Whole blood immune repertoire detection method based on high-flux sequencing technology
An immunological library and detection method technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as inconvenience and directness, and achieve the effect of low cost and good sequencing time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
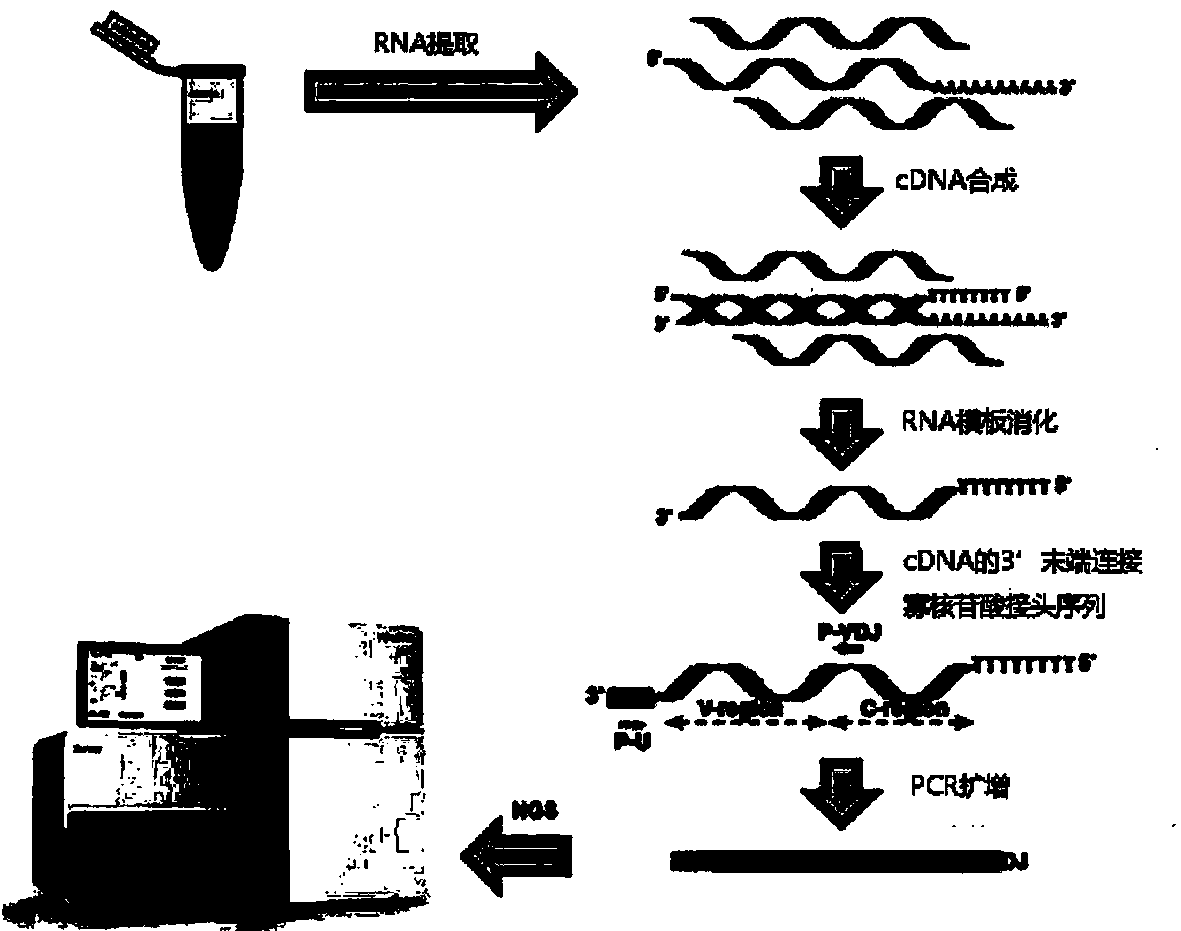
[0022] The whole blood immune repertoire detection method based on high-throughput sequencing technology provided by the present invention comprises the following steps:
[0023] (1) RNA is directly extracted from whole blood samples;
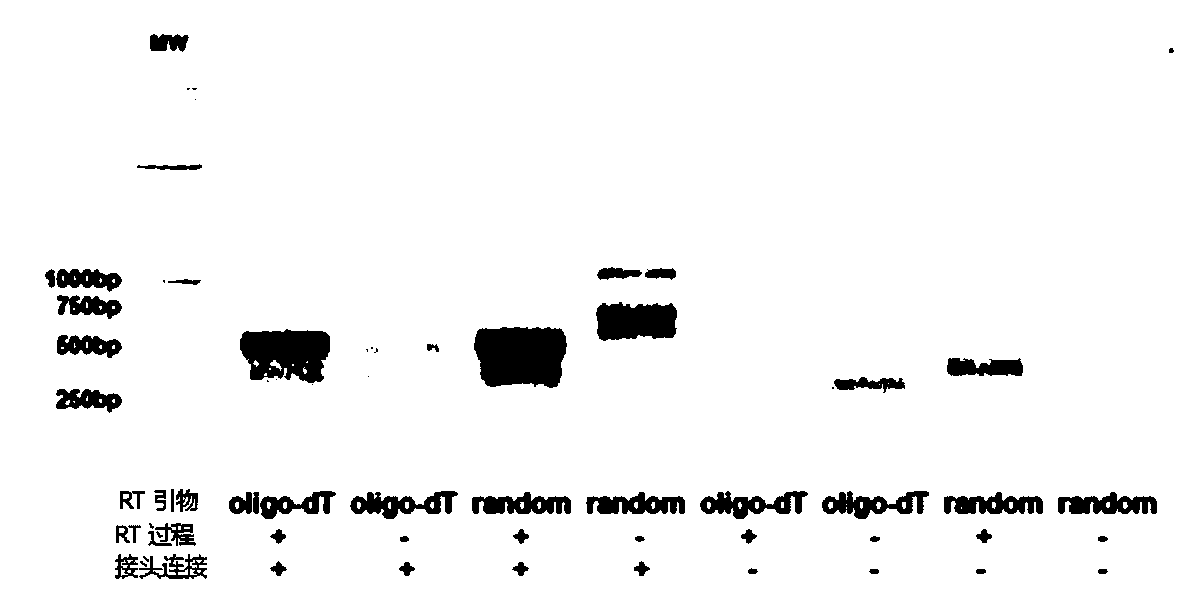
[0024] (2) Generate single-stranded cDNA from RNA reverse transcription, connect the oligonucleotide linker sequence to the 3' end of the single-stranded cDNA, use general primers and special targeting primer P-VDJ whose sequence is GAC CTC GGG TGG GAA CAC The special primer pair for PCR reaction and the single-stranded cDNA connected with the oligonucleotide linker sequence are used as PCR templates for PCR reaction to amplify the immune receptor genome;
[0025] (3) High-throughput sequencing.
[0026] The method provided by the present invention will be further described below in conjunction with examples.
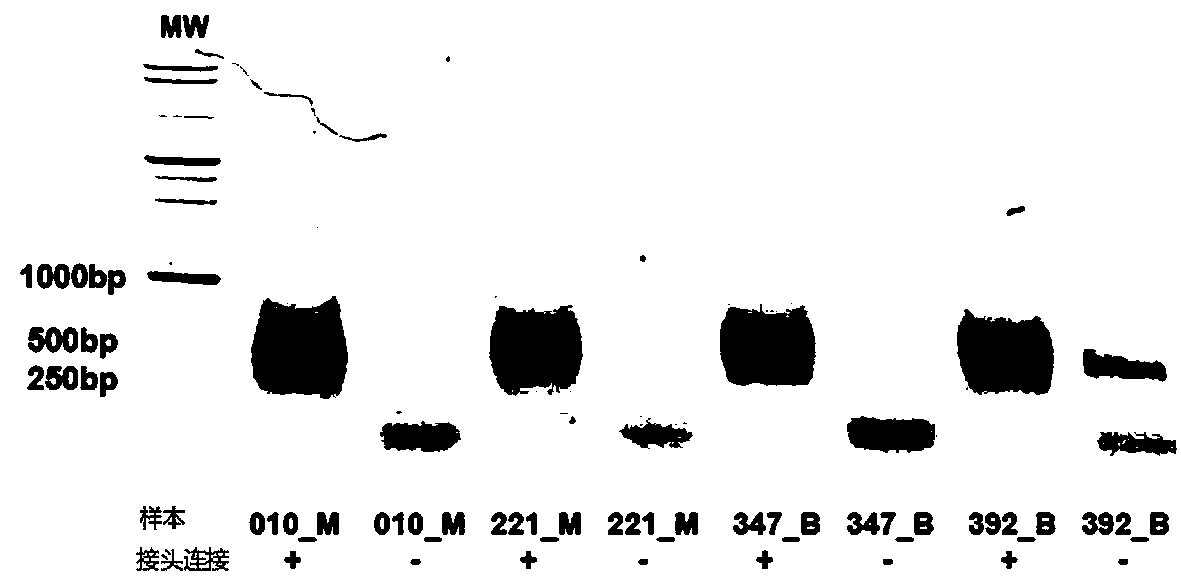
[0027] Using previously collected whole blood samples from two malignant tumor patients and two benign patients, which were stored at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com