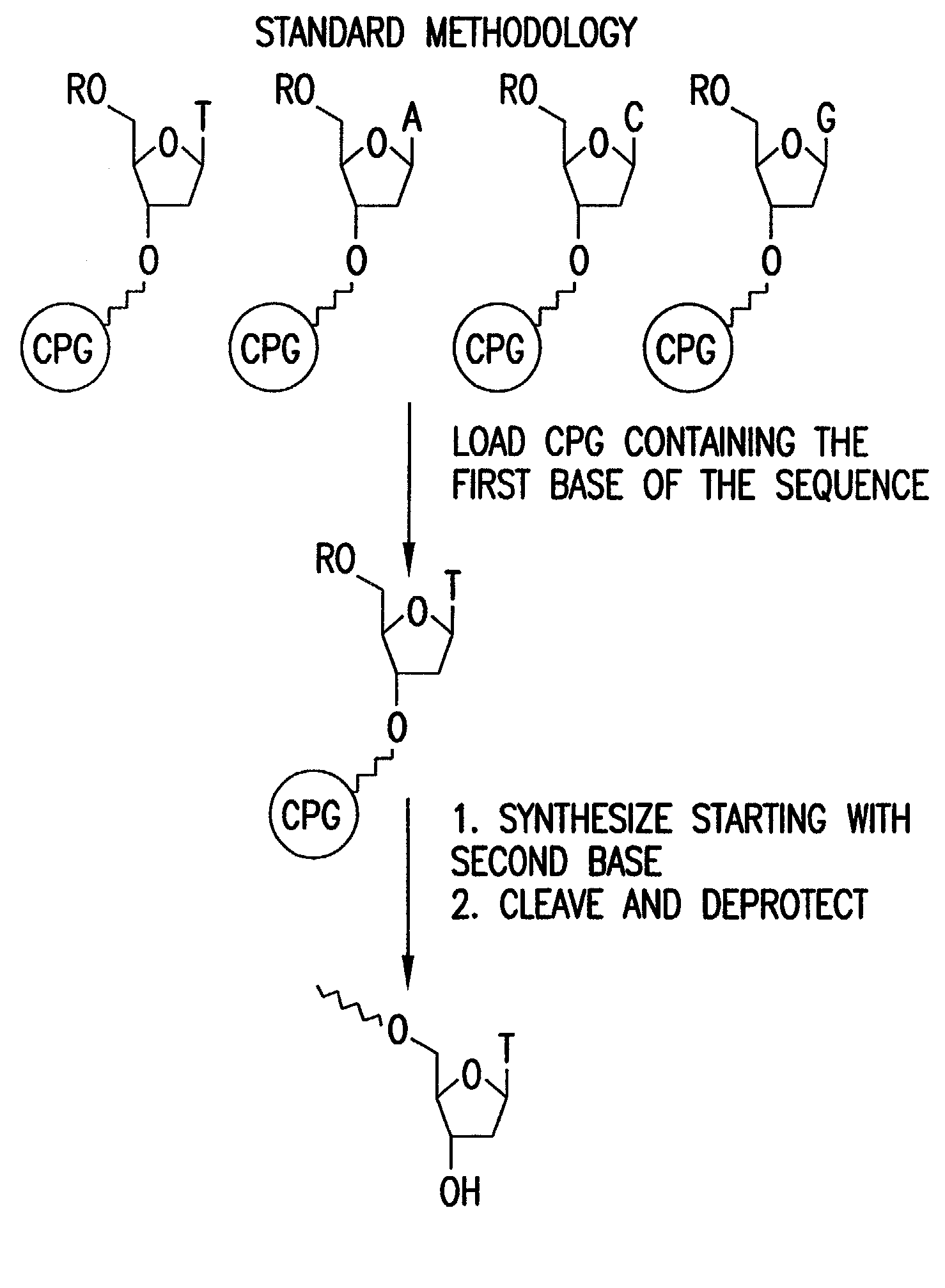
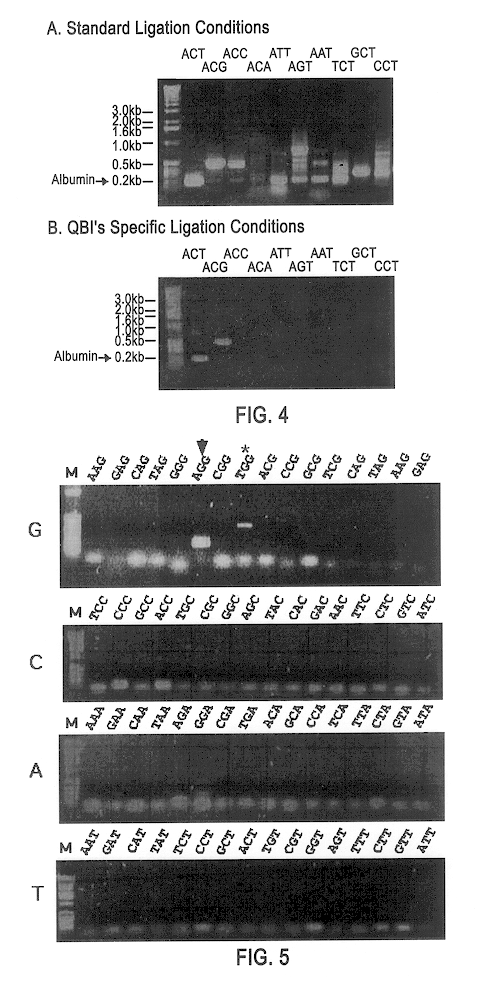
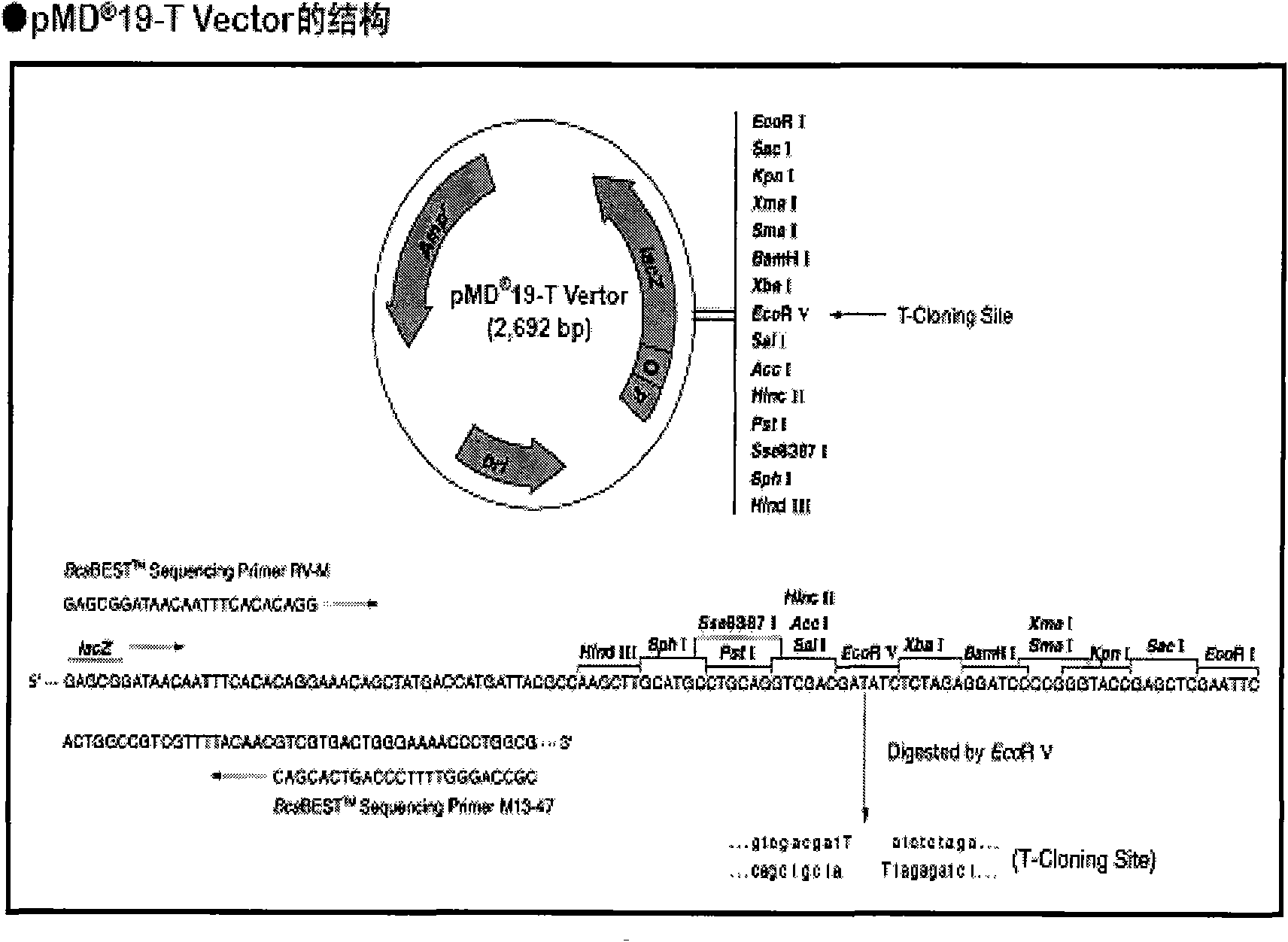
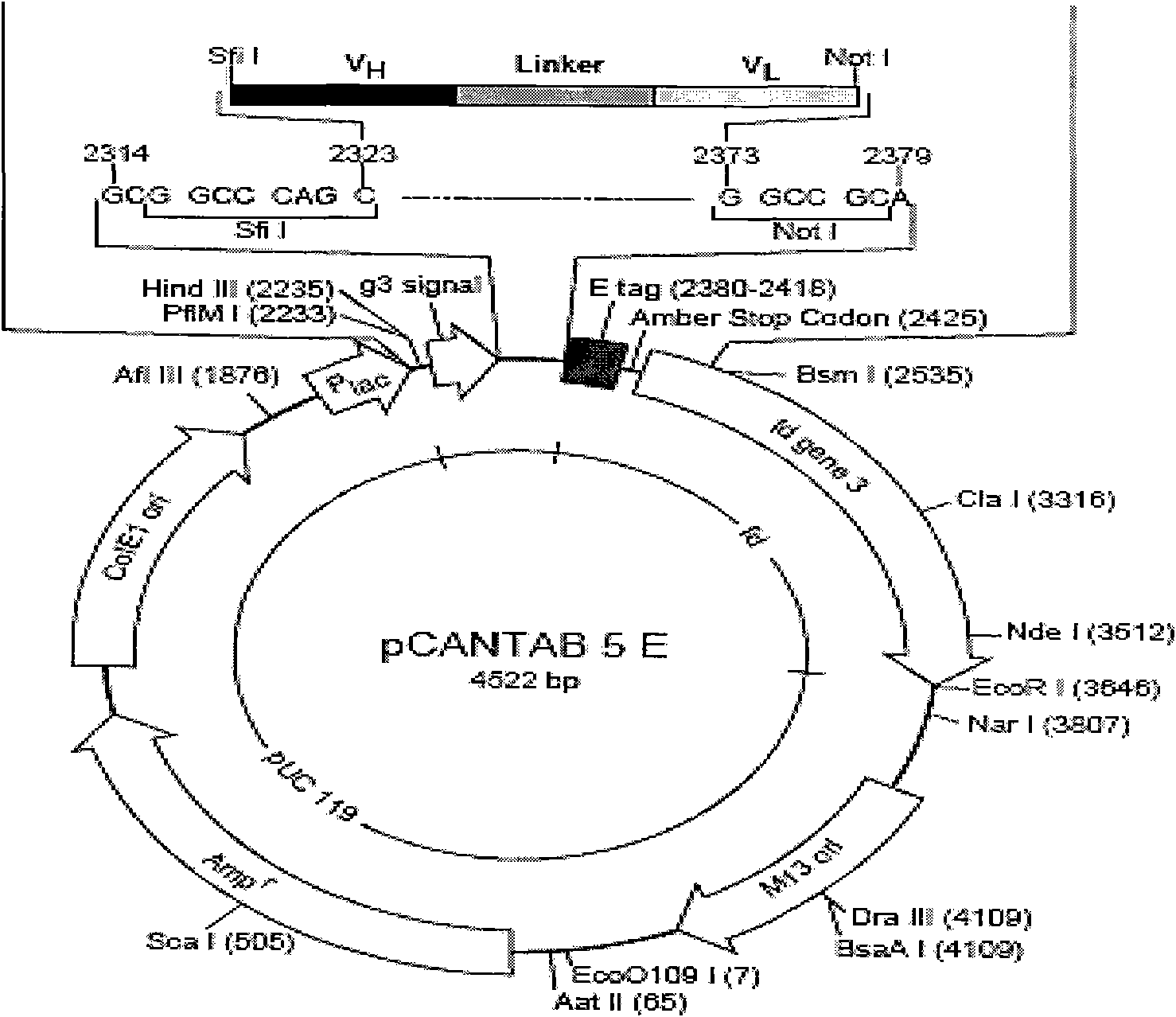
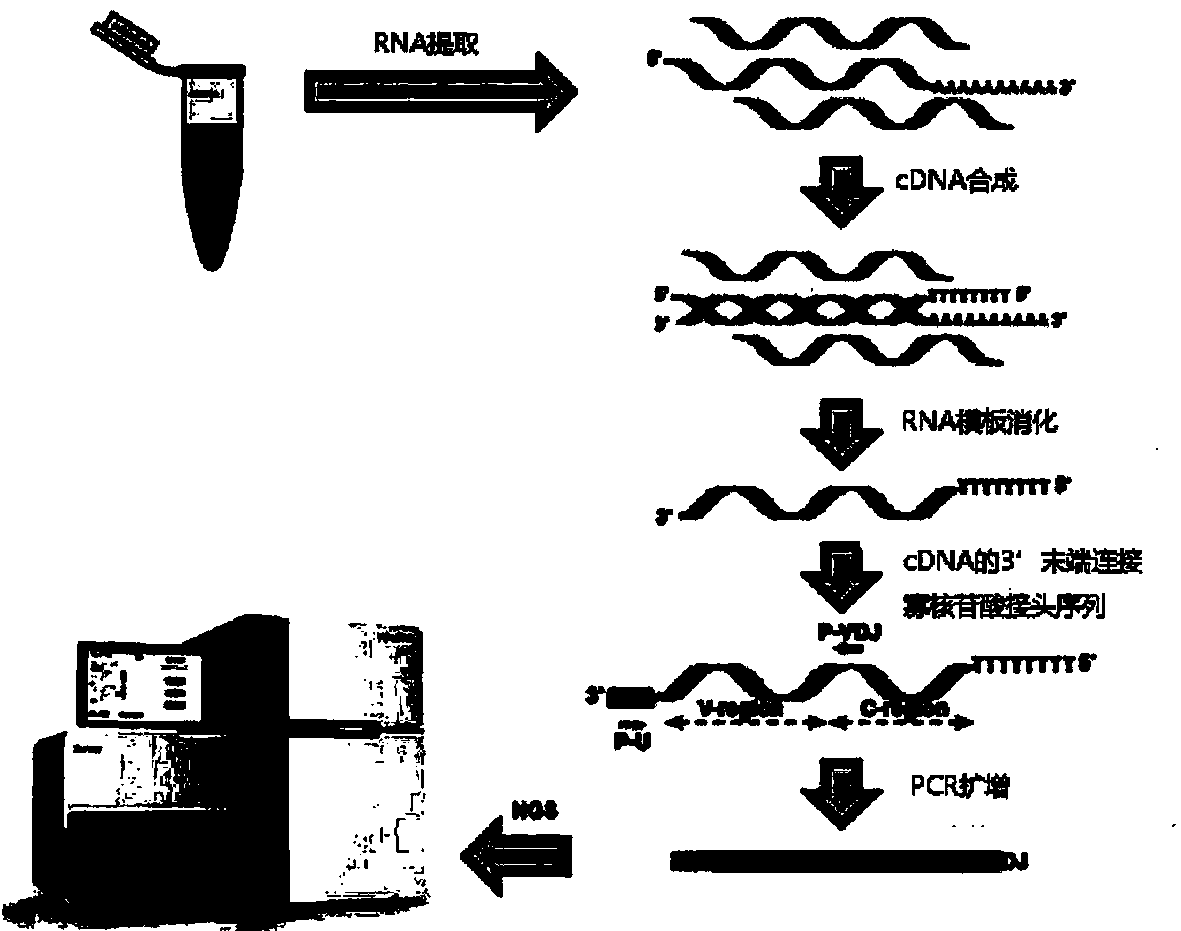
Patents
Literature
34 results about "Oligonucleotide Linker" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
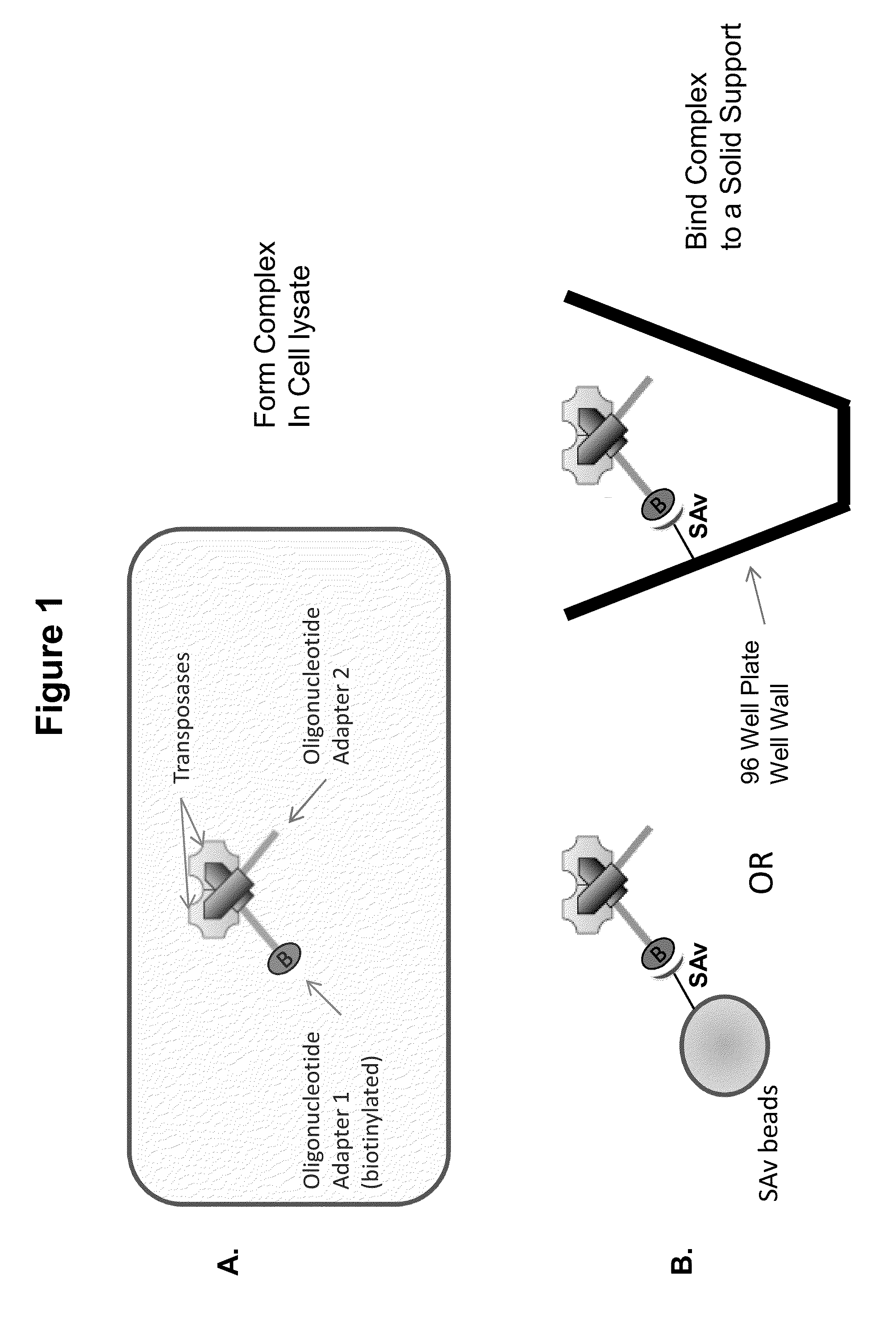
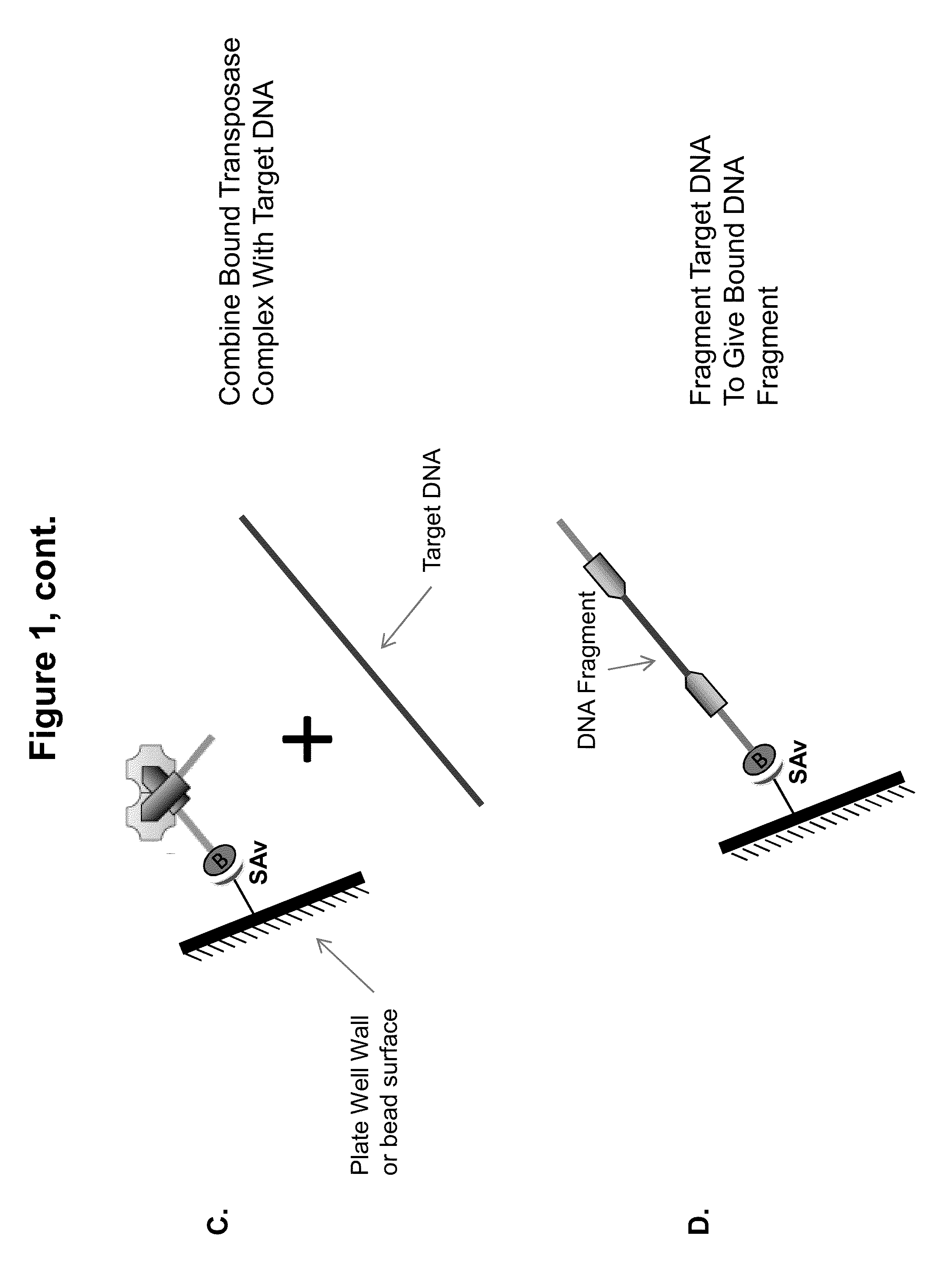
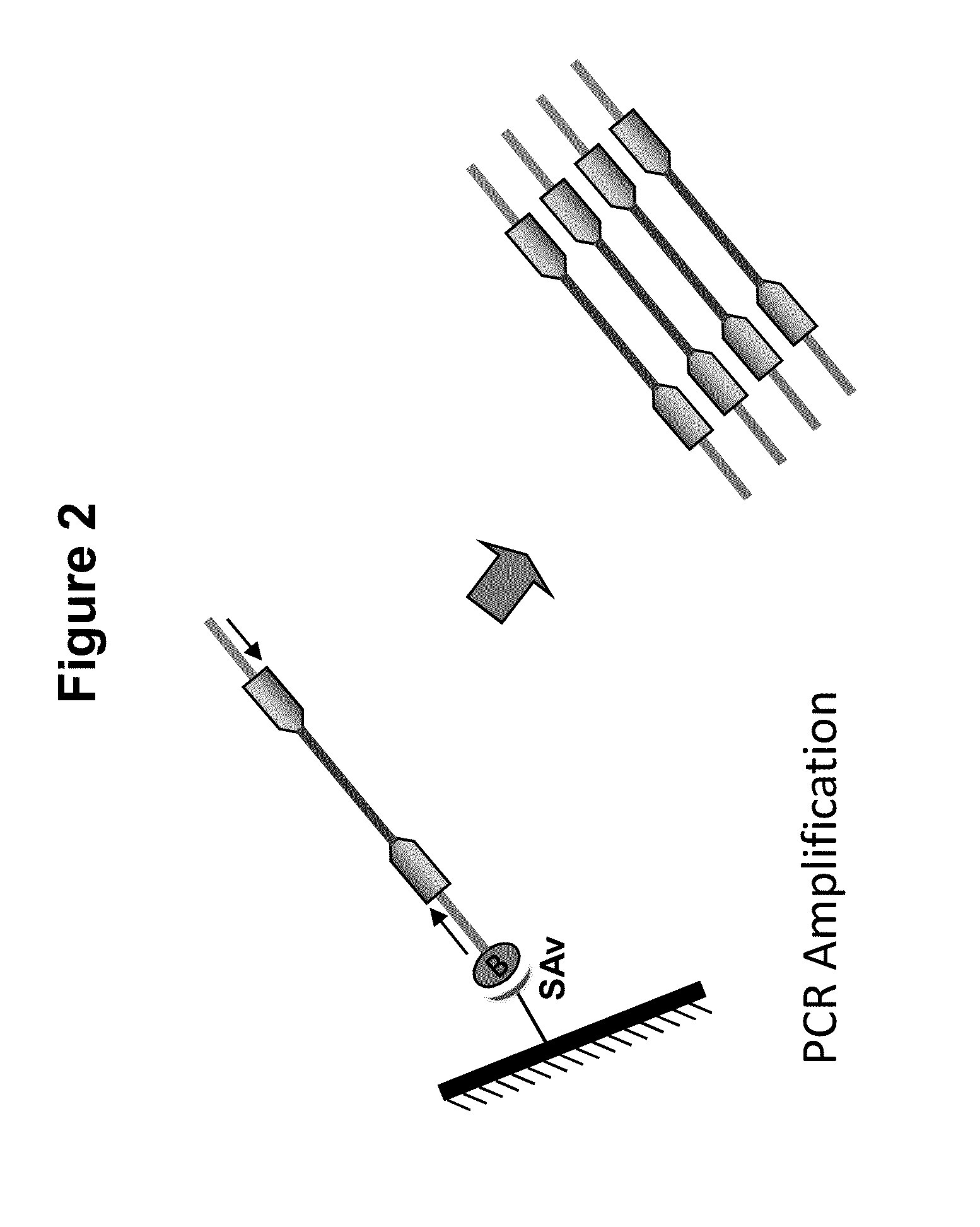
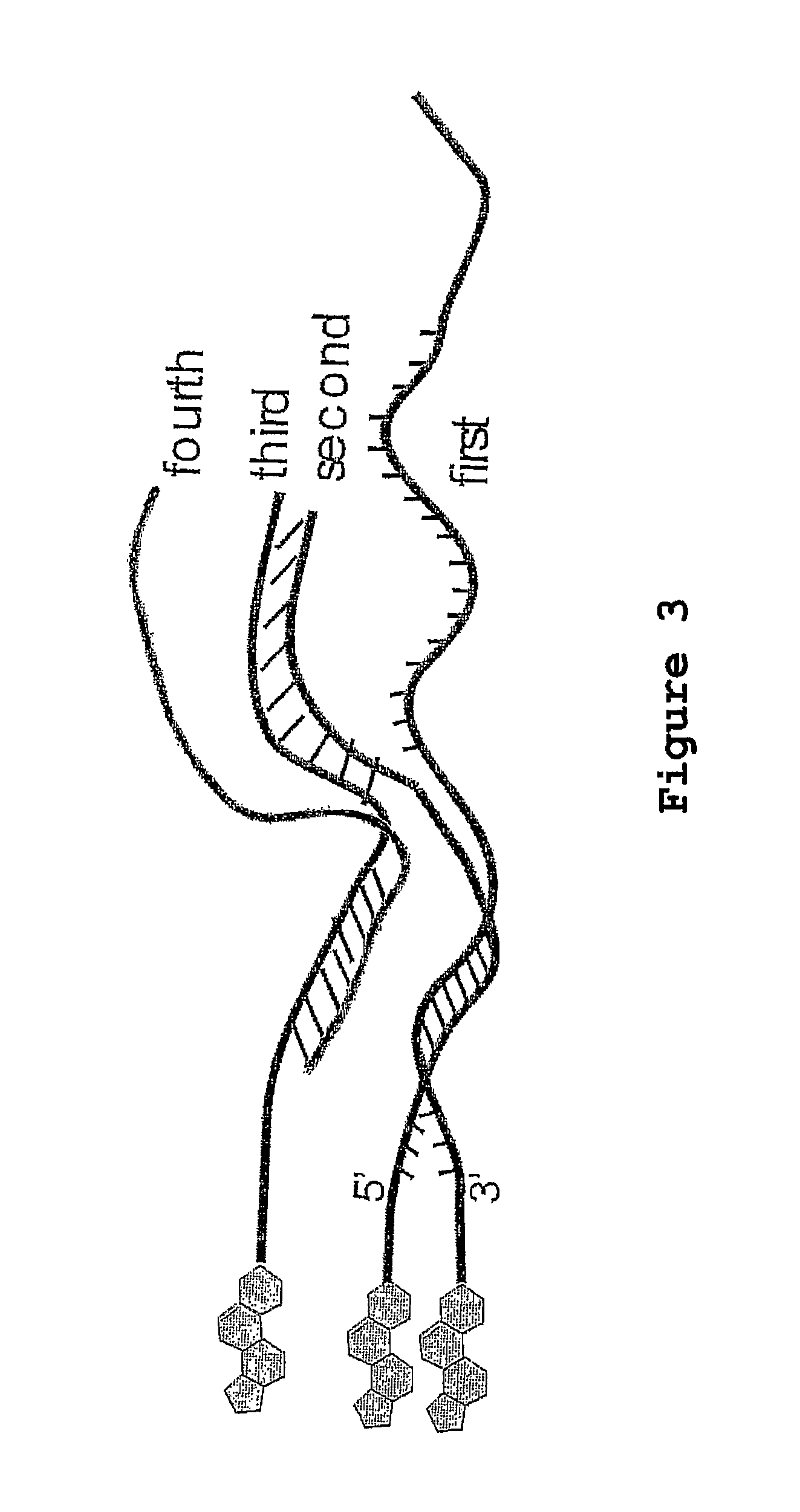
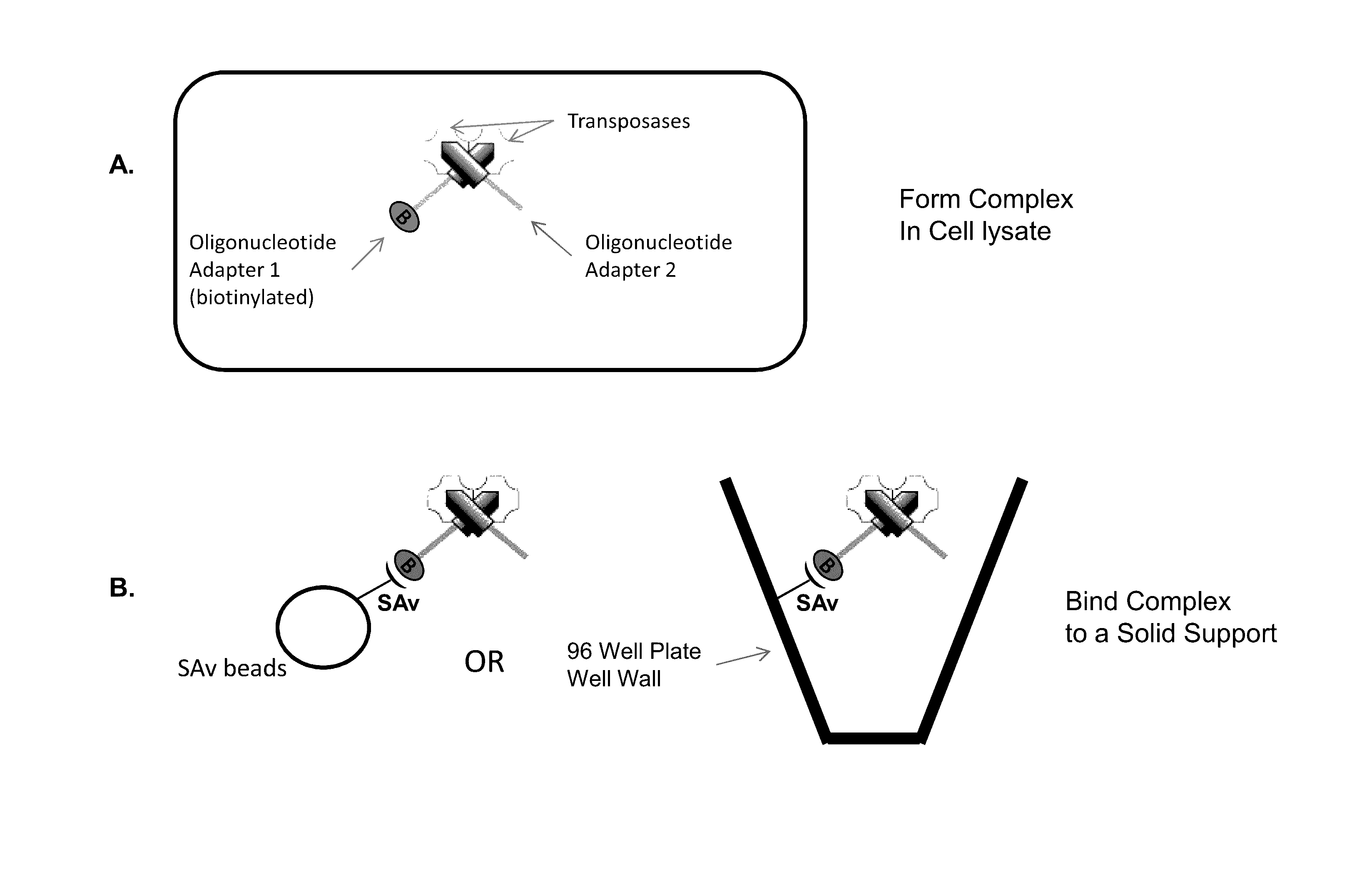
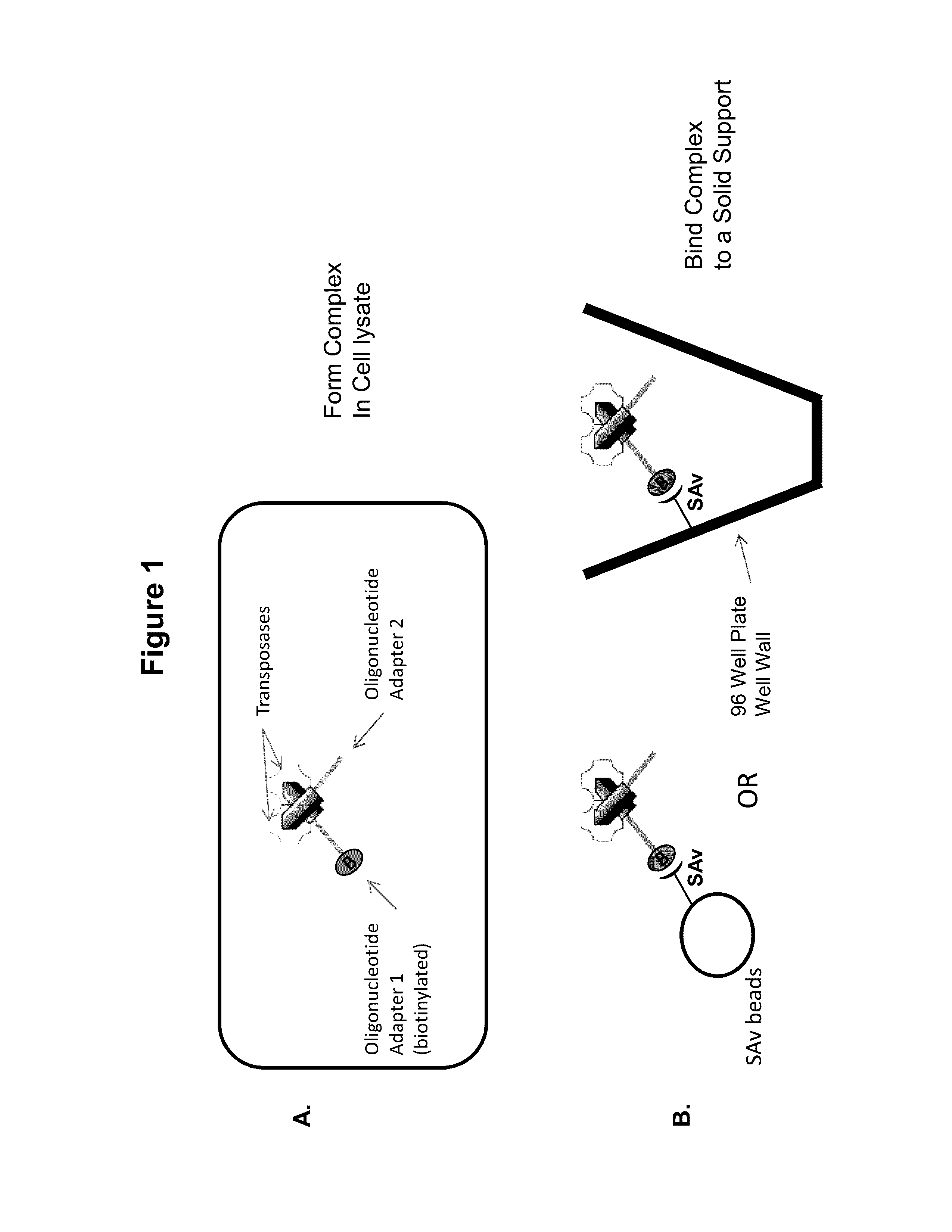
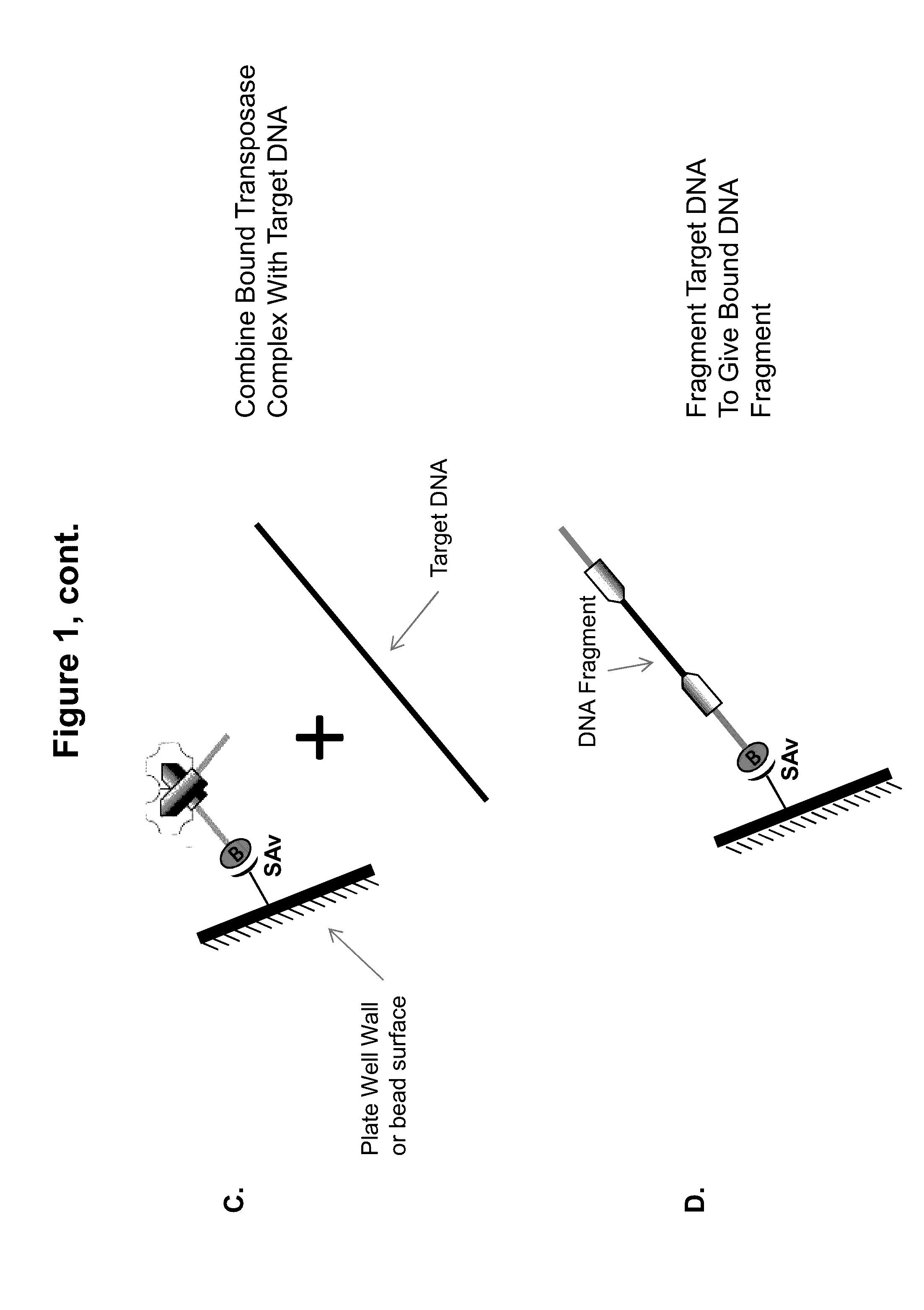
Immobilized transposase complexes for DNA fragmentation and tagging
ActiveUS20140093916A1Maintain good propertiesExcellent characteristicsHydrolasesMicrobiological testing/measurementDNA fragmentationOligonucleotide Linker
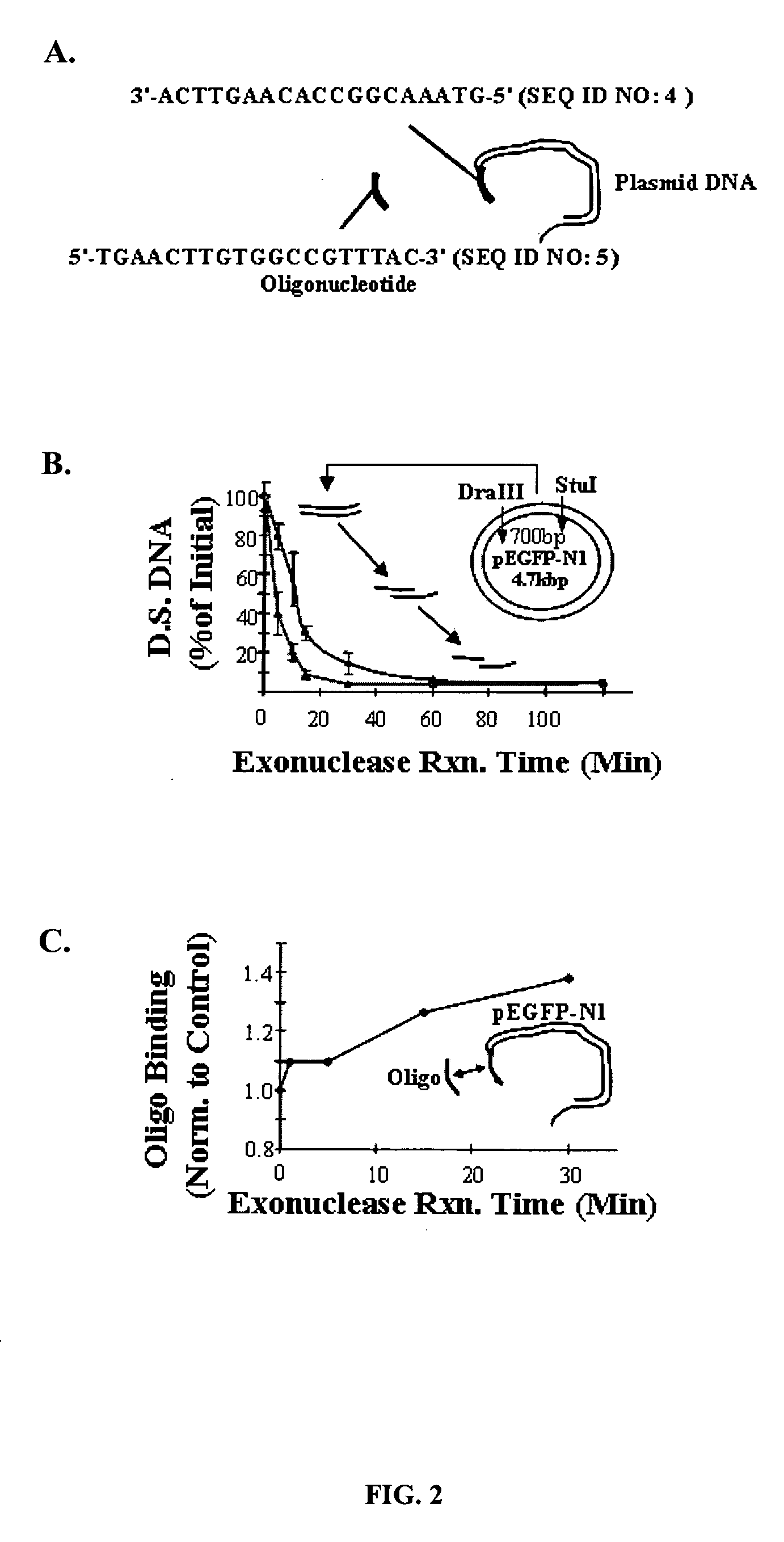
The present invention provides a simple and rapid method for preparing purified transposase complexes that are highly suited for fragmenting DNA. The method includes forming transposase complexes with oligonucleotide adapters in cell lysate, then purifying the complexes from the other substance in the cell lysate. Purification is accomplished using a specific binding pair, in which one member of the pair is bound to an oligonucleotide adapter of the complex and the other member of the pair is bound to a solid substrate. The bound complexes can be immediately used in DNA fragmentation reactions to produce solid substrate-bound DNA fragments, which can be used for any number of purposes, including as templates for amplification and sequencing.
Owner:AGILENT TECH INC
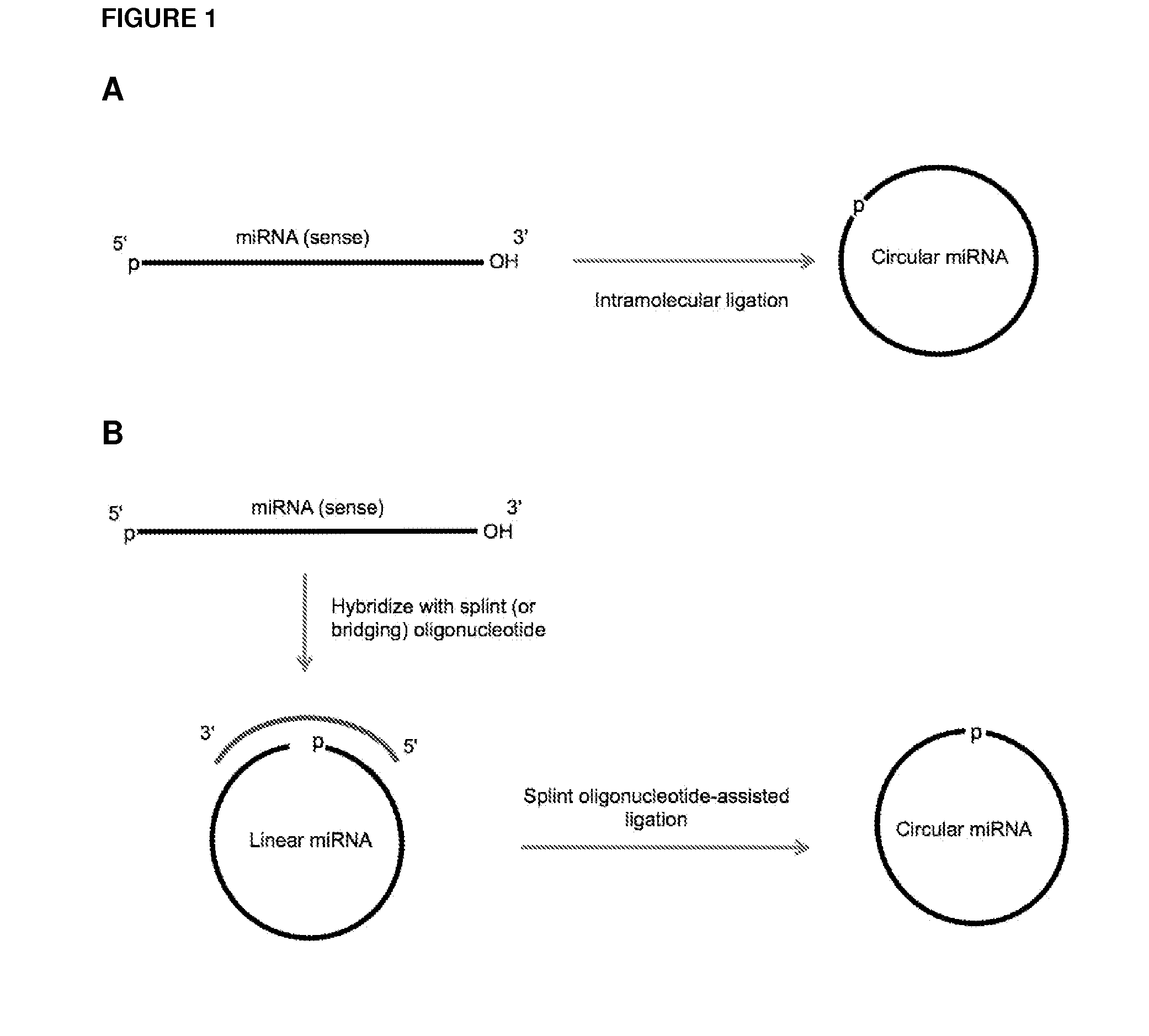
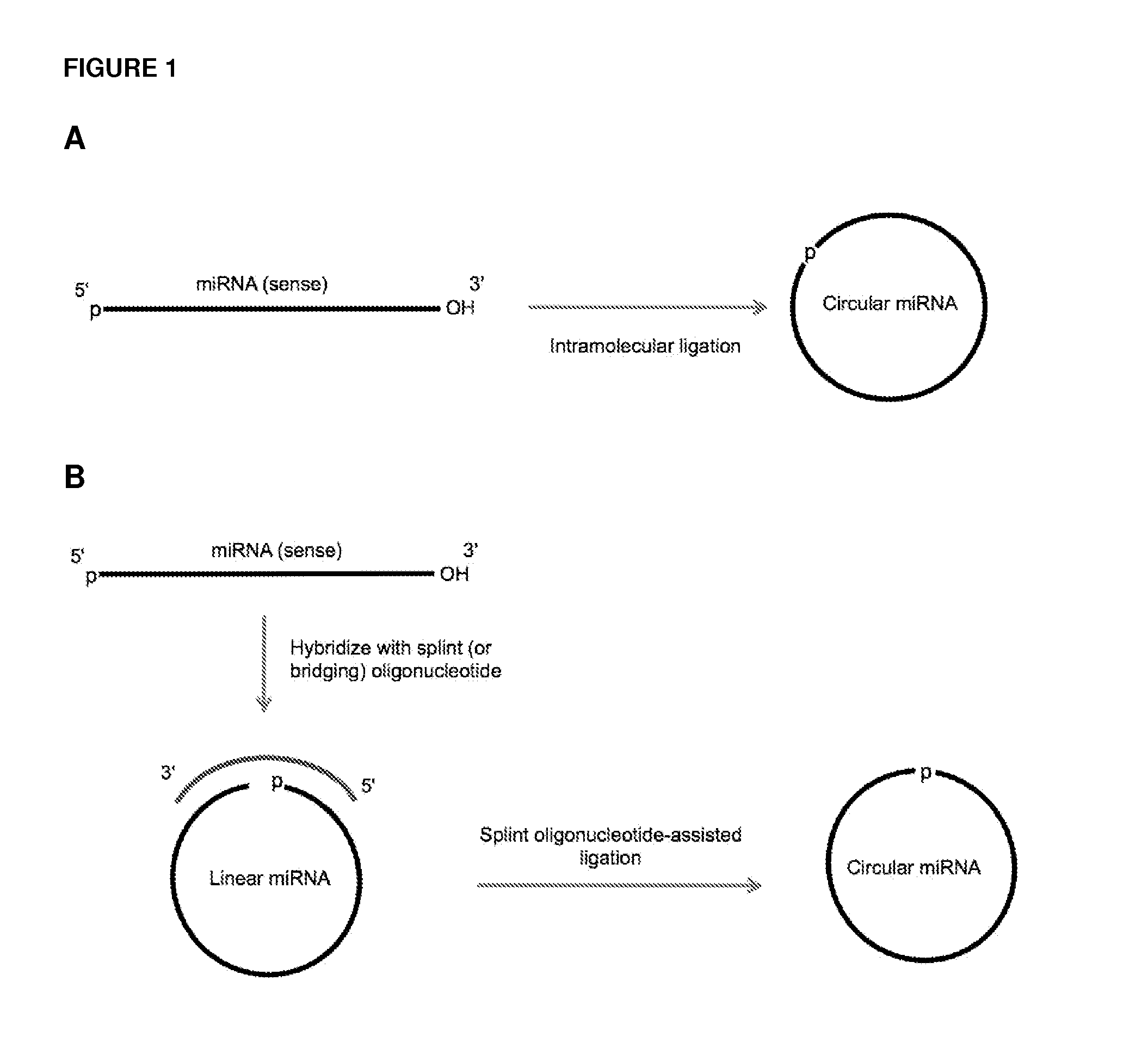
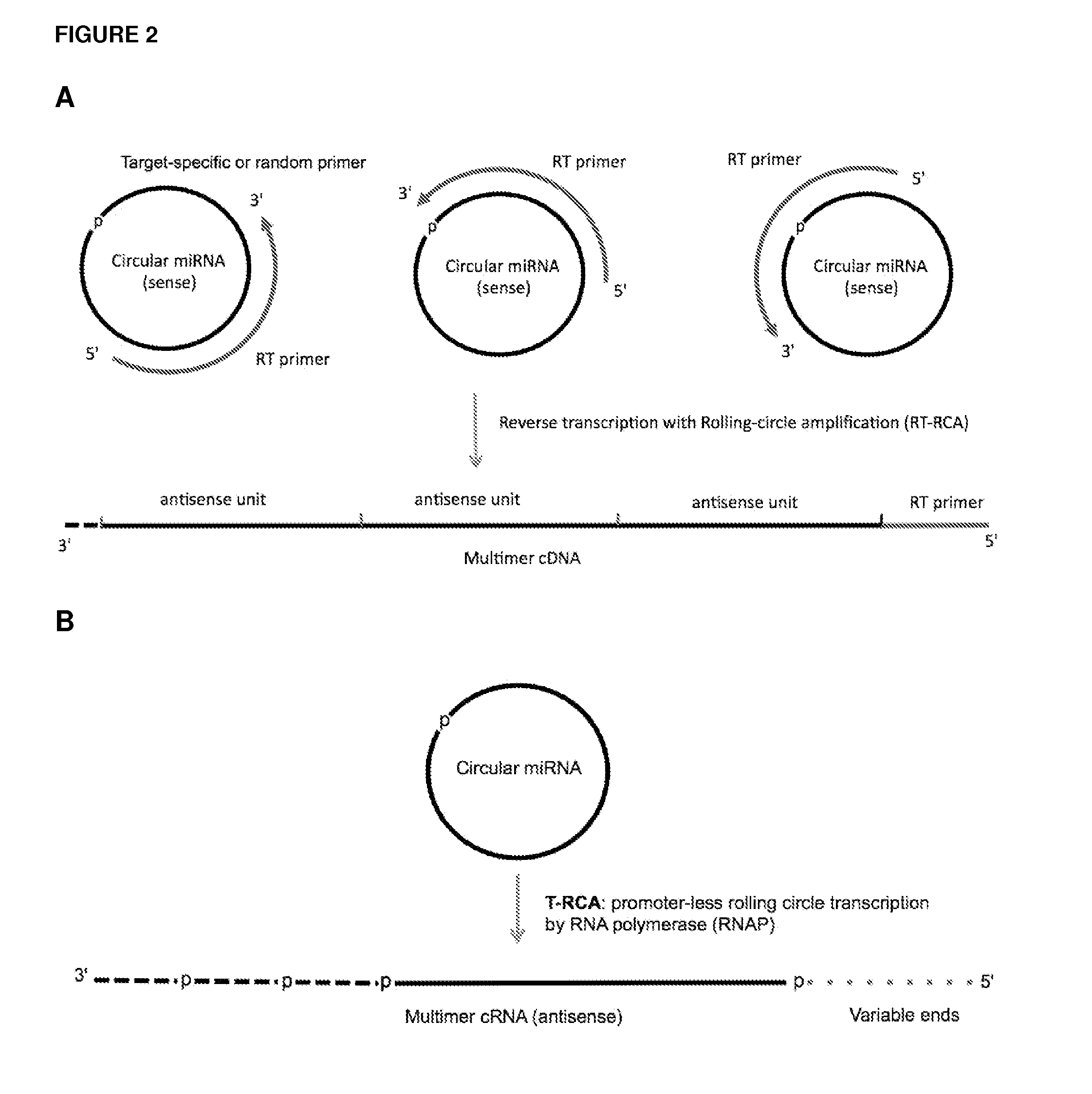
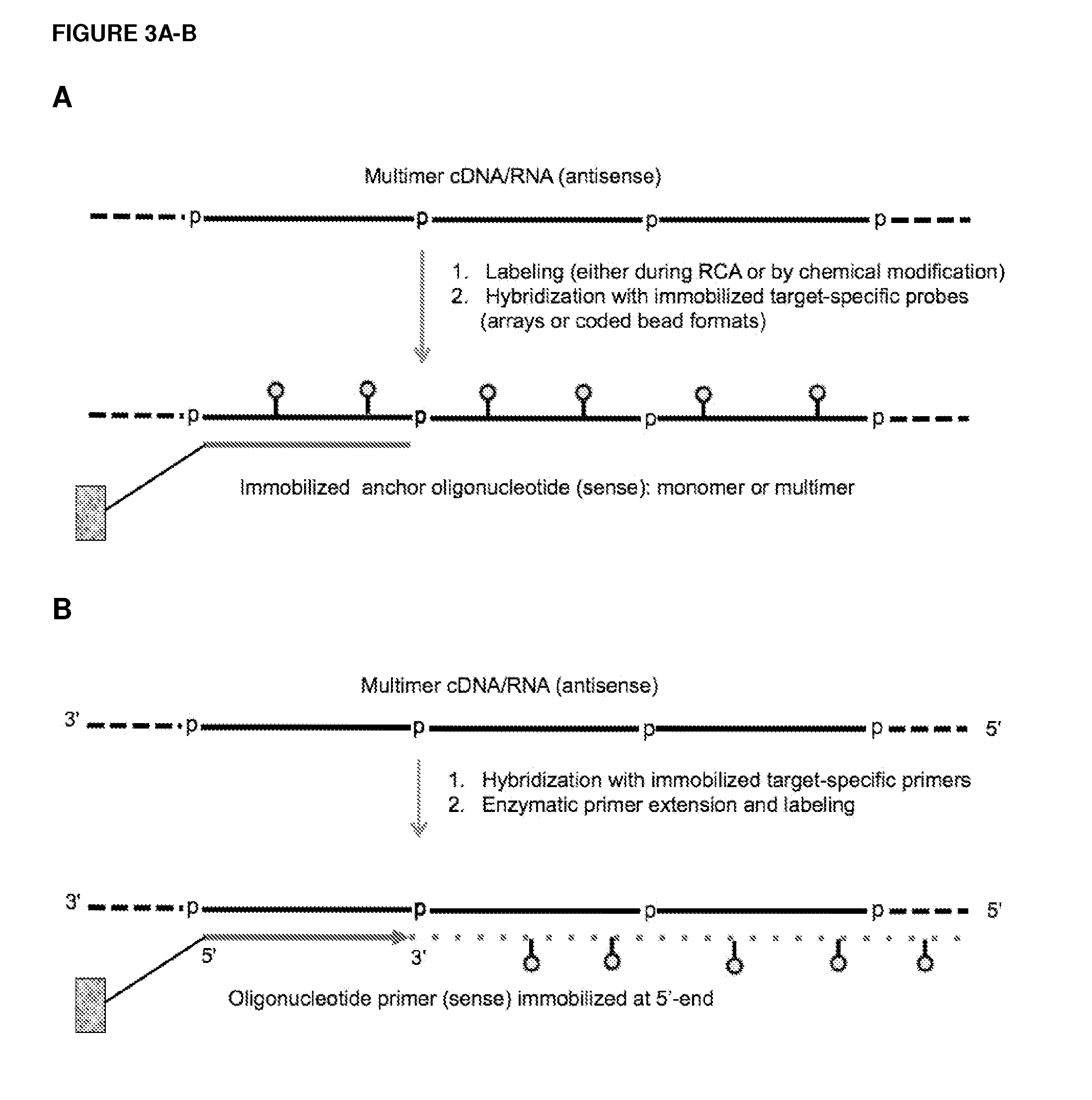
Methods and compositions for detection of small rnas
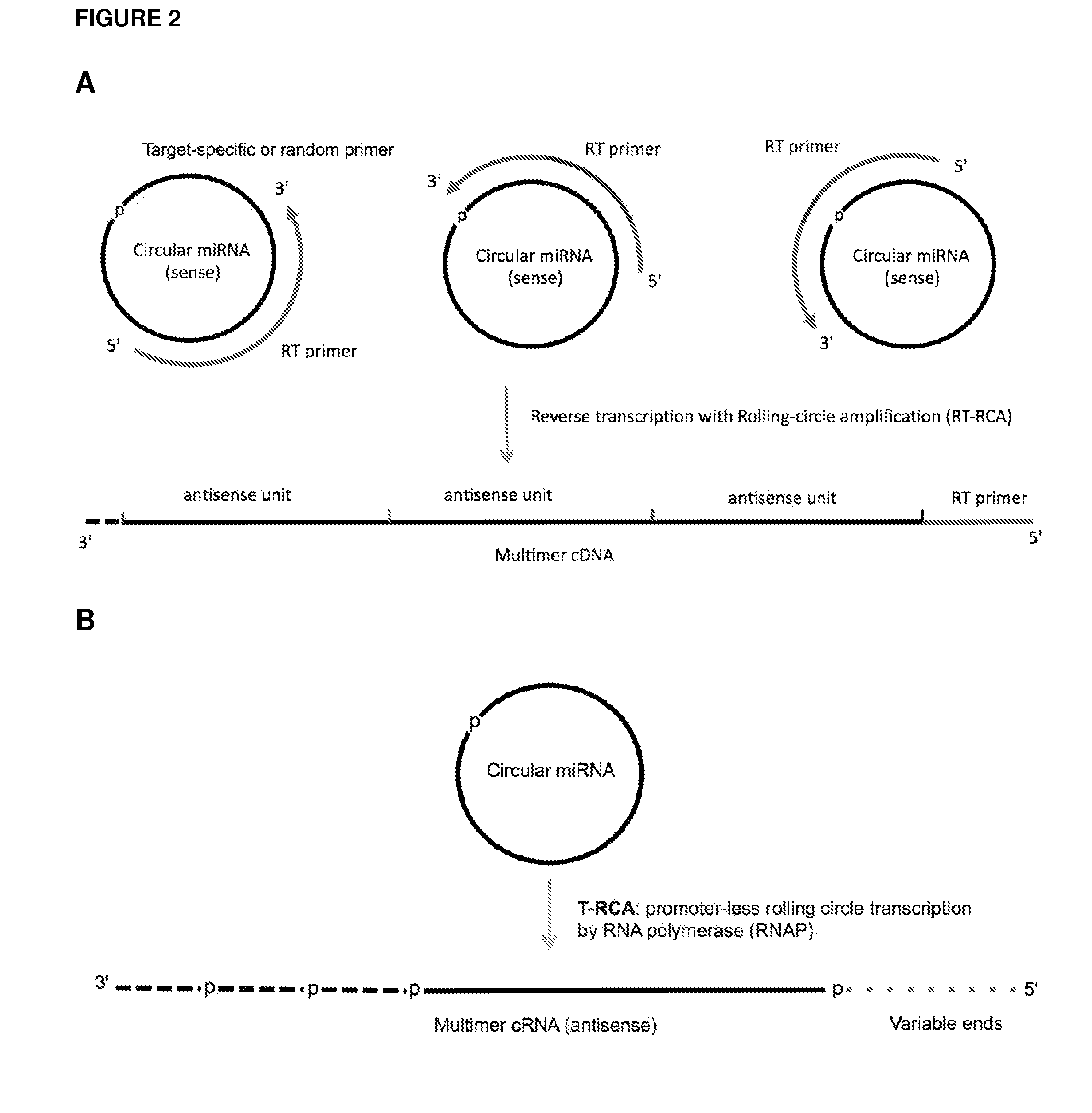
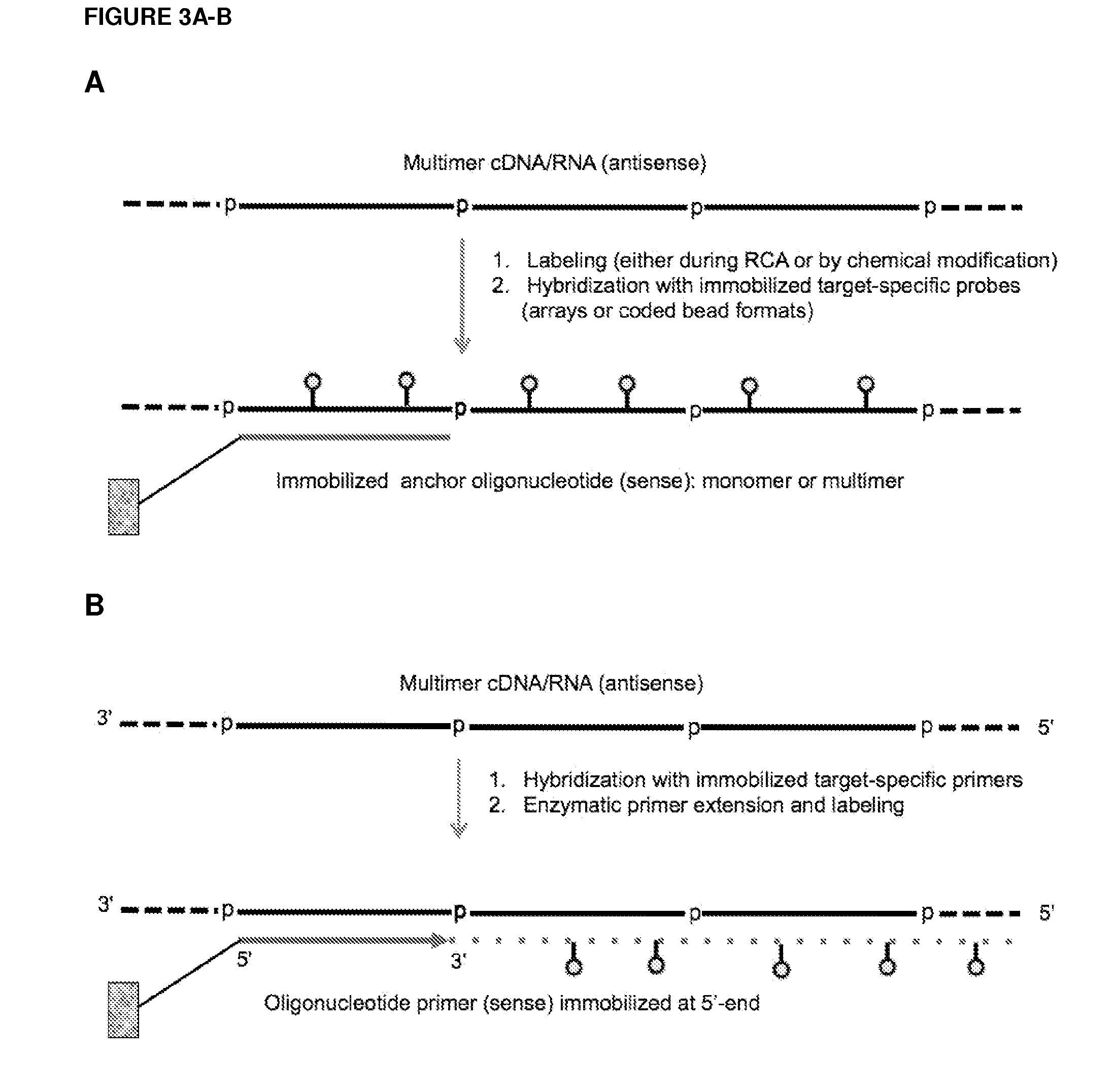
ActiveUS20120164651A1Improve efficiencySimilar efficiencyMicrobiological testing/measurementTrue positive rateCircular RNA
Currently, the circularization of small RNAs is broadly regarded as an obstacle in ligation-related assays and explicitly avoided while short lengths of linear RNA targets is broadly recognized as a factor limiting use of conventional primers in PCR-related assays. In contrast, the disclosed invention capitalizes on circularization of small RNA targets or their conjugates with oligonucleotide adapters. The circular RNA templates provide amplification of the target sequences via synthesis of multimer nucleic acids that can be either labeled for direct detection or subjected to PCR amplification and detection. Structure of small circular RNAs and corresponding multimeric nucleic acids provide certain advantages over current methods including flexibility in design of conventional RT and PCR primers as well as use of 5′-overlapping dimer-primers for efficient and sequence-specific amplification of short target sequences. Our invention also reduces number of steps and reagents while increasing sensitivity and accuracy of detection of small RNAs with both 2′OH and 2′-OMe at their 3′ ends. Our invention increase sensitivity and specificity of detection of microRNAs and other small RNAs with both 2′OH and 2′-OMe at their 3′ ends while allowing us to distinguish these two forms from each other.
Owner:REALSEQ BIOSCI INC
Method for removing a universal linker from an oligonucleotide
InactiveUS20020143166A1Sugar derivativesRecombinant DNA-technologyOligonucleotide LinkerOligonucleotide
The invention relates to a method for cleavage of a linker from an oligonucleotide comprising contacting an oligonucleotide-linker conjugate with a gaseous nucleophilic cleavage reagent under conditions that result in the cleavage of the linker from the oligonucleotide.
Owner:INVITROGEN
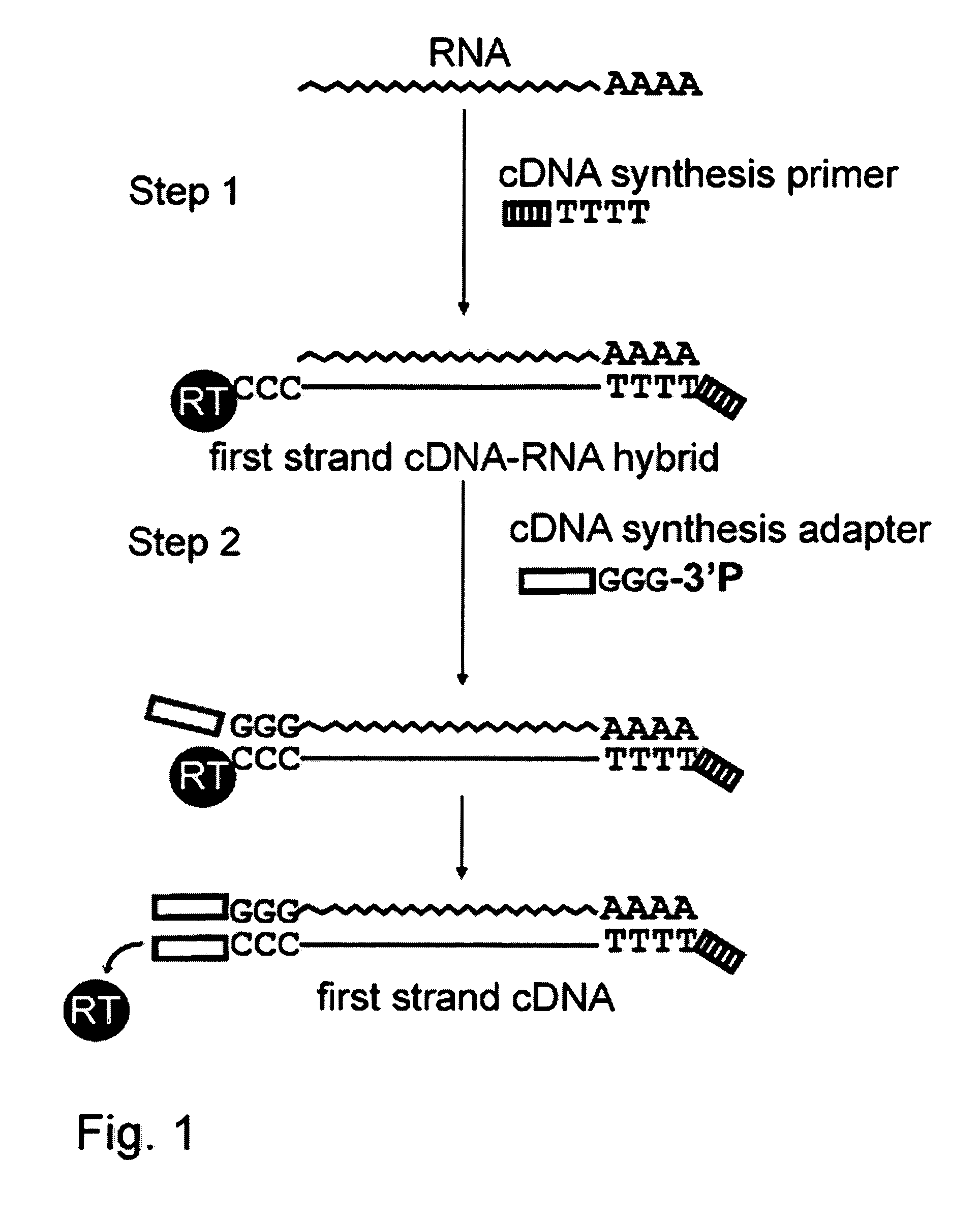
Methods of cDNA preparation
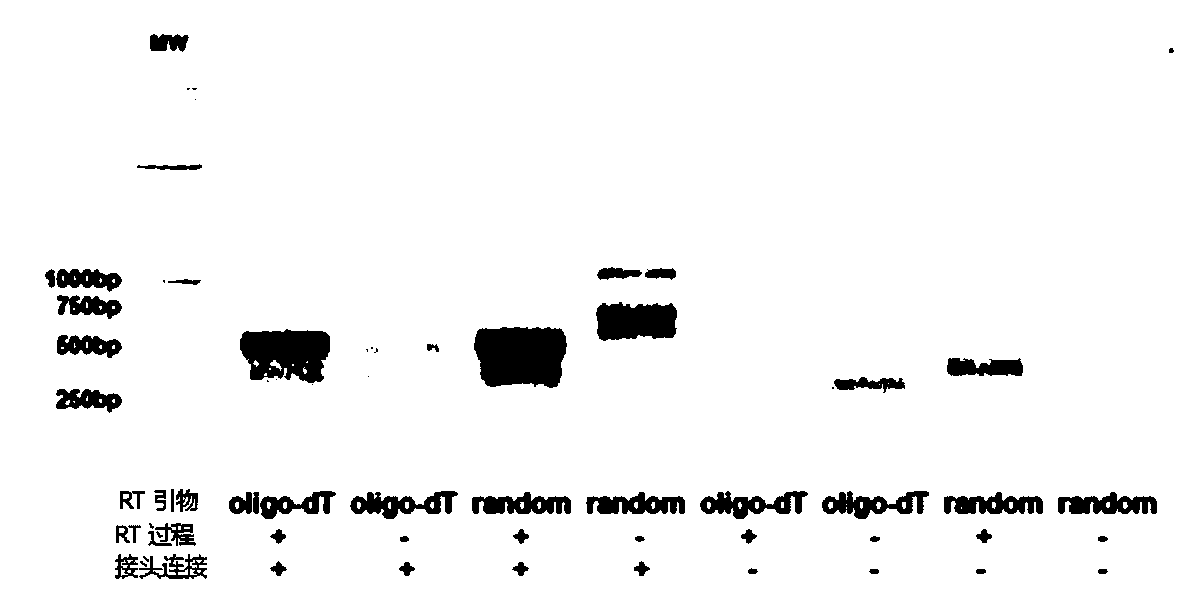
InactiveUS20080145844A1Microbiological testing/measurementFermentation3-deoxyriboseReverse transcriptase activity
The present invention provides an improved method for cDNA preparation. The method of the present invention comprises the following steps: (1) contacting mRNA with a cDNA synthesis primer which can anneal to RNA and a suitable enzyme which possesses reverse transcriptase activity under conditions sufficient to permit the template-dependent extension of the primer to generate an mRNA-cDNA intermediates; (2) contacting a mixture from step 1 with a deoxyribonucleotide adapter in the presence of Mn2+-ions, wherein said oligonucleotide adapter has a pre-selected arbitrary nucleotide sequence at its 5′-end, and a short dG stretch at its 3′-end. The 3′-end nucleotide of the adapter is a terminator nucleotide, e.g., a nucleotide with a modified 3′-OH group of a deoxyribose residue.
Owner:EVROGEN
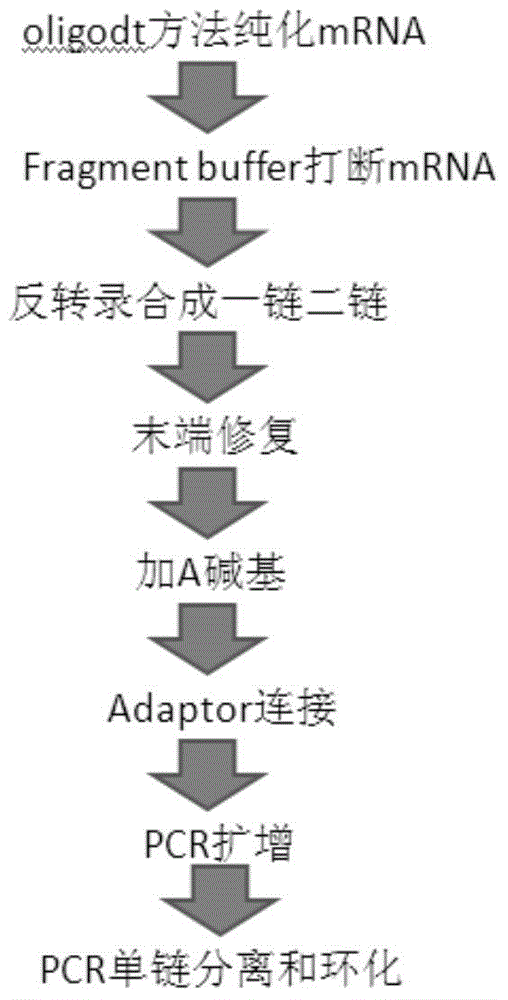
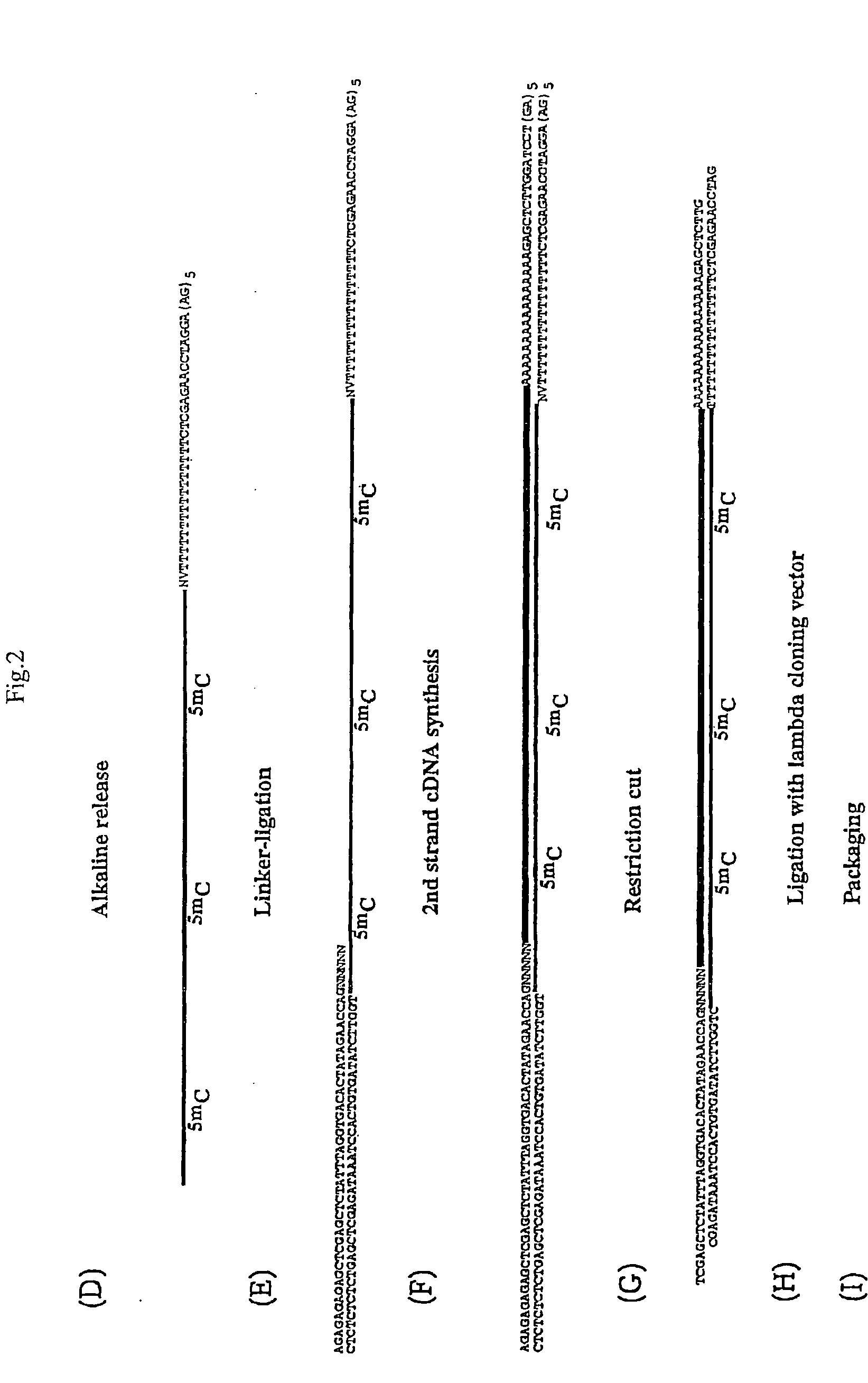
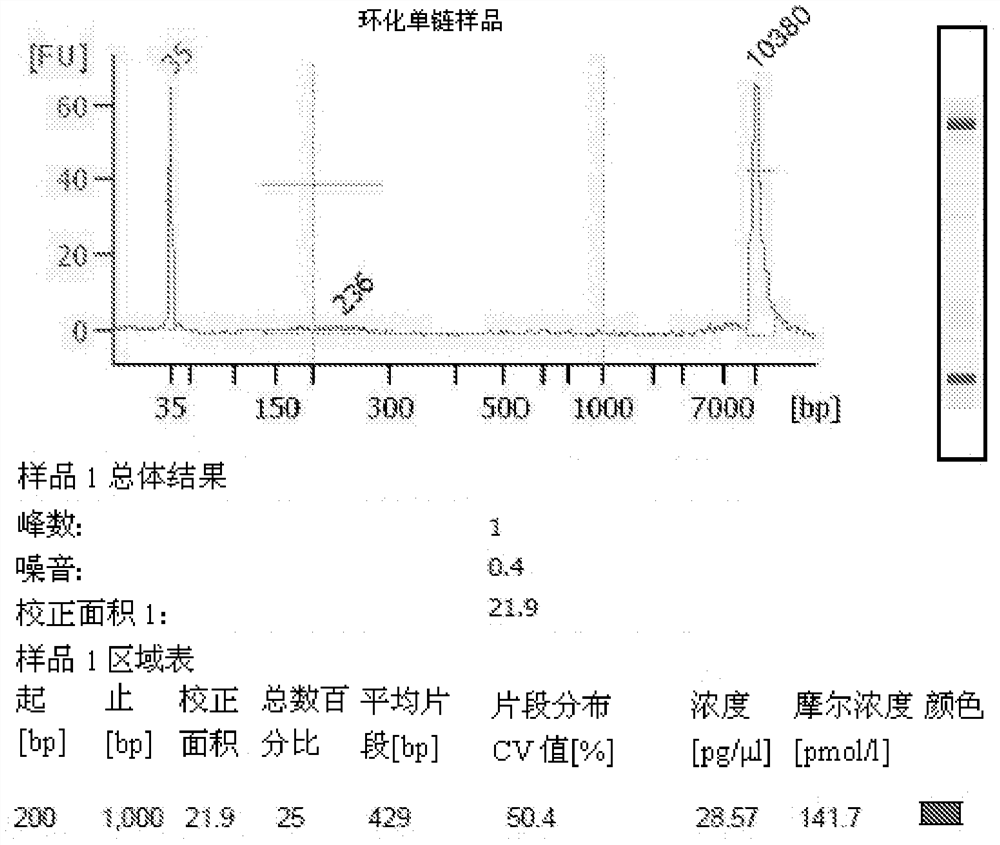
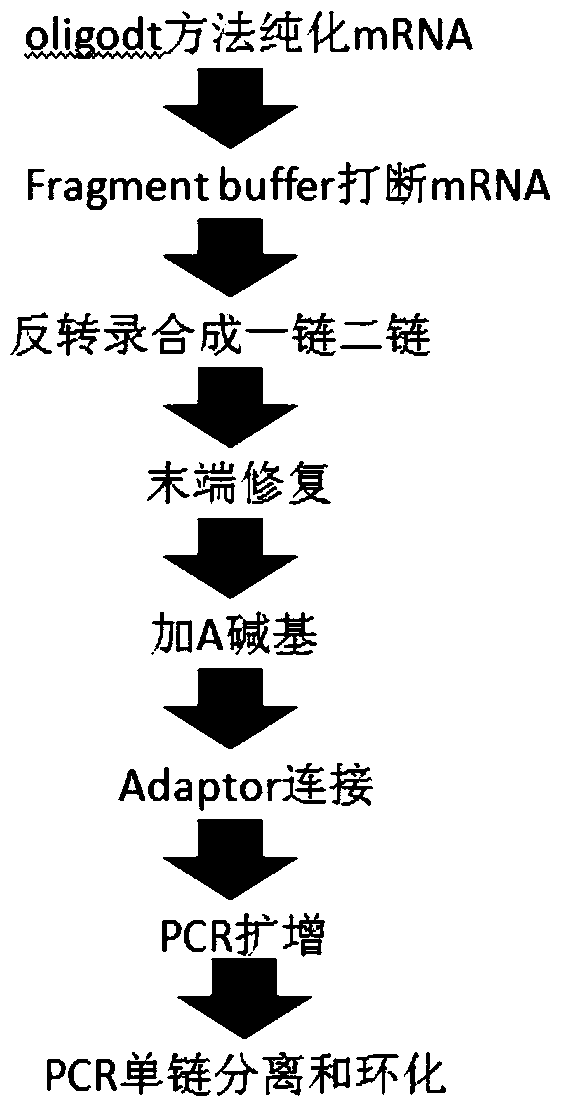
Oligonucleotide linker and applications of oligonucleotide linker in construction of nucleic acid sequencing single-chain cyclic library
ActiveCN105400776AAvoid digestionNucleotide librariesLibrary creationNucleic acid sequencingOligonucleotide Linker
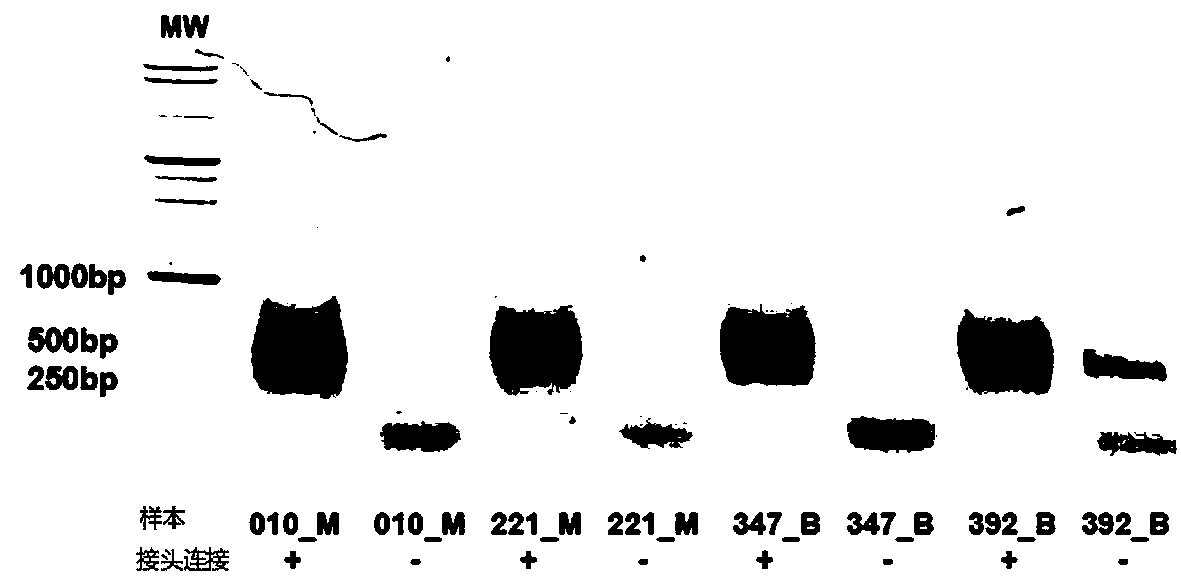
The present invention provides an oligonucleotide linker, and particularly relates to uses of the oligonucleotide linker in construction of a single-chain cyclic library in nucleic acid sequencing, and further in sequencing platforms requiring single-chain cyclic libraries. The oligonucleotide linker of the present invention has advantages of high sequencing throughput, high accuracy, and easy operation.
Owner:MGI TECH CO LTD
Sequence-dependent gene sorting techniques
InactiveUS6468749B1Without redundancySugar derivativesMicrobiological testing/measurementGeneComputational biology
This invention provides a method of sorting genes comprising: (1) preparing ds cDNA molecules from mRNA molecules (2) digesting the ds cDNA molecules (3) ligating to the digested cDNA molecules a set of dsDNA oligonucleotide adaptors; (4) amplifying the ligated cDNA molecules; and (5) sorting the amplified cDNA molecules into nonoverlapping groups. This invention also provides two additional methods of sorting genes. This invention further provides a method of making sub-libraries of ligation sets.
Owner:QUARK FARMACUITIKALS INC
Single-chain antibody of broad-spectrum anti-p21ras protein and preparation method thereof
The invention discloses a single-chain antibody of broad-spectrum anti-p21ras protein and a preparation method thereof. The gene segment of the single-chain antibody is constructed by connecting light and heavy chain variable region gene segments of monoclonal antibody hybridoma cell strains of the broad-spectrum anti-p21ras protein through flexible oligonucleotide linkers; the gene segment of the single-chain antibody is connected with a phagemid expression vector to construct recombinant phagemid; the recombinant phagemid is converted into amber inhibiting colon bacillus TG1 to perform fusion expression; the positive recombinant phagemid is identified by using indirect ELISA screening through phage library exhibition; the positive recombinant phagemid is converted into non-amber inhibiting colon bacillus BL21 (DE3) to perform soluble expression of the single-chain antibody; indirect enzyme-linked immunosorbent assay and immune cell and tissue chemical experiments prove that the single-chain antibody for the soluble expression has strong binding specificity with three p21ras proteins of H-ras, K-ras and N-ras and high affinity; and the single-chain antibody is used for constructing small molecular antibodies such as intracellular antigens and the like and used for diagnosis and research of ras gene related tumors.
Owner:成都军区昆明总医院
Whole blood immune repertoire detection method based on high-flux sequencing technology
ActiveCN104263818ALow costReduce sequencing timeMicrobiological testing/measurementComplementary deoxyribonucleic acidT cell
The invention provides a whole blood immune repertoire detection method based on high-flux sequencing technology. The method comprises the following steps: (1) directly extracting RNA (ribonucleic acid) from a whole blood sample; (2) carrying out reverse transcription on the RNA to generate single chain cDNA (complementary deoxyribonucleic acid), connecting an oligonucleotide joint sequence to the tail end of the single chain cDNA, carrying out PCR (polymerase chain reaction) reaction on the single chain cDNA subjected to oligonucleotide joint sequence connection by using a universal primer and a special targeted primer P-VDJ (of which the sequence is GAC CTC GGG TGG GAA CAC) as a special primer pair for PCR reaction, and amplifying the immune receptor genome; and (3) carrying out high-flux sequencing. The method of directly extracting the RNA from the whole blood sample is more suitable for clinic; the single target specific primer is utilized to uniformly amplify the variable region of the subtype T cell receptor gene; and a Hi-SEQ 2500 of the Illumina corporation and a bar code are utilized for sequencing, thereby ensuring the lower cost, the sequencing time of less than 48 hours and favorable sequencing depth.
Owner:武汉希望组生物科技有限公司
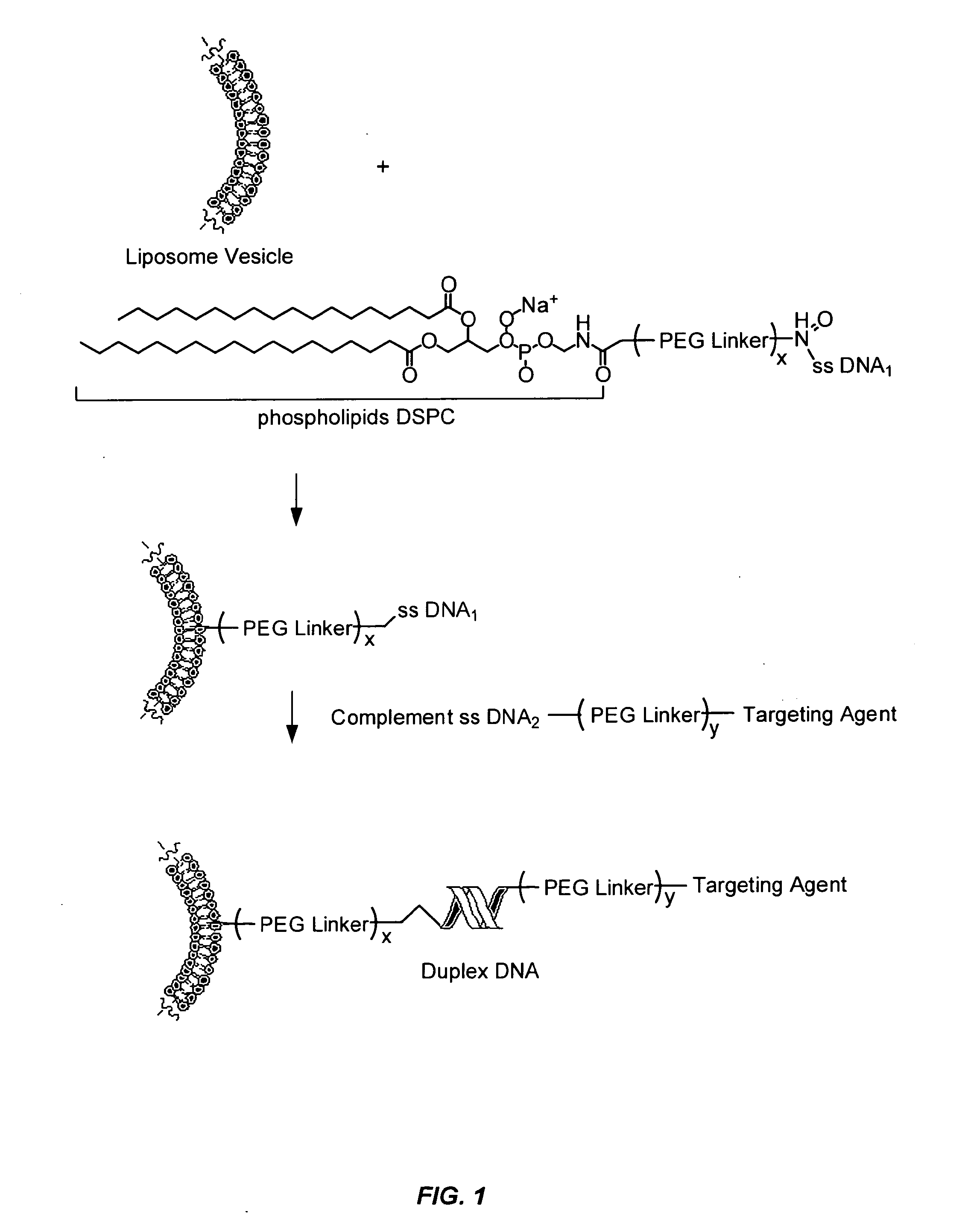
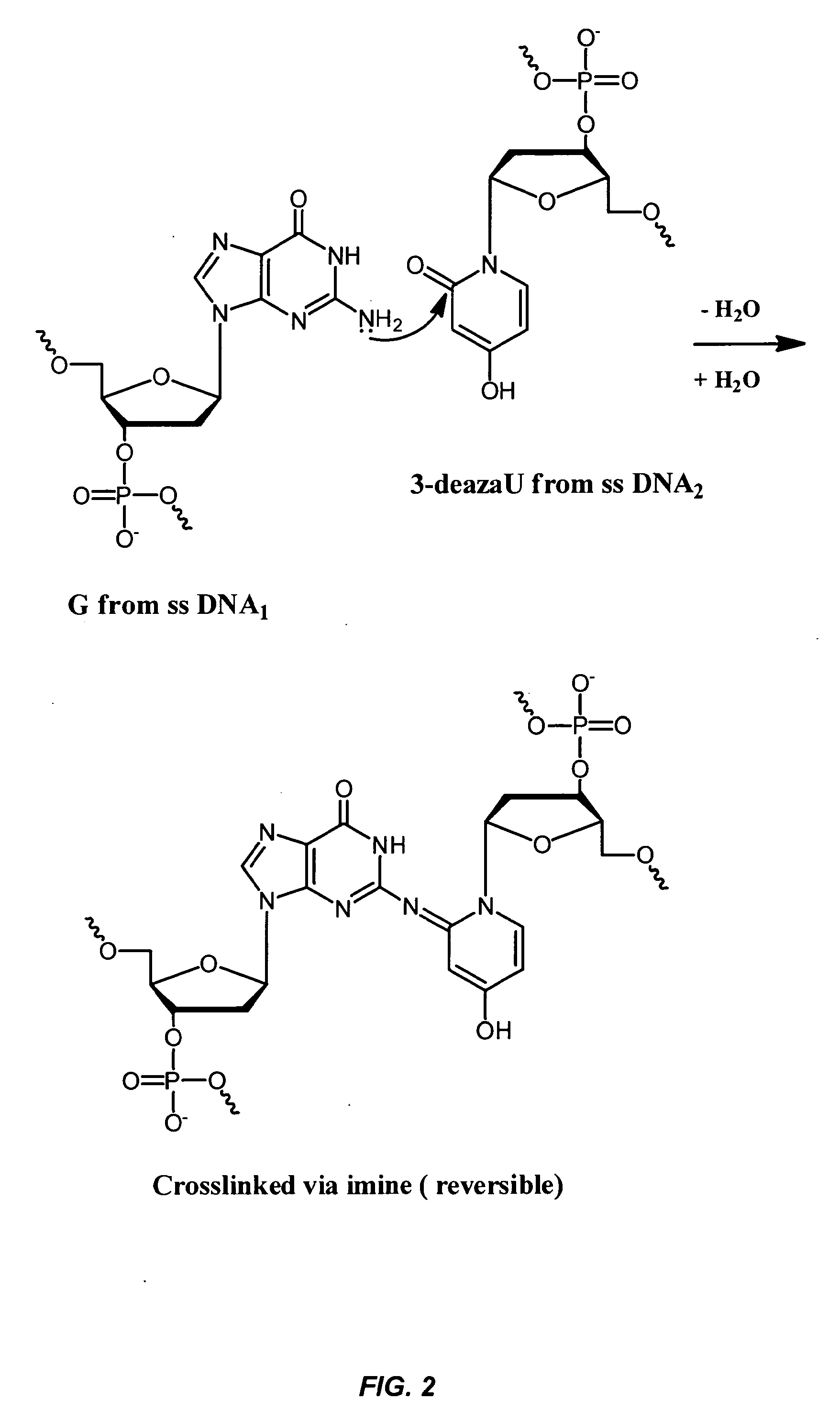
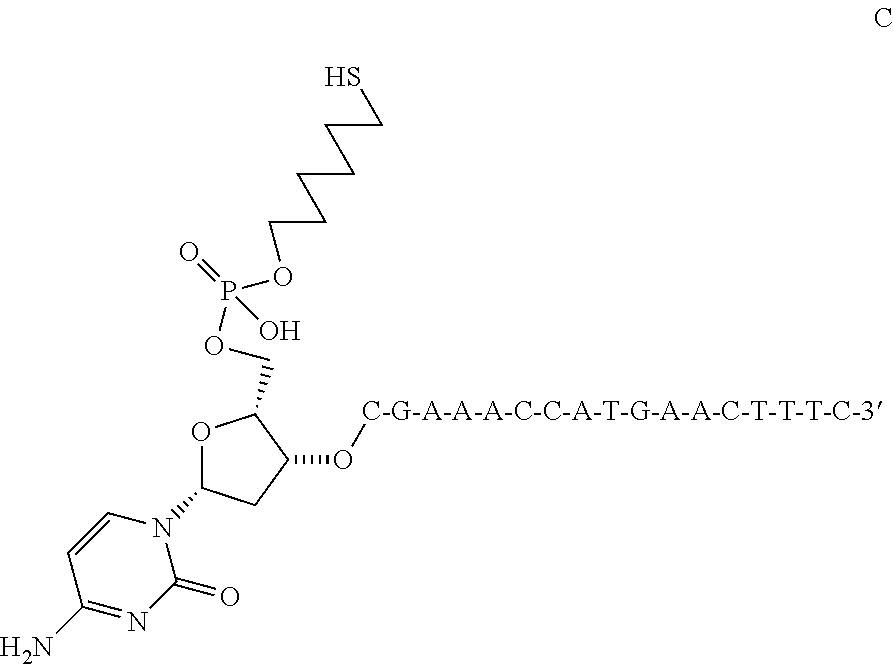
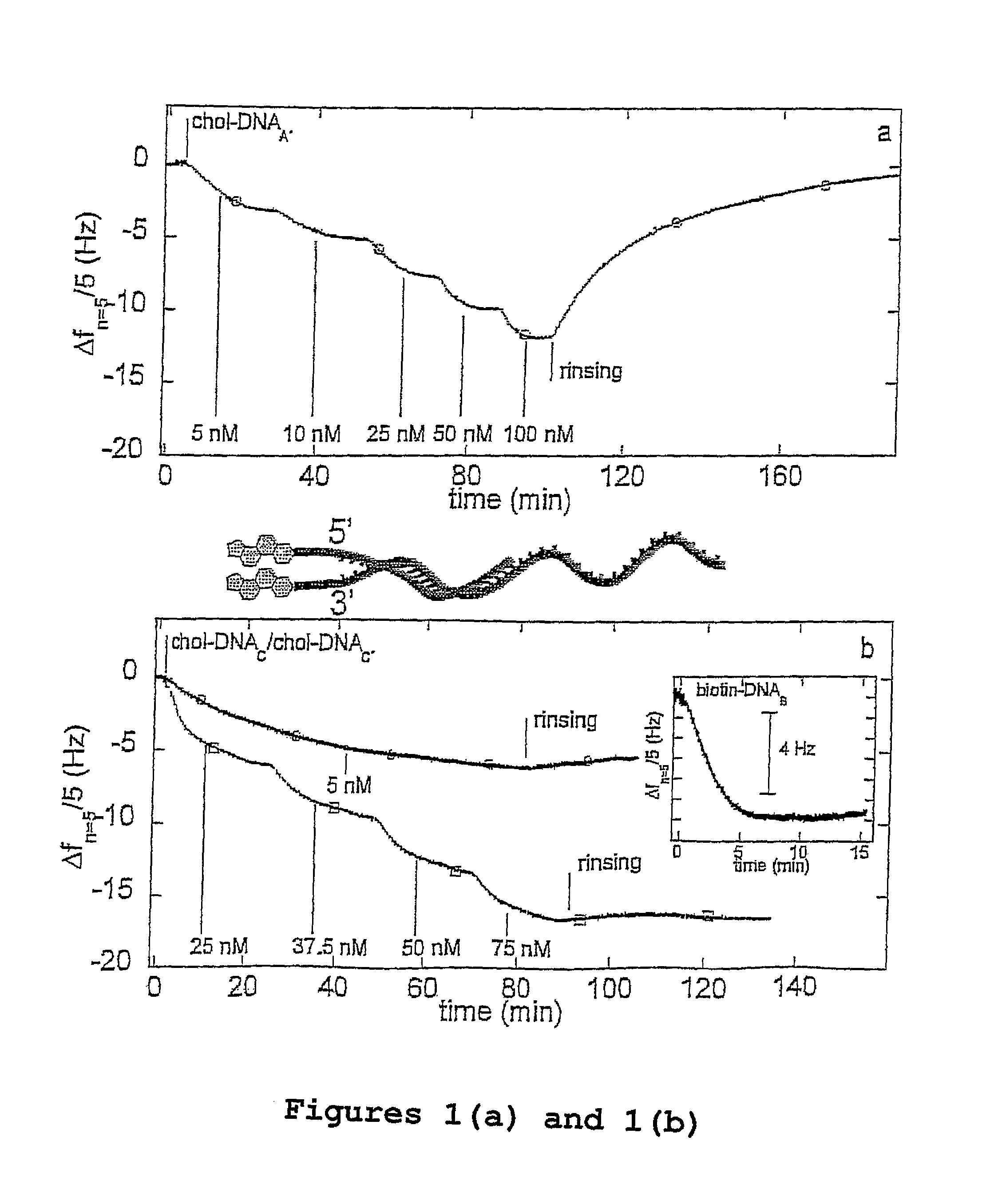
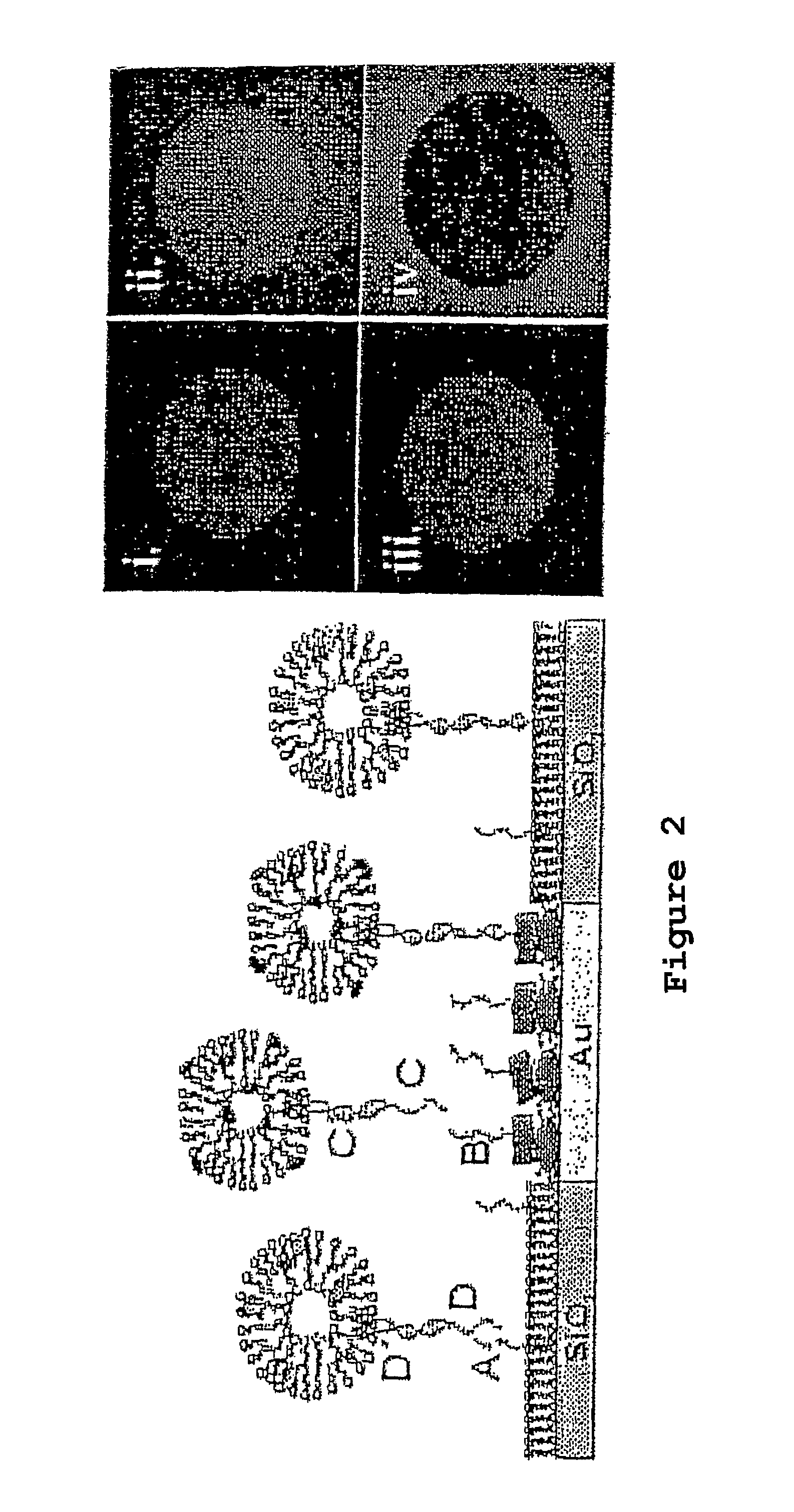
Oligonucleotides Related to Lipid Membrane Attachments
ActiveUS20070275047A1Strong couplingBioreactor/fermenter combinationsSequential/parallel process reactionsLipid formationCell Membrane Proteins
Oligonucleotide structures are provided that are capable of forming more stable bonds to a lipid membrane and thereby generate an improved control of the process whereby oligonucleotide linkers are introduced to lipid membranes. Methods of forming lipid membrane oligonucleotide attachments are provided including lipid vesicles. The oligonucleotides typically comprise at least two hydrophobic anchoring moieties capable of being attached to a lipid membrane. Said moieties may be attached at the terminal ends of an oligonucleotide or, in the case of a first and second strand forming a duplex, at the same terminal end one of the strands other end not being part of the duplex leaving it free to hybridize to additional strands. The lipid vesicles attached with the oligonucleotide can be used in biosensors and may contain membrane proteins.
Owner:BIO RAD LAB INC
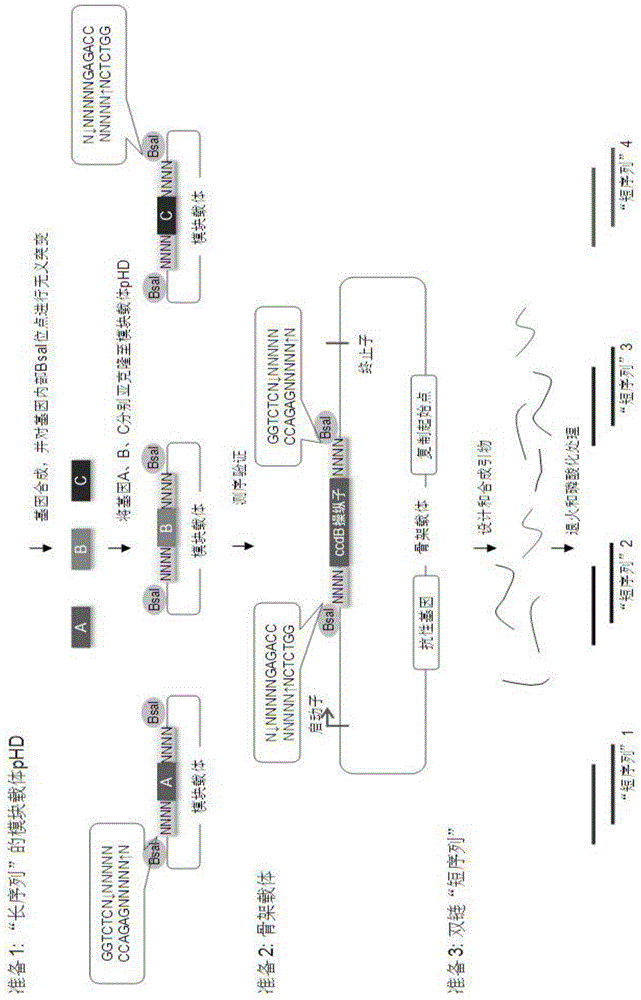
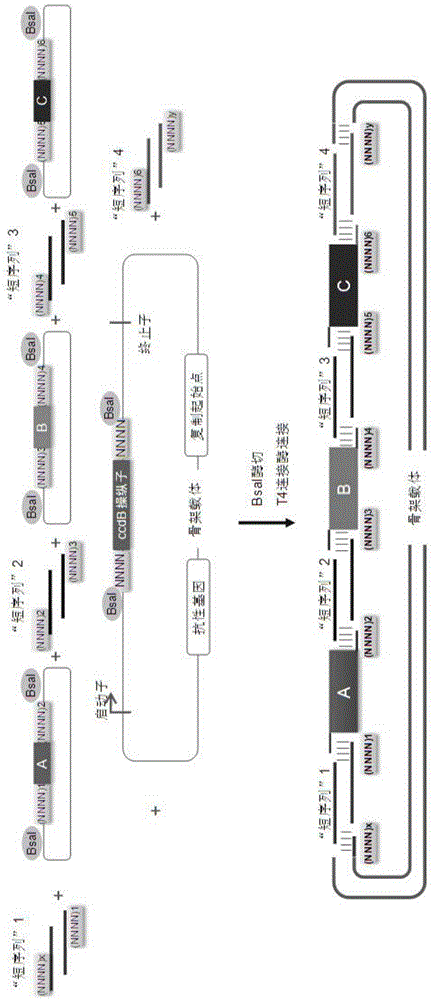
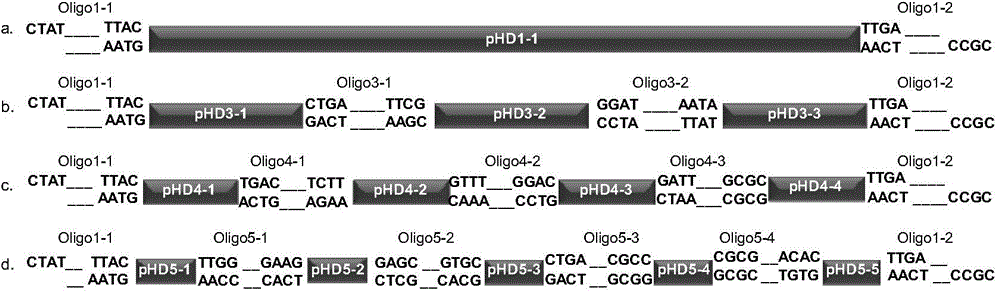
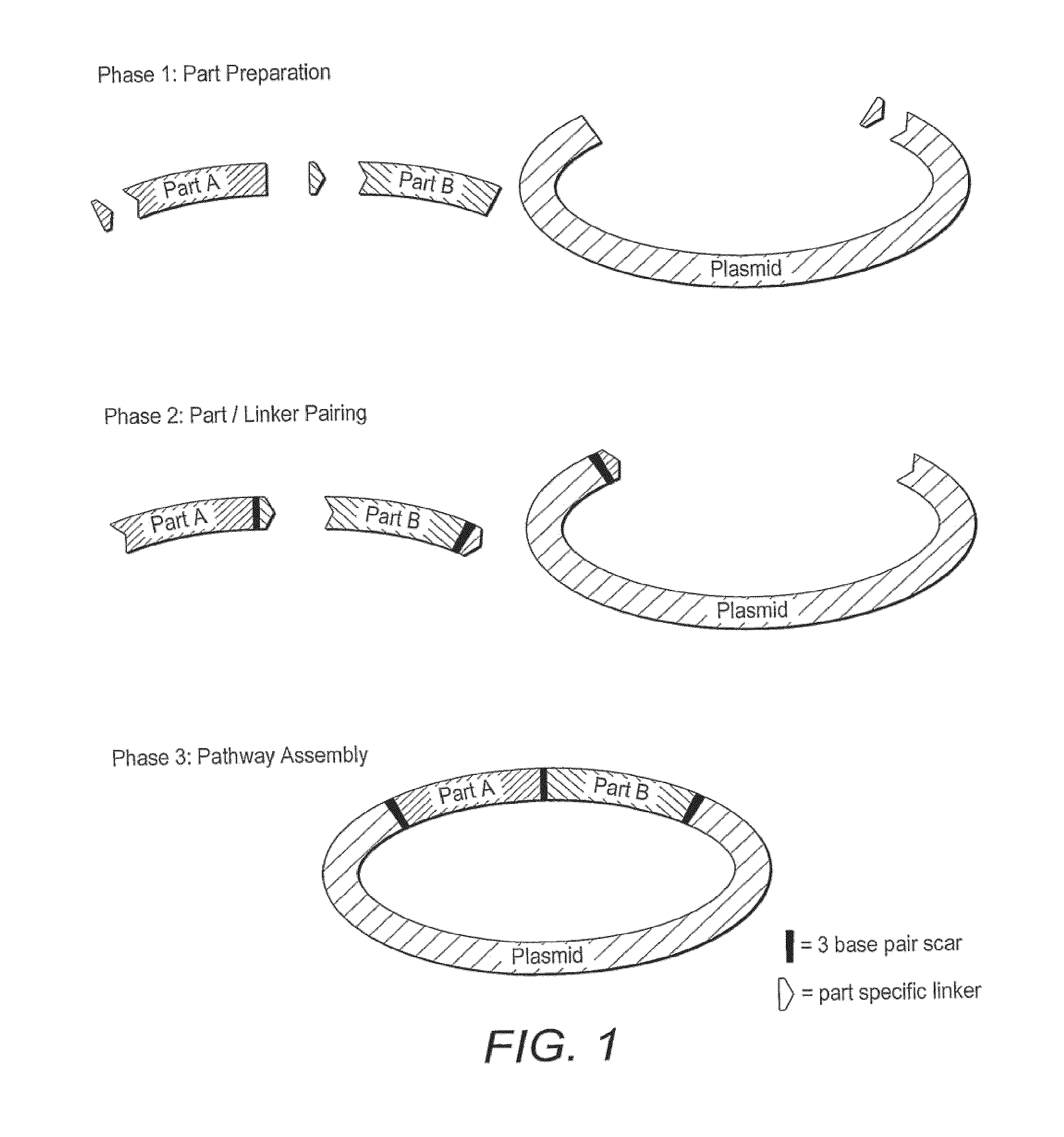
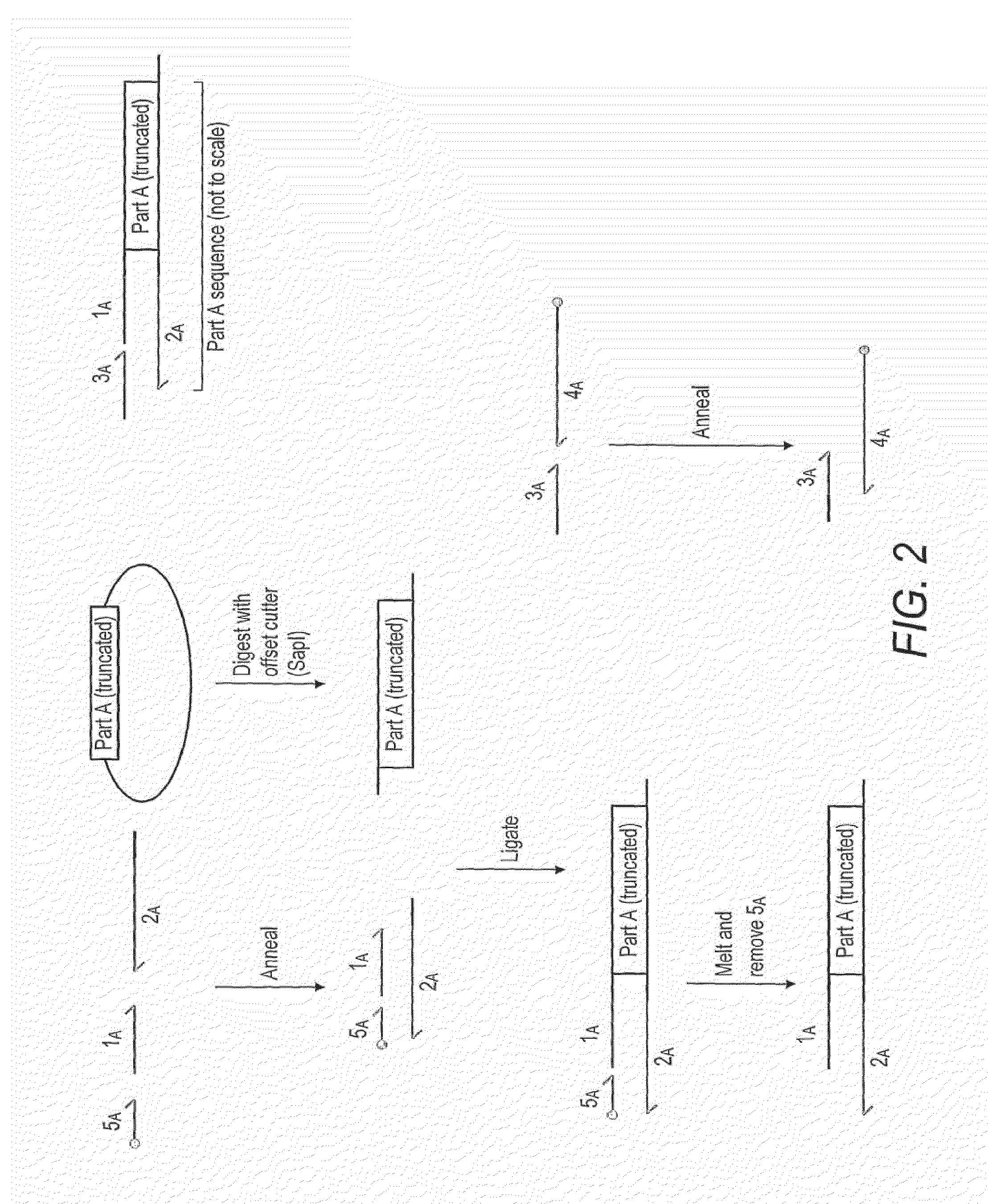
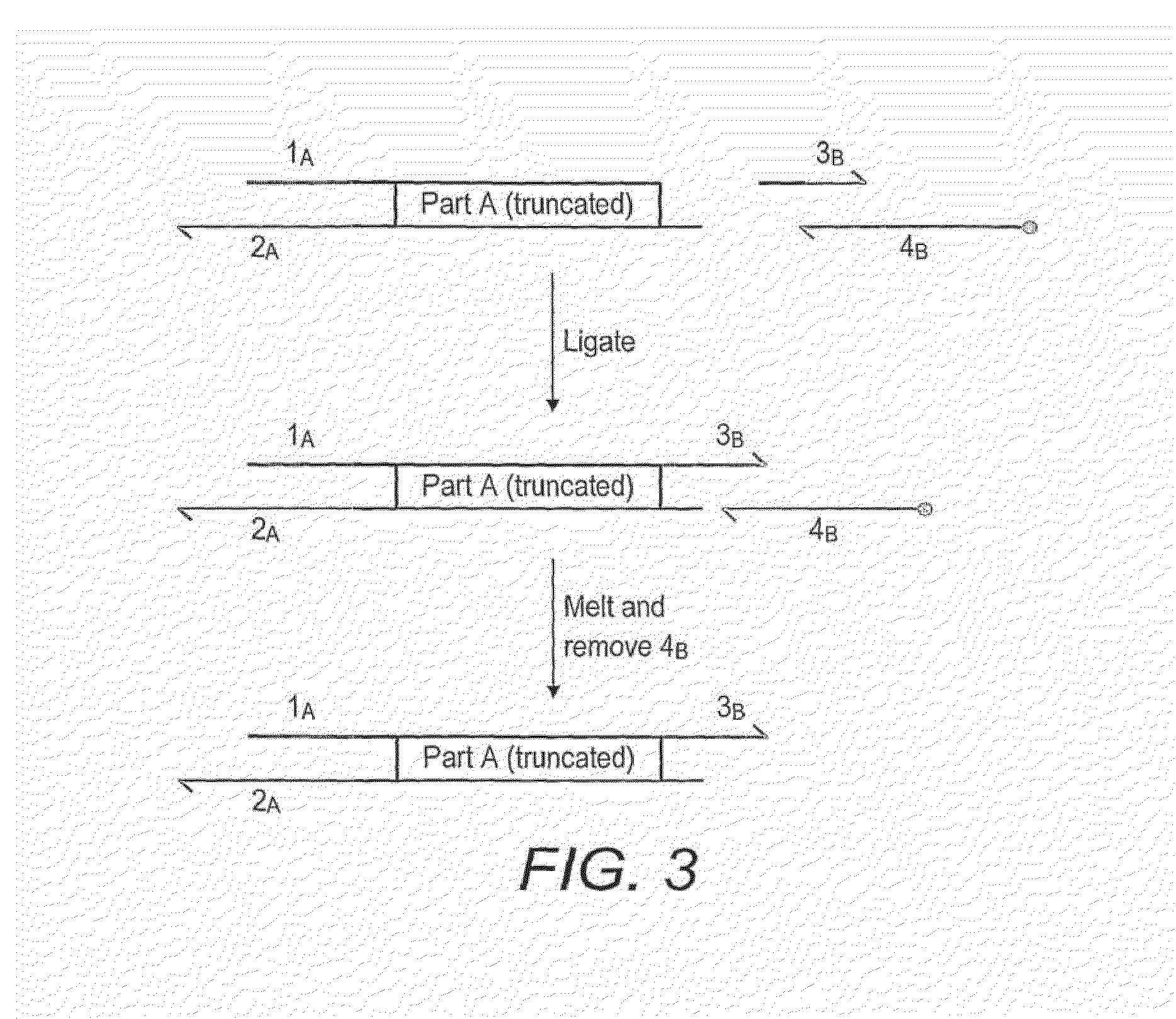
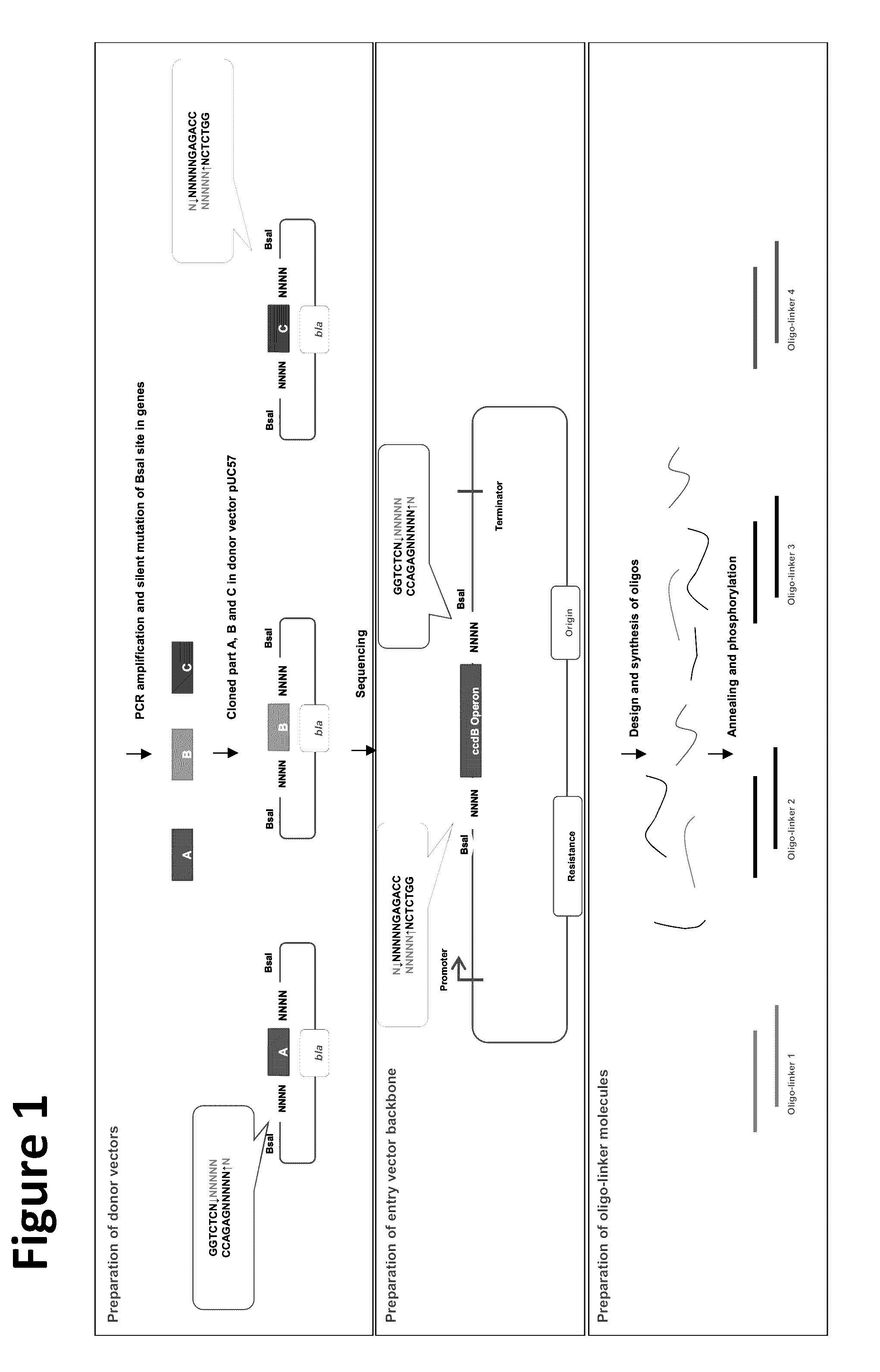
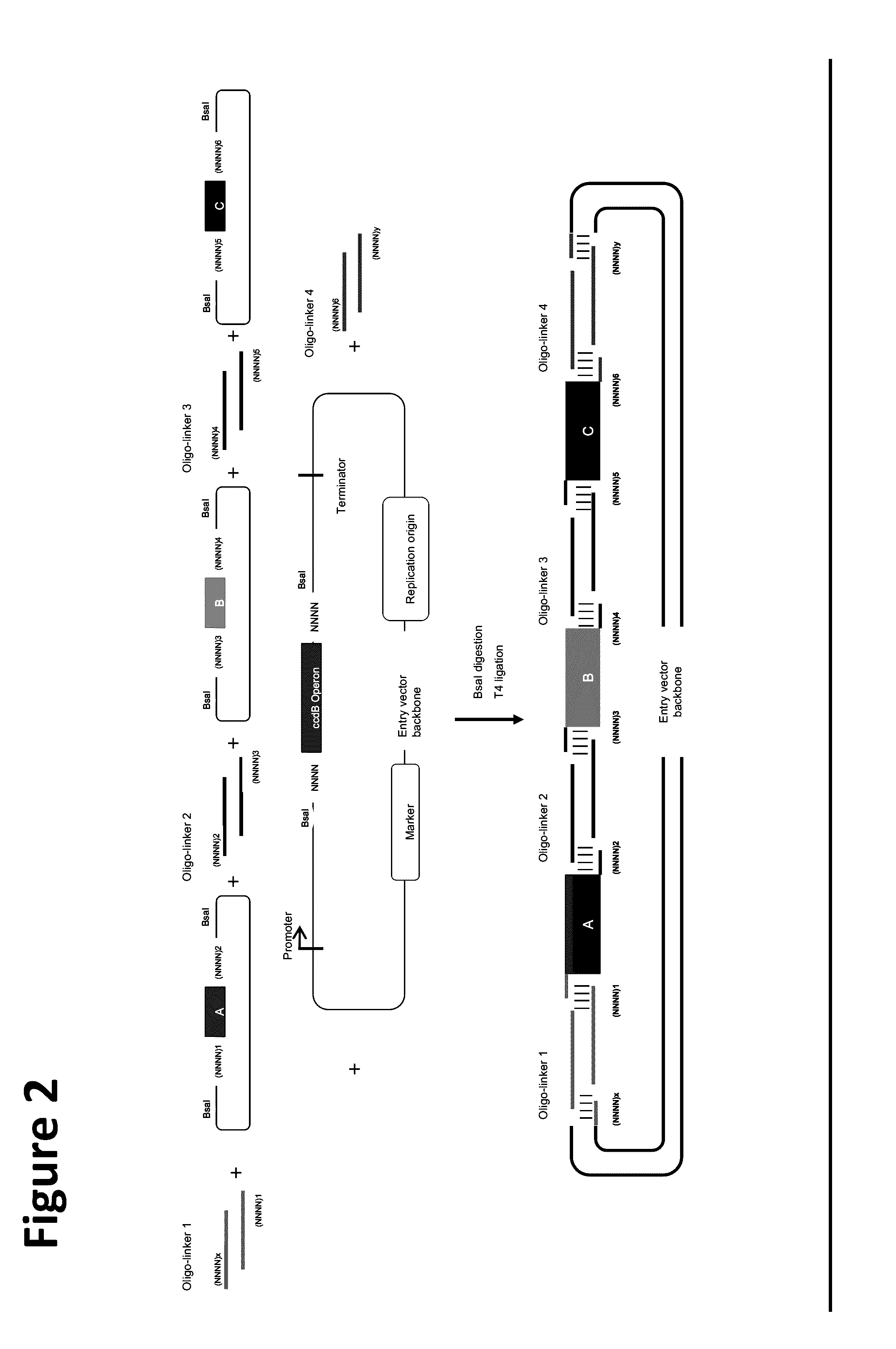
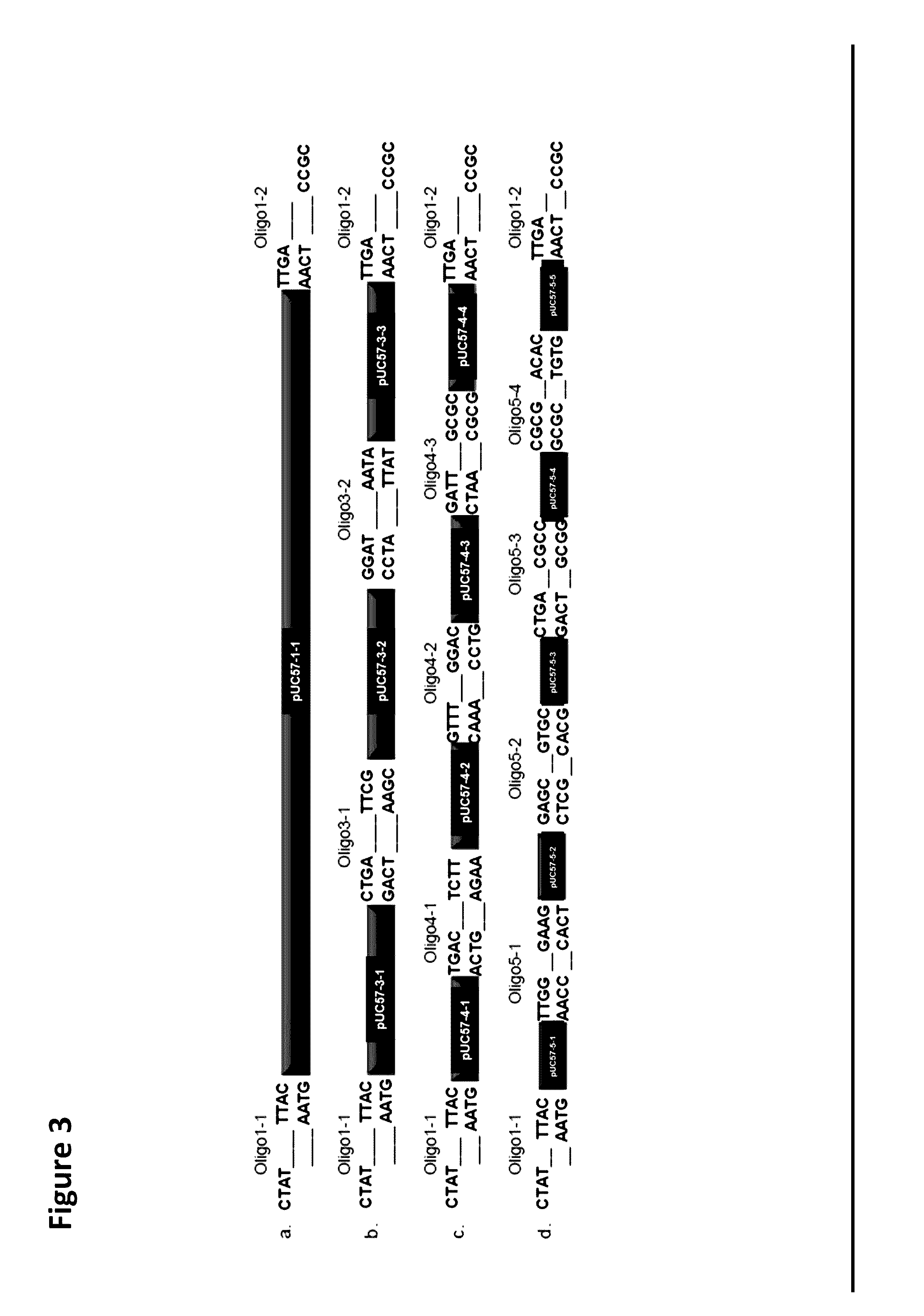
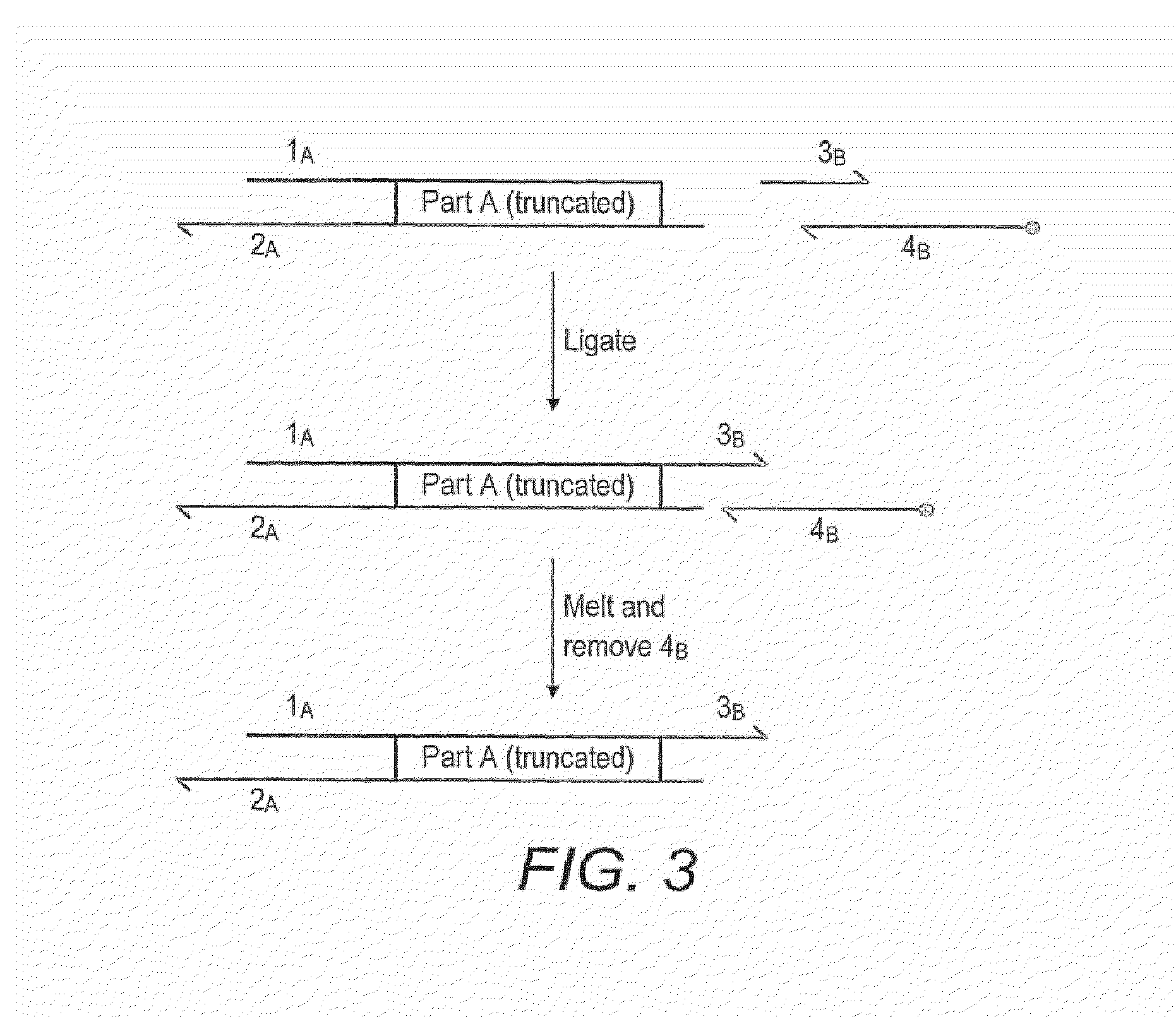
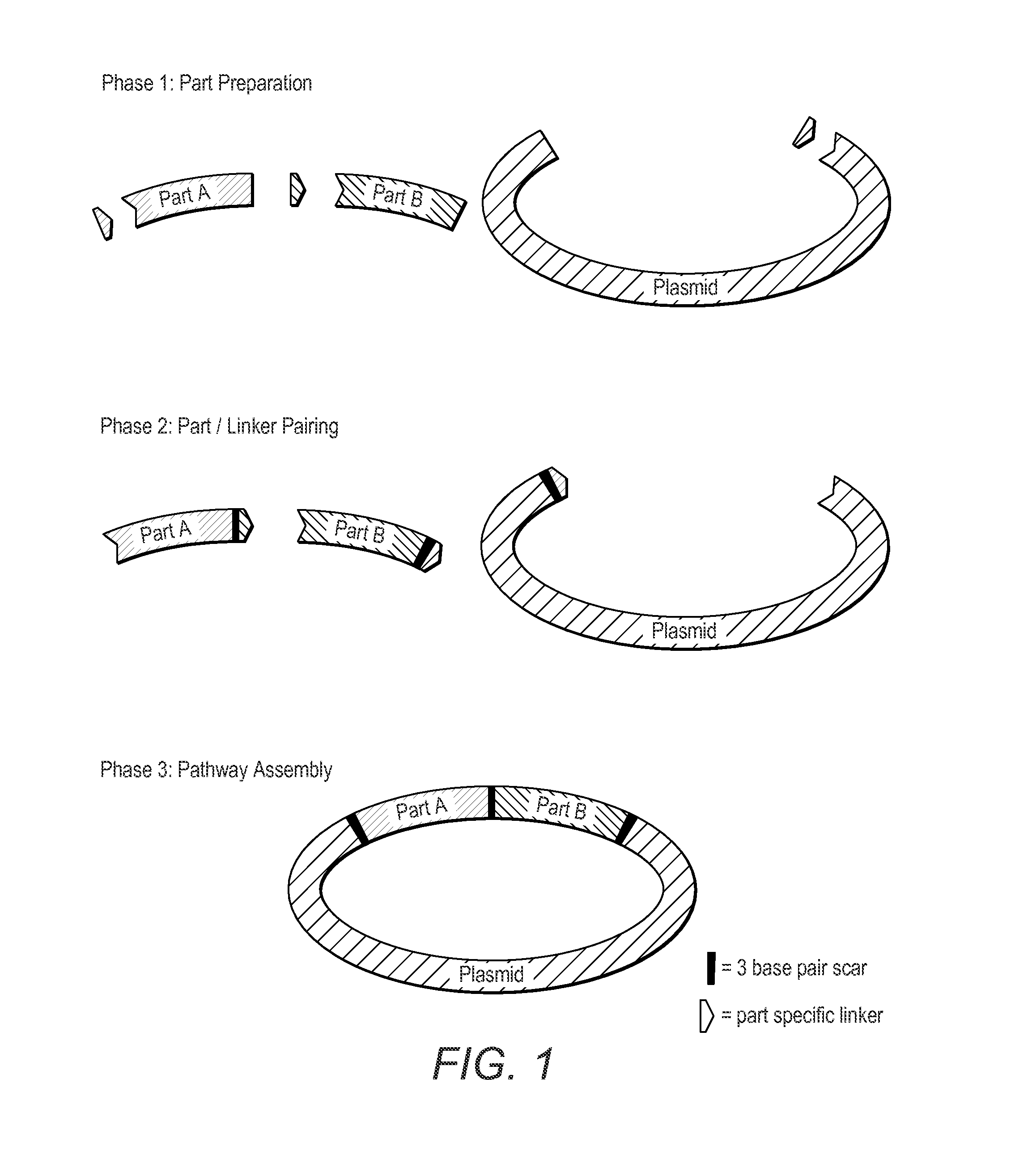
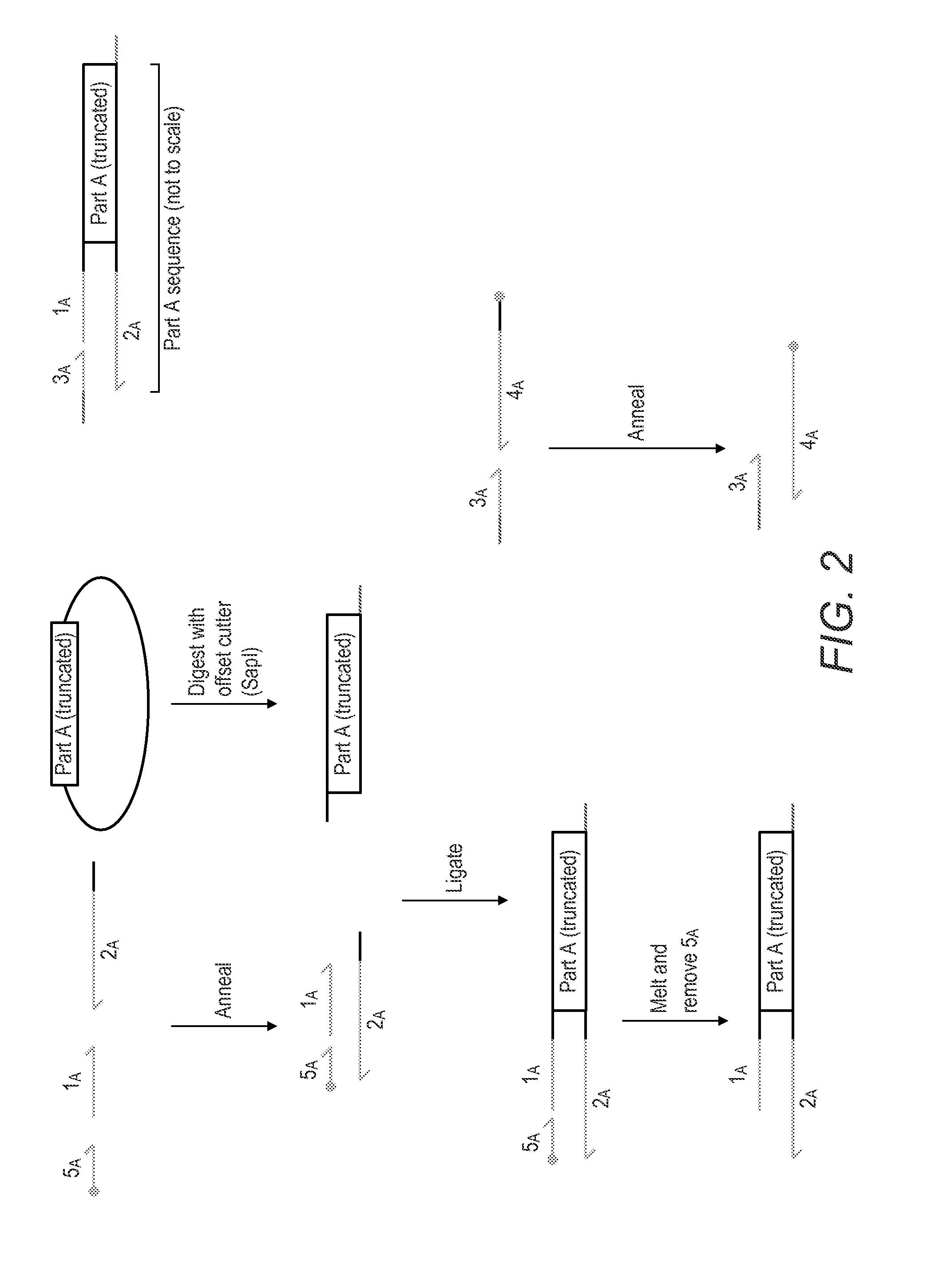
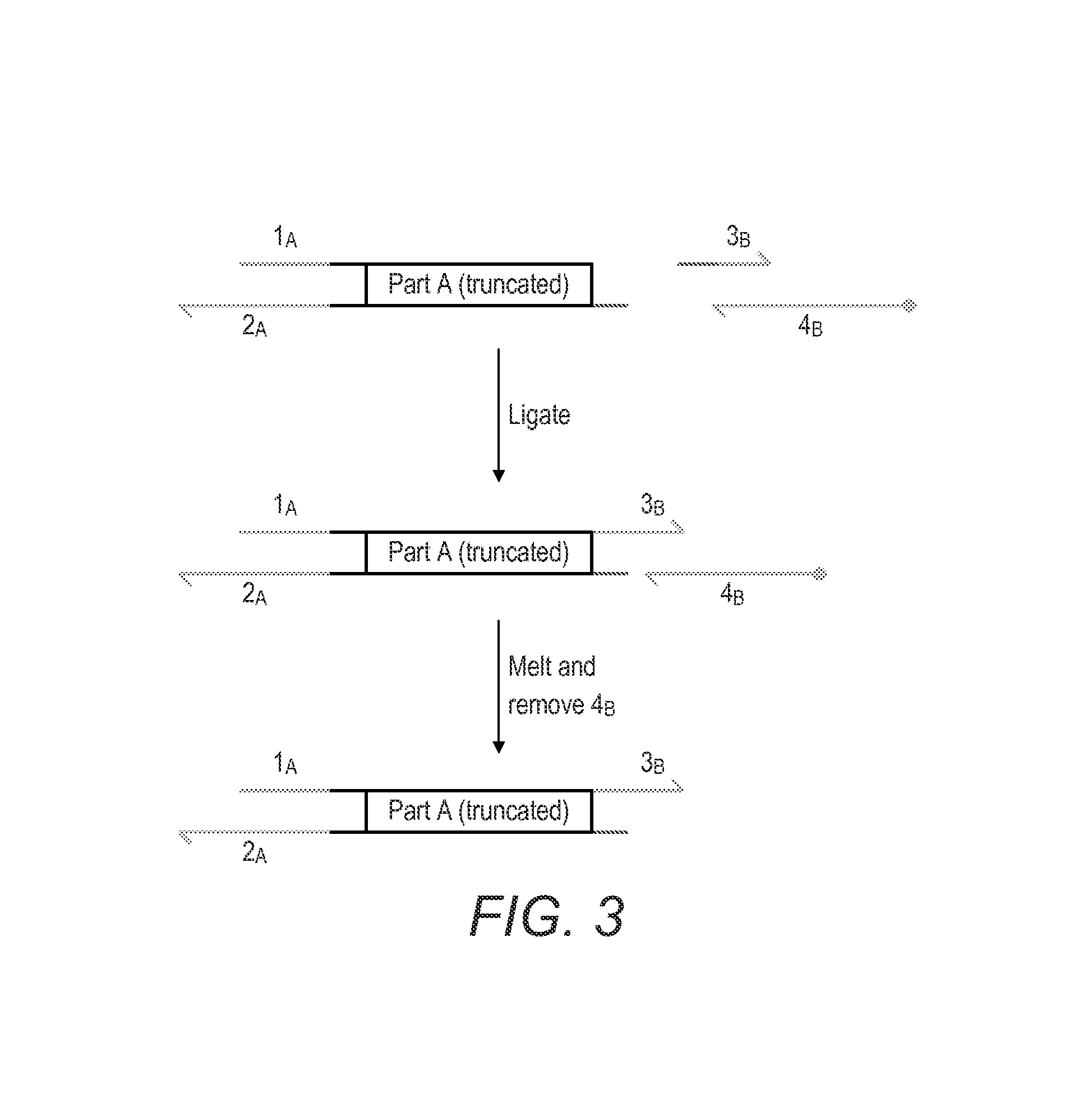
Oligonucleotide-linker-mediated DNA assembly method and application thereof
ActiveCN106282157ATo achieve the purpose of flux optimization pathwayAvoid residueVector-based foreign material introductionDNA preparationOperonBarcode
The invention discloses an oligonucleotide-linker-mediated DNA assembly method and an application thereof. The assembly method comprises steps of synthesis of a long sequence, preparation and assembly of a module carrier, remolding and assembly of a skeleton carrier, preparation of a short sequence, and enzyme-cut and link-up integration of the module carrier, the remolded skeleton carrier and the short sequence. According to the method, the short sequence is a regulating element such as a promoter and an RBS, and the long sequence is a replication initiation point of a gene or a carrier. The short sequence is designed to synthesize a double-stranded DNA chain, and can connect the long sequence and be used as a barcode sequence to determine the assembly sequence in an assembly process. The gene sequence in an operon is achieved by synthesizing short sequence with different cohesive ends. According to the method, multiple factors are allowed to participate in gene regulation at the same time. The method can be used to construct a combinatorial library of a biosynthesis channel, and a high-flux optimized channel is achieved.
Owner:NANJING GENSCRIPT BIOTECH CO LTD
Method and system for delivering nucleic acid into a target cell
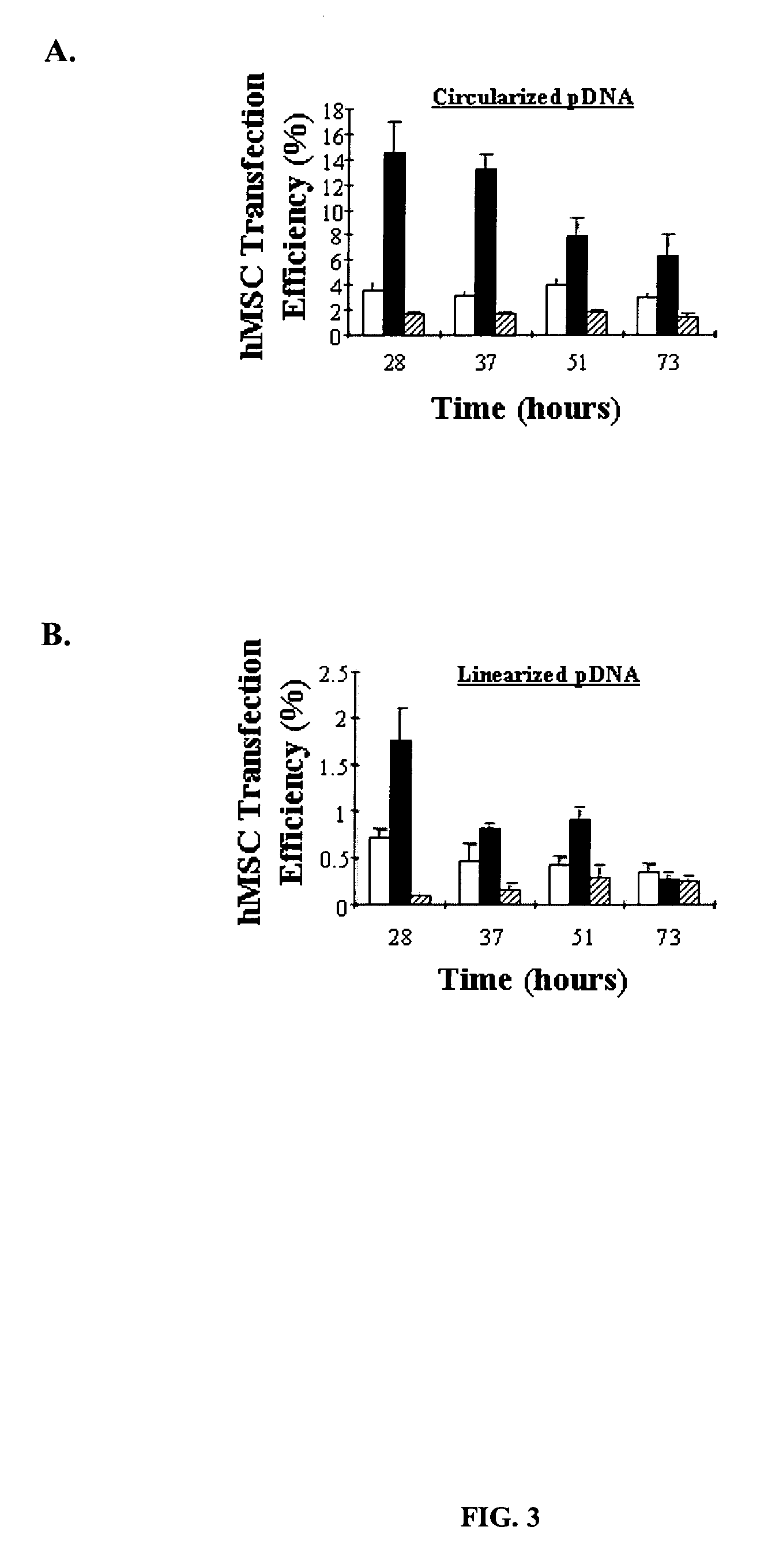
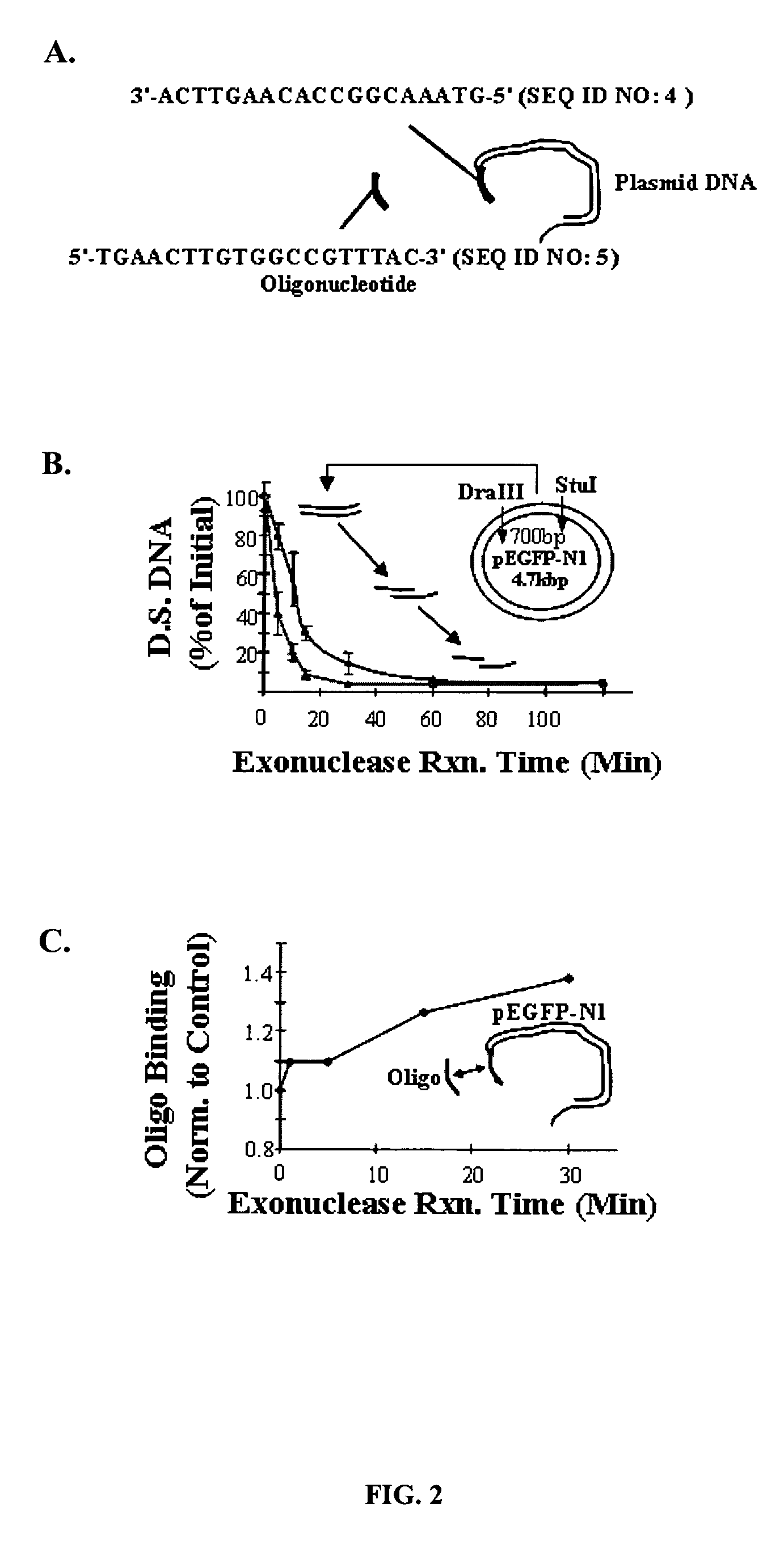
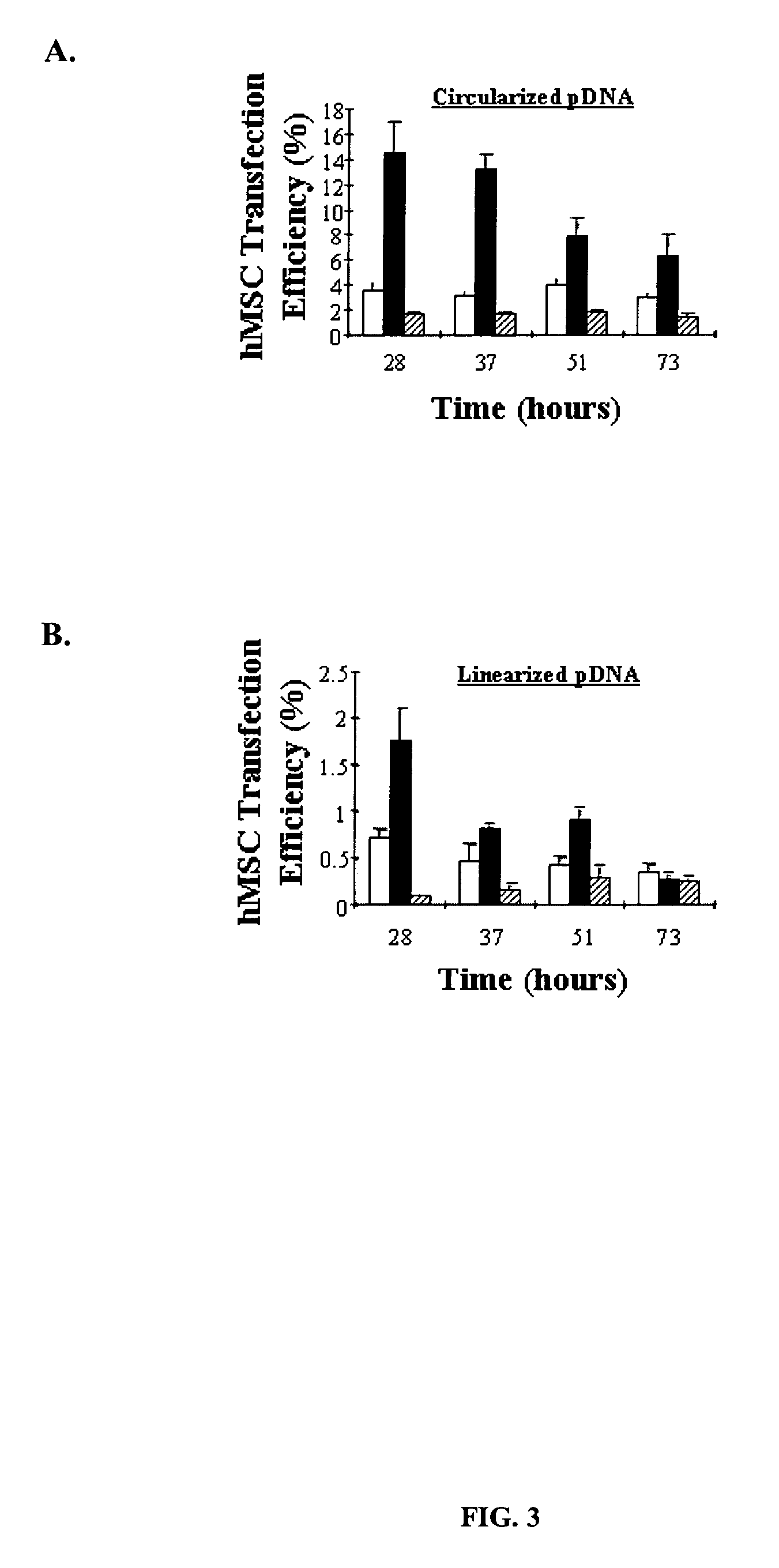
ActiveUS20070092906A1Easy to controlHigh transfection efficiencyBioreactor/fermenter combinationsMaterial nanotechnologyOligonucleotide LinkerOligonucleotide
Controlled delivery of nucleic acid into adhered of cells is achieved by immobilizing the nucleic acid and the cells to a substrate. Improved control over delivery is achieved by immobilizing the nucleic acid to the substrate via complimentary DNA binding interactions with an oligonucleotide linker.
Owner:WISCONSIN ALUMNI RES FOUND
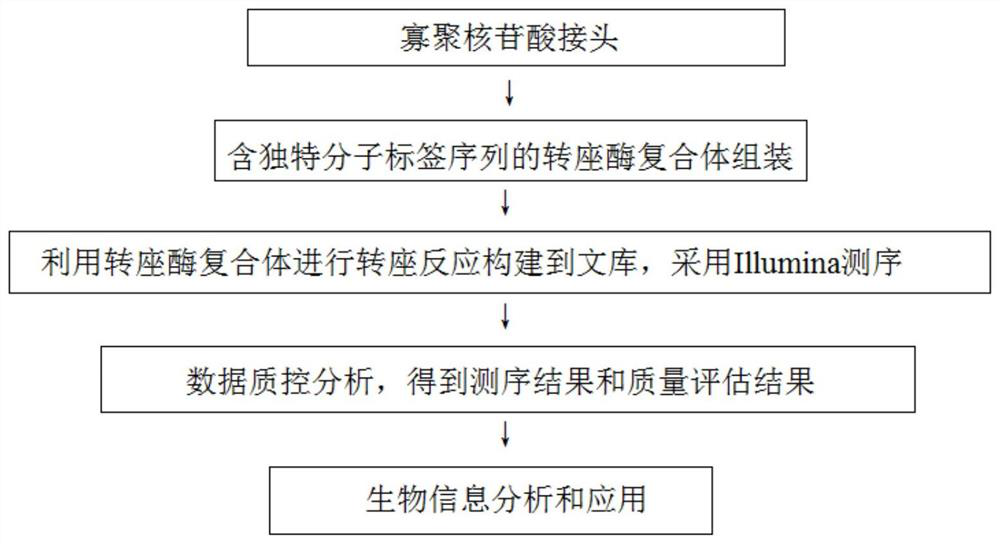
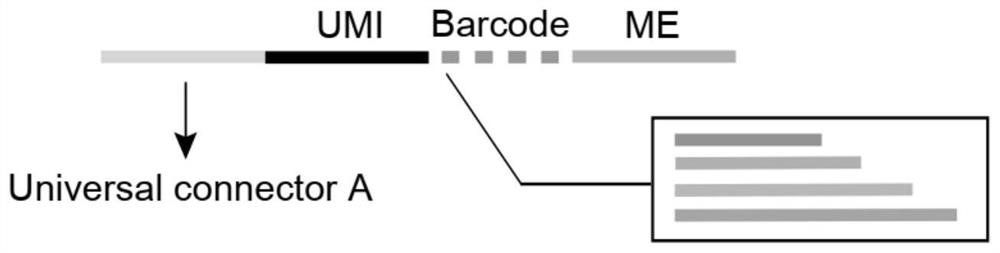
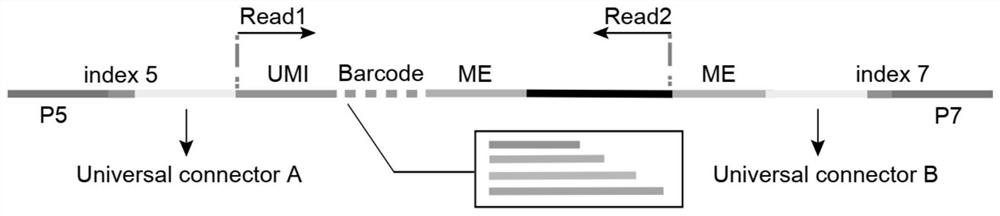
Transposase complex containing unique molecular identifier sequence and application of transposase complex
PendingCN112251422AIncrease profitEfficiently Distinguishing Repeated ReadsMicrobiological testing/measurementTransferasesInsertionProtide
The invention discloses a transposase complex containing a unique molecular identifier sequence and application of the transposase complex. The transposase complex is a complex formed by assembling oligonucleotide linkers and transposase. A next-generation sequencing library constructed by utilizing the transposase complex containing the UMI can effectively distinguish transposase insertion repeatevents and repeat reads caused by PCR, so that the utilization rate of sequencing data, the reliability of sequence variation and frequency identification of the sequence variation, the accuracy of gene expression and chromatin accessibility quantification, and the identification sensitivity of the footprint of the DNA binding protein are improved.
Owner:HUAZHONG AGRI UNIV
Method for assembly of polynucleic acid sequences
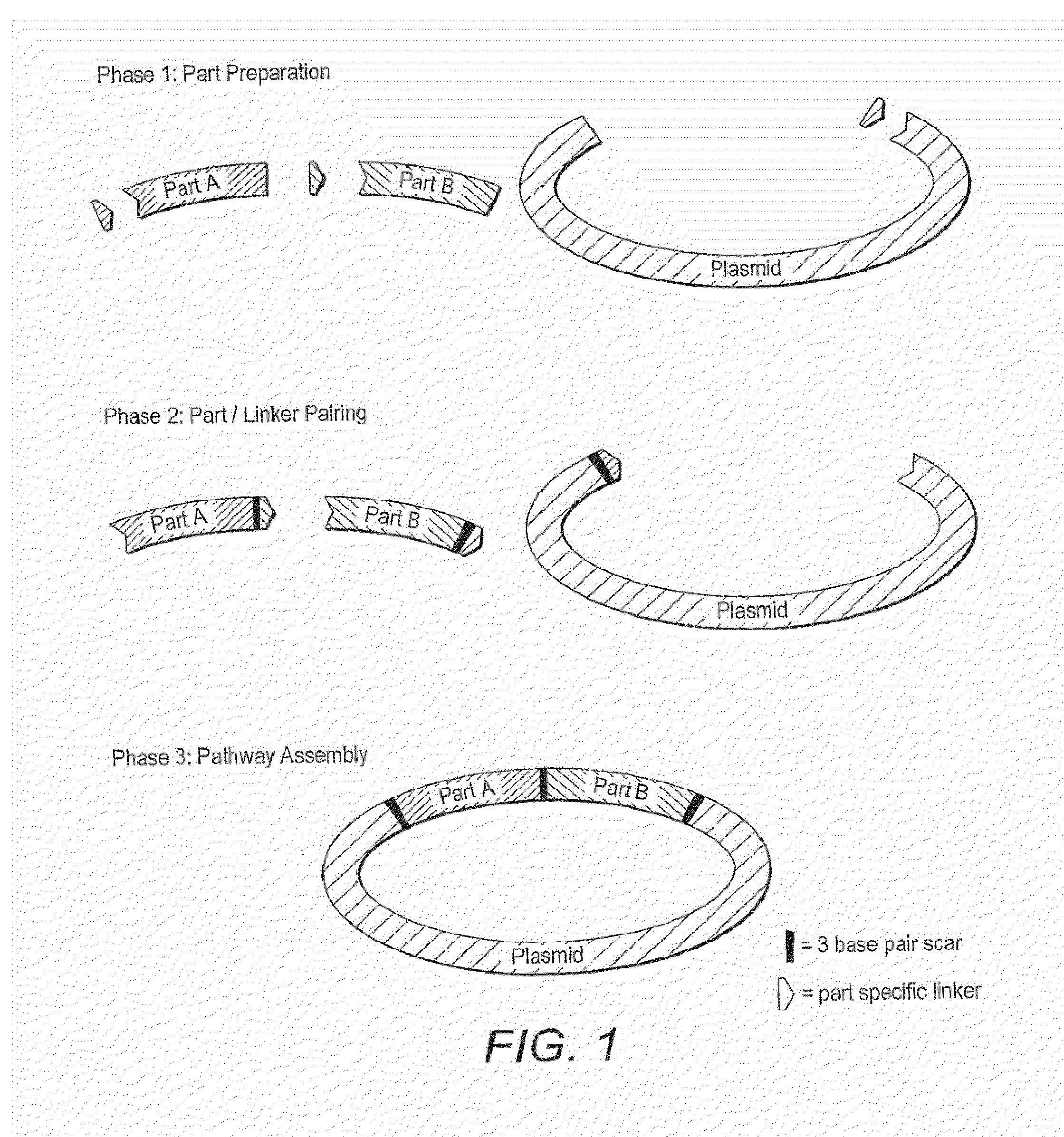
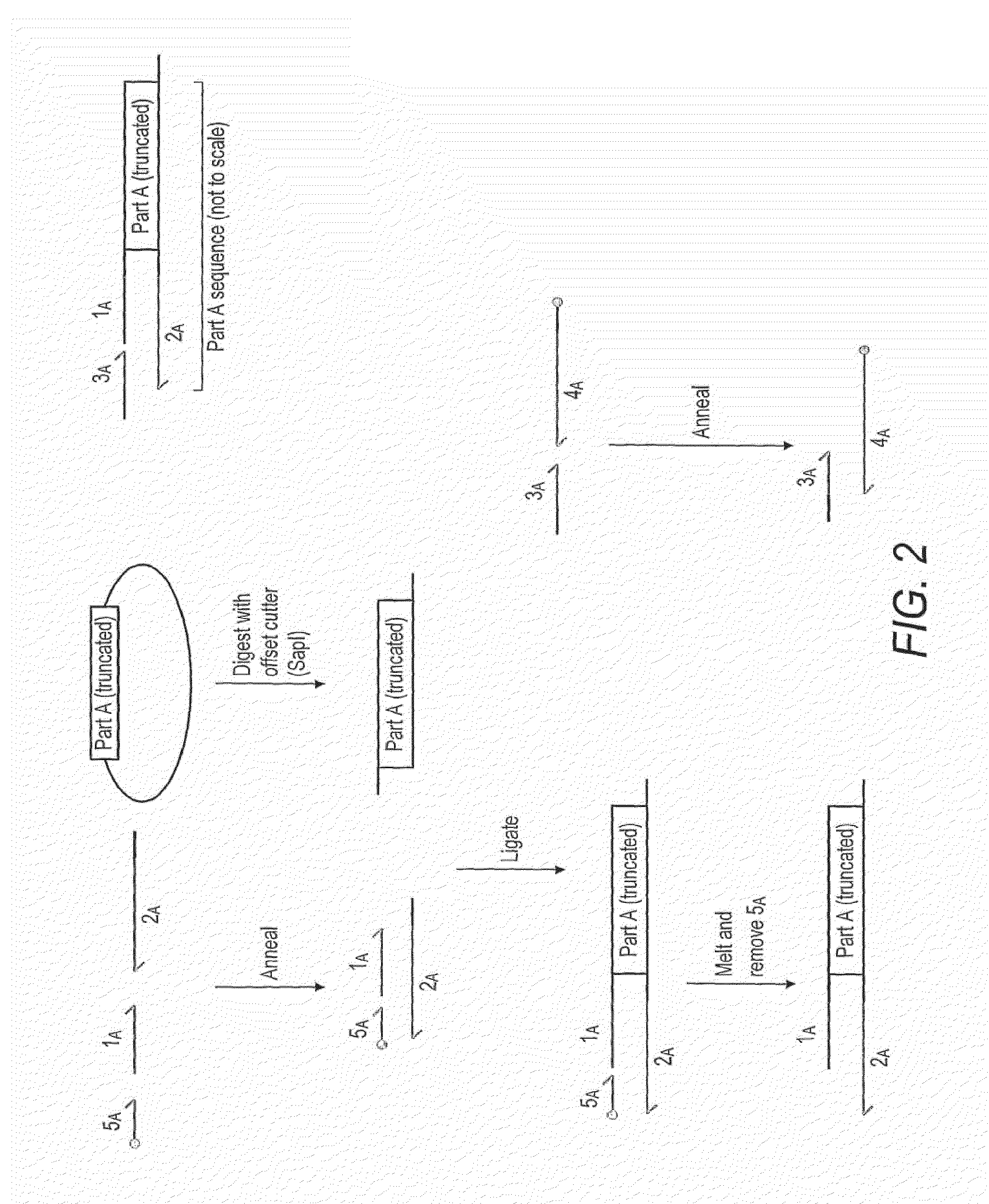
InactiveUS8999679B2Quicker and convenient to useSimple and quick assemblyTransferasesFermentationNucleic acid sequencingOligonucleotide Linker
The present invention provides a method for the assembly of a polynucleic acid sequence from a plurality of nucleic acid sequences in which the polynucleic acid sequence is of a formula Nn+1, in which N represents a nucleic acid sequence and where n is 1 or greater than 1 and each N may be the same or a different nucleic acid sequence, in which the method comprises: (i) providing a first nucleic acid sequence N1 which has an oligonucleotide linker sequence L13 at the 3′-end of the nucleic acid sequence; (ii) providing a second nucleic acid sequence N2 which optionally has an oligonucleotide linker sequence L23′ at the 3′-end of the nucleic acid sequence and which has an oligonucleotide linker sequence L25′ at the 5′-end of the nucleic acid sequence, wherein the 5′-end linker sequence L25′ of nucleic acid sequence N2 is complementary to the 3′-end linker sequence L13′ of nucleic acid sequence N1; (iii) optionally providing one or more additional nucleic acid sequences N, wherein nucleic acid sequence N2 has an oligonucleotide linker sequence L23′ at the 3′-end of the nucleic acid sequence, and wherein said one or more additional nucleic acid sequences N comprises a terminal additional nucleic acid sequence NZ, and wherein each additional nucleic acid sequence N has an oligonucleotide linker sequence at its 3′-end, wherein said terminal additional nucleic acid sequence NZ optionally lacks an oligonucleotide linker sequence at its 3′-end and wherein each additional nucleic acid sequence N has an oligonucleotide linker sequence at its 5′-end, wherein for the first additional nucleic acid sequence N3 the 5′-end linker sequence L35′ is complementary to the 3′-end linker sequence L23′ of nucleic acid sequence N2 and for each second and subsequent additional nucleic acid sequence N the 5′-end linker sequence is complementary to the 3′-end linker sequence of the respective preceding additional nucleic acid sequence; and (iv) ligating said nucleic acid sequences to form said polynucleic acid sequence.
Owner:ITI SCOTLAND
Methods and compositions for detection of small RNAs
Currently, the circularization of small RNAs is broadly regarded as an obstacle in ligation-related assays and explicitly avoided while short lengths of linear RNA targets is broadly recognized as a factor limiting use of conventional primers in PCR-related assays. In contrast, the disclosed invention capitalizes on circularization of small RNA targets or their conjugates with oligonucleotide adapters. The circular RNA templates provide amplification of the target sequences via synthesis of multimer nucleic acids that can be either labeled for direct detection or subjected to PCR amplification and detection. Structure of small circular RNAs and corresponding multimeric nucleic acids provide certain advantages over current methods including flexibility in design of conventional RT and PCR primers as well as use of 5′-overlapping dimer-primers for efficient and sequence-specific amplification of short target sequences. Our invention also reduces number of steps and reagents while increasing sensitivity and accuracy of detection of small RNAs with both 2′OH and 2′-OMe at their 3′ ends. Our invention increase sensitivity and specificity of detection of microRNAs and other small RNAs with both 2′OH and 2′-OMe at their 3′ ends while allowing us to distinguish these two forms from each other.
Owner:REALSEQ BIOSCI INC
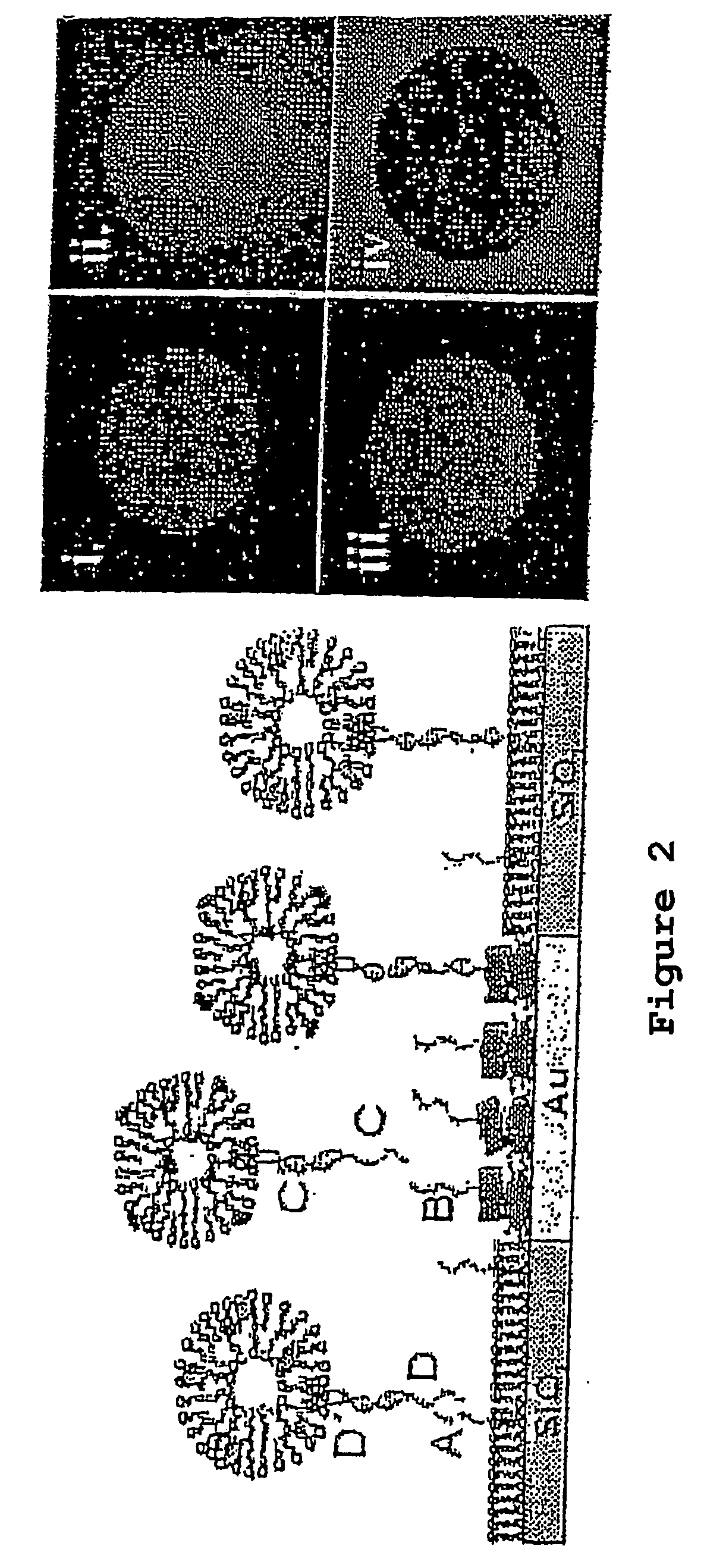
Remote assembly of targeted nanoparticles using complementary oligonucleotide linkers
InactiveUS20140234217A1Ultrasonic/sonic/infrasonic diagnosticsGenetic material ingredientsDiseaseOligonucleotide Linker
The present invention provides targeted delivery compositions and their methods of use in treating and diagnosing a disease state in a subject. Components of the targeted delivery compositions are put together through duplex formation between oligonucleotides.
Owner:MALLINCKRODT INC
Oligonucleotides related to lipid membrane attachments
ActiveUS8389707B2Strong couplingSequential/parallel process reactionsSugar derivativesLipid formationOligonucleotide Linker
Oligonucleotide structures are provided that are capable of forming more stable bonds to a lipid membrane and thereby generate an improved control of the process whereby oligonucleotide linkers are introduced to lipid membranes. Methods of forming lipid membrane oligonucleotide attachments are provided including lipid vesicles. The oligonucleotides typically comprise at least two hydrophobic anchoring moieties capable of being attached to a lipid membrane. Said moieties may be attached at the terminal ends of an oligonucleotide or, in the case of a first and second strand forming a duplex, at the same terminal end one of the strands other end not being part of the duplex leaving it free to hybridize to additional strands. The lipid vesicles attached with the oligonucleotide can be used in biosensors and may contain membrane proteins.
Owner:BIO RAD LAB INC
Immobilized Transposase Complexes For DNA Fragmentation And Tagging
ActiveUS20170058326A1Superior property and characteristicHigh activityHydrolasesMicrobiological testing/measurementDna breakageDNA fragmentation
Owner:AGILENT TECH INC
Novel oligo-linker-mediated DNA assembly method and applications thereof
InactiveUS20160340670A1Efficient assemblyAvoid introducingNucleic acid vectorVector-based foreign material introductionBiological pathwayOligonucleotide Linker
A method for generating a library of expression vectors comprising a plurality of donor sequences and a plurality of oligo-linker nucleic acids, termed Oligonucleotide Linker-Mediated DNA Assembly (OLMA), is described. Also described are applications of the OLMA method, including the simultaneous tuning of several factors in metabolic and biological pathways, and the combinatorial high throughput optimization of metabolic and biological pathways.
Owner:NANJING GENSCRIPT BIOTECH CO LTD
High throughput digital karyotyping for biome characterization
InactiveUS20130267428A1Nucleotide librariesMicrobiological testing/measurementA-DNAOligonucleotide Linker
The invention herein describes a method for identifying a DNA sequence, and oligonucleotide adaptors used in the identification of a DNA sequence.
Owner:WASHINGTON UNIV & ST LOUIS OFFICE OF TECH MANAGEMENT +1
Method For Assembly Of Polynucleic Acid Sequences
ActiveUS20120040870A1Quicker and convenient to useSimple and quick assemblyFermentationVector-based foreign material introductionOligonucleotideOligonucleotide Linker
The present invention provides a method for the assembly of a polynucleic acid sequence from a plurality of nucleic acid sequences in which the polynucleic acid sequence is of a formula Nn+1, in which N represents a nucleic acid sequence and where n is 1 or greater than 1 and each N may be the same or a different nucleic acid sequence, in which the method comprises: (i) providing a first nucleic acid sequence N1 which has an oligonucleotide linker sequence L13 at the 3′-end of the nucleic acid sequence; (ii) providing a second nucleic acid sequence N2 which optionally has an oligonucleotide linker sequence L23′ at the 3′-end of the nucleic acid sequence and which has an oligonucleotide linker sequence L25′ at the 5′-end of the nucleic acid sequence, wherein the 5′-end linker sequence L25′ of nucleic acid sequence N2 is complementary to the 3′-end linker sequence L13′ of nucleic acid sequence N1; (iii) optionally providing one or more additional nucleic acid sequences N, wherein nucleic acid sequence N2 has an oligonucleotide linker sequence L23′ at the 3′-end of the nucleic acid sequence, and wherein said one or more additional nucleic acid sequences N comprises a terminal additional nucleic acid sequence NZ, and wherein each additional nucleic acid sequence N has an oligonucleotide linker sequence at its 3′-end, wherein said terminal additional nucleic acid sequence NZ optionally lacks an oligonucleotide linker sequence at its 3′-end and wherein each additional nucleic acid sequence N has an oligonucleotide linker sequence at its 5′-end, wherein for the first additional nucleic acid sequence N3 the 5′-end linker sequence L35′ is complementary to the 3′-end linker sequence L23′ of nucleic acid sequence N2 and for each second and subsequent additional nucleic acid sequence N the 5′-end linker sequence is complementary to the 3′-end linker sequence of the respective preceding additional nucleic acid sequence; and (iv) ligating said nucleic acid sequences to form said polynucleic acid sequence.
Owner:ITI SCOTLAND
Method And Device
ActiveUS20140030766A1To overcome the large delayReduce error rateBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid sequencingOligonucleotide Linker
The present invention provides a method for the assembly of a polynucleic acid sequence from a plurality of nucleic acid sequences in which the polynucleic acid sequence is of a formula Nn+1, in which N represents a nucleic acid sequence and where n is 1 or greater than 1 and each N may be the same or a different nucleic acid sequence, in which the method comprises:(i) providing a first nucleic acid sequence N1 which has an oligonucleotide linker sequence L13′ at the 3′-end of the nucleic acid sequence;(ii) providing a second nucleic acid sequence N2 which optionally has an oligonucleotide linker sequence L23′ at the 3′-end of the nucleic acid sequence and which has an oligonucleotide linker sequence L25′ at the 5′-end of the nucleic acid sequence,wherein the 5′-end linker sequence L25′ of nucleic acid sequence N2 is complementary to the 3′-end linker sequence L13′ of nucleic acid sequence N1;(iii) optionally providing one or more additional nucleic acid sequences N, wherein nucleic acid sequence N2 has an oligonucleotide linker sequence L23′ at the 3′-end of the nucleic acid sequence, and wherein said one or more additional nucleic acid sequences N comprises a terminal additional nucleic acid sequence NZ, and wherein each additional nucleic acid sequence N has an oligonucleotide linker sequence at its 3′-end, wherein said terminal additional nucleic acid sequence NZ optionally lacks an oligonucleotide linker sequence at its 3′-end and wherein each additional nucleic acid sequence N has an oligonucleotide linker sequence at its 5′-end,wherein for the first additional nucleic acid sequence N3 the 5′-end linker sequence L35′ is complementary to the 3′-end linker sequence L23′ of nucleic acid sequence N2 and for each second and subsequent additional nucleic acid sequence N the 5′-end linker sequence is complementary to the 3′-end linker sequence of the respective preceding additional nucleic acid sequence; and(iv) ligating said nucleic acid sequences to form said polynucleic acid sequence;wherein at least step (iv) is carried out on a microfluidic device.
Owner:ITI SCOTLAND
Epigenetic profiling using targeted chromatin ligation
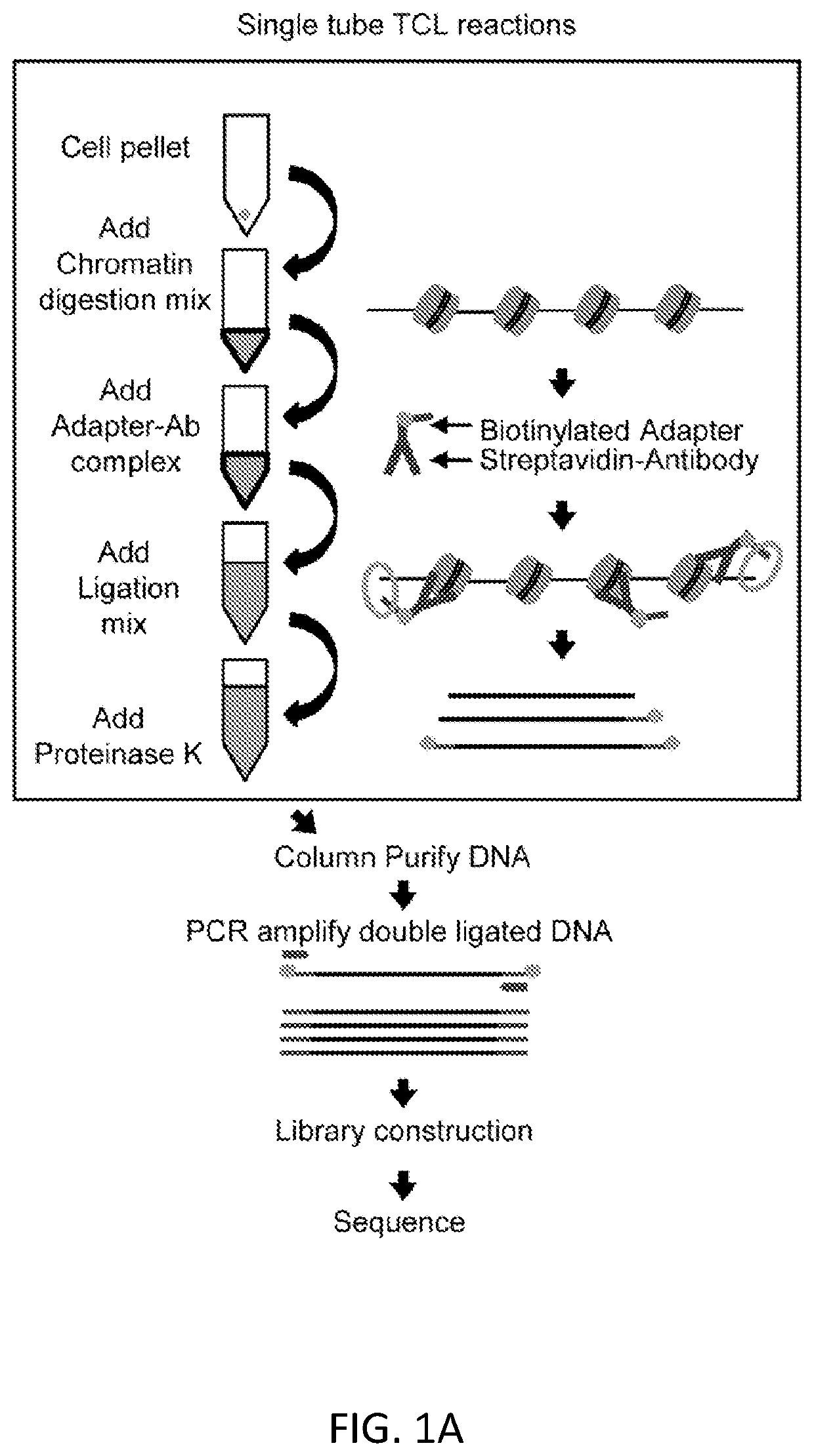
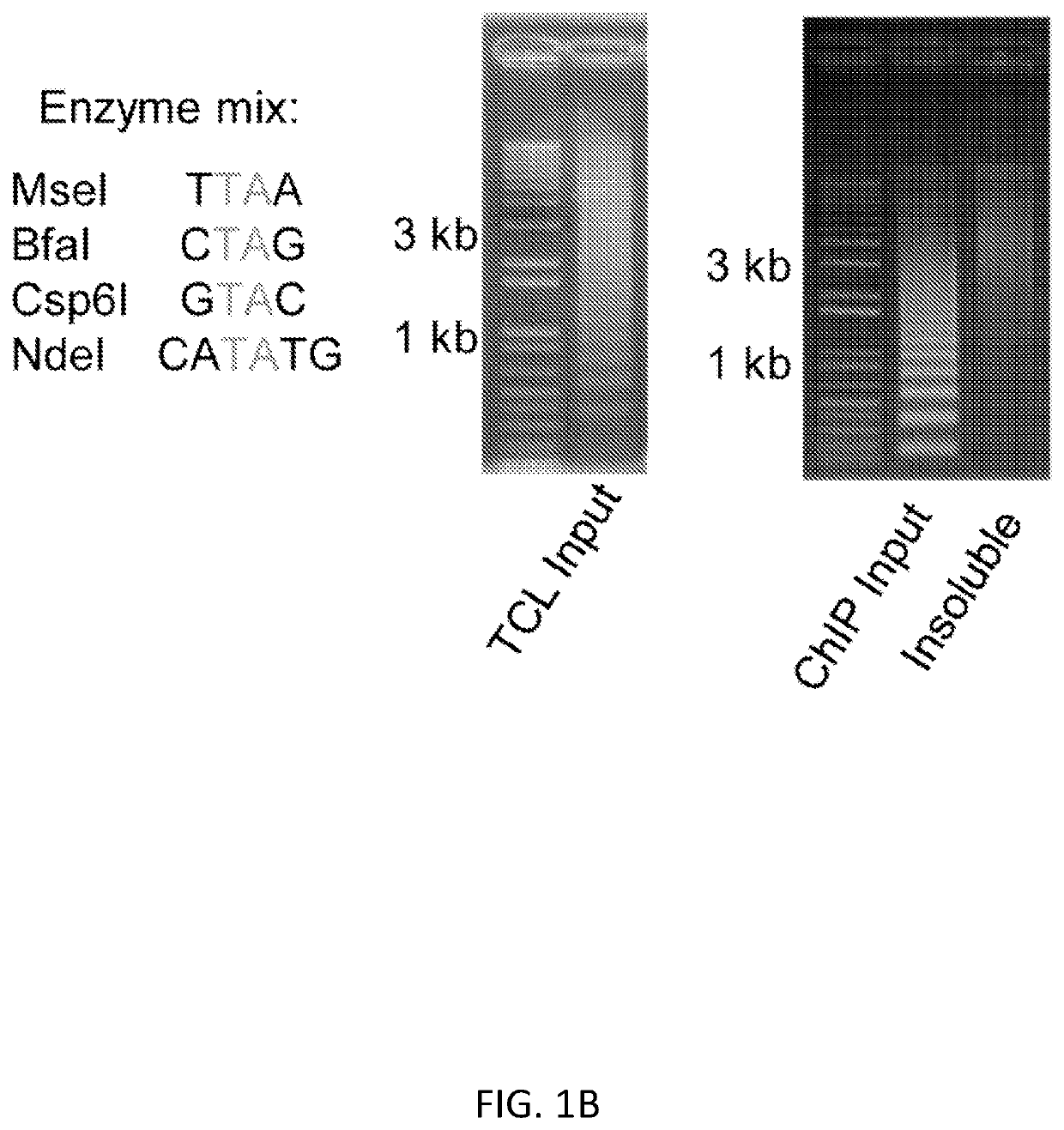
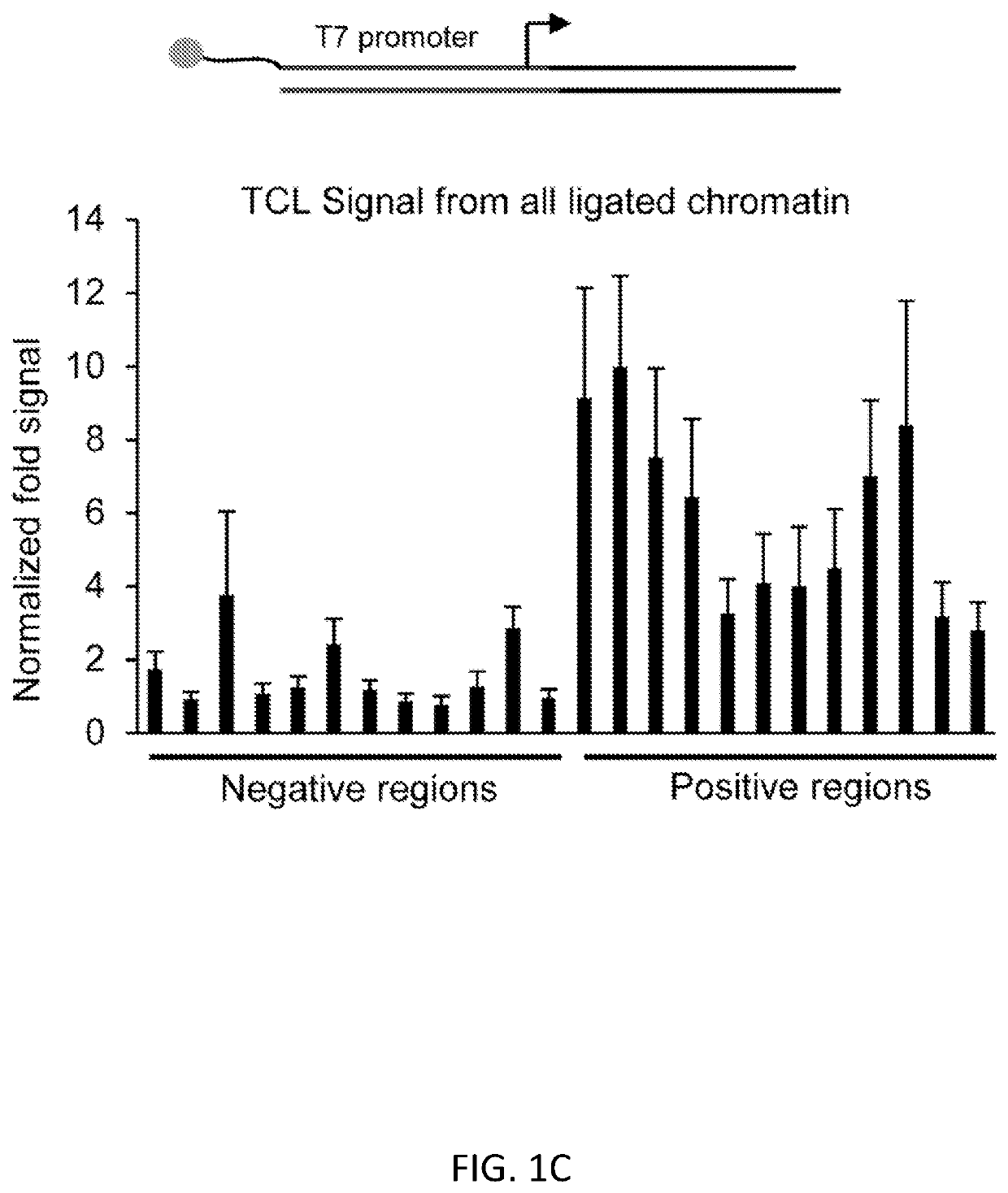
InactiveUS20200123591A1Modify structureEasy to produceCompound screeningApoptosis detectionOligonucleotide LinkerHistone protein
Reagents and methods for epigenetic profiling using targeted chromatin ligation are disclosed. The method utilizes oligonucleotide adapters complexed with antibodies specific for DNA-binding proteins of interest and proximity ligation to tag fragmented chromatin with the adapters. Chromatin fragments having ligated adapters are amplified and sequenced with primers that hybridize to the adapters. This method can be used in epigenetic profiling, for example, for mapping histone modification patterns as well as transcriptional regulatory sites.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Methylation detection composition, kit and method
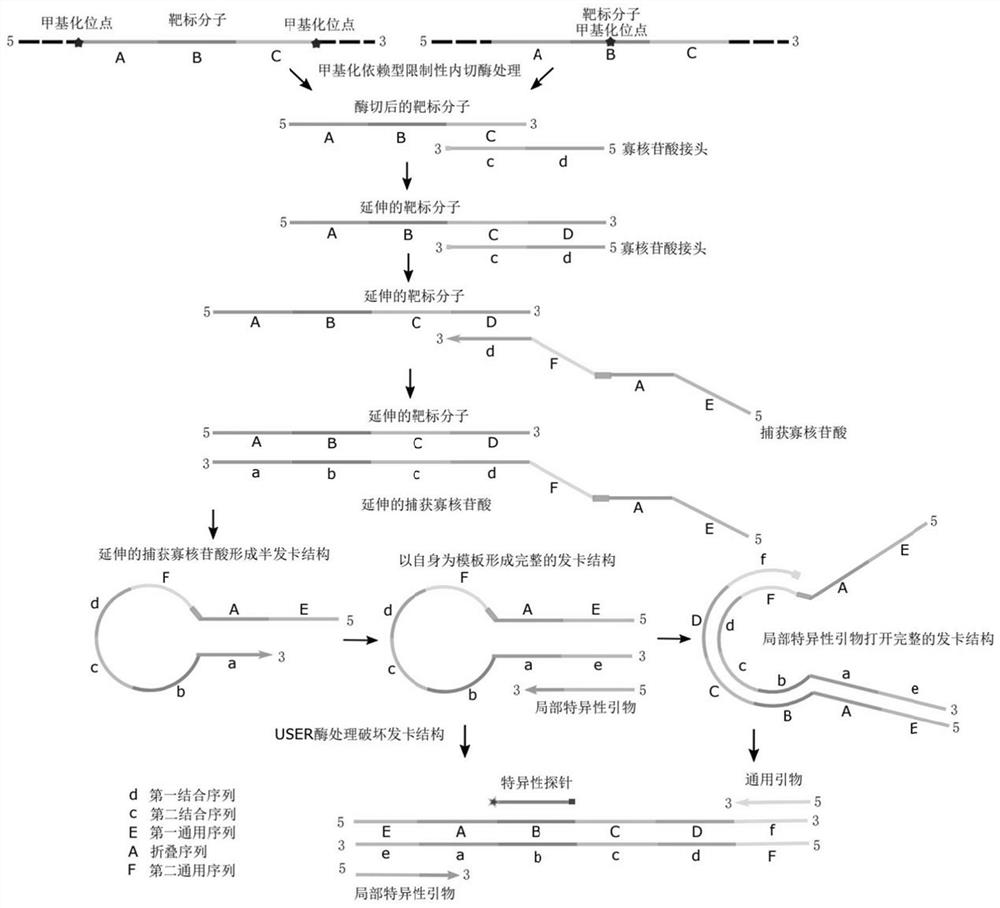
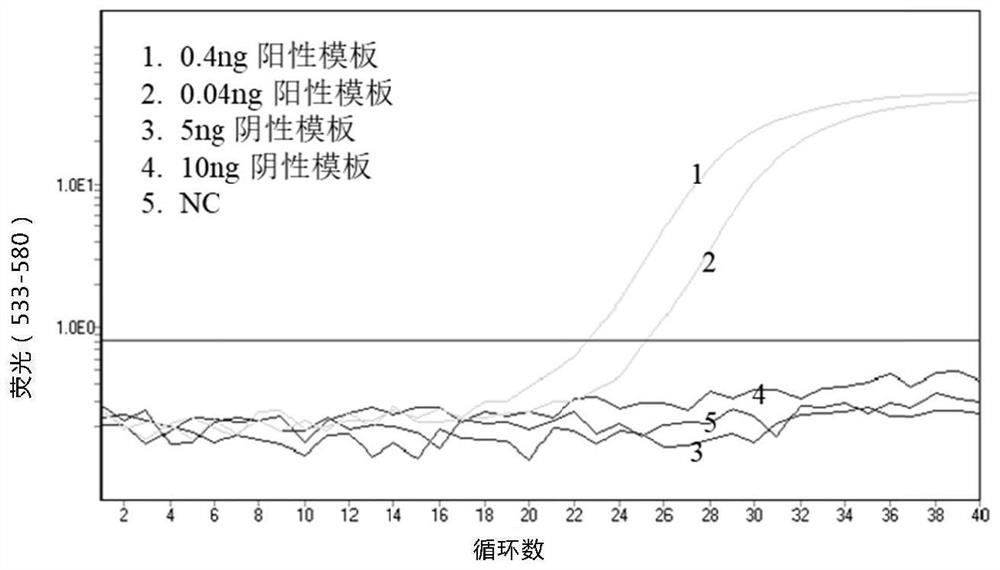
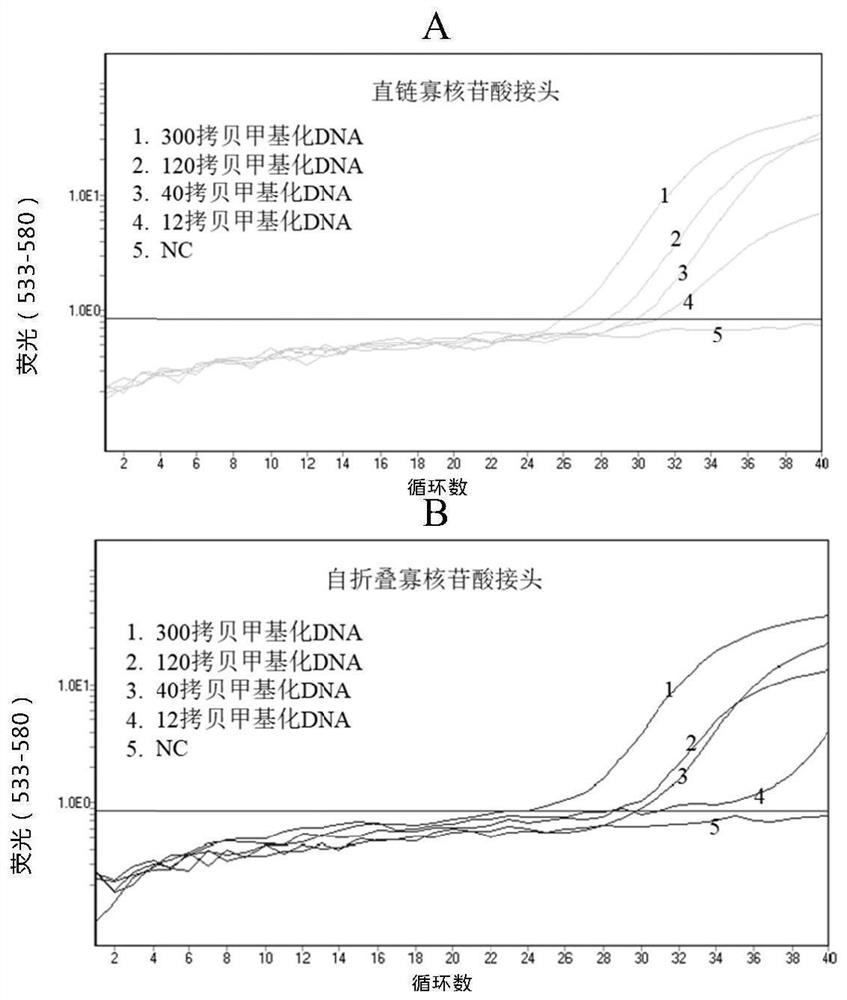
PendingCN114075595AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationDNA methylationOligonucleotide Linker
The invention relates to a methylation detection composition, kit and method, and specifically provides oligonucleotide linkers and a capture oligonucleotide for nucleic acid amplification, use method and use thereof. The oligonucleotide linkers comprise a first binding sequence and a second binding sequence, wherein the first binding sequence comprises a sequence which is the same as a 3' terminal sequence of the capture oligonucleotide, the second binding sequence is complementary with a target molecule, and the capture oligonucleotide comprises a first universal sequence, a folding sequence and a binding capture sequene. According to the invention, the product and the method can realize DNA methylation detection with good specificity and high sensitivity.
Owner:SHANGHAI JIAO TONG UNIV +1
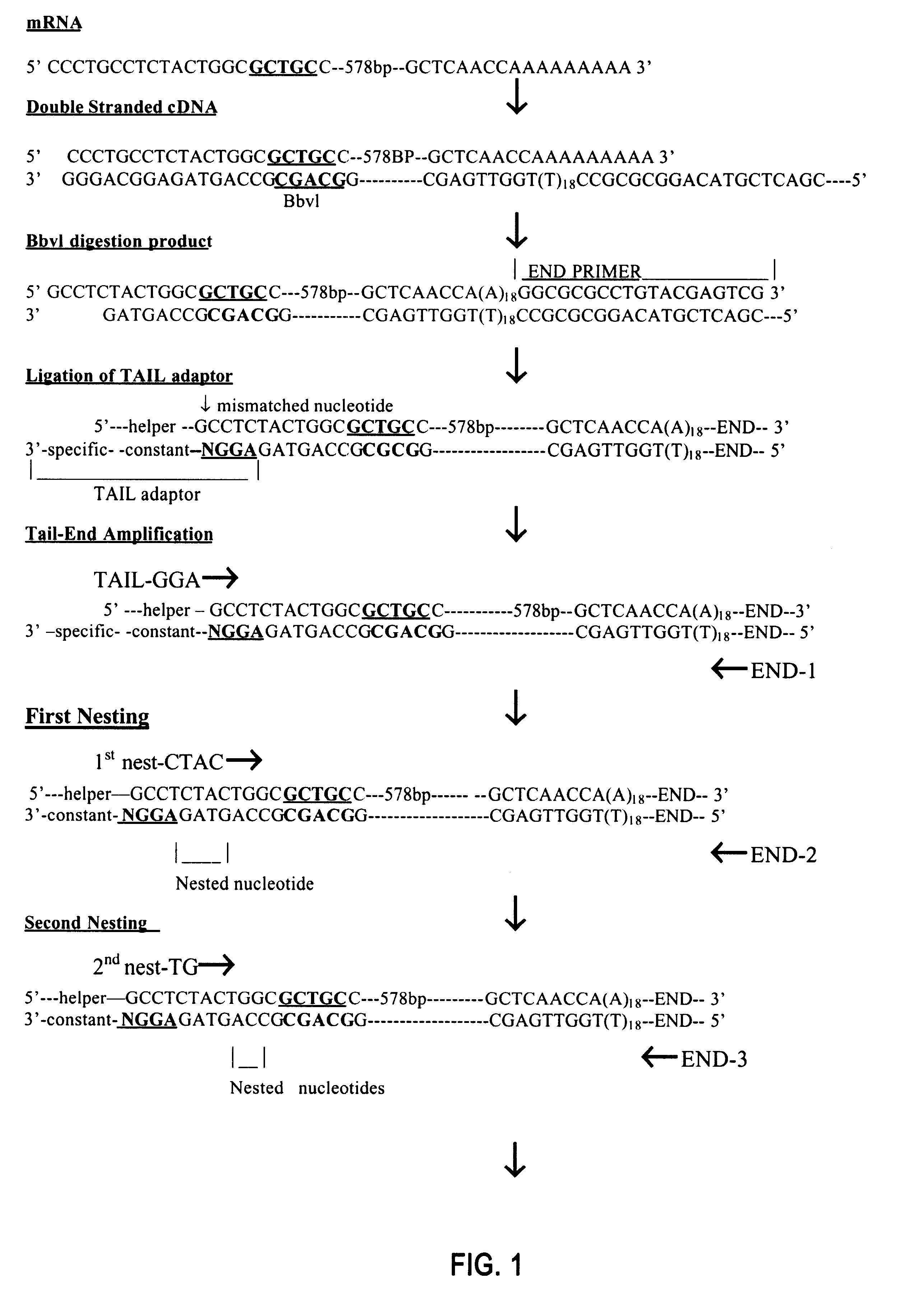
Oligonucleotide linkers comprising a variable cohesive portion and method for the preparation of polynucleotide libraries by using said linkers
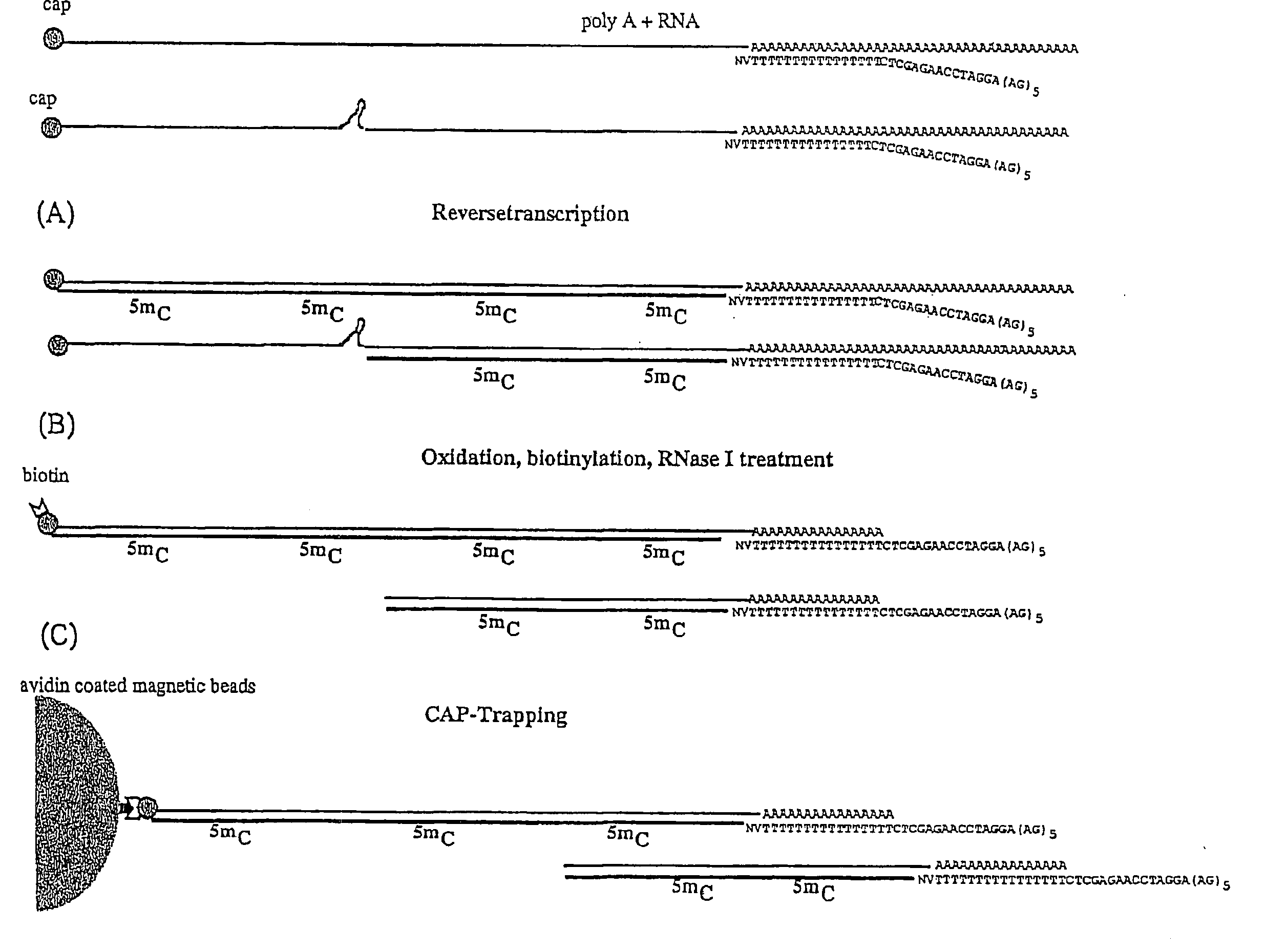
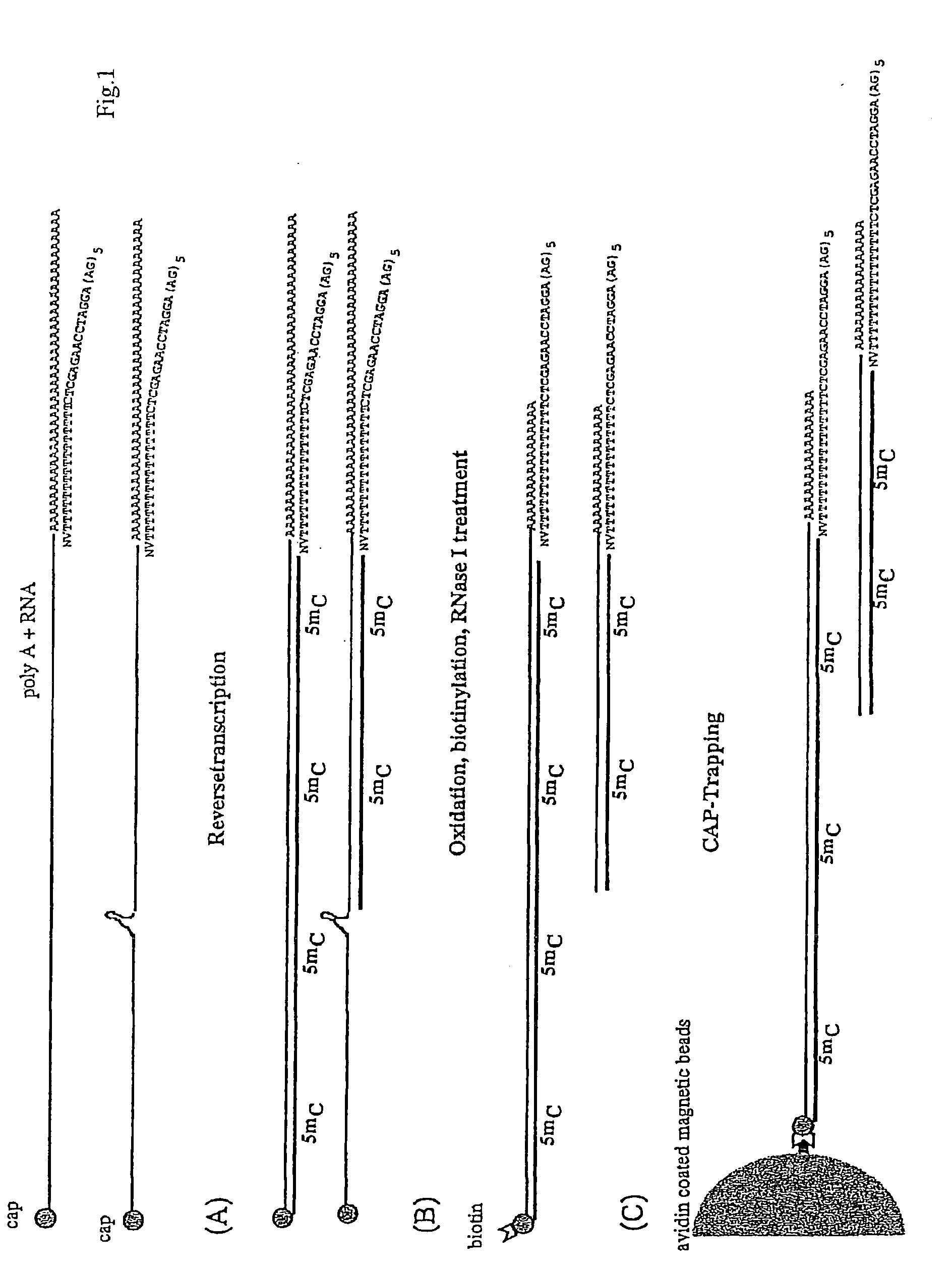
The present invention relates to a linker or population of linkers that include an oligonucleotide fixed portion and an oligonucleotide variable portion represented by formula (N)n, wherein N is A, C, G, T or U, or their derivatives, and n is an integer equal to or higher than 1. A linker-polynucleotide or a population of linker-polynucleotides of the invention may be constituted by said linker or population of linkers and a target first strand polynucleotide bound to said linker. The invention also encompasses a method of preparing said linker or population of linkers and a method of preparing a linker-polynucleotide using said linker or population of linkers. The linkers or polynucleotide-linkers of the invention can be used in a method of preparing a cDNA library.
Owner:HAYASHIZAKI YOSHIHIDE
Novel quencher and reporter dye combinations
Disclosed is a probe for use in biological assays. The probe includes a fluorescent dye bound to a quencher compound through an oligonucleotide linker. Also disclosed are methods of using the probe, such as for a polymerase chain reaction (PCR), such as in a quantitative PCR reaction (qPCR), as well as kits including the probe.
Owner:LIFE TECH CORP
Method and system for delivering nucleic acid into a target cell
ActiveUS8187883B2Low specificityControl strengthMaterial nanotechnologyBioreactor/fermenter combinationsOligonucleotide LinkerOligonucleotide
Controlled delivery of nucleic acid into target cells is achieved by immobilizing the nucleic acid and the cells to a substrate. Improved control over delivery is achieved by immobilizing the nucleic acid to the substrate via complementary DNA binding interactions with an oligonucleotide linker.
Owner:WISCONSIN ALUMNI RES FOUND
Method and reagent for constructing nucleic acid single-stranded circular library
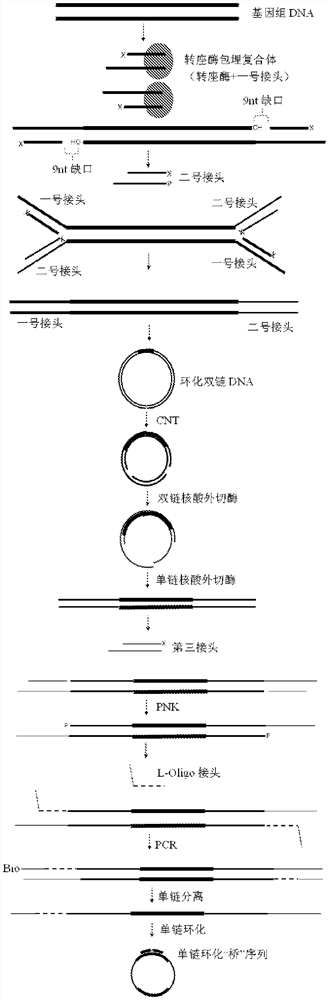
ActiveCN108138364BFlexible length controlThe process is simple and convenientMicrobiological testing/measurementVector-based foreign material introductionSingle strandOligonucleotide Linker
The invention discloses a method and reagents for constructing a nucleic acid single-strand circular library. The method comprises the following steps: using a transposase embedding complex to randomly interrupt the nucleic acid to form a first joint at both ends and a gap. Fragment; connect the second linker at the gap; perform the first PCR reaction to obtain the product of connecting the first linker and the second linker sequence at both ends; enzyme digestion to generate a nick, and then perform double-stranded circularization to generate a circular nucleic acid molecule; from Carry out restriction nick translation reaction at the nick; digest the part that has not occurred restriction nick translation reaction; connect the third adapter and oligonucleotide adapter sequence; perform the second PCR reaction to obtain the third adapter and oligonucleotide at both ends respectively The product of the acid linker sequence; isolate the single-stranded nucleic acid; circularize the single-stranded nucleic acid to obtain a single-stranded circular library. The method of the present invention realizes simple and rapid construction of a nucleic acid single-stranded circular library through the combination of transposase breaking and restriction gap translation reaction.
Owner:MGI TECH CO LTD
Oligonucleotide adapters and their application in the construction of single-strand circular libraries for nucleic acid sequencing
ActiveCN105400776BAvoid digestionNucleotide librariesLibrary creationSingle strandNucleic acid sequencing
The present invention provides an oligonucleotide linker, and particularly relates to uses of the oligonucleotide linker in construction of a single-chain cyclic library in nucleic acid sequencing, and further in sequencing platforms requiring single-chain cyclic libraries. The oligonucleotide linker of the present invention has advantages of high sequencing throughput, high accuracy, and easy operation.
Owner:MGI TECH CO LTD
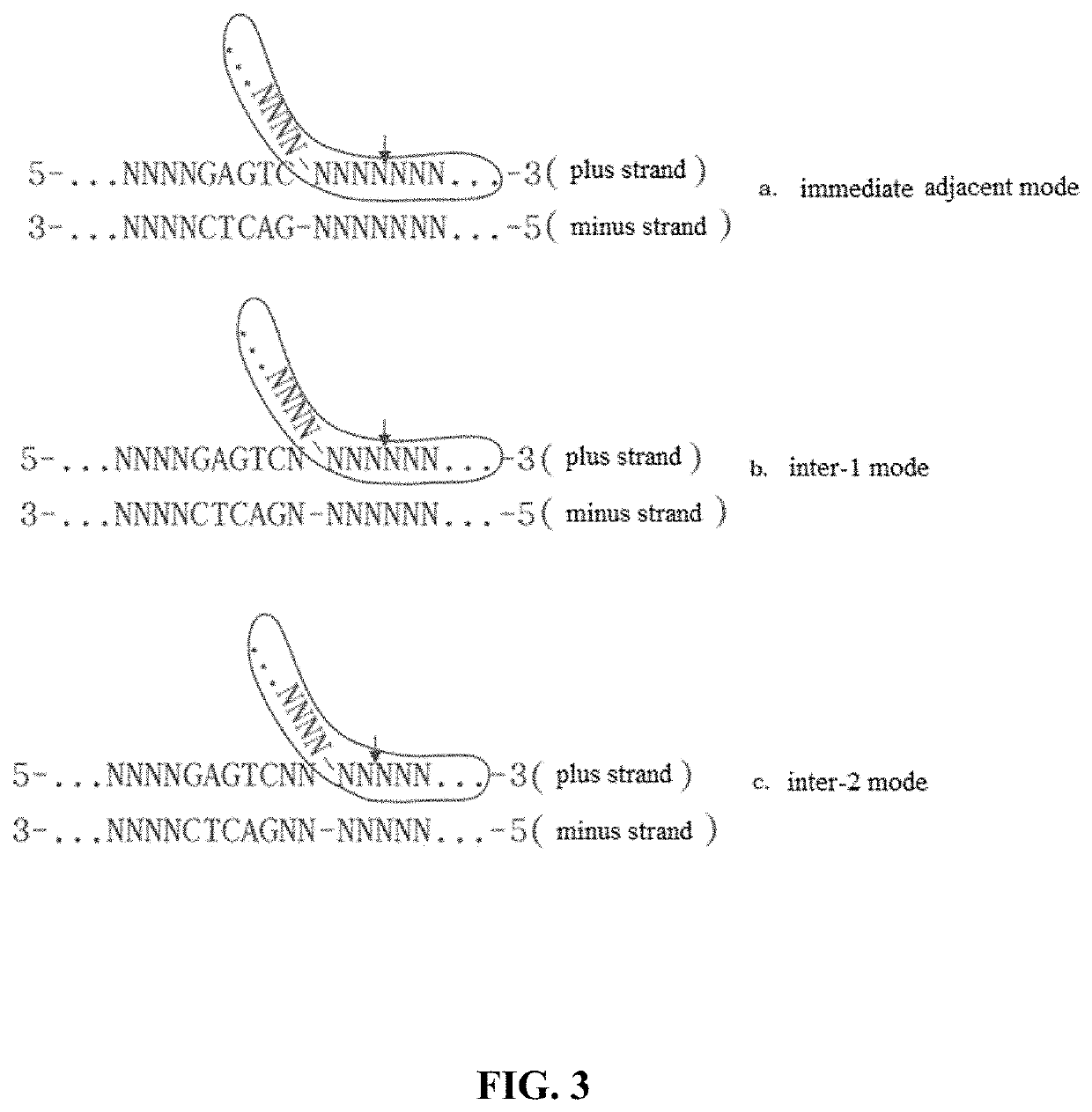
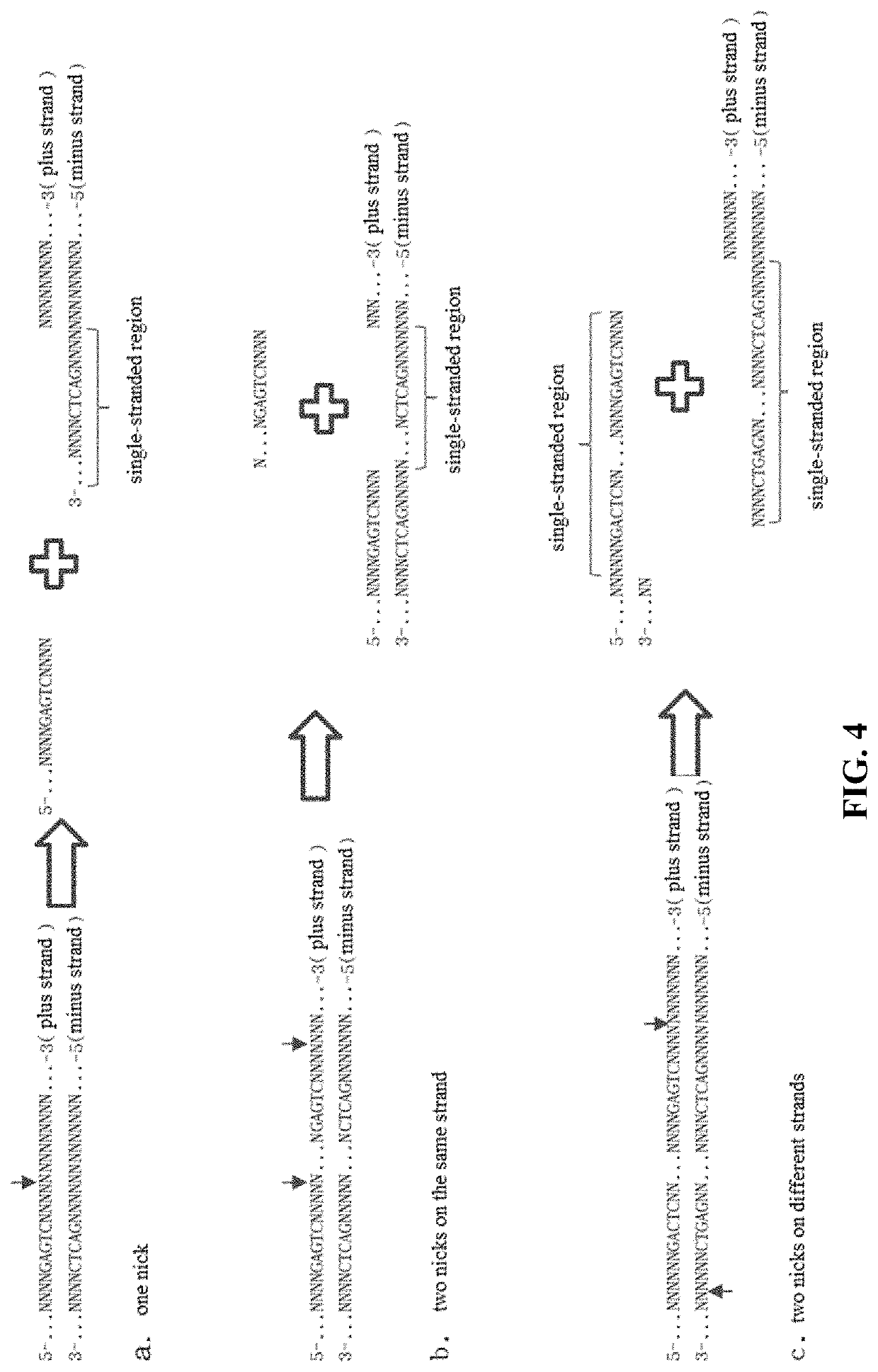
Method for manipulating terminals of double stranded DNA
PendingUS20220290163A1Eliminate needBulky assemblyVector-based foreign material introductionDNA preparationOligonucleotide LinkerDouble strand
A method for manipulating the terminals of a double-stranded DNA. The principle thereof is using a restriction nicking enzyme to first generate one or more nicks on one strand of a double-stranded DNA, then using an oligonucleotide adaptor to bind the same or a different restriction nicking enzyme to generate cleavage on the other strand of the double-stranded DNA, the position of cleavage being determined by the design of the oligonucleotide adapter, and eventually cleaving the double-stranded DNA of interest and generating various lengths of 5′ io protruding terminals and various lengths of 3′ protruding terminals or blunt terminals at the nicks. The bases at the terminals of a double-stranded DNA generated by means of this method can be designed at will, and such terminals can be used for double-stranded DNA splicing, particularly seamless splicing of double-stranded DNA.
Owner:WUXI QINGLAN BIOLOGICAL SCI & TECH
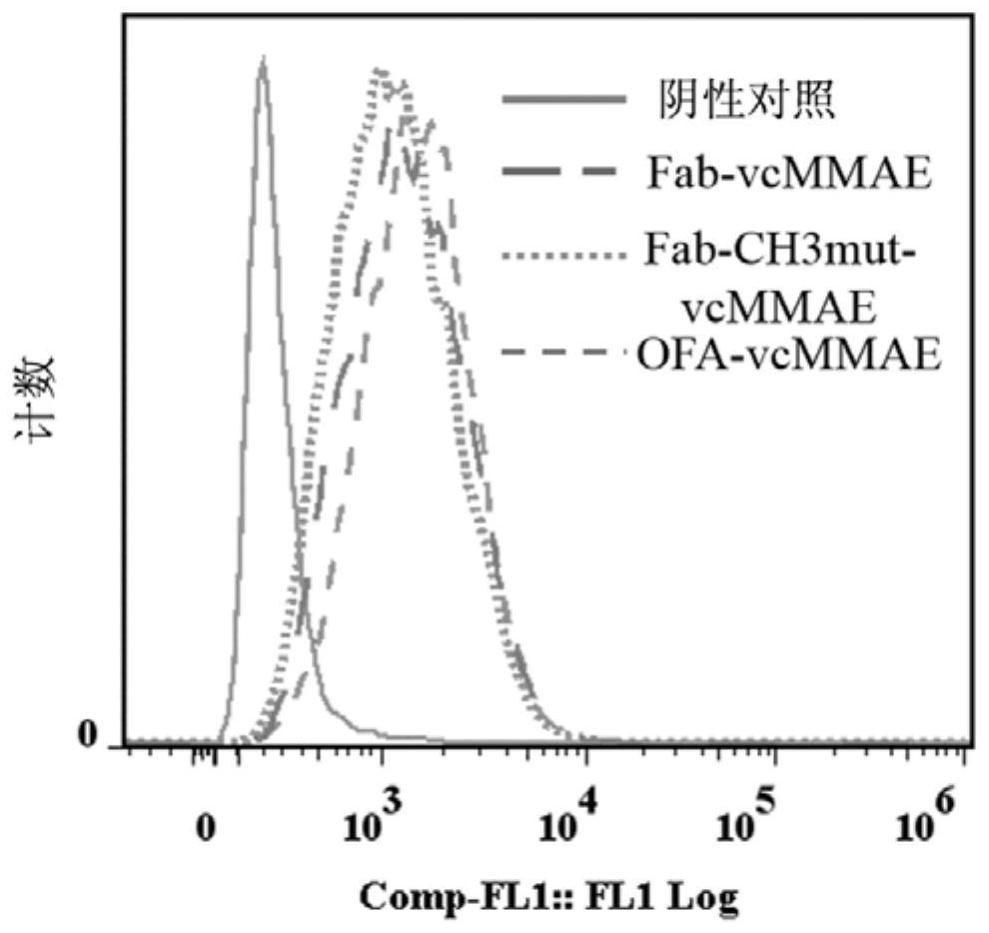
Antigen-binding fragment of an antibody-dolastatin conjugate and its preparation method and application
ActiveCN108084267BRetention of lethalityFast tumor penetration rateAntibody mimetics/scaffoldsTetrapeptide ingredientsCD20Glycine
Owner:ZHEJIANG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com