Methylation detection composition, kit and method
A detection method and sequence-combining technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of reducing sequence complexity, difficult primer design, labor-intensive and time-consuming, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment
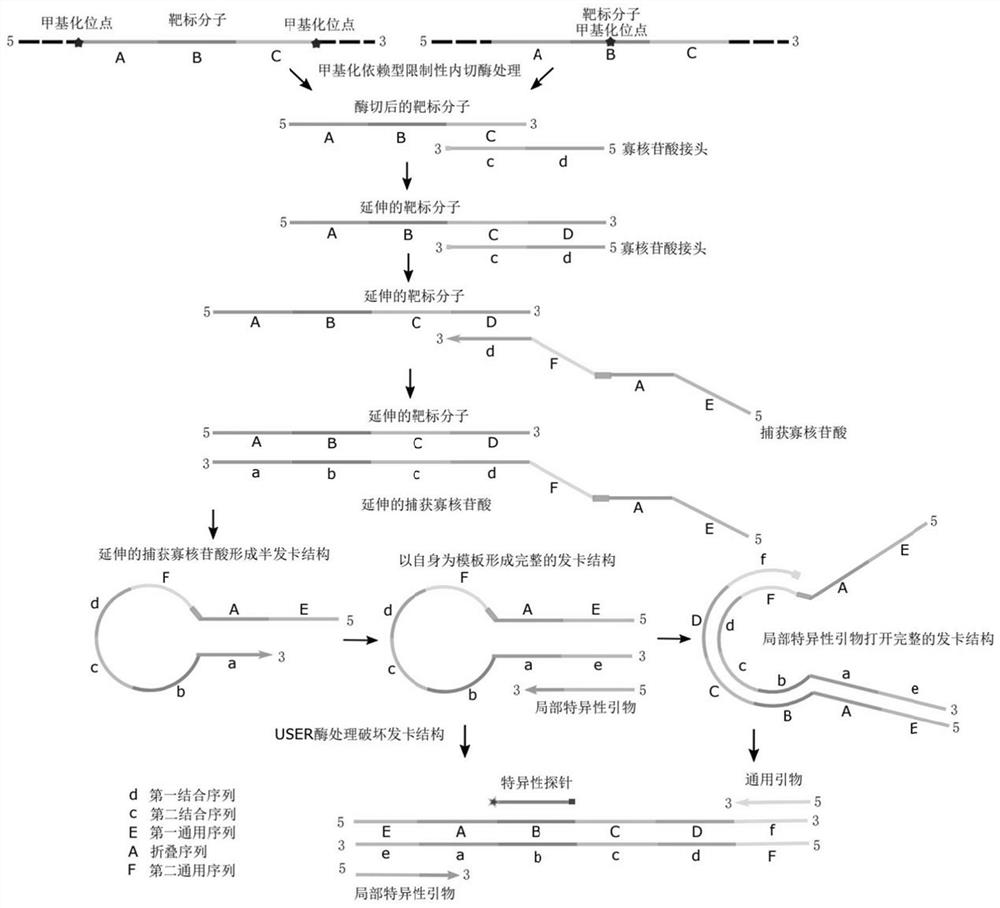
[0142] Specific examples The methylation site in the sequence described in SEQ ID NO: 10 of the Septine 9 gene is detected by the following method:
[0143]Use the oligonucleotide linker (1) shown in SEQ ID NO:1 or 6 to contact with the cfDNA digested by GlaI, the oligonucleotide linker (1) and the target molecule (2) shown in SEQ ID NO:10 Complementary binding; the target molecule (2) is extended using SEQ ID NO: 1 or 6 as a template, and a sequence complementary to SEQ ID NO: 1 or 6 is added to the 3' end of the target molecule (2) to obtain an extended target molecule (3 ); the extended target molecule (3) is complementary to the capture oligonucleotide (4) shown in SEQ ID NO: 2 or 7; the capture oligonucleotide (4) is carried out with the extended target molecule (3) as a template An extension reaction, adding a sequence complementary to the extended target molecule (3) at the 3' end of the capture oligonucleotide (4) to obtain an extended capture oligonucleotide (5); the ...
Embodiment 1
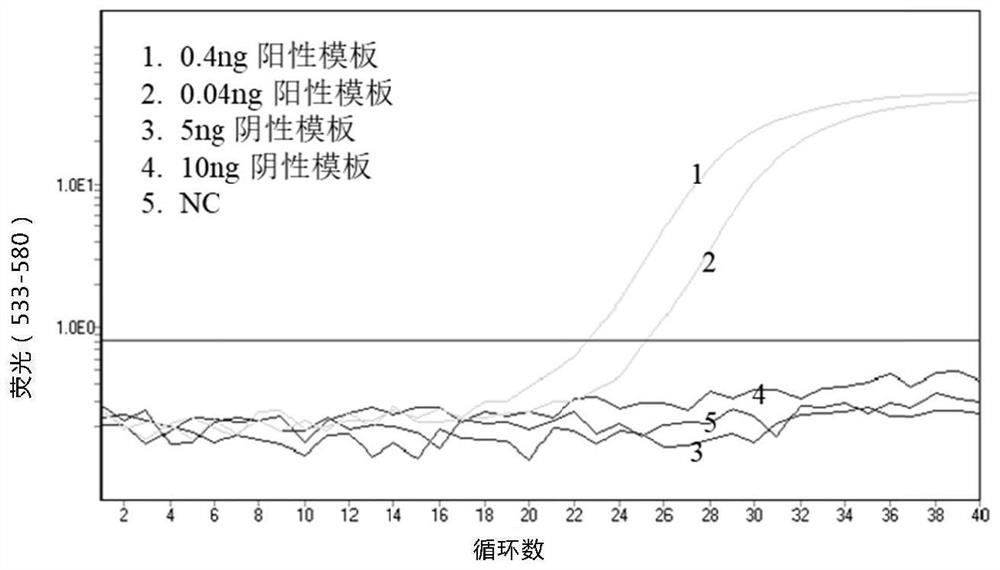
[0152] Example 1: Methylation detection of ultrasonically fragmented DNA of human Septine 9 gene
[0153] (1) Genomic DNA of Jurkat cell line and Hela cell line were extracted respectively, and the methylation status of Septine9 gene was identified by sequencing.
[0154] (2) Ultrasonic treatment of the above two genomic DNAs to obtain fragmented DNA with a fragment size of about 150 bp.
[0155] (3) Use the methylation-dependent restriction endonuclease GlaI to carry out enzyme digestion reaction on the above-mentioned ultrasonically fragmented DNA. The reaction system is: 1× enzyme digestion buffer, 10U GlaI, ultrasonically fragmented DNA at different concentrations, and the total volume is 10 μL; the reaction condition is to incubate at 37°C for 1 hour; after the enzyme digestion reaction, heat the system to 85°C and incubate for 10 minutes to heat inactivate GlaI.
[0156] (4) Add the Septine 9 gene oligonucleotide linker and capture oligonucleotide to the above enzyme di...
Embodiment 2
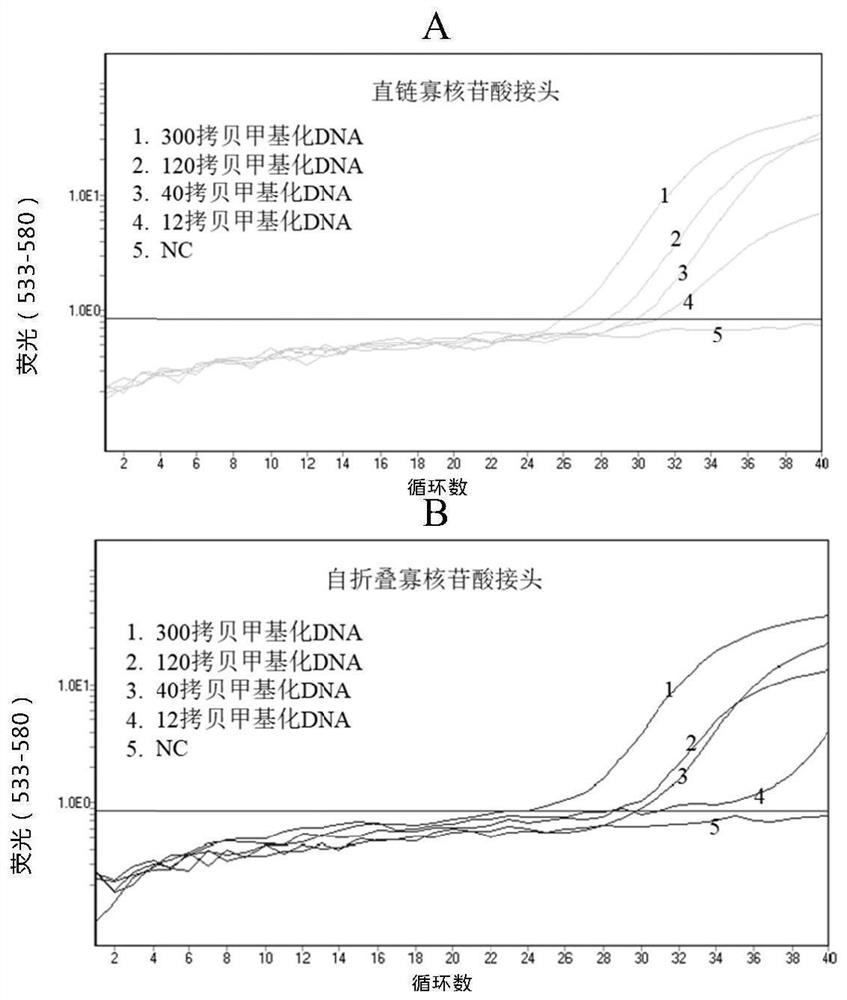
[0175] Embodiment 2: Experiment of different structural oligonucleotide linkers
[0176] In order to reduce the competition between oligonucleotide adapters, capture oligonucleotides and target molecules, in this example, oligonucleotide adapters with different structures were selected for detection. The capture oligonucleotides, specific primers, universal primers and specific probes used are the same as those in Example 1, and the restriction conditions and amplification conditions are the same as those in Example 1.
[0177] Oligonucleotide adapters used include:
[0178] Linear Oligonucleotide Adapter
[0179] 5-TGCCGTCAGAGTCCTGTCTCGAGCGACCCGCTGCCCACCAG (SEQ ID NO: 1)
[0180] Self-folding oligonucleotide adapters (underlined are self-folding regions)
[0181] 5- TCGAGACAGGAC TGCCGTCAGA GTCCTGTCTCGA GCGACCCGCTGCCCAC (SEQ ID NO: 6)
[0182] The partial sequence of the human Septine 9 gene is shown in Example 1.
[0183] Such as image 3As shown, using straight-cha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com