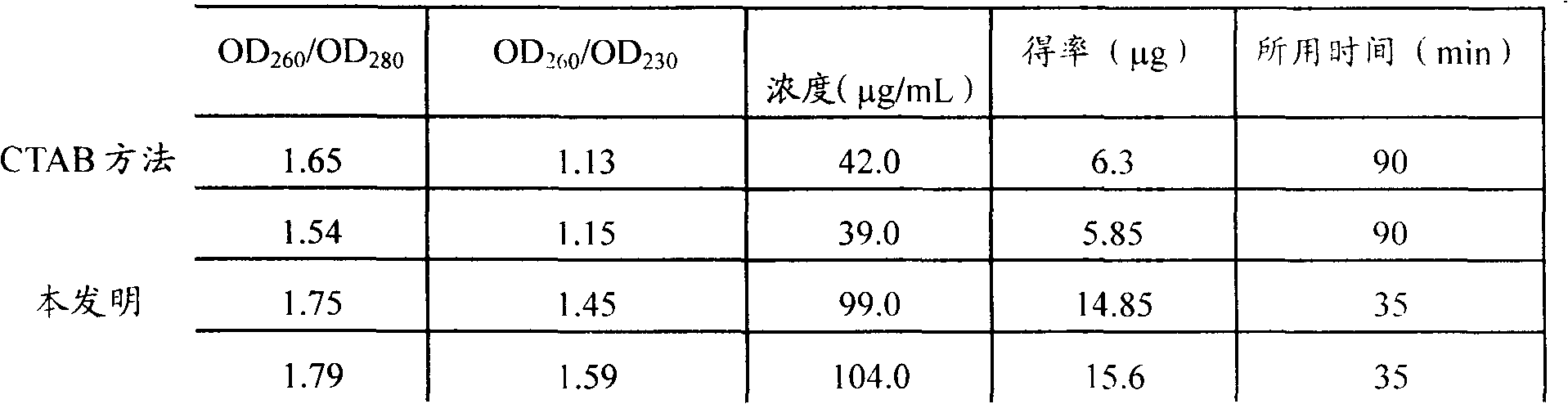
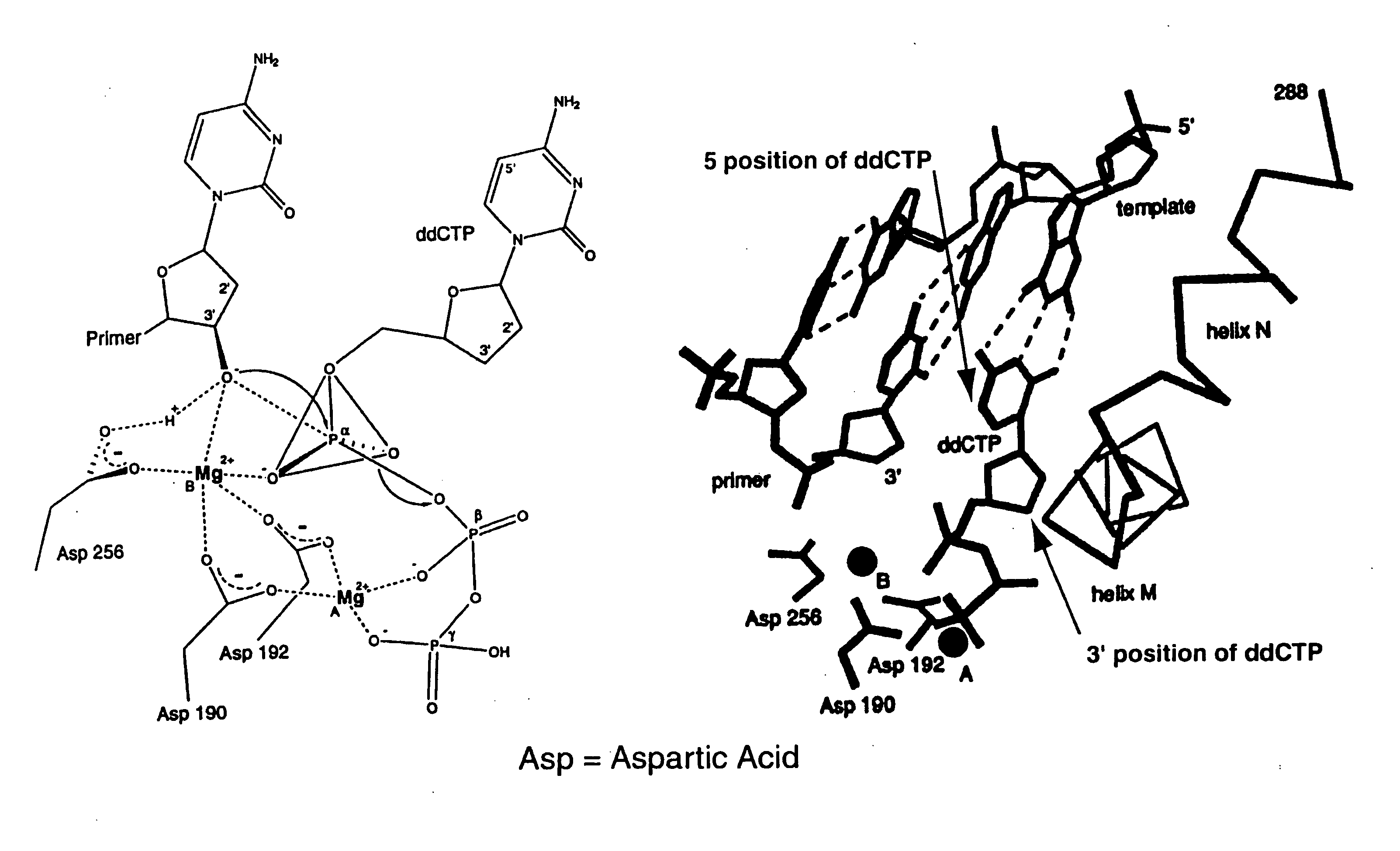
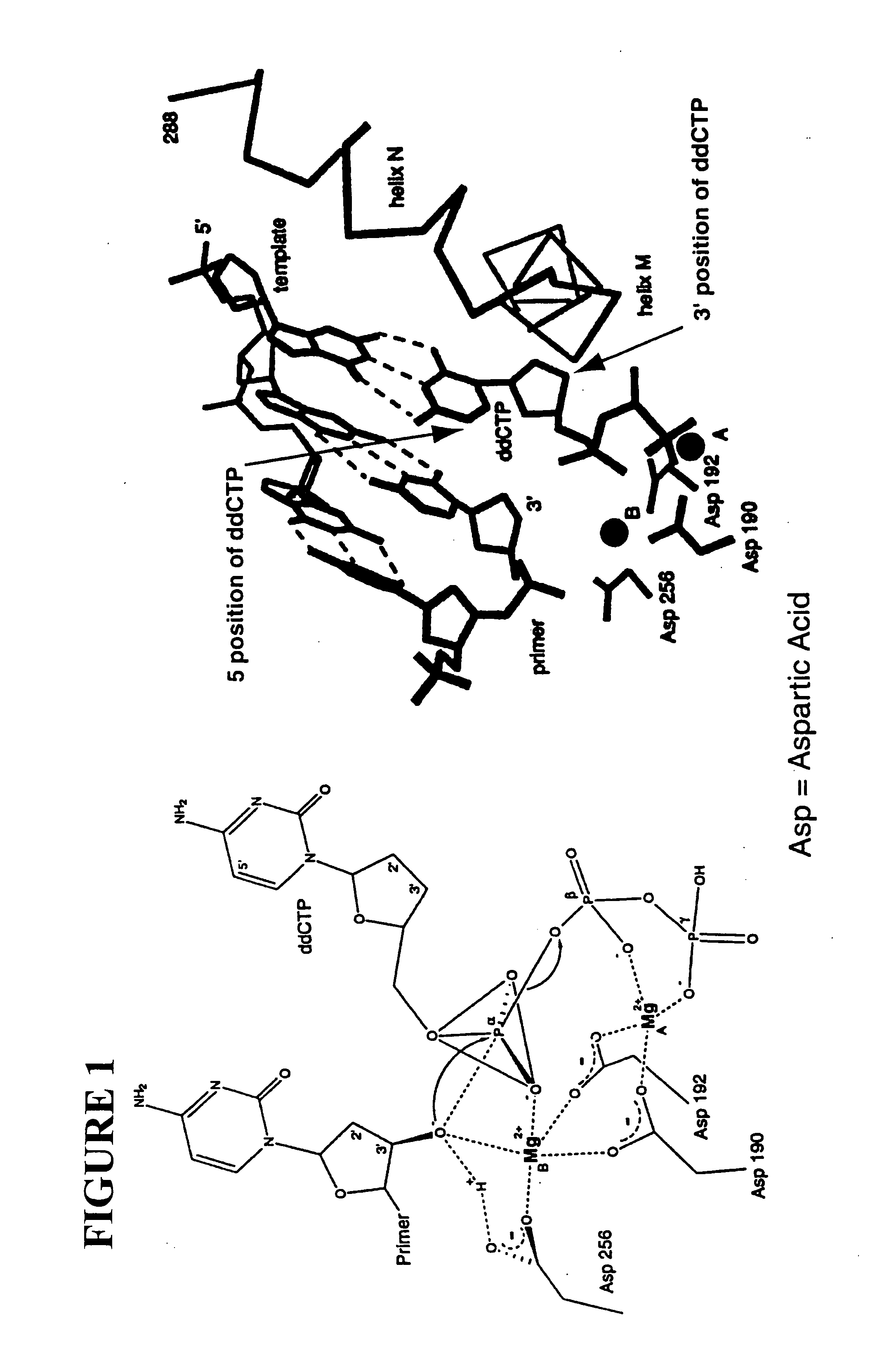
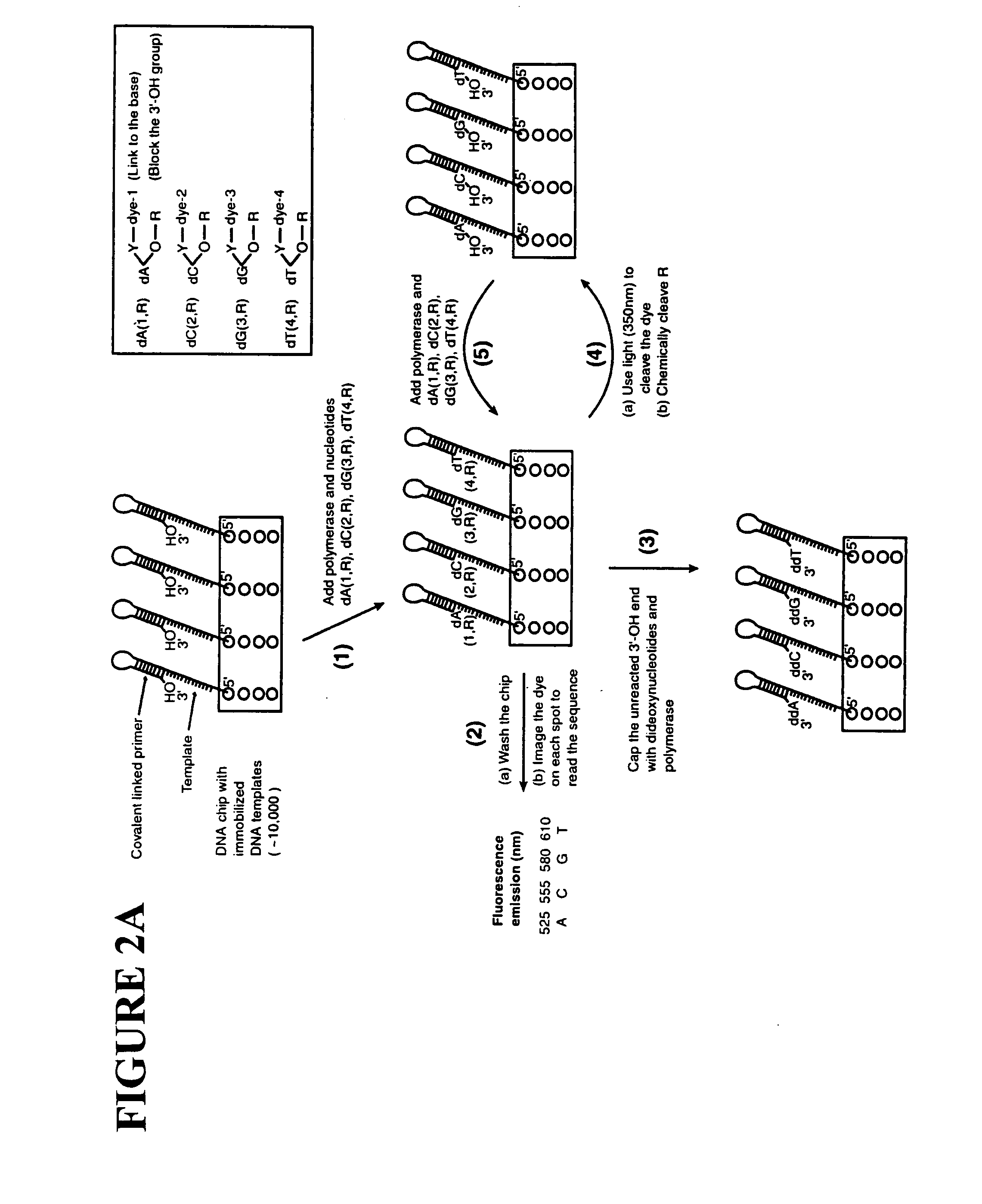
Patents
Literature
639 results about "3-deoxyribose" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of an oxygen atom.
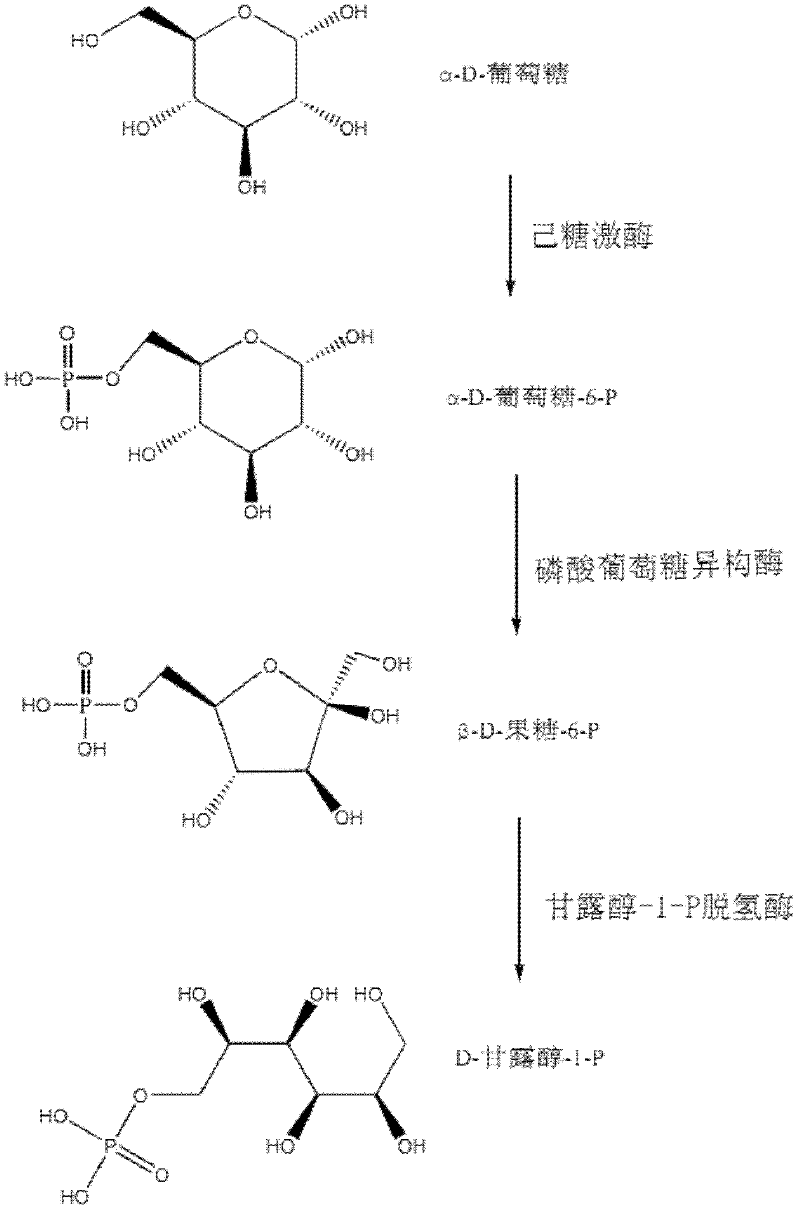
Relevant enzymes for preparing mannitol by performing anabolism on Chinese caterpillar fungus and hirsutella sinensis, gene and application thereof
The invention provides a group of relevant enzymes for preparing mannitol by performing anabolism on Chinese caterpillar fungus serving as a multifunctional production fungus and hirsutella sinensis based on glucose, a gene for encoding these enzymes and application thereof. The relevant enzymes include (1) hexokinase: manA1-A6 proteins of which the sequences are SEQ ID No.1-6, (2) phosphoglucoisomerase: manB1-B3 proteins of which the sequences are SEQ ID No.7-9, and (3) mannitol-1-P dehydrogenase: manC protein of which the sequence is SEQ ID No.10. In the invention, detailed researches are performed on the metabolic pathway of mannitol synthesized by using Chinese caterpillar fungus serving as a multifunctional production fungus, hirsutella sinensis and glucose on the aspect of principle, cloned DNA (Deoxyribose Nucleic Acid) comprising a nucleotide sequence provided by the invention can be transferred into engineering bacteria with transduction, conversion and conjugal transfer methods, and host mannitol is endowed with high expression by regulating the expression of a biosynthetic gene of the mannitol.
Owner:ZHEJIANG UNIV OF TECH +1
DNA (Deoxyribose Nucleic Acid) sequencer
ActiveCN102703312AImprove sequencing efficiencyBioreactor/fermenter combinationsBiological substance pretreatments3-deoxyriboseBiochemical engineering
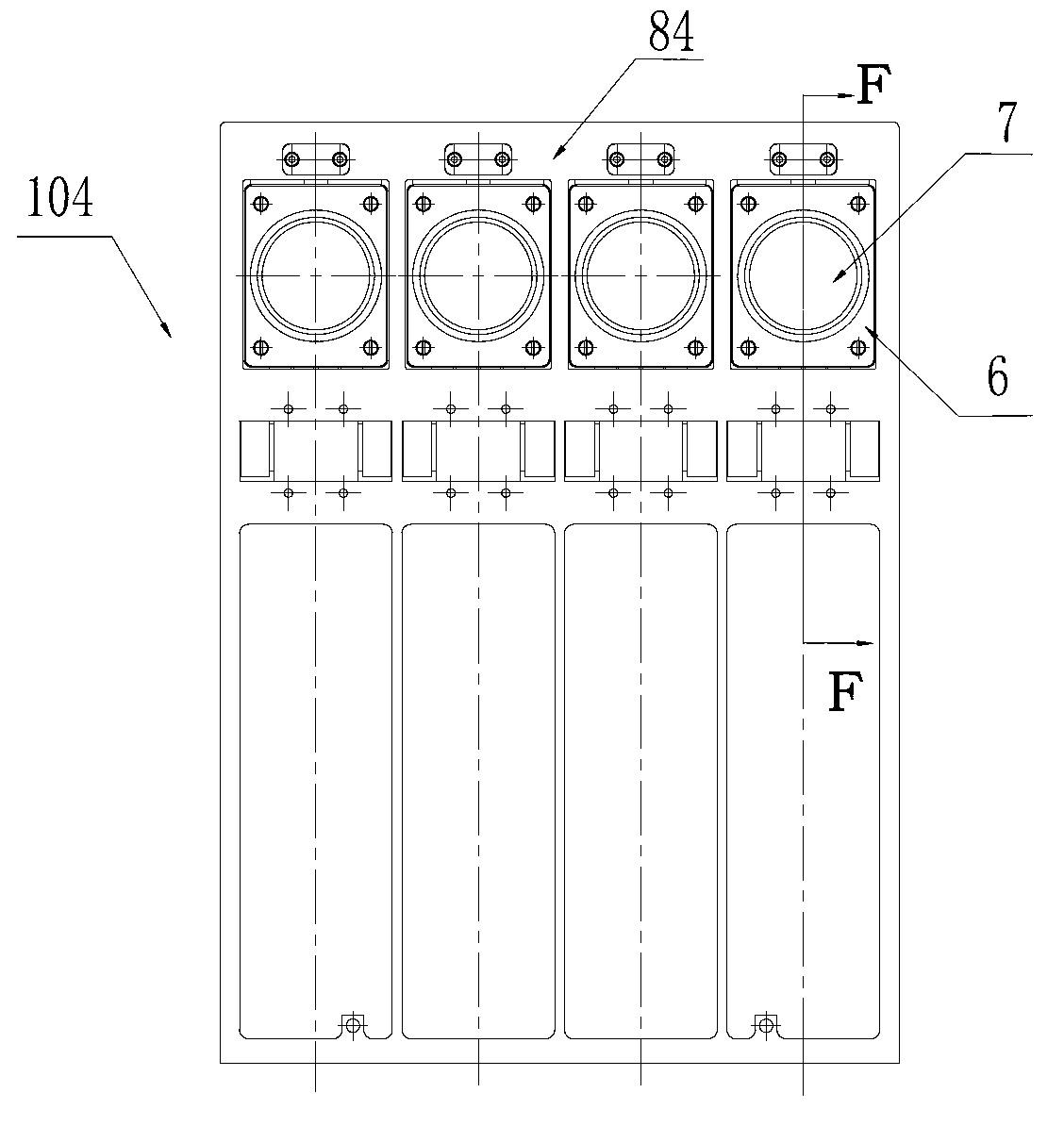
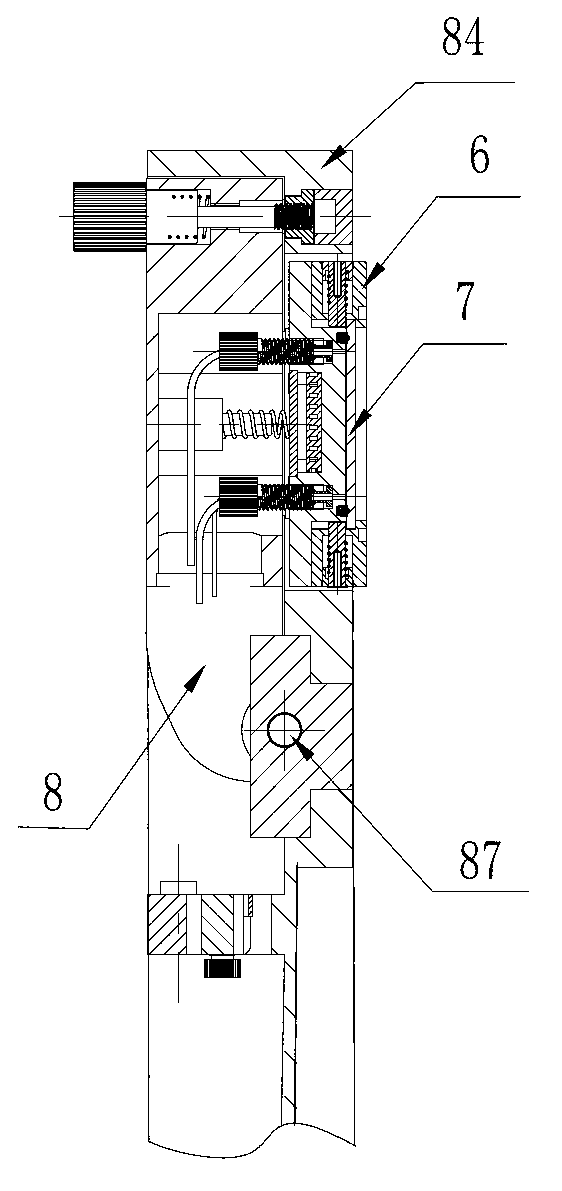
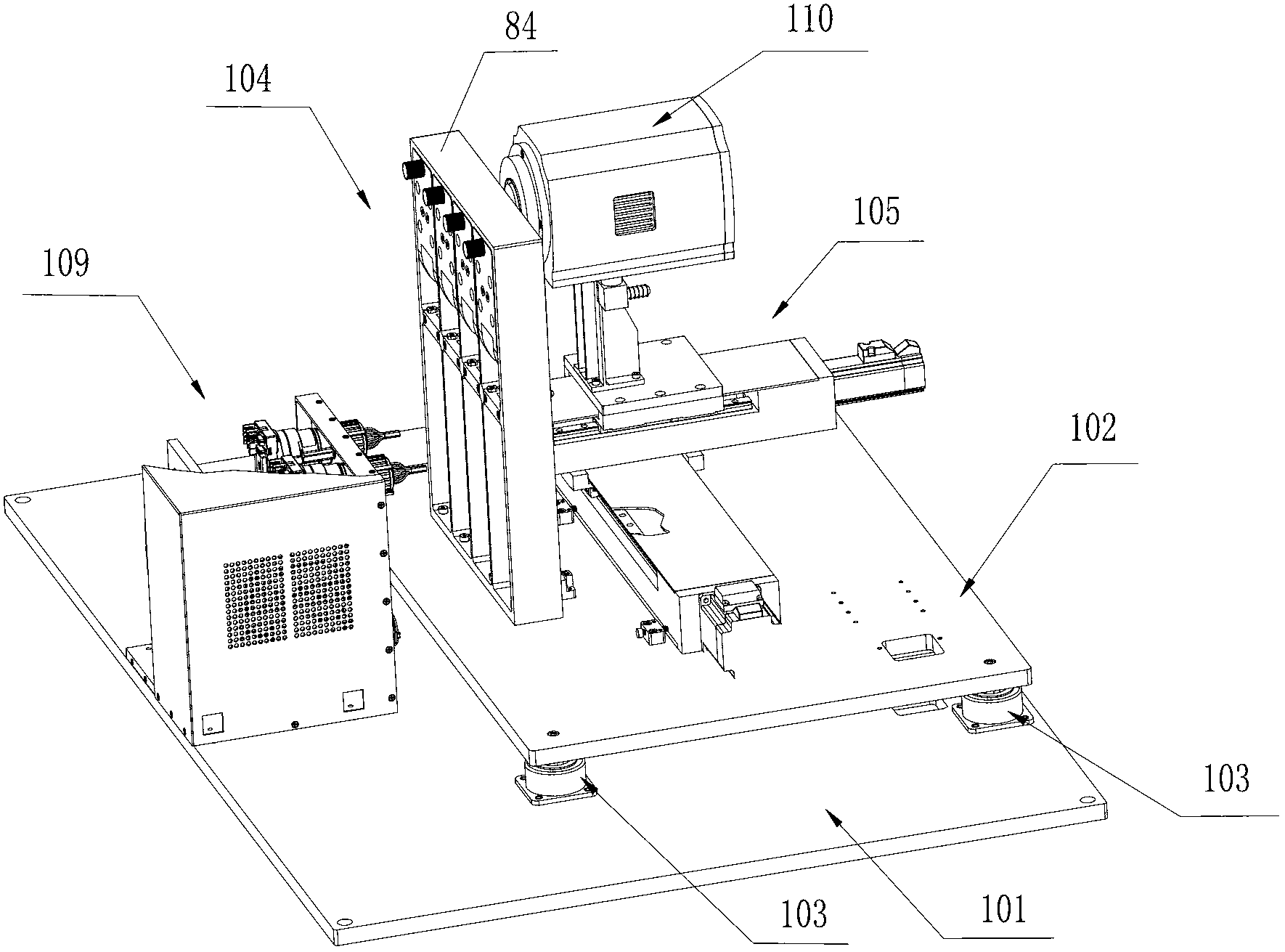
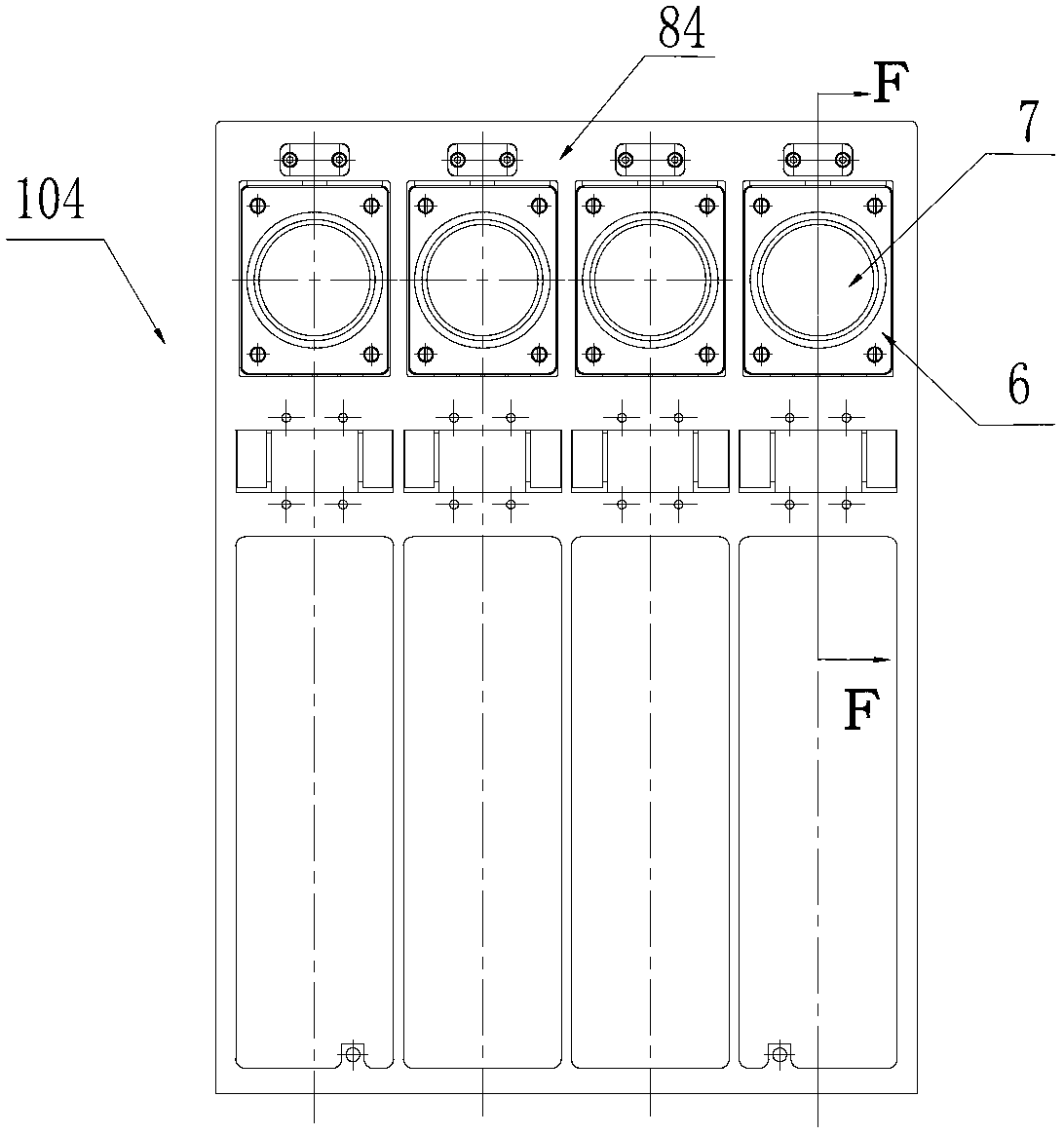
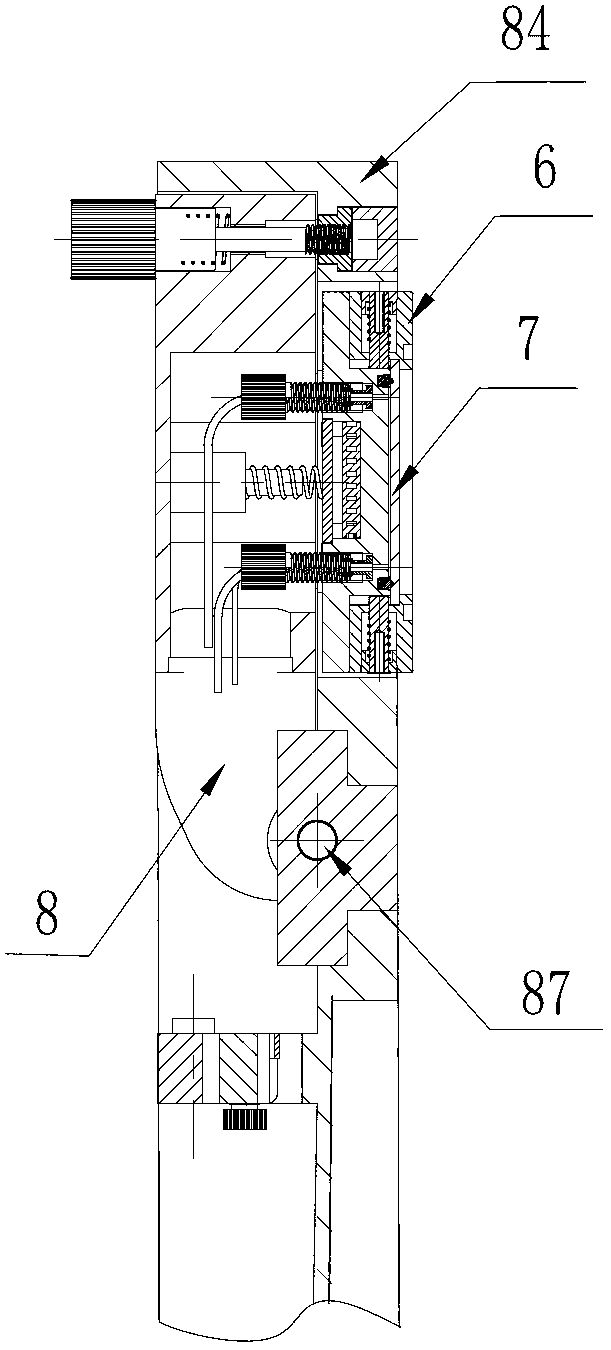
The invention discloses a DNA (Deoxyribose Nucleic Acid) sequencer which comprises a supporting table, a plurality of vibration dampers, a vibration damping plate, a reaction bin assembly, a CCD (Charge Coupled Device) camera, a two-dimensional regulation supporting device and a medicament supply assembly, wherein the vibration damping plate is connected with the supporting table by a plurality of vibration dampers; the reaction bin assembly is fixedly arranged on the vibration damping plate and is used for performing the DNA sequencing reaction; the CCD camera is used for acquiring an optical signal; the two-dimensional regulation supporting device is used for supporting the CCD camera; and the medicament supply assembly is arranged on the supporting table and is used for providing reagents and buffer solution for the reaction bin assembly. According to the DNA sequencer disclosed by the invention, by the arrangement of a plurality of reaction bins and the matching of the two-dimensional regulation supporting device capable of carrying out two-dimensional regulation and the medicament supply assembly capable of supplying the reagents for a plurality of reaction bins, the aim of simultaneously performing a plurality of reactions is fulfilled, a plurality of samples can be simultaneously sequenced and the DNA sequencing efficiency is greatly improved.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION +1
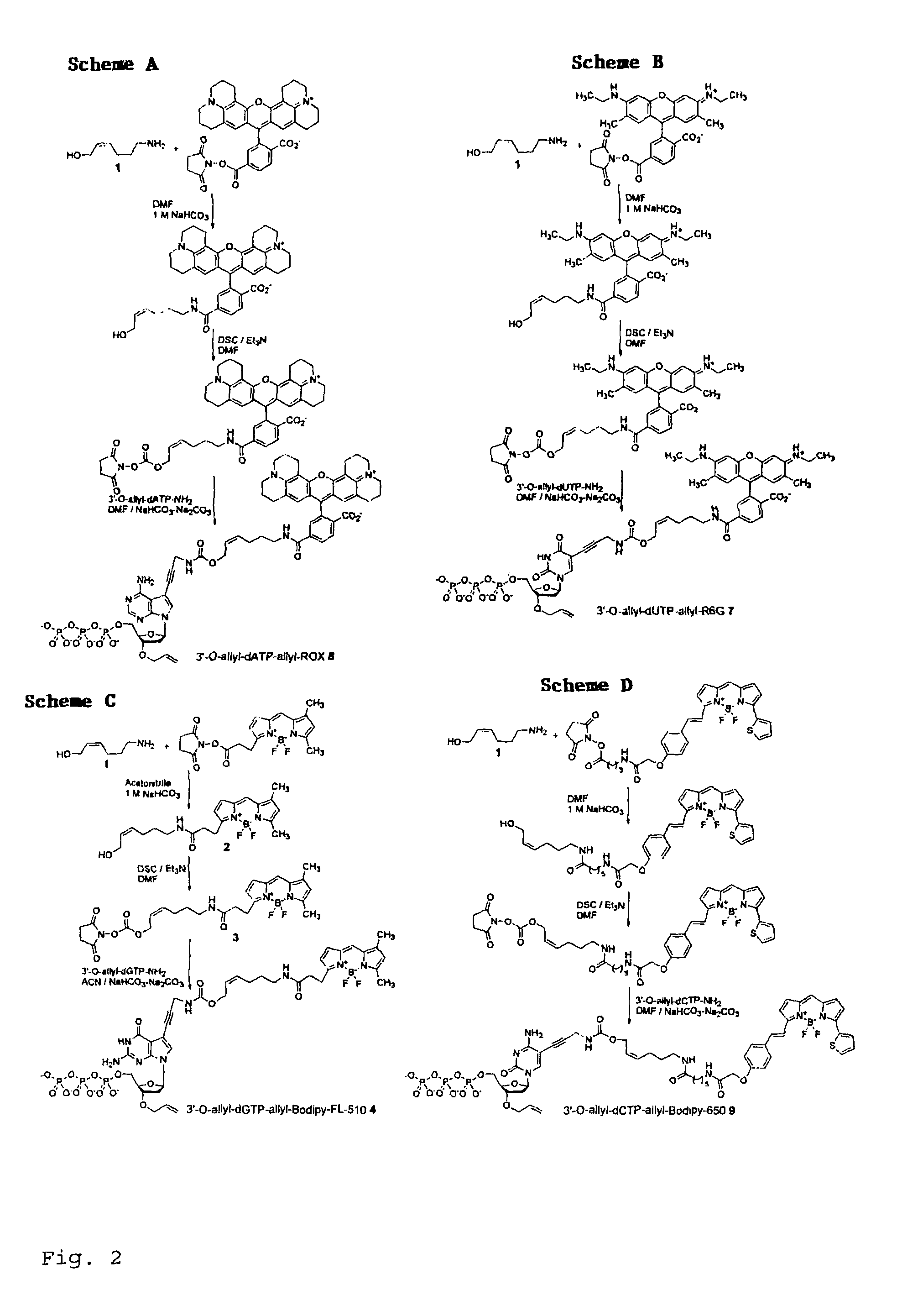
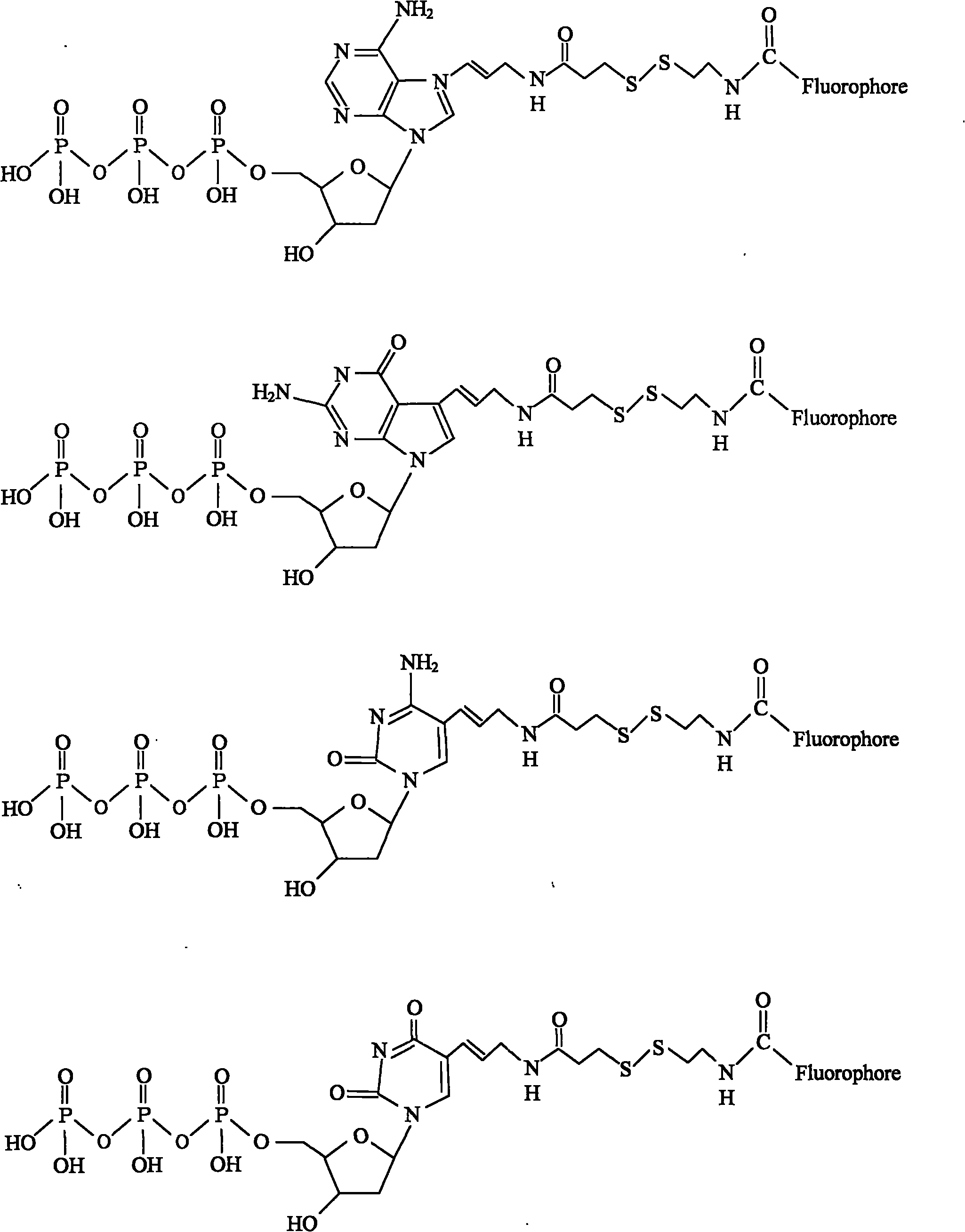
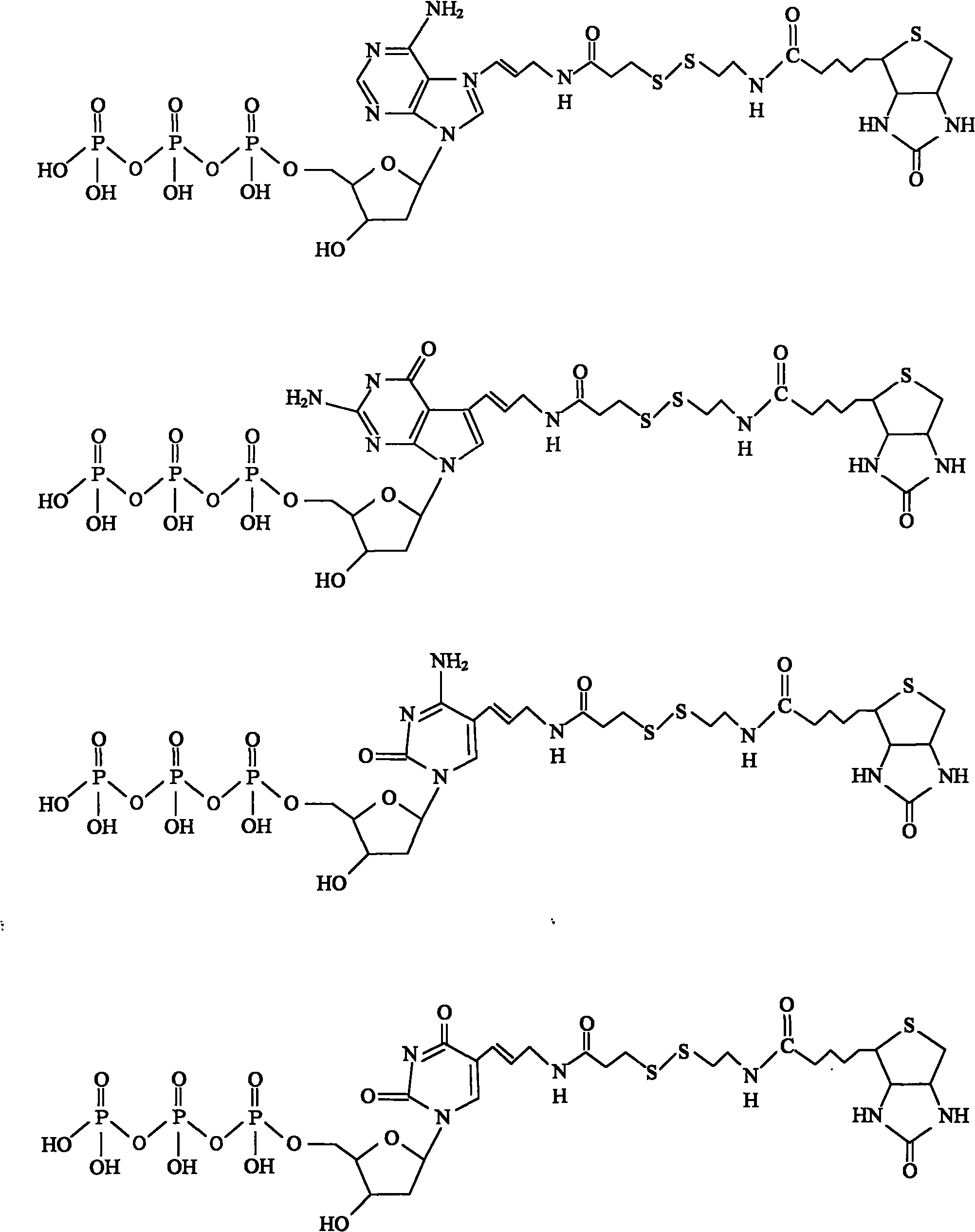
Chemically cleavable 3'-o-allyl-DNTP-allyl-fluorophore fluorescent nucleotide analogues and related methods
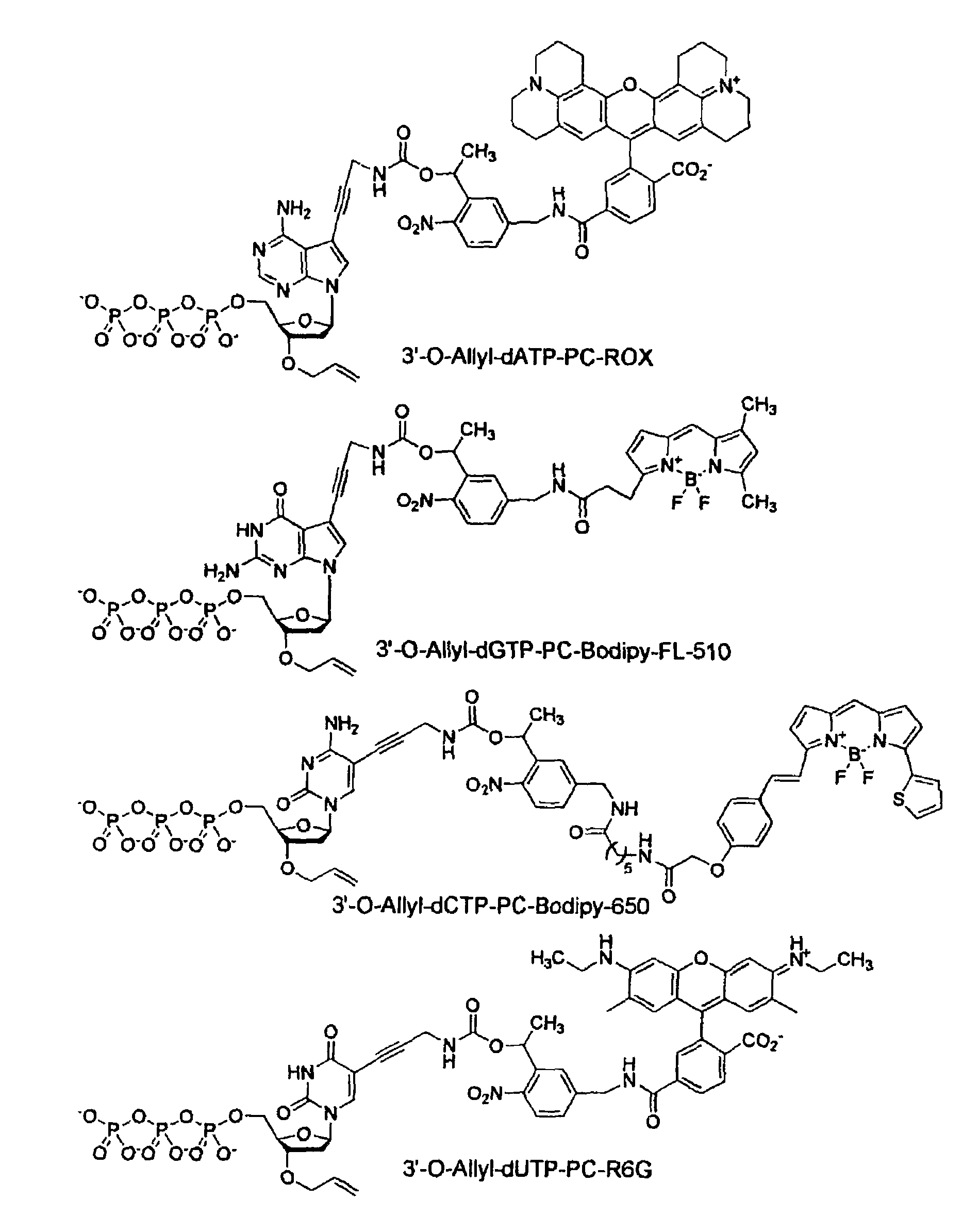
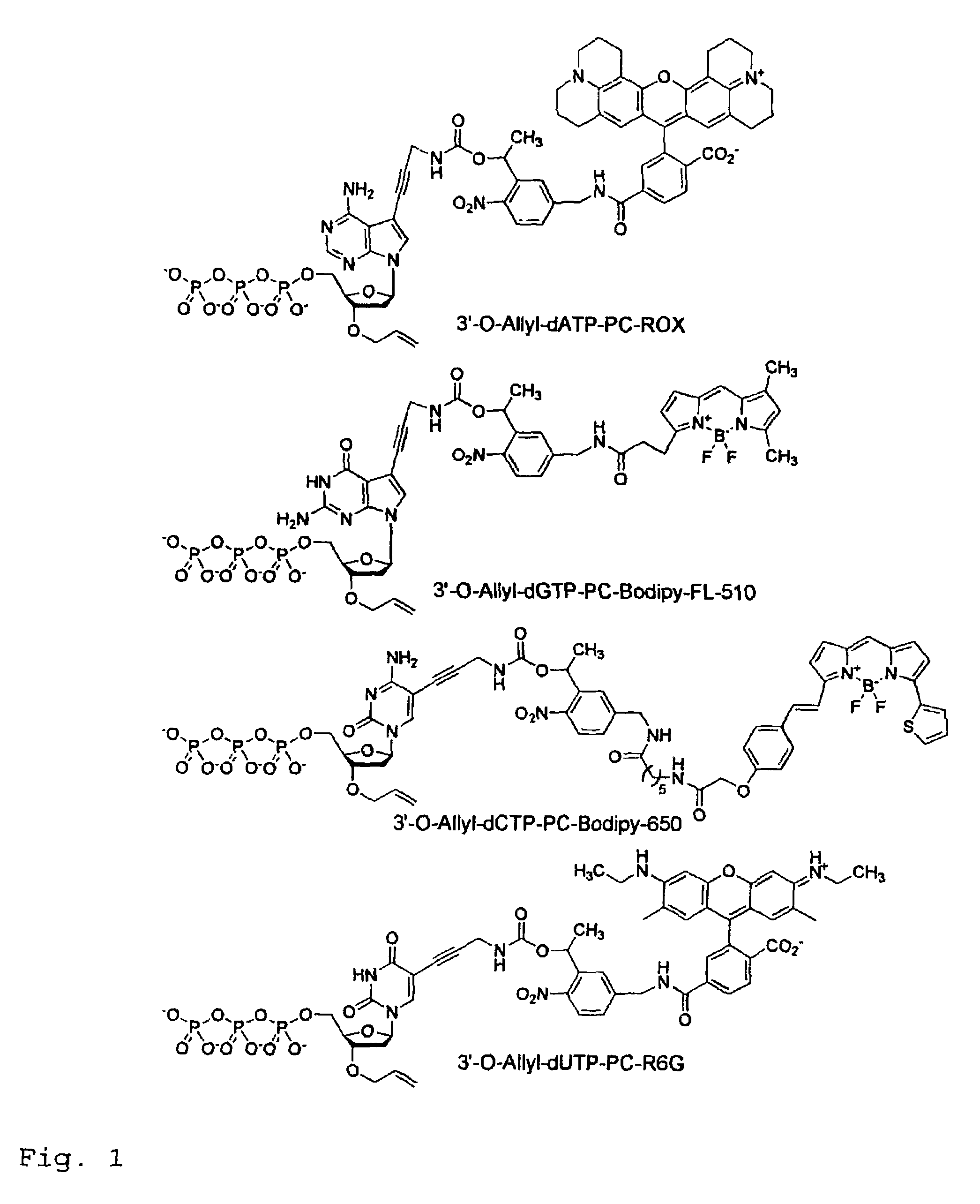
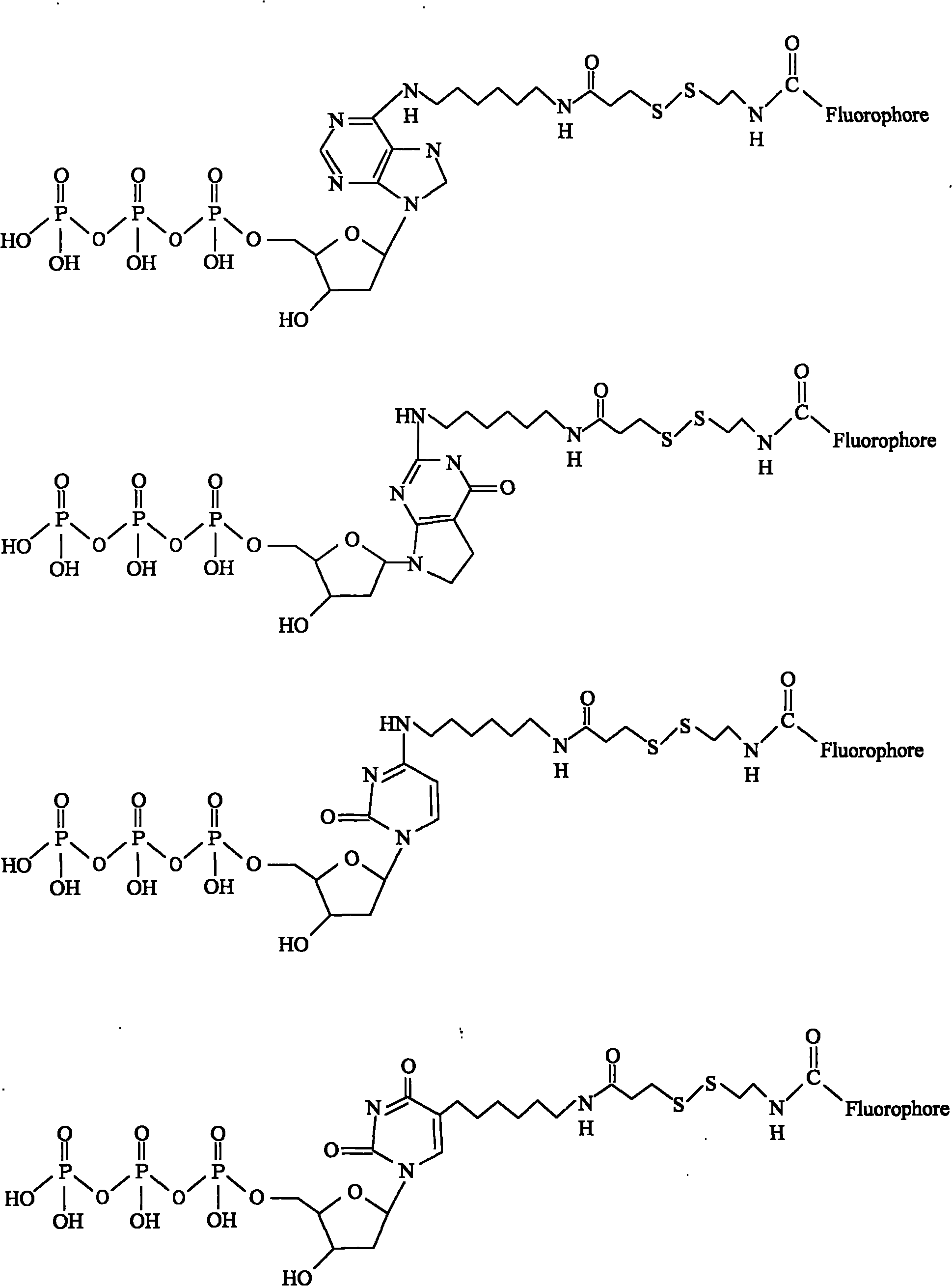
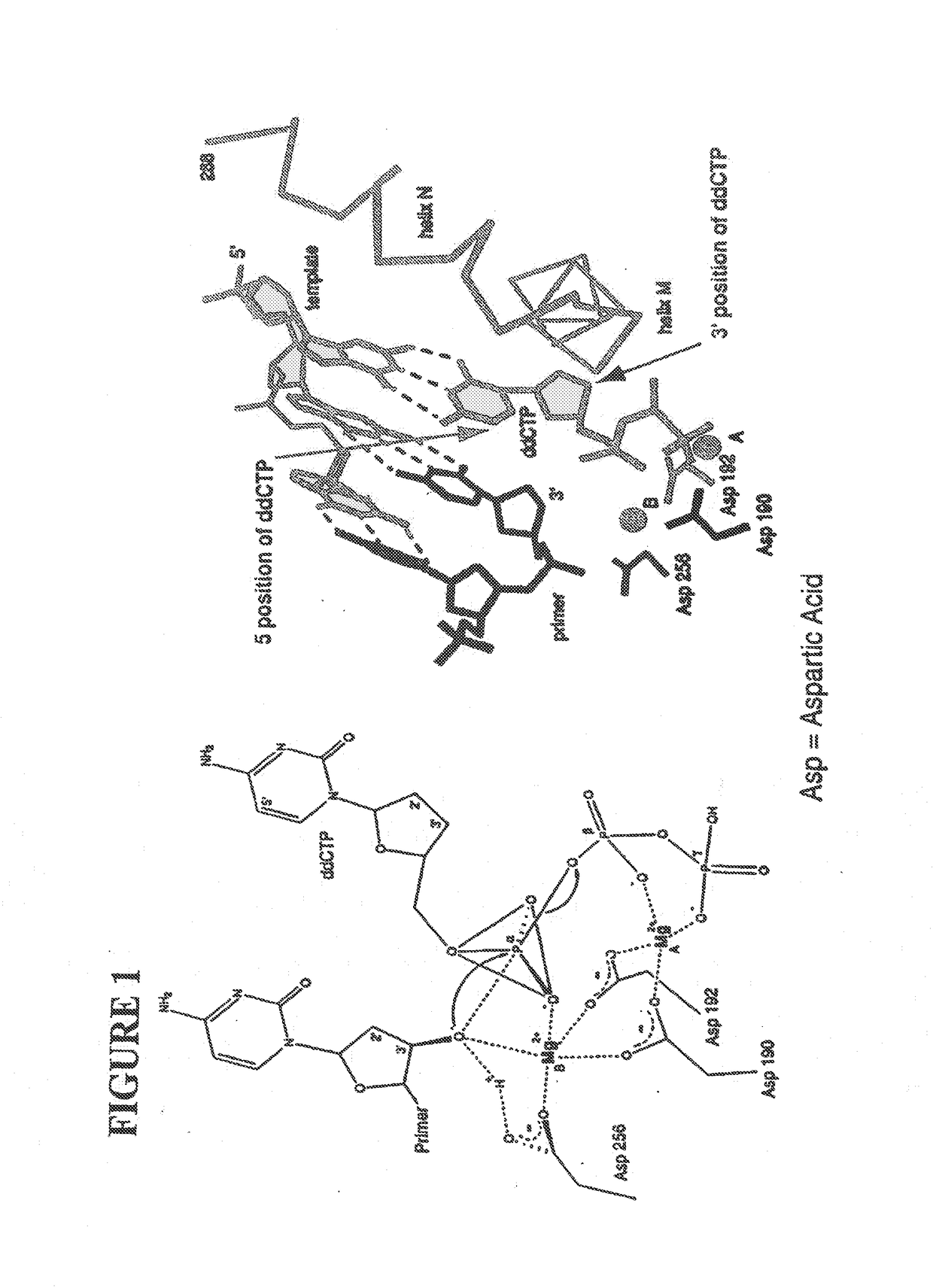
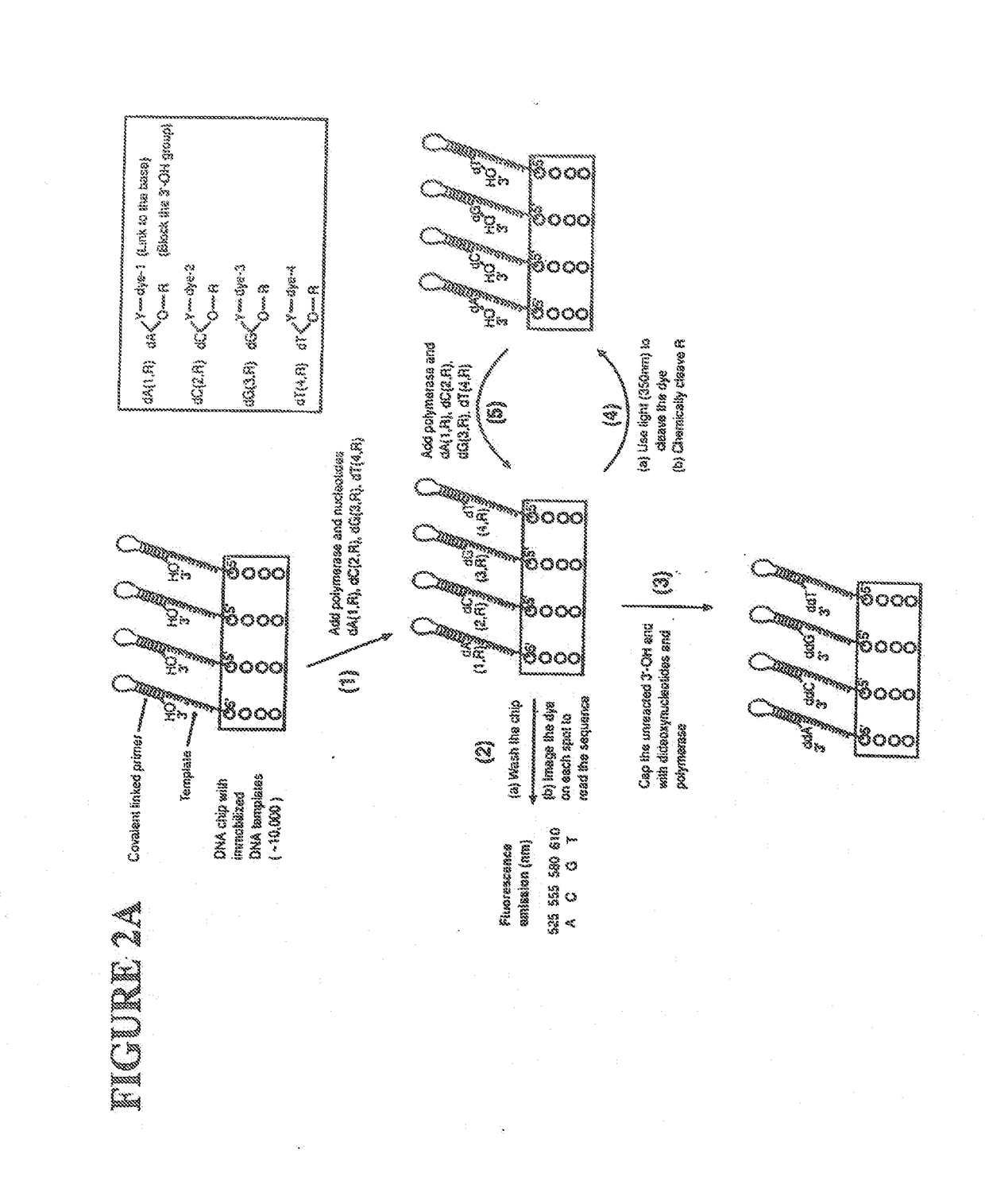
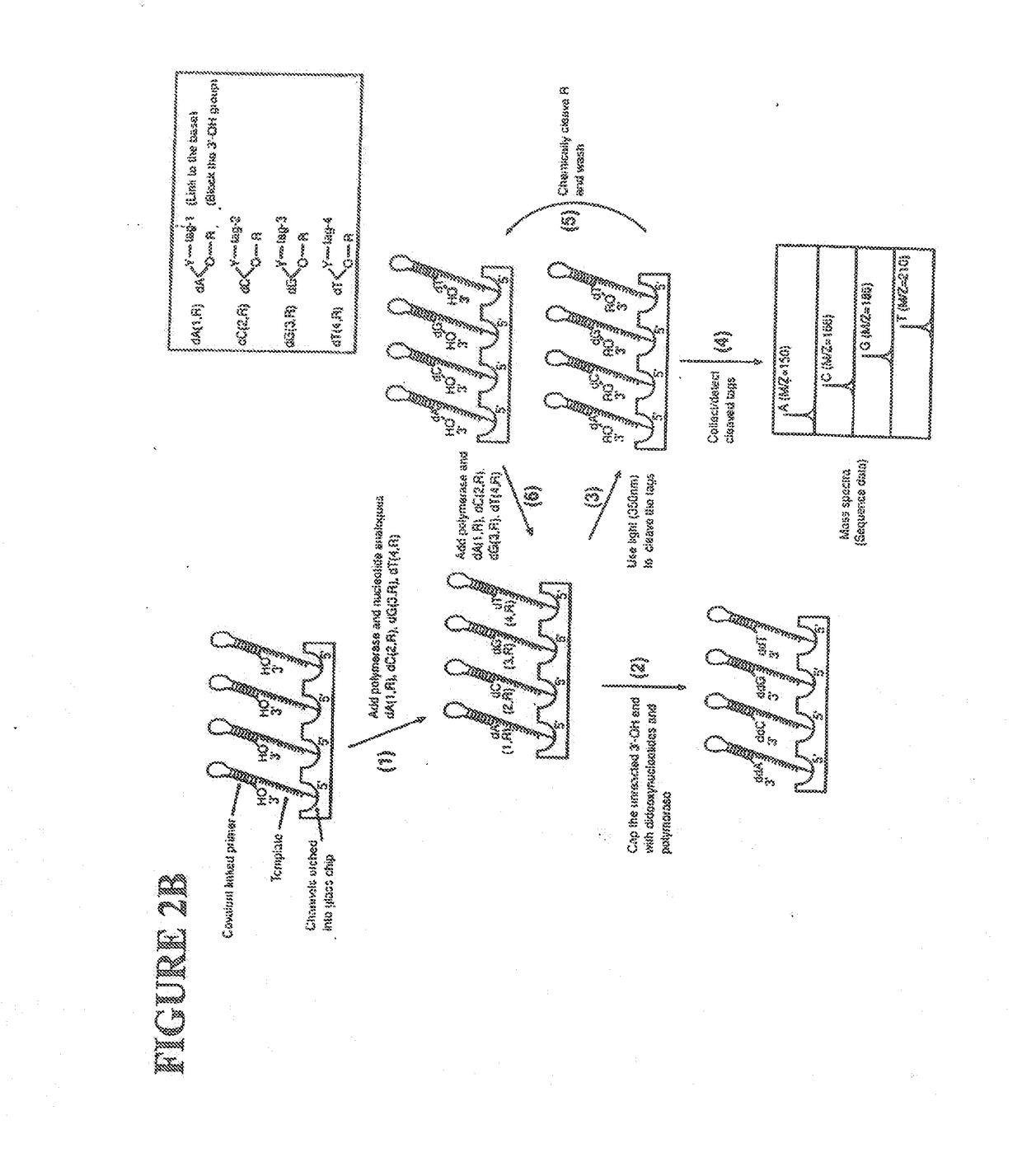
This invention provides a nucleotide analogue comprising (i) a base selected from the group consisting of adenine, guanine, cytosine, thymine and uracil, (ii) a deoxyribose, (iii) an allyl moiety bound to the 3′-oxygen of the deoxyribose and (iv) a fluorophore bound to the base via an allyl linker, and methods of nucleic acid sequencing employing the nucleotide analogue.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Control system for DNA (Deoxyribose Nucleic Acid) sequencer
ActiveCN102703314AImprove sequencing efficiencyTimely and accurate supplyBioreactor/fermenter combinationsBiological substance pretreatmentsPeristaltic pumpControl signal
The invention discloses a control system for a DNA (Deoxyribose Nucleic Acid) sequencer, which comprises a PLC (Programmable Logic Controller), controllers of first and second servo motors, first and second peristaltic pumps and driving motors of first and second multipass reversing valves, wherein the controllers of the first and second servo motors are electrically connected with the PLC; the first and second peristaltic pumps are respectively and electrically connected with the PLC and are used for receiving control signals of the PLC; a CCD (Charge Coupled Device) camera is electrically connected with the PLC and is used for receiving a control signal of the PLC; and a plurality of sensors are respectively and electrically connected with the PLC and are used for sending position signals of the CCD camera to the PLC. According to the control system disclosed by the invention, a reagent supply assembly of the DNA sequencer with a plurality of reaction bins timely and accurately supplies reagents and buffer solution for a plurality of reaction bins and the CCD camera can be ensured to timely read an optical signal in each reaction bin, and therefore, the aim of simultaneously performing a plurality of reactions is fulfilled, so that a plurality of samples can be simultaneously sequenced and the DNA sequencing efficiency is greatly improved.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION +1
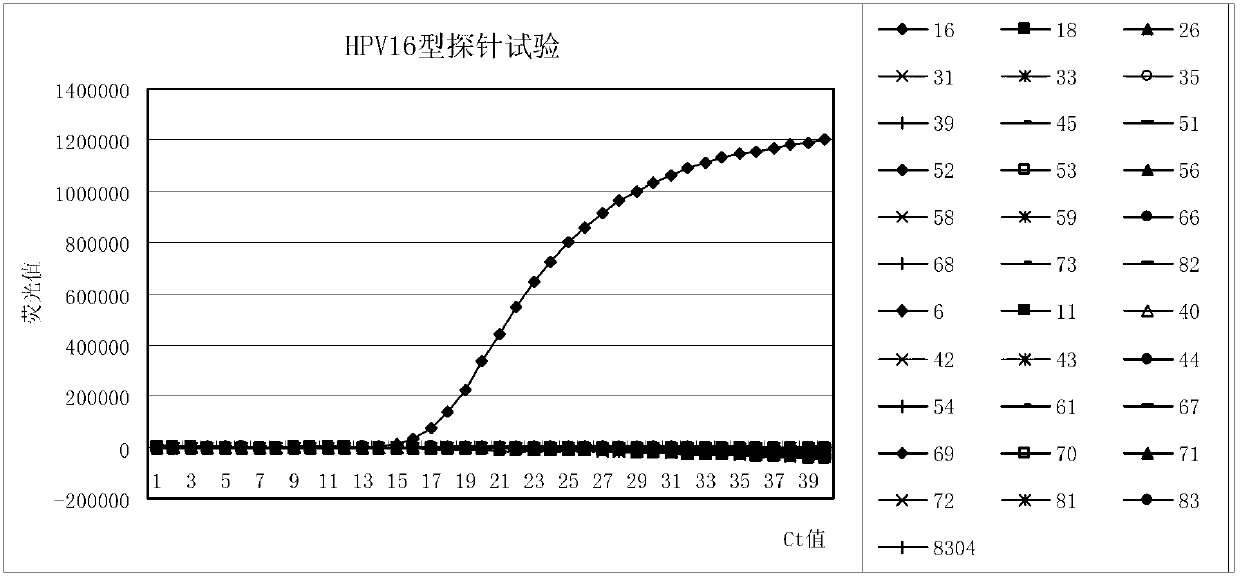
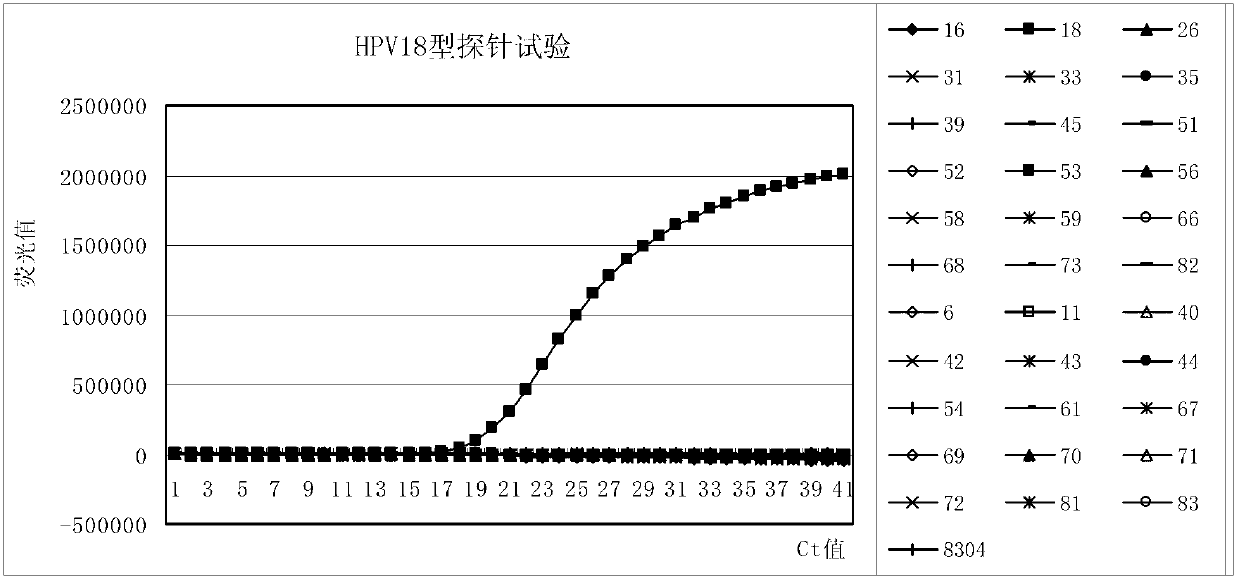
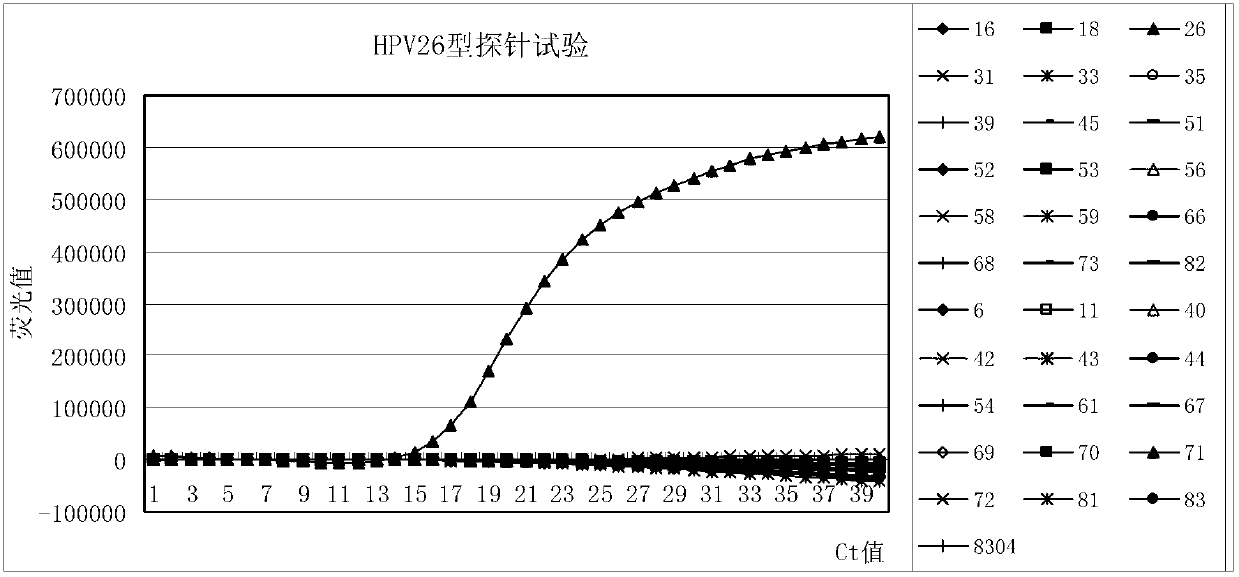
Primer, probe and kit for fluorescence PCR (Polymerase Chain Reaction) detection of 18 high-risk human papilloma viruses
ActiveCN102994651AMicrobiological testing/measurementFluorescence/phosphorescenceHuman papillomavirus DNAFluorescence
The invention discloses a primer, a probe and a kit for fluorescence PCR (Polymerase Chain Reaction) detection of 18 types of high-risk human papilloma viruses. Different typing kits are detected by adding different specificity probes; the DNAs (Deoxyribose Nucleic Acids)) of 18 types of common high-risk human papilloma viruses internationally recognized and closely related to the cervical cancer can be detected once; and typing detection is carried out on HPV (Human Papilloma Virus)16 and 18. The application provides 6 universal primers and 18 specific molecular beacon probes. The DNAs of the 18 types of common high-risk human papilloma viruses can be amplified by the 6 universal primers; and meanwhile, the 18 probes are the specific molecular beacon probes designed by aiming at the 18 high-risk types; different types of probes are added according to the detection need and combined as the kits aiming at the detection need of the different high-risk types; and at the same time, the added probes are marked by different report genes, so that the purpose of carrying out typing detection on the high-risk HPV is achieved.
Owner:英科新创(苏州)生物科技有限公司
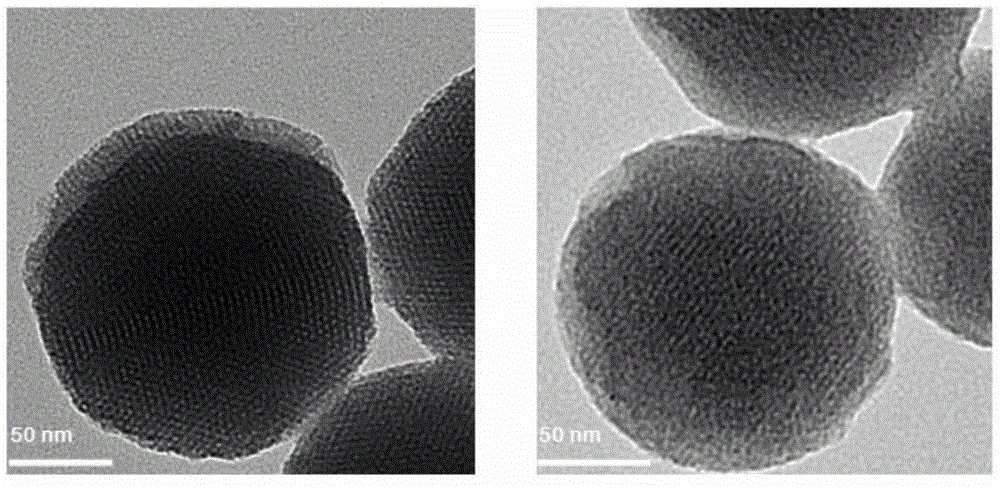
Method for preparing meso-porous silicon nano medicine carrier with cell specificity target, reduction responsiveness and triple anticancer treatment effects
InactiveCN104013965AOvercome toleranceEasy to operatePowder deliveryMacromolecular non-active ingredientsCell membraneCarrier system
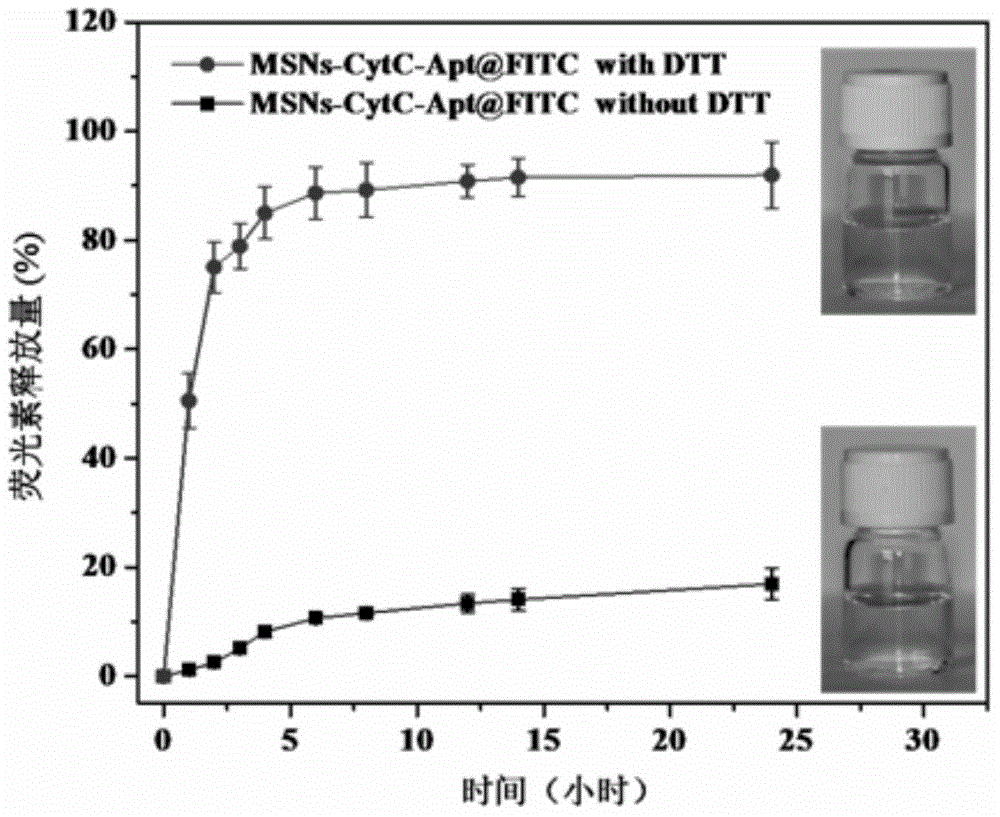
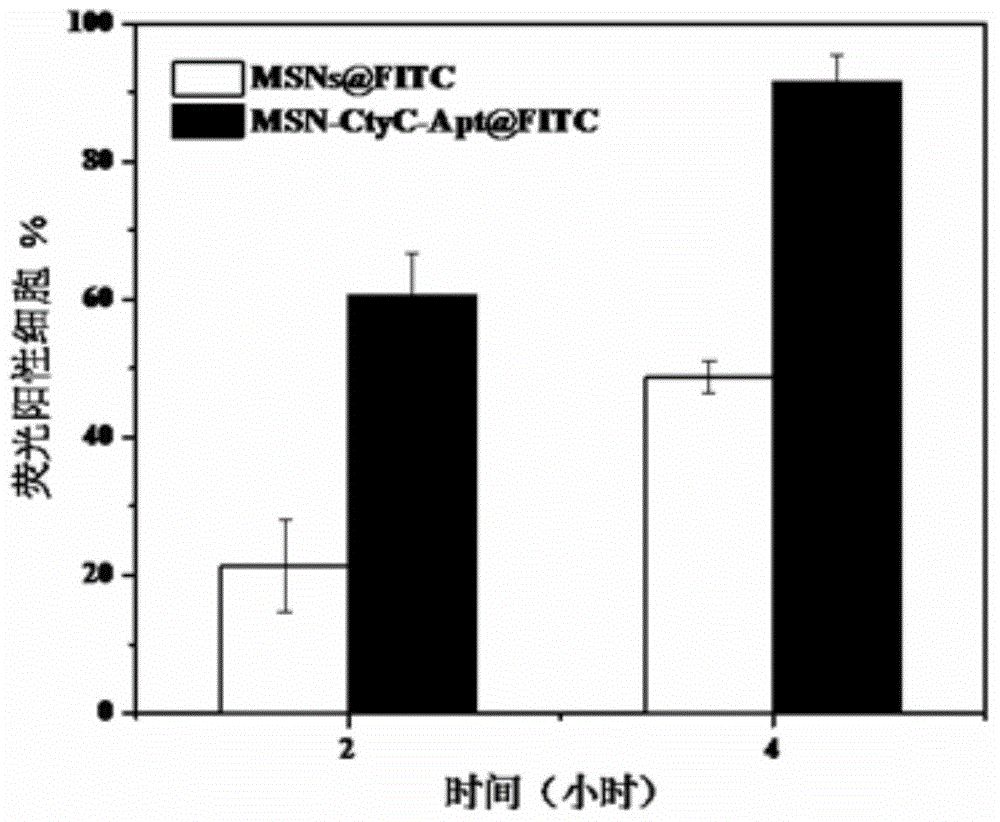
The invention discloses a method for preparing a meso-porous silicon nano medicine carrier with cell specificity target, reduction responsiveness and triple anticancer treatment effects. The method comprises the following steps: firstly, synthesizing meso-porous silicon nano particles by using a gel dissolution method, subsequently, introducing a disulfide bond onto the surface of a meso-porous silicon nano reservoir by using a chemical modification method, innovatively fixing cytochrome C with an apoptosis-inducing function onto the surface of the meso-porous silicon nano reservoir, blocking meso-porous channels with medicines, finally modifying DNA (Deoxyribose Nucleic Acid) aptamer single chain molecules (AS1411, with a cancer cell apoptosis-inducing function) onto the surface of a meso-porous silicon / cytochrome C nano composite system, and taking the system as specificity ligand of a receptor (nucleolin protein) which is overexpressed on the surface of liver cancer cytomembrane, thereby establishing a multifunctional composite type nano medicine carrier system for achieving triple anticancer treatment under combined action of medicines, blocking substances and target molecules inside meso-pores.
Owner:CHONGQING UNIV
Gene-specific molecular marker Pi2SNP of rice blast-resistant gene Pi2 as well as preparation method and application thereof
ActiveCN103320437AStrong specificityHigh sequence identityMicrobiological testing/measurementDNA preparationAgricultural scienceEnzyme digestion
The invention discloses a gene-specific molecular marker Pi2SNP of a rice blast-resistant gene Pi2 as well as a preparation method and application thereof. The molecular marker Pi2SNP is a nucleotide fragment II in a specific banding pattern with the rice blast-resistant gene Pi2 obtained by amplifying F1 and R1 from the total DNA (Deoxyribose Nucleic Acid) of the rice blast-resistant variety carrying rice blast-resistant gene Pi2 by use of a primer to obtain a nucleotide fragment I and then performing enzyme digestion of the nucleotide fragment I by use of restriction endonuclease Pst I or Hinf I. The molecular marker is the first Pi2 gene-specific SNP marker developed for the sequence in the Pi2 gene, and has the advantages of high specificity and low cost and high flux in practical application, and can be widely applied to the colonies with different inheritance backgrounds. By adopting the molecular marker, the utilization efficiency of the gene in molecular marker-assisted selective breeding, gene pyramiding breeding and transgenic breeding can be improved.
Owner:PLANT PROTECTION RES INST OF GUANGDONG ACADEMY OF AGRI SCI
Live attenuated mycoplasma strains
InactiveUS20090068231A1Reduce expressionReduced virulenceAntibacterial agentsBacterial antigen ingredientsBacteroidesVaccination
The present invention provides live, attenuated Mycoplasma bacteria that exhibit reduced expression of one or more proteins selected from the group consisting of pyruvate dehydrogenase, phosphopyruvate hydratase, 2-deoxyribose-5-phosphate aldolase, and ribosomal protein L35, relative to a wild-type Mycoplasma bacterium of the same species. Also provided are vaccines and vaccination methods involving the use of the live, attenuated Mycoplasma bacteria, and methods for making live attenuated Mycoplasma bacteria.
Owner:WYETH LLC
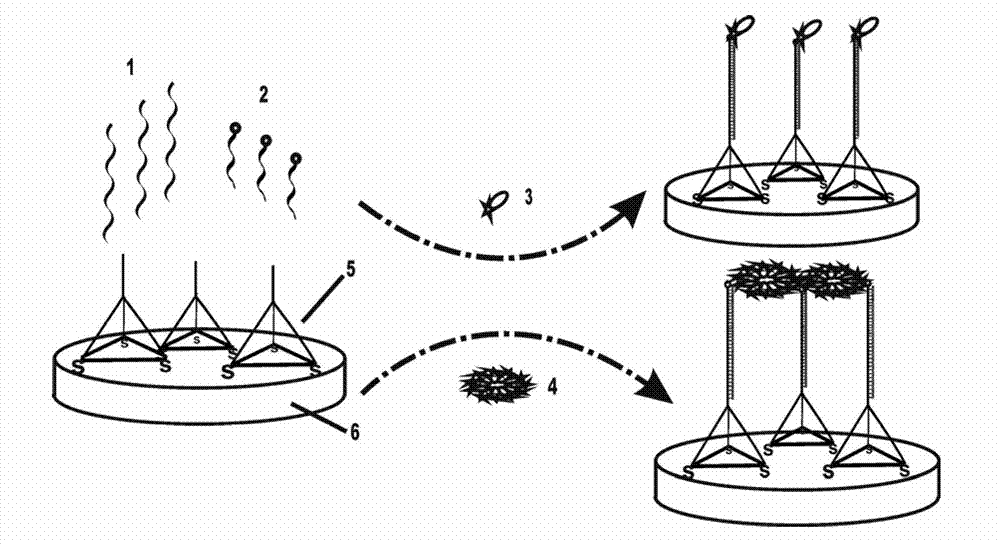
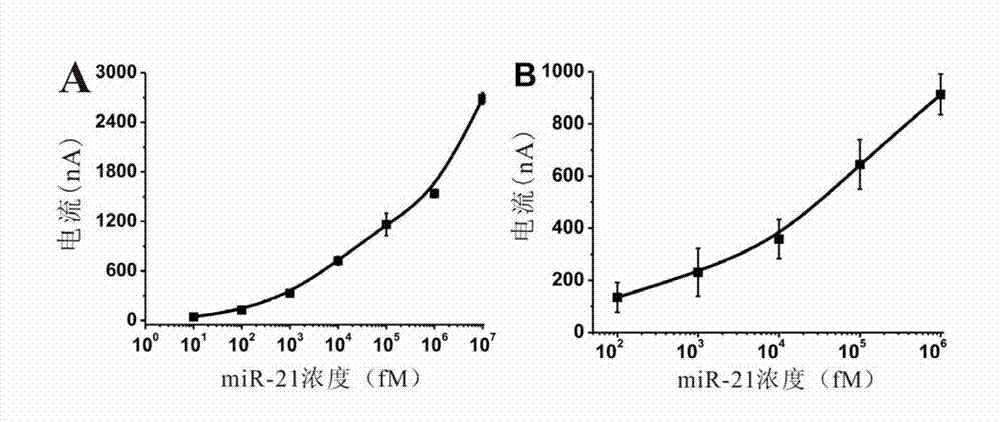
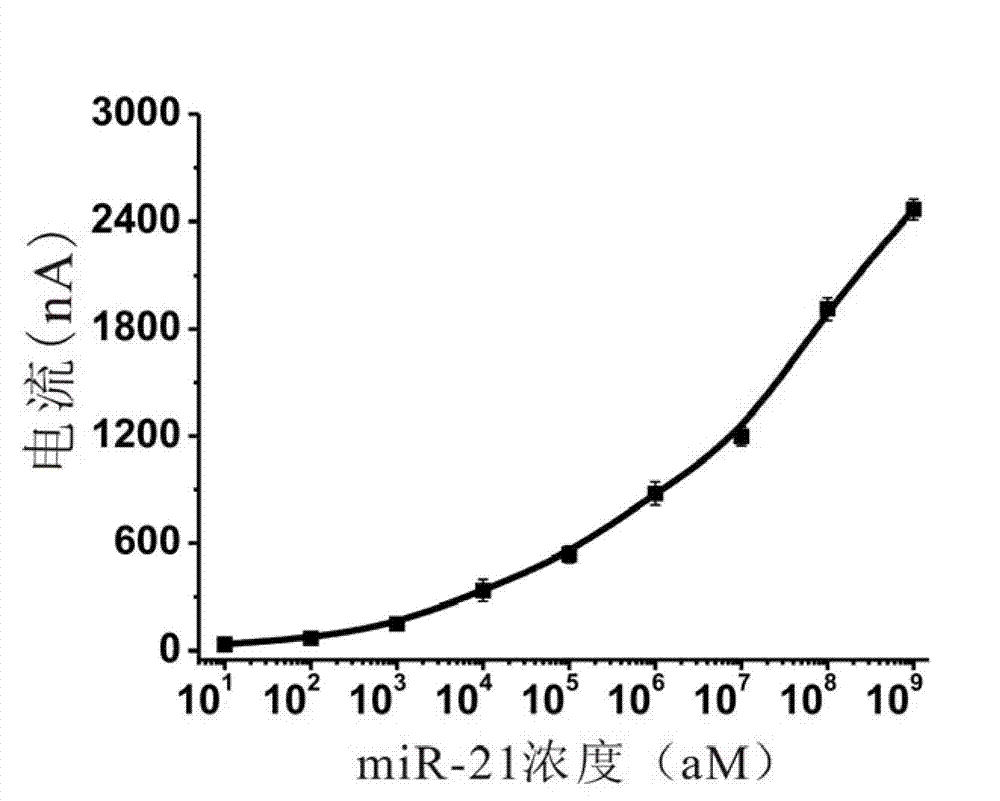
Electrochemical miRNA (micro Ribose Nucleic Acid) detection method based on DNA (Deoxyribose Nucleic Acid) three-dimensional nano structure probe
The invention provides an electrochemical miRNA (micro Ribose Nucleic Acid) detection method based on a DNA (Deoxyribose Nucleic Acid) three-dimensional nano structure probe. The electrochemical miRNA detection method comprises the steps of: synthesizing a DNA three-dimensional nano structure probe through a self-assembly method, wherein the DNA three-dimensional nano structure probe comprises one section of extended recognition sequence; assembling the DNA three-dimensional nano structure probe on the surface of a working electrode of an electrochemical device; hybridizing a target miRNA with the DNA three-dimensional nano structure probe on the surface of the working electrode; and adding oxidordeuctase and a corresponding substrate, and carrying out electrochemical detection by using the electrochemical device. The method can be used for detecting the miRNA of 10aM, therefore, the problem of the great demand on test samples in the detection method in the prior art is solved. In addition, the method has strong specificity selection and can be well used for distinguishing base pair mismatching of same family of miRNA. Compared with the method using the single-chain DNA probe, the electrochemical miRNA detection method is higher in stability.
Owner:SHANGHAI INST OF APPLIED PHYSICS - CHINESE ACAD OF SCI
DNA and fingerprint authentication of mobile devices
ActiveUS9307407B1Expand coverageImprove performanceModulated-carrier systemsUnauthorised/fraudulent call preventionCode division multiple accessControl signal
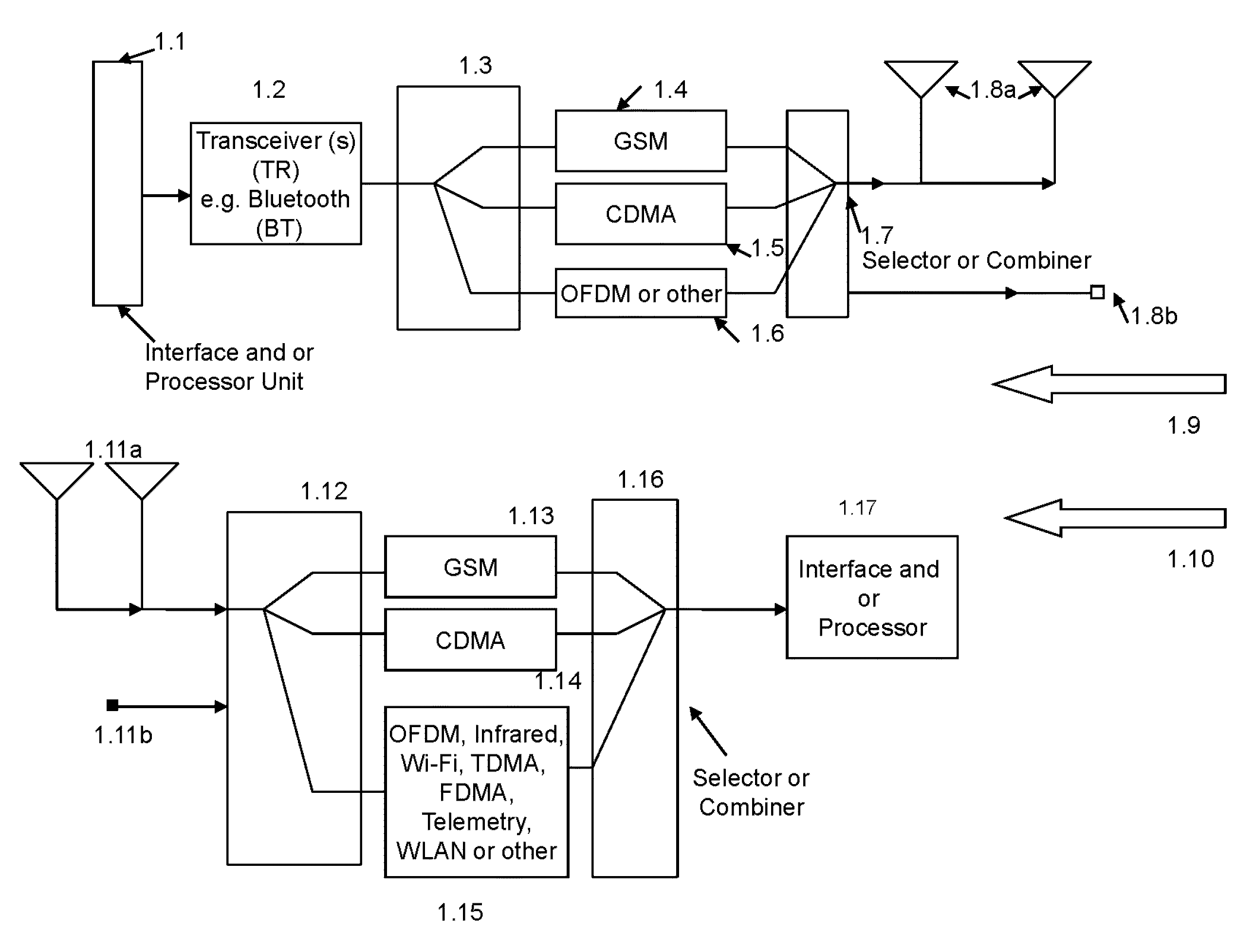
User generated and processed Deoxyribose Nucleic Acid (DNA) or fingerprint signal is used for authentication of a mobile device. A touch screen generated processed control signal is used for control of mobile device operated in a cellular system and a Wi-Fi network. The mobile device receives, demodulates and processes a location finder signal and provides processed location finder signal to an interface unit of mobile device. From a cellular base station the mobile device receives, demodulates and processes a modulated spread spectrum signal into a received, demodulated and processed baseband spread spectrum signal and processes, modulates and transmits in cascade the processed spread spectrum signal in a Wi-Fi system. The mobile device operates in a repeater mode. In one of the embodiments the spread spectrum signal uses a Code Division Multiple Access (CDMA) modulated signal and the Wi-Fi system uses OFDM modulated signal. The mobile device comprises step of processing a video signal into baseband in-phase and a quadrature-phase cross-correlated spread spectrum or Time Division Multiple Access (TDMA) signal. The video TDMA signal is modulated and transmitted as a Gaussian Minimum Shift Keying (GMSK) modulated signal in said cellular system and wherein said video signal comprises a three dimensional (3D) image.
Owner:FEHER KAMILO
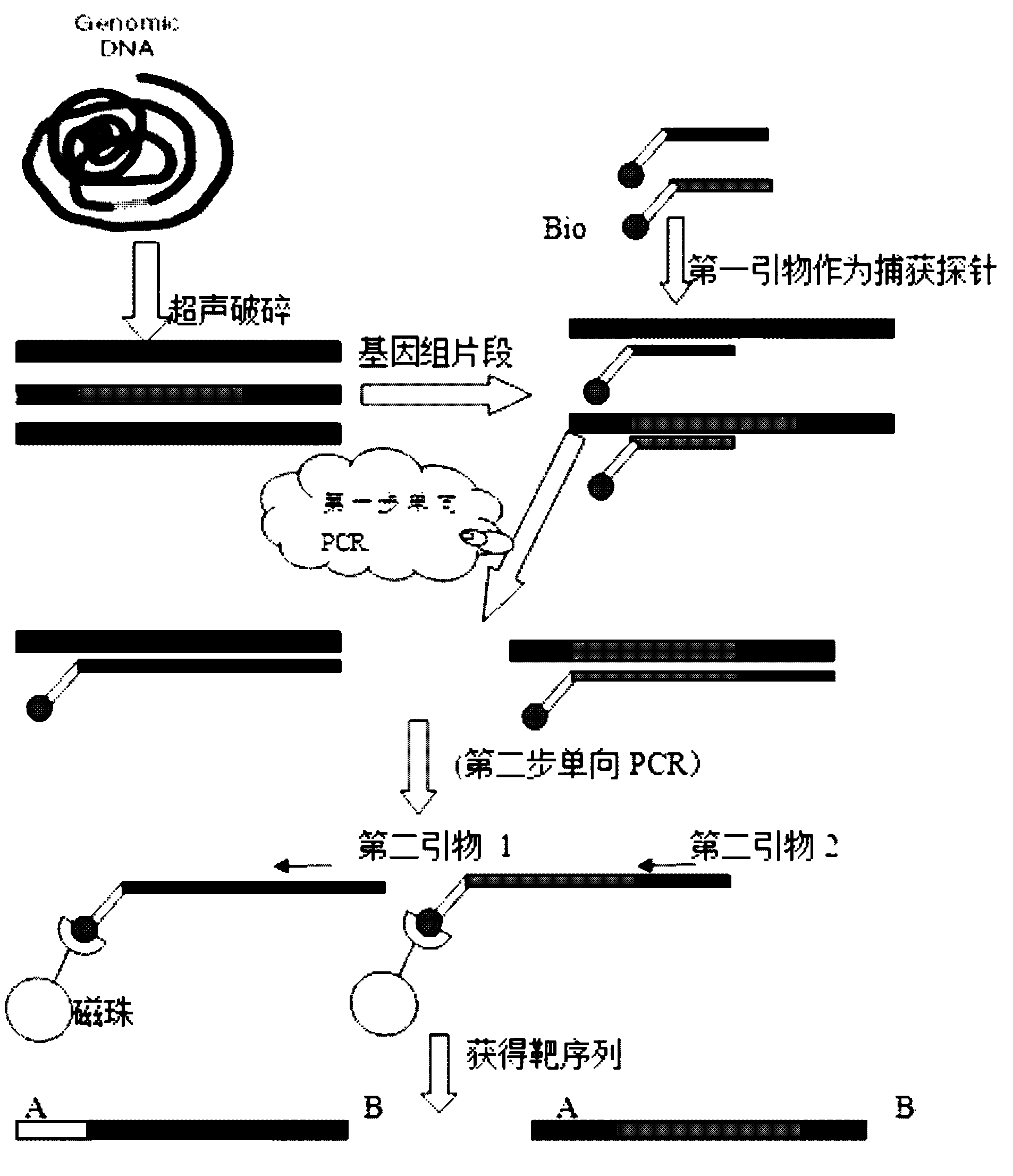
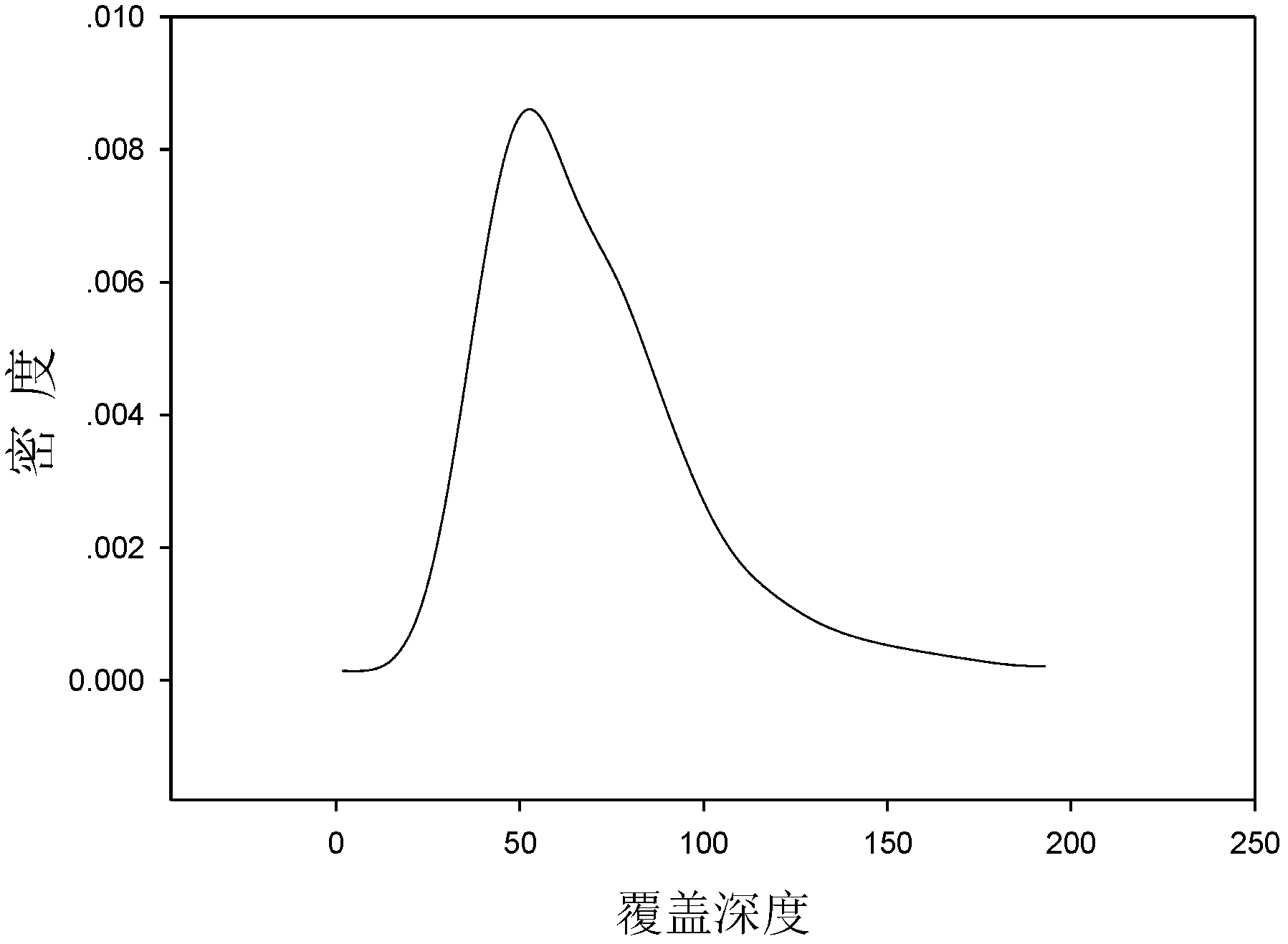
Preparation method of genome mixing sequencing library
InactiveCN105671644AImprove connection efficiencyImprove the efficiency of database constructionMicrobiological testing/measurementLibrary creation3-deoxyriboseDeoxyribose
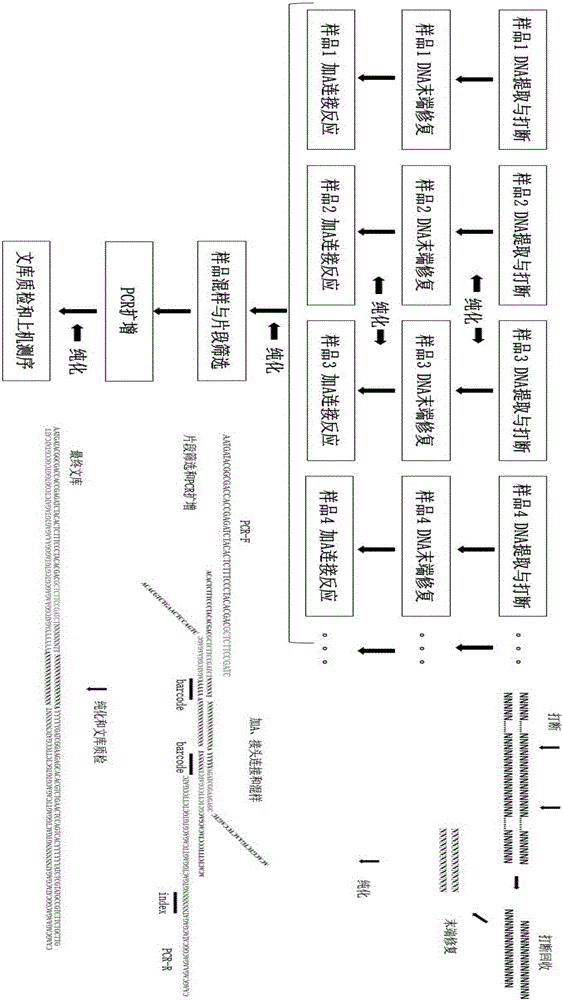
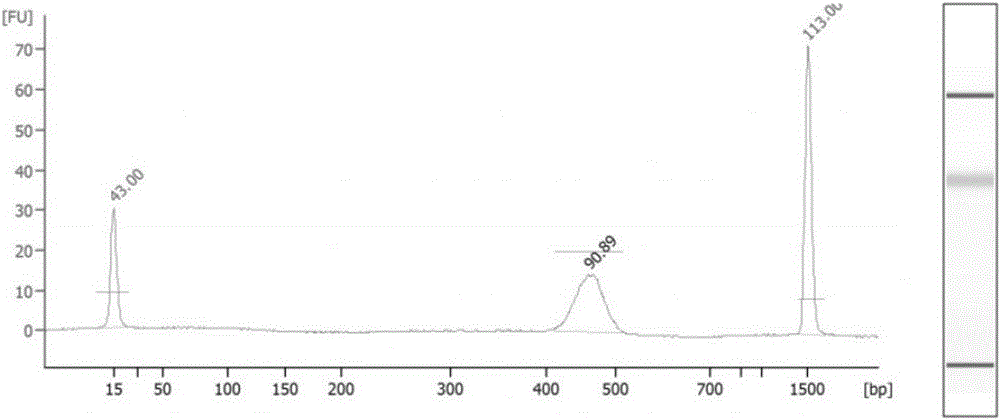
The invention relates to the technical field of molecular biology, and discloses a method for preparing a genome mixed-sample sequencing library, which comprises the following steps: (1) ultrasonic fragmentation of genomic DNA; (2) purification and end repair of fragmented products; (3) ) repair product for purification and adapter ligation; (4) ligation product purification recovery and concentration determination; (5) sample mixing and fragment screening; (6) PCR amplification of the product after fragment screening; (7) purification of the PCR product to obtain Sequencing library; (8) library quality inspection and on-machine sequencing. The present invention provides an adapter compatible with the Illumina next-generation sequencer in the above step (2), a method for purifying the product in the step (2) (3) (4) (7), and a method for mixing samples in the step (4) And the PCR amplification system and program setting in the step (6) ensure that the library building method provided by the present invention can quickly and smoothly carry out multiple sample mixed sample library building, and the method provided by the present invention can be used to obtain good data uniformity. and high-quality sequencing data.
Owner:WUHAN BINGGANG BIOTECH CO LTD
Test strip for detecting aflatoxin B1 or M1 by utilizing aptamer
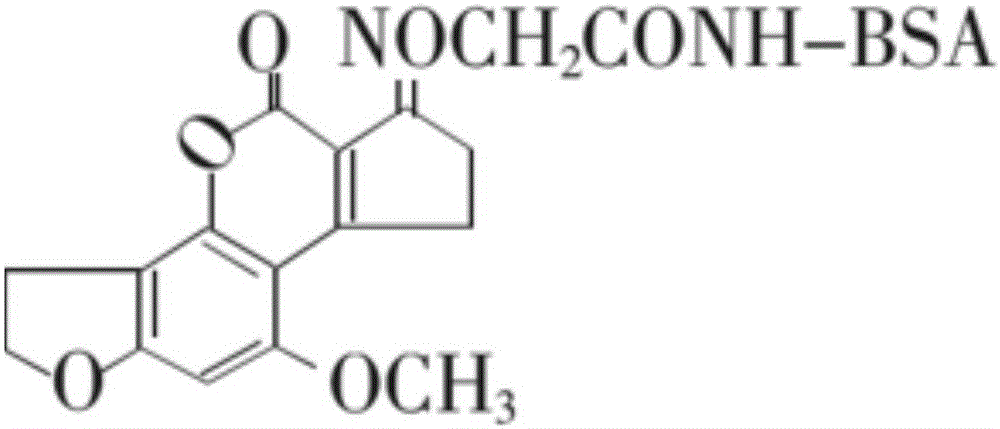
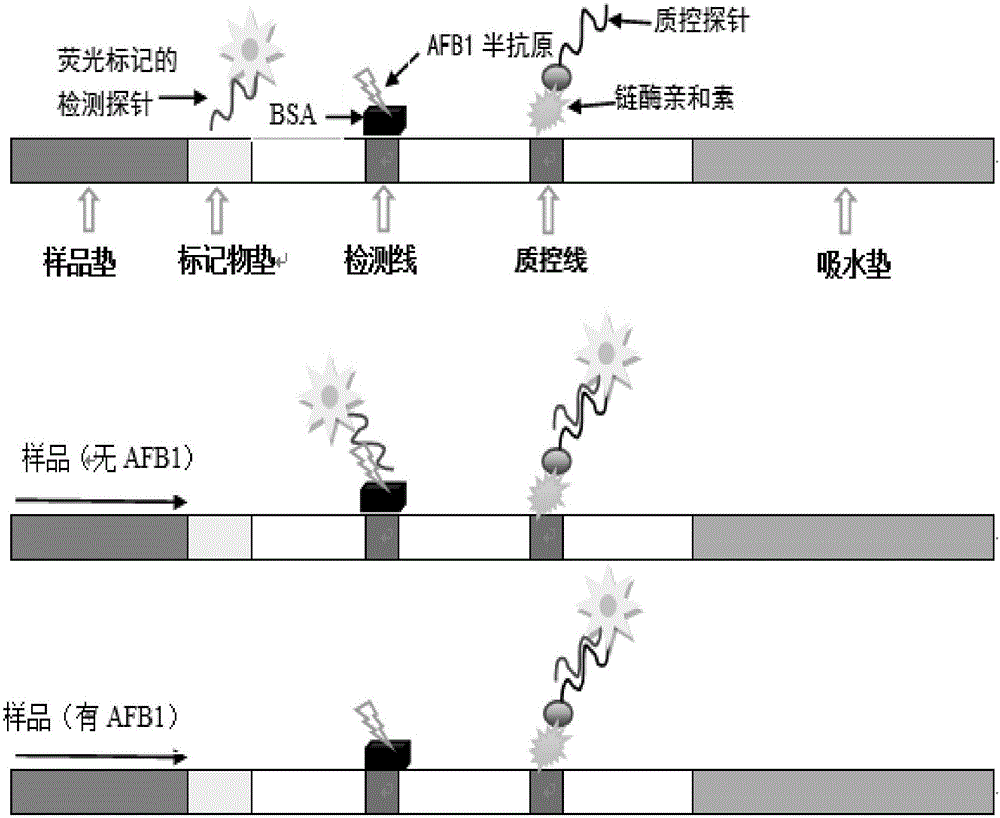
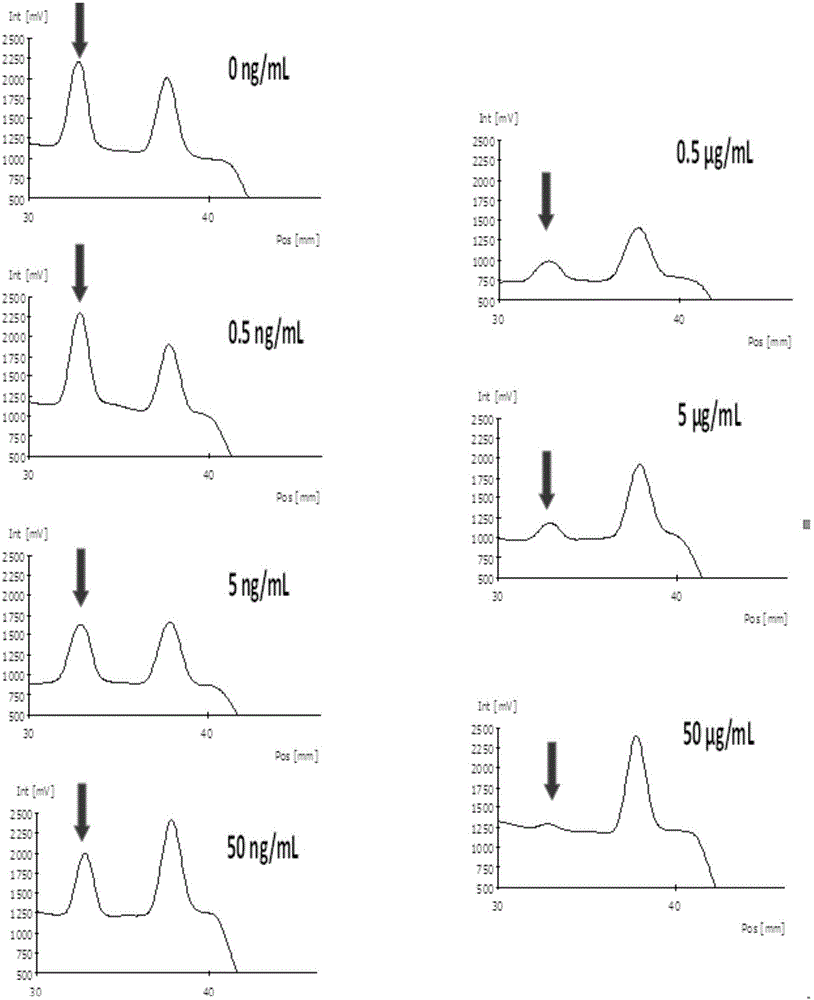
The invention discloses a test strip for detecting aflatoxin B1 or M1 by utilizing an aptamer. The test strip for detecting the aflatoxin B1 or M1 by utilizing the aptamer, provided by the invention, comprises a sample absorption pad, a marker pad, a reaction film and a water absorption pad, wherein the marker pad is coated with a detection probe; the detection probe is an aflatoxin B1 aptamer marked by fluorescein; the nucleotide sequence of the aflatoxin B1 aptamer is sequence 1; the reaction film comprises a detection region and a quality control region; the detection region is coated with a conjugate formed by an aflatoxin B1 hapten and a carrier protein; and the quality control region is coated with a quality control probe, and the quality control probe is a conjugate formed by avidin conjugation probe marked aflatoxin B1 complementary single strand DNA (Deoxyribose Nucleic Acid)) molecules. The test strip for detecting the aflatoxin B1 or M1, provided by the invention, has the advantages of high sensitivity, accuracy in quantifying, strong specificity, simplicity and convenience, and short detection time.
Owner:INST OF QUALITY STANDARD & TESTING TECH FOR AGRO PROD OF CAAS
Method for detecting surviving gene based on graphene-gold composite material electrochemical DNA (Deoxyribose Nucleic Acid) biosensor
InactiveCN103063715AImprove conduction abilityImprove electrocatalytic activityMaterial electrochemical variables3-deoxyriboseSulfur
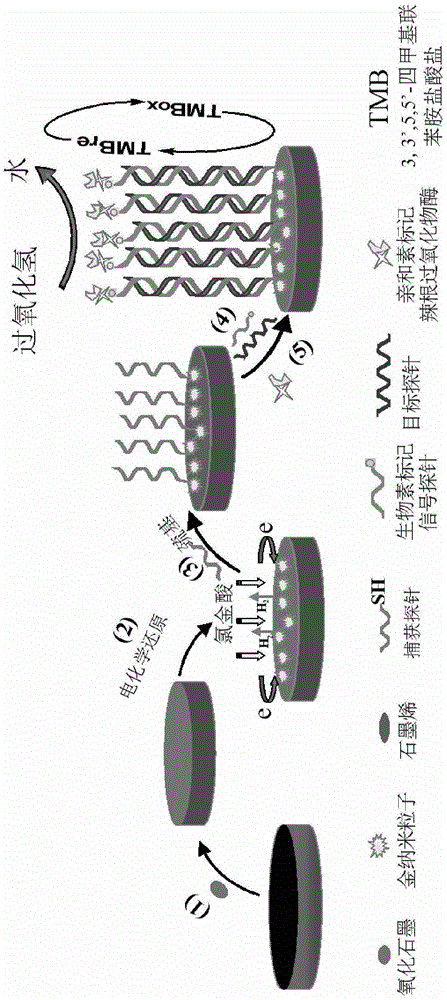
The invention discloses a method for detecting a surviving gene based on a graphene-gold composite material electrochemical DNA (Deoxyribose Nucleic Acid) biosensor. The method comprises the following steps: a specific probe is designed according to a gene segment to be detected; a capture probe is self-assembled on the surface of G-3DAu / GCE through a gold-sulfur bond; the capture probe and a signal probe with the tail end marked by biotin are respectively combined with a target DNA to form a 'sandwich' model in the presence of the target DNA; a horse radish peroxidase marked by avidin can be combined with the biotin marked by the signal probe, so that the HRP (Horse Radish Peroxidase) can be fixed on the surface of an electrode; the electrode is placed in a base solution of 3, 3', 5, 5'-tetramethyl benzidine (TMB) and H2O2, and the H2O2 can oxidize TMB to generate a bidiazotizedbenzidine material under the catalyzing of the HRP, so that an electrochemical signal is generated. The method has the advantages of being simple, quick and green in preparation process, and high in selectivity and sensitivity.
Owner:FUJIAN MEDICAL UNIV
Methods of cDNA preparation
InactiveUS20080145844A1Microbiological testing/measurementFermentation3-deoxyriboseReverse transcriptase activity
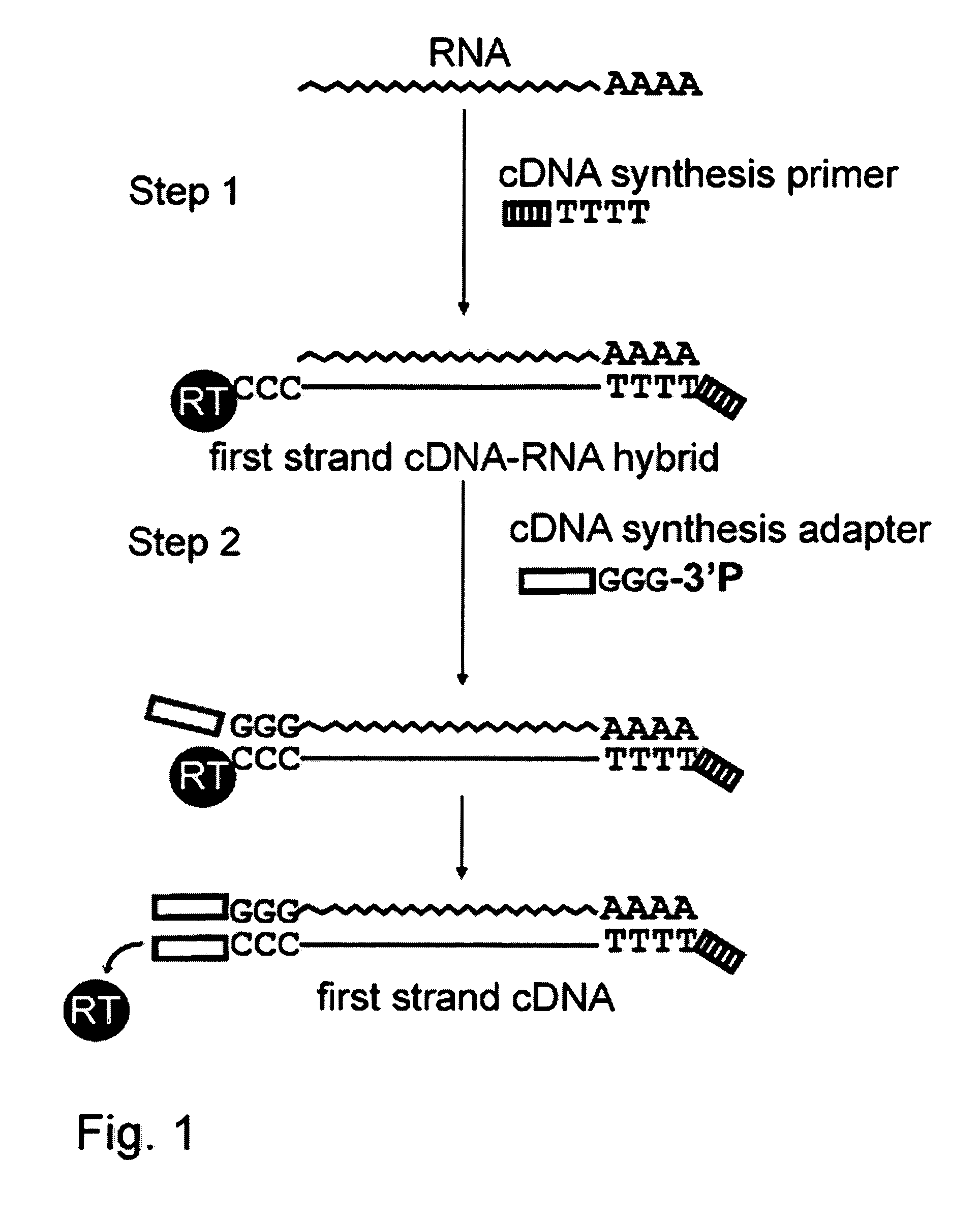
The present invention provides an improved method for cDNA preparation. The method of the present invention comprises the following steps: (1) contacting mRNA with a cDNA synthesis primer which can anneal to RNA and a suitable enzyme which possesses reverse transcriptase activity under conditions sufficient to permit the template-dependent extension of the primer to generate an mRNA-cDNA intermediates; (2) contacting a mixture from step 1 with a deoxyribonucleotide adapter in the presence of Mn2+-ions, wherein said oligonucleotide adapter has a pre-selected arbitrary nucleotide sequence at its 5′-end, and a short dG stretch at its 3′-end. The 3′-end nucleotide of the adapter is a terminator nucleotide, e.g., a nucleotide with a modified 3′-OH group of a deoxyribose residue.
Owner:EVROGEN
Method for synthesizing fluorescent silver nano clusters by taking general DNA (Deoxyribose Nucleic Acid) as stabilizer
The invention belongs to the field of chemical engineering, and particularly relates to a method for large-scale synthesization of fluorescent silver nano clusters by taking general modified DNA (Deoxyribose Nucleic Acid) as a stabilizer. The method mainly comprises the following steps: (1) mixing the general modified DNA with a silver ion solution and reacting in a refrigerator of 4 DEG C for 24 h; and (2) adding a NaBH4 solution into a mixed solution in the step (1) and reacting for 3 h at the room temperature of 10-25 DEG C to obtain a silver nano cluster solution. According to the method, the general modified DNA is taken as the stabilizer for the convenience of large-scale synthesization of the fluorescent silver nano clusters; and the synthetic method is economic, the particle size is smaller, the particle distribution is uniform, and strong fluorescent light can be generated.
Owner:HUNAN UNIV OF SCI & TECH
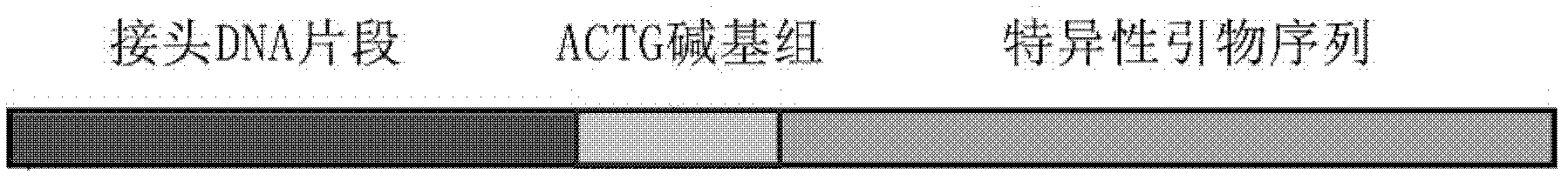
Asymmetric deoxyribose nucleic acid (DNA) artificial adapters by using second-generation high-throughput sequencing technology and application thereof
InactiveCN102061335AImprove connection efficiencyEasy to storeMicrobiological testing/measurement3-deoxyriboseElectrophoresis
The invention provides asymmetric deoxyribose nucleic acid (DNA) artificial adapters by using a second-generation high-throughput sequencing technology, which is composed of two DNA oligonucleotide single strands. The preparation method comprises the following steps: dissolving the two DNA oligonucleotide single strands into a quenching solution; regulating the final concentration to 2mM and volume to 20microlitre; carrying out a quenching reaction to form the DNA artificial adapters which are locally and mutually complemented at a temperature of 95 DEG C for 5 minutes; reducing the temperature of 95 DEG C to 12 DEG C at the speed of 0.1 DEG C per second; keeping the temperature of 12 DEG C, and diluting an adapter solution after quenching to 500 mu M at a ratio of 1:4; and storing at the temperature of minus 20 DEG C. An application of the asymmetric DNA artificial adapters by using the second-generation high-throughput sequencing technology is as follows: A, asymmetric DNA adapters are in ligation with DNA samples; B, non-connected DNA artificial adapters are purified and isolated by using electrophoresis tapping; C, judgment is carried out; and D, a polymerase chain reaction (PCR) amplification reaction is performed based on an asymmetric sequence. The asymmetric DNA artificial adapters by using the second-generation high-throughput sequencing technology has the obvious advantages that: 1) the usage of original DNA samples for establishing a library is reduced to 50 nanogram and the sensitivity of the original DNA samples is improved by 100 times; and 2) 100% of effective sequencing samples are produced in the process of adapter connection for establishing the library.
Owner:SUZHOU ZHONGXIN BIOTECH
Molecule marking method of rice blast-resisting gene
InactiveCN102162011APredicting Rice Blast ResistanceAccurate genetic lociMicrobiological testing/measurementDiseaseAgricultural science
The invention discloses a molecule marking method of a rice blast-resisting gene, belonging to the field of crop molecule heredity and breeding. The method comprises the following steps: (1) taking a rice sample and extracting a genome DNA (Deoxyribose Nucleic Acid) of the rice sample; and (2) carrying out PCR (Polymerase Chain Reaction) amplification on the genome DNA of the rice sample by utilizing any one pair of molecule-marked primers in RM6091 and RM26632, carrying out electrophoresis detection on a PCR amplification product, and if a molecule-marked DNA segment with corresponding size is amplified, showing that a Pi-bdl(t) gene exists. Through rice blast-resisting gene Pi-bdl(t) molecule marking in the invention, whether thin rice as well as crossbred descendants, backcross descendants and multiple cross descendants thereof contain the gene can be detected, the resistance level of the gene on rice blast can be forecasted, the selecting efficiency of rice blast resistant materials can be greatly increased, and the breeding process for disease resistance can be accelerated.
Owner:NANJING AGRICULTURAL UNIVERSITY
Establishment method of long mate pair library
InactiveCN102864498AQuick sortingEfficient screeningNucleotide librariesMicrobiological testing/measurement3-deoxyriboseDeoxyribose
The invention relates to an establishment method of a long mate pair library. The establishment method comprises the steps as follows: 1) randomly interrupting DNA (deoxyribose nucleic acid), and completing and repairing the gaps at the tail end; 2) preparing A-Fragment; 3) carrying out cohesive end connection to A-Fragment and LMP Adaptor; 4) digesting the LMP-Adaptor-Fragment with at least two restriction enzymes which can distinguish four basic groups; 5) cyclizing the fragments in the obtained enzyme digesting library again; and 6) amplifying the purified ring molecule through primer of the LMP Adaptor; and recovering the amplified products, thus finishing the establishment of LMP (long mate pair) library. According to the library establishment method provided by the invention, only two 63bp oligonucleotides sequences are needed to be synthetized, and simple experiments such as molecular biology connection and amplification are carried out, and the two ends of the obtained LMP library have the sequencing primer sequences which can be directly applied to next generation sequencing; and moreover, the fragment subjected to secondary cyclizing has the known restriction enzyme site, and the sequences of two ends of the to-be-tested fragments can be quickly sorted, the sequence testing data can be effectively sieved, and the mosaic sequence can be removed.
Owner:TIANJIN INST OF IND BIOTECH CHINESE ACADEMY OF SCI
Method for quickly identifying pseudo-ginseng
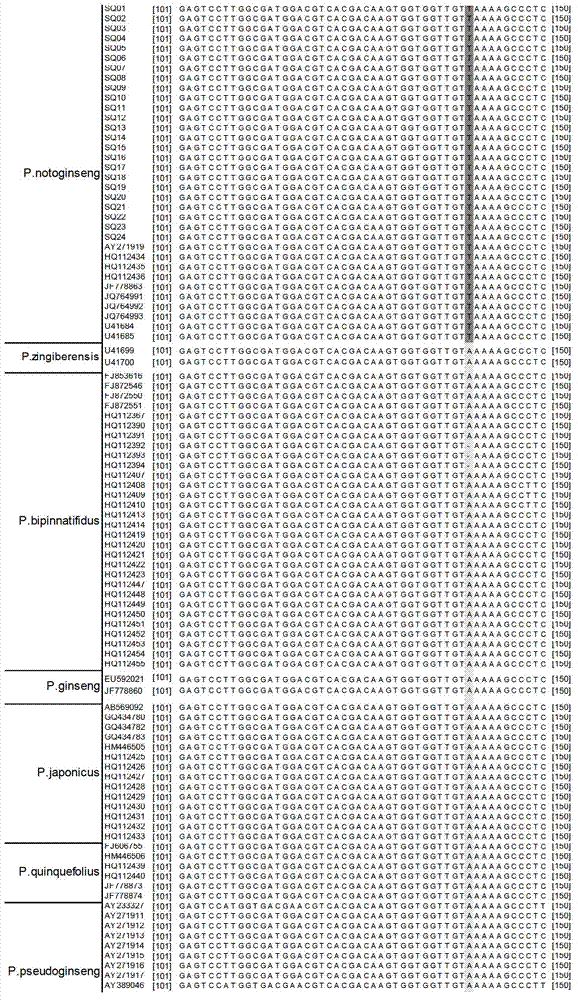
InactiveCN102888456ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentation3-deoxyriboseAdulterant
The invention discloses a method for quickly identifying pseudo-ginseng. The method comprises steps as follows: 1) getting DNA (Deoxyribose Nucleic Acid) of a sample to be detected as a template, and amplifying the fragments containing the sequences shown as SEQ ID No.1 (Sequence Identification Number); and 2) sequencing the amplified products, analyzing the sequences as shown in SEQ ID No.1; if the bit 140 beginning from 5' end in the sequences as shown in SEQ ID No.1 is T or the bit 207 is T, identifying that the sample is pseudo-ginseng; and if the bit 140 and the bit 207 beginning from the 5' end in the sequence as shown in SEQ ID No.1 are not T, identifying that the sample is not pseudo-ginseng. With the adoption of the method disclosed by the invention, the raw pseudo-ginseng, the sibling species of the raw pseudo-ginseng, the crude medicine, the raw medicinal slices, the powder and the adulterant can be quickly and accurately identified.
Owner:INST OF MEDICINAL PLANT DEV CHINESE ACADEMY OF MEDICAL SCI
Reagent kit for quantitatively assessing long-term recurrence risks of breast cancer
InactiveCN102586410AEasy to operateTest results are stableMicrobiological testing/measurementFluorescence/phosphorescenceGenomicsBreast cancer metastasis
The invention relates to the functional genomic and gene expression detection technology and the analysis technology, and discloses a reagent kit for quantitatively assessing long-term recurrence risks of breast cancer. Particularly, a type of genes capable of being used for breast cancer metastasis and prognostic molecular classification is screened within a human functional genome expression profile range, the detection technology is created, and the reagent kit is prepared and applied to breast cancer metastasis and prognostic assessment for a patient. The reagent kit for quantitatively assessing long-term recurrence risks of breast cancer comprises 21 pairs of primers, 21 specific taqman fluorescent probes, 10XRT-PCR (reverse transcription-polymerase chain reaction) buffer solution, 2.5mM of dNTP (diethyl-nitrophenyl thiophosphate) mixed liquor, reverse transcriptase, DNA (deoxyribose nucleic acid) polymerase, 10XPCR buffer solution and RNA (ribonucleic acid) enzyme inhibitor. The reverse transcription PCR technology is combined with the taqman fluorescent quantitative PCR technology, reverse transcription primers, real-time PCR primers and the taqman fluorescent probes are self-designed and optimized, reverse transcription PCR reagent and taqman fluorescent quantitative PCR reagent are integrated to prepare the detection reagent kit, operation is simple and fast, detection results are more stable, and detection cost is lower.
Owner:苏州科贝生物技术有限公司
Kit for extracting genome DNA (Deoxyribose Nucleic Acid) from plant leaves based on paramagnetic particle method and extracting method thereof
The invention belongs to the technical field of molecular biology, and particularly relates to a kit for extracting genome DNA (Deoxyribose Nucleic Acid) from plant leaves based on a paramagnetic particle method and an extracting method thereof. The kit for extracting genome DNA from plant leaves based on the paramagnetic particle method comprises a pretreatment solution, a rapid cracking liquid, a DNA binding liquid, a magnetic bead suspension and an eluent, wherein cell walls and cell membranes can be effectively cracked through the pretreatment solution; and the interference of impurities such as polysaccharides, polyphenol, tannin and the like on genome DAN extraction can be effectively eliminated through the pretreatment solution. In the rapid cracking liquid, guanidine hydrochloride and Tween 20 are taken as main components, so that plant cells can be fully cracked at the normal temperature within 2 minutes, only two minutes are required in a subsequent centrifuging step, and the operation time is greatly saved. The guanidine hydrochloride is a strong denaturant, has a good cell cracking effect. Moreover, organic solvents such as chloroform and the like are not required to be added, so that damages to operating personnel are avoided, and safety and reliability are achieved.
Owner:苏州启巧生物科技有限公司
Massive parallel method for decoding DNA and RNA
This invention provides methods for attaching a nucleic acid to a solid surface and for sequencing nucleic acid by detecting the identity of each nucleotide analogue after the nucleotide analogue is incorporated into a growing strand of DNA in a polymerase reaction. The invention also provides nucleotide analogues which comprise unique labels attached to the nucleotide analogue through a cleavable linker, and a cleavable chemical group to cap the —OH group at the 3′-position of the deoxyribose.
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Method for qualitatively and quantitatively detecting nucleic acid based on high-flux sequencing technology
ActiveCN102628082AStrong specificityEasy to operateMicrobiological testing/measurement3-deoxyriboseHigh flux
The invention discloses a method for qualitatively and quantitatively detecting nucleic acid based on a high-flux sequencing technology. The method comprises the following steps of: preparing a single-chain DNA (Deoxyribose Nucleic Acid) library with a method for capturing a specific nucleic acid target through multi-gene multi-region two-step and one-direction amplification; sequencing the obtained single-chain DNA library based on a high-flux sequencing technology; and qualitatively and quantitatively analyzing nucleic acid according to a sequencing result to obtain a qualitative and quantitative detection result of nucleic acid. Compared with an ordinary real-time Q-PCR (Quantitative Polymerase Chain Reaction) quantifying method, the method has the advantages of lower cost, wider application, increase in the quantifying sample flux of nucleic acid and capability of accurately qualifying and quantifying the series of nucleic acids.
Owner:ANHUI ANKE BIOTECHNOLOGY (GRP) CO LTD
Preservative agent for DNA of saliva
The invention discloses a preservative agent for DNA (Deoxyribose Nucleic Acid) of saliva. The preservative agent comprises the following components in percentage by volume: 5-10 g / 100mL of trihytdroxy methyl-aminomethane, 10-25 g / 100mL of ethylene diamine tetraacetic acid, 7-17 g / 100mL of sugar, 1-10 g / 100mL of sodium chloride and 0.5-5 g / 100mL of surfactant. The solvent of the preservative agent is water; the pH value of the preservative agent is 7.0-9.5; the DNA in a saliva sample can be effectively preserved at room temperature for a long time; and the defects in the prior art that the preservative time of the DNA acquired from the saliva sample is short and the DNA is required to be preserved at low temperature are effectively overcome. The preservative agent disclosed by the invention is low in cost, simple in preparation method and suitable for industrial production.
Owner:XIAMEN ZEESAN BIOTECH
Pseudovirion vector and preparation method and application thereof
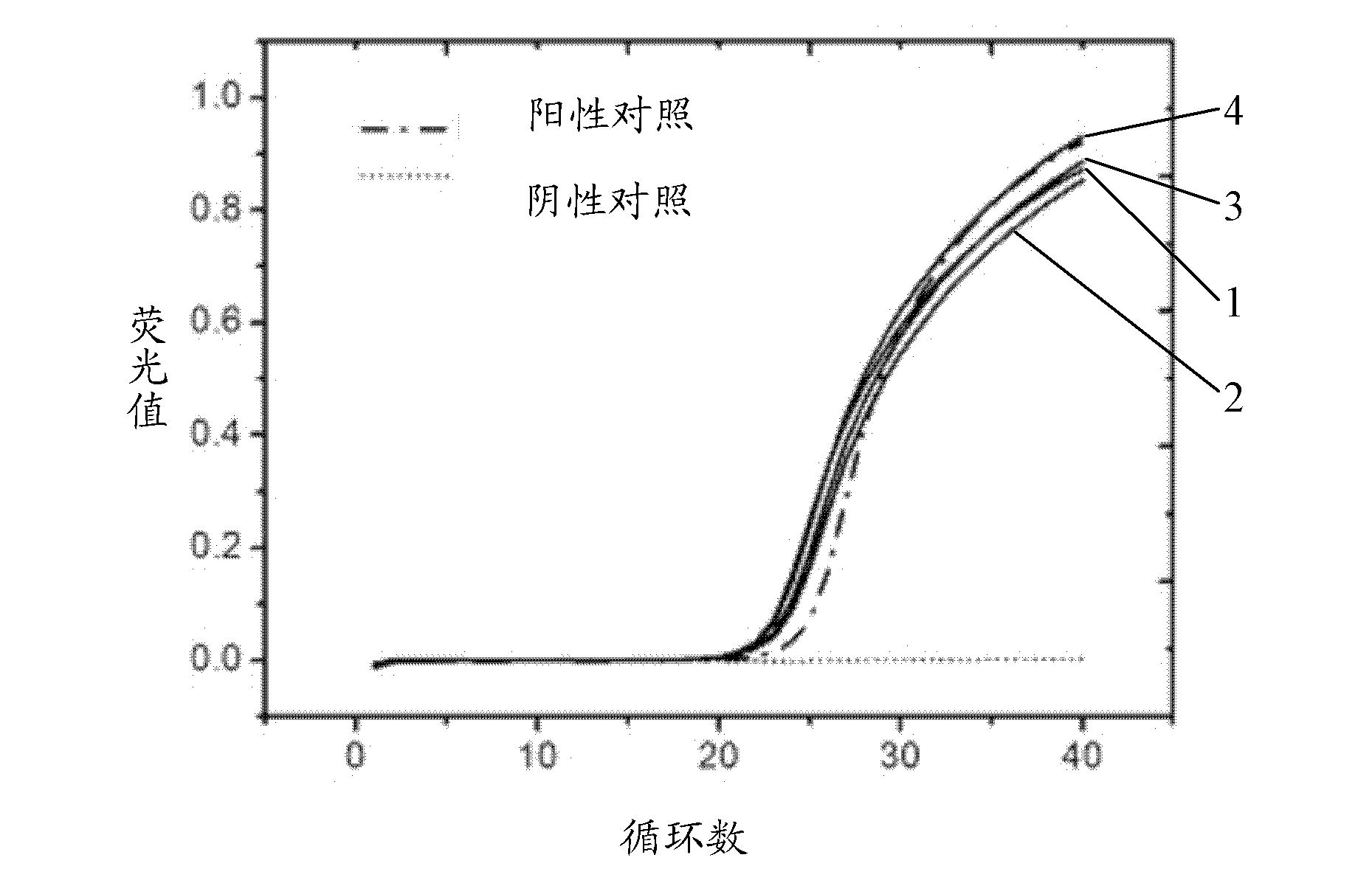
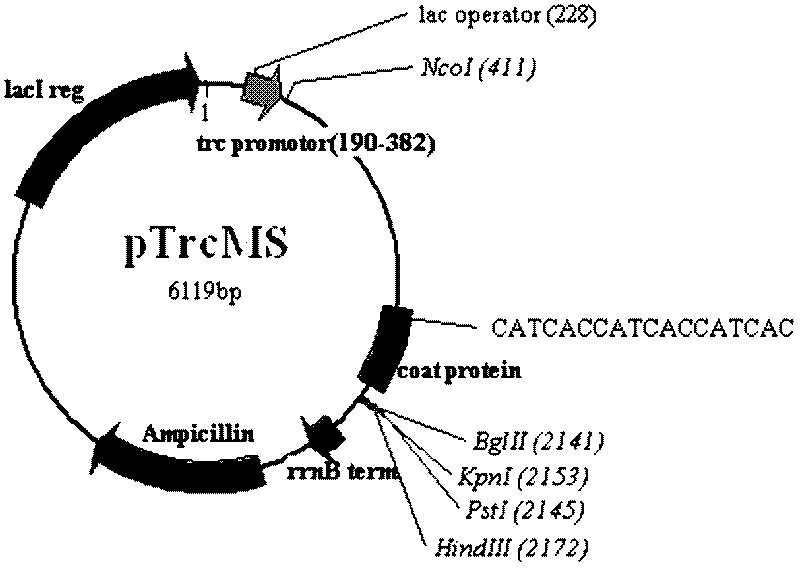
ActiveCN102559731AReduce cumbersome operationsQuality improvementMicrobiological testing/measurementMicroorganism based processesProtein containing complexNucleotide
The invention discloses an animal pathogen microbial pseudovirion vector and a preparation technology thereof. The gene coding sequences of a maturase protein and a coat protein of an MS2 phage and a cDNA (complementary Deoxyribose Nucleic Acid) sequence which corresponds to a 5' non-coded sequence of a gene regulating element sequence included by a genome part are connected to the downstream of a pTrcHis2A vector promoter, so that a pseudovirion vector is constructed. A 6His purification tag is added between the fifteenth and sixteenth amine acids of the coat protein in the MS2 phage througha gene insertion method, a pseudovirion particle comprising an exogenous gene is expressed into an RNA (Ribonucleic Acid)-protein complex, and the pseudovirion particle is captured with a protein purifying method, so that the complex operating process for preparing the pseudovirion particle can be simplified, and the purification quality of the pseudovirion particle is enhanced simultaneously. The pseudovirion vector is pTrcMS of which the nucleotide sequence is shown as SEQ ID No.1.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE
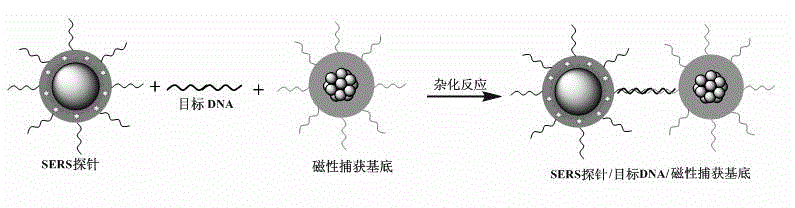
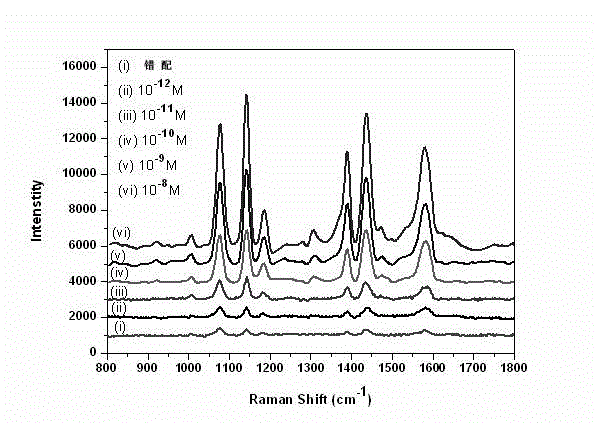
Method for high-sensitivity detection for t-DNA (transfer-deoxyribose nucleic acid) by aid of SERS (surface enhanced Raman spectroscopy) liquid chip
InactiveCN102721680AOvercome efficiencyOvercoming associativityRaman scatteringSurface-enhanced Raman spectroscopyMedical diagnosis
The invention belongs to the technical field of biological molecule detection, and particularly discloses a method for high-sensitivity detection for t-DNA (transfer-deoxyribose nucleic acid) by the aid of an SERS (surface enhanced Raman spectroscopy) liquid chip. In the method, surface enhanced Raman spectroscopy (SERS) is used for coding, and the SERS liquid chip with magnetic composite nanoparticles as a capturing substrate is used for detecting the t-DNA in a high-sensitivity manner. The method includes chemically bonding the nanoparticles (SERS labels) comprising SERS codes with p-DNA (phosphorous-deoxyribose nucleic acid) of a probe to prepare a high-sensitivity SERS probe at first; capturing the c-DNA (complementary-deoxyribose nucleic acid) by the magnetic composite nanoparticles with surfaces richly containing carboxyl in a chemical bonding manner to prepare the magnetic capturing substrate; and constructing the SERS liquid chip by the SERS probe and the magnetic capturing substrate to detect the t-DNA. The method is simple, speedy and sensitive in operation, can be used for high through-put, quantitative and multi-element detection for DNA (deoxyribose nucleic acid), can be widely used in fields such as food safety monitoring, medical diagnosis and forensic examination, and has an important application prospect and a development value.
Owner:FUDAN UNIV
Specific primer pair for identification of spermatophyte species and applications of specific primer pair
InactiveCN102978208AEfficient identificationImprove developmentMicrobiological testing/measurementDNA/RNA fragmentation3-deoxyriboseDNA barcoding
The invention discloses a specific primer pair for identification of spermatophyte species and applications of the specific primer pair. The invention provides a pair of specific primers consisting of a single stranded DNA (Deoxyribose Nucleic Acid) A and a single stranded DNA B. The single stranded DNA A is 15-40bp and has a DNA fragment same as the DNA fragment shown as a sequence I in a sequence table. The single stranded DNA B is 15-40bp and has a DNA fragment same as the DNA fragment shown as a sequence II of the sequence table. In the invention, with the ycflb gene of a plant to be tested as a template, the DNA fragment obtained by PCR (Polymerase Chain Reaction) amplification with the specific primer pair is also protected. The DNA fragment can be used for assisting identification of the spermatophyte species. The specific primer pair can be used for developing general kits, effectively identifying land plant species, and promoting the development of plant DNA bar codes, contributing to social progress.
Owner:INST OF BOTANY CHINESE ACAD OF SCI
Nucleotide carrying modifiers and preparation method thereof and method for gene sequencing
InactiveCN101942000AActive connectionImprove accuracySugar derivativesMicrobiological testing/measurement3-deoxyriboseNucleotide
The invention relates to the gene engineering field, specifically to a nucleotide carrying modifiers and a preparation method thereof and a method for gene sequencing. The structural general formula of the nucleotide carrying modifiers is dNTP-R; in the formula, the dNTP is nucleotide; the modifier -R is connected on a position in the nucleotide except from -OH on a deoxyribose and comprising at least a cutting site and at least a marker. The preparation method thereof comprises the following steps: the position except from the -OH on a deoxyribose is connected with the nucleotide of a first modifier -R1 to react with a second modifier -R2 with the cutting site to combine the nucleotide dNTP-R carrying the modifier. Furthermore, the invention provides a method for gene sequencing using the nucleotide carrying modifiers. When the nucleotide carrying modifiers provided by the invention carries out the gene sequencing, the sequencing accuracy rate is improved and the longer gene sequence is measured.
Owner:SHENGZHEN CHINA GENE TECH COMPANY
Gene editing technique taking Argonaute nuclease as core
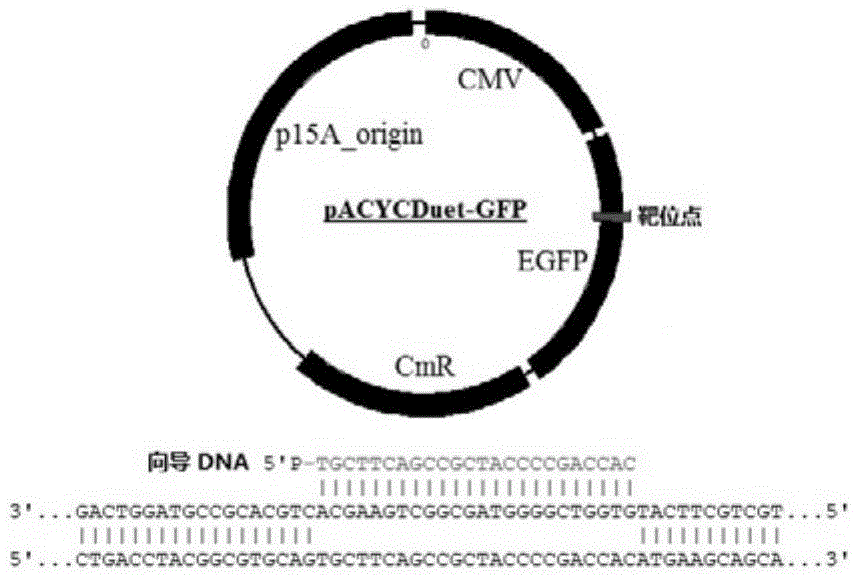
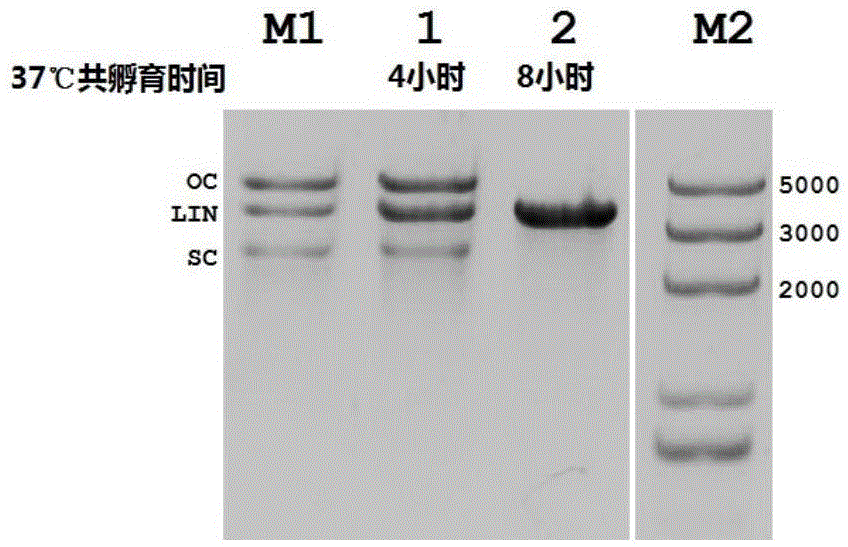
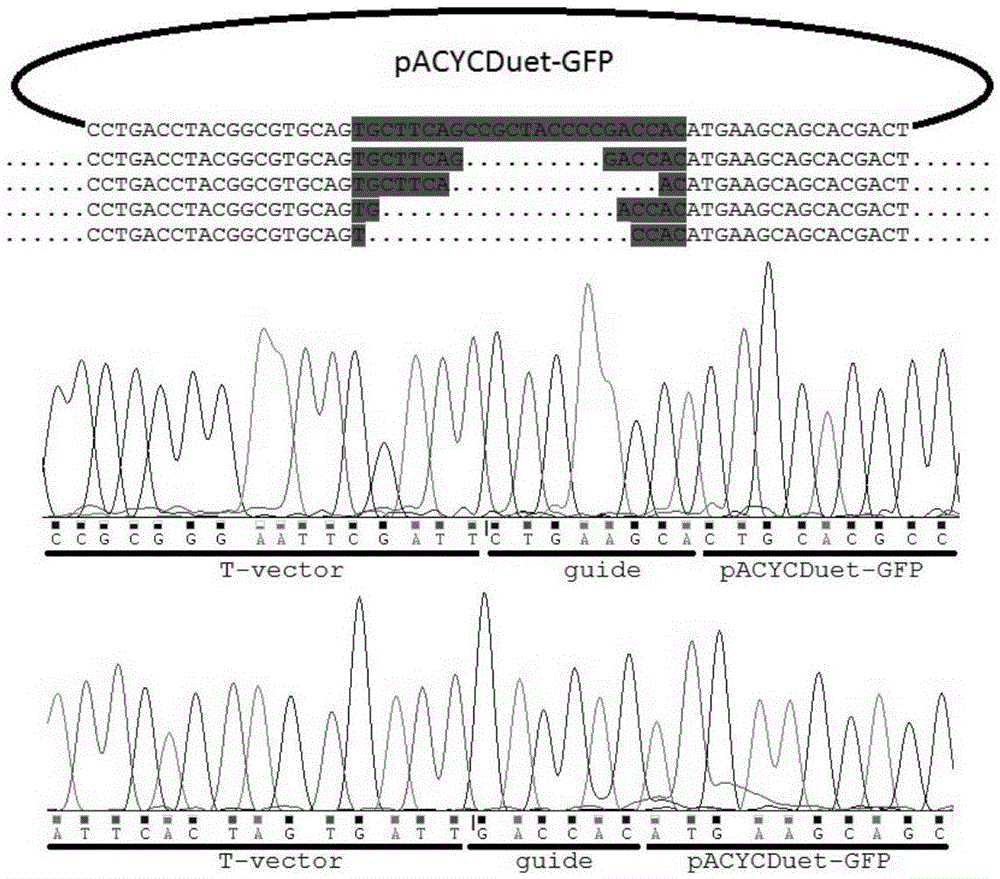
InactiveCN105483118ASimple designThe dosage is easy to controlDNA preparationGenome editing3-deoxyribose
The invention discloses a gene editing technique taking Argonaute nuclease as a core. In the presence of guide DNA (Deoxyribose Nucleic Acid), Argonaute endonuclease can cut any locus of a targeted DNA sequence, including a genome of a mammal to cause breakage of DNA double chains, thereby fulfilling an editing aim. Editing means include incision, deletion, mutagenesis induction, exogenous sequence insertion, fragment substitution and the like. Editing effects include gene inactivation, gene mutation, exogenous gene induction and the like. The protein can serve as an important tool for genome editing. A gene editing tool taking the protein as the core has the characteristics of easiness in operation, high efficiency, low target missing rate and high fidelity. Moreover, almost any genome locus can be effectively targeted and cut by the Argonaute nuclease. By using the technique, high-specificity and high-efficiency genome targeted editing is helpfully realized.
Owner:ZHEJIANG UNIV +1
Massive parallel method for decoding DNA and RNA
Owner:THE TRUSTEES OF COLUMBIA UNIV IN THE CITY OF NEW YORK
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com