Gene editing technique taking Argonaute nuclease as core
A gene editing and nuclease technology, applied in the field of targeted gene editing technology, can solve the problems of low system efficiency and off-target, and achieve the effects of controllable dose, simple transfection of cells, and simple design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: DNA plasmid endolysis in vitro
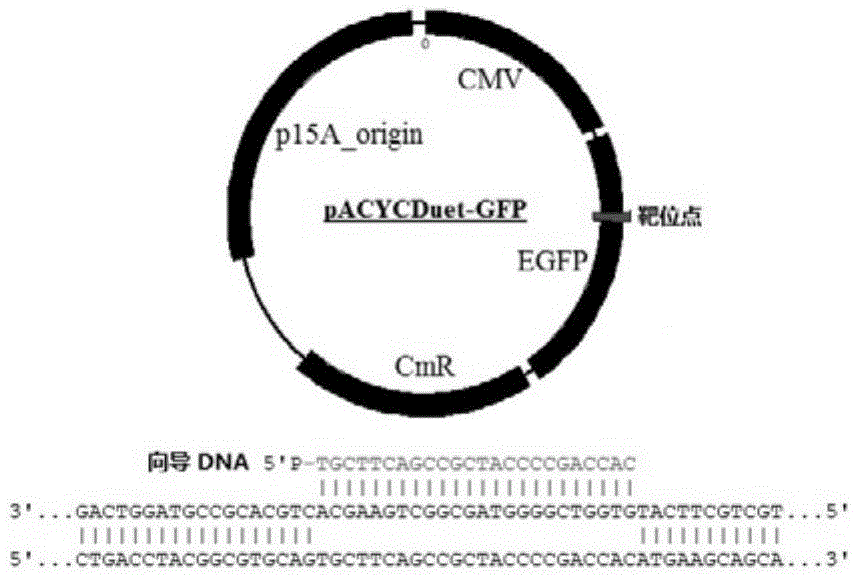
[0030] 1. The complete reading frame of Argonaute (NgAgo for short) obtained from Natronobacterium gregoryi SP2 bacteria is cloned on the pcDNA3.1 plasmid, and its 5' end and 3' end are accompanied by FLAG and HA tag sequences respectively. The amino acid sequence of NgAgo is shown in SEQ ID NO:1.
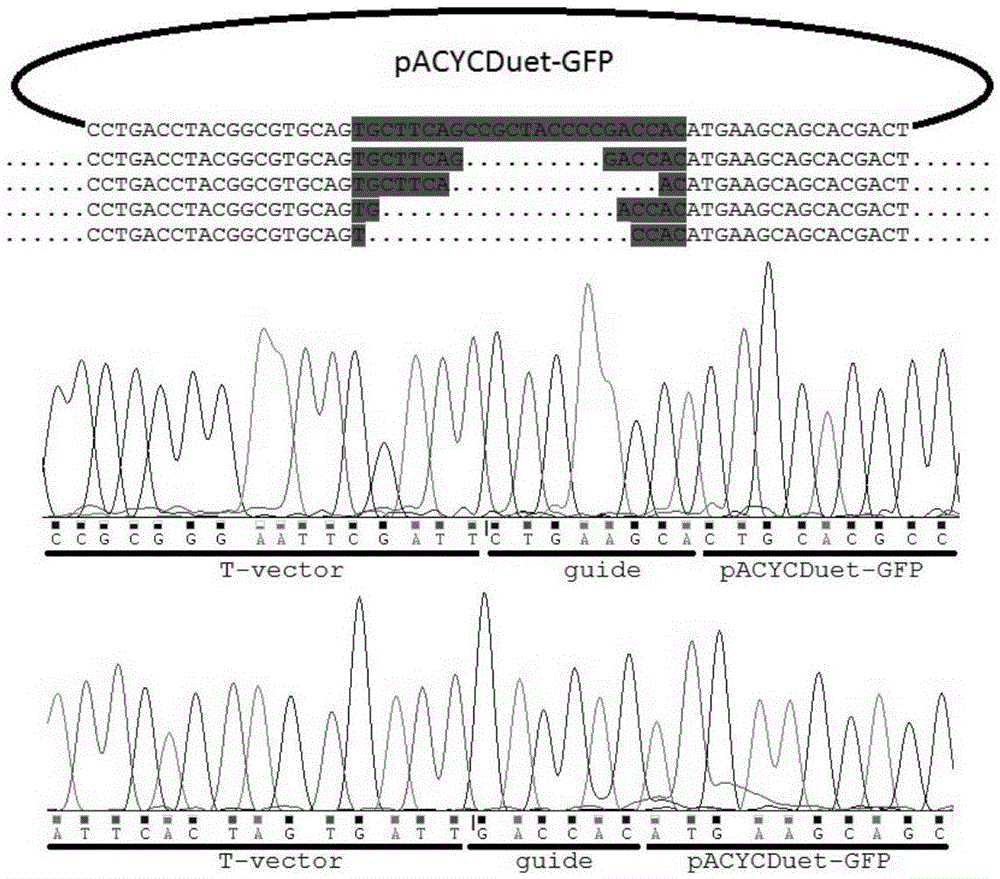
[0031] 2. synthesize a section of 5' phosphorylated guide DNA 5'P-TGCTTCAGCCGCTACCCCGACCAC-3', which is complementary to a section of the GFP gene on the pACYCDuet-GFP targeting plasmid ( Figure 1a ).
[0032] 3. Co-transfect the constructed FLAG-NgAgo-HA-pcDNA3.1 plasmid and the guide fragment in step 2 above into human 293T cells.
[0033] 4. Use FLAG, HATandemAffinityPurificationKit Kit (Sigma-Aldrich Company) to purify the FLAG-NgAgo-HA protein bound to the guide DNA from the transfected cells.
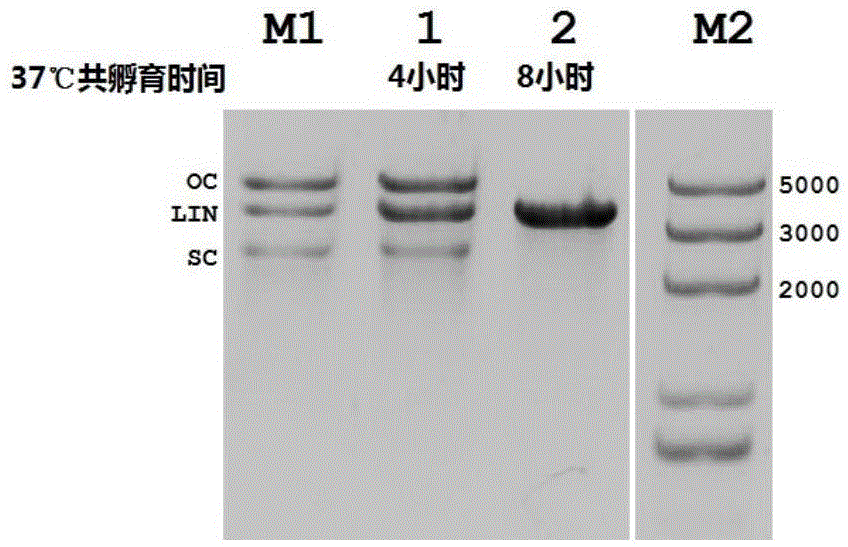
[0034] 5.5 μg of purified NgAgo protein combined with guide DNA and 400ngpACYCDuet-GFP plasmid were placed in a 50ul reaction syst...
Embodiment 2
[0037] Example 2: Argonaute-mediated plasmid cleavage in mammalian cells and comparison with CRISPR / Cas9 system-mediated cleavage efficiency
[0038] 1. A nuclear localization signal (NLS) is added to the N-terminus of the NgAgo protein, and the NLS-NgAgo gene sequence of the improved protein is constructed on the pcDNA3.1 plasmid. NLS-NgAgo-pcDNA3.1 mainly distributed in the nucleus after expressed in Hela cells.
[0039] 2. Hela cells were co-transfected with EGFP-N1 target plasmid and NLS-NgAgo-pcDNA3.1 expression plasmid. In addition, the cells were also transfected with 4 guide DNAs (G1-G4) for the target plasmid CMV promoter region and GFP coding region ( Figure 2a ). Transfection conditions: 200ngNLS-NgAgo-pcDNA3.1 plasmid and 100ng guide DNA were transfected into 2×10 5 Intracellular (in 24-well plate).
[0040] 3. Another group of cells were co-transfected with the EGFP-N1 target plasmid, the Cas9 expression plasmid SpCas9-pCDNA3.1, and the guide RNA (sgRNA1) tar...
Embodiment 3
[0050]Example 3: Argonaute-mediated cleavage of GC-rich sequences in the genome and comparison with CRISPR / Cas9 system-mediated cleavage efficiency in mammalian cells
[0051] 1.293T cells were transfected with the NLS-NgAgo expression plasmid and the guide DNA for the GC-rich region in the HBA2 gene and GATA4 gene ( Figure 3a ).
[0052] 2. As a comparison, some 293T cells were transfected with Cas9 expression plasmid and guide RNA targeting the same region.
[0053] 3. Because the double-strand breaks in the mammalian genome are mainly repaired by the cell's spontaneous indel-forming non-homologous endjoining (NHEJ) pathway, we use the T7 endonuclease I (T7EI) detection method to detect the cleavage efficiency of the genomic target sequence. In each T7EI reaction, 500 ng of PCR product was reacted with T7 endonuclease I, which cleaves unpaired DNA sequences. The products were detected by denaturing PAGE electrophoresis and silver staining. Band density was analyzed by Im...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com