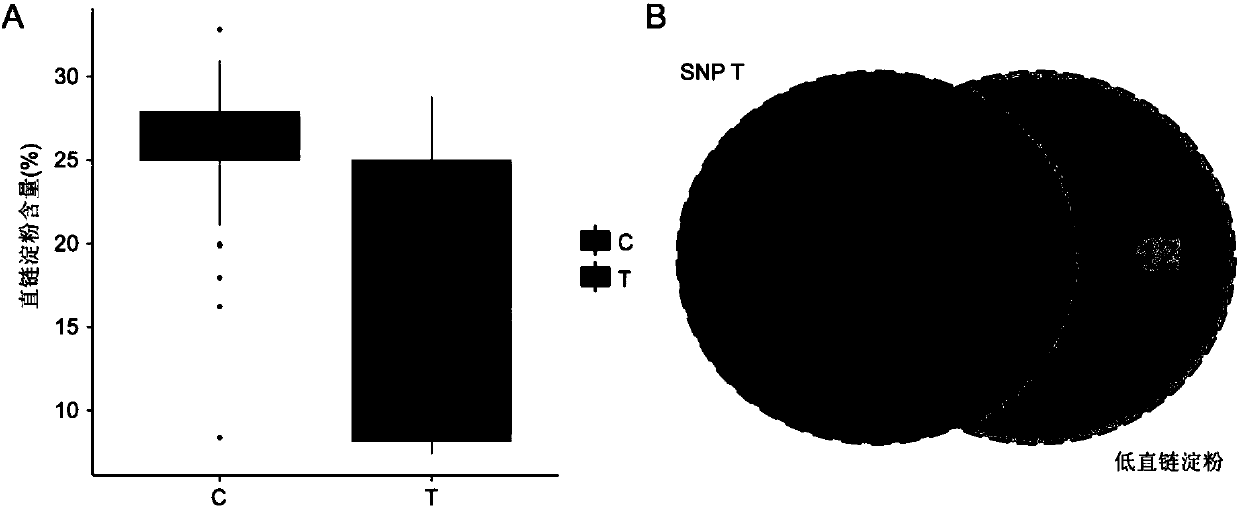
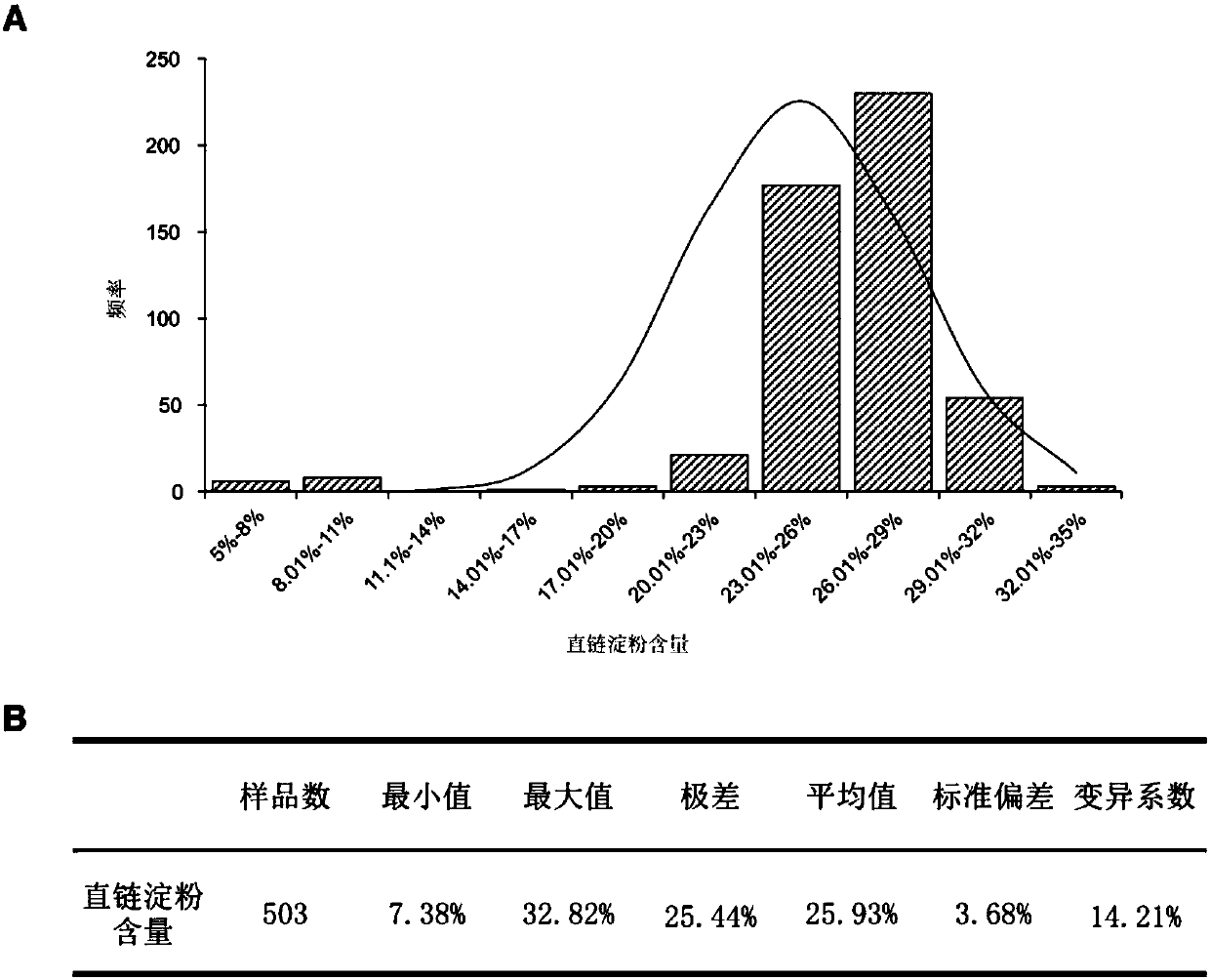
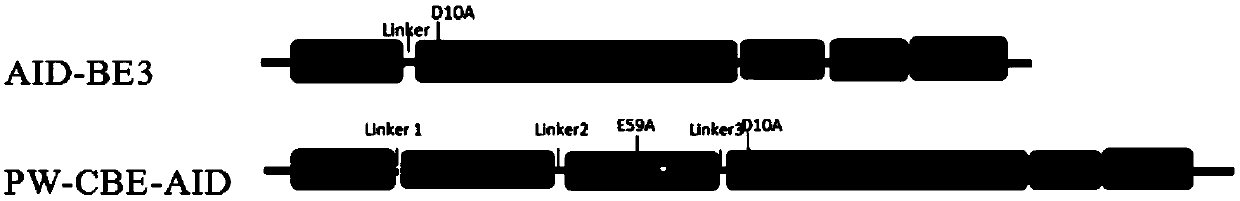
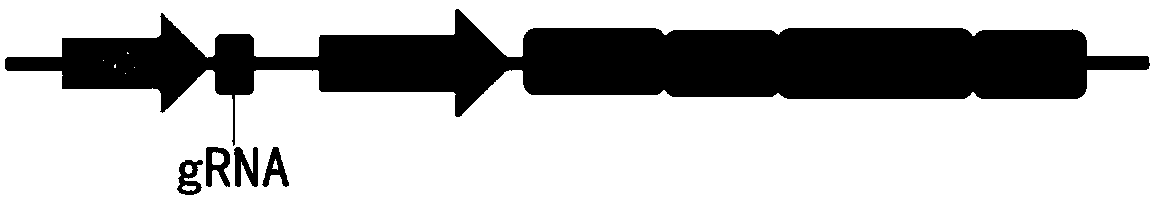
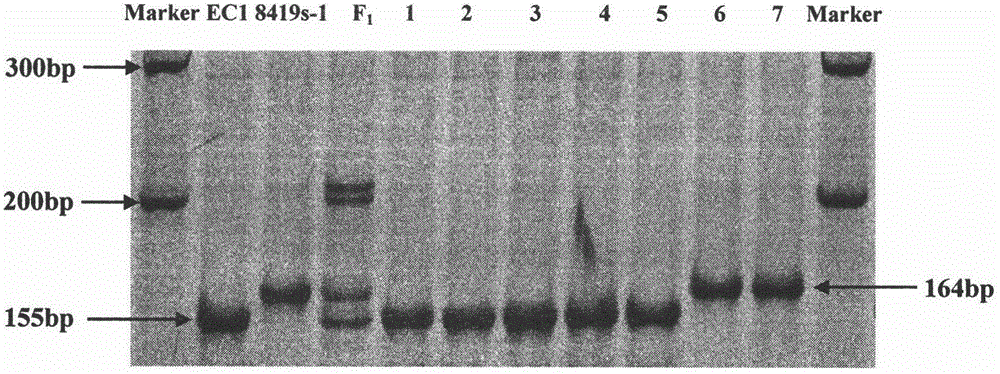
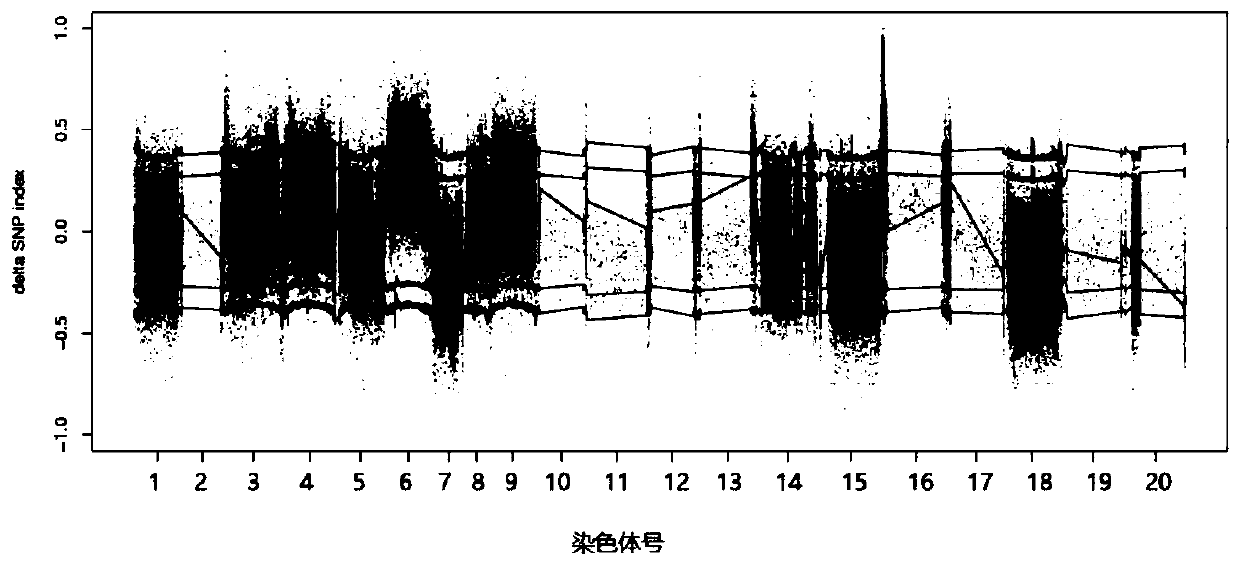
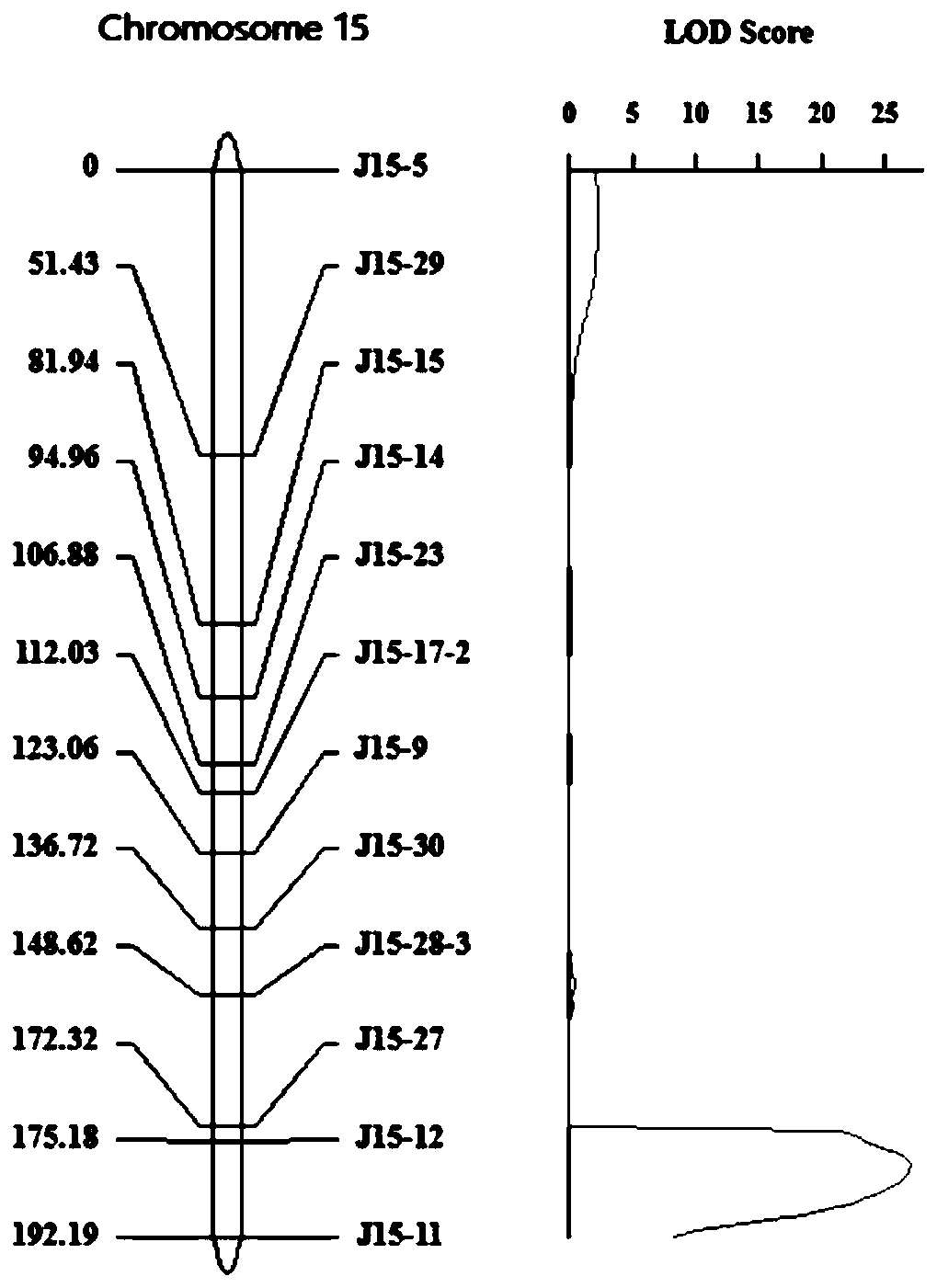
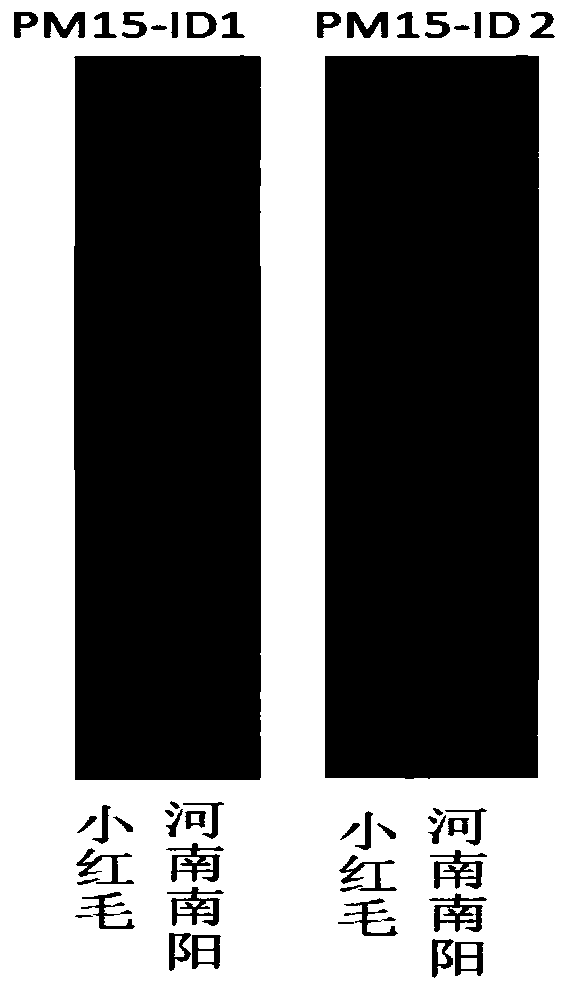Patents
Literature
255 results about "Indel" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Indel is a molecular biology term for an insertion or deletion of bases in the genome of an organism. It is classified among small genetic variations, measuring from 1 to 10 000 base pairs in length, including insertion and deletion events that may be separated by many years, and may not be related to each other in any way. A microindel is defined as an indel that results in a net change of 1 to 50 nucleotides.
Methods, cells & organisms
InactiveUS20150079680A1Reduce chanceGenetically modified cellsStable introduction of DNABiological bodyDNA fragmentation
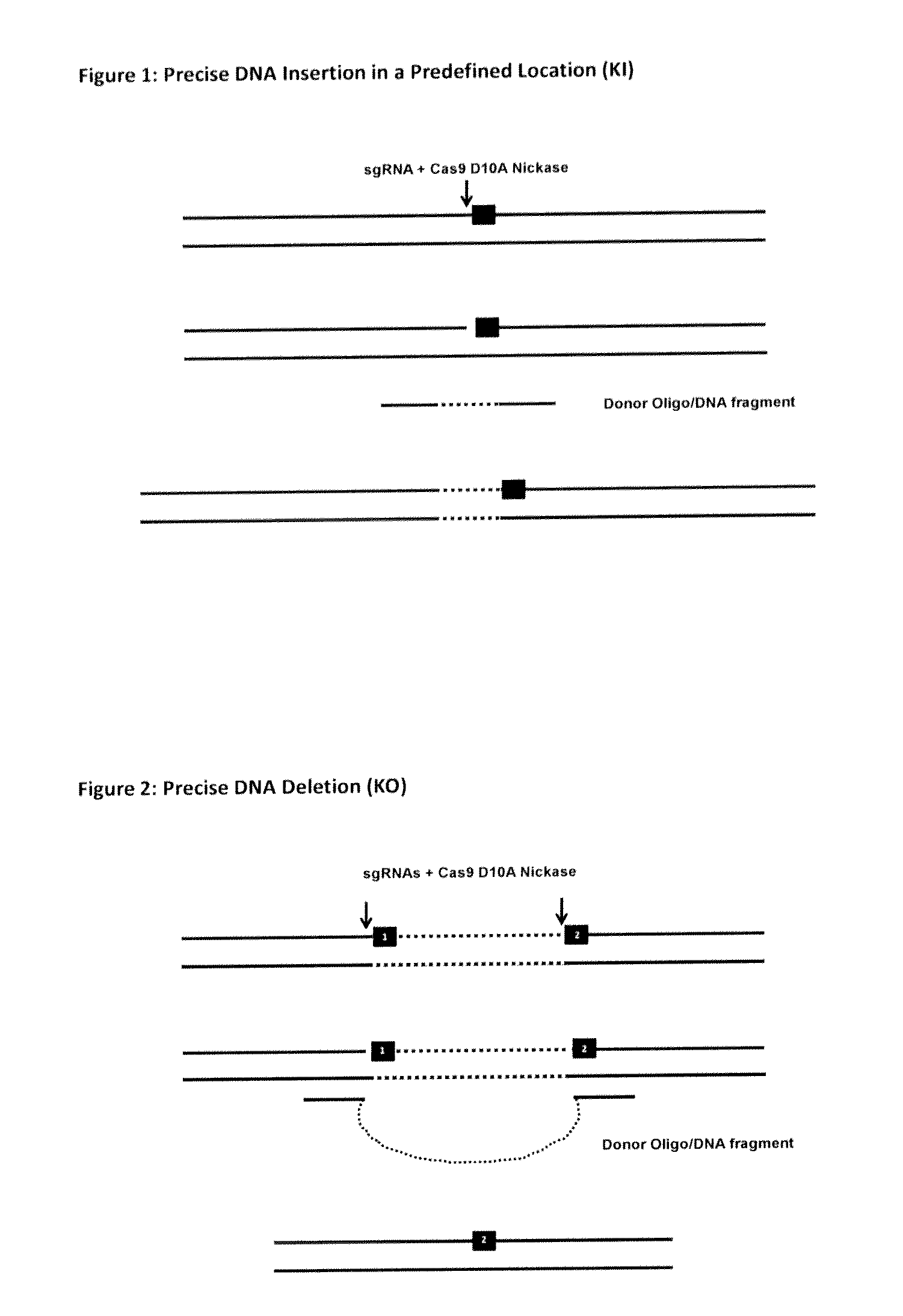
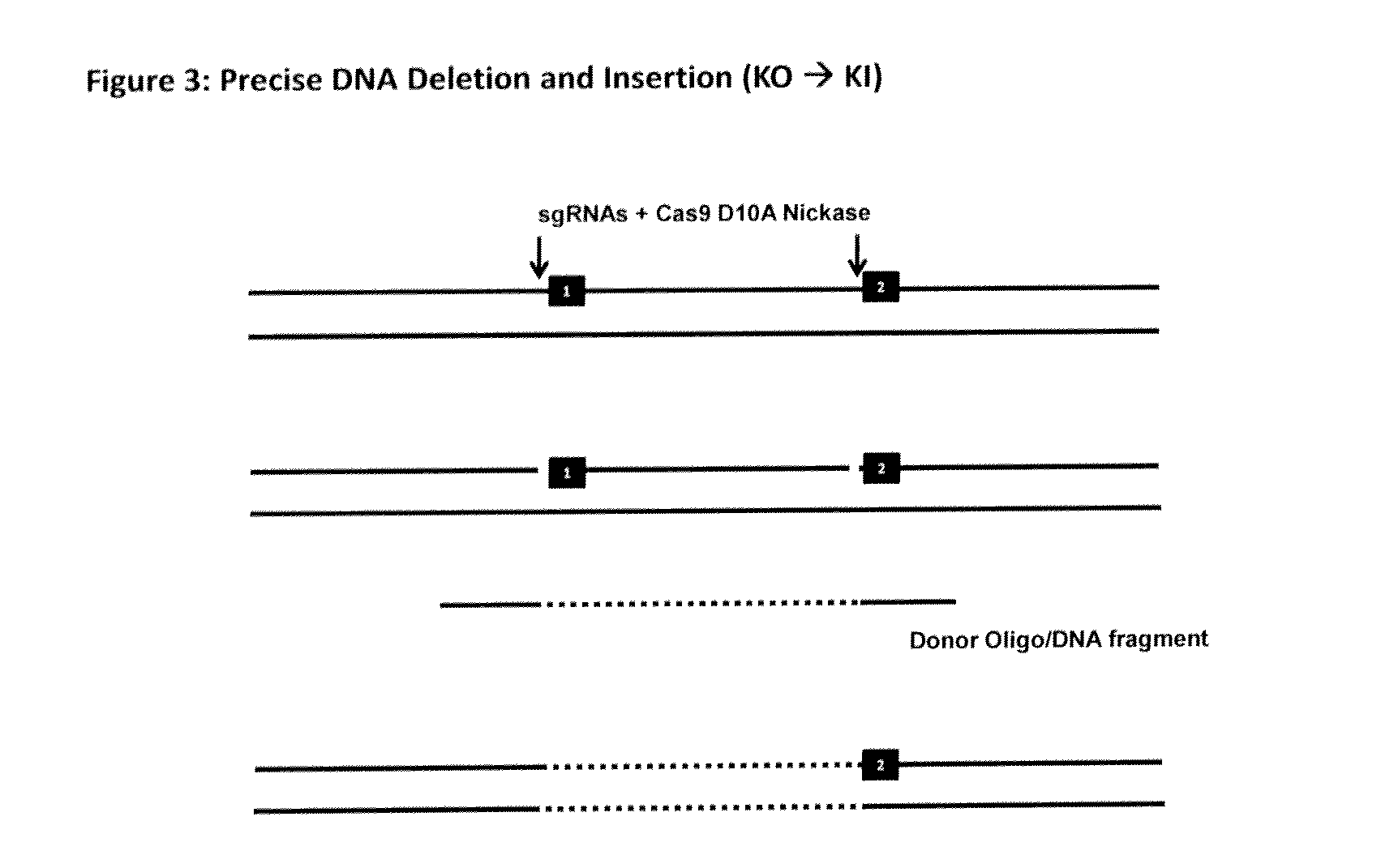
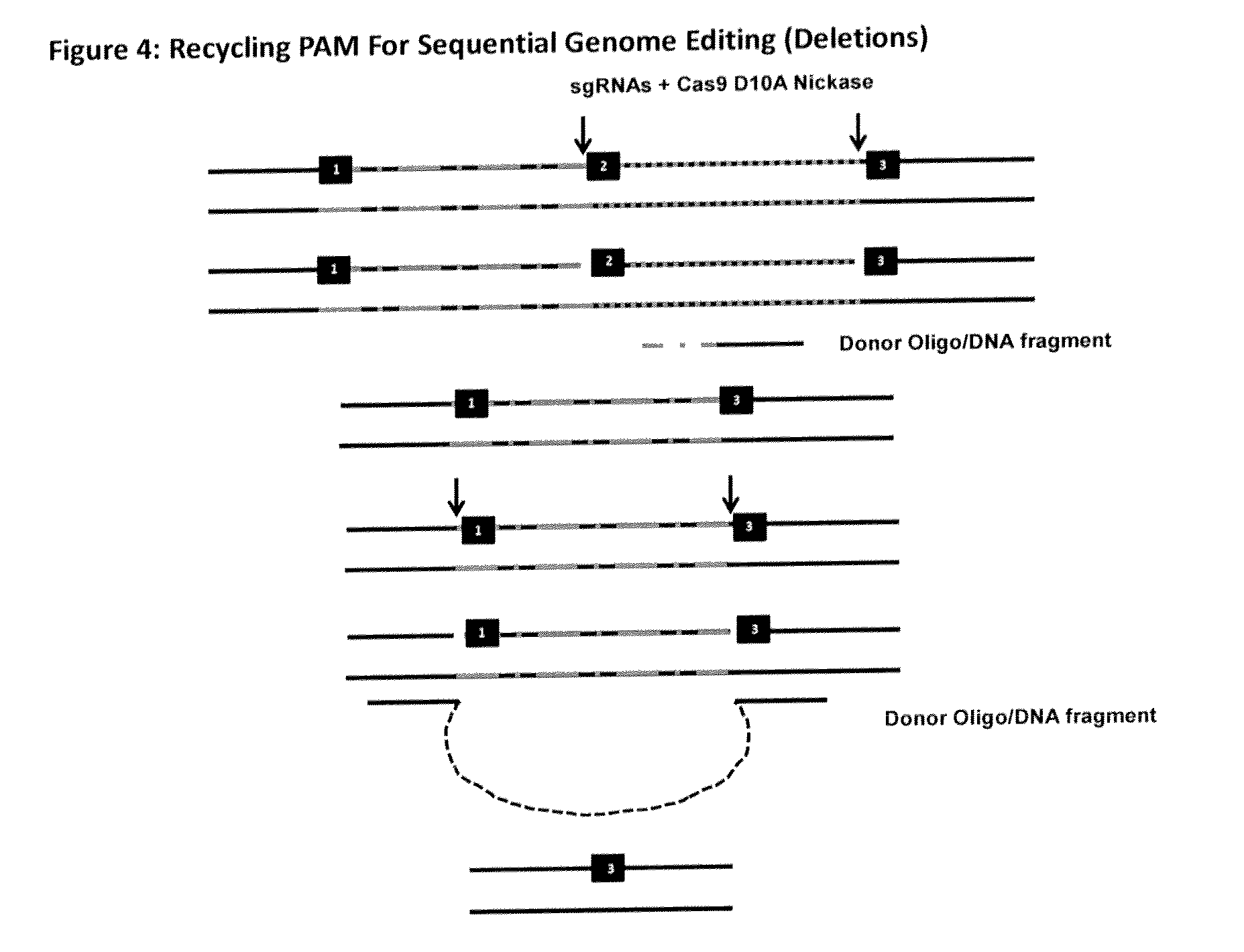
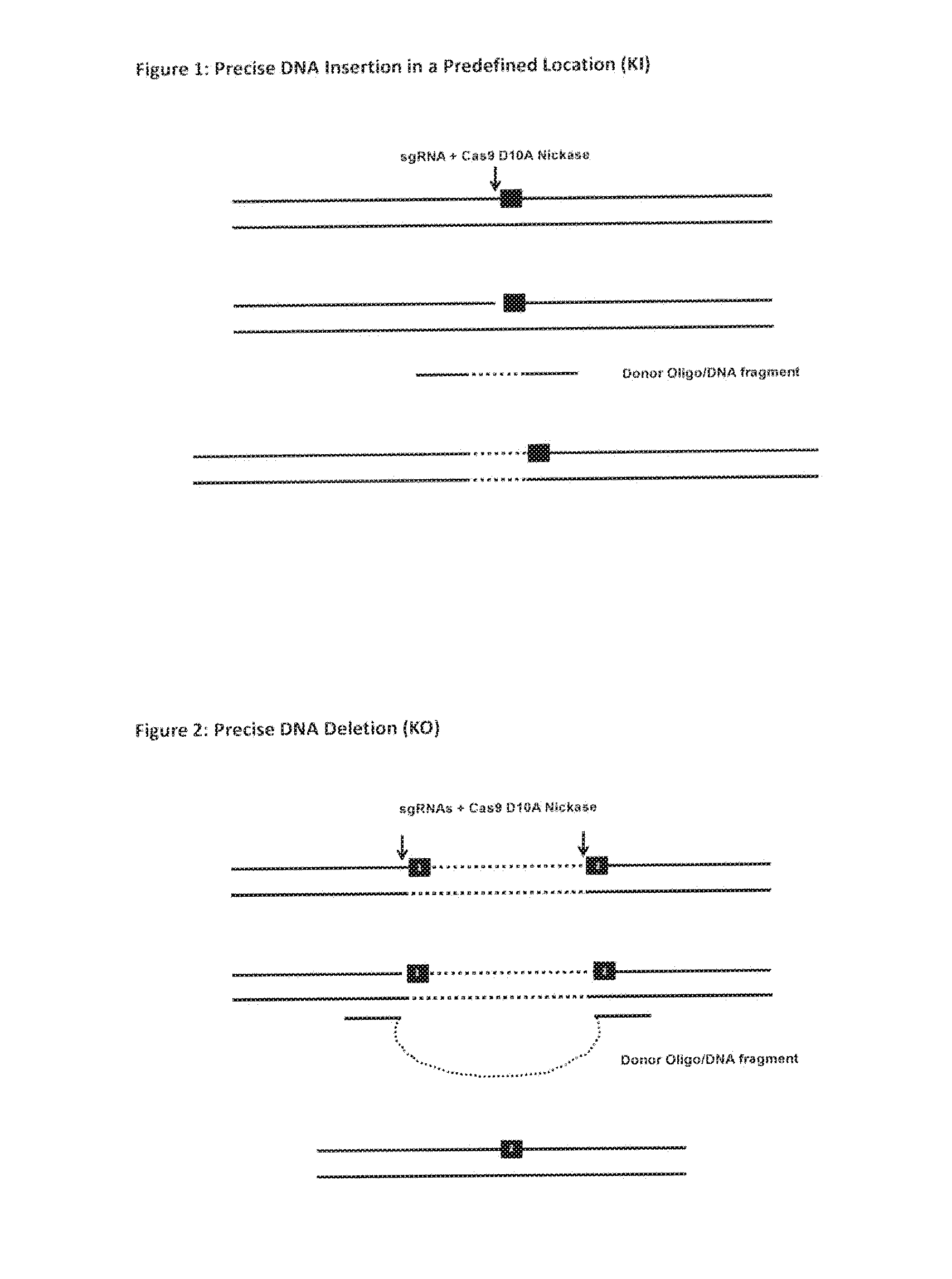
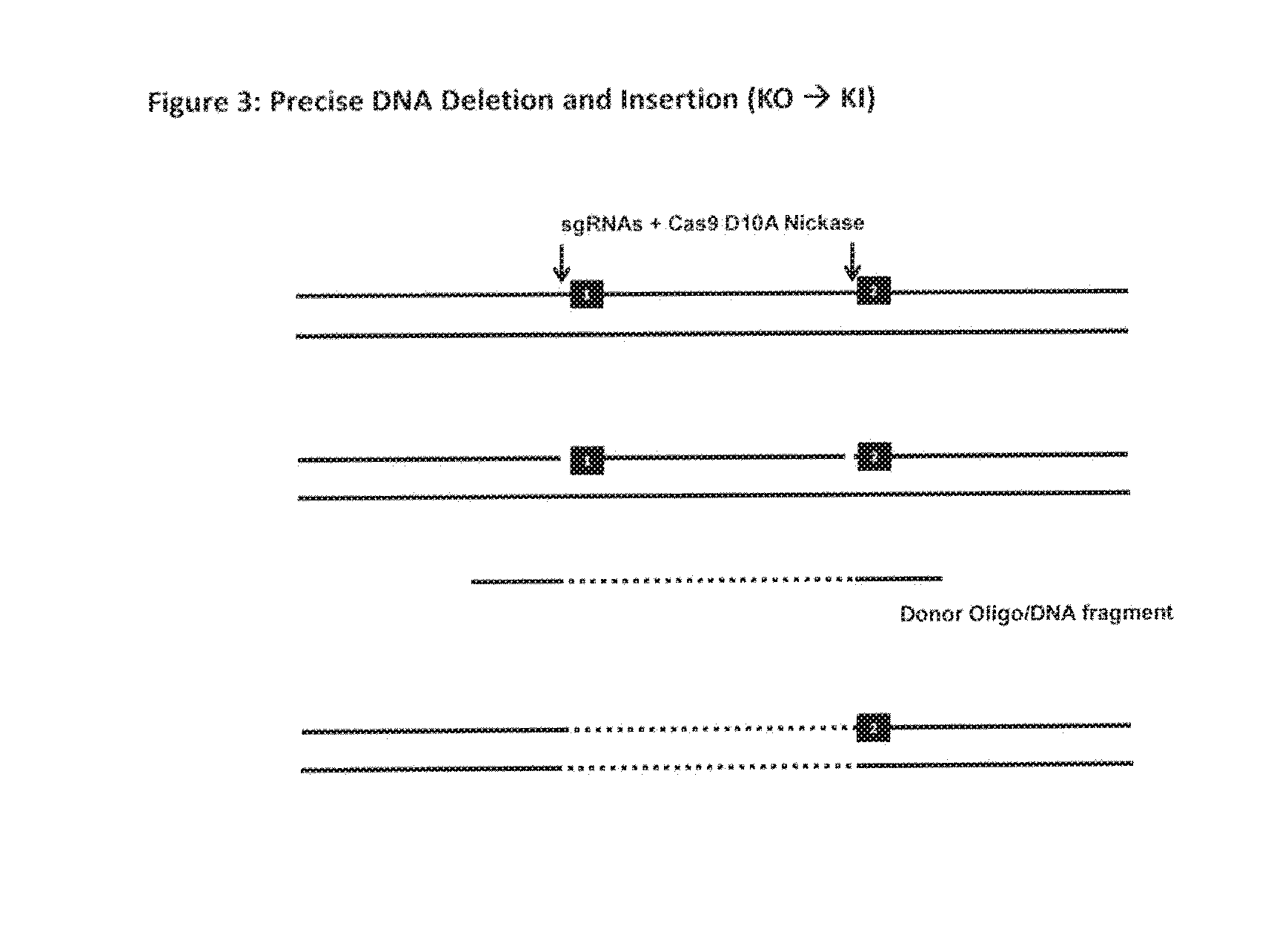
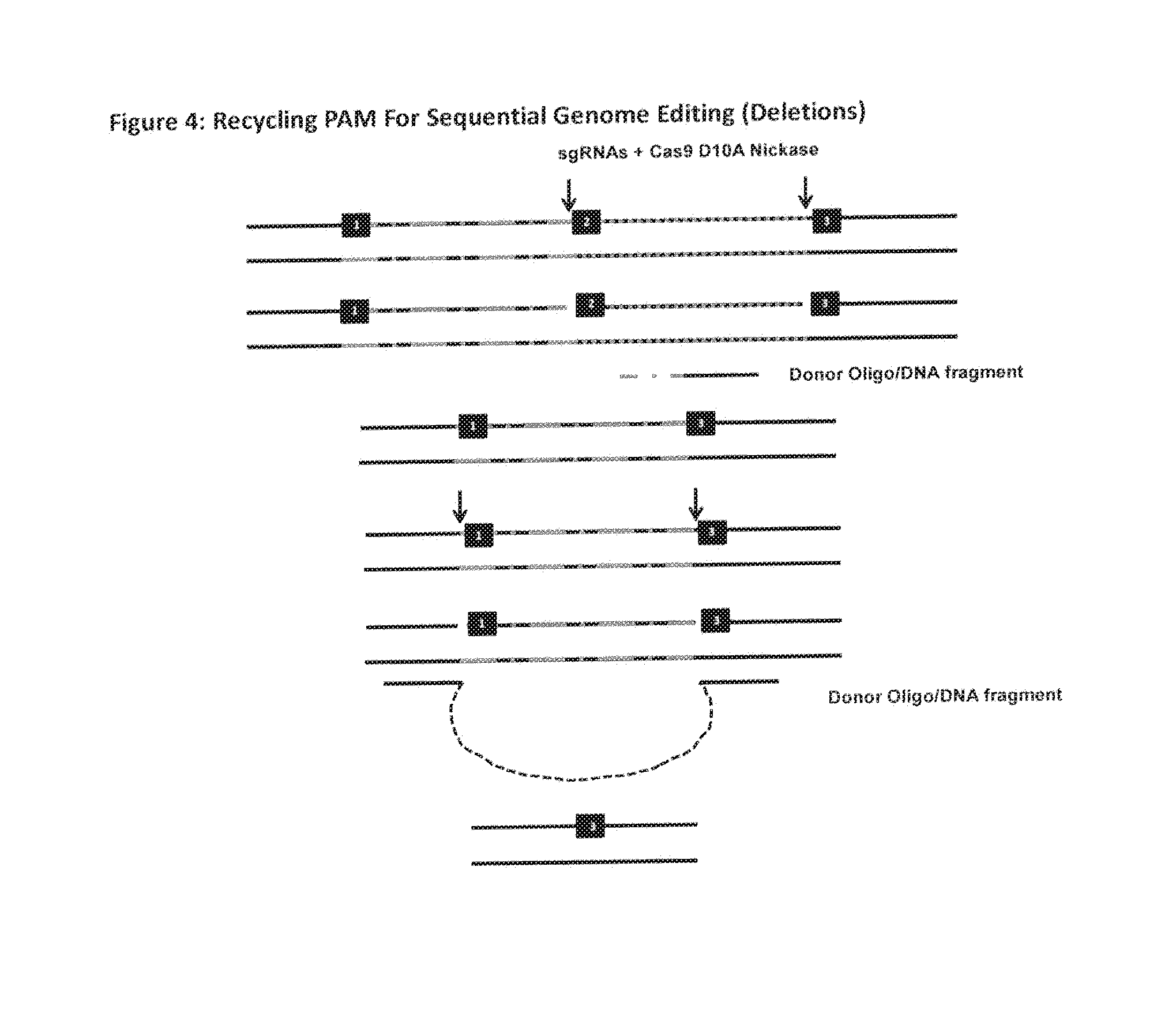
The invention relates to an approach for introducing one or more desired insertions and / or deletions of known sizes into one or more predefined locations in a nucleic acid (eg, in a cell or organism genome). They developed techniques to do this either in a sequential fashion or by inserting a discrete DNA fragment of defined size into the genome precisely in a predefined location or carrying out a discrete deletion of a defined size at a precise location. The technique is based on the observation that DNA single-stranded breaks are preferentially repaired through the HDR pathway, and this reduces the chances of indels (eg, produced by NHEJ) in the present invention and thus is more efficient than prior art techniques. The invention also provides sequential insertion and / or deletions using single- or double-stranded DNA cutting.
Owner:KIMAB LTD
Method for predicating homologous recombination deficiency mechanism and method for predicating response of patients to cancer therapy
InactiveCN107287285AInnovativeOvercoming the pitfalls of inaccurate forecastsMicrobiological testing/measurementSequence analysisAbnormal tissue growthPolymerase L
The invention discloses a method for predicating a homologous recombination deficiency (HRD) mechanism and a method for predicating response of patients to cancer therapy and relates to the field of biological information predication. The method comprises the step of judging whether a tumor sample has homologous recombination deficiency or not according to one or more comprehensive values in a large-segment INDEL (Insertion / Deletion) fraction, a copy number variation fraction and a tumor mutation load fraction, wherein the comprehensive values can also comprise a loss of heterozygosity variation fraction. By adopting the method disclosed by the invention, predication of a chromosome large-segment structure, a chromosome gene type number, a chromosome gene copy number, a chromosome variation interval and abnormal loss of heterozygosity and chromosome telomeric imbalance is realized, so that an evaluation range is more complete and HRD can be accurately predicated; the comprehensive values also can be used for determining whether the patients have response to a therapeutic regimen containing one or more of a PARP (Poly Adenosine Diphosphate Ribose Polymerase) inhibitor, an DNA (Deoxyribonucleic Acid) injury inhibitor, a topoisomerase II / II+inhibitor, a topoisomerase I inhibitor and radiotherapy; the method is simple and has wide general applicability.
Owner:SHANGHAI ORIGIMED CO LTD
Methods and kits for multiplex amplification of short tandem repeat loci
InactiveUS20120122093A1Reduce the possibilityImprove efficiencyMicrobiological testing/measurementAmelogeninIndel
Compositions, methods and kits are disclosed for use in simultaneously amplifying at least 20 specific STR loci of genomic nucleic acid in a single multiplex reaction, as are methods and materials for use in the analysis of the products of such reactions. Included in the present invention are materials and methods for the simultaneous amplification of 23 and 24 specific loci in a single multiplex reaction, comprising the 13 CODIS loci, the Amelogenin locus, an InDel and at least six to ten additional STR loci, including methods, kits and materials for the analysis of these loci.
Owner:LIFE TECH CORP
Compound amplification kit for InDel genetic polymorphic sites of human euchromosome and Y chromosome and application thereof
ActiveCN106868150AIncrease the number of detectionsShort ampliconMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceTyping
The invention discloses a compound amplification kit for InDel genetic polymorphic sites of human euchromosome and Y chromosome and application thereof. The kit comprises 47 pairs of euchromosome InDel site loca, 2 pairs of Y chromosome InDel site loca and a pair of specific amplification primers of a sex determination gene. The kit can be used for human individual recognition, paternity identification and degraded detection material recognition. The kit comprises 49 InDel sites which are relatively balanced in types and one sex determination gene. By adopting a six-colored STR fluorescence detection system, the kit is higher than the disclosed legal medical InDel detection kit. The kit disclosed by the invention is suitable for detecting Chinese groups. An amplification system of 50 sites is short in amplification fragment, and the amplicon is controlled at 200bp, so that the system is suitable for InDel typing of high degraded detection material; the amplification system improves the detection numbers of sites in the degraded detection material; the two Y-InDel sites introduced into the system plays an auxiliary judging role on the sex determination gene Amelogenin.
Owner:GUANGZHOU CRIMINAL SCI & TECH RES INST +2
Fluorescently-labeled insertion/deletion (InDel) genetic polymorphism locus composite amplification system and application thereof
ActiveCN101956005AHigh sensitivityHigh individual recognition rateMicrobiological testing/measurementFluorescence/phosphorescenceDiseaseFluorescein
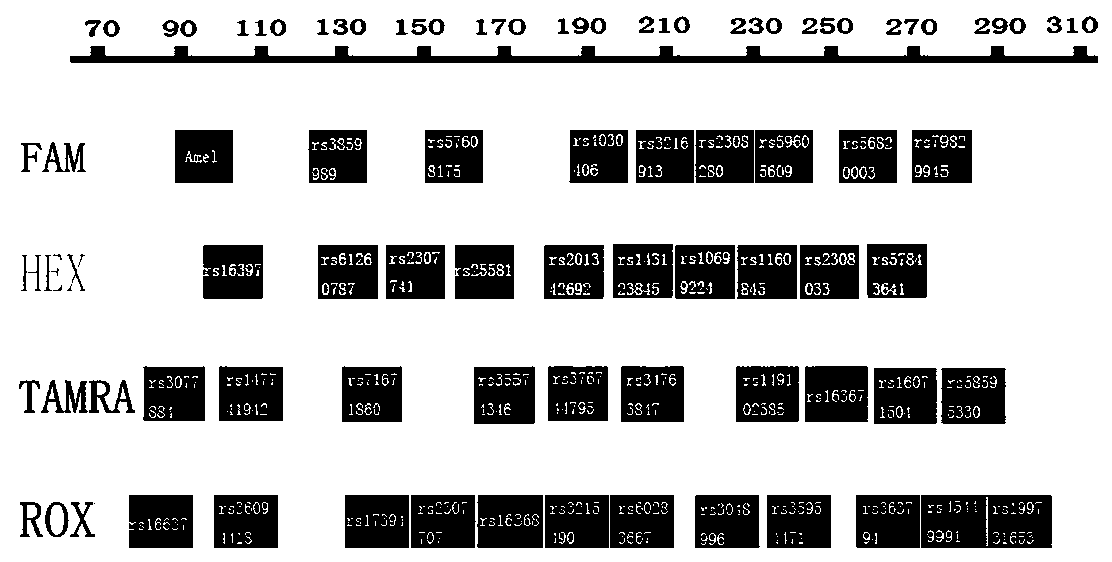
The invention belongs to the technical field of biological detection, and in particular relates to a fluorescently-labeled insertion / deletion (InDel) genetic polymorphism locus composite amplification system and application thereof. The application comprises the following steps of: selecting 35 InDel loci, designing polymerase chain reaction (PCR) amplification primers of the 35 InDel loci, matching fluorescein labels of the PCR primers, compositely amplifying and detecting the 35 InDel loci, and the like. The system can analyze 35 InDel genetic polymorphism loci by the composite amplification. The 35 InDel loci are labeled by FAM, HEX, TAMRA and ROX fluorescein with four different colors respectively. The composite amplification system based on the invention can be made into a kit, serves as an InDel locus fluorescence composite amplification kit with Chinese characteristics, and is applied to the fields, such as triplet paternity test, doublet paternity test, grandparent and grandchild test, sibling test, individual recognition, disease diagnosis, anthropology and the like.
Owner:ACADEMY OF FORENSIC SCIENCE
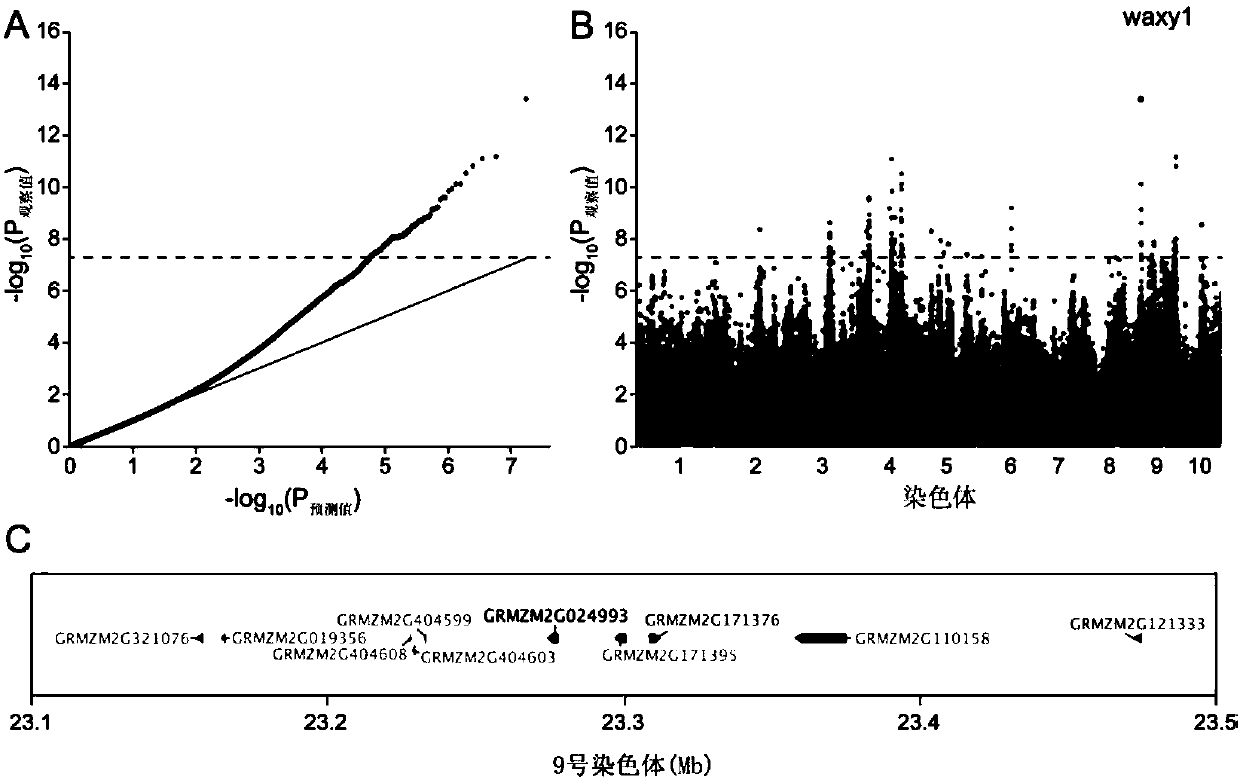
Method for developing waxy1-gene internal molecular markers on basis of association analysis and KASP
ActiveCN107619873AQuick digRapid phenotypeMicrobiological testing/measurementDNA/RNA fragmentationAmylaseCandidate Gene Association Study
The invention discloses a method for developing waxy1-gene internal molecule markers on the basis of association analysis and KASP. One indel and two snap markers located in waxy1 gene and remarkablyassociated with content of amylase are found out mainly through genome-wide association study and candidate gene association analysis techniques; the detected indel is marked as a major mutant type ofglutinous maize in China; glutinous maize materials can be identified through agarose gel by the markers developed on the basis of the indel, and convenience and rapidness is achieved as compared with that of original polyacrylamide gel identification; meanwhile, paired linkage disequilibrium test of 59 parts of waxy1-gene sequence regions of maize hybrid line materials is performed, close linkage of the indel marker and the two snp markers is tested, primers of the two snps are developed on the basis of the KASP technology, the glutinous maize materials can be identified rapidly, efficientlyand accurately, and molecular breeding of glutinous maize is facilitated.
Owner:SHANGHAI JIAO TONG UNIV
Composition for modifying nucleotide sequence, method and application
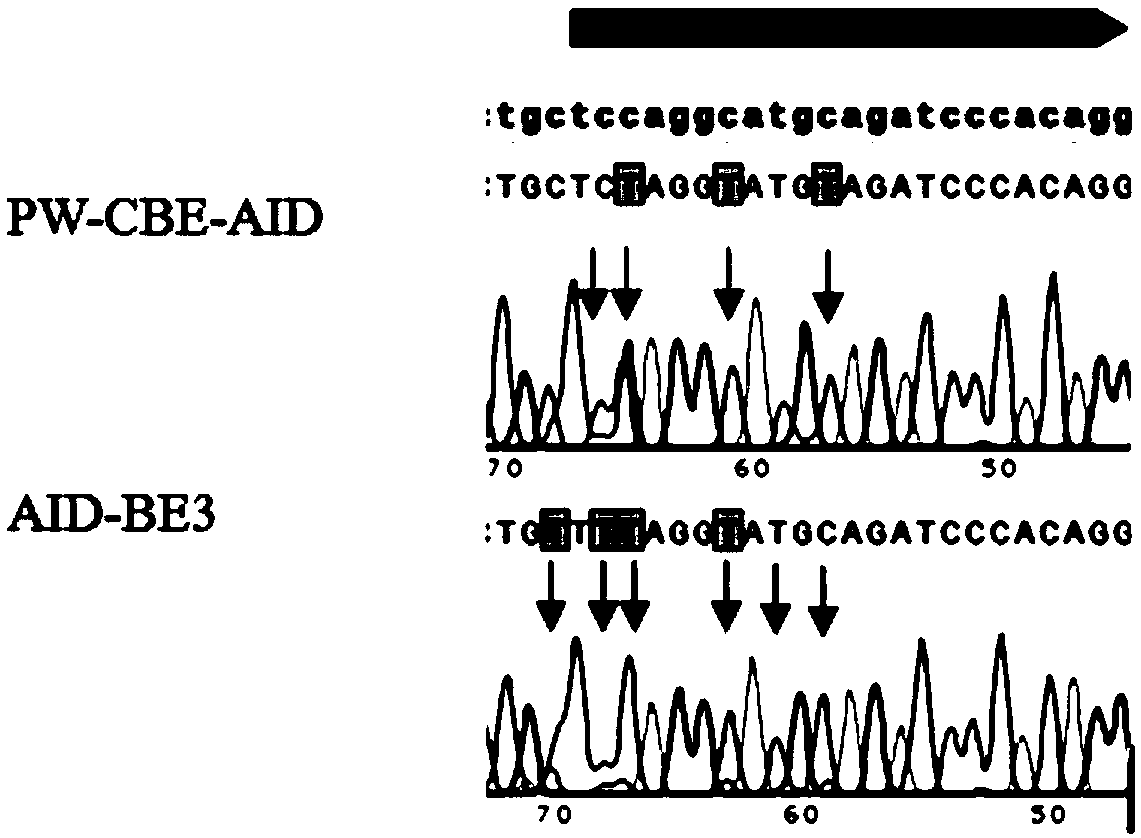
ActiveCN109517841AWide working windowReduce deletionVector-based foreign material introductionDNA preparationCytosine deaminaseNucleotide
The invention discloses a composition for modifying a nucleotide sequence, a method and application, and relates to the technical field of gene editing. The composition comprises a first vector and asecond vector, wherein the first vector is provided with the following expression elements, such as a cytosine deaminase expression element, an adenine deaminase expression element and a mutant Cas enzyme expression element; the second vector is provided with the following expression elements, such as a gRNA (guide ribonucleic acid) expression element and a uracil glycosidase inhibitor expressionelement. The composition is used for modifying the C bases at the 1st to 16th sites at the upstream of PAM (protospacer adjacent motif) of the target nucleotide sequence, so that the transformation ofC / G to T / A is realized. Compared with the existing gene editing technique, the composition and the method have the advantages that the working window is broader; the DSB (double-strand break) and indels (insertion / deletion mutations) are not introduced, the off-target effect is very low, the safety is higher, and the like.
Owner:EAST CHINA NORMAL UNIV +1
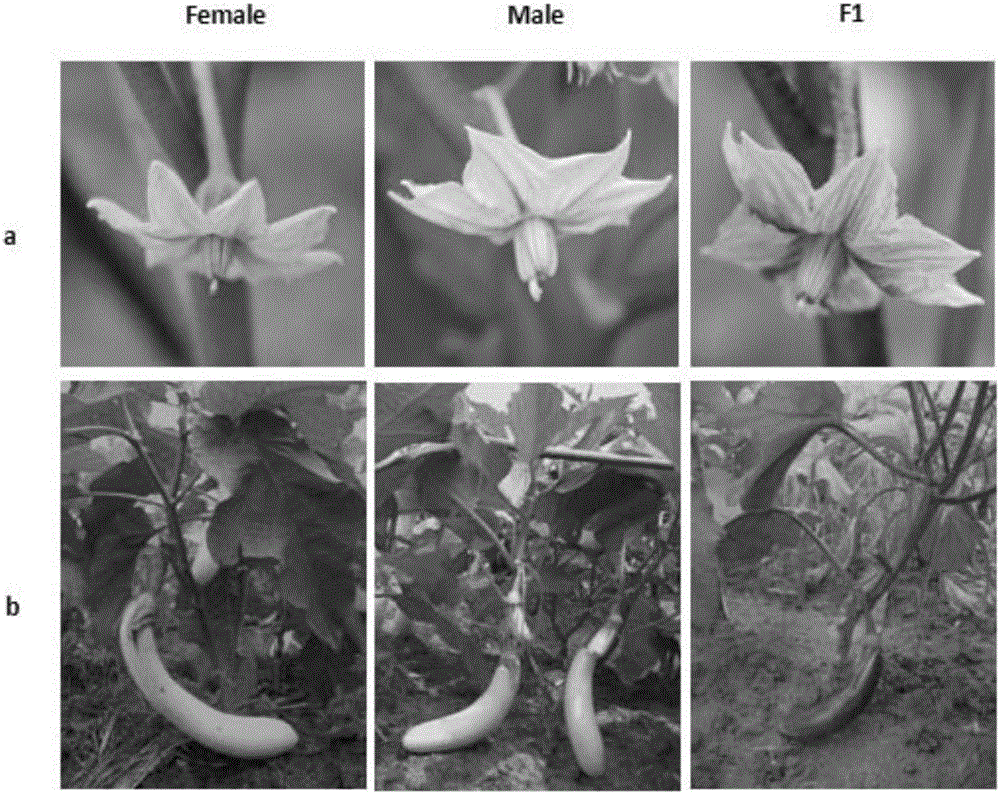
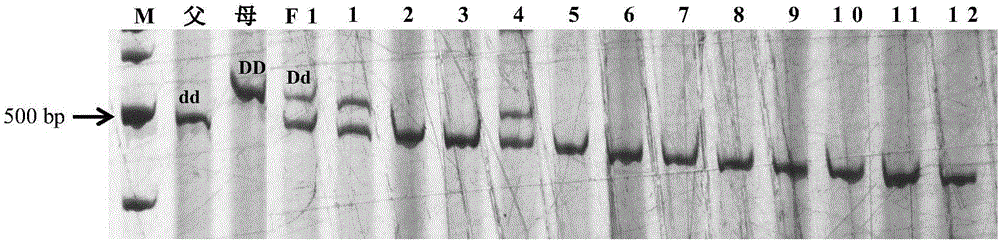
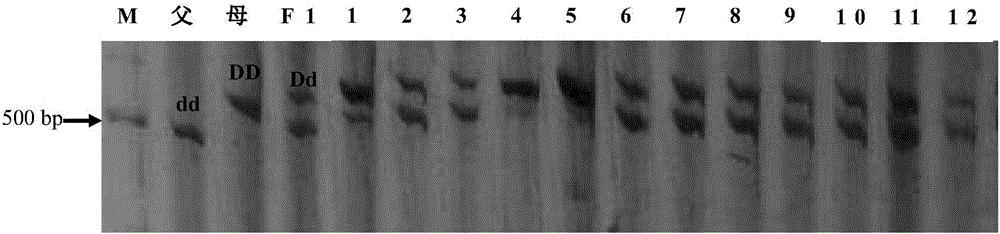
Mapping of eggplant fruit color epistatic gene D, and InDel molecular marker development and application thereof
ActiveCN106701926AImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationRe sequencingAgricultural science
The invention discloses a mapping result of an eggplant fruit color epistatic gene D. The gene D is mapped in a position near a nucleotide sequence Sme2.5_00538.1 on a No.10 chromosome of the eggplant. Based on the re-sequencing result of male and female parents, an InDel molecular marker closely linked with the eggplant fruit color epistatic gene D is obtained near the nucleotide sequence Sme2.5_00538.1 of the No.10 chromosome, and is named as InDel22522; the genetic distance of the marker from the epistatic gene D is 0.61 cM. The nucleotide sequence of the molecular marker InDel 22522 in the female parent has one 29-basic-group InDel more than that in the male parent. The mapping result on the eggplant fruit color epistatic gene D provided by the invention lays the foundation for the precise positioning and cloning of the eggplant fruit color epistatic gene; the InDel 22522 molecular marker and a molecular marker amplification primer can be applied to eggplant fruit color breeding practice in a way of simplicity, convenience, high speed and high flux; the eggplant fruit color property improving process is accelerated.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
Molecular marker interlocked with pepper cucumber mosaic virus disease resistance gene qcmv-2-1 and application of molecular marker
InactiveCN105296475AImprove the detection rateEasy to mark tapeMicrobiological testing/measurementDNA/RNA fragmentationDiseaseResistant genes
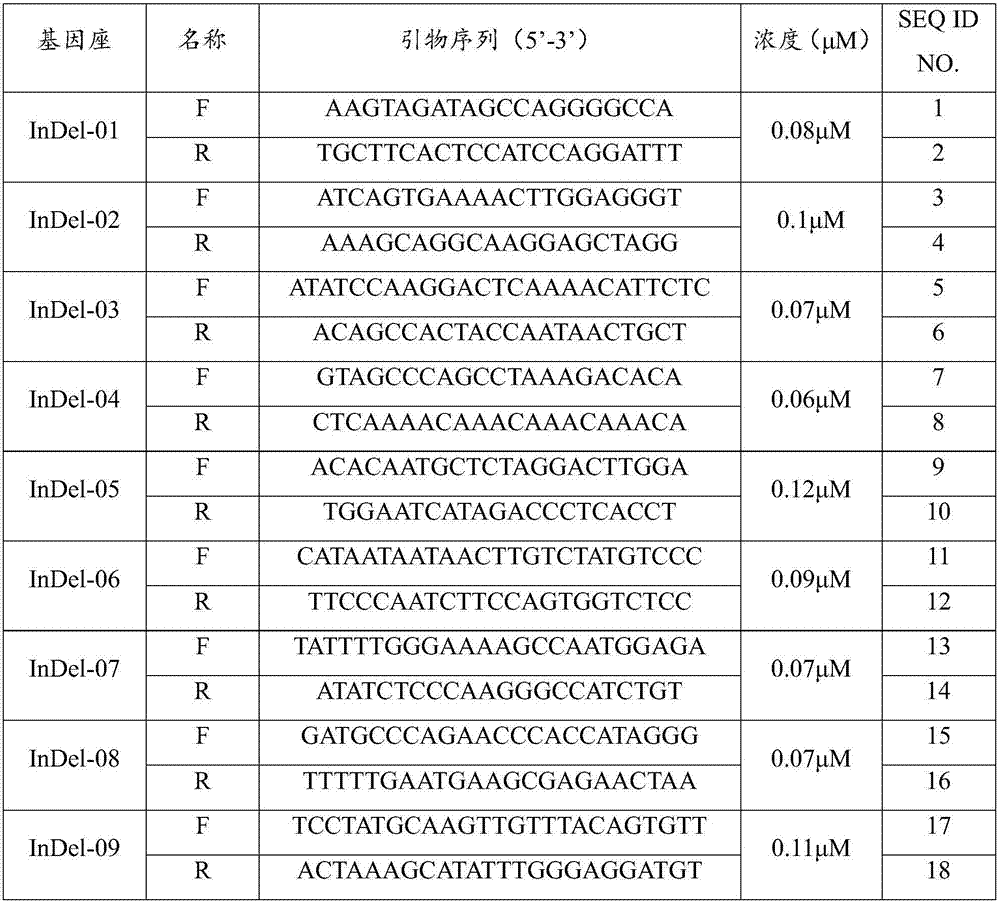
The invention discloses a molecular marker interlocked with a pepper cucumber mosaic virus disease resistance gene qcmv-2-1 and an application of the molecular marker. According to the molecular marker interlocked with the pepper cucumber mosaic virus disease resistance gene qcmv-2-1, pepper genome DNA is amplified through a primer pair InDel-2-134F / InDel-2-134R, and an obtained 230bp amplified fragment is the molecular marker interlocked with the pepper cucumber mosaic virus disease resistance gene qcmv-2-1; the InDel-2-134F sequence is shown as SEQ ID NO.1, and the InDel-2-134R sequence is shown as SEQ ID NO.2. According to the Indel molecular marker and corresponding primers interlocked with the qcmv-2-1 resistance gene and screened through the InDel marker and QTL-Seq technology, marker bands are simple, and the molecular marker and the application of the molecular marker can be directly used for marker-assisted section.
Owner:JIANGSU ACAD OF AGRI SCI
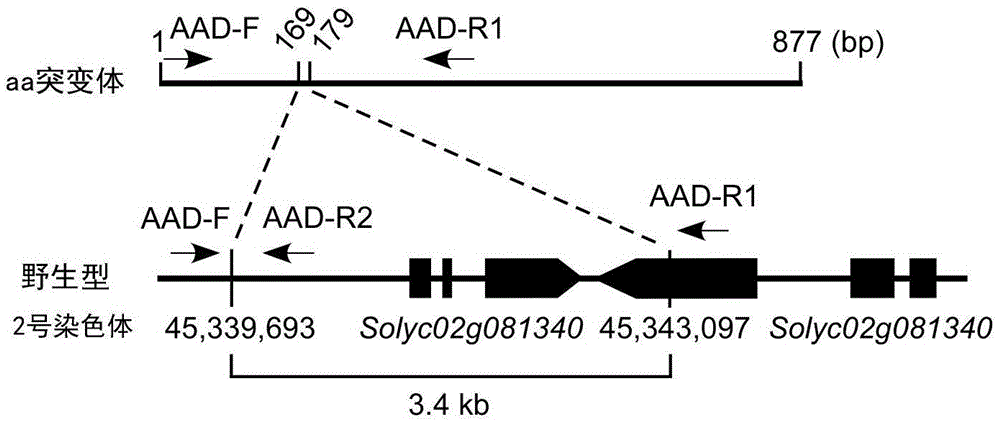
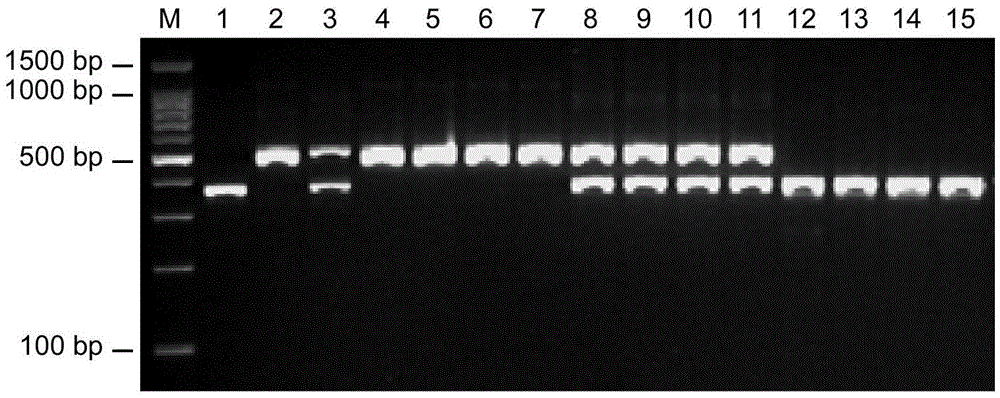
Insertion/deletion (InDel) molecular marker AAD closely-linked to tomato anthocyanin absent character and application thereof
ActiveCN105525016AShorten the breeding cycleImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMolecular breeding
The invention relates to the field of molecular breeding, and in particular to an insertion / deletion (InDel) molecular marker AAD closely-linked to the tomato anthocyanin absent character and application thereof. The physical position of the InDel molecular marker closely-linked to the tomato anthocyanin absent character is No. 45339693 base to No. 45343097 base on chromosome 2 of tomato. The invention can replace a traditional method of selecting a green plant to ensure that a breeding material of tomato contains an anthocyanin absence (AA) mutation site. An AA site hybrid plant in the breeding material with normal phenotype is assist-selected by mean of the molecular marker, the breeding period can be shortened, and the breeding efficiency can be improved.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
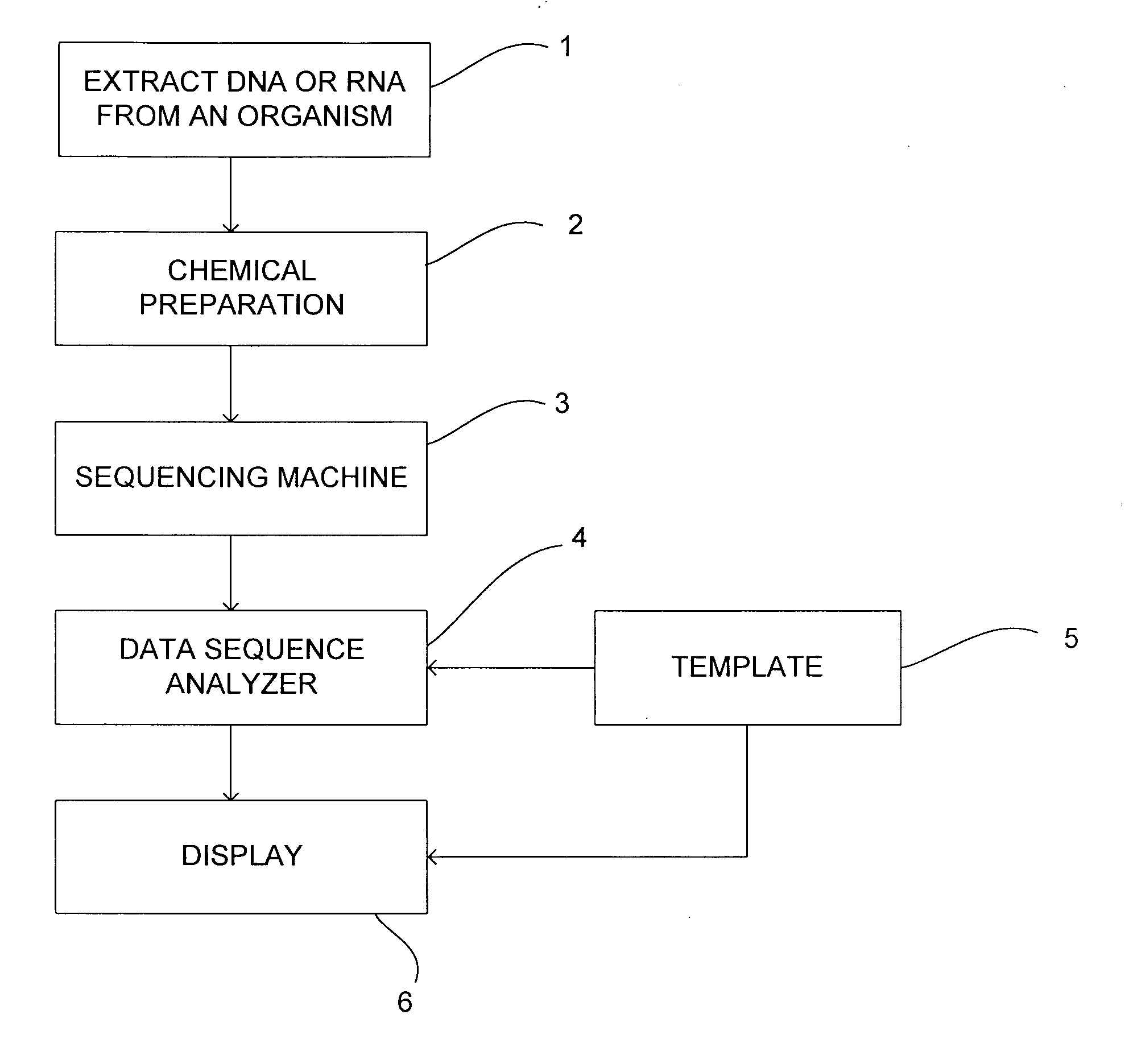
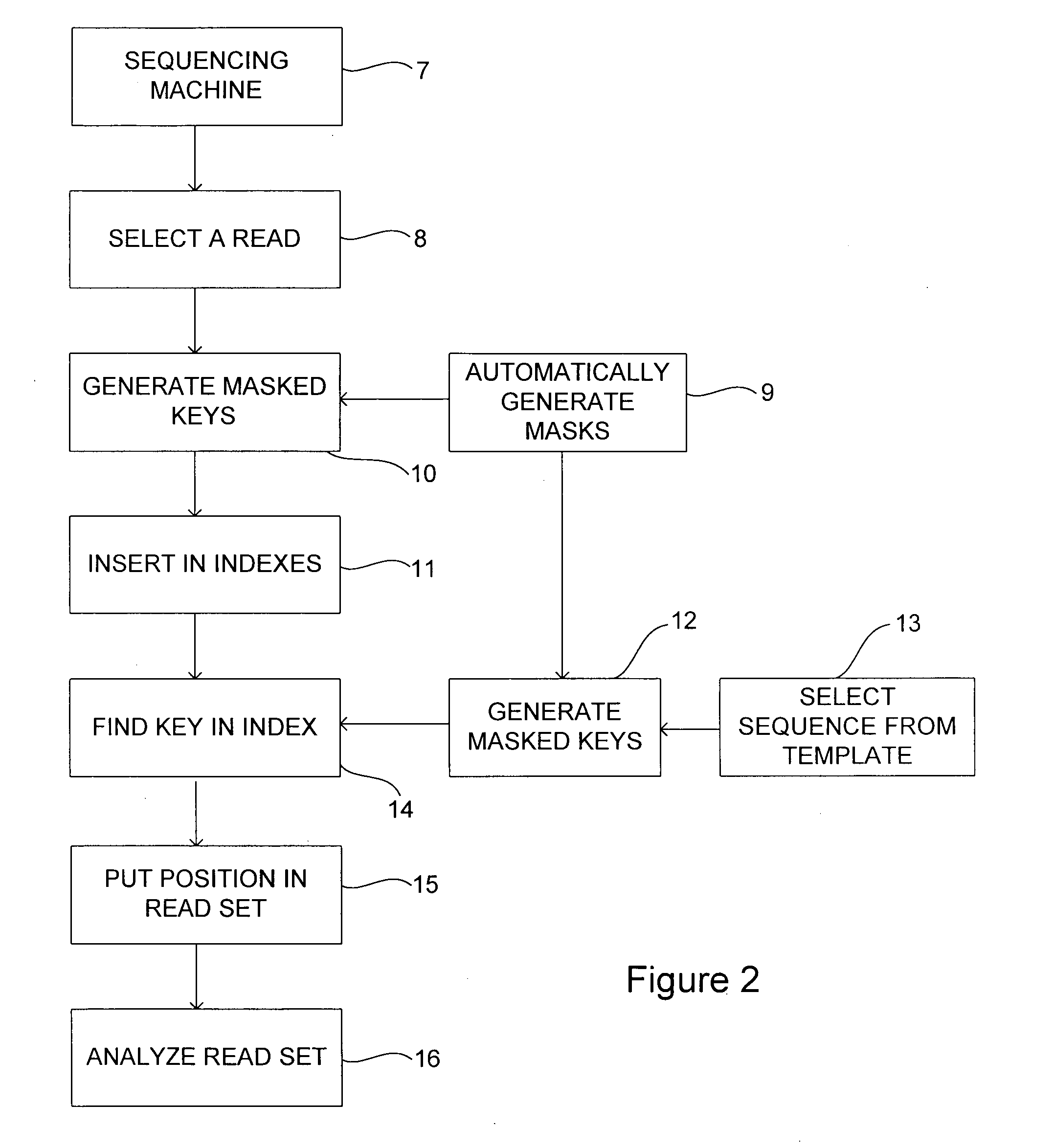
Method and system for analysing data sequences
A sequencing system and method of generating index keys for one or more data sequence based on masked values of reads from a sample data sequence and / or one or more template data sequence. Each index key value may be based upon a concatenated form of each extracted value, although other transformations may be employed. A number of different masks may be applied to the data sequence at a number of locations. At least some of the masks may include indels and / or substitutions. The masks may be manually or computer generated. The data sequence may be one or more reference templates and / or one or more sample sequences, such as DNA or RNA sequences. Sample data may be stored in the one or more index by correlating masked values of reads with index key values and storing an identifier for each read in association with a corresponding index key value. Sample data sequences may be evaluated by comparing sample sequence and template sequences having the same index key value and determining scores for the reads based on the comparison and associating the scores with the reads. Reads may be rejected based upon the comparison. A read may be rejected if there is more than one position at which it has a best score. A read may be rejected if its score falls below a threshold score level.
Owner:REAL TIME GENOMICS
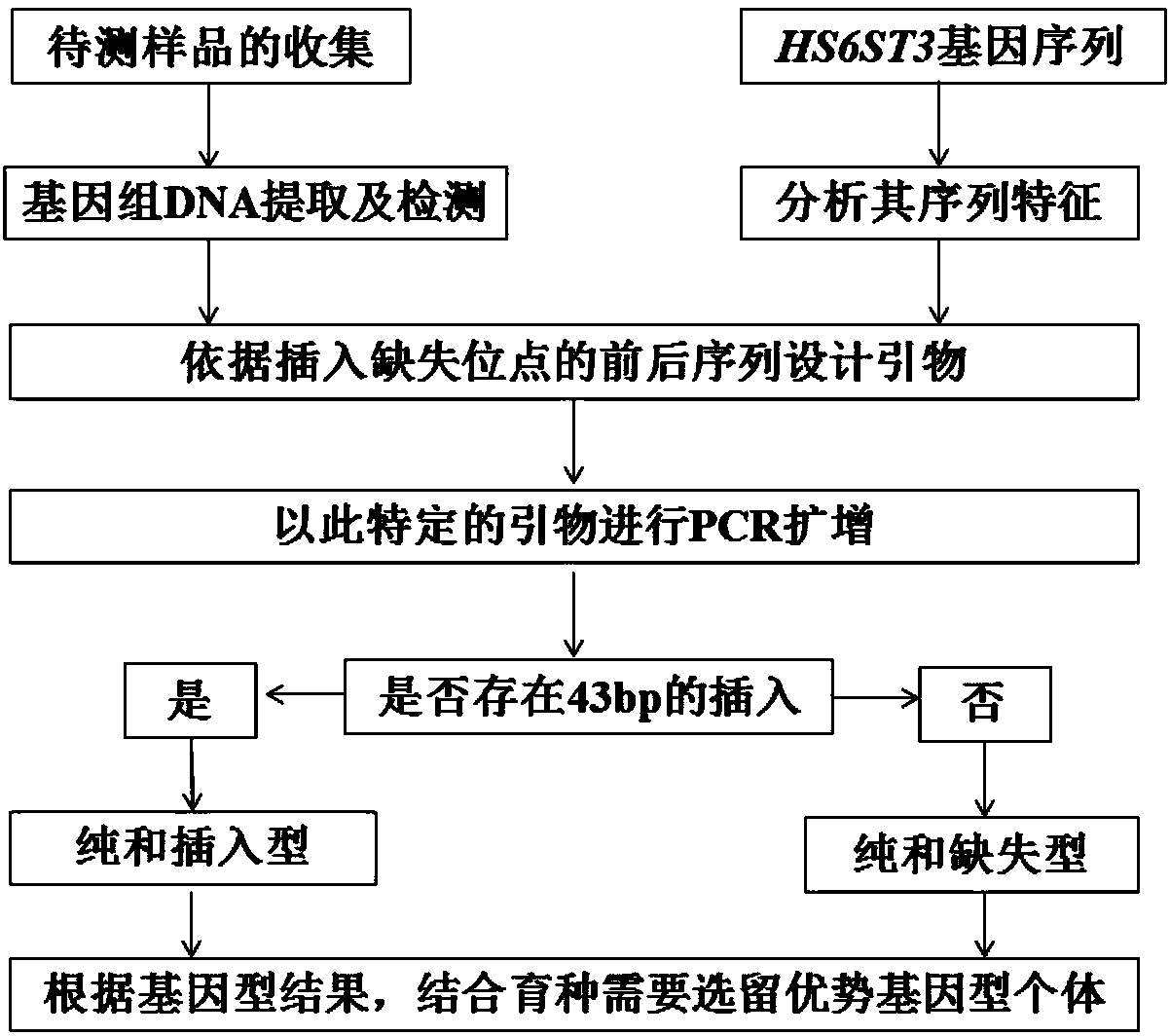
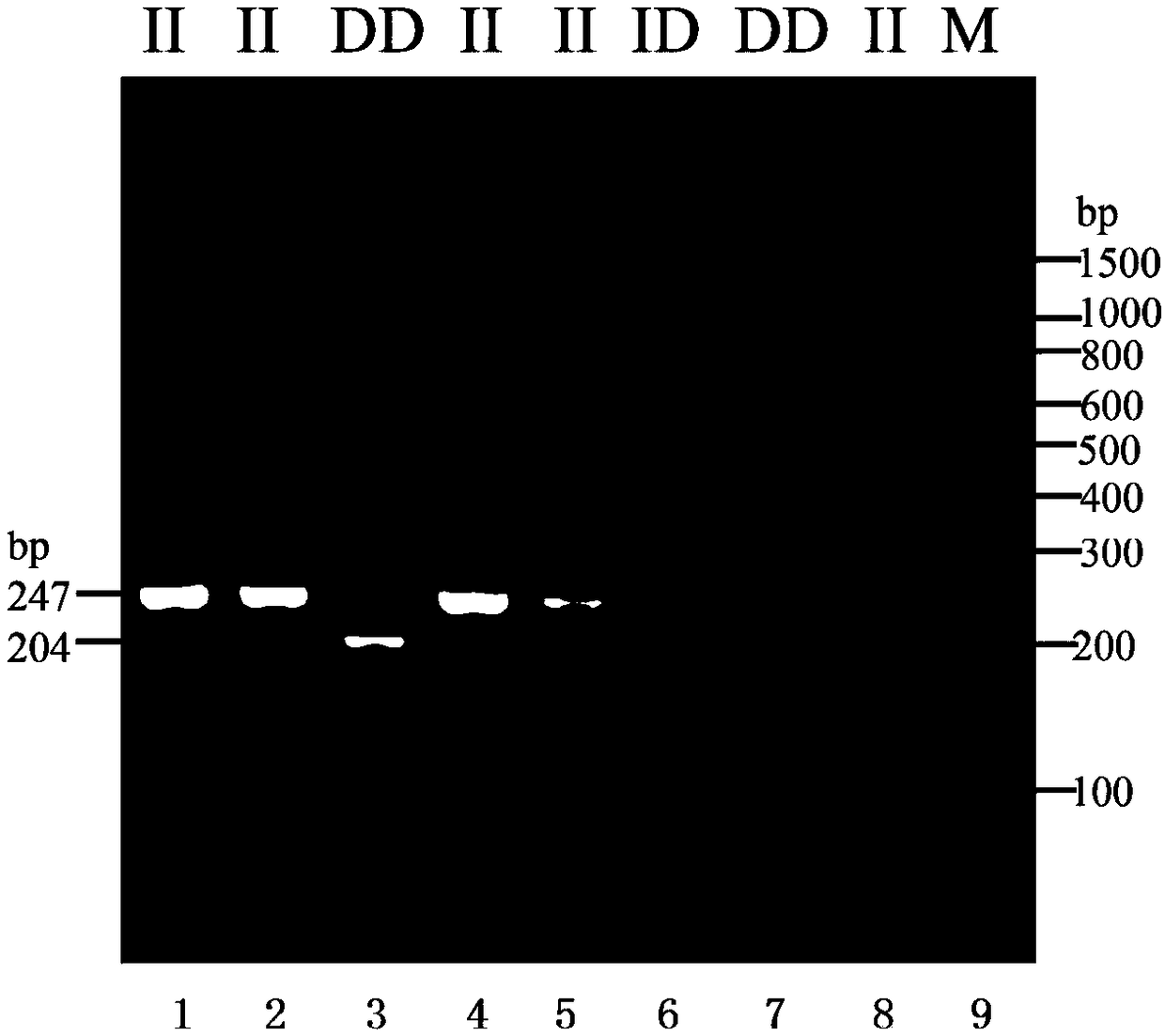
Chicken HS6ST3 gene 43bp indel polymorphic marker, application thereof, detection primer and reagent kit
InactiveCN108531615AReduce manufacturing costAccelerate the progress of genetic selectionMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionIntein
The invention relates to a chicken HS6ST3 gene 43bp indel polymorphic marker, application thereof, a detection primer and a reagent kit, and belongs to the technical field of molecular genetic breeding. The chicken HS6ST3 gene 43bp indel polymorphic marker, the application, the detection primer and the reagent kit have the advantages that the 43bp indel polymorphic marker in first introns of HS6ST3 genes is discovered, is obviously related to diversified economic characters such as 12-week shank circumferences and 6-week body weights of chicken and can be used for assisted selection and molecular breeding on the chicken; particularly, an amplification fragment designed according to a sequence shown as SEQ ID NO.1 contains the primer at 176th-218th bits from a 5' end of the sequence, whether the 176th-218th bits are inserted / deleted can be judged according to the sizes of amplification products, breeding on dominant genotype individuals with 176th-218th bit homozygous deletion can be reinforced, accordingly, the economic benefits of indigenous and commercial chicken can be increased, methods are free of enzyme digestion, accurate in genotype judgment, easy to operate, low in cost and short in period, and discrimination can be facilitated.
Owner:HENAN AGRICULTURAL UNIVERSITY
Molecular breeding method for new rice variety carrying gene Pi65(t) with resistance to rice blast
ActiveCN104774945AMicrobiological testing/measurementPlant genotype modificationDiseaseAgricultural science
The invention belongs to the field of molecular biology and particularly relates to a molecular marker assisted breeding method of a new rice variety with resistance to rice blast and special primers thereof. The molecular mark is a co-dominant molecular mark Indel-1 of the rice gene Pi65(t) with resistance to rice blast and is a nucleotide sequence which is amplified from total DNA of rice by using a primer pair SEQ ID NO: 1 and SEQ ID NO: 2. The method can be applied to molecular marker assisted selection of Pi65(t) in resistance breeding of the resistance to blast of rice, so that the efficiency of the anti-disease variety breeding is improved and the workload of field identification is reduced.
Owner:LIAONING ACAD OF AGRI SCI +2
Fluorescent multiplex amplification system of 37 Y-STR gene locuses and Y-Indel, kit and application thereof
ActiveCN108531610AImprove application efficiencyHigh individual recognition rateMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceBiology
The invention provides a fluorescent multiplex amplification system of 37 Y-STR gene locuses and a Y-Indel, a kit and application thereof, belonging to the technical field of biological detection. Thekit can amplify and analyze 37 Y-STR gene locuses and a Y-Indel at the same time. Six colors of fluorescence are used for marking, the size of the amplified product is 70-490bp, the operation is simple, and very high cumulative individual discernment capability and cumulative probability of paternity exclusion are realized.
Owner:SUZHOU MICROREAD GENETICS
Methods of Identifying Microsatellite Instability
ActiveUS20190194731A1Improve the level ofMicrobiological testing/measurementTumor SampleWilms' tumor
The present invention relates to methods and kits for identifying microsatellite instability (MSI) in a sample. In particular it relates to identifying microsatellite instability in a tumor sample, which may be from a subject suspected of having colorectal cancer or Lynch syndrome. The methods and kits can be used to identify mismatch repair defects. More particularly the invention relates to a panel of markers for a sequencing based MSI test, that can differentiate between MSI-H and MSS CRCs. The invention also allows for determination of biological significance, differentiating between PCR and sequencing errors and MSI induced indels / mutations.
Owner:CANCER RES TECH LTD
Somatic mutation detection method and device for tumor tissue single samples
PendingCN111718982ASolving the Mutation Detection ChallengeEnabling somatic mutation detectionBioreactor/fermenter combinationsBiological substance pretreatmentsMutation detectionHigh flux
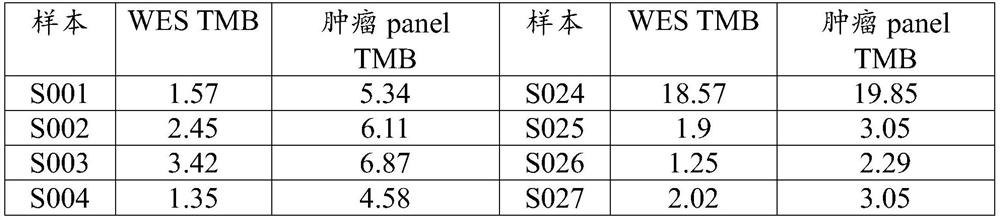
The invention belongs to the technical field of tumor gene detection and provides a high-flux capture sequencing based somatic mutation detection method and device for tumor tissue single samples, andfurther provides a computer readable medium used for operating the method and detection equipment. According to the invention, a normal copy number base line is established by selecting detected normal control samples, and a model capable of accurately screening stable conditions of single sample microsatellite sites is established by selecting the detected normal control samples; and compared with the prior art, the problem that tumor tissue mutation detection is carried out under the condition of no control samples can be solved, besides, multiple types of mutation such as SNV, INDEL, CNV,Fusion and MSI can be detected better, and calculation of tumor mutation burden (TMB) can be performed. The somatic mutation detection of the tumor tissue single samples can be realized under the condition of no control samples.
Owner:SHENZHEN HAPLOX BIOTECH
Kit used for diagnosing/predicting sudden cardiac death
ActiveCN107858423ASusceptibility predictionMicrobiological testing/measurementDNA/RNA fragmentationGenotypeSudden cardiac death
The invention relates to a kit used for diagnosing / predicting sudden cardiac death (SCD). The kit comprises one or more of a specific primer pair (SEQ ID No. 1-SEQ ID No. 6) used for detecting the number rs3917 insertion / deletion polymorphic site on a COL1A2 gene, a specific primer pair (SEQ ID No. 7-SEQ ID No. 12) used for detecting the number rs72014506 insertion / deletion polymorphic site on anMIR155HG gene, a specific primer pair (SEQ ID No. 13-SEQ ID No. 18) used for detecting the number rs113044851 insertion / deletion polymorphic site on a CTH gene and a specific primer pair (SEQ ID No. 19-SEQ ID No. 24) used for detecting the number rs397729601 insertion / deletion polymorphic site on a DSG2 gene. The kit has the advantages that the SCD risks of a patient are evaluated through the insertion / deletion polymorphic evaluation, the associated analysis of the genotype of the four sites can favorably provide molecular-genetics support and help for the diagnosis and prediction of the SCD,and the kit has high application value in the risk evaluation and diagnosis of the SCD; in addition, the kit is good in detection specificity, high in sensitivity and good in accuracy.
Owner:SUZHOU UNIV
InDel molecular marker closely linked with epistatic gene P of eggplant fruit color and application of InDel molecular marker
ActiveCN108034752AImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceHigh flux
The invention discloses an InDel molecular marker closely linked with an epistatic gene P of eggplant fruit color and application of the InDel molecular marker. The genetic distance between the InDelmolecular marker and the epistatic gene P is 0.5cM, and InDel of specific insertion / deletion fragments at 50bp and 35bp in nucleotide sequences of the male parent is not found from the female parent.The InDel molecular marker closely linked with the epistatic gene P of the eggplant fruit color, disclosed by the invention, lays a foundation for fine mapping and cloning of the epistatic gene of theeggplant fruit color. An InDel 5513 molecular marker and a molecular marker amplification primer can be simply and rapidly applied in eggplant fruit color breeding practice at high flux, so that theprogress of improving fruit color characters of eggplants is accelerated.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
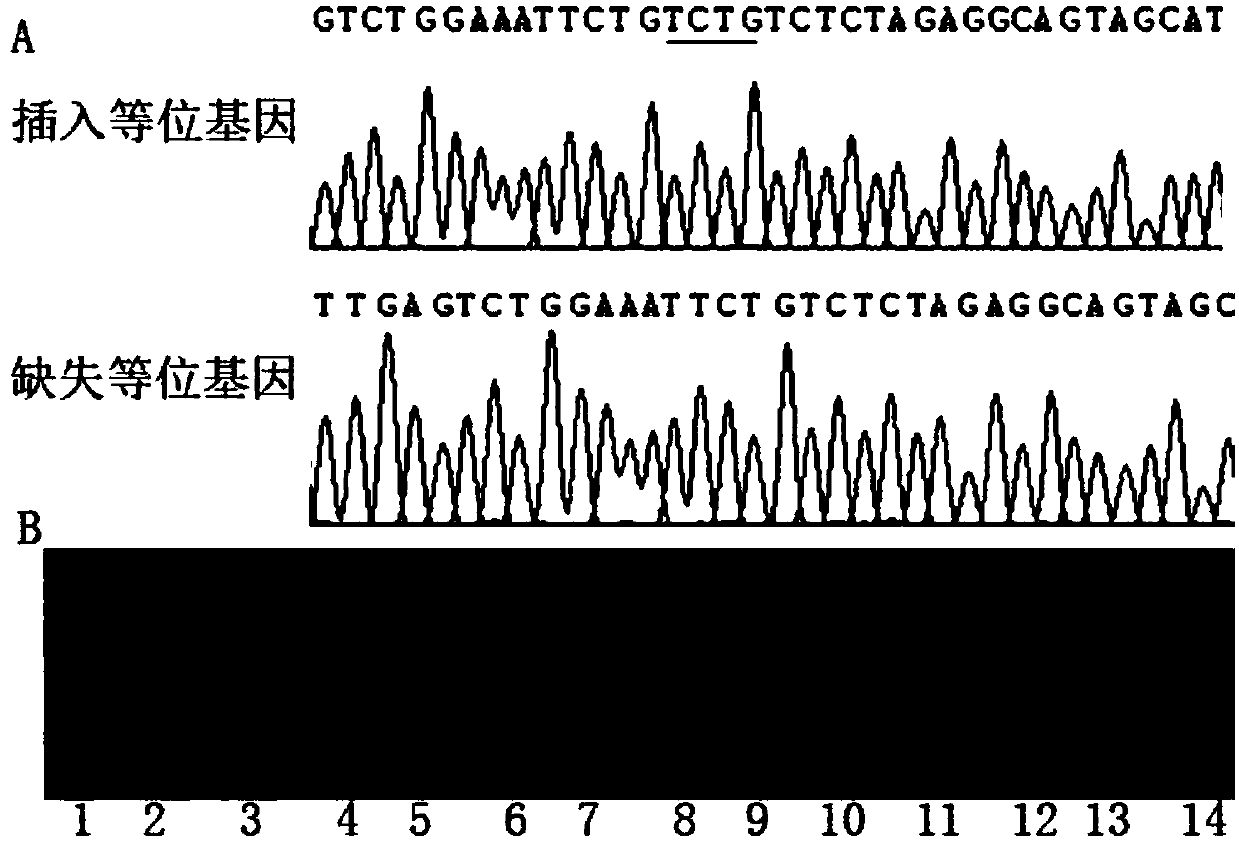
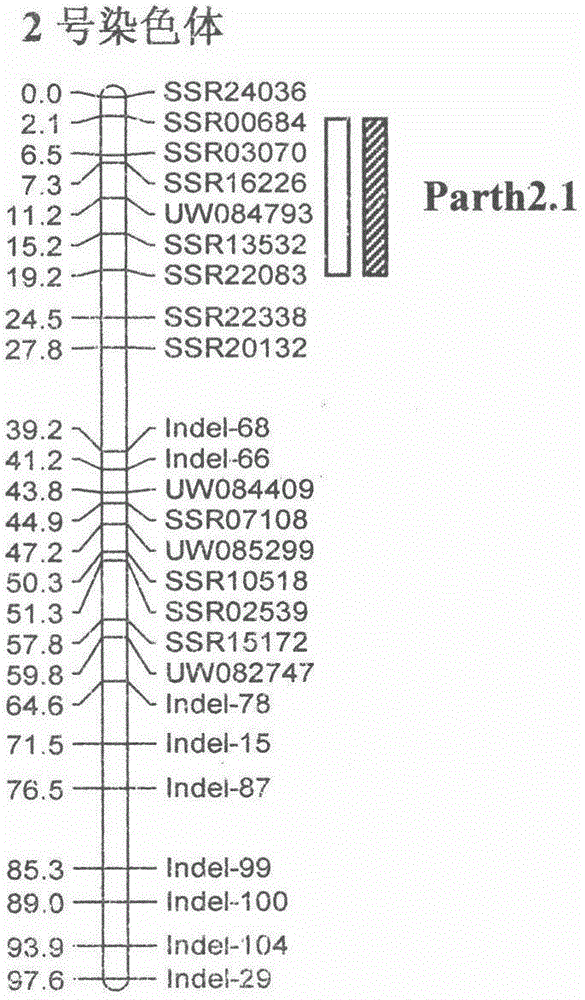
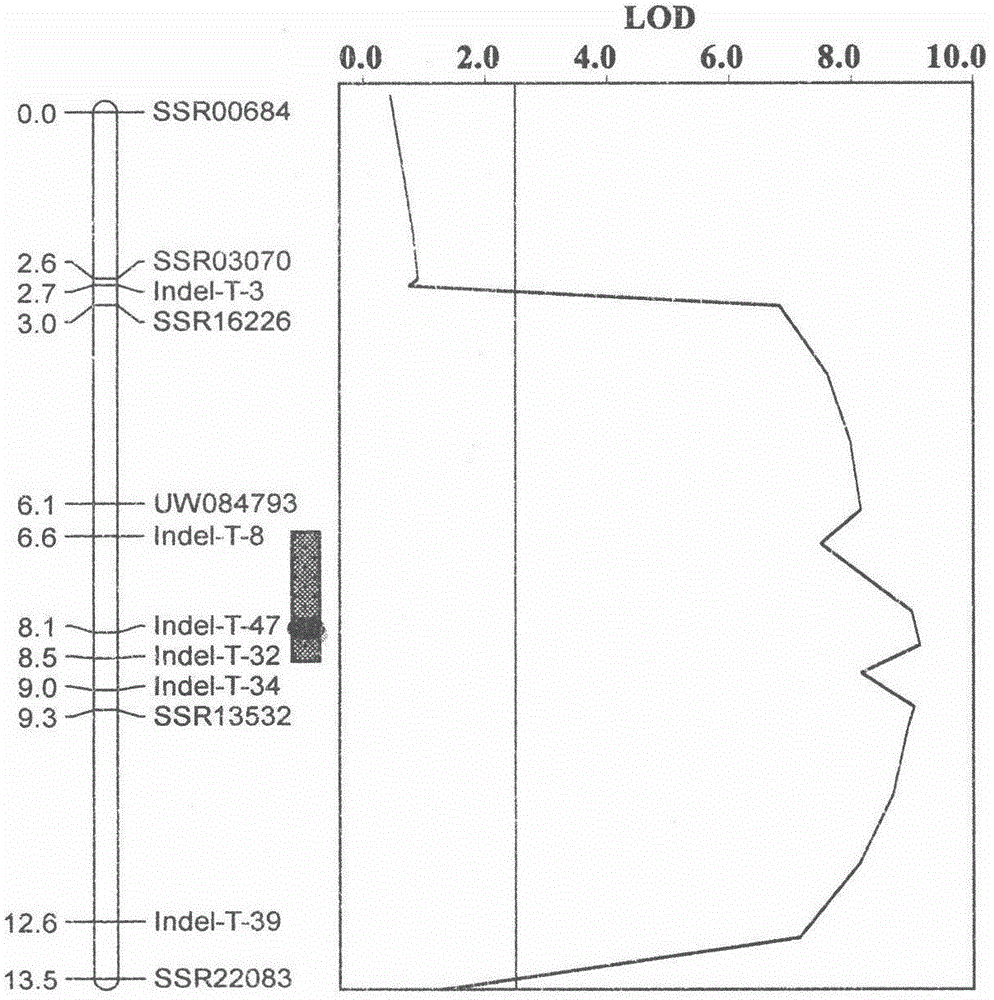
Molecular marker Indel-T-47 for close linkage with cucumber parthenocarpic main-effect QTL (quantitative trait loci)
InactiveCN104789562ASimple methodStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceQuantitative trait locus
The invention discloses a molecular marker Indel-T-47 for close linkage with cucumber parthenocarpic main-effect QTL (quantitative trait loci), and belongs to the technical field of biology. The invention firstly discloses a molecular marker Indel-T-47 for close linkage with QTL Parth2.1, and further discloses a primer Indexl-T-47F / Indel-T-47R of the marker. In a segregating population containing Parth2.1, the marker is significantly related to parthenocarpy; in an all-female selfing line material containing the Parth2.1, a 155bp product can be amplified from a strong parthenocarpy strain by the marker primer Index-T-47F / Index-T-47R; the marker primer Index-T-47F / Index-T-47R is a peak marker of the Parth2.1, and is closely linked with the Parth2.1. The molecular marker for close linkage with the Parth2.1 disclosed by the invention can be applied to assistant breeding of an all-female cucumber parthenocarpy molecular marker.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecular marking method of peanut plant type-related gene loci and application thereof
InactiveCN110592264AClear locationFine locationMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionMarker-assisted selection
The invention belongs to the field of molecular genetic breeding science, and provides a molecular marking method of gene loci related to peanut lateral branch angle, growth habit and plant types andapplication thereof. By designing a CAPS molecular marker, and by using a method combining primer amplification, enzyme digestion and agarose gel electrophoresis, the judgment of allelic variation ofgene loci related to the lateral branch angle, the growth habit and the plant types is realized. By labeling PM15-ID1 and PM15-ID2 with InDel, and by using the method of primer amplification and polyacrylamide gel electrophoresis, the marker-assisted selection of offspring with allelic variation of gene loci related to lateral branch angle, growth habit and plant types. The molecular marking method provided by the invention can be used for detecting the offspring of a breeding group constructed by using peanut creeping germplasm and upright germplasm as parents, can be used for rapidly identifying the allelic variation of gene loci related to lateral branch angle, growth habit and plant types of the offspring, can improve the selection efficiency of the gene loci, and can provide referencefor peanut plant type improvement and high-yield breeding.
Owner:QINGDAO AGRI UNIV
InDel marker of cucumber anti-cucumber-mosaic-virus-disease gene cmv and its application
ActiveCN107674922AReduce restrictionsMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCucumber mosaic virus
The invention provides an InDel marker of a cucumber anti-cucumber-mosaic-virus-disease gene cmv and its application, belonging to the field of biotechnological assistant breeding. The nucleotide sequence of a primer InDel-cmv3-F / InDel-cmv3-R for the InDel marker closely linked with the cucumber anti-cucumber-mosaic-virus-disease gene cmv is as described in the specification; a characteristic band amplified by the InDel marker and closely linked with the anti-cucumber-mosaic-virus-disease gene cmv has a length of 223 bp; and a characteristic band amplified by the InDel marker and closely linked with a cucumber-mosaic-virus-disease susceptibility gene CMV has a length of 245bp. The marker provided by the invention has the advantages of high efficiency and little limitation, improves the efficiency of breeding of anti-cucumber-mosaic-virus-disease cucumber materials and shortens a breeding cycle.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Method for analyzing mixed sample DNA
PendingCN110157786ASuitable for forensic testingProtection securityMicrobiological testing/measurementOrgan transplantationBone marrow transplantations
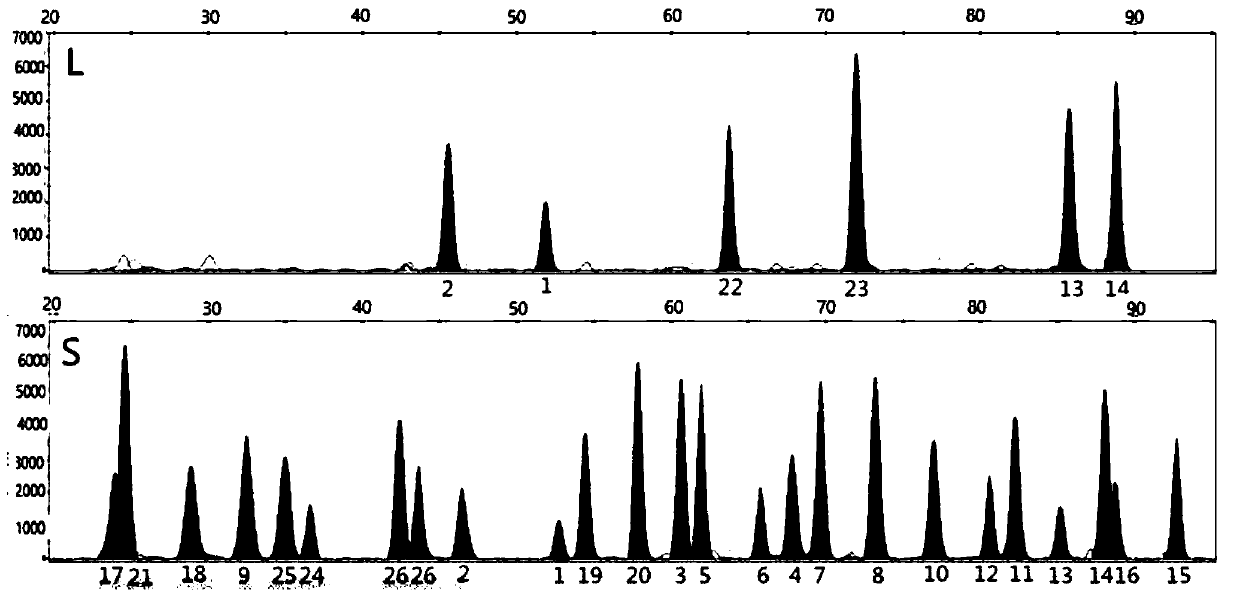
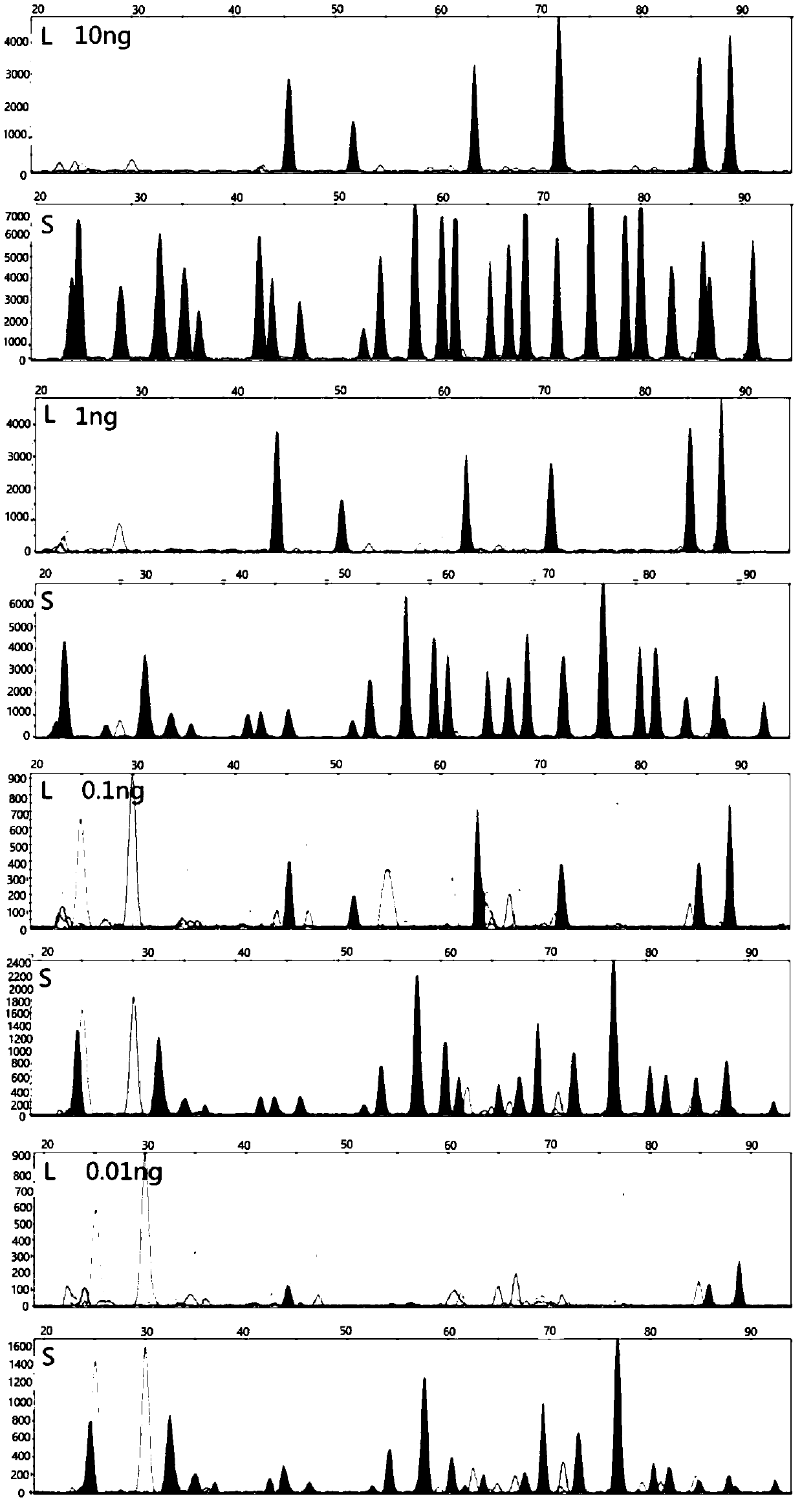
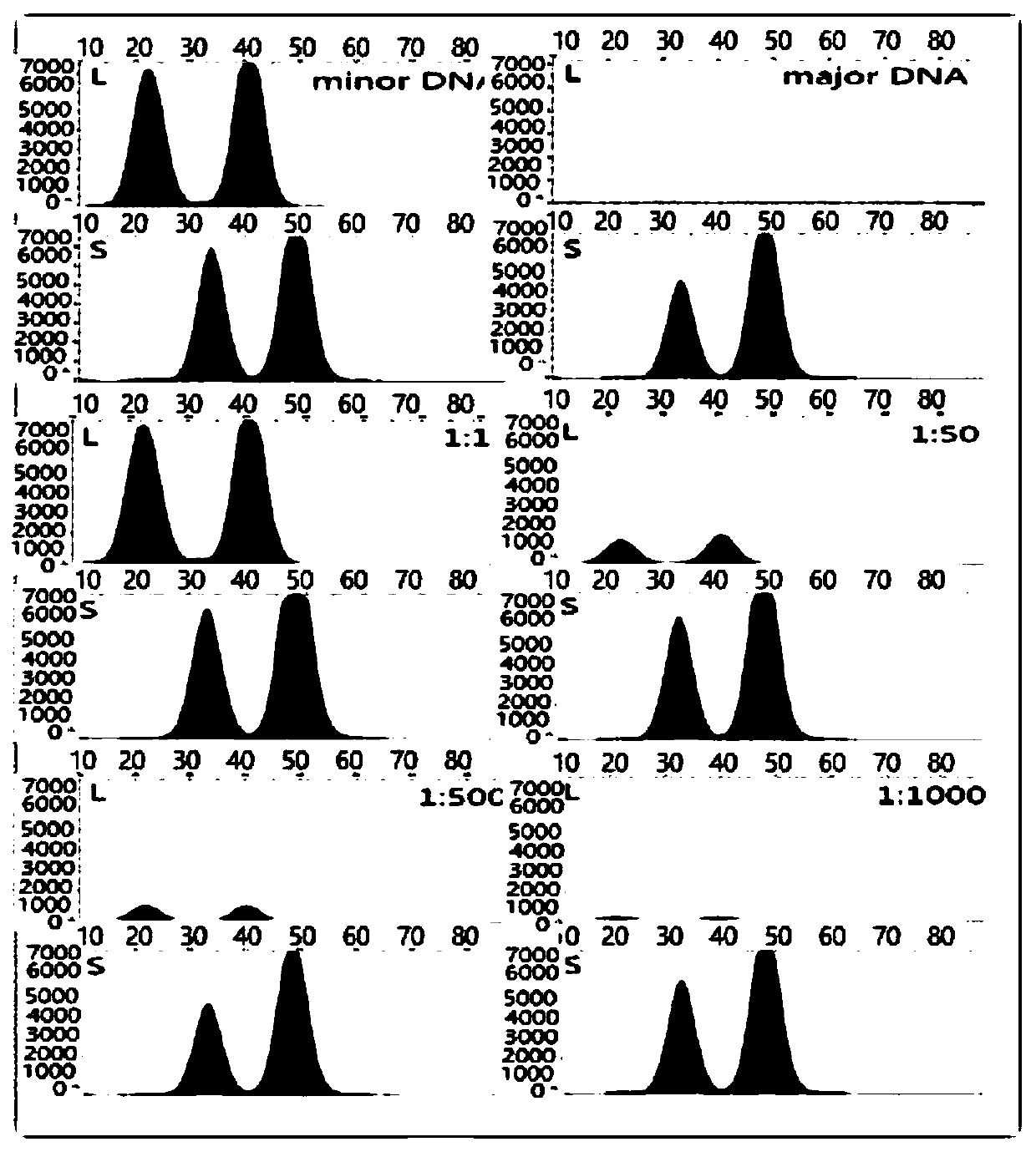
The invention discloses a method for analyzing mixed sample DNA. The mixed sample DNA is analyzed with a method combining INDEL with Microhaplotypes; in other words, father source or mother source DNAof an individual is sorted through differential amplification of the INDEL, and then individual distinguishing is conducted through the Microhaplotypes on DNA molecules; the method is named a DIP-Microhaplotypes method. The method has the advantages that analysis of a mixed DNA sample of two samples and detection of mixed stain sensitivity mainly depend on the INDEL, following detection of SNP isrelated to a detection means, and mixed stains can be successfully analyzed and used for excluding or confirming the individual; an INDEL-SNP primer has species specificity and is suitable for forensic medicine detection; the method is possibly used for monitoring transplant ingredients in peripheral blood after a bone marrow transplantation operation or other organ transplantation operations; the method is possibly used for all cases where individuals need to be distinguished when the mixed sample is involved.
Owner:SHANXI MEDICAL UNIV
Methods, Cells & Organisms
InactiveUS20160177340A1Genetically modified cellsStable introduction of DNASingle strandDouble strand
The invention relates to an approach for introducing one or more desired insertions and / or deletions of known sizes into one or more predefined locations in a nucleic acid (eg, in a cell or organism genome). They developed techniques to do this either in a sequential fashion or by inserting a discrete DNA fragment of defined size into the genome precisely in a predefined location or carrying out a discrete deletion of a defined size at a precise location. The technique is based on the observation that DNA single-stranded breaks are preferentially repaired through the HDR pathway, and this reduces the chances of indels (eg, produced by NHEJ) in the present invention and thus is more efficient than prior art techniques. The invention also provides sequential insertion and / or deletions using single- or double-stranded DNA cutting.
Owner:KIMAB LTD
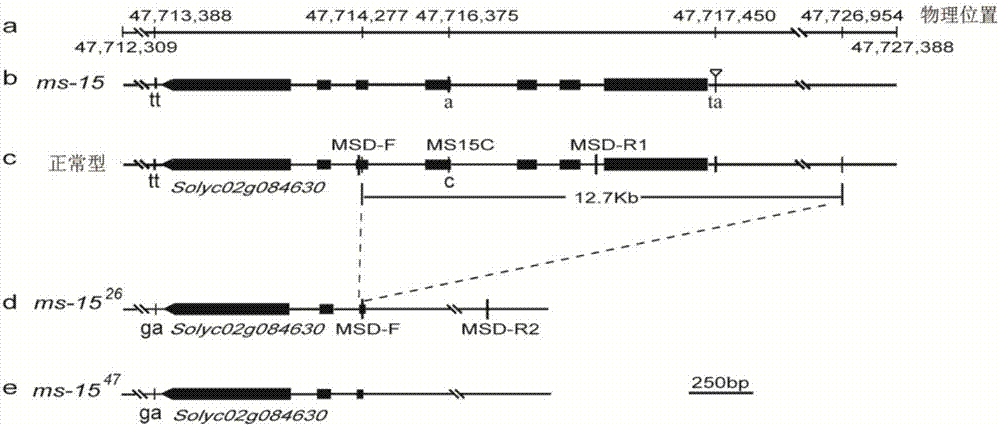
Molecular markers associated with male sterile mutation sites ms-15, ms-26 and ms-47 of tomatoes and application
ActiveCN107881251AShorten the breeding cycleImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceIndel
The invention belongs to the fields of agricultural biotechnology engineering and genetic breeding of vegetables, and particularly relates to molecular markers associated with male sterile mutation sites ms-15, ms-26 and ms-47 of tomatoes, a specific primer and application. The molecular markers comprise the co-dominance molecular marker MS15C used for detecting the SNP in the site ms-15, and theco-dominance molecular marker MS26D used for detecting InDels in the sites ms-26 and ms-47. The molecular markers can be employed to replace the traditional method that by selecting male sterile plants in the flowering stage, it is ensured that a tomato breeding material contains the male sterile mutation sites ms-15 or ms-26 or ms-47. The molecular markers can assist in selecting homozygous plants and hybrid plants containing the mutation sites ms-15 or ms-26 or ms-47 in the seedling stage, so that the breeding period is shortened, and the breeding efficiency is improved.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
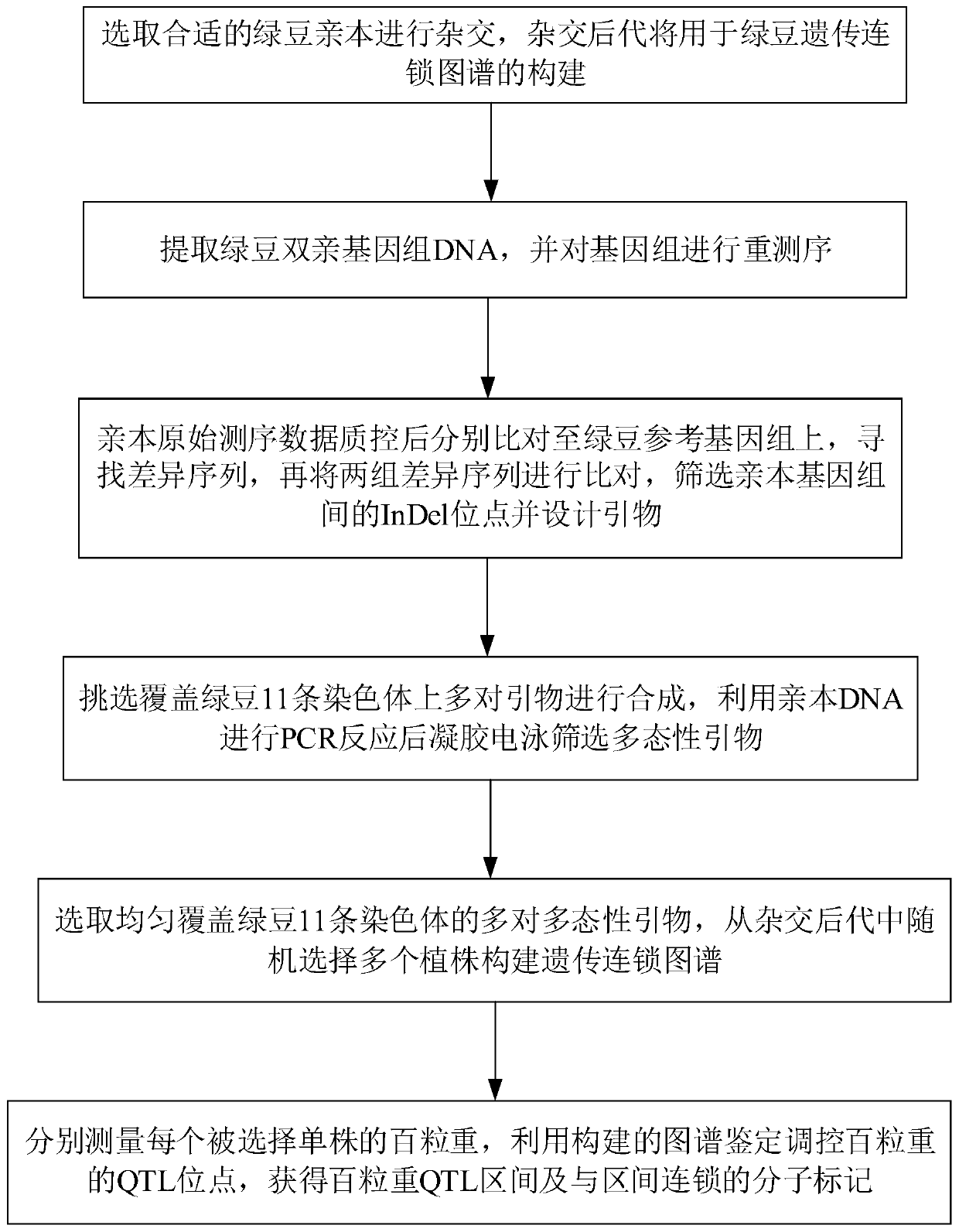
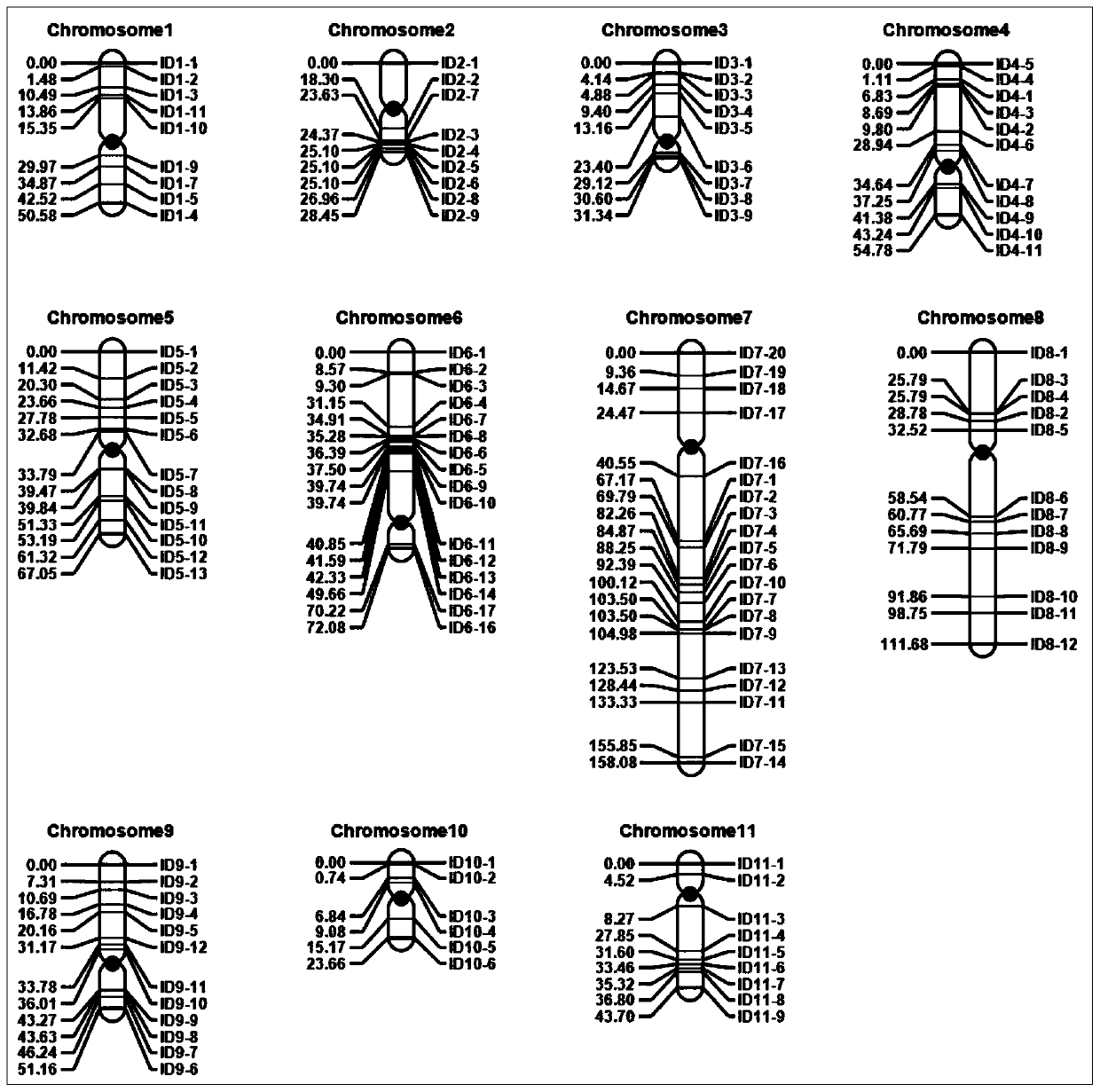
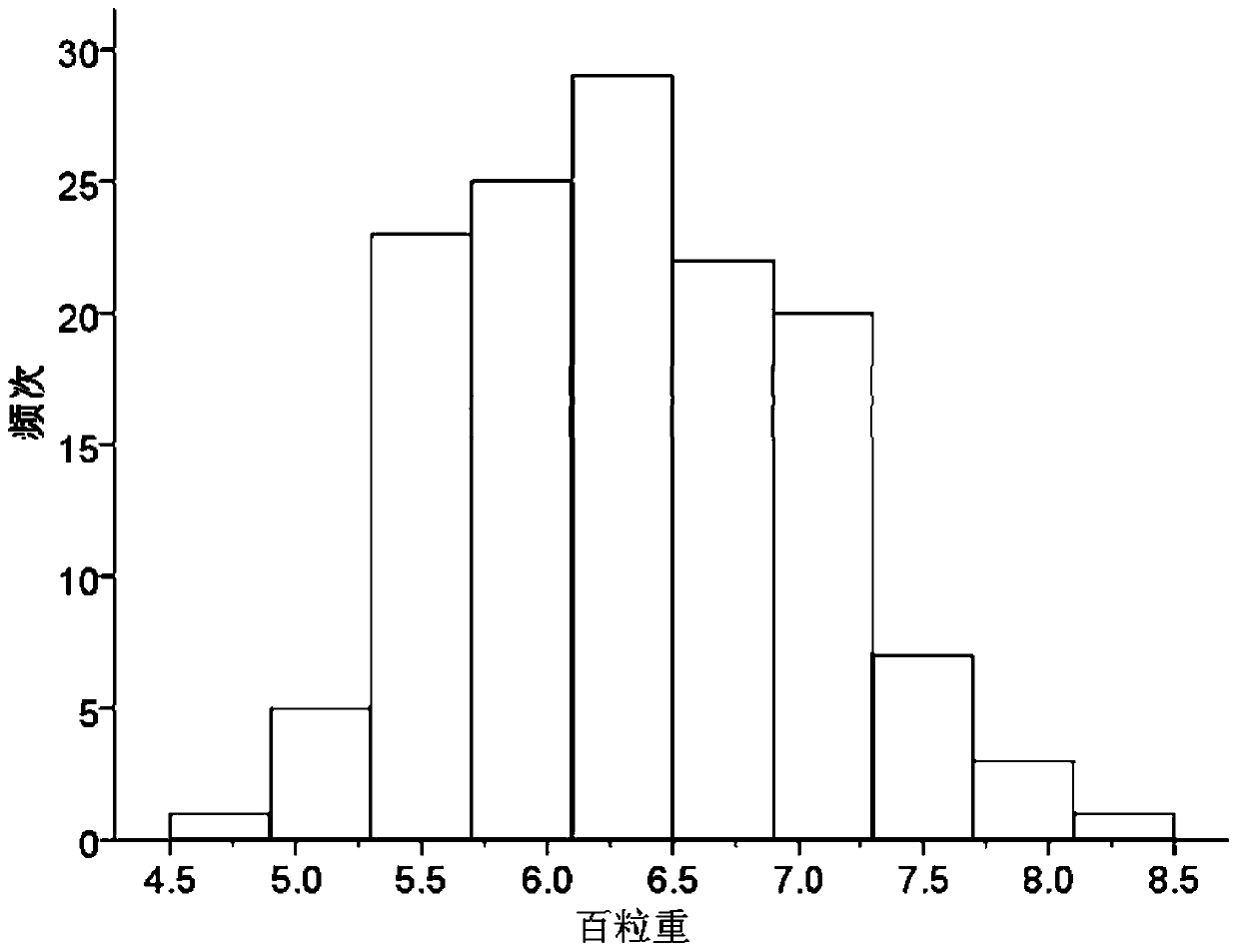
Set of mung bean InDel molecular markers and development method thereof
ActiveCN110656200AIncrease success rateRaise the ratioMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyPlantlet
The invention discloses a set of mung bean InDel molecular markers and a development method thereof. The development method comprises the steps: mung bean parents are selected to be hybridized so as to construct a mung bean genetic mapping population; parent genomes are re-sequenced; parental sequencing data are subjected to quality control and then compared to mung bean reference genomes to screen differential sequences, then InDel loci between the parental genomes are screened, finally primers are designed for the InDel loci, and the multiple pairs of primers are obtained; the polymorphic primers evenly covering 11 chromosomes of mung beans are selected, and a plurality of filial generation plants are selected at random to construct a mung bean genetic linkage map; and the hundred-grainweight per plant is measured, quantitative trait loci (QTL) for controlling and adjusting the hundred-grain weight are identified, and QTL intervals and the linked markers are obtained. The success rate and the polymorphic primer ratio of primer design are increased, polymorphism is better, and the genetic linkage map constructed through the InDel markers can be used for identification of the QTLof quantitative traits and is beneficial to important gene identification of the mung beans and assistant breeding of the molecular markers.
Owner:CROP INST ANHUI PROV ACAD OF AGRI SCI
Compound amplification detection kit containing forty InDel genetic polymorphic sites of human X chromosome
ActiveCN110578009ALow mutation rateHigh sensitivityMicrobiological testing/measurementInsertion deletionX chromosome
The invention discloses a compound amplification detection kit containing forty InDel genetic polymorphic sites of human X chromosome. The forty InDel sites of the X chromosome and an individual identification location point are amplified simultaneously by a primer group. Compared with a traditional STR genetic marker, an insertion deletion genetic marker used in the compound amplification detection kit has the advantages of low mutation rate and high sensitivity; and the polymorphism detection sites involved in the compound amplification detection kit combine the advantages of an InDel genetic marker and the special genetic characteristics of the X chromosome, and effective supplementary means can be provided for identification of difficult or special cases.
Owner:GUANGDONG HUAMEI ZHONGYUAN BIOLOGICAL SCI & TECH +2
Systems and methods for detecting genetic alterations
ActiveUS20190071732A1High sensitivityStrong specificityNucleotide librariesMicrobiological testing/measurementGene splicingAndrogen Receptor Gene
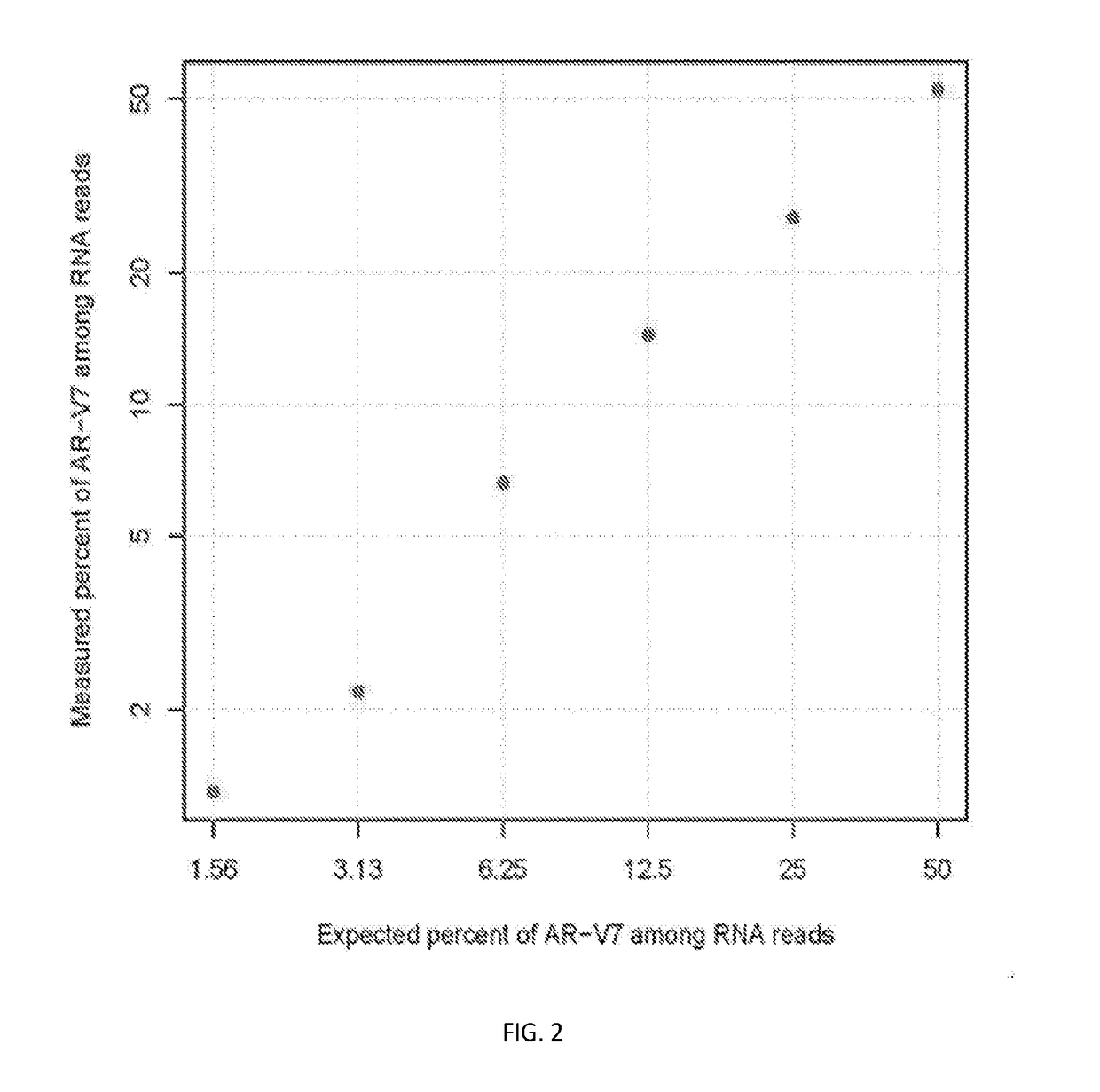
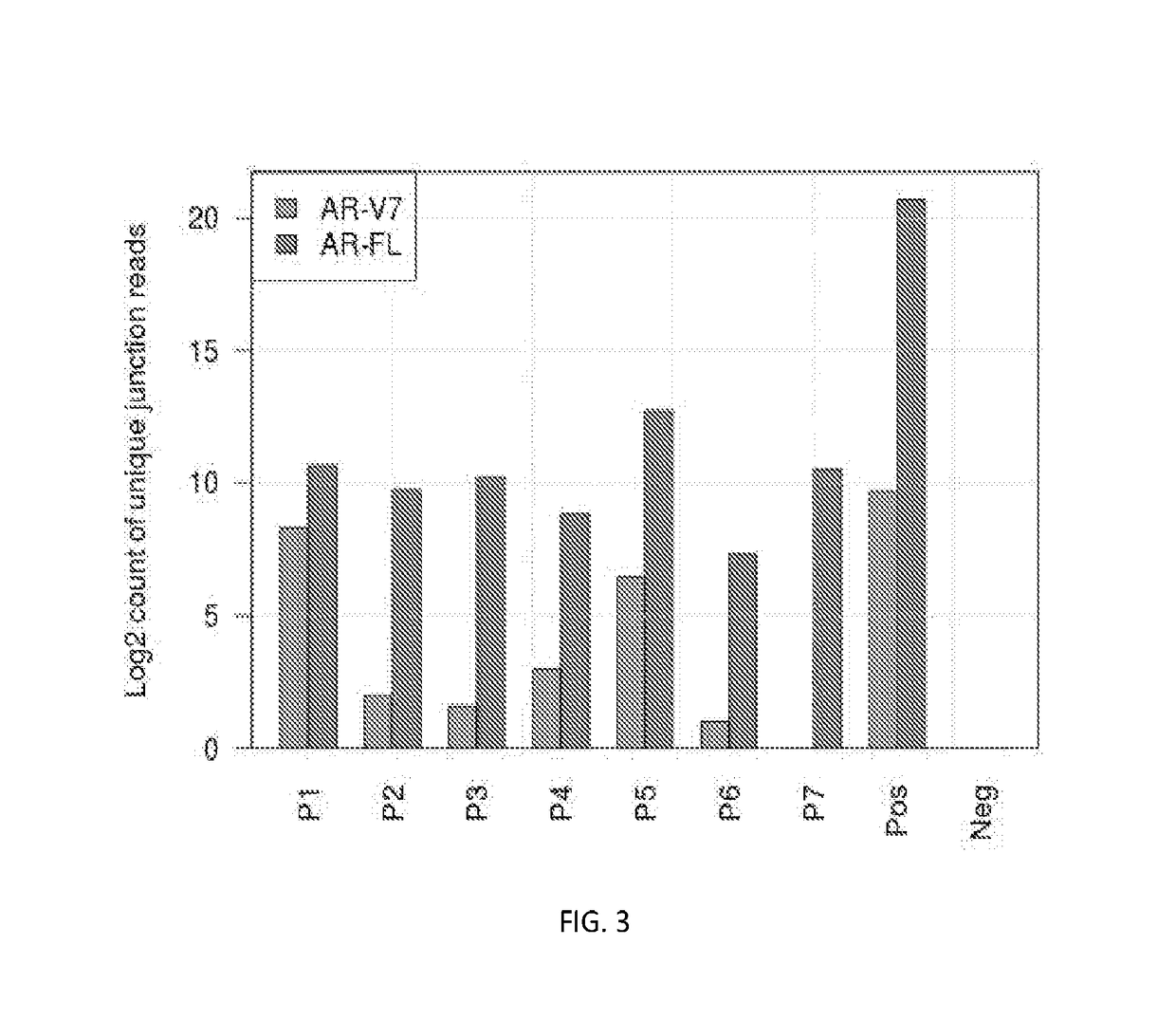
Disclosed are systems and methods for detecting genetic alterations comprising androgen receptor gene splice variants (AR-Vs), mutations, indel, copy number changes, fusion and combination thereof, in a biofluid sample from the patient. The systems and methods are similarly applicable to the detection of gene alterations comprising gene splicing variants, mutations, indel, copy number changes, fusion and combination thereof of other genes of interest. The streamlined methods improve the consistency and simplicity of non-invasive detections of biomarkers.
Owner:PREDICINE INC
Fluorescently-labeled X-InDel locus composite amplification system and application thereof
ActiveCN104131067AHigh sensitivityHigh individual recognition rateMicrobiological testing/measurementFluorescence/phosphorescenceInsertion deletionDNA paternity testing
The invention provides a fluorescently-labeled X-InDel locus composite amplification system. By means of the system, eighteen insertion-deletion genetic polymorphism loci on an X chromosome can be compositely amplified, wherein the eighteen loci are respectively labeled with three fluoresceins FAM, HEX and TAMRA. The composite amplification system is high in sensitivity and individual identification rate, is high in polymorphism, is good in stability and repeatability, is accurate in typing results and can satisfy a practical requirement. A kit can be manufactured on the basis of the composite amplification system, can be used for paternity test, dyad paternity test, grandparent and grandchild test, sibling test and individual identification. New technologies can be provided for the fields such as anthropology, medical genetics and the like.
Owner:ACADEMY OF FORENSIC SCIENCE
Lymphoma gene capture chip and applications thereof
PendingCN109385666AAchieve early screeningRealize Auxiliary DiagnosisNucleotide librariesMicrobiological testing/measurementNucleotideSignalling pathways
The invention proposes a lymphoma gene capture chip, which comprises: a substrate and a probe, wherein the probe is arranged on the substrate, and specifically recognizes at least one of genes represented by table 1. According to the present invention, the chip comprises the related Driver Genes, the high frequency mutation genes, the important genes in five cancer-related signaling pathways, thetargeted drug-sensitive and chemotherapeutic drug-sensitive genes and the drug-resistant related genes in various subtypes of common highly-incident lymphoma, can be used for the detection of the single-nucleotide site variation (SNV), the Indel insertion or deletion (Indel), the gene copy number variation (CNV) and the chromosome structural variation (SV) of lymphoma-associated genes, can achievethe detection of various subtypes of lymphoma, and can be effectively used for the early screening, the assisted diagnosis, the medication guidance and the recurrence monitoring of lymphoma.
Owner:BGI GENOMICS CO LTD +2
Molecular marker of major QTL, qTLA-9 for regulating angle of tilt of paddy rice leaves, and application thereof
InactiveCN109777886AHigh LOD valueHigh spectral densityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceIndel
The invention discloses a major QTL, qTLA-9, which is used for regulating the angle of tilt of paddy rice leaves, and locates between the mark Indel Tal-1 and on Indel Tal-2 on the ninth chromosome, and the genetic distance is 74.80 cm-98.33 cm, the physical distance is 17448080 bp-2293 8953 bp. 2293 8953 bp. The invention also provides a mark Indel of the major QTL, qTLA-9, which is used for regulating the angle of tilt of paddy rice leaves; the invention also provides the application of the major QTL: by developing the molecular marker closely linked to the major QTL, the test result that whether there are leaf inclination-related QTLs in rice varieties or strains can be attained, thereby speeding up the breeding process of excellent rice varieties.
Owner:ZHEJIANG NORMAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com