Molecular marking method of peanut plant type-related gene loci and application thereof
A gene locus and molecular marker technology, applied in the field of molecular genetics and molecular breeding, to achieve the effect of cost saving, quick and easy identification method and high selection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
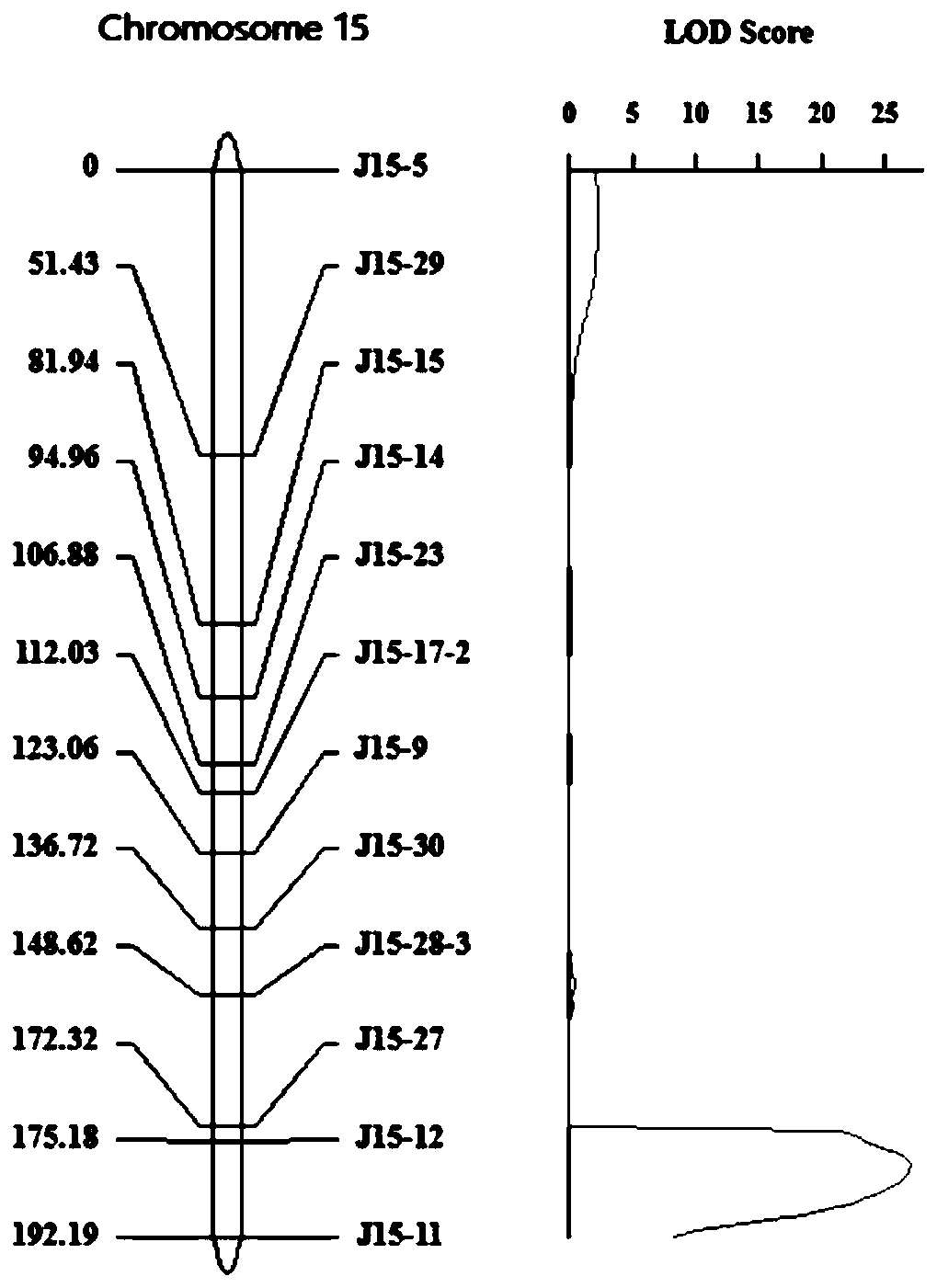
[0034] Example 1: Genetic Mapping of Peanut Side Branch Angle, Growth Habit and Plant Type-Related Gene Loci
[0035] (1) Materials and methods:
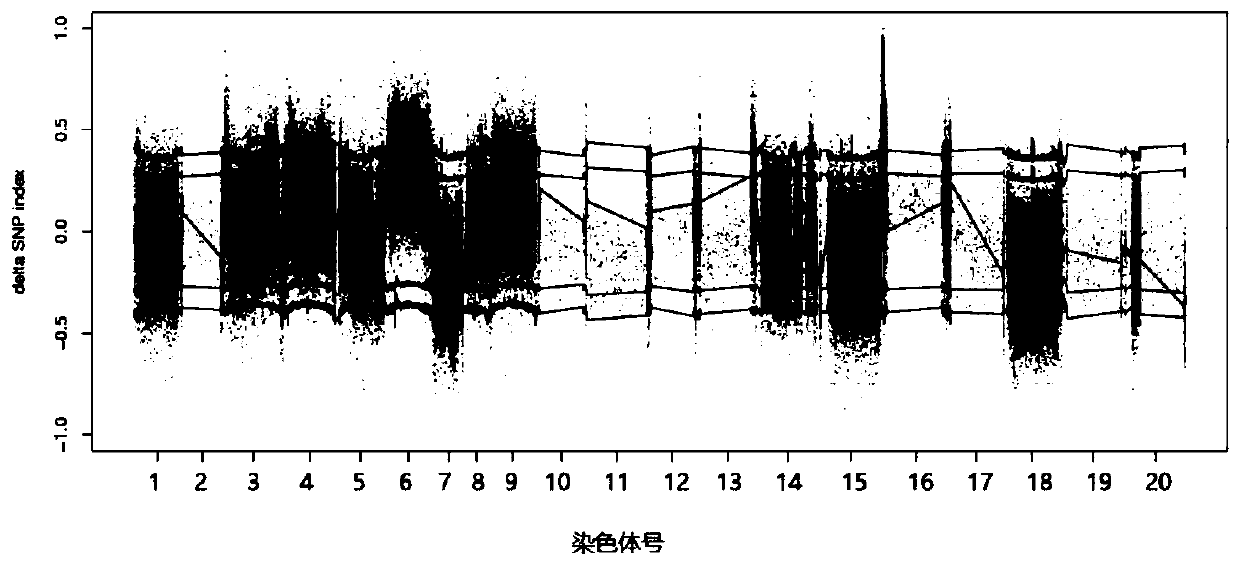
[0036] (1) Construction of plant materials and genetic populations: the creeping cultivated peanut Xiaohongmao was crossed with the erect cultivated peanut Henan Nanyang peanut, the hybrids were planted, and F 2 Separation of populations, continuous self-breeding of multiple generations of strains, and finally the formation of F 6 Recombinant inbred line HN-F 6 . It is used for the detection and location of the angle-related gene loci between the main effect and side branches.
[0037] (2) Extract individual DNA by CTAB method (refer to "Molecular Cloning Experiment Guide" for details).
[0038] (3) Development of polymorphic InDel markers: According to the comparison between the resequencing data of the parental genome and the reference genome (Tifrunner genome data: https: / / www.peanutbase.org / peanut_genome), 31 pairs of InDel ...
Embodiment 2
[0049] (1) Development and application of co-segregation marker LBA5b-caps for lateral branch angle, growth habit and plant type related gene loci
[0050] According to the result of embodiment 1, the difference of TA insertion between parents on the candidate gene LBA5b is developed as a CAPS marker, which is named as LBA5b-caps (see Figure 4 ). Amplify HN-F using the upstream and downstream primers of CAPS-labeled LBA5b-caps 6 The genomic DNA of each individual in the population is digested by restriction endonuclease SacⅠ, and then detected by electrophoresis on 1% agarose gel electrophoresis. If the PCR product is cut into two small fragments, it means that the individual has an LBA5b potentiation allele If the PCR product cannot be cut at the mutated gene locus (from Xiaohongmao), it means that the individual does not have the LBA5b synergistic variant gene, but has a depleted allelic variant lba5b (from Nanyang peanut). Allelic variation analysis and phenotypic analysi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com