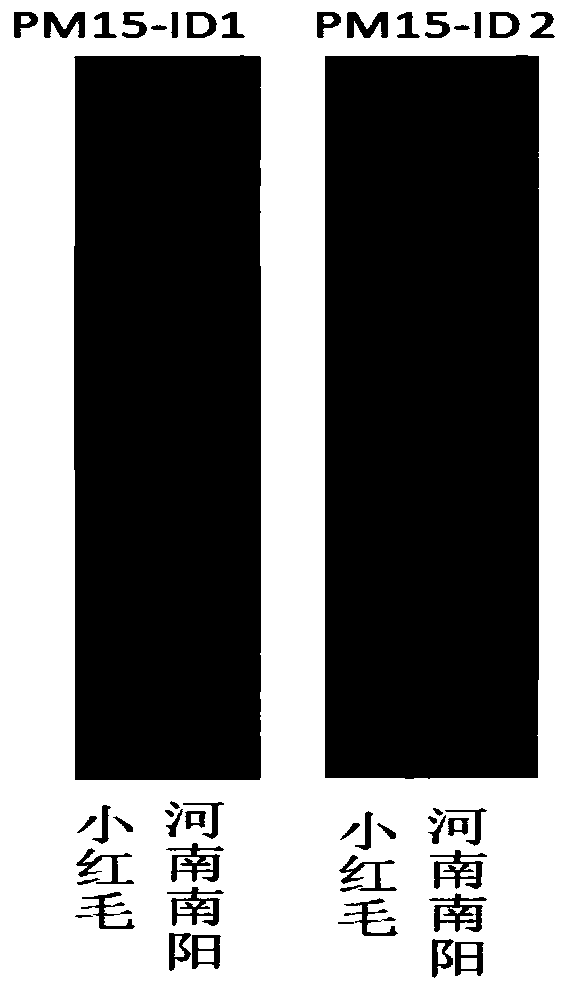Patents
Literature
109results about How to "Identification method is simple" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
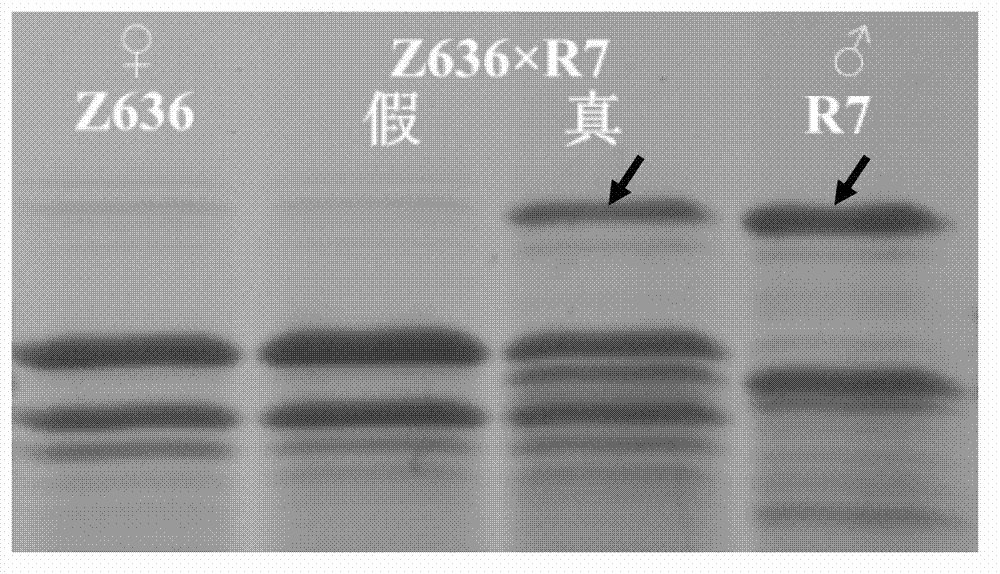
Real-time fluorescence quantification PCR detection primer for quickly identifying genotypes of duck circovirus and detection method thereof
PendingCN106065419AIdentification method is simpleImprove shortcutMicrobiological testing/measurementMicroorganism based processesFluorescencePcr method
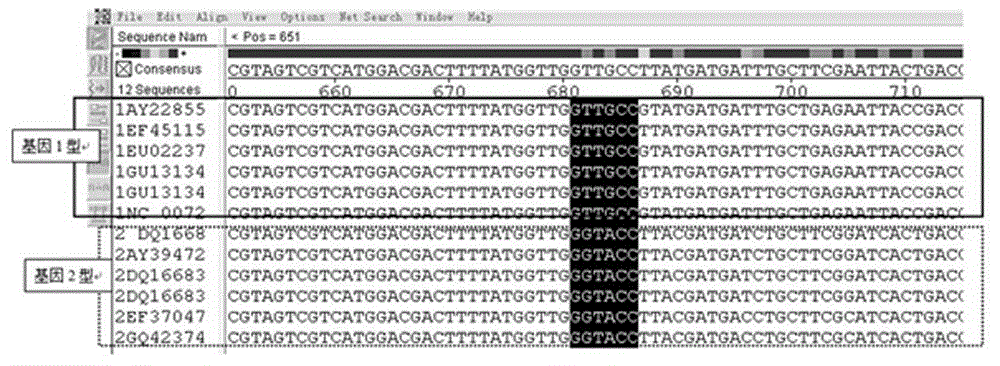
The invention relates to a real-time fluorescence quantification PCR detection primer for quickly identifying genotypes of duck circovirus and a detection method thereof. The sequences of the primer are described as follows: an upstream primer P1: 5'-CAGATCCCCGGGCACGAGA-3'; a downstream primer P2: 5'-CCTCACCTTCAGGGATTC-3'. The detection method includes the steps of: establishing the real-time fluorescence quantification PCR method based on SYBR Green I with the primer, wherein difference of temperature of a melting curve exists due to the difference of GC content of a nucleotide in a specific gene fragment region of gene 1-type duck circovirus and gene 2-type duck circovirus which are amplified by the primer, so that infection of the gene 1-type duck circovirus and the gene 2-type duck circovirus can be directly carried out according to the difference of Tm value in the melting curve. The method can distinguish the infection caused by the duck circovirus in different genotypes just with one group of primer, thereby providing technical fundament for healthy breeding in duck breeding industry.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
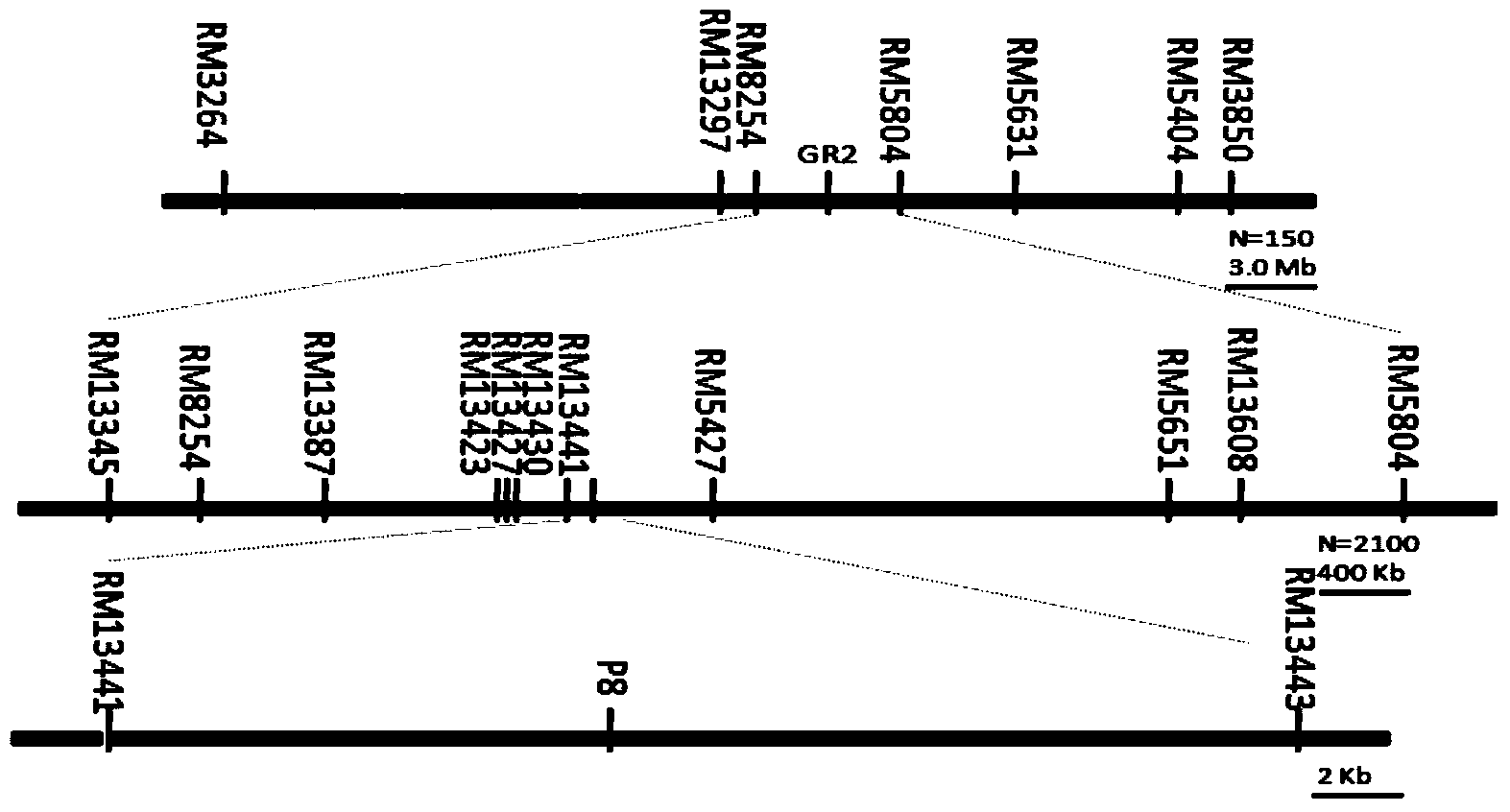
Molecular marker of rice seed salt tolerant germination major QTL qGR2 and its application
ActiveCN103642801AQuick filterClear locationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceElectrophoresis
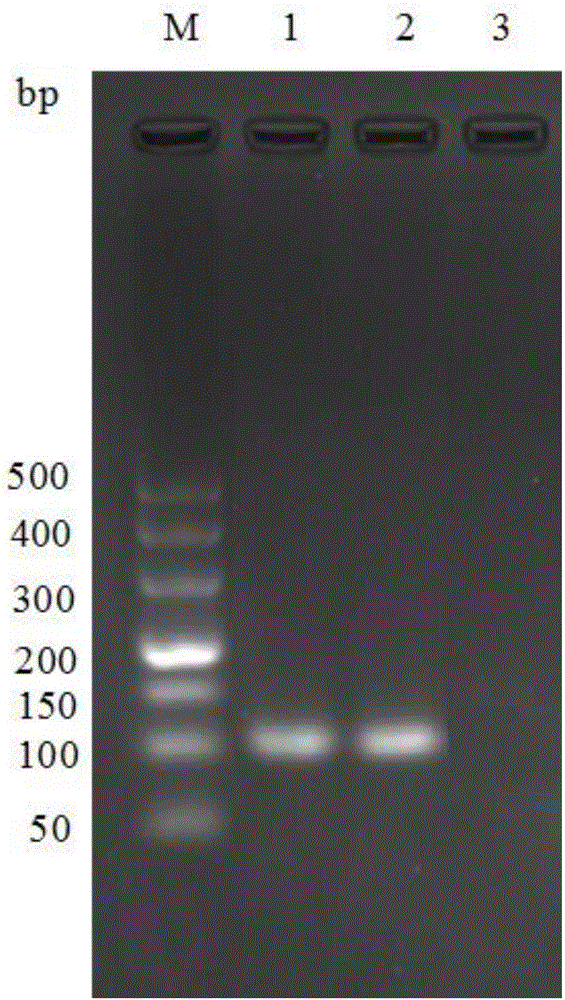
Belonging to the fields seed science and technology application, the invention relates to a molecular marker of a rice seed salt tolerant germination major QTL qGR2 and its application. One or more pairs of rice genome DNA in SSR marked primers are employed to conduct PCR amplification, and the PCR amplification product is subjected to electrophoresis detection on 8% non-denaturing polyacrylamide gel. If a DNA fragment with a corresponding size is amplified, major QTL qGR2 synergetic allele for controlling rice seed salt tolerant germination can exist. By means of a molecular marker method in coseparation and close linkage with the seed salt tolerant germination gene qGR2, the salt tolerance level during rice germination can be predicted, and the salt rice variety in a rice germination period can be screened out rapidly.
Owner:NANJING AGRICULTURAL UNIVERSITY
High-efficiency identification method for salt resistance of cotton germplasms
ActiveCN103364383AStrong salt toleranceThe result is accurate and reliableFluorescence/phosphorescenceSalt resistanceAgricultural science
The invention provides a high-efficiency identification method for salt resistance of cotton germplasms. By utilizing a flower pot soil seedling cultivation method, cotton seedings are cultivated to a three-leaf-and-one-leaflet stage. The materials with normal and consistent growth vigour are selected and subjected to salt stress treatment and clear water treatment with the same volume respectively. The second true leaves are selected, the PSII maximum photochemical efficiencies of the true leaves with different treatments are measured respectively, the relative values are calculated, and brought into the standard equation of salt resistant indexes: D=1.943X-0.882 to calculate the salt resistant indexes and determine the salt resistance, wherein X is for a relative value of a PSII maximum photochemical efficiency and D is for a salt resistant index. The larger the salt resistant index, the stronger the salt resistance. When the method is compared to the prior art, the results of the obtained standard equation of the method are accurate and credible. The measurement method of PSII maximum photochemical efficiencies is simple and fast, can be performed in vivo, does not harm plants, and can ensure normal running of downstream experiments. The measurement time is the cotton seedling stage, so the identification time can be shortened effectively, the efficiency is raised, and the method is suitable for large scale germplasm resource identification.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Primer and probe for detecting echinococcus granulosus in canine feces, and kit of primer and probe
InactiveCN107365862AQuick checkIncreased sensitivityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFeces
The invention provides a primer for detecting echinococcus granulosus in canine feces, wherein the sequence of an upstream primer is as shown in SEQ ID No. 1; and the sequence of a downstream primer is as shown in SEQ ID No. 2. The invention also provides a probe for detecting the echinococcus granulosus in the canine feces, wherein the sequence is as shown in SEQ ID No. 3. The invention further provides a kit for detecting the echinococcus granulosus in the canine feces; the kit comprises the primer for detecting the echinococcus granulosus in the canine feces and the probe for detecting the echinococcus granulosus in the canine feces, wherein the sequence of the upstream primer is as shown in SEQ ID No. 1; the sequence of the downstream primer is as shown in SEQ ID No. 2; and the sequence of the probe is as shown in SEQ ID No. 3. In the invention, by using the primer, the probe and the kit, a target gene DNA can be rapidly and specifically detected with high sensitivity; even when a specimen contains the trace amount of echinococcus DNA, the target gene DNA can be detected.
Owner:STATION OF VIRUS PREVENTION & CONTROL CHINA DISEASES PREVENTION & CONTROL CENT
PCR-RFLP primer and method for rapidly identifying genotypes of duck circoviruses
InactiveCN105177186AIdentification method is simpleMicrobiological testing/measurementMicroorganism based processesRestriction Enzyme Cut SiteDistinctin
The invention discloses a PCR-RFLP primer and method for rapidly identifying the genotypes of duck circoviruses. The primer comprises an upstream primer body P1:5'-TGCGCCAAAGAGTCGACATAC-3' and a downstream primer body P2:5'-CAAATTGGTCTCAGTAGTTTATT-3'. According to the method, distinguishing is carried out through the difference of restriction enzyme cutting sites contained in coding regions of copied protein genes of the duck circoviruses in different genotypes (the first genotype and the second genotype). The method includes the steps that DNA is extracted, Kpn I restriction enzyme cutting is carried out after PCR amplification is carried out to obtain corresponding target fragments, and then RFLP analysis is carried out. The identification method is rapid and easy to operate and high in accuracy.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
Molecular marking method of rice stigma exsertion major QTL sites
InactiveCN104178560AAccurate locationFine locationMicrobiological testing/measurementAgricultural scienceElectrophoresis
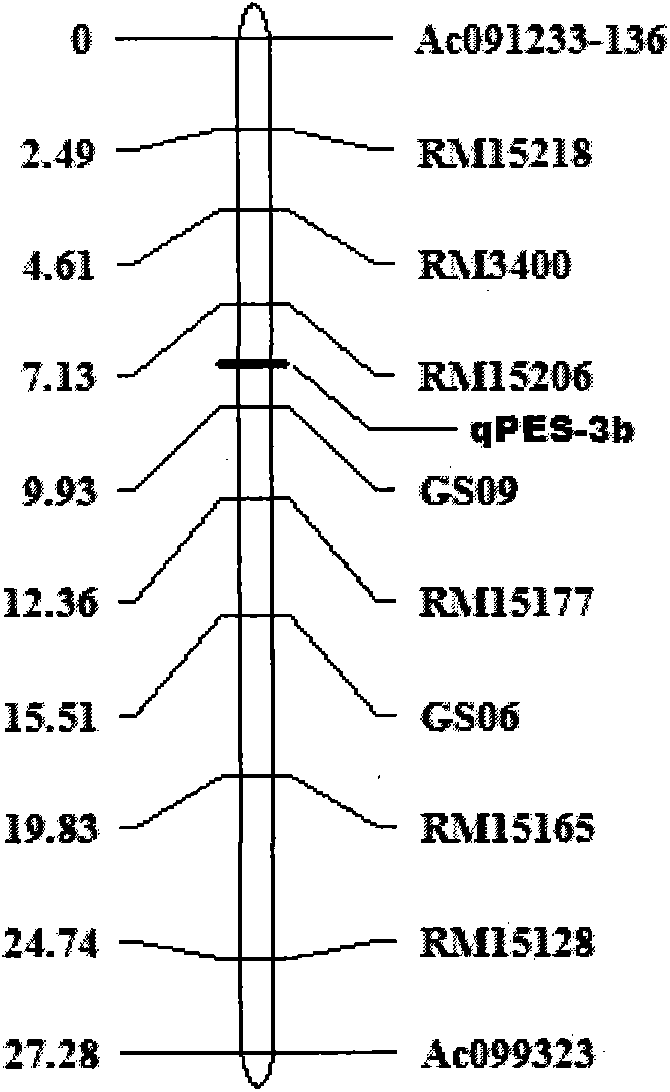
The invention belongs to the field of molecular genetics and breeding, and discloses a molecular marking method of rice stigma exsertion major QTL sites. Development of a new molecular marker and fine mapping of rice stigma exsertion rate QTL are carried out for a target zone of third chromosome, PCR amplification of rice genome DNA is carried out by using one or two pairs of primers of Indel marked primer GS09 and SSR marked primer RM15206, and the above obtained PCR amplification products are detected through electrophoresis on 8% native polyacrylamide gel. A qPES-3b synergistic site exists if a corresponding size of a fragment is amplified. The rice high stigma exsertion germplasm and breeding populations constructed by all parents can be detected through tightly linked molecular markers controlling the stigma exsertion major QTL sites, so the major QLT controlling the stigma exsertion can be rapidly introduced, the selection efficiency of major sites is improved, and the improvement of hybrid rice sterile line stigma exsertion and the improvement of the seed yield are benefited.
Owner:国家粳稻工程技术研究中心 +1
Universal primers for identifying bacillus and classification method using same
ActiveCN102226216APolymorphism richThe classification result is accurateMicrobiological testing/measurementDNA/RNA fragmentationClassification methodsSubspecies
The invention discloses universal primers for identifying bacillus. The universal primers comprise a forward degenerate primer composed of a nucleotide sequence shown as SEQID NO:1 and a reverse degenerate primer composed of a nucleotide sequence shown as SEQID NO:2. The invention also discloses a method for classifying and identifying bacillus, which comprises the following steps: utilizing the universal primers to perform PCR (Polymerase Chain Reaction) amplification; sequencing the PCR amplification product; utilizing sequence comparison software to perform sequence comparison in a Genbank data base; and acquiring the variety of a known strain having the highest sequence similarity, namely the variety of the strain. All varieties of bacillus can be classified by the universal primers, and the classification result is accurate and reliable; and the universal primers can be used for classifying bacillus on a variety level and can be also used for classifying bacillus on a subvariety level. Besides, the classification and identification method is simple and has a high identification speed.
Owner:南京科绿丰科技有限公司
Identification method for clubroot disease resistance of high mountain radishes in field
InactiveCN106508336AAccurate Disease IndexIdentification method is simpleCultivating equipmentsPlant cultivationDiseaseBlack spot
The invention discloses an identification method for clubroot disease resistance of high mountain radishes in a field. The identification method comprises the following steps: selecting experimental radish varieties, enriching bacterium sources of a clubroot disease nursery in a high mountain region, and sowing radishes; grading clubroot disease morbidity degree of full-grown radishes to obtain disease condition grades of the radishes; calculating radish variety disease condition indexes according to the disease condition grades of the radishes and a radish variety disease condition index general formula; and evaluating the resistance type of each radish variety to the clubroot disease by combining a radish disease resistance evaluation standard according to a calculation result. According to the identification method, the disease condition grading of the radishes which are infected with a disease is simplified; the influence on radish clubroot disease condition grading caused by black spots and black scars on the radishes is excluded; the existing grading mode is changed essentially; the overall identification process and the result processing analysis become simple, and meanwhile, the clubroot disease resistance of different radish varieties is judged rigorously and accurately.
Owner:INST OF ECONOMIC CROP HUBEI ACADEMY OF AGRI SCI
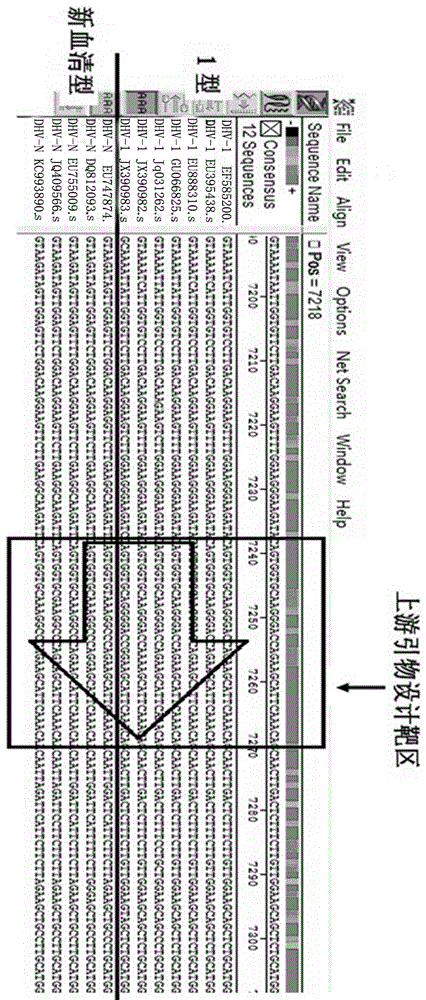
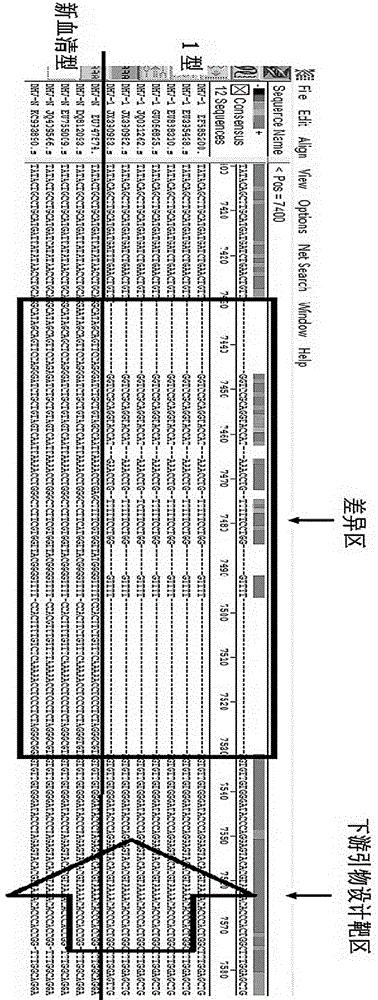
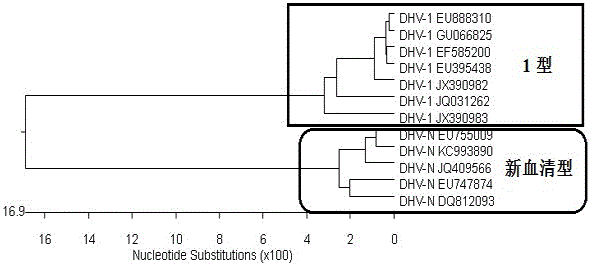
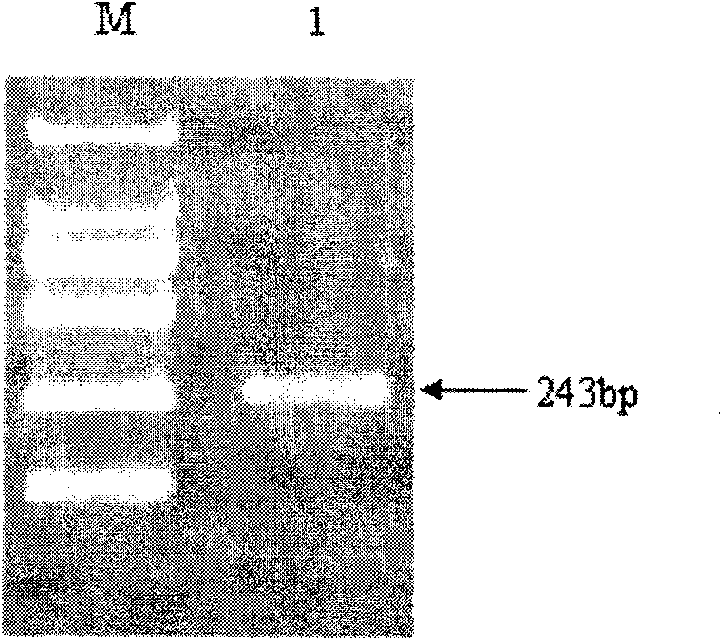
RT-PCR discriminating diagnosis primers for type 1 and new serotype duck hepatitis virus
InactiveCN106011315AIdentification method is simpleFast and Accurate Differential Diagnostic TestingMicrobiological testing/measurementMicroorganism based processesDuck hepatitis A virusNucleotide
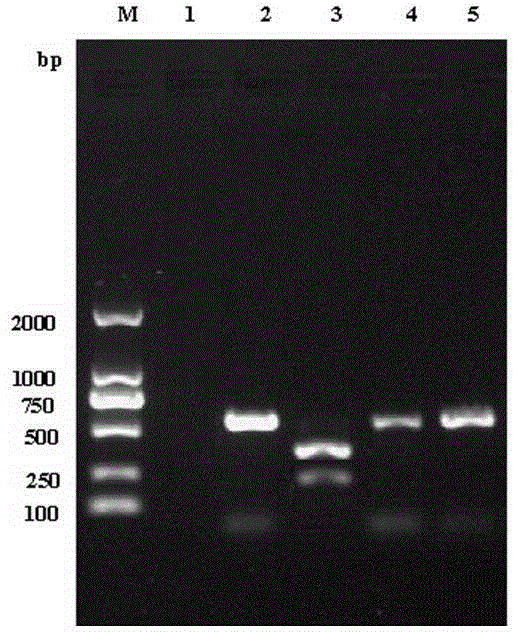
The invention discloses a group of RT-PCR discriminating diagnosis primers for discriminating type 1 and new serotype duck hepatitis virus. A primer sequence is shown as a SEQ ID NO. 1-2. The primer is designed according to existed difference of a 3'-terminal nucleotide sequence of a type 1 and new serotype duck hepatitis virus whole-genome sequence, The group of the primers performs discriminating diagnosis in a specific amplification area for amplifying the type 1 and new serotype duck hepatitis virus according to the length difference of the nucleotide sequences, wherein size of a type 1 duck hepatitis virus amplification fragment is about 258 bp, and the size of a new serotype duck hepatitis virus amplification fragment is about 321 bp. The identification method of the primer has the advantages of easy and rapid operation, and high accuracy, only one group of the primers can discriminate the type 1 and new serotype duck hepatitis virus types, and the primers provide technical guarantee for aquatic bird healthy culture.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
Kit and special primer for detecting H1N1 influenza A virus and target sequence
InactiveCN101591715AIdentification method is simpleShort detection timeMicrobiological testing/measurementMicroorganism based processesH1n1 virusElectrophoresis
The invention discloses a special primer for detecting H1N1 influenza A virus, a kit comprising the primer for detecting H1N1 influenza A virus and a target sequence corresponding to the primer, and belongs to the field of quick diagnosis of pathogen genes. The base sequence of the target sequence corresponding to the special primer is shown as SEQ ID No.1; and the kit utilizes the technical principles of genetic reverse transcription and isothermal amplification to quickly detect H1N1 influenza A virus nucleic acid, and the detection result can be judged by naked eyes and also can be analyzed by using electrophoresis. The kit has the characteristics of quickness, sensitivity, specificity and convenient operation.
Owner:INST OF BASIC MEDICAL SCI ACAD OF MILITARY MEDICAL SCI OF PLA
Molecular marking method of peanut plant type-related gene loci and application thereof
InactiveCN110592264AClear locationFine locationMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionMarker-assisted selection
The invention belongs to the field of molecular genetic breeding science, and provides a molecular marking method of gene loci related to peanut lateral branch angle, growth habit and plant types andapplication thereof. By designing a CAPS molecular marker, and by using a method combining primer amplification, enzyme digestion and agarose gel electrophoresis, the judgment of allelic variation ofgene loci related to the lateral branch angle, the growth habit and the plant types is realized. By labeling PM15-ID1 and PM15-ID2 with InDel, and by using the method of primer amplification and polyacrylamide gel electrophoresis, the marker-assisted selection of offspring with allelic variation of gene loci related to lateral branch angle, growth habit and plant types. The molecular marking method provided by the invention can be used for detecting the offspring of a breeding group constructed by using peanut creeping germplasm and upright germplasm as parents, can be used for rapidly identifying the allelic variation of gene loci related to lateral branch angle, growth habit and plant types of the offspring, can improve the selection efficiency of the gene loci, and can provide referencefor peanut plant type improvement and high-yield breeding.
Owner:QINGDAO AGRI UNIV
Molecular identification method for cotton restoring line containing sterile cytoplasm of gossypium harknessii
InactiveCN102618647AThe results are clearAccelerateMicrobiological testing/measurementDNA/RNA fragmentationCytoplasmic StructureMolecular level
The invention belongs to the field of biological technique, relates to a molecular identification method for the purity of a cotton restoring line, and particularly provides a primer of a SSR (Simple Sequence Repeat) marker for identifying the cotton restoring line containing the sterile cytoplasm of gossypium harknessii. The primer is as shown in nucleotide sequences such as SEQ ID No.1 and 2 and / or SEQ ID No. 3 and 4. The SSR marker is adopted for carrying out polymorphism detection of molecular level on the cotton restoring line material containing the sterile cytoplasm of the gossypium harknessii. The molecular identification method is fast in identification speed, high in accuracy and simple in operation and is very suitable for the indoor identification of the cotton restoring line and the assisted-selective breeding of molecular markers.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Application of cabbage type rape floral leaf mutant
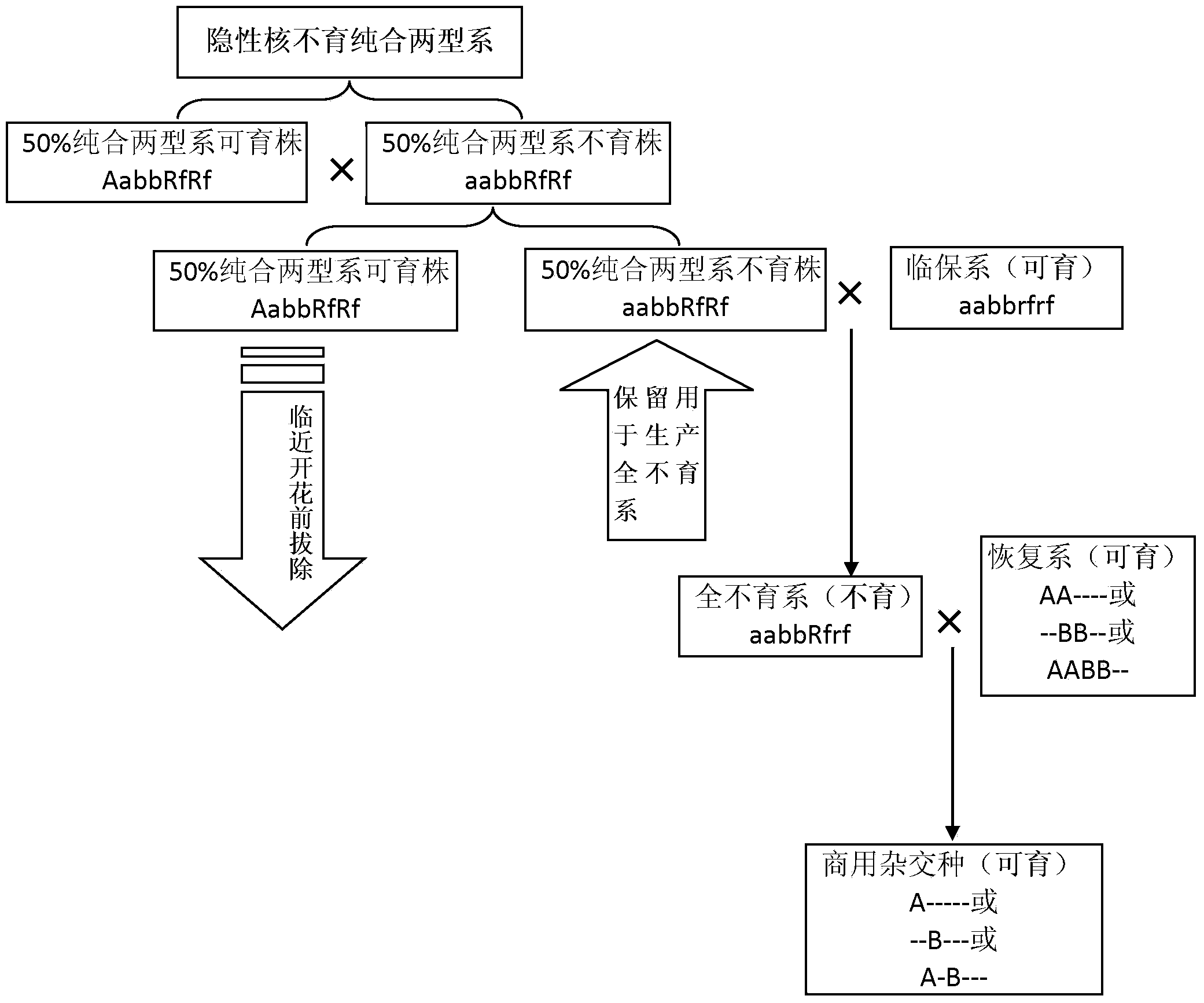
ActiveCN103931486AEfficient identificationEfficient removal of impuritiesPlant genotype modificationBiotechnologyHybrid seed
The invention relates to an application of a cabbage type rape floral leaf mutant. The cabbage type rape floral leaf mutant is applied by the following steps: hybridizing a sterile plant in a cabbage type rape recessive genic male sterile homozygous two-type line 20118AB serving as a female parent with a cabbage type rape floral leaf mutant serving as a male parent, bagging and performing inbreeding to obtain a floral leaf sterile plant; hybridizing the floral leaf sterile plant with a fertile plant in 20118AB to breed a new homozygous two-type line, namely, a floral leaf recessive genic male sterile homozygous two-type line; hybridizing the floral leaf sterile plant with a temporary maintaining line to obtain a new temporary maintaining line, namely, a floral leaf temporary maintaining line; and hybridizing the cabbage type rape floral leaf recessive genic male sterile homozygous two-type line with the floral leaf temporary maintaining line to produce a cabbage type rape floral leaf complete sterile line. Purity identification and impurity removal at the seedling stage of the complete sterile line are realized by using the character of a floral leaf, the purity of the complete sterile line is increased, the genic male sterile seed production procedure is simplified, and the labor amount of field hybrid seed production is reduced.
Owner:SHANGHAI ACAD OF AGRI SCI +1
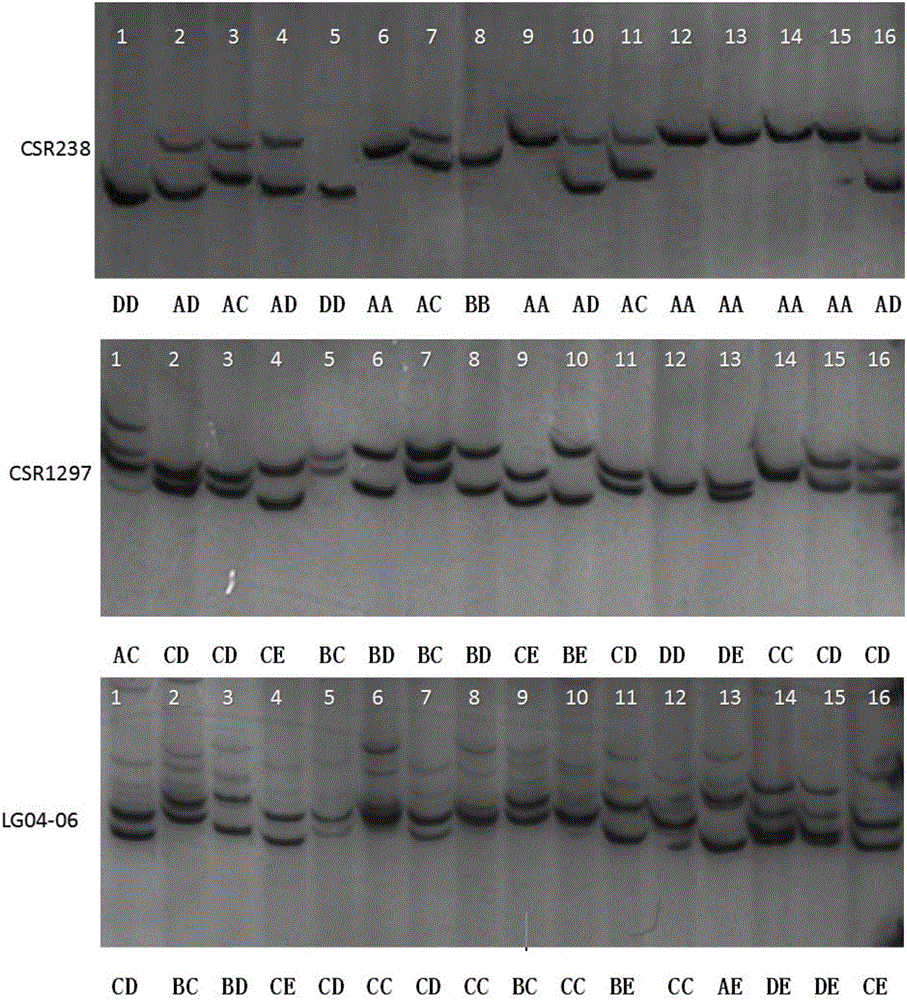
Molecular marker combination for rapidly identifying different albino tea tree varieties, method and application
ActiveCN106480224ASolve mixed problemsEasy to confirmMicrobiological testing/measurementDNA/RNA fragmentationTea leafMolecular marker
The invention discloses a molecular marker combination for rapidly identifying different albino tea tree varieties. The molecular marker combination adopts CSR238, CSR 1297 and LG04-06 primer combinations; the invention further discloses a method for rapidly identifying different albino tea tree varieties by using the molecular marker combination and a kit containing the molecular marker. The molecular marker combination can distinguish and identify the albino tea tree varieties on the market, and the identifying method is simple and fast, and can solve the problem of confused names of albino tea tree varieties on the market for a long time; the invention further discloses a finger-print of the albino tea tree varieties; tea selling enterprises can apply the two-dimensional code albino tea tree varieties generated by the invention to mark the product formed by processing different albino tea tree varieties, thus consumers can more conveniently learn about the product information.
Owner:TEA RES INST CHINESE ACAD OF AGRI SCI
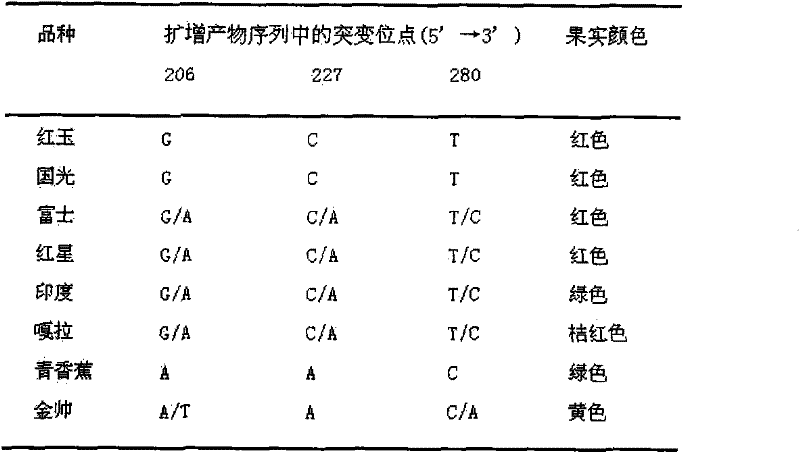
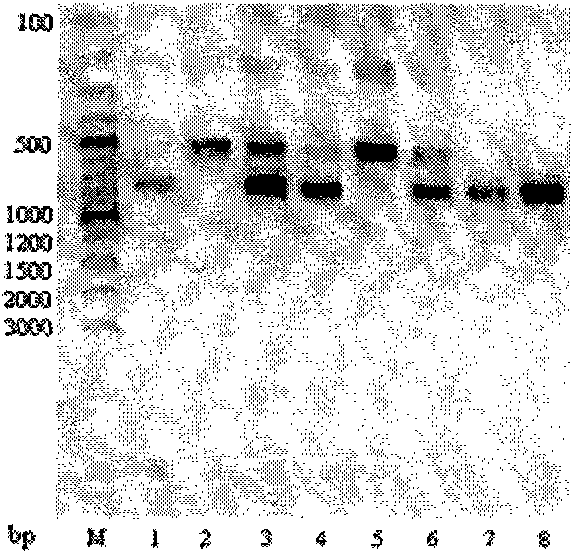
Two color-related mutations in apple MdMYB1 gene and detection method thereof
InactiveCN102242186AIdentification method is simpleShorten electrophoresis timeMicrobiological testing/measurementSequence analysisAgarose electrophoresis
Two color-related mutations in apple MdMYB1 gene and a detection method thereof. The invention belongs to the field of biotechnology and provides a low-cost molecular breeding means for the improvement of apple color traits. A pair of primers is designed by the use of apple color related gene MdMYB1, and apple genome DNA is taken as a template to carry out PCR amplification so as to obtain a PCR amplification segment. Sequence analysis is carried out on PCR amplification segments of 8 apple varieties to find out two gene mutations related with the apple color traits, wherein one gene mutationis Pm1 Iendonuclease site. The enzyme site is used to develop a CAPS marker Mb2P for detecting the mutation. The substantial progress lies in that common agarose gel electrophoresis can be used for the detection of the developed marker so as to greatly reduce experimental cost, in comparison with expensive NuSieve GTG agarose gel electrophoresis which is needed for the detection of the dCAPS marker in the prior art. By the use of the CAPS marker or the sequencing method to detect mutations, apple varieties and the color traits of hybrid posterity fruits can be identified. The color traits of hybrid posterity apples are selected at an early stage to abandon parts of plants, so as to save the management and screening cost of progeny plants.
Owner:SHANDONG INST OF POMOLOGY
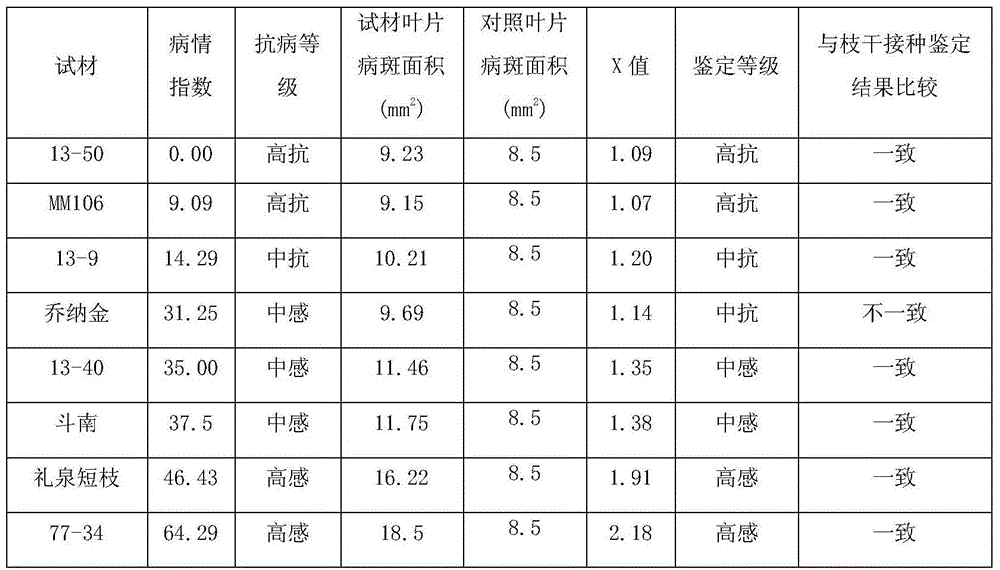
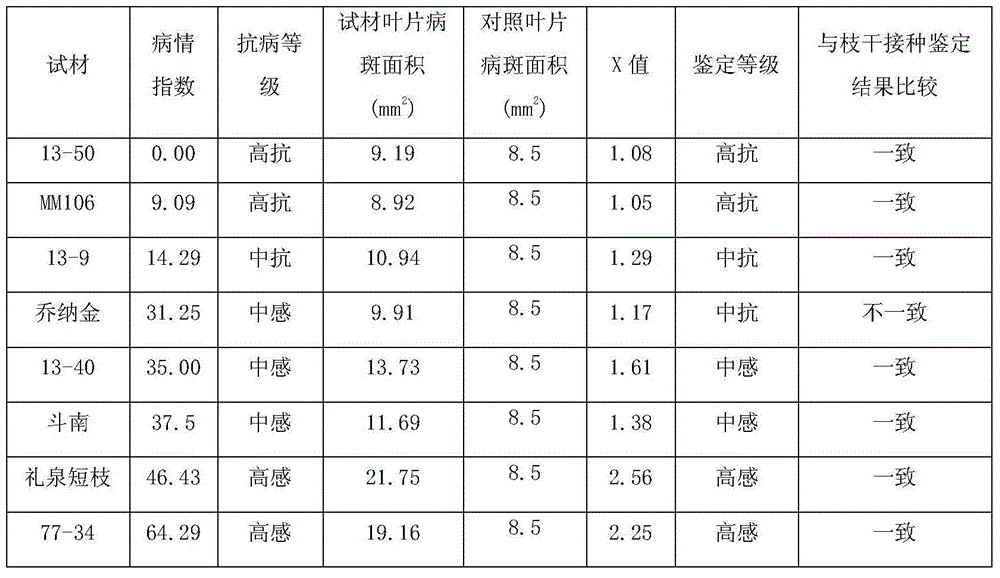
Method for rapidly identifying resistance of apple branches and trunks to physalospora piricola nose
The invention relates to a physalospora piricola nose resistance identification method, in particular to a method for rapidly identifying resistance of apple branches and trunks to the physalospora piricola nose. The physalospora piricola nose is inoculated to a PDA culture medium to be cultured so that a physalospora piricola nose culture medium bacterial colony can be obtained; pseudo identified varieties or rootstock young sprout leaves without plant diseases or insect pests at the top end, the front faces of the leaves are stabbed by an inoculating needle, and then the physalospora piricola nose is inoculated; after the bacteria-bearing leaves are cultured for six days, the diameters of disease spots of the leaves are measured according to a crossing method, the disease spot area of the leaves are worked out and compared with the disease spot areas of known resistance grade materials, and the grade of resistance of varieties to be identified or rootstocks to the branch and trunk physalospora piricola nose is determined. According to the method for rapidly identifying the resistance of apple branches and trunks to the physalospora piricola nose, leaves are inoculated, infection conditions suitable for the physalospora piricola nose are created, physalospora piricola nose resistance identification is performed on the infection result, few identification materials are needed, identification time is short, repeatability is high, and the identification method is simple, easy to master and popularize, and low in cost.
Owner:HEBEI AGRICULTURAL UNIV.
Composition based on stem cells and application thereof in preparing preparation for coronary heart disease
InactiveCN104928235AImprove systolic functionPromote diastolic functionDead animal preservationMammal material medical ingredientsCoronary heart diseaseBiochemistry
The invention discloses a composition based on stem cells and application thereof in preparing preparation for the coronary heart disease. The composition based on the stem cells includes the stem cells prepared through an optimizing method and cell preserving liquid serving as a solvent. Firstly, proper stem cells are selected; secondly, the selected stem cells are further processed through the optimizing method; lastly, the stem cells are collected and resuspended in the cell preserving liquid. A various cell implanting method is adopted in the application of the composition based on the stem cells in preparing the preparation for the coronary heart disease, the time nodes before an operation, during the operation and after the operation are combined, and it is ensured that the transplanted stem cells can give the full play. The optimizing method is used for preparing the stem cells and the stem cell composition in the cell preserving liquid, the ventricle function can be extremely obviously enhanced after cell transplantation, and the left ventricle reconstitution is restrained. The optimal cell composition is provided for treating the coronary heart disease, the source is wide, preparation is simple, toxic and side effects are avoided, and the transplanting effect is good.
Owner:奥思达干细胞有限公司
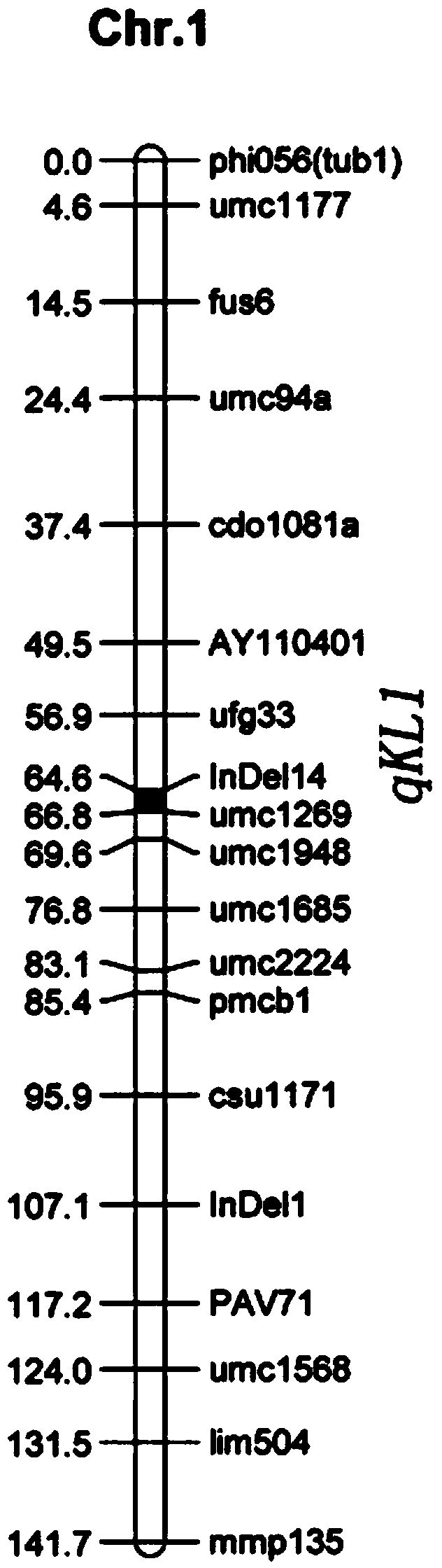
Molecular marker for regulating main effect QTL of grain length of corn and application of molecular marker
ActiveCN104212801AIdentification method is simpleImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationMolecular breedingAgricultural science
The invention relates to the field of molecular breeding of plants and particularly relates to a molecular marker for regulating main effect QTL of grain length of corn and an application of the molecular marker. The molecular marker for regulating the main effect QTL of grain length of corn consists of two pairs of primers InDel14 and umc1269. A method for assisted selection of long grain corn comprises the following steps: extracting genome DNA of corn to be detected; carrying out PCR amplification on the two pairs of primers InDel14 and umc1269; and if amplification products with the lengths of 236bp and 350bp are obtained, determining the corn to be detected is candidate long grain corn, and applying the candidate long grain corn to breeding. By carrying out assisted selection of long grain corn by means of the molecular marker provided by the invention, the length of the grain can be predicted just by detecting the characteristic amplification strip of the molecular marker, so that the molecular marker is simple in identification method and high in selecting efficiency.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Method for identifying seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin
InactiveCN103293213AHigh priceIdentification method is simpleMaterial analysis by electric/magnetic meansBiotechnologyDismutase
The invention discloses a method for identifying the seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin, which comprises the following steps of: preparation of enzyme liquid, preparation of a gel plate, sample application and electrophoresis, SOD (superoxide dismutase) isozyme dyeing process, variety conformation and the like. The method mainly makes use of the physiological and biochemical difference between the seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin, and takes the SOD isozyme zymogram as an index for identifying the seeds of glycyrrhiza uralensis and glycyrrhiza inflata batalin. If the SOD isozyme zymogram of the seed to be detected is the same as the two lanes on the left side of a standard zymogram, the seed is the glycyrrhiza uralensis seed; if the SOD isozyme zymogram of the seed is the same as the two lanes on the right side of the standard zymogram, the seed is the glycyrrhiza inflata batalin seed. The identifying method disclosed by the invention has the advantage of simple, quick and accurate identification.
Owner:TIANJIN NORMAL UNIVERSITY
Kit and method for identifying cortex berberis origins
InactiveCN106086197AIdentification method is simpleGuarantee the quality of medicinal materialsMicrobiological testing/measurementDNA/RNA fragmentationMedicinal herbsClinical efficacy
The invention discloses a kit for identifying cortex berberis origins. The kit contains a reagent for amplifying four gene segments containing nucleotide sequences shown as SEQIDNO: 3-6. The invention further discloses a method for identifying the cortex berberis origin. The kit and the method can effectively and accurately distinguish various origins of cortex berberis, and the identifying method is simple and stable, has the important significance on usage of standard cortex berberis and can effectively guarantee the quality of medicinal materials and a clinical curative effect, and the application prospect is good.
Owner:CHENGDU UNIV OF TRADITIONAL CHINESE MEDICINE
Transparent liquid for rapidly identifying oocyte nuclei of shrimps and crabs
InactiveCN104818314AQuick observationRapid germinal vesicles observed in shrimp and crab oocytesMicrobiological testing/measurementAcetic acidAlcohol
The invention relates to the field of aquaculture, and provides a transparent liquid which can be used for rapidly transparentizing oocytes of shrimps and crabs and a method for treating and detecting maturity conditions of the oocytes of the shrimps and crabs by using the transparent liquid. The transparent liquid is a mixture which is prepared from anhydrous ethyl alcohol, glacial acetic acid and formaldehyde at a volume ratio of 60:1:30. The transparent liquid provided by the invention can be used for treating the oocytes of the shrimps and crabs, can be used for effectively transparentizing the oocytes of the shrimps and crabs, and can be used for rapidly identifying germinal vesicles of the oocytes of the shrimps and crabs.
Owner:SHANGHAI OCEAN UNIV
Method for detecting X-type HMW-GS (High Molecular Weight Glutenin Subunit) gene in distant hybridization wheat crossbreed
ActiveCN103114134AIdentification method is simpleReliable resultsMicrobiological testing/measurementDNA/RNA fragmentationCrossbreedPolymerase chain reaction
The invention provides a method for detecting an aegilops sharonensis X-type HMW-GS (High Molecular Weight Glutenin Subunit) gene in a distant hybridization wheat crossbreed. The invention further provides a primer for detecting the aegilops sharonensis X-type HMW-GS gene. The nucleotide sequence of the primer is shown in SEQ ID NO. 1-2. Whether the distant hybridization wheat crossbreed contains an aegilops sharonensis X-type HMW-GS genetic locus can be rapidly and accurately detected by carrying out PCR (Polymerase Chain Reaction) through using the primer. By utilizing the method, the transfer and existence of the aegilops sharonensis X-type HMW-GS gene can be accurately detected, the gene can be conveniently tracked, the working efficiency in breeding is greatly improved, and the progress in wheat quality breeding is accelerated, so that the method has good application prospects in the field of crossbreeding of wheat species.
Owner:SICHUAN AGRI UNIV
Identification and evaluation method for canker resistance of kiwi fruit
InactiveCN107083416AIncrease awarenessPromote healthy and sustainable developmentMicrobiological testing/measurementMicroorganism based processesHigh resistanceHigh humidity
The invention discloses an identification and evaluation method for the canker resistance of a kiwi fruit. The condition that in vitro branch culture after inoculation treatment is carried out at a relatively low temperature and relatively high humidity is ensured through determining reasonable in vitro branch sampling time of the kiwi fruit and innovating an in vitro branch inoculation culture mode; a branch peeling mode is utilized for resistance identification; the canker resistance of the kiwi fruit is identified and evaluated through the branch browning degree, so that the identification method is simple and intuitive; the identification result and the expression of field canker resistance of the kiwi fruit have relatively high consistency; and the method is suitable for most of kiwi fruit varieties. According to the method, recognition on the canker resistance of an existing kiwi fruit variety and other wild germplasm resources is greatly promoted, a producer can select a variety with high resistance for planting according to the requirements, further development and utilization of excellent resistant germplasm resources are facilitated, and healthy and sustainable development of the whole kiwi fruit industry is promoted.
Owner:GUANGXI INST OF BOTANY THE CHINESE ACAD OF SCI
Quick screening method of peanut mutant and application
InactiveCN109156352AIdentification method is simpleReduce invalid laborPlant tissue cultureHorticulture methodsMutagenic ProcessAgricultural science
The invention discloses a quick screening method of a peanut mutant and application, and belongs to the field of peanut seed breeding. The quick screening method of the peanut mutant comprises the steps as follows: identifying genotype mutagenicity of regeneration seedlings obtained by in-vitro mutagenesis of peanuts with an SSR (simple sequence repeat) method, weeding out the regeneration seedlings without genotype mutation, and identifying the regeneration seedlings with the genotype mutation as peanut mutants. The identification method of the mutagenicity of tissue culture seedlings is simple, can effectively weed out the regeneration seedlings without the mutation, saves ineffectual work of grafting, transplanting and subsequent mutagenicity identification of the regeneration seedlingswithout the mutation, and saves manpower, material resources and financial resources. The SSR method for screening the peanut genotype mutants is combined with an offspring character screening method, so that the seed breeding time can be shortened by 2-3 years. Some mutants without obvious phenotypic variation and with quality character variation can also be identified with the SSR method.
Owner:QINGDAO AGRI UNIV
Molecular marker tightly in linkage with corn line grain number main effect QTL and application thereof
InactiveCN106906301ANot affectedIdentification method is simpleMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenotype
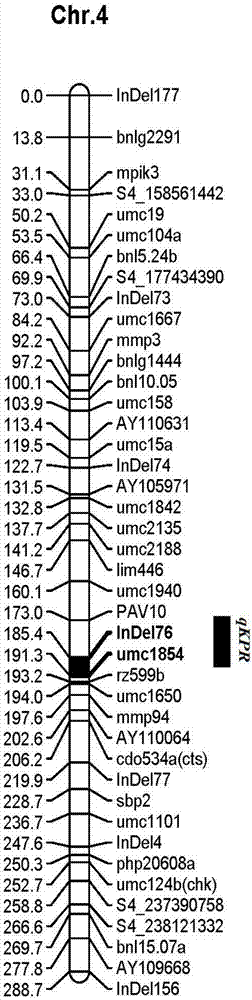
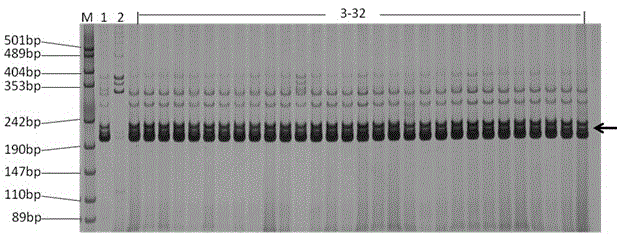
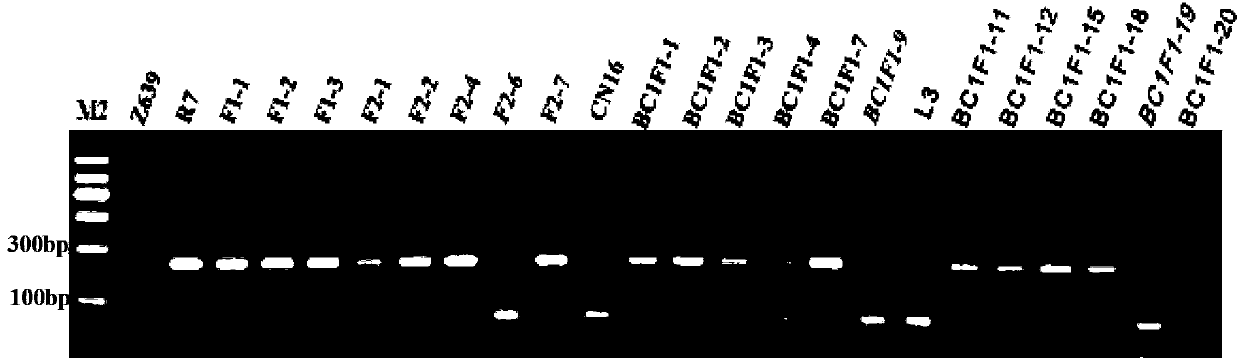
The invention discloses a molecular marker tightly in linkage with main effect QTL of corn line grain number and application thereof. The provided QTL is positioned on a first chromosome 4.09bin, and tightly linked molecular marker consists of primers InDe176 and umc1854. A genome DNA of a triple test-crossed group of corn to be tested is extracted, PCR amplification is carried out by using the primers InDe176 and umc1854, and amplification products having the lengths of 174bp and 142bp are obtained and can be used for predicting the corn line grain number. The molecular marker is used for genotype detection of the corn and its derived varieties or strains to judge the line grain number of the varieties or the strains and improve the selection accuracy and breeding efficiency.
Owner:JIANGSU ACAD OF AGRI SCI
Molecular Identification Method of Cotton Null Fruiting Branch Gene
ActiveCN104561284BSpeed up breedingImprove convenienceMicrobiological testing/measurementMolecular identificationAgricultural science
The invention belongs to the field of biotechnology, relates to a molecular identification method for zero-type fruit branches of cotton, and particularly provides a gene specific primer marker used for identifying a cotton strain with zero-type fruit branch genes. Nucleotide sequences are as shown in SEQ ID No.1 and SEQ ID No.2. An individual strain carrying the zero-type fruit branch genes in a segregation population is identified by the gen specific marker. The method is high in identification speed, high in accuracy, simple to carry out and very suitable for indoor identification and molecular marker-assisted selection of the zero-type branches of the cotton.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Method for detecting X-type HMW-GS (High Molecular Weight Glutenin Subunit) gene in distant hybridization wheat crossbreed
ActiveCN103114134BAchieve genetic improvementMake up for cumbersome deficienciesMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGlutenin
The invention provides a method for detecting an aegilops sharonensis X-type HMW-GS (High Molecular Weight Glutenin Subunit) gene in a distant hybridization wheat crossbreed. The invention further provides a primer for detecting the aegilops sharonensis X-type HMW-GS gene. The nucleotide sequence of the primer is shown in SEQ ID NO. 1-2. Whether the distant hybridization wheat crossbreed contains an aegilops sharonensis X-type HMW-GS genetic locus can be rapidly and accurately detected by carrying out PCR (Polymerase Chain Reaction) through using the primer. By utilizing the method, the transfer and existence of the aegilops sharonensis X-type HMW-GS gene can be accurately detected, the gene can be conveniently tracked, the working efficiency in breeding is greatly improved, and the progress in wheat quality breeding is accelerated, so that the method has good application prospects in the field of crossbreeding of wheat species.
Owner:SICHUAN AGRI UNIV
Molecular marker for Y-type high molecular weight glutelin subunit gene in distant hybridization generation of wheat and aegilops sharonensis and application of molecular marker
ActiveCN103397025BIdentification method is simpleReliable resultsMicrobiological testing/measurementPlant genotype modificationAgricultural scienceTriticeae
The invention provides a molecular marker for a Y-type high molecular weight glutelin subunit (HMW-GS) gene in a distant hybridization generation of wheat and aegilops sharonensis, wherein the molecular marker is obtained by amplifying a primer with a nucleotide sequence shown in SEQ ID NO.1-2. PCR (polymerase chain reaction) is carried out by utilizing the primer, so whether the distant hybridization generation of wheat and aegilops sharonensis contains an aegilops sharonensis Y-type HMW-GS gene locus or not can be rapidly and accurately detected, and whether the distant hybridization generation of wheat and aegilops sharonensis contains a Y-type HMW-GS gene or not is judged. By adopting the molecular marker method, whether the aegilops sharonensis Y-type HMW-GS gene transfers or exists in a wheat hybrid can be accurately detected, the aegilops sharonensis Y-type HMW-GS gene can be conveniently tracked, breeding working efficiency is greatly improved, a wheat quality breeding course is sped up, and the molecular marker for the Y-type HMW-GS gene in the distant hybridization generation of wheat and aegilops sharonensis has a good application prospect in the field of wheat crossbreeding.
Owner:SICHUAN AGRI UNIV
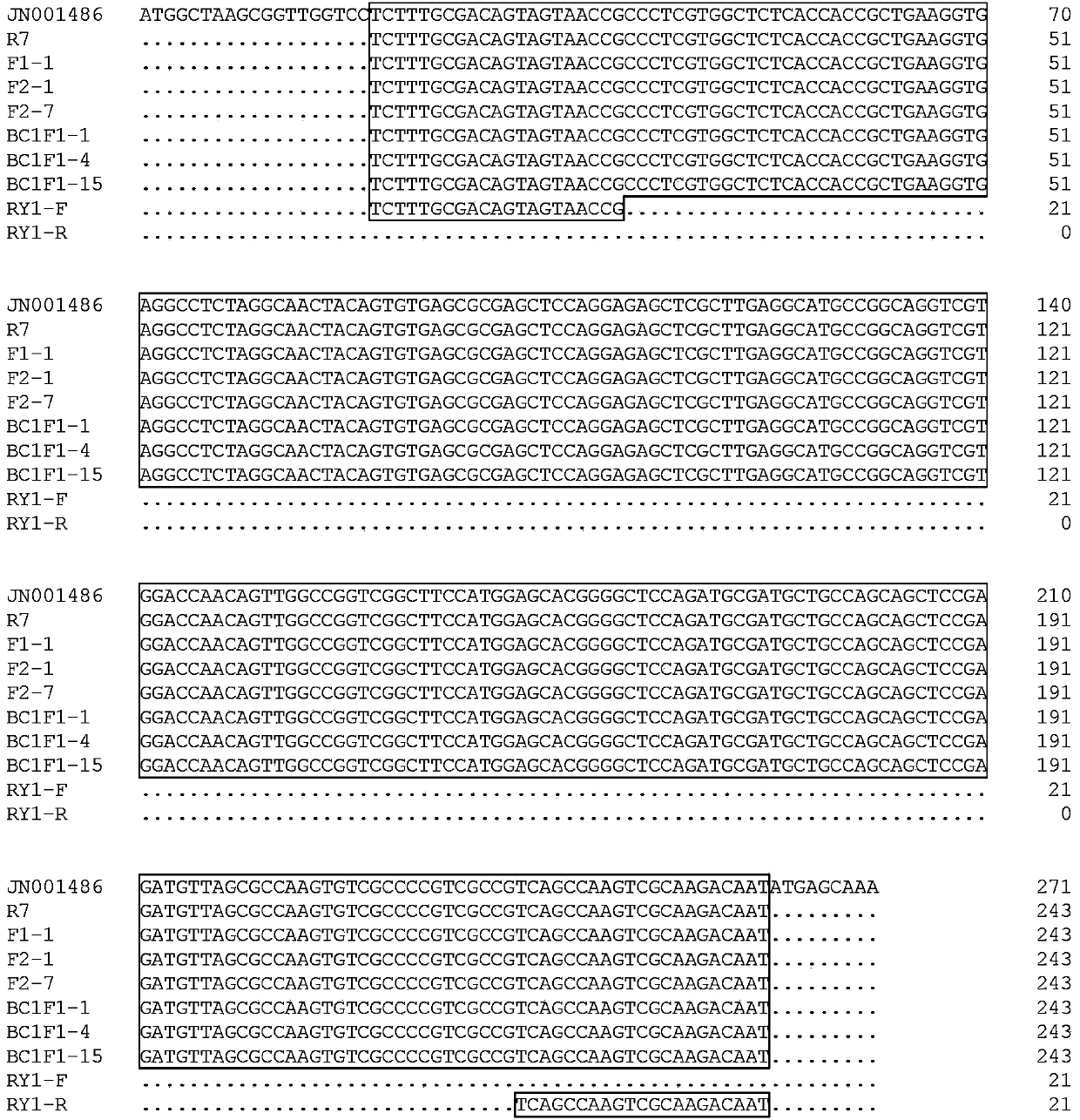
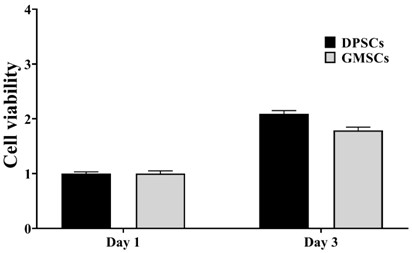
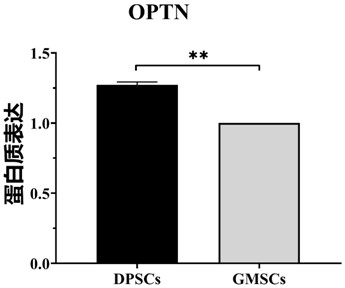
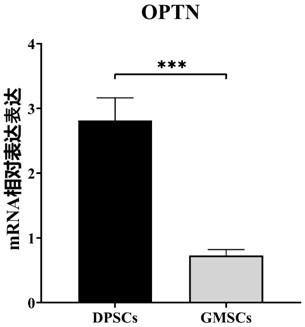
Application of OPTN gene in identification of dental pulp stem cells for identifying dental pulp stem cells and method thereof and product
ActiveCN111926089AImprove proliferative abilityEasy identificationMicrobiological testing/measurementDNA/RNA fragmentationBiologic markerDentistry
The invention relates to the technical field of biology, and particularly provides an application of an OPTN gene in identification of dental pulp stem cells for identifying the dental pulp stem cells, and a method thereof and a product. The biomarker OPTN gene capable of being used for identifying the dental pulp stem cells is discovered for the first time through tests, and the high-purity dental pulp stem cells can be identified and separated through differential expression of the gene.
Owner:KYBIOSTEM CO LTD
Method of preparing alpha-chlorothalonil
ActiveCN1752069AQuick transitionTransformation GuaranteeBiocideAnimal repellantsBoiling pointSolvent
A process for preparing alpha-chlorothalonil includes such steps as loading the raw chlorthalonil into a container, filling solvent, stirring or adding more solvent, adding alpha-chlorothalonil powder as crystal seeds, transform reaction at temp lower than 150 deg.C, and vacuum drying.
Owner:溧阳常大技术转移中心有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com