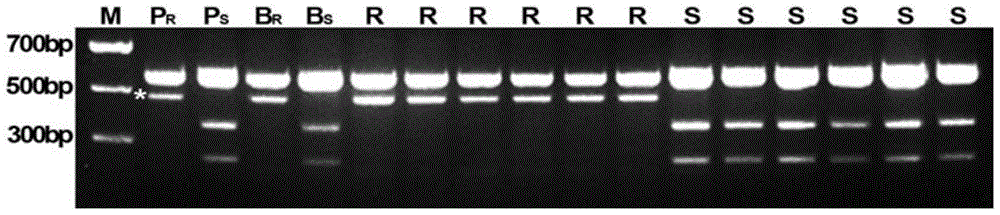
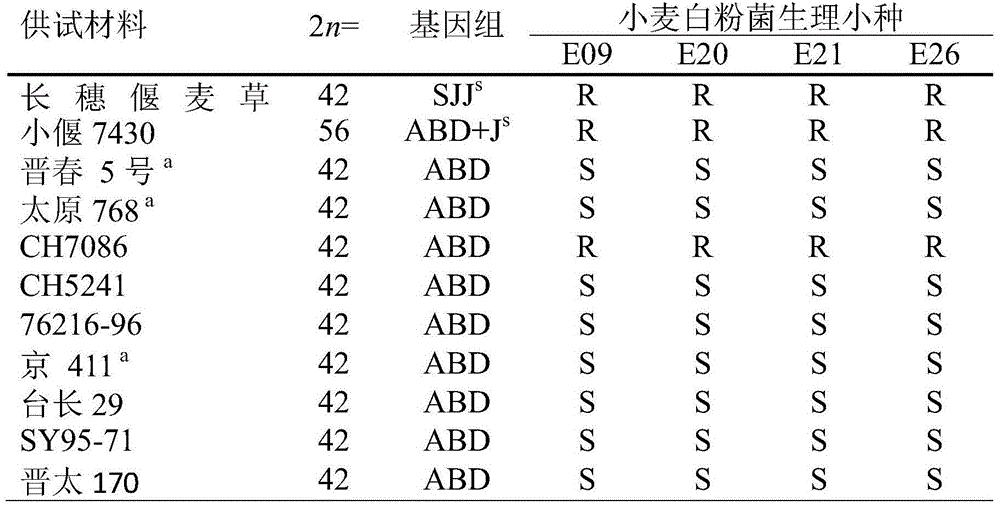
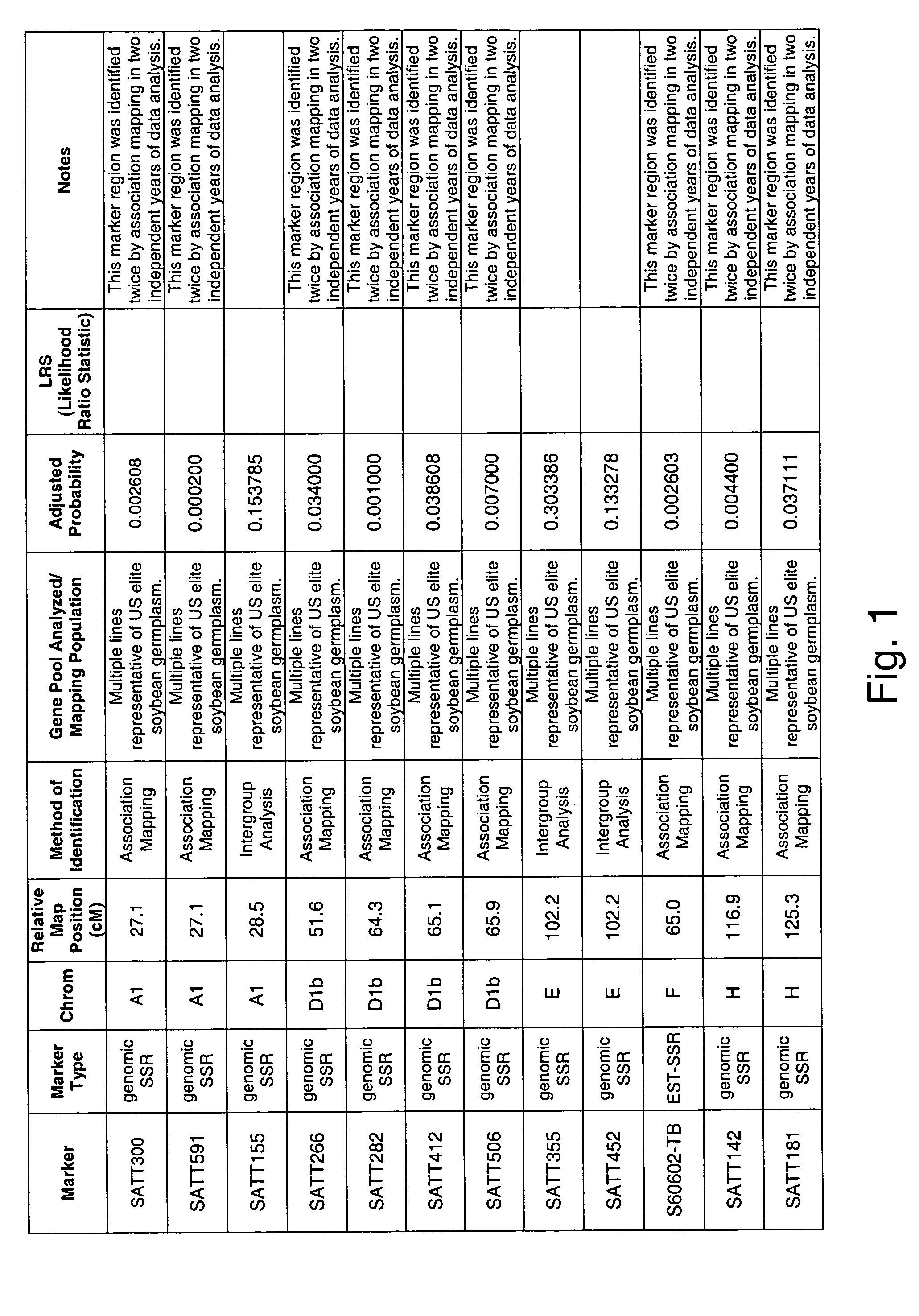
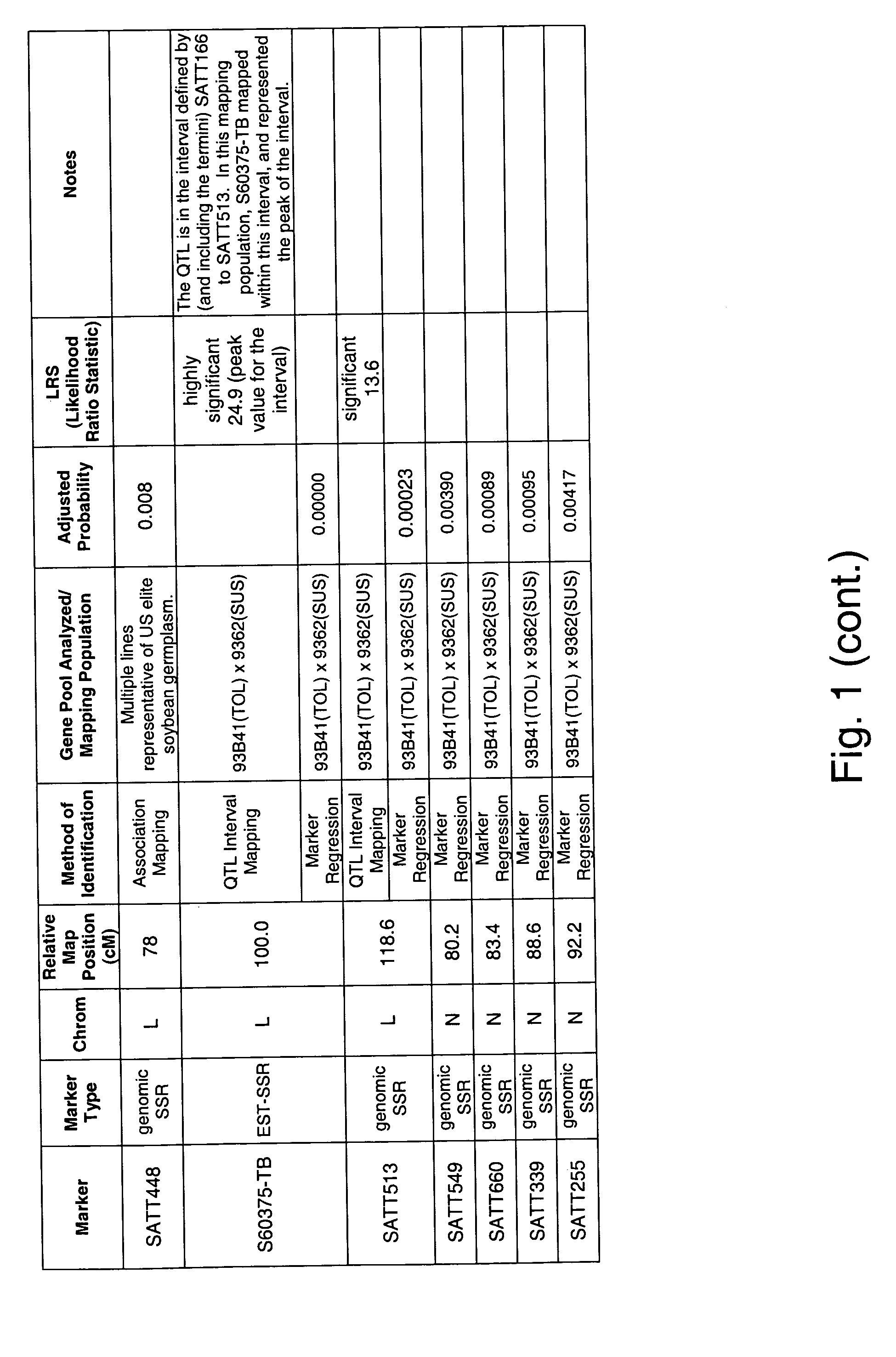
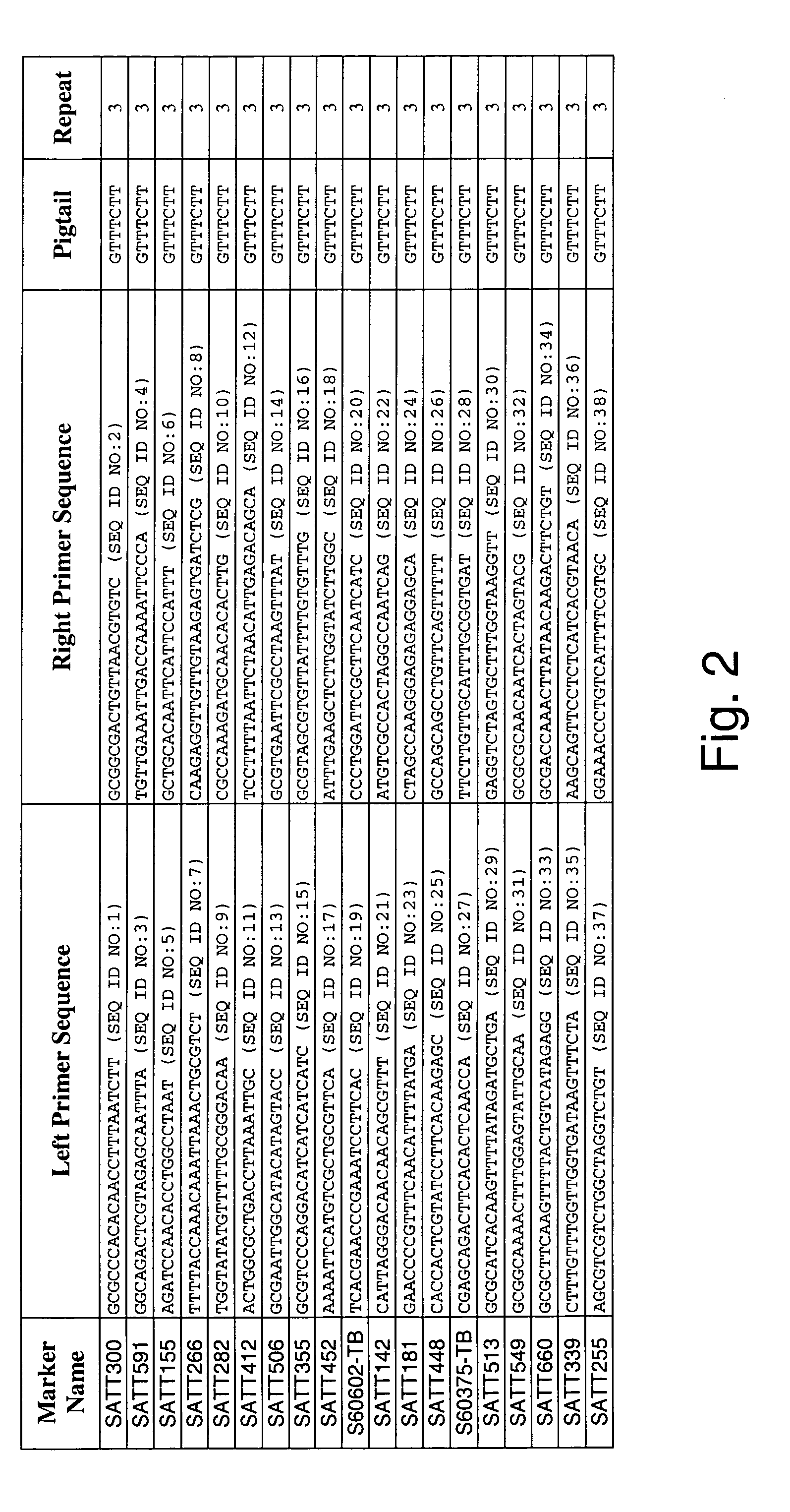
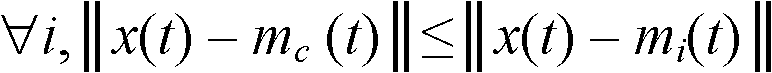Patents
Literature
905 results about "Molecular genetics" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Molecular genetics is the field of biology that studies the structure and function of genes at a molecular level and thus employs methods of both molecular biology and genetics. The study of chromosomes and gene expression of an organism can give insight into heredity, genetic variation, and mutations. This is useful in the study of developmental biology and in understanding and treating genetic diseases.
Device and method for detecting analytes
InactiveUS6548311B1Influence is negligibleRelative density is smallBioreactor/fermenter combinationsBiological substance pretreatmentsElectricityAnalyte
The invention relates to a method for detecting analytes and to a device for carrying out the method, for use for analysis or diagnosis in the fields of chemistry, biochemistry, molecular genetics, food chemistry, biotechnology, the environment and medicine. Marker particles (5) with different electrical properties or a different relative permeability to those of the measuring solution (3) surrounding them are used to detect the analytes (8). The marker particles (5) either bond specifically to the analytes (8) or to a base (2) in competition with the analyte. The analytes (8) are detected by the changes in an electrical field or an electrical current generated by electrodes (2) or in an electrical voltage applied to an electrode or in a magnetic field, said changes being caused by marker particles which have bonded with the analytes or by marker particles which have instead bonded to the base in an electrical field.
Owner:KNOLL MEINHARD
Method for modifying genetic characteristics of an organism
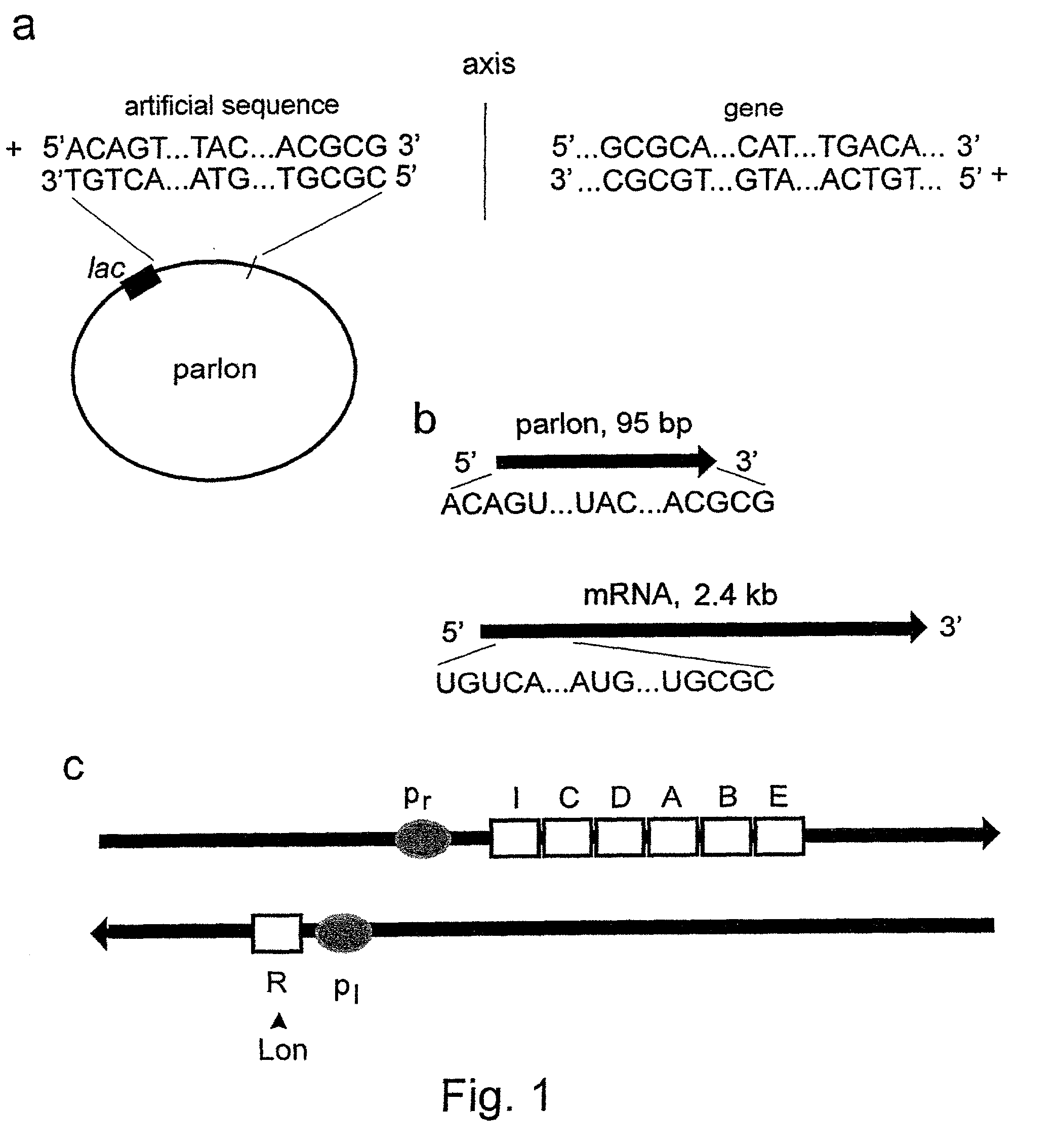
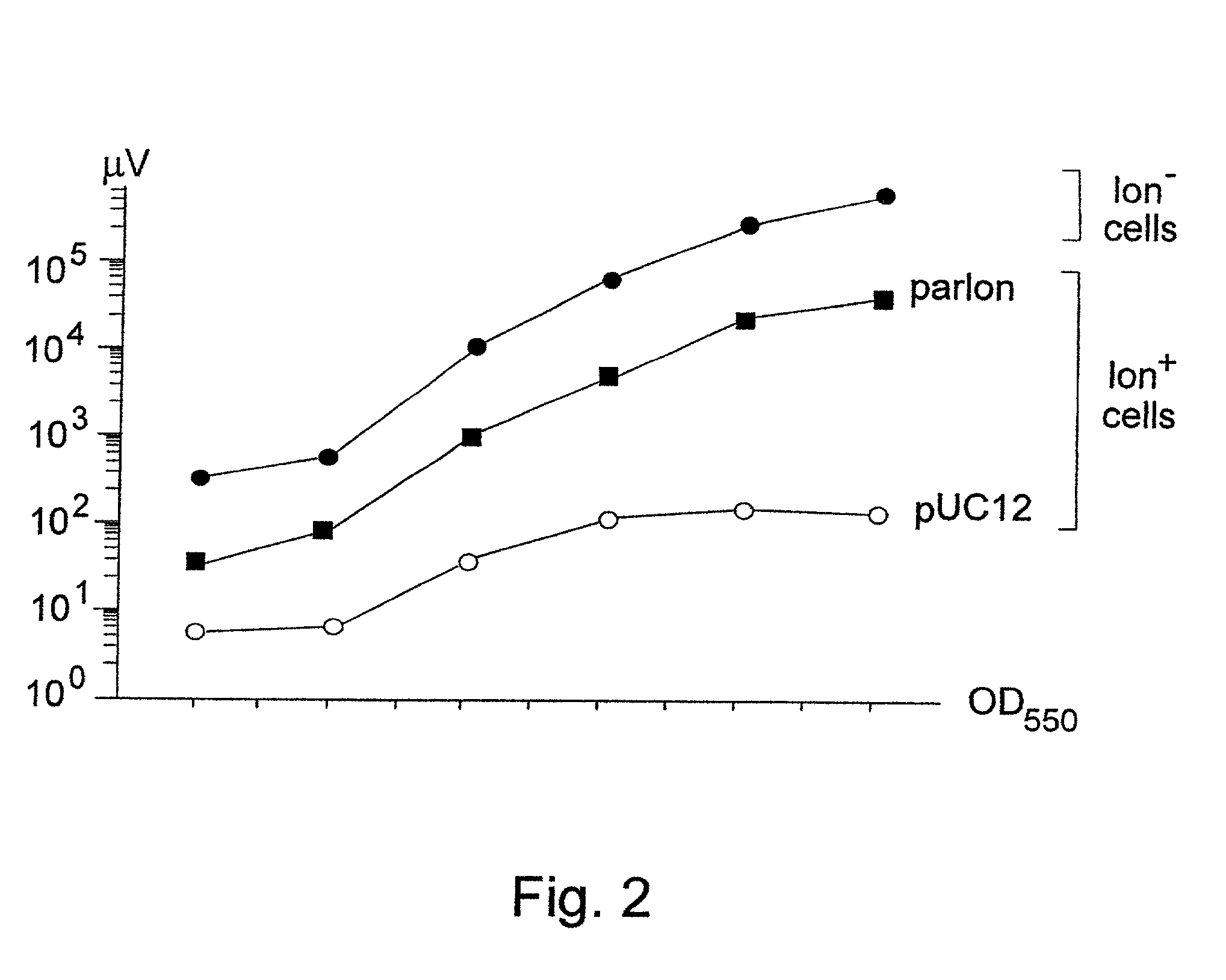
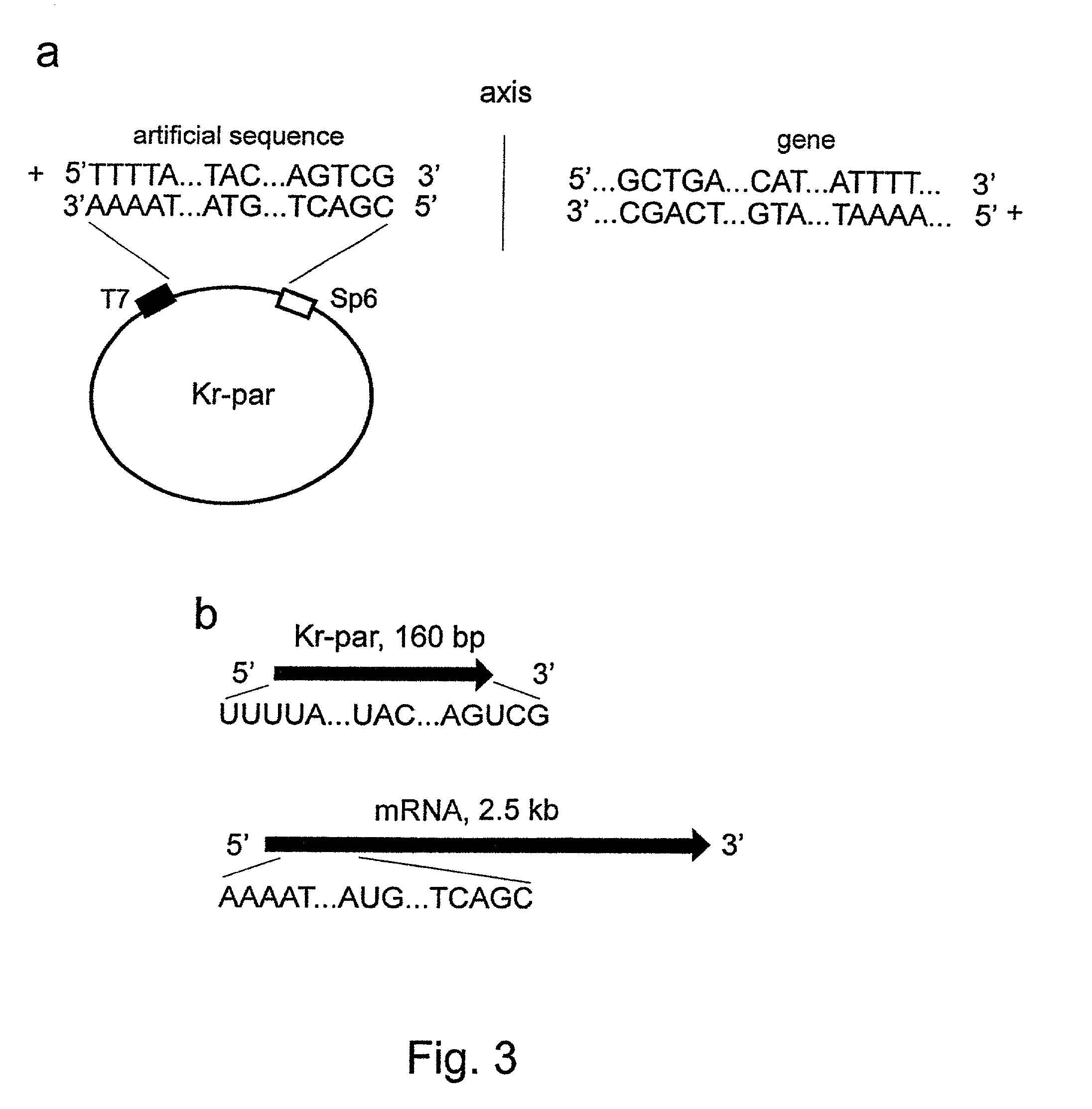
The invention concerns with the molecular biology, molecular genetics and biotechnology and can be used in the gene-therapy in the medicine and the agriculture or in the industrial biotechnology for a gene-specific silencing of the disease-related genes or the genes interfering a buildup of a product, respectively. The approach is suggested for changing of genetic properties of an organism by RNA interference leading to gene-specific silencing of a selected gene by RNA molecules, that are complementary in a parallel orientation (pcRNA) to mRNA of the selected gene; pcRNA are synthesized in vivo or in vitro on the artificial DNA sequence possessing symmetrical nucleotide ordering (mirror inversion) in respect to the nucleotide sequence of the gene. The invention suggests the general approach for changing of genetic properties of an organism and is based on the biological properties of mirror inversions of nucleotide sequences that are realized in RNA interference and gene-specific silencing.
Owner:INST MOLEKULJARNOJ BIOLOGII IM V A EHNGELGARDTA RAN
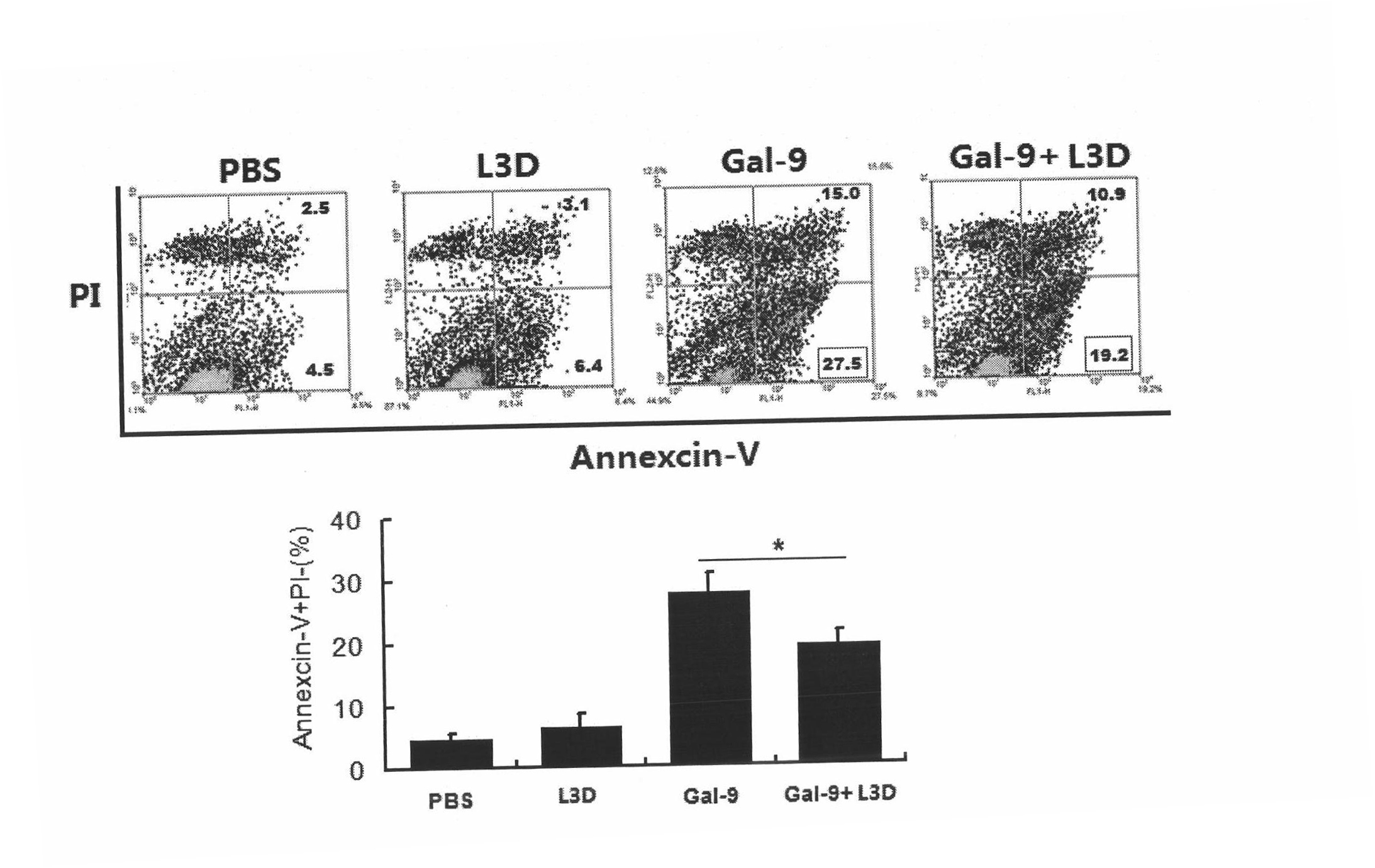
Anti-human Tim-3 neutralized monoclonal antibody L3D and application thereof
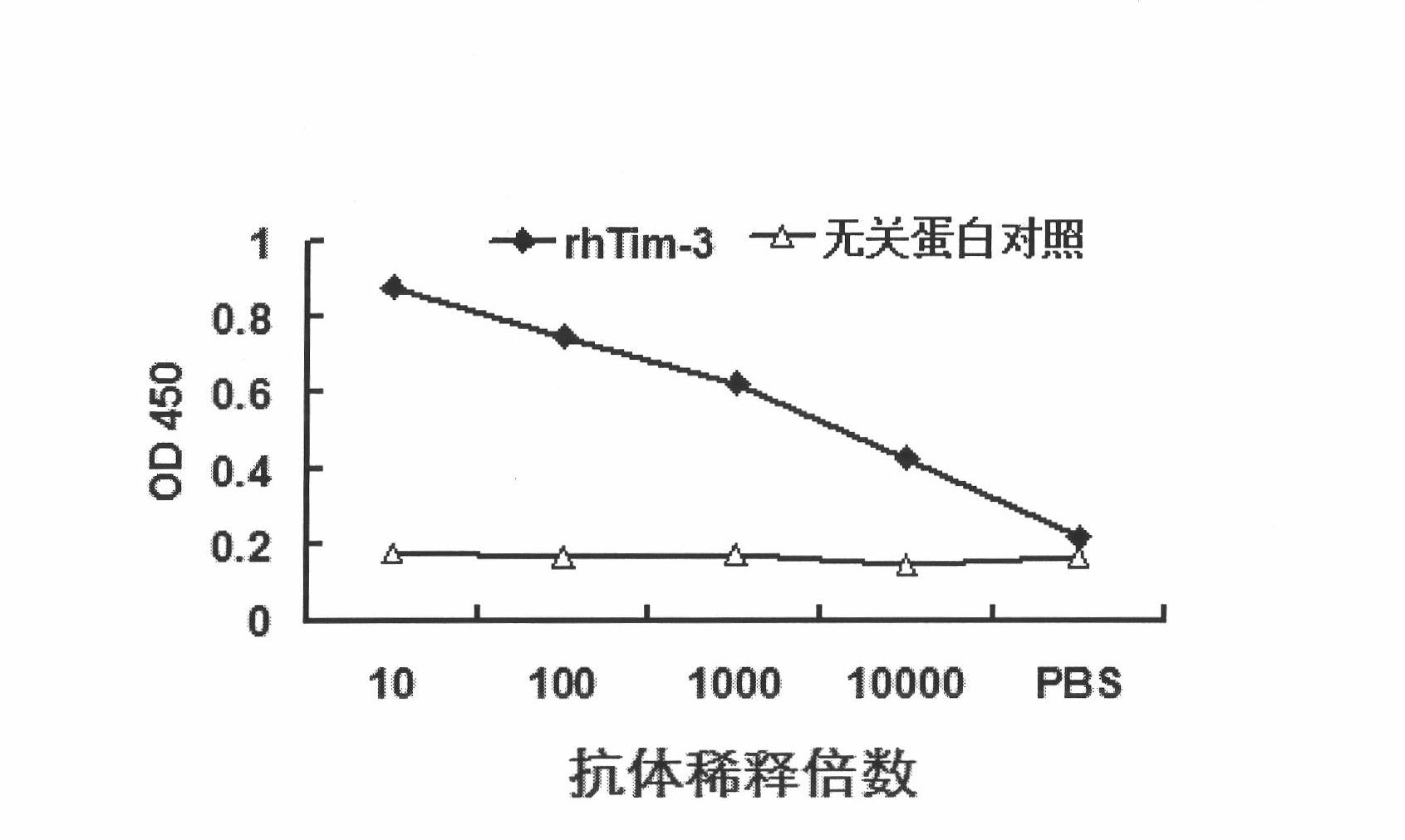
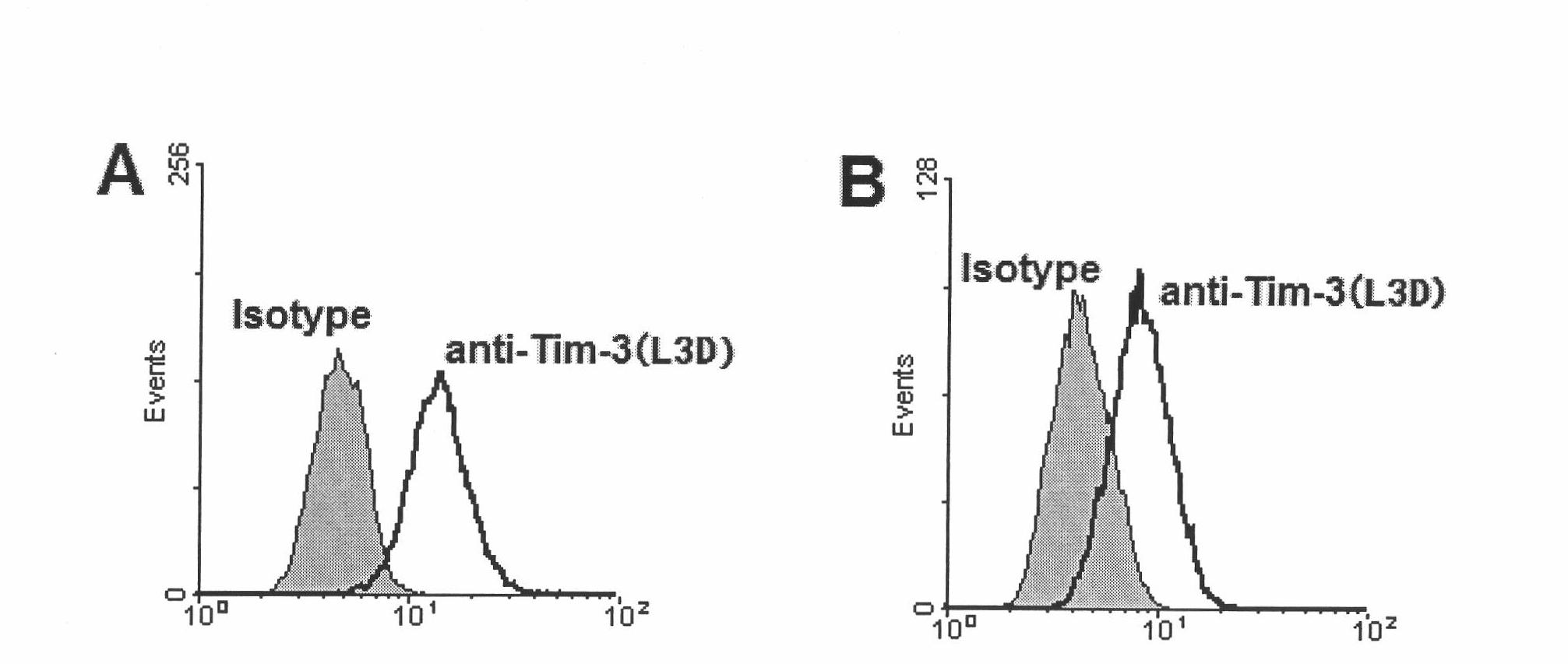
The invention discloses an anti-human Tim-3 neutralized monoclonal antibody L3D and an application thereof. Antibody light and heavy chain variable region genes are cloned from a prepared anti-human Tim-3 neutralized monoclonal antibody L3D hybridoma cell; and the obtained light chain and heavy chain variable region genes can be used for encoding a monoclonal antibody variable region. Based on the light and heavy chain variable region genes of the monoclonal antibody, a plurality of small molecular genetic engineering antibodies can be constructed and expressed. Polypeptides or proteins encoded on the basis of the genes can be cross-linked with a plurality of bioactive molecules, so that a Tim-3 expression level detection reagent is prepared for diagnosing and treating diseases caused by abnormal expression of Tim-3.
Owner:INST OF BASIC MEDICAL SCI ACAD OF MILITARY MEDICAL SCI OF PLA
Functional molecular marker for rice anti-blast gene Pi9 and application thereof
ActiveCN103146695AImprove breeding efficiencyImprove accuracyMicrobiological testing/measurementPlant genotype modificationBiotechnologyNucleotide
The invention provides a functional molecular marker for rice anti-blast gene Pi9 and application thereof, belonging to the field of crop molecular genetic breeding science. The invention finds out that the rice anti-blast gene Pi9 has a 10bp insertion / deletion site positioned between the gene promoter -516 and -517, wherein the nucleotide sequence is disclosed as SEQ ID NO.1; and on such basis, the invention develops a method of the functional molecular marker for gene Pi9. By detecting the functional molecular marker, the invention can accurately detect whether genomes of different rice species contain the Pi9 functional gene and detect the homozygotic state, and can be used for screening rice hybrid transformed descendant plants to enhance the breeding efficiency of the rice anti-blast material, thereby effectively controlling the size of the breeding population, obviously saving the breeding and screening cost and obtaining the anti-blast rice species containing the Pi9 functional gene.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
Molecular improvement method for lowering rice grain seed holding
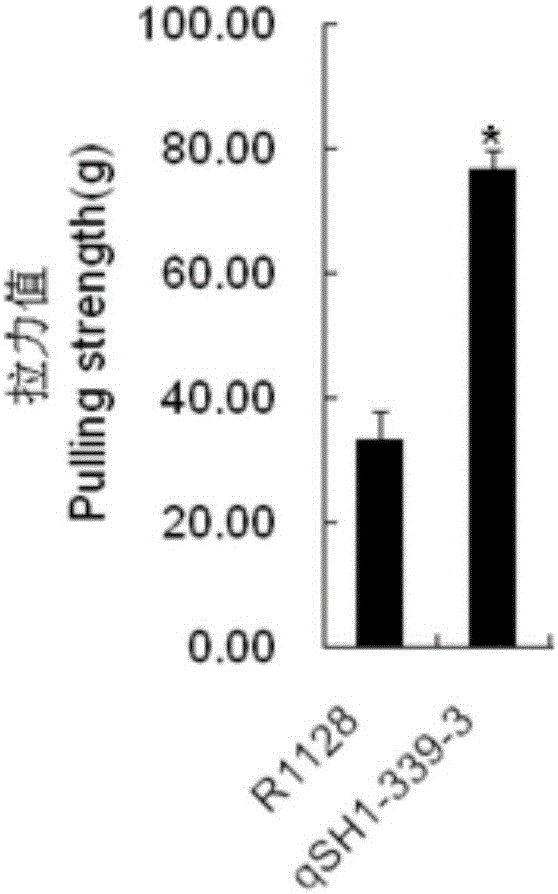
ActiveCN106191107AReduce shatteringImprove work efficiencyPlant peptidesFermentationTransgenesisMolecular genetics
The invention discloses a molecular genetic improvement method for lowering rice seed holding by targeted modification on rice seed holding gene qSH1 by using a CRISPR (clustered regularly interspaced short palindromic repeats) / Cas9 system. The method comprises the following steps: 1) selecting an appropriate target; 2) establishing a vector containing the target spot sequence; 3) establishing a recombinant vector containing the target spot sequence by utilizing the vector; 4) introducing the recombinant vector into receptor rice to obtain a transgenic positive plant; 5) obtaining a targeted-mutation mutant plant by utilizing the transgenic positive plant; 6) carrying out additive-generation growth on the mutant plant to obtain a transgenic-component-free homozygous mutant plant; and 7) carrying out seed holding investigation by using the homozygous mutant plant to obtain the plant with obviously lower seed holding as the seed-holding-improved plant. The method has the advantages of high directionality, small genetic background changes and the like, can avoid the risks caused by genetic transformation, and can culture the new species and new combination of transgenic-component-free rice with obviously lower seed holding.
Owner:HUNAN AGRICULTURAL UNIV +1
Molecular mark method for rice variety anti-brownspot gene site
InactiveCN1896281AEasy to detectNot affectedMicrobiological testing/measurementDNA/RNA fragmentationBrown planthopperGenotype
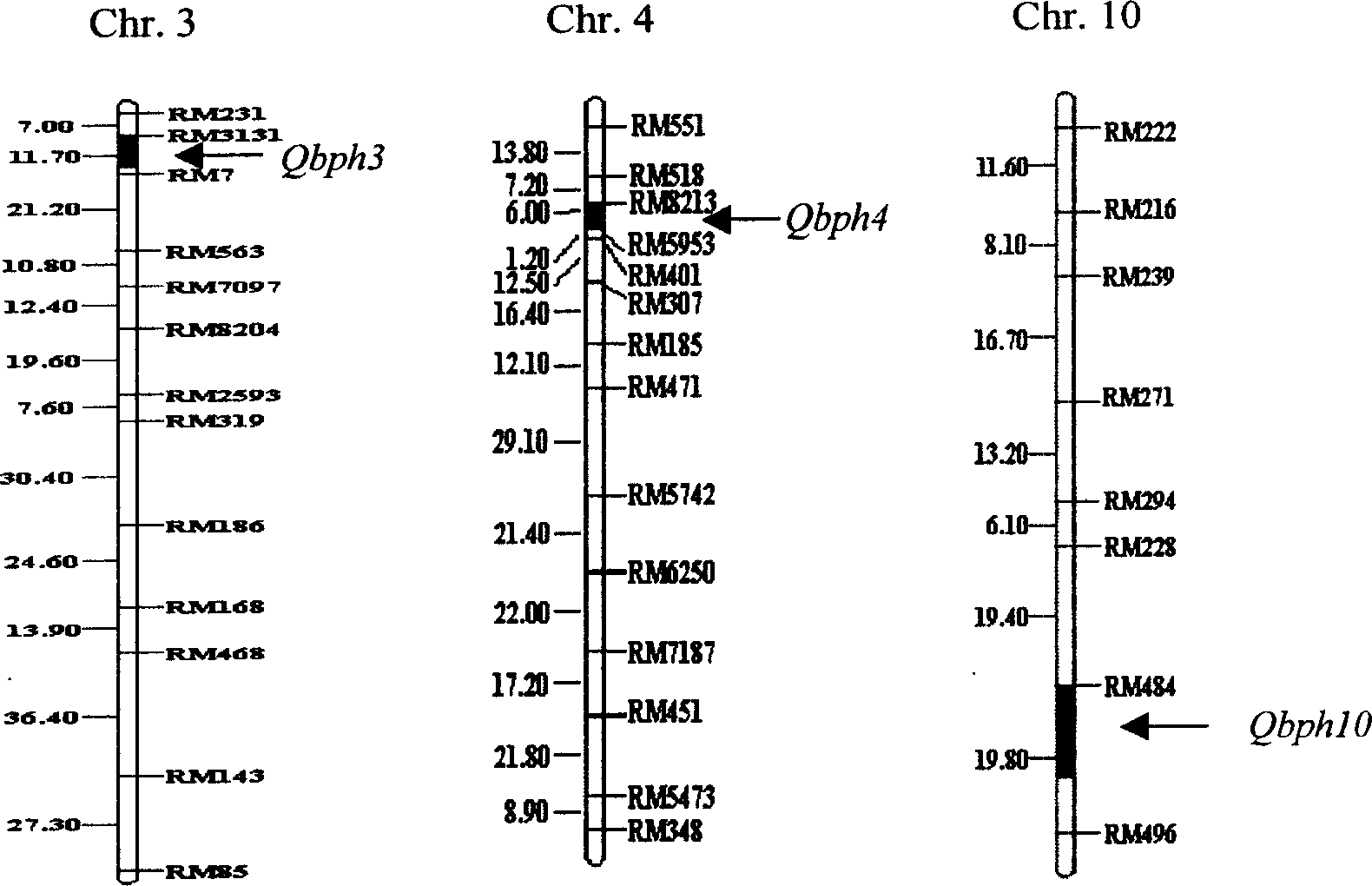
The present invention belongs to the molecular genetics field and relates to the molecular marker method of the major gene for brown planthopper resistance in the rice variety Rathu Heenati. Via genetic linkage analysis of the individual F2 plant genotype and the brown planthopper resistance level of each F2:3 family produced by hybridizing the insect-resistant variety Rathu Heenati with insect-susceptible variety 02428, the molecular markers RM8213 and RM5953 of the major gene loci Qbph4(Bph3) in the insect-resistant variety Rathu Heenati are obtained, the loci Qbph4(Bph3) is 3.6cM and 3.2cM distant from the markers RM8213 and RM5953 respectively. Detection of this major gene loci in Rathu Heenati and its derivatives via these molecular markers will increase the selection efficiency of brown plant hopper resistant rices.
Owner:NANJING AGRICULTURAL UNIVERSITY
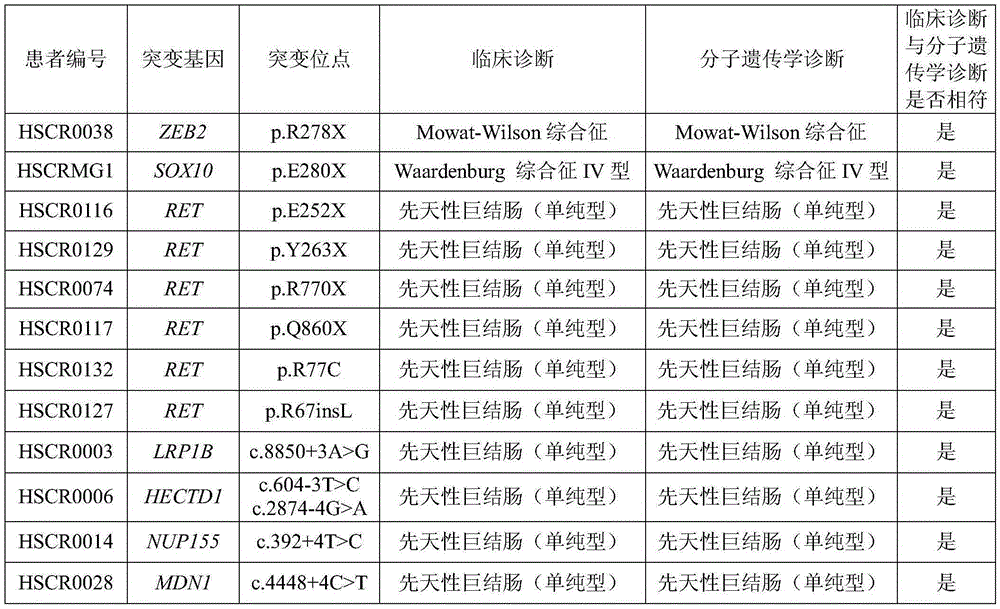
Probe set and reagent kit used for detecting pathopoiesia/susceptibility genes of congenital megacolon and relative syndromes
ActiveCN105256051AStrong targetingReduce or avoid birthsMicrobiological testing/measurementDNA/RNA fragmentationDiseaseMass survey
The invention provides a probe set and a reagent kit used for detecting pathopoiesia / susceptibility genes of congenital megacolon and relative syndromes. At most 172 pathopoiesia / susceptibility genes related to diseases can be detected at the same time, and reference can be provided for affected individual molecular genetics diagnosis, mass survey of congenital megacolon high-incidence areas, screening of high-risk affected groups in congenital megacolon family members and corresponding genetic counseling or prenatal intervention.
Owner:首都儿科研究所 +1
Bovine germline D-genes and their application
ActiveUS7196185B2Large capacitySugar derivativesMicrobiological testing/measurementDiseaseImmunocompetence
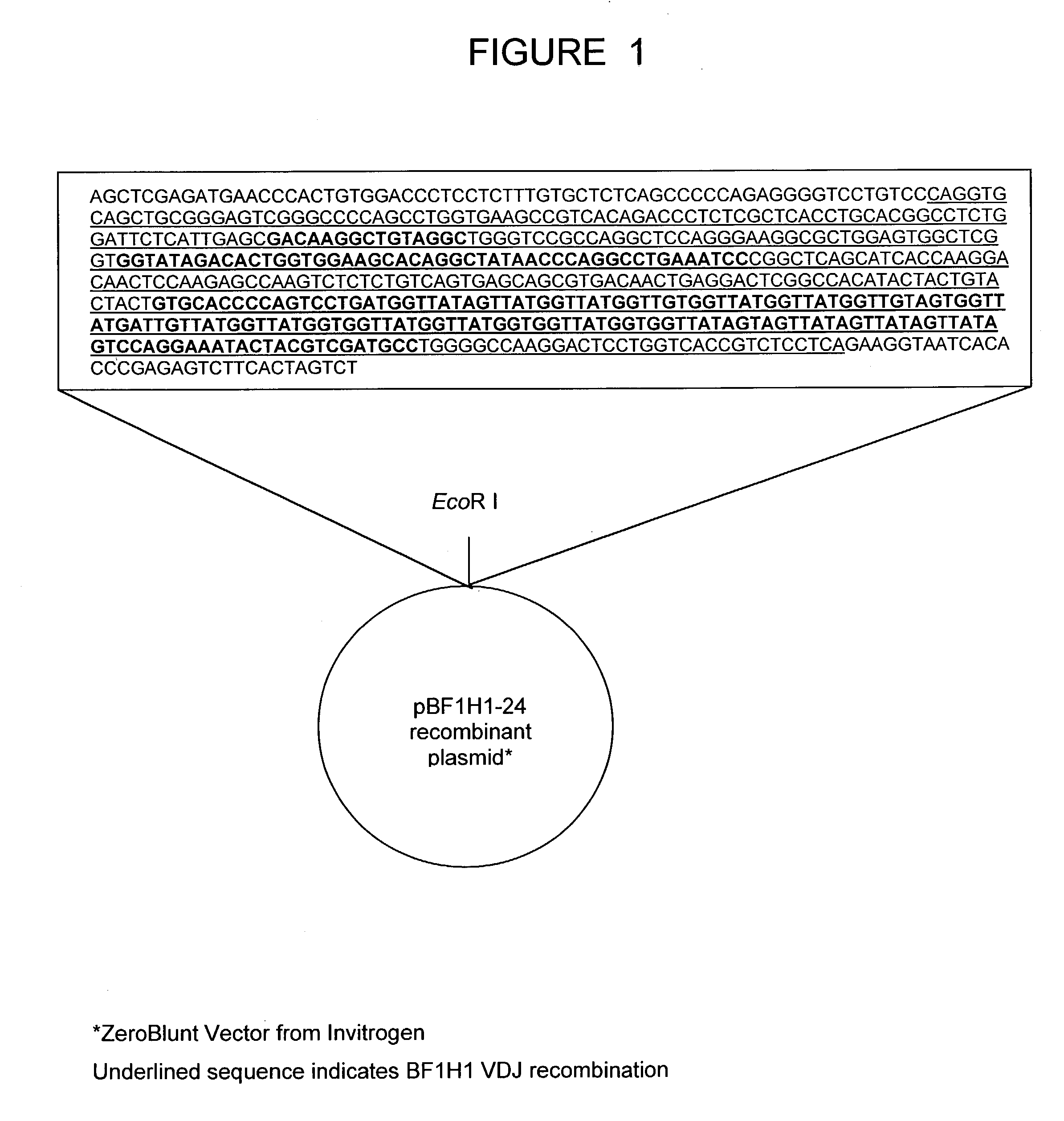
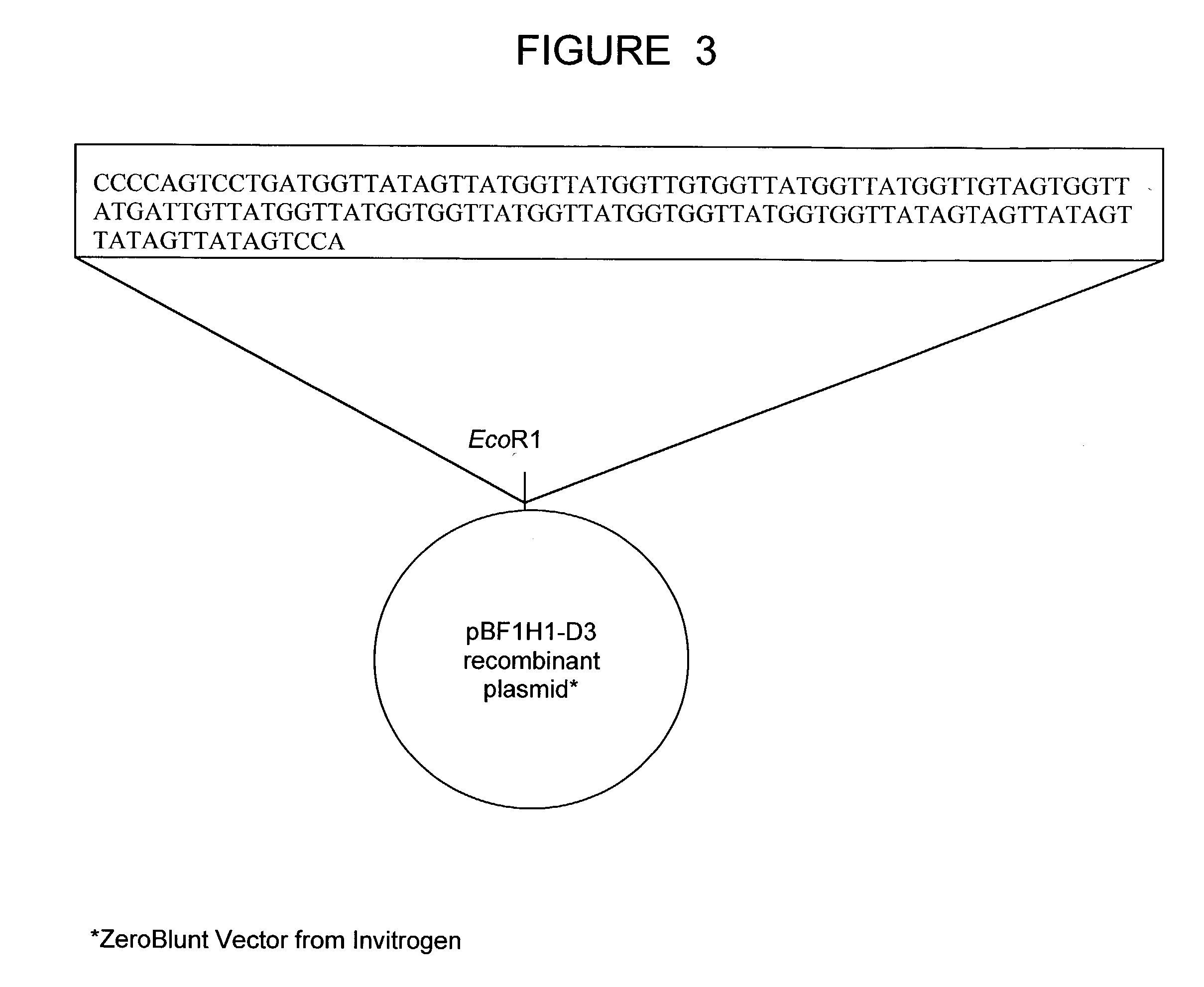
The present invention relates to a bovine VDJ cassette (BF1H1) that provides the novel ability to develop chimeric immunoglobulin molecule capable of incorporating both linear T cell epitope(s) (CDR1H and CDR2H) as well as conformational B cell epitope(s) (exceptionally long CDR3H). Further, multiple epitopes can be incorporated for development of multivalent vaccine by replacing at least a portion of an immunoglobulin molecule with the desired epitope such that functional ability of both epitope(s) and parent VDJ rearrangement is retained. The antigenized immunoglobulin incorporating both T and B epitopes of interest is especially useful for development of oral vaccines for use in humans apart from other species including cattle. The long CDR3H in BF1H1 VDJ rearrangement originates from long germline D-genes. The novel bovine germline D-genes provide unique molecular genetic marker for sustaining the D-gene pool in cattle essential for immunocompetence via selective breeding. D-gene specific DNA probe permits typing and selection of breeding cattle stock for maximum gemline D gene pool for better health and disease prevention. The bovine D-genes are unique to cattle and, therefore, provide sensitive and specific forensic analytical tool using molecular biology techniques to determine tissues suspected of bovine origin.
Owner:KAUSHIK AZAD KUMAR +2
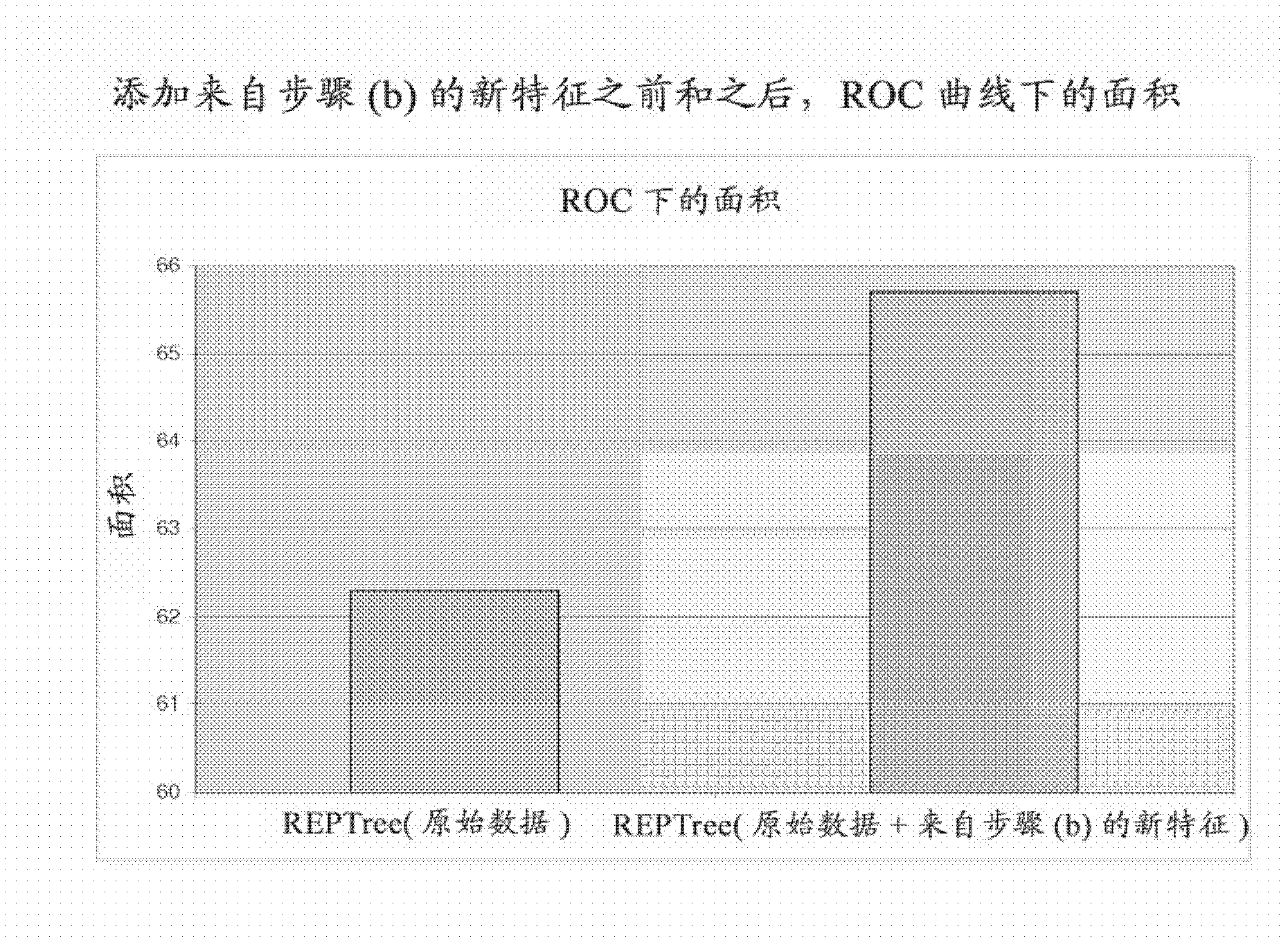
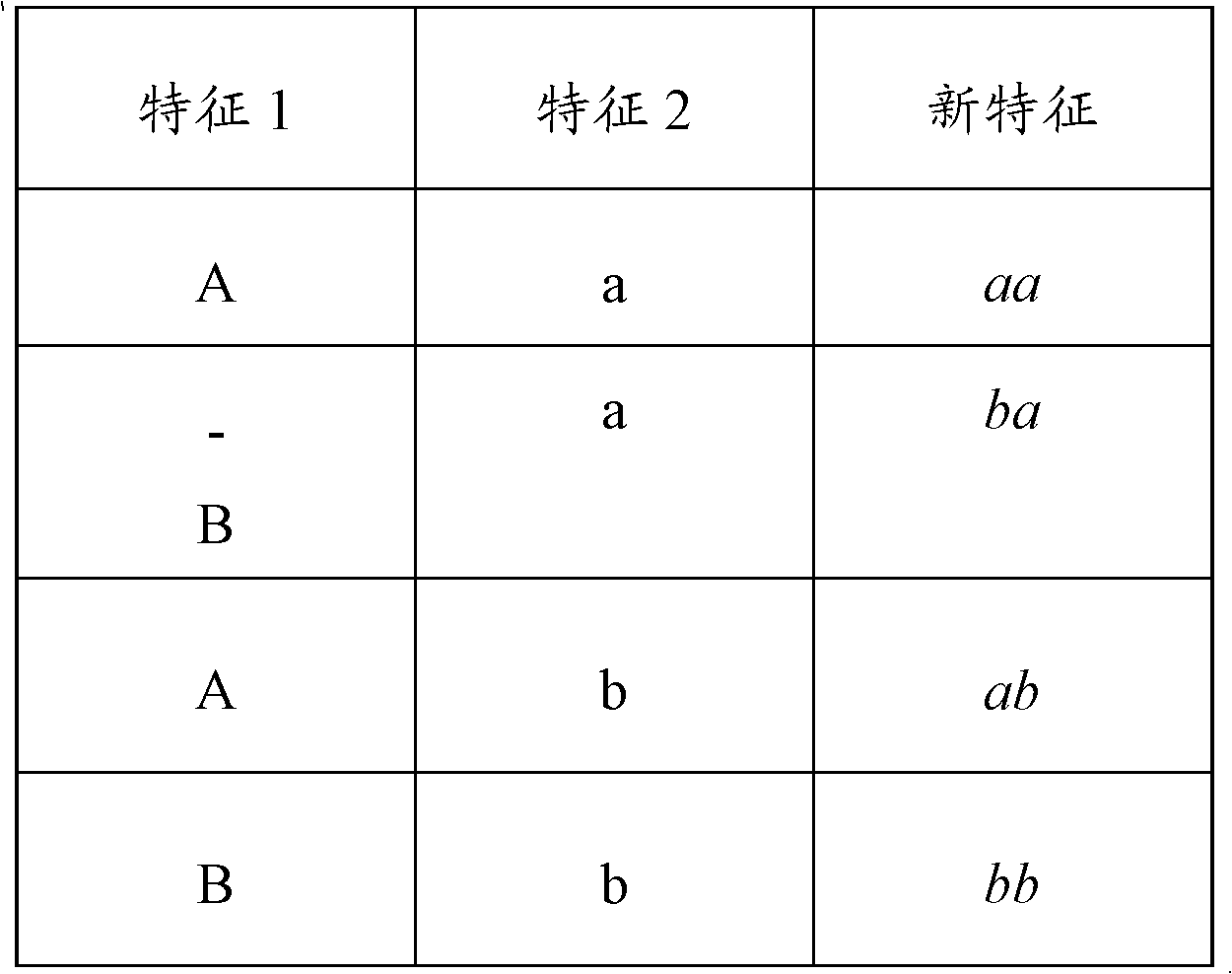
Application of machine learning methods for mining association rules in plant and animal data sets containing molecular genetic markers, followed by classification or prediction utilizing features created from these association rules
The disclosure relates to the use of one or more association rule mining algorithms to mine data sets containing features created from at least one plant or animal-based molecular genetic marker, find association rules and utilize features created from these association rules for classification or prediction.
Owner:DOW AGROSCIENCES LLC
SSR molecular marker A002 for identifying males and females of kiwi fruit hybrid populations
ActiveCN104313019AQuick digImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMarker-assisted selection
The invention discloses an SSR molecular marker A002 for identifying males and females of kiwi fruit hybrid populations, relating to the technical field of molecular genetic breeding. The male plant specific fragment nucleotide sequence of the molecular marker A002 is shown in SEQ.ID No.1; the female plant specific fragment nucleotide sequence is shown in SEQ.ID No.2; and the primer sequence is shown in SEQ.ID No.3 and SEQ.ID No.4. The molecular marker A002 can be used for the early identification detection of males and females of kiwi fruits so as to avoid the great waste caused by abundant male plants in the breeding and production; by utilizing a high-density genetic map, sex related molecular markers can be rapidly explored to assist the breeding and important practical experience is provided for the development of the functional markers; and being used for marker assisted selection of males and females, the molecular marker A002 is capable of greatly improving the selection efficiency of the female plants in the breeding and shortening the breeding period, and has great application potential and relatively high economic value.
Owner:WUHAN BOTANICAL GARDEN CHINESE ACAD OF SCI
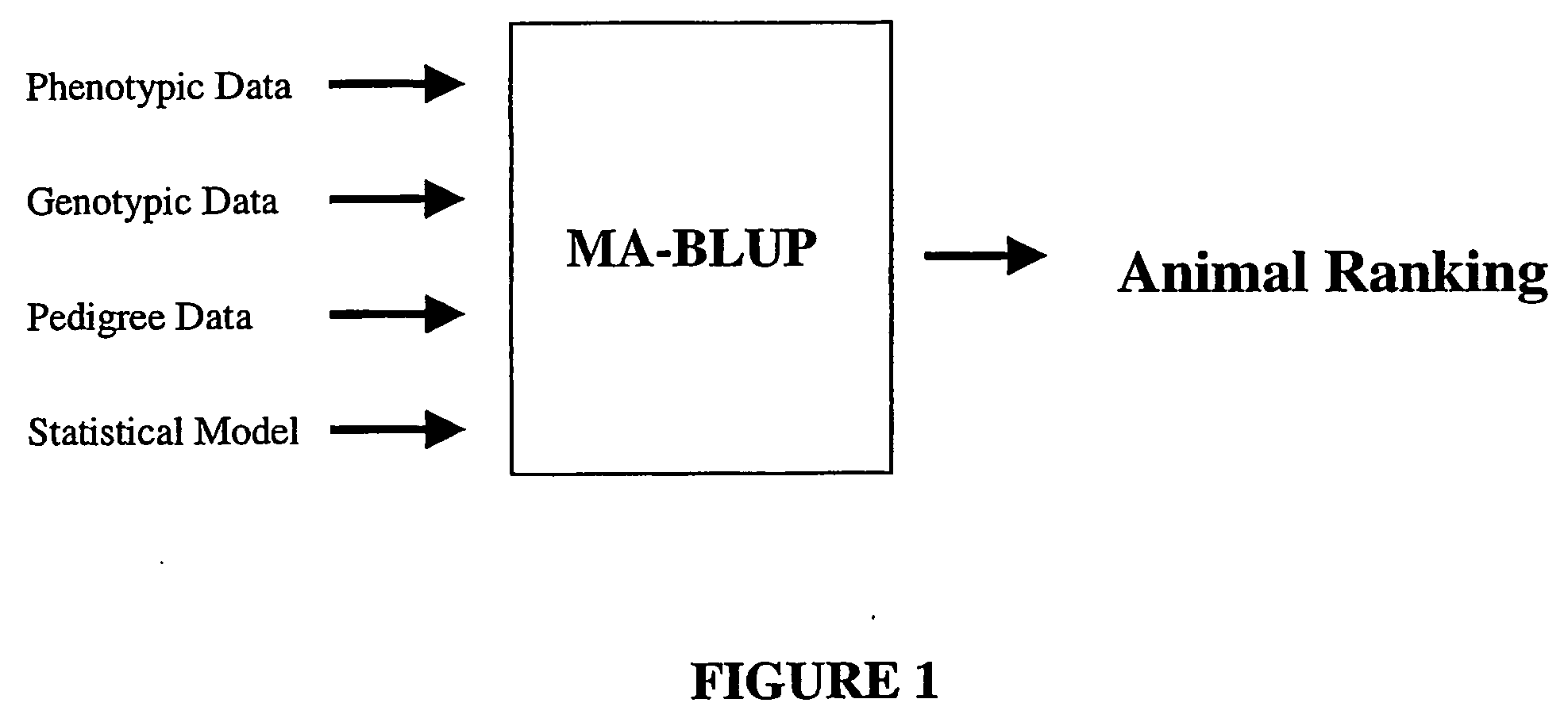
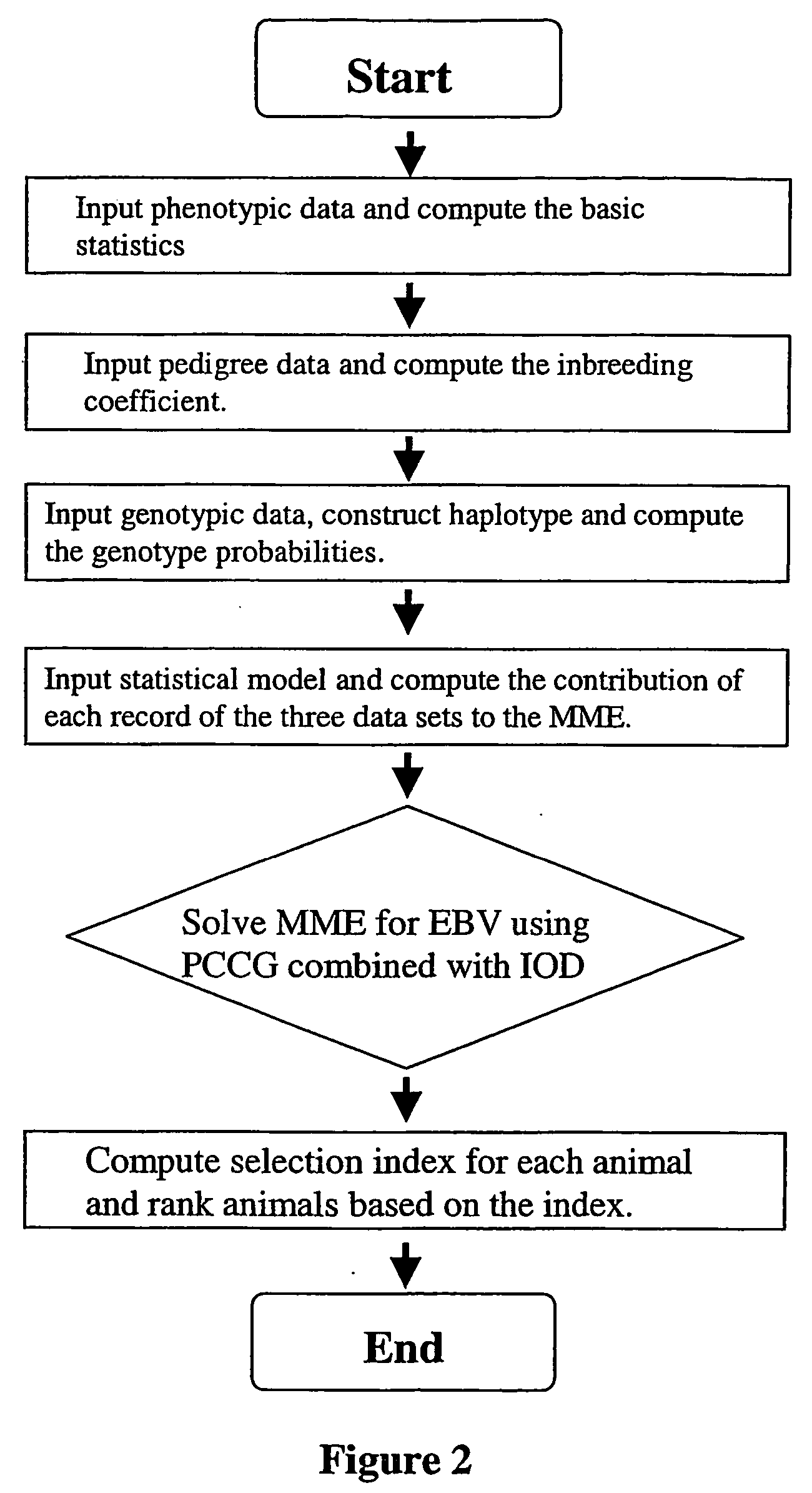
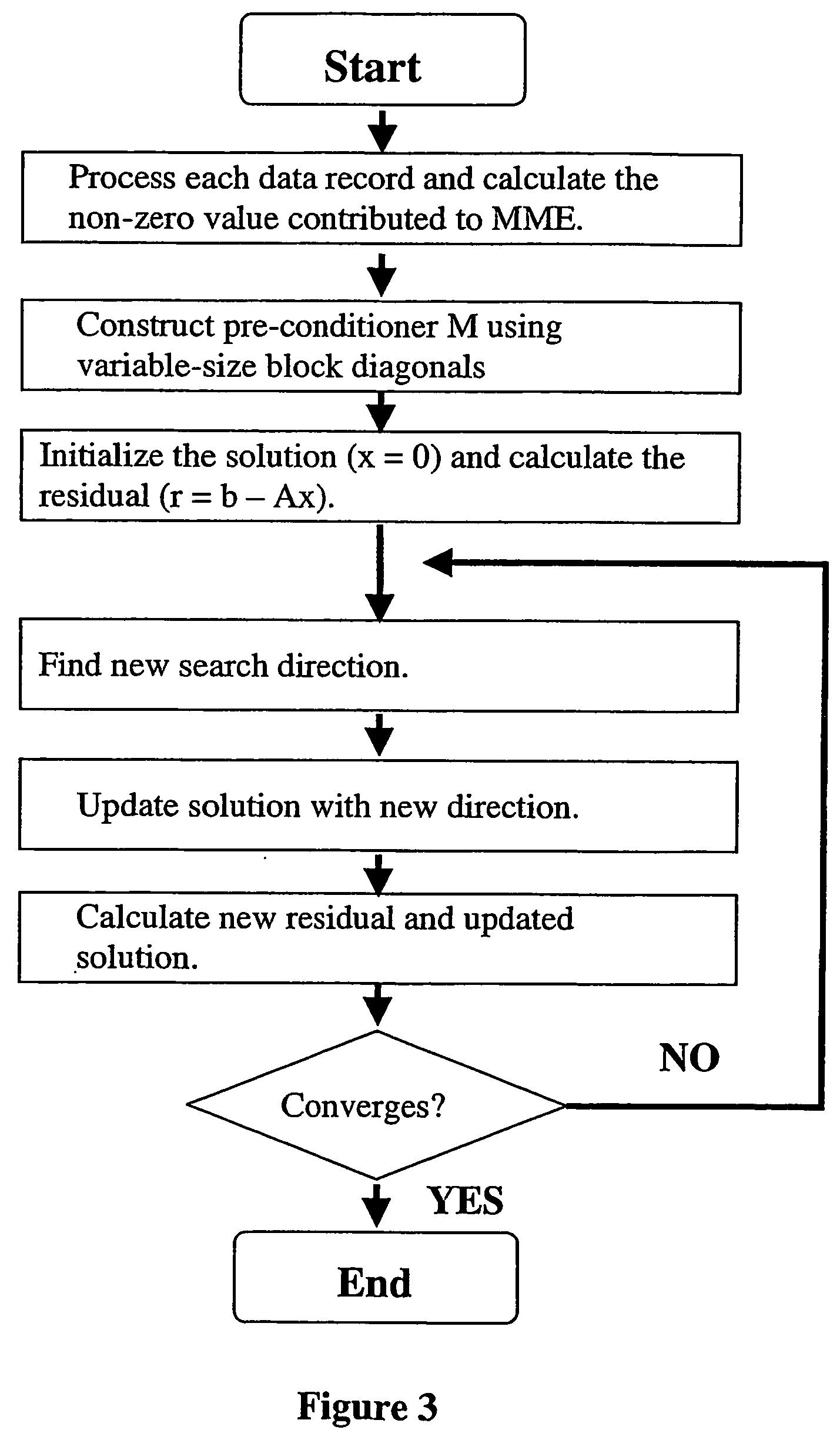
Marker assisted best linear unbiased prediction (ma-blup): software adaptions for large breeding populations in farm animal species
InactiveUS20070105107A1Genetic responseReduce memory requirementsMicrobiological testing/measurementProteomicsQuantitative trait locusHerd
The invention provides methodologies for improved molecular genetic analysis of individual animals and animal populations. The invention includes methods and systems for identifying those animals in a population that are most likely to heritably pass on desirable traits. Provided are means for evaluating the estimated breeding values and increasing the average genetic merit for animals in a population. For each trait, the instant invention provides methods for evaluating the relative effect of one or more quantitative trait loci (QTL) and three or more molecular genetic markers for each QTL The relationship between these various markers and the pre-selected trait and QTL is calculated, along with the contribution of other factors such as pedigree and known measures with respect to quantitative trait, and these data are used to calculate estimated breeding values for the animals in the herd and to rank the animals according to these estimated breeding values.
Owner:SCIDERA
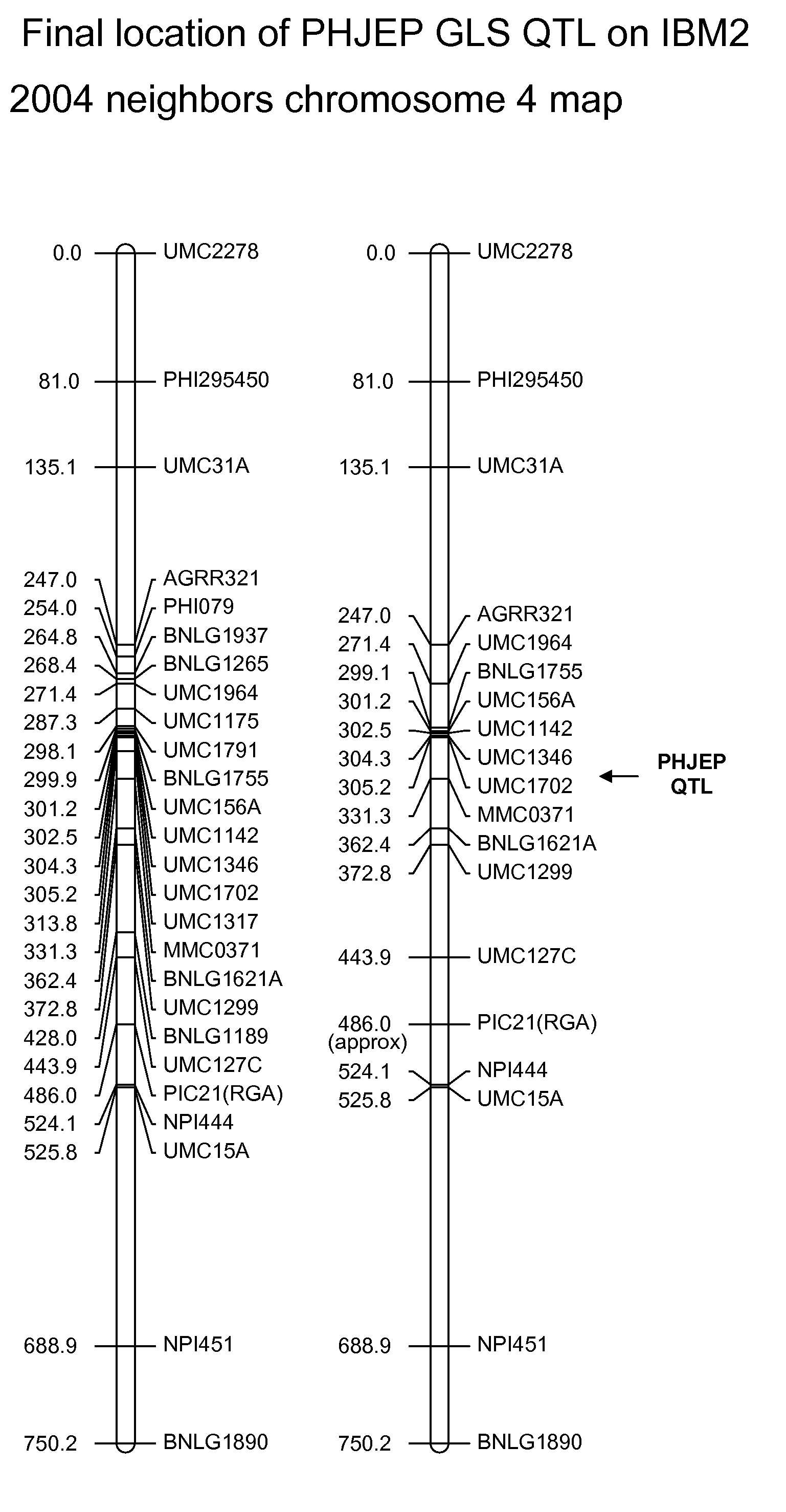
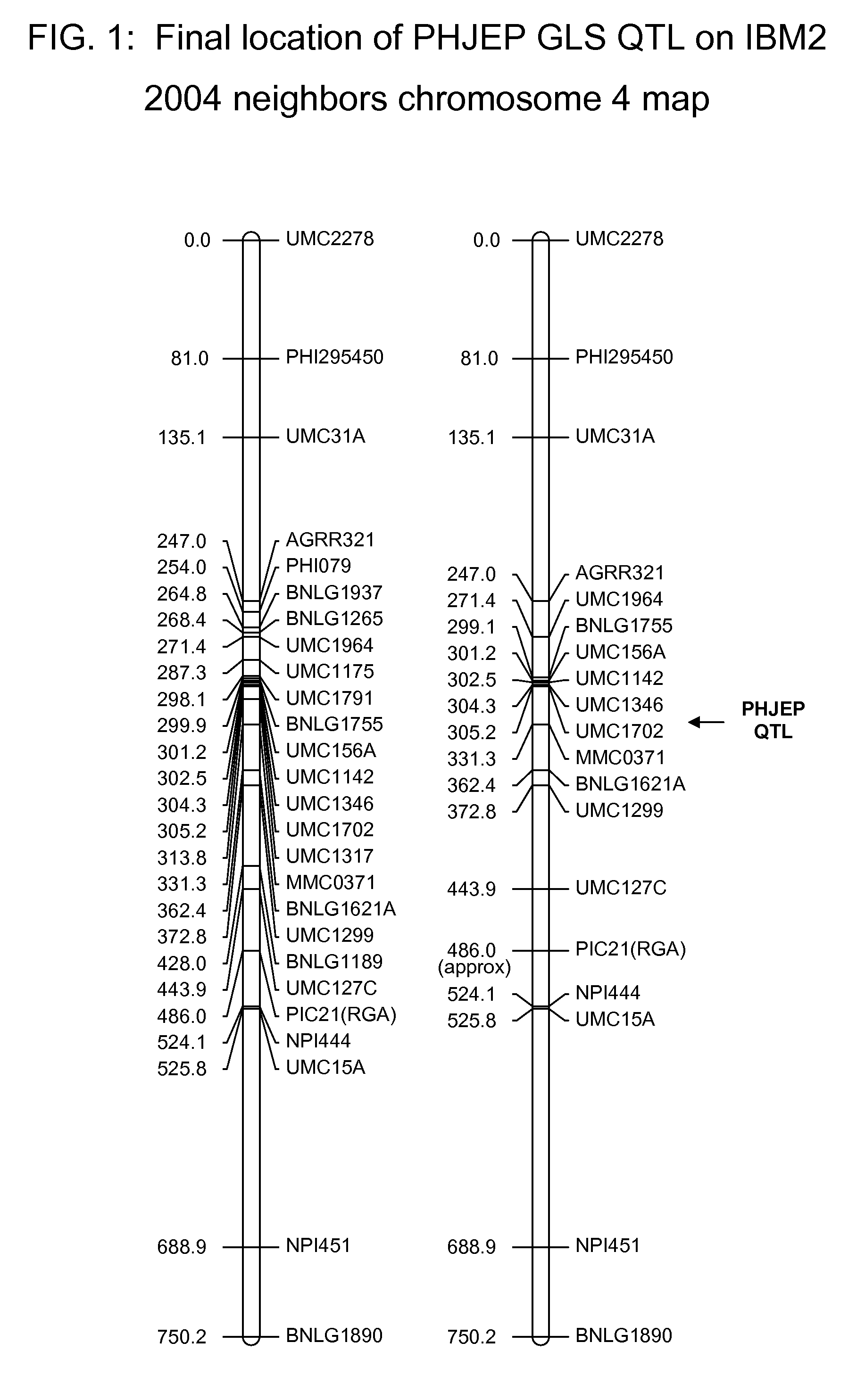
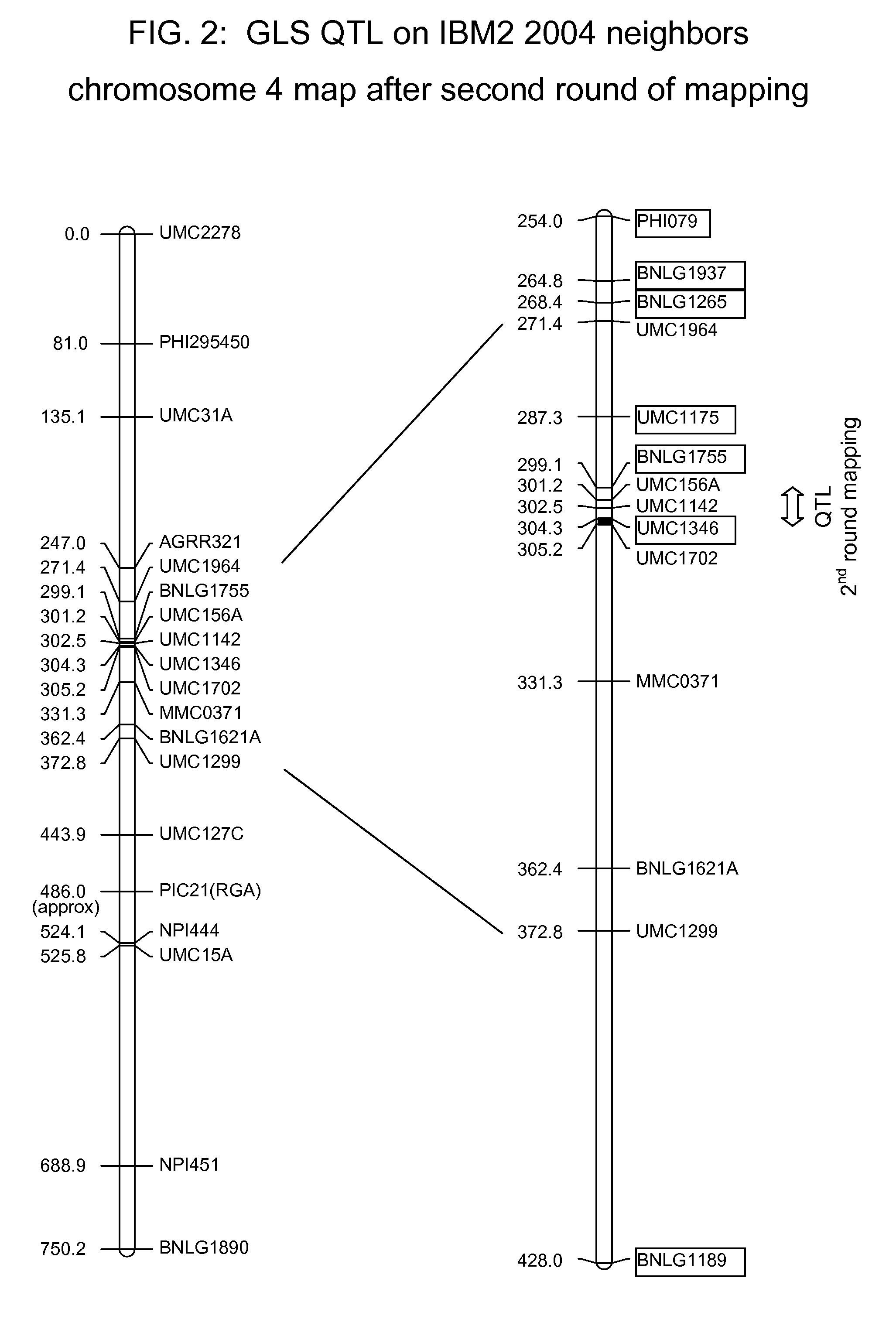
Gray leaf spot tolerant maize and methods of production
ActiveUS20090172845A1Less affectedHigh yieldMicrobiological testing/measurementLibrary member identificationBiotechnologyMolecular genetics
The invention relates to methods and compositions for identifying maize plants that have newly conferred tolerance or enhanced tolerance to, or are susceptible to, Gray Leaf Spot (GLS). The methods use molecular genetic markers to identify, select and / or construct tolerant plants or identify and counter-select susceptible plants. Maize plants that display newly conferred tolerance or enhanced tolerance to GLS that are generated by the methods of the invention are also a feature of the invention.
Owner:CORTEVA AGRISCIENCE LLC +1
Molecular marker method for anti-rice blast gene Pi-hk1 (t) of paddy
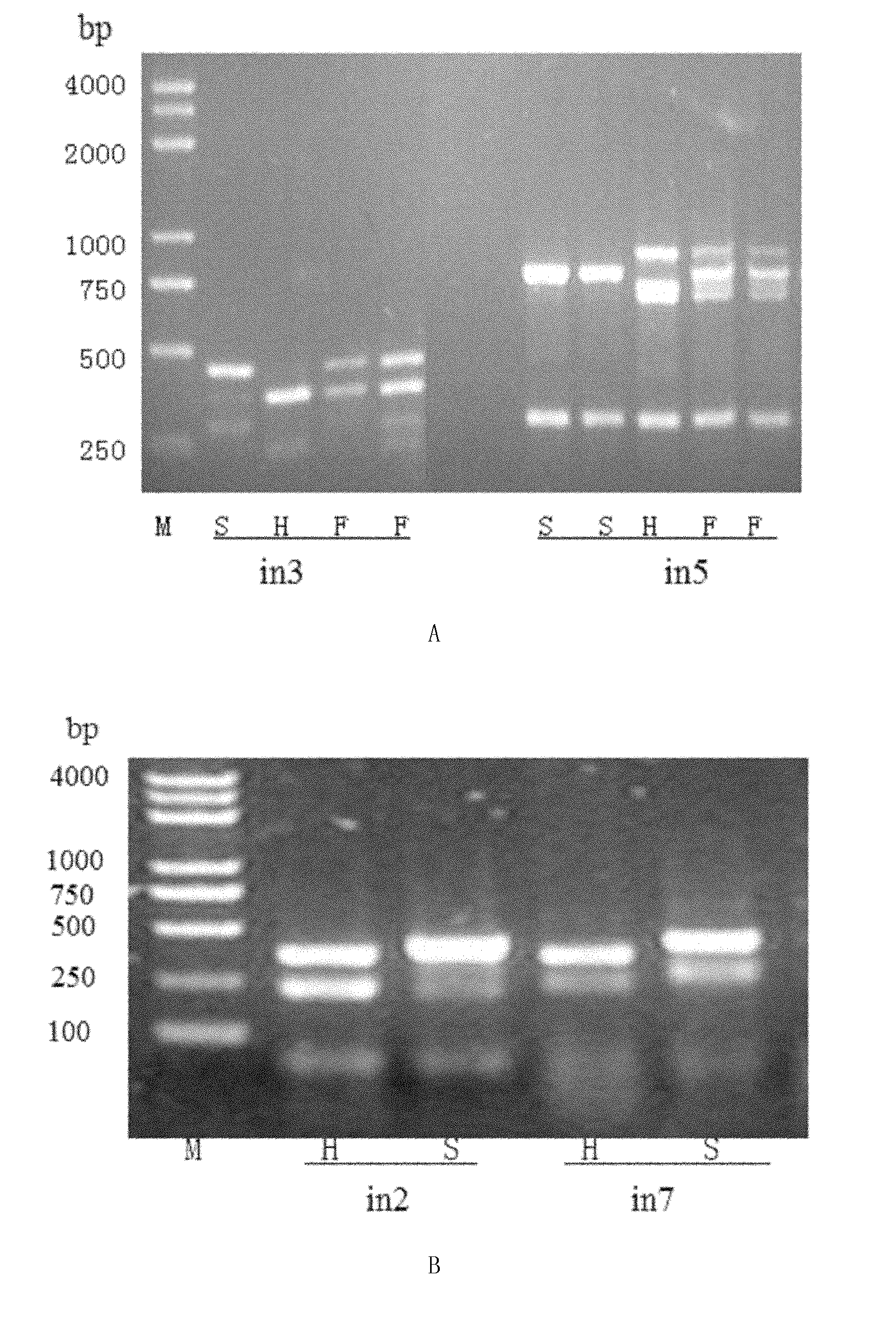
InactiveCN102154470APredicting Rice Blast ResistanceAccurate genetic lociMicrobiological testing/measurementDiseaseElectrophoresis
The invention belongs to the field of molecular genetics, and discloses a molecular marker method for anti-rice blast gene Pi-hk1 (t) of paddy. The method comprises the following steps of: (1) selecting a paddy sample, and extracting genome deoxyribonucleic acid (DNA) of the paddy sample; and (2) performing polymerase chain reaction (PCR) amplification on the genome DNA of the paddy sample by utilizing any one, two or three pairs of molecular marker primers in s01, in5 and in3, performing electrophoretic detection on a PCR amplification product, and if corresponding DNA fragments of molecularmarkers are amplified, indicating that the Pi-hk1 (t) gene exists. In the method, the selection efficiency of a single marker is over 99.92 percent, and the selection efficiency of any double markersis 100 percent. The resistance level of the gene on rice blast can be forecasted by detecting whether the paddy contains the gene or not by the molecular marker primers provided by the invention, so the selection efficiency of the rice blast resistance of the paddy is improved greatly, and the process for breeding for disease resistance is accelerated.
Owner:NANJING AGRICULTURAL UNIVERSITY
Method of screening silky character of chicken by SNP (Single Nucleotide Polymorphism) detection
ActiveCN103667429AImprove the efficiency of breeding and conservationEasy to operateMicrobiological testing/measurementAnimal husbandryGeneticsA-DNA
The invention discloses a method of screening silky character of chicken by SNP (Single Nucleotide Polymorphism) detection. The method comprises the following steps: 1) extracting a DNA (Deoxyribonucleic Acid) sample of chicken to be detected, and carrying out PCR (Polymerase Chain Reaction) amplification by using DNA specific primer pairs to obtain a PCR amplification product; and 2) carrying out SNP detection on the third chromosome 70486623bp site of chicken in the PCR amplification product. The invention provides an efficient, accurate, simple and quick chicken breeding assistant selection molecular genetic mark and provides an efficient molecular mark breeding means for seed selection and retention of silky character of chicken. The detection method disclosed by the invention is simple to operate, low in cost and high in accuracy, and can realize automatic detection.
Owner:CHINA AGRI UNIV
Method for extensively screening scallop SNP
InactiveCN101845489AThe principle is safe and reliableReliable Large-Scale Screening RunsMicrobiological testing/measurementMolecular geneticsPhenol
The invention belongs to a method for extensively screening scallop SNP in the technical field of the scallop DNA molecular genetic marker, which comprises the following steps that: solution D and phenol-chloroform are firstly used for extracting a plurality of Chlamys farreri RNA, and a ultraviolet specrophotometer is used for measuring the concentration of the mixture of solution D and the phenol-chloroform which are uniformly mixed; a scallop full-length cDNA sample is re-established and comprises the synthesis of a cDNA first chain and the synthesis and augmentation of a cDNA second chain; then homogenization of full-length cDNA is performed, and ultrasonic breaking is performed; then sequence measuring joints are connected, and terminal repair, joint connection and sample augmentation are included; finally a biological software is used for analyze the data to obtain an SNP marker; then an SNP marker to be screened is selected to design a primer; and DNA of individual scallop is extracted to be equivalently mixed to be used as a template, conventional colony sequence measuring is undertaken after the PCR augmentation, and the position point with the occurring frequency of single base difference being 10 percent or more is defined as an SNP marker. The method has safe and reliable principle, simple and controllable flow procedures, reliable running of large-scale screening, excellent effect and strong practicability.
Owner:OCEAN UNIV OF CHINA
SNP (single nucleotide polymorphism) marker for evaluating growth performance of ctenopharyngodonidella, primer and evaluation method
ActiveCN106755527AEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationGenetic exchangeIntein
The invention provides an SNP (single nucleotide polymorphism) marker (comprising SNP1 and SNP4) for evaluating growth performance of ctenopharyngodonidella and a primer pair for amplifying a gene segment where an SNP locus is positioned by virtue of methods of molecular genetics and molecular biology. SNP1 is positioned at the 940th locus in a promoter of a complete sequence of a MyoD (myogenic determining factor) gene of the ctenopharyngodonidella, and a base T is inserted or deficient at the position; SNP4 is positioned in a 107th locus in a first intron of the complete sequence of the MyoD gene of the ctenopharyngodonidella, and a base at the position is A or T. The growth performance of the ctenopharyngodonidella is determined by detecting haplotypes of the two SNP loci. The adopted haplotypes are mutated according to bases generated in the MyoD gene, so that genetic exchange and further phenotype verification are avoided. By virtue of the haplotypes of the SNP marker, rapidly growing ctenopharyngodonidella can be simply and rapidly identified, and rapidly growing ctenopharyngodonidella of a new line can also be guided to be bred.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
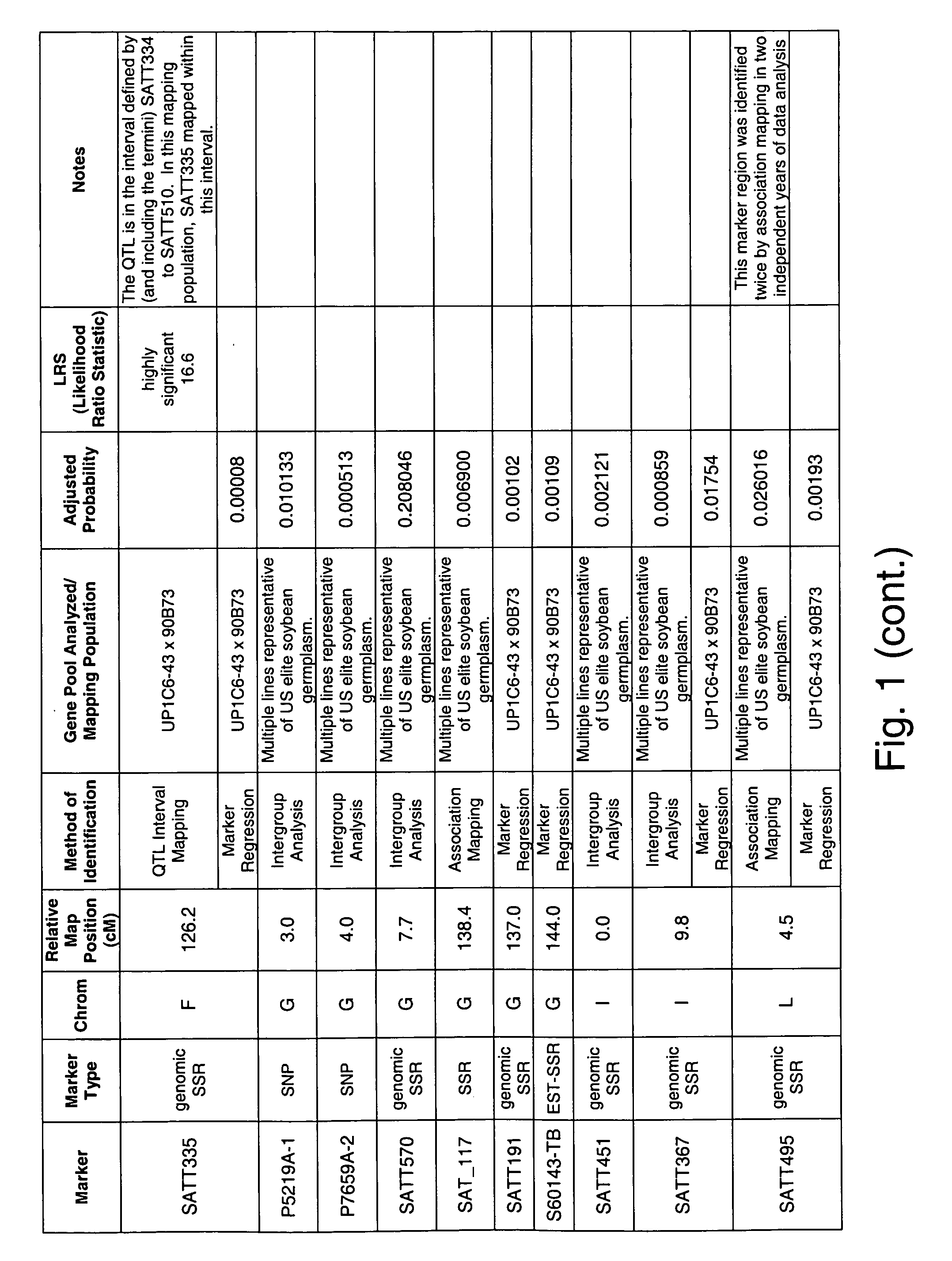
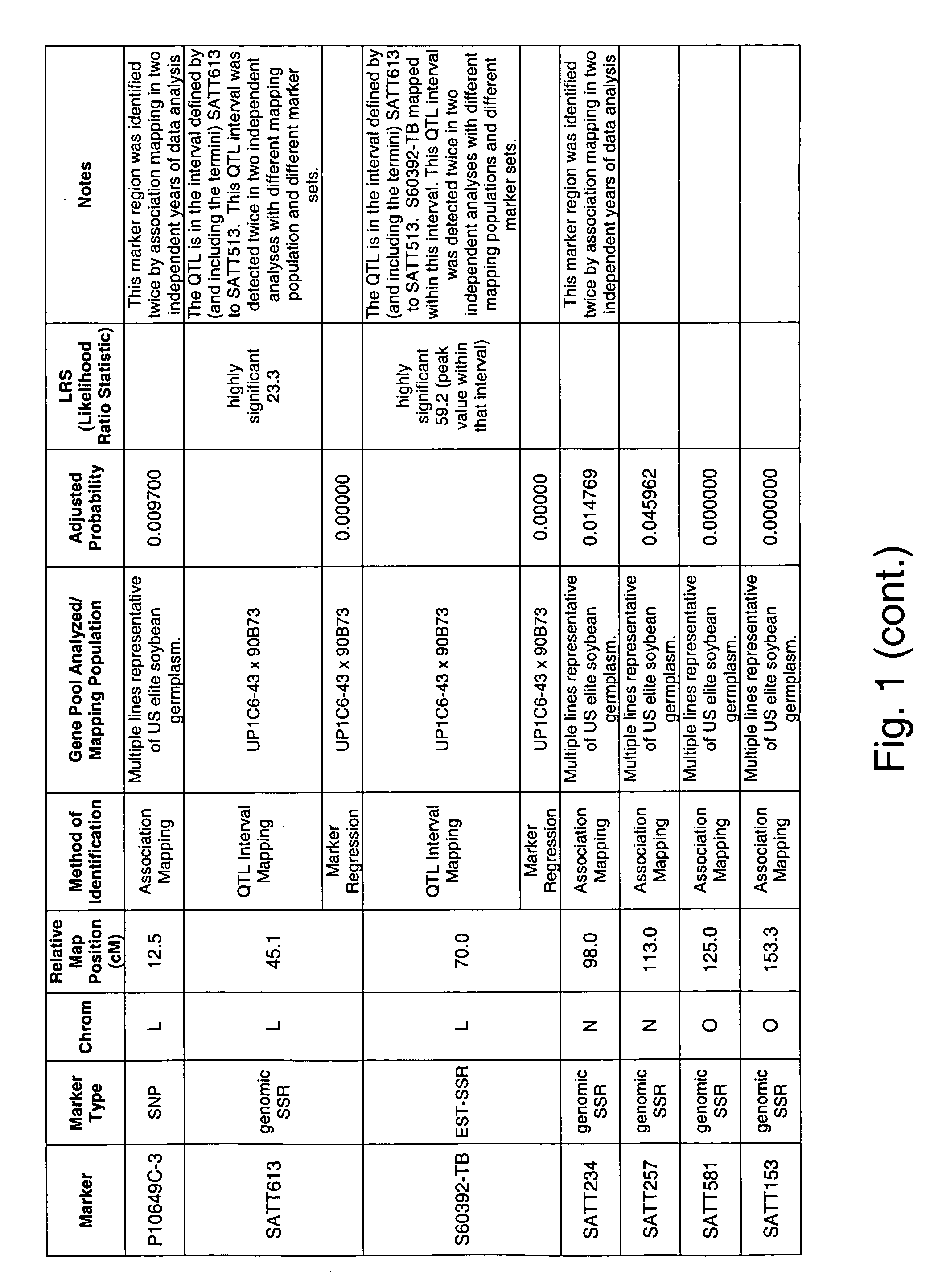
Genetic loci associated with iron deficiency tolerance in soybean
InactiveUS20060041951A1Improve toleranceIncreased susceptibilityMicrobiological testing/measurementOther foreign material introduction processesIron deficientPhytophthora sp.
The invention relates to methods and compositions for identifying soybean plants that are tolerant, have improved tolerance or are susceptible to iron deficient growth conditions. The methods use molecular genetic markers to identify, select and / or construct disease-tolerant plants or identify and counterselect disease-susceptible plants. Soybean plants that display tolerance or improved tolerance to Phytophthora root rot infection that are generated by the methods of the invention are also a feature of the invention.
Owner:PIONEER HI BRED INT INC
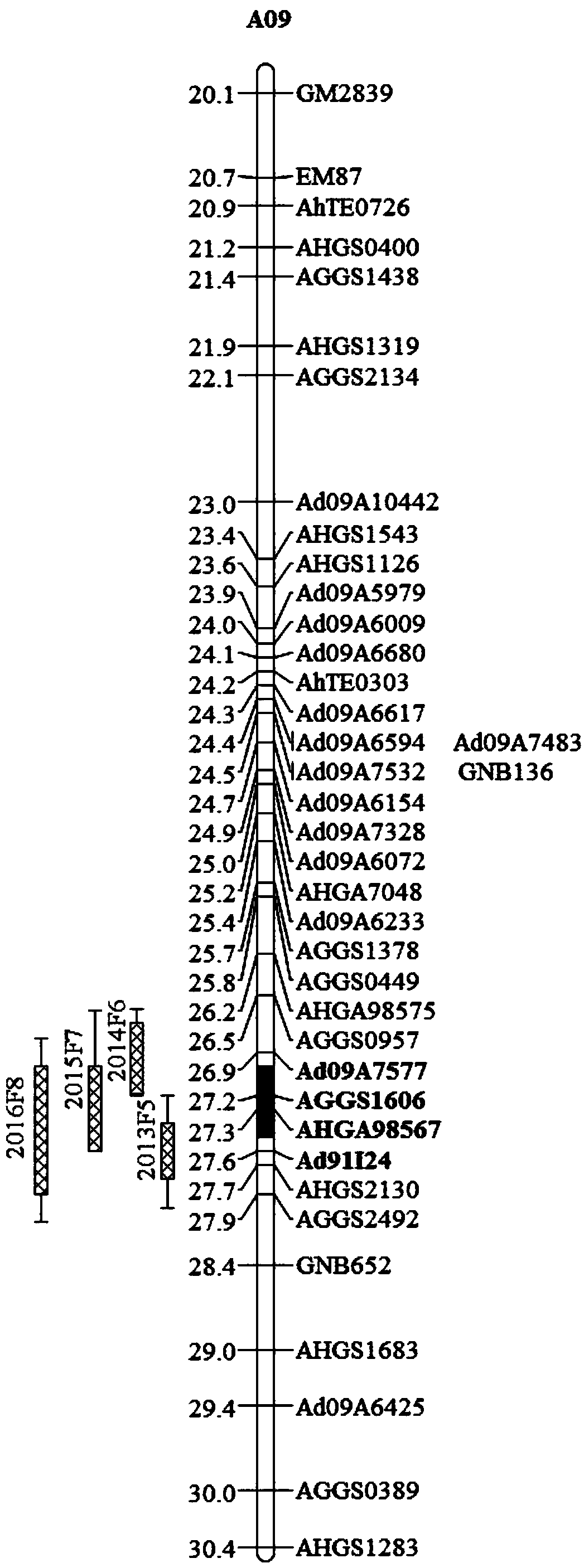
Molecular markers for peanut kernel yield major QTL (quantitative trait locus) and application thereof
ActiveCN108374053AConducive to the promotion of breedingImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationQuantitative trait locusElectrophoresis
The invention belongs to the field of molecular genetic breeding and particularly relates to molecular markers for peanut kernel yield major QTL (quantitative trait locus) and application thereof. Themolecular markers include Ad09A7577, AGGS1606, AHGA98567 and Ad91I24; one or more of primer pairs of the molecular markers Ad09A7577, AGGS1606, AHGA98567 and Ad91I24 are used to perform PCR (polymerase chain reaction) amplification on DNA of peanut material under identification, the amplified product is subjected to electrophoretic detection; if a DNA fragment of corresponding size is obtained byamplifying, it is shown that kernel yield major QTL cqSPA09 is present, and pods have high kernel yield.
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
Molecular genetic maker for egg quality of laying hen and application of molecular genetic marker
InactiveCN106701919AHigh precisionEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionNucleotide
The invention belongs to the technical field of laying hen molecular genetic marker assisted selecting, and particularly relates to a molecular genetic marker for egg quality of a laying hen and application of the molecular genetic marker. The molecular genetic marker is a partial sequence of genes VLDLR and CTSD of the laying hen, and a VLDLR nucleotide sequence is as shown in SEQ ID No.1, and VLDLR nucleotide sequences are as shown in SEQ ID No.2-4. The molecular making method is utilized for selecting egg quality characters, so that a molecular marking method which is more effective, simpler and more convenient to implement is provided for marker assisted selecting in chicken breeding work; meanwhile, the molecular genetic marker can overcome the defects that the normal egg quality character selecting method is great in workload, is easily affected by the environment, is long in breeding period and the like, and provides an effective molecular marker breeding means for egg quality character improving, so that genetic progress of chicken is quickened.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
SNP (single nucleotide polymorphism) marker and method for identifying MC1R (melanocortin receptor 1) genes of pig breeds and colors of pig breeds as well as application
PendingCN105648071AThe screening method is accurateEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationUnknown SourceScreening method
The invention discloses an SNP (single nucleotide polymorphism) marker for identifying MC1R (melanocortin receptor 1) genes of pig breeds and colors of the pig breeds and further discloses an SNP marker primer for identifying the MC1R genes of the pig breeds and the colors of the pig breeds as well as an SNP marker method for identifying the MC1R genes of the pig breeds and the colors of the pig breeds. The molecular genetic marker is not limited by ages, sexes and the like of pigs, can be applied to early breeding of pure breeds of local black hogs and can further be used for performing pig breed and color identification and genetic relationship deduction on pork samples from unknown sources. A screening method is accurate, operation is simple, the cost is low, and the efficiency is high.
Owner:KUNMING UNIV +1
Method for detecting chicken fatty character
InactiveCN1900316AEasy to breedImprove conversion rateMicrobiological testing/measurementMarker-assisted selectionMolecular breeding
The method of detecting chicken fatty character is to detect whether the 646-th base from the 5' end of chicken PGC-1alpha gene cDNA is A or G. The present invention provides one effective, accurate, simple and feasible molecular genetic marker for the auxiliary molecular mark selection of meat chicken breeding and the improvement of chicken's abdomen fatty character. The detection method is simple, low in cost and accurate.
Owner:CHINA AGRI UNIV
Methods and compositions for diagnosis of hyperhomocysteinemia
InactiveUS6132965AReduce potential steric hindrancePromote resultsSugar derivativesMicrobiological testing/measurementHyperhomocysteinemiaMolecular genetics
Owner:HAMILTON CIVIC HOSPITALS RESARCH DEV
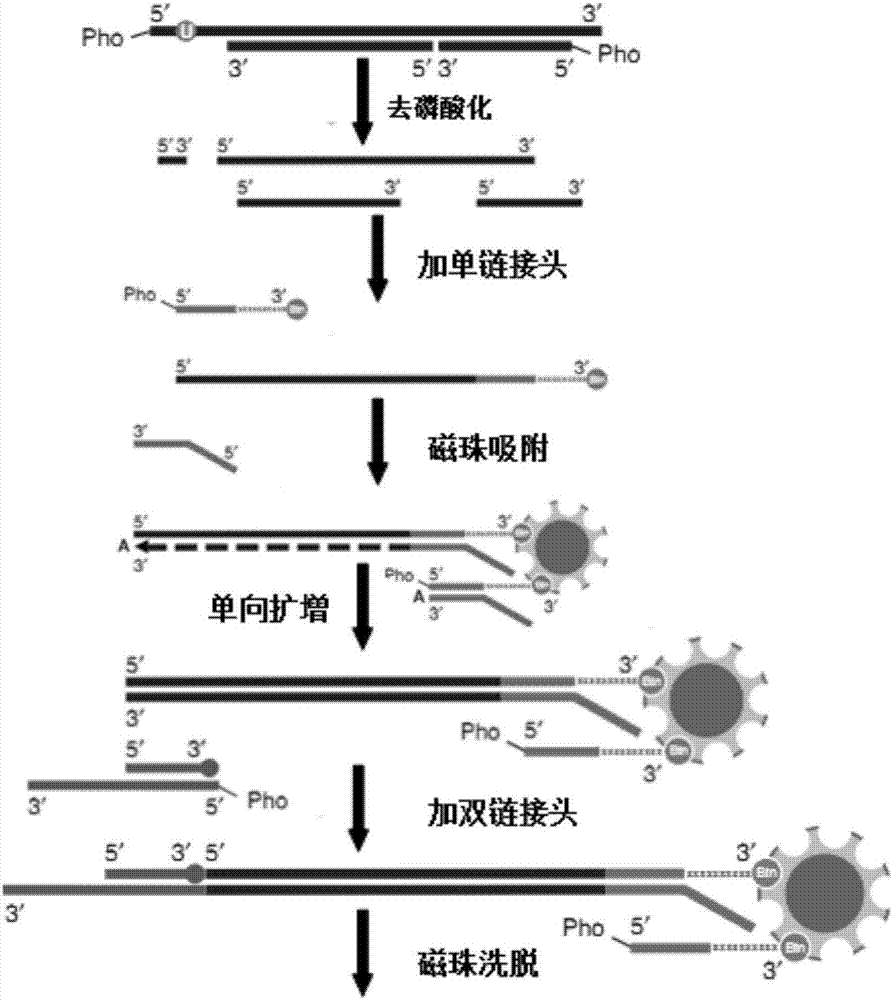
Linker, primer group and kit for cfDNA library construction and library construction method
InactiveCN106867995AAvoid lossEasy to operateNucleotide librariesMicrobiological testing/measurementPhosphorylationMolecular genetics
The invention belongs to field of molecular genetics, and particularly relates to a linker, a primer group and a kit for cfDNA library construction and a library construction method. The linker comprises a single-chain linker A and a double-chain linker B; the primer group comprises a sequence shown as SEQ ID NO.4-6; the kit comprises the linker and the primer group; the library construction method comprises the following steps: (1) dephosphorylating and denaturating cfDNA, to obtain the single-chain cfDNA; (2) connecting the single-chain cfDNA with a single-chain linker A with a modified 3'terminal, to obtain a single-chain connecting product with a modified 3'terminal; (3) unidirectionally amplifying a first-time connecting product, to obtain a double-chain cfDNA; (4) connecting the double-chain cfDNA with a double-chain linker B, to obtain a double-chain connecting product; (5) removing one chain with modified 3'terminal in the double-chain connecting product; and (6) performing library quality inspection. According to the linker, the primer group and the kit for cfDNA library construction and library construction method, the cfDNA is firstly denaturated so that the double chain is changed to single chain, then the single chain is connected with the linker through a single-chain connecting enzyme so as to cause almost all cfDNA to be captured, the loss of cfDNA can be avoided, and further the detection sensitivity can be improved.
Owner:ANHUI ANKE BIOTECHNOLOGY (GRP) CO LTD
Bovine serum microRNA molecular marker of milk cow fatty liver disease and milk cow perinatal period-associated metabolic disease
ActiveCN105420405AAid in early detectionAids in diagnosisMicrobiological testing/measurementDNA/RNA fragmentationFatty liverAnimal science
The invention relates to molecular genetics, and provides a bovine serum microRNA molecular marker of a milk cow fatty liver disease and a milk cow perinatal period-associated metabolic disease. The molecular marker is used for early detection and diagnosis of a diseased cow in production practice of the milk cow, thus greatly improving diagnosis accuracy and greatly avoiding economic loss to the great extent. The marker can be used for variety breeding and idioplasm improvement of the milk cow.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Molecular marker of wheat powdery mildew disease-resistant genes Pm51 and application of molecular marker
ActiveCN104313021AImprove utilizationRapid identificationMicrobiological testing/measurementDNA/RNA fragmentationDiseaseBiotechnology
The invention relates to plant molecular genetics and wheat breeding and in particular relates to a molecular marker of novel wheat powdery mildew disease-resistant genes Pm51 and application of the molecular marker. A functional molecular marker BQ246670 linked with powdery mildew disease-resistant genes Pm51 is obtained. The marker is a co-dominant marker, the genetic distance between the marker and the Pm51 is 1.5cM, the existence and existential state of wheat genes Pm51 can be rapidly and accurately identified, and the resistance of wheat powdery mildew disease is predicted, so that the utilization of the wheat powdery mildew disease-resistant genes Pm51 is accelerated.
Owner:SHANXI UNIV OF CHINESE MEDICINE
Genetic loci associated with Fusarium solani tolerance in soybean
ActiveUS7767882B2Rapid lossLosses in viabilityMicrobiological testing/measurementGenetic engineeringBiotechnologyFUSARIUM INFECTIONS
The invention relates to methods and compositions for identifying soybean plants that are tolerant, have improved tolerance or are susceptible to Fusarium solani infection (the causative agent of sudden death syndrome or SDS). The methods use molecular genetic markers to identify, select and / or construct disease-tolerant plants or identify and counterselect disease-susceptible plants. Soybean plants that display tolerance or improved tolerance to Fusarium solani infection that are generated by the methods of the invention are also a feature of the invention.
Owner:PIONEER HI BRED INT INC
Multiple controls for molecular genetic analyses
InactiveUS20050227229A1Less stringentMinimize the numberMicrobiological testing/measurementFermentationPositive controlGenomic DNA
A method for constructing multiple nucleic acid sequences for use as positive controls in a genetic test is described. Compositions according to the invention including multiple nucleic acid sequences constructed as described are the optimal controls for simultaneously testing multiple variable nucleic acid sequences at one or more DNA or RNA sites in a subject or subjects. Sequences according to the invention can be prepared chemically and / or by PCR amplification for use directly or after cloning and propagation. At the same time, some sequences can be PCR amplified and / or cloned directly from total genomic DNA obtained from an individual carrying the mutation or variant. Alternatively, the normal sequence to be changed can be cloned and then modified by site directed mutagenesis. Several single mutant or polymorphic sequences that together comprise a panel of multiple control sequences can be added individually to single site tests or mixed together or ligated together by further PCR or by cloning into vectors prior to use in individual or multiplex tests. Controls sequences constructed according to the invention can be used when testing any genetically transmitted nucleic acid sequence by organizations testing quality assurance and by companies maintaining quality control of manufactured genetic test kits.
Owner:CHILDRENS HOSPITAL MEDICAL CENT OF AKRON
Molecular genetic marker associated with egg-laying traits of ducks and application of molecular genetic marker
ActiveCN106755326AImplement early selectionFast detection methodMicrobiological testing/measurementDNA/RNA fragmentationTypingLaying duck
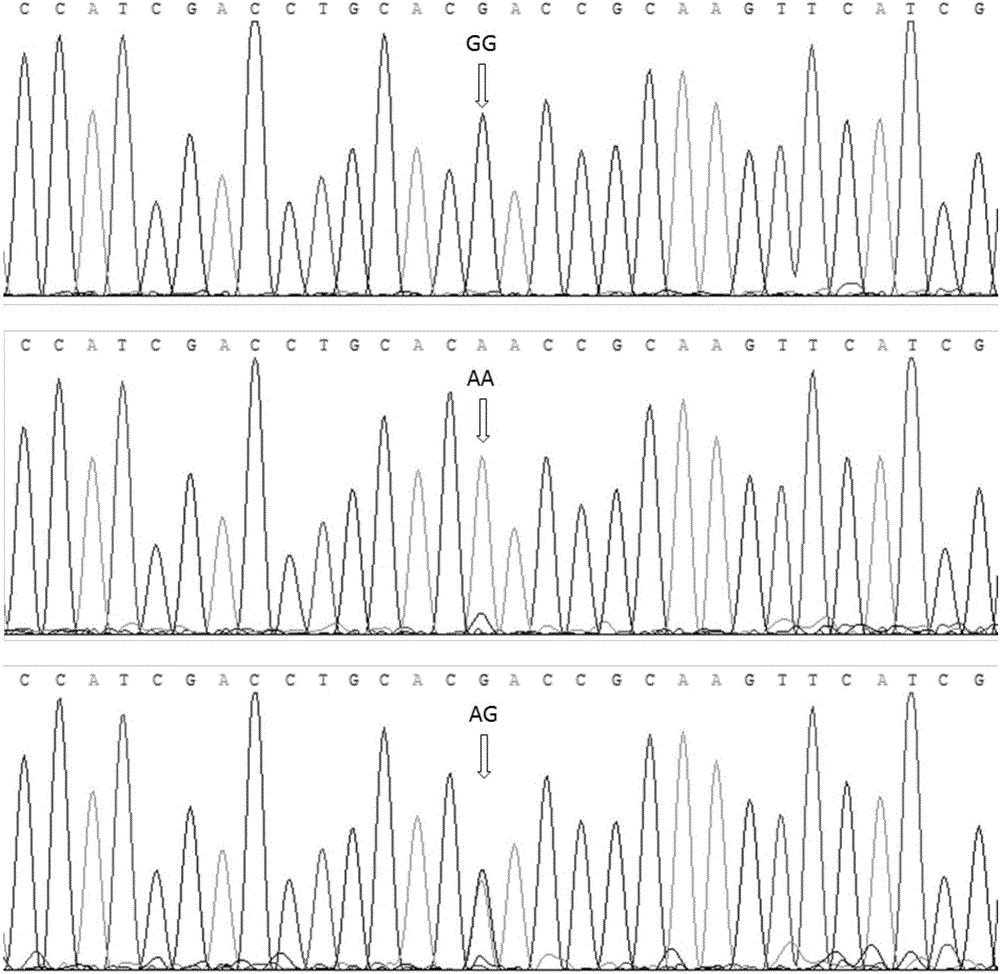
The invention relates to a molecular genetic marker associated with egg-laying traits of ducks and application of the molecular genetic marker, belongs to the field of poultry breeding, and particularly relates to application of a G575A site of an AMH (anti-Mullerian hormone) gene, associated with egg-laying traits of ducks, serving as a molecular genetic marker in duck production performance research. A specific DNA segment is obtained from the AMH gene to serve as a molecular marker associated with the egg-laying traits of the ducks, one G575-A575 basic group in a sequence of the molecular marker mutates, the mutation site is significantly associated with the 43-week-old egg production and the 72-week-old egg production of the ducks (P smaller than 0.05), and by performing PCR (polymerase chain reaction) amplification and performing sequencing-based typing on a PCR product, a GG genotype is selected as an laying duck. A detection method provided by the invention is significant in effect, is simple and quick, and is not influenced by an external environment.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
Method for screening novel restoring gene for HL cytoplasm male sterile rice
InactiveCN101899522AFast wayAccurate methodMicrobiological testing/measurementPlant genotype modificationBand patternMolecular genetics
The invention discloses a method for screening a novel restoring gene for HL cytoplasm male sterile rice. The method comprises the following steps of: firstly, designing a primer of a restoring gene coseparation molecular marker and establishing a standard polymerase chain reaction (PCR) amplification system; secondly, comparing and analyzing a molecular marker amplification banding pattern and screening a rice variety or an ecotype having different molecular markers; and finally, verifying a new restoring gene locus by using genetic analysis and determining the novel restoring gene. In the method, the conventional genetic analysis is combined with molecular genetic marker technology, and a new gene resource is developed and utilized purposefully. The method has the characteristics of simple and convenient operation, rapidness, correctness and accurate selection of a novel restorer and is favorable for expanding restoring spectrum, improving the polymerization selection efficiency of excellent recovering genes and promoting the maturing rate, yield and sustainable development of hybrid rice.
Owner:WUHAN UNIV
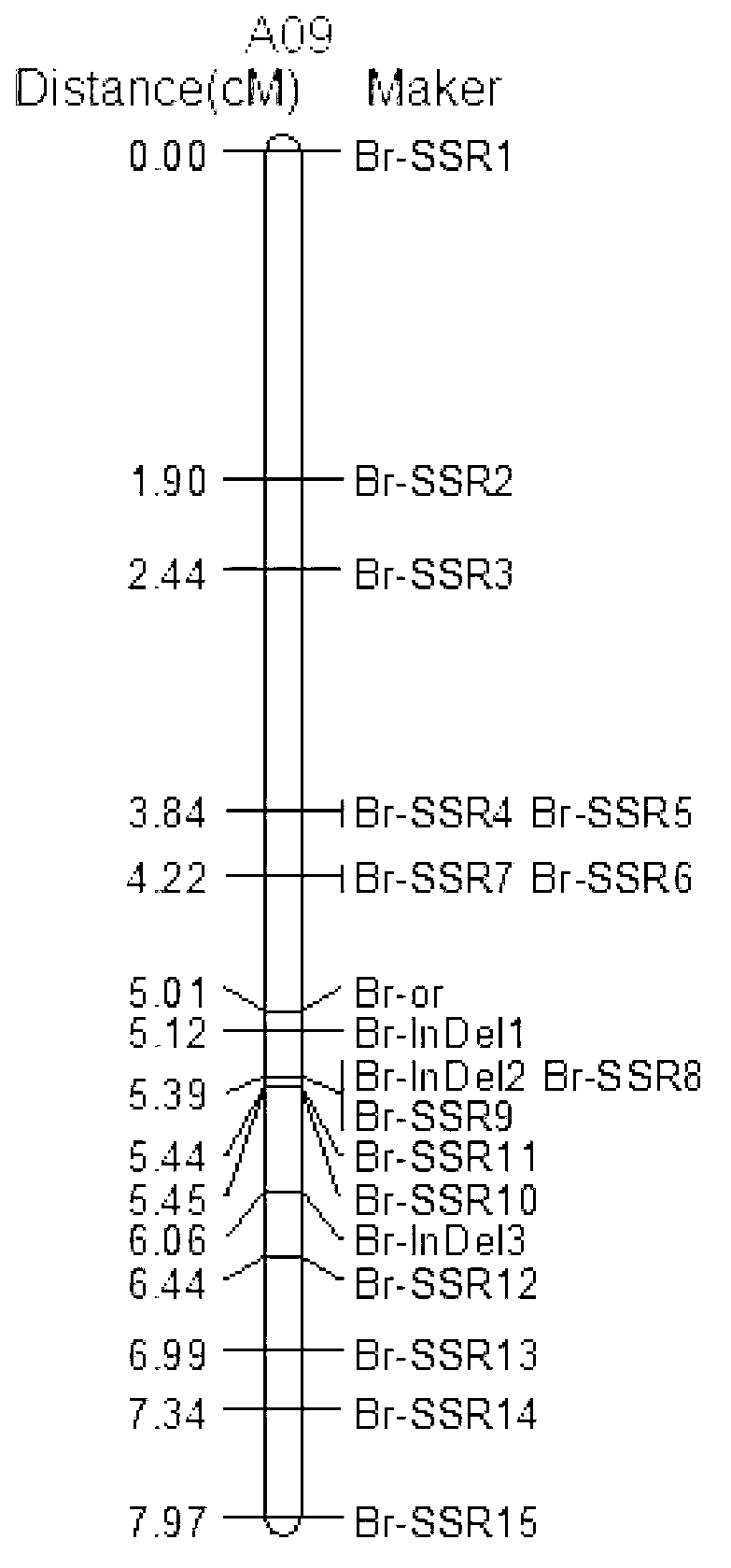
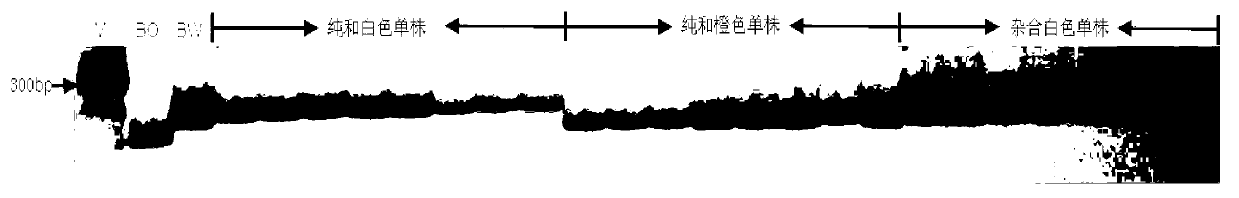
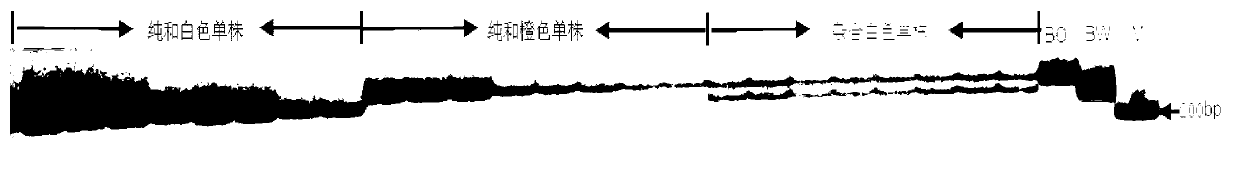
SSR (Simples sequence repeats) and InDel (insertion/deletion) molecular marker primer linked with brassica campestris orange head gene Br-or, and application thereof
InactiveCN103103184AEasy to detectStable amplificationMicrobiological testing/measurementDNA/RNA fragmentationBrassicaAgricultural science
The invention discloses an SSR (simples sequence repeats) and InDel (insertion / deletion) molecular marker primer linked with brassica campestris orange head gene Br-or and an application of the molecular marker primer. The marker primer is obtained by the steps of: based on the genome DNA (deoxyribonucleic acid) of orange brassica campestris and normal white brassica campestris and F2S4 separation group as a template, carrying out polymorphism primer screening to orange head and white head single strain DNA mixed tank in F2S4 by using 480 pairs of SSR and InDel primer pairs, then analyzing single strain of F2S4 group, and screening the molecular marker linked with the brassica campestris orange head gene Br-or to finally obtain fifteen Br-SSR and three Br-InDel molecular markers linked with the Br-or gene, wherein a molecular genetic map of the brassica campestris orange head gene Br-or is created on the ninth link group (A09). According to the link analysis, the genetic distance of two markers most tightly linked with both sides of the Br-or gene are 0.11cM and 0.79cM; and the molecular marker primer has the advantages of being convenient for detection, stable in amplification, and high in repeatability and accuracy, and thus a basis is provided to the molecule assistant selective breeding of brassica campestris orange head character and the breeding process is accelerated.
Owner:NORTHWEST A & F UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com