Linker, primer group and kit for cfDNA library construction and library construction method
A kit and primer set technology, applied in the field of molecular genetics, can solve the problems of limited detection sensitivity, cfDNA cannot be filled with junctions, low cfDNA content, etc., and achieve the effects of improving detection sensitivity, stable sequencing results, and low sequencing costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
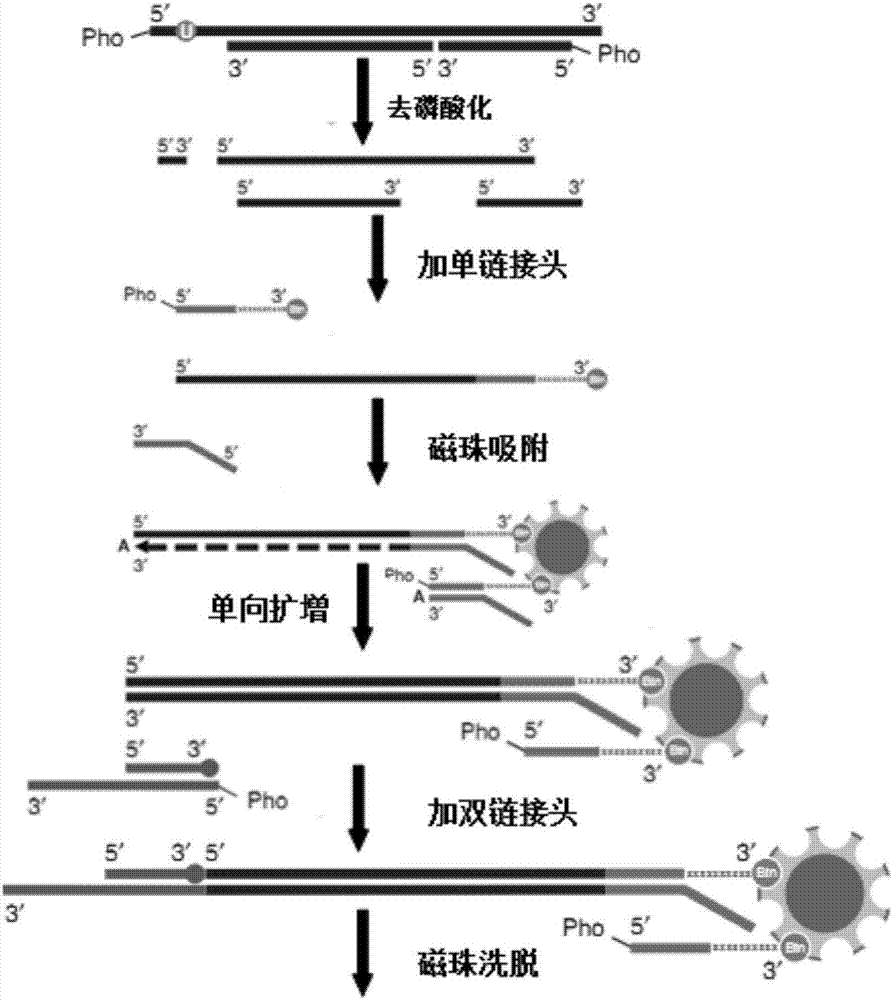
[0023] The invention provides a cfDNA library construction kit product, which includes:
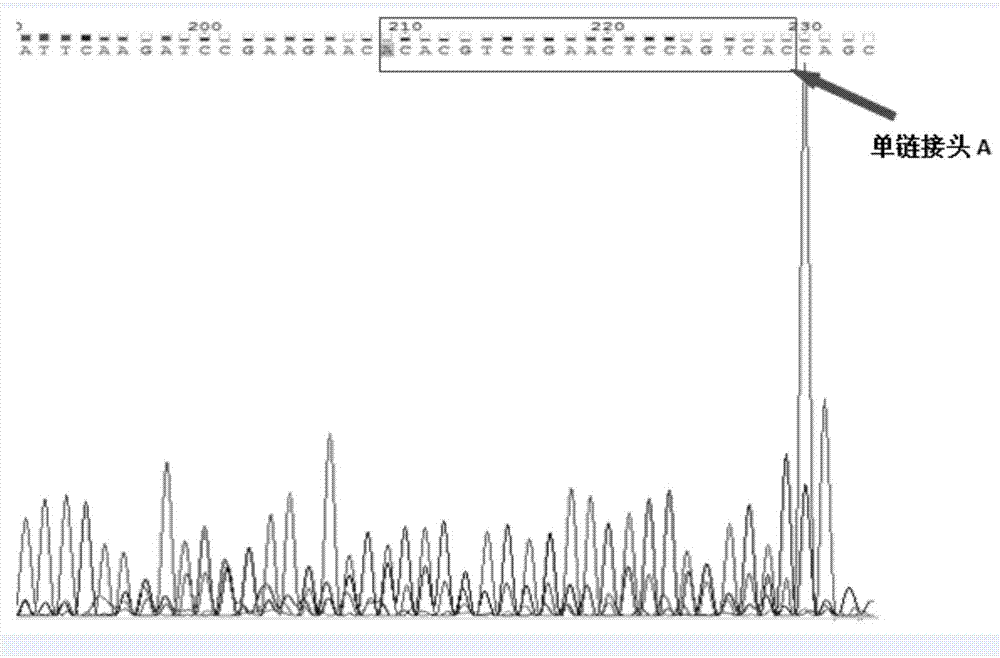
[0024] A single-stranded linker A for connecting and labeling the denatured single-stranded cfDNA, the 3' end of the single-stranded linker A is modified with biotin;
[0025] Single-stranded ligase for ligation of single-stranded cfDNA and single-stranded adapter A and buffer for single-stranded ligase;
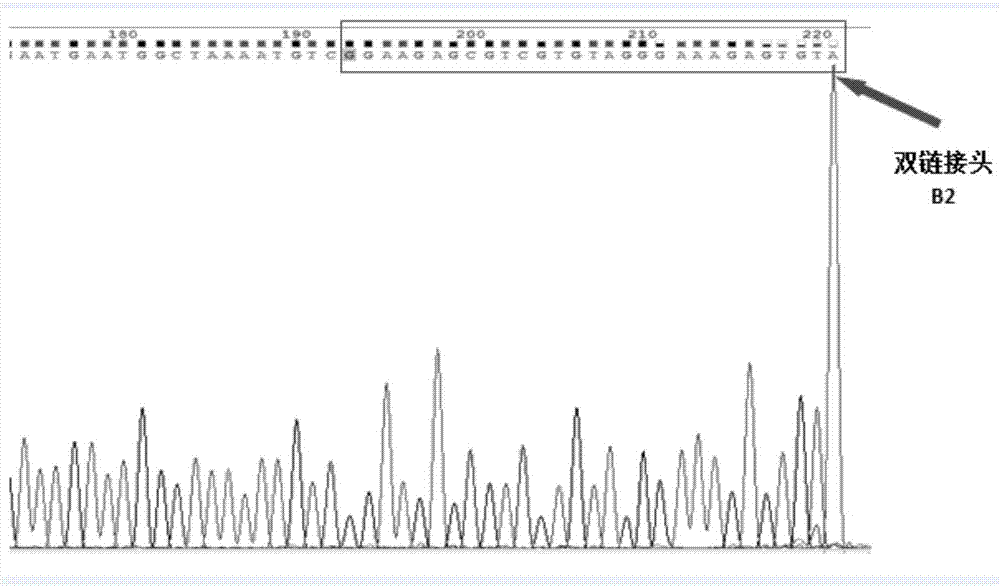
[0026] A double-stranded adapter B for connecting double-stranded cfDNA, said double-stranded adapter B comprising adapter B1 and adapter B2;
[0027] T4 DNA ligase for ligation of double-stranded adapter B of double-stranded cfDNA and buffer for T4 DNA ligase;
[0028] Primer CL9 for unidirectional amplification of single-stranded cfDNA and primers IS7 and IS8 for amplifying library molecules for library detection:
[0029]
[0030] Wherein, the double connector B is made by the following method:
[0031] Perform PCR on the following system, react at 95°C for 10S, and then cool d...
Embodiment 2
[0054] A method for building a cfDNA library, comprising the steps of:
[0055] (1) Dephosphorylate and denature cfDNA to obtain single-stranded cfDNA;
[0056] (a1): According to the instructions for use of the QIAamp Circulating Nucleic Acid Kit (50) kit provided by QIAGEN, use the kit to extract free cfDNA in plasma;
[0057] (b1): Add the following components to the PCR tube:
[0058]
[0059] Using a PCR instrument, first react at 37°C for 10 minutes, then react at 95°C for 2 minutes;
[0060] Take out the PCR tube, quickly put it into the ice-water mixture, keep it in the ice bath for at least 1min, take it out, centrifuge it, and store it at room temperature.
[0061] (2) Ligate the single-stranded cfDNA and the single-stranded linker A modified at the 3' end to obtain the single-stranded ligation product modified at the 3' end
[0062] (a2): Add the following components to the PCR tube:
[0063]
[0064]
[0065] Use a PCR instrument, react at 60°C for 1 h...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com