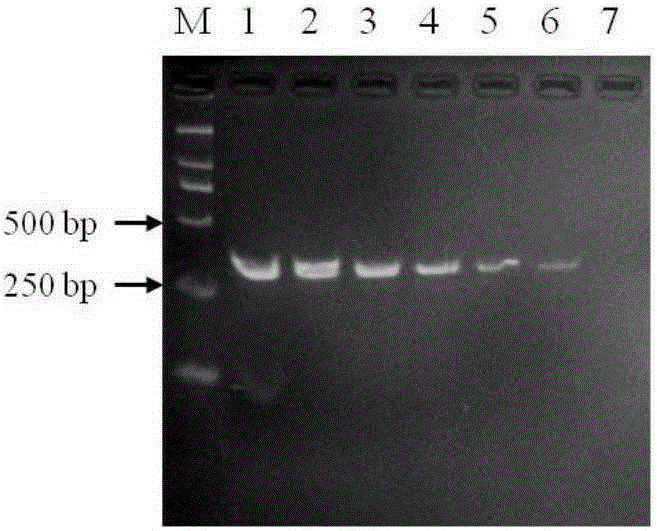
Patents
Literature
282results about How to "Shorten identification time" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
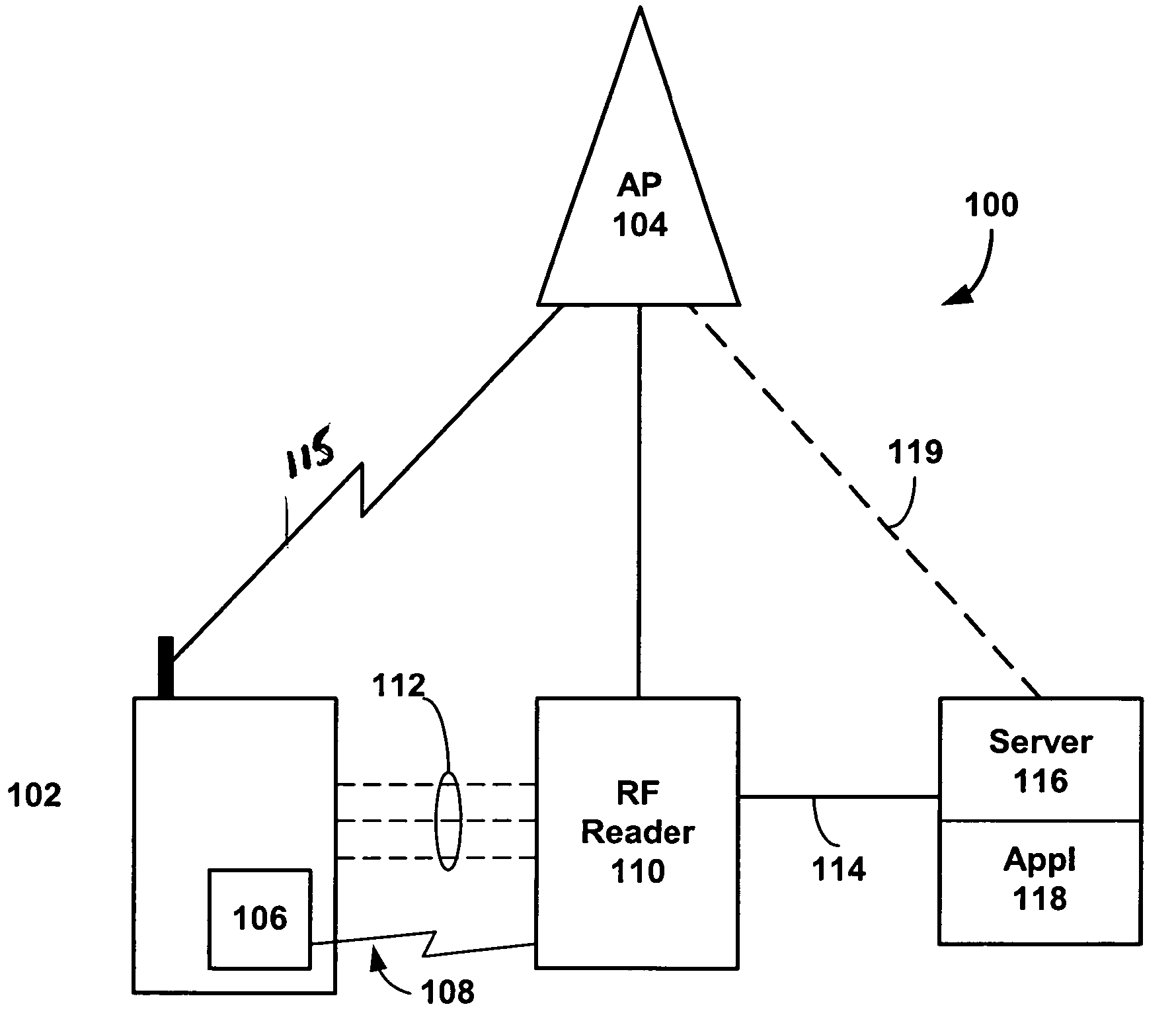
Radio frequency identification (rf-id) based discovery for short range radio communication
InactiveUS20050079817A1Shorten identification timeShorten session set-up timeConnection managementData switching by path configurationRf fieldConnectionless communication
A RF-ID based wireless terminal has shortened session set-up and user identification time for conducting transactions with interactive service applications. The wireless terminal includes a terminal identification number and a user identification as a RF-ID tag. A RF-ID reader transmits a RF field for detecting the RF-ID tag in the terminal and provides an output signal when the terminal is within the reader field. The output signal establishes a connectionless communication to an access point or other terminal which initiates a wireless paging operation, in lieu of conducting a terminal discovery process, based upon the content of the RF-ID tag. The terminal initiates a wireless session between the terminal and the access point or terminal for conducting transactions with a service application linked to the access point or terminal.
Owner:NOKIA TECHNOLOGLES OY
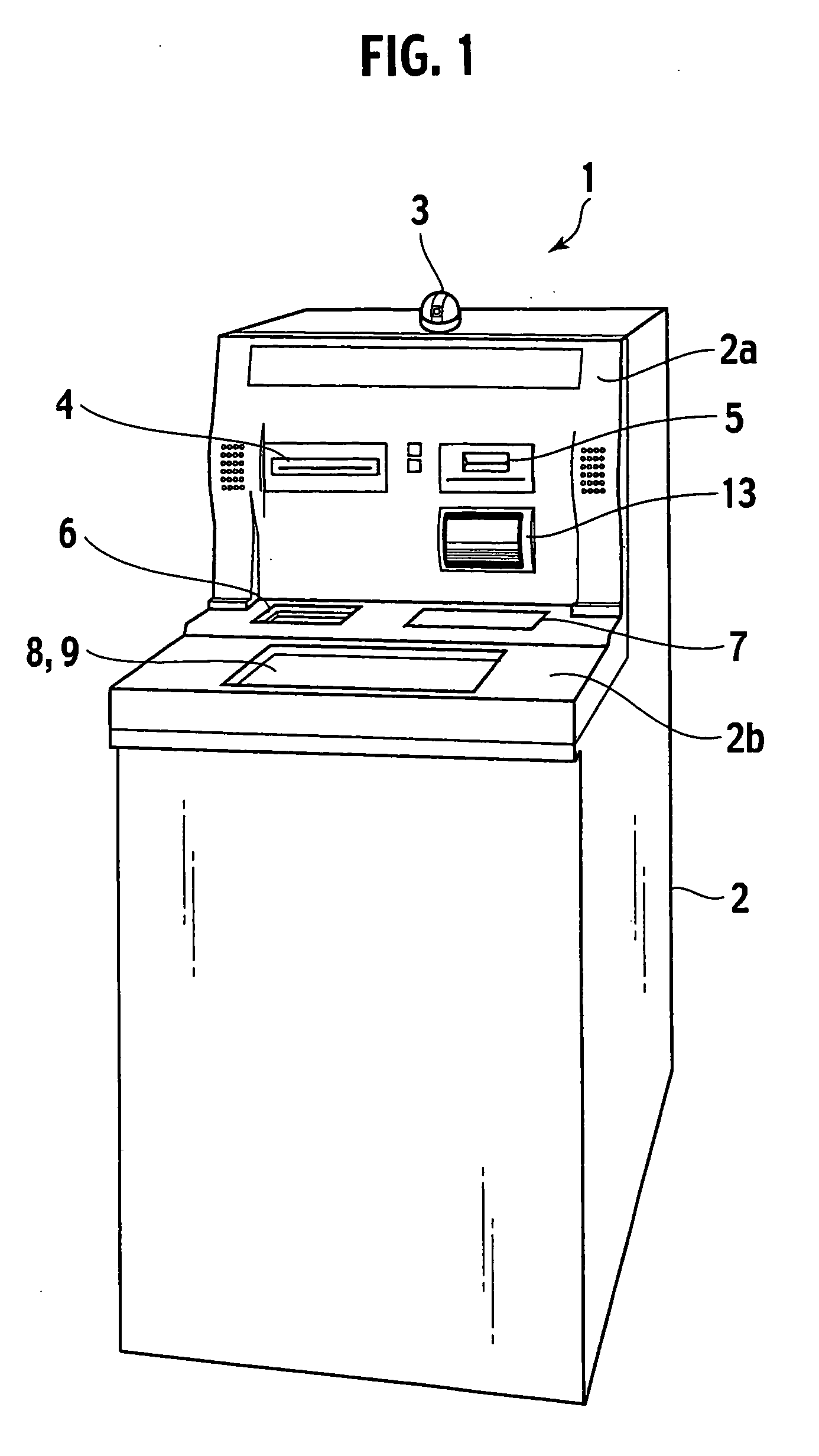
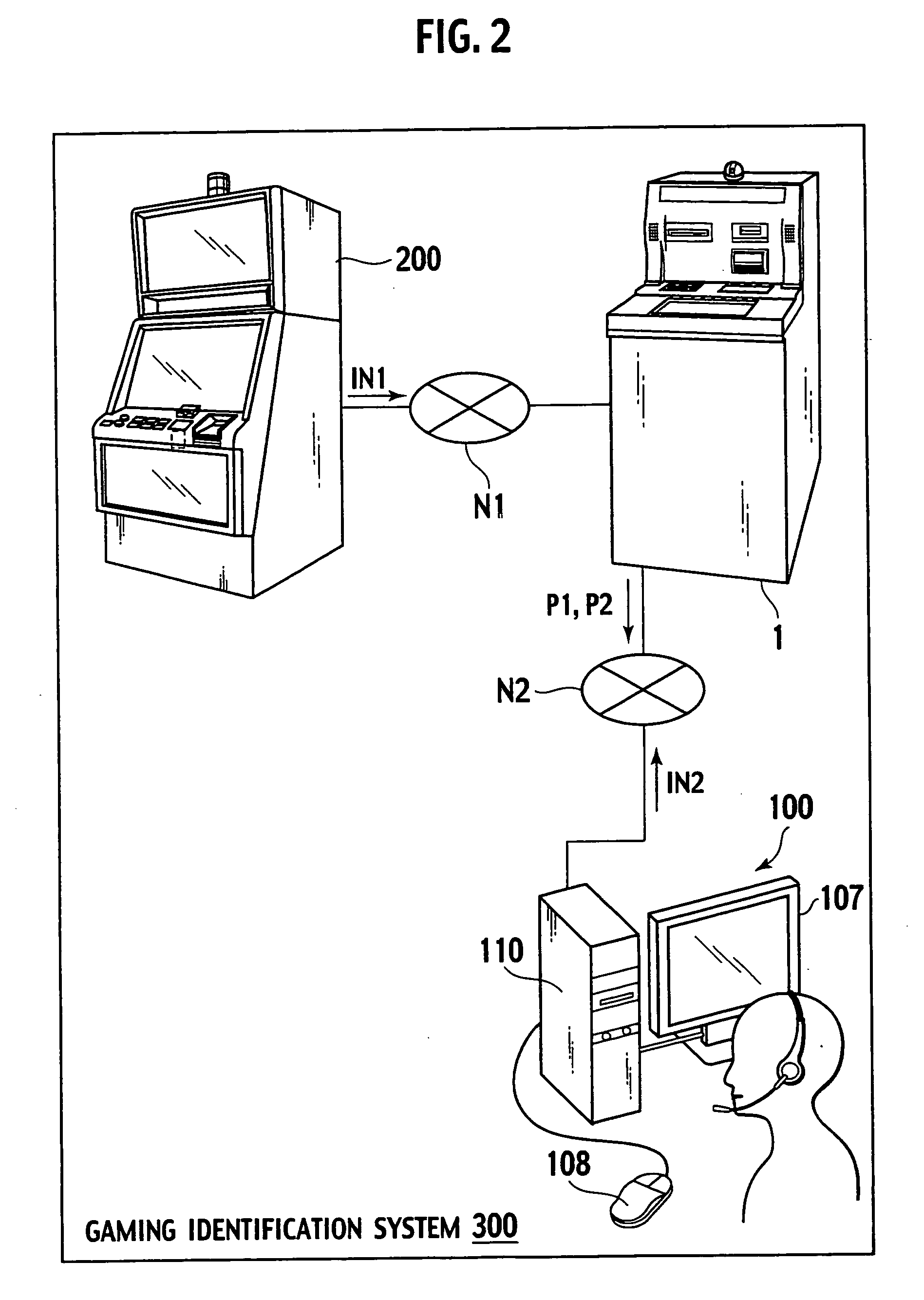
Gaming settlement machine, gaming terminal machine and gaming identification system
InactiveUS20060205494A1Shorten identification timeReduce power consumptionApparatus for meter-controlled dispensingVideo gamesImaging dataIdentification system
A gaming identification system comprises a gaming settlement machine and a gaming terminal machine connected to the gaming settlement machine. The gaming settlement machine generates a player image data for identifying a player who will obtain a payout, and then sends the player image data to the gaming terminal machine. The gaming settlement machine executes the payout according to a payout order data for ordering whether or not the gaming settlement machine is permitted to execute the payout to the player who is identified, when receiving it from the gaming terminal machine. The gaming terminal machine displays a player's image according to the player image data sent from the gaming settlement machine. The gaming terminal machine generates the payout order data according to the displayed player's image, and then sends the payout order data to the gaming settlement machine.
Owner:UNIVERSAL ENTERTAINMENT CORP
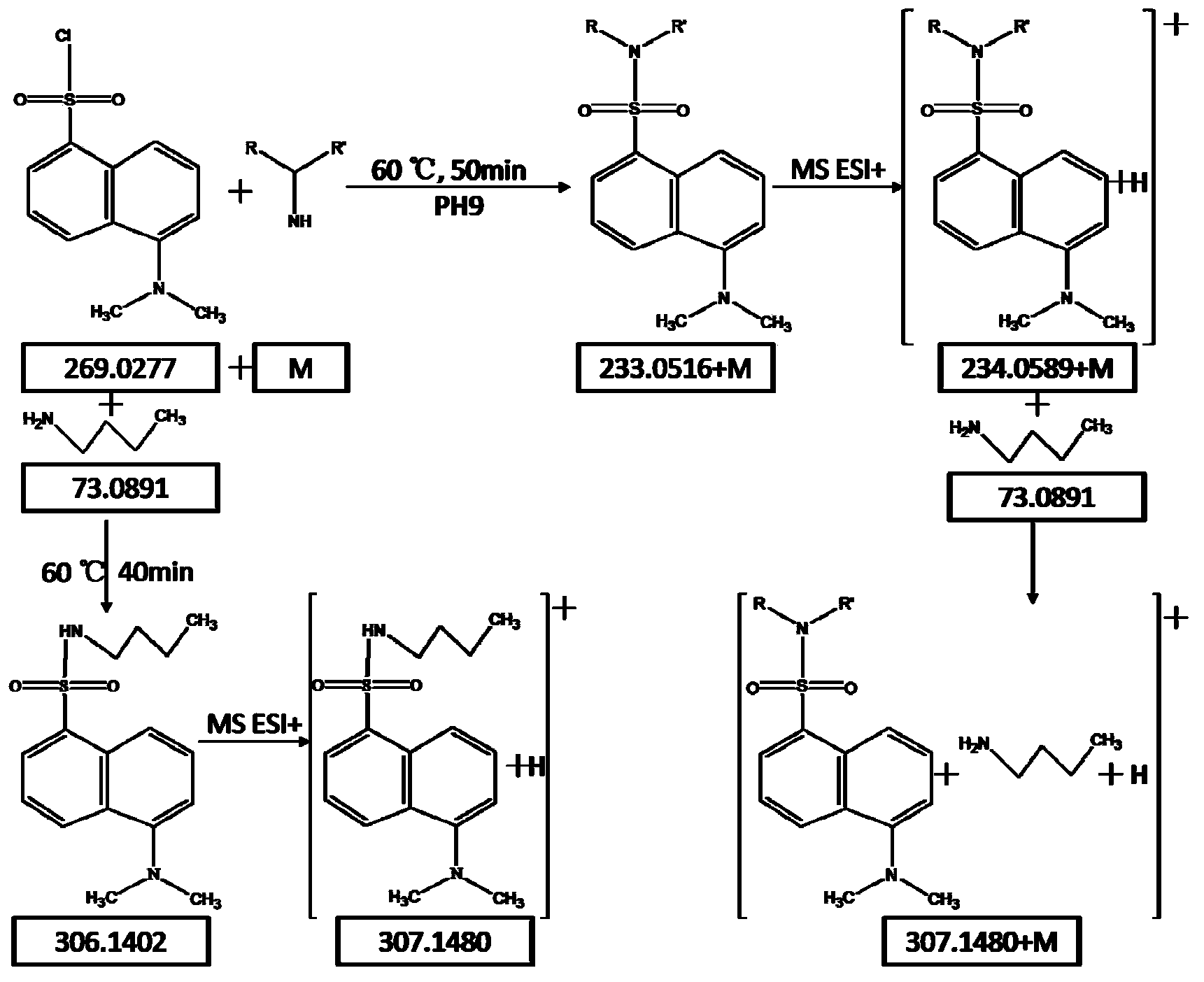
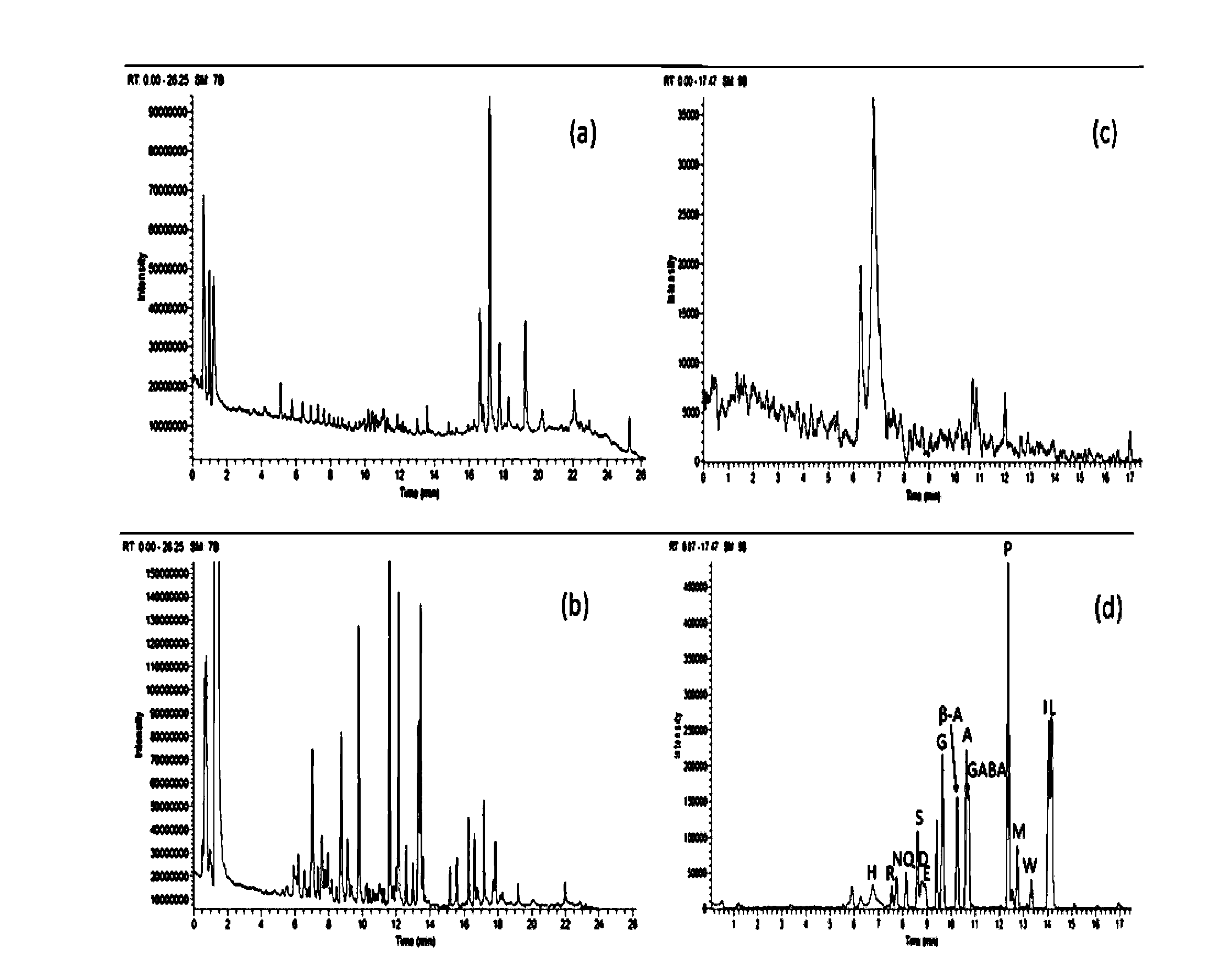
Method for analyzing amine substances in dansyl chloride derived-plasma based on liquid chromatography mass spectrometry
InactiveCN103822998AReduce consumptionStrong specificityComponent separationDipeptideGas chromatography–mass spectrometry
The invention discloses a method for analyzing amine substances in dansyl chloride derived-plasma based on liquid chromatography mass spectrometry. The method is characterized in that dansyl chloride is used as a derivatization reagent to perform derivatization on a metabolite containing primary amine and secondary amine, therefore the retention capability of the amine substances in reversion phase chromatography is increased, the ionization efficiency in mass spectrometric detection is increased, and the purpose of increasing the quantitative accuracy and detection sensitivity of the amine substances is achieved. The method has wide coverage for amine metabolites (comprising amino acid, methylation products, acetylation products, dipeptide and the like), can utilize 100 [mu]L of plasma samples to detect 113 amine substances, supplies the outline information of metabolome of the amine substances, and has the characteristics of good specificity, high accuracy and repeatability, less reagent consumption and the like.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
Method for studying metabolic difference of transgenic rice and non-transgenic rice
InactiveCN102478563AThe pre-processing process is simpleImprove reliabilityComponent separationBiotechnologyGenetically modified rice
The invention discloses a method for studying metabolic difference of transgenic rice and non-transgenic rice. The method analyzes rice seed extract with liquid chromatography-mass spectrometry (LC-MS) technology to obtain metabolic profiling of rice, studies integral difference of metabolic profiling data between transgenic rice and non-transgenic rice by multivariate statistics method, and finds out compound with significant contribution to metabolic phenotype difference, to provide a basis for evaluation of unintended effect and safety of transgenic rice. The simple and rapid analysis method with good repeatability is suitable for batch analysis of practical samples.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
Crop seed variety authenticity identification method based on near infrared spectrum
InactiveCN104062262AImprove stabilityImprove adaptabilityMaterial analysis by optical meansInfraredFeature extraction
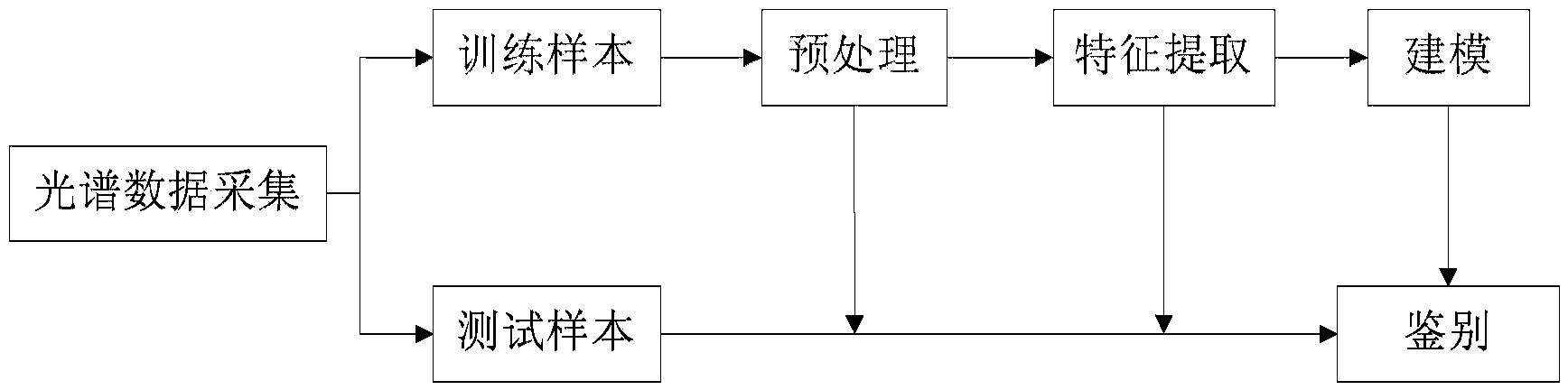
The invention discloses a crop seed variety authenticity identification method based on near infrared spectrum. The method comprises the following steps of 1, acquiring near infrared spectrum data of crop seed samples, and preprocessing the near infrared spectrum data which is taken as a training sample; 2, selecting the needed spectrum data from the preprocessed near infrared spectrum data, and establishing a qualitative analysis model of the crop seeds by a feature extraction method and a modeling method; and 3, carrying out variety authenticity identification on the spectrum of the crop seeds which are taken as test samples and about to be identified. The crop seed variety authenticity identification method is based on the near infrared spectrum, the general near infrared spectrum qualitative analysis model can be established by a series of operation such as spectrum preprocessing, feature extraction, modeling and identification, and the method is capable of rapidly and accurately identifying the crop seed variety authenticity.
Owner:INST OF SEMICONDUCTORS - CHINESE ACAD OF SCI
A DNA bar code standard gene sequence for Taiwan Lasiohelea
ActiveCN102690829AFacilitate the realization of molecular identificationShorten identification timeMicrobiological testing/measurementFermentationRapid identificationLasiohelea
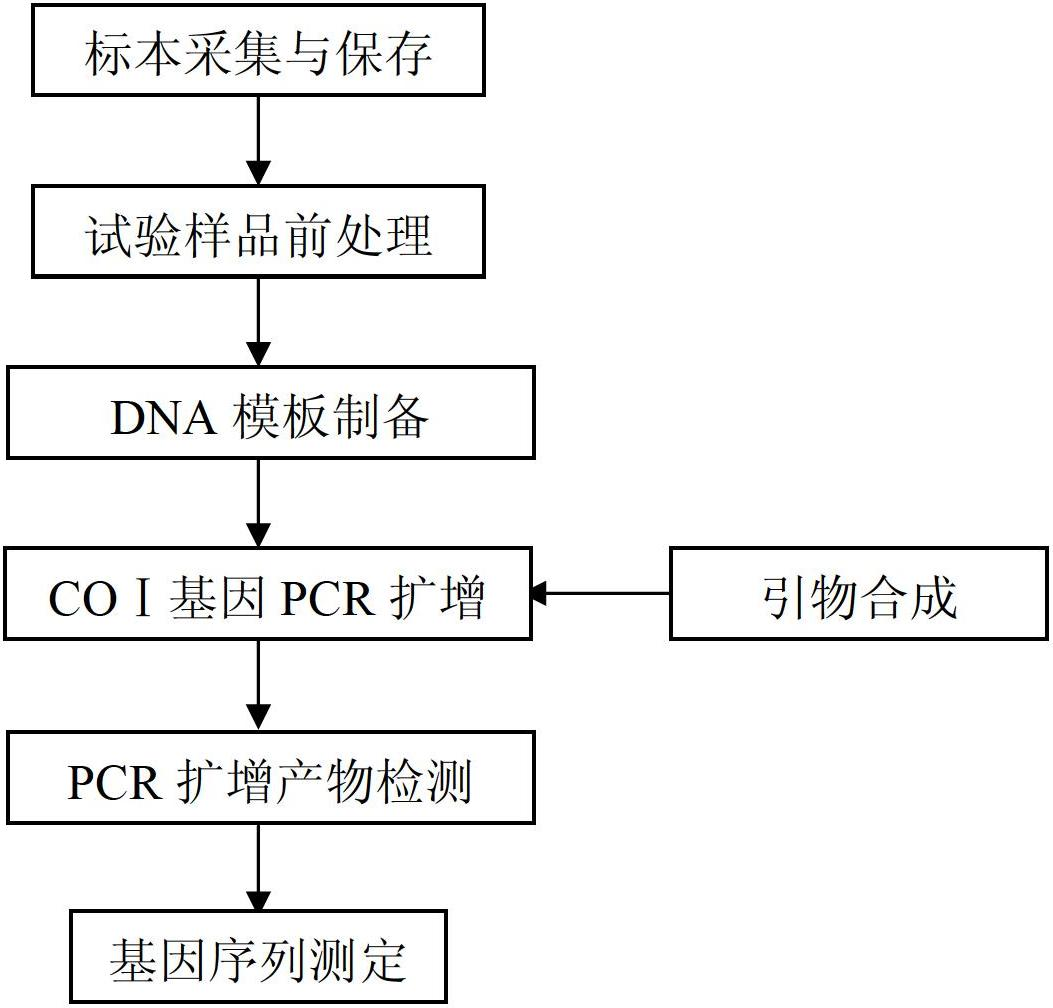
The invention discloses a DNA bar code standard gene sequence for Taiwan Lasiohelea, which is characterized in that a bar code standard detection gene is COI gene, possessing gene sequence SEQ ID NO.1. Manual proofreading and sequence assembly are carried out to the sequencing result and Blast similarity search is carried out in NCBI to ensure that the obtained sequence is the target sequence. Species identification of Taiwan Lasiohelea is realized through morphological identification, thereby result reliability is guaranteed. The DNA bar code standard gene sequence for Taiwan Lasiohelea provided in the invention is in favor of realizing rapid identification of Taiwan Lasiohelea, and reduces identification time.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Molecular identification method for lasiohelea taiwana
ActiveCN103805609AAccurate identificationShorten identification timeMicrobiological testing/measurementFermentationBiotechnologyMolecular identification
The invention discloses a molecular identification method for lasiohelea taiwana. The method is characterized in that two different genes, namely a COI gene and an ITS2 gene are adopted to be combined, and the lasiohelea taiwana is subjected to species identification by comparison of the similarity of the gene sequences. By adopting the molecular identification method for lasiohelea taiwana provided by the invention, accurate and rapid identification of the lasiohelea taiwana is facilitated. A basis is provided for molecular identification of the lasiohelea taiwana. The new variety can play a certain role in transmission of plant pollen and environment harmless reaction.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
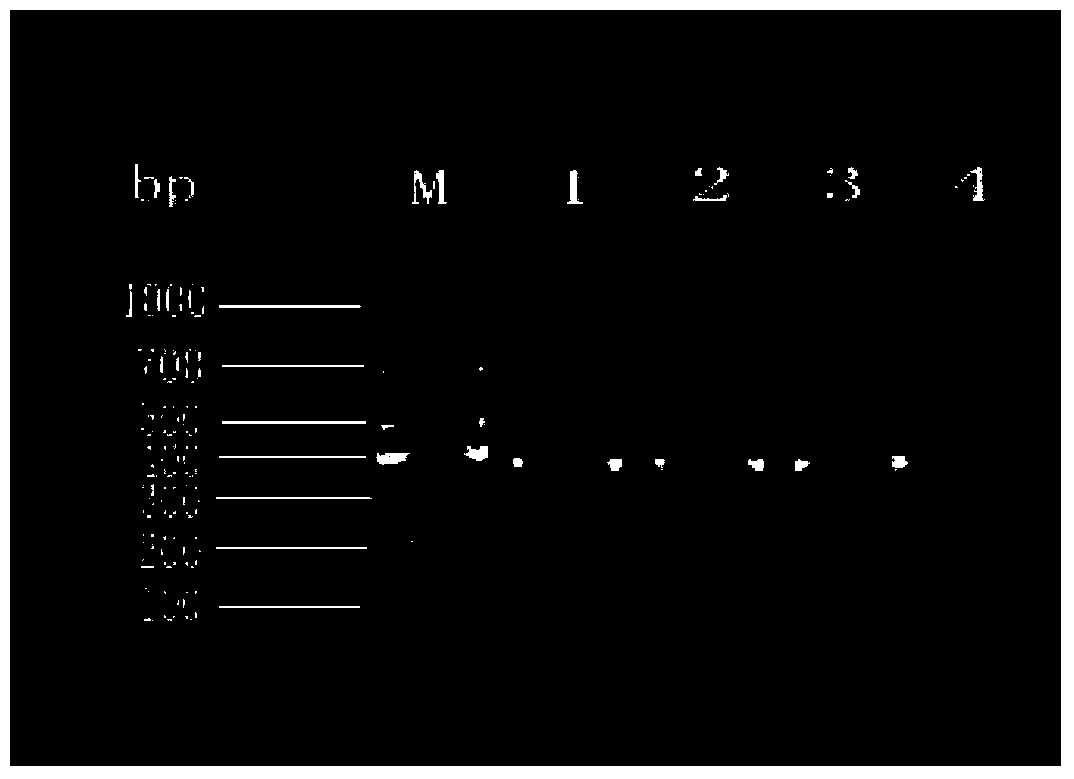
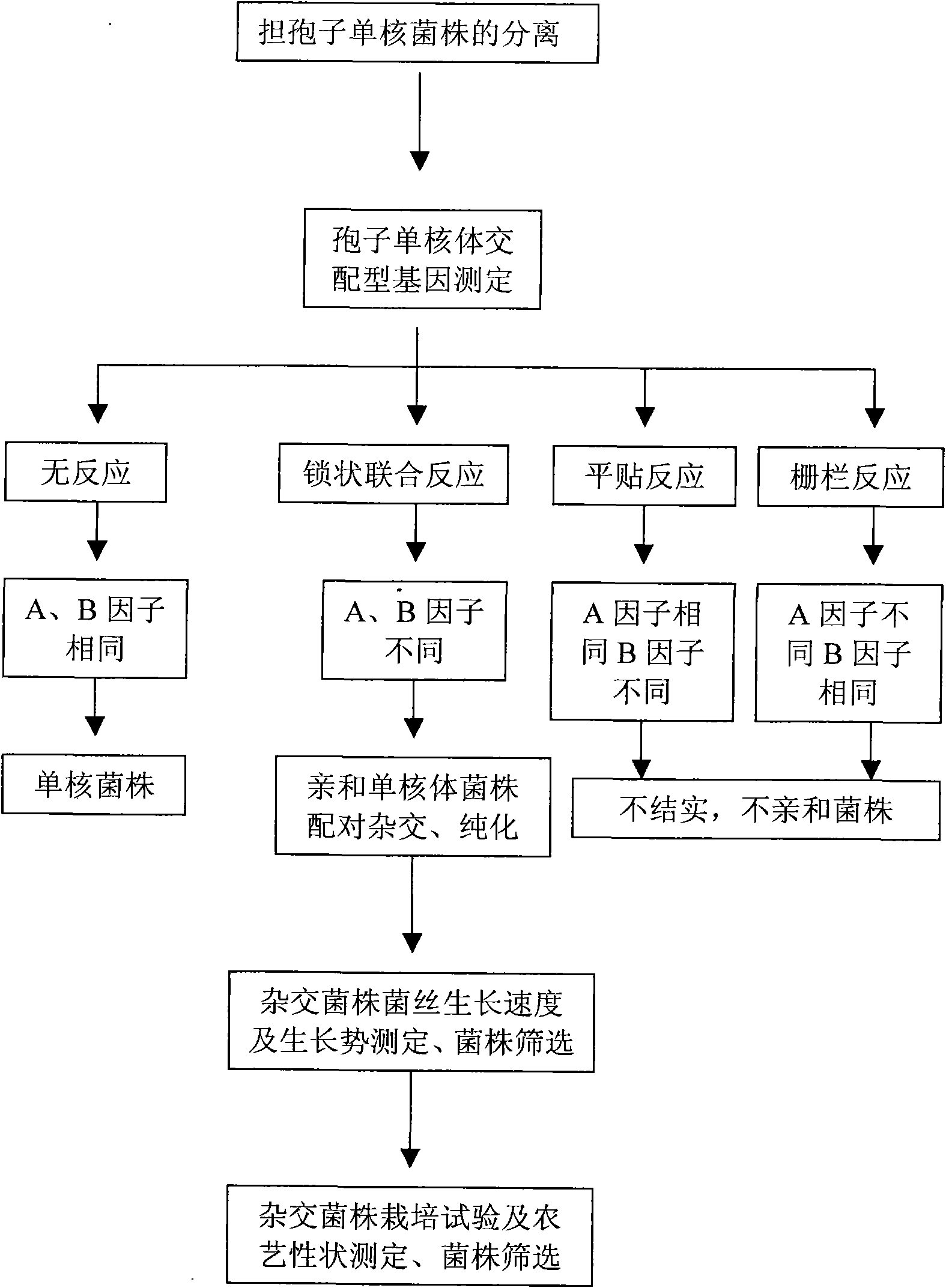
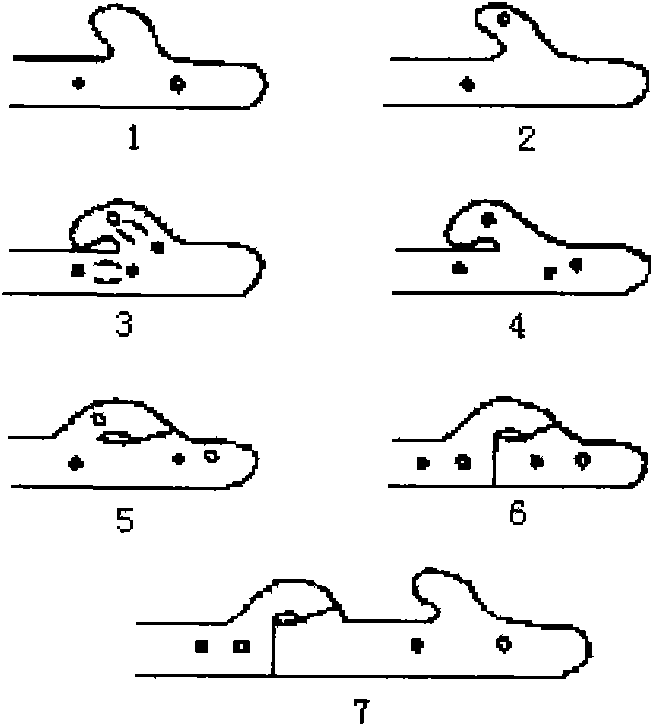
Method for identifying hypsizigus marmoreus mating type gene
InactiveCN101608213AReduced number of contrast screensShorten identification timeMicrobiological testing/measurementPlant genotype modificationBasidiosporeClamp connection
The invention provides a method for performing mating type gene identification on a basidiospore monocaryon strain which is separated from different hypsizigus marmoreus strains. In the identifying process, in particular application of a flushing reaction, a palisade reaction and transplanting, the method can primarily identify the mating type gene of the hypsizigus marmoreus intuitively and quickly without other instruments and equipment. When the method is applied in cross breeding, the mating type gene of basidiospore monocaryon parent and progeny strains can be identified conveniently so as to improve the purposiveness and the effectiveness of the cross breeding process. The adopted method can shorten the time for identifying the mating type gene for 5 to 10 days, reduce the number of comparing-screening strains by 75 percent theoretically in cross breeding, improve the fecundity, and is more convenient and efficient particularly in application of multiple cross breeding of multiple strains. Then, the method selects the compatible basidiospore monocaryon strain having clamp connection according to the identification result to carry out biparental crossing, and screens hybrid strains according to growth speed, growth potential and economical character index, thereby obtaining stable, high yield and high quality hybrid strains.
Owner:SHANGHAI ACAD OF AGRI SCI
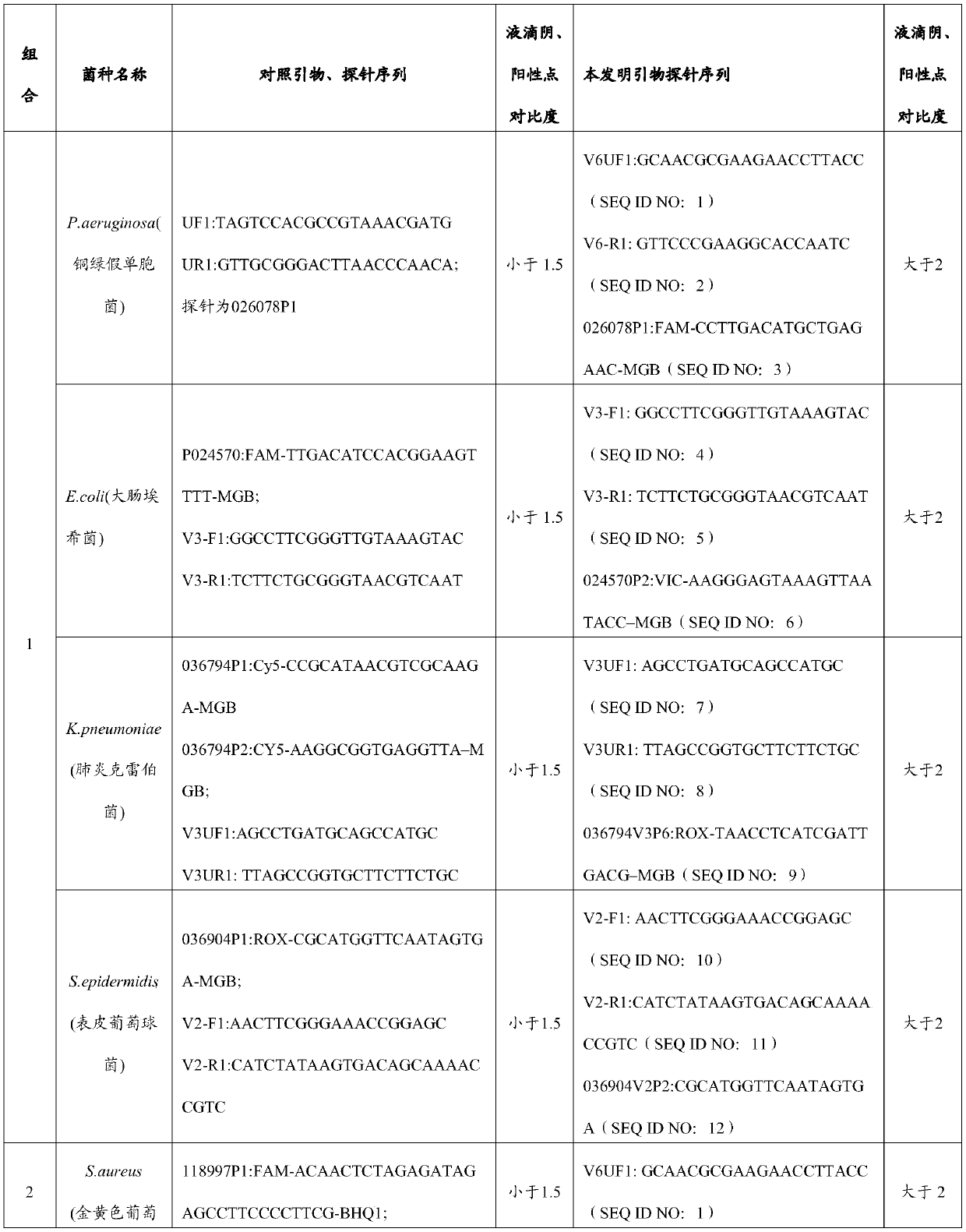
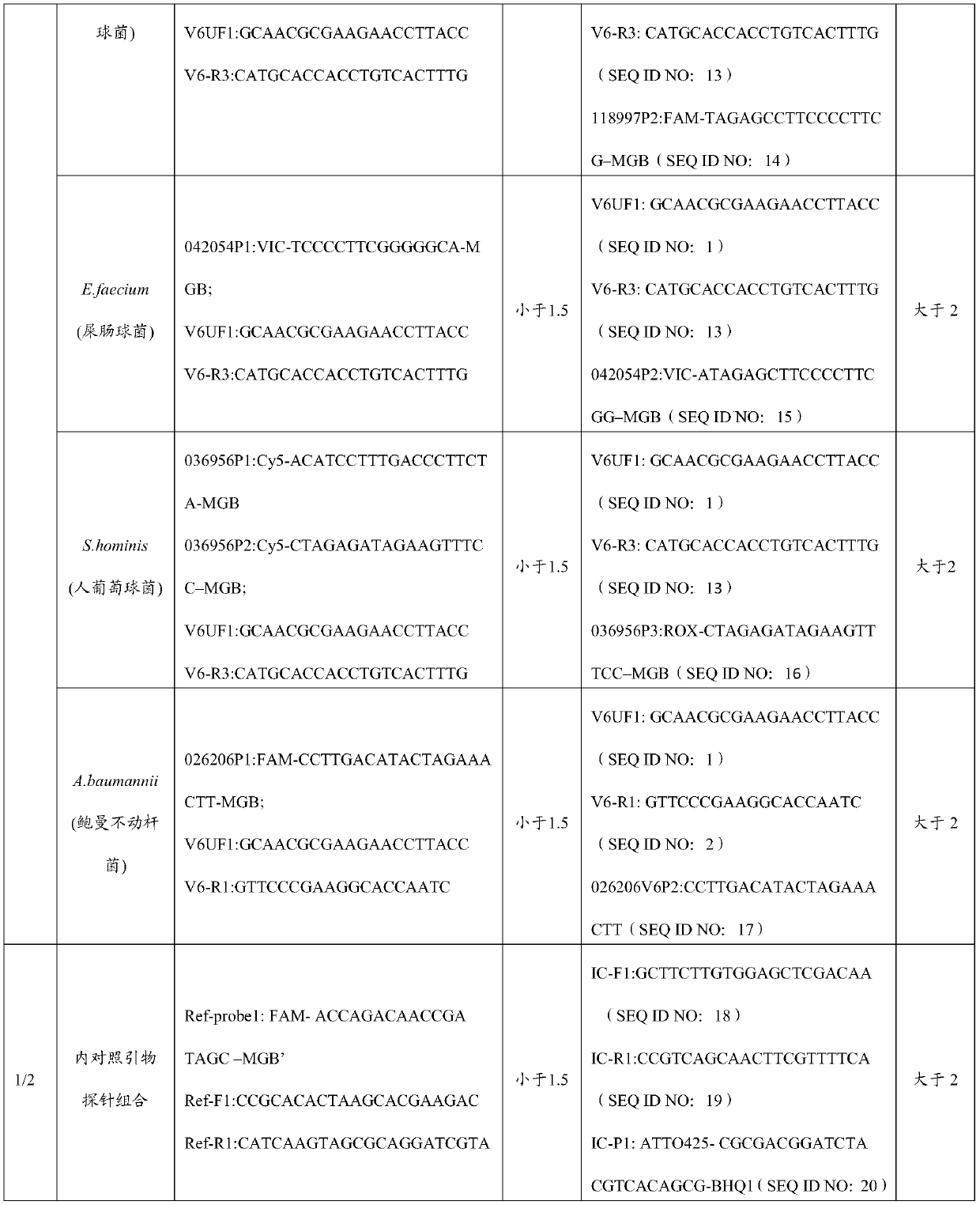
Primer and probe combination and kit for detecting human pathogenic bacteria
PendingCN110564824AShorten identification timeImprove positive detectionMicrobiological testing/measurementDNA/RNA fragmentationInfected patientTreatment effect
The invention relates to the technical field of medical detection, and in particular, relates to a primer and probe combination and a kit for detecting human pathogenic bacteria. The kit comprises theprimer and probe combination for detecting pseudomonas aeruginosa, escherichia coli, klebsiella pneumoniae, staphylococcus epidermidis, staphylococcus aureus, enterococcus faecium, human staphylococcus and acinetobacter baumannii. According to the kit, the primer and probe combination is adopted, multiple digital PCR is combined, the pathogenic bacteria types and the target nucleic acid copy number in the detection range can be rapidly and accurately identified, the detection method has high sensitivity, and positive detection of low-copy nucleic acid fragments of pathogenic bacteria is guaranteed; meanwhile, the variety of the pathogenic bacteria or the change of the copy number of the target nucleic acid fragment in the body of a pathogenic bacteria infected patient can be dynamically monitored, the treatment effect is assisted and evaluated in time, and reference is provided for optimization of a doctor clinical scheme.
Owner:PILOT GENE TECH HANGZHOU CO LTD
Primers used for rapid detection of Neofusicoccum parvum and application thereof
InactiveCN102747078AIncreased sensitivityShorten the identification timeMicrobiological testing/measurementMicroorganism based processesGenomic DNADisease
The invention discloses a pair of primers used for rapid detection of Neofusicoccum parvum strains and application thereof. Nucleotide sequences of the pair of the primers are respectively represented by sequence 1 and sequence 2 in a sequence table, and the primers can specifically amplify the product of 370bp in Neofusicoccum parvum and have a sensitivity of 10 pg to genomic DNAs of the Neofusicoccum parvum strain. It is proved that the pair of the primers can be used for rapid reliable detection and identification of the Neofusicoccum parvum strain in disease samples of grapes in the field, and the problems of cumbersome steps, a long identification period and the like in traditional identification methods are overcome.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
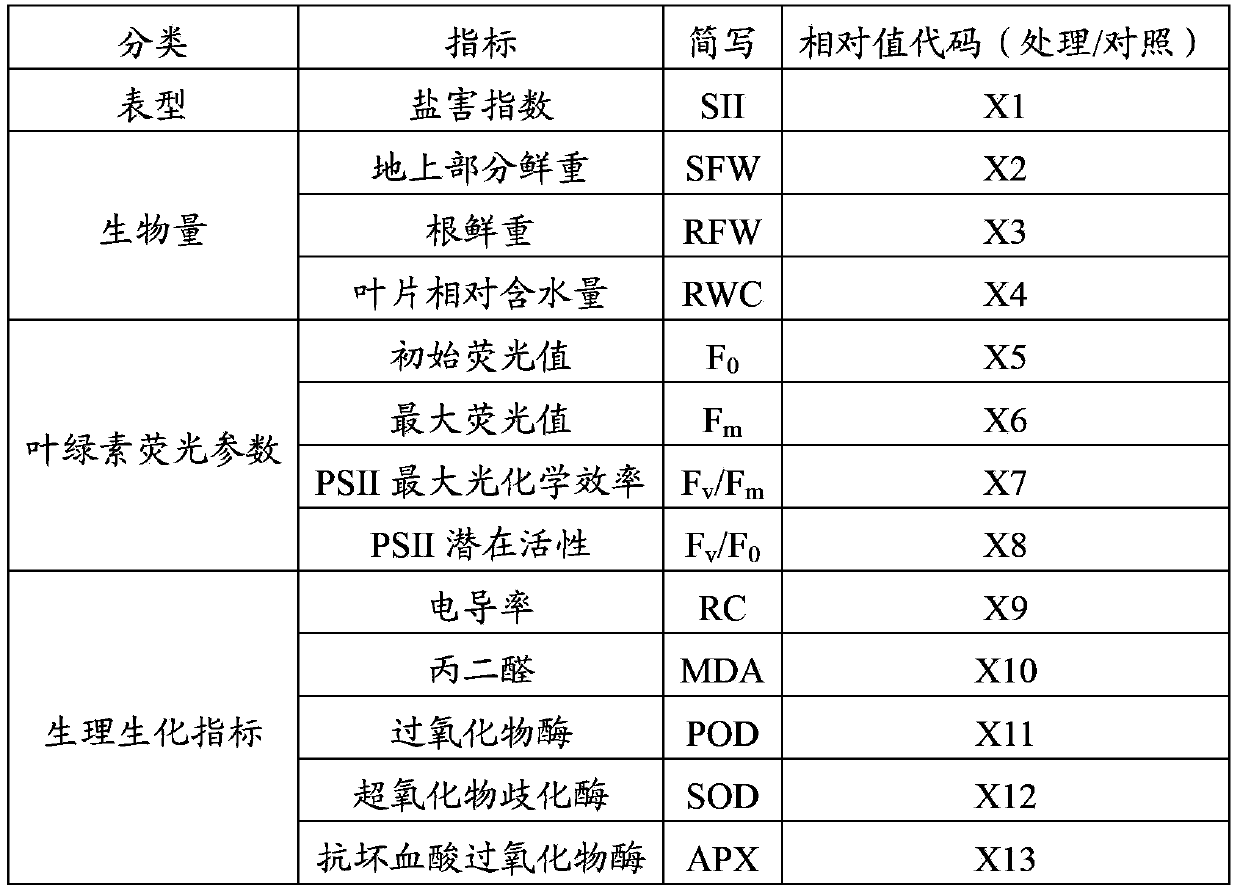
High-efficiency identification method for salt resistance of cotton germplasms
ActiveCN103364383AStrong salt toleranceThe result is accurate and reliableFluorescence/phosphorescenceSalt resistanceAgricultural science
The invention provides a high-efficiency identification method for salt resistance of cotton germplasms. By utilizing a flower pot soil seedling cultivation method, cotton seedings are cultivated to a three-leaf-and-one-leaflet stage. The materials with normal and consistent growth vigour are selected and subjected to salt stress treatment and clear water treatment with the same volume respectively. The second true leaves are selected, the PSII maximum photochemical efficiencies of the true leaves with different treatments are measured respectively, the relative values are calculated, and brought into the standard equation of salt resistant indexes: D=1.943X-0.882 to calculate the salt resistant indexes and determine the salt resistance, wherein X is for a relative value of a PSII maximum photochemical efficiency and D is for a salt resistant index. The larger the salt resistant index, the stronger the salt resistance. When the method is compared to the prior art, the results of the obtained standard equation of the method are accurate and credible. The measurement method of PSII maximum photochemical efficiencies is simple and fast, can be performed in vivo, does not harm plants, and can ensure normal running of downstream experiments. The measurement time is the cotton seedling stage, so the identification time can be shortened effectively, the efficiency is raised, and the method is suitable for large scale germplasm resource identification.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Method for rapidly identifying black shank resistance of tobaccos
InactiveCN104745672ARapid identificationEasy to breedFungiMicrobiological testing/measurementDiseaseAnimal science
The invention belongs to the field of crop disease resistance identification and especially a method for rapidly identifying the black shank resistance of tobaccos. The method for rapidly identifying the black shank resistance of the tobaccos is capable of effectively overcoming the defects of exiting identification, and comprises the following steps: step 1, culturing tobacco black shank bacteria; step 2, taking tobacco seeds to be tested and sterilizing the surface; and step 3, performing germination rate and morbidity statistics and performing disease resistance evaluation. The method is capable of saving manpower and material resources and also greatly reducing the identification time; the resistance level to the black shank is not limited by the growth seasons; besides, the method is simple in identification operation steps and relatively low in test room requirement conditions; according to the method, the purpose of rapidly identifying the black shank resistance of the tobaccos at low cost and high flux is achieved; the method is suitable for identifying the black shank resistance of a large batch of germplasm materials in tobacco breeding study; and as a result, the anti-disease germplasm resource screening, the resistance hereditary character study and the anti-disease variety breeding and popularization / application can be accelerated.
Owner:TOBACCO RES INST HENAN ACADEMY OF AGRI SCI
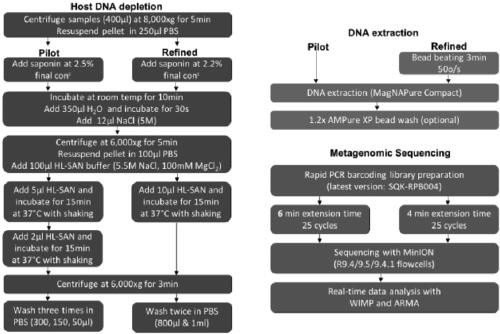
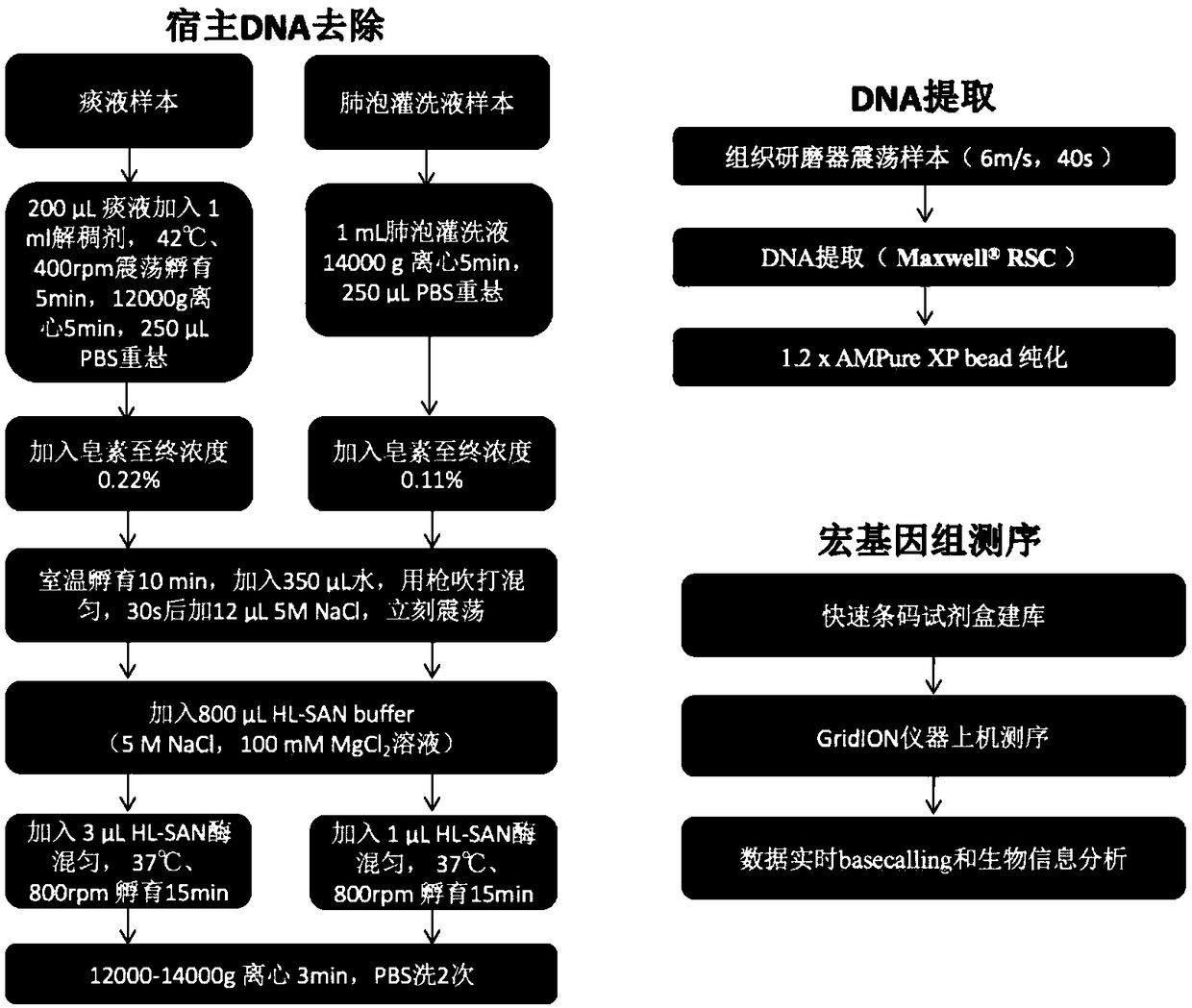
Metagenome sample library construction method and identification method based on nanopore sequencing platform and kit
ActiveCN109487345AShorten the timeTrue reflection compositionMicrobiological testing/measurementLibrary creationComputational biologyBuilding construction
The invention relates to a metagenome sample library construction method and an identification method based on a nanopore sequencing platform and a kit. The method comprises host removal pre-treatmentand sample library construction on human and animal metagenome samples by combining incomplete host removal based on the nanopore sequencing platform so as to sequence and identify the metagenomes. Compared with a conventional process flow, the method is simpler and rapider, meets the reasonable detection demands, omits flows such as library construction PCR in the identifying process, extrudes deviation caused by PCR amplification, and can be used for identifying metagenome microorganisms more truly and accurately.
Owner:BEIJING SIMCERE MEDICAL LAB CO LTD +2
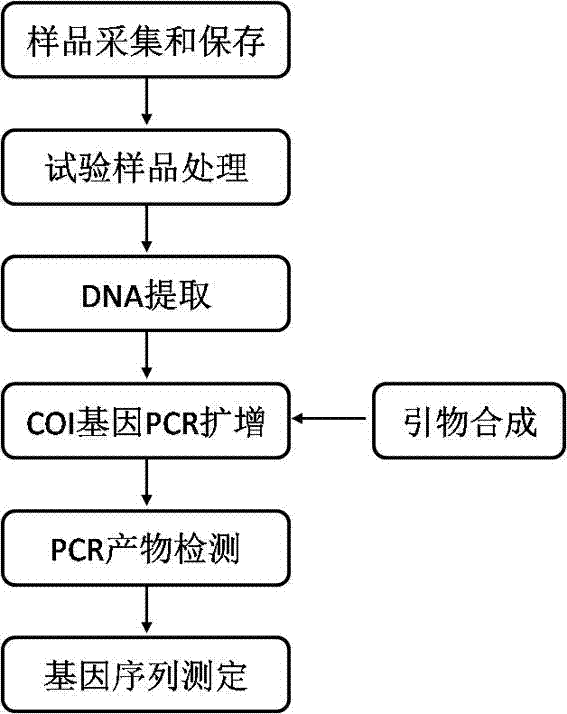
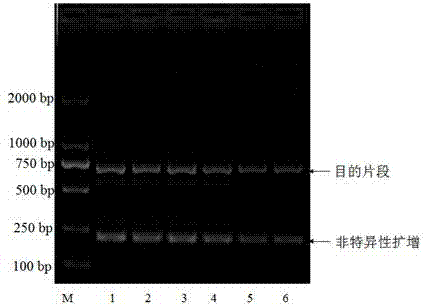
DNA barcoding standard gene sequence of Rizhao Blepharipoda liberata and species identification method based thereon
InactiveCN103937802AFacilitate the realization of molecular identificationAchieve molecular identificationMicrobiological testing/measurementFermentationMolecular identificationForward primer
Relating to the technical field of species identification, the invention specifically relates to a DNA barcoding standard gene sequence of Rizhao Blepharipoda liberata and species identification method based on the gene sequence. The DNA barcoding standard gene is COI gene, which has a gene sequence shown as SEQIDNO.1. The identification method includes: conducting extraction separation of DNA on a to-be-tested tissue, and amplifying a target gene by polymerase chain reaction. A pair of primers used in the amplification reaction are shown as the following: a forward primer 5'-GGTCAACAAATCATAAAGATATTGG-3', and a reverse primer 5'-TAAACTTCAGGGTGACCAAAAAATCA-3'. The gene sequence obtained in the invention is in favor of realizing molecular identification of the Rizhao Blepharipoda liberata, and can effectively shorten the identification time of the Rizhao Blepharipoda liberata.
Owner:刘丰铭
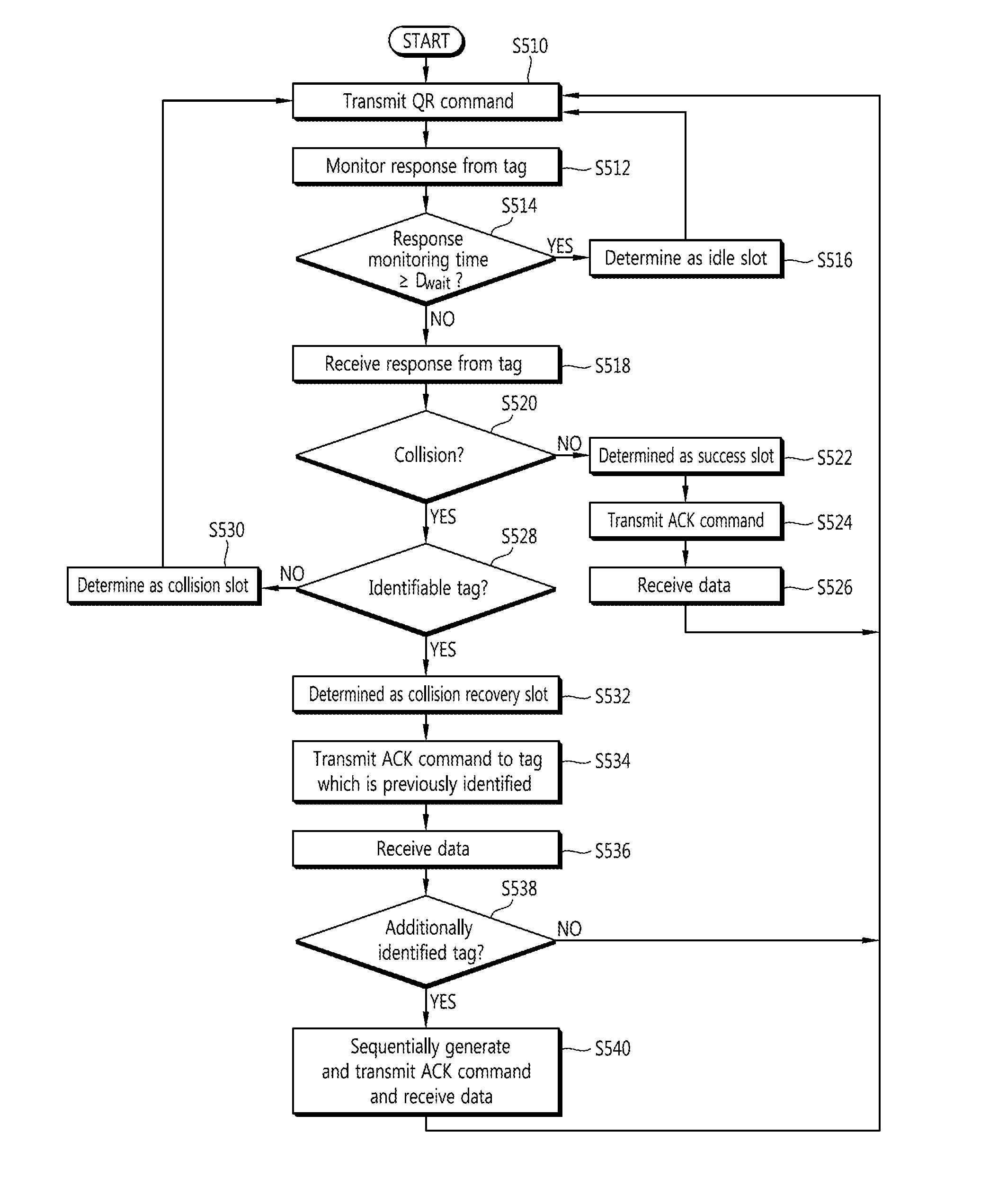
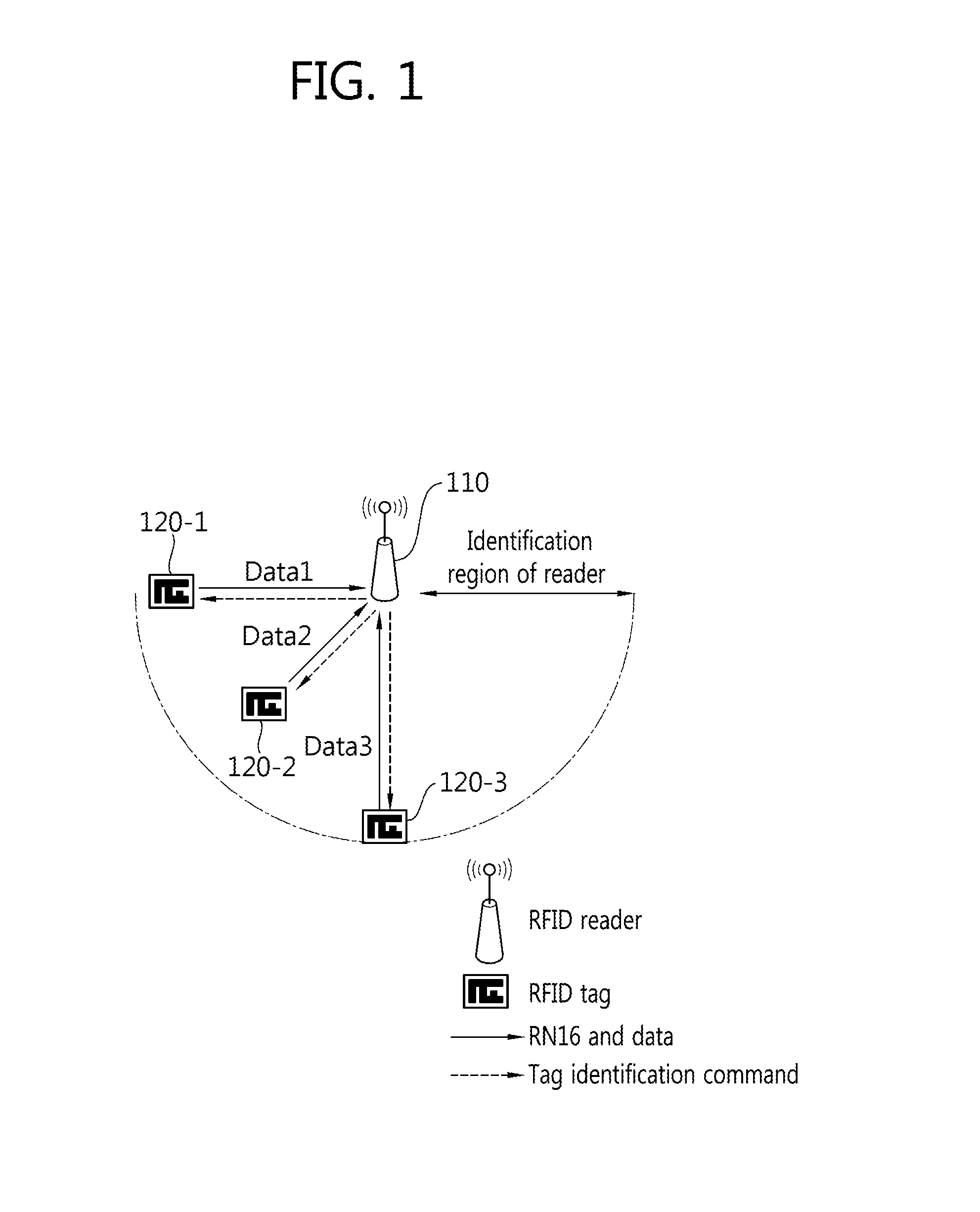
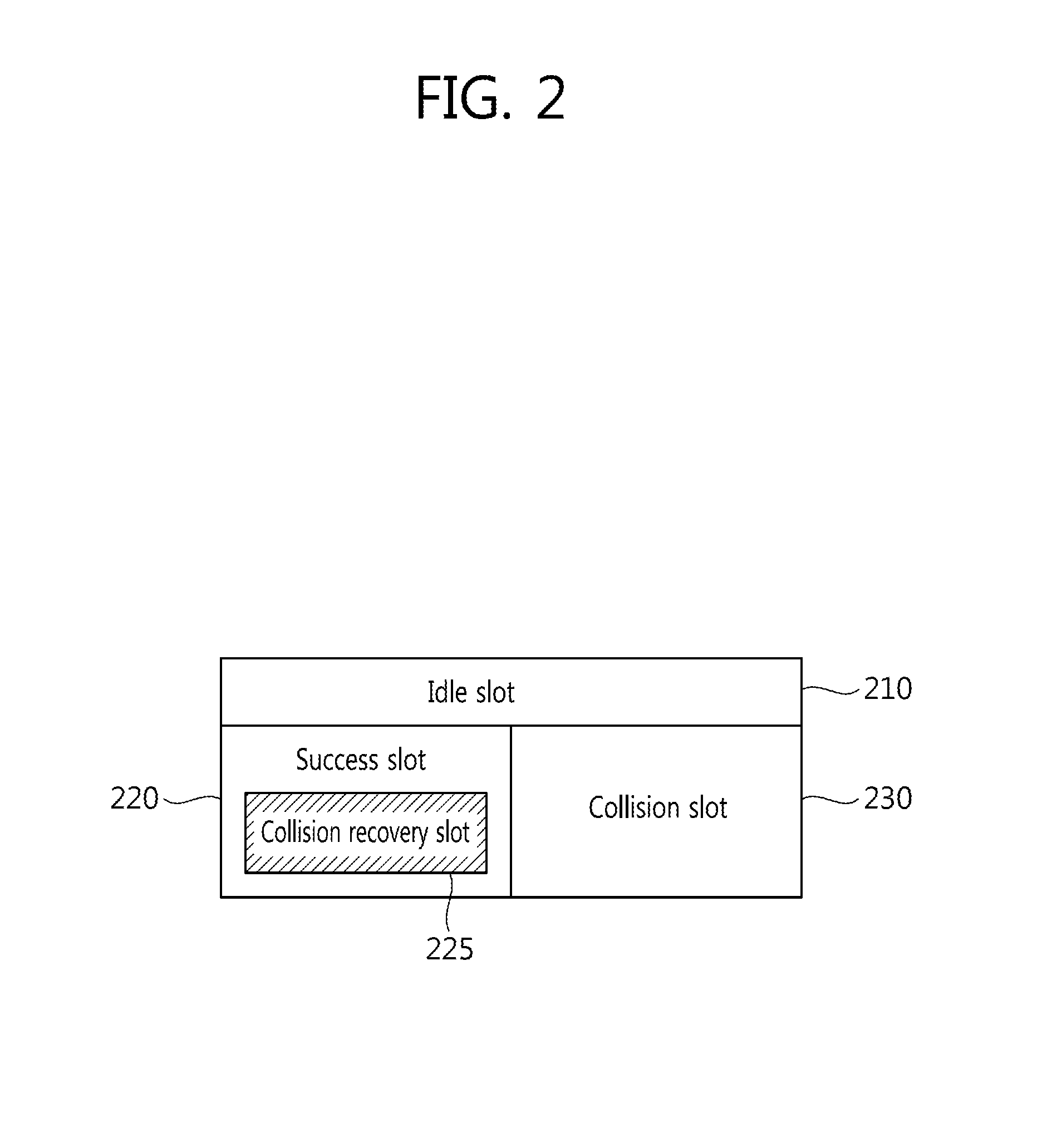
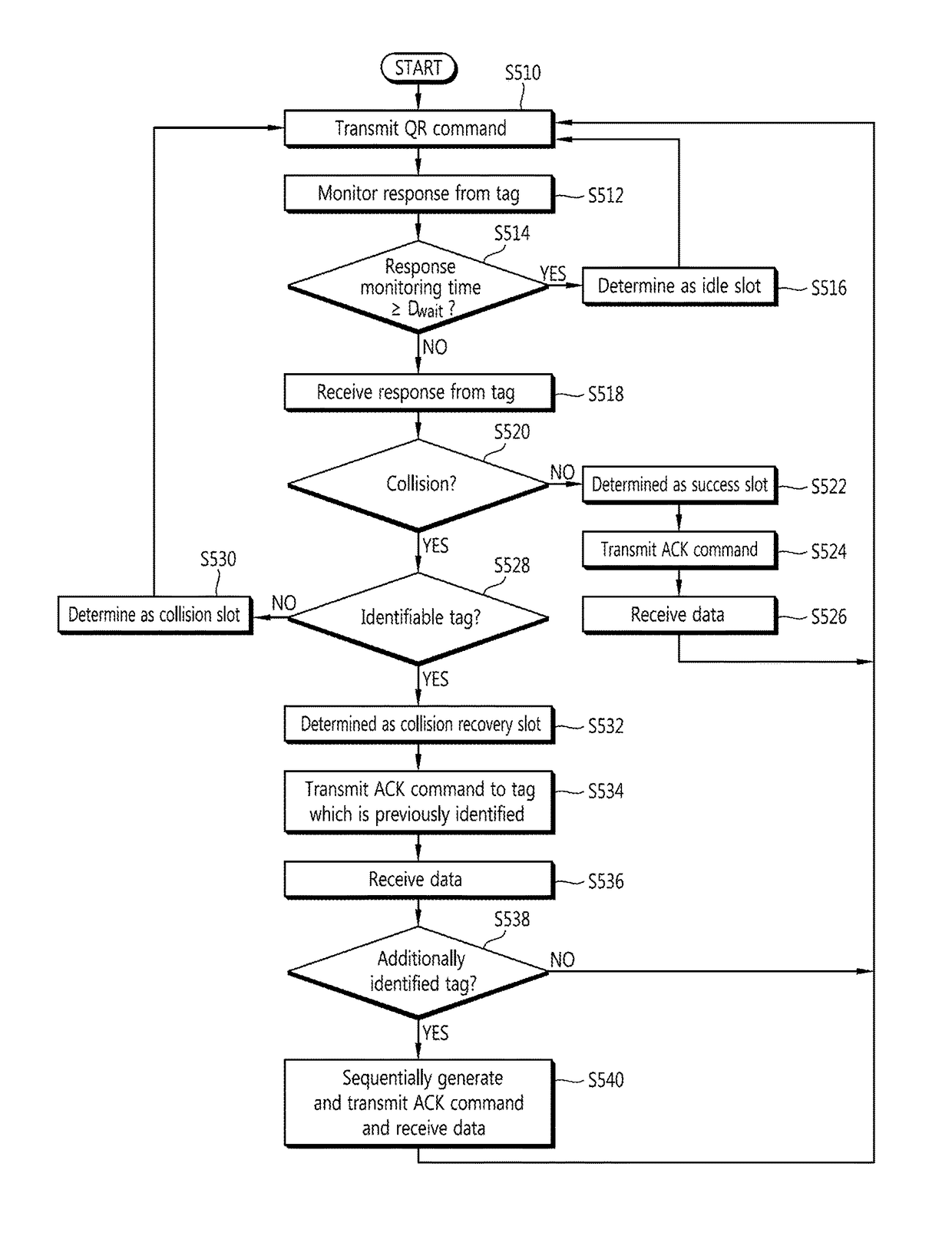
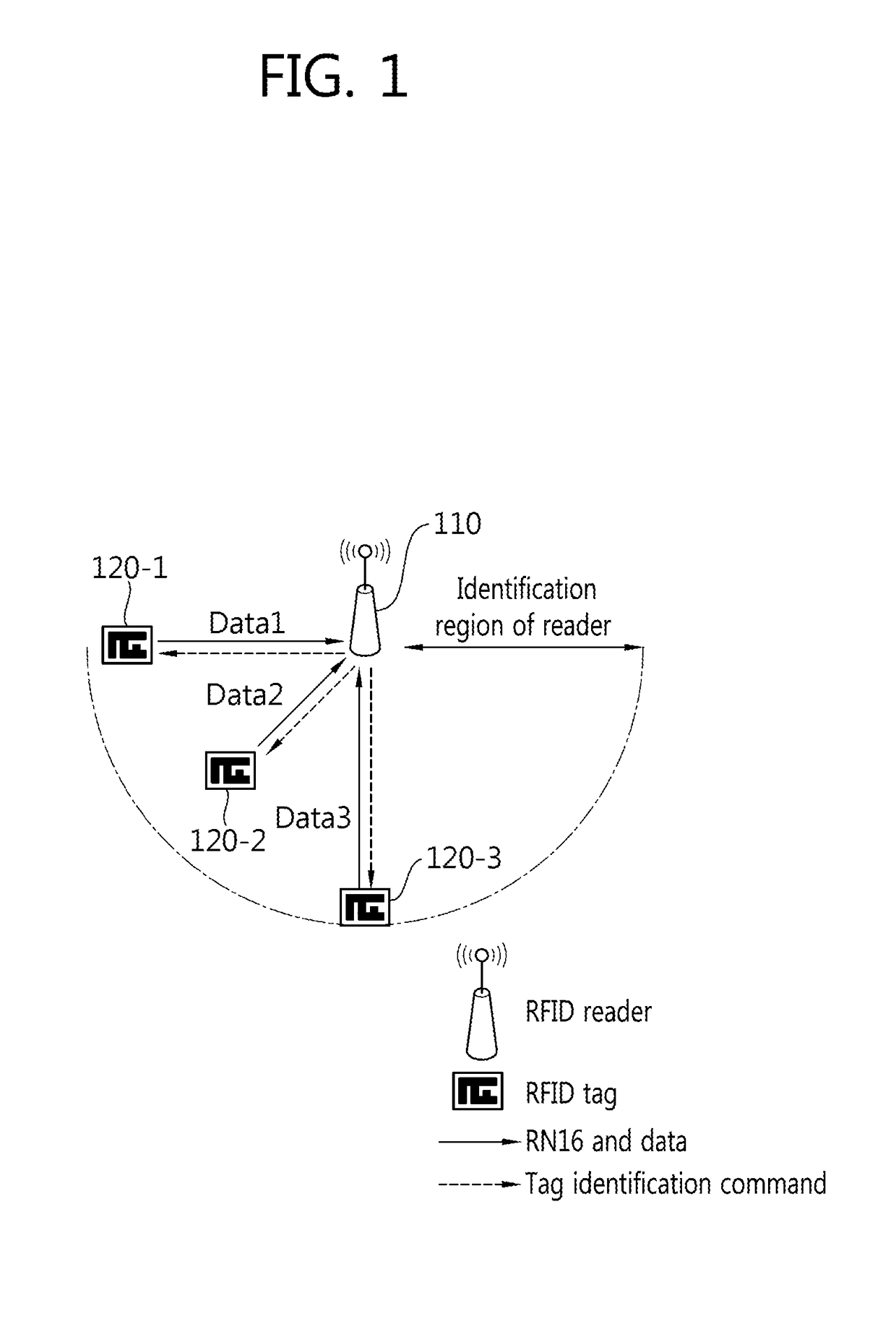
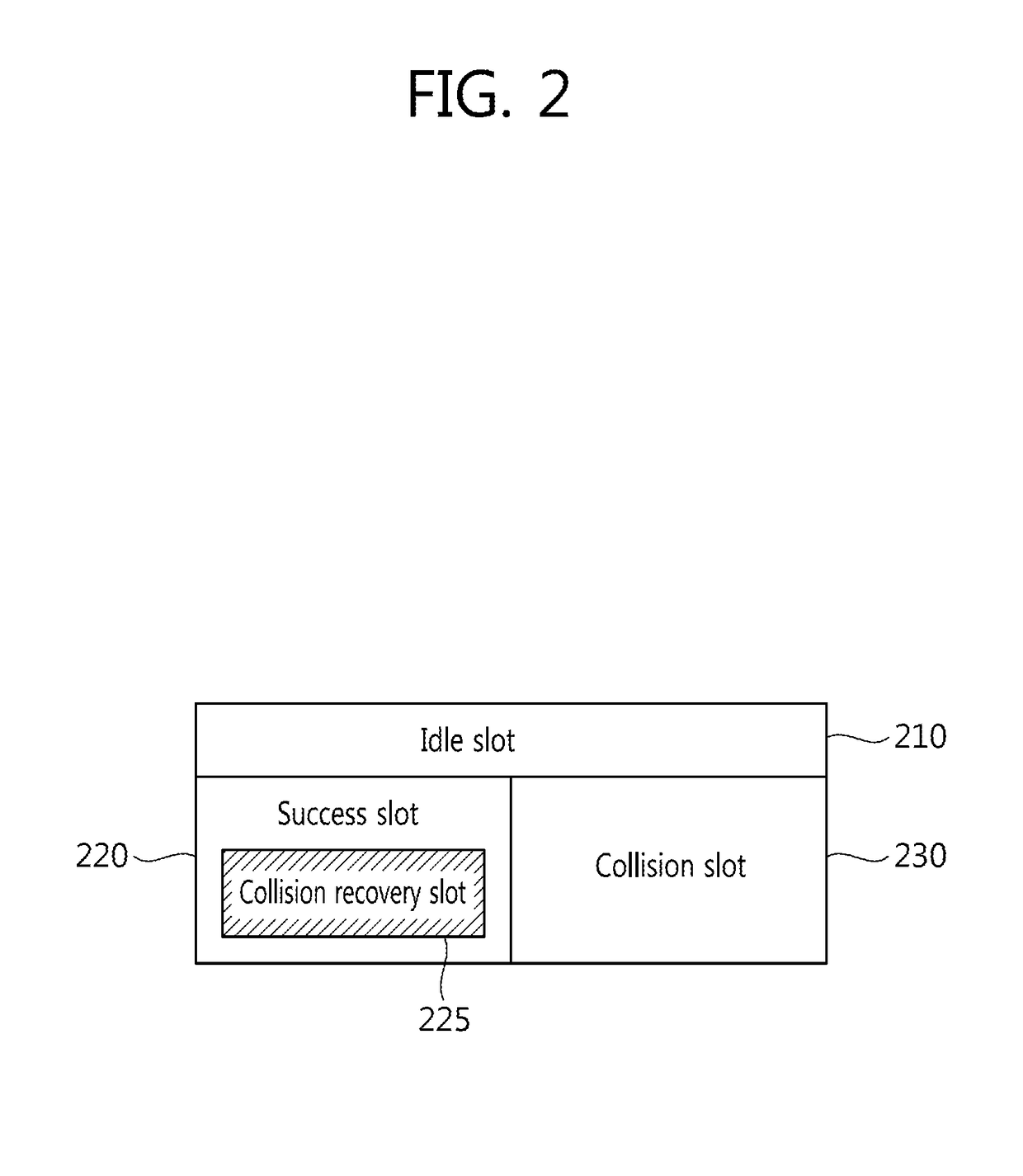
Tag Anti-collision method, reader apparatus and system for RFID systems with multi-packet reception capability
ActiveUS20160239692A1Improve recognition rateShorten identification timeTesting sensing arrangementsMemory record carrier reading problemsData transmissionEmbedded system
Provided is a method of preventing collision between a plurality of RFID tags by an RFID reader. An anti-collision method by a tag reader apparatus in an RFID system including a plurality of RFID tags, includes changing a length of a collision recovery slot in the collision recovery slot which successfully identifies a plurality of tags even though at least two tags simultaneously replied in one slot to cause collision so that data transmission of at least one identified tag is achieved.
Owner:RES & BUSINESS FOUND SUNGKYUNKWAN UNIV
Culicoides bellulus specific gene and molecular identification method thereof
InactiveCN105543249AGuaranteed accuracyEnsure reliabilityMicrobiological testing/measurementEnzymesMolecular identificationCulicoides
The invention discloses a culicoides bellulus specific gene and a molecular identification method thereof. The culicoides bellulus specific gene is characterized in that a molecular biology method is adopted to obtain the COI gene sequence of culicoides, and the species of the culicoides is identified by comparing gene sequence similarity. The ulicoides bellulus molecular identification method is beneficial for accurately and fast identifying culicoides bellulus.
Owner:FUJIAN INT TRAVEL HEALTH CARE CENT
Tag anti-collision method, reader apparatus and system for RFID systems with multi-packet reception capability
ActiveUS9773132B2Improve throughput and tag identification rateImprove recognition rateTesting sensing arrangementsMemory record carrier reading problemsData transmissionEmbedded system
Provided is a method of preventing collision between a plurality of RFID tags by an RFID reader. An anti-collision method by a tag reader apparatus in an RFID system including a plurality of RFID tags, includes changing a length of a collision recovery slot in the collision recovery slot which successfully identifies a plurality of tags even though at least two tags simultaneously replied in one slot to cause collision so that data transmission of at least one identified tag is achieved.
Owner:RES & BUSINESS FOUND SUNGKYUNKWAN UNIV
PCR-RFLP method for rapidly identifying radix tetrastigme and various counterfeits and adulterants of radix tetrastigme
ActiveCN104611424ARapid identificationImprove accuracyMicrobiological testing/measurementMedicinal herbsEnzyme digestion
The invention belongs to the field of molecular markers and discloses a PCR-RFLP method for rapidly identifying radix tetrastigme and various counterfeits and adulterants of the radix tetrastigme. The method comprises the following steps: 1, extracting the NDA of the medicinal material (namely the radix tetrastigme); 2, carrying out PCR amplification by forward and reverse primers of internal transcribed spacer ITS2 sequences of a pair of amplified ribosomal DNAs; 3, digesting the PCR product by restriction enzyme NCO I; 4, carrying out agarose gel electrophoresis analysis. After the enzyme digestion of amplified products of the DNA of the radix tetrastigme, two DNA fragments with sizes about 335 bp and 200 bp are generated, and the various counterfeits and adulterants are not identified by the NCO I enzyme. By utilizing the differences between the DNA sequences of radix tetrastigme and the counterfeits and adulterants of the radix tetrastigme, a quick, convenient and reliable PCR-RFLP identification method is established and used for identifying whether counterfeits and adulterants are mixed in the radix tetrastigme or not, so that the technical problem that the truth and false cannot be identified by sensory or physical and chemical analysis methods is solved, and the medication safety of the radix tetrastigme is ensured.
Owner:ZHEJIANG PHARMA COLLEGE
Dual PCR primer for detecting two types of pathogens of grape canker at same time and application of dual PCR primer
ActiveCN105177148AShorten identification timeTo achieve the purpose of early targeted disease preventionMicrobiological testing/measurementMicroorganism based processesNucleotide compositionBioinformatics
The invention discloses dual PCR primer for detecting two types of pathogens of grape canker at the same time and application of the dual PCR primer. The dual PCR primer is composed of nucleotides shown in the first sequence in the sequence list, nucleotides shown in the second sequence in the sequence list, nucleotides shown in the third sequence in the sequence list and nucleotides shown in the fourth sequence in the sequence list. The primer can detect existence of the two types of pathogens of the grape canker, namely, Neofusicoccum parvum and Botryosphaeria dothidea in a dual PCR reaction, it shows that the primer can rapidly, accurately and flexibly determine the two types of pathogens of the grape canker in grape nursery stock and field grape disease samples, and foundation is provided for nursery stock bacterium-carrying detection and specially preventing and controlling the grape canker caused by the two types of pathogens.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Rapid Fungus detection kit and detection method using same
ActiveCN101974640AShorten identification timeSave rescue timeMicrobiological testing/measurementMicroorganismMagnetic bead
The invention relates to the field of clinical microorganism identification, and provides a rapid fungus detection kit and a detection method using the same. The kit comprises two universal primers for amplifying an internal transcribed spacer (ITS1) of a fungus 18S, 5.8S, a sequencing primer for determining a sequence of a reverse section of the ITS1, a magnetic bead marked by an avidin, and a sequencing result and database comparison and judgment species. By using the kit, the fungus in a clinical sample can be distinguished by one-time sequencing, so the kit can be used for clinical diagnosis. The kit not only gains precious rescue time for clinical diagnosis and treatment, but also has the advantages that: the cost is low, the kit is convenient to operate and the specificity is high.
Owner:呼和浩特迪安医学检验所有限公司
Dual-PCR method for detecting tomato TY-1 gene and Mi gene at the same time
InactiveCN101638683AAccelerate the aggregation breeding processShorten identification timeMicrobiological testing/measurementEnzymePcr method
The invention provides a dual-PCR method for detecting a tomato TY-1 gene and an Mi gene at the same time, which comprises the following steps: (1) performing PCR reaction on a PCR system comprising the sequences of a Ty-1 primer and an Mi primer to obtain a PCR product; (2) keeping an enzyme cutting system at a temperature of 65 DEG C for 1.5 hours to obtain an enzyme cutting product; and (3) detecting the PRC product and the enzyme cutting product, using EB for dyeing the final result, adopting a Bio-RAD gel imaging system for taking pictures, and analyzing the result. The dual-PCR techniquehas the advantages that: 1, two genes are detected at the same time in the PCR, and the used time is just one half of that for detecting the two genes respectively so that the identification time isgreatly saved; and 2, the medicines used by the PCR are greatly saved, and the experimental cost is reduced.
Owner:SHANGHAI ACAD OF AGRI SCI
Specific gene and molecular identification method of culicoides cyancus
ActiveCN105483261AGuaranteed accuracyEnsure reliabilityMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationCulicoides
The invention discloses a specific gene and a molecular identification method of culicoides cyancus. The molecular identification method is characterized in that a COI gene sequence of the culicoides cyancus is acquired by adopting a molecular biology method, and species identification is performed on the culicoides cyancus through comparison of the gene sequence similarity. According to the molecular identification method of the culicoides cyancus, achievement of accurate and rapid identification on the culicoides cyancus is facilitated.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE
Identification method for ginkgo seedling male and female plants
InactiveCN103149159ASimple and fast operationEasy to operateMaterial analysis by observing effect on chemical indicatorColor/spectral properties measurementsSeedlingHorticulture
The invention provides an identification method for ginkgo seedling male and female plants, and the method is mainly directed at identification of the seedling stages of ginkgo seedling male and female plants in urban garden greening. By preparing a phosphate buffer solution, utilizing the chromogenic reaction of a bromothymol blue solution, and a proper treatment during a seedling stage, various conditions needed by early identification of ginkgo seedling male and female plants can be guaranteed. Comprising the steps of grinding, extraction, centrifugation and the like on the ginkgo seedling male and female plants, the provided method can be applied in large areas during production practice of urban garden greening, and can solve the identification problem of ginkgo seedling male and female plants. Being effective, the method has the advantages of simple operation, high accuracy, reduction of identification time, and cost saving.
Owner:LIAONING UNIVERSITY
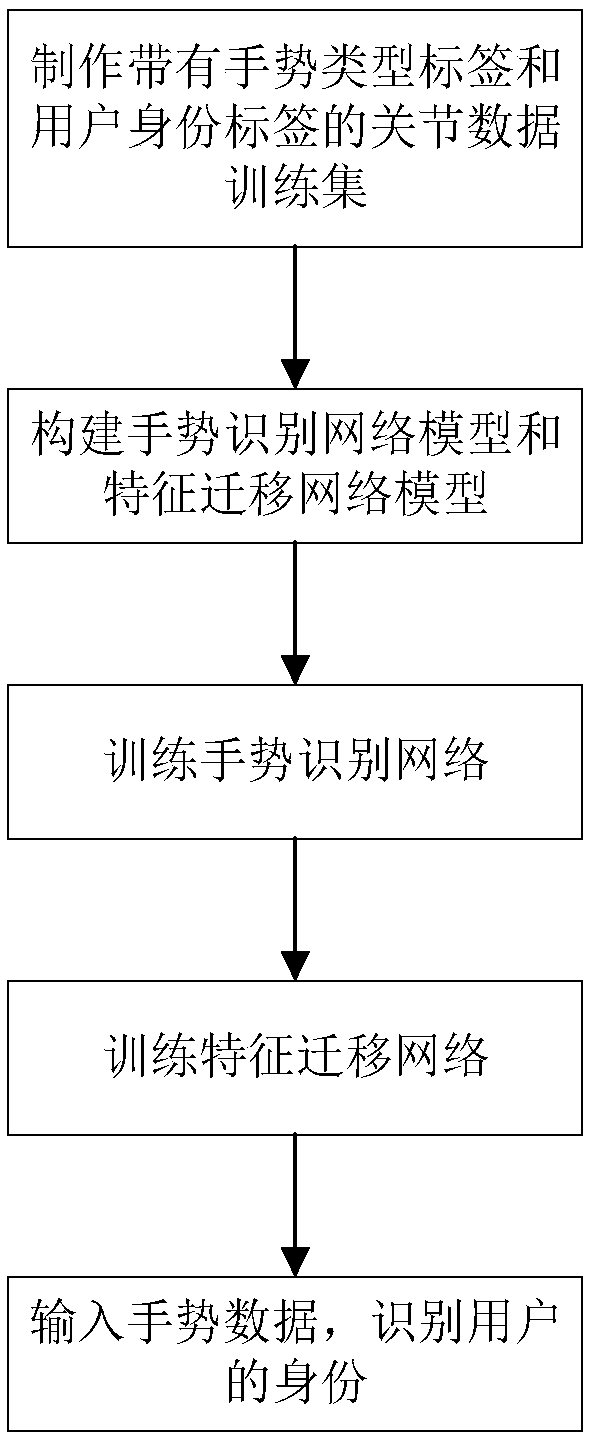
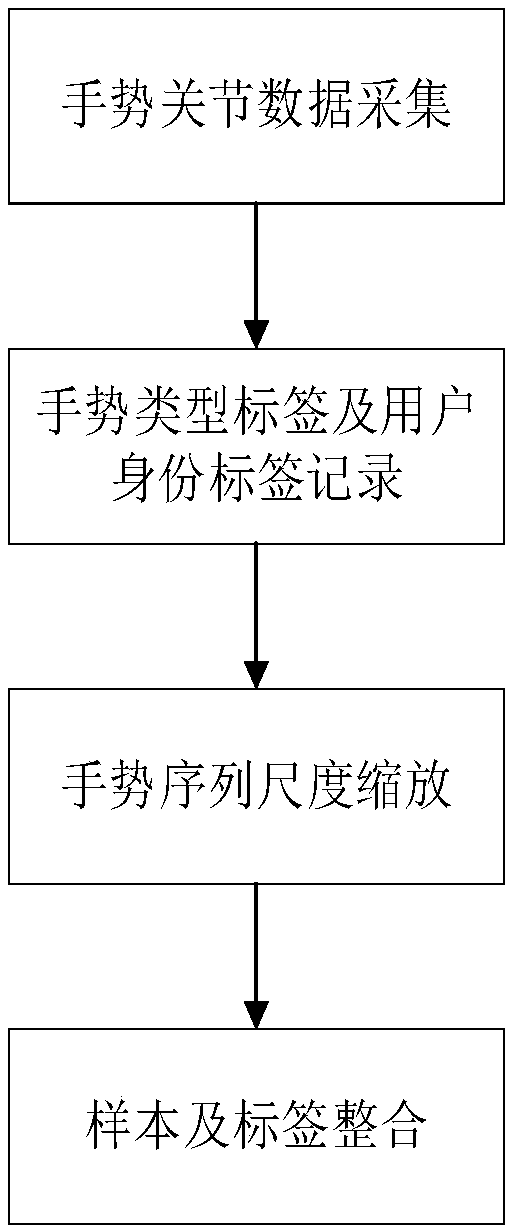
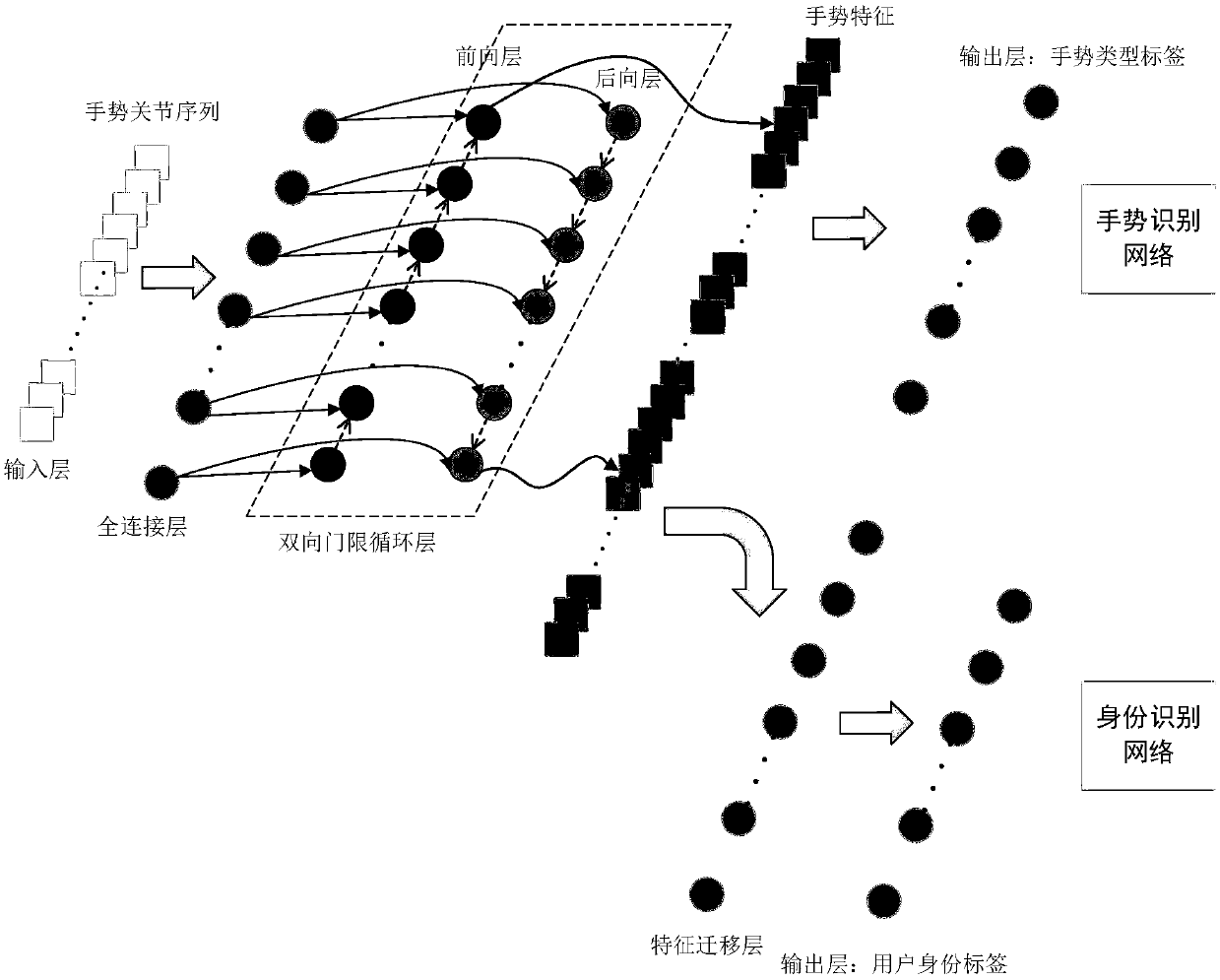
Method of converting gesture recognition to identification recognition on the basis of feature transfer learning
ActiveCN108960171AImprove performanceReduced user identification timeCharacter and pattern recognitionNeural architecturesNetwork modelNetwork on
The invention discloses a method of converting gesture recognition to identification recognition on the basis of feature transfer learning, belonging to the technical field of pattern recognition andbiometric recognition. The method includes the following main steps: step 1, making a gesture training set including both a gesture type tag and a user identity tag; step 2, constructing a gesture recognition network and a feature transfer network model; step 3, training the gesture recognition network on the basis of the made data set; step 4, training the feature transfer network model on the basis of the made data set; step 5, inputting a dynamic gesture on the basis of parameters of the learned feature transfer network model, and verifying a corresponding user identity. The invention provides a gesture recognition network based on a two-way threshold circulating network, adopts the feature transfer network to convert gesture recognition into identity recognition, and has a wide application prospect in the field of information security, medical dust prevention and the like.
Owner:ANHUI UNIVERSITY OF TECHNOLOGY
SSR (simple sequence repeat) molecular marker for detecting fertility of stamens of beta vulgaris and application of SSR molecular marker
ActiveCN107267660ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFertility
The invention relates to an SSR (simple sequence repeat) molecular marker for detecting fertility of stamens of beta vulgaris and application of the SSR molecular marker, and can be applied to detecting fertility of stamens of beta vulgaris. By the aid of the SSR molecular marker and the application, the problems of deficiency of molecular markers related to fertility of stamens of beta vulgaris and difficulty in identifying the fertility of the stamens of the beta vulgaris can be solved. Primer pairs for carrying out PCR (polymerase chain reaction) amplification on the molecular marker are BvRE051-S and BvRE051-A. The SSR molecular marker can be used for identifying the fertility of the stamens of the beta vulgaris. The SSR molecular marker and the application have the advantages that the consistency of detection results obtained by the aid of the SSR molecular marker and the performance of traits of the fertility of the stamens of the beta vulgaris can reach 70% and can reach 63.33% and 76.67% in fertile and infertile individuals; chain relations between the SSR molecular marker and the fertility of the stamens are shown, and accordingly the SSR molecular marker can be used for detecting the fertility of the stamens.
Owner:HEILONGJIANG UNIV +1
Method for identifying authenticity of pepper varieties and special SSR primer combination thereof
ActiveCN111270004AGuarantee authenticityImprove representationMicrobiological testing/measurementDNA/RNA fragmentationNucleotideGenetics
The invention belongs to the field of molecular markers and detecting thereof, and particularly relates to a method for identifying the authenticity of pepper varieties and a special SSR primer combination thereof. SSR loci for identifying the authenticity of the pepper variety are obtained by performing data mining according to a reference genome Capsicum.annum.L of pepper and 40 parts of germplasm resource whole genome re-sequencing data; the SSR primer combination is selected from the group consisting of the first SSR primer pair to the twenty-second SSR primer pair which are used for PCR amplification of the first SSR site to the twenty-second SSR site respectively, and preferably, the first SSR primer pair to the twenty-second SSR primer pair are nucleotide sequences with the homologyof 85%-100% with the nucleotide sequences shown as SEQ ID NO: 1-44 in the sequence table respectively. The method can be used for identifying the authenticity of the pepper variety in the full life cycle from seeds, and technical support is provided for pepper germplasm resource and new variety protection.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Method for identifying authenticity of brassica rapa variety and special SSR (simple sequence repeat) primer combination of method
ActiveCN111235302AGuarantee authenticityProtection of rights and interestsMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention belongs to the field of molecular markers and detection thereof and in particular relates to a method for identifying authenticity of a brassica rapa variety and a special SSR (simple sequence repeat) primer combination of the method. An SSR locus for identifying authenticity of a brassica rapa variety is obtained through data digging according to a reference genome Brassica_rapa V1.5 of brassica rapa and a reference genome Brassica olerecea V2.1 of brassica oleracea together with whole genome resequencing data of 40 genetic resources; the SSR primer combination is selected froma first SSR primer pair to a twenty-third SSR primer pair which are respectively applied to PCR (polymerase chain reaction) amplification on a first SSR locus to a twenty-third SSR locus, and are respectively optimally selected as nucleotide sequences which have 85-100% of homology with nucleotide sequences of SEQ ID NO:1-46 in a sequence table. The method can be applied to authenticity identification on the brassica rapa variety for a whole life cycle from seeds, and technical support is provided for protection on genetic resources and new varieties of brassica rapa.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
A calligraphy and painting record authentication device based on multistage magnification times
PendingCN106529506AEasy to adjustImprove accuracyTelevision system detailsColor television detailsMagnificationImage acquisition
The invention provides a calligraphy and painting record authentication device based on multistage magnification times. The device comprises a housing, a main board, a display screen, a storage unit and image acquisition devices. The technical key points are that the number of the image acquisition devices, of which the relative distance is fixed and magnification times are same or different, is N, N>=2; and the number of the image acquisition devices, of which the magnification times are different, is M, M>=2, and N>=M. The calligraphy and painting record authentication device has the advantages of simple system, high record and authentication efficiency, reliable authentication result, low authentication cost, high anti-counterfeiting capability and easy popularization and the like.
Owner:沈阳毓沣恒装饰材料有限公司
Standard detection sequence for DNA bar code of Leuciscus waleckii and application of standard detection sequence
ActiveCN107354226AAccurate identificationAccurate identification methodMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationDNA barcoding
The invention relates to a standard detection sequence for a DNA bar code of Leuciscus waleckii and application of the standard detection sequence. The invention aims at providing the standard detection sequence for the DNA bar code of the Leuciscus waleckii and application of the standard detection sequence in species identification. The standard detection sequence is represented by SEQ ID NO:1 in a sequence table. By utilizing the standard detection sequence for the DNA bar code of the Leuciscus waleckii, the molecular identification of the Leuciscus waleckii can be realized, and the identification time can be effectively shortened. The standard detection sequence is used for identifying the Leuciscus waleckii.
Owner:HEILONGJIANG RIVER FISHERY RES INST CHINESE ACADEMY OF FISHERIES SCI
Citrus mitochondrion InDel molecular markers and application thereof
ActiveCN110669863AHigh resolutionImprove stabilityMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyCitrus volkameriana
The invention discloses a set of citrus mitochondrion InDel molecular markers and application thereof, and belongs to the technical field of plant molecular marker assisted germplasm resource identification. Primer sequences of the 5 pairs of the citrus mitochondrion InDel molecular markers are shown as SEQ ID NO.1-10. Germplasms of citrus, poncirus and fortunella hindsii swingle can be rapidly and accurately identified by using primers of the citrus mitochondrion InDel molecular markers, the mitochondrial origin of a citrus protoplast fusion progeny can be identified, and the female parent origin of parent-unknown citrus cultivars can be analyzed. The citrus mitochondrion InDel molecular markers are of great significance in protecting and utilizing wild citrus resources.
Owner:HUAZHONG AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com