PCR-RFLP method for rapidly identifying radix tetrastigme and various counterfeits and adulterants of radix tetrastigme
A PCR-RFLP, cloverleaf technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of effective identification, high stability and versatility, and shortened identification time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Extraction of medicinal material DNA
[0043] Collect samples of Clover and various medicinal materials, and use the common CTAB (cetyltrimethylammonium bromide) method for extracting plant DNA or use the plant genome DNA extraction kit to extract sample DNA.
[0044] 1. Take about 0.5g of the sample in a mortar, add liquid nitrogen to grind it into a fine powder, and carefully transfer it to a 2ml centrifuge tube;
[0045] 2. Add 700 μL of 2% CTAB extract (CTAB 4g, NaCl 16.364g, 1M Tris-HCl (pH8.0), 0.5M EDTA 8ml) preheated at 65°C, dissolve it with 70ml ddH2O, and then dilute to 200ml Sterilize, add 0.2-1% β-mercaptoethanol after cooling), mix thoroughly, keep warm in 65°C water bath for 40min, and shake gently from time to time;
[0046] 3. Take out the centrifuge tube, centrifuge at 12000r / min for 10min, use a micropipette to suck out the supernatant, put it into a clean centrifuge tube, add an equal volume of chloroform-isoamyl alcohol (24:1), mix well, ...
Embodiment 2
[0051] Embodiment two PCR amplification of medicinal material DNA
[0052] 1. Design the primer sequence according to the internal transcribed spacer region of ribosomal DNA, the forward primer has the nucleotide sequence described in SEQID NO.1: 5'ATGCGATACTTGGTGTGAAT 3'; the reverse primer has the nucleotide sequence described in SEQID NO.2 Acid sequence: 5'GACGCTTCTCCAGACTACAAT 3'.
[0053] 2. PCR amplification is a 25 μL reaction system, which consists of the following components:
[0054]
[0055] 3. Amplification program: pre-denaturation at 95°C for 5 minutes; then cycle, denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 40 sec, 35 cycles; final extension at 72°C for 10 min.
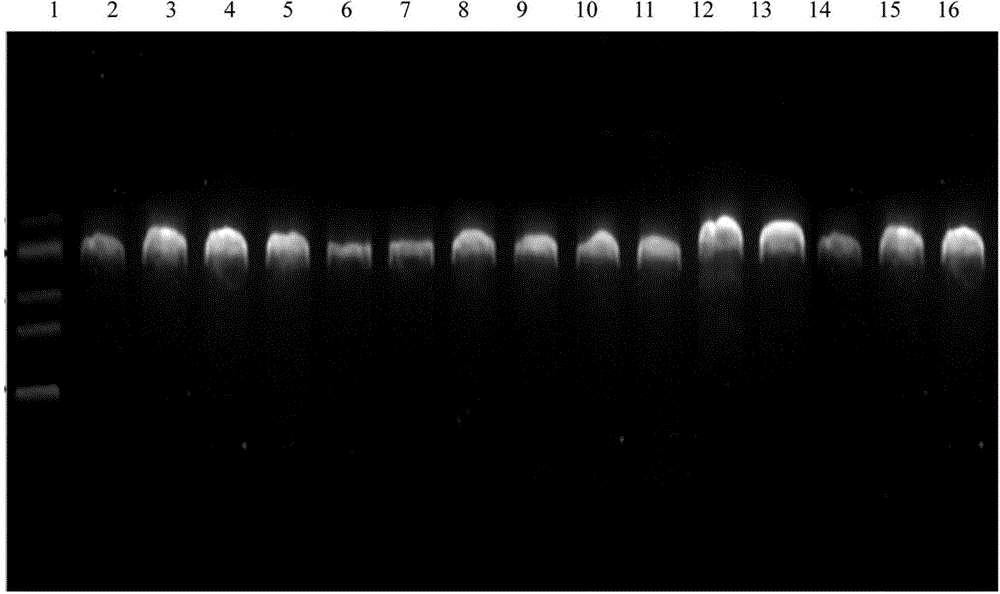
[0056] 4. Quality inspection of PCR amplification products: take 5 μL of DNA sample, electrophoresis on 2% agarose gel, voltage 120V, electrophoresis time is about 30min, after electrophoresis, the gel is stained with 0.5μg / mL EB, and run on a UV gel imaging ...
Embodiment 3
[0057] Example 3 Sequence Analysis of PCR Products
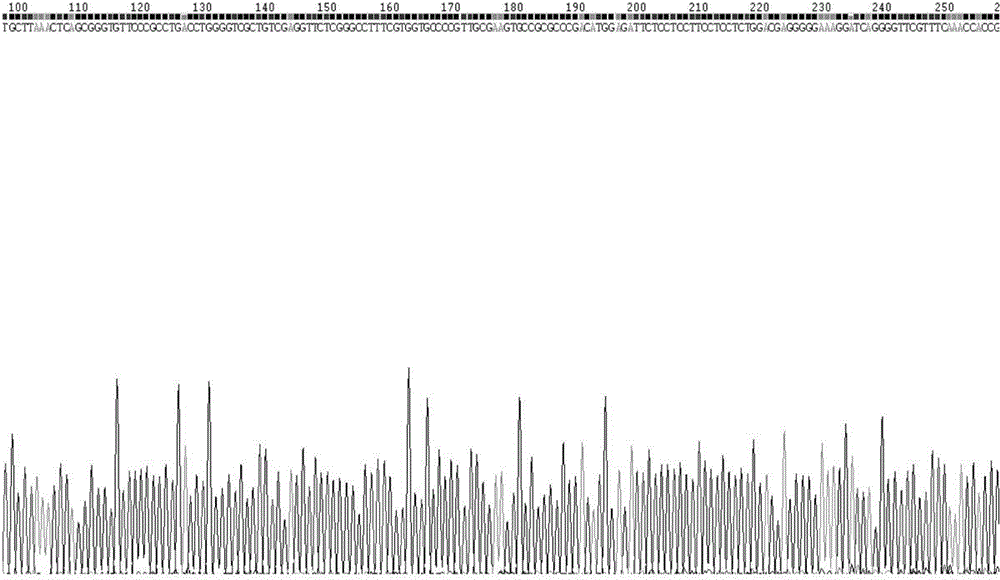
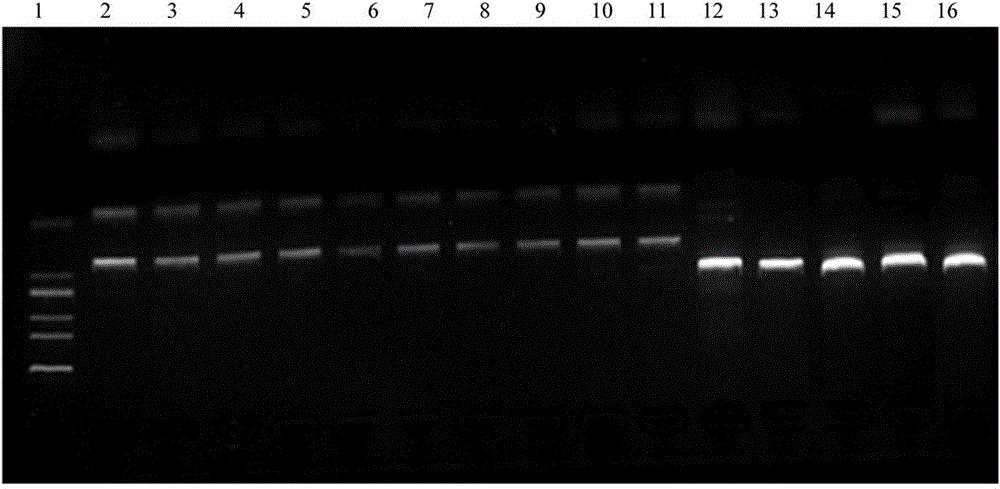
[0058] The PCR product was taken for DNA sequencing, using bidirectional sequencing, and part of the sequencing peaks are shown in figure 2 , the sequence determination result has the nucleotide sequence described in SEQID NO.3, which does not include the primer sequences at the 5' and 3' ends. The samples from Lishui, Wenling, Xiangshan in Zhejiang, Tianlin and Leye in Guangxi, Shaoyang, Yiyang, Yuanling in Hunan, Wanan and Shangrao in Jiangxi and other closely related varieties Tuqier, Aconitum, Daqingteng, The sequence of the PCR amplification products of the close relatives of A. clover and Crawling twig, after sequence analysis, the sample sequence of A. clover has the restriction endonuclease Nco I recognition sequence CCATGG, while other closely related samples are around 315bp There is no recognition site for this enzyme.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com