Patents
Literature
118 results about "Internal transcribed spacer" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
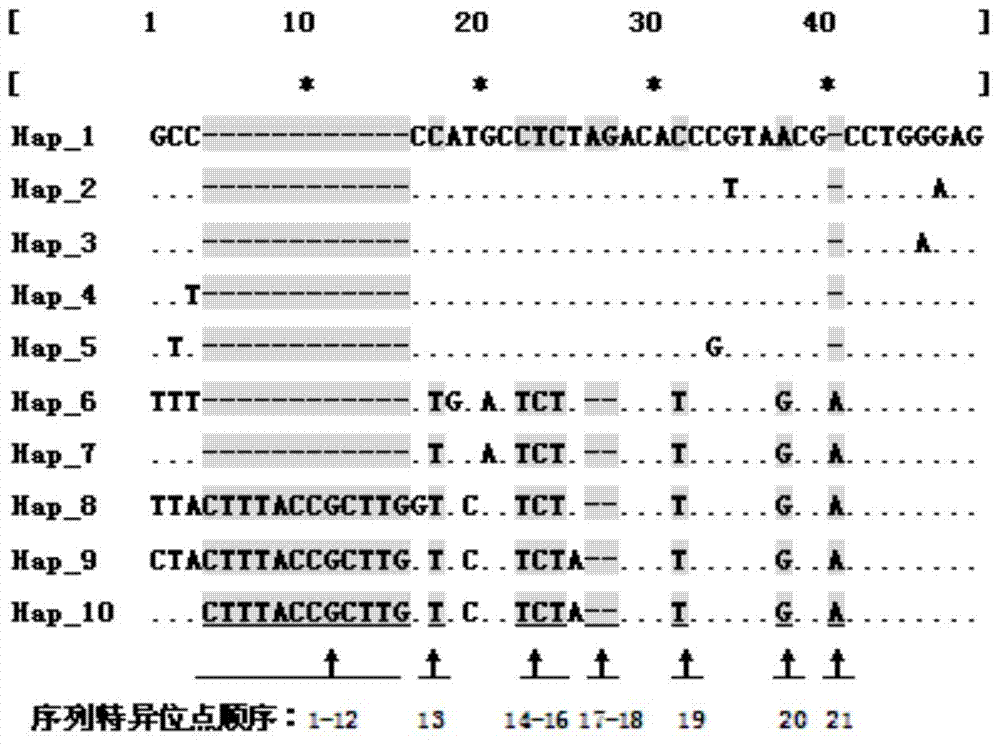
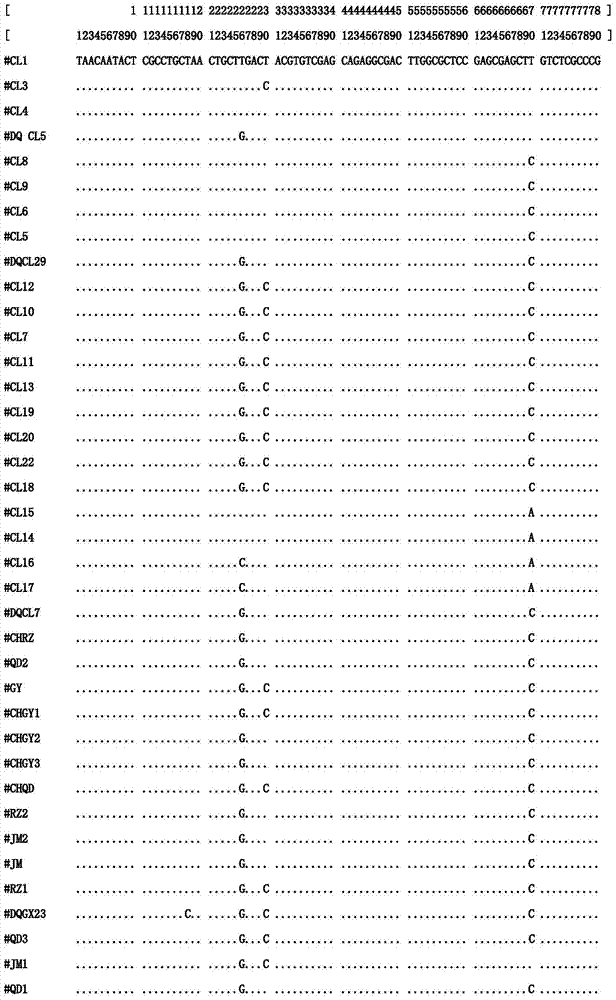
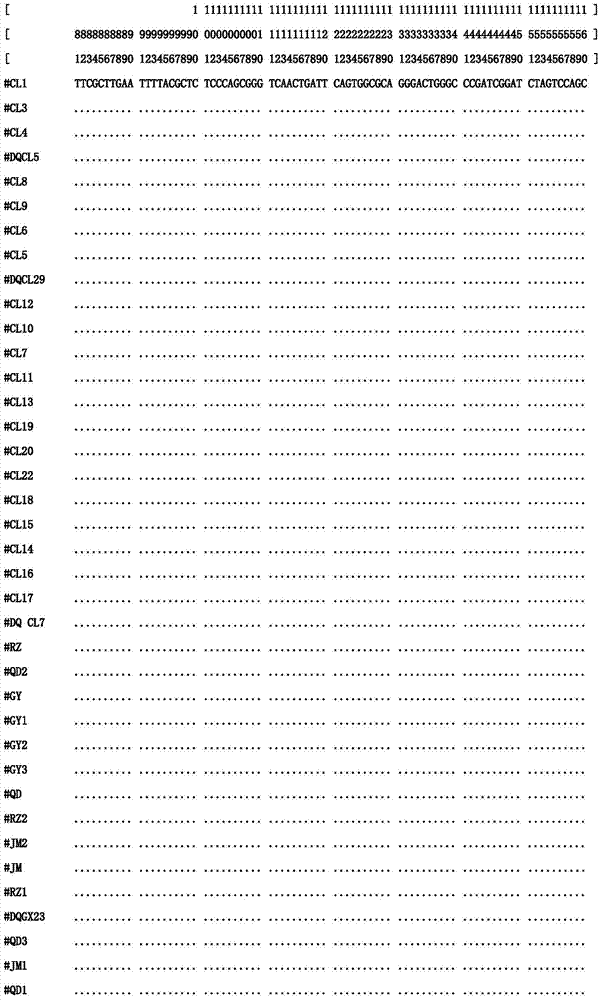
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript.
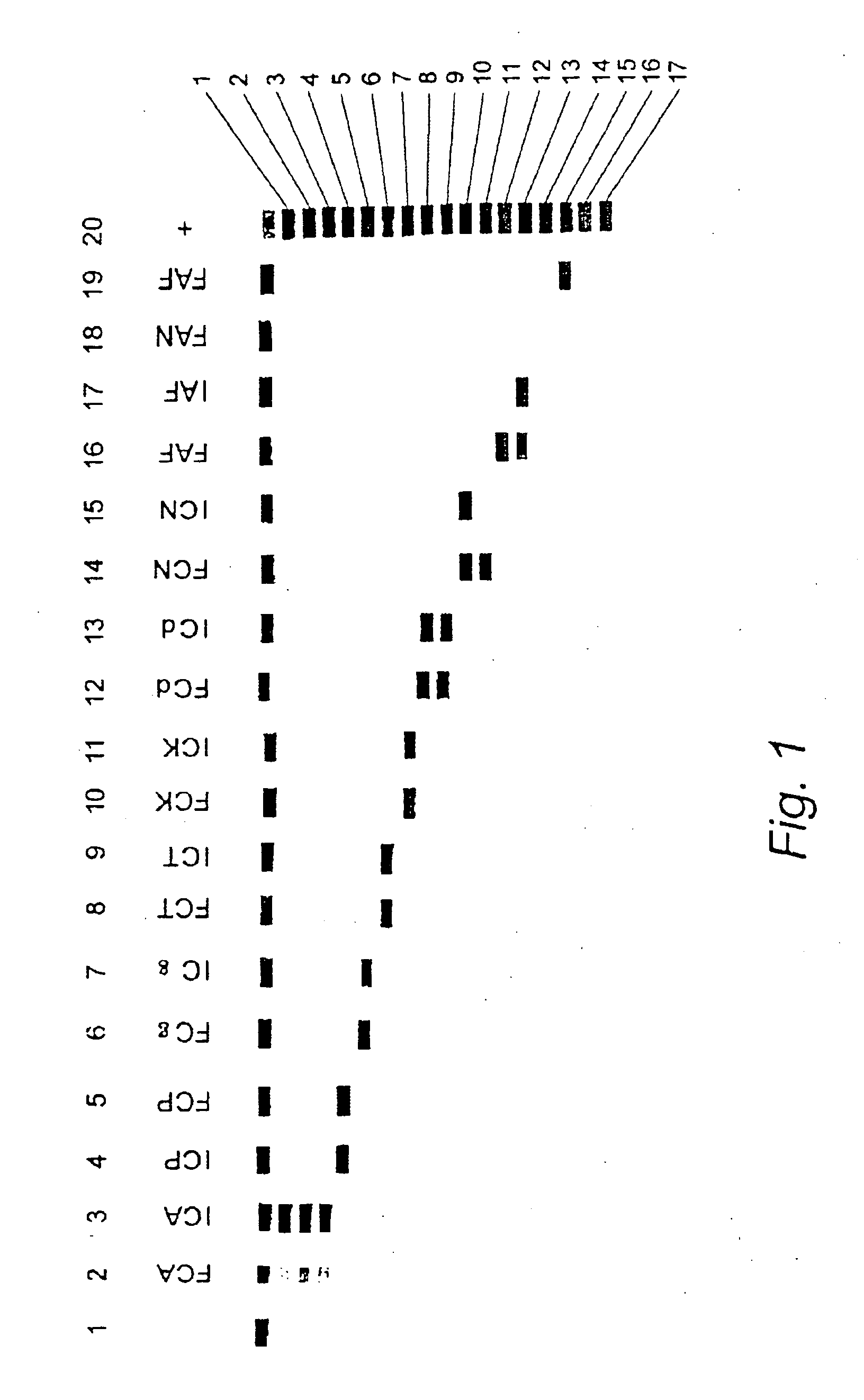
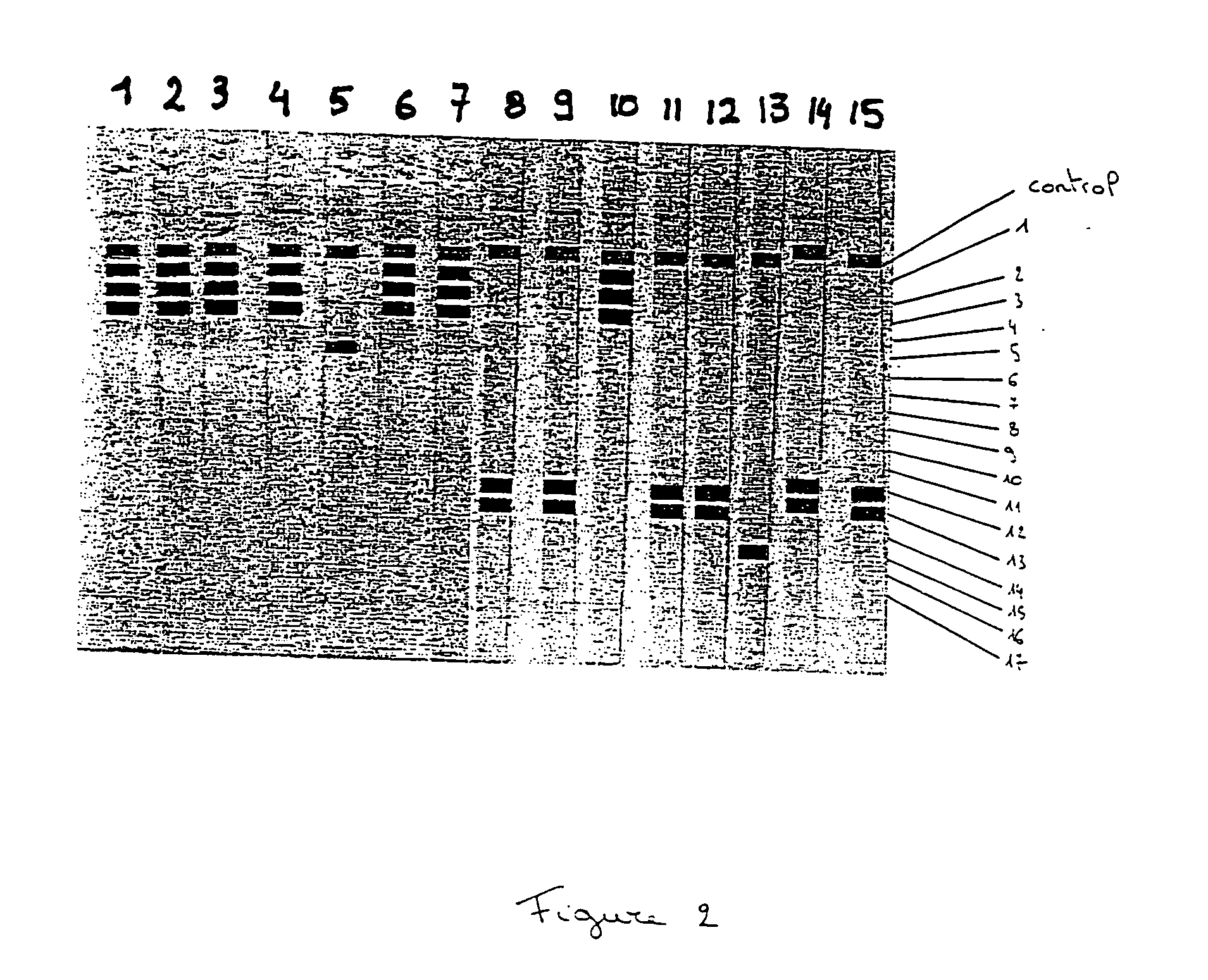
Nucleic acid probes and methods for detecting clinically important fungal pathogens
InactiveUS6858387B1Function increaseAttached with easeSugar derivativesMicrobiological testing/measurementNucleic Acid ProbesAspergillus flavus
The current invention relates to the field of detection and identification of clinically important fungi. More particularely, the present invention relates to species specific probes originating from the Internal Transcribed Spacer (ITS) region of rDNA for the detection of fungal species such as Candida albicans, Candida parapsilosis, Candida tropicalis, Candida kefyr, Candida krusei, Candida glabrata, Candida dubliniensis, Aspergillus flavus, Aspergillus versicolor, Aspergillus nidulans, Aspergillus fumigatus, Cyptococcus neoformans and Pneumocystis carinii in clinical samples, and methods using said probes.
Owner:ENTERPRISE IRELAND +2
Detection of fungal pathogens
Unique DNA sequences are provided which are useful in identifying different pathogenic fungi, such as those which infect grape plants. These unique DNA sequences can be used to provide oligonucleotide primers in PCR based analysis for the identification of fungal pathogens. The DNA sequences of the present invention include the internal transcribed spacer (ITS) of the ribosomal RNA gene regions of particular fungal pathogens, as well as oligonucleotide primers which are derived from these regions which are capable of identifying the particular pathogen.
Owner:E & J GALLO WINERY
Detection of fermentation-related microorganisms
InactiveUS6248519B1Sugar derivativesMicrobiological testing/measurementMicroorganismOligonucleotide primers
DNA sequences are provided which are useful in identifying different fermentation-related microorganisms, such as those involved in fermentations. These DNA sequences can be used to provide oligonucleotide primers in PCR based analysis for the identification of fermentation-related microorganisms. The DNA sequences of the present invention include the internal transcribed spacer (ITS) of the ribosomal RNA gene regions of particular fermentation-related microorganisms, as well as oligonucleotide primers which are derived from these regions which are capable of identifying the particular microorganism.
Owner:E & J GALLO WINERY
Microbial population analysis
ActiveUS20170159108A1Increase diversityMicrobiological testing/measurementICT adaptationMicroorganismIts region
The present invention relates to a method of typing a microbiome for having a desirable or undesirable signature, comprising analyzing the composition of the population of microorganisms in said microbiome based on taxonomic variation in the DNA sequence of the microbial 16S-23S rRNA internal transcribed spacer (ITS) regions in the genomic DNA of said microorganisms, wherein the sequences of conserved DNA regions comprised in the 16S and 23S rRNA sequences flanking said ITS region in the genome of said microorganisms comprise primer binding sites for amplification of said ITS regions.
Owner:IS DIAGNOSTICS
Application of nucleotide sequence of rDNA (recombinant deoxyribonucleic acid) ITS (internal transcribed spacer)-D3 region in establishment of DNA (deoxyribonucleic acid) bar code identification system for medicinal plants
InactiveCN102191318AImprove versatilitySequence alignment is accurateMicrobiological testing/measurementSpecial data processing applicationsNucleotideDNA barcoding
The invention relates to a method for identifying medicinal plants by utilizing nucleotide sequences, in particular to an application of a nucleotide sequence of an rDNA (recombinant deoxyribonucleic acid) ITS (internal transcribed spacer)-D3 region in establishment of a DNA (deoxyribonucleic acid) bar code identification system for medicinal plants. The application comprises the following steps of: firstly, detecting the nucleotide sequences of the ITS-D3 regions of medicinal plants, and establishing a DNA bar code database; then, detecting the nucleotide sequence of the ITS-D3 region of a sample to be identified; constructing a clustering tree by the detected nucleotide sequence and the nucleotide sequences of the ITS-D3 regions in the DNA bar code database; determining the medicinal plant having the closest genetic relationship with the sample to be identified in the DNA bar code database according to the clustering tree, and comparing the difference between the nucleotide sequences of the ITS-D3 regions of the medicinal plant and the sample to be identified; and then, by taking the name of the species of the medicinal plant having the closest genetic relationship with the sample to be identified in the database as a reference and combining information such as morphological characteristics and the like, determining the name of the species of the sample to be identified. In the invention, the DNA bar codes have the characteristics of good primer universality, accurate sequence comparison and strong species distinguishing capability.
Owner:GUANGZHOU UNIVERSITY OF CHINESE MEDICINE +1
Nucleic acid probes and methods for detecting clinically important fungal pathogens
InactiveUS20050164243A1Faster and simpler to performSuitable for automationSugar derivativesMicrobiological testing/measurementNucleic Acid ProbesAspergillus flavus
The current invention relates to the field of detection and identification of clinically important fungi. More particularely, the present invention relates to species specific probes originating from the Internal Transcribed Spacer (ITS) region of rDNA for the detection of fungal species such as Candida albicans, Candida parapsilosis, Candida tropicalis, Candida kefyr, Candida krusei, Candida glabrata, Candida dubliniensis, Aspergillus flavus, Aspergillus versicolor, Aspergillus nidulans, Aspergillus fumigatus, Cryptococcus neoformans and Pneumocystis carinii in clinical samples, and methods using said probes.
Owner:INNOGENETICS NV +2
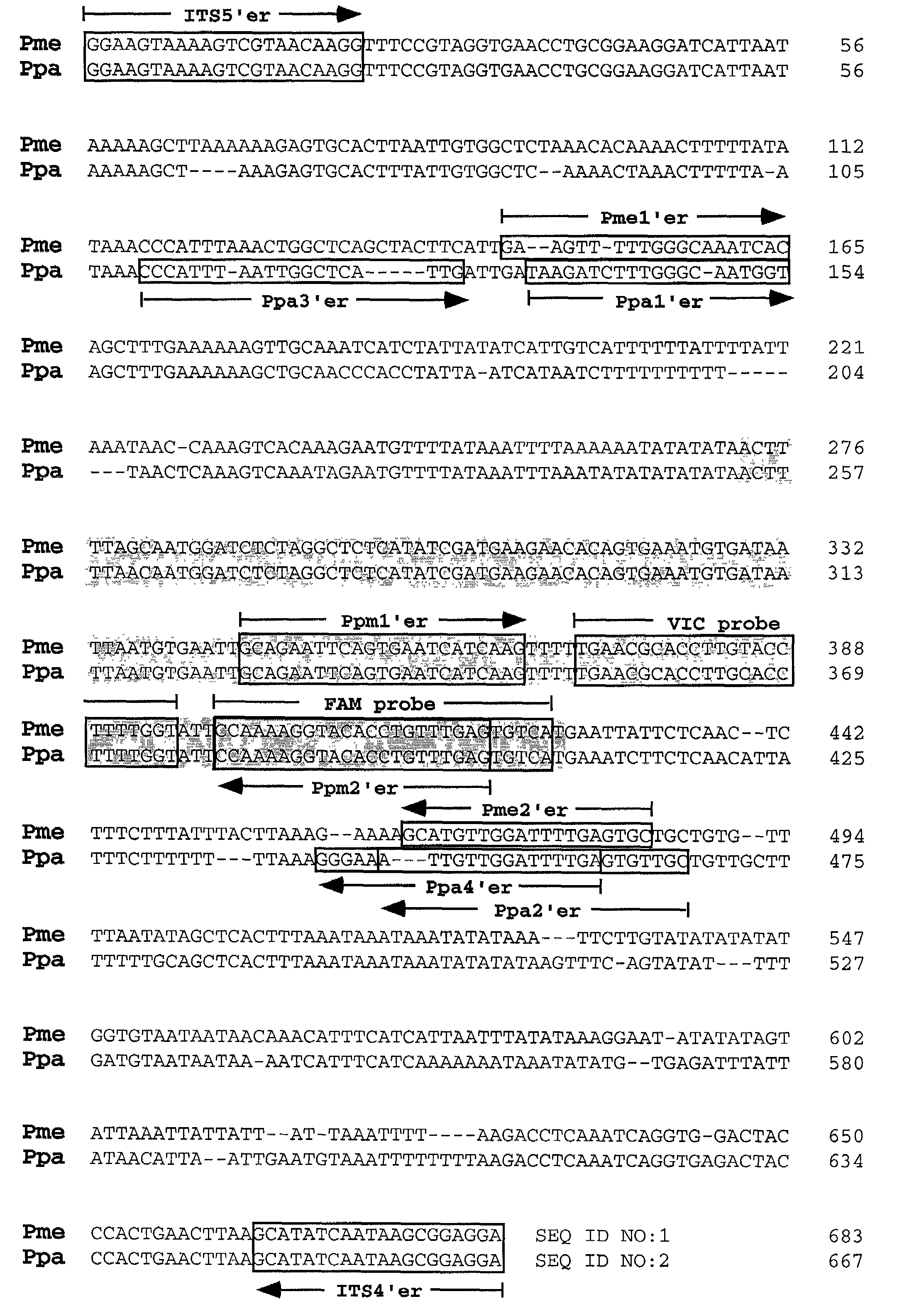
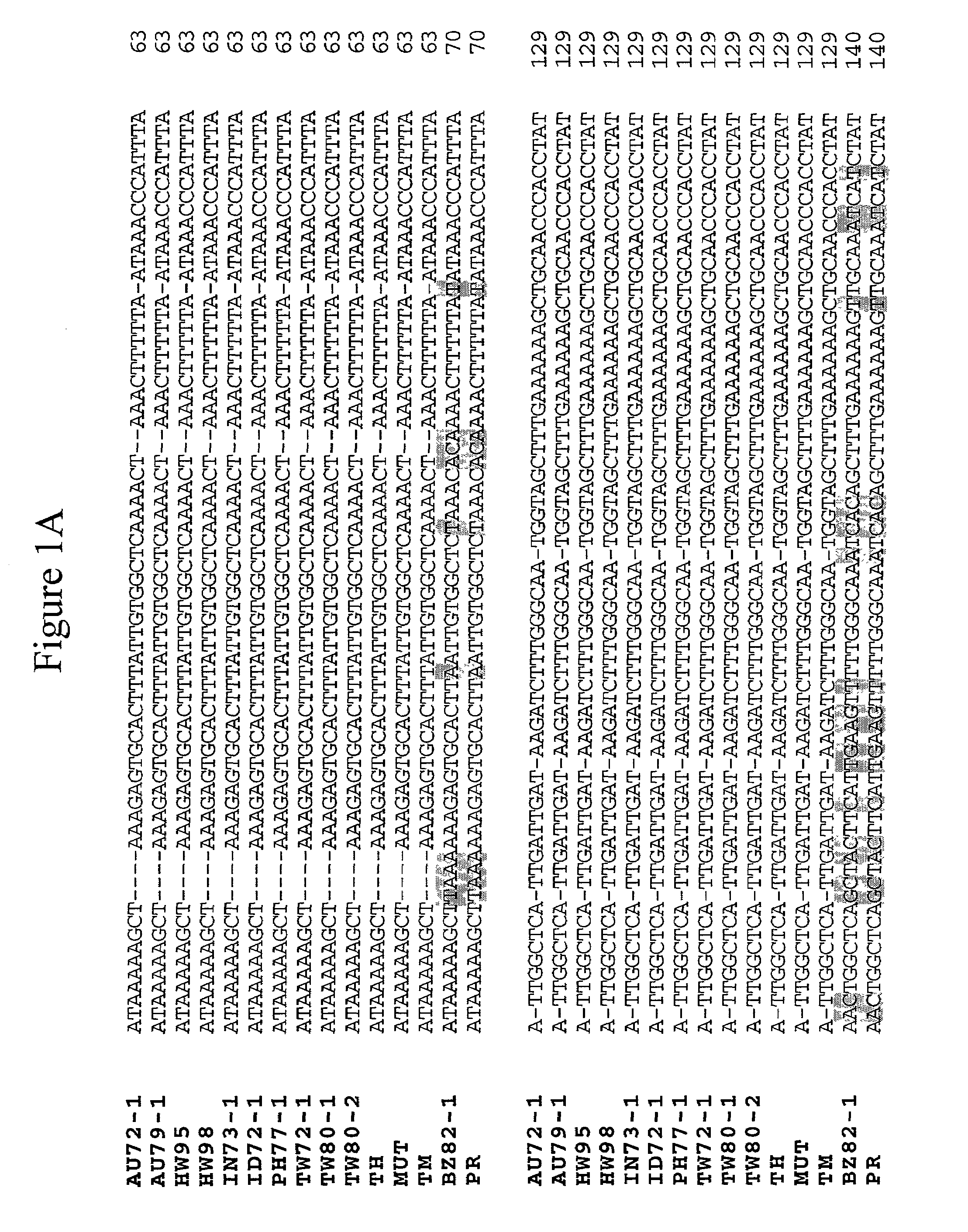
PCR methods for the identification and detection of the soybean rust pathogen Phakopsora pachyrhizi
InactiveUS7097975B1Readily apparentSugar derivativesMicrobiological testing/measurementPhakopsora pachyrhiziIts region
Soybean rust occurs in many countries throughout Asia, Australia, Africa, and South America. The causal agents of soybean rust are two closely related fungi, Phakopsora pachyrhizi and P. meibomiae, which are differentiated based upon morphological characteristics of the telia. Determination of the nucleotide sequence of the internal transcribed spacer (ITS) region revealed greater than 99% / 95% nucleotide sequence similarity among isolates of either P. pachyrhizi or P. meibomiae, but only 80% sequence similarity between the two species. Utilizing differences within the ITS region, four sets of PCR primers were designed specifically for P. pachyrhizi, and two sets of PCR primers were made specific to P. meibomiae. Classical and real-time fluorescent PCR assays were developed to identify and differentiate between P. pachyrhizi and P. meibomiae.
Owner:US SEC AGRI
Detection, identification and differentiation of eubacterial taxa using a hybridization assay
InactiveUS20060115819A1Rapid and reliable hybridization method for detectionSugar derivativesMicrobiological testing/measurementSpecific detectionIts region
The present invention relates to a method for the specific detection and / or identification of Staphylococcus species, in particular Staphylococcus aureus, using new nucleic acid sequences derived from the ITS (Internal Transcribed Spacer) region. The present invention relates also to said new nucleic acid sequences derived from the ITS region, between the 16S and 23S ribosomal ribonucleic acid (rRNA) or rRNA genes, to be used for the specific detection and / or identification of Staphylococcus species, in particular of S. aureus, in a biological sample. It relates also to nucleic acid primers to be used for the amplification of said spacer region of Staphylococcus species in a sample.
Owner:INNOGENETICS NV +1
High through-put detection of pathogenic yeasts in the genus trichosporon
InactiveUS20060216723A1Rapid and simple to performSugar derivativesMicrobiological testing/measurementSequence analysisPresent method
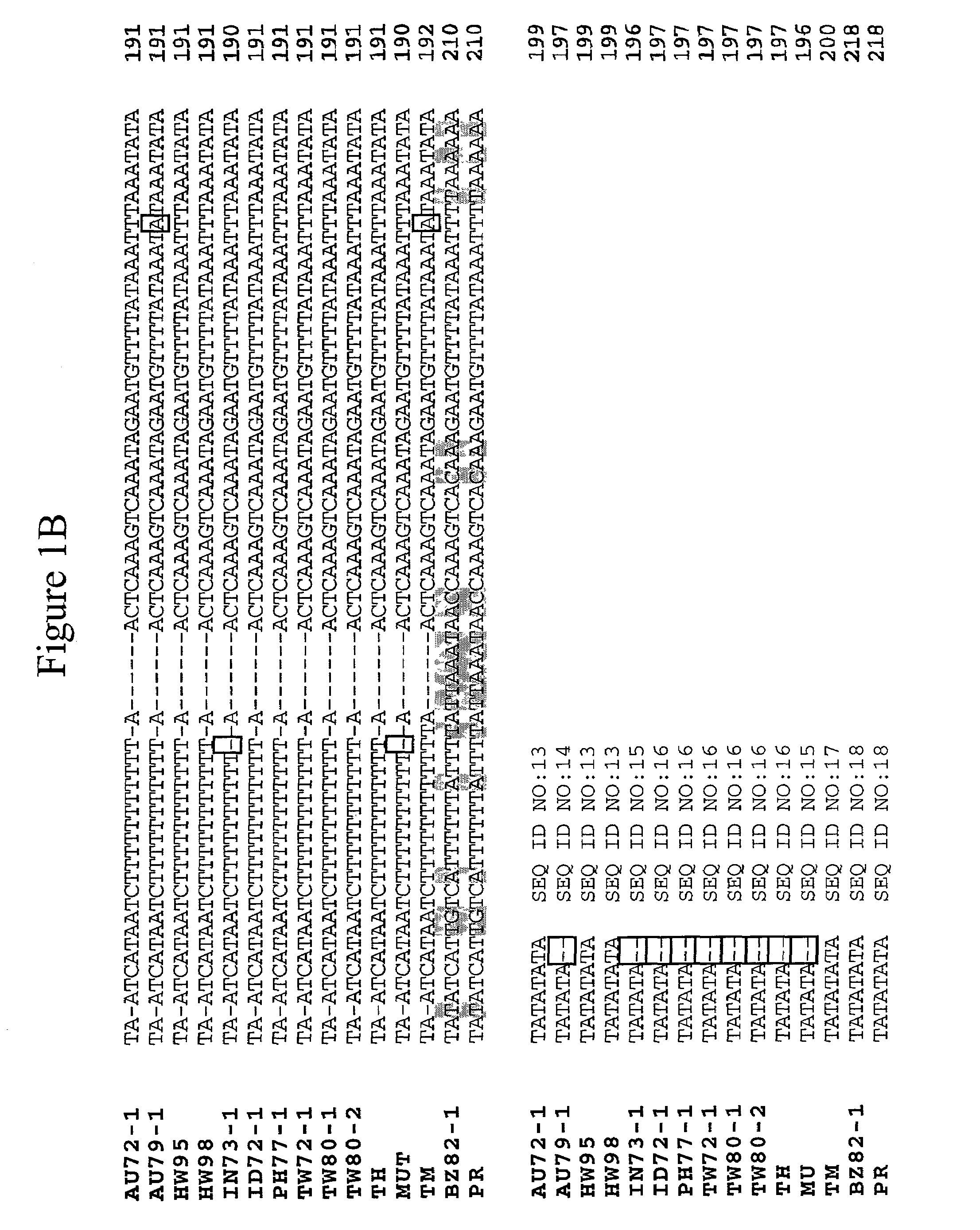
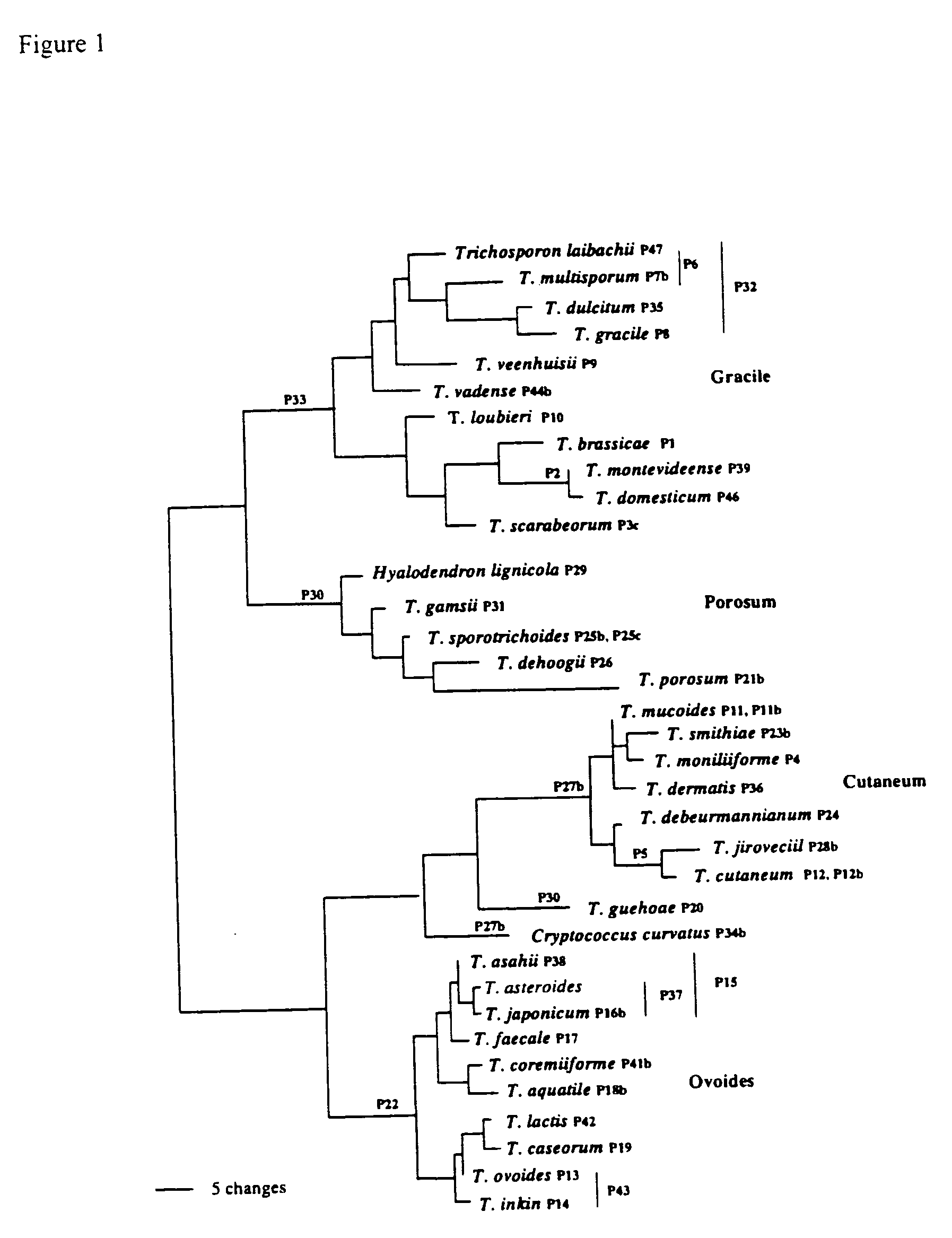
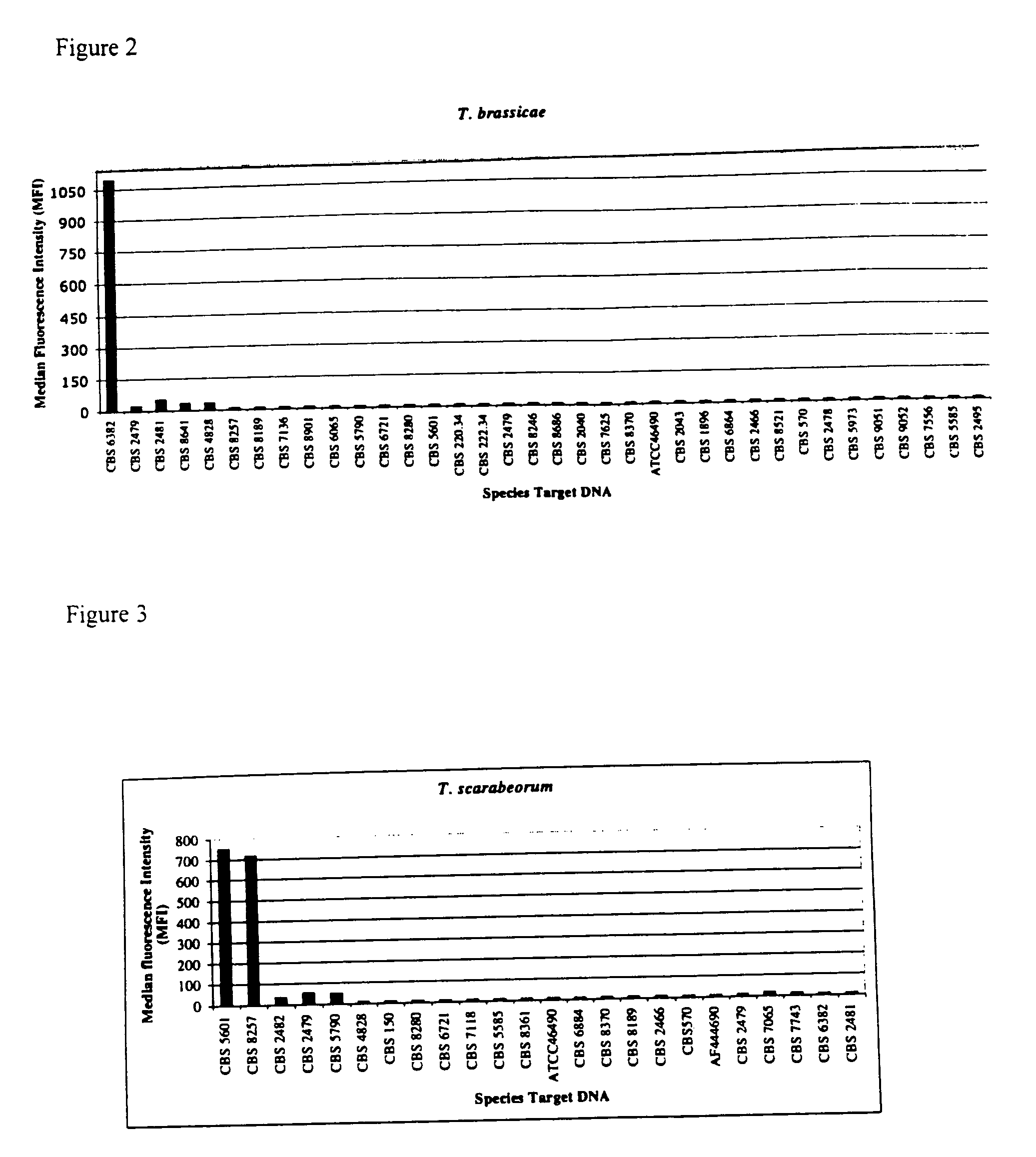
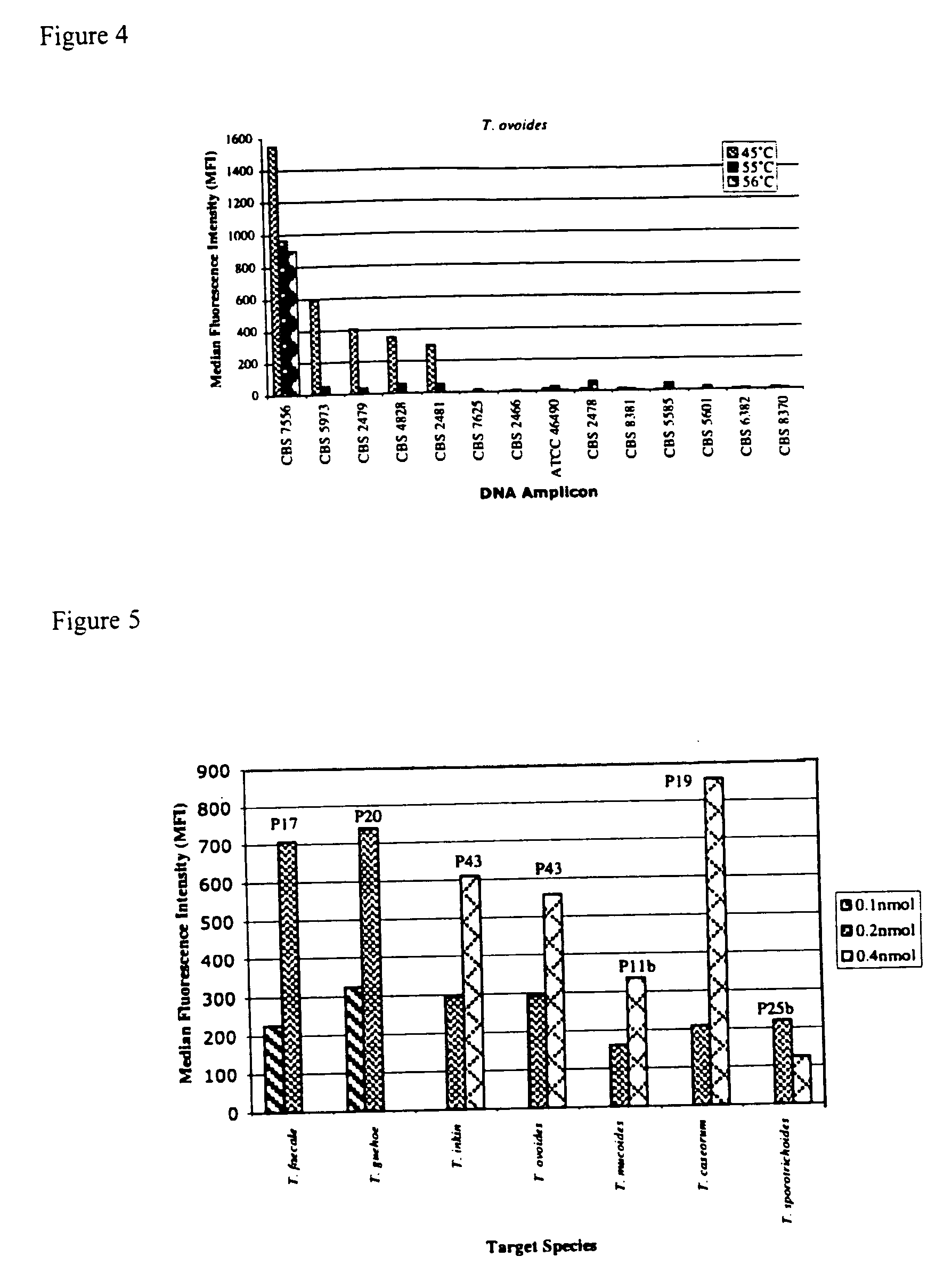
The emergence of opportunistic and antifungal resistant strains has given rise to an urgent need for a rapid and accurate method for the detection of fungal pathogens. In this application, we demonstrate the detection of medically important fungal pathogens at the species level. The present method, which is based on a nucleotide hybridization assay, consists of a combination of different sets of fluorescent beads covalently bound to species specific capture probes. Upon hybridization, the beads bearing the target amplicons are classified by their spectral addresses with a 635 nm laser. Quantitation of the hybridized biotinylated amplicon is based on the fluorescent detection with a 532 nm laser. Using this technology we designed and tested various multiplex formats, the performance of forty eight species specific and group specific capture probes designed from sequence analysis in the D1 / D2 region of ribosomal DNA, internal transcribed spacer regions (ITS), and intergenic spacer region (IGS). Species-specific biotinylated amplicons (>600 bp) were generated with three sets of primers to yield fragments from the three regions. The developed assay was specific and relatively fast, as it discriminated species differing by one nucleotide and required less than 50 min following amplification to process a 96 well plate with the capability to detect up to 100 species per well. The sensitivity of the assay allowed the detection as low as 102 genome molecules in PCR reactions and 107 to 108 molecules of biotinylated amplification product. This technology provided a rapid means of detection of Trichosporon species and had the flexibility to identify species in a multiplex format by combining different sets of beads. The assay can be expanded to include all known pathogenic fungal species.
Owner:MIAMI UNIVERISTY OF
Rapid molecular detection method and application for plasmopara viticola
ActiveCN103243166AEfficient detectionReliable detectionMicrobiological testing/measurementPlasmopara viticolaGenome
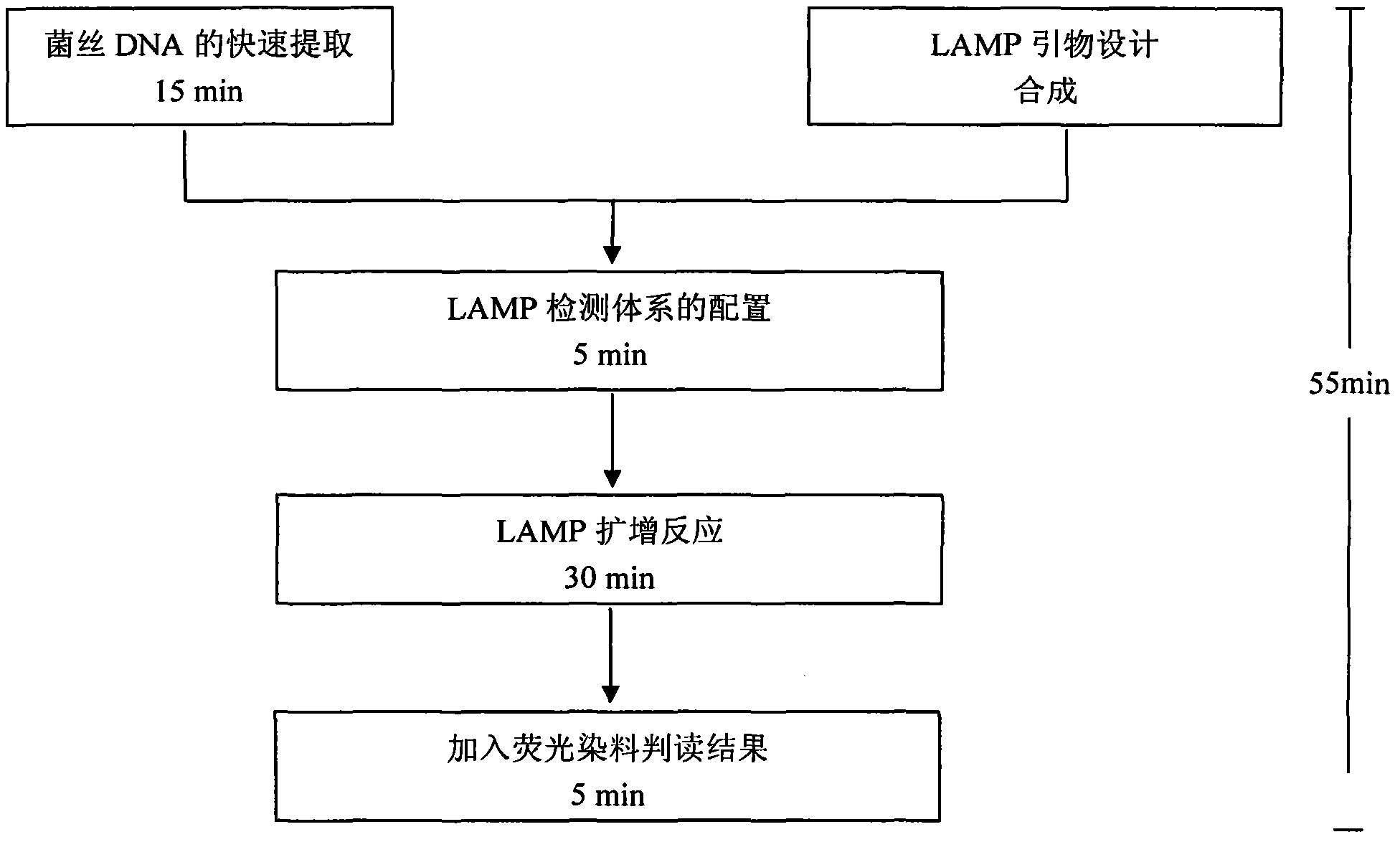
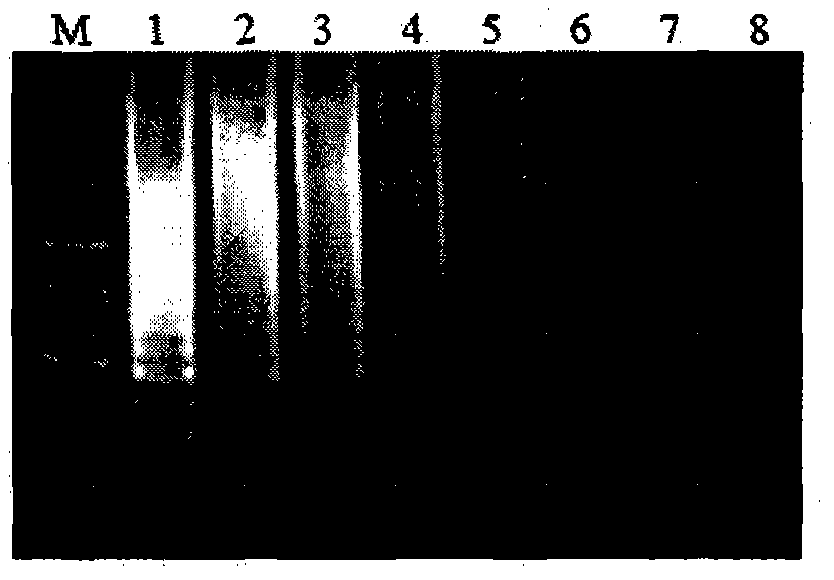
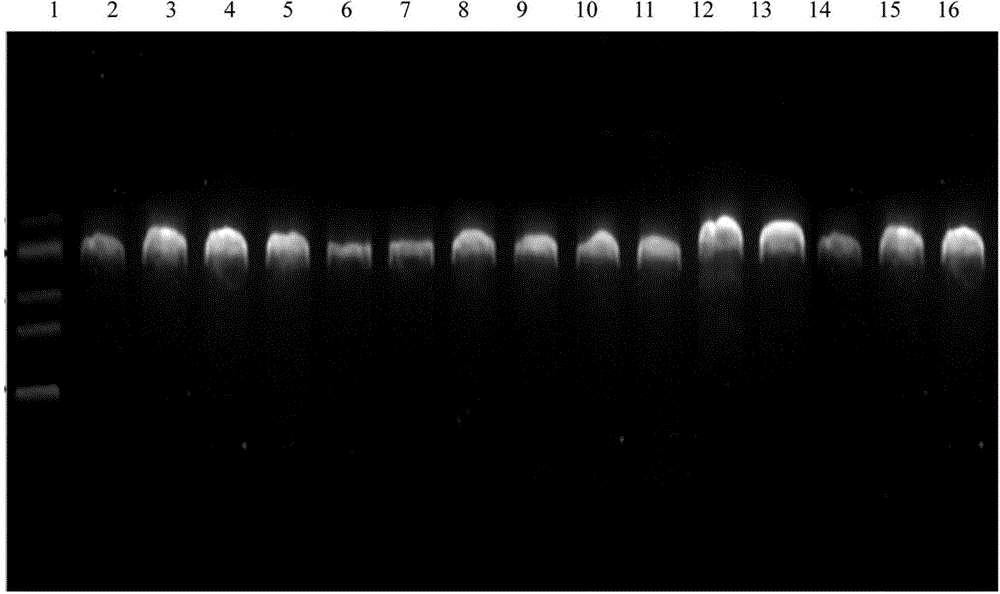
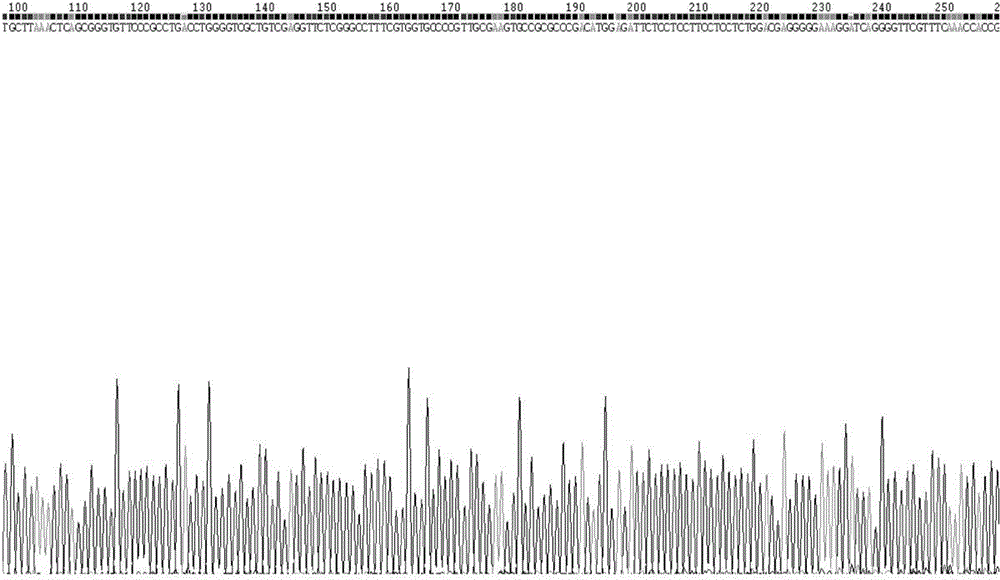
The invention belongs to the technical field of crop disease diagnosis and control, and in particular relates to a molecular detection method and application of plasmopara viticola. According to the molecular detection method, a group of looped-mediated isothermal amplification (LAMP) primers are self-designed through bioinformatic analysis according to the internal transcribed spacer (ITS) sequence information of a variety of disclosed peronosporales fungi and other common plant pathogenic fungi; and the existence of the deoxyribonucleic acid (DNA) of a plasmopara viticola genome at the level of 33fg / mu l can be rapidly detected from the plasmopara viticola samples of different varieties in different grape growing regions within only 55 minutes by combining a rapid extraction technology for the genome of the plasmopara viticola and adopting a one-step LAMP reaction. The rapid molecular detection method has high specificity, sensitivity and detection accuracy, and wide application range, and can be popularized and applied in various places as a rapid detection technology for the plasmopara viticola.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
PCR-RFLP method for rapidly identifying radix tetrastigme and various counterfeits and adulterants of radix tetrastigme
ActiveCN104611424ARapid identificationImprove accuracyMicrobiological testing/measurementMedicinal herbsEnzyme digestion
The invention belongs to the field of molecular markers and discloses a PCR-RFLP method for rapidly identifying radix tetrastigme and various counterfeits and adulterants of the radix tetrastigme. The method comprises the following steps: 1, extracting the NDA of the medicinal material (namely the radix tetrastigme); 2, carrying out PCR amplification by forward and reverse primers of internal transcribed spacer ITS2 sequences of a pair of amplified ribosomal DNAs; 3, digesting the PCR product by restriction enzyme NCO I; 4, carrying out agarose gel electrophoresis analysis. After the enzyme digestion of amplified products of the DNA of the radix tetrastigme, two DNA fragments with sizes about 335 bp and 200 bp are generated, and the various counterfeits and adulterants are not identified by the NCO I enzyme. By utilizing the differences between the DNA sequences of radix tetrastigme and the counterfeits and adulterants of the radix tetrastigme, a quick, convenient and reliable PCR-RFLP identification method is established and used for identifying whether counterfeits and adulterants are mixed in the radix tetrastigme or not, so that the technical problem that the truth and false cannot be identified by sensory or physical and chemical analysis methods is solved, and the medication safety of the radix tetrastigme is ensured.
Owner:ZHEJIANG PHARMA COLLEGE
Nucleotide specific to ITS of Balcillus proteus mirabilis and use thereof
ActiveCN101469326AQuick checkSensitive detectionSugar derivativesMicrobiological testing/measurementSocial benefitsNucleotide
The invention relates to nucleotide special to an Internal transcribed spacer (ITS for short) of a 16S rRNA-23S rRNA gene in Proteus mirabilis, in particular to oligonucleotide special to the ITS in the Proteus mirabilis and application thereof. The invention also provides a PCR detection reagent taking an oligonucleotide pair as a primer and a detection method thereof. The detection method utilizes the PCR reagent to detect the Proteus mirabilis in human body and the environment, is simple, convenient and quick, has good specificity and high sensitivity, can be used in the fields of supervision and detection of food and clinical samples, detection of pathogen in the food, microbial classification and epidemiological investigation and so on, and has deep social benefit and large economic benefit.
Owner:TIANJIN BIOCHIP TECH CO LTD
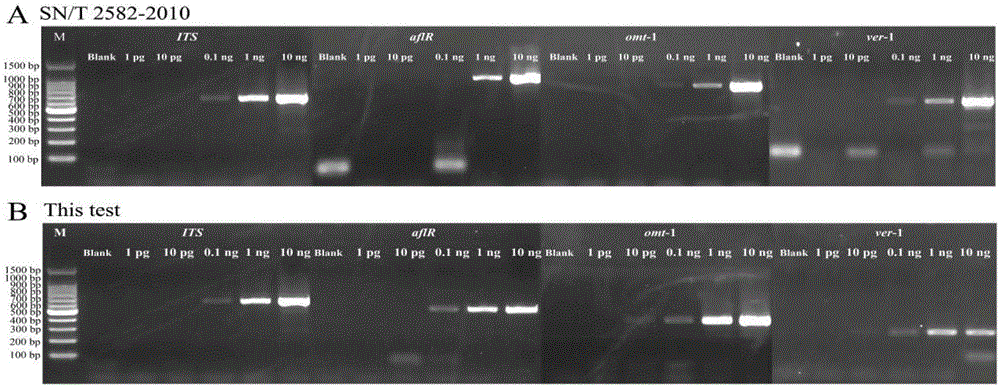
Multiple PCR detection primer group, kit and detection method for fungus producing aflatoxin
ActiveCN106318939AHigh detection throughputEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationMultiplex pcrsBiology
The invention relates to the field of biotechnology and provides a multiple PCR detection primer group, a kit and a detection method for fungus producing aflatoxin. According to the invention, through combined utilization of a fungus universal molecular marker sequence (internal transcribed spacer, ITS) and three key genes, comprising an aflatoxin production regulating gene (aflR), a sterigmatocystin-o-methoxy transferase gene (omt-1), and versicolorin A dehydrogenase gene (ver-1), in the biochemically synthetic process of the aflatoxin, the multiple PCR detection primer group, the kit and the detection method have the technical effects of wide coverage range, strong specificity and high sensitivity during the detection of the fungus producing aflatoxin. The primer group, the kit and the detection method are short in detection time, are simple in operation and are low in device requirement, can save large amount of manpower, materials and costs, are suitable for quick detection and are easy to promote in practical production.
Owner:COFCO GROUP +1
Rapid Fungus detection kit and detection method using same
ActiveCN101974640AShorten identification timeSave rescue timeMicrobiological testing/measurementMicroorganismMagnetic bead
The invention relates to the field of clinical microorganism identification, and provides a rapid fungus detection kit and a detection method using the same. The kit comprises two universal primers for amplifying an internal transcribed spacer (ITS1) of a fungus 18S, 5.8S, a sequencing primer for determining a sequence of a reverse section of the ITS1, a magnetic bead marked by an avidin, and a sequencing result and database comparison and judgment species. By using the kit, the fungus in a clinical sample can be distinguished by one-time sequencing, so the kit can be used for clinical diagnosis. The kit not only gains precious rescue time for clinical diagnosis and treatment, but also has the advantages that: the cost is low, the kit is convenient to operate and the specificity is high.
Owner:呼和浩特迪安医学检验所有限公司
Culture method for inducing differentiation of umbilical cord mesenchymal stem cell into cartilage and culture medium used in method
InactiveCN109456939ATight textureFast inductionCulture processSkeletal/connective tissue cellsCartilage cellsDexamethasone
The invention relates to the technical field of cell induction culture, and particularly relates to a culture method for inducing the differentiation of an umbilical cord mesenchymal stem cell into acartilage, and a culture medium used in the method. The culture medium is obtained by at least additionally adding the following components in a basal culture medium: 7 ng / mL to 13 ng / mL of TGF (Transforming Growth Factor)-beta 3, 0.07 mumol / L to 0.13 mumol / L of dexamethasone, 40 mug / mL to 60 mug / mL of ascorbate, 0.7 w / v% to 1.3 w / v% of 1*ITS (Internal Transcribed Spacer), 30 mug / mL to 50 mug / mL of L-proline and 0.7 mmol / L to 1.3 mmol / L of sodium pyruvate. According to the culture medium, through optimizing an induction component and matching a specific culture method, the induction velocity is improved; the hard and compact cartilage can be formed after 7 days; all inoculated mesenchymal stem cells can be differentiated into cartilage cells, and the differentiation rate is improved. An induced cartilage globule is compact in texture, and displayed through specific staining, and all mesenchymal stem cells are sufficiently differentiated into the cartilage cells.
Owner:BEIJING TAIDONG BIOTECH CO LTD
Primer and detection kit for detecting seven types of chicken eimeria tenella
ActiveCN103451288AEasy to operateRealize epidemiological investigationMicrobiological testing/measurementDNA/RNA fragmentationEimeria maximaIntergenic spacer
The invention belongs to the technical field of biological detection and particularly discloses a primer and a detection kit for detecting seven types of chicken eimeria tenella. The primer is obtained by designing IGS (Intergenic Spacer) and ITS (Internal Transcribed Spacer) specific sequences of the seven types of eimeria tenella, namely eimeria tenella, eimeria necatrix, eimeria maxima, eimeria acervulina, mild eimeria tenella, eimeria praecox and eimeria brunette. A fluorescence quantitative PCR (Polymerase Chain Reaction) kit prepared by the seven pairs of primers can be used for accurately, objectively and quantitatively detecting the seven types of chicken eimeria tenella; the detection kit has very high specificity and sensitivity.
Owner:ZHAOQING INST OF BIOTECHNOLOGY CO LTD
Nucleotides with specificity to internal transcribed spacer(ITS) of neisseria meningitidis and application thereof
ActiveCN102154466APracticalThe preparation method is simple and easyMicrobiological testing/measurementMicroorganism based processesSocial benefitsNucleotide
The invention relates to nucleotides SEQ ID NO: 1 to SEQ ID NO: 3 with specificity to the 16S rRNA-23S rRNA internal transcribed spacer (ITS) of neisseria meningitidis and application thereof, and provides a fluorescent quantitative PCR(polymerase chain reaction) detection kit and a detection method by using oligonucleotide as a primer and a probe. The fluorescent quantitative PCR detection kit has the advantages of simple operation, quick result, good specificity and high sensitivity when used for detecting the neisseria meningitidis in a human body and environment, can be used in fields such as supervision and detection of foods and clinical samples, detection of pathogenic bacteria of foods, bacteriological classification and epidemiological study and has significant social benefit andgreat economic benefit.
Owner:TIANJIN BIOCHIP TECH CO LTD
Method for extracting total DNA (deoxyribonucleic acid) of fungal hyphae
InactiveCN103820434AEasy to useReduce usageMicroorganism based processesDNA preparationWater bathsFreeze thawing
The invention belongs to the field of molecular biology study of fungi, and in particular relates to a method for extracting total DNA (deoxyribonucleic acid) of fungal hyphae, which has the advantages of avoiding liquid nitrogen grinding and relatively thoroughly breaking cell walls to extract the total DNA of the fungal hyphae. The method comprises the following steps: (1) collecting the fungal hyphae; (2) putting into a DNA extracting solution, and mashing; (3) repeatedly freeze-thawing for 2 to 3 times in a low temperature environment of -80 DEG C and a water bath environment of 65 DEG C; (4) adding protease K and lysozyme for treating; (5) removing the enzymes and purifying the DNA. The treatment method is simple, and the required agents are common; a repeated freeze-thawing and mashing mode is adopted, so that a sample is prevented from liquid nitrogen grinding, and then the use in places where liquid nitrogen can not be obtained easily is facilitated; a mortar is not used, so that cross contamination is prevented; toxic reagents such as phenol, beta-mercaptoethanol and the like are not used; the whole extracting process is mild, the quality of the extracted total DNA of the fungal hyphae is high, and subsequent molecular biology studies such as PCR (polymerase chain reaction) amplification and sequencing of an ITS (internal transcribed spacer) sequence and the like can be performed.
Owner:INST OF SOIL FERTILIZER SICHUAN ACAD OF AGRI SCI +2
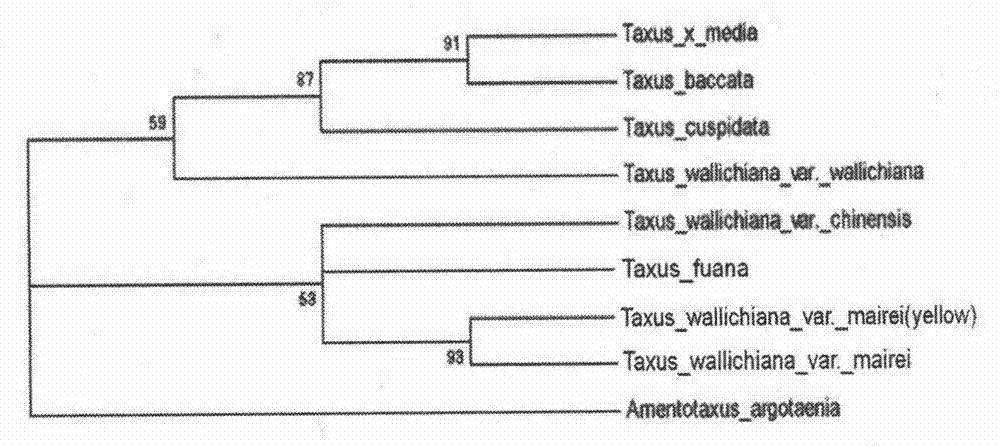
Nucleotide sequence and method used for identifying and differentiating species and varieties of Taxus chinensis
The invention discloses a nucleotide sequence used for identifying and differentiating species and varieties of Taxus chinensis. The sequence is originated from an internal transcribed spacer (ITS1) sequence of ribosome DNA (rDNA) of a proto-variety of Taxus wallichiana, and can be used for contrast and identification of the species and varieties of Chinese yew; and through analysis of the genetic characteristics of the germplasm resource of Taxus chinensis at a DNA level, effective molecular marks are provided for differentiation of the species and varieties of Taxus chinensis, and a good base is laid for identification, differentiation and screening of excellent germplasm of the species and varieties of Taxus chinensis. Meanwhile, the invention provides a method for identifying the species of Taxus chinensis by using the nucleotide sequence. The method is simple and practical to operate and can realize convenient, rapid, objective and accurate differentiation of different species and varieties of Taxus chinensis.
Owner:INST OF BOTANY JIANGSU PROVINCE & CHINESE ACADEMY OF SCI
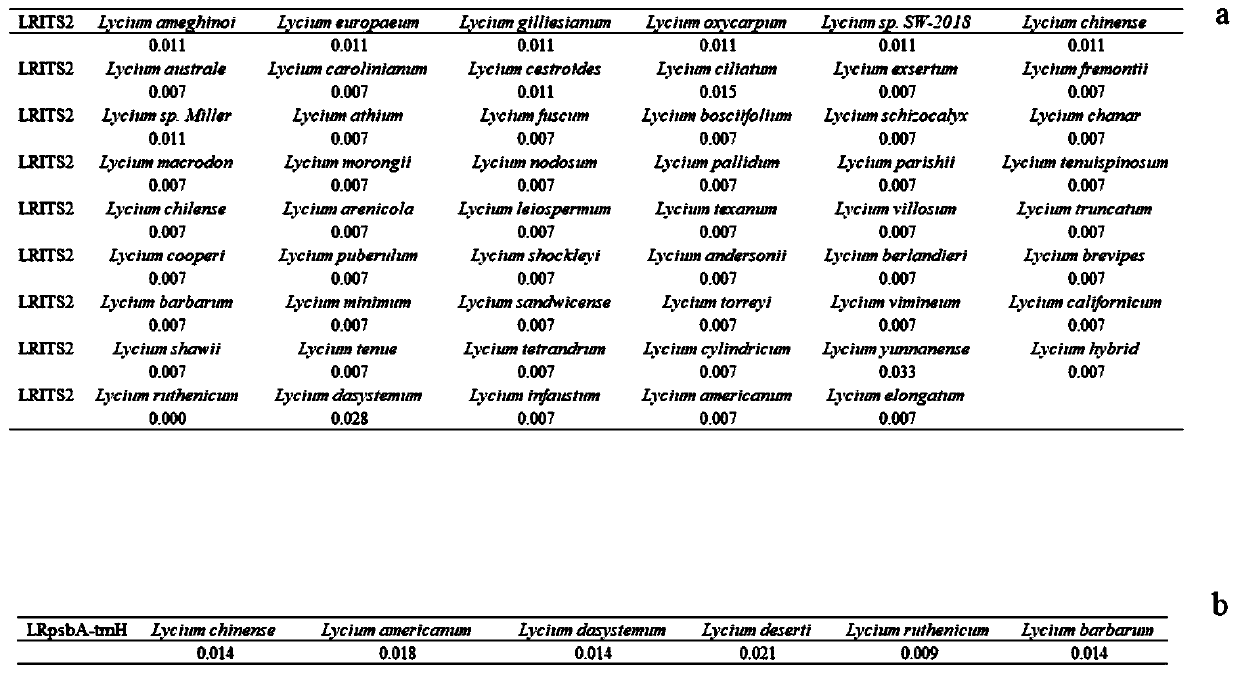
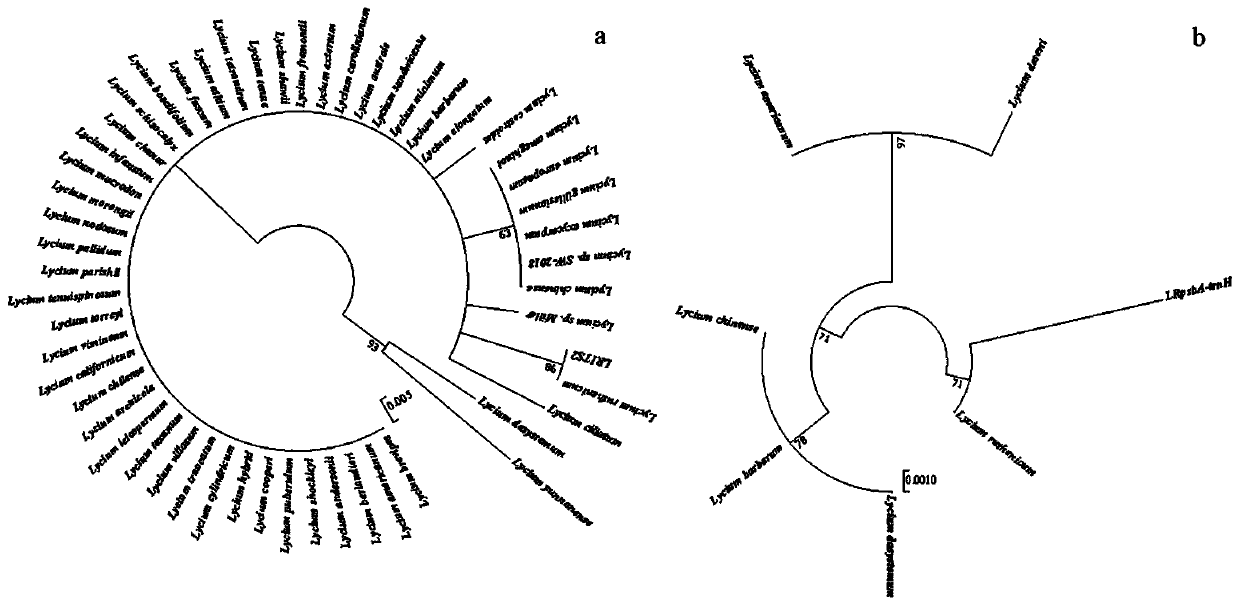
Method for identifying lycium ruthenicum based on DNA bar code and application thereof
PendingCN110229927AEfficient identificationAccurate identificationMicrobiological testing/measurementSocial benefitsDNA barcoding
The invention provides a method for identifying lycium ruthenicum based on a DNA bar code. The DNA bar code gene sequences for identifying the lycium ruthenicum include LRITS2 (the second internal transcribed spacer) and LRpsbA-trnH (a non-coding region between the chloroplast genes psbA and trnH). The DNA bar code sequences LRITS2 / LRpsbA-trnH for identifying lycium ruthenicum can be used simultaneously or optionally. The method can efficiently and accurately identify the lycium ruthenicum raw materials, prevents similar confusion products or counterfeit products, simultaneously can be used for identifying fruit powder, fruit fragments and the like, and has important application value and great social benefit for guaranteeing food safety and consumer rights and interests.
Owner:SHANGHAI NOVANAT BIORESOURCES CO LTD
Method for rapidly identifying coelomactra antiquate colony by rDNA ITS2 (recombinant Deoxyribose Nucleic Acid Internal Transcribed Spacer) sequence tag
InactiveCN103497995AAccurate identificationAvoid confusionMicrobiological testing/measurement3-deoxyriboseCoelomactra
The invention relates to a method for rapidly identifying a coelomactra antiquate colony by an rDNA ITS2 (recombinant Deoxyribose Nucleic Acid Internal Transcribed Spacer) sequence tag. The method comprises the steps of (1), collecting a coelomactra antiquate individual, and extracting DNA; (2), synthesizing an ITS2 sequence tag primer of the rDNA; (3), carrying out PCR (Polymerase Chain Reaction) amplification of the ITS2 sequence on a coelomactra antiquate colony sample; (4), purifying PCR products; and (5), carrying out sequencing and sequence alignment on the purified PCR products to obtain a specific locus, and judging the type of the sample. The method is stable in result, high in repeatability and simple in operation, and is of great importance in genetic resource conservation and scientific utilization of shellfish.
Owner:INST OF OCEANOLOGY & MARINE FISHERIES JIANGSU
Application of ITS (internal transcribed spacer) sequence as DNA barcode in identifying Jiaxing Zuili (Prunus salicina lindl.) and identifying method
ActiveCN105671185AImprove detection accuracyGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationPrunus salicina
The invention discloses application of ITS (internal transcribed spacer) sequence as a DNA barcode in identifying Jiaxing Zuili (Prunus salicina lindl.) and an identifying method and belongs to the technical field of molecular identification. The identifying method comprises: amplifying ITS sequence of a sample to be tested by using PCR (polymerase chain reaction) and carrying out sequencing; results show that if both basic group 165bp and basic group 195bp of the ITS sequence are 'T', it is determined that the sample to be tested is Jiaxing Zuili. The invention also discloses a method for identifying early maturing and late maturing varieties of Jiaxing Zuili; if the basic group sequence of the ITS sequence is shown as in SEQ ID No. 1, it is determined that the sample to be tested is an early maturing variety of Jiaxing Zuili; if the basic group sequence of the ITS sequence is shown as in SEQ ID No. 2, it is determined that the sample to be tested is a late maturing variety of Jiaxing Zuili. The ITS sequence is used herein as the DNA barcode to identify Jiaxing Zuili, detection accuracy is high, detection repeatability is high, and identifying time is short.
Owner:ZHEJIANG JIAXING AGRI SCI ACADEMY INST
New combination of universal amplification primers for plant parasitic nematode ribosome ITS (internal transcribed spacer) and application method thereof
InactiveCN104032002ASolve many problems of expansionBroad SpectrumMicrobiological testing/measurementDNA/RNA fragmentationPlant nematodeEnzyme digestion
The invention provides a new combination of two pairs of universal amplification primers for plant parasitic nematode ribosome ITS (internal transcribed spacer) and an application method thereof for PCR (polymerase chain reaction) amplification. The key innovation point of the method is that after the new primer combination is applied to the PCR amplification of the plant parasitic nematode ITS, the plant nematode type which can be amplified by the new PCR amplification system has broader spectrum, the amplification efficiency and success rate are remarkably improved, and the problems of the ITS amplification in the plant nematode identification before are effectively solved. The PCR amplification product amplified by the method provided by the invention meets the needs of the molecular detection technologies such as nematode DNA (deoxyribonucleic acid) barcode identification and RFLP (restricted fragment length polymorphism) enzyme digestion identification, and can be popularized to the quarantine identification of the port and agriculture / forestry departments in China.
Owner:ANIMAL & PLANT & FOOD INSPECTION CENT OF TIANJIN ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Identifying method for pleurotus nebrodensis-pleurotus eryngii interspecific hybrid
InactiveCN102618663AShort detection timeImprove accuracyMicrobiological testing/measurementPleurotus nebrodensisBio engineering
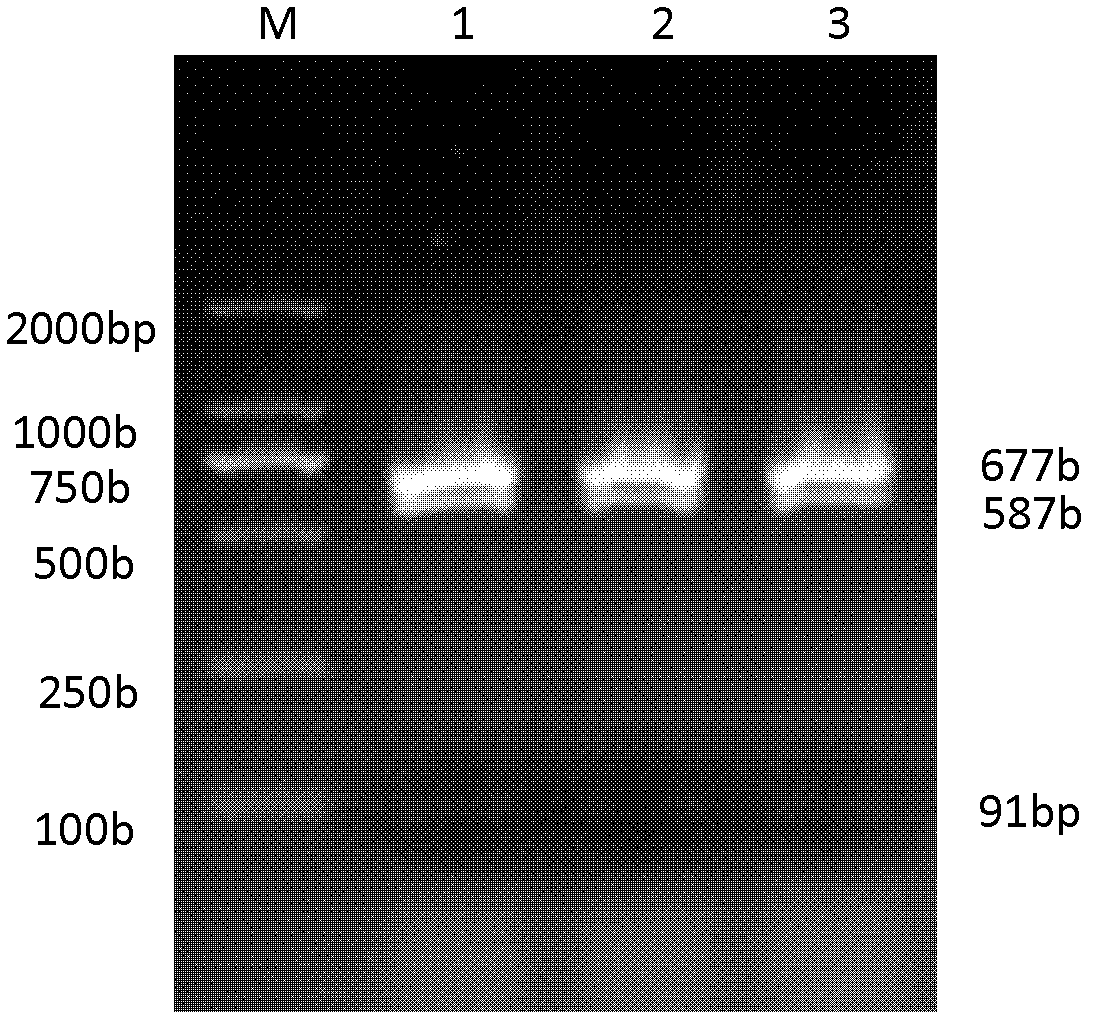
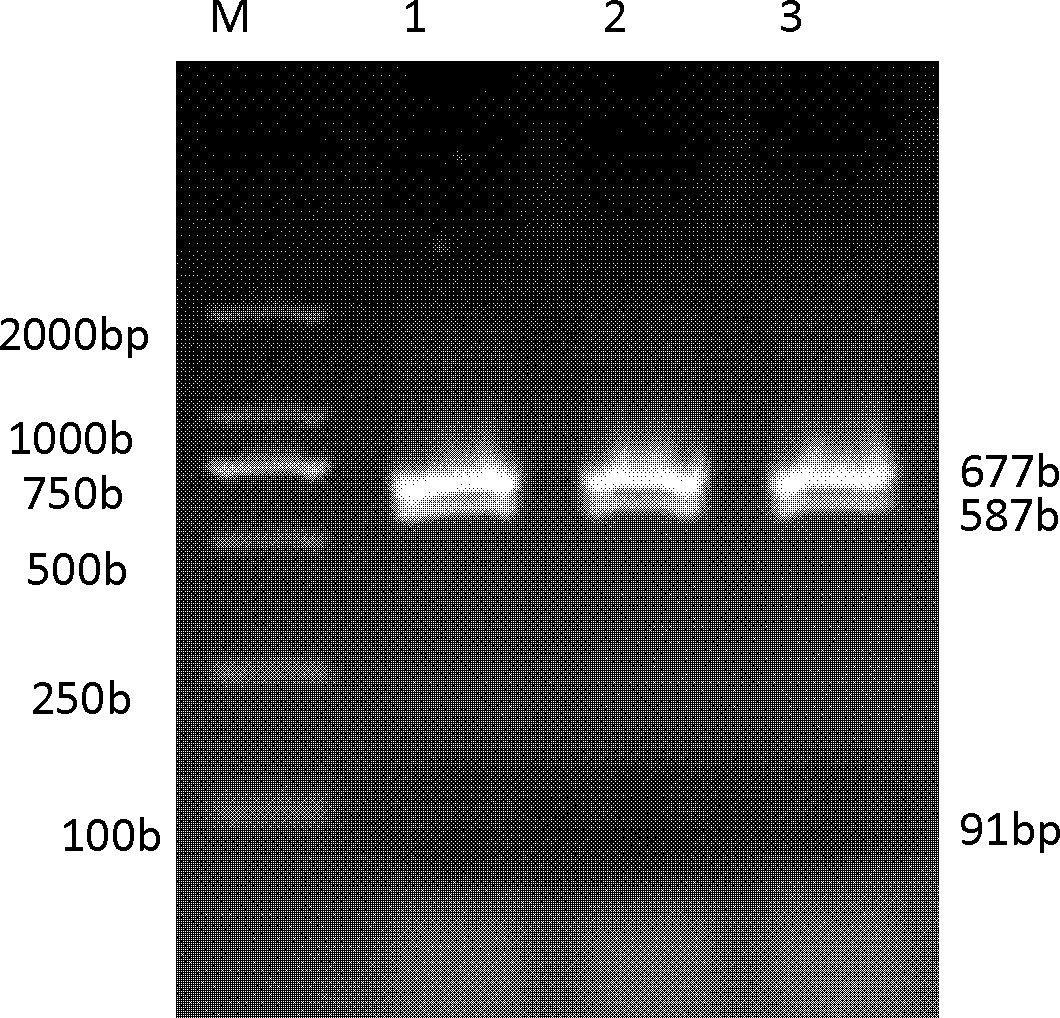
The invention belongs to the field of biological engineering, and is used for solving the technical problem of difficulty in screening a pleurotus nebrodensis-pleurotus eryngii interspecific hybrid in the prior art. The invention provides an identifying method of the pleurotus nebrodensis-pleurotus eryngii interspecific hybrid. The method comprises the following steps of: preparing a strain protoplast; culturing the protoplast; culturing and collecting hyphae in the protoplast; extracting genome DNAs (Deoxyribonucleic Acids) in the hyphae; performing ITS-PCR (Internal Transcribed Spacer-Polymerase Chain Reaction) amplification on the extracted DNAs; performing specific enzyme splicing on an amplification product; and performing electrophoresis on an enzyme splicing product. If DNA marking strips of which the molecular weights are 675-685bp, 585-595bp and 85-95bp are generated according to electrophoresis detection, the strain is an interspecific hybrid of pleurotus nebrodensis and pleurotus eryngii. The identifying method disclosed by the invention has the advantages of short detection time, high accuracy, easiness for judging and high stability.
Owner:SHANGHAI BAIRONG VELVET MUSHROOM
ITS (internal transcribed spacer) sequence of dalbergia odorifera and method for identifying dalbergia odorifera by ITS sequence
InactiveCN106434645AEasy to identifyEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationNucleotide sequencing
The invention discloses an ITS (internal transcribed spacer) sequence of dalbergia odorifera and a method for identifying the dalbergia odorifera by the ITS sequence. The ITS sequence of the dalbergia odorifera is characterized in that a nucleotide sequence is shown in SEQ ID NO:1. The method comprises the following steps of using primers ITS4 and ITS5 to perform PCR (polymerase chain reaction) proliferation on DNA (deoxyribonucleic acid) of a to-be-tested wood sample, so as to obtain ITS sequences; matching and comparing the two ITS sequences, and checking basic group information of ten sites, so as to judge whether the to-be-tested wood sample is the dalbergia odorifera or not. The method has the advantages that the dalbergia odorifera can be simply, quickly and efficiently subjected to molecular identification at the molecular biological level; the cost is low, the operation is simple, and the identification accuracy is high.
Owner:GUANGDONG PHARMA UNIV
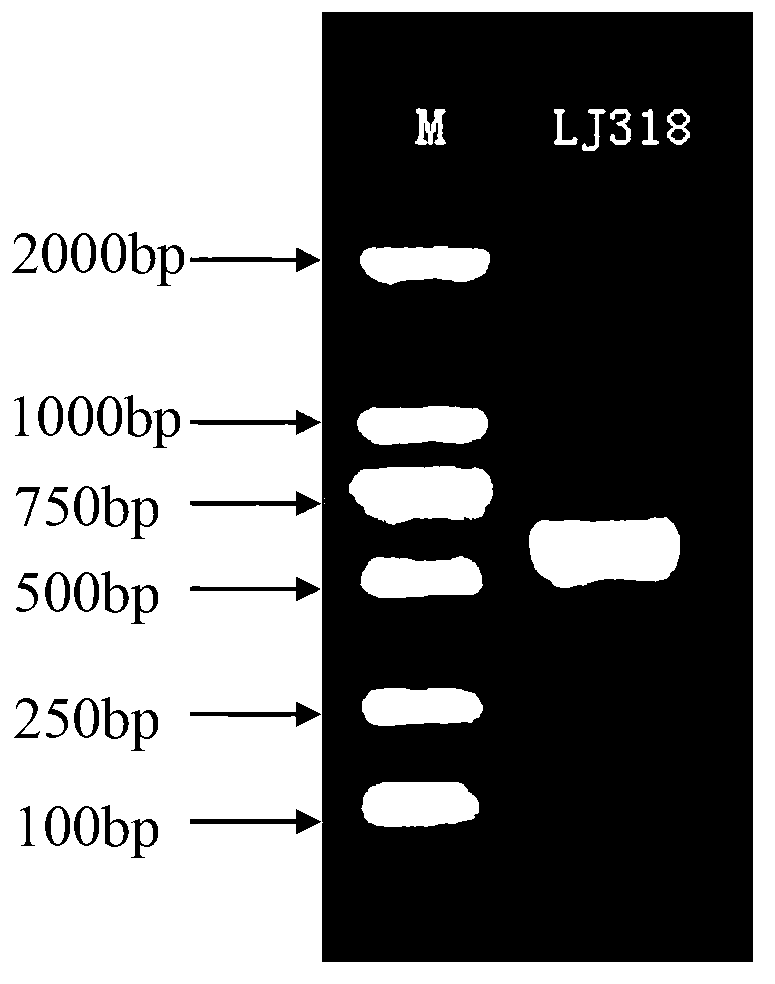
Penicillium citrinum LJ318 for degrading chlortetracycline
InactiveCN103289904APromote degradationGrow fastFungiMicroorganism based processesMicroorganismRibosomal DNA
The invention relates to a strain of Penicillium citrinum LJ318 capable of degrading chlortetracycline and belongs to the technical field of microorganisms. The strain is preserved in China General Microbiological Culture Collection Center (CGMCC) in March 3rd, 2012, and the preservation number of the strain is CGMCC No.5850. An rDNA-ITS (Ribosomal DNA internal transcribed spacer) gene sequence of the chlortetracycline degrading strain LJ318 consists of 544 basic groups. The strain is wide in chlortetracycline-tolerating range and has relatively good degradation effects on the residual chlortetracycline in both liquid and solid wastes.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY +1
Single PCR detection primer pair group for fungus producing aflatoxin, detection method and application thereof
ActiveCN106318938AWide range of applicationsComprehensive species coverageMicrobiological testing/measurementMicroorganism based processesMicrobiologyDehydrogenase
The invention relates to the field of biotechnology and provides a single PCR detection primer pair group aiming to a fungus producing aflatoxin, a detection method, and an application thereof in detection on the fungus producing aflatoxin. In the invention, the single PCR detection primer pair group is composed of primer pairs that aim to a fungus universal molecular marker sequence (internal transcribed spacer, ITS) and three key genes, comprising an aflatoxin production regulating gene (aflR), a sterigmatocystin-o-methoxy transferase gene (omt-1), and versicolorin A dehydrogenase gene (ver-1), in the biochemically synthetic process of the aflatoxin. Through the primer pair group, technical effects of wide coverage range, strong specificity and high sensitivity during the detection of the fungus producing aflatoxin are achieved. The primer pair group is short in detection time, is simple in operation and is low in device requirement, can save large amount of manpower, materials and costs, is suitable for quick detection and is easy to promote in practical production.
Owner:COFCO GROUP +1
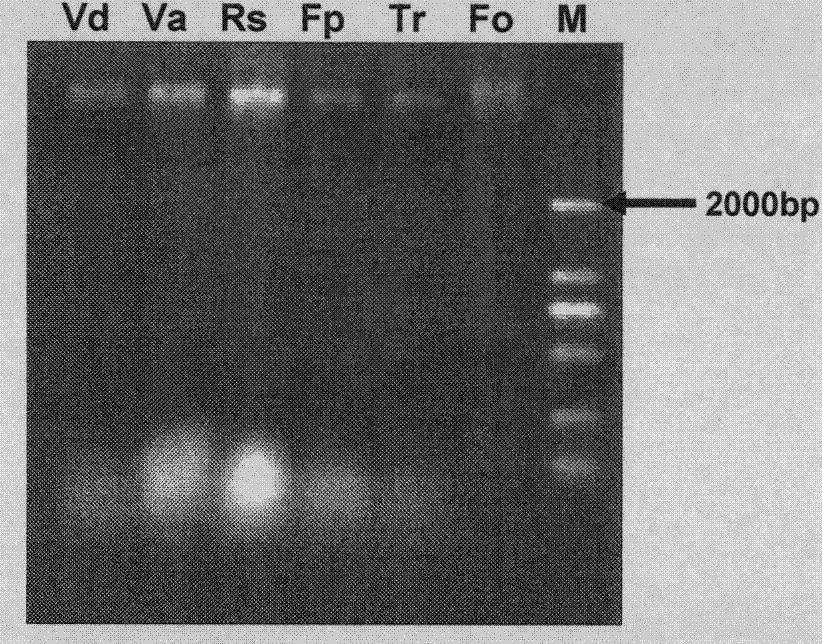
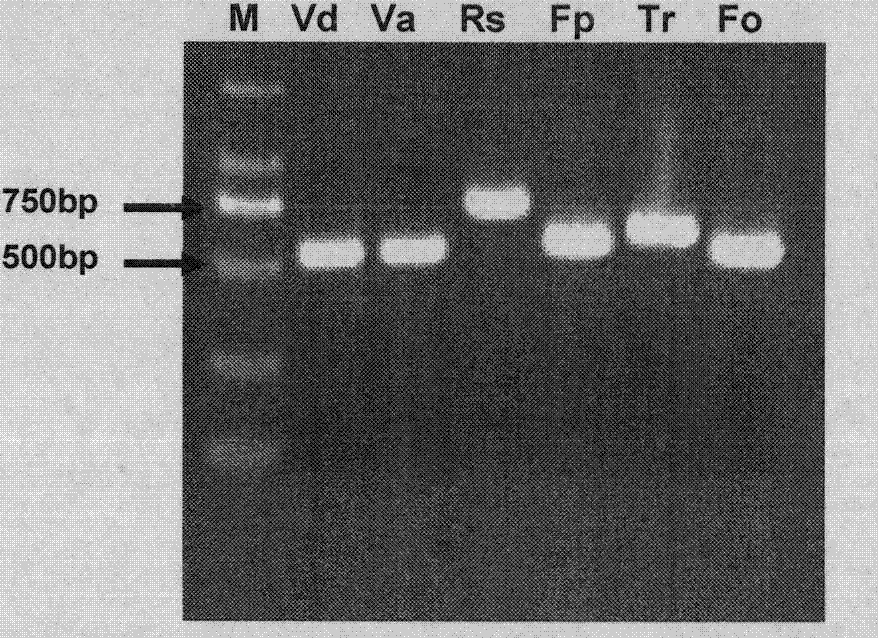
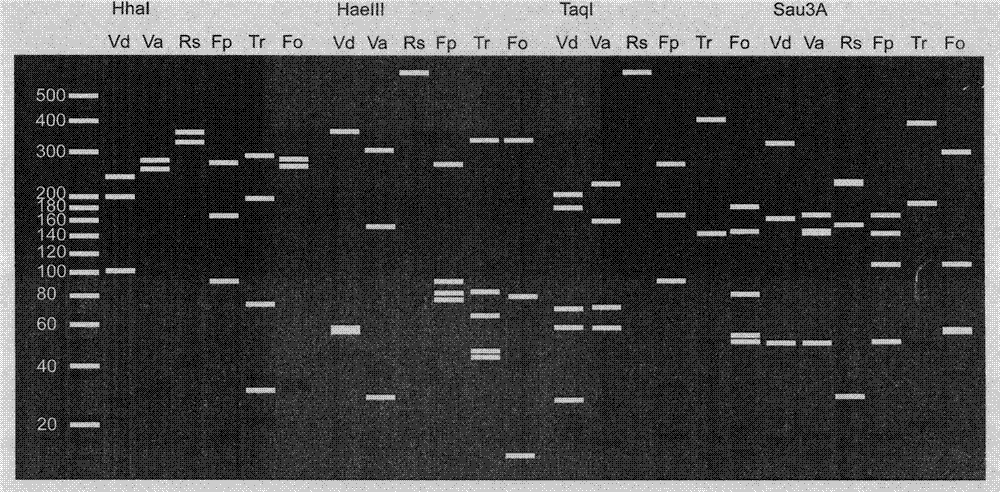
ITS-RFLP method for rapidly identifying main cotton pathogenic fungus
InactiveCN103695533ARapid identificationEasy to operateMicrobiological testing/measurementMicroorganism based processesEnzyme digestionDigestion
The invention discloses an ITS-RFLP method for rapidly identifying main cotton pathogenic fungus, which mainly comprises the following steps: 1) extracting cotton main pathogenic fungus mycelia genome DNA, which comprises Verticillium dahliae, Verticillium alboatrum, Rhizoctonia solani, Fusarium moniliforme, Trichothecium roseum and Fusarium oxysporum; 2) amplifying ribosome internal transcribed spacer (ITC) using PCR: carrying out rapid amplification using general primer ITS4 and ITS5 of fungi ITS; 3) carrying out enzyme digestion reaction using restriction enzymes and identifying band spectrums: carrying out single digestion reaction of ITS products of cotton main pathogenic fungus randomly using restriction enzymes HhaI, HaeIII, TaqI and Sau3A whose recognition sites are four basic groups, and comparing ITS-RFLP band spectrum correlation tables and standard electronic maps, thereby rapidly identifying classes of pathogens. Compared with traditional time-consuming and labor-consuming morphological and physiological identification as well as ITS clone sequencing identification method of pathogen with high cost, the invention has the advantages of rapid, accuracy and economy, and simultaneous identifications of a plurality of pathogenic fungus.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
LAMP (loop-mediated isothermal amplification) primer group for quickly detecting sporisorium scitamineum, kit and detection method thereof
ActiveCN104031997ALow costEasy to operateMicrobiological testing/measurementMicroorganism based processesBiotechnologySugar cane
The invention relates to the field of biotechnology and particularly discloses an LAMP (loop-mediated isothermal amplification) primer group for quickly detecting sporisorium scitamineum, a kit and a detection method thereof. In the invention, a primer group F3 / B3 and FIP / BIP is designed according to the ribosome gene internal transcribed spacer (ITS) sequence of sporisorium scitamineum, the sporisorium scitamineum is detected by the primer group or a kit containing the primer group through LAMP and the detection has the advantages of low cost, easy operation, simplicity, high speed, visual detection result, judgment with naked eyes, high sensitivity, high specificity and the like; and the LAMP primer group, the kit and the detection method disclosed by the invention are applicable to the sugarcane introduction quarantine in the basic-level departments, quality detection on detoxified seedlings, field early detection of culmicolous smut and the like, and has extensive and practical application value.
Owner:SOUTH CHINA AGRI UNIV
Primer pair, kit and PCR (Polymerase Chain Reaction) detection method for distinguishing octodonta nipae from brontispa longissima
InactiveCN103540664AImprove accuracyImprove efficiencyMicrobiological testing/measurementDNA/RNA fragmentationGel electrophoresisOctodonta nipae
The invention discloses a method for quickly distinguishing octodonta nipae from brontispa longissima through a PCR (Polymerase Chain Reaction) detection technology. The method comprises the following steps: enabling specific primers of COI (Cytochrome Oxidase Subunit I) and ITS (Internal Transcribed Spacer) gene segments of the octodonta nipae to be subjected to PCR reaction with DNA (Deoxyribose Nucleic Acid) templates of the octodonta nipae and the brontispa longissima, placing reaction products on an electrophoresis apparatus, and performing agarose gel electrophoresis, thus distinguishing the octodonta nipae from the brontispa longissima through electrophoretic bands. The method is simple, quick and high in accuracy, and plays a significant role in intensive study of the octodonta nipae and the brontispa longissima.
Owner:FUJIAN AGRI & FORESTRY UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com