ITS-RFLP method for rapidly identifying main cotton pathogenic fungus
A technology of pathogenic fungi and identification method, applied in the field of plant pathology, to achieve the effect of simple reaction procedure, low false positive rate and accurate identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
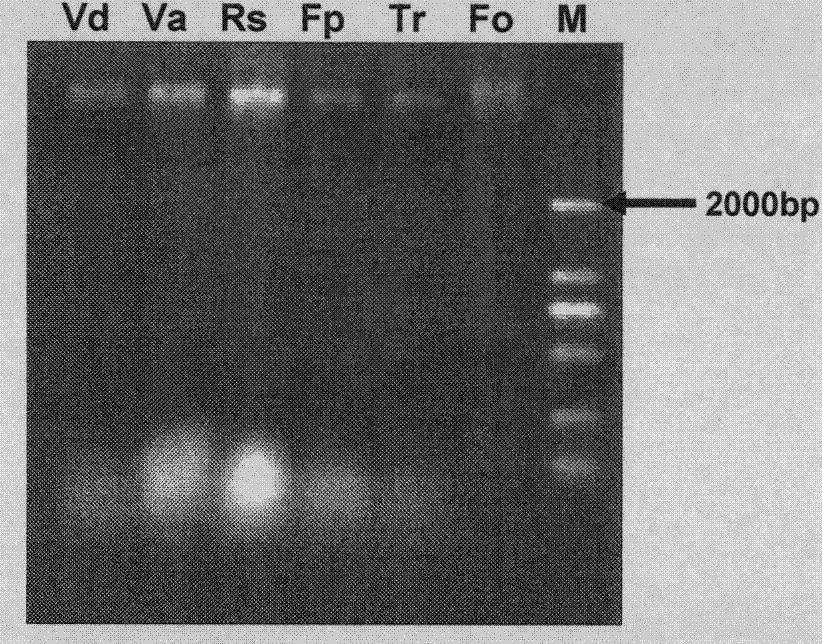
[0032] The extraction of embodiment 1 cotton pathogenic fungus genomic DNA
[0033] 1) Scrape 50-100 mg of fresh mycelium from the PDA solid medium, put it in a 1.5 ml centrifuge tube, add 200 μl of preheated DNA extraction buffer [recipe: 50 mmol / L Hris-HCl (pH=7.5), 50mmol / L EDTA (pH=8.0), 1.4mol / L Nacl, 20g / L CTAB], grind it with a micro-grinding rod, then add 450μl extraction buffer, bathe in 65°C water for 30min, reverse every 10min;
[0034] 2) Add an equal volume of 1:1 saturated phenol and chloroform, mix well by inverting up and down, and centrifuge at 12000g for 15min at room temperature;
[0035] 3) Carefully pipette the supernatant into a new 1.5ml centrifuge tube, add an equal volume of ice-cold absolute ethanol to precipitate, place at room temperature for 10min, centrifuge at 12000g for 10min to collect the precipitate, discard the supernatant, and dry in an oven at 37°C;
[0036] 4) Add 30μl of ddH 2Dissolve genomic DNA in O and store in -20°C refrigerator fo...
Embodiment 2
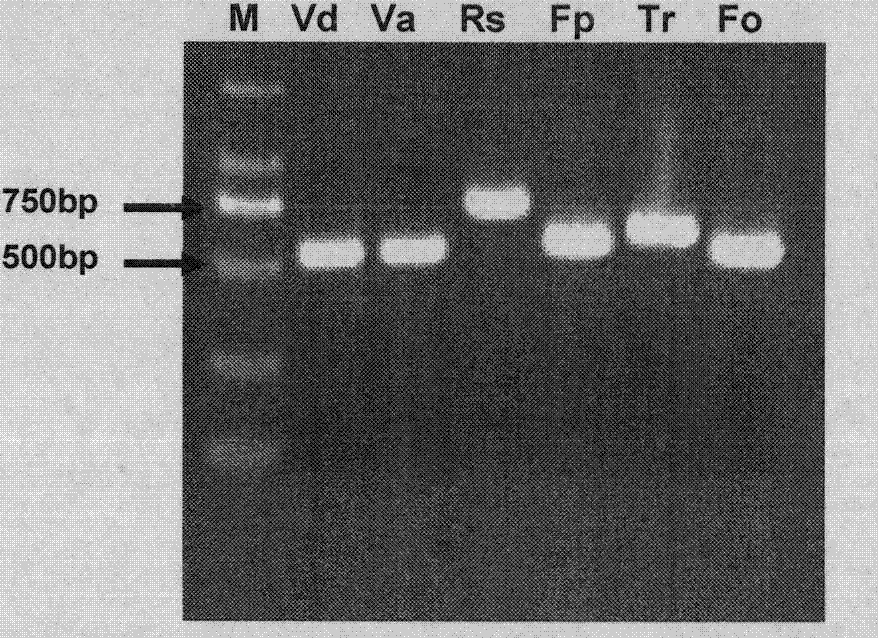
[0038] Embodiment 2 Cotton pathogenic fungus ITS amplification
[0039] 1) Amplification reaction system: using the genomic DNA of the above-mentioned pathogenic fungi as a template, using the primer pair ITS4 5'-TCC TCC GCT TAT TGA TAT GC-3' and ITS5 5'-GGA AGT AAA AGT CGT AAC AAG G-3' Perform PCR amplification. 20μl PCR reaction system: including 10×PCR buffer (Mg 2+ ) 2 μl, 1.6 μl of dNTP solution with a concentration of 10 mM, 1 μl of each primer ITS4 / ITS5 with a concentration of 10 uM, 1 μl of genomic DNA, 0.2 μl of Taq enzyme with a concentration of 5 U / μl, and 13.2 μl of ddH 2 O.
[0040] 2) The amplification reaction program is: pre-denaturation at 94°C for 2 minutes, followed by denaturation at 94°C for 30s, annealing at 50°C for 30s, extension at 72°C for 40s, a total of 25 cycles, and finally extension at 72°C for 10 minutes to obtain the PCR product of the internal transcriptional spacer .
[0041] 3) For electrophoresis detection, take 5 μl of ITS PCR product,...
Embodiment 3
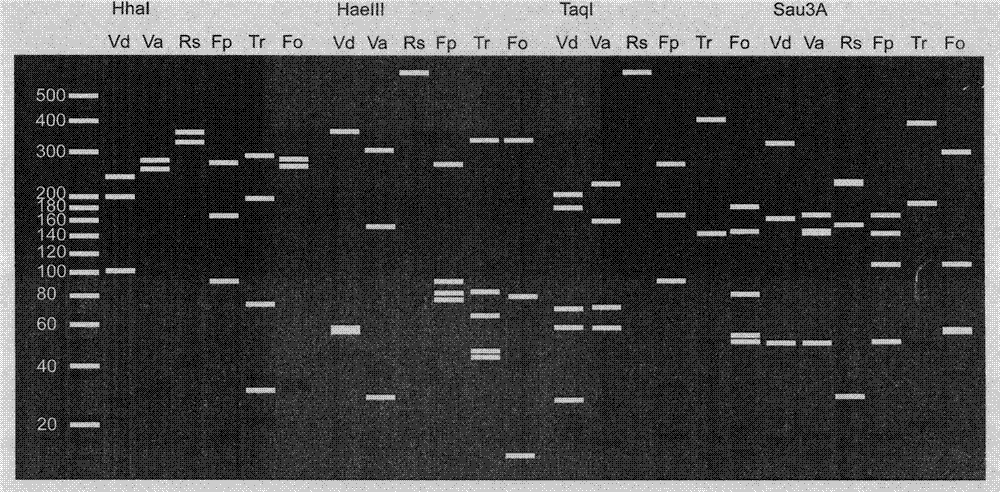
[0042] The selection of embodiment 3 restriction endonucleases
[0043] Screen for suitable four-base restriction enzymes. Based on the ITS sequence determination results of six cotton pathogenic fungi, the recognition of nine four-base restriction endonucleases (Acc II, AfaI, AluI, HaeIII, HhaI, Sau3A, MspI, TaqI and XspI) on different ITSs were analyzed one by one site, calculate the length of the product, fully consider the characteristics of polymorphism and fungal species specificity, and finally select four common endonucleases HhaI, HaeIII, TaqI, Sau3A.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com