Method for rapidly identifying coelomactra antiquate colony by rDNA ITS2 (recombinant Deoxyribose Nucleic Acid Internal Transcribed Spacer) sequence tag
A Xishi tongue and population technology, which is applied in the field of rapid identification of Xishi tongue population by rDNA ITS2 sequence markers, can solve the problems of lack of molecular marker identification methods, difficulties in group identification, adverse effects, etc., and achieve rapid, efficient and accurate identification, and avoid confusion and variation. The effect of fewer sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
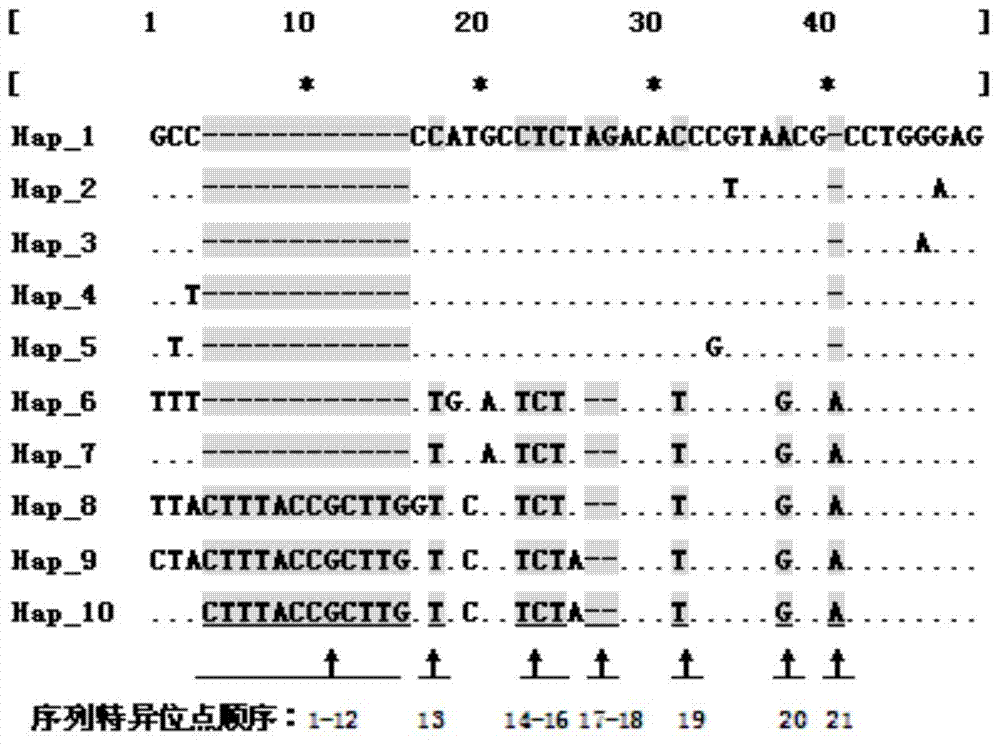
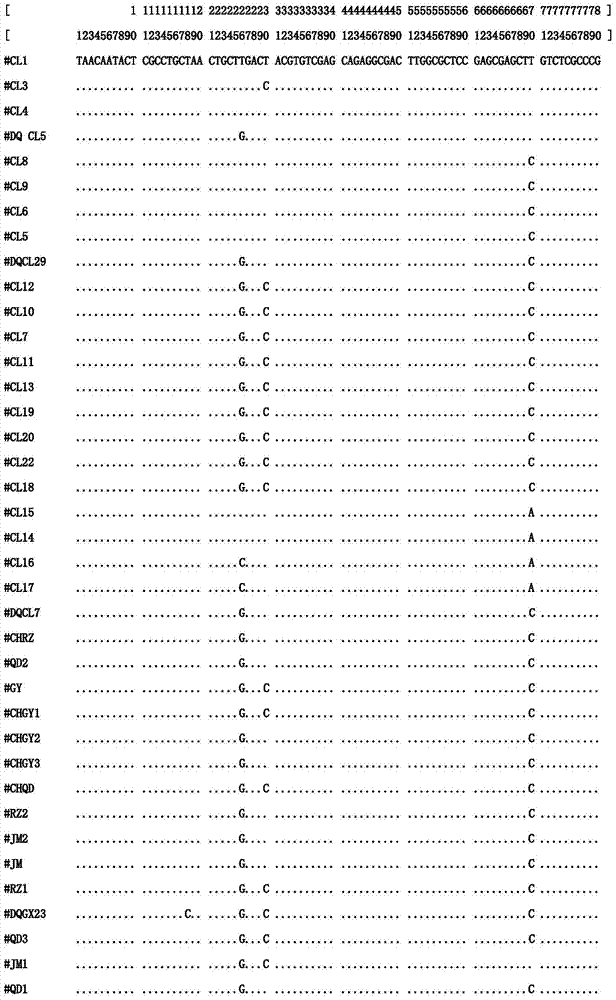
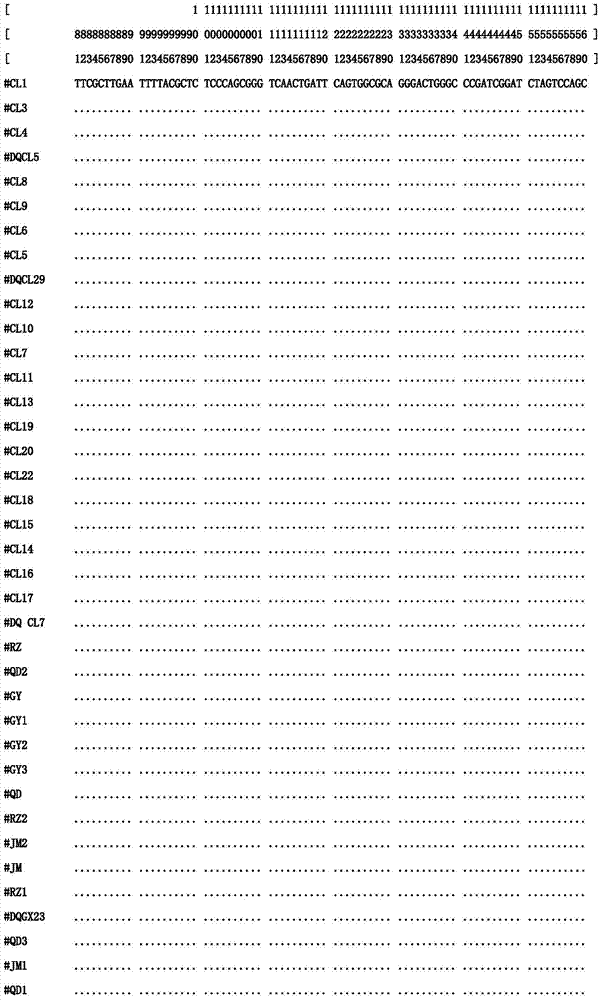
[0023] 1) Sample collection and DNA extraction from different sources:
[0024] Naturally grown Xishi tongue individuals were collected from sea areas such as Jimo (JM) and Rizhao (RZ) in Shandong Province, Ganyu (GY) and Qidong (QD) in Jiangsu Province, and Changle (CL) in Fujian Province. The adductor muscle tissue of the sample was taken with a sterilized scalpel and forceps. The total genomic DNA can be extracted by DNA extraction kit or phenol-chloroform extraction method. Finally, the OD260 and OD280 values of the sample DNA were measured by UV spectrophotometer to determine its concentration and purity, and the mother solution was stored at -20°C. The DNA concentration of 100ng / ul was stored at 4°C for use.
[0025] 2) PCR amplification conditions:
[0026] The primer base sequences are:
[0027] ITS2-F(2A): 5′-GGG TCG ATG AAG AAC GCA G-3′
[0028] ITS2-R(2B): 5′-GCT CTT CCC GCT TCA CTC G-3′
[0029] PCR reaction system is 30μL, each reaction system contains: 50...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com