Molecular marker of rice seed salt tolerant germination major QTL qGR2 and its application
A technology of molecular markers and rice seeds, which is applied in the determination/testing of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of few researches on seed germination period, and achieve fast and simple identification methods, high selection efficiency, Choose a well-targeted effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] (1) Materials and methods:
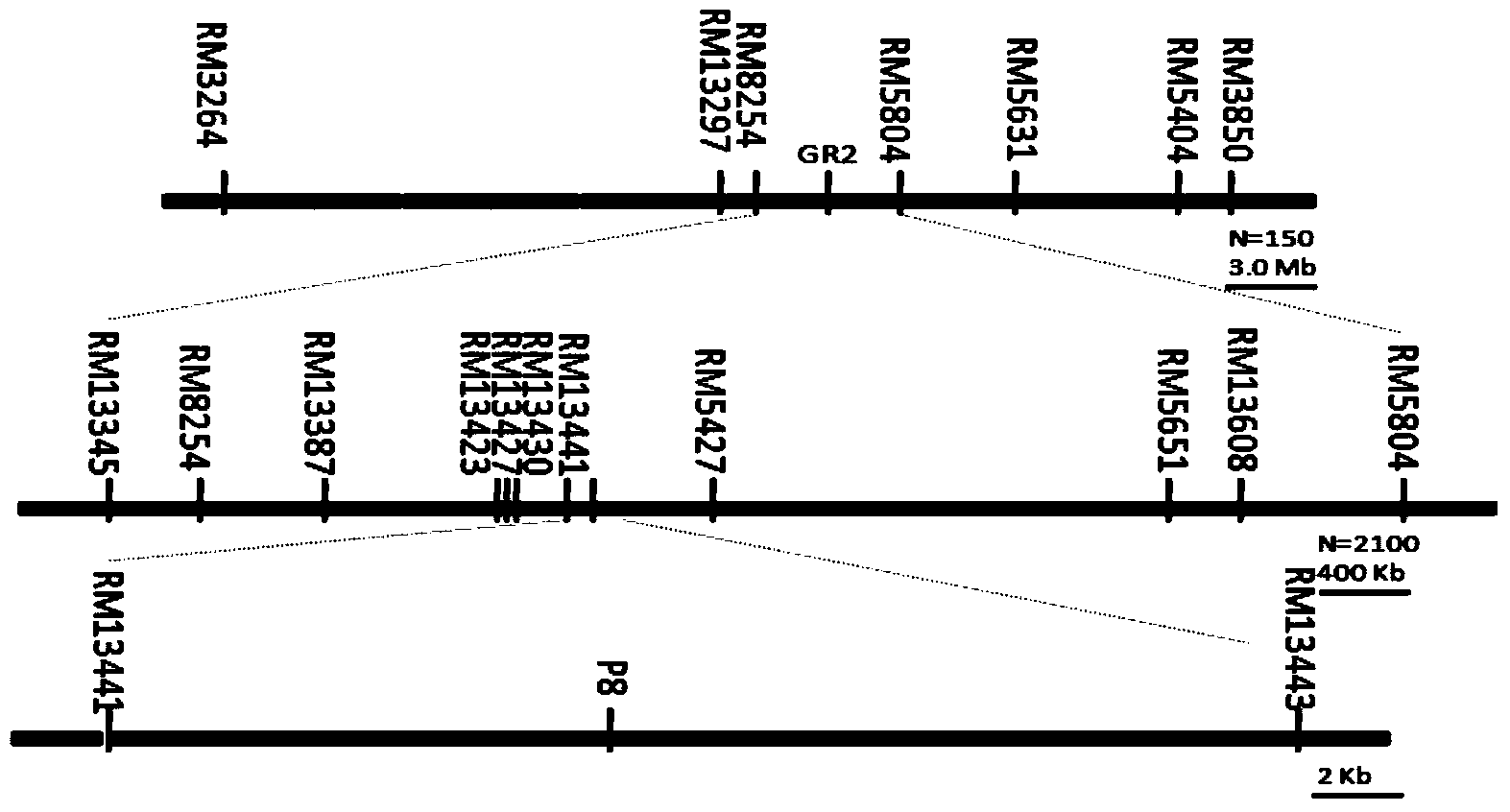
[0032] 1. Material: strong salt-tolerant variety Chive Green and IR 26 Hybrid ( figure 1 ), to obtain 150 RIL families F 2:9 , for initial positioning, and then select the background-like IR containing the qGR2 locus 26 The families were backcrossed and selfed to obtain segregation BC 2 f 2 Populations are isolated, fine-mapped, and linked markers identified.
[0033] 2. Extract individual DNA by SDS method.
[0034] 3. Screening of polymorphic markers: select 900 pairs of SSR primers, and use leek green and IR 26 As a template, carry out PCR amplification, and screen for polymorphic markers.
[0035] 4. PCR reaction system: the volume is 25 microliters, including 2.5 microliters of 10×buffer, 25mM MgCl 2 1.5 microliters, 2.5 microliters of 4 pmol / microliter primer pair, 2 microliters of 2.5mM dNTPs, 0.2 microliters of 5 units / microliter Taq enzyme, 20 nanograms of template DNA, and add water to 25 microliters. The reaction program wa...
Embodiment 2
[0045] (1) Materials and methods:
[0046] 1. Material: rice variety Leek Green and IR 26 .
[0047] 2. Extract individual DNA by SDS method.
[0048] 3. Marking: RM13441, P8, RM13443.
[0049] 4. PCR reaction system: the volume is 25 microliters, including 2.5 microliters of 10×buffer, 25mM MgCl 2 1.5 microliters, 2.5 microliters of 4 pmol / microliter primer pair, 2 microliters of 2.5mM dNTPs, 0.2 microliters of 5 units / microliter Taq enzyme, 20 nanograms of template DNA, and add water to 25 microliters. The reaction program was DNA pre-denaturation at 95°C for 5 min; pre-denaturation at 95°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 30 s, and 35 cycles; finally, extension at 72°C for 10 min. PCR amplification was carried out on a biometer amplification instrument, and the amplified products were separated by electrophoresis on 8% non-denaturing polyacrylamide gel (containing 7.6g acrylamide and 0.4g methylenebisacrylamide in 100ml polyacrylamide solution...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com