Molecular marker tightly in linkage with corn line grain number main effect QTL and application thereof
A technology of molecular markers and row kernel number, which is applied in the field of corn breeding and molecular biology, can solve the problems of inconsistent results, etc., and achieve the effect of simple identification method, clear selection target and high selection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] A molecular marker of the main QTL controlling the number of kernels in a row of maize. The molecular marker is composed of two pairs of primers, InDe176 and umc1854. The sequence of the primer InDel14 is:
[0022] Forward: 5'-TCAGTGCAAAAAGGAATCTCAA-3'
[0023] Reverse: 5'-TGAACGTCTCCCCTTACAATGC-3'
[0024] The sequence of the primer umc1269 is:
[0025] Forward: 5'-TCTAAAGTATGGCTGGAAACGAGC-3'
[0026] Reverse: 5'-GCAACGATAAGTACCAGAATATGGC-3'.
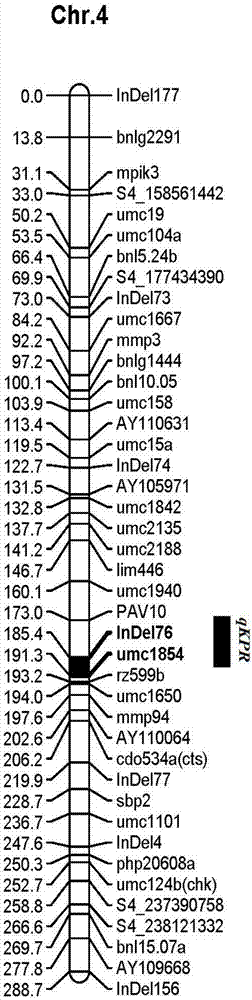
[0027] Extract the genomic DNA of the corn triple testcross population to be tested, and use primers InDe176 and umc1854 for PCR amplification to obtain amplified products with a length of 174bp and 142bp, and the amplified products are passed through 6% polyacrylamide gel and 1% agarose gel. After separation by gel electrophoresis, InDe176 and umc1854 were obtained respectively, and the corn to be tested was the corn with a large number of rows.
[0028] The described maize varieties are B73 and Mo17 and the IBM groups comp...
Embodiment 2
[0032] A detailed step for obtaining the main effect QTL molecular markers for controlling the number of grains in a row of corn is as follows:
[0033] (1) Construction of corn triple test cross (TTC) population and measurement of row kernel number
[0034] 82 high genetic exchange rate IBM recombinant inbred lines were crossed with their parents B73, Mo17 and F1 respectively to obtain TC(B73), TC(Mo17) and TC(F1) triple testcross populations. 82 IBM populations, TC(B73), TC(Mo17) and TC(F1) triple test cross populations were planted in Liuhe Experimental Base of Jiangsu Academy of Agricultural Sciences in 2012 according to random block design, with three replicates. Each genotype material was planted in a single row, with a plant spacing of 30 cm and a row spacing of 60 cm, and normal field management. After emergence, the leaf tissue of the IBM population was collected to extract DNA. After the grains were physiologically mature, the mature ears of IBM recombinant inbred ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com