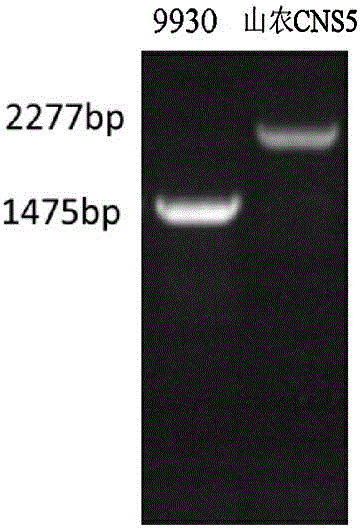
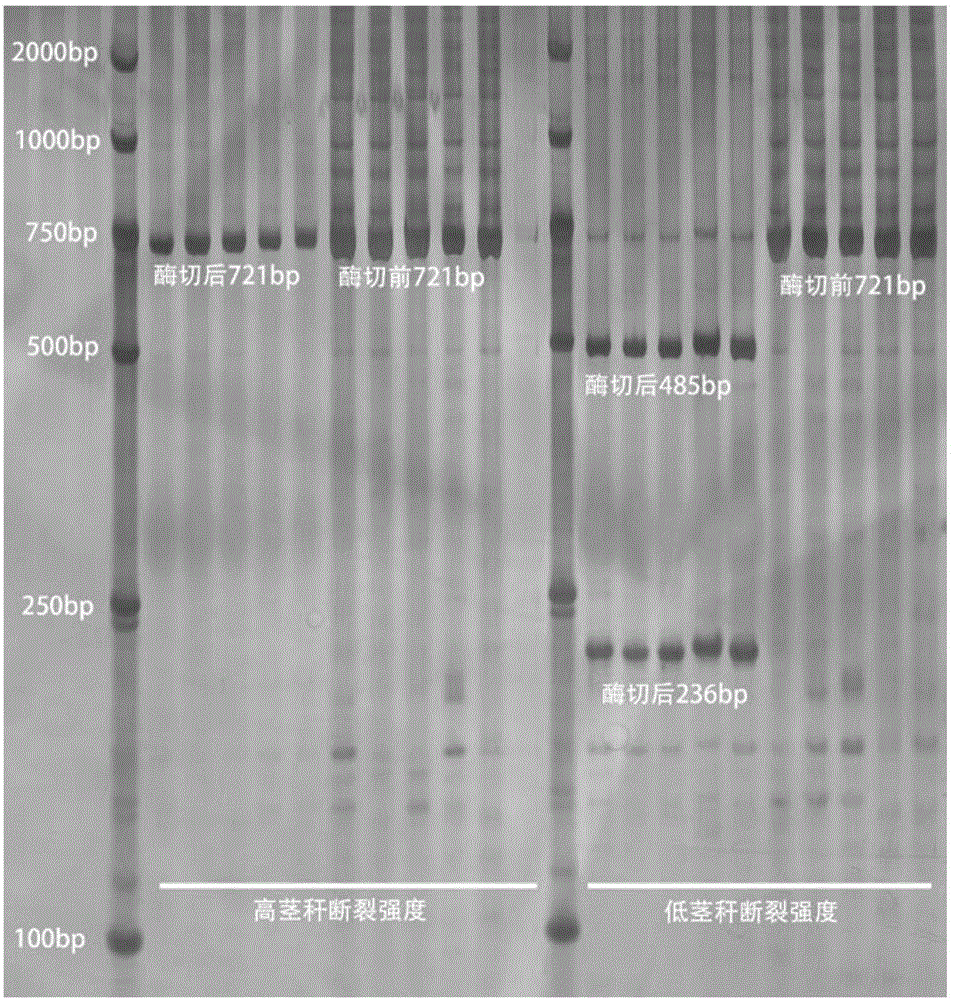
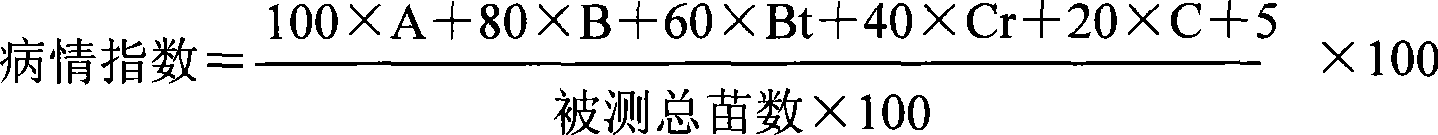Patents
Literature
106results about How to "Choose a clear goal" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Functional molecular marker for rice anti-blast gene Pi9 and application thereof
ActiveCN103146695AImprove breeding efficiencyImprove accuracyMicrobiological testing/measurementPlant genotype modificationBiotechnologyNucleotide
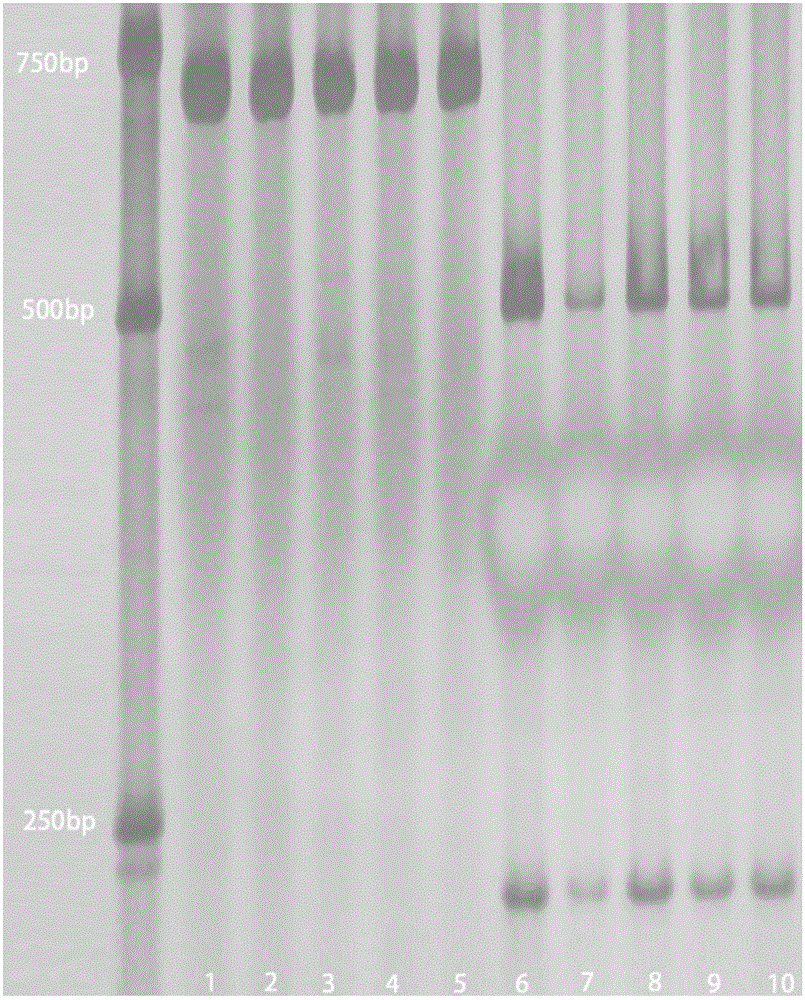
The invention provides a functional molecular marker for rice anti-blast gene Pi9 and application thereof, belonging to the field of crop molecular genetic breeding science. The invention finds out that the rice anti-blast gene Pi9 has a 10bp insertion / deletion site positioned between the gene promoter -516 and -517, wherein the nucleotide sequence is disclosed as SEQ ID NO.1; and on such basis, the invention develops a method of the functional molecular marker for gene Pi9. By detecting the functional molecular marker, the invention can accurately detect whether genomes of different rice species contain the Pi9 functional gene and detect the homozygotic state, and can be used for screening rice hybrid transformed descendant plants to enhance the breeding efficiency of the rice anti-blast material, thereby effectively controlling the size of the breeding population, obviously saving the breeding and screening cost and obtaining the anti-blast rice species containing the Pi9 functional gene.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
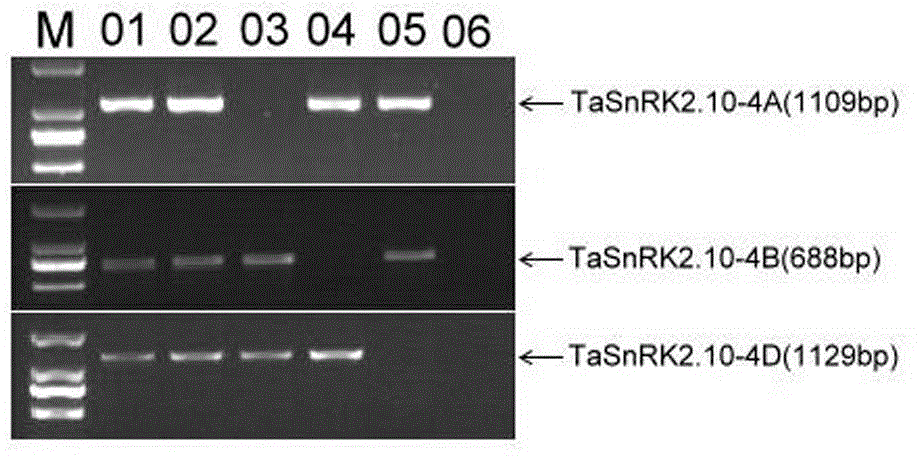
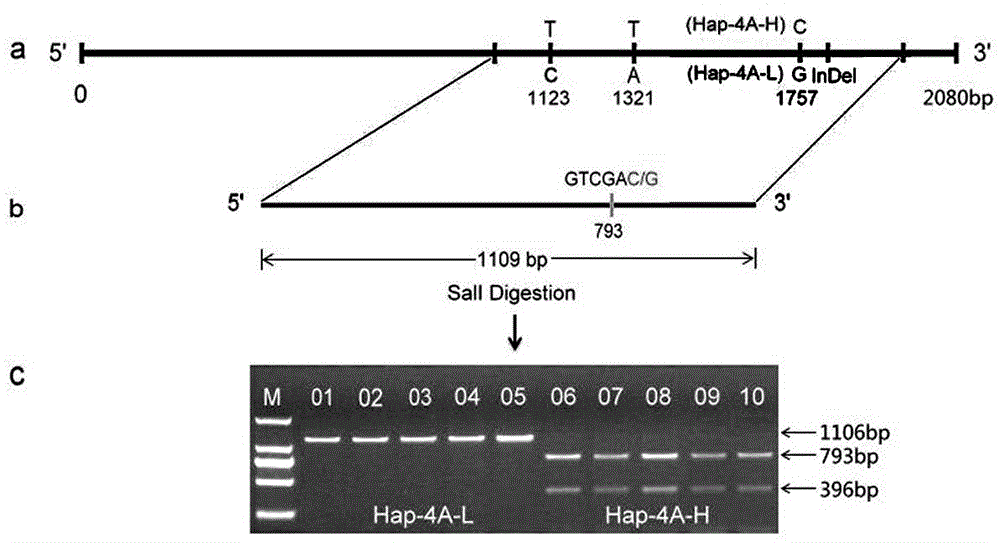
Gene relevant to wheat thousand seed weight, functional marker and application thereof
ActiveCN103820476ANot affectedImprove selection efficiencyMicrobiological testing/measurementFermentationBiotechnologyTriticeae
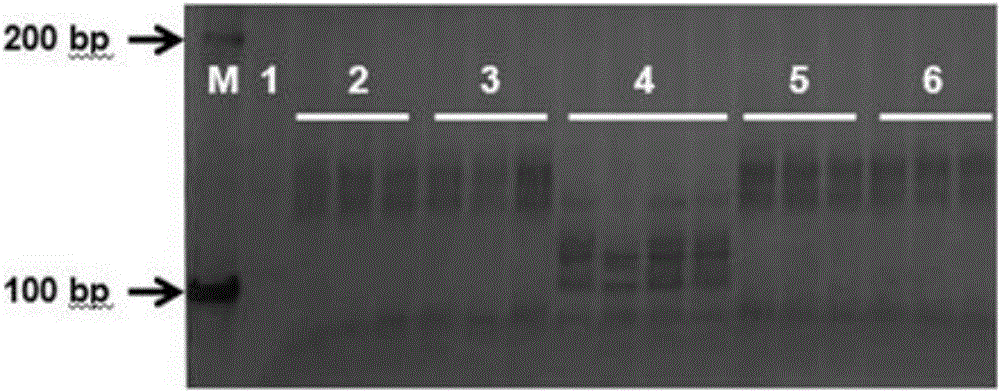
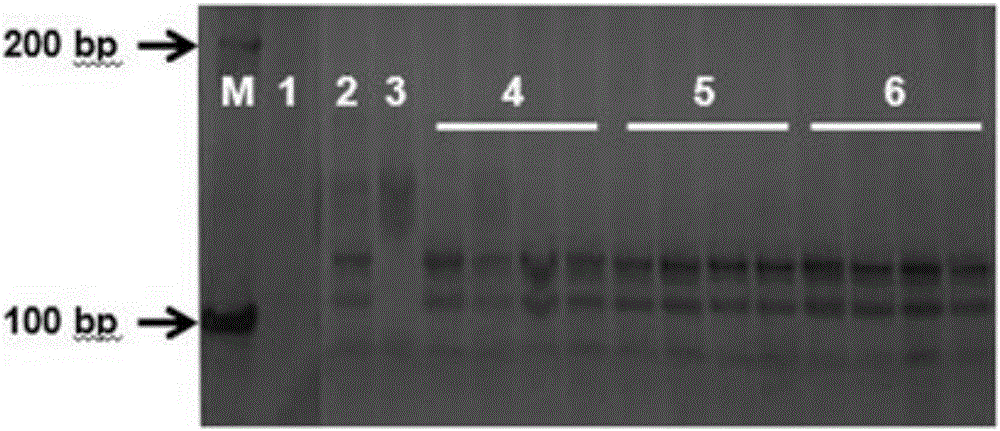
The invention discloses a gene TaSnRK2.10 relevant to the wheat thousand seed weight, a molecular marker TaSnTK2.10-4A-caps relevant to the gene and application of the marker. The DNA of a wheat variety to be detected is subjected to PCR (polymerase chain reaction) multiplication through a marker primer TaSnTK2.10-4A-caps; a multiplication product is subjected to cleavage through SalI incision enzyme and is also subjected to electrophoretic separation; for example, PCR products are two bands with the sizes of 793bp and 316bp; the wheat variety is a variety with haplotype with high thousand grain weight of the gene; the PCR product is only a band with the size of 106bp; the wheat variety is a variety which is not provided with the haplotype with high thousand grain weight of the gene. According to the gene TaSnRK2.10 relevant to the wheat thousand seed weight and the molecular marker TaSnTK2.10-4A-caps thereof, the wheat variety or strain with high thousand grain weight can be conveniently detected or screened, and the selection process of the high-yield variety of wheat can be greatly accelerated.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Molecule marking method of rice blast-resisting gene
InactiveCN102162011APredicting Rice Blast ResistanceAccurate genetic lociMicrobiological testing/measurementDiseaseAgricultural science
The invention discloses a molecule marking method of a rice blast-resisting gene, belonging to the field of crop molecule heredity and breeding. The method comprises the following steps: (1) taking a rice sample and extracting a genome DNA (Deoxyribose Nucleic Acid) of the rice sample; and (2) carrying out PCR (Polymerase Chain Reaction) amplification on the genome DNA of the rice sample by utilizing any one pair of molecule-marked primers in RM6091 and RM26632, carrying out electrophoresis detection on a PCR amplification product, and if a molecule-marked DNA segment with corresponding size is amplified, showing that a Pi-bdl(t) gene exists. Through rice blast-resisting gene Pi-bdl(t) molecule marking in the invention, whether thin rice as well as crossbred descendants, backcross descendants and multiple cross descendants thereof contain the gene can be detected, the resistance level of the gene on rice blast can be forecasted, the selecting efficiency of rice blast resistant materials can be greatly increased, and the breeding process for disease resistance can be accelerated.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecular marker-assisted selection primer and method for wilt resistance and gynoecious line of cucumis melon
InactiveCN103866005ALow costReduce workloadMicrobiological testing/measurementDNA/RNA fragmentationEnzymePolymerase chain reaction
The invention discloses a molecular marker-assisted selection primer and method for the wilt resistance and gynoecious line of cucumis melon. According to the method, CAPS (cleaved amplified polymorphic sequence) primers A-as and A-s are designed according to single-gene mutation between a 6045th basic group on the DNA (deoxyribonucleic acid) sequence of a gene A of the cucumis melon and a corresponding locus of a gene a, and an SCAR (sequence characterized amplified region) primer pair G-as and G-s and an SCAR primer pair g-as and g-s are respectively designed according to genes G and g of the cucumis melon and are used for differentiating and labeling sex-determining genes of the cucumis melon; and meanwhile, a comparison result of genes MR-1 and WI998Fom-2 at an SCOP functional domain shows that only a 481th basic group can be developed as a CAPS-marked site, and a primer pair F-s and F-as is designed according to the comparison result. The polymorphism that the digestion is carried out after amplification by virtue of a PCR (Polymerase Chain Reaction) can be applied to the molecular marker-assisted breeding of the wilt resistance and gynoecious line of the cucumis melon. The primer and the method are used for carrying out assisted breeding and have the advantages of definite target selection, cost conservation and the like.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular marker closely linked with major QTL of wheat height and uppermost internode length as well as acquisition method and application of molecular marker
InactiveCN104372003AImprove selection efficiencyQuality improvementMicrobiological testing/measurementDNA preparationBiotechnologyGenetic stock
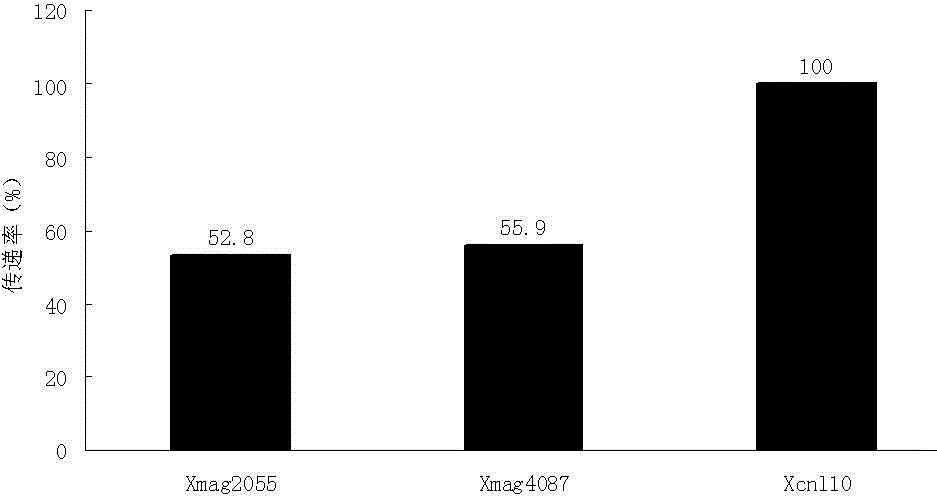
The invention discloses a molecular marker closely linked with the major QTL of wheat height and uppermost internode length as well as an acquisition method and application of the molecular marker. The acquisition method comprises the following steps: carrying out PCR amplification on DNA of a wheat variety Kenong9204 by adopting a marked primer Xcnl10; carrying out electrophoretic separation on the amplified product on 6.0% native polyacrylamide gel to obtain the amplified product with molecular weight of 700bp, namely the molecular marker of major QTL of wheat height and uppermost internode length. The molecular marker can be used for detecting whether a wheat variety or line contains QTL capable of reducing plant height and uppermost internode length, or not, so that the wheat variety or line containing QTL capable of reducing the plant height and uppermost internode length can be quickly screened for breeding, and the breeding progress for excellent wheat plant variety can be greatly accelerated.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
Molecular marker of TaGS2 gene related with wheat plant height, obtaining method and application thereof
ActiveCN104846099ASpeed up shapingQuality improvementMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyEnvironment effect
The invention discloses a molecular marker IN10 of a TaGS2 gene related with the wheat plant height, and the application in the field of screening wheat variety or strain plant height by using the molecular marker; in particular, a marker primer of the molecular marker IN10 is used for performing PCR (polymerase chain reaction) amplification on a wheat genome DNA (deoxyribonucleic acid) to be tested, and the product of amplification is subjected to electrophoresis separation; the obtained corresponding product of amplification has bands in 555bp, 466bp and 426bp of molecular weights, and is used for judging the change of plant height of the variety, which is used as a theoretical guide for breeding new wheat variety of different plant height types. The screening method provided by the invention is quick and accurate and is not affected by environment, the selection target is definite, a reasonable plant height group can be quickly produced, and the selection efficiency and quality of high-yield wheat variety or strain can be improved greatly.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI
Molecule label linked with rice anti-rice blast gene
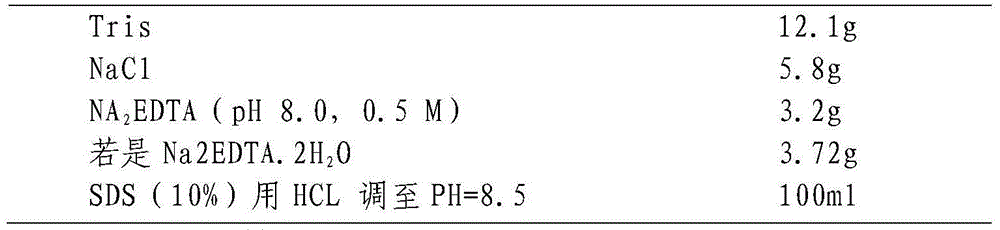
InactiveCN1648253AChoose a clear goalSave time and costMicrobiological testing/measurementGenetic engineeringJaponica riceAgricultural science
The present invention relates to molecular label linked with rice blast resisting gene and specially for the screening of rice blast resisting gene. Single plant DNA's of black hull japonica rice, Suyu glutinous rice and their F1 and F2 separated colony are extracted in SDS process; 350 pairs of SSR molecular labels are adopted in polymorphism screening of the amplified products of two parents and their F1; and the individuals of F2 anti-infective pond and F2 colony are screened for rice blast resisting gene linked label. It is found that the SSR labels RM144H240+260, RM7654H120, RM7212H120 and RM5923H150 in the long arm end of 11-th chromosome are linked with rice blast resisting gene Pi-hkl(t); and the SSR labels RM277H70, RM511H80+90, and RM2972H90 in the 12-th chromosome are linked with rice blast resisting gene Pi-hk2(t). The present invention can target the rice blast resisting gene accurately and raise the rice blast resisting selectively breeding efficiency greatly.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecule making method for gene locus for preventing mycosphaerella melonis of muskmelon
InactiveCN101173311AClear locationEasy to identifyMicrobiological testing/measurementMelon (food)Cultivar
The invention relates to a molecular marker method for a melon blight resistance gene, belonging to the field of biotechnology. There are four AFLP molecular markers that are linked to the gene for resistance to blight blight, the marker EcoRI-TG / MseI-CTC200 is a 200bp band; the marker EcoRI-AT / MseI-CTG90 is a 90bp band; the EcoRI-TC / MseI-CAG60 is a 60bp band band; the marker EcoRI-TG / MseI-CTA70 is a 70bp band; the linkage distances are 2.0, 6.0, 5.4 and 6.0cM, respectively. Detect whether PI 420145 and its derivative varieties (lines) contain the gene through the molecular marker method provided by the present invention, predict its resistance level to vine blight, improve the selection efficiency, and help to accelerate the selection of excellent vine blight-resistant melon varieties in my country It is used in the production process to identify the purity of muskmelon varieties resistant to vine blight.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecular marker of rice blast resisting gene and application of molecular marker
ActiveCN106555001AImprove selection efficiencyLow breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationGenomicsAgricultural science
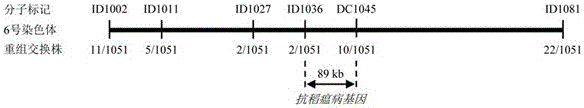
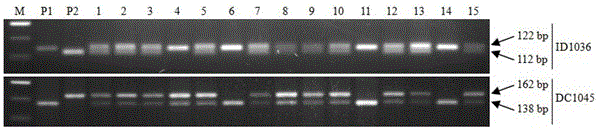
The invention discloses a molecular marker of a rice blast resisting gene and application of the molecular marker and belongs to the field of crop molecular genetics breeding science. The study means of phytopathology, molecular genetics and genomics are utilized, the rice blast resisting gene of the local rice variety of indica type rice in the northern Fujian province is located between the six chromosome molecular marker ID1036 and the six chromosome molecular marker DC1045. The molecular marker combination of ID1036 and DC1045 is utilized, plants, carrying the anti-disease gene, in the hybrid transform breeding group of the rice blast resisting variety of the indica type rice in the northern Fujian province can be efficiently screened, and the efficiency of breeding the rice blasting rice variety comprising the gene is improved.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
Eriocheir-sinensis-growth-trait-related EST (expressed sequence tag)-SSR (simple sequence repeat) molecular marker and application thereof
InactiveCN103667272AChoose a clear goalChoose accuratelyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceEriocheir
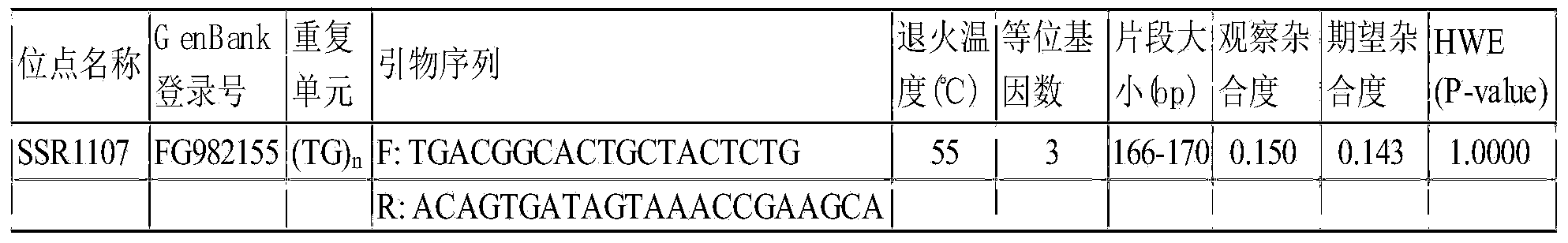
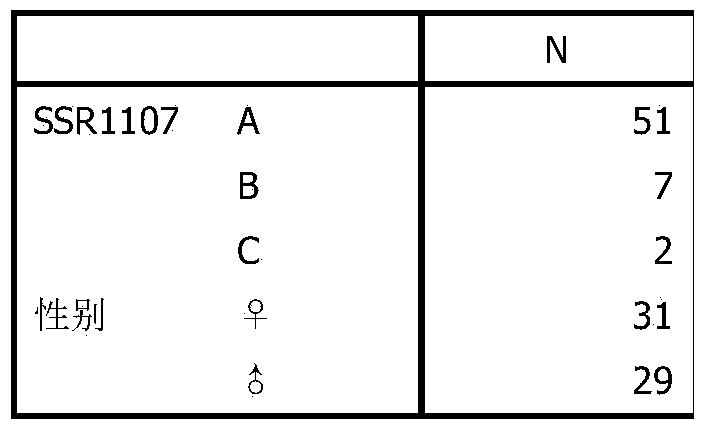
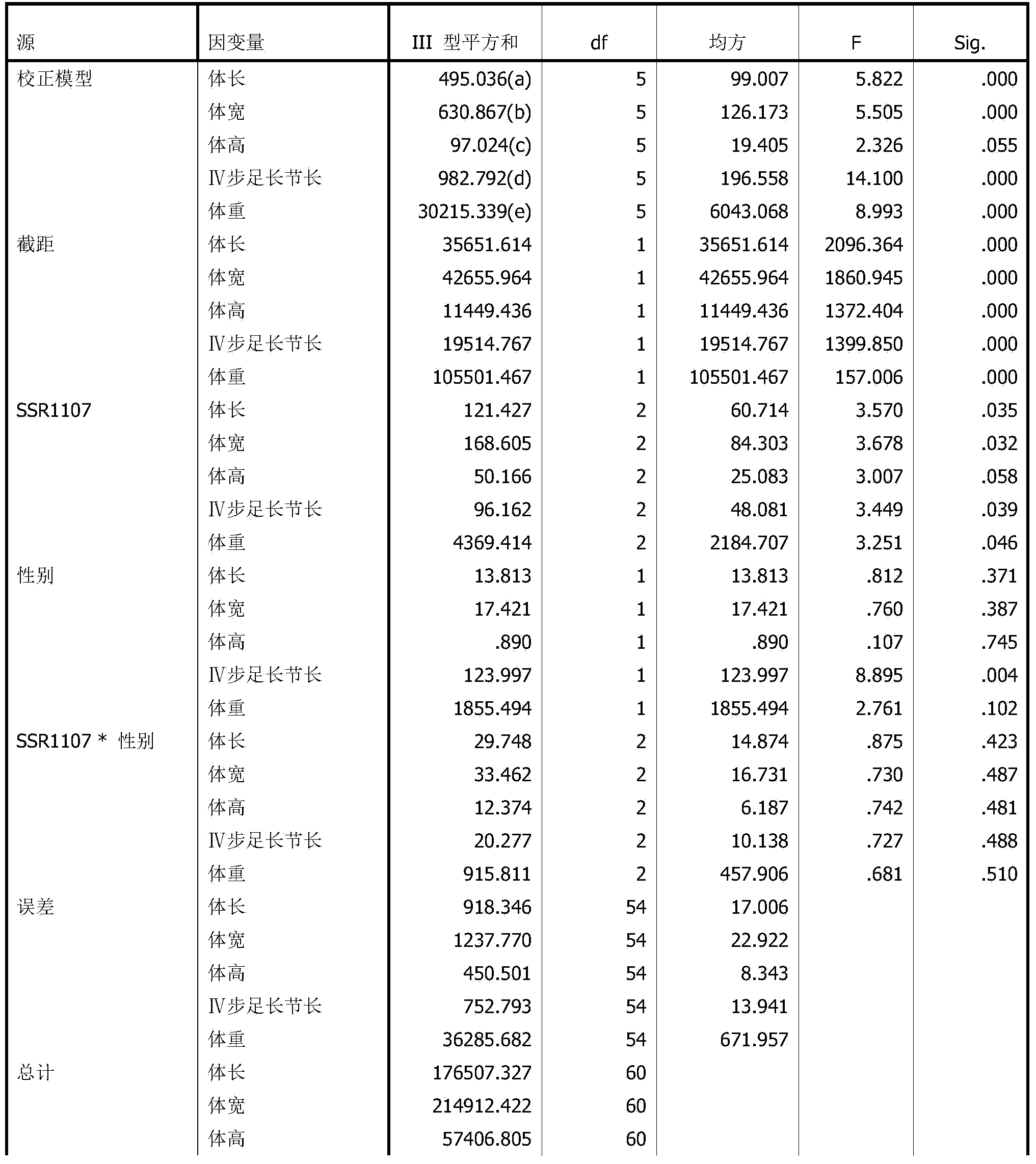
The invention discloses an eriocheir-sinensis-growth-trait-related EST (expressed sequence tag)-SSR (simple sequence repeat) molecular marker, the molecular marker is named as SSR1107, and comprises a core sequence (TG) n, an upstream primer and a downstream primer, the upstream primer is shown in SEQ ID NO.1, the downstream primer is shown as SEQ ID NO.2, and n is 7-9. The molecular marker SSR1107 overcomes the defects that a traditional breeding method mainly relies on phenotypic trait identification, and is greatly affected by environmental factors, and the growth trait breeding is difficult and inefficient. Through use of the eriocheir-sinensis-growth-trait-related EST-SSR molecular marker, biological parents excellent in growth trait can be fast and accurately screened for river crab breeding, the selection target is clear, and the time and labor are saved.
Owner:天津市水生动物疫病预防控制中心
Molecular marker closely linked with wheatear grain number major QTL and application of molecular marker
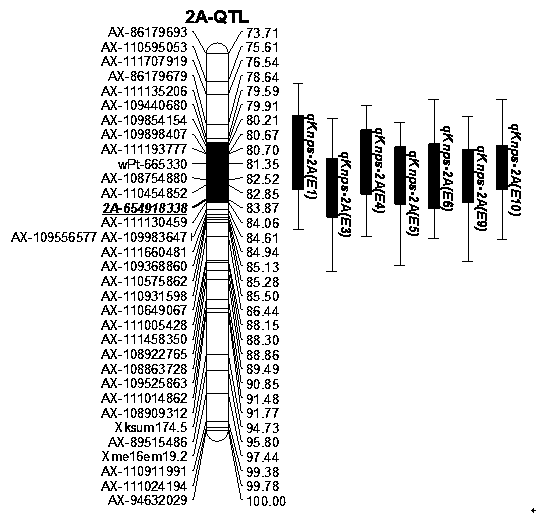
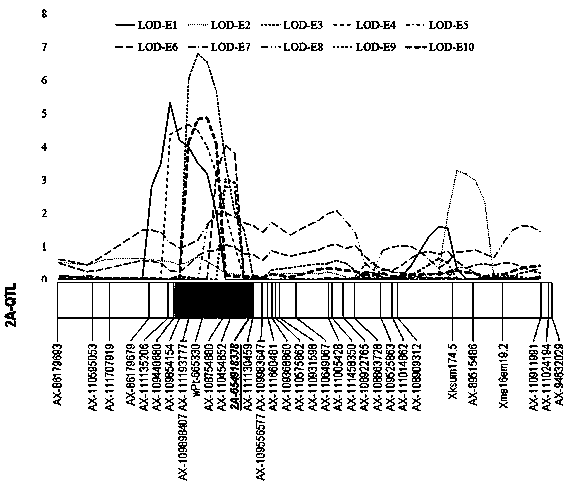
ActiveCN109825639AQuick judgmentSpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGermplasm
The invention discloses a molecular marker closely linked with wheatear grain number major QTL. Wheat genome DNA serves as a template, a PCR primer pair is adopted to conduct PCR amplification, the amplificated product is subjected to ionophortic separation through polyacrylamide gel to obtain the molecular marker 2A-654918338. The invention further discloses application of the molecular marker 2A-654918338 in detecting whether a wheat variety or line contains the QTL for increasing the wheatear grain number and application of the molecular marker 2A-654918338 in molecular breeding of wheat. By means of the molecular marker, the genetic improvement progress of the wheatear grain number is accelerated, the selection efficiency and quality of the wheat variety or line are greatly improved, identification of the target gene in wheat genetic resources and breeding progenies is directly achieved, and more new wheatear grain number genetic resources are developed for wheat breeding.
Owner:LUDONG UNIVERSITY
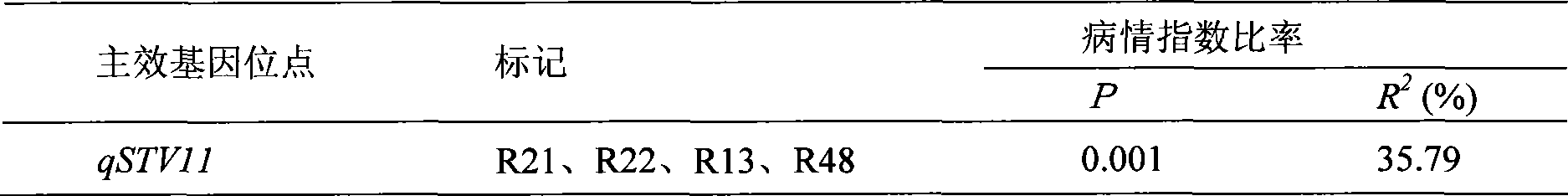
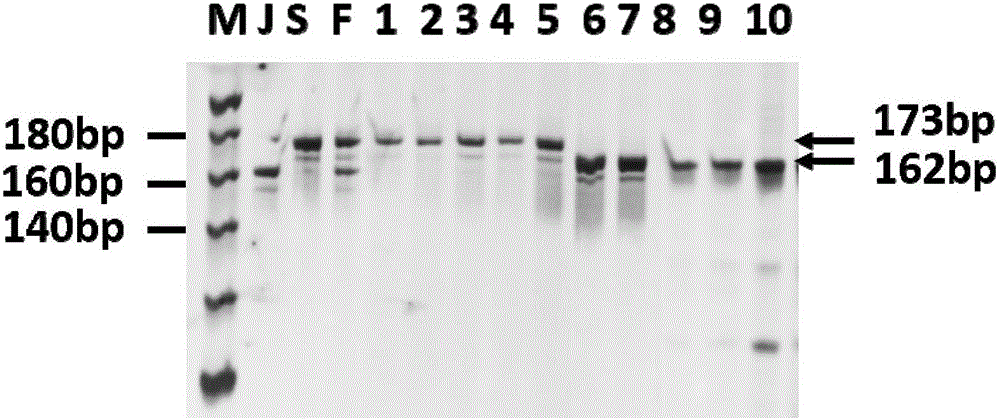
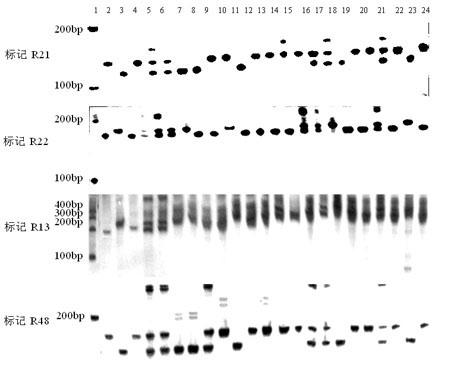
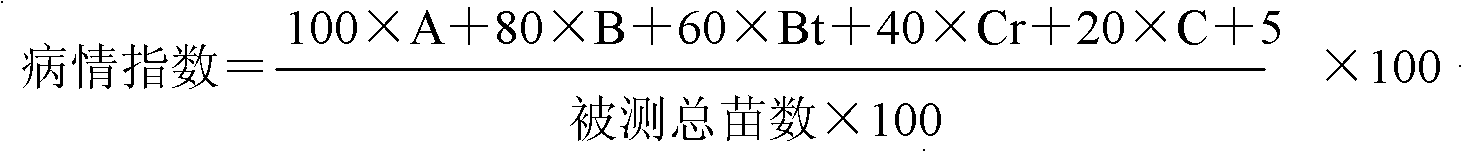
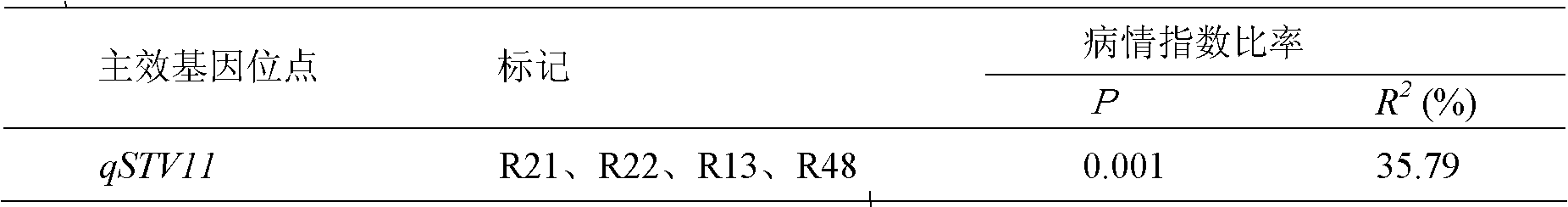
Molecular marker method for rice anti-rice stripe major gene loci qSTV11
InactiveCN101487050AImprove utilizationEasy to detectMicrobiological testing/measurementDiseaseBiotechnology
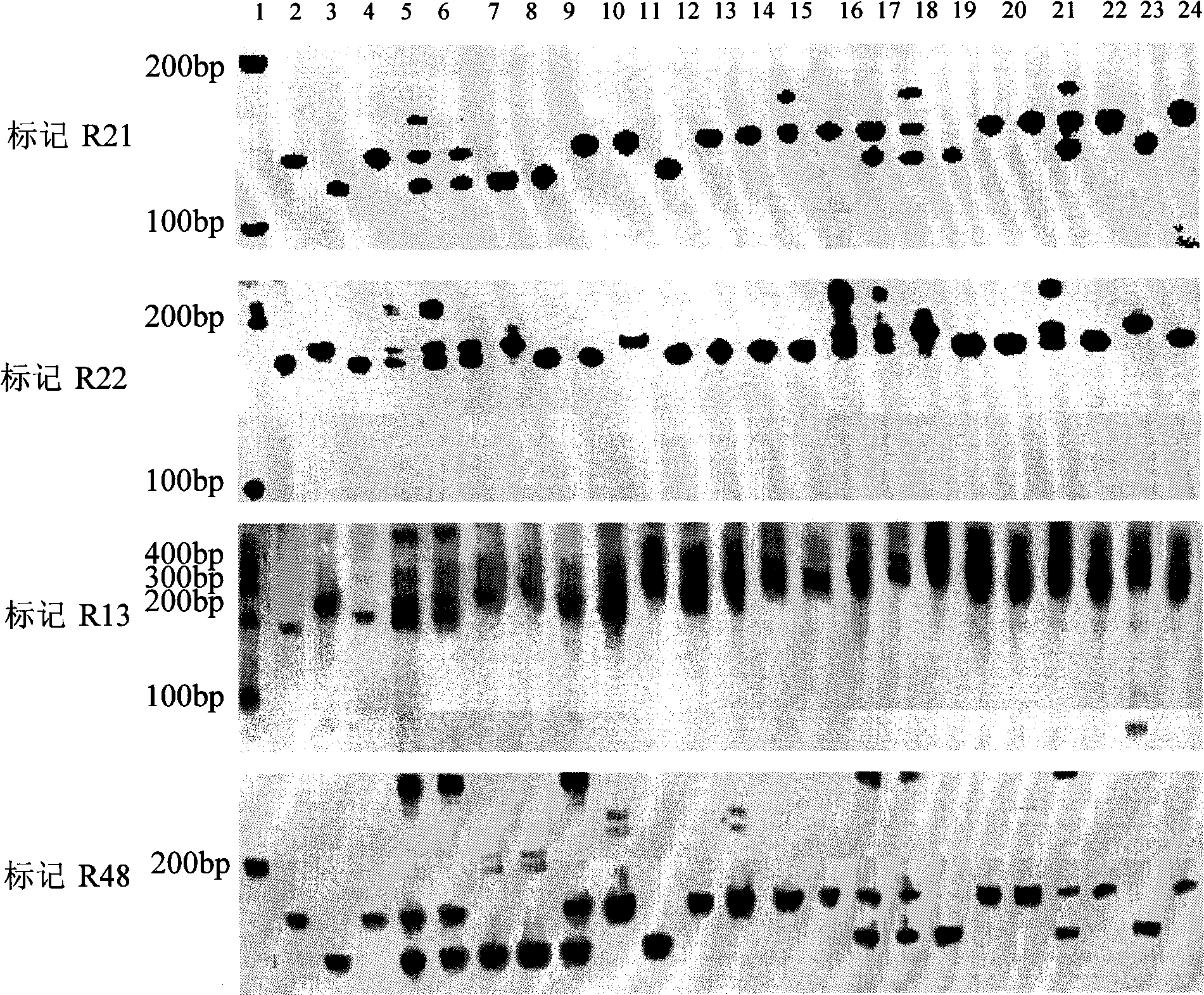
The invention relates to a molecular marking method of a major gene locus qSTV11 with rice stripe and leaf blight resistance and belongs to the field of molecular genetics. Hybridization with female plant of rice Nipponbare and male plant of Kasalath, backcross with Nipponbare and derivation are orderly realized, thus obtaining a backcross recombinant inbred system, genetic linkage analysis between genotypes and corresponding disease indexes and ratios in each family is realized, the resistant effects of the major gene locus qSTV11 with rice stripe and leaf blight resistance and allelic genes from Kasalath are detected, and molecular markers R21, R22, R13 and R48 that can be utilized in breeding are obtained through filtering. The four molecular markers are utilized for detecting whether Kasalath and derived varieties thereof contain the major gene locus, and consequently the rice stripe and leaf blight resistant level of the varieties can be predicted, and the selection efficiency of rice varieties with rice stripe and leaf blight resistance can be remarkably raised.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecular mark method for rice anti-leaf drop streak site
InactiveCN1896283AImprove utilizationEasy to detectMicrobiological testing/measurementDNA/RNA fragmentationDiseaseAgricultural science
The present invention belongs to the molecular genetics field and relates to the the molecular marker method of the major gene loci conferring rice stripe disease resistance. The rice variety Nipponbare is hybridized with Kasalath and then backcrossed with Nipponbare. Via genetic linkage analysis of the genotype and disease index ratio of each family in aquired backcross inbred lines, the molecular marker BJ11-8 of the major rice stripe disease resistance gene loci qSTVll is obtained. Detection of this major gene locus in Kasalath and its derivatives via the molecular marker BJ11-8 will forecast the resistance level to the rice stripe disease and increase the selection efficiency of rice stripe disease resistant rices significantly.
Owner:NANJING AGRICULTURAL UNIVERSITY
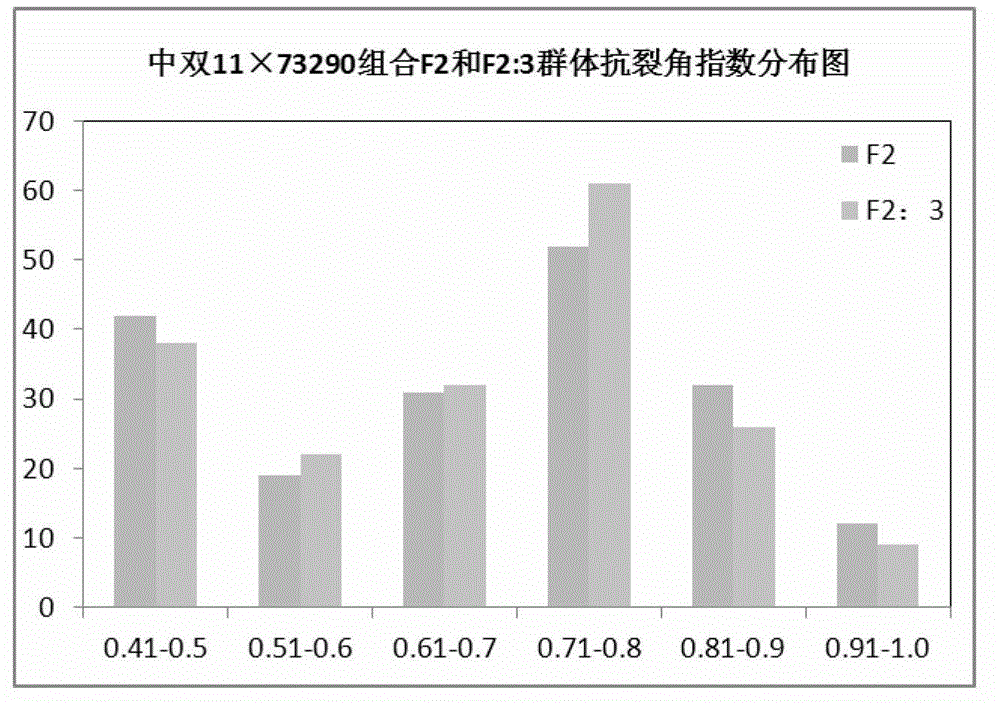
Molecular marker of pot shattering resistance trait major gene locus of rapes and application
ActiveCN102747080AImprove accuracySelection inefficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceField experiment
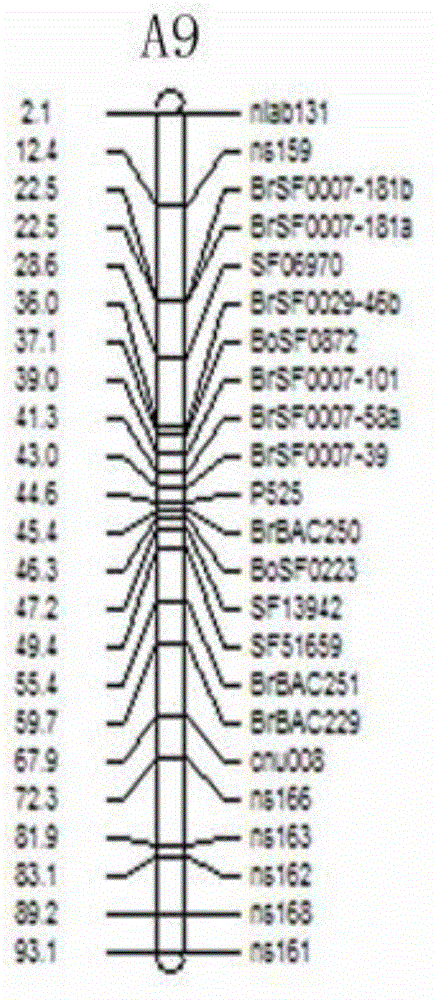
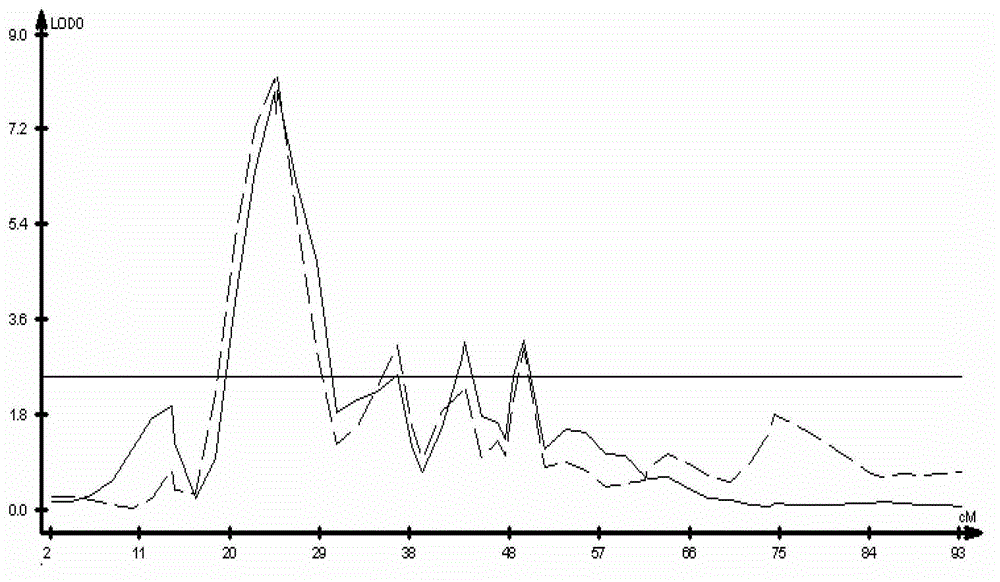
The invention discloses a molecular marker of a pot shattering resistance trait major gene locus of rapes and an application. The steps are as follows: 1) hybridizing double 11 variety and 73290 variety (which have obvious differences in the pot shattering resistance traits) in a cabbage type rape varieties, and carrying out selfing on progenies to obtain F2 and F2:3 segregation populations with the segregated pod shattering resistance traits; 2) carrying out polymorphic screening on parent DNA (deoxyribonucleic acid) by utilizing an SSR (simple sequence repeat) primer, and establishing a genetic linkage map by carrying out SSR molecular marker genotyping on the F2 progeny segregation populations; 3) carrying out field experiments and plant inquisition on the F2 and F2:3 segregation populations so as to obtain phenotype data of the pod shattering resistance traits; and 4) carrying out QTL (quantitative trait locus) detection by combining with the developed high-density molecular marker genetic linkage map and genotype and the phenotype data of the segregation populations, utilizing QTL Cart2.5 software to obtain the major gene locus Psr.A9 for controlling the pod shattering resistance of the rapes on a cabbage type rape A9 linkage group and also obtain the molecular marker BrSF0007-39 closely linked with the major gene locus. The selection efficiency of breeding with the pod shattering resistance can be greatly improved by utilizing the marker for assistant selection.
Owner:INST OF OIL CROPS RES CHINESE ACAD OF AGRI SCI
Molecular marker for rapidly identifying density of spines of cucumber line, identification method and application
ActiveCN104805216APrecise screeningNot affectedMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide
The invention discloses a molecular marker for rapidly identifying the density of spines of a cucumber line. The molecular marker is a molecular marker MM3, a molecular marker MM1 or a molecular marker MM2; the molecular marker MM3 consists of a nucleotide sequence shown in SEQ ID NO:9 and a nucleotide sequence shown in SEQ ID NO:10, wherein the nucleotide sequence shown in the SEQ ID NO:9 is totally linked with a cucumber sparse spine gene FS1; the nucleotide sequence shown in the SEQ ID NO:10 is totally linked with a cucumber sparse spine gene fs1; meanwhile, the invention also provides a method for rapidly identifying a cucumber variety or the density of the spines of the line by utilizing the molecular marker MM3. The molecular marker and the identification method provided by the invention are beneficial for building a molecular marking assisting breeding system for sparse / dense spine cucumbers and applying the molecular marking assisting breeding system to cucumber breeding practices and have the advantages of being rapid and accurate in screening, not affected by environment, definite in selected target, capable of lowering the production cost and greatly improving the selection efficiency and quality of a target cucumber variety or the spine type of the line, and quite high in social and economic values.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Wheat grain weight molecular marker and its use in breeding
ActiveCN104164428AImprove selection efficiencyQuality improvementMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceChromosome
The invention relates to the technical field of gene engineering and provides a wheat grain weight molecular marker. The wheat grain weight molecular marker can be used in the field of wheat breeding. The wheat grain weight molecular marker is located between wheat 6A chromosome BOBWHITE_C20706_135 and RA_C28538_971. Through use of the wheat grain weight molecular marker, it is detected if the wheat variety or line comprises QGW6A-164 for increasing thousand seed weight so that wheat high-yield variety breeding is accelerated. Through use of the special-purpose grain weight molecular marker, screening is fast and accurate, is not influenced by the environment and has a clear selection object, a production cost is greatly reduced and high-yield wheat variety or line selection efficiency and quality are greatly improved.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Wheat stalk breaking strength molecular marker QWQD4B.4-13 and application thereof
ActiveCN106480180AImprove qualitySpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationBreaking strengthAgricultural science
A wheat stalk breaking strength molecular marker is disclosed, the wheat stalk breaking strength molecular marker is located between wheat 4B chromosome TDURUM-CONTIG63670-287 and IACX557, and named as QWQD4B.4-13; through application of the molecular marker, whether QWQD4B.4-13 gene loci for increasing stalk breaking strength exist in wheat varieties or lines can be detected, the breeding process of lodging-resistant wheat varieties can be accelerated, by use of the special stalk breaking strength molecular marker, the screening is fast and accurate, and is not affected by the environment, a target is clear to select, cost of production is saved, the selection efficiency and quality of the lodging-resistant high yield wheat varieties or lines are greatly improved, QTL detection effect and precision can be improved, the effective molecular marker is developed, a stalk breaking strength molecular assisted breeding system is developed, and the stalk breaking strength molecular assisted breeding system can be programmatically applied in breeding practices.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Special primer for identifying gummy stem blight resistance of muskmelon and molecular marking method
InactiveCN106701917AChoose a clear goalReduce workloadMicrobiological testing/measurementDNA/RNA fragmentationElectrophoresisPathogenic bacteria
The invention provides a special primer for identifying gummy stem blight resistance of muskmelon and a molecular marking method. Specific to the problems existing in gummy stem blight resisting breeding work of the muskmelon, such as the defects of inaccuracy and large workload in manual inoculation of pathogenic bacteria and identification of gummy stem blight resistance, the method comprises the following steps: extracting a genome DNA (Deoxyribonucleic Acid) of a muskmelon sample to be detected to serve as a template; performing polymerase chain reaction (PCR) amplification by using a primer special for molecular marking; detecting an amplification product through electrophoresis. By adopting the SSR (Simple Sequence Repeat) molecular marker for identifying the gummy stem blight resistance and the special primer thereof, the aim of screening muskmelon materials resisting to the gummy stem blight and not resisting to the gummy stem blight can be fulfilled at the early growth period of muskmelon plants, and the molecular marker and the special primer have the advantages of accurate identification, freeness from limitations by infection conditions, adoption of readily-available raw materials and the like. Muskmelon auxiliary breeding is performed, so that the special primer has the advantages of definite target selection, saving in breeding time and labor cost, and increase of the breeding efficiency.
Owner:TIANJIN UNIV
SSR mark linked with pseudoperonospora cubensis resistance main effect QTL and application of SSR mark
ActiveCN105734057AShorten the breeding cycleImprove selection accuracyMicrobiological testing/measurementDNA/RNA fragmentationPopulationResistant strain
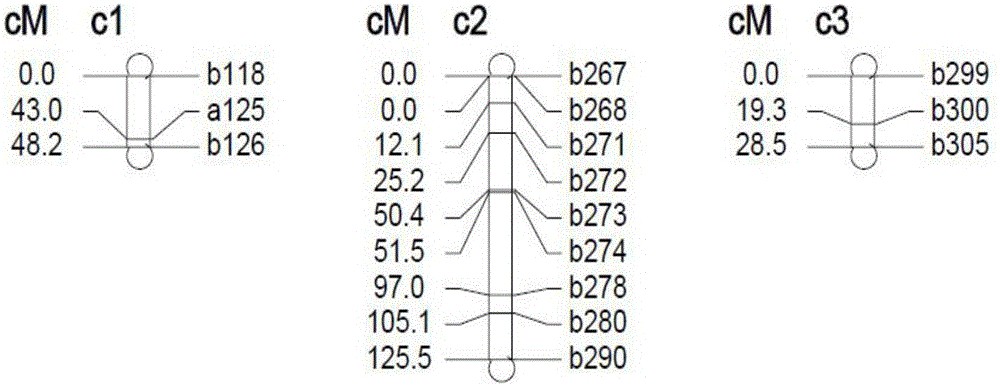
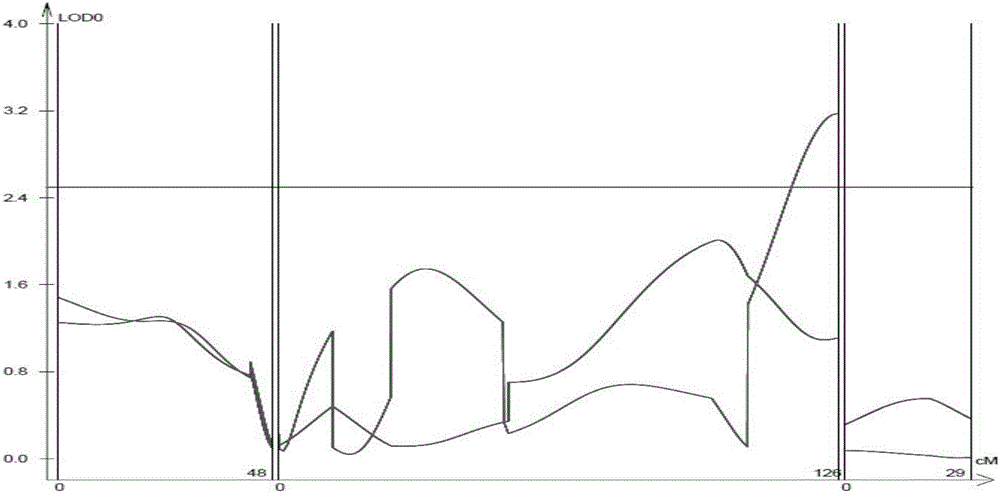
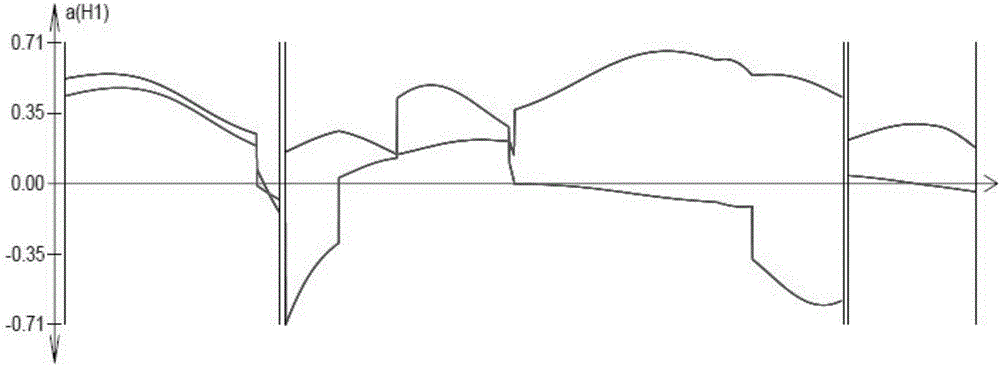
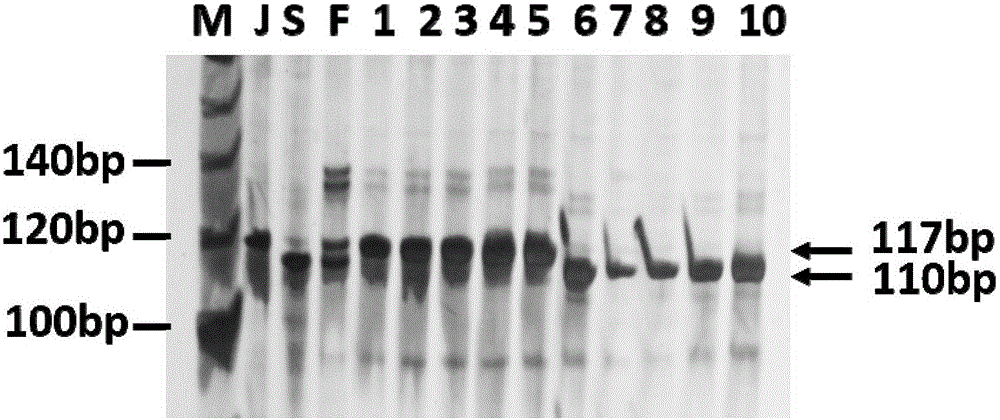
The invention discloses an SSR mark linked with a pseudoperonospora cubensis resistance main effect QTL and an application of the SSR mark. A high-resistance resource PI390452 and an infected farm variety 'Kelakesai' construct a pseudoperonospora cubensis infection-resistant F2 generation population, and well-known genetic software is used for performing linkage disequilibrium analysis and establishing a genetic linkage graph, the LOD result analysis shows a peak value at b290, and LOD is more than 3.0. According to b290, well-known SSR search software is used for re-searching an SSS site, and well-known PCR primer design software is used for re-designing an SSR primer. According to a phenotype and a genotype, well-known genetic software is finally used for finding the molecular mark SSR-pc283 linked with the pseudoperonospora cubensis resistance main effect QTL, the genetic distance is 0.5CM, and the SSR mark can be used for molecular mark assisted breeding, so that the good technical effects of shortening the breeding cycle and improving the selection accuracy in the process of breeding a pseudoperonospora cubensis resistant strain are achieved.
Owner:XINJIANG AGRI SCI ACAD CANTALOUPE RES CENT
Molecular marker closely interlocked with neck blast resistance gene of paddy rice Jiangnan lateness and application thereof
ActiveCN105713983AAccurate genetic lociEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyElectrophoreses
The invention belongs to the field of crop molecular genetic thremmatology, and discloses a molecular marker interlocked with a neck blast resistance gene of paddy rice Jiangnan lateness and an application thereof. The molecular marker is prepared by the following steps: (1) acquiring a paddy rice sample, and extracting a genome DNA of the paddy rice sample; (2) performing PCR amplification for the genome DNA of the paddy rice sample by utilizing a primer pair of any pair of molecular markers in RM27273 and BS33, and performing the electrophoresis detection on a PCR amplification product, wherein if a molecular marker DNA fragment of the corresponding size is amplified, the existence of the Pb-jn1 gene is marked. By virtue of the molecular marker of the paddy rice neck blast resistance gene Pb-jn1, whether the paddy rice Jiangnan lateness as well as the hybrid, the backcross and the multiple-cross later generations of the paddy rice Jiangnan lateness contain the gene can be detected, the resistance level to the paddy neck blast can be predicted, the selection efficiency of the paddy rice neck blast resistance material can be greatly improved, and the anti-disease breeding process can be accelerated.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecular markers of rice stripe virus disease-resistant major gene locus qSTV11
InactiveCN102021244AEasy to detectQuick filterMicrobiological testing/measurementAgricultural scienceGenotype
The invention relates to molecular markers of a rice stripe virus disease-resistant major gene locus qSTV11, which belongs to the field of molecular genetics. The molecular markers are obtained by the following steps of: hybridizing a rice variety Nipponbare serving as a female parent with kasalath serving as a male parent; performing back crossing on a hybrid product and the Nipponbare; deriving to obtain a back crossing recombinant inbred line; performing genetic linkage analysis on the genotype of each family and corresponding disease index ratio; detecting the stripe virus disease-resistant major gene locus qSTV11, wherein alleles from kasalath have resistance effects; and screening to obtain molecular markers R21, R22, R13 and R48 which can be bred and utilized. The four molecular markers are used for detecting whether the kasalath and a derivative variety (line) contain the major gene locus or not, so that the rice stripe virus disease resistance level can be predicted and the selection efficiency of a stripe virus disease-resistant rice variety is improved remarkably.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecular marker for co-separating from aroma character
InactiveCN101921761AEasy to identifyAccurate and convenient identificationMicrobiological testing/measurementDNA/RNA fragmentationWater bathsNose
The invention relates to a molecular marker for co-separating from aroma characters, belonging to the field of molecular genetics. 1-2 pieces of spires at the upper part of individual rice are taken at the tillering stage and are reserved in a refrigerator with the temperature of below 80 DEG C for later use. Rice leaves with the length of 2-4cm are taken to be placed in a centrifugal tube of 1.5ml, liquid nitrogen is added, grinding is carried out, 900 mu l of TPS solution is added, and water bath is performed for 20min at the temperature of 75 DEG C. A cover is opened to smell the odour by a nose to determine whether aroma exists. A specific PCR primer L01 or L02 is utilized to obtain the molecular marker by PCR amplification. The invention can be specially used for seed selection and genetic research of the rice aroma variety and can be used for cloning aroma genes.
Owner:SHANDONG RICE RES INST
Molecular marker closely linked with melon pseudoperonospora cubensis resisting gene and application thereof
ActiveCN105821135AShorten the breeding cycleImprove selection accuracyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceHigh resistance
The invention discloses a molecular marker closely linked with a melon pseudoperonospora cubensis resisting gene and the application thereof. Based on the seedling-stage resource pseudoperonospora cubensis resistance identification and selection principle and the prior art, a melon resource PI438685 with high pseudoperonospora cubensis resistance is selected as the high-resistance resource, a melon pseudoperonospora cubensis anti-fever F2-generation group is established by means of the high-resistance resource PI438685 and the susceptible farm variety Clarke, the molecular marker SSR-666230 linked with the PI438685 pseudoperonospora cubensis resisting gene is found by means of well-known heredity software, and the genetic distance between the marker and the resistance gene is 2.66 cm. By extracting genome DNA of a single melon plant and conducting PCR amplification with the marker SSR-666230, whether a unique 230bp specific band appears is detected, it is predicted that the single melon plant has pseudoperonospora cubensis resistance if yes, the technical effects of shortening breeding period and improving selection accuracy during pseudoperonospora cubensis resisting strain breeding are realized, and practical value is high.
Owner:XINJIANG AGRI SCI ACAD CANTALOUPE RES CENT
InDel molecular marker of rice blast-resistant gene Pid3-A4, and detection method and application thereof
ActiveCN108913809AImprove breeding efficiencyEasy accessMicrobiological testing/measurementPlant genotype modificationResistant genesAgricultural science
The invention discloses an InDel molecular marker of a rice blast-resistant functional gene Pid3-A4, wherein the InDel molecular marker includes an InDel molecular marker PA4-C, and the InDel molecular marker PA4-C is used for identifying and screening a rice material with a functional gene Pid3-A4 having 9 bp deletion at first 1815 bases of a coding region from rice materials; a 9 bp deletion sequence is CCCTGAAGA. The functional molecular marker can accurately detect whether a Pid3-A4 functional gene is contained in genomes of different rice varieties and detect the homozygous status, can beapplied for screening rice hybridized and transferred offspring plants, improves the breeding efficiency of the rice blast-resistant material of rice, and obtains rice blast-resistant rice varietiescontaining the functional gene Pid3-A4.
Owner:HUNAN HYBRID RICE RES CENT
Grain weight molecular marker for wheat and application of grain weight molecular marker in breeding
ActiveCN104087586AImprove selection efficiencyQuality improvementMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGrain weight
The invention relates to the technical field of genetic engineering and can be applied to the field of wheat breeding, and particularly provides a grain weight molecular marker for wheat. The marker is located between RFL-CONTIG4632-1512 and TaGW2-CAPS of 6A chromosome of wheat. By applying the molecular marker, whether a wheat variety or strain has QGW6A-232 for increasing thousand seed weight or not can be detected so as to accelerate the breeding progress of high-yield variety of wheat. As the special grain weight molecular marker is adopted, screening becomes quick and accurate without being affected by the environment, the selected target is clear, moreover, the production cost is saved, so that the selecting efficiency and quality of the high-yield variety or strain of wheat are greatly improved.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Molecular marker tightly in linkage with corn line grain number main effect QTL and application thereof
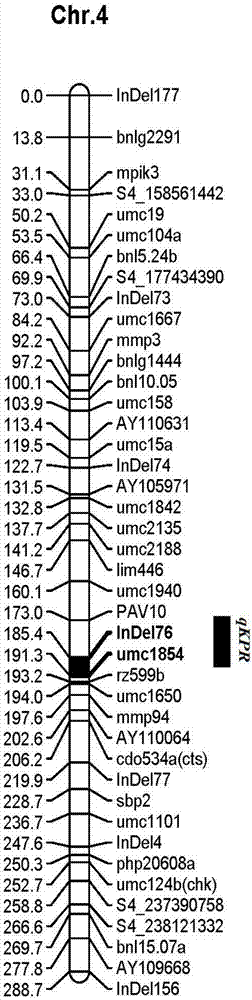
InactiveCN106906301ANot affectedIdentification method is simpleMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenotype
The invention discloses a molecular marker tightly in linkage with main effect QTL of corn line grain number and application thereof. The provided QTL is positioned on a first chromosome 4.09bin, and tightly linked molecular marker consists of primers InDe176 and umc1854. A genome DNA of a triple test-crossed group of corn to be tested is extracted, PCR amplification is carried out by using the primers InDe176 and umc1854, and amplification products having the lengths of 174bp and 142bp are obtained and can be used for predicting the corn line grain number. The molecular marker is used for genotype detection of the corn and its derived varieties or strains to judge the line grain number of the varieties or the strains and improve the selection accuracy and breeding efficiency.
Owner:JIANGSU ACAD OF AGRI SCI
Molecular marker interlocked with cotton fiber length major QTL and application thereof
ActiveCN109055591AChoose a clear goalLow costMicrobiological testing/measurementDNA/RNA fragmentationMolecular breedingBiology
The invention relates to the technical field of molecular breeding and specifically relates to molecular marker which is from sea island cotton 1 and related to fiber length and application thereof. The molecular marker is SSRN106270 and SSRN189240. The molecular marker is favorable for overcoming the defect of an existing breeding technology for identifying the fiber length, can improve fiber length selection efficiency and can quicken a high-quality novel variety breeding process.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Molecular marker for regulating corn single spike weight main effect QTL and application method thereof
ActiveCN107058590AIdentification method is simpleImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMolecular breeding
The invention belongs to the field of crop molecular breeding, particularly relates to a molecular marker for regulating corn single spike weight main effect QTL, and also relates to a method for assisting the selection of corn single spike weight. The molecular marker for regulating the corn single spike weight main effect QTL is composed of two pairs of SSR markers of umc1120 and umc2134. The method for assisting the selection of the corn single spike weight comprises the following steps that the genome DNA of corns to be tested is extracted and PCR amplification is performed by using the umc1120 and umc2134 primers, when amplified products with the length of 338bp and 234bp are obtained, the corns to be tested are candidate large-spike-type corns. When the method is used to assist the selection of large-spike-type corn materials, only does the characteristic strip of the molecular marker need to be detected to predict the single spike weight size. The method is simple and feasible, and the selection efficiency is high. A large-spike-type corn individual plant can be identified during the corn growth early period, and the selection object is clear and not affected by the environment. A high-yield corn hybrid combination is designed accordingly.
Owner:安徽英诺管理咨询有限公司
Molecular marker of pear PpAIV2 gene locus and screening method of molecular marker
ActiveCN106701911ASpeed up the selection processQuick filterMicrobiological testing/measurementDNA/RNA fragmentationScreening effectSaccharum
The invention relates to the field of molecular genetics, discloses a molecular marker of a pear PpAIV2 gene locus and further discloses a screening method of the molecular marker of the pear PpAIV2 gene locus. A genome simple sequence repeat (SSR) locus adjacent to pear soluble acid invertase gene PpAIV2 is predicated and a locus specific primer is designed; one SSR allelic variation locus which is tightly interlocked with the PpAIV2 is obtained through amplification in germplasms with different genetic backgrounds and F1-generation population genomes; the SSR allelic variation locus is named as P.zaas-1. The pear soluble acid invertase gene PpAIV2 is a key gene for accumulating pear fruit sucrose; the P.zaas-1 has potential identification and screening effects on materials with remarkable differences on accumulation of the fruit sucrose.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Functional molecular marker for rice anti-blast gene Pi9 and application thereof
ActiveCN103146695BImprove breeding efficiencyImprove accuracyMicrobiological testing/measurementPlant genotype modificationBiotechnologyNucleotide
The invention provides a functional molecular marker for rice anti-blast gene Pi9 and application thereof, belonging to the field of crop molecular genetic breeding science. The invention finds out that the rice anti-blast gene Pi9 has a 10bp insertion / deletion site positioned between the gene promoter -516 and -517, wherein the nucleotide sequence is disclosed as SEQ ID NO.1; and on such basis, the invention develops a method of the functional molecular marker for gene Pi9. By detecting the functional molecular marker, the invention can accurately detect whether genomes of different rice species contain the Pi9 functional gene and detect the homozygotic state, and can be used for screening rice hybrid transformed descendant plants to enhance the breeding efficiency of the rice anti-blast material, thereby effectively controlling the size of the breeding population, obviously saving the breeding and screening cost and obtaining the anti-blast rice species containing the Pi9 functional gene.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com