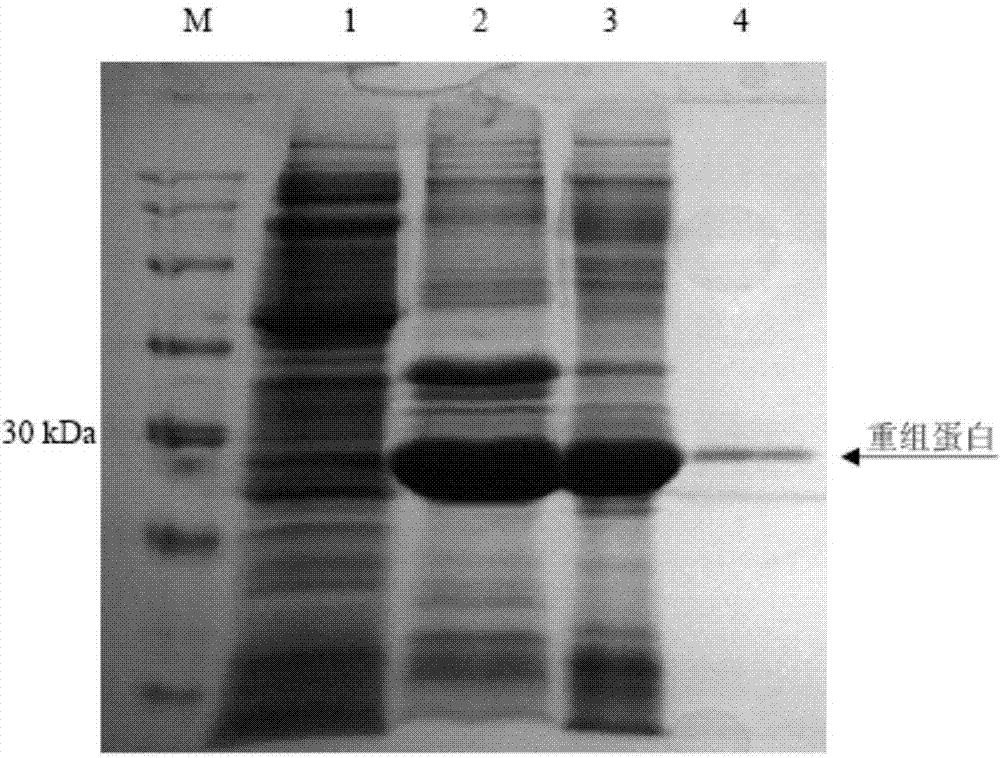
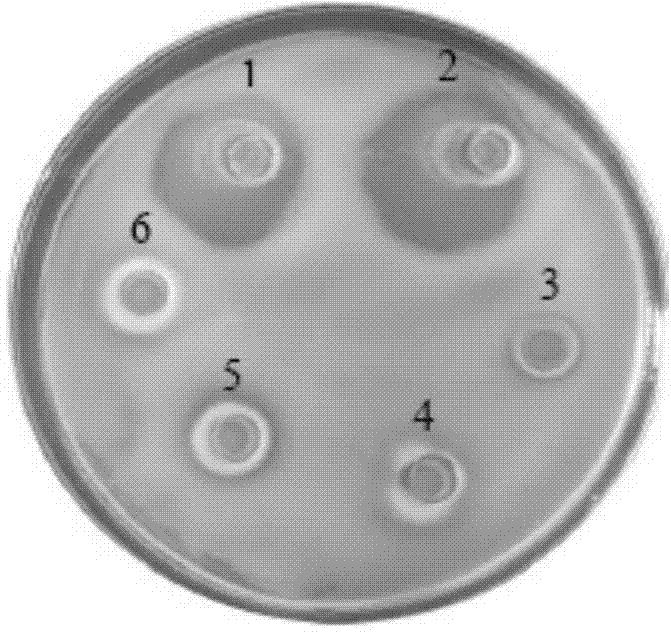
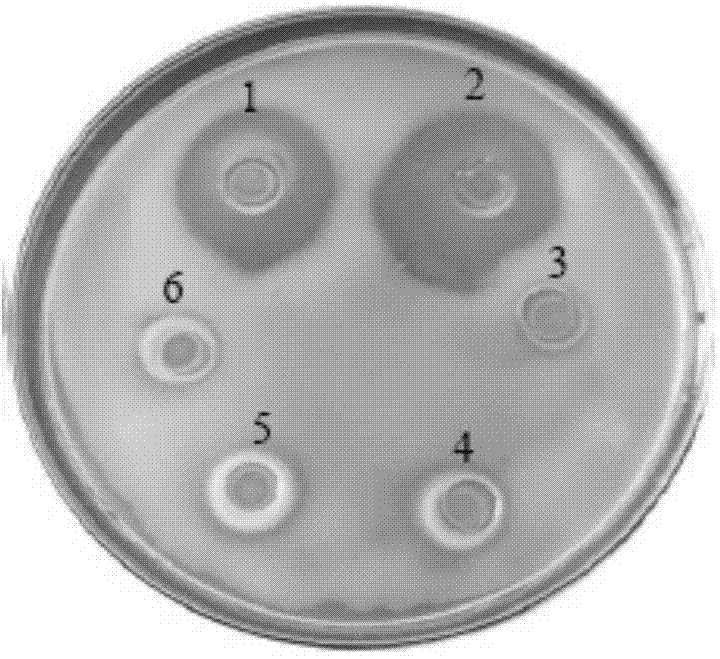
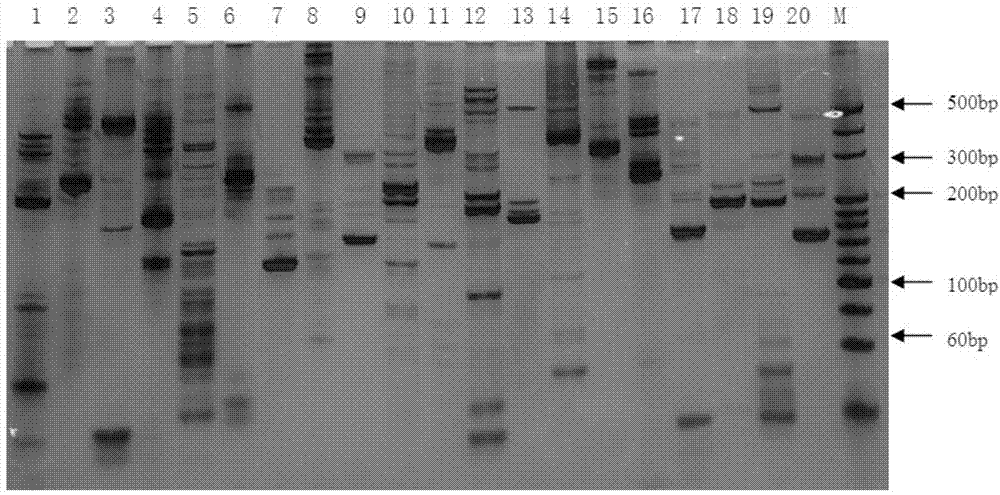
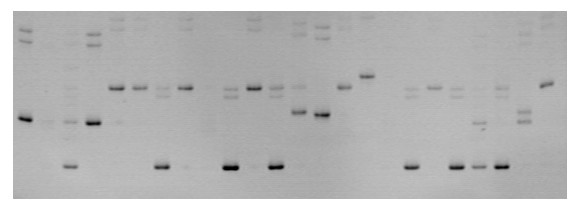
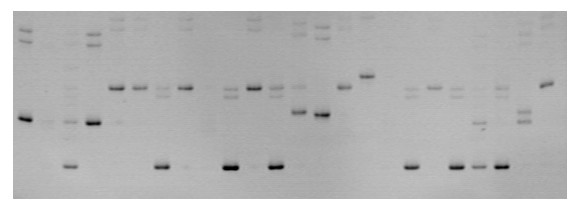
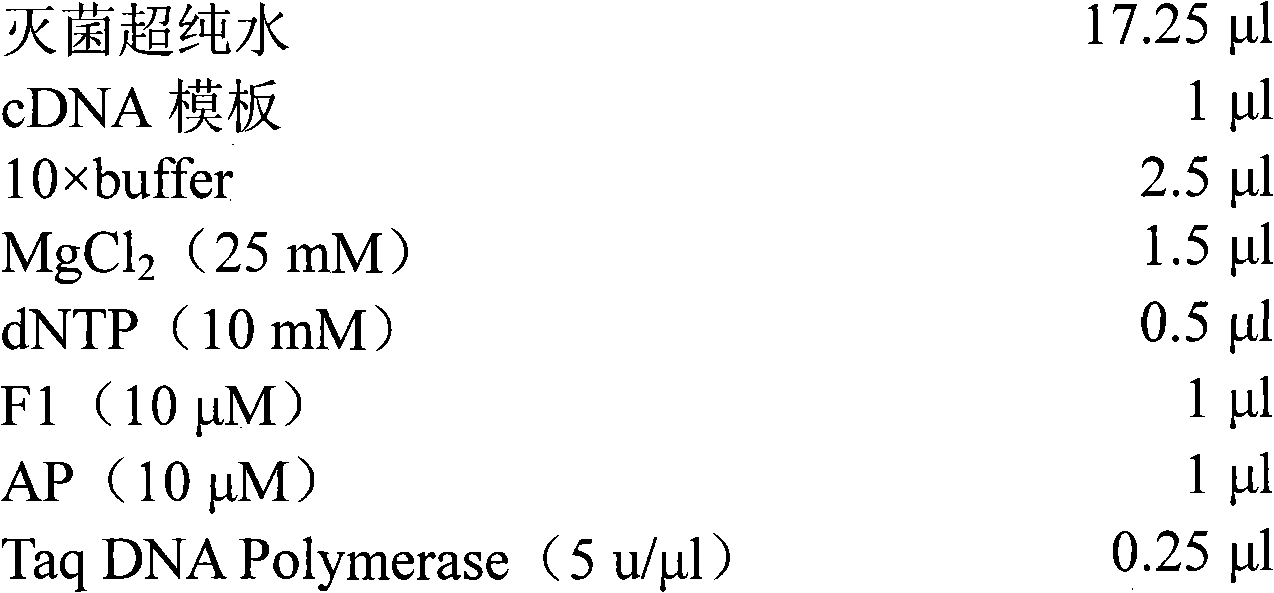Patents
Literature
169 results about "Expressed sequence tag" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and are instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proceeded rapidly, with approximately 74.2 million ESTs now available in public databases (e.g. GenBank 1 January 2013, all species).
PRO362 polypeptides
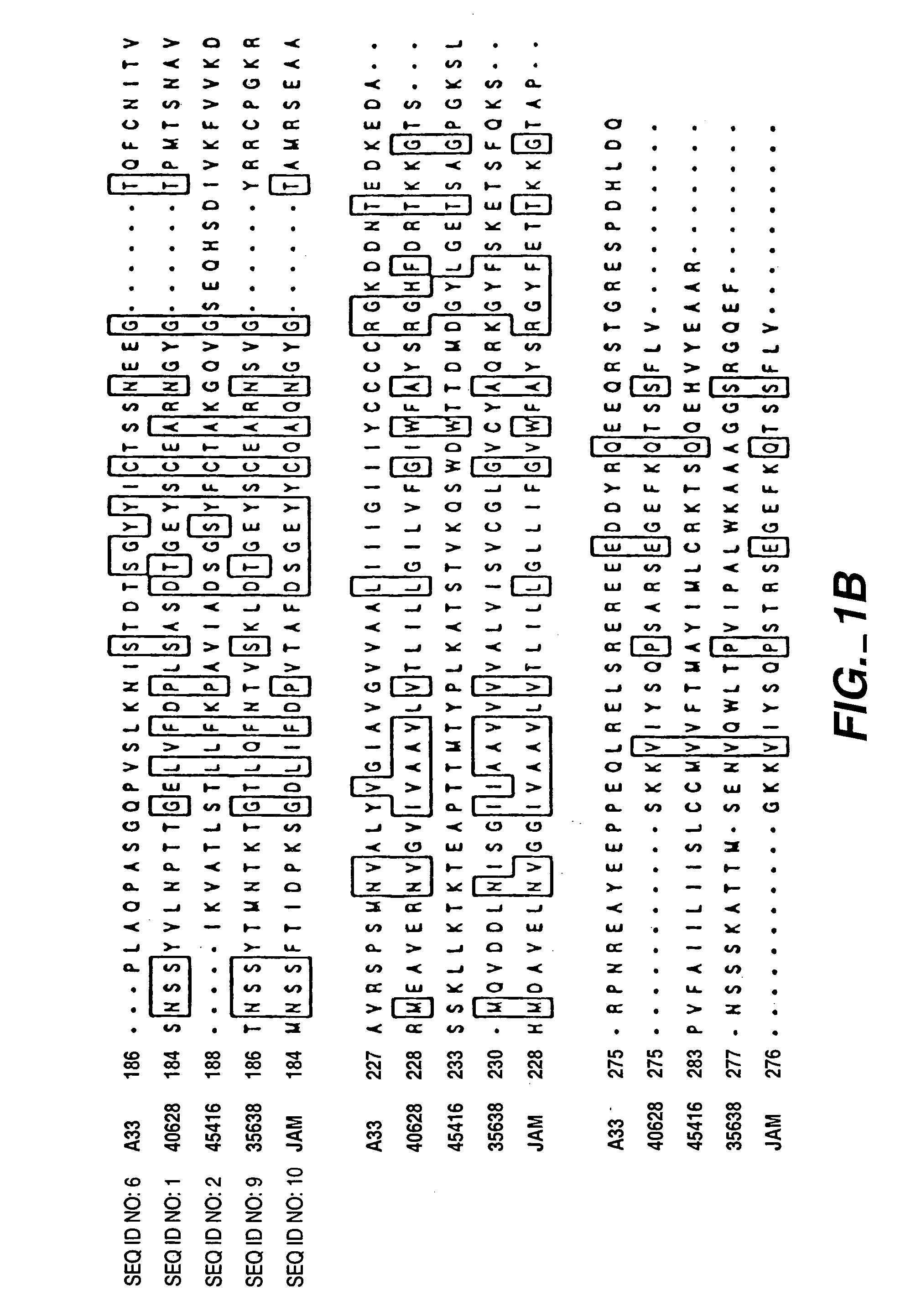
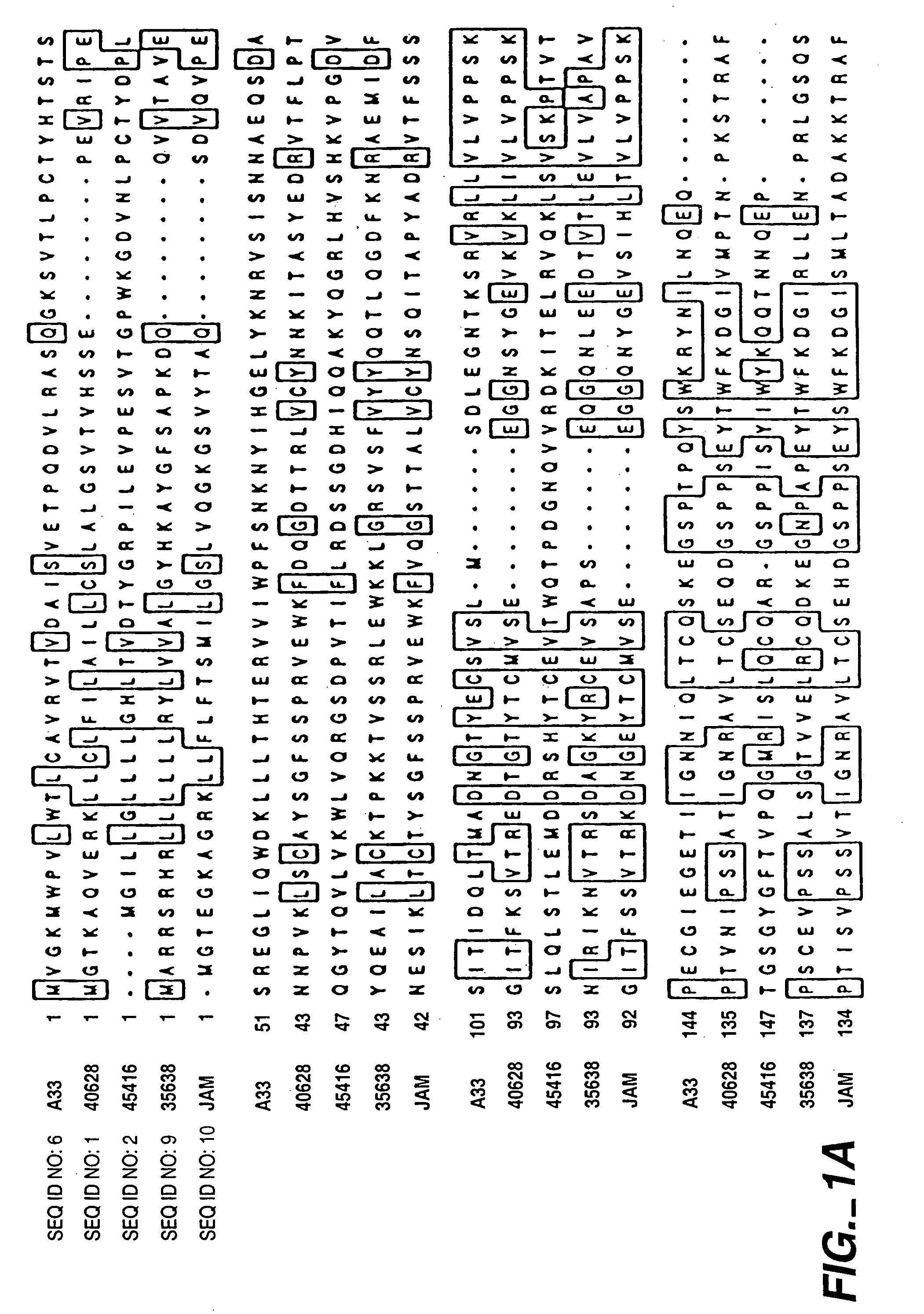
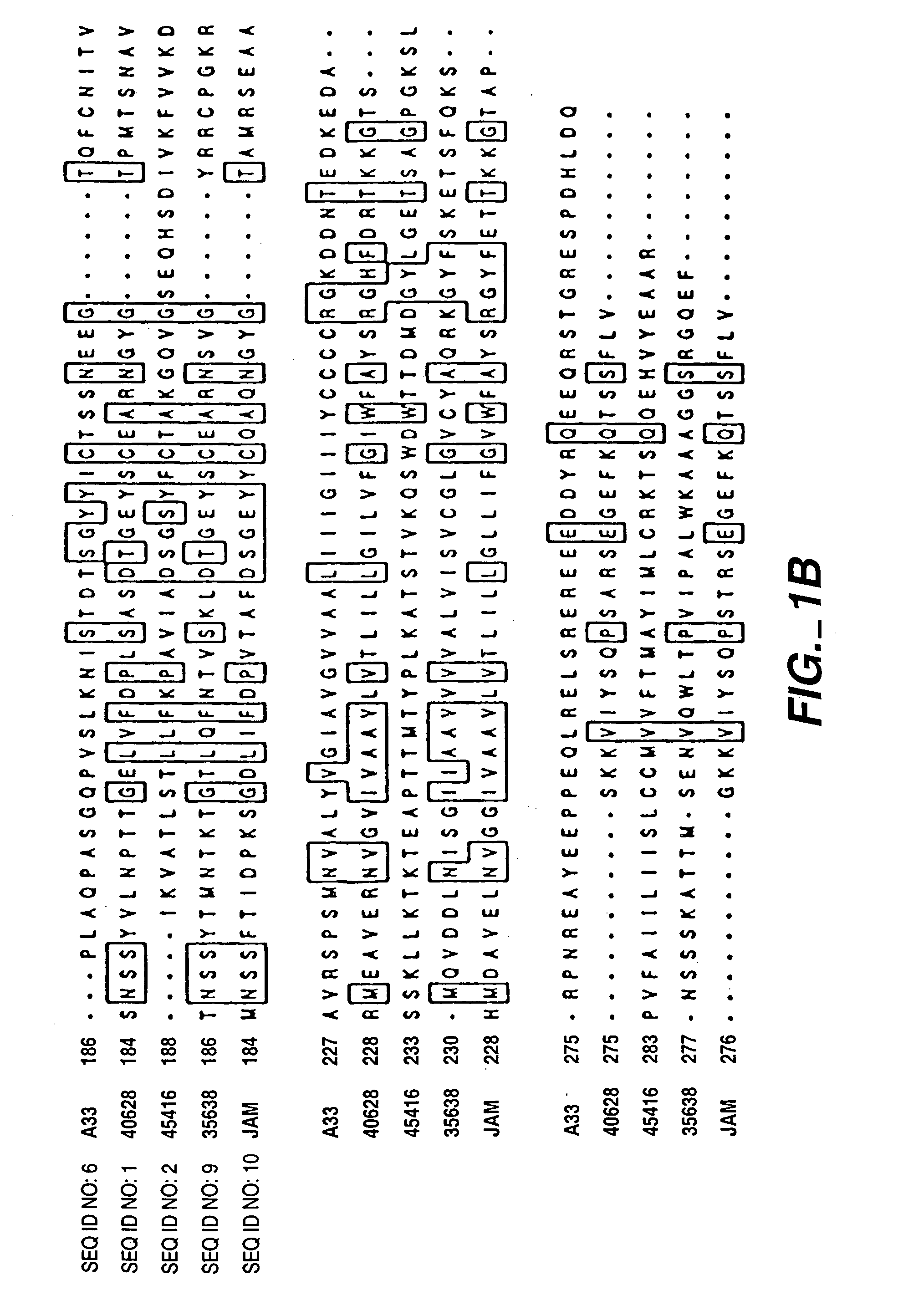
InactiveUS7282565B2Cell receptors/surface-antigens/surface-determinantsAntibody mimetics/scaffoldsAntigenInflammatory bowel disease
The present invention relates to compositions and methods of treating and diagnosing disorders characterized the by the presence of antigens associated with inflammatory diseases and / or cancer, and nucleotide sequences, including expressed sequence tags (ESTs), oligonucleotide probes, polypeptides, vectors and host cells expressing such antigens PRO301, PRO362 or PRO245.
Polynucleotides encoding GFRα3
InactiveUS7026138B1Facilitating identification and characterizationEasy to detectSugar derivativesTissue cultureNucleotideReceptor activation
The present invention relates to nucleotide sequences, including expressed sequence tags (ESTs), oligonucleotide probes, polypeptides, vectors and host cells expressing, and immunoadhesions and antibodies to mammalian GFRα3, a novel α-subunit receptor of the GDNF (i.e. GFR) receptor family. It further relates to an assay for measuring activation of an α-subunit receptor by detecting tyrosine kinase receptor activation (i.e., autophosphorylation) or other activities related to ligand-induced α-subunit receptor homo-dimerization or homo-oligomerization.
Owner:GENENTECH INC
Compounds, compositions and methods for the treatment of diseases characterized by A-33 related antigens
InactiveUS7211400B2Cell receptors/surface-antigens/surface-determinantsPeptide/protein ingredientsAntigenNucleotide sequencing
The present invention relates to compositions and methods of treating and diagnosing disorders characterized the by the presence of antigens associated with inflammatory diseases and / or cancer, and nucleotide sequences, including expressed sequence tags (ESTs), oligonucleotide probes, polypeptides, vectors and host cells expressing such antigens PRO301, PRO362 or PRO245.
Owner:GENENTECH INC
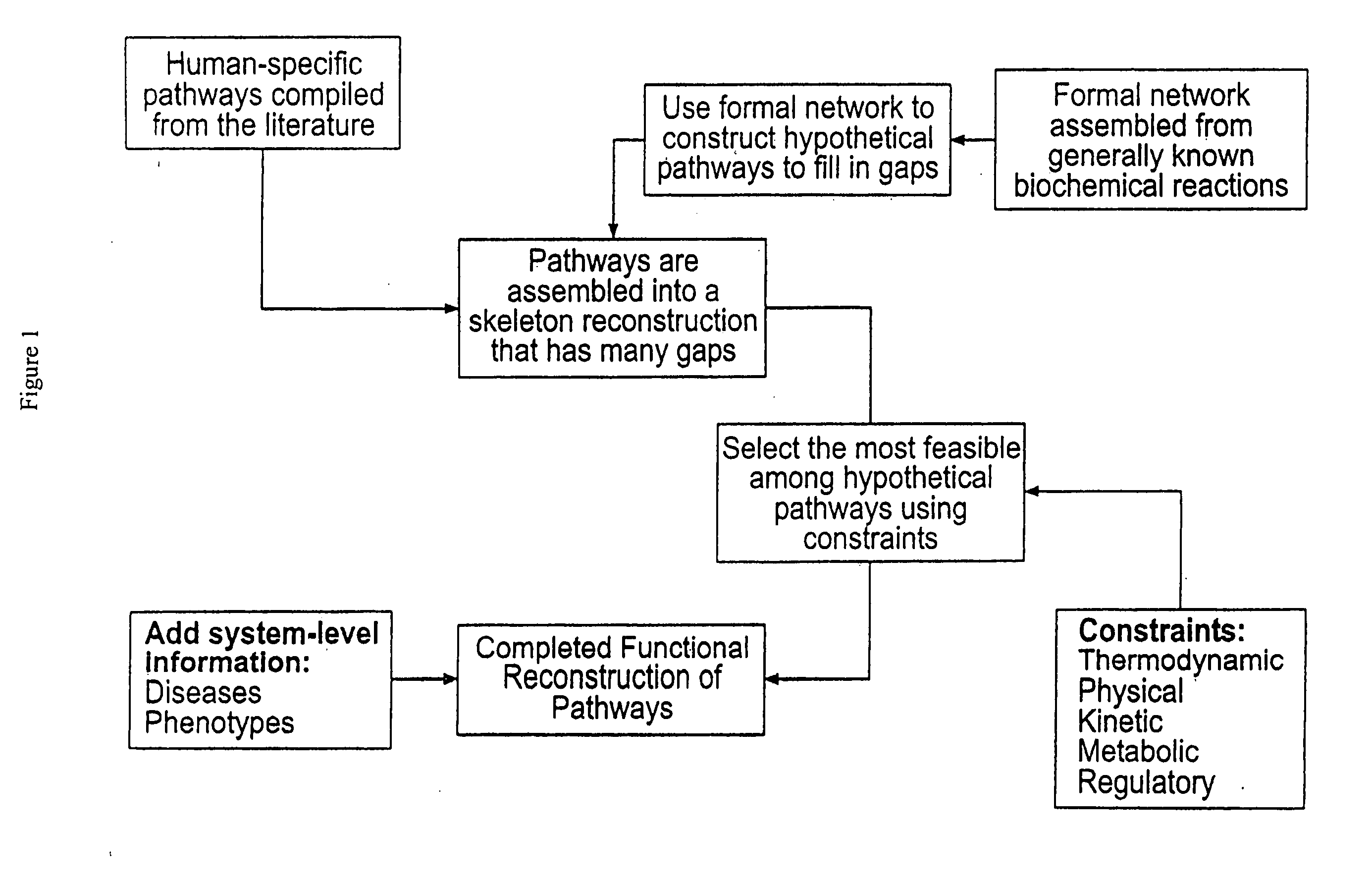
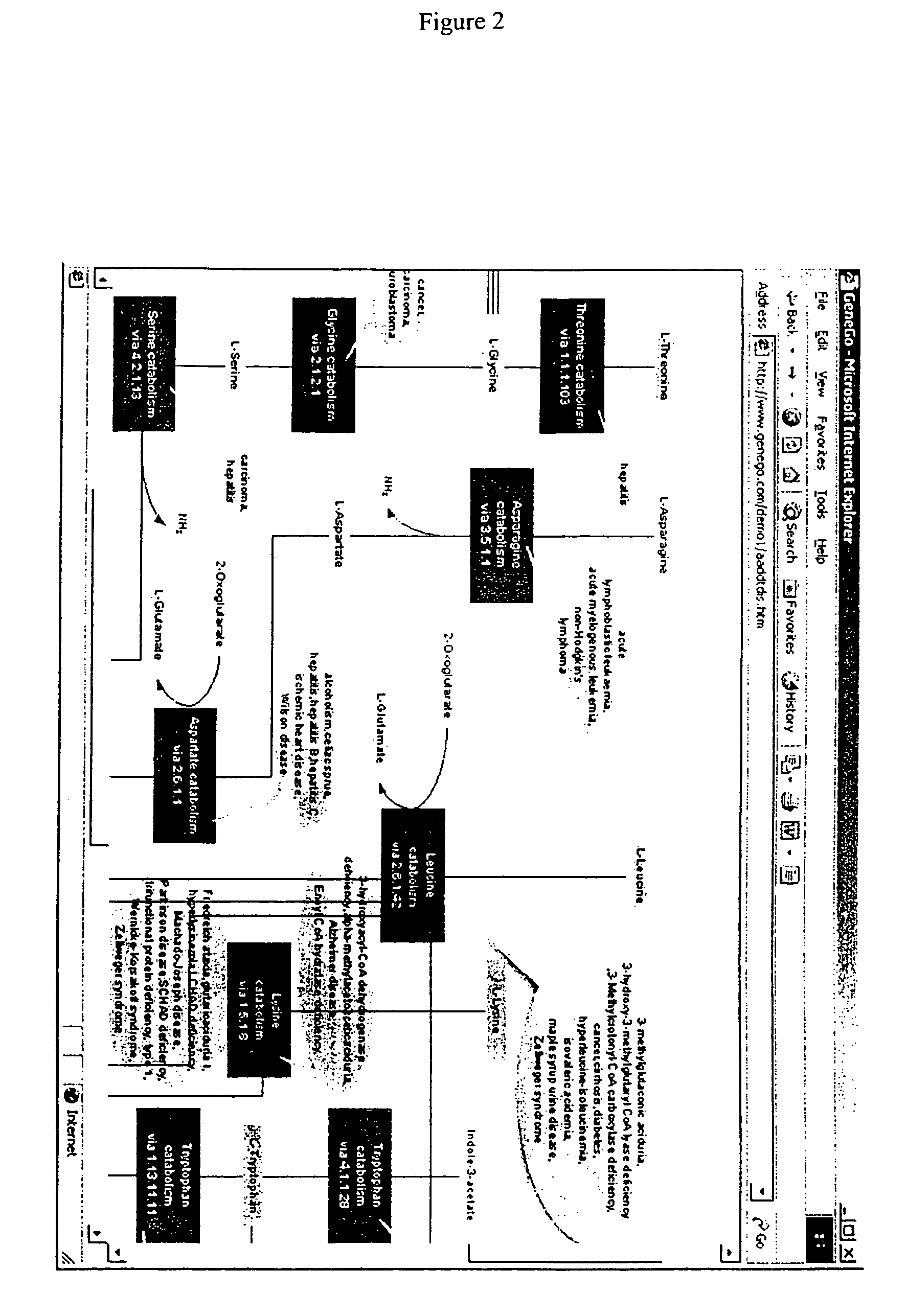
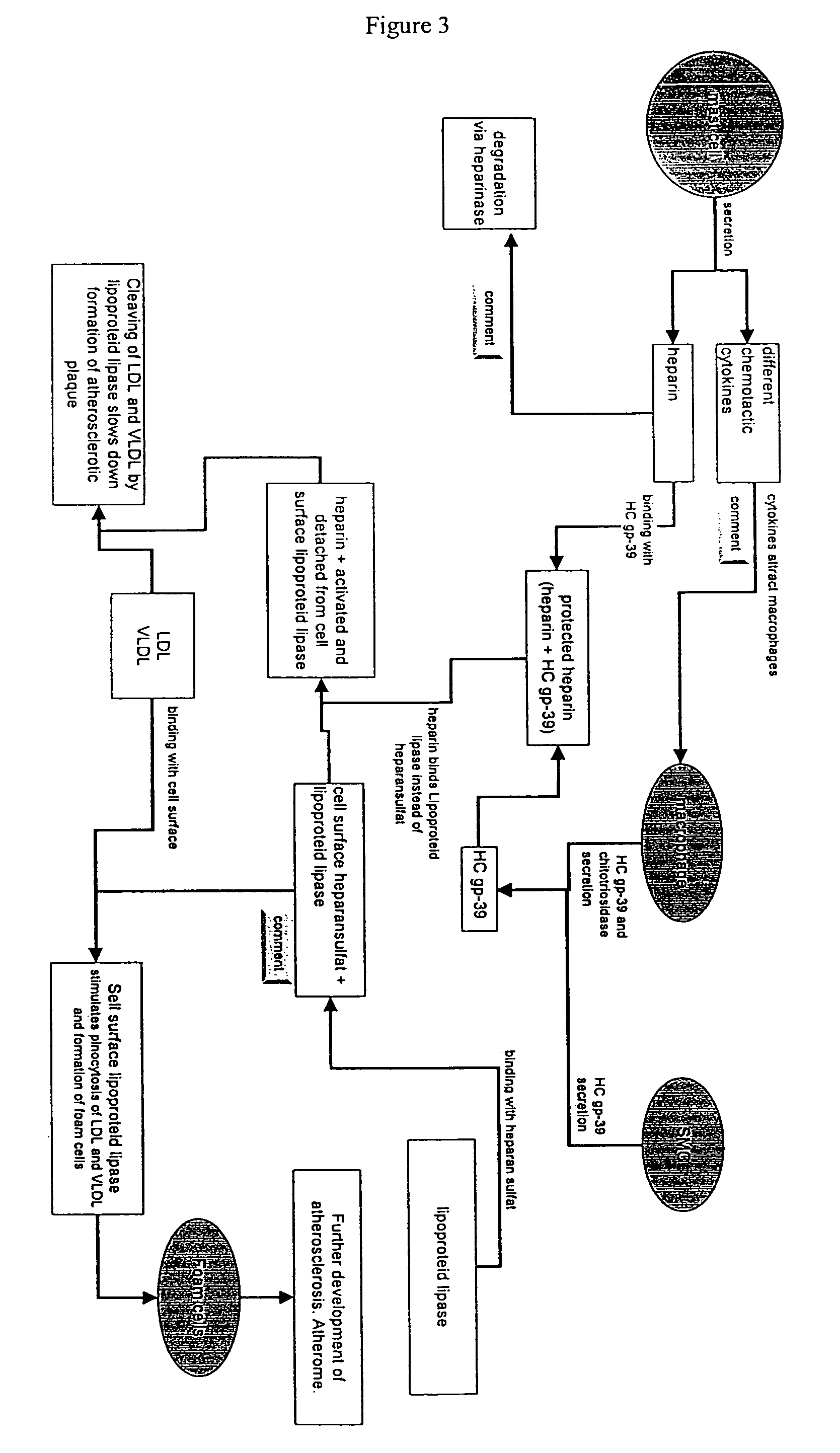
Methods for identification of novel protein drug targets and biomarkers utilizing functional networks
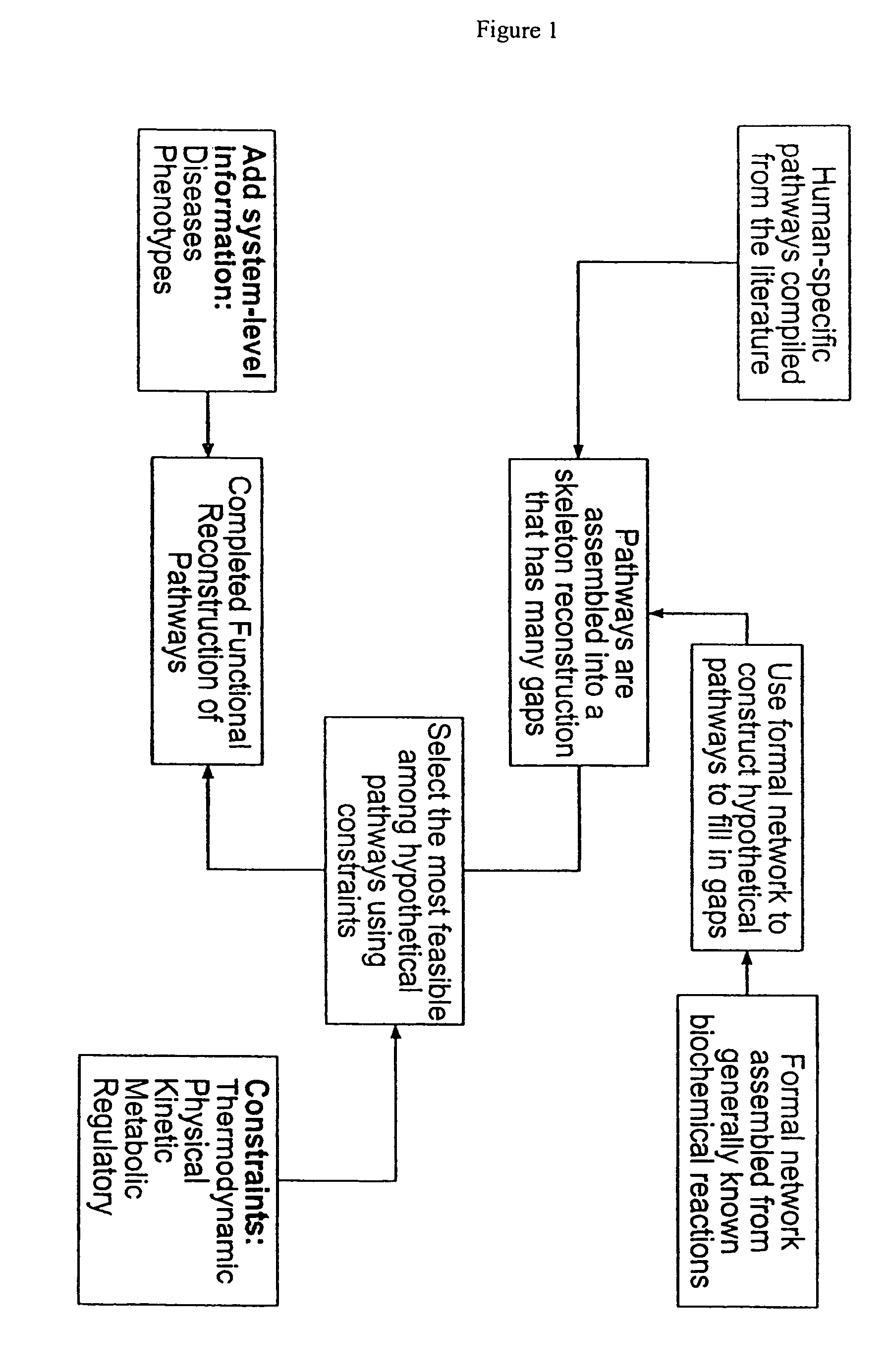
The process of System Reconstruction is used to integrate sequence data, clinical data, experimental data, and literature into functional models of disease pathways. System Reconstruction models serve as informational skeletons for integrating various types of high-throughput data. The present invention provides the first metabolic reconstruction study of a eukaryotic organism based solely on expressed sequence tag (EST) data. System Reconstruction also provides a method for the identification of novel therapeutic targets and biomarkers using network analysis. The initial seed networks are built from the lists of novel targets for diseases with the high-throughput experimental data being superimposed on the seed networks to identify specific targets.
Owner:CAMELOT UK BIDCO LTD
GFRalpha3 and its uses
InactiveUS20060216289A1Facilitating identification and characterizationEasy to detectFungiSenses disorderAutophosphorylationReceptor activation
The present invention relates to nucleotide sequences, including expressed sequence tags (ESTs), oligonucleotide probes, polypeptides, vectors and host cells expressing, and immunoadhesions and antibodies to mammalian GFRα3, a novel α-subunit receptor of the GDNF (i.e. GFR) receptor family. It further relates to an assay for measuring activation of an α-subunit receptor by detecting tyrosine kinase receptor activation (i.e., autophosphorylation) or other activities related to ligand-induced α-subunit receptor homo-dimerization or homo-oligomerization.
Owner:DE SAUVAGE FREDERIC +3
Nucleic acid molecules and other molecules associated with plants
InactiveUS20080263730A1Reduces and depresses expression of proteinReduce protein expressionImmunoglobulinsFermentationNovel genePlant genomics
Expressed Sequence Tags (ESTs) isolated from maize are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:MONSANTO TECH LLC
Methods and Compositions for Homozygous Gene Inactivation Using Collections of Pre-Defined Nucleotide Sequences Complementary Chromosomal Transcripts
Methods and compositions for performing homozygous gene inactivation assays are provided. A feature of the subject methods is the use of a library of constructs that synthesize predefined nucleic acids, where each constituent predefined nucleic acid of the library is of known sequence that corresponds to a sequence of a chromosomal transcript, e.g., where a representative embodiment of a predefined nucleic acid is an expressed sequence tag (i.e., EST). In certain embodiments, the subject libraries are produced using an amplification protocol that preserves the sequence representation profile of the template nucleic acids. The subject methods and compositions find use in a variety of different applications, including the identification of novel diagnostic and therapeutic genetic targets.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
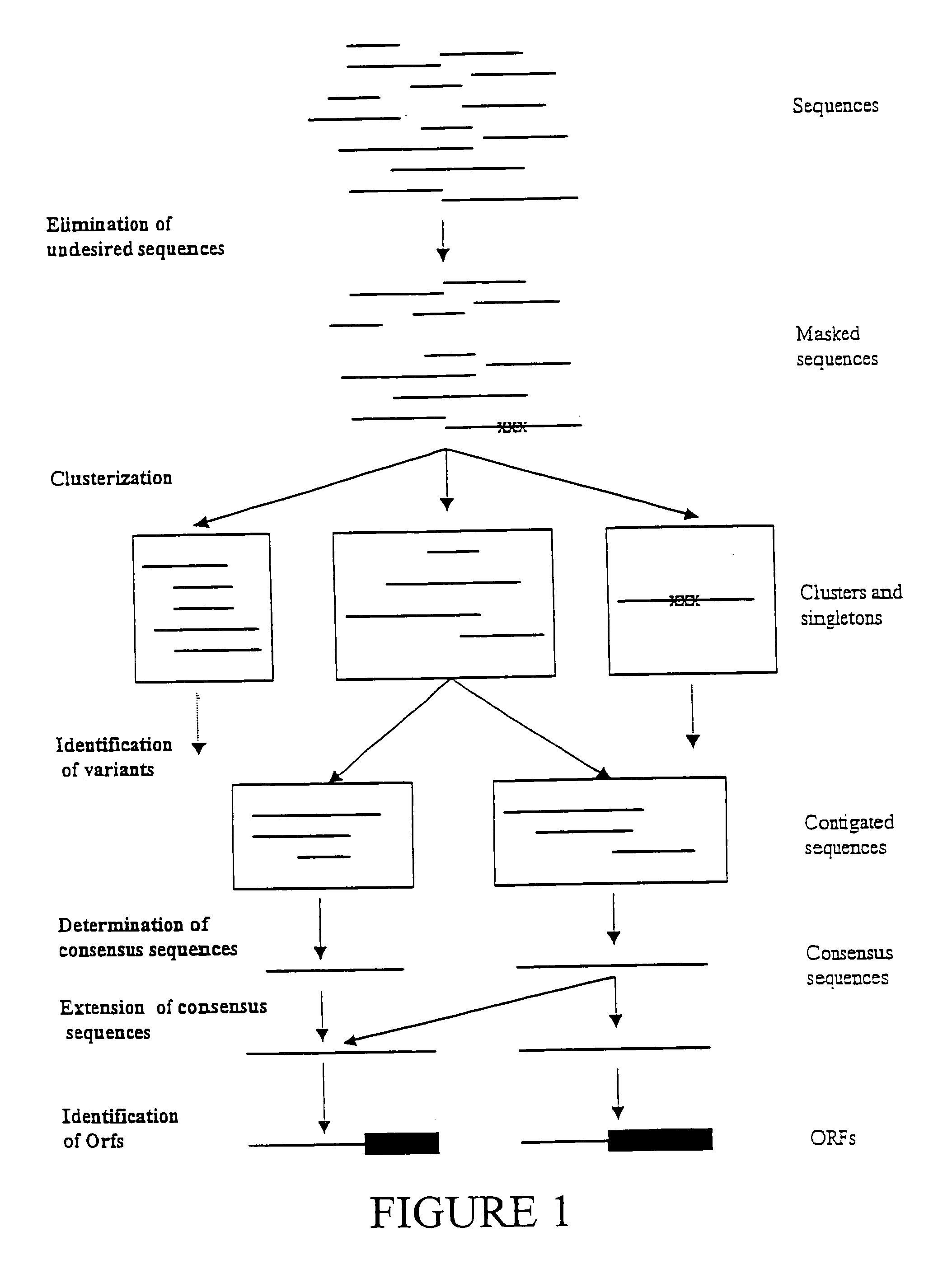
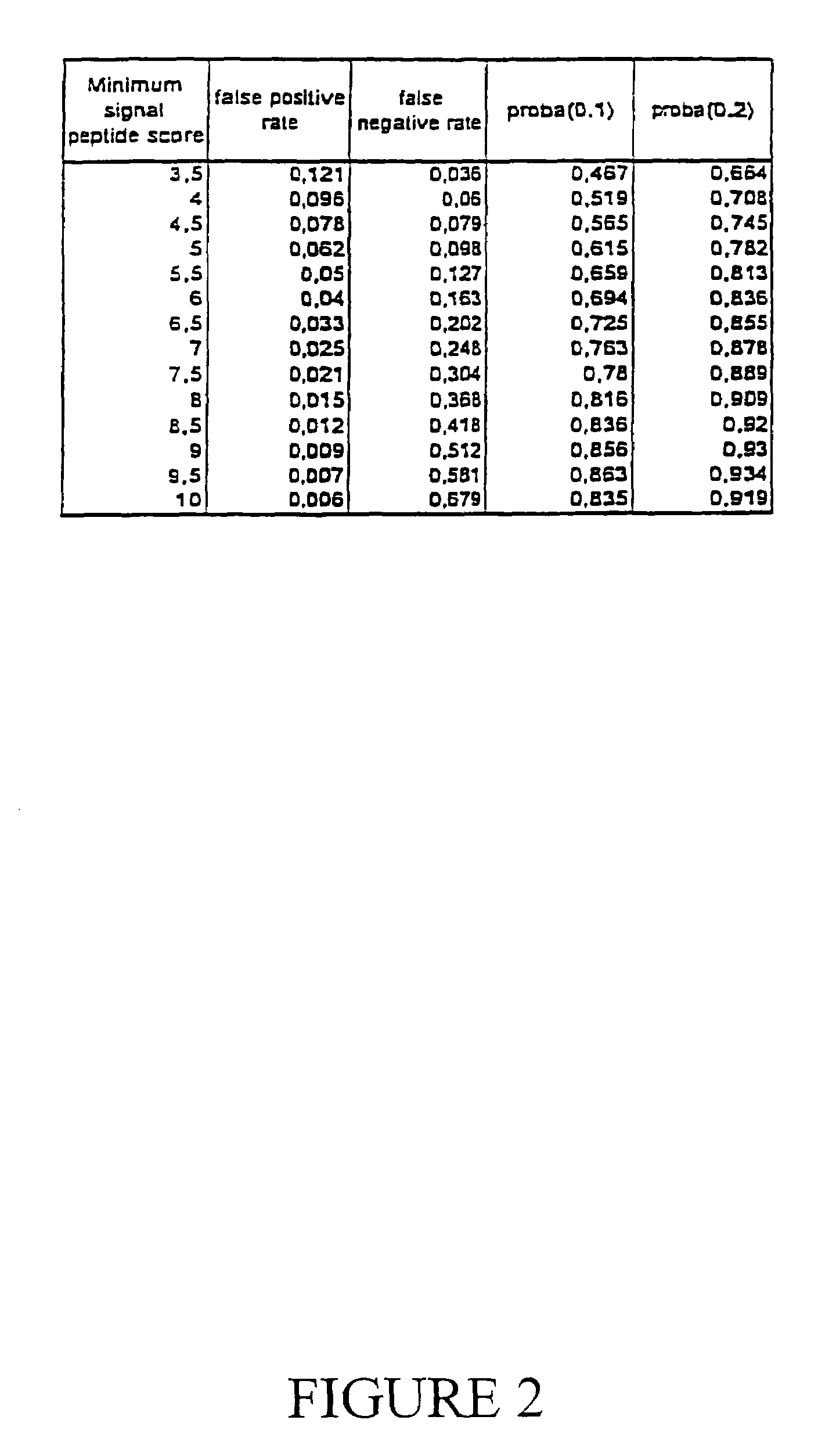
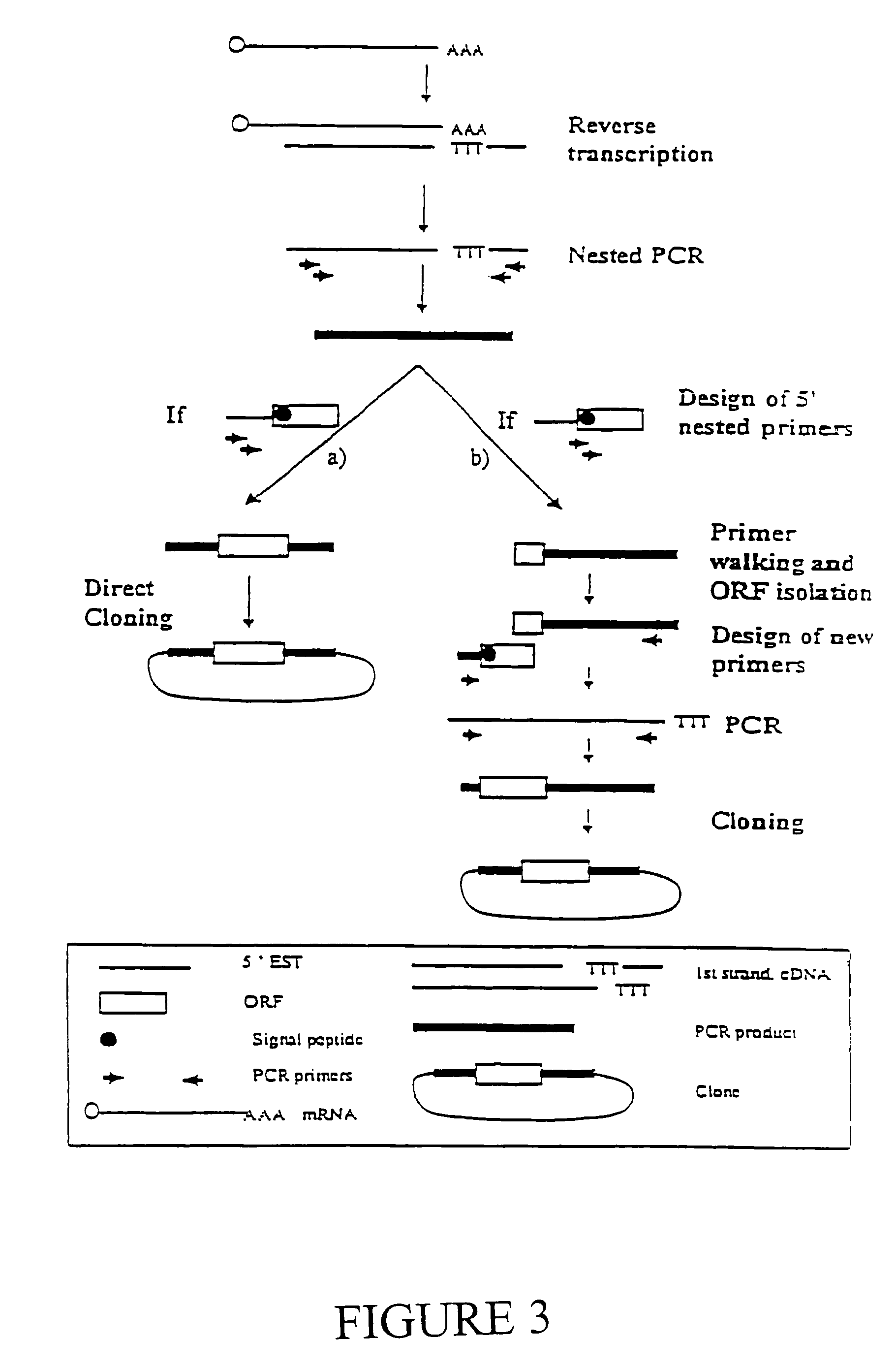
Expressed sequence tags and encoded human proteins
The sequences of 5′ ESTs derived from mRNAs encoding secreted proteins are disclosed. The 5′ ESTs may be to obtain cDNAs and genomic DNAs corresponding to the 5′ ESTs. The 5′ ESTs may also be used in diagnostic, forensic, gene therapy, and chromosome mapping procedures. Upstream regulatory sequences may also be obtained using the 5′ ESTs. The 5′ ESTs may also be used to design expression vectors and secretion vectors.
Owner:SERONO GENETICS INST SA
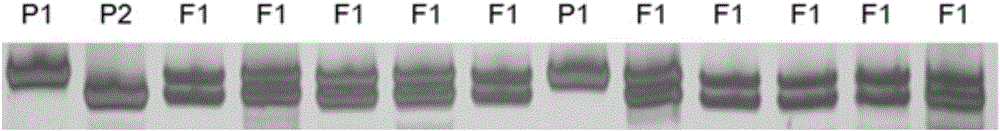
Method for identifying seed purity of muskmelon hybrid variety based on EST-SSR (expressed sequence tag-simple sequence repeat) marker
InactiveCN104059992AImprove stabilityLow costMicrobiological testing/measurementDNA/RNA fragmentationField testsGenetic stock
The invention discloses a method for identifying seed purity of a muskmelon variety namely green angel hybrid variety based on an EST-SSR (expressed sequence tag-simple sequence repeat) marker. The method comprises the following steps: using the muskmelon variety namely green angel hybrid variety and parental genome DNA (deoxyribonucleic acid) of the muskmelon variety namely green angel hybrid variety as a template; designing 57 pairs of EST-SSR primers by using Primer 3.0 online primer design through 214 muskmelon EST sequences published by a GeneBank database; obtaining a pair of complementary band type primers of which the hybrid variety band type is parental by screening. By virtue of a field verification test, the primer is good in stability, is relatively matched with results of the field test, and can be used for performing purity identification on the muskmelon hybrid variety. A detection method disclosed by the invention can be used for completing seed purity identification within 3 hours, and has the advantages of high speed, low cost, convenience in operation and the like.
Owner:TIANJIN RES INST OF VEGETABLE +1
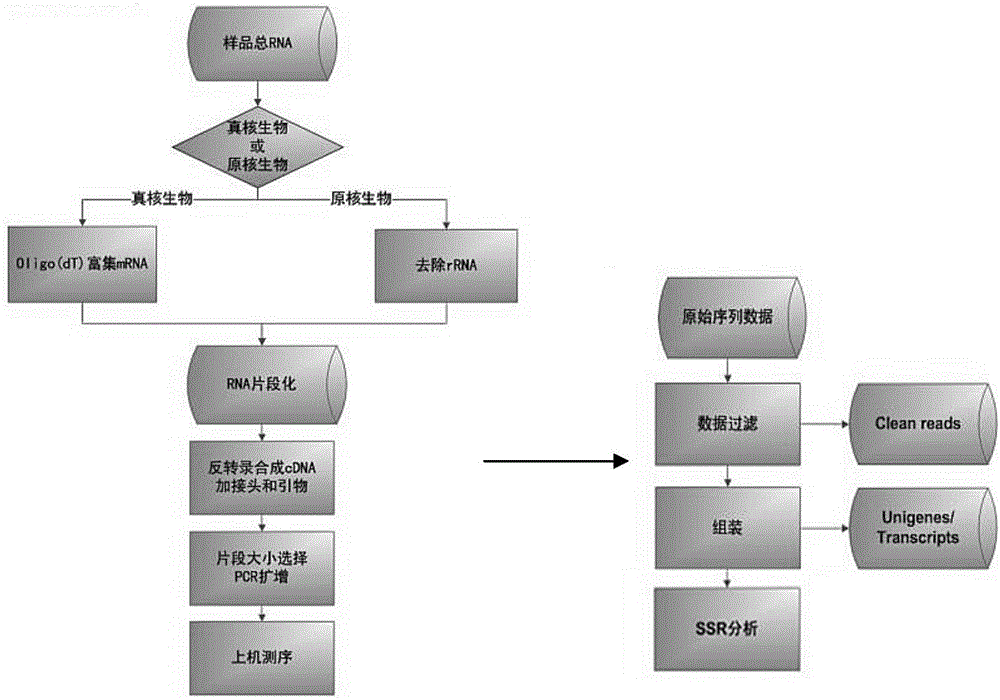
Method for developing dendranthema SSR (Simple Sequence Repeat) primer based on transcriptome sequencing
ActiveCN103233075AAdd raw dataOvercoming access difficultiesMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceLymphatic Spread
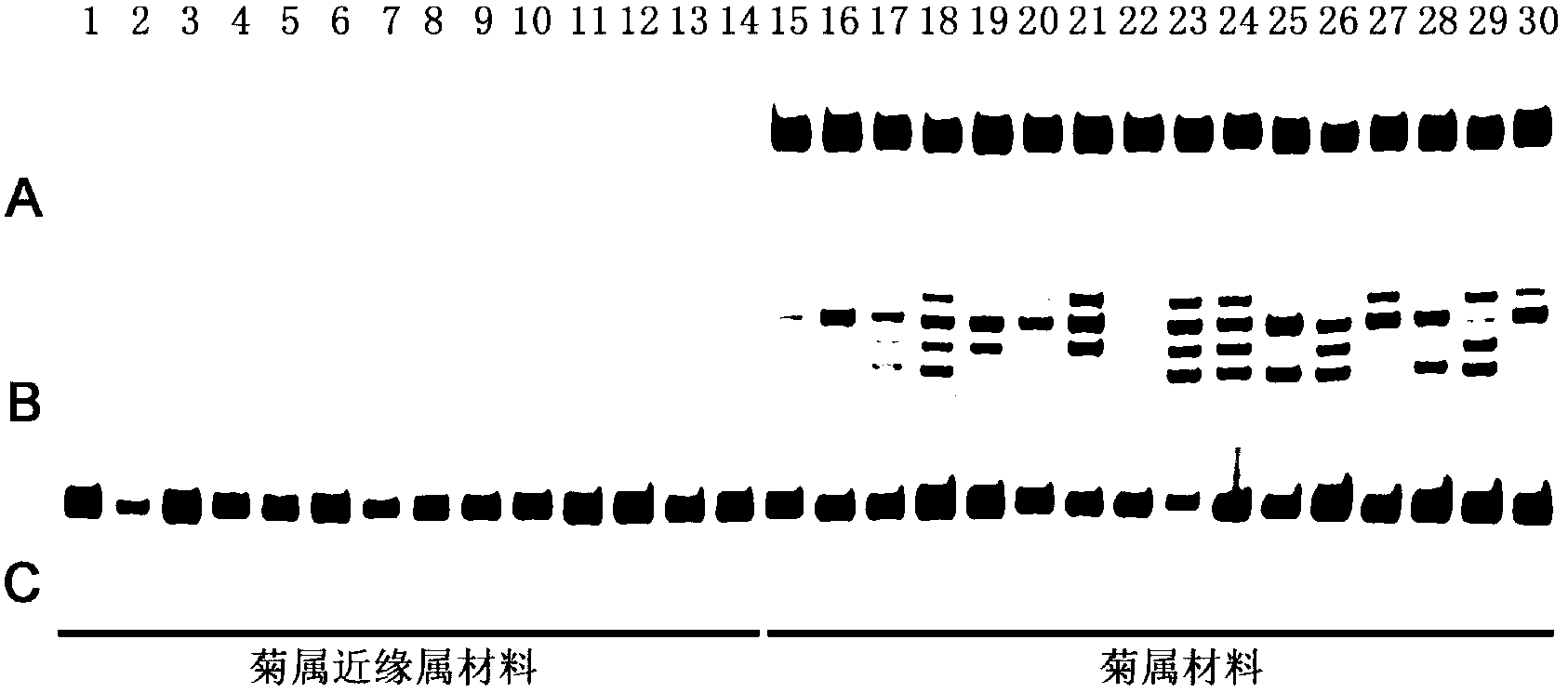
The invention belongs to biotechnology field, and relates to a method for developing a dendranthema SSR (Simple Sequence Repeat) primer based on transcriptome sequencing. A lot of sequence information is processed in batch for searching an SSR sequence and designing an SSR labeled primer based on the transcriptome sequencing by utilizing EST-SSR (Expressed Sequence Tags-Simple Sequence Repeat) interspecific metastasis and using a Perl (Practical Extraction and Reporting Language) programming language method in combination, so that the defects of low efficiency, long time consumption, high cost and the like of the SSR development are conquered. Different dendranthema materials are selected for verifying the designed SSR primer, and the primer is a successful SSR primer if detected out by any strip. By adopting the method, 1788 pairs of SSR primers are successively designed, so that a novel method and thinking are provided for the development of the dendranthema SSR primer to further achieve molecular marker assistant selection breeding and comparative genomics research.
Owner:NANJING AGRICULTURAL UNIVERSITY
Nucleic acid molecules and other molecules associated with plants
InactiveUS20070016974A1Desirable effectIncrease profitSugar derivativesOther foreign material introduction processesNovel genePlant genomics
Expressed Sequence Tags (ESTs) isolated from rice are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:BYRUM JOSEPH +2
Method for generating pterostilbene by utilizing grape resveratrol-oxygen-methyl transferase to catalyze resveratrol
InactiveCN102120996AMicrobiological testing/measurementFermentationRapid amplification of cDNA endsBiotechnology
The invention discloses a method for generating pterostilbene by utilizing grape resveratrol-oxygen-methyltransferase to catalyze resveratrol, grape ROMT (resveratrol-oxygen-methyl transferase) genes are cloned by utilizing 5' and 3' RACE (rapid amplification of cDNA ends) full-length gene cloning technology according to EST (expressed sequence tag) sequence of the ROMT genes of Chinese wild grapes in East China, and the full length of an open reading frame of the genes is 1074bp; and the cloned grape STS (steroid sulfatase) and ROMT genes can be simultaneously transferred into model plant tobacco, tobacco plant analysis is utilized for analyzing the biological synthesis path of generating the pterostilbene by utilizing the grape ROMT to catalyze the resveratrol, a grape ROMT gene sequence is obtained for orientation, and the ROMT genes can catalyze the resveratrol to generate the pterostilbene in the transgenic tobacco, thereby providing the sequence of the ROMT genes and the method for synthesizing the pterostilbene by utilizing plants and providing a basis for getting high-quality, disease-resistant and health care crops.
Owner:NORTHWEST A & F UNIV
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from soybean are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:BUEHLER ROBERT E +3
Methods for identification of novel protein drug targets and biomarkers utilizing functional networks
ActiveUS8000949B2Analogue computers for chemical processesSequence analysisDrug targetBiomarker (petroleum)
The process of System Reconstruction is used to integrate sequence data, clinical data, experimental data, and literature into functional models of disease pathways. System Reconstruction models serve as informational skeletons for integrating various types of high-throughput data. The present invention provides the first metabolic reconstruction study of a eukaryotic organism based solely on expressed sequence tag (EST) data. System Reconstruction also provides a method for the identification of novel therapeutic targets and biomarkers using network analysis. The initial seed networks are built from the lists of novel targets for diseases with the high-throughput experimental data being superimposed on the seed networks to identify specific targets.
Owner:CAMELOT UK BIDCO LTD
Task scheduling method and device
ActiveCN104915251ATake advantage ofImprove task execution efficiencyMultiprogramming arrangementsResource utilizationExpressed sequence tag
The invention discloses a task scheduling method and a task scheduling device. The method comprises the following steps of creating a task priority queue for a task cluster formed by networked task nodes; selecting a task from the task priority queue as a current task according to a priority sequence, and calculating a resource utilization rate of the current task; judging whether certain resources remain or not according to the resource utilization rate of the current task, and if certain resources remain, selecting at least one task which requires resources fewer than the remaining resources and of which an EST (Expressed Sequence Tag) is 0 from the task priority queue as the current task; simultaneously executing each current task; subtracting 1 from the EST of a parent task of each level of each current task, and returning to execute the operation of selecting the current task from the task priority queue. Compared with the prior art, the task scheduling method and the task scheduling device have the advantage that the remaining resources of the cluster are fully utilized, so that task execution efficiency is improved.
Owner:厦门见福连锁管理有限公司
Nucleic acid molecule SEQ ID NO. 68811 and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from maize are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:MONSANTO TECH LLC
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from cotton are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:MONSANTO TECH LLC
Nucleic acid molecules and other molecules associated with plants
InactiveUS20080168583A1Reduces and depresses expression of proteinReduce protein expressionPeptide/protein ingredientsImmunoglobulinsNovel genePlant genomics
Expressed Sequence Tags (ESTs) isolated from cotton are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:FINCHER KAREN L +3
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from cotton are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:FINCHER KAREN L +3
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from soybean are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:BUEHLER ROBERT E +3
Method for quickly identifying purity of watermelon variety Zhentian 1217 by using EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker
The invention relates to the field of molecular markers, and provides a method for quickly identifying purity of a watermelon variety Zhentian 1217 by using an EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker. According to the method, a specially designed EST-SSR primer is adopted, a genome DNA of the Zhentian 1217 is used as a template to perform amplification, and a specific band of an amplification result is used for detecting the variety purity of the Zhentian 1217 to be detected. By adopting the method, the identification can be performed at any period of the growth of a Zhentian 1217 plant, and hybrid seeds can be differentiated from female parent selfed seeds and male parent selfed seeds; and the method is high in accuracy and broad in application popularization prospect, and provides a scientific basis for carrying out watermelon seed preparation and selling high-quality seeds timely.
Owner:华盛农业集团股份有限公司
Method for obtaining SSR (Simple Sequence Repeat) primer of lycium ruthenicum based on transcription sequencing
ActiveCN106754886AMicrobiological testing/measurementDNA preparationGenetic diversityMolecular breeding
The invention relates to a method for obtaining an SSR (Simple Sequence Repeat) primer of lycium ruthenicum based on transcription sequencing, and belongs to the technical field of molecular marking techniques of molecular genetics of biology. The method comprises the following steps of (1), carrying out transcriptome sequencing; (2), screening an EST-SSR (Expressed Sequence Tag-Simple Sequence Repeat) site; (3), carrying out primer design; (4), extracting DNA (Deoxyribonucleic Acid); (5), carrying out primer screening; (6), carrying out polymorphic primer screening by using 8-percent native polyacrylamide gel electrophoresis. The method for obtaining the SSR primer of the lycium ruthenicum based on the transcription sequencing, which is provided by the invention, can be used as a method for the correction of a linkage map of a genome between remote species and comparative mapping, and has higher utilization value in two aspects. The greatest advantages of the method are that the development is simple and quick and the expense is low. The general characteristic of the SSR of the lycium ruthenicum is specified by utilizing an SSR molecular marking technique; the SSR primer of the lycium ruthenicum is developed; the foundation is laid for carrying out the research on the genetic diversity, the linkage map construction and the affinity relationship of species resources of the lycium ruthenicum by utilizing an SSR molecular marker, so that the method is better applied to molecular breeding.
Owner:BEIJING FORESTRY UNIVERSITY
Nucleic acid molecules and other molecules associated with plants
InactiveUS20070283459A1Desirable effectIncrease profitImmunoglobulinsFermentationNovel genePlant genomics
Expressed Sequence Tags (ESTs) isolated from soybean are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:BYRUM JOSEPH +2
Stichopus japonicas BPI gene, encoded protein, cloning method of stichopus japonicas BPI gene, and method for constructing recombinant stichopus japonicas BPI genetically engineered bacterium
ActiveCN104745595AEfficient killingAntibacterial agentsPeptide/protein ingredientsBPI proteinVibrio parahaemolyticus
The invention discloses a stichopus japonicas BPI gene, an encoded protein, a cloning method of the stichopus japonicas BPI gene, and a method for constructing a recombinant stichopus japonicas BPI genetically engineered bacterium. The stichopus japonicas BPI gene is characterized in that a stichopus japonicas BPI gene sequence is as shown in SEQ ID NO.1; the cloning method comprises the step of designing a nested primer of RACE according to an expressed sequence tag EST sequence which is homologous to the BPI gene; a full-length gene is expanded by employing an RACE technique; a stichopus japonicas BPI protein sequence is as shown in SEQ ID NO.2; an N-terminal protein structure domain sequence is as shown in SEQ ID NO.3; an N-terminal structure domain of the stichopus japonicas BPI protein is amplified by employing primers which respectively comprise BamH I sites and Not I sites; the cloned target gene is inserted into a vector to obtain recombinant plasmids; and the recombinant plasmids are subjected to induced expression, and purification and renaturation, so as to obtain the genetically engineered bacterium. The stichopus japonicas BPI gene has the advantage of having obvious sterilization effect on vibrio parahaemolyticus, vibrio harveyi and micrococcus luteus.
Owner:NINGBO UNIV
Brassica juncea EST-SSR (expressed sequence tag-simple sequence repeat) marker primer group based on development of transcriptome sequence
ActiveCN104846093AOvercoming the Rare ProblemAdd raw dataMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMarker-assisted selection
The invention discloses a brassica juncea EST-SSR (expressed sequence tag-simple sequence repeat) marker primer group based on development of a transcriptome sequence, and belongs to the technical field of biologics. The brassica juncea EST-SSR marker primer group has the advantages that the primer group is obtained on the basis of development of the transcriptome sequence; on the basis of transcriptome sequencing, a large amount of sequence information is processed by batches, and the finding of an SSR sequence and the primer design of an SSR marker are performed, so as to overcome the defects of fewer SSR markers, low development efficiency and the like in the present brassica juncea; as the brassica juncea and brassica campestris are different materials, the effectiveness and universality of SSR primer are verified, and a foundation is laid for the development of the brassica juncea SSR primer, the auxiliary selecting of the molecule marker, the building of the genetic linkage map, the evaluation of genetic diversity, and the like.
Owner:ZHEJIANG UNIV
Clam ferroprotein gene as well as coding protein and application of in vitro recombinant expression product of same
InactiveCN102051363APeptide/protein ingredientsImmunological disordersRapid amplification of cDNA endsBiology
The invention relates to the technologies of clam ferroprotein gene cloning and in vitro recombinant expression in molecular biology. In the invention, the expression sequence tag (EST) technology and rapid amplification of cDNA ends (RACE) 3' and 5' are utilized to clone ferroprotein gene cDNA with the total length of 798bp from a clam larva, the cloned ferroprotein gene contains an open reading frame with the length of 513bp, 171 amino acids are coded, the length of a non-coding region 5' is 119bp, the length of a non-coding region 3' is 154bp, a tailing signal is contained, and the gene plays an important role in formation of a shell during development of the clam larva. In the invention, the in vitro prokaryotic recombinant expression technology is utilized to obtain recombinant clam ferroprotein; and the ferroprotein has antioxidant activity, can be used for regulating and controlling formation and growth of shells of shellfish larvae such as clams and the like, and can be applicable to development regulation and immunological enhancement in the shellfish larva production process.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Detection method for Apostichopus japonicas AjE101 micro-satellite DNA label
InactiveCN102140522AGood polymorphismImprove stabilityMicrobiological testing/measurementGerm plasmGenetic diversity
The invention belongs to the field of molecular biology DNA labeling technology and application, and in particular relates to Apostichopus japonicas express sequence tag EST micro-satellite label screening development and application. The invention provides Apostichopus japonicas AjE101 micro-satellite specific DNA primers, a polymerase chain reaction (PCR) reaction system by utilizing the primers, and a detection method for an Apostichopus japonicas AjE101 micro-satellite DNA label. The detection method comprises the following steps of: extracting genome of Litopenaeus vannamei Boone; designing the specific primers at two ends of an Apostichopus japonicas AjE101 micro-satellite DNA core sequence; and performing PCR amplification on genome DNA of different groups of the Litopenaeus vannamei Boone or individuals in the group by using the primers, analyzing products, and determining the genome of each individual so as to obtain an Apostichopus japonicas polymorphism genetic variation map. The invention is mainly applied to Apostichopus japonicas germ plasm resource and genetic diversity analysis, cular population genetics, construction of genetic maps and research of functional genes.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
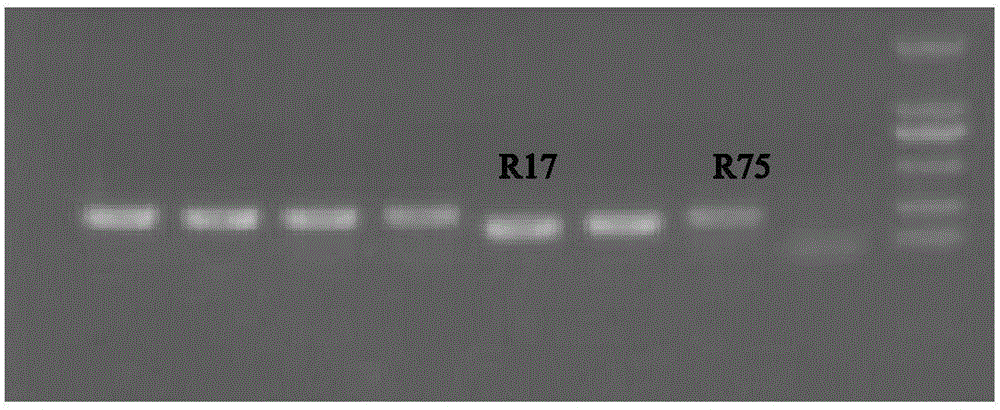
EST-SSR (expressed sequence tag-simple sequence repeats) molecular markers for heat-resistant radish, primers of molecular markers and application thereof
InactiveCN106636403AHigh purityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceNucleotide sequencing
The invention provides EST-SSR (expressed sequence tag-simple sequence repeats) molecular markers for heat-resistant radish. The EST-SSR molecular markers comprise a type of or two types of SSR 17 and SSR 75. Nucleotide sequences of the SSR 17 are shown as SEQ ID No.1 in sequence tables; nucleotide sequences of the SSR 75 are shown as SEQ ID No.2 in the sequence tables. The EST-SSR molecular markers have the advantages that heat-resistant radish varieties, namely Summer Resistance 40-day, are screened to obtain the two EST-SSR molecular markers, the SSR 17 and the SSR 75 have codominant expression characteristics, the population purity of the commodity heat-resistant radish varieties, namely the Summer Resistance 40-day, is detected by the aid of the two EST-SSR molecular markers in codominant relationships, the purity can reach 92% as shown by results and is consistent with the purity of field phenotype, and accordingly the two EST-SSR molecular markers can be used for detecting the purity of the heat-resistant radish varieties.
Owner:HUANGGANG NORMAL UNIV
CRIg polypeptide for prevention and treatment of complement-associated disorders
The present invention relates to compositions and methods of treating and diagnosing disorders characterized by the presence of antigens associated with inflammatory diseases and / or cancer, and nucleotide sequences, including expressed sequence tags (ESTs), oligonucleotide probes, polypeptides, vectors and host cells expressing such antigens PRO301, PRO362 or PRO245.
Owner:GENENTECH INC
Specific primer system of EST (expressed sequence tag)-SSR (simple sequence repeat) molecular markers for Pleurotus ostreatus and application of specific primer system
InactiveCN102181559AEasy to manageImprove the level of comprehensive utilizationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceResearch efficiency
The invention relates to a specific primer system of EST (expressed sequence tag)-SSR (simple sequence repeat) molecular markers for Pleurotus ostreatus and application of the specific primer system, belonging to the technical field of molecular biology. The specific primer system of EST-SSR molecular markers for Pleurotus ostreatus comprises 4 pairs of specific primers for EST-SSR molecular markers. The invention also provides the application of the specific primer system of EST-SSR molecular markers in the genetic diversity analysis and the germplasm resource identification of the Pleurotusostreatus. The specific primer system disclosed by the invention is used for establishing a core germplasm bank of the Pleurotus ostreatus, has convenience for use, can simply and rapidly carry out the genetic diversity analysis of germplasm resources, germplasm resource identification, genetic map construction and research on functional genes; and compared with a traditional method, the invention can greatly shorten an analytical cycle and increase research efficiency.
Owner:INST OF AGRI RESOURCES & ENVIRONMENT SHANDONG ACADEMY OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com