Patents
Literature
209 results about "Sequence homology" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
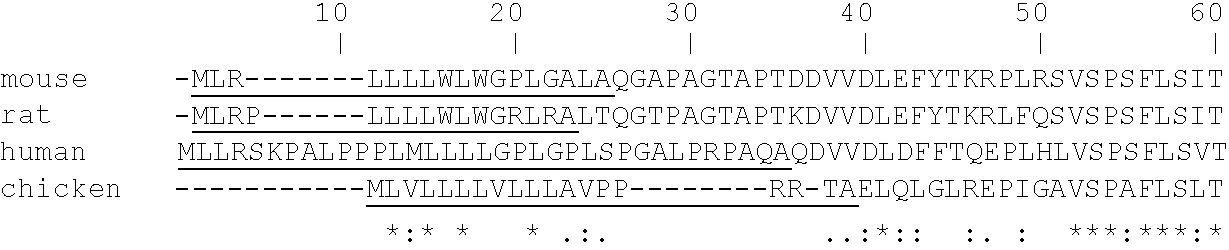
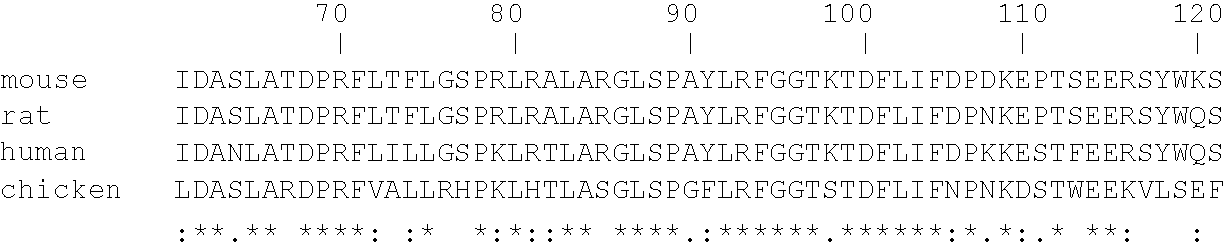
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs).
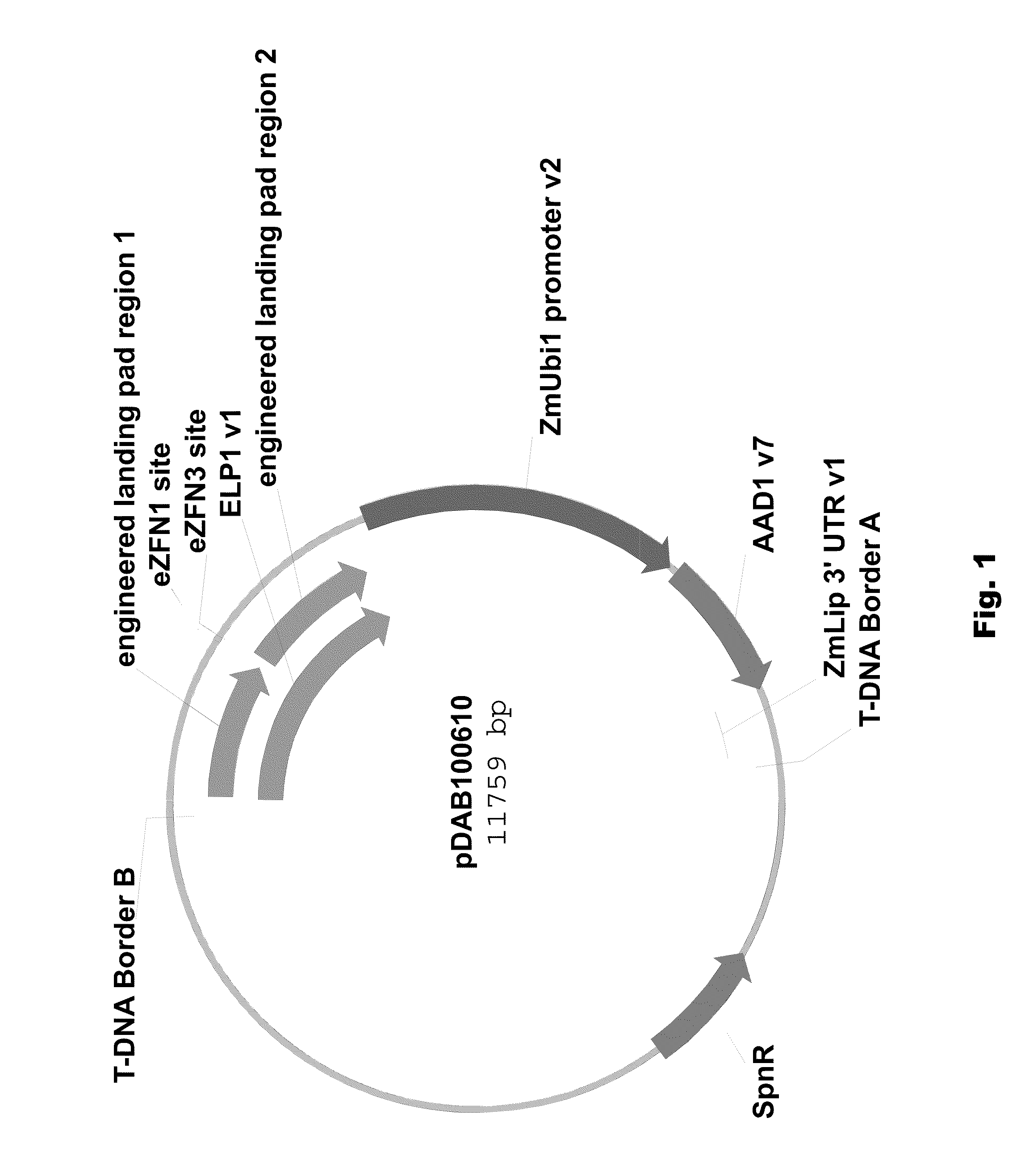
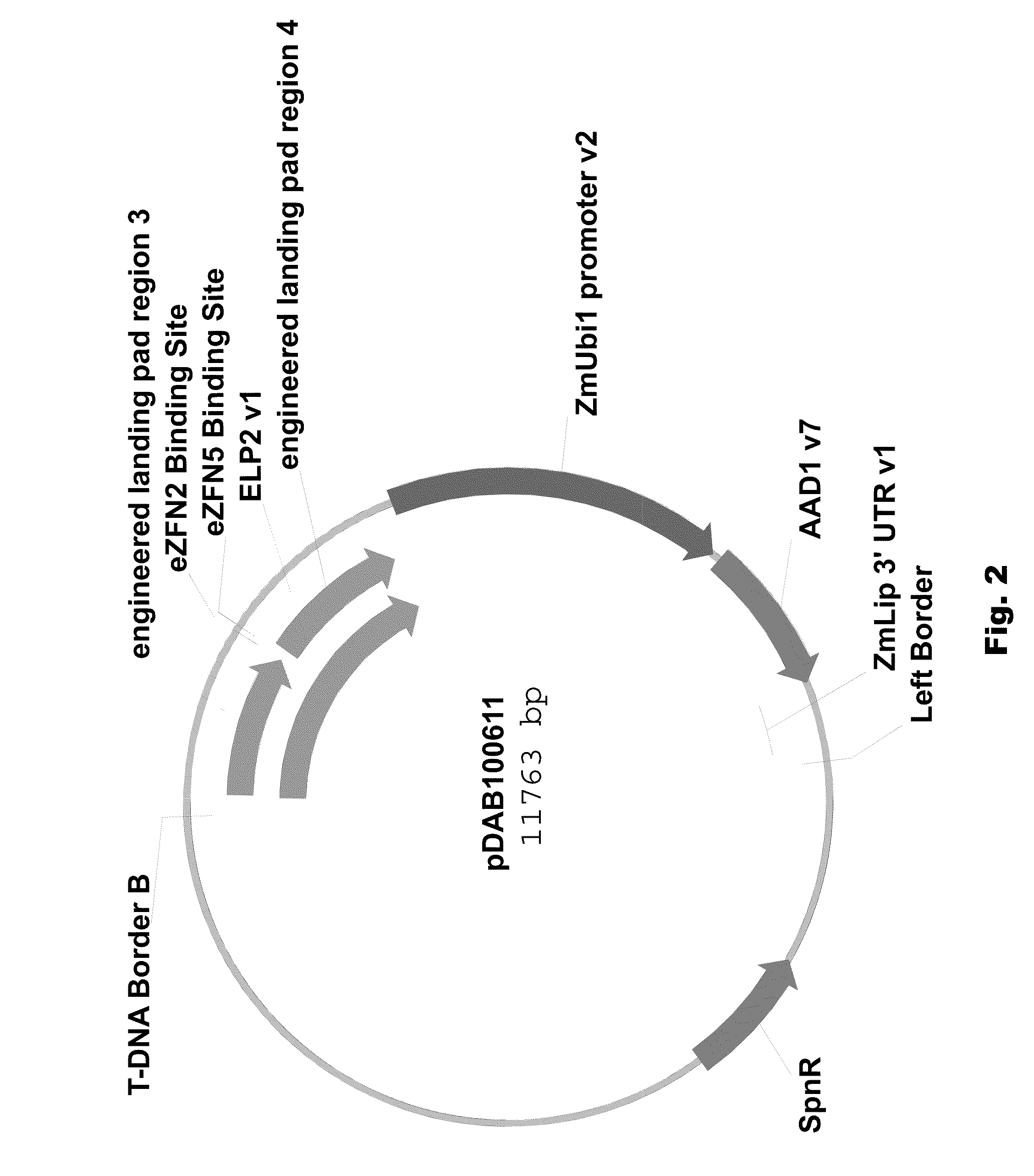
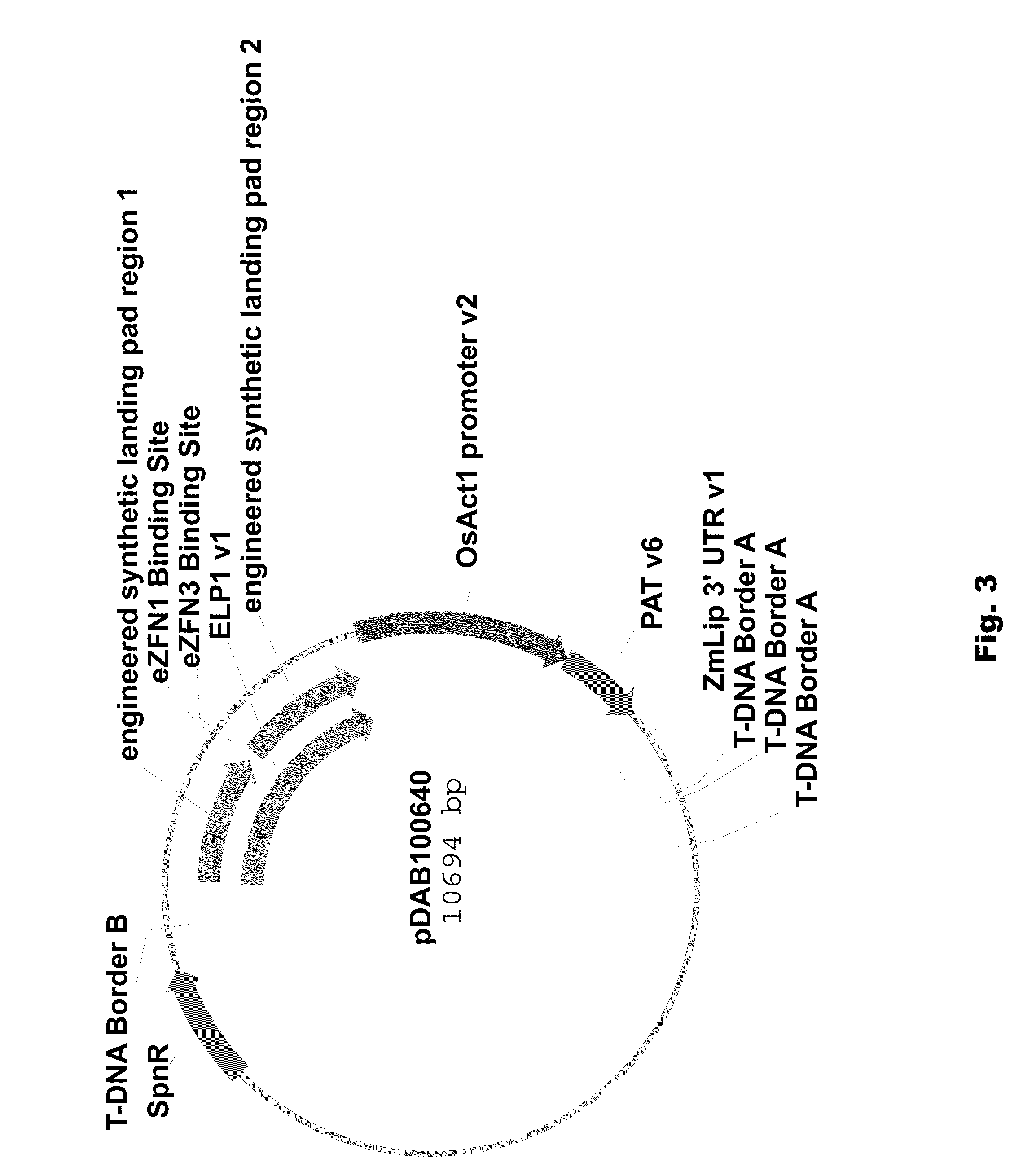
Engineered landing pads for gene targeting in plants
InactiveUS20110191899A1Reduce and eliminate needEasy to reorganizeOther foreign material introduction processesFermentationZinc finger nucleaseGene targets
A method for producing a transgenic plant includes providing a nucleic acid molecule comprising at least two regions of nucleic acid sequence that lack sequence homology with genomic DNA of the plant cell, and at least two zinc finger nuclease recognition sites, wherein the at least two regions of nucleic acid sequence that lack sequence homology with genomic DNA of the plant cell flank the at least two zinc finger nuclease recognition sites. A plant cell or tissue having the nucleic acid molecule stably integrated into the genome of the plant cell is transformed. A plant is regenerated from the plant cell. Transgenic plants are produced by the method. Seeds are produced by the transgenic plants.
Owner:CORTEVA AGRISCIENCE LLC
Dideoxynucleotide-triphosphate utilization by the hyper-thermophilic DNA polymerase from the archaeon Pyrococcus furiosus
InactiveUS6333183B1High sensitivityImprove thermal stabilityBacteriaSugar derivativesDideoxynucleotide TriphosphatesBinding site
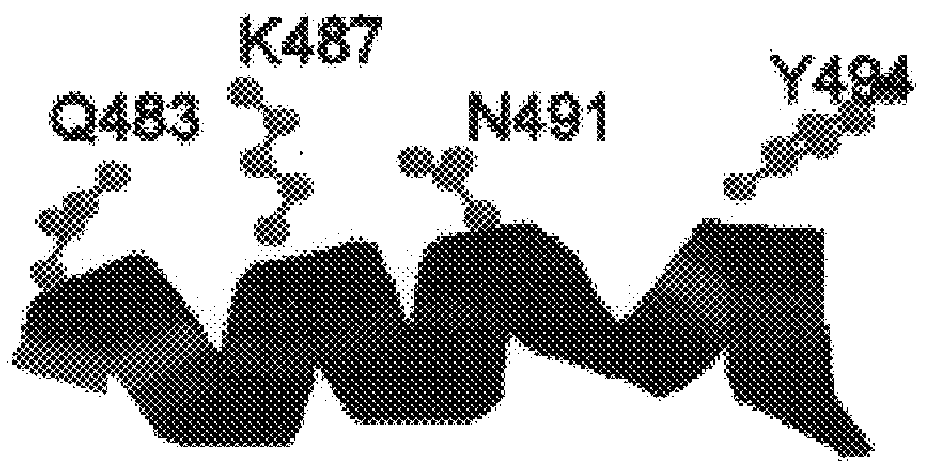
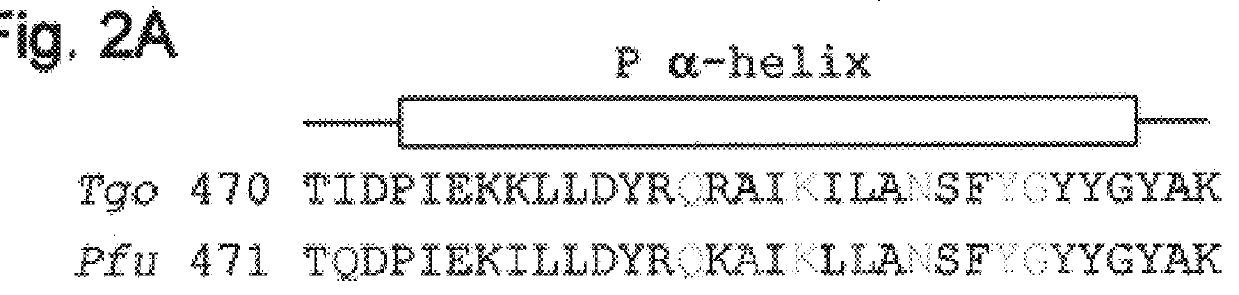
Polymerases from the Pol I family which are able to efficiently use ddNTPs have demonstrated a much improved performance when used to sequence DNA. A number of mutations have been made to the gene coding for the Pol II family DNA polymerase from the archaeon Pyrococcus furiosus with the aim of improving ddNTP utilisation. "Rational" alterations to amino acids likely to be near the dNTP binding site (based on sequence homologies and structural information) did not yield the desired level of selectivity for ddNTPs. However, alteration at four positions (Q472, A486, L490 and Y497) gave rise to variants which incorporated ddNTPs better than the wild type, allowing sequencing reactions to be carried out at lowered ddNTP:dNTP ratios. Wild type Pfu-Pol required a ddNTP:dNTP ratio of 30:1; values of 5:1 (Q472H), 1:3 (L490Y), 1:5 (A486Y) and 5:1 (Y497A) were found with the four mutants; A486Y representing a 150-fold improvement over the wild type. A486, L490 and Y407 are on an alpha-helix that lines the dNTP binding groove, but the side chains of the three amino acids point away from this groove; Q472 is in a loop that connects this alpha-helix to a second long helix. None of the four amino acids can contact the dNTP directly. Therefore, the increased selectivity for ddNTPs is likely to arise from two factors: 1) Small overall changes in conformation that subtly alter the nucleotide triphosphate binding site such that ddNTPs become favoured; 2) interference with a conformational change that may be critical both for the polymerisation step and discrimination between different nucleotide triphosphates.
Owner:GE HEALTHCARE BIO SCI CORP
Novel peptide and use thereof
InactiveUS20090305973A1Promoting fibroblast proliferationPromoting wound healingPeptide/protein ingredientsMuscular disorderWound healingAmino acid
The present invention relates to a novel peptide and use thereof, more particularly to an isolated peptide comprising 21-41 contiguous amino acids selected from the amino acid sequence of SEQ ID NO: 1 or the amino acid sequence having at least 90% sequence homology to the amino acid sequence of SEQ ID NO: 1 and methods for promoting fibroblast proliferation and wound healing, which comprise administering to a subject in need thereof an effective amount of the peptide.
Owner:ATYR PHARM INC
TGF-Beta Modulators and Use Thereof
InactiveUS20090036382A1Cell receptors/surface-antigens/surface-determinantsPeptide/protein ingredientsHypertension medicationsMRNA synthesis
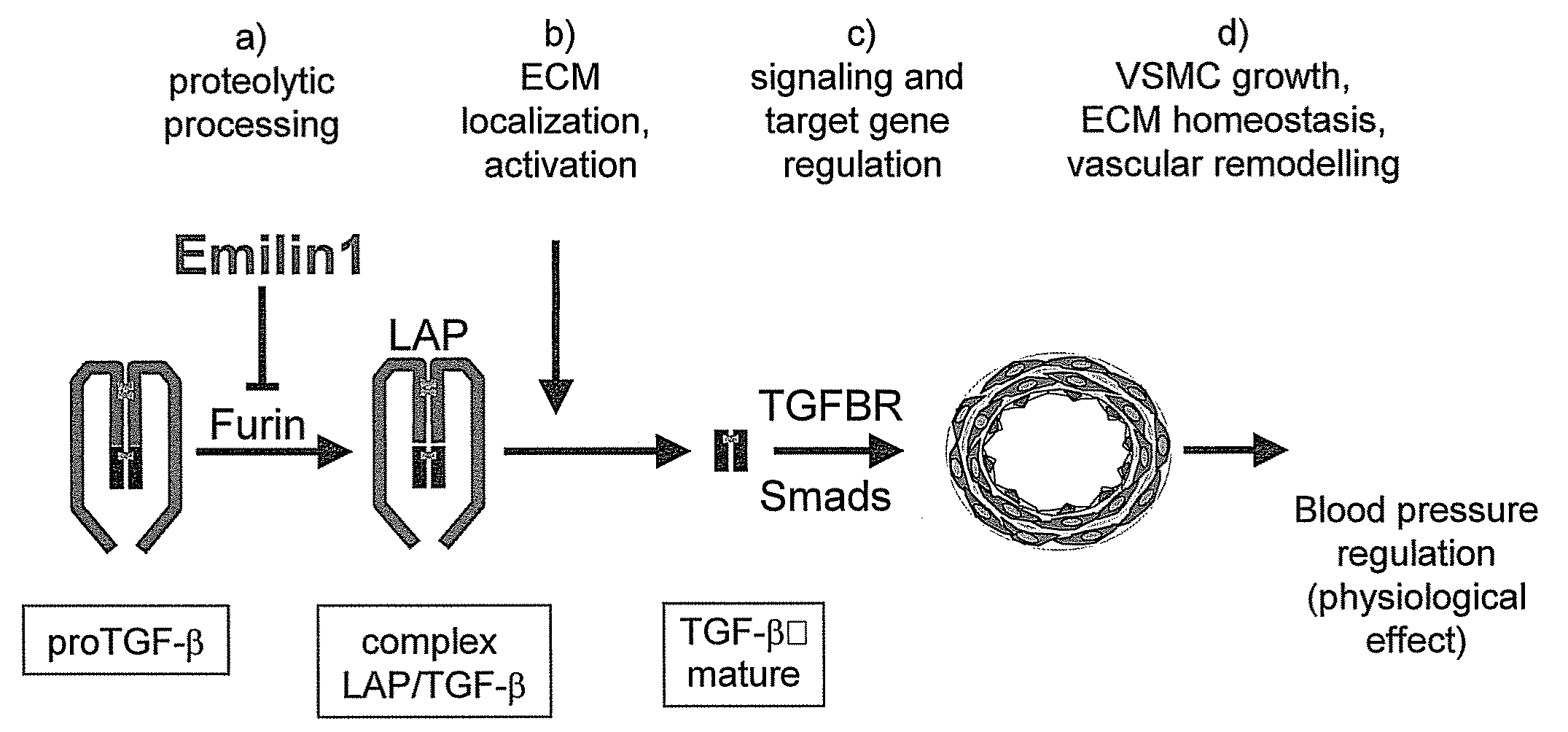
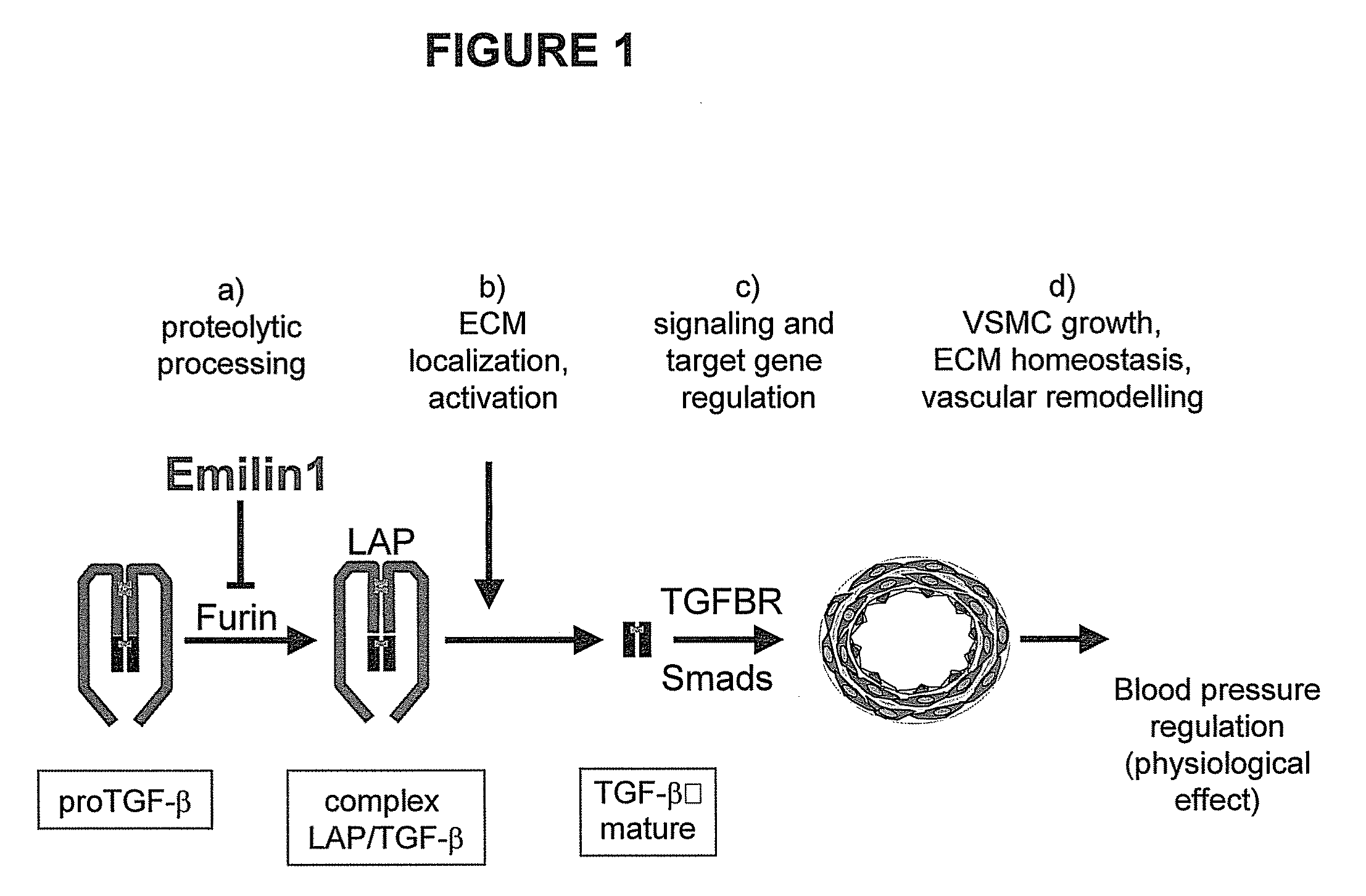
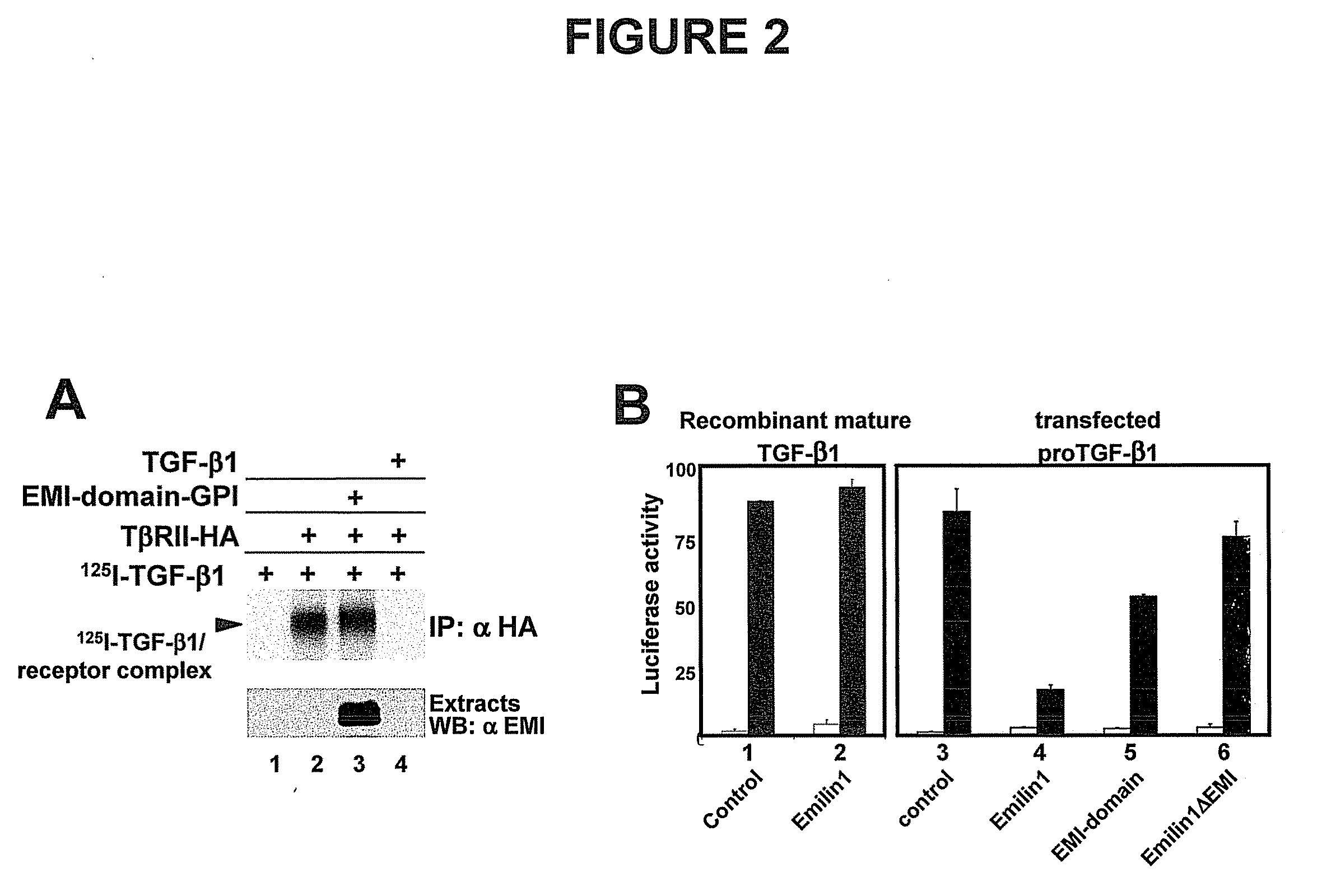
The present invention relates to molecules preferably of polypeptide nature with negative regulatory activity on the amount or activity of TGF-β through direct interaction with pro-TGF-β, and containing as active region a cysteine-rich polypeptide sequence defined as “EMI domain”, or its subfragments, wherein said “EMI domain” has at least 25% sequence homology to the ID NO2 sequence for pharmaceutical use. Even more preferably said polypeptide sequence consists of the EMI domain of the following proteins: emilin-1, emilin-2 and / or multimerin-2 or their subfragments having a length of at least 6 amino acids, capable of inhibiting the conversion of pro-TGFβ to mature TGFβ as anti-hypertensive drugs and polypeptides active on the cardiovascular system. The invention extends to the use of molecules which are known to negatively regulate TGF-β and to molecules which interfere with TGF-β binding to its receptors, or to inhibitors of TGF-β mRNA synthesis or TGF-β expression for the same therapeutic uses as anti-hypertensive drugs and polypeptides active on the cardiovascular system.
Owner:UNIV DEGLI STUDI DI PADOVA +1
Novel polypeptide having Anti-tumor activity
InactiveUS20110124582A1Prevent proliferationEasy to useBacteriaPeptide/protein ingredientsApoptosisPolynucleotide
The present invention relates to a novel polypeptide having anti-tumor activity through inducing apoptosis of endothelial cell and use thereof. More particularly, the present invention relates to a method for inducing apoptosis of endothelial cell, and for preventing or treating cancer, comprising administering to a subject in need thereof an effective amount of (a) an isolated polypeptide having the amino acid sequence of SEQ ID NO: 9 or the amino acid sequence having at least 90% sequence homology to the amino acid sequence of SEQ ID NO: 9; or (b) an isolated polynucleotide encoding the polypeptide of (a).
Owner:ATYR PHARM INC
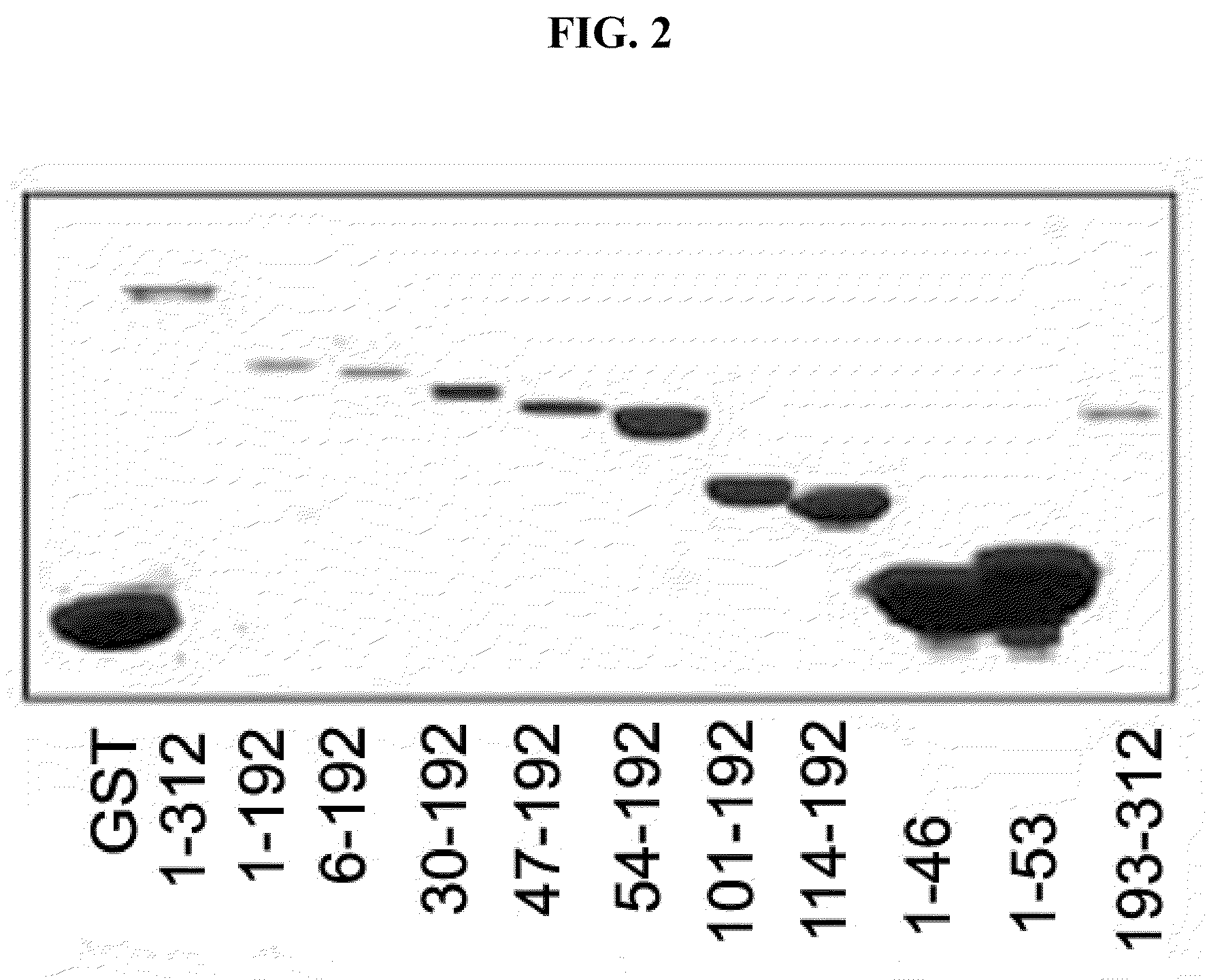
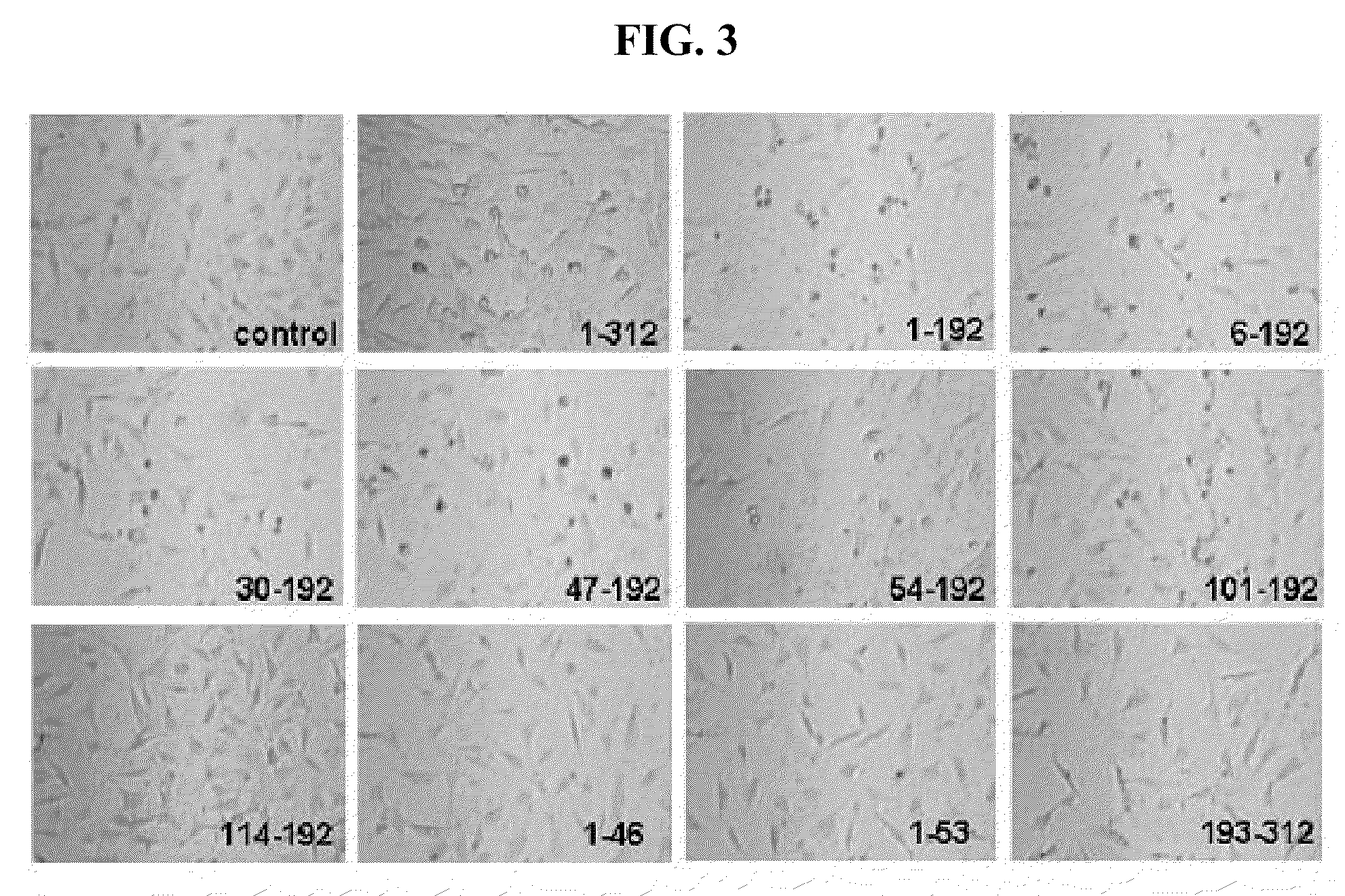
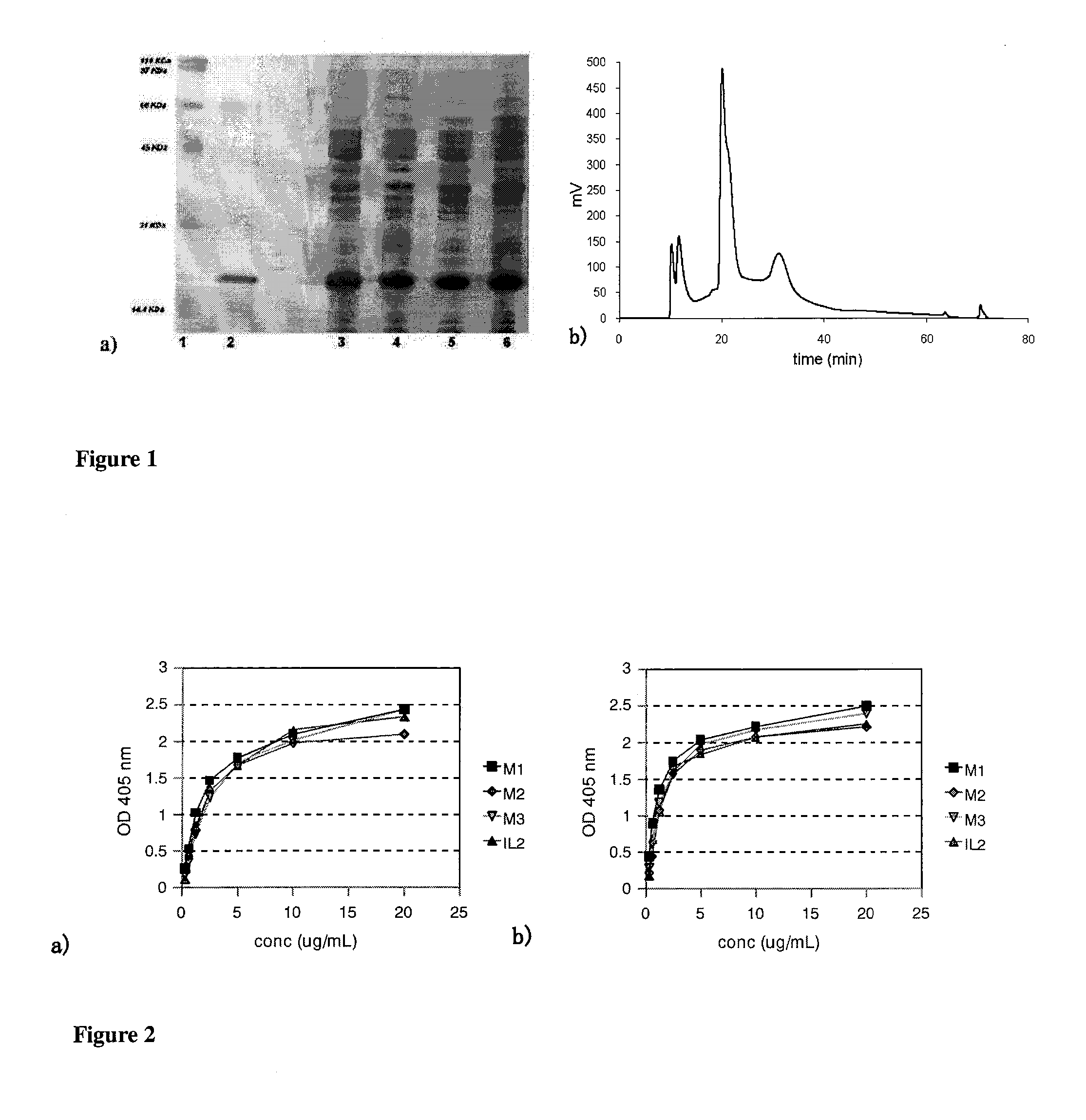
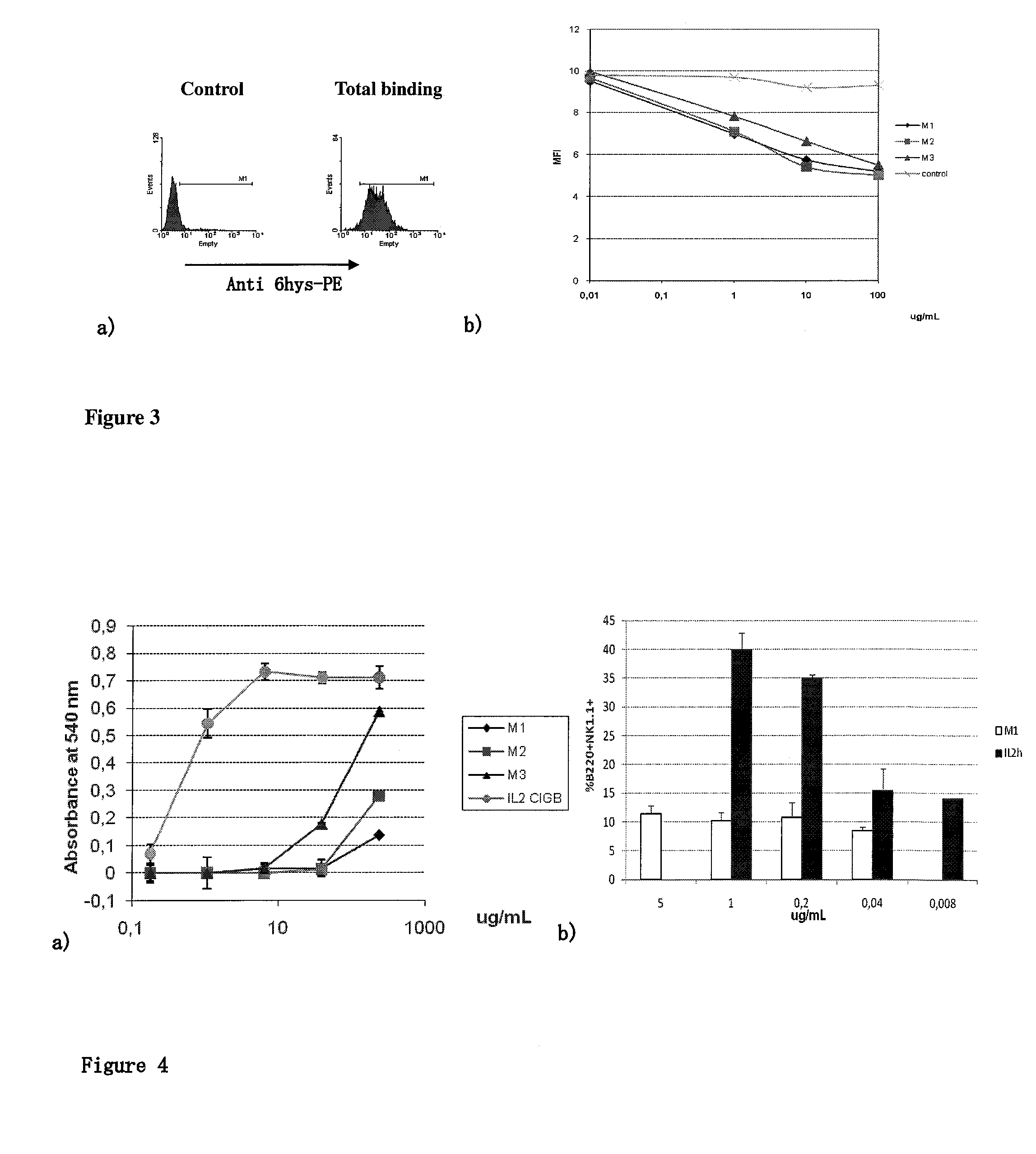
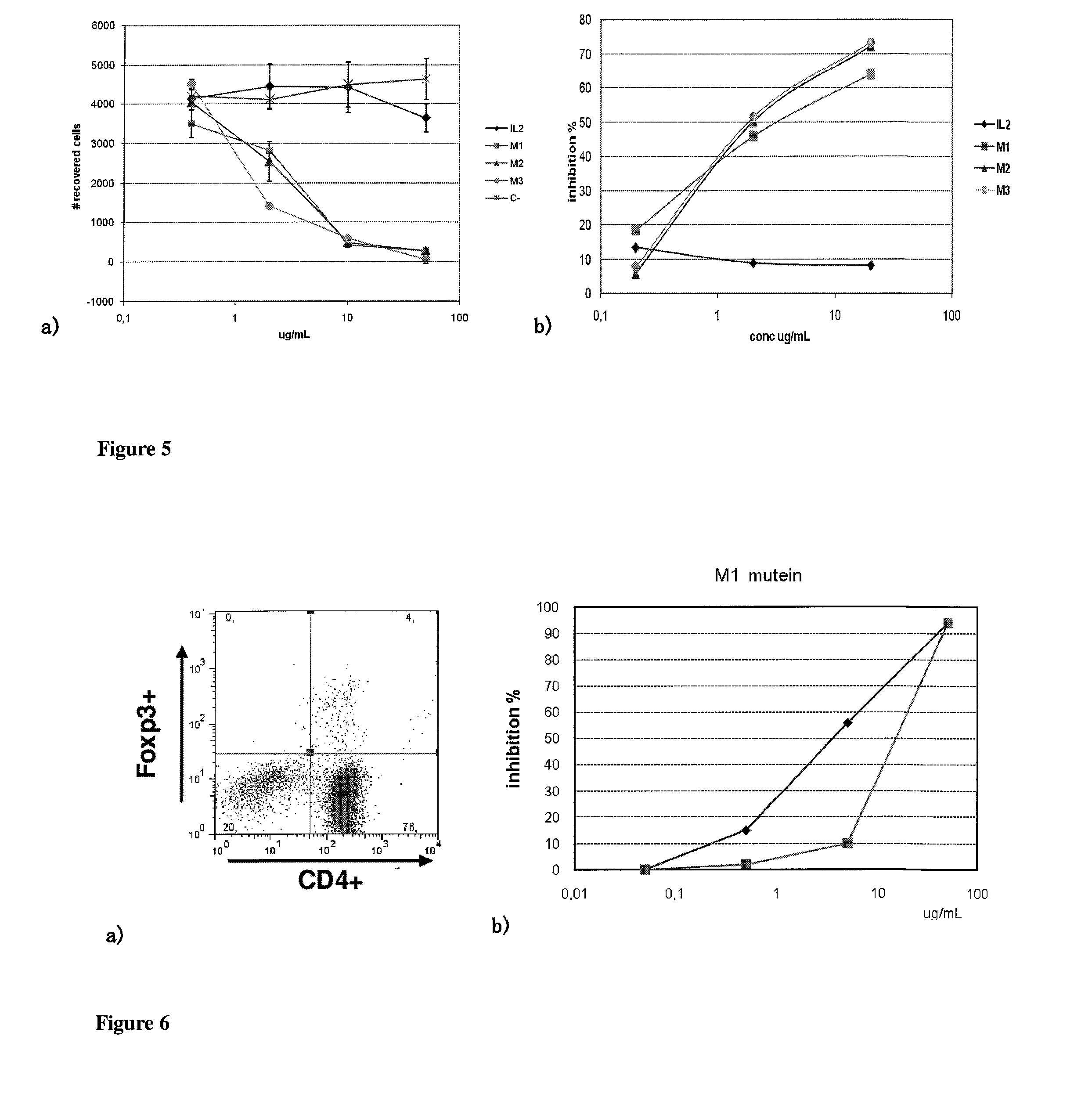
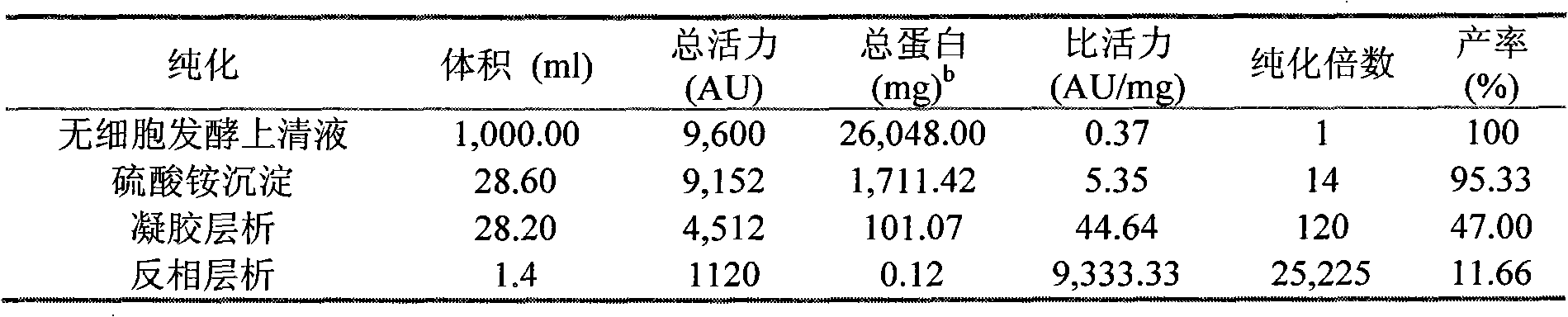
Immunomodulatory interleukin-2 polypeptides and methods of treating melanoma
The present invention relates generally to polypeptides whose primary sequence has high sequence homology with human interleukin 2 (IL-2) with some punctual mutations in the sequence of native IL-2. The polypeptides of the present invention have an immunomodulatory effect on the immune system, which is selective / preferential on regulatory T cells. The present invention also relates to specific polypeptides whose amino acid sequence is disclosed herein. In another aspect the present invention relates to pharmaceutical compositions comprising as active ingredient the polypeptides disclosed. Finally, the present invention relates to the therapeutic use of the polypeptides and pharmaceutical compositions disclosed due to their immune modulating effect on diseases such as cancer and chronic infectious diseases.
Owner:CENT DE INMUNOLOGIA MOLECULAR CENT DE INMUNOLO
Lactobacillus plantarum and bacteriocins produced by lactobacillus plantarum and capable of inhibiting Gram negative bacteria
InactiveCN101812414AHigh activityBroad antibacterial spectrumBacteriaMicroorganism based processesAlcohol sugarsFermentation
The invention relates to a strain of lactobacillus plantarum and bacteriocins produced by the lactobacillus plantarum and capable of inhibiting Gram negative bacteria. The bacteriocins have the advantages of broad antibacterial spectrum, thermal stability and stable pH, degradability by protease, no residue in a human body and high safety. A lactobacillus plantarum strain is preserved on June 29, 2009 with a preservation number of CGMCC No.3151. A production strain is obtained by separating 'Jiaoke', a conventional dairy product in Inner Mongolia; on a MRS culture medium, colonies are ivory and round with a protruded center and orderly edges; the strain has two blunt round ends and a short and straight stem; and the size of the strain is 0.4 to 0.7 mu m *2 to 3 mu m. The strain is a non-spore Gram positive bacillus, and is identified as the lactobacillus plantarum through an API50CHL sugar alcohol fermentation test, a 16S rRNA sequence homology analysis test and a recA gene multiplex PCR method. The lactobacillus plantarum KLDS1.0391 is used as the production strain and is fermented and purified by using the improved MRS culture medium to obtain the bacteriocins of the lactobacillus plantarum. The bacteriocins are used in food preservatives.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Method for the prediction of an epitope
The invention provides methods for the prediction of an epitope in a target protein. An epitope of the target protein can be bound by a given molecule, e.g., by an antibody. In particular, the methods of the invention comprise (i) identifying a plurality of cross-reactive proteins, i.e., proteins that can be bound by the same molecule, e.g., by the same antibody, as the target protein using, e.g., protein microarrays; and (ii) comparing the amino acid sequences of the target protein and the cross-reactive proteins with each other to identify windows of sequence homology, wherein the windows of sequence homology correspond to the epitope.
Owner:PROTOMETRIX
Heparanase activity neutralizing anti-heparanase monoclonal antibody and other anti-heparanase antibodies
InactiveUS20040170631A1Peptide/protein ingredientsBiological material analysisMonoclonal antibodyDrug development
Specific anti-heparanase antibodies which bind specifically to heparanase having sequence homology to human heparanase, which can be used to treat and diagnose conditions associated with heparanase catalytic activity, for purification of heparanase, and for drug development in heparanase associated conditions are disclosed.
Owner:YACOBY ZEEVI ORON +9
Urocortin-III and uses thereof
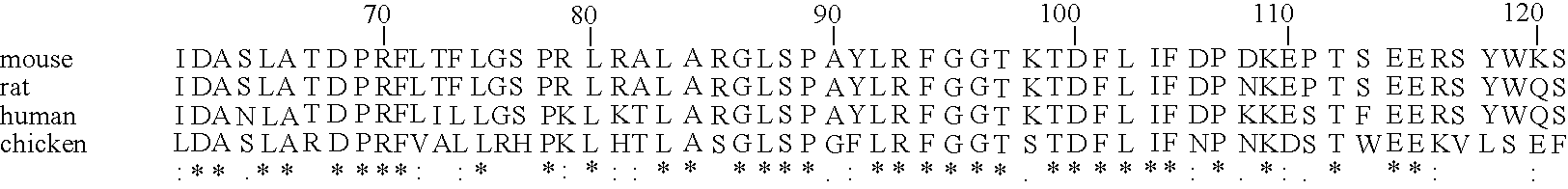
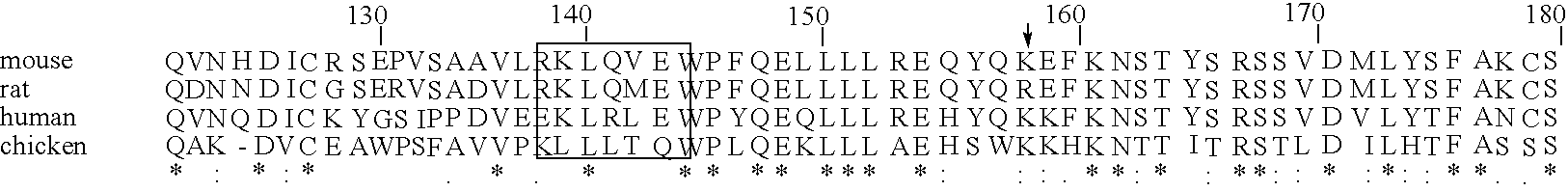
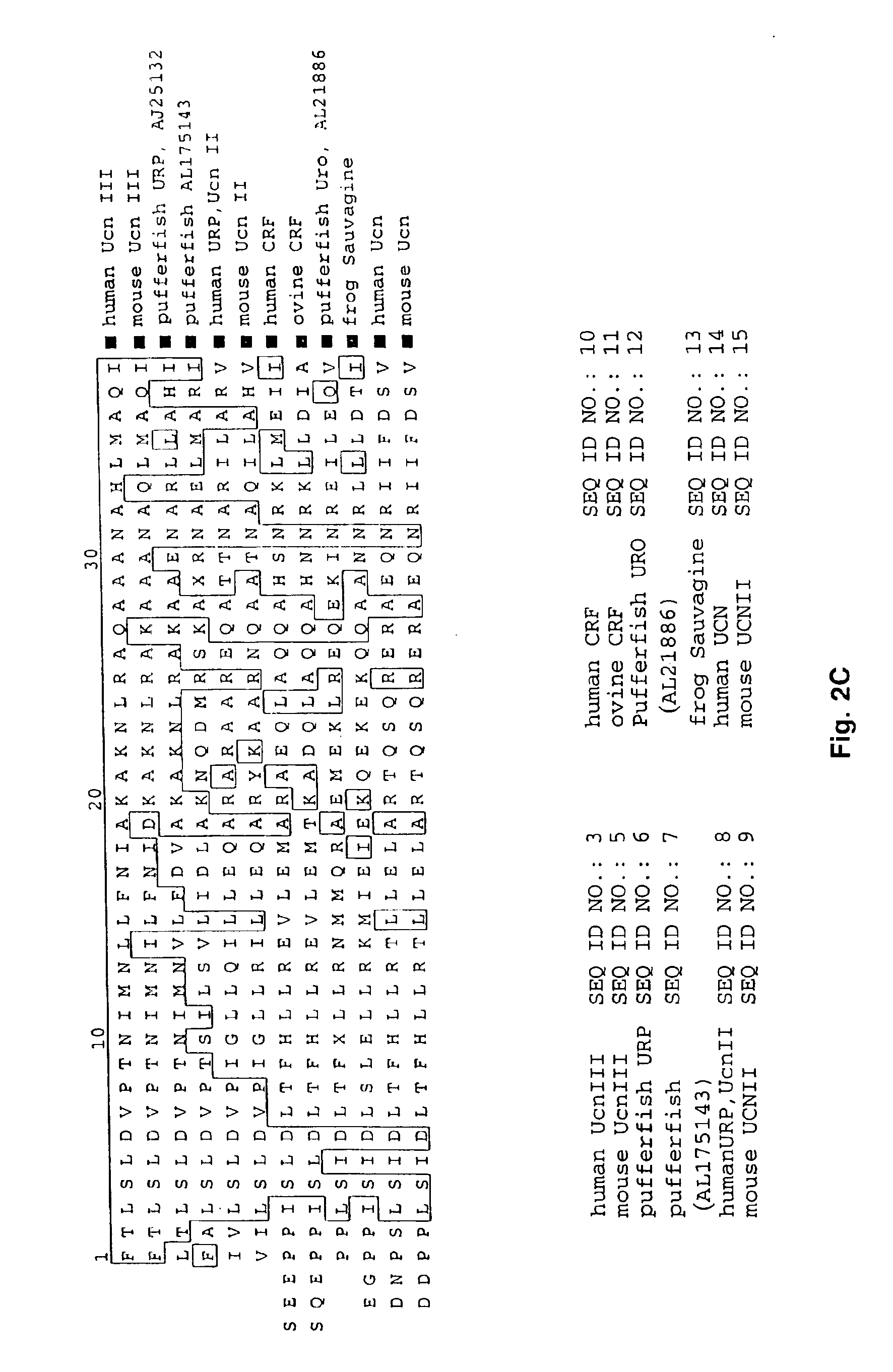
A search of the public human genome database identified a human EST, GenBank accession number AW293249, which has high homology to known pufferfish urocortin sequences. The full length sequence was amplified from human genomic DNA and sequenced. Sequence homology comparisons of the novel sequence with human urocortin I and urocortin II revealed that the sequence encoded a novel human urocortin, which was designated urocortin III (UcnIII). While urocortin III does not have high affinity for either CRF-R1 or CRF-R2, the affinity for CRF-R2 is greater than the affinity for CRF-R1. Urocortin III is capable stimulating cyclic AMP production in cells expressing CRF-R2α or β. Thus, the affinity is high enough that urocortin III could act as a native agonist of CRF-R2. However, it is also likely that urocortin III is a stronger agonist of a yet to be identified receptor.
Owner:RES DEVMENT FOUND
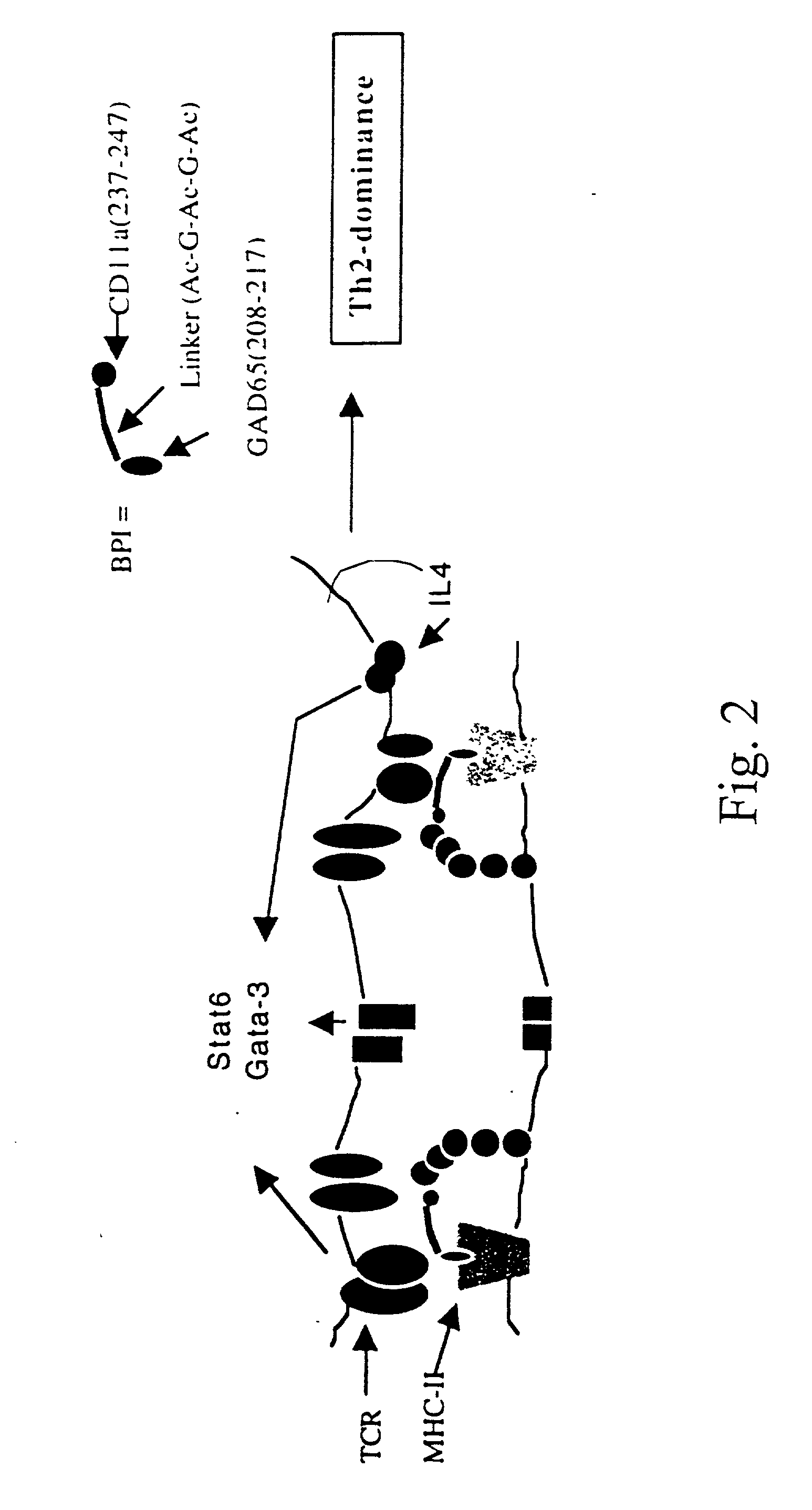
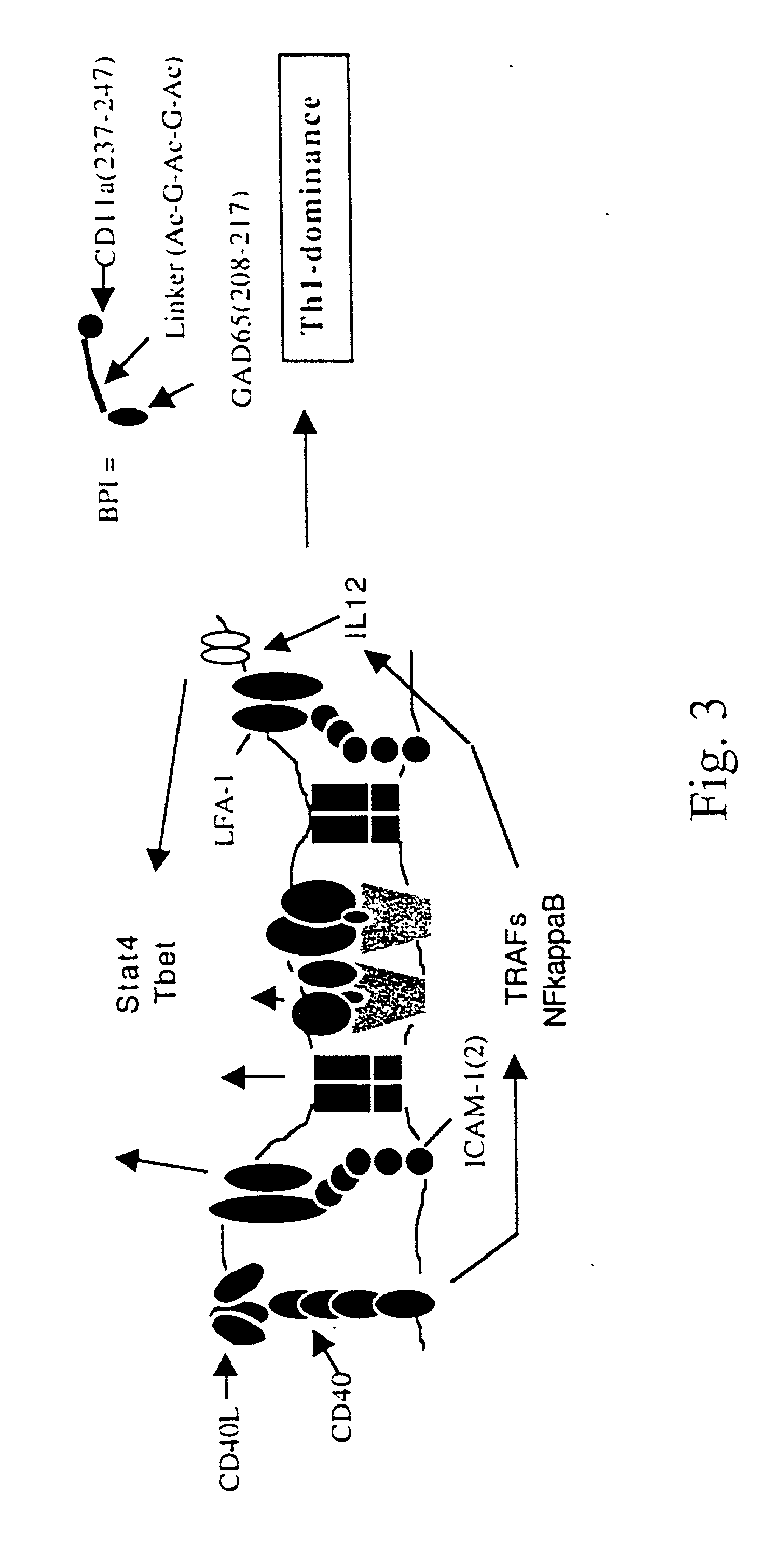
Signal-1/signal-2 bifunctional peptide inhibitors
InactiveUS20050107585A1Decreased LFA-1 : ICAM- signalingPreventing translocation stepCell receptors/surface-antigens/surface-determinantsPeptide/protein ingredientsMajor histocompatibilityPeptide sequence
A novel peptide sequence having the general formula AB wherein each of A and B represent a chain of amino acid residues and wherein said A chain is capable of binding with a major histocompatibility complex on an antigen presenting cell, and wherein said B chain is capable of binding with a Signal-2 receptor on an antigen presenting cell. Preferred forms of the peptide sequence further include an X chain positioned intermediate the A chain and the B chain. Moreover, preferred forms include an A chain which has at least about 10% sequence homology with a Signal-1 moiety, or is a peptidomimetic of a Signal-1 moiety, said B chain has at least 10% sequence homology with a Signal-2 receptor moiety, or is a peptidomimetic of a Signal-2 receptor moiety, and wherein the X chain has at least one amino acid residue, or is a peptidomimetic of that amino acid residue. Advantageously, the novel peptide sequence is capable of shifting a type-1 immune response to a type-2 immune response or from a type-2 immune response to a type-1 immune response.
Owner:KANSAS UNIV OF
Bactrian camel heavy-chain (HC) variable-domain antibody resisting porcine circovirus 2 as well as preparation method and application thereof
ActiveCN102766207AMicrobiological testing/measurementMicroorganism based processesSingle-domain antibodyAmino acid
The invention discloses a bactrian camel heavy-chain (HC) variable-domain antibody resisting a porcine circovirus 2 as well as a preparation method and an application thereof, and belongs to the field of a biotechnology. A PCV2 (Porcine Circovirus Virus) inactivated vaccine immunized bactrian camel is firstly utilized and a phage display technology is utilized to establish a PCV2 immune VHH antibody base; and a high-specificity variable domain antibody of heavy chains of HCAbs (VHH) which has a good affinity with the PCV, particularly a PCV2 virus antigen; and the antibody disclosed by the invention has an amino acid sequence shown as SEQ (sequence) ID NO.1 or an amino acid sequence which is shown as SEQ ID NO.2 and has the sequence isogeny being more than 95%. The PCV2 heavy-chain variable-domain antibody disclosed by the invention has the very important meanings on researching porcine circovirus, particularly researching a porcine circovirus 2-type infection mechanism and establishing rapid antigen detection with high sensitivity and specificity or diagnosing a reagent.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
Nucleic acid molecules and other molecules associated with plants
InactiveUS20080263730A1Reduces and depresses expression of proteinReduce protein expressionImmunoglobulinsFermentationNovel genePlant genomics
Expressed Sequence Tags (ESTs) isolated from maize are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:MONSANTO TECH LLC
Heparanase activity neutralizing anti-heparanase monoclonal antibody and other anti-heparanase antibodies
InactiveUS20040213789A1Peptide/protein ingredientsBiological material analysisAntiendomysial antibodiesPharmaceutical drug
Specific anti-heparanase antibodies which bind specifically to heparanase having sequence homology to human heparanase, which can be used to treat and diagnose conditions associated with heparanase catalytic activity, for purification of heparanase, and for drug development in heparanase associated conditions are disclosed.
Owner:INSIGHT STRATEGY & MARKETING +1
Nucleic acid molecules and other molecules associated with plants
InactiveUS20070016974A1Desirable effectIncrease profitSugar derivativesOther foreign material introduction processesNovel genePlant genomics
Expressed Sequence Tags (ESTs) isolated from rice are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:BYRUM JOSEPH +2
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from soybean are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:BUEHLER ROBERT E +3
Thermostable Alcohol Dehydrogenase Derived From Thermococcus Guaymasensis
An alcohol dehydrogenase (ADH) from hyperthermophilic archaeon Thermococcus guaymasensis was purified to homogeneity and was found to be a homotetramer with a subunit size of 40±1 kDa. The gene encoding the enzyme was cloned and sequenced, and found to have significant sequence homology to known zinc-containing ADHs and L-threonine dehydrogenases with both binding motifs of catalytic zinc and NADP+. The wild-type enzyme is a primary-secondary ADH that exhibits a substrate preference for secondary alcohols and corresponding ketones, and exhibits unusual stereoselectivity. The wild-type enzyme was found to have outstanding thermostability, demonstrating 60% activity after incubation at 80° C. for 40 hours. Site-directed mutagenesis was used to substitute the cyteine residue at position 56 with a serine, to provide the TgADH(C56S) mutant. In the assays that we carried out, we found virtually no difference in enzyme activity and oxygen-sensitivity between the mutant TgADH (C56S) and wild type TgADH.
Owner:MA KESEN +1
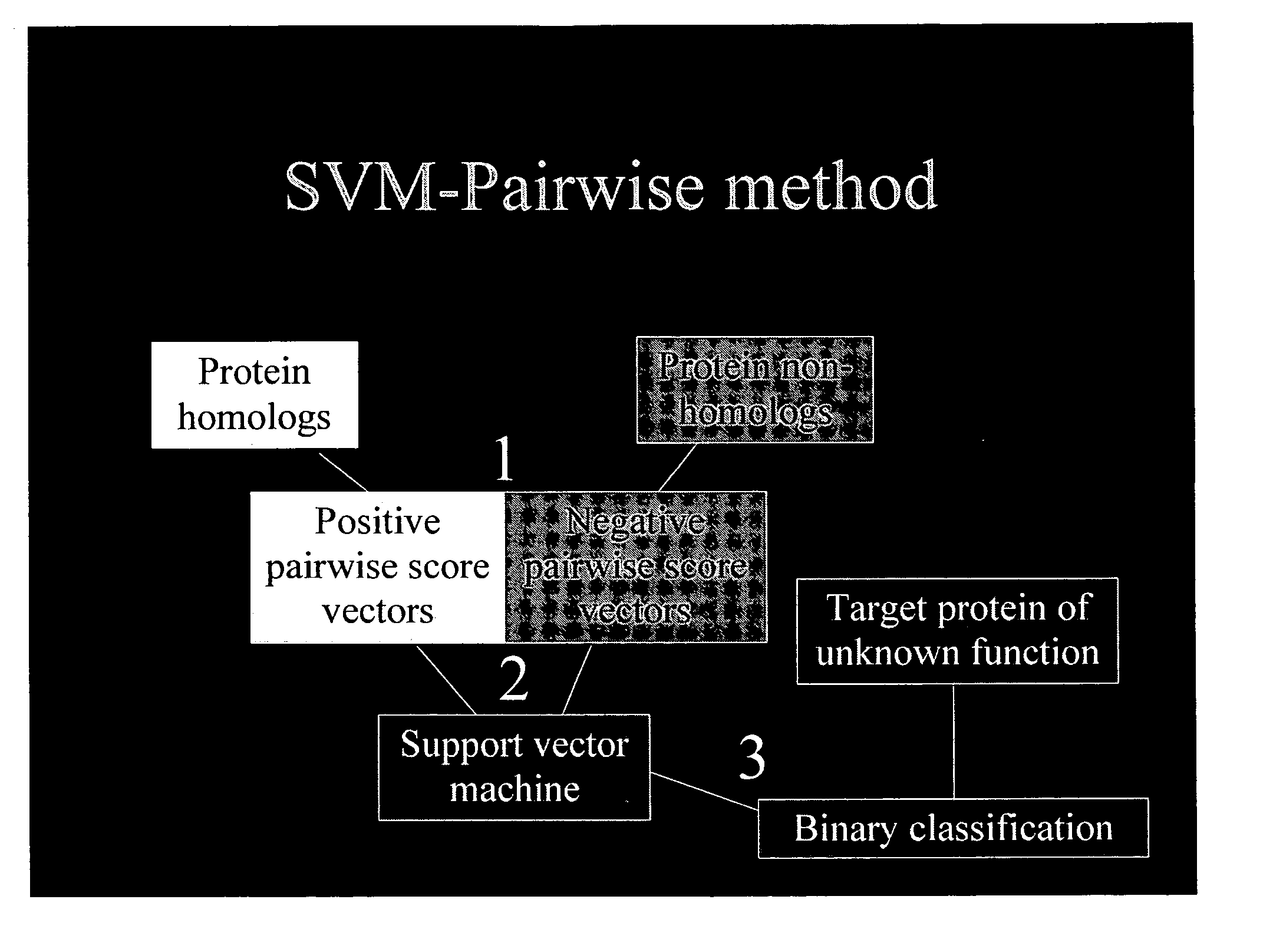
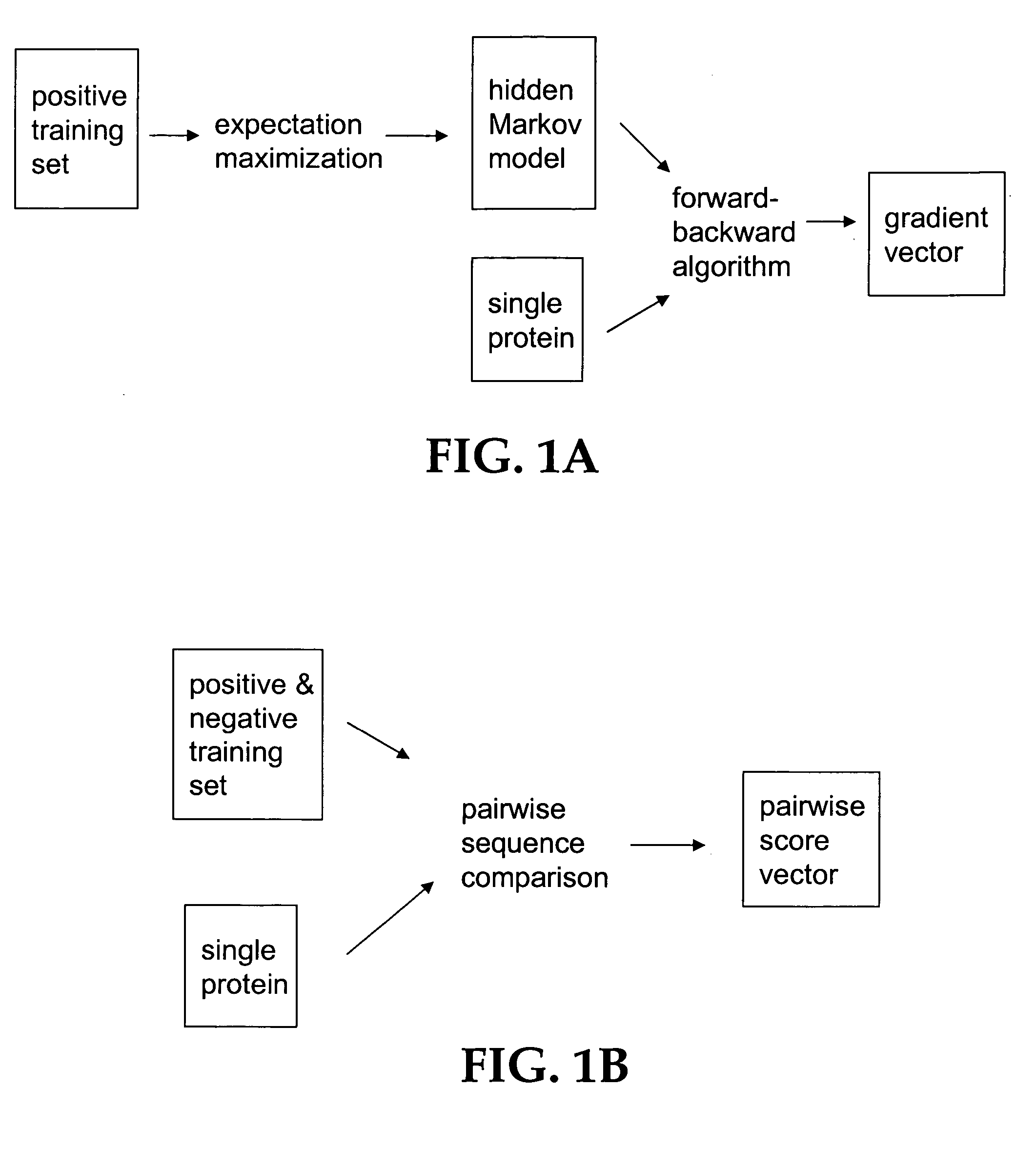
Computational method for detecting remote sequence homology
InactiveUS20050100992A1Enhanced remote detectionAccurate detectionBiostatisticsSequence analysisAlgorithmCalculation methods
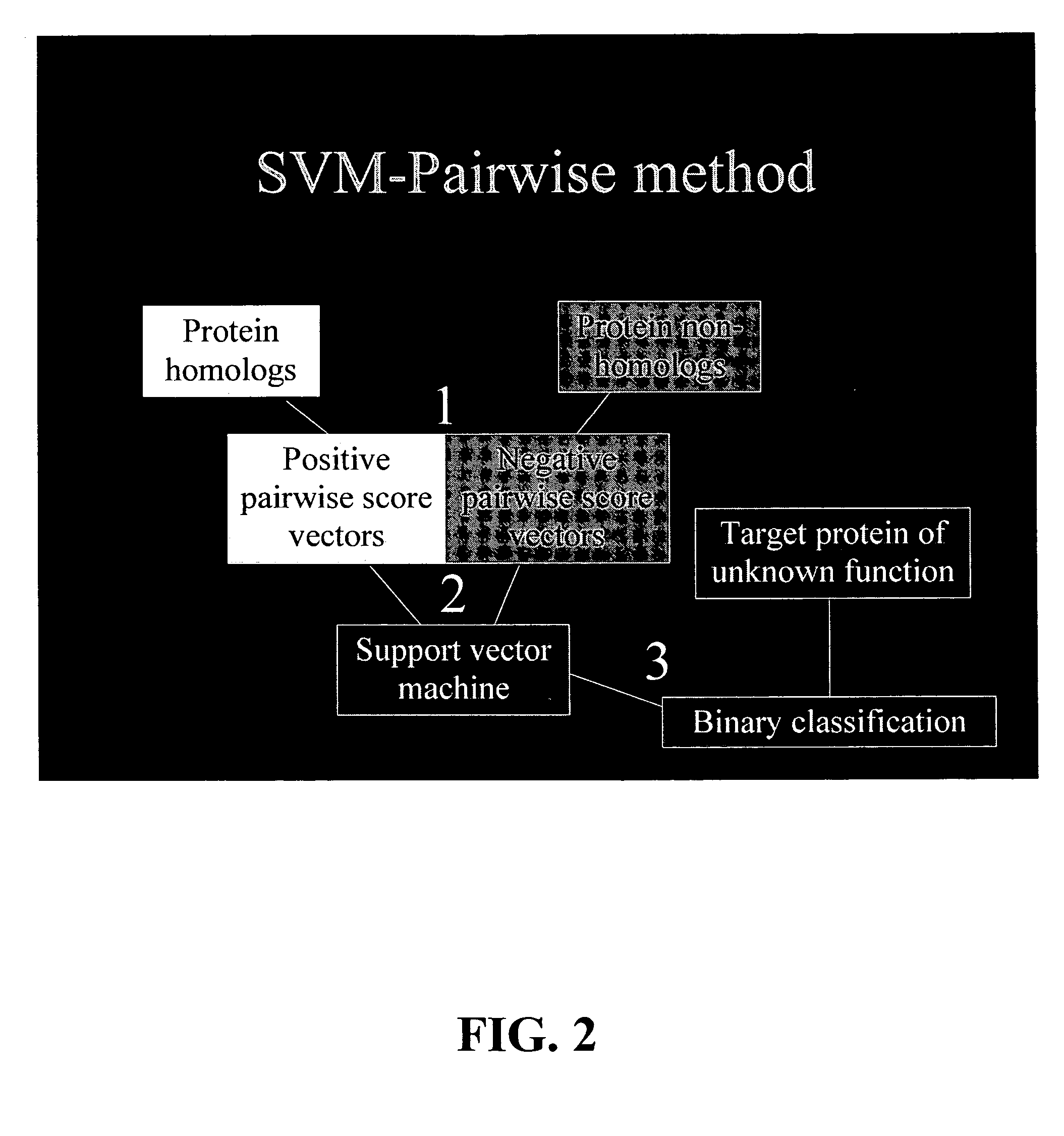
The present invention relates to a computation method for detecting remote sequence homologies. The method comprises the following steps: First, a training sequence set of positive and negative examples each having a corresponding binary label is provided together with a database query sequence set (typically large) of unlabeled sequences. Second, each sequence in the training set is converted into a fixed-length vector of real values by computing pairwise sequence similarity scores with respect to the vectorization set to obtain vectorized training sequences each having corresponding binary labels. Third, the vectorized training sequences (along with their binary labels) are used to train a discriminative classification algorithm to obtain a trained discriminative classification algorithm. Fourth, the the database of unlabeled sequences are converted into pairwise score vectors, using the vectorization set to obtain vectorized database sequences. Finally, each vectorized database query sequence is presented to the trained discriminative classification algorithm to produce predicted classifications for the database query sequence.
Owner:NOBLE WILLIAM STAFFORD
Composition for Organ, Tissue, or Cell Transplantation, Kit, and Transplantation Method
ActiveUS20170058001A1Facilitate functioningImprove survivabilityPeptide/protein ingredientsSurgical drugsBULK ACTIVE INGREDIENTActive ingredient
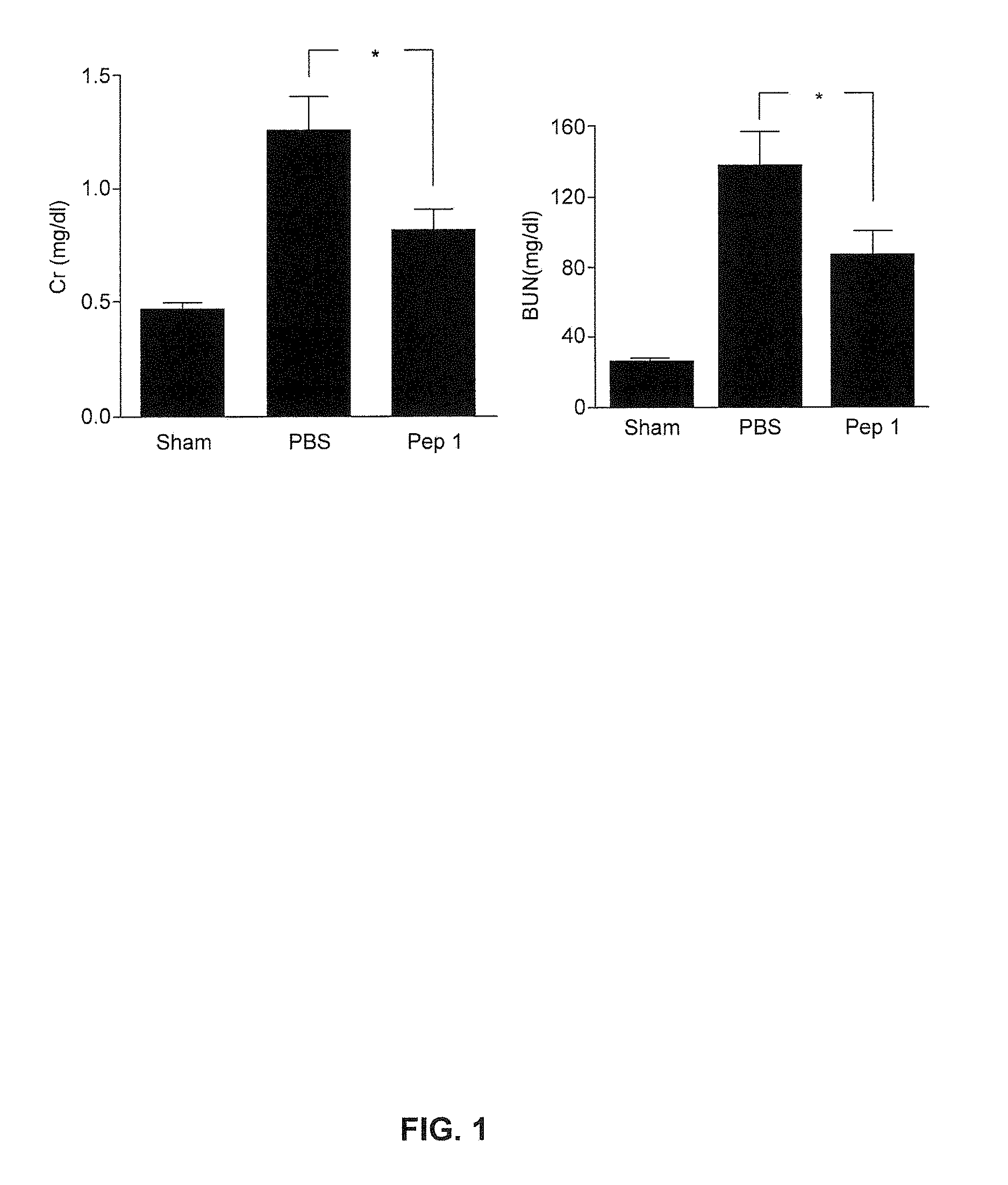
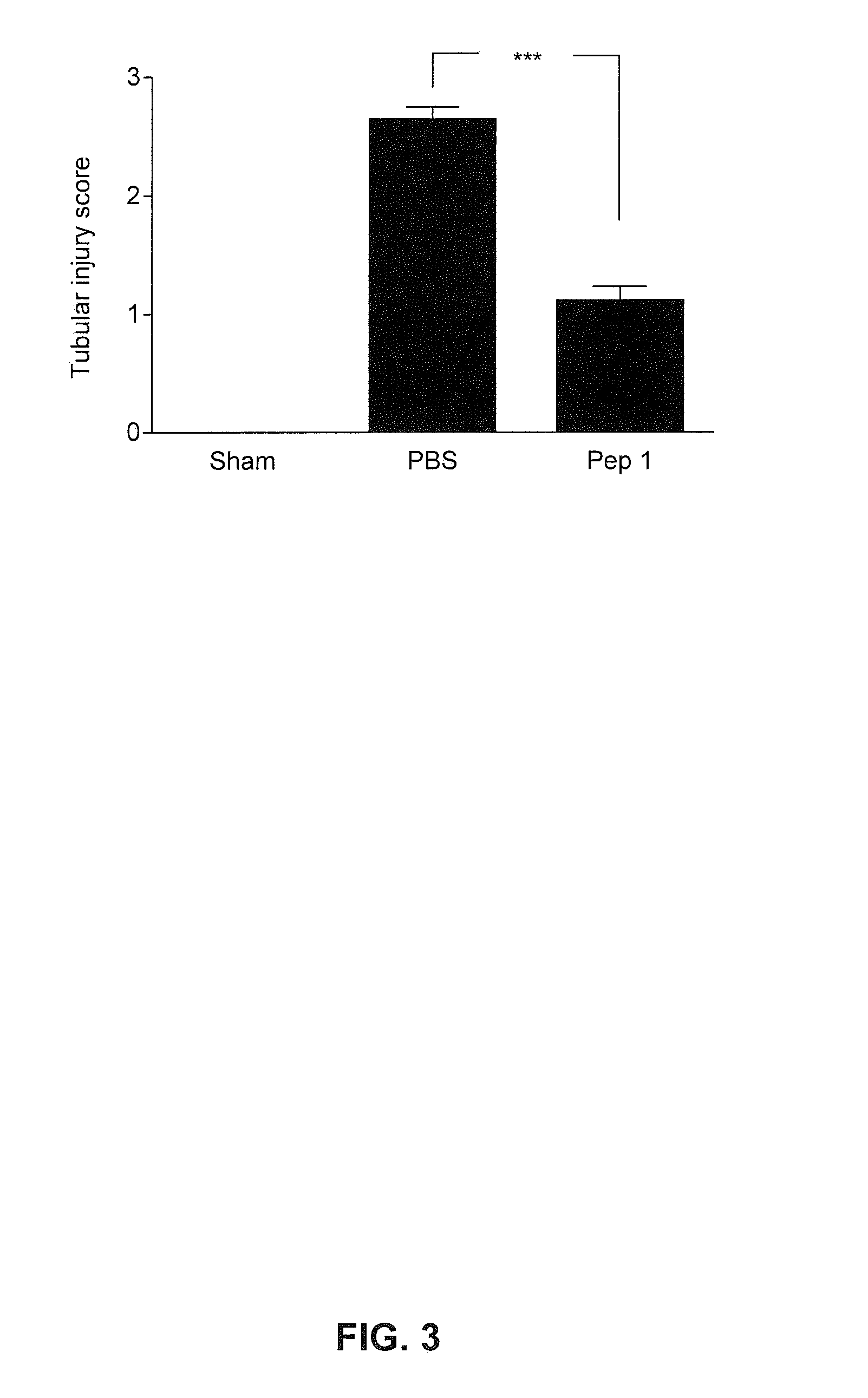
Disclosed are: a composition for organ, tissue or cell transplantation, containing, as an active ingredient, a peptide comprising the amino acid sequence of SEQ ID NO: 1, a peptide having at least 80% sequence homology with the amino acid sequence, or a peptide which is a fragment thereof; a kit comprising the composition; or a method using the composition. By using the composition, the kit, or the method, the viability and / or function of an organ, tissue, or cell after transplantation are strengthened and an organ, tissue or cell isolated from a living body is preserved temporarily without damage.
Owner:GEMVAX & KAEL
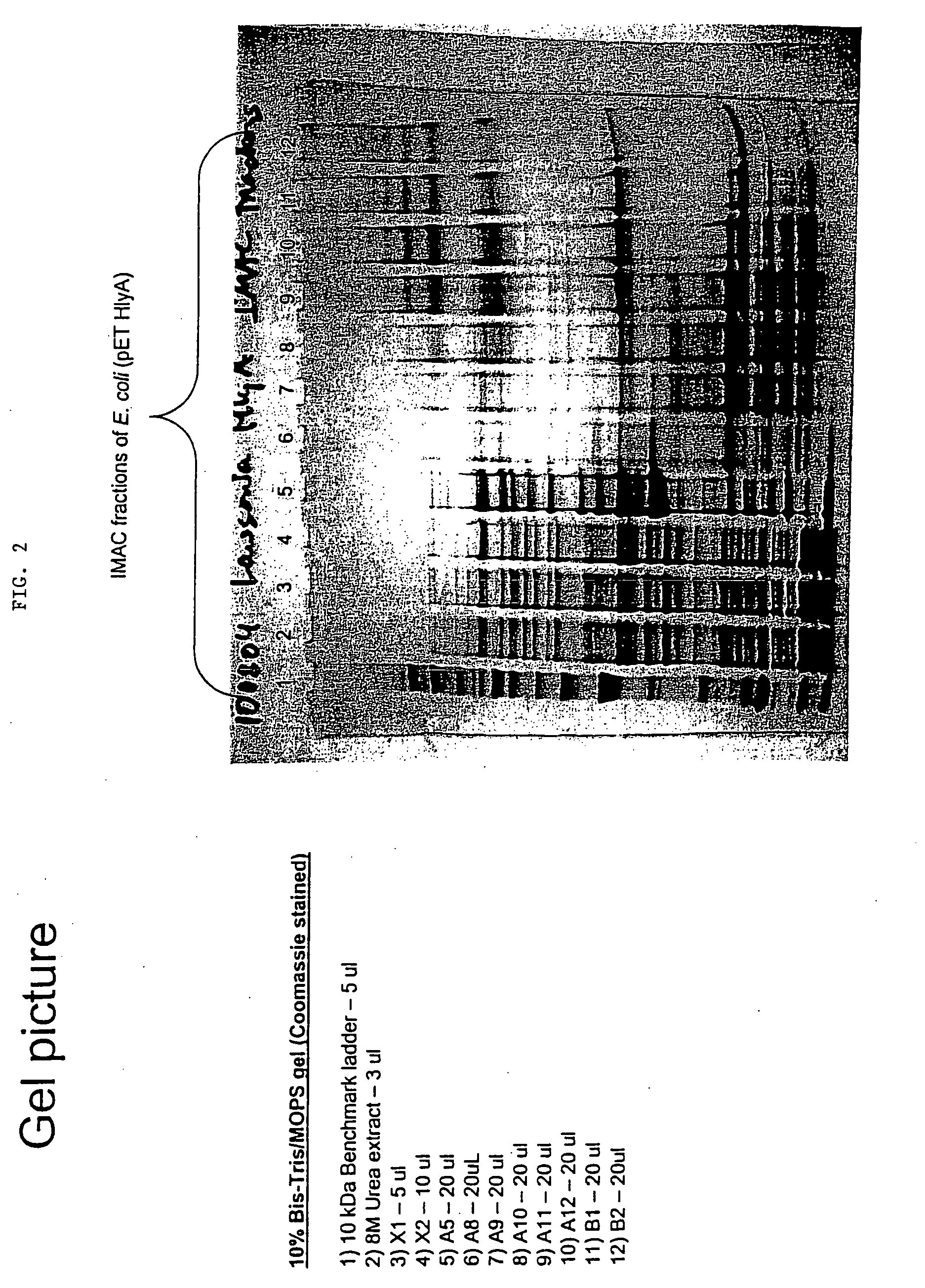
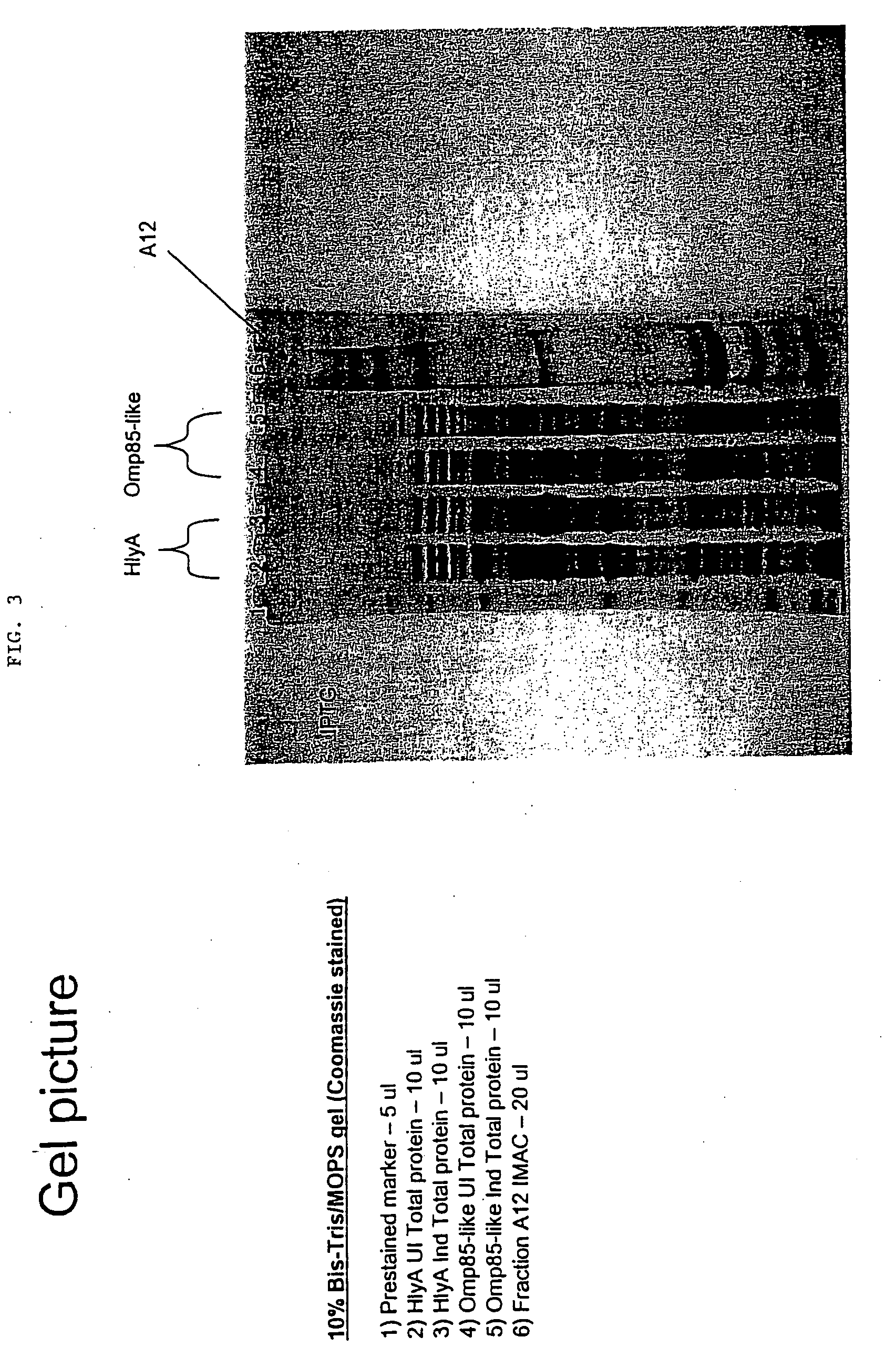
Lawsonia intracellularis immunological proteins
InactiveUS20080279893A1Improve the immunityReduce the severity of the diseaseAntibacterial agentsPeptide/protein ingredientsA-DNAImmunogenicity
The present invention provides nucleic acid and amino acid sequences useful as the immunogenic portion of vaccines or immunogenic compositions effective for lessening the severity of the clinical symptoms associated with Lawsonia intracellularis infection or conferring protective immunity to an animal susceptible to such infection. Preferred amino acid sequences are selected from the group consisting of 1) a polypeptide comprising a sequence selected from the group consisting of SEQ ID Nos.: 1-455, SEQ ID No 466, or the polypeptide encoded by SEQ ID No: 456, SEQ ID No: 457 or SEQ ID No: 466; 2) any polypeptide that has at least 85% sequence homology, more preferably at least about 90% sequence homology, still more preferably at least about 95% sequence homology, even more preferably at least about 97% sequence homology, still even more preferably at least about 98% sequence homology, and even more preferably at least about 99% sequence homology to the polypeptide of 1); 3) any immunogenic portion of the polypeptides of 1) and / or 2) 4) the immunogenic portion of 3), comprising at least 300, 290, 280, 270, 260, 250, 240, 230, 220, 210, 200, 190, 180, 170, 160, 150, 140, 130, 120, 110, 100, 90, 80, 70, 60, 50, 45, 40, 35, 30, 25, 20, 18, 15, 13, 10, or most preferably 9 contiguous amino acids included in the sequences of SEQ ID No: 1-455, SEQ ID No: 456, or the amino acid sequence encoded by SEQ ID No: 457 or SEQ ID No: 466; and / or 5) a polypeptide that is encoded by a DNA that codes for a peptide comprising the sequence of SEQ ID No: 1-455 or SEQ ID No: 466. Thus, the nucleic acid sequences encoding such proteins, or the proteins themselves are included in vaccine compositions, together with veterinary-acceptable carrier and administered to an animal in need thereof.
Owner:BOEHRINGER LNGELHEIM VETMEDICA GMBH
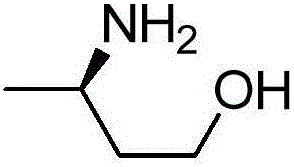
Improved transaminase and application thereof in preparation of (R)-3-aminobutanol
ActiveCN106754806AHigh catalytic activityHigh chiral purityBacteriaTransferasesSubstrate concentrationAmino acid
The invention relates to improved transaminase, an encoding gene and application thereof in preparation of (R)-3-aminobutanol. The improved transaminase is higher than wild transaminase in enzymatic activity. The amino acid sequence of the wild transaminase is shown in SEQ ID No.2 and is rooted in Aspergillus terreus NIH2624. The improved transaminase comprises an amino acid sequence with at least 95% sequence homology as the amino acid sequence shown in the SEQ ID No.2, and the 215th bit corresponding to the amino acid sequence shown in SEQ ID No.2 is a proline residue. The reaction conditions are mild when the improved transaminase is used for preparing (R)-3-aminobutanol, the product is high in chirality purity and yield, the concentration of a substrate reaches up to 50 g / L, and the good industrial application prospect is achieved.
Owner:SYNCOZYMES SHANGHAI
Recombinant human-like collagen protein-human cell growth factor fusion protein and preparation method and application thereof
The invention discloses a recombinant human-like collagen protein-human original cell growth factor fusion protein and a preparation method and application thereof. The prepared fusion protein is provided with a special fibrous structure and good biological tissue compatibility, and can promote proliferation and differentiation of epidermis cells, endothelial cells and fibroblast cells. The preparation method comprises the following steps: obtaining encoded DNA sequence of the fusion protein, reconstructing and recombining fusion protein expressed by an expression vectors in escherichia coli, pichia pastoris or chinese hamster ovary cells. The fusion protein comprises a first zone and a second zone, wherein the first zone has at least 85% of sequence homology with human collagen protein and has the function of the human collagen protein, and the second zone has at least 85% of human original cell growth factor and has the function of the human original cell growth factor; connecting peptide is arranged between the first zone and the second zone; the general formula of the connecting peptide is (GGGS)n; preferably, the amino acid sequence of the fusion protein is SEQ ID NO. 15.
Owner:浙江肽源生物科技有限公司
Nucleic acid molecule SEQ ID NO. 68811 and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from maize are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:MONSANTO TECH LLC
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from cotton are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:MONSANTO TECH LLC
Nucleic acid molecules and other molecules associated with plants
InactiveUS20080168583A1Reduces and depresses expression of proteinReduce protein expressionPeptide/protein ingredientsImmunoglobulinsNovel genePlant genomics
Expressed Sequence Tags (ESTs) isolated from cotton are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:FINCHER KAREN L +3
Rapid Genomic Sequence Homology Assessment Scheme Based on Combinatorial-Analytic Concepts
InactiveUS20120215463A1Reduce in quantityEfficient identificationBiological testingSequence analysisNucleotideGenome
The present invention relates to methods and apparatus for rapid assessment of genomic sequences using the difference set model. The invention provides methods to determine the presence and identity of similarities and differences in genomic sequences. In particular, the invention provides methods and apparatus to assess homology, the presence and identity of insertion and deletion segments and the presence and identity of single nucleotide polymorphisms in genomic sequences.
Owner:MITRE SPORTS INT LTD
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from cotton are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:FINCHER KAREN L +3
Nucleic acid molecules and other molecules associated with plants
Expressed Sequence Tags (ESTs) isolated from soybean are disclosed. The ESTs provide a unique molecular tool for the targeting and isolation of novel genes for plant protection and improvement. The disclosed ESTs have utility in the development of new strategies for understanding critical plant developmental and metabolic pathways. The disclosed ESTs have particular utility in isolating genes and promoters, identifying and mapping the genes involved in developmental and metabolic pathways, and determining gene function. Sequence homology analyses using the ESTs provided in the present invention, will result in more efficient gene screening for desirable agronomic traits. An expanding database of these select pieces of the plant genomics puzzle will quickly expand the knowledge necessary for subsequent functional validation, a key limitation in current plant biotechnology efforts.
Owner:BUEHLER ROBERT E +3
High-throughput active peptide screening method based on tandem mass spectrum and molecular docking
InactiveCN107132360AQuick searchEasy to operateBiological testingProtein targetHigh-Throughput Screening Methods
The invention relates to the field of medicine, and discloses a high-throughput screening method for active polypeptides based on tandem mass spectrometry and molecular docking. The invention uses modern biochemical technology to extract and prepare mixed polypeptides from marine molluscs, fish and other animals. After preliminary activity separation, the active site / component group is determined, and the amino acid sequence of all polypeptides in the active site / component group is identified by LC-MS / MS method. In the molecular docking software, all polypeptide sequences are homologously modeled, and then imported Molecular docking software docks with the target protein, analyzes the binding rate of the polypeptide and the target protein, and evaluates its inhibitory / activating activity, and screens the confirmed active polypeptide through solid-phase synthesis combined with in vitro activity evaluation to verify the accuracy of the screening. The invention has the advantages of fast and efficient search for marine active polypeptides, strong operability and important application value.
Owner:NANJING UNIVERSITY OF TRADITIONAL CHINESE MEDICINE
Improved ketoreductase polypeptide and coding gene thereof, and cell for expressing polypeptide
The invention relates to an improved ketoreductase polypeptide. The enzymatic activity of the improved ketoreductase polypeptide is higher than that of a wild type ketoreductase polypeptide; the enzymatic activity refers to the enzymatic activity for catalyzing a ketone compound to join in a reduction reaction to generate a chiral alcohol; the wild type ketoreductase polypeptide has an amino acidsequence shown in SEQ ID No. 2 and is from Lactobacillus kefiri DSM 20587; the improved ketoreductase polypeptide contains an amino acid sequence having at least 90% of sequence homology with the amino acid sequence shown in SEQ ID No. 2; and a residue in the amino acid sequence of the improved ketoreductase polypeptide in relation to the 94th residue in the amino acid sequence shown in SEQ ID No. 2 is a tyrosine residue.
Owner:SYNCOZYMES SHANGHAI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com