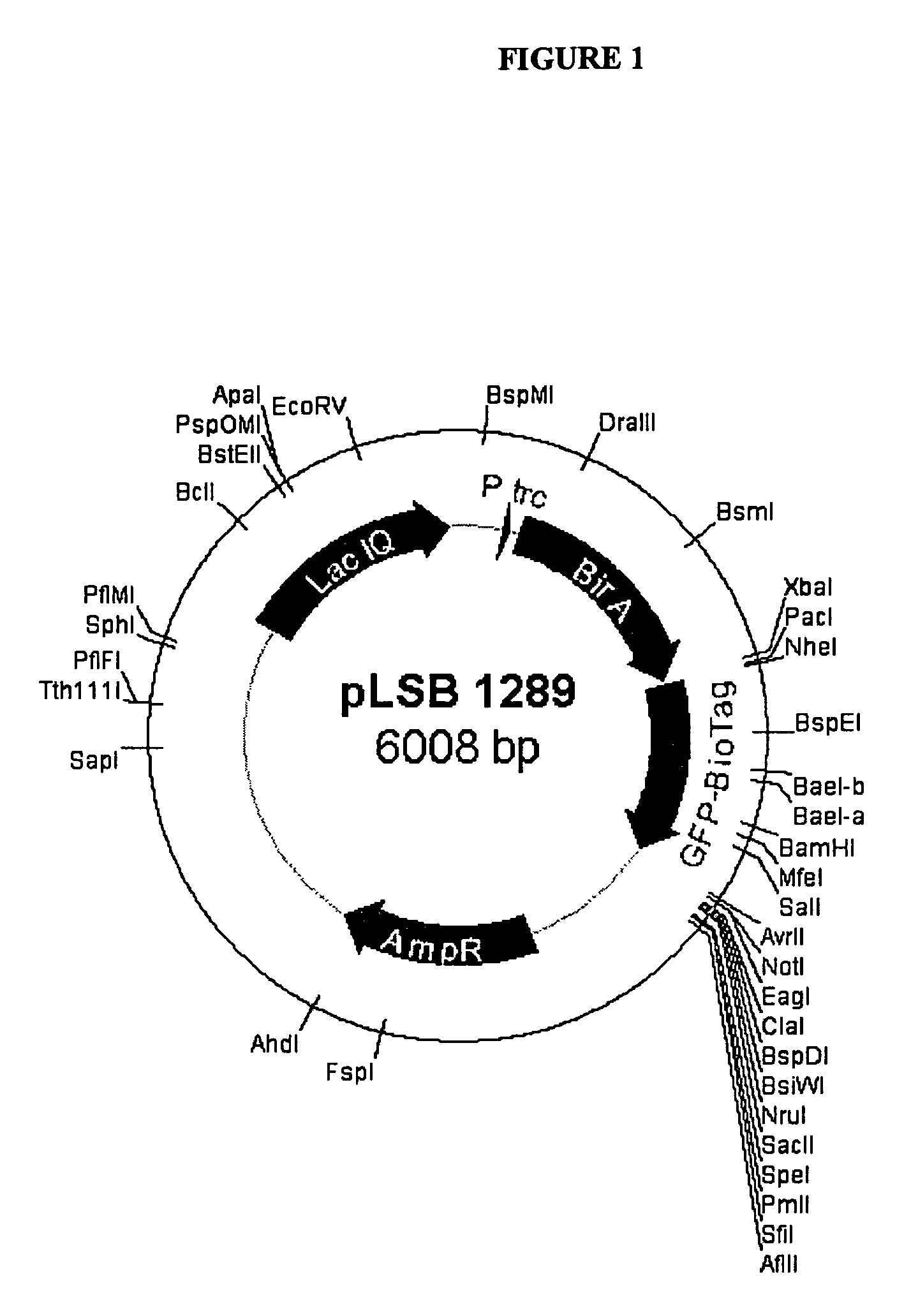
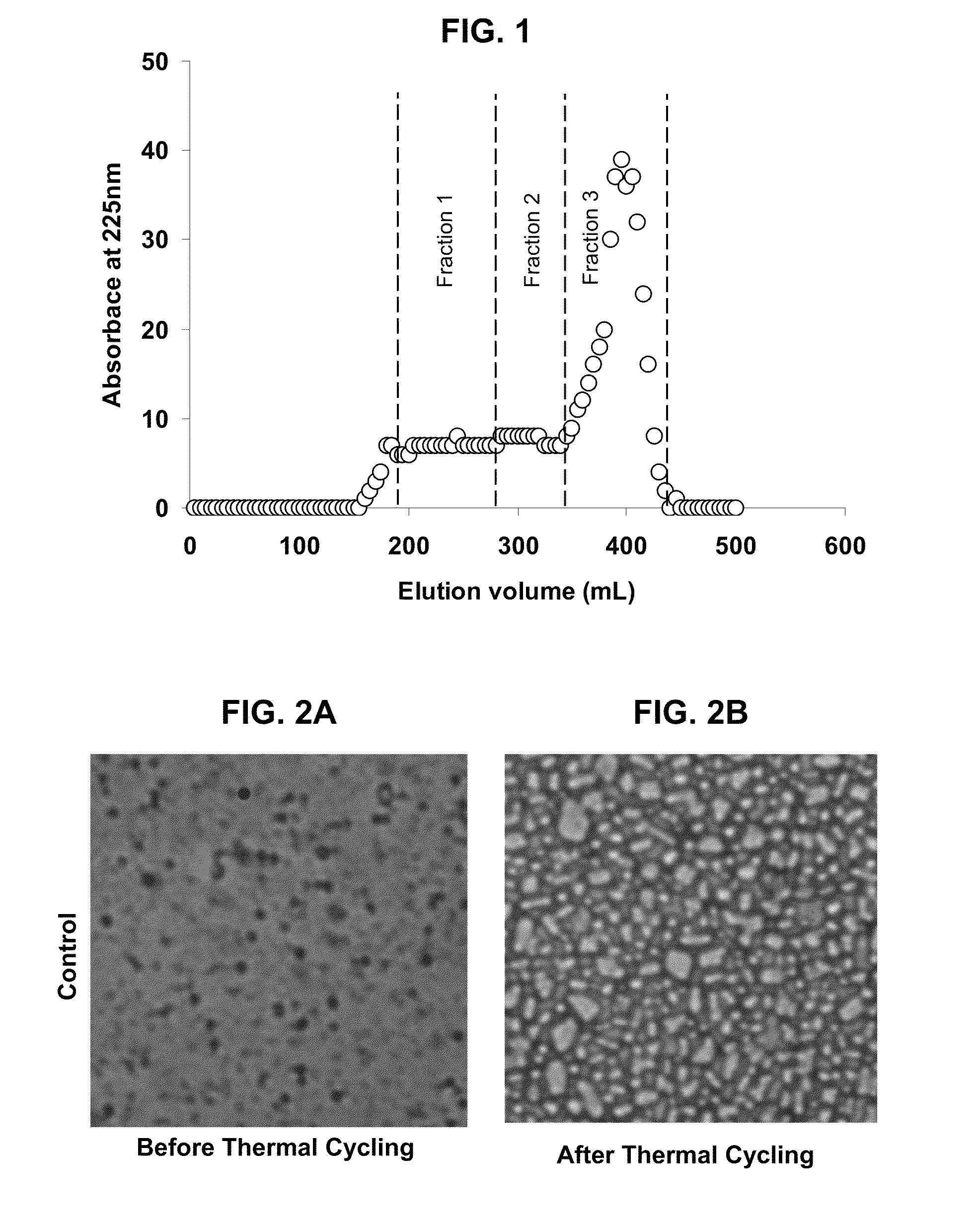
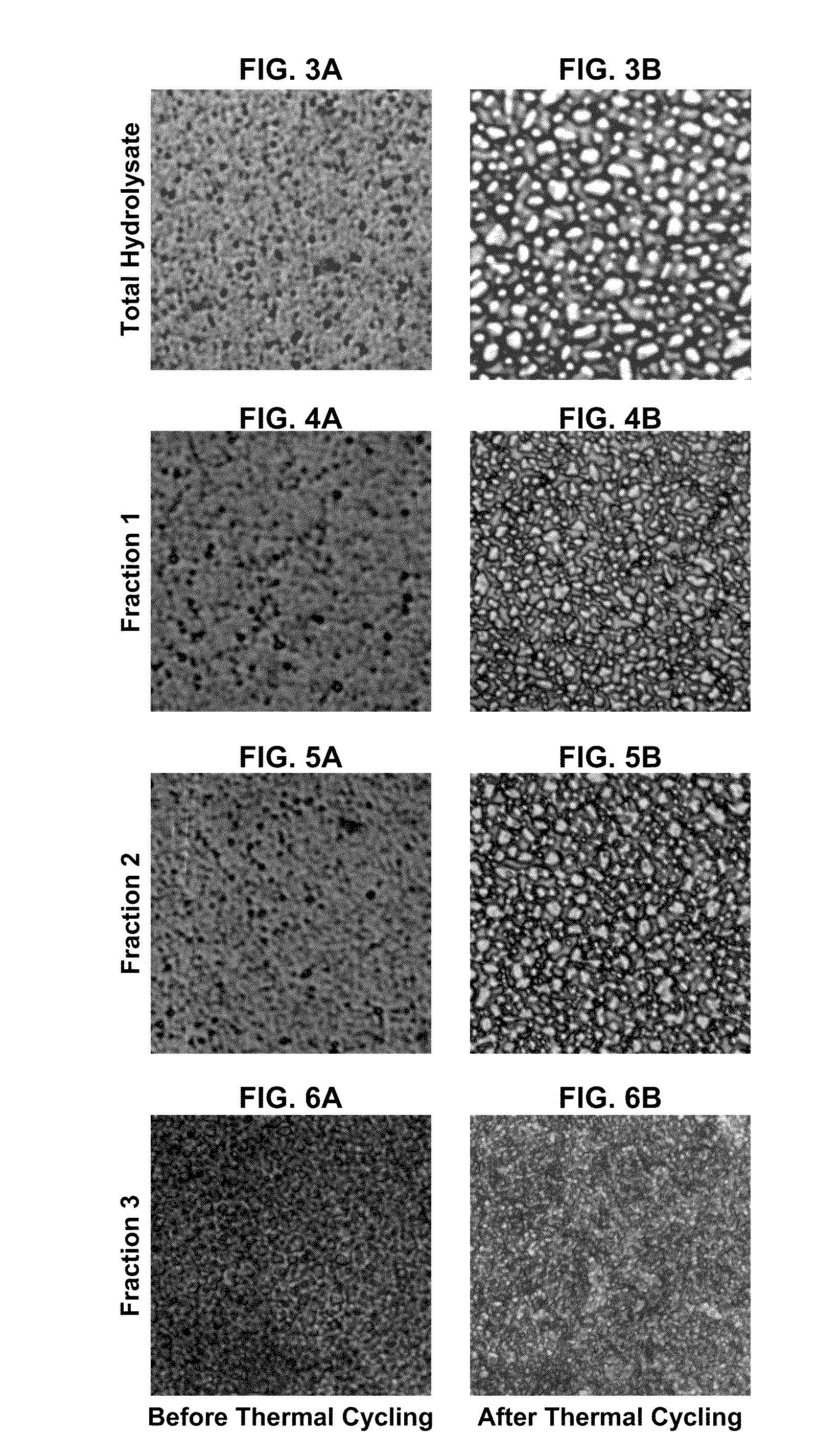
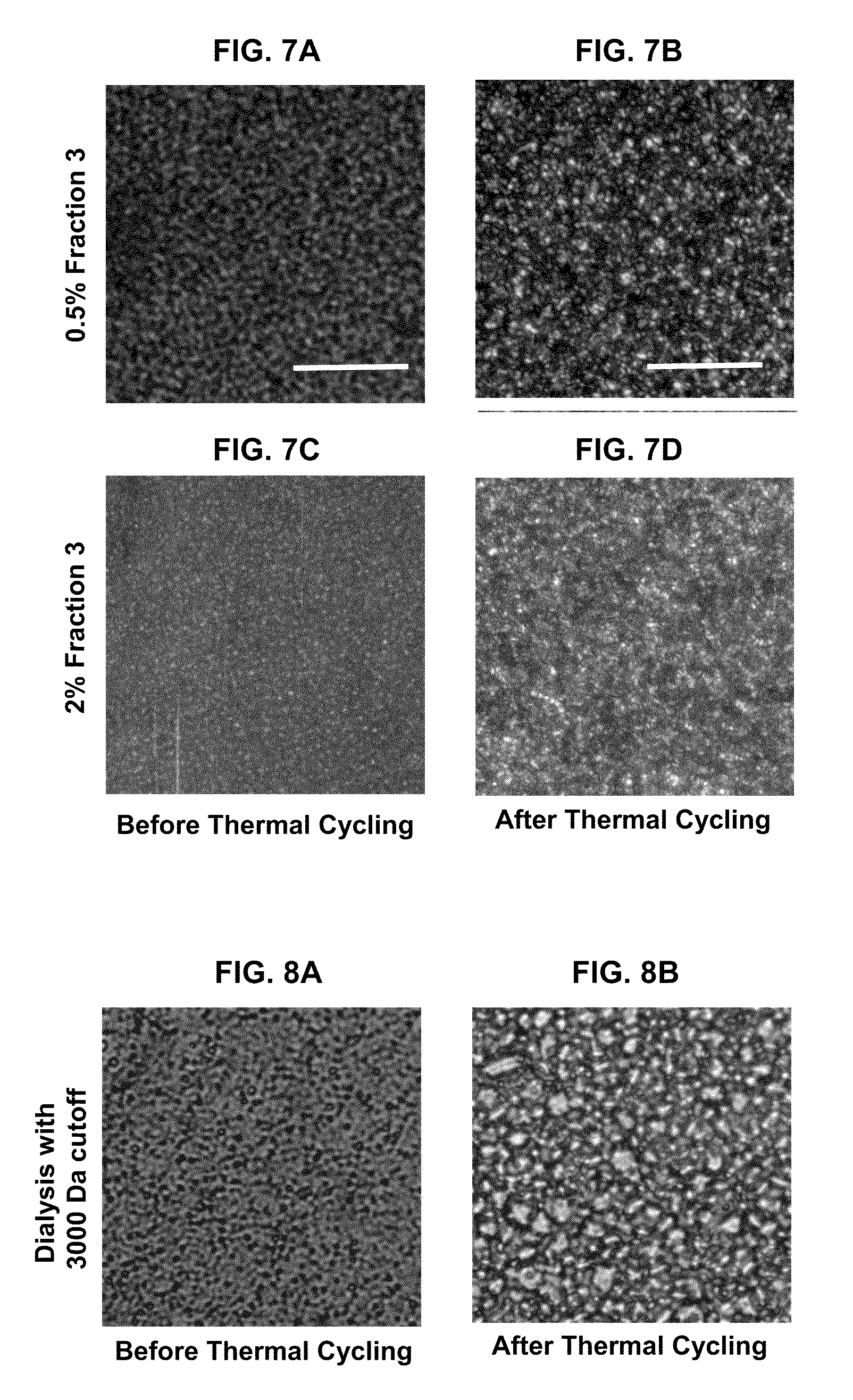
Patents
Literature
71 results about "Primary sequence" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
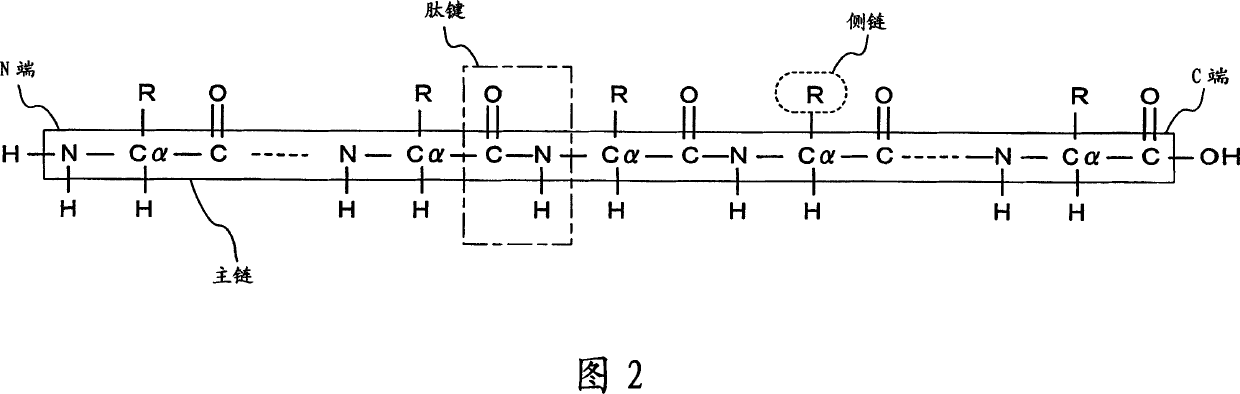
The primary structure (or sequence) of a peptide or protein is always written starting with the amino terminus on the left and progressing towards the carboxy terminus.
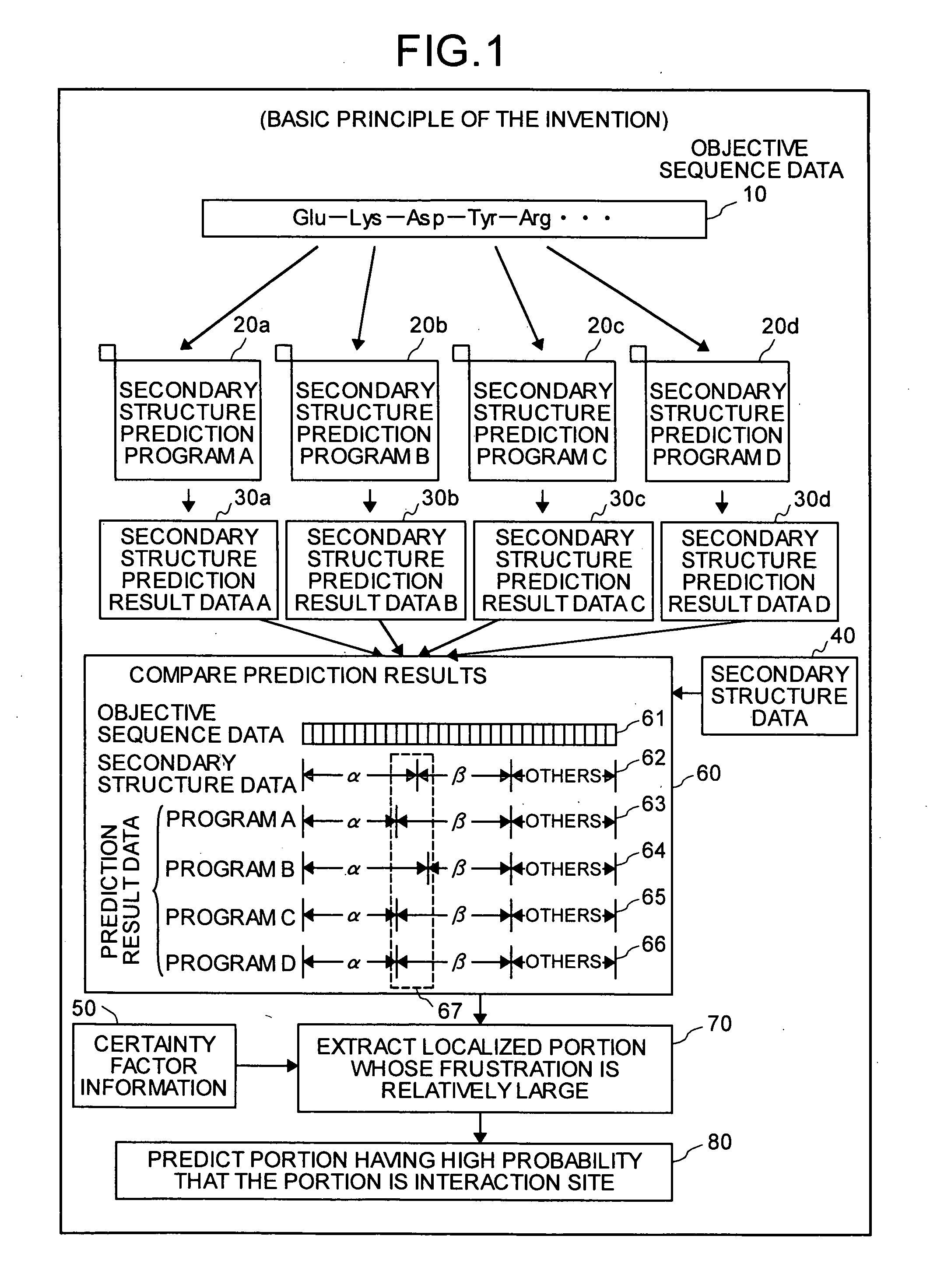
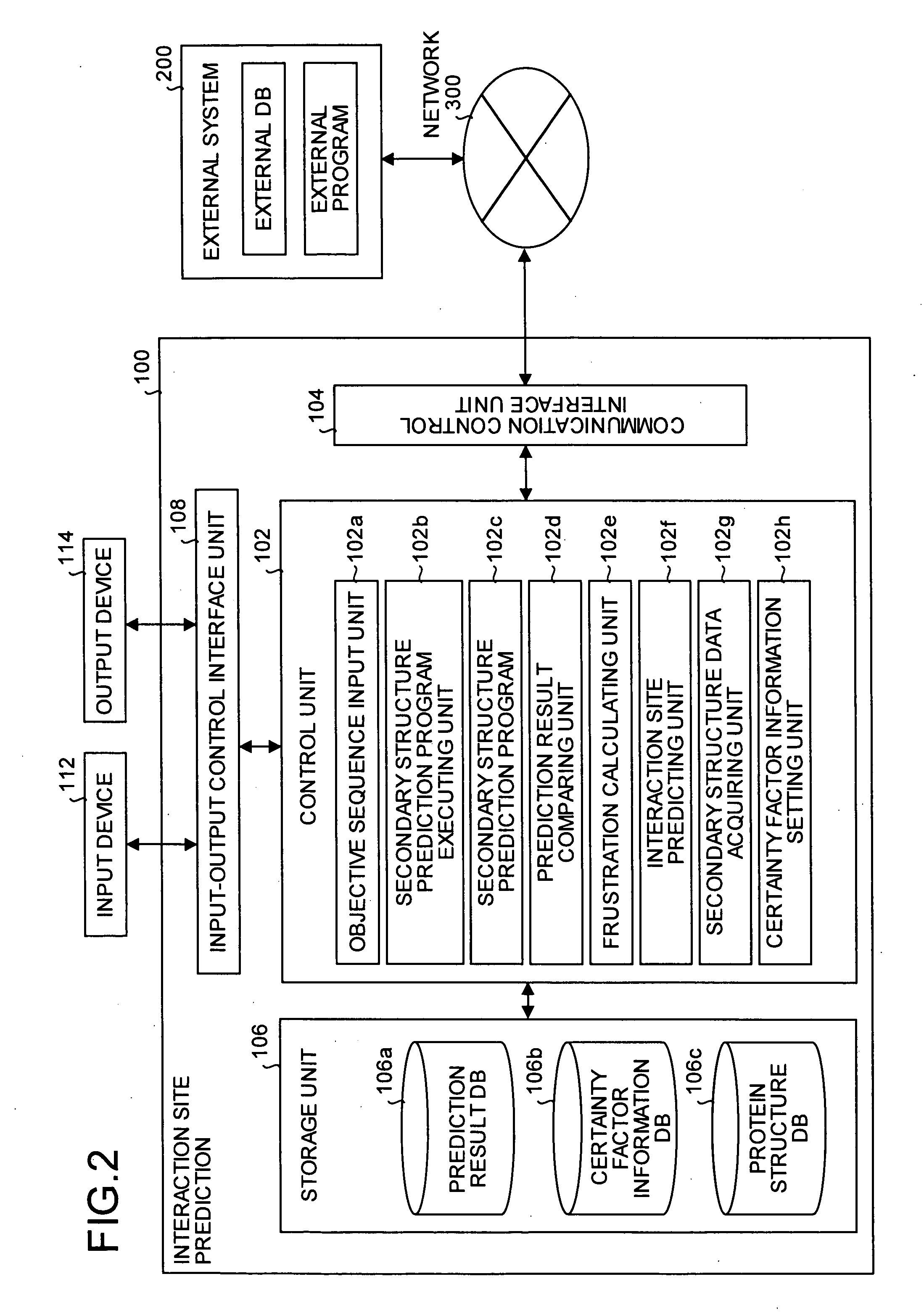
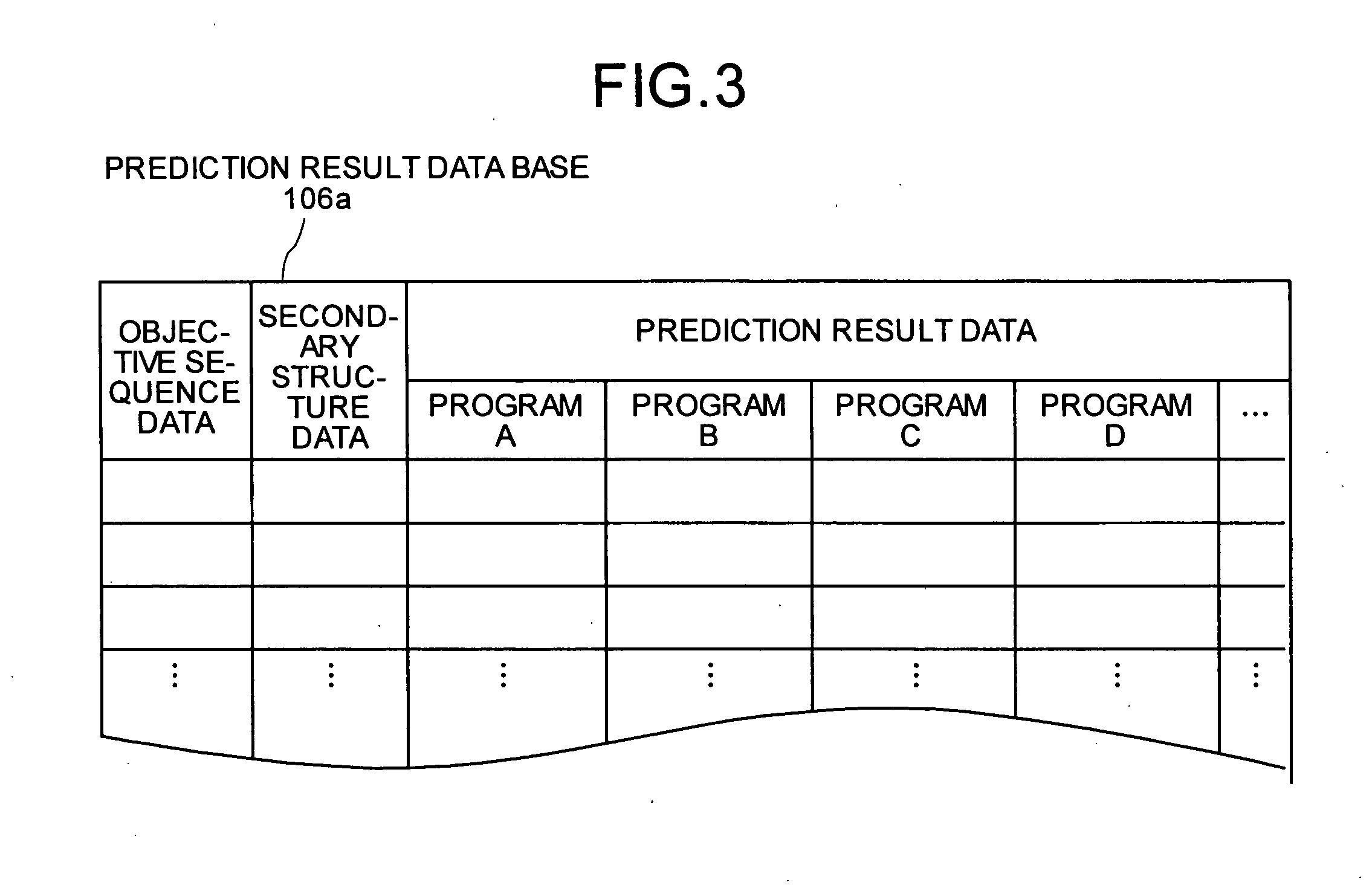
Interaction predicting device
InactiveUS20050130224A1Predict interactionAnalogue computers for chemical processesPeptide preparation methodsFrustrationInteraction site
Objective sequence data (10) which is primary sequence information on an objective protein is entered in an interaction site predicting device by the user. A secondary structure prediction simulation is executed on the objective sequence data (10) entered for secondary structure prediction programs (20a to 20d) that predict a secondary structure of a protein from primary sequence information of the protein. Results of secondary structure prediction (30a to 30d) from the respective secondary structure prediction programs (20a to 20d) are compared (60). Based on the comparison result, frustration of a local portion in the primary sequence information of the objective protein is calculated (70). An interaction site of the objective protein is predicted from the calculated frustration of the local portion (80).
Owner:CELESTAR LEXICO SCI
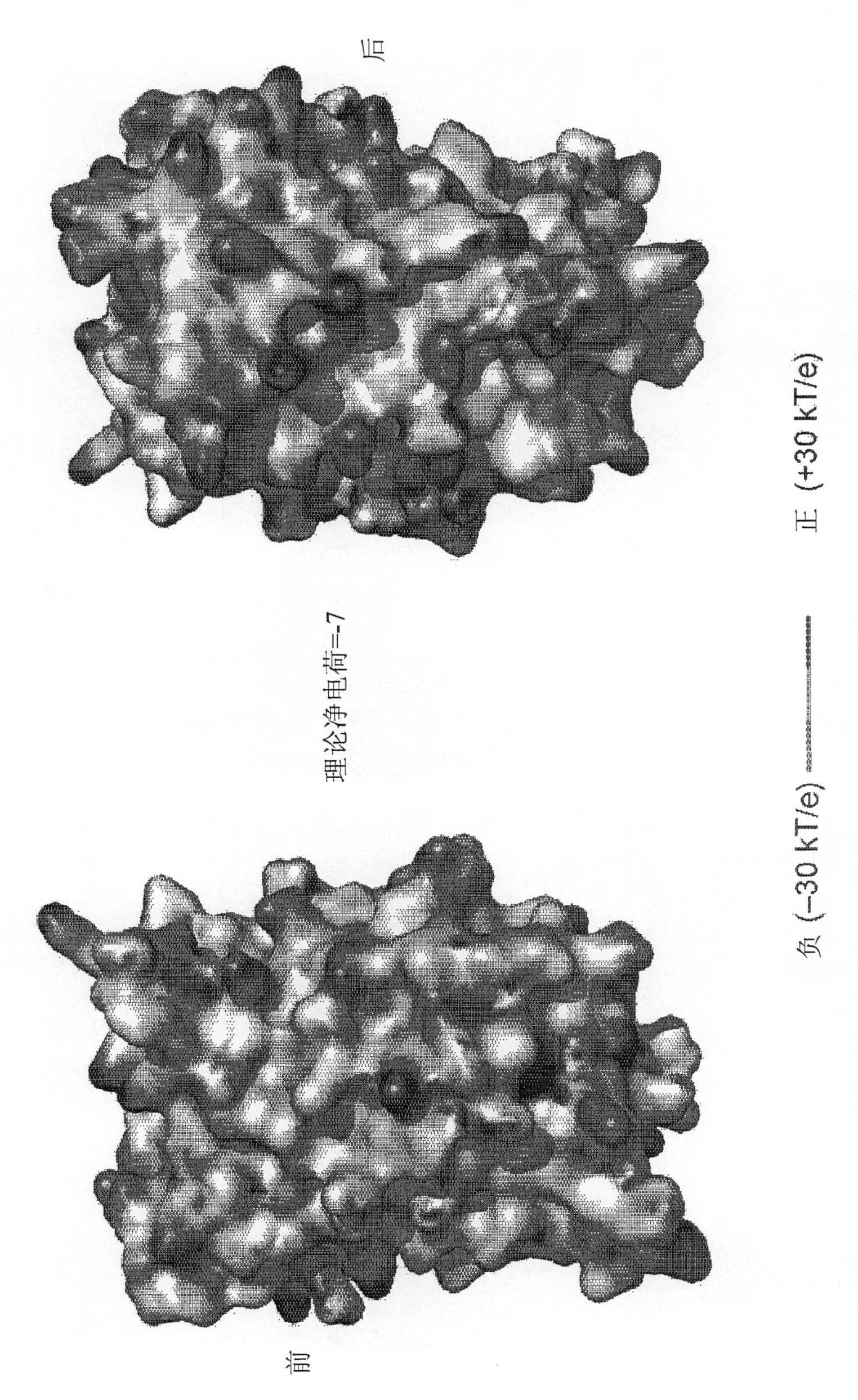
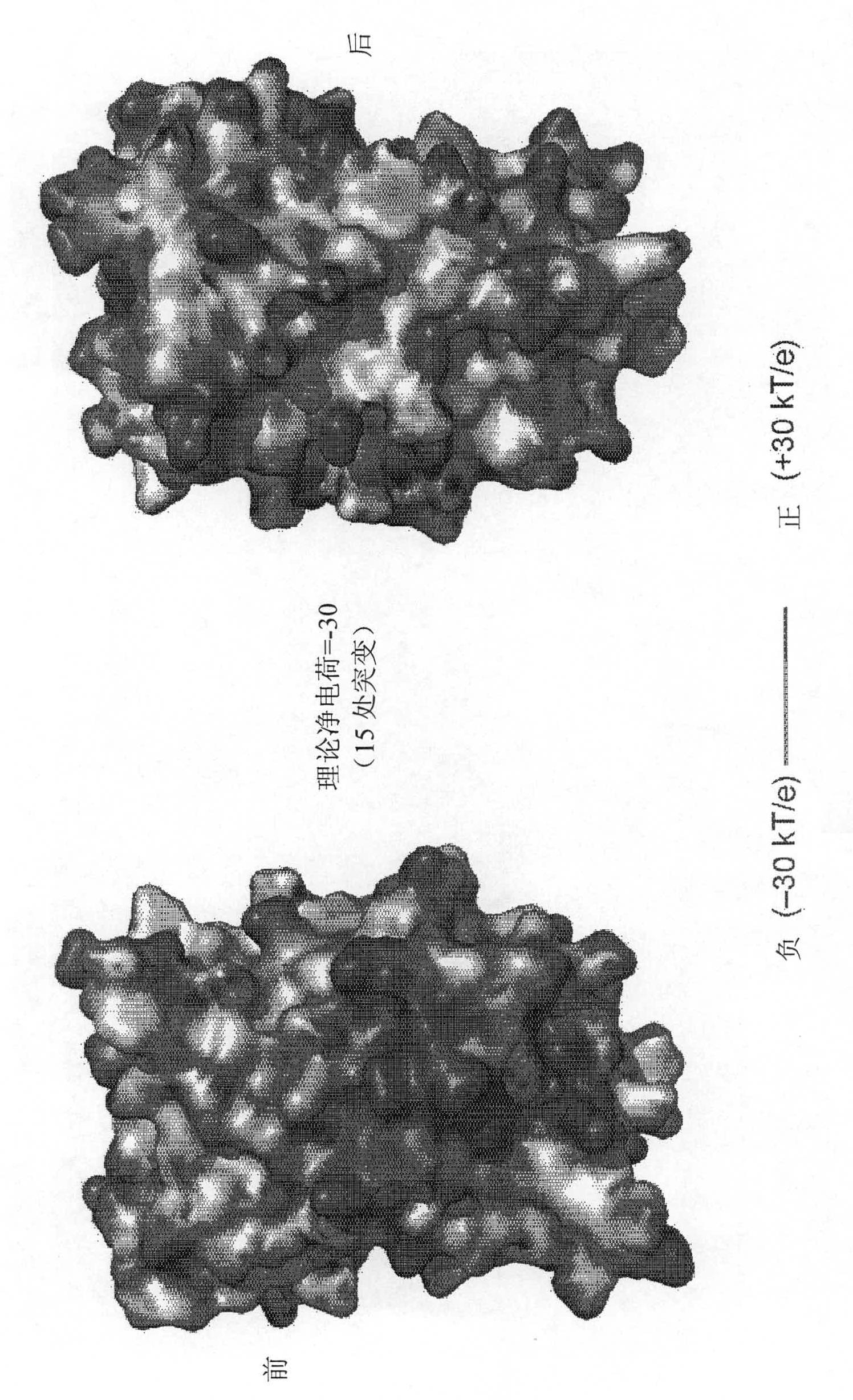
Supercharged proteins for cell penetration
InactiveUS20110112040A1Improved cell penetrationReduce biological activityMicrobiological testing/measurementSaccharide peptide ingredientsInfective disorderDisease cause
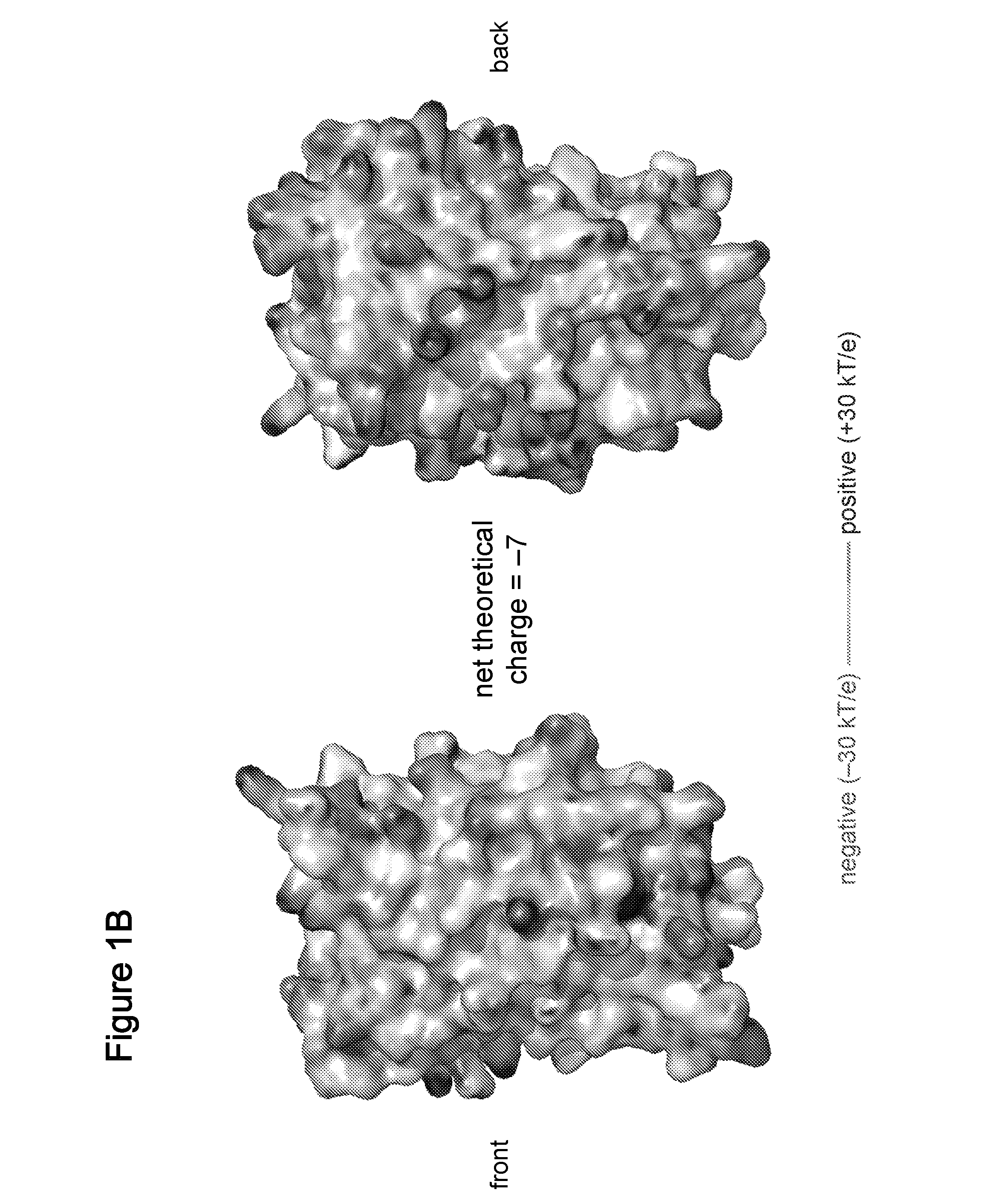
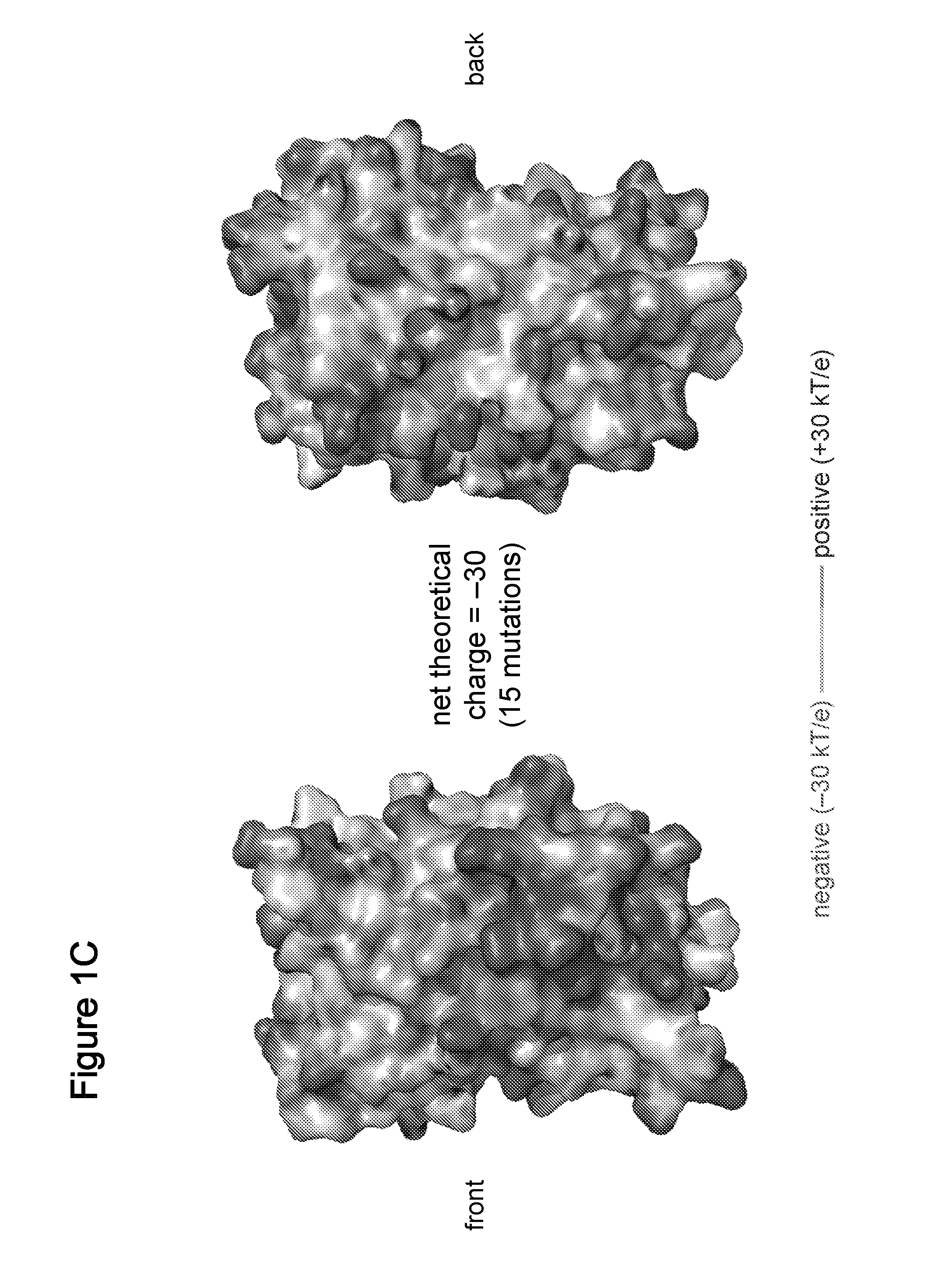
Compositions, systems and related methods for delivering a supercharged protein or a complex of a supercharged protein and therapeutic agent (e g, nucleic acid, peptide, small molecule) to cells are disclosed. Superpositively charged proteins may be associated with nucleic acids (which typically have a net negative charge) via electrostatic interactions. The systems and methods may involve altering the primary sequence of a protein in order to “supercharge” the protein (e g, to generate a superpositively-charged protein). The compositions may be used to treat proliferative diseases, infectious diseases, cardiovascular diseases, inborn errors in metabolism, genetic diseases, etc.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
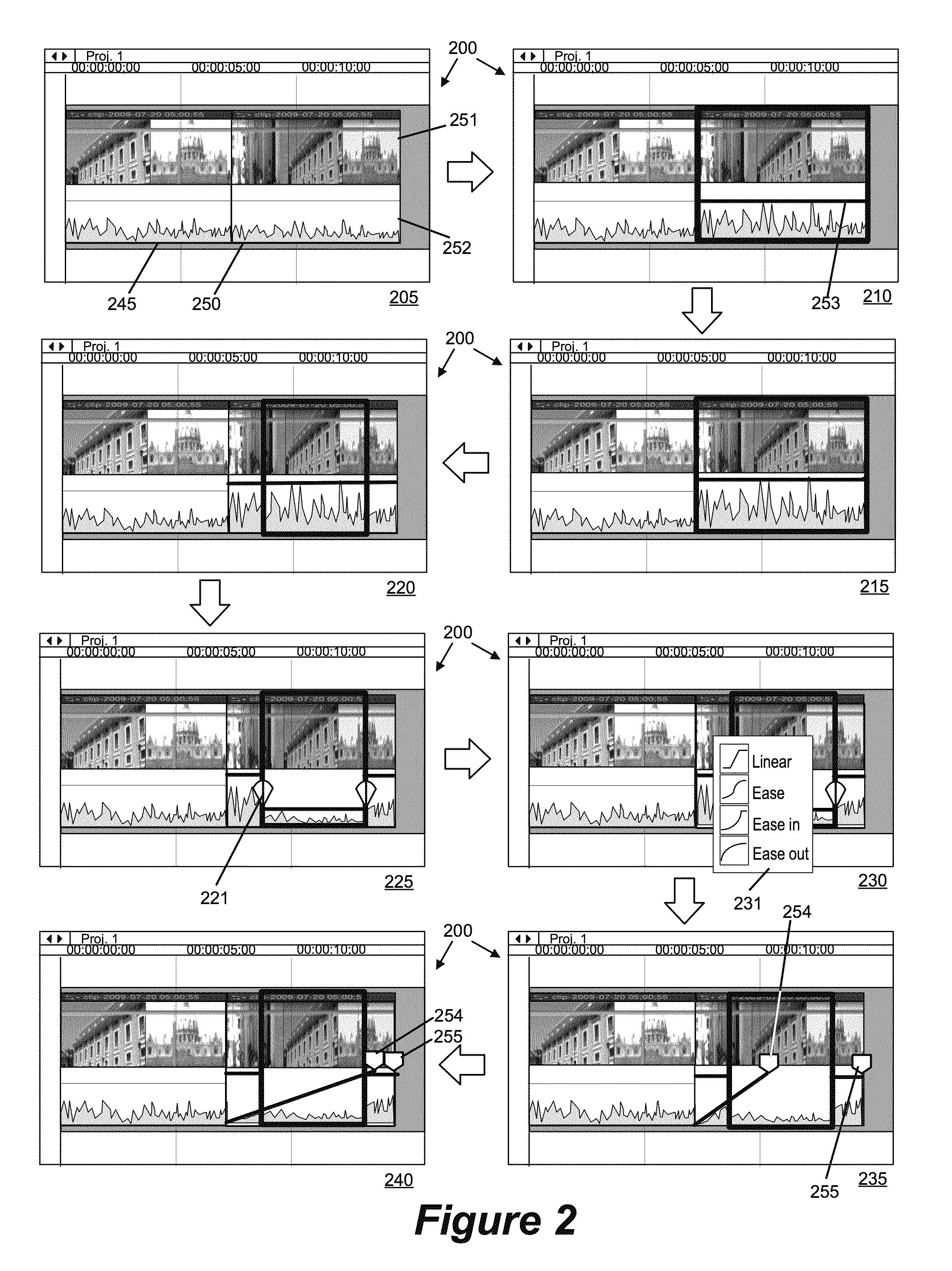
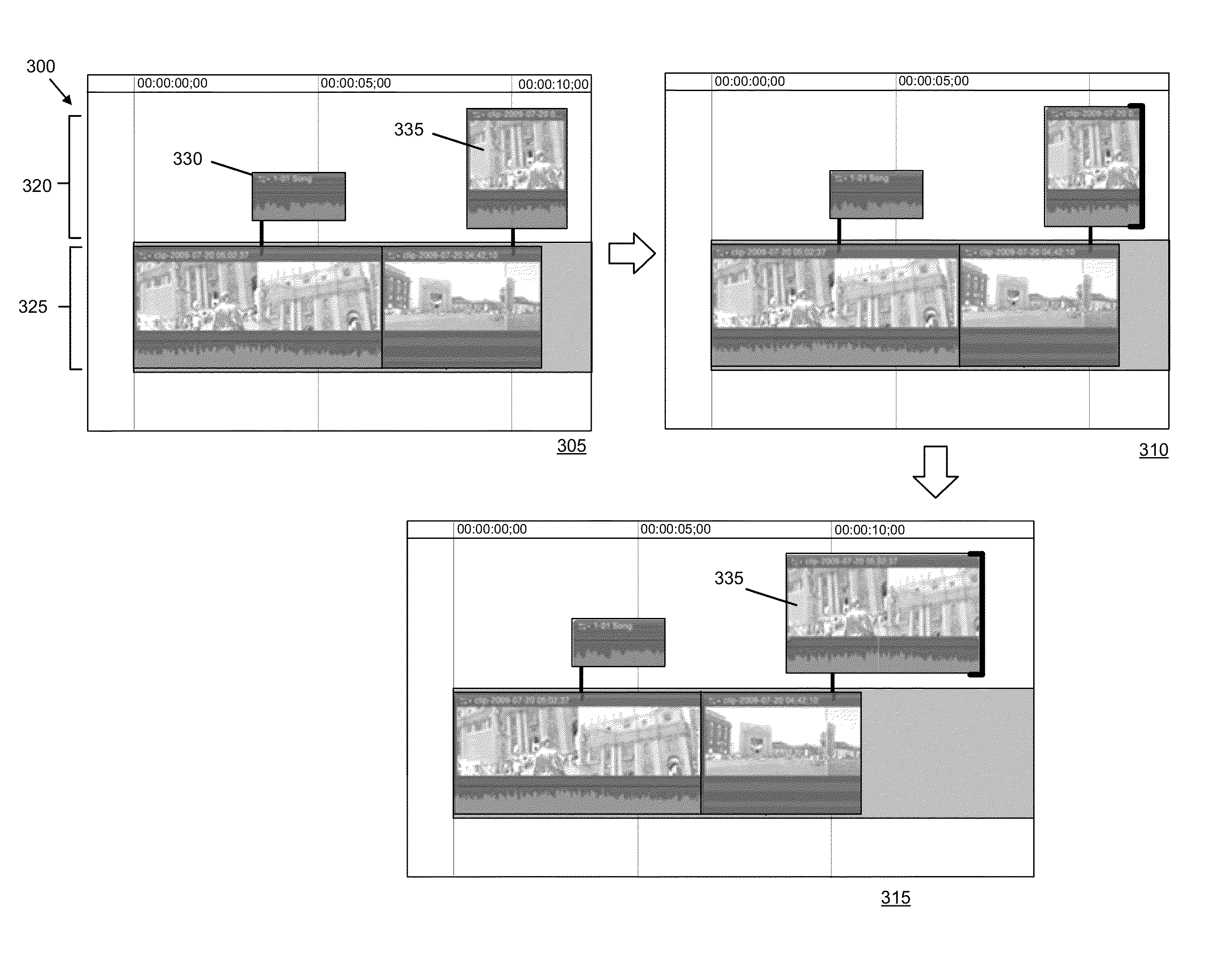
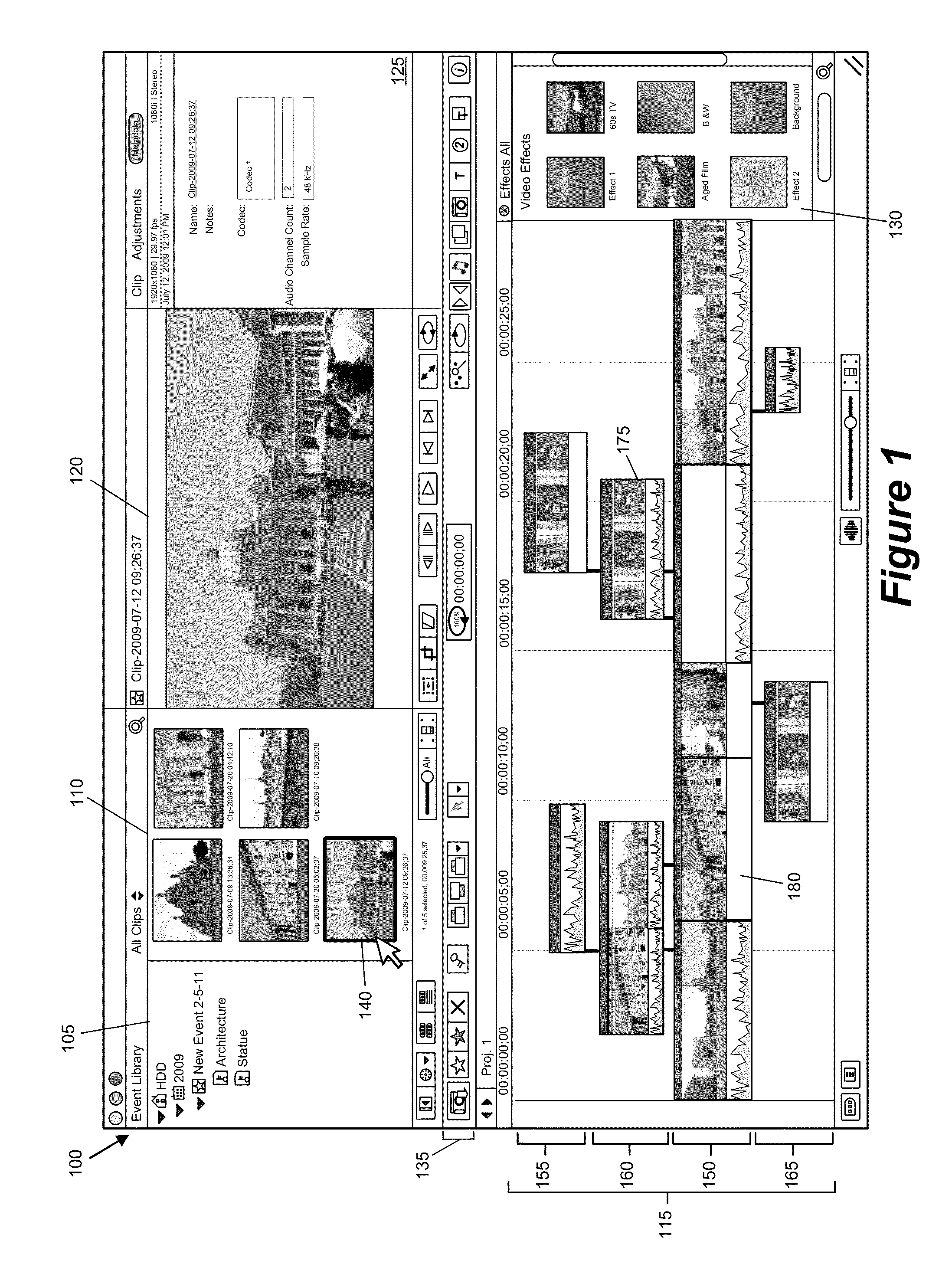
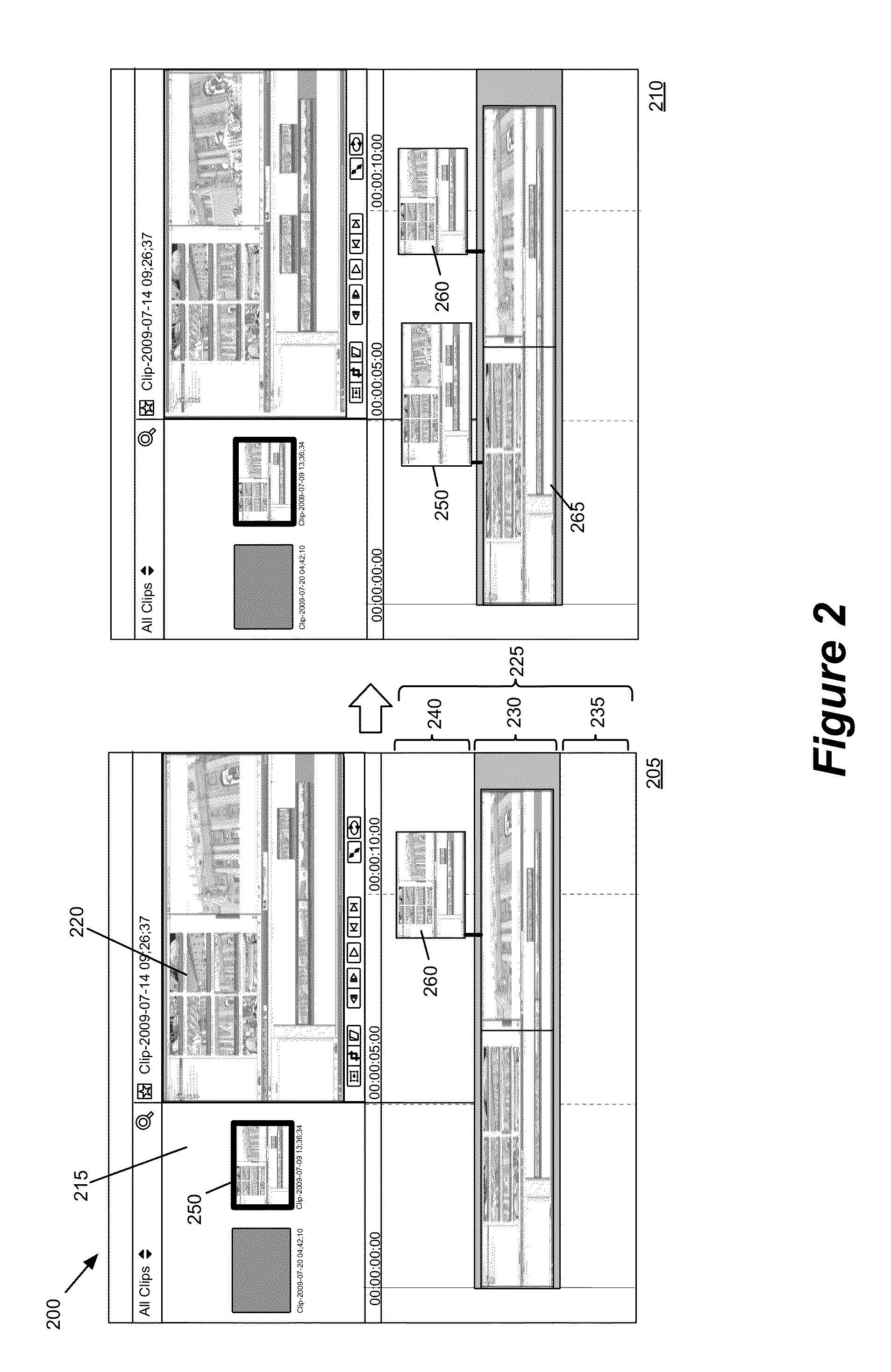
Media-Editing Application with Anchored Timeline
ActiveUS20120210230A1Direct accessDown playback speedTelevision system detailsElectronic editing digitised analogue information signalsComposite mediaComputer graphics (images)
Owner:APPLE INC
Engineered binding proteins
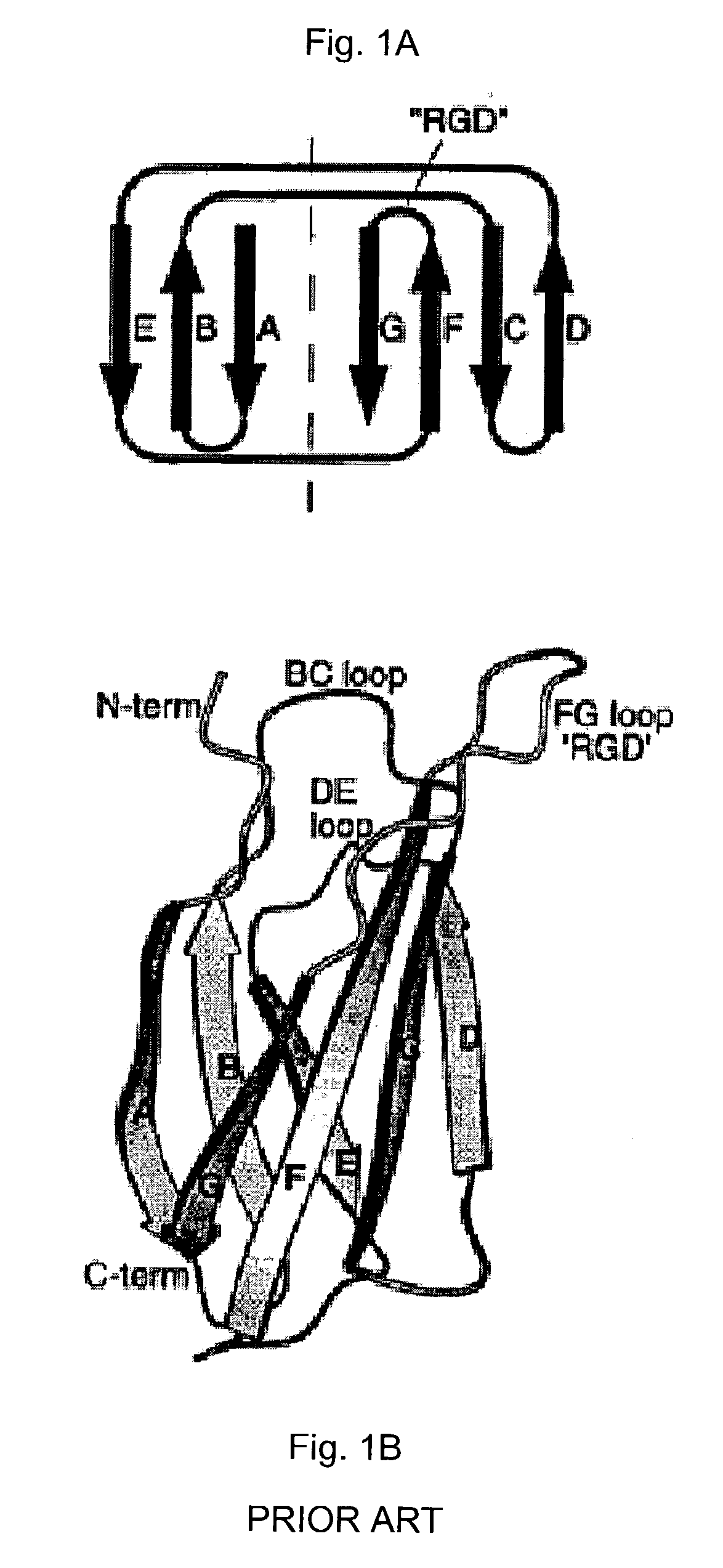
InactiveUS20040009530A1Unusual thermal stabilityMaterial nanotechnologyPeptide librariesADAMTS ProteinsRubredoxin
Engineered binding proteins are provided. In some cases, the parent protein corresponding to the engineered protein has a three-layer swiveling beta / beta / alpha domain. In other cases, the parent protein corresponding to the engineered protein has a rubredoxin-like fold. At least one portion of the primary sequence of the engineered protein is determined by an engineering scheme. In some case, the engineered protein is characterized by an ability to bind to a compound that the parent protein does not bind. In some cases, the parent protein is derived from a domain of a chaperonin or a rubredoxin. One form of engineering scheme used is a randomization scheme. A method for making libraries of engineered proteins, all based on a single parent protein is provided. Methods to identify proteins that bind to compounds of interest in libraries of engineered libraries is provided. An array of engineered proteins immobilized on a support is provided. Each engineered protein in the array is a chaperonin domain or a rubredoxin that has been subjected to an engineering scheme.
Owner:WILSON DAVID S +1
Hco32 and hco27 and related examples
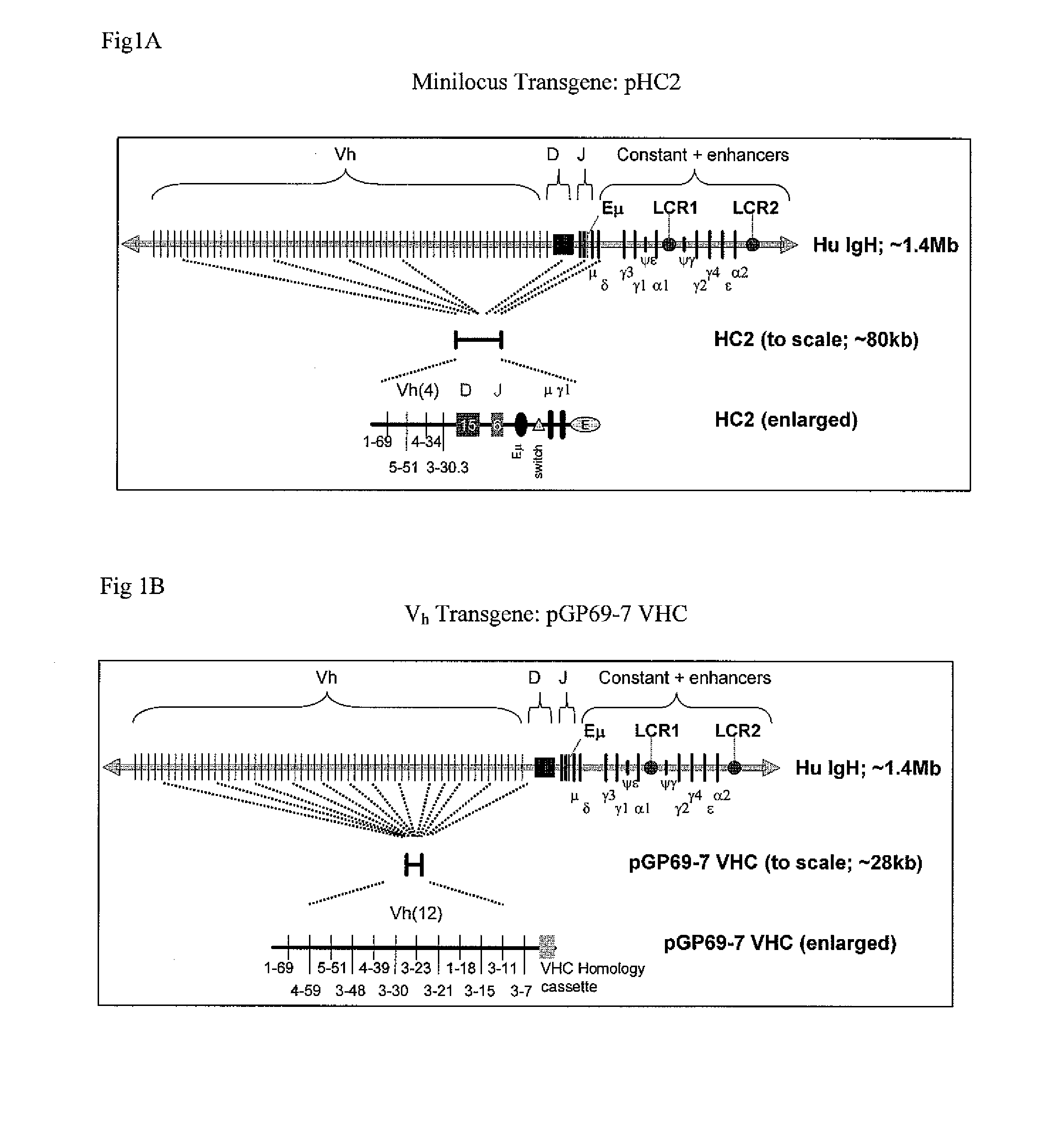
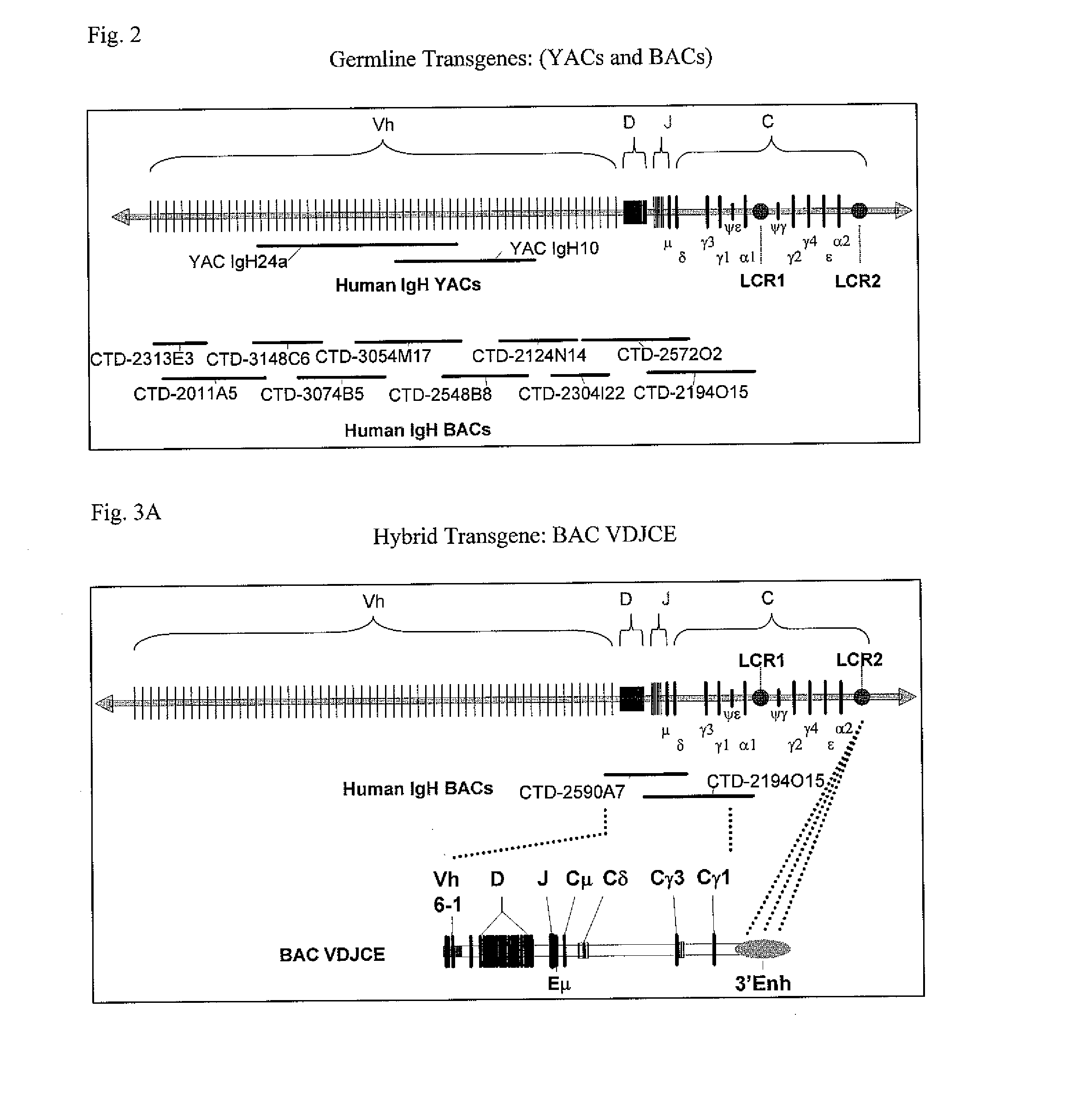
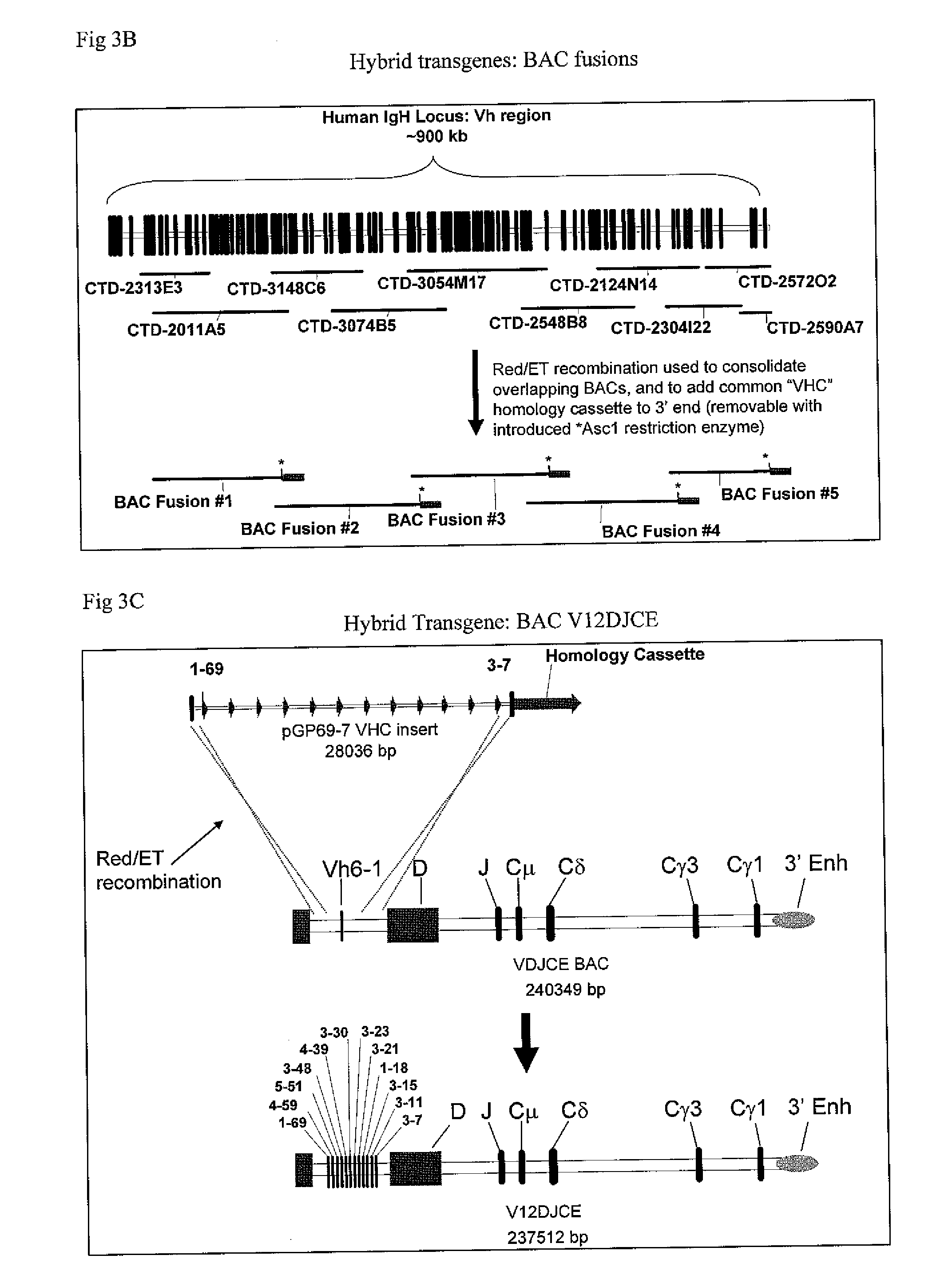
The instant invention relates to transgenic non-human animals capable of producing heterologous antibodies, transgenes used to produce such transgenic animals, transgenes capable of functionally rearranging a heterologous D gene in V-D-J recombination, immortalized B-cells capable of producing heterologous antibodies, methods and transgenes for producing heterologous antibodies of multiple isotypes, methods and transgenes for producing heterologous antibodies wherein a variable region sequence comprises somatic mutation as compared to germline rearranged variable region sequences, transgenic nonhuman animals which produce antibodies having a human primary sequence and which bind to human antigens, hybridomas made from B cells of such transgenic animals, and monoclonal antibodies expressed by such hybridomas.
Owner:ER SQUIBB & SONS INC
Techniques for Reducing a Cell Identification Falsing Rate in a Wireless Communication System
A technique of operating a wireless communication device includes selecting, from a primary sequence group that includes respective primary sequences, one of the respective primary sequences as a first portion of a cell identification (ID). In this case, the respective primary sequences are each associated with respective secondary sequence subgroups included in a secondary sequence group. Each of the respective secondary sequence subgroups include secondary sequences. One of the secondary sequences is selected (from one of the respective secondary sequence subgroups that is associated with the selected one of the respective primary sequences) for a second portion of the cell ID. At least some of the secondary sequences are only included in one of the respective secondary sequence subgroups. The first portion of the cell ID is encoded on a first downlink waveform that is to be transmitted and the second portion of the cell ID is encoded on a second downlink waveform that is to be transmitted. The first and second downlink waveforms are then transmitted.
Owner:APPLE INC
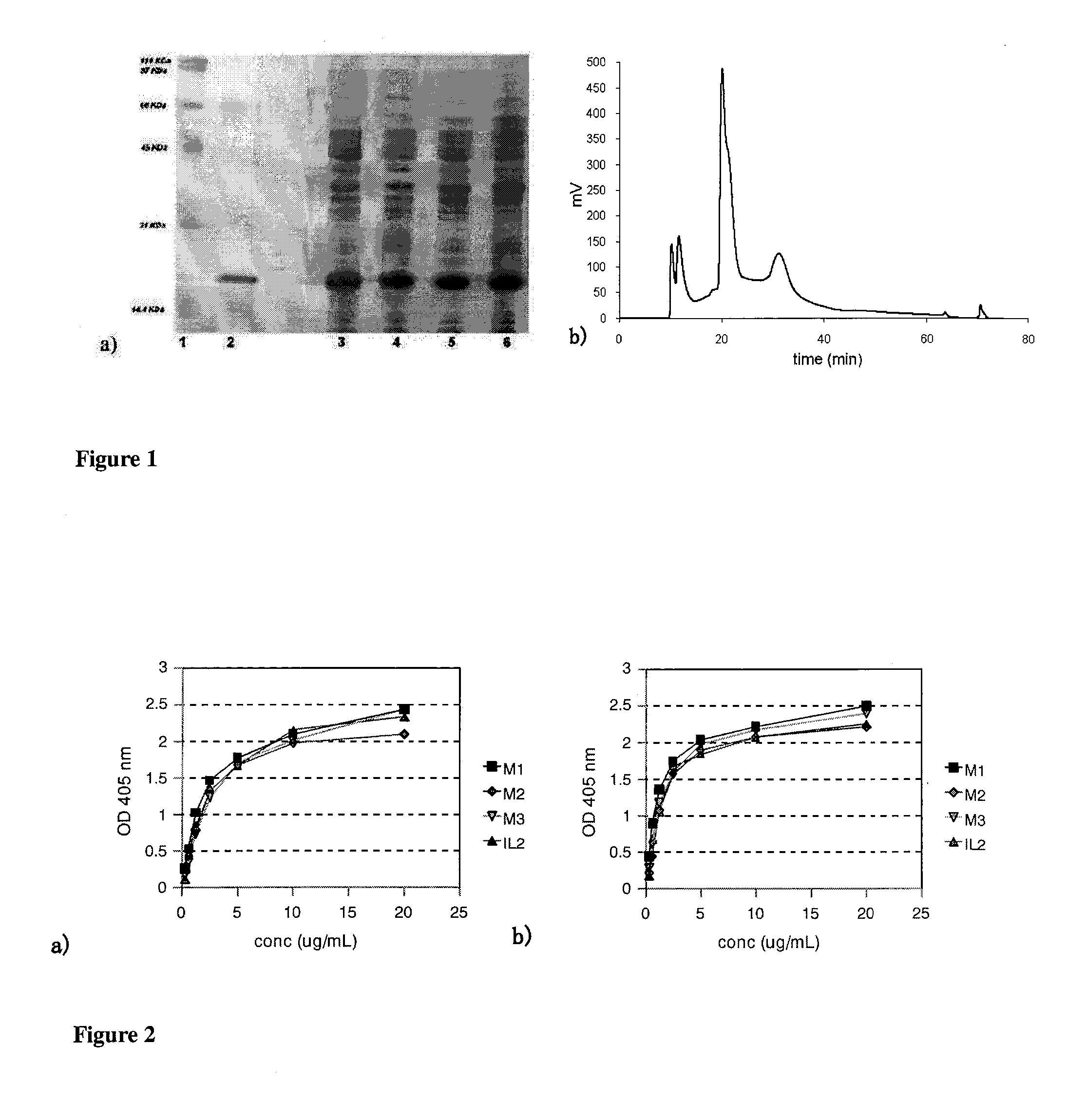
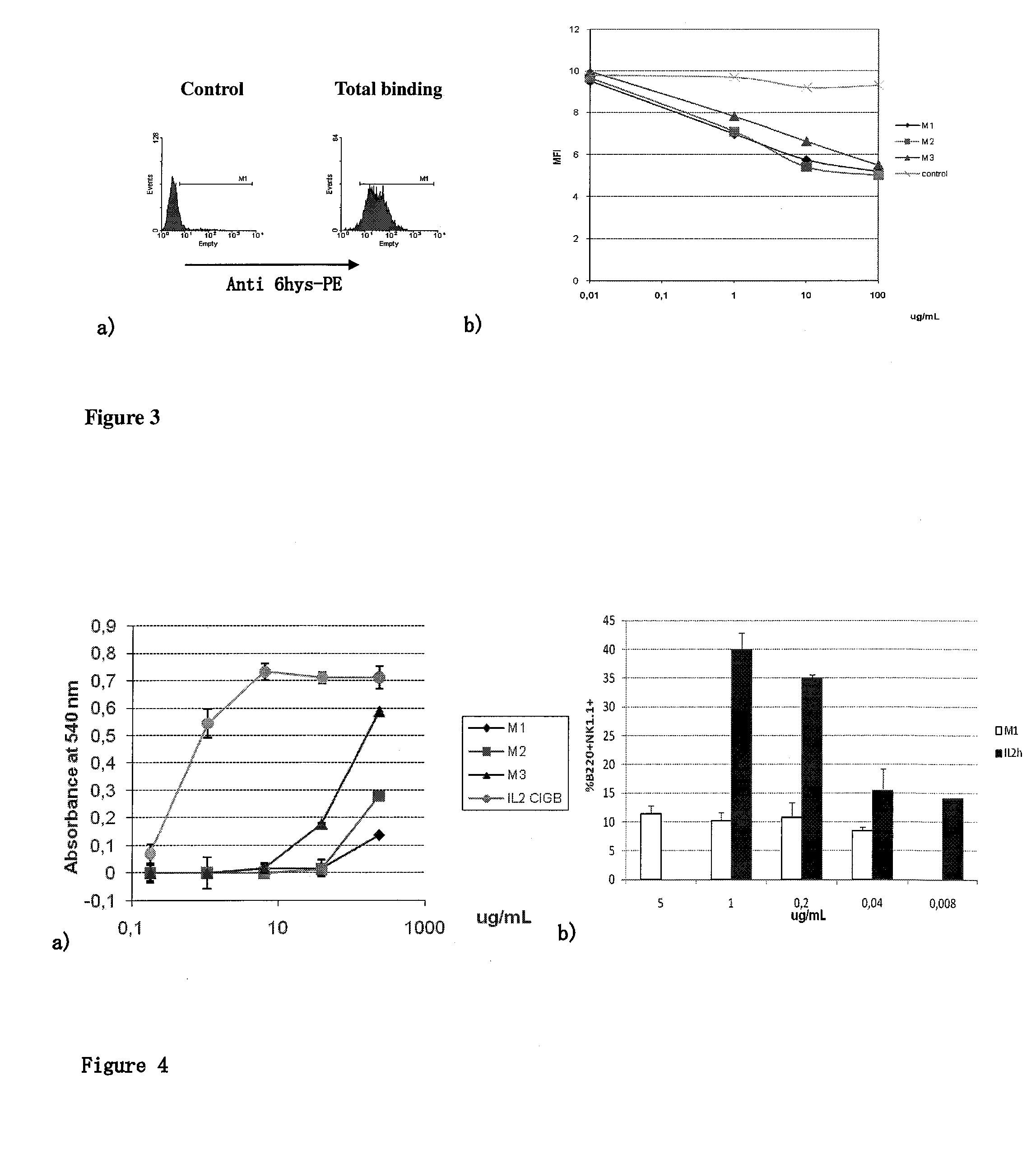
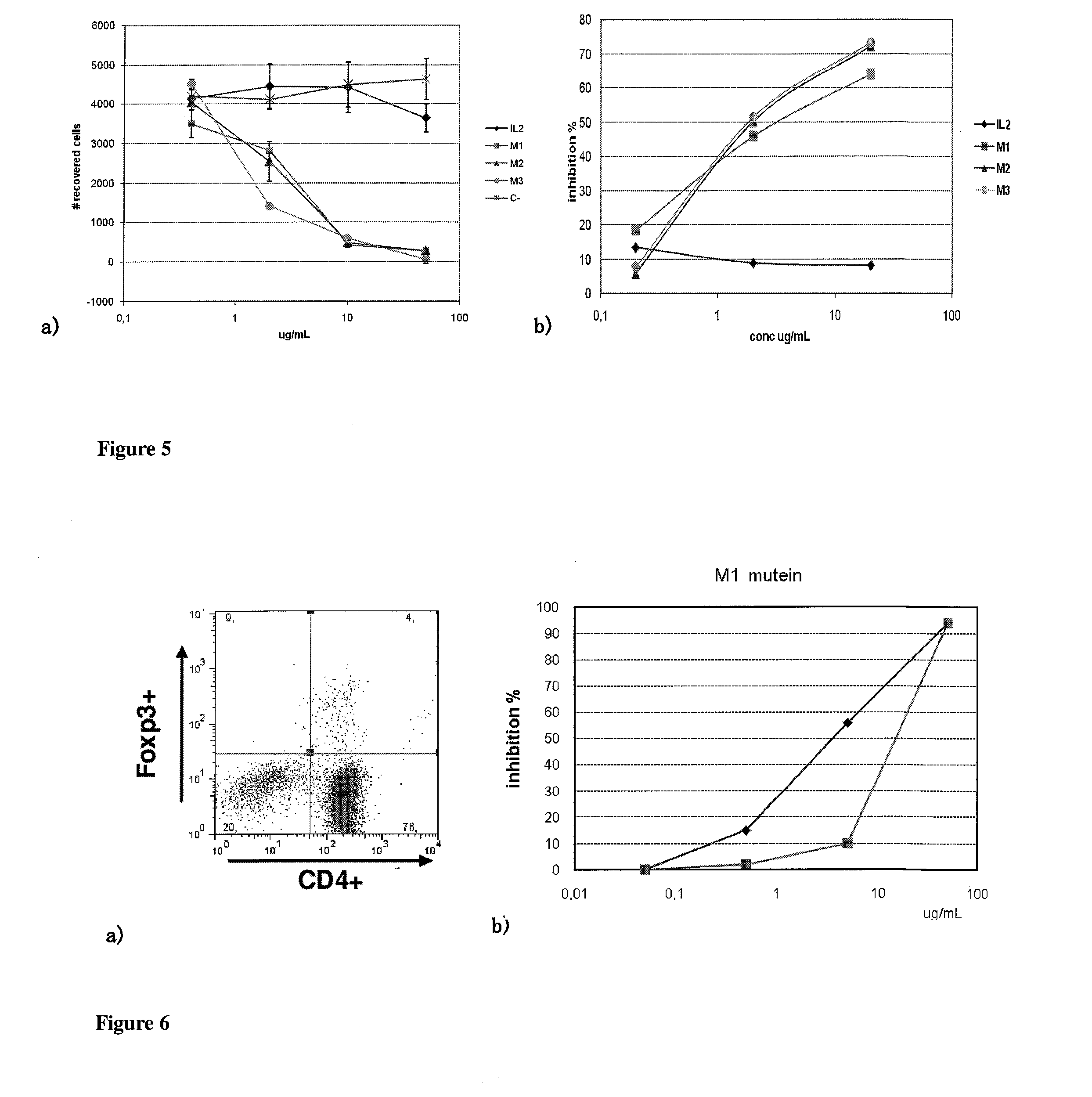
Immunomodulatory interleukin-2 polypeptides and methods of treating melanoma
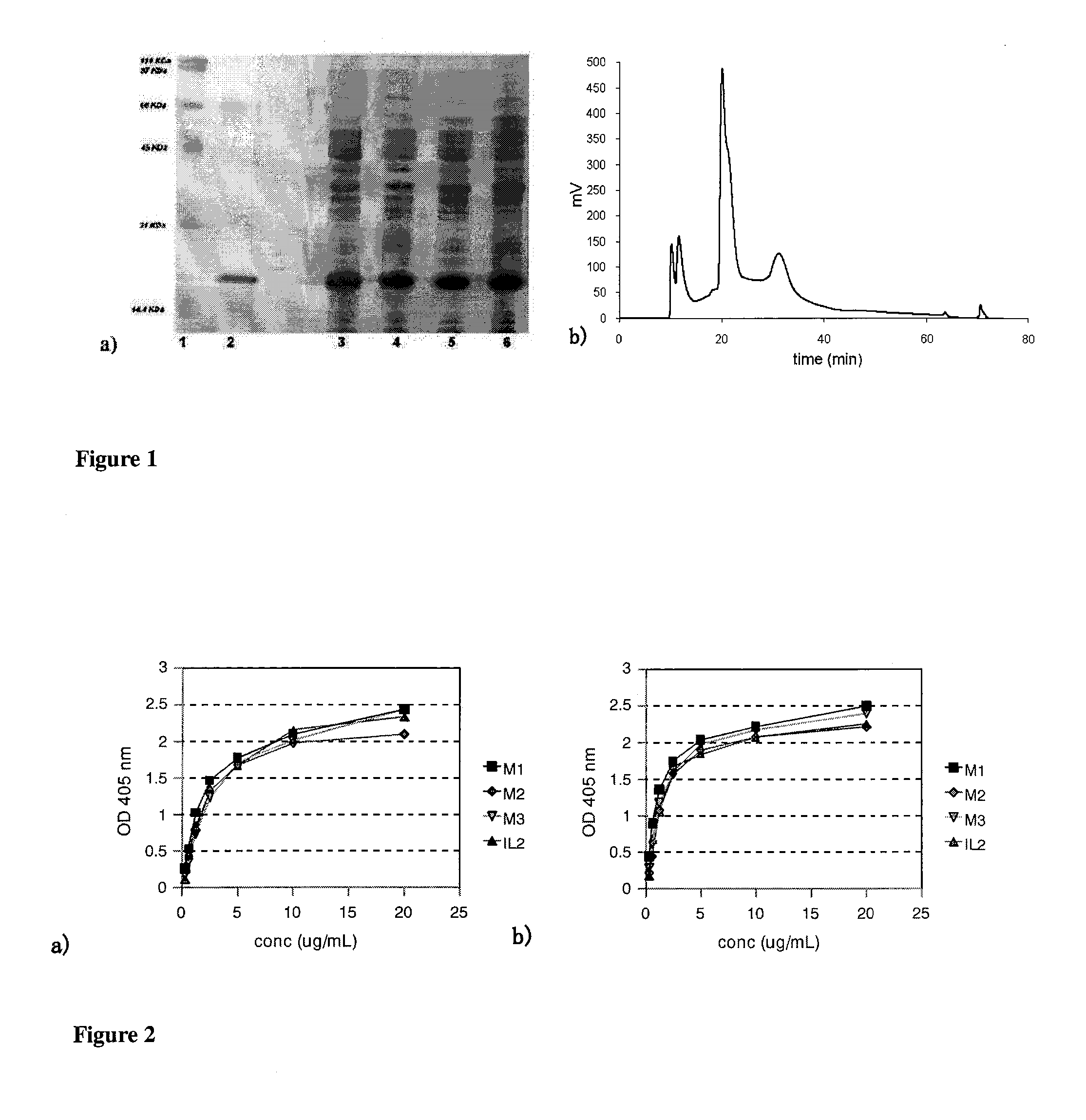
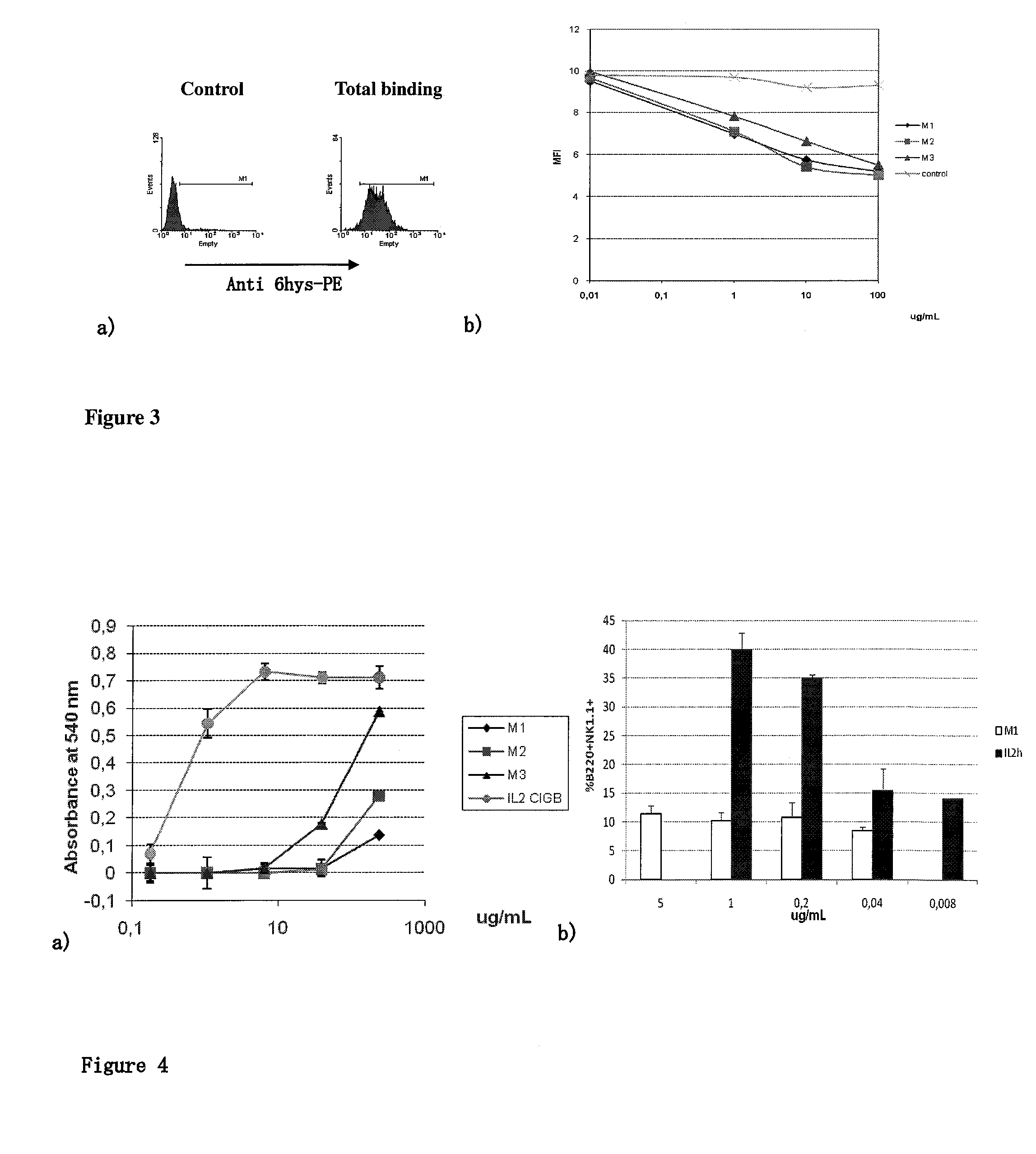
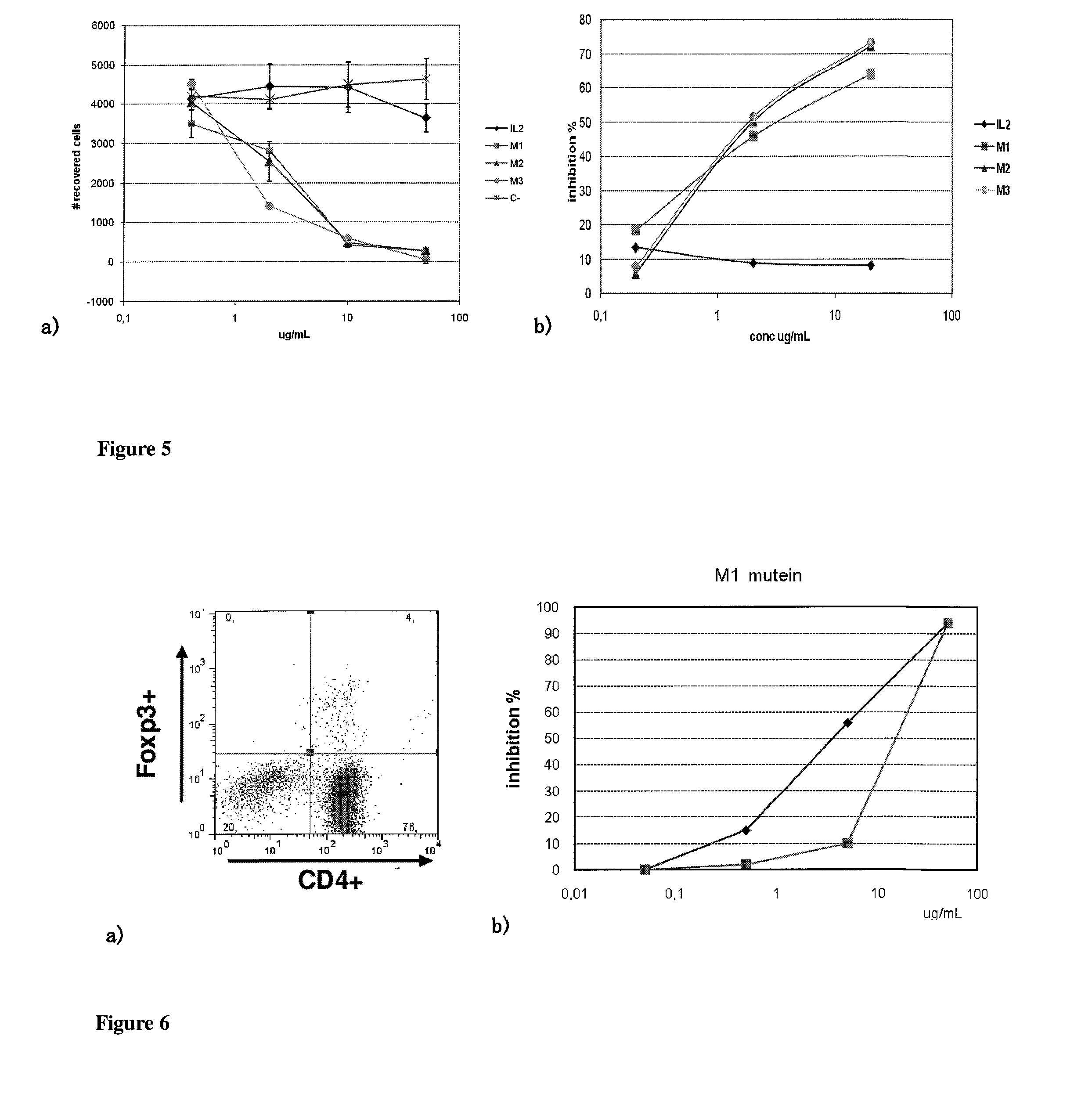
The present invention relates generally to polypeptides whose primary sequence has high sequence homology with human interleukin 2 (IL-2) with some punctual mutations in the sequence of native IL-2. The polypeptides of the present invention have an immunomodulatory effect on the immune system, which is selective / preferential on regulatory T cells. The present invention also relates to specific polypeptides whose amino acid sequence is disclosed herein. In another aspect the present invention relates to pharmaceutical compositions comprising as active ingredient the polypeptides disclosed. Finally, the present invention relates to the therapeutic use of the polypeptides and pharmaceutical compositions disclosed due to their immune modulating effect on diseases such as cancer and chronic infectious diseases.
Owner:CENT DE INMUNOLOGIA MOLECULAR CENT DE INMUNOLO
Bone polypeptide-1
The present invention relates to a bone polypeptide, particularly bone polypeptide-1 and nucleic acid molecules encoding the same. The present invention provides the human primary sequence of bone polypeptide-1 as well as sequences of vertebrate homologs. The invention also provides cell lines engineered to express the cDNA, antibodies to detect its translation products, and recombinant adenoviruses to deliver an expressible cDNA into a host. Bone polypeptide-1 can be utilized to treat diseases affecting renal and bone functions.
Owner:ALEXION PHARMA INC
Polypeptides having a functional domain of interest and methods of identifying and using same
InactiveUS6309820B1Specificity is easedEffect is exertedCompound screeningApoptosis detectionPrimary sequenceDNA
Novel polypeptides having functional domains of interest are described, along with DNA sequences that encode the same. A method of identifying these polypeptides by means of a sequence-independent (that is, independent of the primary sequence of the polypeptide sought), recognition unit-based functional screen is also disclosed. Various applications of the method and of the polypeptides identified are described, including their use in assay kits for drug discovery, modification, and refinement.
Owner:CYTOGEN CORP +1
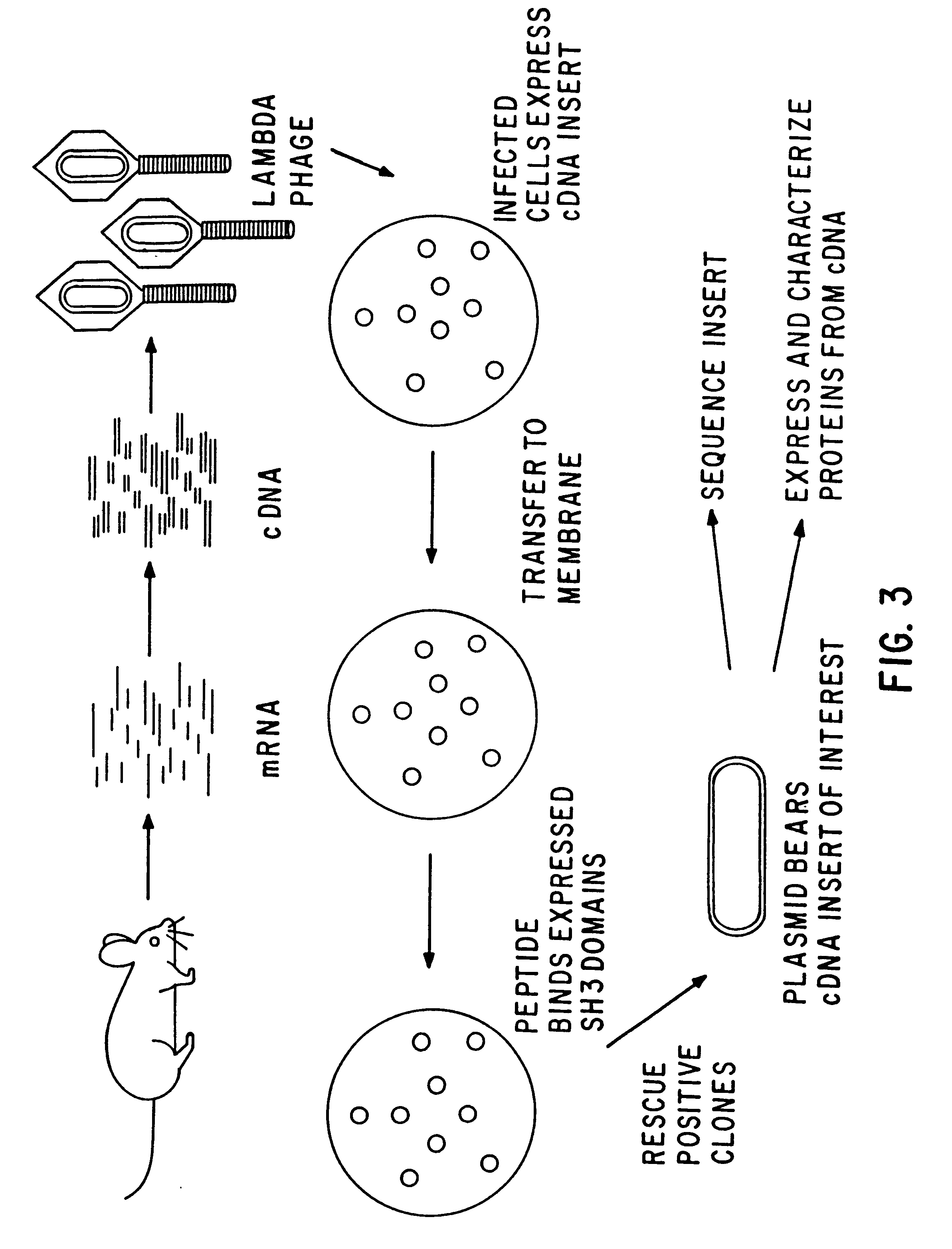
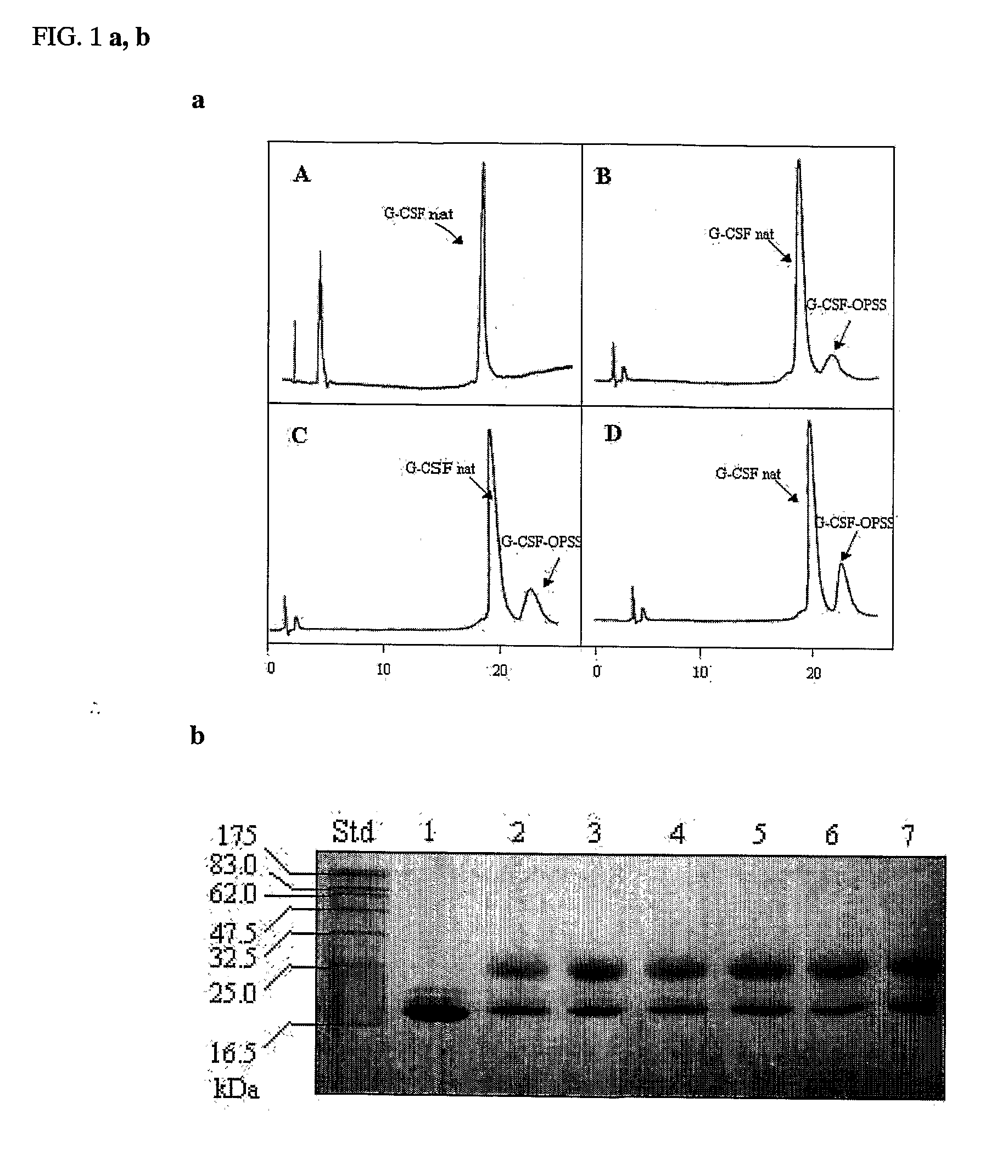
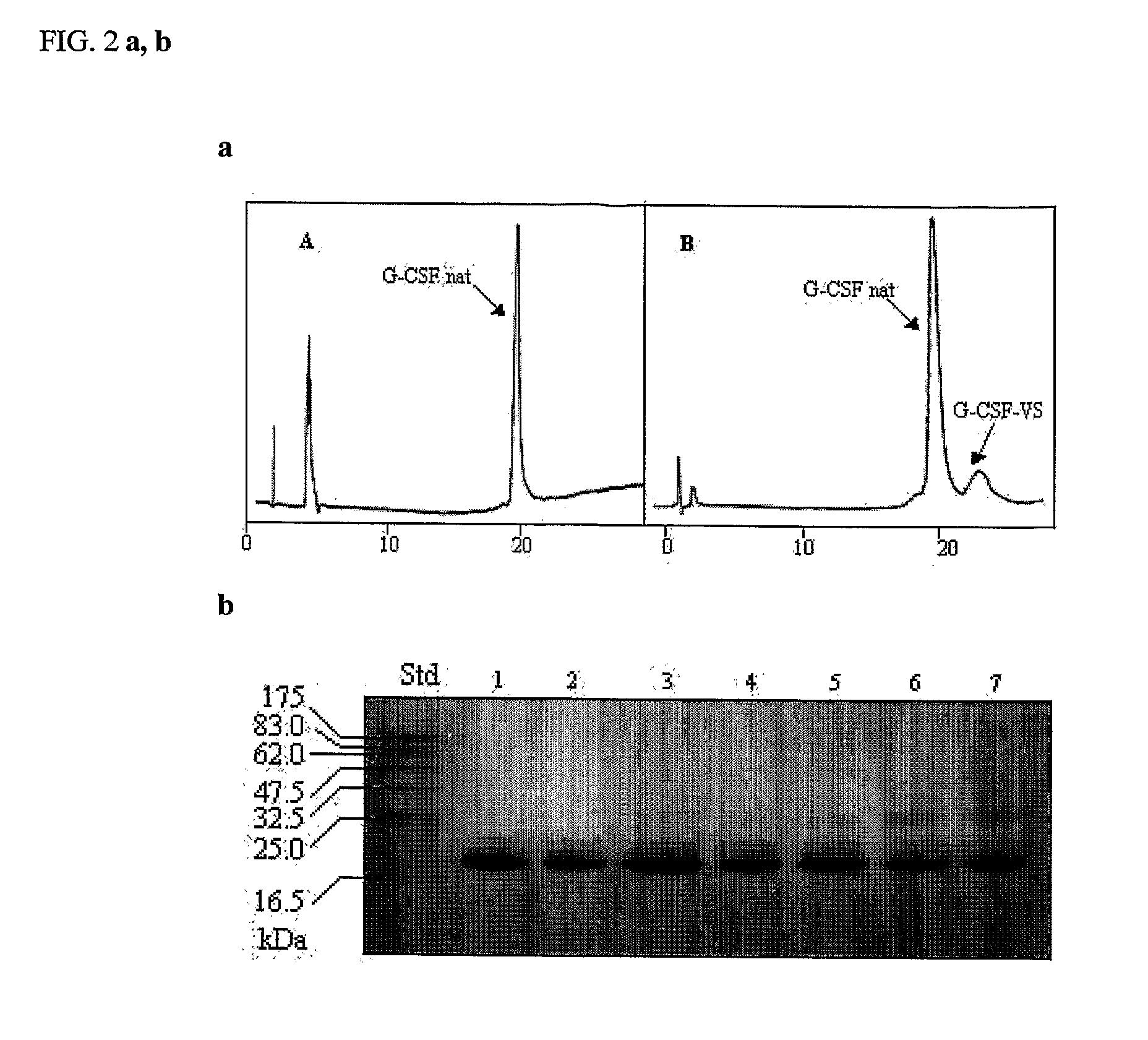
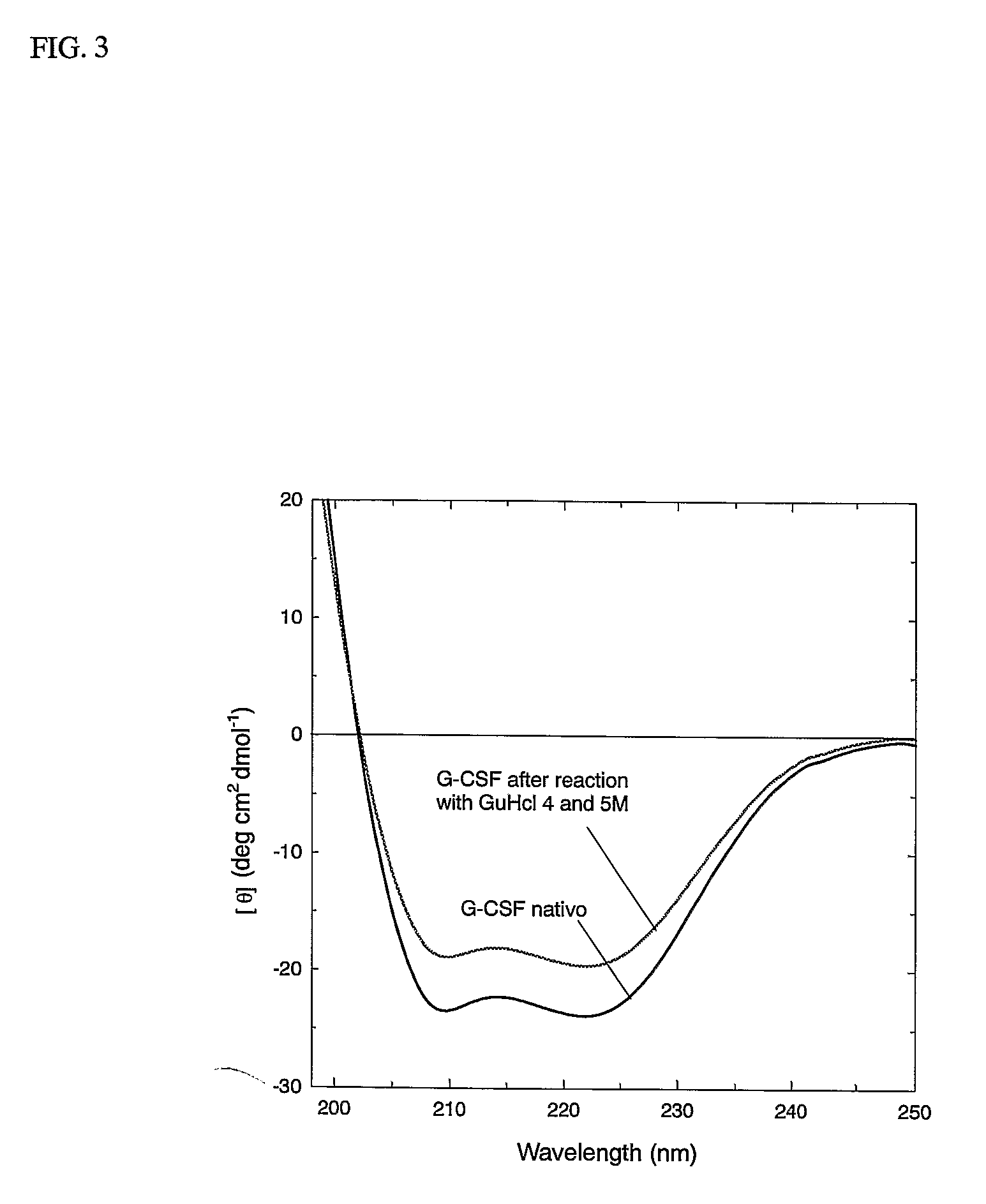
Novel g-csf conjugates
InactiveUS20070166278A1Simplified mixtureSelectivity in conjugationPeptide/protein ingredientsColony-stimulating factorThiolCysteine thiolate
The present application relates to novel PEG-G-CSF conjugates in which a PEG molecule is linked to the cyteine residue in position 17 of native G-CSF primary sequence or to the cysteine residue of the corresponding position of a G-CSF analogue. The present application also describes a process for the manufacture of such conjugates, such process comprising the following steps: (i) subjecting the G-CSF protein to conditions inducing reversible denaturation of the protein, (ii) conjugation of the denatured protein obtained is step i with a thiol-reactivc PEG under denaturing conditions, (iii) subjecting the conjugates obtained in step ii to conditions promoting renaturation of the conjugate yielding biologically active G-CSF-PEG conjugate.
Owner:NEKTAR THERAPEUTICS INC
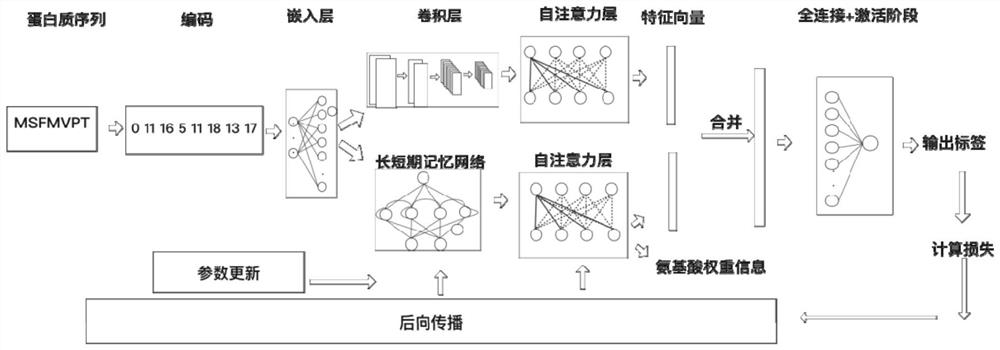
DNA binding protein identification and function annotation deep learning method based on self-attention mechanism
The invention discloses a DNA binding protein identification and function annotation deep learning method based on a self-attention mechanism. The method comprises the following steps: selecting a data set from a protein database, training and testing a constructed deep learning model by using the selected data set, and predicting whether protein can be combined with DNA by using the trained deeplearning model, wherein the deep learning model comprises a coding layer, an embedding layer, a long short-term memory neural network layer (LSTM), a convolutional neural network layer (CNN) and a self-attention layer (Self-Attention). Through a deep learning model based on a self-attention mechanism, whether the protein has a function of combining with DNA or not can be predicted according to primary sequence information of the protein, and an area with a combining function is found out.
Owner:TIANJIN UNIV
Anchor override for a media-editing application with an anchored timeline
ActiveUS8966367B2Electronic editing digitised analogue information signalsCarrier indicating arrangementsGraphicsGraphical user interface
Owner:APPLE INC
Supercharged proteins for cell penetration
Compositions, systems and related methods for delivering a supercharged protein or a complex of a supercharged protein and therapeutic agent (e g, nucleic acid, peptide, small molecule) to cells are disclosed. Superpositively charged proteins may be associated with nucleic acids (which typically have a net negative charge) via electrostatic interactions. The systems and methods may involve altering the primary sequence of a protein in order to ''supercharge'' the protein (e g, to generate a superpositively-charged protein). The compositions may be used to treat proliferative diseases, infectious diseases, cardiovascular diseases, inborn errors in metabolism, genetic diseases, etc.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
System and method for resource reallocation based on ambient condition data
Owner:GM GLOBAL TECH OPERATIONS LLC
Application of scolopendra mutilans neurotoxin peptide omega-SLPTX-Ssmla
InactiveCN102671185AEnhancement effect is goodNervous disorderPeptide/protein ingredientsDiseaseSequence signal
The invention relates to application of scolopendra mutilans neurotoxin peptide omega-SLPTX-Ssmla, belonging to the field of biomedicine. A primary sequence of the omega-SLPTX-Ssmla comprises 83 amino acid residues, wherein three pairs of disulfide bonds are formed by six aminothiopropionic acids, and the molecular weight is 8811.3 Da. The code of the neurotoxin gene comprises 426 nucleotides of which include signal peptides, premise peptides and mature peptides. The scolopendra mutilans neurotoxin omega-SLPTX-Ssml has a quite obvious enhancement effect on calcium ion channel current on a dorsal ganglion root and is a novel calcium channel agonist. The scolopendra mutilans neurotoxin omega-SLPTX-Ssm provided by the invention can be used for increasing the openness of the calcium channel to enable calcium ions to flow internally and preparing medicaments for treating calcium channel diseases and can also be used as a new tool reagent for calcium channel researches.
Owner:KUNMING INST OF ZOOLOGY CHINESE ACAD OF SCI
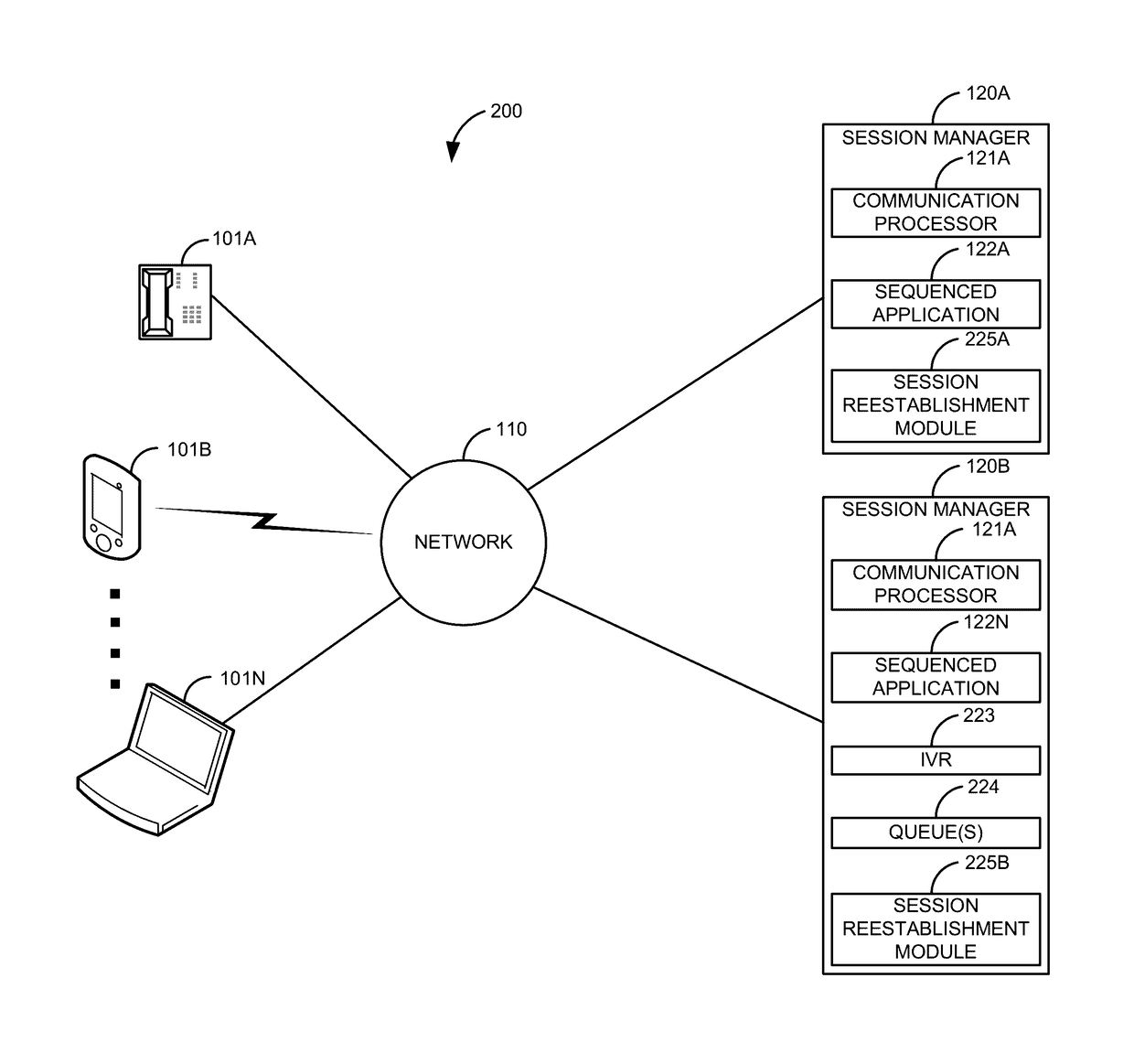
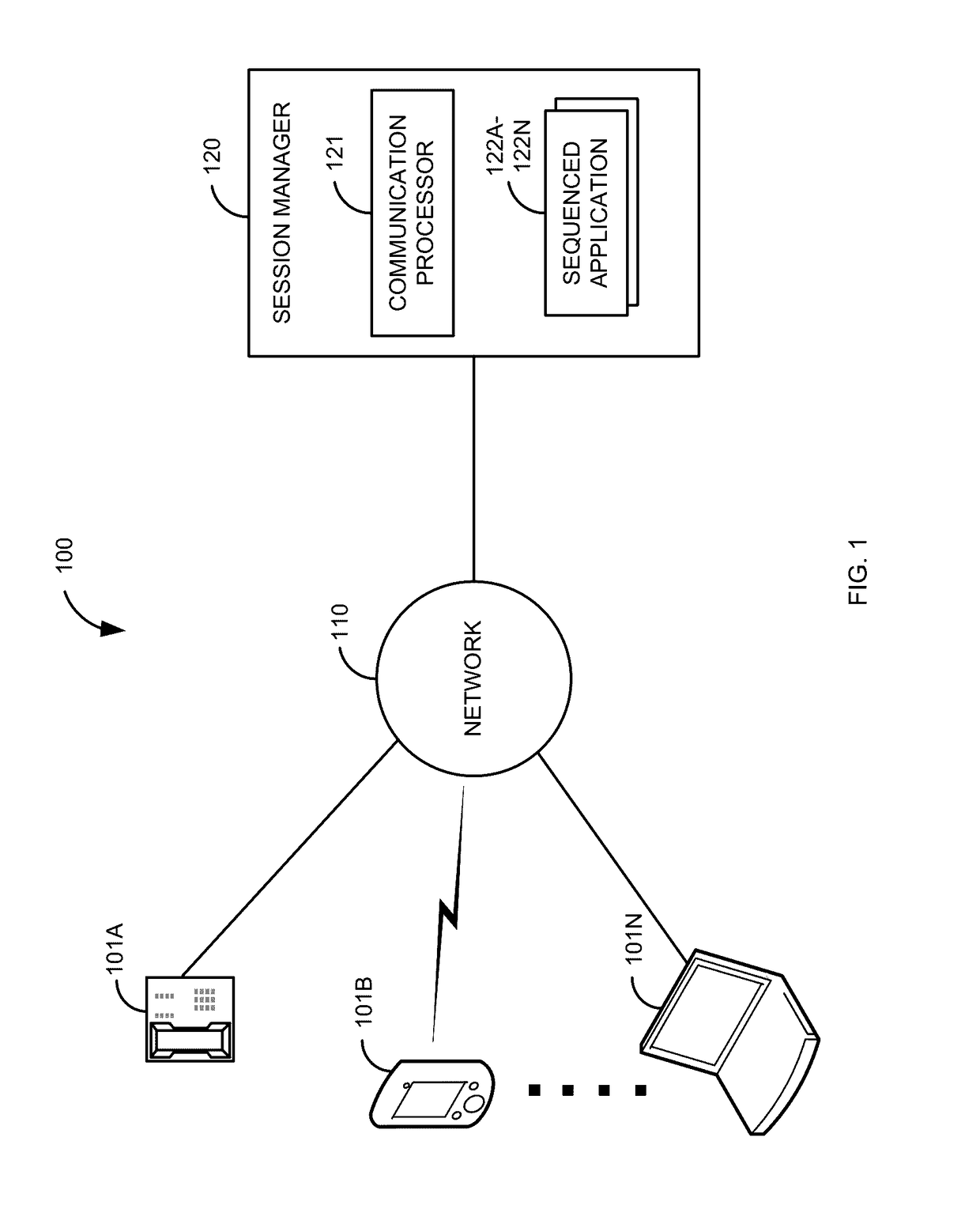
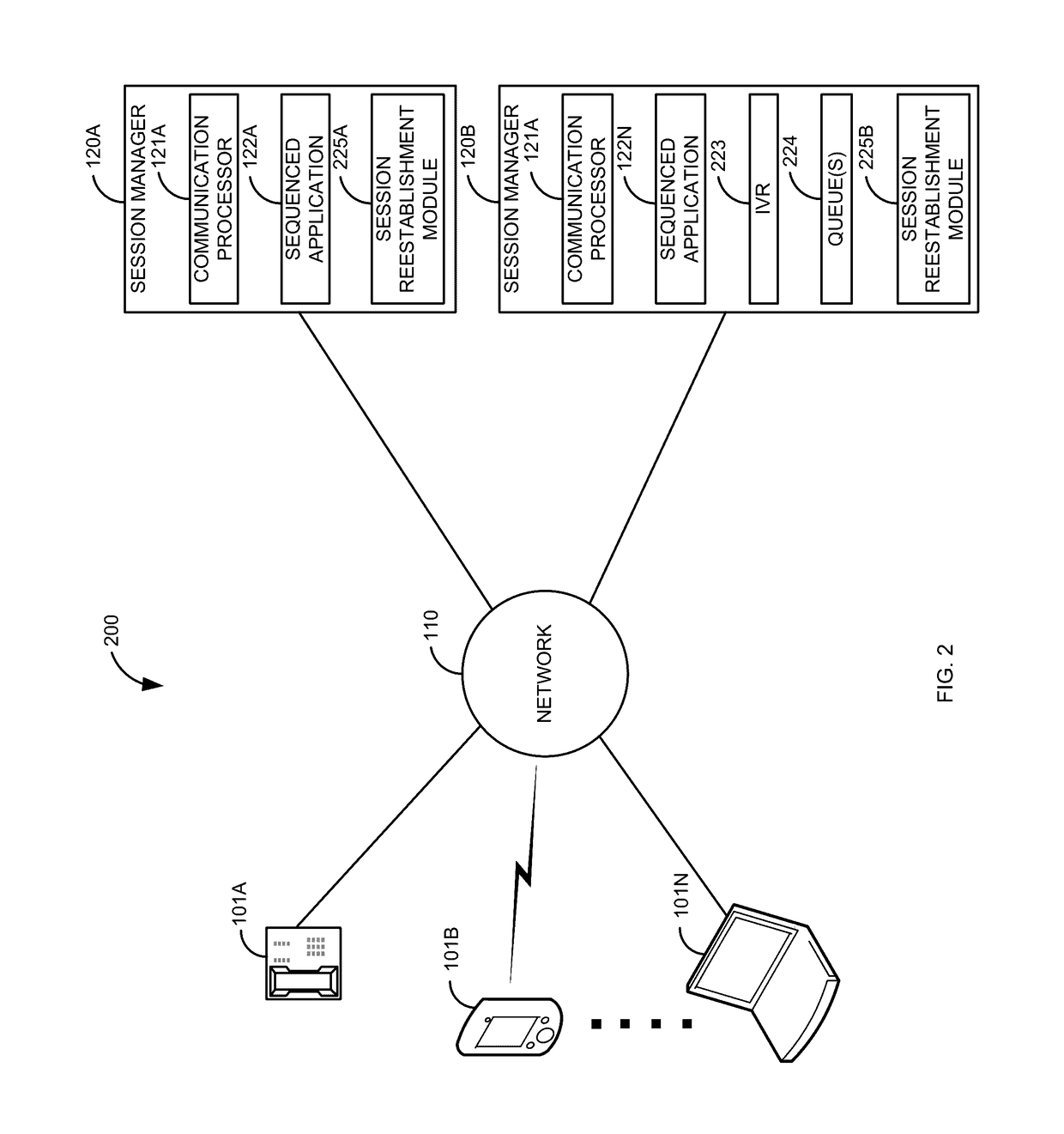
High availability take over for in-dialog communication sessions
When a communication session is established, a primary sequenced application is sequenced into the communication session. At some point, the primary sequenced application fails. Before the communication session has been reestablished, an in-dialog message is received for the first communication session. In response to determining that the primary sequenced application has failed and receiving the in-dialog message for the first communication session, reestablishment of the first communication session is expedited to a backup sequenced application. The communication session is reestablished. This allows a communication session that was normally going to be dropped to stay established, thus providing higher reliability over current systems.
Owner:AVAYA INC
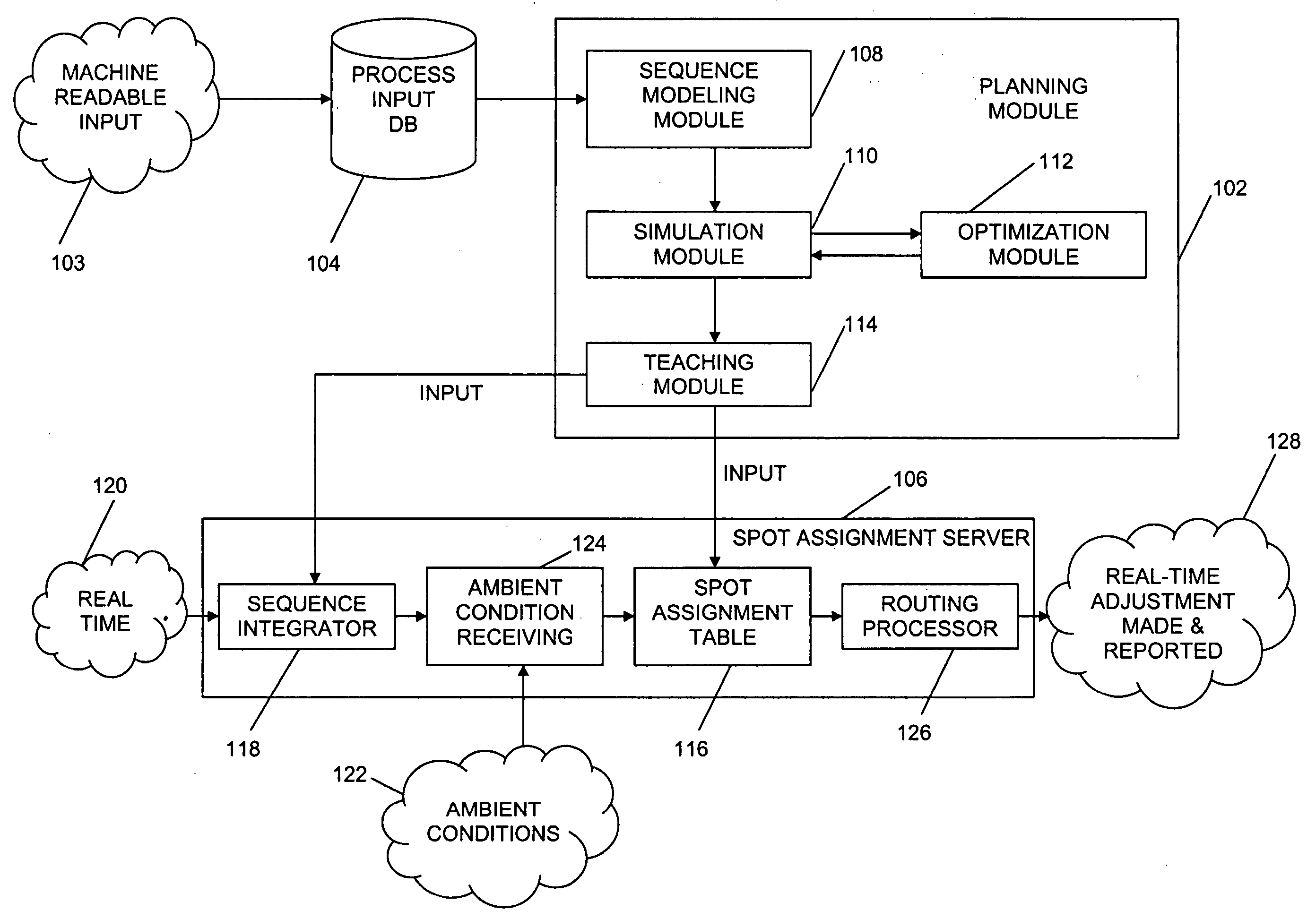
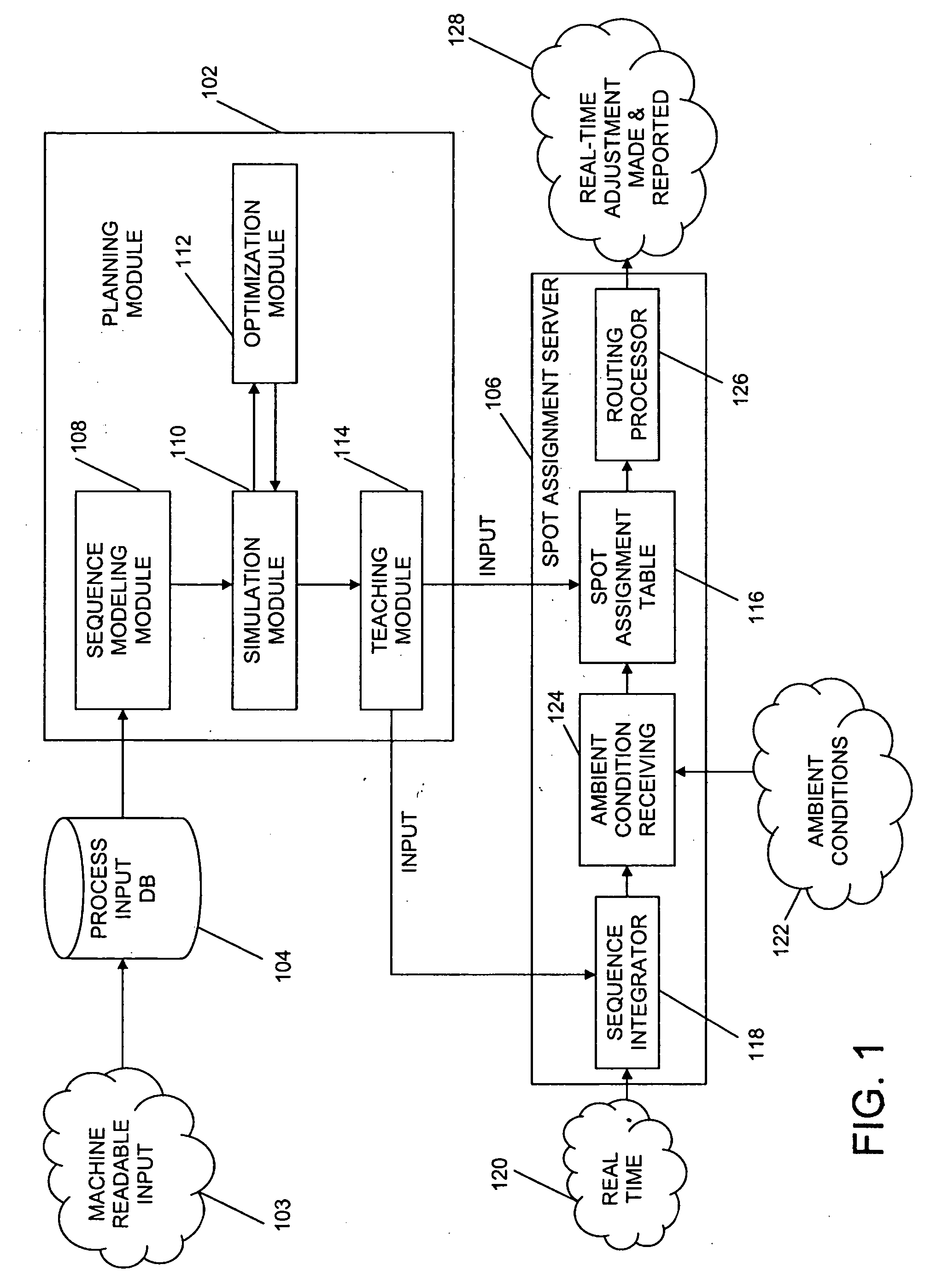
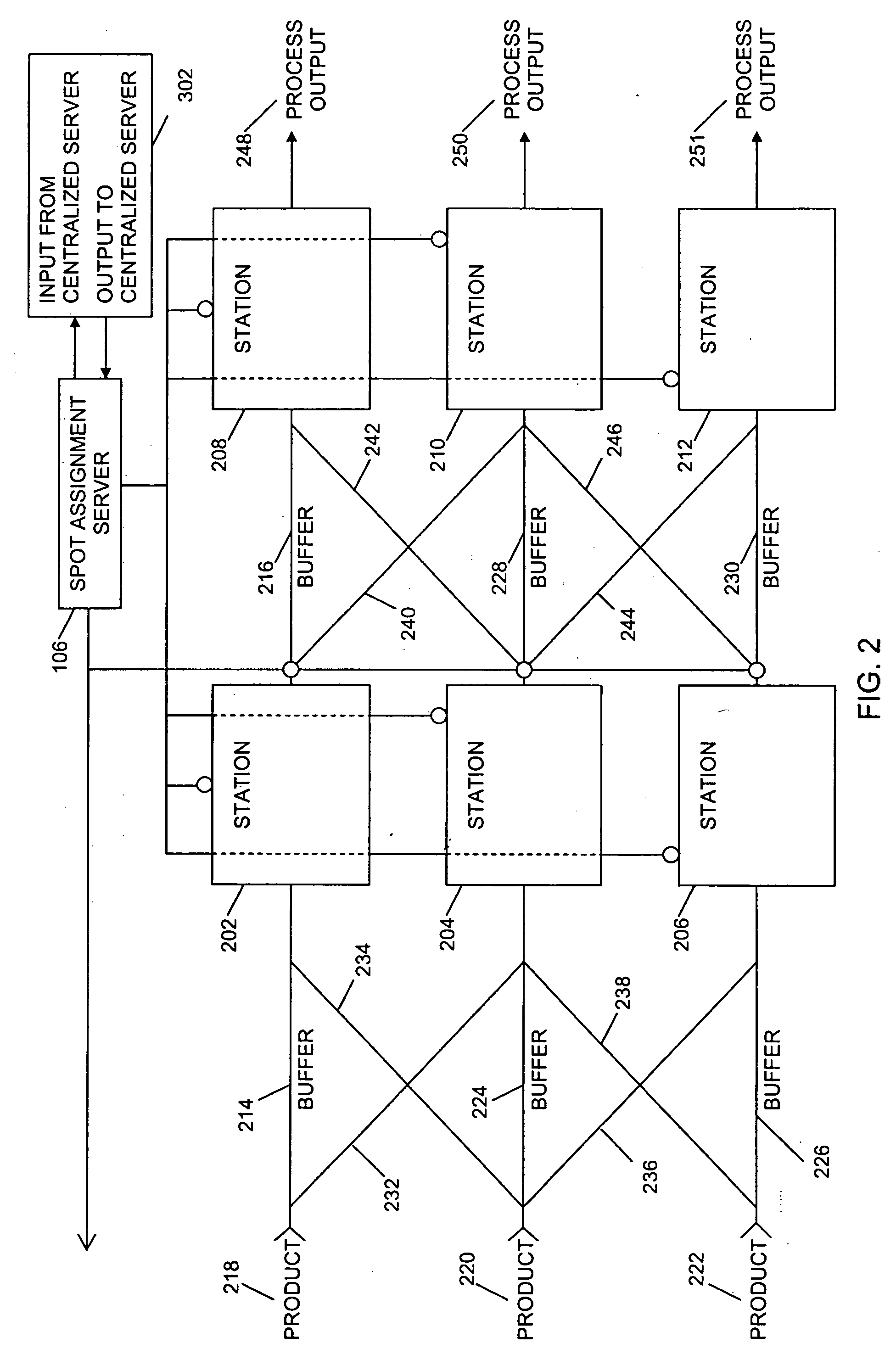
System and method for adaptive machine programming
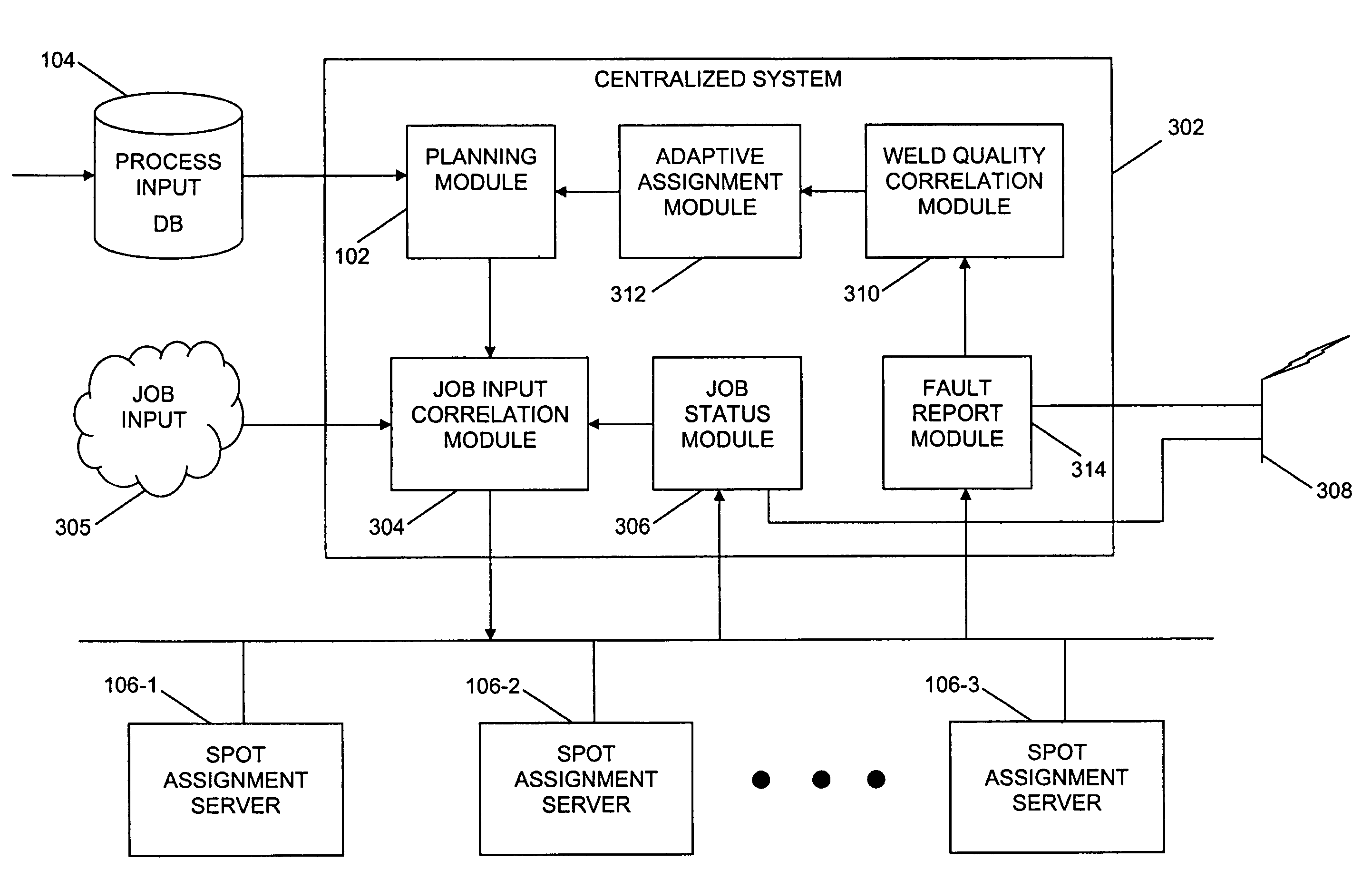
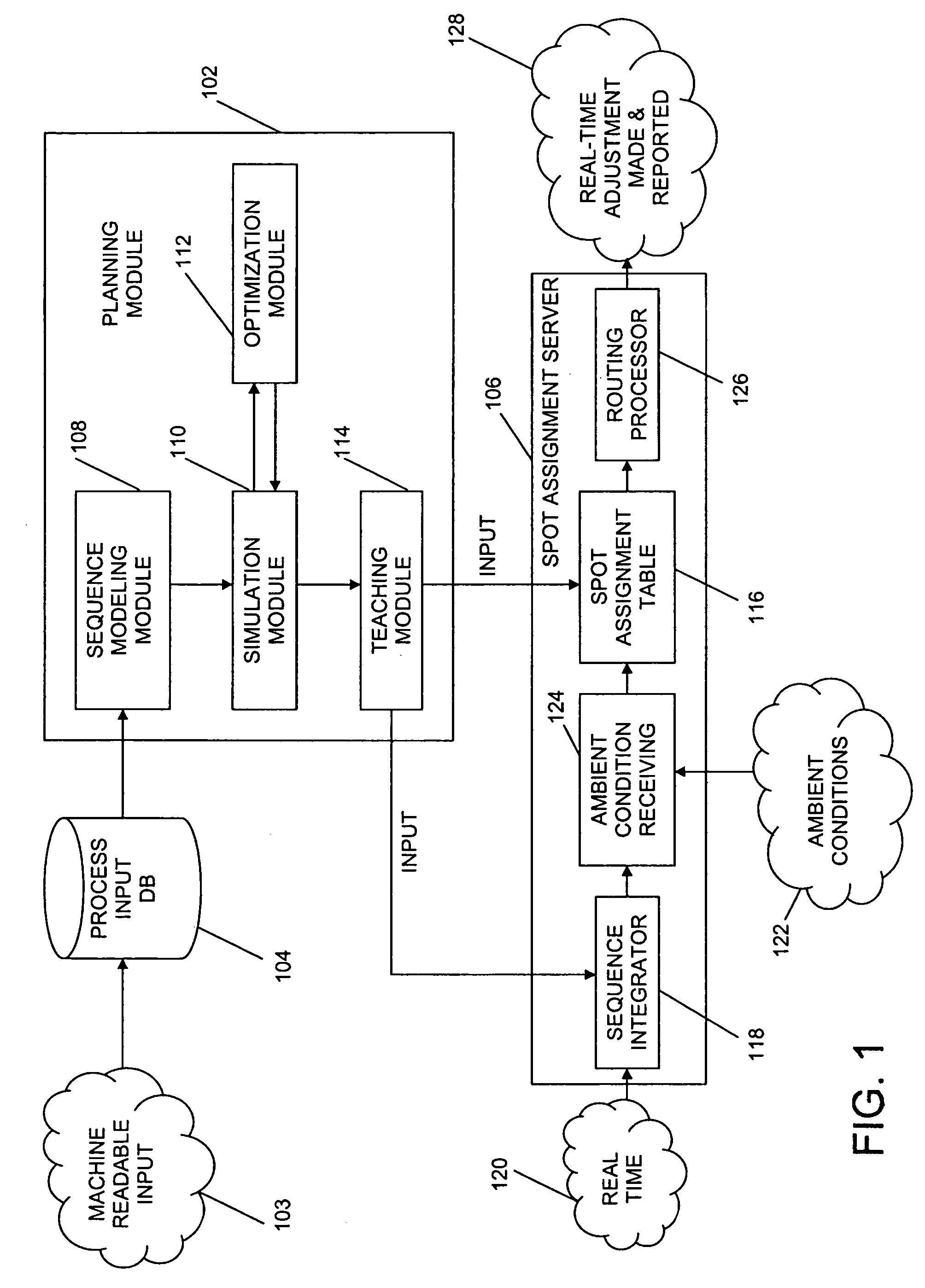
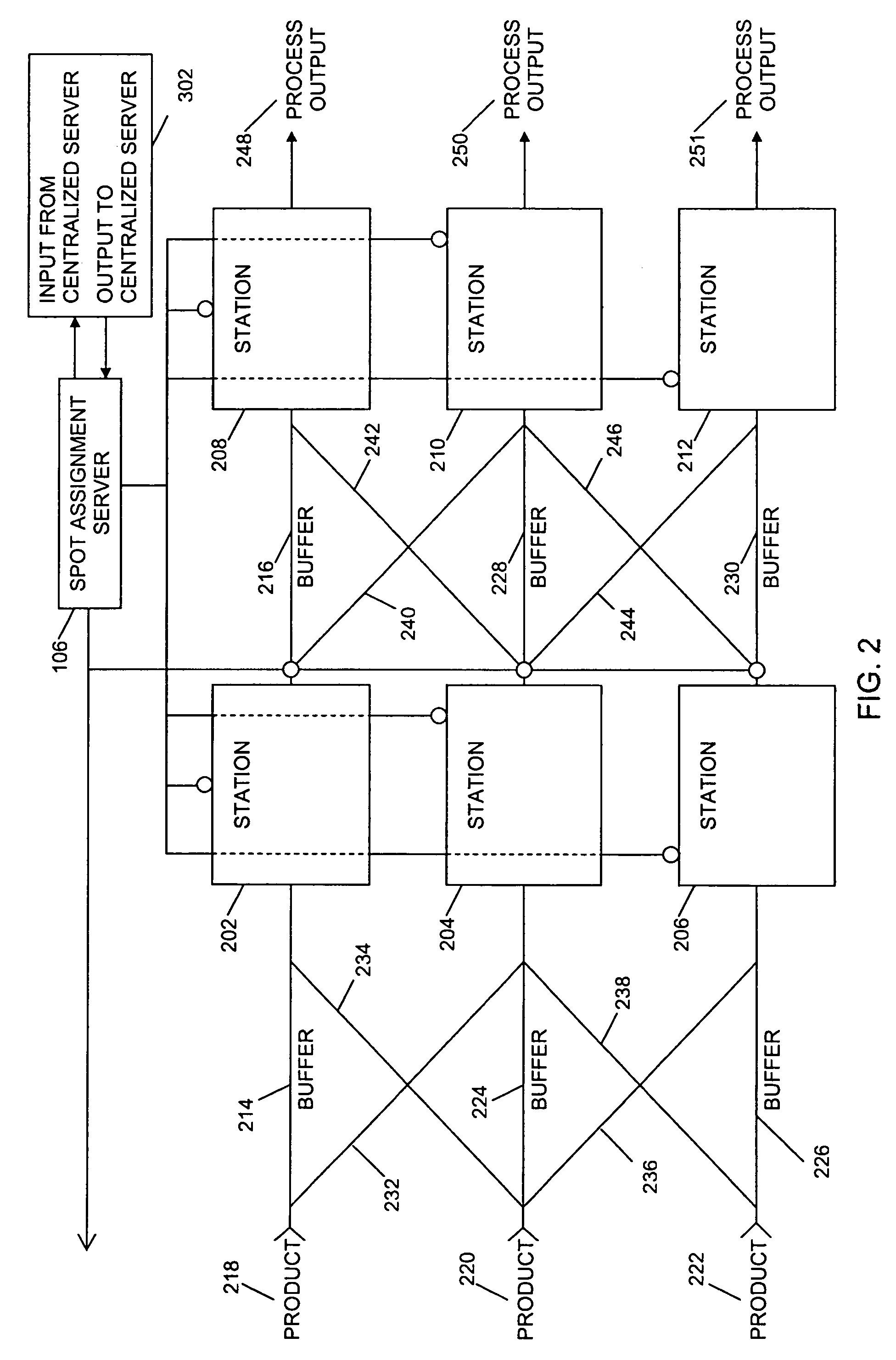
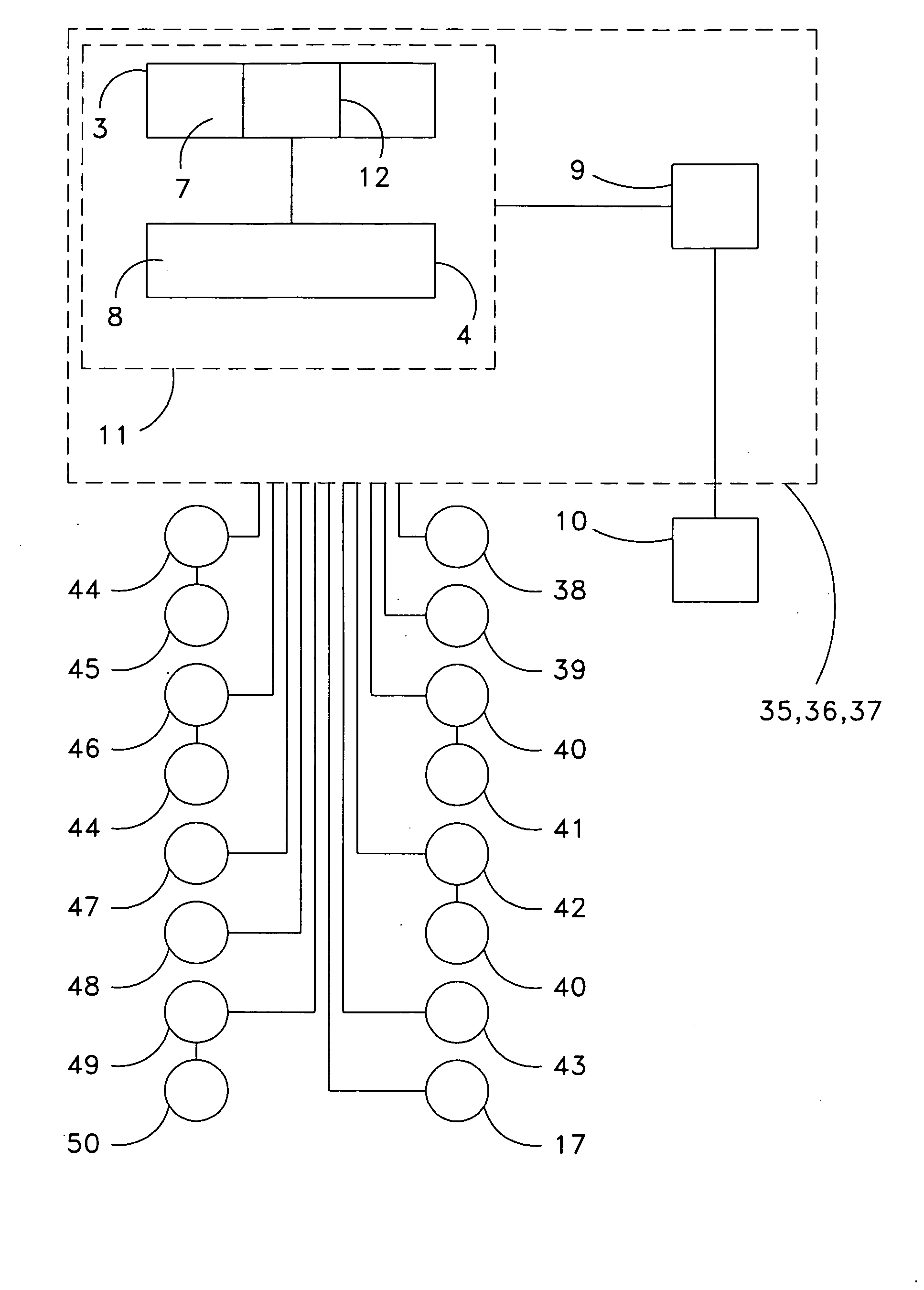
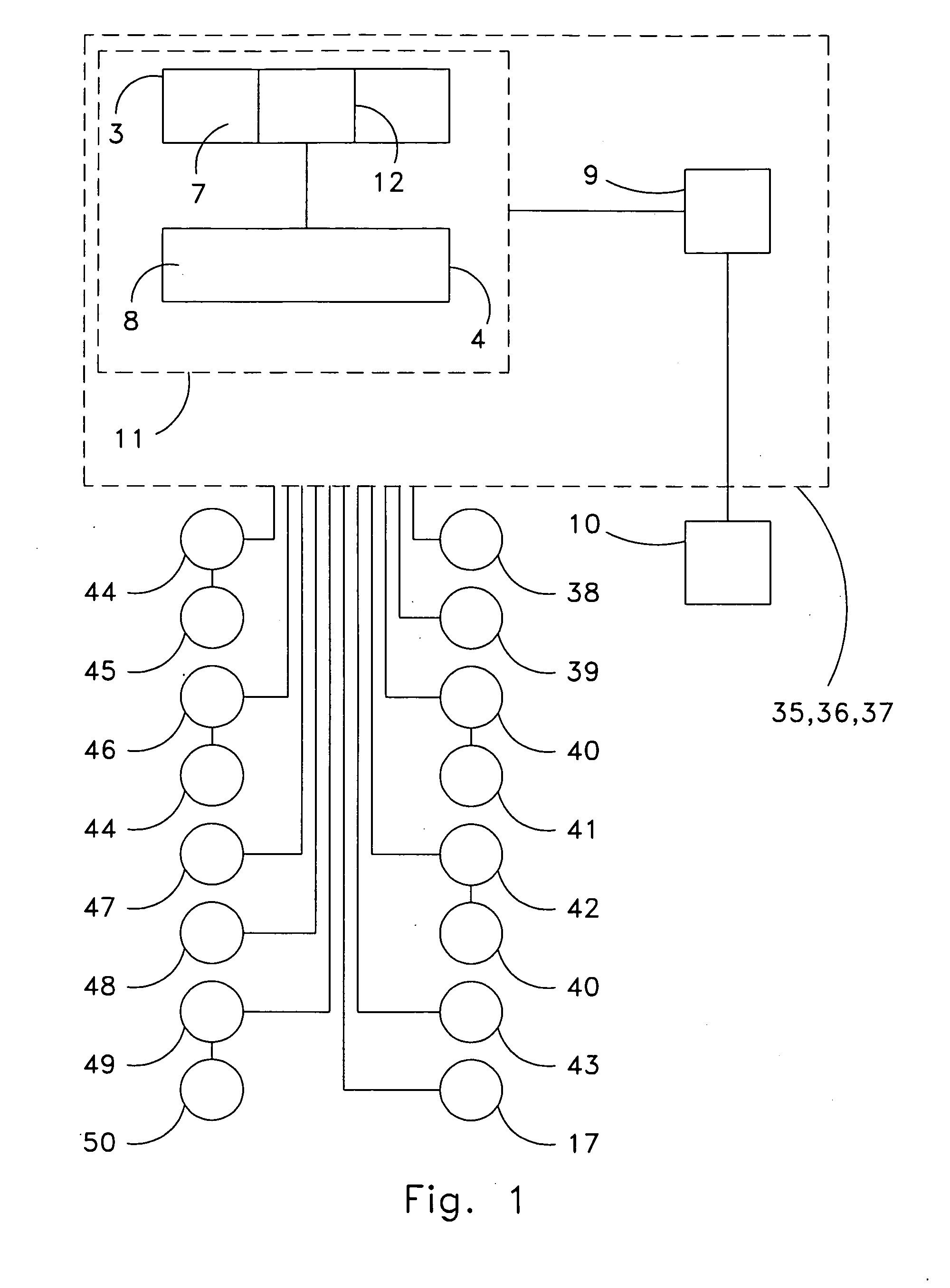
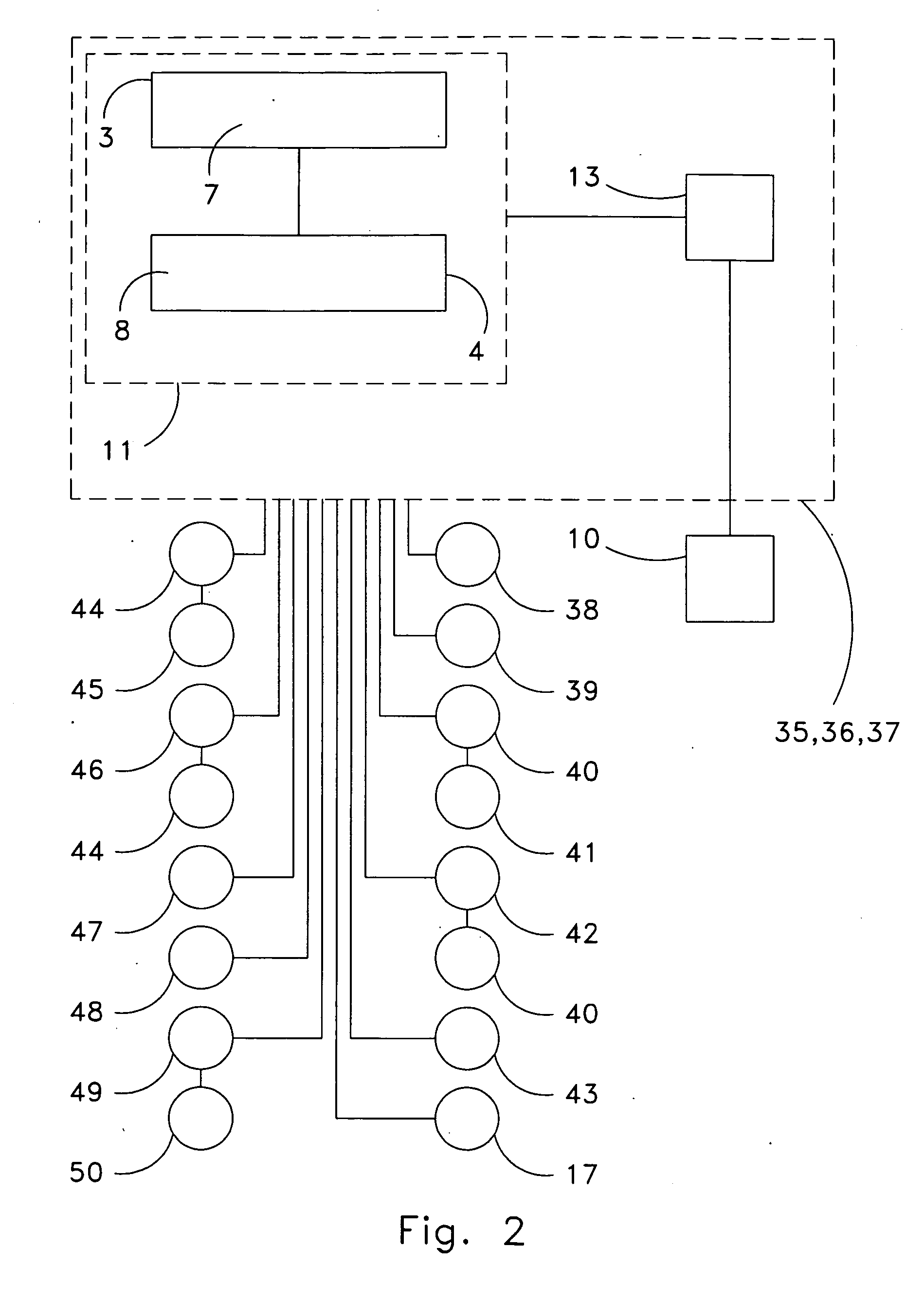
ActiveUS20060190131A1Welding/cutting auxillary devicesTotal factory controlAmbient dataResource reallocation
A system, method and apparatus for resource reallocation based on ambient condition data is disclosed herein. The system includes a planning module (102) for configuring plan data to include a primary sequence and an auxiliary sequence, a rule table (116) for storing the auxiliary sequence, an ambient condition receiver (124) for receiving the ambient condition data, and a routing processor (126) for accessing the rule table (116) to allocate an auxiliary sequence in place of a primary sequence in the event that particular ambient data is received.
Owner:GM GLOBAL TECH OPERATIONS LLC
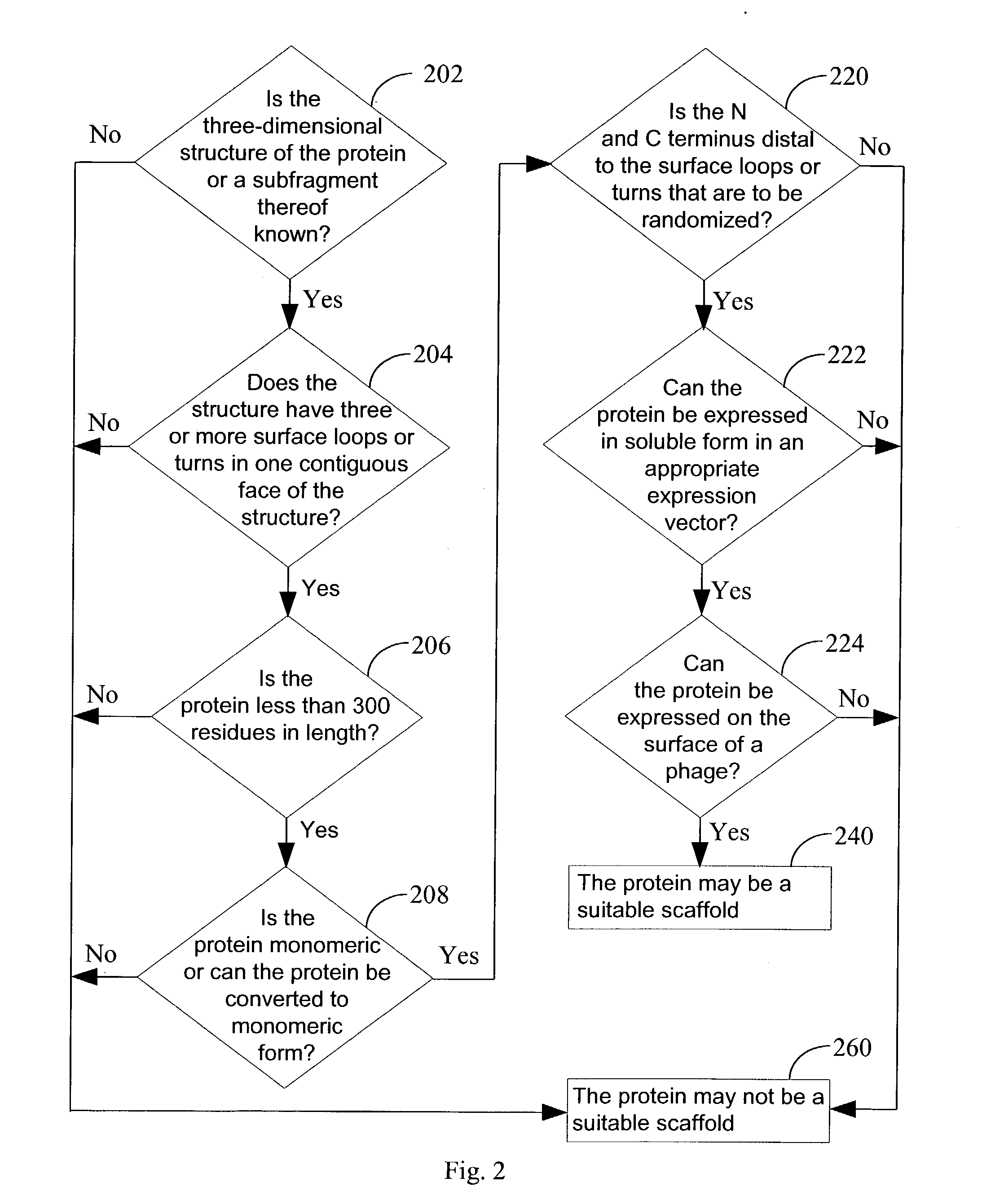
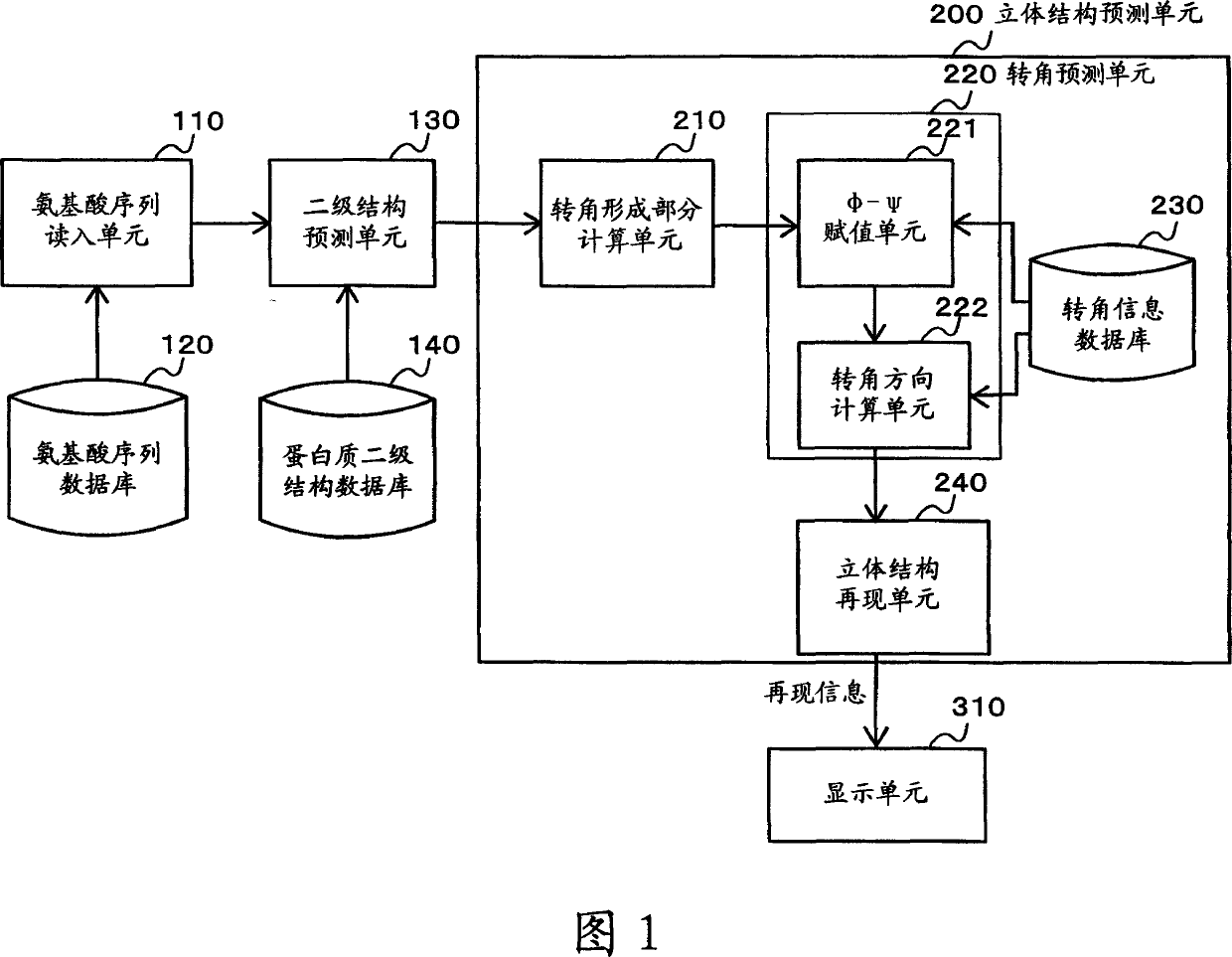
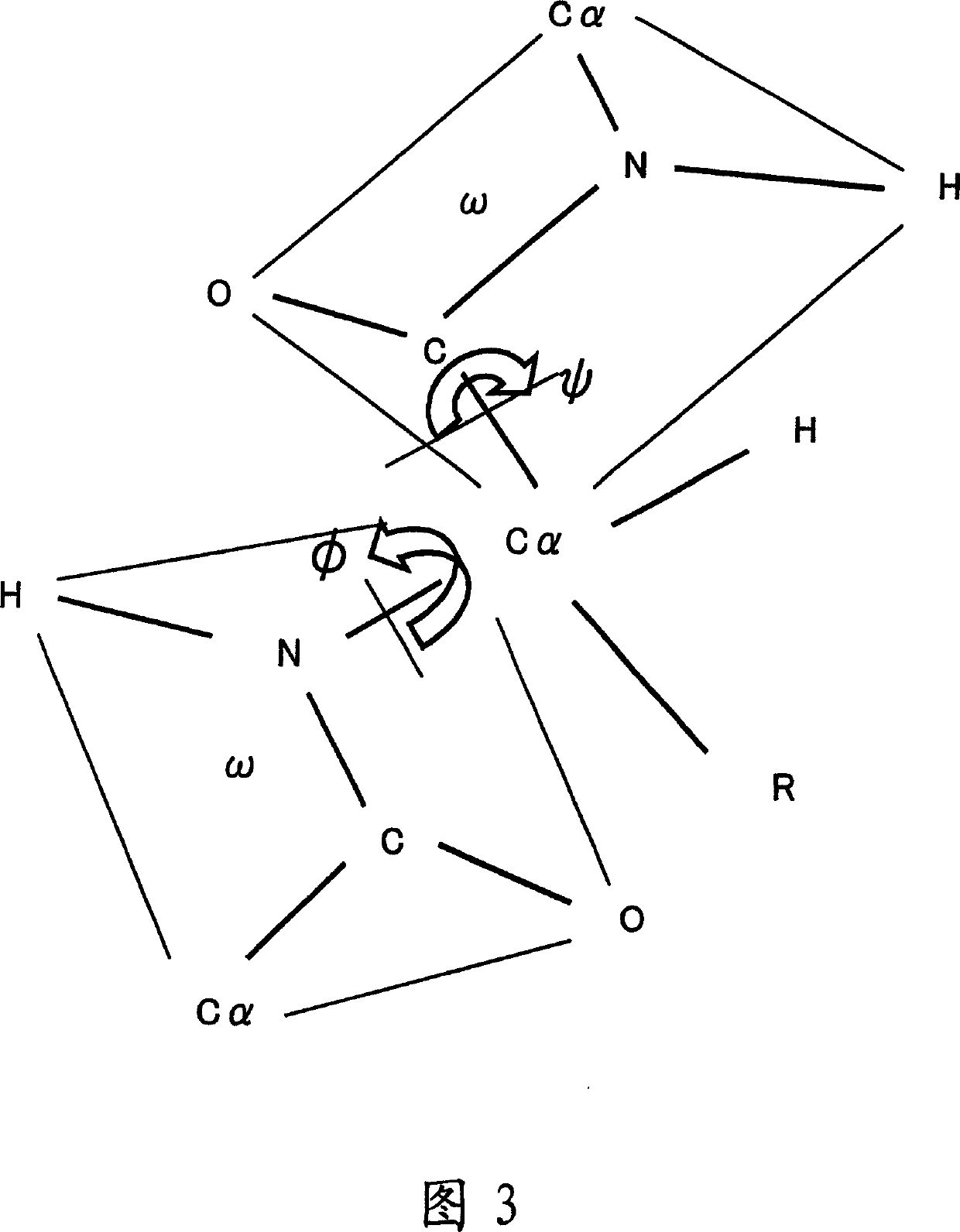
Apparatus for predicting stereostructure of protein and prediction method
A three-dimensional structure of a protein is predicted by reproducing turns. An amino-acid-sequence read-in unit (110) reads in an amino-acid sequence as a primary sequence, and a secondary-structure prediction unit (130) predicts secondary structures corresponding to the amino-acid sequence. In a three-dimensional-structure prediction unit (200), a turn-forming-portion calculation means (210) calculates the number of amino acids forming the turn based on the predicted secondary structures. In a turn-information DB (230), information on structures of probable turns is registered, where the information is obtained according to secondary structures and the numbers of amino acids. The turn prediction means (220) searches for information on a structure of a probable turn, based on the secondary structures and the calculated number of the amino acids, and reproduces the turn based on the information which is searched for. A three-dimensional-structure reproduction means (240) reproduces the three-dimensional structure of the protein by using the reproduced turn.
Owner:FUJITSU LTD
Rice fertility recovery gene auxiliary breeding molecular marker and application thereof
ActiveCN107385024AReduce wasteAchieve pollutionMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFertility
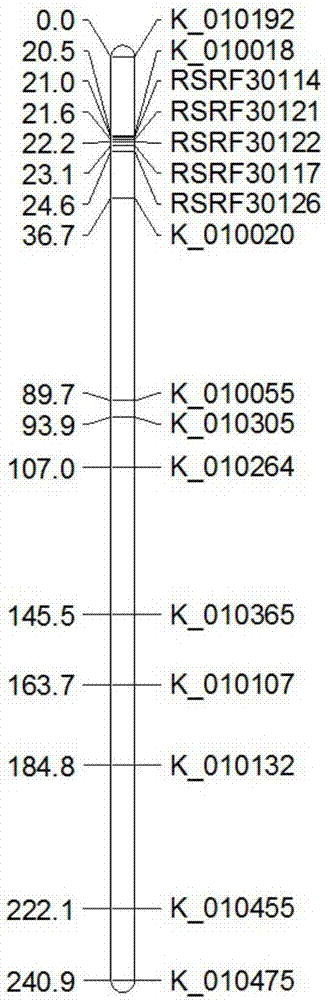
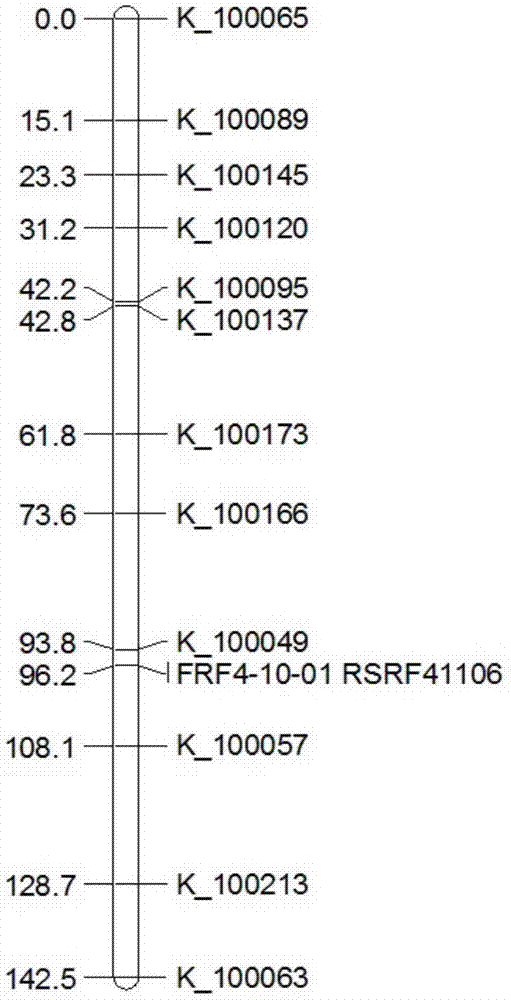
The invention provides a rice fertility recovery gene auxiliary breeding molecular marker, which is SNP markers RSRF30121 and FRF4-10-01 respectively co-separated from rice fertility recovery genes Rf3 and Rf4. The marker RSRF30121 is used for detecting a 5606898-th site basic group of an No.1 rice chromosome; the marker FRF4-10-01 is used for detecting a 18838172-th site basic group of an No.10 rice chromosome; primer sequences of the marker RSRF30121 developed on the basis of KASP technology are shown as SEQ ID NO:1-3. Primary sequences of the marker FRF4-10-01 developed on the basis of the KASP technology are shown as SEQ ID NO:4-6. The invention also provides application of the SNP markers RSRF30121 and FRF4-10-01 to rice fertility recovery gene auxiliary breeding. By using the marker combination, whether the rice has the fertility recovery gene or not can be fast identified; important significance is realized on promoting the application of the Rf3 and Rf4 fertility recovery genes to commercial breeding.
Owner:HUAZHI RICE BIO TECH CO LTD
Process and device for synchronization and codegroup identification in cellular communication systems and computer program therefor
ActiveUS7386006B2Simple processing circuitReduce computational complexityTime-division multiplexRadio transmissionCDMA2000Cellular communication systems
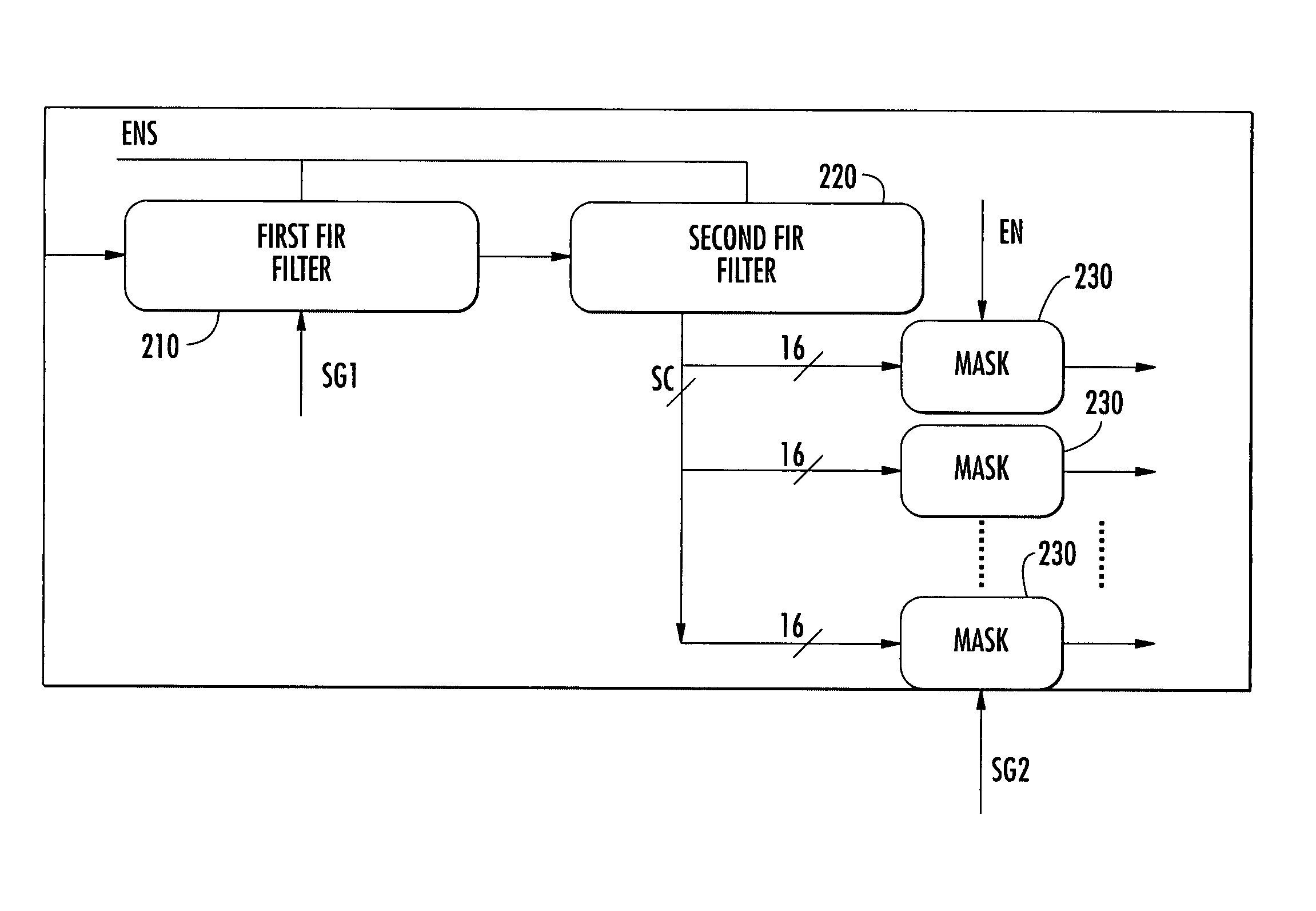
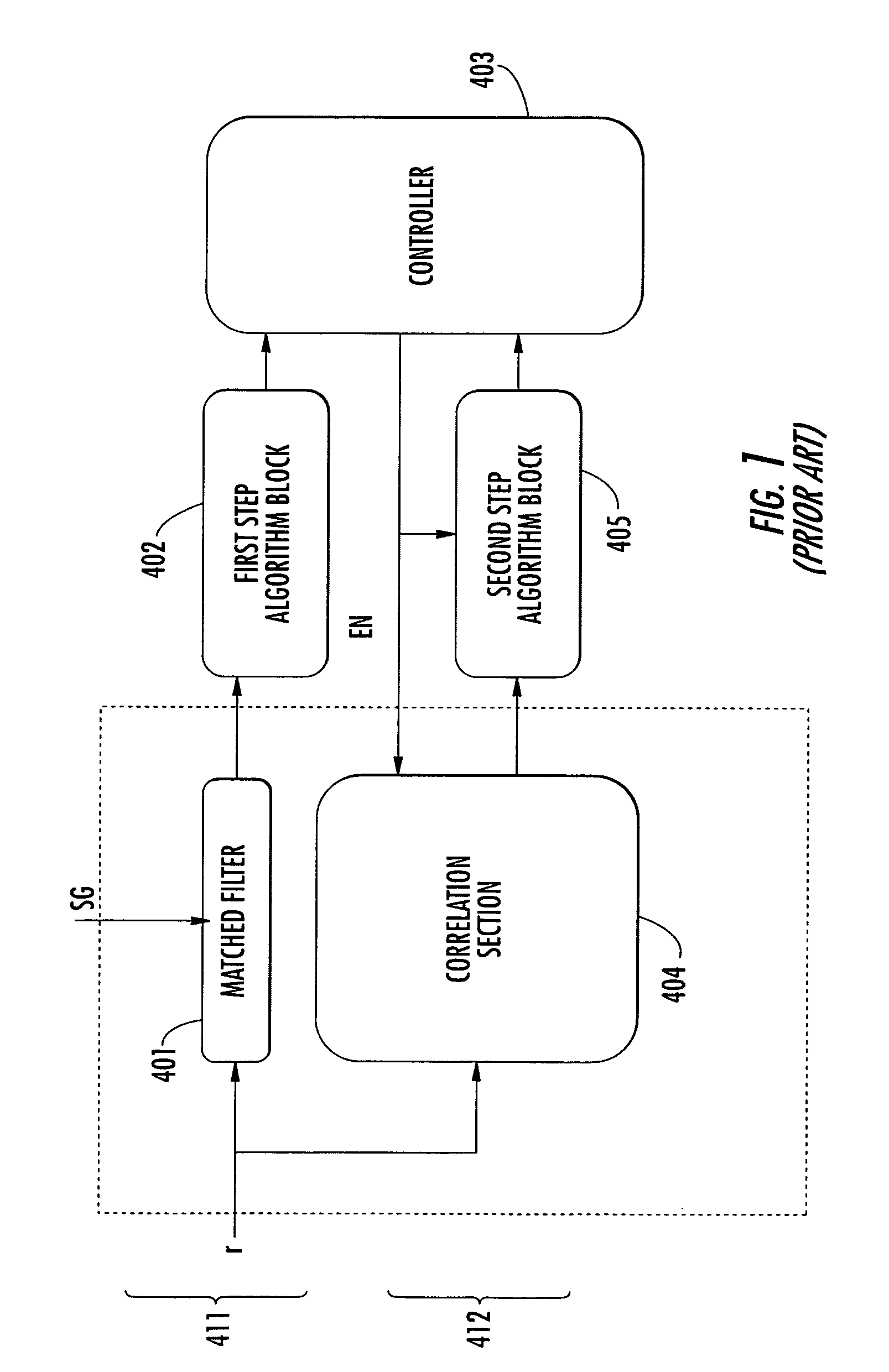
In a first step, slot synchronization may be obtained by setting in correlation the received signal with a primary sequence, which represents the primary channel, and storing the received signal. During a second step, the correlator may be re-used for correlating the received signal with a secondary sequence corresponding to the secondary synchronization codes. The correlator may include a first filter and a second filter connected in series, which receive a first secondary sequence and a second secondary sequence, which may include Golay sequences. Architectures of parallel and serial types, as well as architectures designed for reusing further circuit parts are also disclosed. The invention is particularly applicable in mobile communication systems based upon standards such as UMTS, CDMA2000, IS95, and WBCDMA.
Owner:MICROELECTRONIC INNOVATIONS LLC
Moving image encoding method, moving image decoding method, moving image encoding device, moving image decoding device, program, and integrated circuit
InactiveCN102246525AImprove coding efficiencyEasy to changePulse modulation television signal transmissionDigital video signal modificationDecoding methodsValue assignment
Owner:PANASONIC CORP
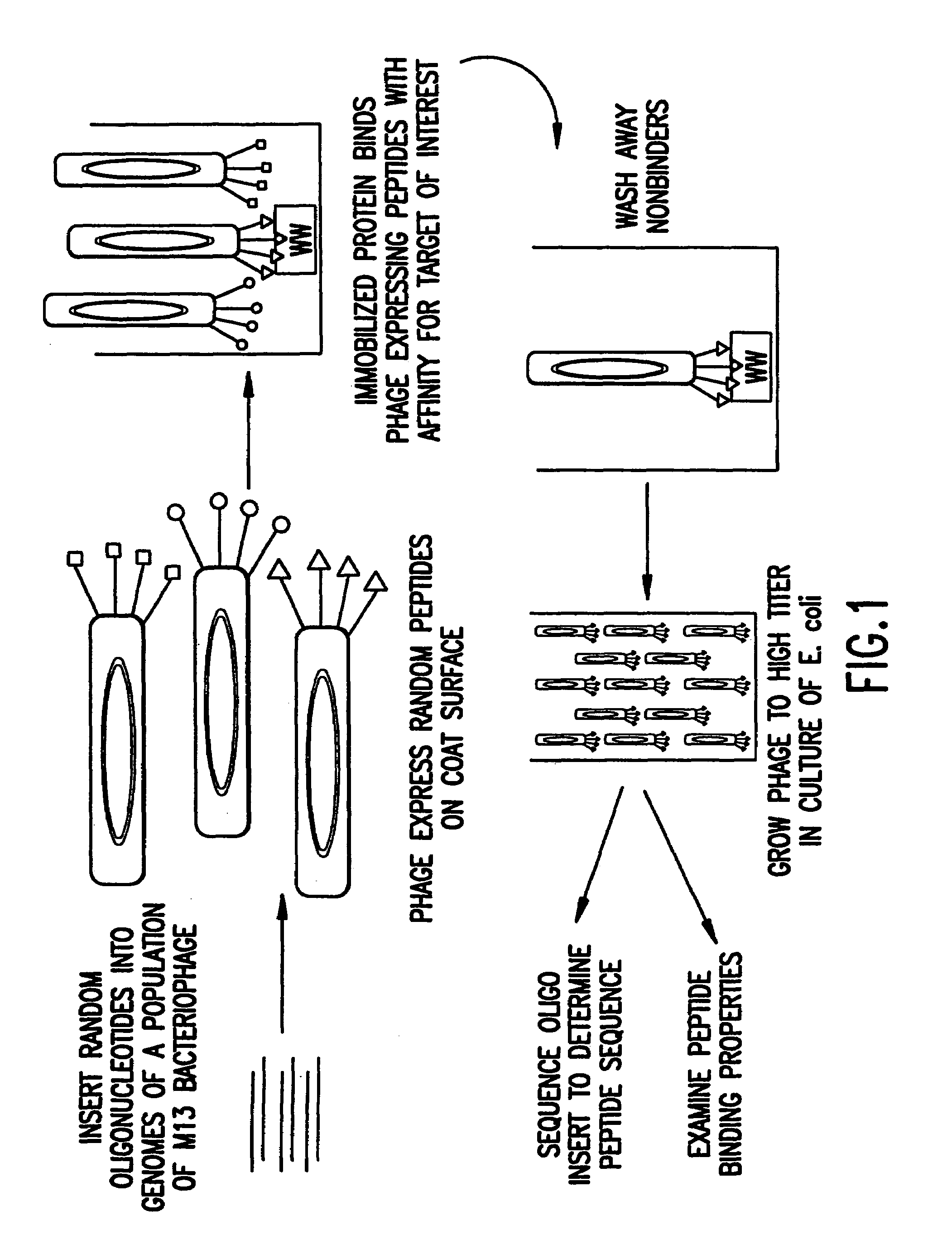
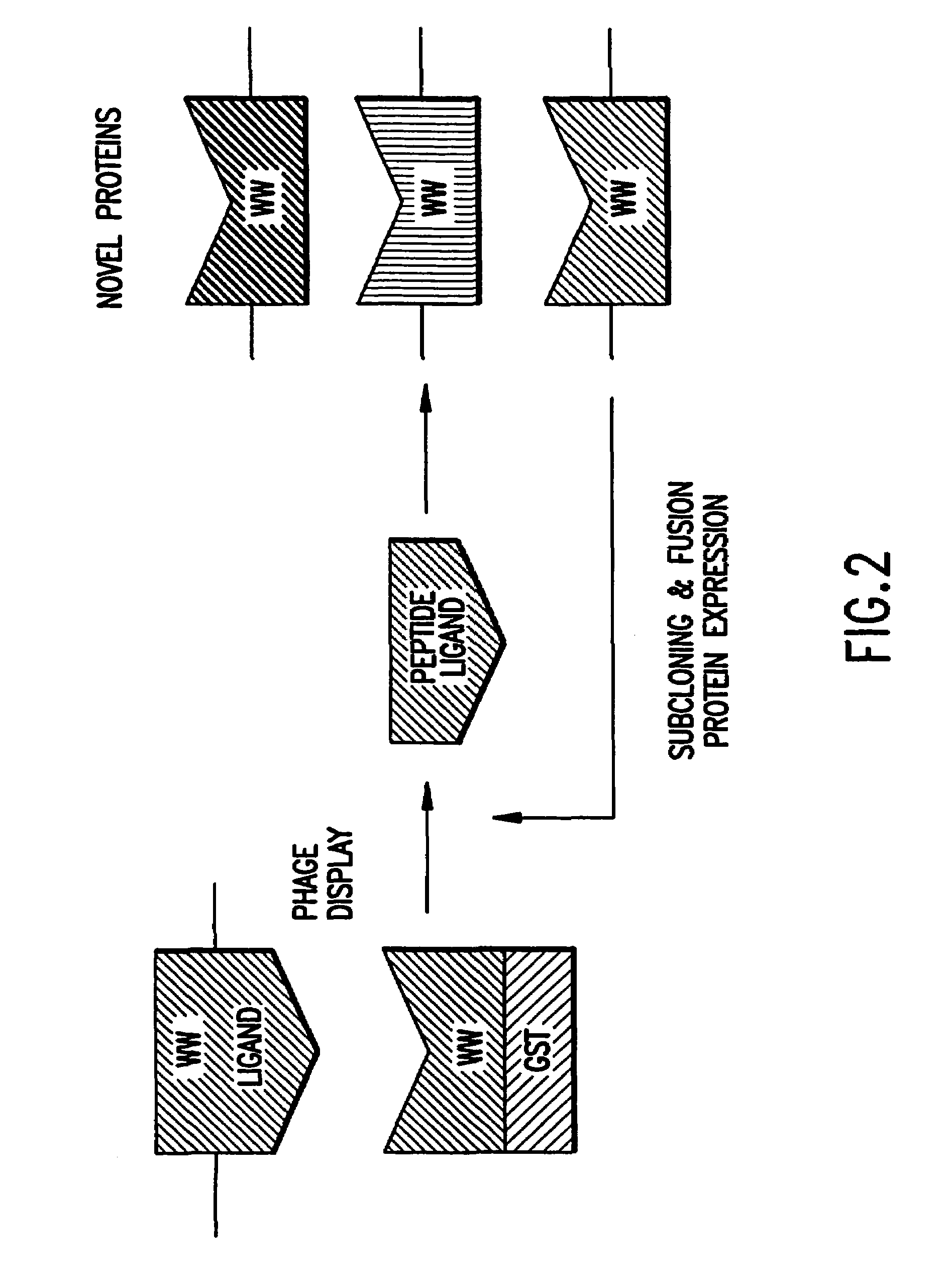
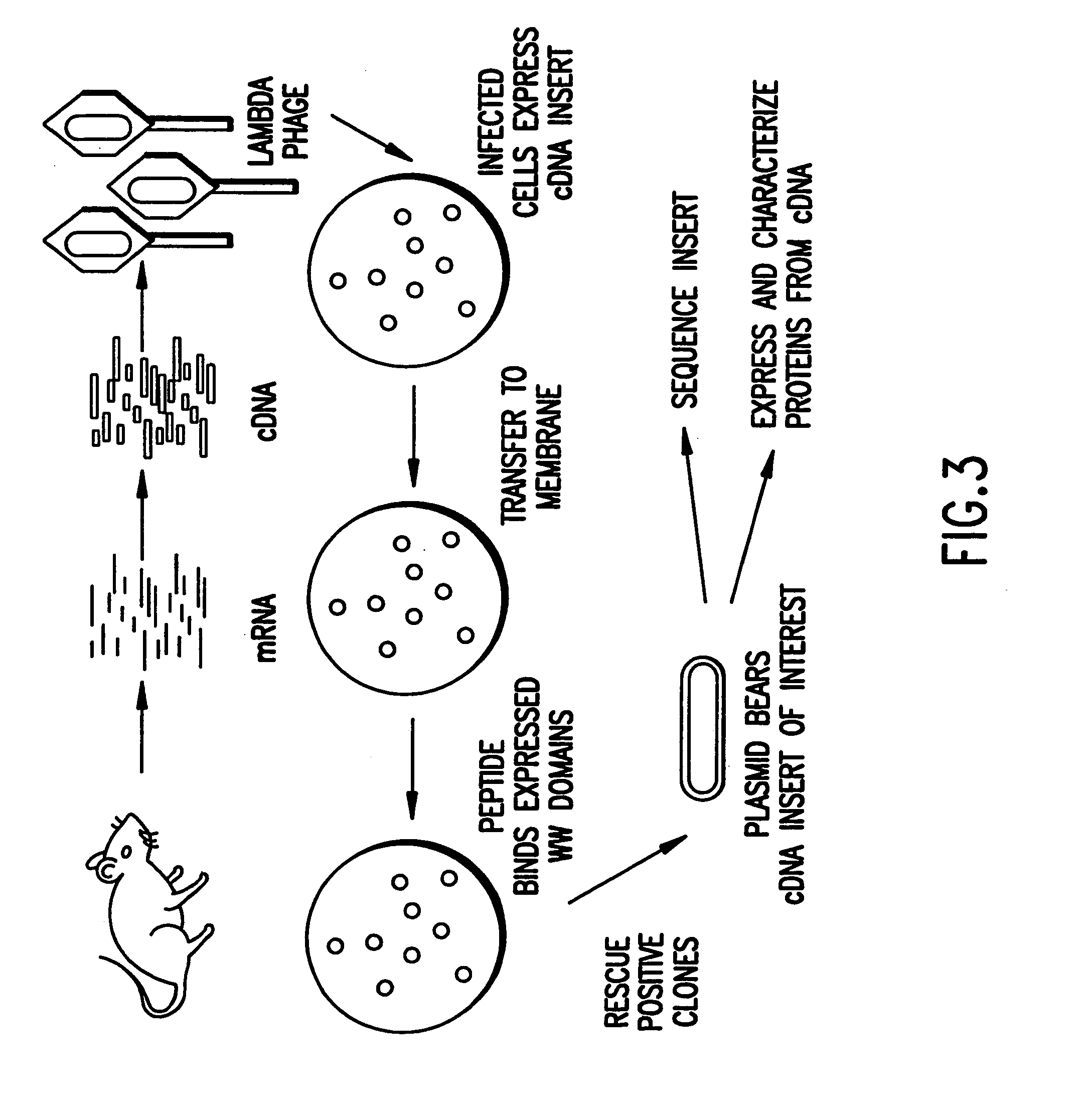
Identification and isolation of novel polypeptides having WW domains and methods of using same
InactiveUS7279548B2Specificity is easedPeptide/protein ingredientsAntibody mimetics/scaffoldsWW domainPrimary sequence
Novel polypeptides having WW domains of interest are described, along with DNA sequences that encode the same. A method of identifying these polypeptides by means of a sequence-independent (that is, independent of the primary sequence of the polypeptide sought), recognition unit-based functional screen is also disclosed. Various applications of the method and of the polypeptides identified are described, including their use in assay kits for drug discovery, modification, and refinement.
Owner:CYTOGEN CORP +1
Modified tobacco mosaic virus particles as scaffolds for display of protein antigens for vaccine applications
InactiveUS20090053261A1Many of characteristicSsRNA viruses positive-senseAntibody mimetics/scaffoldsVaccinationADAMTS Proteins
Display of peptides or proteins in an ordered, repetitive array, such as on the surface of a virus-like particle, is known to induce an enhanced immune response relative to vaccination with the “free” protein antigen. The 2100 coat proteins comprising the rod-shaped capsid of Tobacco mosaic virus (TMV) can accommodate short peptide insertions into the primary sequence, but the display of larger protein moieties on the virion surface by genetic fusions to the capsid protein has not been possible. Since TMV lacks surface exposed residues compatible with commonly available linker chemistries, we employed a randomized library approach to introduce a reactive lysine at the externally located at the amino-terminus of the coat protein. We found that we could easily control the extent of virion conjugation and demonstrated stoichiometric biotinylation of the introduced lysine. To characterize this modular platform for the display of heterologous proteins, we bound a model antigen (streptavidin (SA)-green fluorescent protein (GFP), expressed and purified from plants) to the surface of TMV, creating a GFP-SA decorated virus particle. Rapid and quantitative determination of the level of TMV capsid decoration was accomplished by subjecting the complex to amino acid analysis and solving the family of linear equations relating the pmoles of each residue to the known amino acid composition of the complex components. We obtained a GFP-SA tetramer loading of 26%, which corresponds to display of approximately 2200 GFP moieties per intact virion. We evaluated the immunogenicity of GFP decorated virions in both mice and guinea pigs, and found augmented humoral IgG titers in both species, relative to unbound GFP-SA tetramer. In mice, we observed a detectable humoral immune response after only a single immunization with the TMV-protein complex. By demonstrating the presentation of whole proteins, this study expands the utility of TMV as a vaccine scaffold beyond that which is possible by genetic manipulation.
Owner:KENTUCKY BIOPROCESSING
Inhibition of ice crystal growth
Antifreeze polypeptides, antifreeze compositions including the polypeptides, nucleotides encoding the antifreeze polypeptides, methods of making antifreeze compositions, and methods of inhibiting ice crystal growth are provided herein. The peptides are based on the primary sequence of collagen and include those having a molecular weight between about 500-7000 Da. The peptides preferably include cationic polypeptides. The methods of making antifreeze compositions include digesting collagen or gelatin into hydrolysates with peptides having molecular weights between about 500-7000 Da. The digestions are performed with proteases and / or non-enzymatic hydrolysis. The methods of inhibiting ice crystal growth include adding the antifreeze polypeptides or compositions described herein to a composition to be frozen. The methods may be used to inhibit ice crystal growth in frozen food products.
Owner:DAMODARAN SRINIVASAN
Immunomodulator polypeptides derived from il-2 and their use thereof in the therapeutic of cancer and chronic infections
ActiveUS20120315245A1Peptide/protein ingredientsAntibody mimetics/scaffoldsRegulatory T cellBULK ACTIVE INGREDIENT
The present invention relates generally to polypeptides whose primary sequence has high sequence homology with human interleukin 2 (IL-2) with some punctual mutations in the sequence of native IL-2. The polypeptides of the present invention have an immunomodulatory effect on the immune system, which is selective / preferential on regulatory T cells. The present invention also relates to specific polypeptides whose amino acid sequence is disclosed herein. In another aspect the present invention relates to pharmaceutical compositions comprising as active ingredient the polypeptides disclosed. Finally, the present invention relates to the therapeutic use of the polypeptides and pharmaceutical compositions disclosed due to their immune modulating effect on diseases such as cancer and chronic infectious diseases.
Owner:CENT DE INMUNOLOGIA MOLECULAR CENT DE INMUNOLO
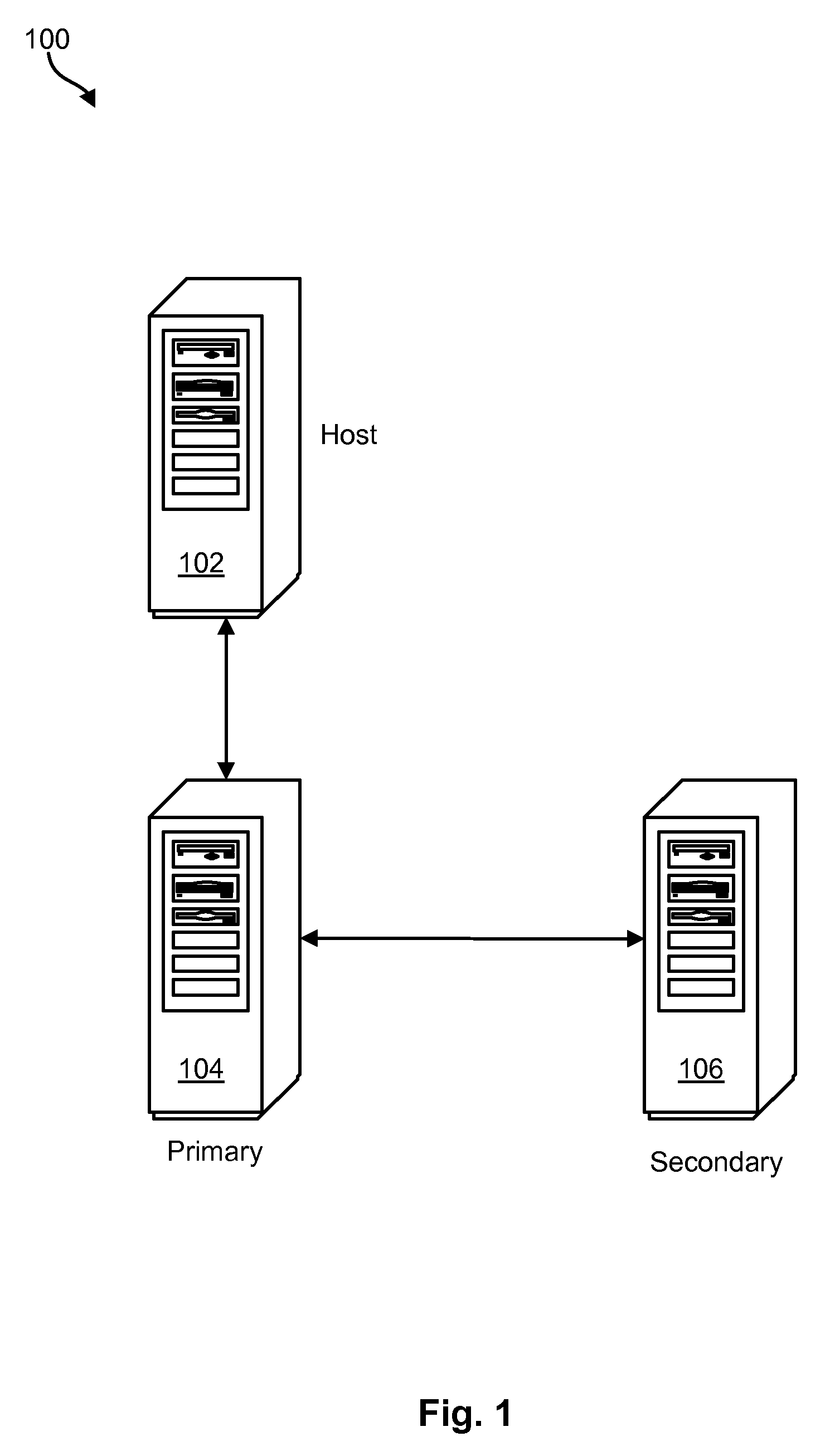
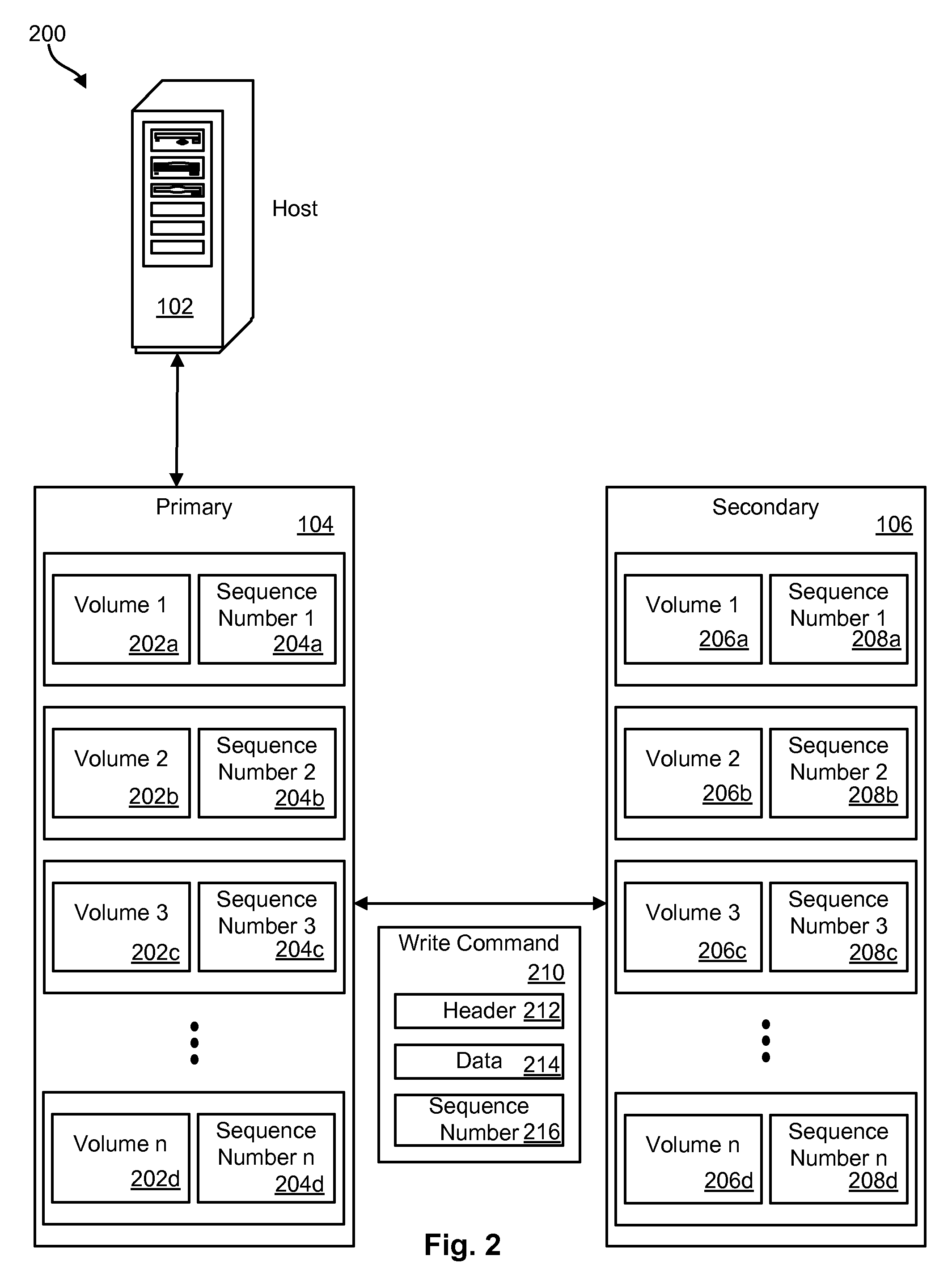
Command sequence numbering apparatus and method
InactiveUS20100049928A1Memory loss protectionError detection/correctionComputer hardwareData integrity
A method, system, and computer program product to preserve data integrity in a mirror and copy environment is disclosed herein. In one embodiment, a method may include receiving a write command and data from a host device. The method may further include writing the data to a primary storage device and attaching a primary sequence number associated with the primary storage device to the write command, thereby providing a numbered write command with a command sequence number. The numbered write command may then be transmitted to a secondary storage device. The method may further include comparing the command sequence number to a secondary sequence number associated with the secondary storage device. If the command sequence number matches the secondary sequence number, then the command may be executed. Otherwise, it may be ignored.
Owner:IBM CORP
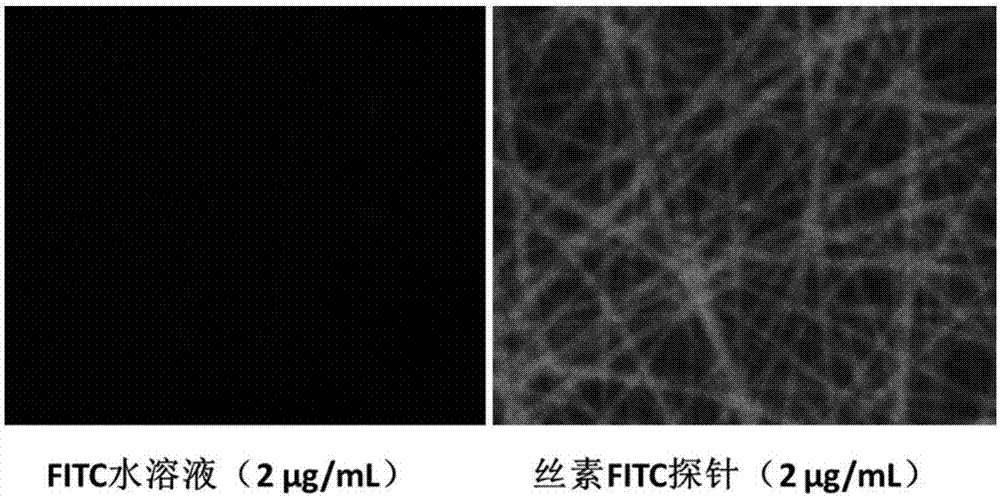
Preparation method of fibroin fluorescent probe
ActiveCN107057683AHigh sensitivityThe detection process is fastPeptide preparation methodsFluorescence/phosphorescenceFluoresceinBacteriophage
The invention discloses a preparation method of a fibroin fluorescent probe. The method comprises the following steps: screening a phage library by using a fibroin material to obtain a phage specifically bonded with fibroin proteins; carrying out gene sequencing; using an amino acid sequence corresponding to the gene obtained by sequencing as a fibroin affinity peptide primary sequence; carrying out polypeptide synthesis on the fibroin affinity peptide primary sequence by adopting a polypeptide synthesis technology to obtain fibroin affinity peptide powder; and bonding the fibroin affinity peptide powder with fluorescein to obtain a fibroin fluorescent probe. The phage screened by the invention has very high affinity to the fibroin material, is high in probe sensitivity, high in detection speed and wide in application prospect, and provides more choices for high-efficient targeting and identification of the fibroin protein and the fibroin material.
Owner:ZHEJIANG UNIV
Methods and Apparatus for the Manipulation of Conferenced Data
InactiveUS20090232032A1Multiplex system selection arrangementsSpecial service provision for substationElectronic communicationData filling
Methods and apparatus may permit the manipulation of conferenced data. Primary sequenced audio-optical data structures may be populated with conferenced data, and secondary sequenced audio-optical data structures may be populated with derivative conferenced data. Electronic identity characteristics may be utilized to selectively deny access of conference participants to an exchange of conferenced electronic communications, verify the identity of prospective conference participants to an exchange of conferenced electronic communications, and verify the identity of known and unknown individuals in conjunction with electronic identity signatures. Conferenced data tags may be associated to conferenced data elements to create derivative conferenced data. Derivative conferenced data may be utilized in conjunction with primary conferenced data structures and secondary conferenced data structures to create augmented functionalities for conferenced data elements.
Owner:VERBAL WORLD
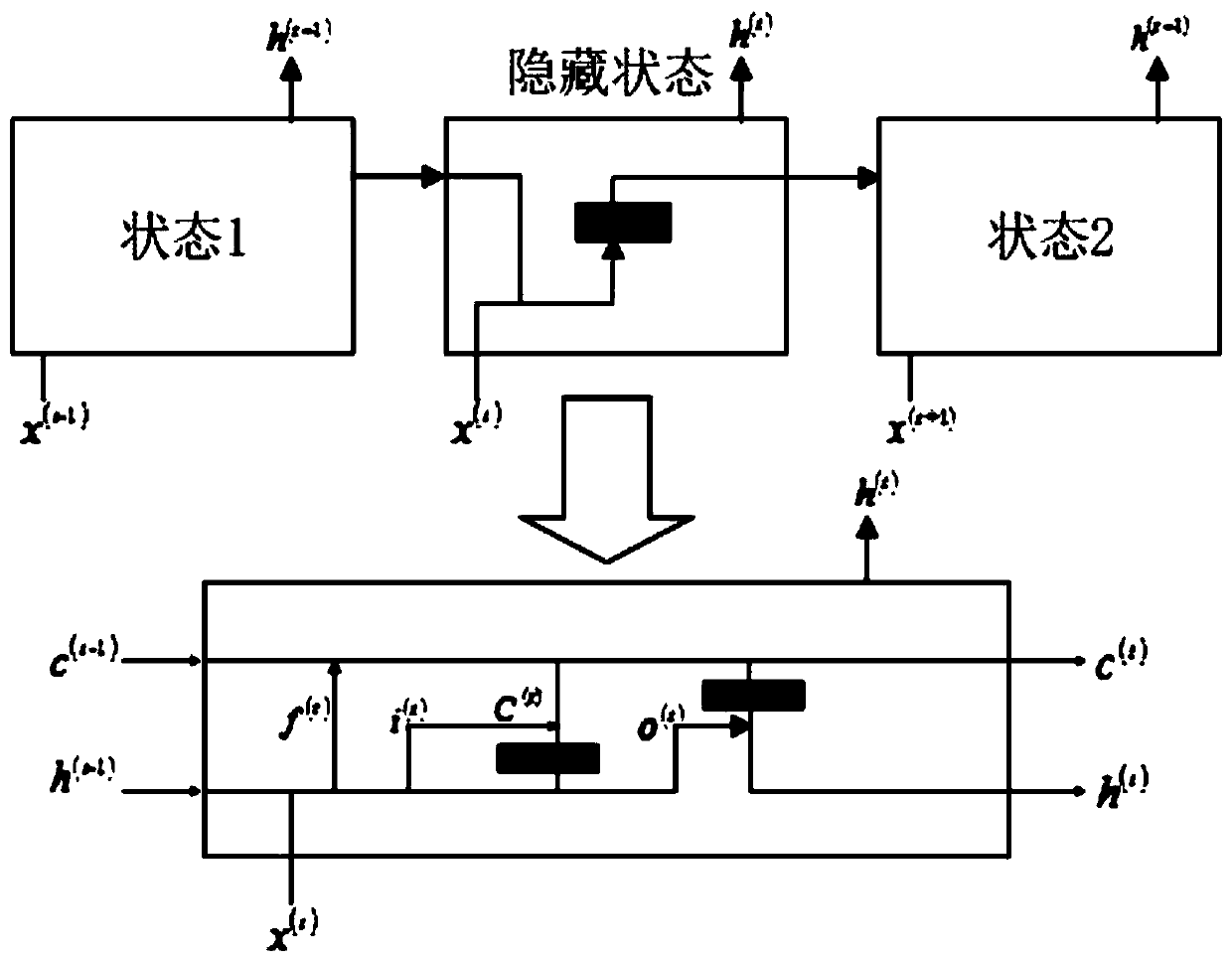
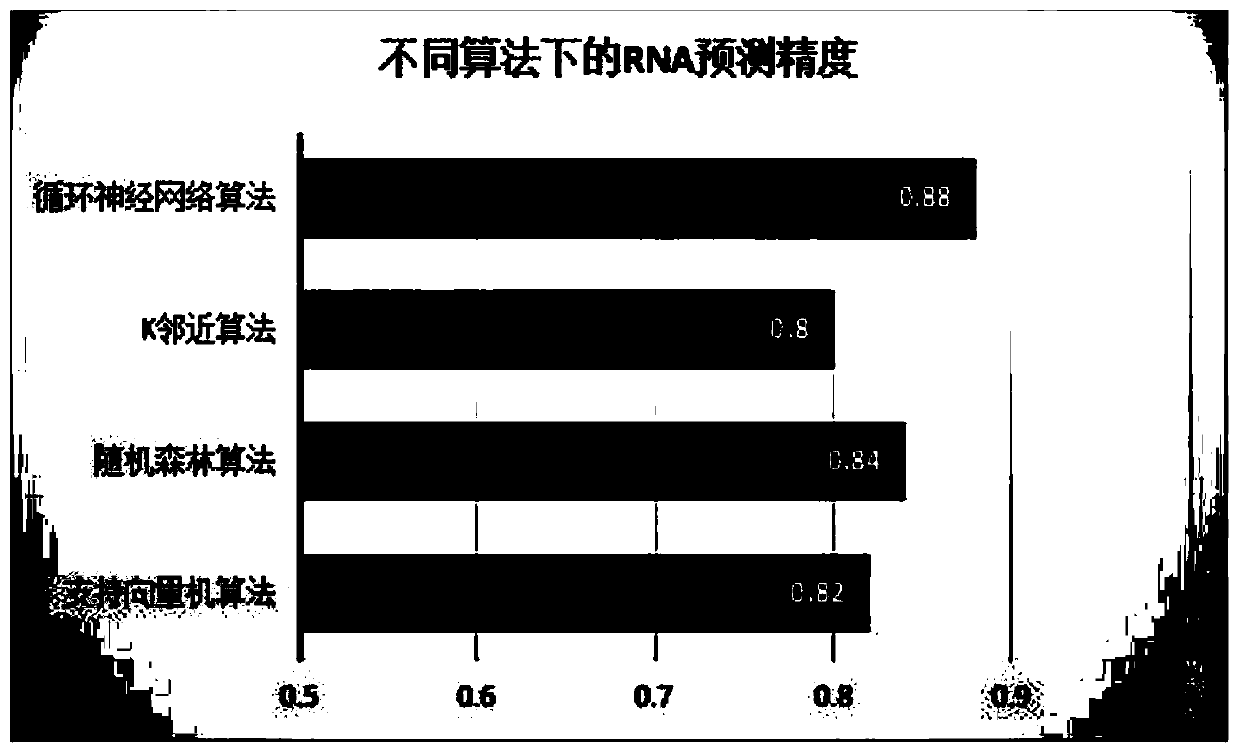
RNA secondary structure prediction method based on recurrent neural network
PendingCN110060728AEasy to findImprove forecast accuracyInstrumentsMolecular structuresData setAlgorithm
The invention discloses an RNA secondary structure prediction method based on a recurrent neural network. The RNA secondary structure prediction method includes the steps: carrying out data preprocessing on an RNA primary sequence data set in a PDB data set; dividing the RNA primary sequence into a long sequence, a medium sequence and a long sequence according to the length; vectorizing the sequence information to obtain feature information expressed in a matrix form, and filling the feature information of the sequence samples unsatisfying the standard by taking the longest sequence information of the long sequence, the medium sequence and the short sequence as the standard to obtain a feature matrix with a fixed dimension; and inputting the feature matrix into an LSTM model established based on a recurrent neural network, and performing RNA secondary structure prediction by using the LSTM model. The RNA secondary structure prediction method based on a recurrent neural network can predict the RNA secondary structure, and the prediction result is relatively accurate, and the implicit features of the RNA sequence can be further mined, and the more accurate RNA secondary structure canbe predicted.
Owner:ZHEJIANG UNIVERSITY OF SCIENCE AND TECHNOLOGY
Method for Derivatization of Proteins Using Hydrostatic Pressure
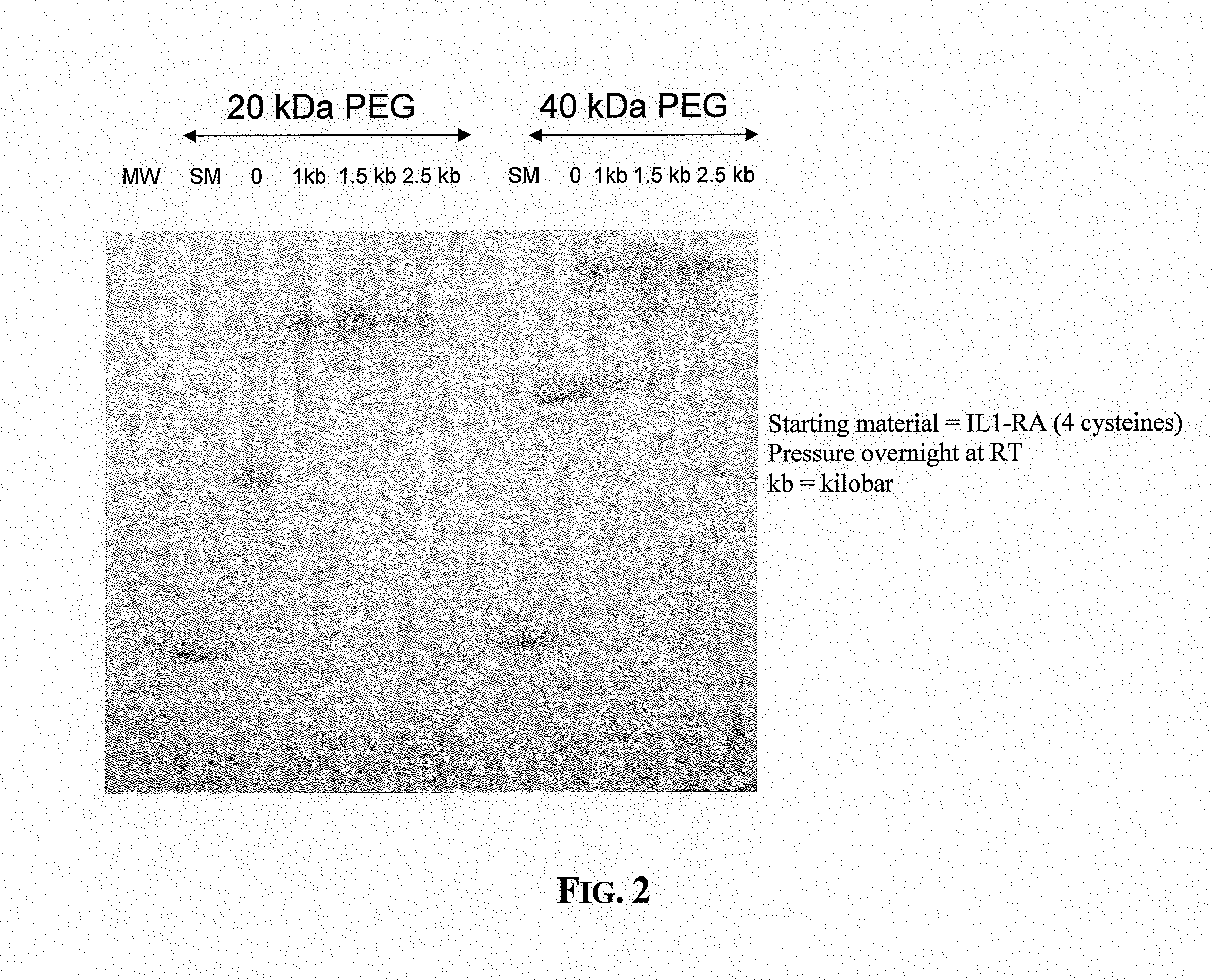
The present invention provides an effective method for derivatization of proteins using hydrostatic pressure to reversibly perturb the native conformation of a protein such that a normally buried functional group on the protein, such as an amino acid residue, or a ligand or cofactor associated with the protein, is exposed and available for derivatization by a polymer molecule or a cytotoxic agent. The methods described herein do not require use of chaotropes, changes in pH, changes in temperature, or genetic modification of the native primary sequence of the protein and are applicable to substantially all proteins.
Owner:BARFOLD INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com