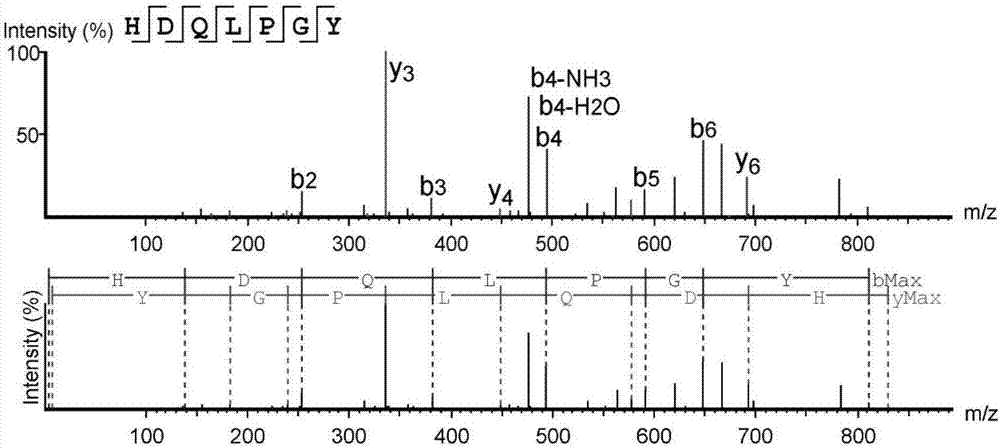
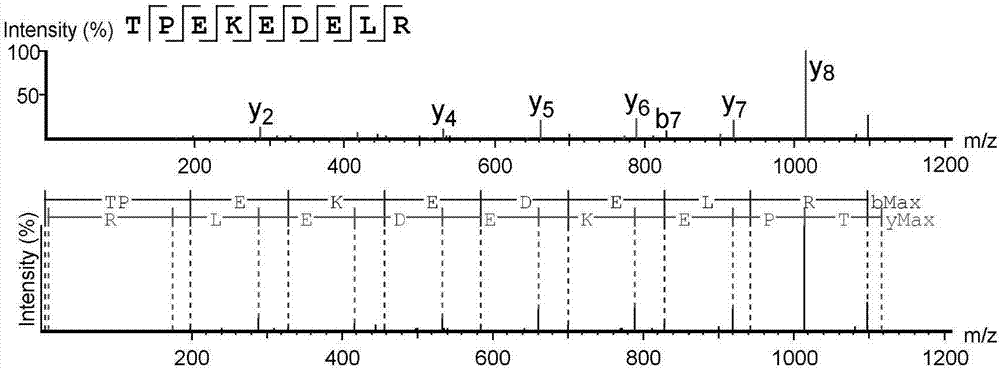
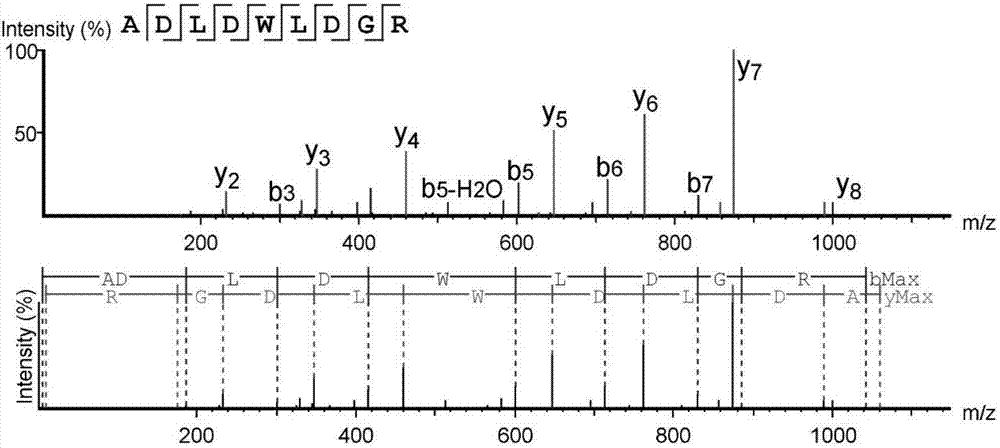
High-throughput active peptide screening method based on tandem mass spectrum and molecular docking
A technology of molecular docking and tandem mass spectrometry, applied in measurement devices, analytical materials, material inspection products, etc., can solve problems such as laboriousness, lack of separation, analysis, screening, identification solutions, and low throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Screening of dipeptidyl peptidase-IV (DPP-IV) inhibitors in variegated clams:
[0051] 1. Preparation of clam hydrolyzate:
[0052] Wash the variegated clam soft and silt, add 3 times the amount of water to decoct twice, 40 minutes each time, separate the decoction and meat dregs, drain; take the meat dregs, add 3 times the amount of water to homogenize, add enzyme activity 50000U / g of papain enzymolysis, the weight of papain added is 1% of the weight of meat dregs, the temperature of enzymolysis is 55°C, the pH of enzymolysis is 7, and the reaction time of enzymolysis is 4h; after the end of enzymolysis, Inactivate in a boiling water bath, then add a certain amount of absolute ethanol for graded alcohol precipitation, and finally prepare 30% ethanol precipitation site (alcohol precipitation site 1), 70% alcohol precipitation site (alcohol precipitation site 2) and alcohol precipitation site The supernatant (alcohol precipitation part 3) was concentrated and freeze-dri...
Embodiment 2
[0071] Screening of angiotensin-converting enzyme (ACE) inhibitory peptides from clam extracts:
[0072] 1. Preparation of clam extract
[0073] Wash the clam software from the sediment, add 3 times the amount of 60% ethanol to decoct twice, each time for 30 minutes, combine the two extracts, concentrate, and freeze-dry to obtain freeze-dried powder.
[0074] 2. Separation of clam extract by reverse phase column chromatography
[0075]The clam extract freeze-dried powder was reconstituted, and the Waters preparative HPLC system (2545-2489HPLC system) was used to separate the clam extract, using a C18 chromatographic column (5μm, 19×150mm), a binary gradient pump, and a detection wavelength of 220nm; Mobile phase A: 0.1% TFA, mobile phase B: methanol; elution conditions: 2% B, 0-3min, 2%-35% B, 3-9min, flow rate 10ml / min. The loading volume is 500 μl. The chromatographic peaks were collected, separated to obtain 5 different parts, concentrated and freeze-dried, and the freez...
Embodiment 3
[0095] Example 3 Screening and discovery of polypeptides with thrombin inhibitor activity in cockles
[0096] 1. Preparation of clam enzymatic hydrolyzate:
[0097] Wash the clam soft body from the sediment, add 3 times the amount of water to decoct twice, 40 minutes each time, separate the decoction liquid and meat dregs, and drain; take the meat dregs, add 3 times the amount of water to homogenize, and then add enzyme activity 10000U / g of neutral protease for enzymolysis, the added weight of neutral protease is 2% of the weight of meat dregs, the temperature of enzymolysis is 45°C, the pH of enzymolysis is 7, and the reaction time of enzymolysis is 4h; the end of enzymolysis Afterwards, it was inactivated in a boiling water bath, and freeze-dried to obtain a freeze-dried powder.
[0098] 2. Molecular exclusion separation of clam enzymatic hydrolyzate
[0099] Redissolve the lyophilized cockles enzymatic hydrolyzate, centrifuge and load the supernatant in batches on a Sepha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com