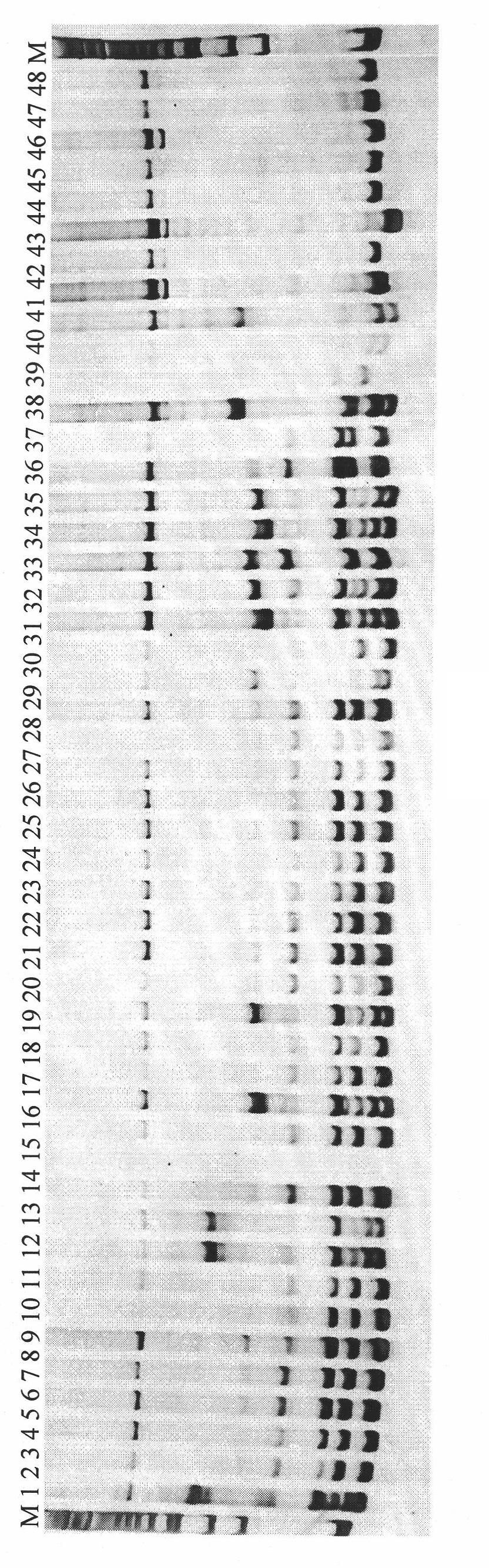
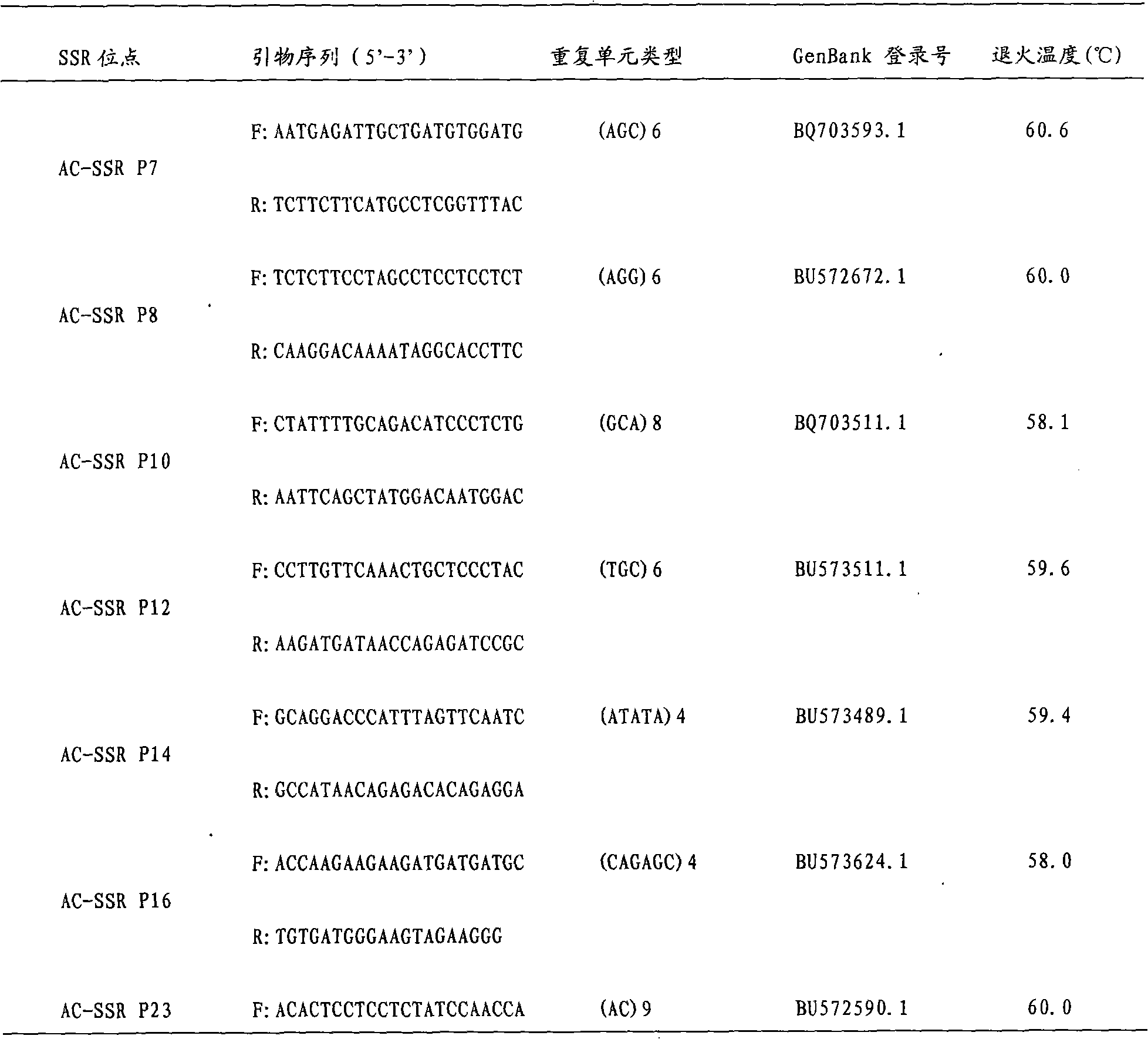
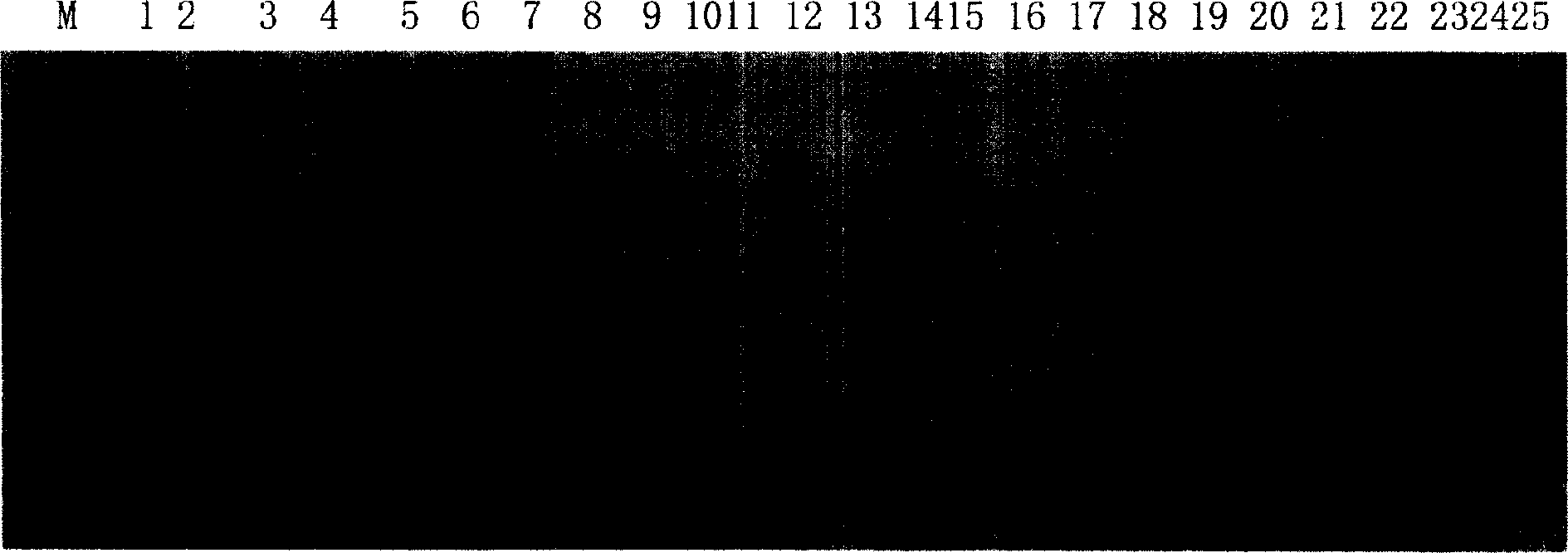Patents
Literature
58 results about "Satellite DNA" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
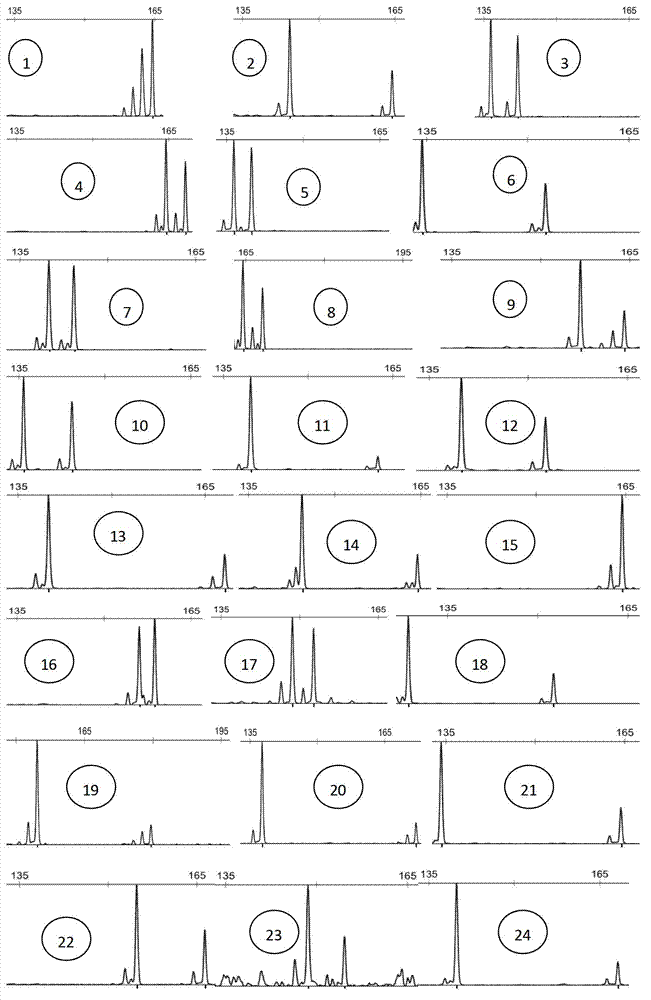
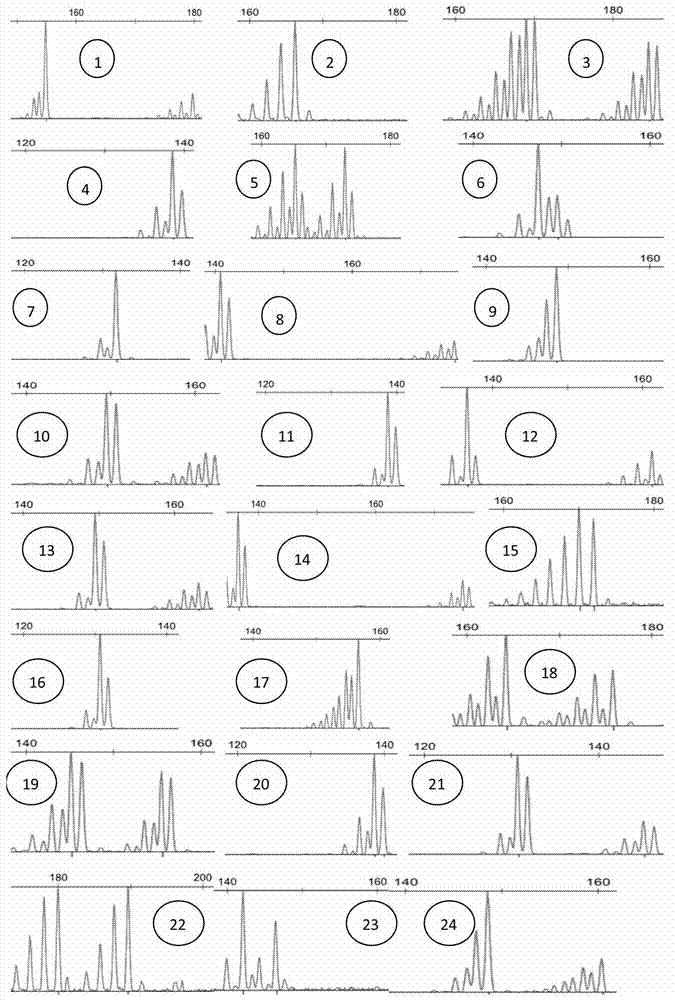
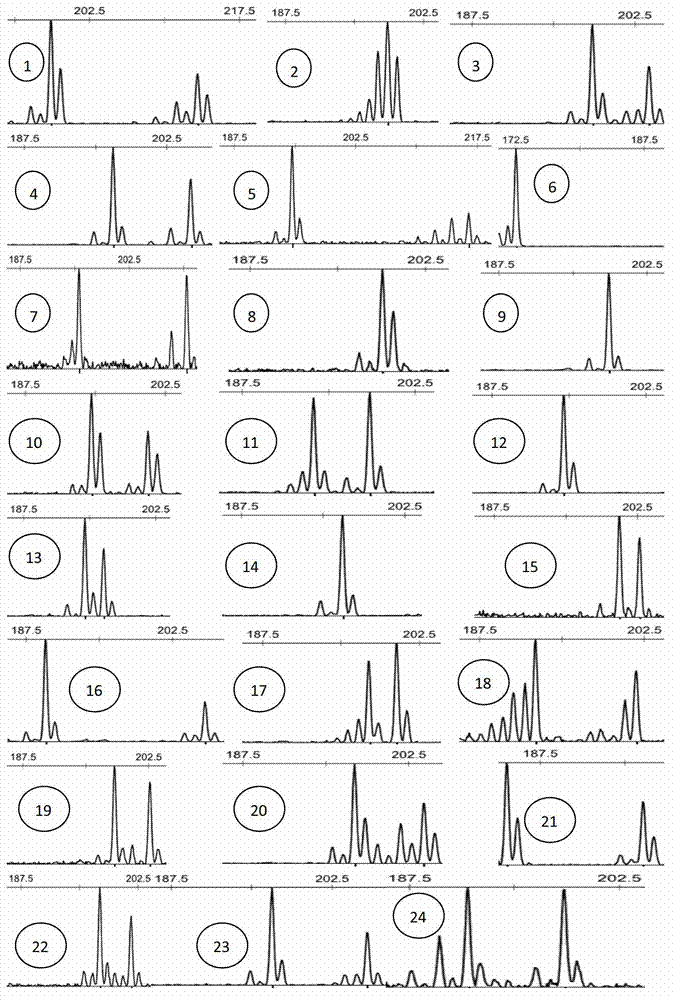
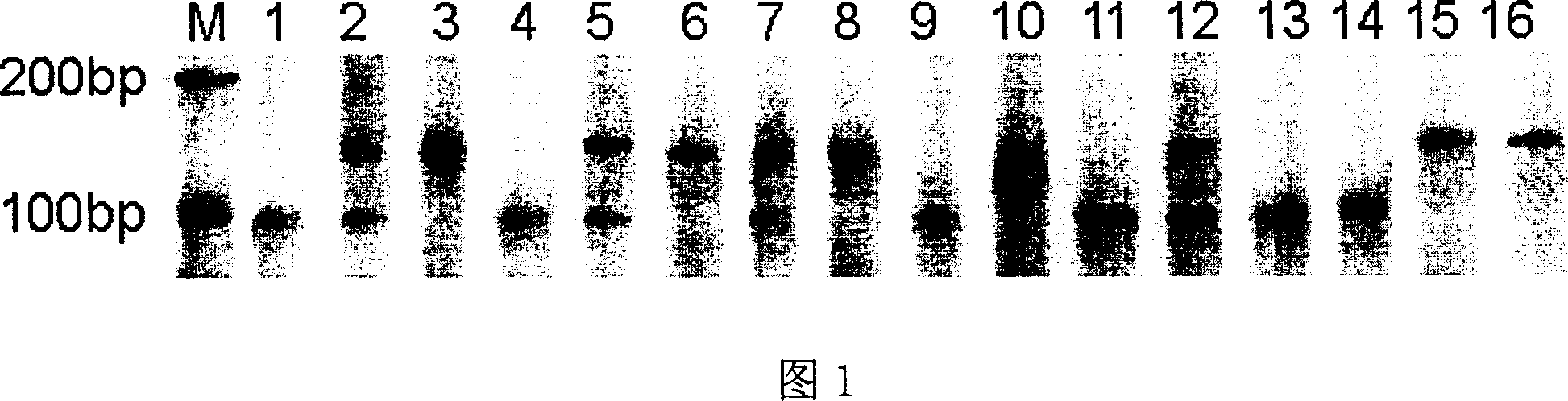
Satellite DNA consists of very large arrays of tandemly repeating, non-coding DNA. Satellite DNA is the main component of functional centromeres, and form the main structural constituent of heterochromatin.
Method for detecting cell proliferative disorders
InactiveUS6291163B1Fast and reliable and sensitive and non-invasive screeningHigh copy numberSugar derivativesMicrobiological testing/measurementCervical tissuePolymerase L
The present invention relates to the detection of a cell proliferative disorder associated with alterations of microsatellite DNA in a sample. The microsatellite DNA can be contained within any of a variety of samples, such as urine, sputum, bile, stool, cervical tissue, saliva, tears, or cerebral spinal fluid. The invention is a method to detect an allelic imbalance by assaying microsatellite DNA. Allelic imbalance is detected by observing an abnormality in an allele, such as an increase or decrease in microsatellite DNA which is at or corresponds to an allele. An increase can be detected as the appearance of a new allele. In practicing the invention, DNA amplification methods, particularly polymerase chain reactions, are useful for amplifying the DNA. DNA analysis methods can be used to detect such a decrease or increase. The invention is also a method to detect genetic instability of microsatellite DNA. Genetic instability is detected by observing an amplification or deletion of the small, tandem repeat DNA sequences in the microsatellite DNA which is at or corresponds to an allele. The invention is also a kit for practicing these methods.
Owner:THE JOHNS HOPKINS UNIVERSITY SCHOOL OF MEDICINE
Preparation method of developing Phaseolus vulgaris SSR primers by magnetic bead enrichment
InactiveCN102533727AImprove efficiencyDecline in clone sequencingMicrobiological testing/measurementDNA preparationAgricultural scienceStreptolydigin
The invention discloses a preparation method of developing Phaseolus vulgaris SSR primers by magnetic bead enrichment. The method comprises the following steps of: (1) digesting Phaseolus vulgaris genome DNA with Msel, and establishing a library by AFLP; (2) hybridizing the genome PCR library with a biotin-labeled repetitive sequence probe; (3) adding the hybridization mixed liquid into streptavidin-coated magnetic beads; (4) amplifying micro-satellite DNA fragments by PCR using eluent-purified micro-satellite DNA as DNA template; (5) linking the purified PCR amplification product on a T vector for cloning to obtain a DNA sequence with inserted fragments; and (6) repeating on the two sides of the core area to design primers, and screening primers to obtain effective microsatellite loci. The method is simple and feasible, increases efficiency of the primers, screens 20 pairs of effective primers from 48 pairs of primers, and provides guarantee for large-batch development of SSR primers.
Owner:WUHAN VEGETABLE RES INST
Microsatellite DNA markers for Trionyx sinensis
The invention discloses microsatellite DNA markers for Trionyx sinensis, which is characterized in that an enriched library of microsatellites (CA)n, (AAC)n, (ACAG)n and (GATA)n from the Trionyx sinensis is constructed, and positive clones of microsatellite sequences are screened and sequenced to obtain 63 clones containing microsatellite repetitive sequences, thus determining 15 microsatellite markers with rich polymorphism, namely, PSW01, PSW02, PSW03, PSW04, PSW05, PSW06, PSW07, PSW08, PSW09, PSW10, PSW11, PSW12, PSW13, PSW14 and PSW15. The microsatellite DNA markers for the Trionyx sinensis of the invention can be applied to researching genetic diversity and paternity test for the Trionyx sinensis in different geographical populations, have the advantage of good repeatability, and are reliable and effective molecular markers.
Owner:ANHUI NORMAL UNIV
Microsatellite molecular marker of lycoris
The invention discloses a microsatellite deoxyribonucleic acid (DNA) molecular marker of lycoris. 33 microsatellite DNA-containing clones are obtained and 10 microsatellite molecular markers with rich polymorphism, namely Lyra-1, Lyra-2, Lyra-3, Lyra-4, Lyra-5, Lyra-6, Lyra-7, Lyra-8, Lyra-9 and Lyra-10, are determined by establishing a lycoris microsatellite (CT)n enrichment library, screening and sequencing the positive clones of the microsatellite. The marker can be used for researching genetic diversity protection of the lycoris and variation and evolution relation between species and populations of lycoris plants and identifying the varieties of the lycoris; and the marker has high repeatability and is a reliable and effective molecular marker.
Owner:ANHUI NORMAL UNIV
Blue crab ptssr17 microsatellite DNA marker testing technique
ActiveCN101294217AEasy to detectSimple methodMicrobiological testing/measurementMicrosatelliteGenotype
The invention relates to a detection technology by ptssr17 microsatellite DNA markers in a blue crab. The technology is characterized in that: first, genome DNA of the blue crab is extracted and diluted to reserve; second, by utilizing a ptssr17 microsatellite core sequence in a genomic library of the blue crab, specificity primers are designed at the two ends of the sequence thereof; third, the genome DNA of different geographic groups of the blue crab or individuals in a blue crab group is processed through the PCR amplification by using the primers, and PCR products are processed through the modified polyacrylamide gel detection; finally, bands which are generated in the products are utilized for analyzing so as to determine the genotype of each individual, thereby obtaining a polymorphic map on the enormous genetic variation of the blue crab in the a ptssr17 core sequence area. The polymorphic map that the ptssr17 genetic mark gene locus of the blue crab shows the enormous genetic variation can be obtained rapidly; the method is simple and convenient; the each individual genotype of the blue crab at the locus can be detected intuitively from the obtained results. The detection technology is mainly used in the genetic marks among the blue crab groups, the genealogical identification, the genetic map construction, etc.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Polymorphism micro-satellite DNA molecular marker for deer and application of polymorphism micro-satellite DNA molecular marker
ActiveCN107164468AClear bandHigh polymorphismMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityGenetics
The invention relates to a micro-satellite DNA molecular marker for cervine animals. The molecular markers can be used for identifying allelic polymorphism, identify same or related cervine animals, distinguish the cervine animals and research the genetic diversity of the species group. The molecular marker can also be used for the genetic and phenotype research utilizing a statistics method such as linkage analysis, association mapping, linkage imbalance. The information can be used for breeding and / or selecting plants.
Owner:INST OF SPECIAL ANIMAL & PLANT SCI OF CAAS
Detection method for Apostichopus japonicas AjE101 micro-satellite DNA label
InactiveCN102140522AGood polymorphismImprove stabilityMicrobiological testing/measurementGerm plasmGenetic diversity
The invention belongs to the field of molecular biology DNA labeling technology and application, and in particular relates to Apostichopus japonicas express sequence tag EST micro-satellite label screening development and application. The invention provides Apostichopus japonicas AjE101 micro-satellite specific DNA primers, a polymerase chain reaction (PCR) reaction system by utilizing the primers, and a detection method for an Apostichopus japonicas AjE101 micro-satellite DNA label. The detection method comprises the following steps of: extracting genome of Litopenaeus vannamei Boone; designing the specific primers at two ends of an Apostichopus japonicas AjE101 micro-satellite DNA core sequence; and performing PCR amplification on genome DNA of different groups of the Litopenaeus vannamei Boone or individuals in the group by using the primers, analyzing products, and determining the genome of each individual so as to obtain an Apostichopus japonicas polymorphism genetic variation map. The invention is mainly applied to Apostichopus japonicas germ plasm resource and genetic diversity analysis, cular population genetics, construction of genetic maps and research of functional genes.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Novel plant DNA virus carrier and its use
InactiveCN1425772AEasy accessClear functionMicrobiological testing/measurementGenetic engineeringViral FunctionForeign protein
The plant DNA virus carrier includes the infectious carrier of Chinese tomato yellowing curly leaf virus DNA-A and DNA-beta, derived plant gene silencing carrier and virus expressing carrier. The present invention also provides one new kind of satellite DNA molecule constructing plant DNA virus carrier and it has at least 72% homology with any one sequence of SEQ ID No.1-12. The present invention also provides the use and method of utilizing plant DNA virus carrier in virus functional genome research, plant functional genome research and foreign protein expression. The present invention provides one important platorm for researching virus genomee structure, function and expression and regulation mechanism and the interaction between virus and plant; provides one excellent tool for researching plant functional gene; and establishing one technological platform for using plant as bioreactor expressing foreign protein.
Owner:ZHEJIANG UNIV
Method and kit for constructing poplar core collection through micro-satellite DNA molecular marking technology
InactiveCN104293895APolymorphism richReduce operating errorsMicrobiological testing/measurementGermplasmGenomic DNA
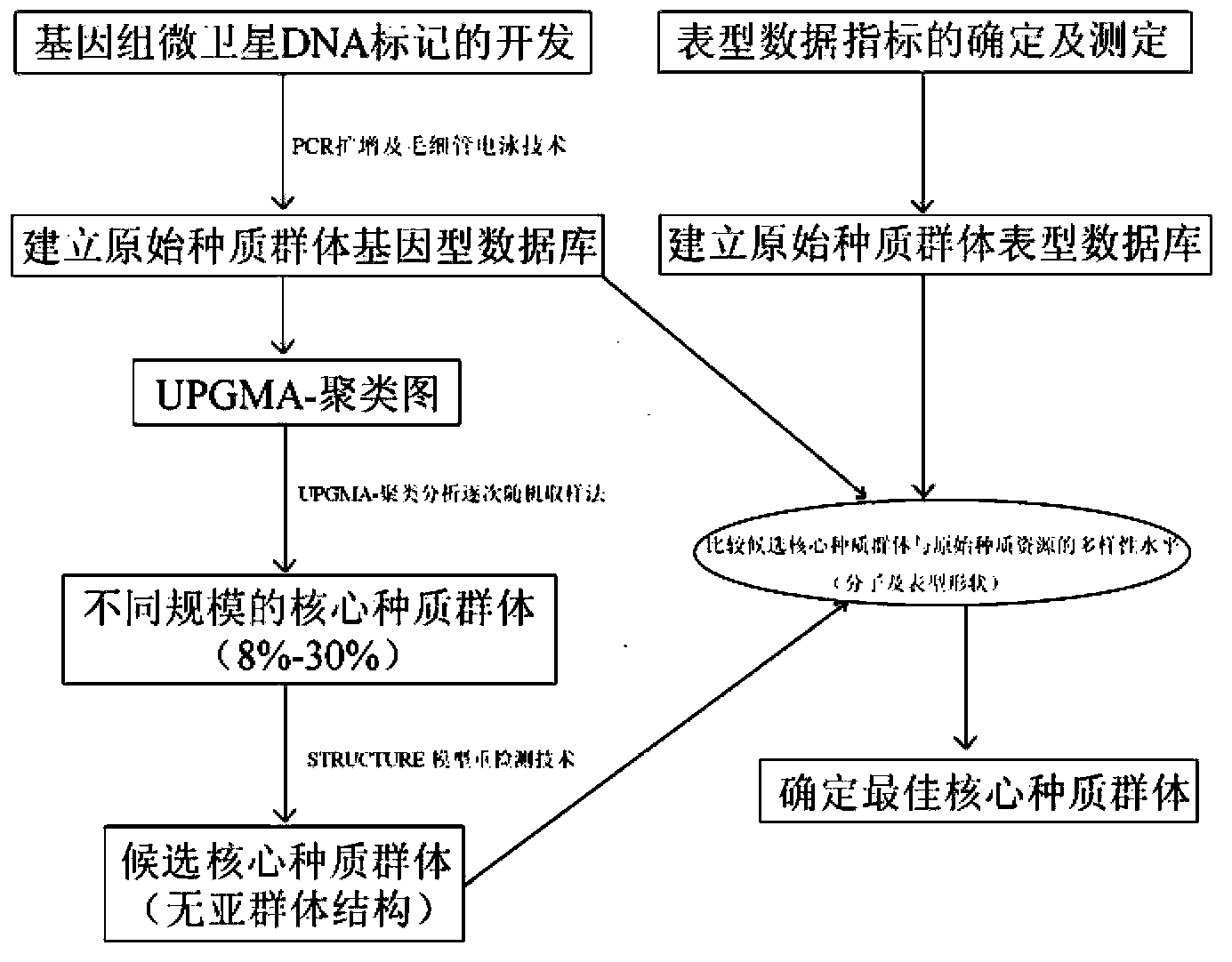
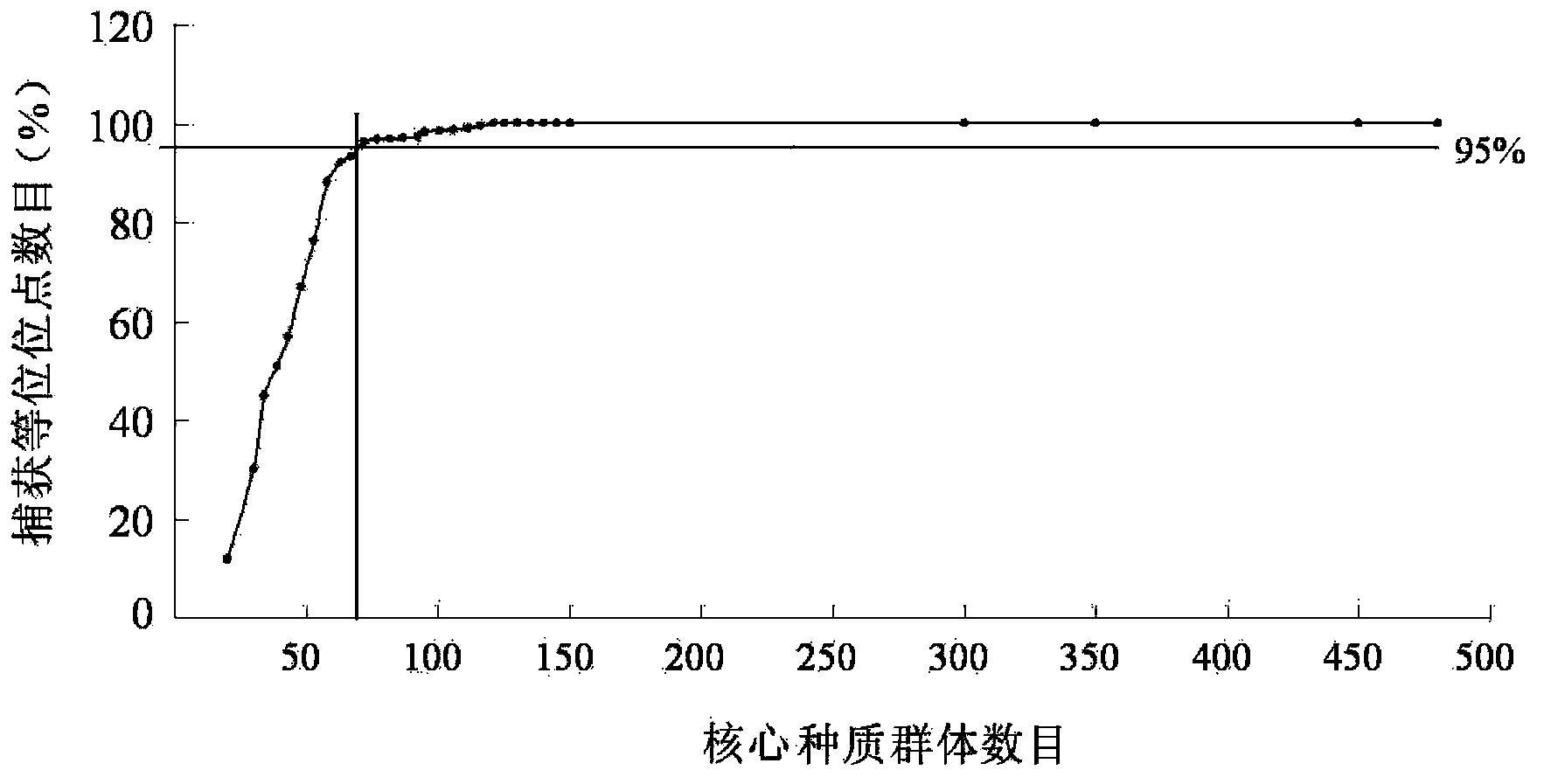
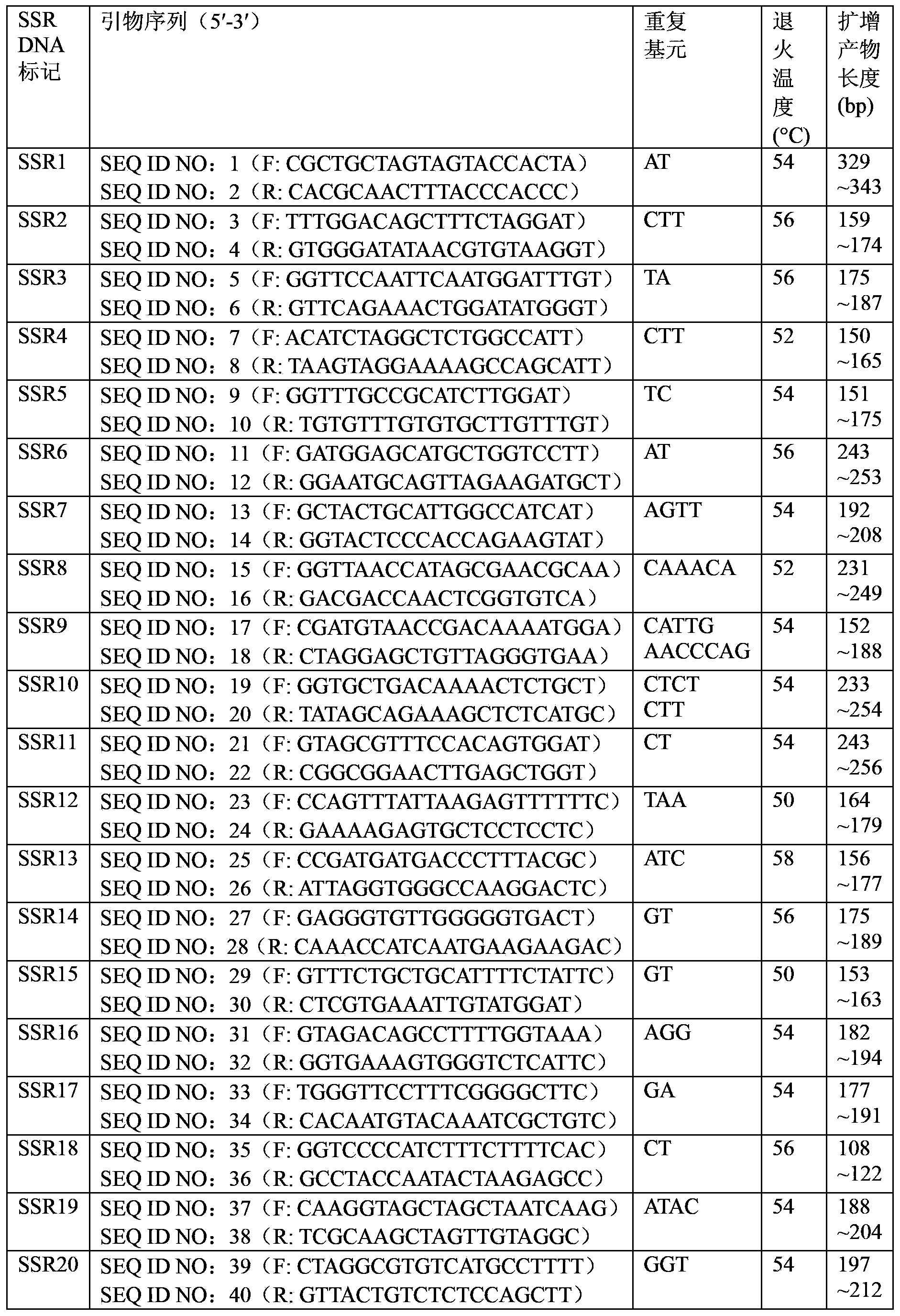
The invention discloses a method and a kit for constructing a poplar core collection through a micro-satellite DNA molecular marking technology. The method includes following steps: (S1) collecting populus tomentosa germplasm resources; (S2) extracting genomic DNA of the populus tomentosa in the step (S1); (S3) performing PCR amplification on a polymorphism micro-satellite DNA marker developed in a whole genome region of the populus tomentosa; (S4) construction a candidate core collection group, having different scales being 8-30%, through UPGMA-cluster analysis gradually random sampling and an STRCUTURE model re-detection combined analytic method; and (S5) determining 15% of the core collection group through comparison between the candidate core collection group and an original group according to differences in molecular genetic diversity and quantitative character variation level. By means of the method and the kit, on one hand, phenotypic character diversity of the species can be reflected well and precious core collection resources are free from being lost; and on the other hand, a good core collection resource material can be provided for developing molecule-marking directional selective breeding in a later period.
Owner:BEIJING FORESTRY UNIVERSITY
Nibea japonica microsatellite DNA markers
ActiveCN104726555AGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityMicrosatellite
The invention relates to the technology of molecular markers, and particularly relates to nibea japonica microsatellite DNA markers. The nibea japonica microsatellite DNA markers have microsatellite numbers of NJB01, NJB04, NJB06, NJB15 and NJB35, and have nucleotide sequences as shown in SEQ ID NO:1-5 sequentially. The six new microsatellite sites of nibea japonica as well as primer sequence and amplification method for amplifying the six microsatellite sites can be applied to studies on genetic diversity and paternity identification of different groups of nibea japonica. The nibea japonica microsatellite DNA markers have good repeatability, and are reliable and effective molecular markers.
Owner:MARINE FISHERIES RES INST OF ZHEJIANG
Chinese alligator microsatellite DNA mark
InactiveCN101368205AMicrobiological testing/measurementFermentationChinese alligatorGenetic diversity
The invention discloses an alligator sinensis microsatellite DNA mark which includes the construction of the enriched libraries of (CT / AG)n, (AC / GT)n and (GATA)n of an alligator sinensis microsatellite, the screening and sequencing of the positive clones of a microsatellite sequence, obtaining 25 clones which comprise the repeated sequence of the microsatellite and conforming 10 microsatellite marks of Asiu1, Asiu2, Asiu4, Asiu20, Asiu44, Asiu45, Asiu84, Asiu96, Asiu122 and Asiu234 of abundant polymorphism. The alligator sinensis microsatellite DNA mark can be used for the studies on genetic diversity of different geographic populations of the alligator sinensis, has good repetitiveness and is a reliable and effective molecule mark.
Owner:ANHUI NORMAL UNIV
Amygdalus communis EST (expressed sequence tag) microsatellite marker screening method
The invention relates to an Amygdalus communis EST (expressed sequence tag) microsatellite marker screening method. For the sequence of ESTs of Amygdalus communis disclosed by GeneBank, microsatellite retrieval software WebSat is used to excavate an EST microsatellite molecular marker and screen out two pairs of high-polymorphism Amygdalus communis microsatellite primer sequences. The Amygdalus communis microsatellite DNA marker is used for researches on the resource evaluation, the identification and the genetic diversity analysis of the Amygdalus communis, the construction of a genetic map, the molecular auxiliary selective breeding and the like, thereby being a reliable and effective molecular marker.
Owner:ZHEJIANG FORESTRY UNIVERSITY
Ramie micro-satellite DNA Label
This invention relates to a DNA molecular genetic marker of ramee and provides 15 DNA sequences of the ramee micro-satellite sites and the primer sequences for expanding the sites and a method, in which, the sites are used in classifying its varieties, its genetic relation analysis and marker assistant breeding and is a reliable and effective molecular marker.
Owner:INST OF BAST FIBER CROPS CHINESE ACADEMY OF AGRI SCI
Method for constructing walnut microsatellite DNA marker fingerprint spectrum and primer pair applied to method
PendingCN108300798AEasy and fast retrievalRapid identificationMicrobiological testing/measurementDNA/RNA fragmentationForward primerAgricultural science
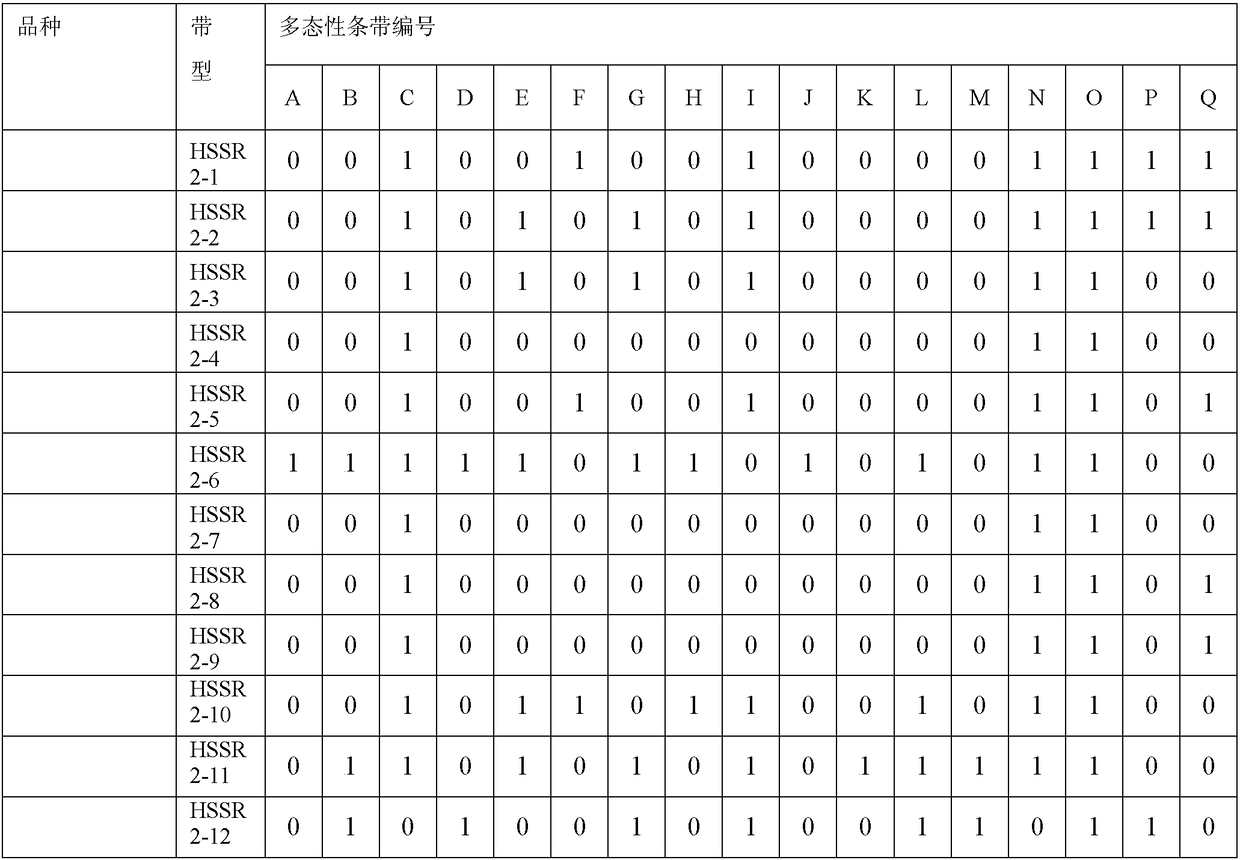
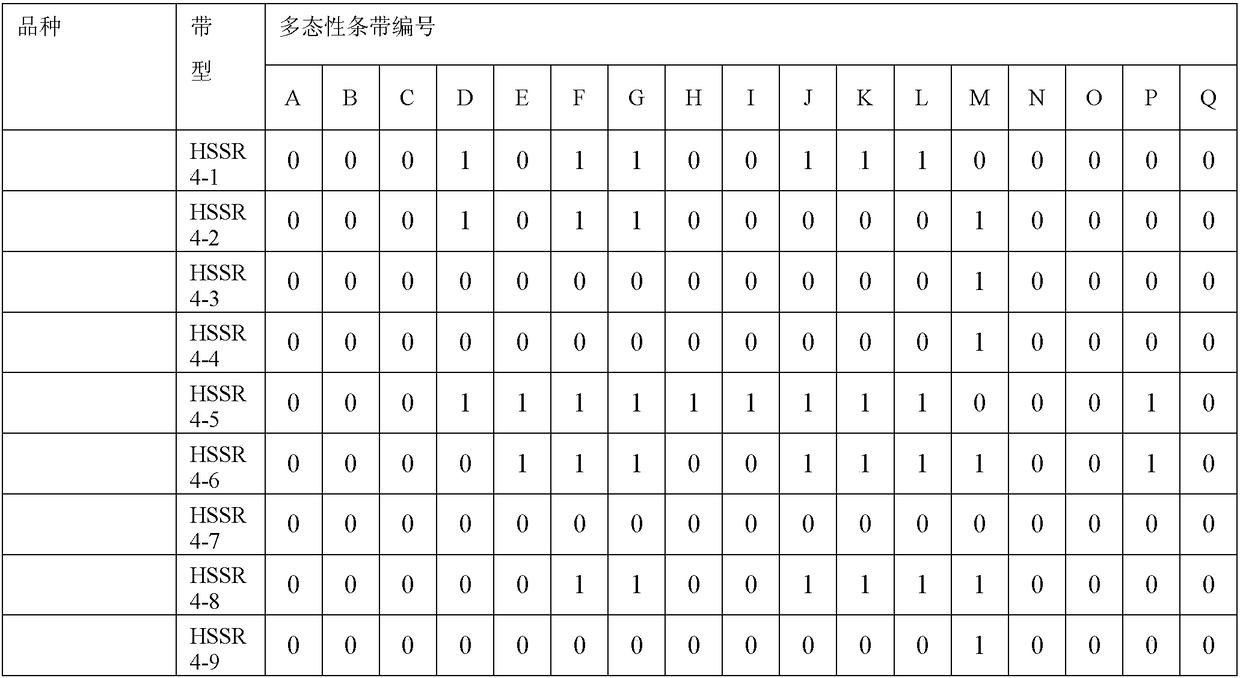
The invention relates to a method for constructing a walnut microsatellite DNA marker fingerprint spectrum and a primer pair applied to the method. A special primer HSSR2 for constructing the walnut microsatellite DNA marker fingerprint spectrum is constructed, the sequence of a forward primer is 5'-GATTTCTCCAGGAAGGCTCC-3', and the sequence of a reverse primer is 5'-AGGAAGAAGAGGAGGGCTTG-3'; a specific primer HSSR4 is constructed, the sequence of a forward primer is 5'-GAATGATGGCCTTGATGATTG-3', and the sequence of a reverse primer is 5'-GCAACCCATCCTCAACTTCT-3'. The constructed DNA marker fingerprint spectrum has high polymorphism, abundant fingerprints and good repeatability; the identification accuracy and efficiency of detecting walnut varieties by using the DNA marker fingerprint spectrum are greatly improved.
Owner:HORTICULTURE INST OF XINJIANG ACAD OF AGRI SCI
Blue crab ptssr36 microsatellite DNA marker testing technique
ActiveCN101294218AEasy to detectSimple methodMicrobiological testing/measurementMicrosatelliteGenotype
The invention discloses a detection technology by ptssr36 microsatellite DNA markers in a blue crab. The detection technology is characterized in that: first, genome DNA of the blue crab is extracted and diluted to reserve; second, by utilizing a ptssr36 microsatellite core sequence in a genomic library of the blue crab, specificity primers are designed at two ends of the sequence thereof; third, the genome DNA of different geographic groups of the blue crab or individuals in a blue crab group is processed through the PCR amplification by using the primers, and PCR products are processed through the modified polyacrylamide gel detection; finally, bands which are generated in the products are utilized for analyzing so as to determine the genotype of each individual, thereby obtaining a polymorphic map on the enormous genetic variation of the blue crab in the a ptssr36 core sequence area. The polymorphic map that the ptssr36 genetic mark gene locus of the blue crab shows the enormous genetic variation can be obtained rapidly; the method is simple and convenient; the each individual genotype of the blue crab at the locus can be detected intuitively from the obtained results. The detection technology is mainly used in the genetic marks among the blue crab groups, the genealogical identification, the genetic map construction, etc.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Northeast China tiger hair DNA extracting method and microsatellite DNA label primer sequence
The present invention is northeast China tiger hair DNA extracting method and microsatellite DNA label primer sequence. The hair DNA extracting process includes flushing the hair root inside sterilized centrifuge tube with double distilled water and alcohol and dry at room temperature; adding mixed solution of Tris-HCl, EDTA and NaCl and containing also SDS, DTT and proteinase K in 0.5 ml to obtain liquid A; twice extracting liquid A and separating for several times to obtain sample E; and obtaining the microsatellite DNA label primer sequence Pti002 of northeast China tiger. The obtained microsatellite DNA label primer sequence Pti002 is shown in the present invention.
Owner:李迪强 +1
Flax micro-satellite DNA mark
The invention discloses a flax micro-satellite DNA mark and provides a DNA sequence of 25 flax micro-satellite loci and an enlarging method for enlarging the primer sequence of the 25 flax micro-satellite loci. The 25 flax micro-satellite loci are used for classifying flax species, analyzing the genetic affinity and marking auxiliary culturing. With favorable repetitiveness, the flax micro-satellite DNA mark is a reliable and effective molecular mark.
Owner:INST OF BAST FIBER CROPS CHINESE ACADEMY OF AGRI SCI
Method for identifying fx151 genetic variation map by using litopenaeus vannamei boone fx151 microsatellite DNA marker
InactiveCN101967519AGood polymorphismImprove stabilityMicrobiological testing/measurementMicrosatelliteGenotype
The invention discloses a method for identifying an fx151 genetic variation map by using a litopenaeus vannamei boone fx151 microsatellite Deoxyribonucleic Acid (DNA) marker, which is characterized by comprising the following steps of: extracting a litopenaeus vannamei boone genome and diluting for later use, designing specific primers at two ends of a sequence by utilizing a litopenaeus vannamei boone fx151 microsatellite DNA core sequence, performing Polymerase Chain Reaction (PCR) amplification on genomes DNA of different groups or individuals in the groups of the litopenaeus vannamei boone by using the primers, and performing denaturing polyacrylamide gel detection on a PCR product; and performing silver staining development by utilizing stripes generated by the product and analyzing, and determining a genotype of each individual to obtain a polymorphic genetic variation map of the litopenaeus vannamei boone at a fx151 genetic marker gene locus. The method can rapidly obtain the polymorphic map of the litopenaeus vannamei boone fx151 genetic marker gene locus which presents high genetic variation, is convenient and simple, and the obtain result can intuitively detect the genotype of each individual litopenaeus vannamei boone.
Owner:SUN YAT SEN UNIV
Detection method of ecssr1024 microsatellite dna marker in white shrimp
ActiveCN102277438ASimple methodAccurate methodMicrobiological testing/measurementExopalaemon carinicaudaMicrosatellite
The invention provides a method for detecting an EcSSR1024 microsatellite DNA marker in exopalaemon carinicauda, which comprises: extracting genomic DNA of the exopalaemon carinicauda and diluting the genomic DNA for later use; designing specific primers at the two ends of the core sequence of an EcSSR1024 microsatellite; performing the polymerase chain reaction (PCR) amplification of the genomicDNA of the exopalaemon carinicauda by using the primers, and detecting the product of the PCR amplification; and analyzing by using strips appeared in the product, and obtaining the polymorphic map of a highly-genetic variation in the core sequence area of the EcSSR1024 in the exopalaemon carinicauda. By using the primers and detection method, which are provided by the invention, the genetic variation in the core area of the EcSSR1024 microsatellite in each individual of exopalaemon carinicauda can be detected, and the polymorphic map of the highly-genetic variation in a genetic marker gene locus in the EcSSR1024 in exopalaemon carinicaud can be obtained quickly. The method is suitable to be used in population genetic marking, genealogical identification, genetic map construction and the like.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Hibiscus cannabinus L. expression sequence tag SSR (Simple Sequence Repeat) DNA markers
The invention relates to a molecular marker technology and specifically relates to hibiscus cannabinus L. expression sequence tag SSR (Simple Sequence Repeat) DNA markers. The following steps are included: new SSR sites are developed based on a hibiscus cannabinus L. transcriptome sequencing sequence, and the primer sequences of the SSR sites are amplified and polymorphic analysis is performed on the SSR sites. The hibiscus cannabinus L. expression sequence tag SSR DNA markers are characterized in that the DNA sequences of 24 SSR markers are separated and identified and the primer sequences of the 24 SSR sites are amplified based on the transcription factor expression sequence tags of hibiscus cannabinus L. transcriptome sequencing. The 24 new hibiscus cannabinus L. SSR DNA markers can be applied to researches such as genetic diversity analysis of the hibiscus cannabinus L. breeds, genetic linkage map establishment and molecular marker assisted selection, and are good in repeatability; as a result, the 24 new hibiscus cannabinus L. SSR DNA markers are reliable and effective molecular markers.
Owner:FUJIAN AGRI & FORESTRY UNIV
Identification method and identification kit for genetic relationships among different species among sections of populus
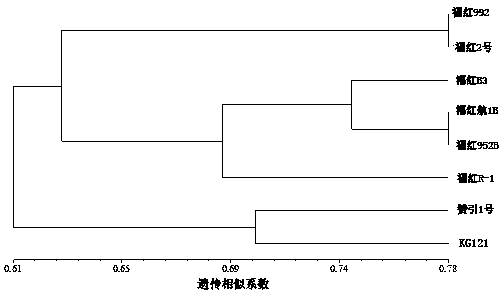
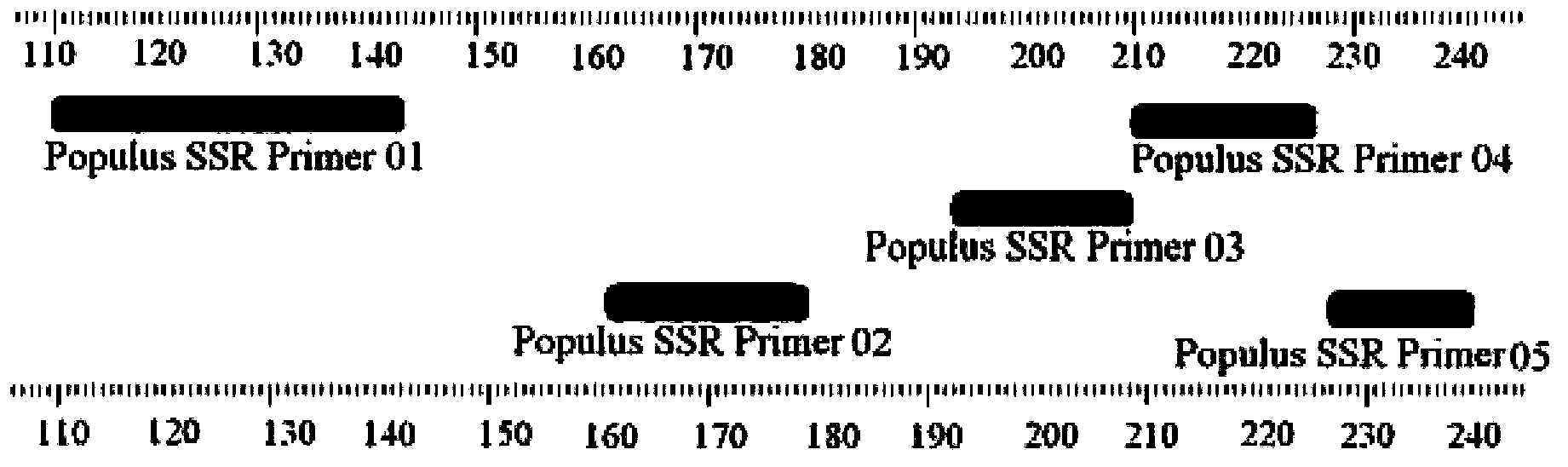
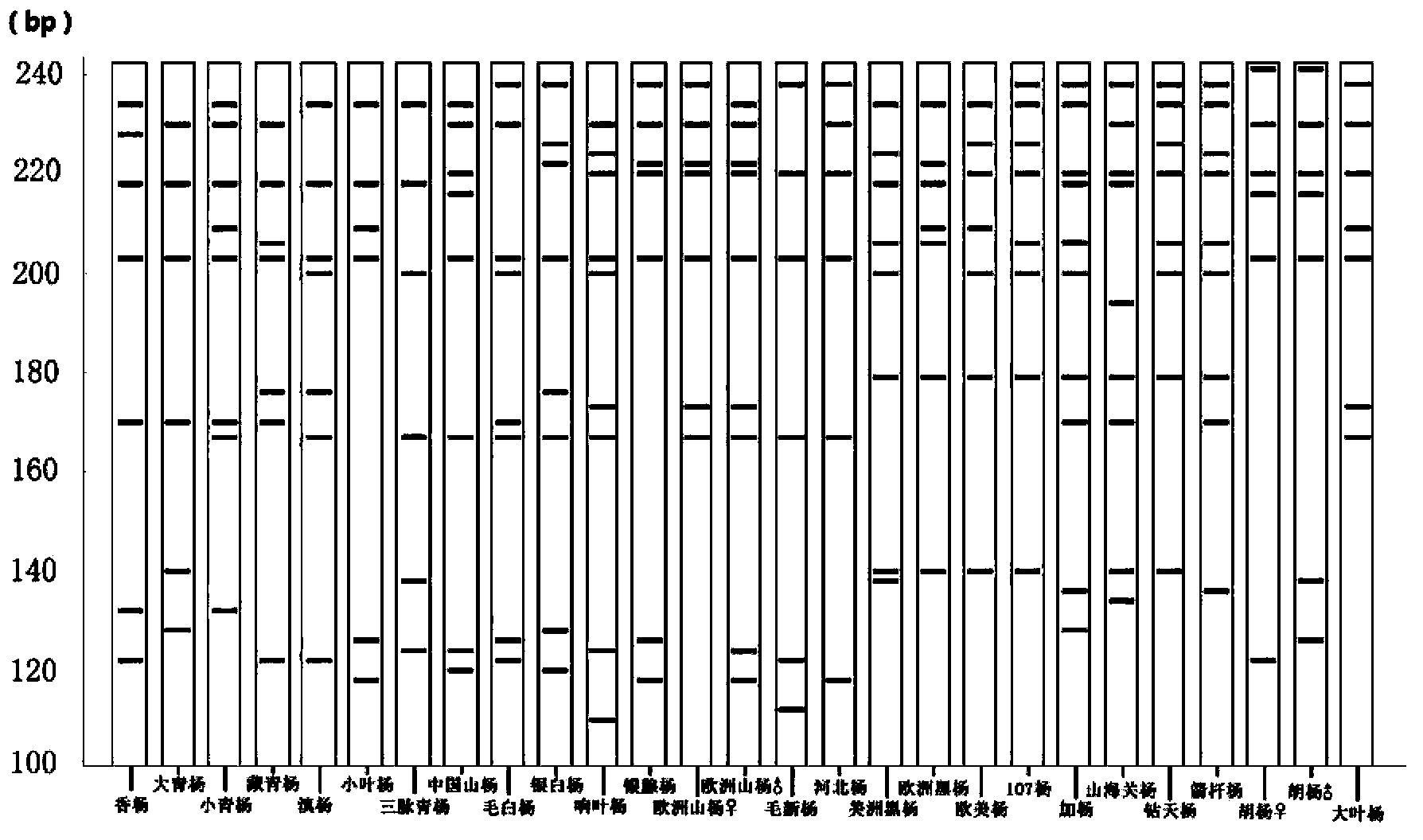
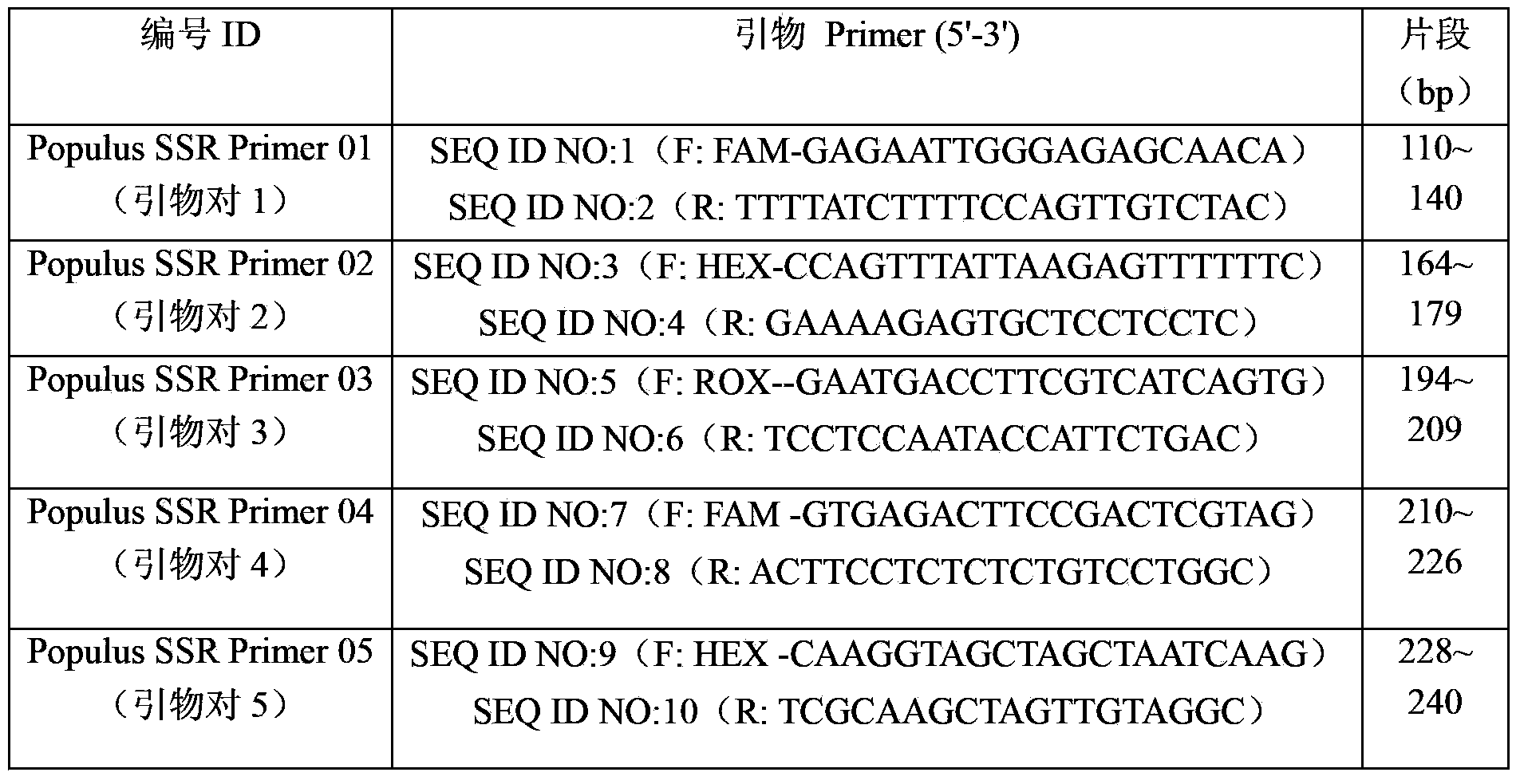
An identification method and an identification kit for genetic relationships among different species among sections of populus are disclosed. The method includes following steps of: S1) subjecting populus to genetic typing by adoption of polymorphic microsatellite DNA primer pairs, and constructing microsatellite DNA finger-prints of individuals in the species; and S2) subjecting populus to be identified to genotype amplification by adoption of the polymorphic microsatellite DNA primer pairs, and determining the genetic relationships of the populus to be identified according to the microsatellite DNA finger-prints, wherein the polymorphic microsatellite DNA primer pairs comprise a primer pair 1, a primer pair 2, a primer pair 3, a primer pair 4 and a primer pair 5. By application of the technical scheme, the microsatellite DNA finger-prints are constructed by utilization of the five pairs of polymorphic microsatellite DNA primers which are high in polymorphism and free of interaction between each two, and then genetic relationships of the populus among sections are determined through the microsatellite DNA finger-prints. The method is rapid, efficient, stable and suitable for different growth periods of the populus.
Owner:BEIJING FORESTRY UNIVERSITY
Larimichthys polyactis microsatellite DNA molecular marker
InactiveCN102876668AMicrobiological testing/measurementDNA/RNA fragmentationLarimichthys polyactisPolymorphic microsatellites
The invention discloses a larimichthys polyactis microsatellite DNA molecular marker. The invention adopts the technical scheme that an enhanced library of larimichthys polyactis microsatellites (GT) 17, (GGT) 9 and (CTAT) 8 is built, larimichthys polyactis polymorphic microsatellite primers are screened, and the genetic polymorphism detection is carried out on larimichthys polyactis microsatellite loca, so that 21 larimichthys polyactis microsatellite markers with rich polymorphism are developed. The larimichthys polyactis microsatellite DNA molecular marker is applicable to researches on the population genetic structure and the genetic variation level of a larimichthys polyactis population.
Owner:SUN YAT SEN UNIV
Bluish dogbean micro satellite DNA label
A DNA molecular genetic marker of kenaf is disclosed. The DNA sequences of 5 polymorphic micro-satellite DNA markers are separated and cloned. Their primer sequences are amplified. Said micro-satellite DNA markers can be used for classifying the varieties of kenaf, discriminating the genetic relation and culturing new variety.
Owner:HUNAN NORMAL UNIVERSITY
Radix polygonati officinalis microsatellite DNA (deoxyribonucleic acid) molecular marker
InactiveCN102703444AHigh polymorphismMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityOfficinalis
The invention discloses a radix polygonati officinalis microsatellite DNA molecular marker, comprising the steps of constructing a radix polygonati officinalis microsatellite (CT) n enrichment library; screening and sequencing microsatellite sequence positive clone; obtaining 55 clones containing the microsatellite repeated sequence; and screening to obtain 9 high polymorphic microsatellite molecular markers Po1, Po2, Po3, Po4, Po5, Po6, Po7, Po8 and Po9. The radix polygonati officinalis microsatellite DNA molecular marker can be used for researching genetic diversity of the radix polygonati officinalis, variation and evolutionary relationship between polygonatum plants and population, has good repeatability and is a reliable and efficient molecular marker.
Owner:ANHUI NORMAL UNIV
Microsatellite DNA (Deoxyribonucleic Acid) marker fingerprint spectrum of red ramies and application of microsatellite DNA marker fingerprint spectrum
The invention relates to a DNA (Deoxyribonucleic Acid) fingerprint spectrum of red ramie germplasm resources. The fingerprint spectrum is a DNA fingerprint spectrum constructed by taking combination of 64 pairs of SSR (Simple Sequence Repeat) primer allelic segments as a foundation and taking 24 red ramie varieties / series as test materials through DNA extracting, PCR (Polymerase Chain Reaction) amplification, identification of a PCR product and data analysis, and finally taking 215 SSR markers. The fingerprint spectrum can be used for distinguishing red ramie varieties / series for testing and can be used for identifying or analyzing the purity of the red ramie varieties / series. Compared with traditional morphological and cytological identification, the fingerprint spectrum has the advantages of short detection time, high accuracy and good repeatability.
Owner:FUJIAN AGRI & FORESTRY UNIV
Method for detecting Jassr131 microsatellite deoxyribonucleic acid (DNA) marker of Charybdis japonica
InactiveCN102162009AEasy to detectSimple methodMicrobiological testing/measurementCharybdis japonicaGenomic DNA
The invention discloses a method for detecting a Jassr131 microsatellite deoxyribonucleic acid (DNA) marker of Charybdis japonica. The method is characterized by comprising the following steps of: extracting genomic DNA of the Charybdis japonica and diluting for later use; designing a specific primer at both ends of a Jassr131 microsatellite core sequence in an enriched microsatellite library of the Charybdis japonica; performing polymerase chain reaction (PCR) amplification on genomic DNA of different geographical populations of the Charybdis japonica or genomic DNA of individuals in the populations by using the primer, and detecting a PCR product by using metamorphic polyacrylamide gel; and analyzing by utilizing stripes of the product, and determining a genotype of each individual so as to obtain a polymorphic map of high genetic variation of the Charybdis japonica in a Jassr131 core sequence region. Sequences of the specific primer of the Jassr131 microsatellite core sequence are that: the sequence of a plus strand is 5'-CCAGGGAATTGAAACACT-3', and the sequence of a minus strand is 3'-TATGAAGGCTCTGCGAAA-5'; and the annealing temperature is 50 DEG C. The method is simple and convenient; and the genotype of each individual of the Charybdis japonica at the site can be intuitively detected according to the obtained results.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Rattus losea microsatellite DNA marker and amplification primer thereof and a detection method and application thereof
ActiveCN108300793AGood repeatabilityMicrobiological testing/measurementDNA/RNA fragmentationNucleotideGenetic diversity
The invention discloses a rattus losea microsatellite DNA marker and an amplification primer thereof and a detection method and application thereof. Through the construction of rattus losea microsatellite (CT) n, (AG) n, (TG) n and (AC) n enriched libraries, screening and sequencing of positive clones containing microsatellite sequences and the cloning containing a microsatellite repeated sequence, eight microsatellite markers chrom1-26, chrom2-2, chrom15-6, chrom16-6, chrom16-12, chrom20-9, chrom20-37 and chrom-16 rich in polymorphism are obtained, and nucleotide sequences thereof are respectively as shown in SEQ ID No. 1-8. The rattus losea microsatellite DNA marker can be applied to researches of study of genetic diversity and the identification of parental relationship of different populations of rattus losea, is good in repeatability and is a reliable and effective molecular marker.
Owner:INST OF ZOOLOGY GUANGDONG ACAD OF SCI
Method for detecting EcSSR0003 microsatellite deoxyribonucleic acid (DNA) markers of palaemon carincauda holthuis
The invention provides a method for detecting EcSSR0003 microsatellite deoxyribonucleic acid (DNA) markers of palaemon carincauda holthuis. The method comprises the following steps of: extracting DNA of a genome of the palaemon carincauda holthuis, and diluting for later use; designing specific primers at two ends of an EcSSR0003 microsatellite core sequence; performing polymerase chain reaction (PCR) amplification on the DNA of the genome of the palaemon carincauda holthuis by using the primers, and detecting a PCR product; and analyzing by utilizing stripes of the product to obtain a polymorphic map of the palaemon carincauda holthuis subjected to high genetic variation in an EcSSR0003 core sequence area. By the primers and the detection method, the genetic variation of each individual of the palaemon carincauda holthuis in the EcSSR0003 microsatellite core area can be detected, and the polymorphic map of EcSSR0003 genetic marker gene loci, which present the high genetic variation, of the palaemon carincauda holthuis can be obtained quickly. The method is suitable for application of genetic markers, genealogy authentication, the construction of genetic maps and the like of palaemon carincauda holthuis groups.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Micro-satellite DNA (Deoxyribose Nucleic Acid) method for identifying reproductive mode of termite nest group
InactiveCN102559896AImprove targetingMicrobiological testing/measurementRepetitive SequencesGenotype
The invention relates to a micro-satellite DNA (Deoxyribose Nucleic Acid) method for identifying a reproductive mode of a termite nest group, which comprises the following steps of: collecting above 20 termite samples from a same nest group in the open air, extracting a sample genome DNA and diluting; searching a micro-satellite site related to a termite reproductive mode on GenBank, designing a primer on the side wing of a micro-satellite repetitive sequence by utilizing software Primer Premier 5.0, carrying out electrophoresis on an automatic analyzer after the termite samples DNA are subjected to PCR (Polymerase Chain Reaction) amplification by selecting a micro-satellite primer and determining the genotype of each strip corresponding sample in a electrophoretic diagram by using SAGAGT software; and carrying out chi-square test on observed genotype frequencies and expected genotype frequencies of the termite samples based on a Mendel's law and judging the reproductive mode of the termite nest group. The micro-satellite DNA identification for reproductive modes of ten reticulitermes chinensis nest groups in Changsha by adopting the micro-satellite DNA method shows that the reticulitermes chinensis nest groups include seven simple nest groups, two amplified nest groups and one mixed nest group. The micro-satellite DNA method has the advantages that the operation is simple and convenient, the results are reliable, the reproductive modes of the termite group can be accurately judged, and the important reference value is provided for the formulation of a termite prevention strategy.
Owner:HUAZHONG AGRI UNIV
Blue crab ptssr17 microsatellite DNA marker testing technique
ActiveCN101294217BEasy to detectSimple methodMicrobiological testing/measurementMicrosatelliteGenotype
The invention relates to a detection technology by ptssr17 microsatellite DNA markers in a blue crab. The technology is characterized in that: first, genome DNA of the blue crab is extracted and diluted to reserve; second, by utilizing a ptssr17 microsatellite core sequence in a genomic library of the blue crab, specificity primers are designed at the two ends of the sequence thereof; third, the genome DNA of different geographic groups of the blue crab or individuals in a blue crab group is processed through the PCR amplification by using the primers, and PCR products are processed through the modified polyacrylamide gel detection; finally, bands which are generated in the products are utilized for analyzing so as to determine the genotype of each individual, thereby obtaining a polymorphic map on the enormous genetic variation of the blue crab in the a ptssr17 core sequence area. The polymorphic map that the ptssr17 genetic mark gene locus of the blue crab shows the enormous geneticvariation can be obtained rapidly; the method is simple and convenient; the each individual genotype of the blue crab at the locus can be detected intuitively from the obtained results. The detectiontechnology is mainly used in the genetic marks among the blue crab groups, the genealogical identification, the genetic map construction, etc.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com