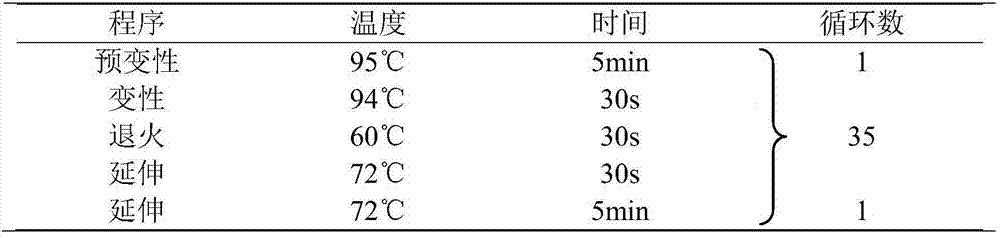
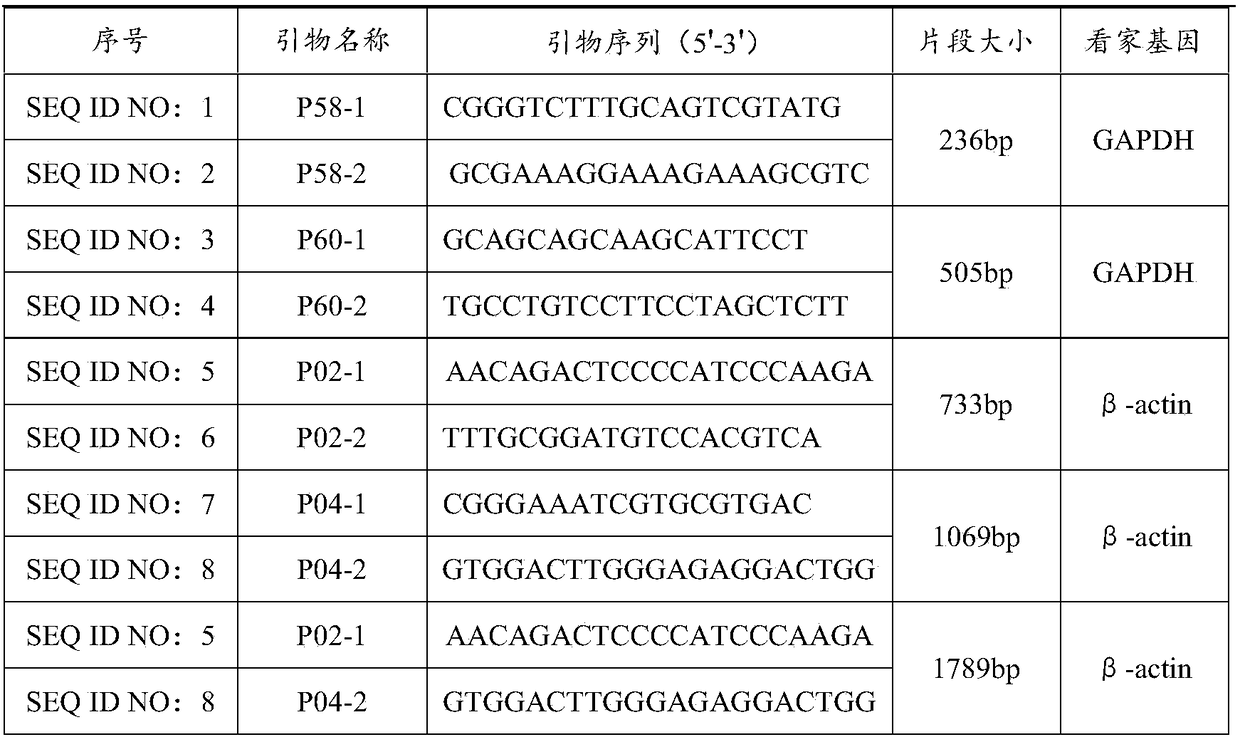
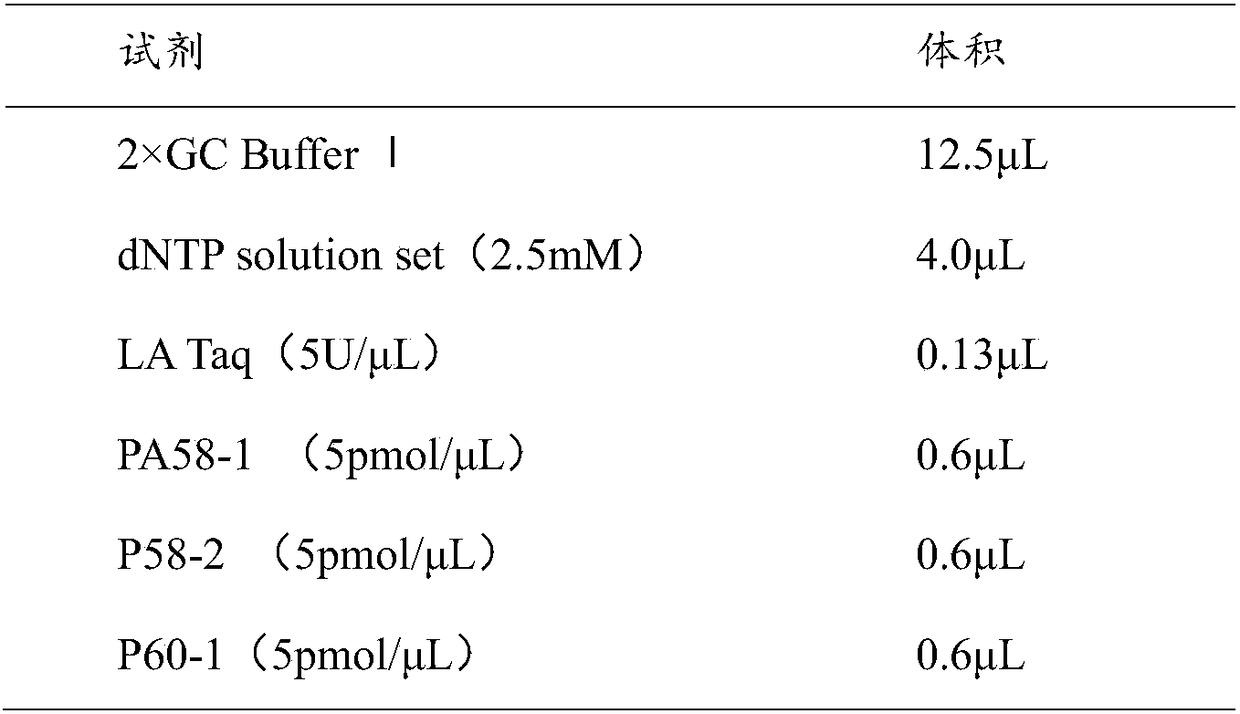
Patents
Literature
158results about How to "Clear band" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
SSR molecular marker method of brassica allohexaploid and primers thereof
ActiveCN104059971AReduce concentrationReduce generationMicrobiological testing/measurementDNA/RNA fragmentationBrassicaAgricultural science
The invention discloses an SSR molecular marker method of a brassica allohexaploid and primers thereof. The SSR molecular marker method of the brassica allohexaploid comprises the steps: hybridizing a brassica allohexaploid parent, and performing microspore culture on the obtained filial generation to obtain a DH colony; extracting genome DNAs of the brassica allohexaploid parent and the DH colony, and performing PCR amplification with all genome DNAs as a template by using the primers; constructing a genetic map of the brassica allohexaploid parent according to a PCR amplification result, wherein 217 pairs of primers are included, with base sequences respectively shown in SEQ ID No. 1 / 2-433 / 434. According to the SSR molecular marker method, a first genetic map of the brassica allohexaploid is constructed, and the basis is provided for further performing QTL location and molecular marker assistant breeding.
Owner:ZHEJIANG UNIV
Purity testing method for tomato hybrid based on PCR technology
InactiveCN101017151AClear bandGood repeatabilityMaterial analysis by observing effect on chemical indicatorMicrobiological testing/measurementRelationship - FatherPcr ctpp
This invention relates to one tomato mixture purification test method based on PCR technique in biological technique field, which comprises the following steps: using product mixture tomato Shu power 8 as subject to extend molecule label filtering to identify one RAPD and one SSR label object of local abnormal band; RAPD label generate mother property label of NAURP409800 and father property label of NAURP4091100; SSR label lead object property mother label of NAUSSRTOM47140 and father label of NAUSSRTOM 47148.
Owner:NANJING AGRICULTURAL UNIVERSITY
Rice blast resistance gene Pi9 functional specificity molecular marker and application thereof
InactiveCN105950743ALower quality requirementsDetection is simple and fastMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionAgricultural science
The invention provides a rice blast resistance gene Pi9 functional specificity molecular marker and application thereof. The molecular marker is named as Pi9InDel, and the nucleotide sequence is shown in SEQ ID NO.1. The molecular marker Pi9InDel being a nucleotide fragment I in a specificity banding pattern with a rice blast resistance gene Pi9 is amplified from total DNA of a rice blast resistant variety carrying a rice blast resistance gene Pi9 through primer pairs of F1 and R1. The molecular marker is a first Pi9 gene specificity InDel marker developed for an internal sequence of the gene Pi9 and has the following advantages that specificity is high, in practical application, cost is low, throughput is high, and the molecular marker can be widely applied to colonies with different genetic backgrounds. By means of the molecular marker, the utilization efficiency of the gene in marker-assisted selection breeding, gene pyramiding breeding and transgenosis breeding can be improved.
Owner:NANJING FORESTRY UNIV
Method for identifying soybean cytoplasmic male sterile line seed purity through molecular marker
InactiveCN103160599ANovel extraction methodQuick extractionMicrobiological testing/measurementBiotechnologyGenomic DNA
The invention provides a method for identifying soybean RN type cytoplasmic male sterile line seed purity through a molecular marker. The method comprises the steps of: extracting genomic DNA of an RN type cytoplasmic male sterile line seed; carrying out PCR (Polymerase Chain Reaction) amplification by taking the genomic DNA as a template and utilizing a primer InDel-cms1, and carrying out gel electrophoresis analysis on an amplification product, wherein the length of a sterile line fragment after amplification is 200bp, and the length of a maintainer line fragment after amplification is 212bp; and accurately distinguishing the sterile line and the maintainer line through the length difference of the fragments, wherein the seed with the amplified fragment different from the amplified sterile line fragment is a false sterile line seed, and the percentage of the number of sterile line fragments with specific length obtained by detection accounting for all the detected seeds is the purity of RN type cytoplasmic male sterile line seeds. The method can be used for rapidly, accurately and reliably identifying the maintainer line mixed in the RN type soybean cytoplasmic male sterile line by a molecular biology experiment means so as to ensure the purity of sterile line seeds.
Owner:JILIN ACAD OF AGRI SCI
Polymorphism micro-satellite DNA molecular marker for deer and application of polymorphism micro-satellite DNA molecular marker
ActiveCN107164468AClear bandHigh polymorphismMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityGenetics
The invention relates to a micro-satellite DNA molecular marker for cervine animals. The molecular markers can be used for identifying allelic polymorphism, identify same or related cervine animals, distinguish the cervine animals and research the genetic diversity of the species group. The molecular marker can also be used for the genetic and phenotype research utilizing a statistics method such as linkage analysis, association mapping, linkage imbalance. The information can be used for breeding and / or selecting plants.
Owner:INST OF SPECIAL ANIMAL & PLANT SCI OF CAAS
Rice blast resistance gene Pi5 function specificity molecular marker and application thereof
ActiveCN106148510AStrong specificityLow costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGermplasm
The invention discloses a rice blast resistance gene Pi5 function specificity molecular marker named as Pi5InDel and application of the rice blast resistance gene Pi5 function specificity molecular marker. The nucleotide sequence of the rice blast resistance gene Pi5 function specificity molecular marker is shown in the sequence SEQ ID NO.1. The rice blast resistance gene Pi5 function specificity molecular marker is high in specificity, is low in cost and high in throughput in practical application, and can be widely applied groups with different genetic backgrounds to screen rice germplasm containing the rice blast resistance gene Pi. By means of the molecular marker, the utilization efficiency of the molecular marker in molecular marker auxiliary selective breeding, gene pyramiding breeding and transgene breeding can be improved.
Owner:苏州顶德生物科技有限公司
Method for identifying soybean male sterile cytoplasm through SNP marks of chloroplast DNA
ActiveCN104004853AMark stableImprove reliabilityMicrobiological testing/measurementBiotechnologyChloroplast
The invention provides a method for identifying soybean male sterile cytoplasm through SNP marks of chloroplast DNA. The method can also be applied to distinction of a soybean cytoplasmic male sterile line and a maintainer line containing normal cytoplasm. The method includes the following steps of extracting the seed genome DNA of the soybean cytoplasmic male sterile line and the seed genome DNA of the maintainer line to serve as amplification templates, selecting four pairs of the SNPs marks of the chloroplast DNA, and designing a pair of primers on the side wing sequences of each pair of SNPs respectively to conduct PCR amplification. The PCR products obtained through amplification conduct identification on the difference between the length of the enzyme-digested products of the sterile line and the length of the enzyme-digested products of the maintainer line through digestion of restriction enzymes. According to the method, the soybean male sterile cytoplasm and normal fertile cytoplasm can be rapidly and accurately identified. The method can also be used for detecting the maintainer line which contains fertile cytoplasm and is mixed in the sterile line containing sterile cytoplasm in the breeding process of the sterile line, and provides guarantees for the purity requirement in the production process of soybean sterile line seeds.
Owner:JILIN ACAD OF AGRI SCI
Method for identifying cherry variety
ActiveCN106957914AThe test result is accurateReduce experiment costMicrobiological testing/measurementDNA/RNA fragmentationS genotypingAgricultural science
The invention discloses a method for identifying a cherry variety, and belongs to the field of molecule identification of cherry varieties. The invention firstly discloses 11 pairs of SSR (Simple Sequence Repeats) primers for identifying the varieties of cherry, wherein the nucleotide sequences of the primers are as shown in SEQ ID No.1 to SEQ ID No.22. By adopting the method, core primer combinations are further screened from the 11 pairs of the SSR primers, 87 cherry germplasms can be distinguished by using 5 core primer combinations, and the obtained primer combinations have the advantages of being high in polymorphism, high in amplification stability, good in repeatability, clear in electrophoretic band and the like. The invention further discloses a cherry variety identification method and a kit based on S genotype information and SSR primer combinations. By adopting the primers, identification upon different cherry varieties can be rapidly and accurately completed. The SSR primer pairs, the cherry variety identification method and the kit disclosed by the invention can be applied to cherry germplasm evaluation, cherry germplasm innovation, parent choice for breeding, and the like.
Owner:ZHENGZHOU FRUIT RES INST CHINESE ACADEMY OF AGRI SCI
Microsatellite primers for identifying crassostrea hongkongensis, crassostrea ariakensis and crassostrea gigas and hybrid thereof and identification method
ActiveCN105624307ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationPacific oceanMicrosatellite
The invention discloses microsatellite primers for identifying crassostrea hongkongensis, crassostrea ariakensis and crassostrea gigas and a hybrid thereof and an identification method.The microsatellite primers are F:5'-CGACTGGTGGGAGTTTCTGAC-3' and R:5'-GCCGCTTCTATCTCCTTTGC-3'.Due to the fact that the external morphology of oysters changes greatly along with different living environments, identification is usually difficult only according to morphological characteristics; the oysters in the larval stage are more difficult to distinguish according to the morphological characteristics.By means of the microsatellite primers and the identification method, crassostrea hongkongensis, crassostrea ariakensis and crassostrea gigas individuals can be identified rapidly and accurately, and identification of the hybrid individual of the crassostrea hongkongensis, crassostrea ariakensis and crassostrea gigas individuals has the potential application value.Meanwhile, the microsatellite primers for identifying the crassostrea hongkongensis, the crassostrea ariakensis and the crassostrea gigas and the hybrid thereof and the identification method have the advantages of being simple in method, visual, accurate and effective in result, free of influences of environment and growth stage, low in cost and the like.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Method for extracting mushroom genome
InactiveCN101434630AQuality improvementAvoid cross contaminationSugar derivativesDNA preparationDissolutionGenome
The invention relates to a method for quickly extracting the genome of lentinus edodes, pertaining to the technical field of molecular biology. The method for extracting the genome of the lentinus edodes comprises the following steps sequentially: cultivation of thalli, washing of the thalli, cellular wall breaking of the thalli, extraction of a genome, removal of RNA, removal of protein and deposition, washing and dissolution of genome DNA; and is characterized in that the purpose of simply and quickly extracting the lentinus-edodes genome DNA with stable effect is achieved by adopting the special steps of acid cleaning of glass beads and cellular wall breaking by oscillation.
Owner:INST OF SOIL & FERTILIZER SHANDONG ACAD OF AGRI SCI
Extraction method of biological membrane proteins and preparation method of electrophoresis sample
The invention provides an extraction method of biological membrane proteins and a preparation method of an electrophoresis sample. The extraction method comprises the following steps: pretreating a biological membrane sample to obtain a precipitate; mixing the precipitate with a pyrolysis solution and a phenylmethylsulfonyl fluoride solution, and performing ultrasonic pyrolysis under an ice bath condition to obtain a sample pyrolysis solution; and shaking and centrifuging the sample pyrolysis solution, taking a supernatant, and treating the supernatant by adopting a TCA-acetone precipitation method, thereby obtaining a protein sample. The preparation method comprises the following steps: dissolving the protein sample by using a dissolving solution, and sequentially performing ultrasonic treatment, vortex oscillation and centrifugation, and taking a supernatant to obtain the electrophoresis sample. According to the methods provided by the invention, the pyrolysis solution containing various detergents is adopted and is combined with the ice bath ultrasound to break walls of microbial cells, and meanwhile, the TCA-acetone precipitation method is adopted to purify the protein, so that whole proteins in the biological membrane sample can be effectively extracted, and the masking action of humus to the proteins can be reduced to ensure that the analysis result of electrophoresis is clear.
Owner:TONGJI UNIV
Method for monitoring Siniperca chuatsi germplasms by virtue of microsatellite genetic markers
PendingCN106222271AHigh amplification efficiencyClear bandData processing applicationsMicrobiological testing/measurementGermplasmPolymorphic microsatellites
The invention discloses a method for monitoring Siniperca chuatsi germplasms by virtue of microsatellite genetic marker. The method comprises genome DNA extraction and detection, microsatellite sequence search and analysis, polymorphic microsatellite primer selection and optimization, polymorphic microsatellite detection and monitoring of different geographic siniperca chuatsi populations and hereditary features thereof. Microsatellite loci selection and polymorphic microsatellite marker development and verification are performed on a large number of EST generated by siniperca chuatsi transcriptome sequencing, so that 15 EST-SSR polymorphic microsatellite markers are developed, and different geographic siniperca chuatsi populations and hereditary features thereof can be monitored; the limitations of fuzzy genetic background and less molecular marker amount in existing germplasm detection, selective breeding and germplasm improvement research of siniperca chuatsi can be broken, and a basis is provided for germplasm detection, selective breeding and germplasm improvement of siniperca chuatsi, so that germplasm detection, auxiliary genetic breeding and genomic research processes of siniperca chuatsi molecular markers are promoted.
Owner:SUZHOU UNIV
DNA extracting method for petroleum microorganism in raw petroleum environmental sample
InactiveCN103667255AIncrease concentrationIncrease brightnessDNA preparationBiotechnologyMicroorganism
The invention relates to the technical field of extraction of DNA in raw petroleum and particularly relates to a DNA extracting method for a petroleum microorganism in a raw petroleum environmental sample. The method comprises the following steps: firstly, adding 10-50mul of raw petroleum into a centrifugal tube, then, adding 100mul of cleaning buffer solution into the centrifugal tube with the raw petroleum, and uniformly mixing to obtain a mixed solution. The concentration of DNA extracted by using the method is increased by 50% as comparison with the concentration of DNA extracted by using a bacterial DNA extraction kit, the DNA extracted by the invention is remarkably brighter than the DNA extracted by using the bacterial DNA extraction kit, and the obtained DNA is clear in strip and free of trailing so as to prove that the DNA extracting method is used for easily extracting the DNA from the raw petroleum, is higher in extraction efficiency and more capable of accurately, comprehensively and truly showing the kinds and amounts of microorganisms in the raw petroleum as comparison with the bacterial DNA extraction kit.
Owner:KARAMAY JINSHAN PETROCHEMICAL LTD CO
Method for separating beta-lactoglobulin from desalted whey powder
InactiveCN103923202AEasy to operateSimple processPeptide preparation methodsAnimals/human peptidesUltrafiltrationSalting out
The invention discloses a method for separating beta-lactoglobulin from desalted whey powder. According to the method, desalted whey powder is used as a treatment object which is subjected to ammonium sulfate salting out, primary ultrafiltration desalting, primary ion-exchange column chromatography, secondary ultrafiltration desalting, secondary ion-exchange column chromatography, gel chromatography and final separation so as to obtain beta-lactoglobulin with relatively high purity. Compared with the prior art, the method has the advantages that firstly, the operation is convenient, the process is simplified, related requirements are lowly required, and the operation can be carried out in a small laboratory; secondly, operators are lowly required, and the popularization and the application are convenient; thirdly, beta-lactoglobulin is relatively high in separation purity, the strip is clear and the separation is thorough; fourthly, the application range is wide, and different varieties of protein can be separated.
Owner:CHINA JILIANG UNIV
Method utilizing environmental sediment samples to monitor freshwater benthic animals
PendingCN111593099AGuaranteed uniformityOvercome the defect of low accuracy of monitoring resultsMicrobiological testing/measurementCentrifugationFishery
The invention belongs to the technical field of benthic animal identification, and discloses a method utilizing environmental sediment samples to monitor freshwater benthic animals. The method includes the following steps: 1) collecting an environmental sediment sample to preserve after cleaning; 2) adding anhydrous ethanol in the preserved environmental sediment sample to perform extraction treatment, taking uniformly mixed extract liquid after treating for a period of time, performing vacuum centrifugation on the extract liquid, and discarding supernatant to obtain dried tissue residues so as to perform DNA extraction; 3) utilizing the extracted DNA barcode fragment to perform amplification so as to obtain an amplified product; and 4) sequencing and analyzing the amplified product. The method performs extraction on the sample by adopting the anhydrous ethanol so as to make the DNA of a benthic animal uniformly released in the solution, so that a homogeneous state can be achieved; andthrough the collection of the ethanol solution containing the DNA sample and the extraction of the DNA after centrifugal concentration, the accurate monitoring of a wide range of environmental samples can be realized.
Owner:南京易基诺环保科技有限公司
Method for evaluating DNA quality of FFPE sample
InactiveCN108103168AEffective assessmentAccurate DNA qualityMicrobiological testing/measurementDNA/RNA fragmentationNucleotideNucleotide sequencing
The invention relates to a method for evaluating DNA quality of a FFPE sample. Generally, primers shown by nucleotide sequences, such as SEQ ID No:1-SEQ ID No:8 are used for carrying out PCR amplification of DNA in the FFPE sample; quality grades of DNA are effectively evaluated according agarose gel electrophoresis results, and accurate grading of DNA quality of the FFPF sample is carried out.
Owner:ANNOROAD GENE TECH BEIJING +2
Identification primer suitable for hippopus and common tridacna varieties in South China Sea and hybrids thereof and method thereof
ActiveCN110760595ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyForward primer
The invention discloses an identification primer suitable for hippopus and common tridacna varieties in South China Sea and hybrids thereof and a method thereof. The identification primer is a TC114 primer pair, and a forward primer and a reverse primer of the identification primer are as follows: F: 5'-AACACTGATTGGGCTCTT-3'R: 5'-TTGACGACGATGACGATA-3'. A pair of microsatellite primer TC114 primerpair is screened from 60 pairs of tridacna crocea microsatellite primers, can be universally used in hippopus and common tridacna varieties (tridacna derasa, tridacna squamosa, tridacna maxima, tridacna crocea and tridacna ovalvata) in South China, and can be distinguished according to the size of an amplified band, meanwhile, the microsatellite primer TC114 primer pair is clear in banding pattern, good in repeatability and high in reliability, and has potential application value in individual identification of hybrids between hippopus and common tridacna in South China Sea.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Polyacrylamide gel and kit thereof
ActiveCN108181369AStable gel solidification speedGood effectMaterial analysis by electric/magnetic meansMultiple componentAmmonium sulfate
The invention relates to a polyacrylamide gel. The polyacrylamide gel is prepared from a separation gel and a concentration gel, wherein the separation gel is prepared from 6 to 15wt% of polyacrylamide, 0.35 to 0.4M of tris(hydroxymethyl)methyl aminomethane, 0.03 to 0.2wt% of tetraethylethylenediamine, 0.1 to 1wt% of sodium hydrogen sulfite and 0.08 to 0.15wt% of ammonium persulfate; the concentration gel is prepared from 4 to 5wt% of polyacrylamide, 0.1 to 0.15M of tris(hydroxymethyl)methyl aminomethane, 0.08 to 0.4wt% of tetraethylethylenediamine, 0.05 to 0.5wt% of sodium hydrogen sulfite and 0.08 to 0.15wt% of ammonium persulfate. The polyacrylamide gel has the advantages that under the synergistic function of multiple components, the stable setting speed of the gel is ensured, and thelong-term stability of the gel is improved; the effect of protein separating by gel electrophoresis is good, and the protein stripes are clear; by not adding the volatile poisonous accelerant TEMED, the injury to the experiment person by the volatile poisonous compound is avoided.
Owner:CHONGQING KANGZHI PHARM
Cotton blastogenesis identification method based on SSR markers and capillary electrophoresis
InactiveCN105734124AWide range of applicationsClear bandMicrobiological testing/measurementGenomic DNAGenetic resources
The invention belongs to thebiotechnology field, and relates to a genetic identification method based on SSR markers and capillary electrophoresis high throughput for cotton. The cotton blastogenesis identification method based on the SSR markers and capillary electrophoresis is characterized by comprising the following steps that 1, a plurality of tender cotton leaves are adopted as research materials, and cotton genomic DNA is extracted; 2, primers are screened and synthesized; 3, PCR amplified reaction is carried out; 4, capillary gel electrophoresis detection is carried out; 5, data statistics and analysis of genetic diversity is carried out. By means of the method, time and labor are saved, and electrophoresis film analysis automation and data processing computerization can be achieved, so that genetic identification analysis is carried out on cotton genetic resource materials with high throughout, large scale, multiple batches and high efficiency, and the result is accurate, reliable, economical and practical.
Owner:XINJIANG ACADEMY OF AGRI & RECLAMATION SCI
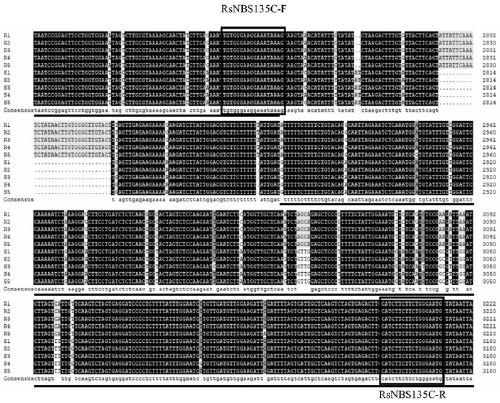
Method for identifying clubroot resistance of radishes
ActiveCN111424111AThe detection method is simple and convenientClear bandMicrobiological testing/measurementDNA/RNA fragmentationResistant genesNucleotide
The invention discloses a method for identifying clubroot resistance of radishes. The turnip clubroot resistance functional marker RsNBS135C is obtained by amplifying turnip genome DNA (deoxyribonucleic acid) by virtue of an upstream primer RsNBS135C-F and a downstream primer RsNBS135C-R. A nucleotide sequence obtained by amplification of the marker in radish genome DNA presents a specific bandingpattern after agarose gel electrophoresis, can quickly and directly identify a target resistance gene locus in radish germplasm, is beneficial to efficient and reliable utilization of a clubroot-resistant target gene, and accelerates a clubroot resistance genetic improvement process.
Owner:NANJING AGRICULTURAL UNIVERSITY +1
Method for screening pollination pear varieties suitable for Dangshan pears
InactiveCN105400887APromote amplificationClear bandMicrobiological testing/measurementPEARGenetic similarity
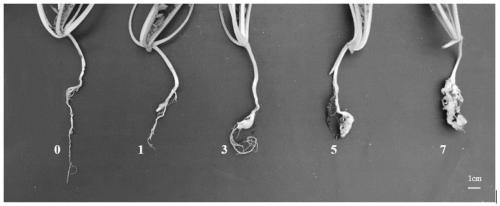
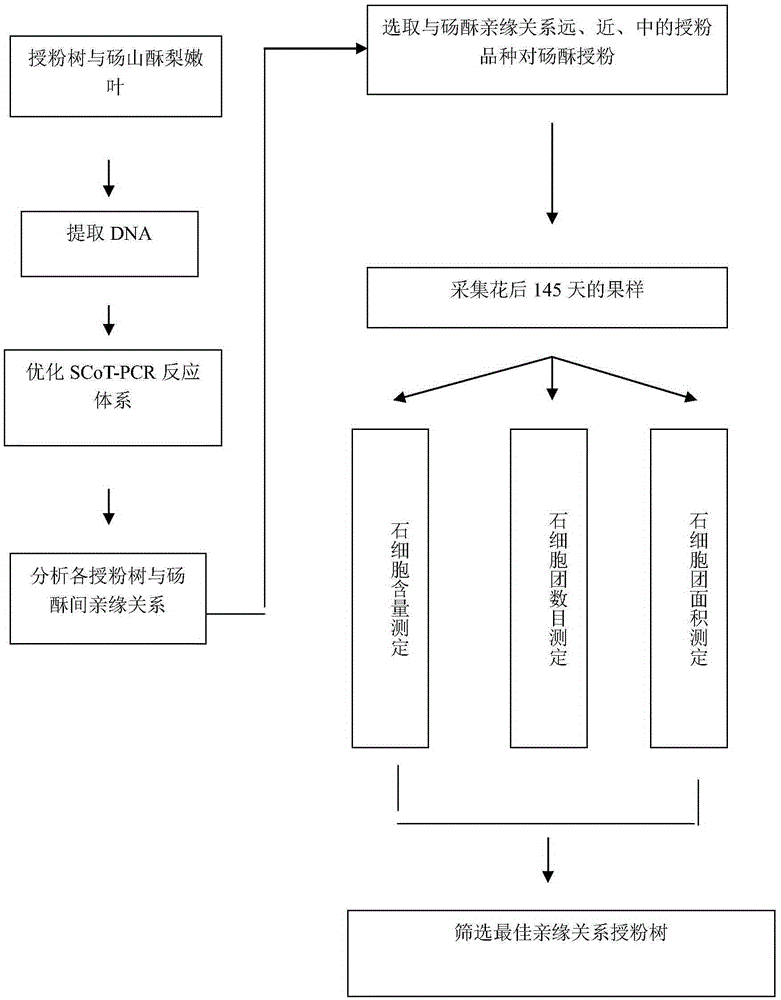
A method for screening pollination pear varieties suitable for Dangshan pears comprises the specific steps that 1, DNA of leaves of the pollination pear varieties is extracted; 2, a SCoT-PCR is carried out, a genetic similarity coefficient is calculated, and a genetic relationship dendrogram between the pollination pear varieties is constructed; 3, according to a clustering analysis result, the pollination pear varieties with the far, intermediate and near genetic relationships are selected to pollinate the Dangshan pears; 4, the number and area of sclereid groups of pulp tissue of mature fruits borne by each pollination pear variety are microscopically observed and calculated, and the content of sclereid is measured; 5, the pollination pear varieties with the optimal generic relationship are screened out through the analysis result of the step 4. According to the method, the genetic relationships between different pear variety resources can be revealed fast and accurately from the molecular level, the influence of the different genetic relation pollination pear varieties on the size and content of the sclereid of Dangshan pear fruits is analyzed, then the excellent pollination varieties with the proper genetic relationship are screened out, the sclereid content of the Dangshan pear fruits is improved, and the quality of the Dangshan pear fruits is improved.
Owner:ANHUI AGRICULTURAL UNIVERSITY
Determining method for phosphorylation level of calpain
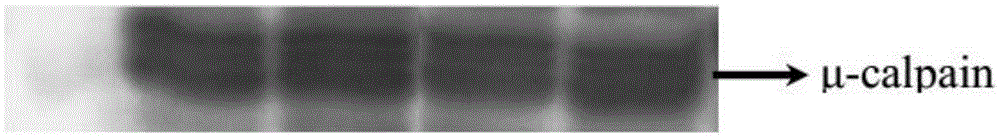
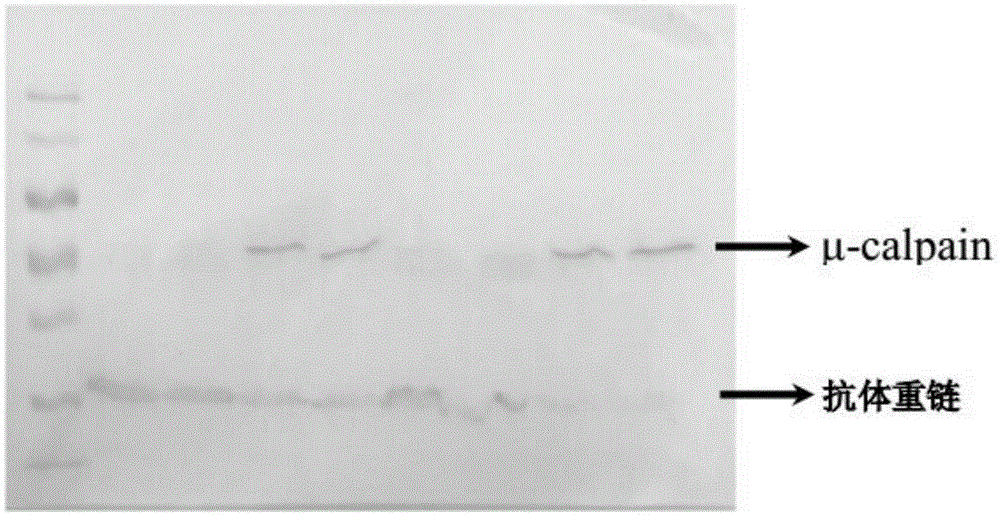
ActiveCN105334329ASmall amount of sampleReduce the potential for damageBiological testingImmune complex depositionPhosphorylation
The invention discloses a determining method for the phosphorylation level of calpain. The determining method includes the steps that 1, the protein concentration of a protein sample containing calpain is adjusted to 5.83-12.5 micrograms / microliter, a phosphorylated antibody is added into the protein sample, the mass ratio of the phosphorylated antibody to the total reacting weight of protein in the protein sample containing calpain is 1 to 1750-2500, then the phophorylated antibody and the protein sample are subjected to vibration hatching at 4 DEG C to form an immune complex, and then through a protein immune precipitation method, a phosphorylated calpain sample is obtained through gathering; 2, through a protein western blot method, the phosphorylation level of the phosphorylated calpain sample obtained in the step1 is detected. Through the protein immune precipitation technology and the western blot technology, phosphorylated calpain in post-slaughter muscles can be detected, the number of needed muscle samples is small, the operation process is simple, repeatability is good, stripes obtained through detection are clear, and the test method is small in potential hurt to the human body.
Owner:INST OF AGRO FOOD SCI & TECH CHINESE ACADEMY OF AGRI SCI
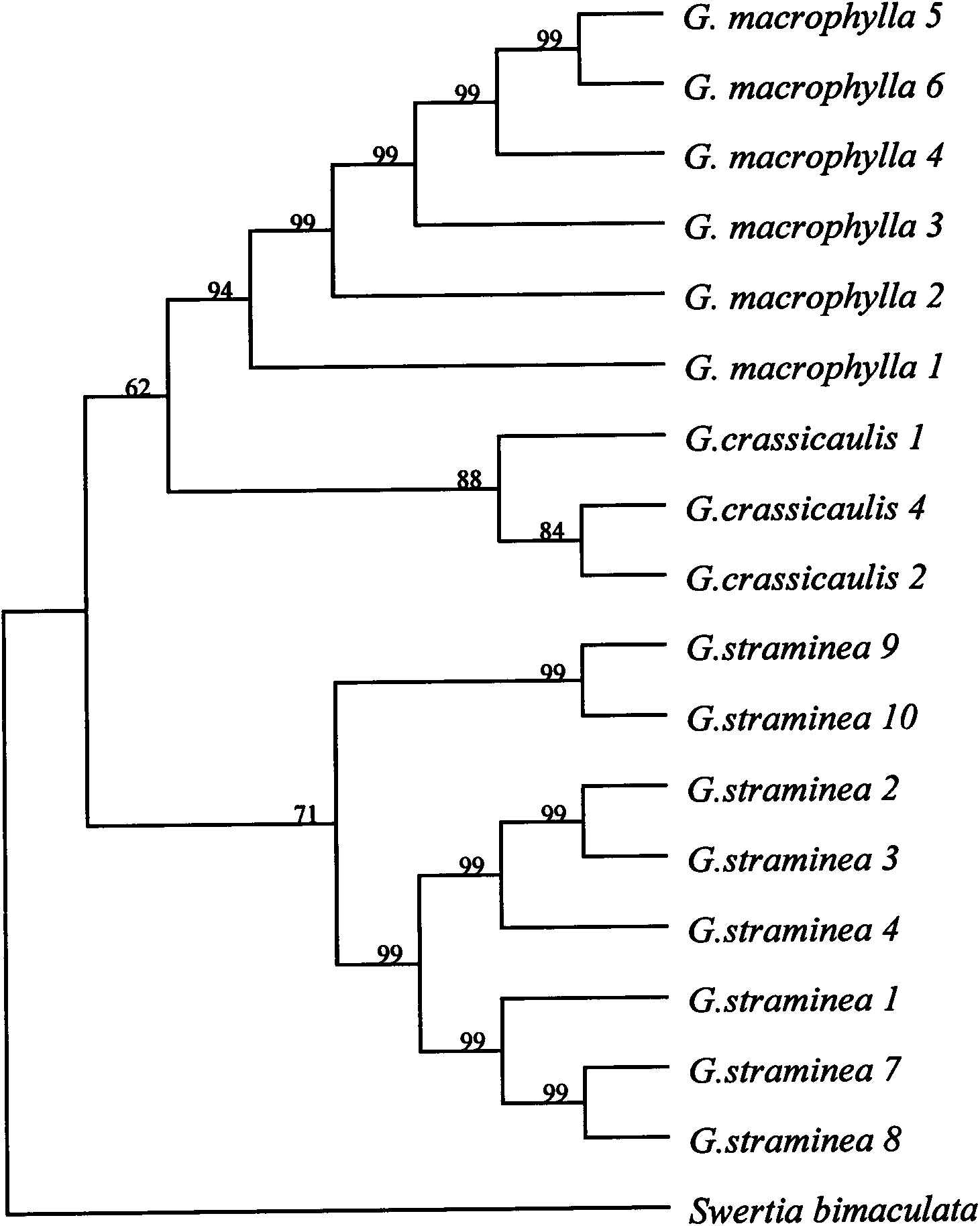
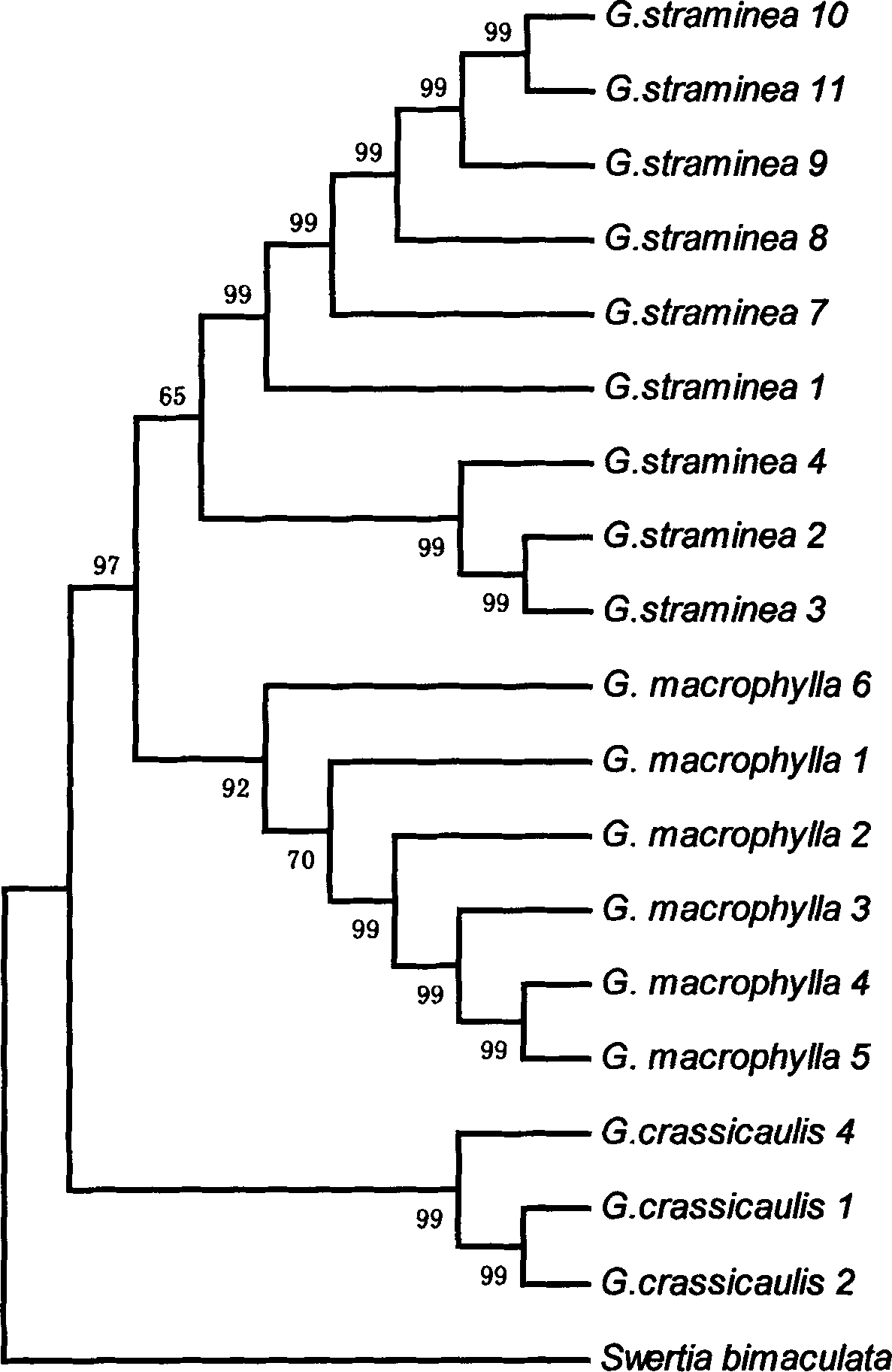
Method for identifying DNA barcodes of three gentiana macrophylla medicinal materials in pharmacopeia
ActiveCN103509871AEasy to proposeAvoid crackingMicrobiological testing/measurementMedicinal herbsHeating time
The invention discloses a method for identifying DNA (Deoxyribose Nucleic Acid) barcodes of three gentiana macrophylla medicinal materials in the pharmacopeia. Three gentian macrophylla medicinal materials in the pharmacopeia are respectively Gentiana macrophylla Pall, G.crassicaulis Duthie, and G.straminea Maxim. The identification method comprises two steps of carrying out DNA extraction and splicing obtained DNA sequences, wherein in the DNA extraction step, a good extraction effect is obtained by prolonging cracking heating time and utilizing phenol and trichloromethane, between which the volume ratio is 1:1, to remove interference of phenol and polysaccharide substances; then the obtained DNA sequences are spliced to identify gentiana macrophylla so as to obtain a good identifying effect.
Owner:ANHUI UNIVERSITY OF TRADITIONAL CHINESE MEDICINE
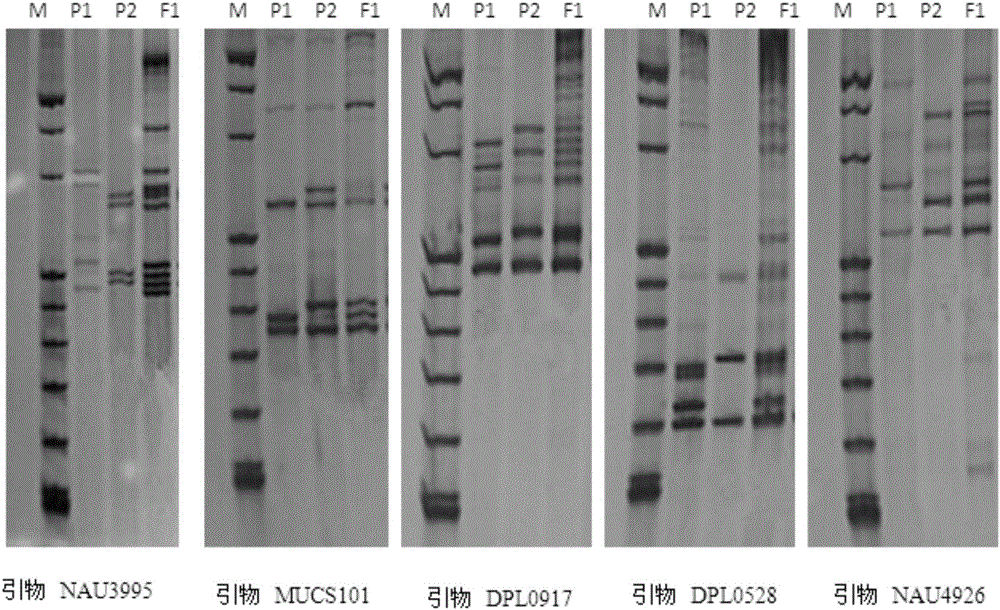
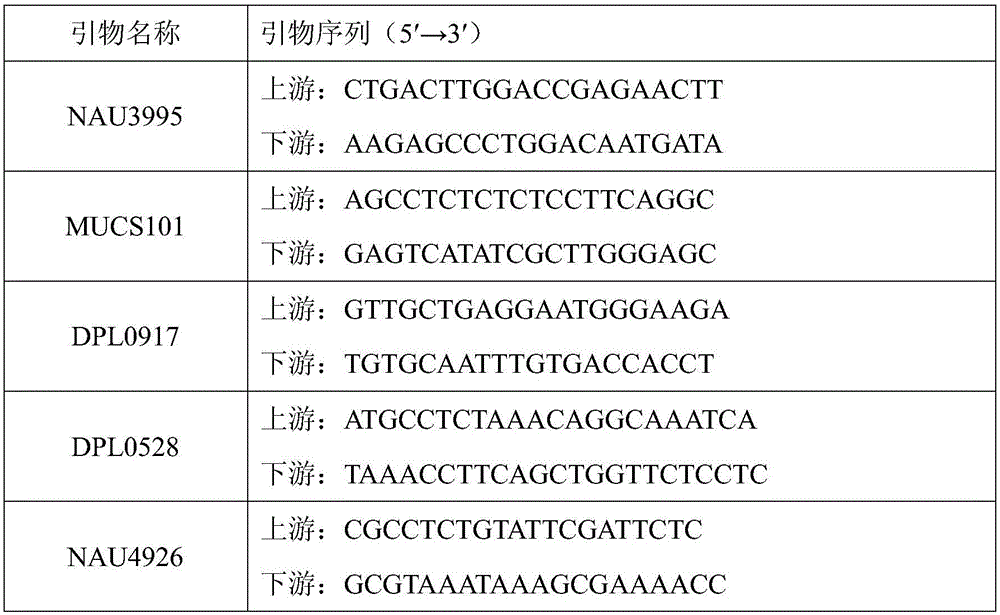
SSR molecular marker primer set for identifying purity of variety Shandong cotton research number 34 and application thereof
InactiveCN105695587AClear bandStrong reliabilityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceBand pattern
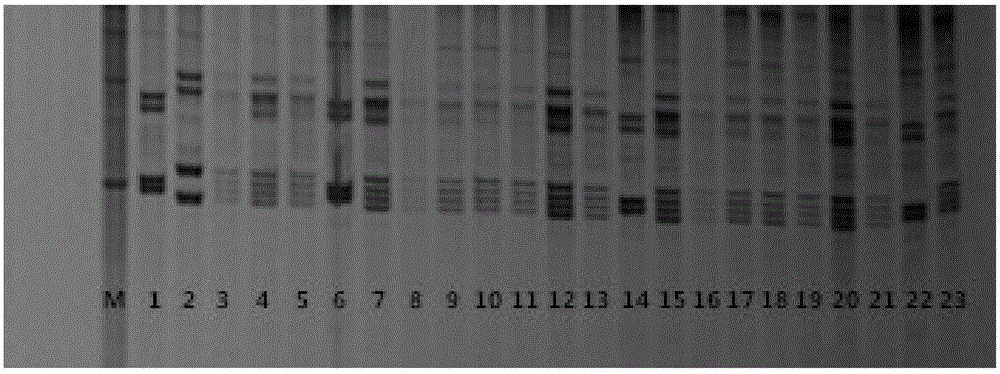
The invention relates to an SSR molecular marker primer set for identifying the purity of Lumianyan No. 34 variety and its application. The primer set consisted of 5 pairs of primers, namely: primers for molecular marker NAU3995, primers for molecular marker MUCS101, primers for molecular marker DPL0917, primers for molecular marker DPL0528, and primers for molecular marker NAU4926. The present invention screens and studies the hybrid Lumianyan 34 and its parental genome DNA characteristic SSR primers, and identifies that the bands with specific markers of the father and mother can be produced simultaneously in the first generation of hybrids, and the band type is clear and repeatable. 5 pairs of characteristic SSR primers of Lumianyan No. 34 with strong reliability constitute the SSR molecular marker primer set for identifying the purity of Lumianyan No. 34 variety. This primer set can be used to quickly detect the seed purity of Lumianyan No. 34 variety.
Owner:SHANDONG COTTON RES CENT
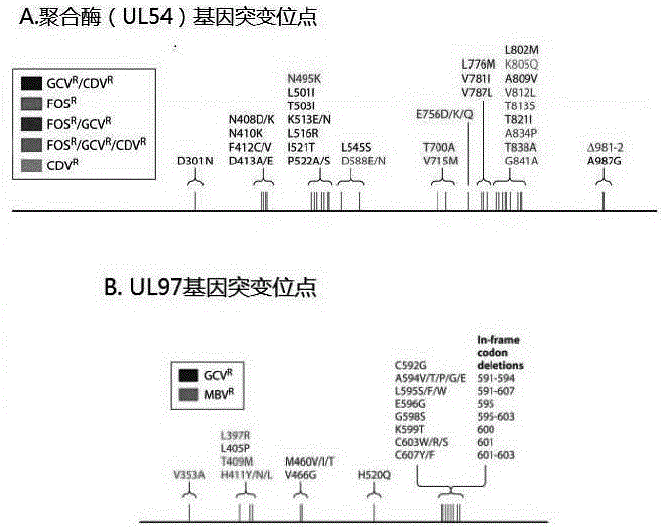
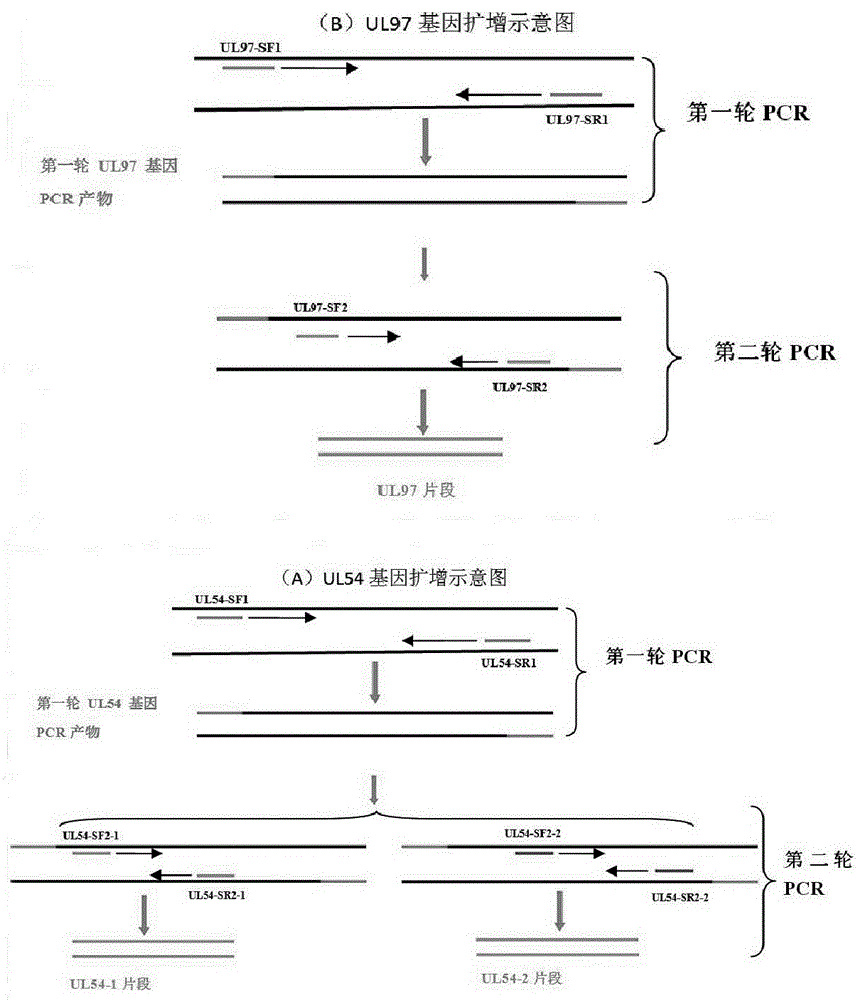
Nested PCR primers and method for detecting drug-resistant mutation of cytomegalovirus UL54 and UL97 genes
InactiveCN106636462AHigh sensitivityEfficient detectionMicrobiological testing/measurementMicroorganism based processesPcr methodDrug resistance
The invention relates to the technical field of detection for virogene mutation, and in particular relates to nested PCR primers and a nested PCR method for detecting drug-resistant mutation of cytomegalovirus UL54 and UL97 genes. Five pairs of primers are adopted, for the UL54 gene, the primers for first round of PCR amplification are the first pair of primers, the primers for second round of PCR amplification are the second and third pairs of primers, and the sequences of the primers are shown as SEQ ID No.1-6; for the UL97 gene, the primers for first round of PCR amplification are the fourth pair of primers, the primers for second round of PCR amplification are the fifth pair of primers, and the sequences of the primers are shown as SEQ ID No.7-10. For the nested PCR primers provided by the invention, the nested PCR method is adopted, the PCR reaction system and reaction procedure are optimized, the amplification for the UL54 and UL97 genes has high sensitivity and high amplification specificity, the primers and the method can be used for analyzing the drug resistance gene mutation, the practicability is strong, and the demands for clinical promotion are satisfied.
Owner:DONGGUAN EIGHTH PEOPLES HOSPITALDONGGUAN CHILDRENS HOSPITAL +1
Molecular marker primer combination for rapid high-throughput identification of sex traits of torreya grandis and application thereof
ActiveCN111304356AHigh amplification efficiencyImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationGermplasmRapid identification
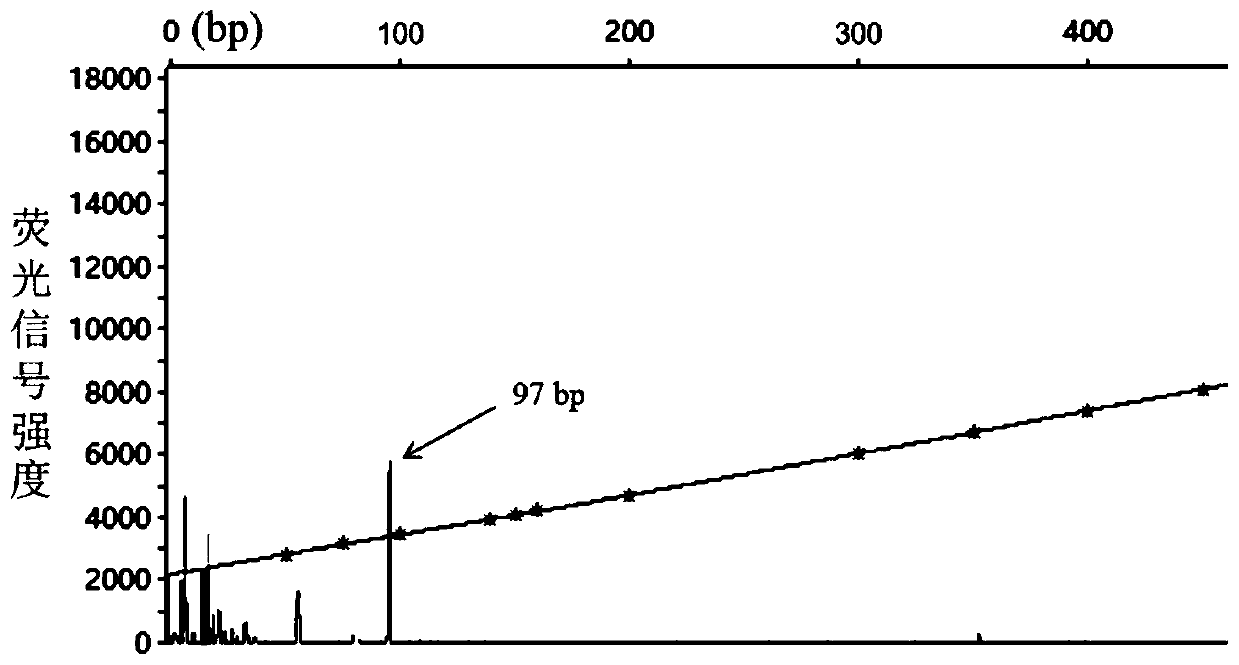
The invention discloses a molecular marker primer combination for rapid high-throughput identification of sex traits of torreya grandis and application thereof, so as to be applied to assisting screening of early molecular markers for torreya grandis breeding. The molecular marker primer combination is JY1754, a PCR amplification product of the JY1754 has a length of only 97 bp, and the JY1754 isparticularly suitable for high-throughput detection platforms such as fluorescent quantitative PCR instruments and the like, and can be used for rapid identification of sex traits of torreya grandis germplasm resources. The invention provides a method for assisting screening of torreya grandis germplasm resources with different sexes based on the developed molecular marker primer combination, hasimportant guiding significance in predicting phenotype of sex traits of torreya grandis, and has advantages of simple operation, high throughput, universal multi-detection analysis platform, accurateresult and the like.
Owner:NINGBO ACAD OF AGRI SCI
Molecule marking method for identifying variety purity of insect resistance hybrid cotton Suza No.3
InactiveCN101104869AAccurate detectionQuick checkMicrobiological testing/measurementMaterial analysis by electric/magnetic meansAgricultural scienceRapid identification
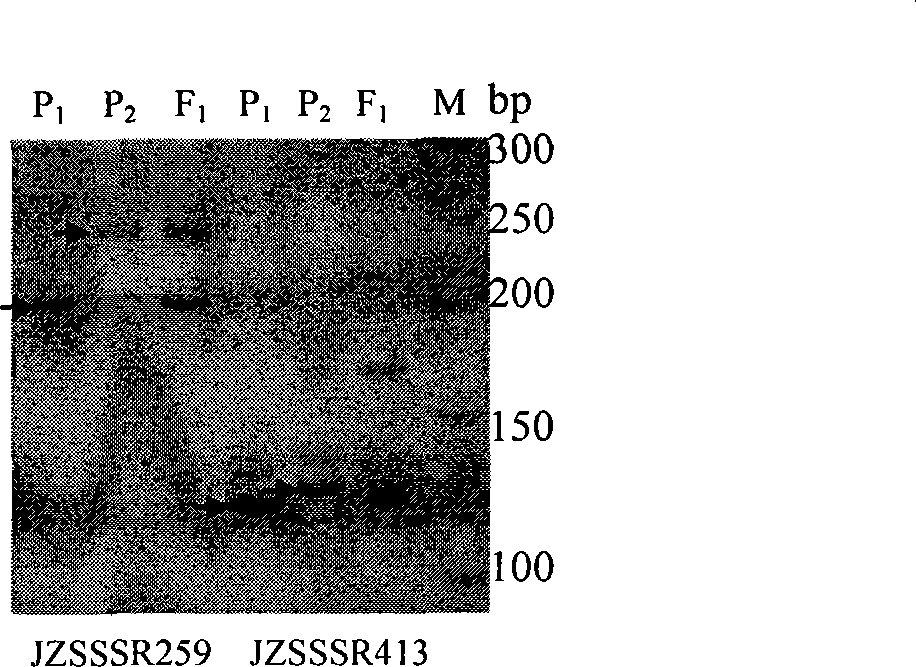
The invention relates to a SSR labeling method to appraisal the authenticity and / or variety purity insect-resistant hybrid cotton suza3, which belongs to biological technology field. The invention takes a commodity of insect-resistant hybrid cotton <SUZA3> and parental DNA as template to screen with SSR molecular marker, which can differentiate two SSR markers of male parent, female parent and hybrid at the same time. A band size of female parent specific marker JZSSSR259205 produced by SSR probe JZSSSR259 is 205bp; female parent specific marker JZSSSR259245 band size is 245bp; a band size of female parent specific marker JZSSSR413125 produced by SSR probe JZSSSR413 is 125bp; male parent specific marker JZSSSR413135 band size is 135bp. The two SSR marker can not only identify the hybrid cotton variety purity and authenticity, but also can do mutual identification which is good for controlling quality accurately and efficiently of the hybrid cotton breed and quicken the quality inspection process to identify quickly to mass sample in short time with a reliable result.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Dye liquid special for preparation of high-resolution chromosome by karyotype analysis
ActiveCN109540633AImprove dispersion characteristicsImprove dyeing qualityPreparing sample for investigationAlcoholKaryotype
The invention relates to a dye liquid special for preparation of high-resolution chromosome by karyotype analysis. The dye liquid is prepared by the following raw materials according to a proportion:1-3g of Giemsa dye power, 66-69ml of glycerinum, 66ml of methyl alcohol and 1-10 ml of dispersing agent. The dye particle is more refined and standard by grinding the dye particle, the dispersing characteristic of the dye particle is improved by addition of a dispersing agent, so that the uniformity and the clearness of dyeing are improved. By the novel dye liquid, the banding dyeing quality of the high-resolution chromosome is obviously improved, the banding belt dyeing effect of the high-resolution chromosome is clearer, the dyeing speed of human chromosome is rapid and controllable, and thebelt is clear. The dye liquid can be applied to large-scale clinic genetic laboratory.
Owner:BEIJING INST OF HUMAN GENETICS & REPRODUCTION MEDICINE LTD CHINA +3
Amaranth EST-SSR marker primer and method for identifying amaranth varieties
InactiveCN108384880AGood repeatabilityClear bandMicrobiological testing/measurementDNA/RNA fragmentationSpectrum bandAgricultural science
The invention provides an amaranth EST-SSR marker primer and a method for identifying amaranth varieties. The nucleotide sequence (CATA)5F of the amaranth EST-SSR marker (CATA)5 is CACGTCCTTCATTCGGATCT, and the nucleotide sequence (CATA)5R of the amaranth EST-SSR marker (CATA)5 is AATCCCGGAGGTAGGATCAC. The amaranth EST-SSR marker (CATA)5 has good polymorphism among the amaranth varieties and is clear in spectrum band and stable and reliable. The amaranth EST-SSR marker (CATA)5 is also applicable to germplasm resource identification and genetic diversity analysis of more amaranth plants.
Owner:FUJIAN AGRI & FORESTRY UNIV
High-quality DNA extraction method of idesia polycarpa
The invention belongs to the technical field of DNA extraction and discloses a high-quality DNA extraction method of idesia polycarpa. According to the method, a CTAB method is taken as a framework, an anti-oxidant is added during grinding, PEG elution is carried out before cracking, subsequent CTAB method operation steps are optimized, so that the high-quality idesia polycarpa DNA extraction method comprising the steps of material collection, material grinding, PEG elution, CTAB cracking, extraction, DNA precipitation, collection and the like is formed. The idesia polycarpa DNA extracted by the method is high in concentration and purity, the quality of the DNA is obviously better than that of an existing method, the average concentration is 438.93 ng / ul, the average concentration is 8.5 times that of a kit method, the OD260 / 280 ratio is 1.94-1.97, the OD260 / 230 ratio is 2.1-2.17, protein contamination and DNA carbohydrate and salt contamination are all well controlled, and the DNA issintact. The examples show that the idesia polycarpa DNA extracted by the method can meet the requirements of molecular tests such as SSR marker typing and genome sequencing simplification; meanwhile,phenol is not used for re-extraction in the method, so that the test steps are simplified, the test time is shortened, and the harm of organic solvents to human health is reduced.
Owner:RES INST OF SUBTROPICAL FORESTRY CHINESE ACAD OF FORESTRY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com