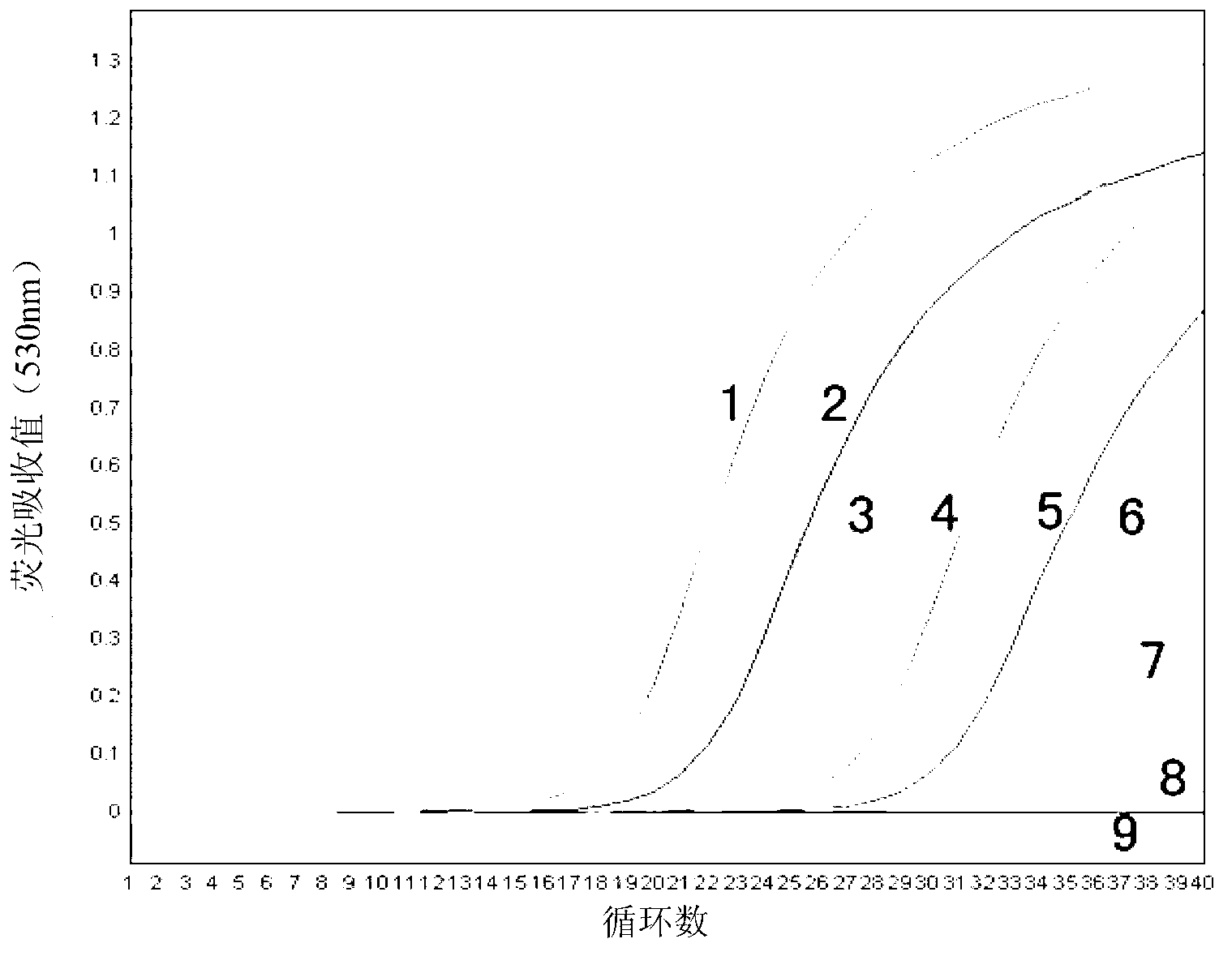
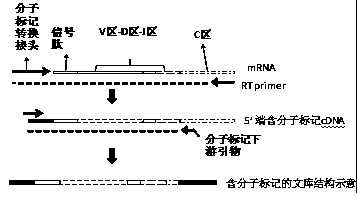
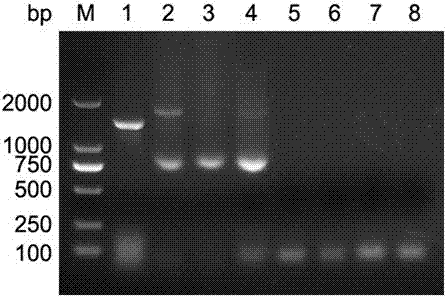
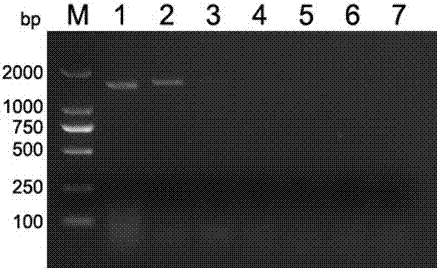
Patents
Literature
75results about How to "Improve amplification specificity" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for semi-quantitatively detecting pathogenic nucleic acid by adding internal control nucleic acid
InactiveCN101957373AAvoid diagnostic problems that are prone to false negativesAvoid problems prone to false negativesMicrobiological testing/measurementMaterial analysisTest sampleQuality control
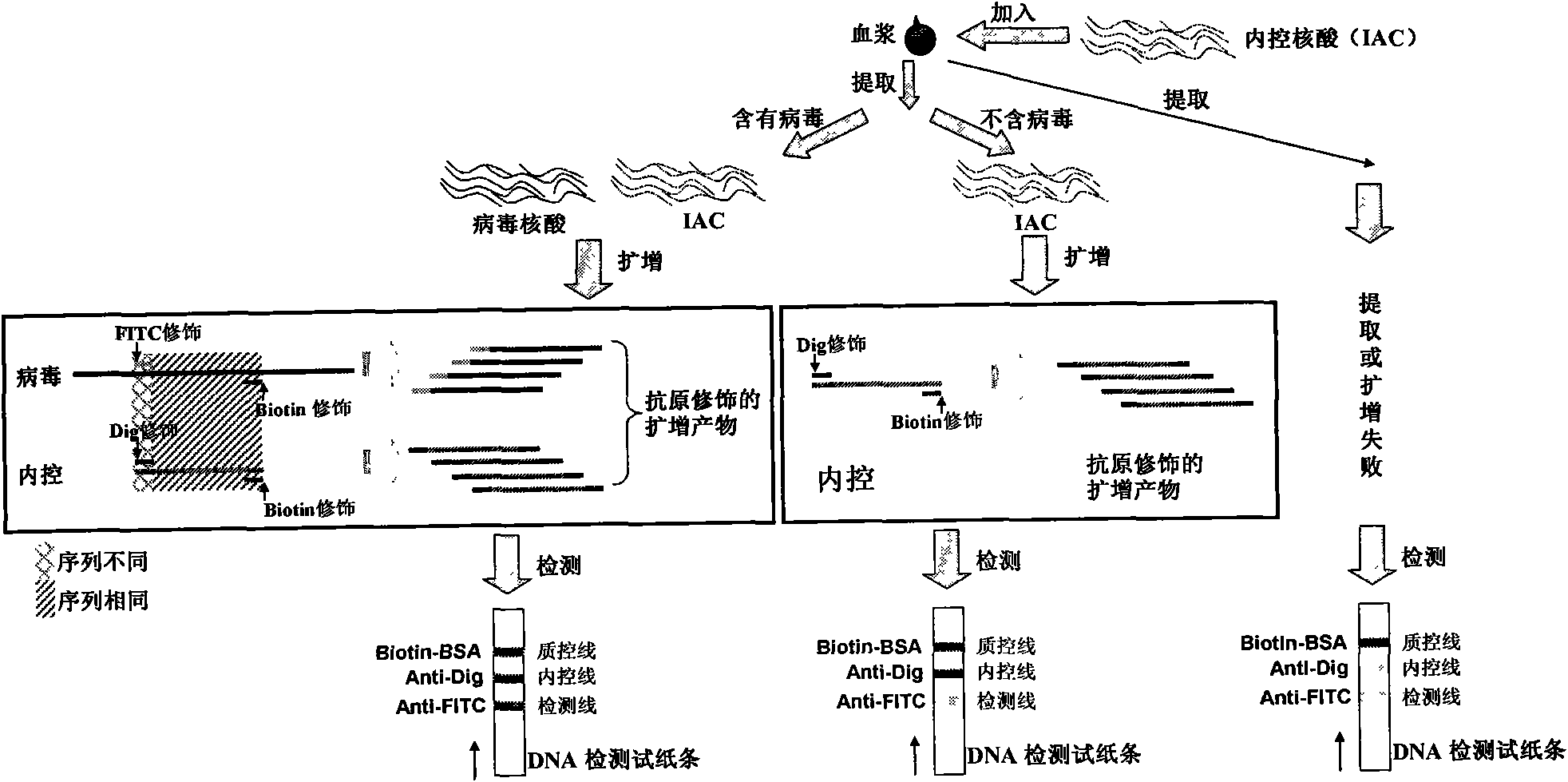
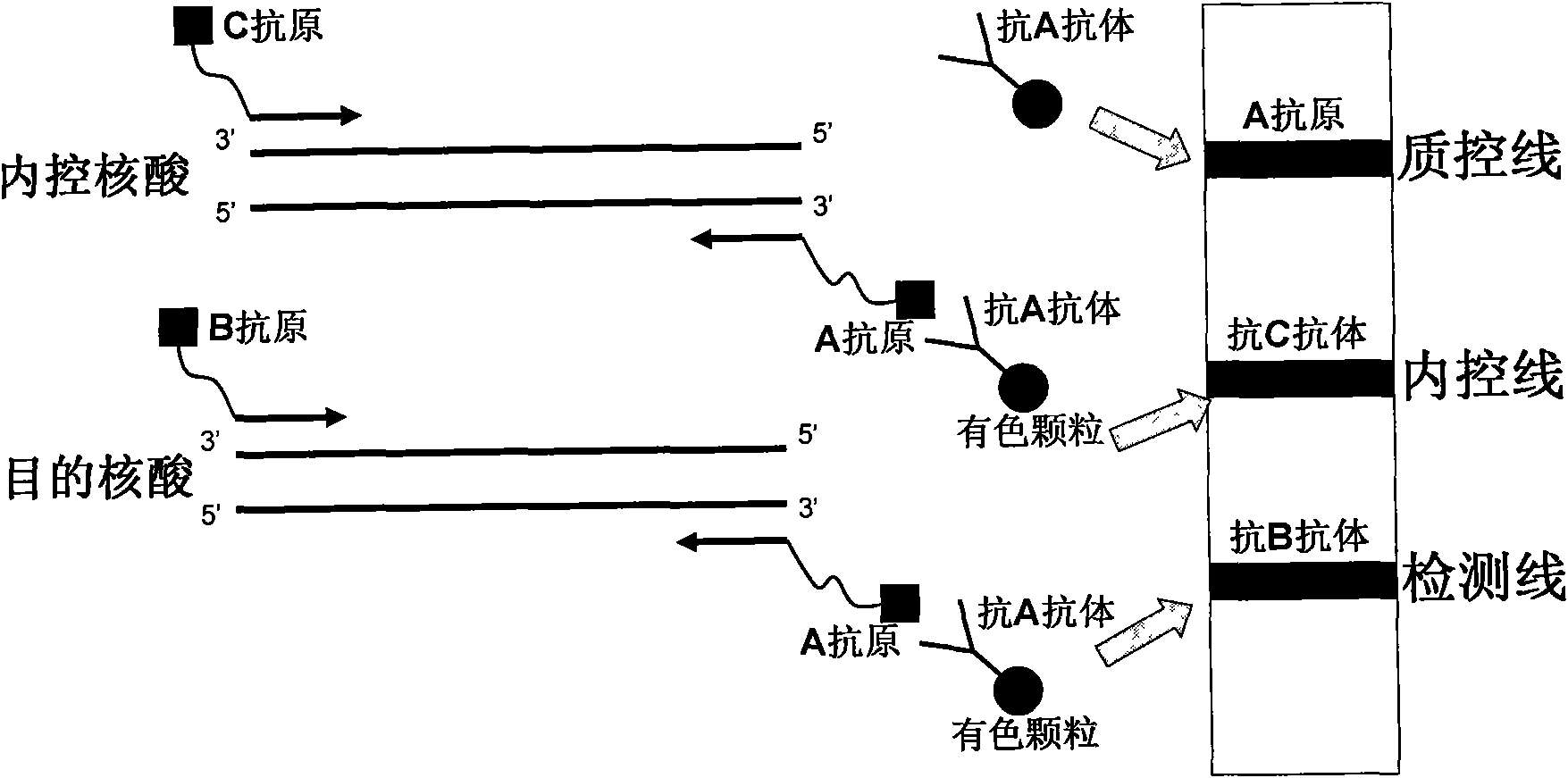
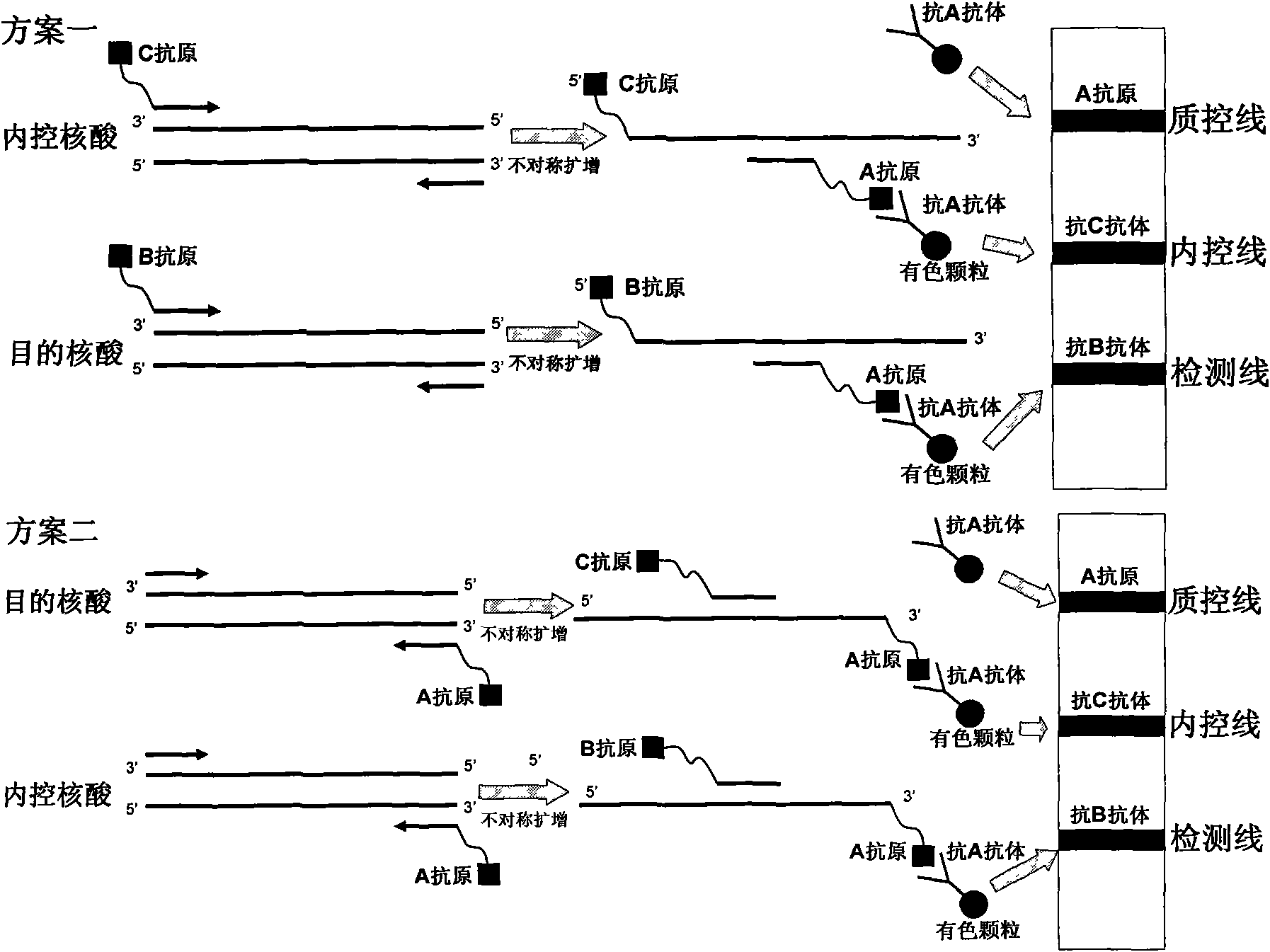
The invention belongs to the field of nucleic acid detection and discloses a method for semi-quantitatively detecting pathogenic nucleic acid by adding internal control nucleic acid. Corresponding internal control is added in the whole process of extracting and amplifying target nucleic acid and testing by using a test paper, so that the internal control and a target segment are parallelly operated, and the semi-quantitative detection is performed finally through color development and intensity contrast of three strips, namely a detection line, an internal control line and a quality control line on the test paper. In the method, in the whole process of processing the target nucleic acid, the corresponding internal control is taken as a positive contrast, and false negative results due to links such as extraction, amplification or sample application errors are avoided in the processing of detecting by using the test paper. Meanwhile, by comparing color development intensity of the internal control line and a sample line and introducing the semi-quantitative function on the basis of the qualitative function of the immunochromatographic test paper to estimate the copy number of tested samples, the detection results are more detailed, accurate and reliable. The method has the advantages of convenient and quick operation and capacity of meeting the actual clinical requirement.
Owner:HUADONG RES INST FOR MEDICINE & BIOTECHNICS
Multiplex amplification detection kit for detecting 60 loca of autosomes and Y-chromosomes simultaneously
ActiveCN109439765ARapid expansionImprove balanceMicrobiological testing/measurementCapillary electrophoresisMagnetic bead
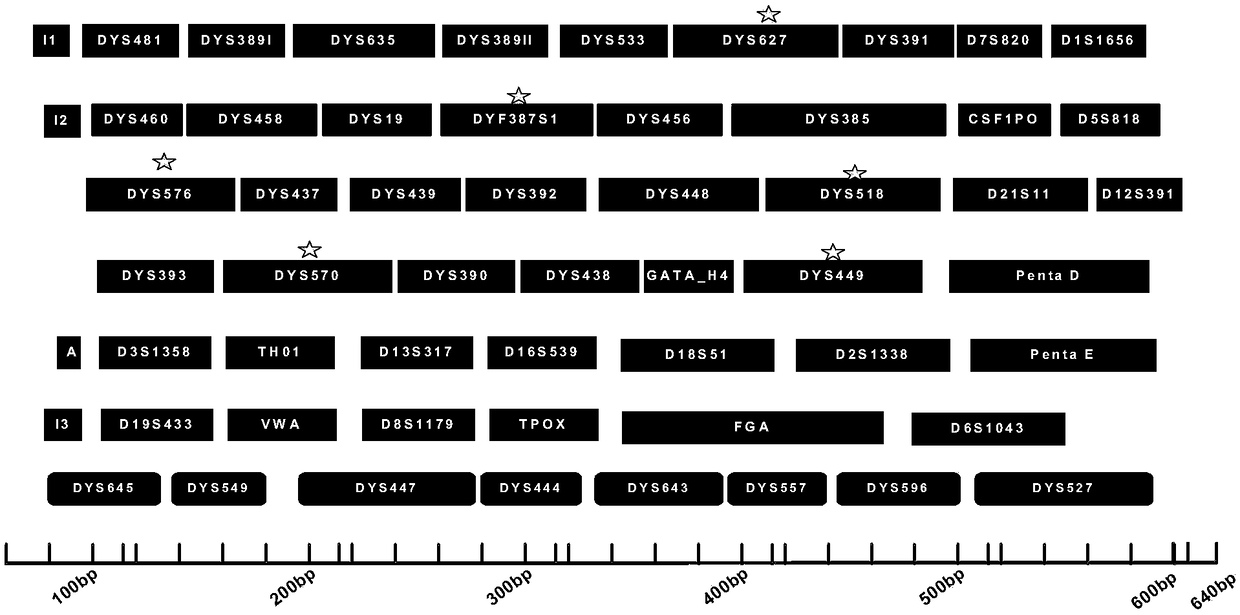
The invention discloses a multiplex amplification detection kit for detecting 60 loca of autosomes and Y-chromosomes simultaneously. The 60 loca are amplified in a multiplex manner through a polymerase chain reaction, and amplification products of the loca are detected by using a gene sequencer. The kit can be used for detecting the 60 loca of the autosomes and the Y-chromosomes simultaneously, which is a case that most STR loca can be detected by a primary reaction with a capillary electrophoresis method at present, and databases can be built for autosome STR and Y-chromosome STR simultaneously by the primary reaction. The kit has strong adaption to biomaterials, namely, one kit can be used for amplifying various biomaterial samples, wherein different biomaterial samples comprise a male genome DNA extracted by a Chelex100 method, a magnetic bead extracting method or an organic extracting method and male blood or oral cells of human, which is / are collected by any one carrier of filterpaper, an FTA card, a cotton bud, gauze and the like. The kit has higher amplification specificity and higher thermal tolerance.
Owner:江苏苏博生物医学科技南京有限公司 +1
Composite amplification kit for 47 human autosome and Y chromosome loci and application thereof
ActiveCN109750110AEffective expansionAmplification specificMicrobiological testing/measurementDNA/RNA fragmentationDNA paternity testingFluorescence
The invention relates to a composite amplification kit of 47 human autosome and Y chromosome loci and an application thereof. The invention provides a composite amplification system of the 47 human autosome and Y chromosome loci, which comprises specific primers for amplifying the 47 loci, wherein the 47 loci comprise 19 autosome STR loci, 27 Y chromosome STR loci and 1 sex recognition locus. The47 pairs of specific primers are subjected to grouping fluorescence labeling by utilizing a six-color fluorescence labeling technology, and the simultaneous high-efficiency, specific and sensitive amplification of the 47 human autosome and Y chromosome loci is achieved through the design and optimization of primer sequences and working concentrations. The detection result of the composite amplification system has high individual identification capability and good data compatibility, and can be used for paternity identification and individual identification in practice, so that the detection cost of human DNA typing is effectively reduced, and the detection working efficiency is improved.
Owner:BEIJING PEOPLESPOT TECH
SSR molecular marker method of brassica allohexaploid and primers thereof
ActiveCN104059971AReduce concentrationReduce generationMicrobiological testing/measurementDNA/RNA fragmentationBrassicaAgricultural science
The invention discloses an SSR molecular marker method of a brassica allohexaploid and primers thereof. The SSR molecular marker method of the brassica allohexaploid comprises the steps: hybridizing a brassica allohexaploid parent, and performing microspore culture on the obtained filial generation to obtain a DH colony; extracting genome DNAs of the brassica allohexaploid parent and the DH colony, and performing PCR amplification with all genome DNAs as a template by using the primers; constructing a genetic map of the brassica allohexaploid parent according to a PCR amplification result, wherein 217 pairs of primers are included, with base sequences respectively shown in SEQ ID No. 1 / 2-433 / 434. According to the SSR molecular marker method, a first genetic map of the brassica allohexaploid is constructed, and the basis is provided for further performing QTL location and molecular marker assistant breeding.
Owner:ZHEJIANG UNIV
PCR (Polymerase Chain Reaction) primer for detecting and identifying porcine circovirus 3 (PCV3) and detection method and detection kit
InactiveCN107012259ASimple and efficient operationStrong specificityMicrobiological testing/measurementMicroorganism based processesConserved sequenceMicrobiology
The invention discloses a PCR (Polymerase Chain Reaction) primer for detecting and identifying porcine circovirus 3 (PCV3) and a detection method and a detection kit. A pair of PCR primers for detecting the PCV3 is designed by performing preference comparison with genomic sequences of the PCV3 according to a conserved sequence on a cap protein of the PCV3. Sequences of the forward and reverse primers PCV3-F / R of the PCR are sequentially shown as SEQ ID NO.1 to SEQ ID NO.2. The invention further provides a PCR detection method and kit for the PCV3. The primer and the detection method disclosed by the invention are capable of rapidly, conveniently and efficiently detecting and identifying the PCV3, the specificity is high, the sensitivity is high, and the detection method is simple and high-efficiency in operation, is convenient for clinical detection and convenient for developing epidemiological investigation and has great application prospects.
Owner:SOUTH CHINA AGRI UNIV
Composite amplification kit for 44 human Y chromosome loci and application thereof
ActiveCN109880912AEfficient amplificationAmplification specificMicrobiological testing/measurementAgainst vector-borne diseasesFluorescenceY-Chromosome Genes
The invention relates to a composite amplification kit for 44 human Y chromosome loci and application thereof. The invention provides a composite amplification system of the 44 human Y chromosome loci. The system includes specific primers for amplifying the 44 loci including 41 Y chromosome STR loci and 3 Y chromosome Indel loci. The 44 pairs of specific primers are subjected to group fluorescencelabeling by the six-color fluorescent labeling technology. Through design and optimization of the primer sequence and the working concentration, simultaneous efficient specific sensitive amplification of the 44 human Y chromosome loci is achieved. A detection result of the composite amplification system has high individual recognition capability and good data compatibility, and can be used in practice for paternity testing and individual recognition, thereby effectively reducing the detection cost of human DNA typing and improving the detection working efficiency.
Owner:BEIJING PEOPLESPOT TECH
Typing detecting kit and method for detecting digital PCR absolute quantification of HBV-B/C
ActiveCN105441595AStrong amplification specificityAccurate Quantification of Copy NumberMicrobiological testing/measurementFluorescenceA-DNA
The invention discloses a typing detecting kit and method for detecting the digital PCR absolute quantification of HBV-B / C. The kit comprises a DNA extraction solution, a digital PCR reaction buffer solution A, a digital PCR reaction buffer solution B, an HBV-B virus gene positive quality control, an HBV-C virus gene positive quality control, a negative quality control and an internal standard solution. The method includes the following steps of firstly, processing a to-be-detected sample; secondly, processing a quality control product; thirdly, preparing a digital PCR reaction mixed solution; fourthly, generating a micro-reaction liquid drop, and conducting digital PCR reaction amplification; fifthly, reading a fluorescent signal, analyzing an HBV type and calculating the number of copying times. By means of the digital PCR technology and the dual-color florescent probe, two subtypes of HBV are detected at the same time; without depending on an external standard object or a standard curve, the kit and method are easy and convenient to operate and can directly detect the HBV precise absolute quantification.
Owner:ANHUI UNIV OF SCI & TECH
Composite amplification kit for 25 human chromosome loci and application thereof
ActiveCN109880911AEfficient amplificationAmplification specificMicrobiological testing/measurementAgainst vector-borne diseasesDNA paternity testingTyping
The invention relates to a composite amplification kit for 25 human chromosome loci and application thereof. The invention provides a composite amplification system of the 25 human chromosome loci. The system includes specific primers for amplifying the 25 loci including 23 autosomal STR loci, 1 gender recognition locus and a Y chromosome Indel locus. The 25 pairs of specific primers are subjectedto group fluorescence labeling by the six-color fluorescent labeling technology. Through design and optimization of the primer sequence and the working primer concentration, simultaneous efficient specific sensitive amplification of the 25 human chromosome loci is achieved. A detection result of the composite amplification system has high individual recognition capability and good data compatibility, and can be used in practice for paternity testing and individual recognition, thereby effectively reducing the detection cost of human DNA typing and improving the detection working efficiency.
Owner:BEIJING PEOPLESPOT TECH
Avian pneumovirus Taqman probe fluorescent quantitative RT-PCR detection kit
ActiveCN103320542AShort detection timeHigh detection specificityMicrobiological testing/measurementFluorescence/phosphorescenceAgricultural scienceSingle strand dna
The invention discloses an avian pneumovirus Taqman probe fluorescent quantitative RT-PCR detection kit and an application thereof. The kit contains a composition for detecting the avian pneumovirus, and the composition is composed of a primer pair and a probe; the primer pair is composed of a primer 1 and a primer 2, the primer is single-stranded DNA represented by sequence 1 in a sequence table, and the primer 2 is single-stranded DNA represented by sequence 2 in the sequence table; and the sequence of the probe is sequence 3 in the sequence table. Experiments prove that the kit has the advantages of short detection time, strong specificity, high sensitivity, simple operation and low price when the kit is used for the avian pneumovirus Taqman probe fluorescent quantitative RT-PCR detection.
Owner:GUANGXI VETERINARY RES INST
Y-STR compound amplification detection kit marked by novel fluorescent labeling method and use method thereof
ActiveCN106834456AHigh fluorescence efficiencyHigh sensitivityMicrobiological testing/measurementEnergy transferBiology
The invention discloses a Y-STR compound amplification detection kit marked by a novel fluorescent labeling method and a use method thereof. The Y-STR compound amplification detection kit performs compound amplification of 27 Y-STR loca with a compound amplification primer by adopting a polymerase chain reaction; the amplification product is detected by means of a gene sequencing instrument of a single channel or multichannel capillary tube; 27 Y-STR loca comprise 20 conventional low mutation rate loca and 7 fast mutant loca. The novel fluorescent labeling method comprises the steps of: marking a first alkaline group at the 5' end of the primer by a conventional dye; and marking the sixth alkaline group with a short excitation wavelength fluorescent dye to realize fluorescent energy transfer. The signal strength generated by the method is 3-11 times higher than that of the conventional method, and an amplification result which is more stable and balanced and higher in sensitivity can be obtained.
Owner:JIANGSU SUPERBIO LIFE SCI CO LTD +1
Construction method of immune repertoire sequencing library and test kit
PendingCN111575343AEffectively eliminate amplification biasRule out amplification biasMicrobiological testing/measurementLibrary creationAmplification biasImmune repertoire
The invention discloses a construction method of an immune repertoire sequencing library and a test kit, belongs to the field of biological medicines, and relates to a detection method of immune repertoire diversity. One of the purposes of the invention is to provide the construction method of the immune repertoire sequencing library; the construction method comprises the following steps of (a) extracting sample RNA, (b) carrying out reverse transcription by using a reverse transcription primer to synthesize cDNA, and adding a specific linker containing a molecular tag sequence at the tail end; (c) by taking the product obtained in the step b as a template, carrying out amplification through a specific linker primer and a gene specific primer containing a molecular tag sequence; and (d) finally, adding a sequencing library linker to an amplification product, so as to complete library construction. When sequencing data is analyzed, original RNA molecules are identified through moleculartag sequences, and amplification bias is excluded. By using the method, all CDR regions of TCR and BCR can be covered, PCR amplification bias interference is effectively eliminated, the specificity is higher, and the analysis result is more accurate. The other purpose of the invention is to provide the test kit for immune repertoire sequencing.
Owner:北京全谱医学检验实验室有限公司
PCR primers, PCR detection method and PCR detection kit for detecting and identifying atypical porcine pestivirus (APPV)
InactiveCN106929606AImprove amplification specificityHigh sensitivityMicrobiological testing/measurementMicroorganism based processesForward primerConserved sequence
The invention discloses PCR primers, a PCR detection method and a PCR detection kit for detecting and identifying atypical porcine pestivirus (APPV). By referring to and comparing the genomic sequence of the atypical porcine pestivirus, based on the conserved sequence of the cap protein of the atypical porcine pestivirus, a pair of PCR primers for detecting the atypical porcine pestivirus is designed, and the sequences of the forward primer and the reverse primer of the PCR primers APPV-F / R are shown as SEQ ID NO.1- SEQ ID NO.2 in sequence; the PCR detection method and the PCR detection kit for detecting the atypical porcine pestivirus are further provided. The primers and the detection method can detect and identify the atypical porcine pestivirus fast, conveniently and efficiently, specificity is good, and sensitivity is high; moreover, operation of the detection method is easy and efficient, and the detection method brings convenience to clinical detection and epidemiological investigation, and has a broad application prospect.
Owner:SOUTH CHINA AGRI UNIV
Semi-random primer based on PCR walking technology, and kit thereof
ActiveCN103397028AEasy to operateImprove amplification specificityMicrobiological testing/measurementDNA/RNA fragmentationMicrobiology
The present invention discloses a semi-random primer based on a PCR walking technology, a kit and a method for performing PCR walking by using the kit. The sequence of the semi-random primer is represented by the SEQ ID NO.7 in the sequence list. The kit comprises the semi-random primer. The method is similar to the ordinary two-primer PCR, at most requires two PCR cycles to obtain a target sequence, and has characteristics of simple, rapid and efficient operation. According to the method, an annealing temperature of the whole PCR cycle process is increased, amplification specificity is increased, the used experimental materials are not limited, and the kit can be used by microorganism samples, animal samples and plant samples. The method can be used in experiments so as to simplify operations, shorten an experiment time, improve experiment efficiency, reduce experiment cost, and provide broad application prospects.
Owner:BRIGHT DAIRY & FOOD
Kit for human papilloma virus E6/E7 gene detection and detection method
ActiveCN105886664AReduce sensitivityReduce background signalMicrobiological testing/measurementMicroorganism based processesPolymerase chain reactionGenotype Analysis
The invention discloses a kit for human papilloma virus E6 / E7 gene detection. The kit comprises 22 pairs of upstream primers and downstream primers designed by aiming at 22 kinds of different subtypes of human papilloma viruses, wherein one of each pair of upstream primer and downstream primer is modified with a fluorophore; each pair of PCR (polymerase chain reaction) primers aims at one kind of HPV E6 / E7 gene. The invention also discloses a method for human papilloma virus E6 / E7 gene detection. The method comprises the following steps that 10uL of PCR reaction systems, 2.35uL of DDH2O, 1uL of 10*PCR buffer, 1uL of 25nM Mg2<+>, 1.5uL of 2nM DNTP, a mixture of 22 pairs of upstream primers and downstream primers designed by aiming at 22 kinds of different subtypes of human papilloma viruses, 0.15uL of 5U / uL FastTaq enzymes and 3uL of reactive templates are used; a genetic analyzer is used for performing capillary electrophoresis and genotype analysis on a PCR product; the HPV subtypes are judged according to the specific color and position peak on the capillary electrophoresis figure. The method provided by the invention adopts a Touchdown multiplex PCR technology, so that the multiplex PCR non-specificity amplification can be effectively reduced.
Owner:SUZHOU MUNICIPAL HOSPITAL
Composite amplification kit for 38 human Y chromosome loci and application thereof
ActiveCN109880913AEfficient amplificationAmplification specificMicrobiological testing/measurementAgainst vector-borne diseasesDNA paternity testingFluorescence
The invention relates to a composite amplification kit for 38 human Y chromosome loci and application thereof. The invention provides a composite amplification system of the 38 human Y chromosome loci. The system includes specific primers for amplifying the 38 loci including 35 STR loci and 3 Indel loci. The 38 pairs of specific primers are subjected to group fluorescence labeling by the six-colorfluorescent labeling technology. Through design and optimization of the primer sequence and the working concentration, simultaneous efficient specific sensitive amplification of the 38 human Y chromosome loci is achieved. A detection result of the composite amplification system has high individual recognition capability and good data compatibility, and can be used in practice for paternity testing and individual recognition, thereby effectively reducing the detection cost of human DNA typing and improving the detection working efficiency.
Owner:BEIJING PEOPLESPOT TECH
Potato low-temperature sweetening resistant molecular marker combination and application thereof in potato low-temperature sweetening resistant breeding
InactiveCN105713991AImprove accuracyEasy to detectMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
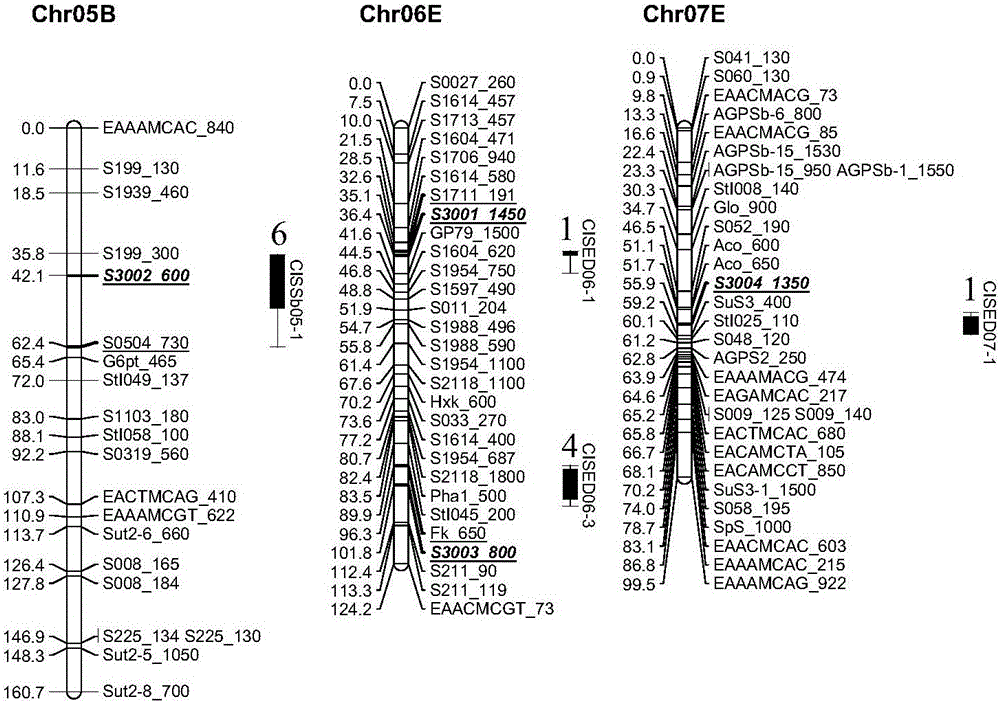
The invention relates to the fields of molecular biology and genetic breeding and in particular relates to a potato low-temperature sweetening resistant molecular marker and application thereof. Potato low-temperature sweetening resistant molecular marker combination is any one or more of nucleotide sequences S3001-S3004; upstream and downstream primer sequences of the nucleotide sequences S3001-S3004 are shown in SEQ ID No.1-8. The potato low-temperature sweetening resistant molecular marker combination provided by the invention has the beneficial effects that related molecular marker polymorphic analysis and low temperature reducing sugar content determination are carried out on varieties (lines) with different low-temperature sweetening resistant capacities, correlation between the molecular marker and low-temperature sweetening resistance is analyzed, screening is carried out for obtaining a molecular maker combination used for establishing a low-temperature sweetening resistant breeding molecular marker-assisted selection system, the molecular maker combination has relatively high accuracy on low-temperature sweetening resistant potato genotype screening, and technical support is provided for identification of a potato low-temperature sweetening resistant strain and low-temperature sweetening resistant breeding.
Owner:HUAZHONG AGRI UNIV
Kit for detecting abundance of common probiotics
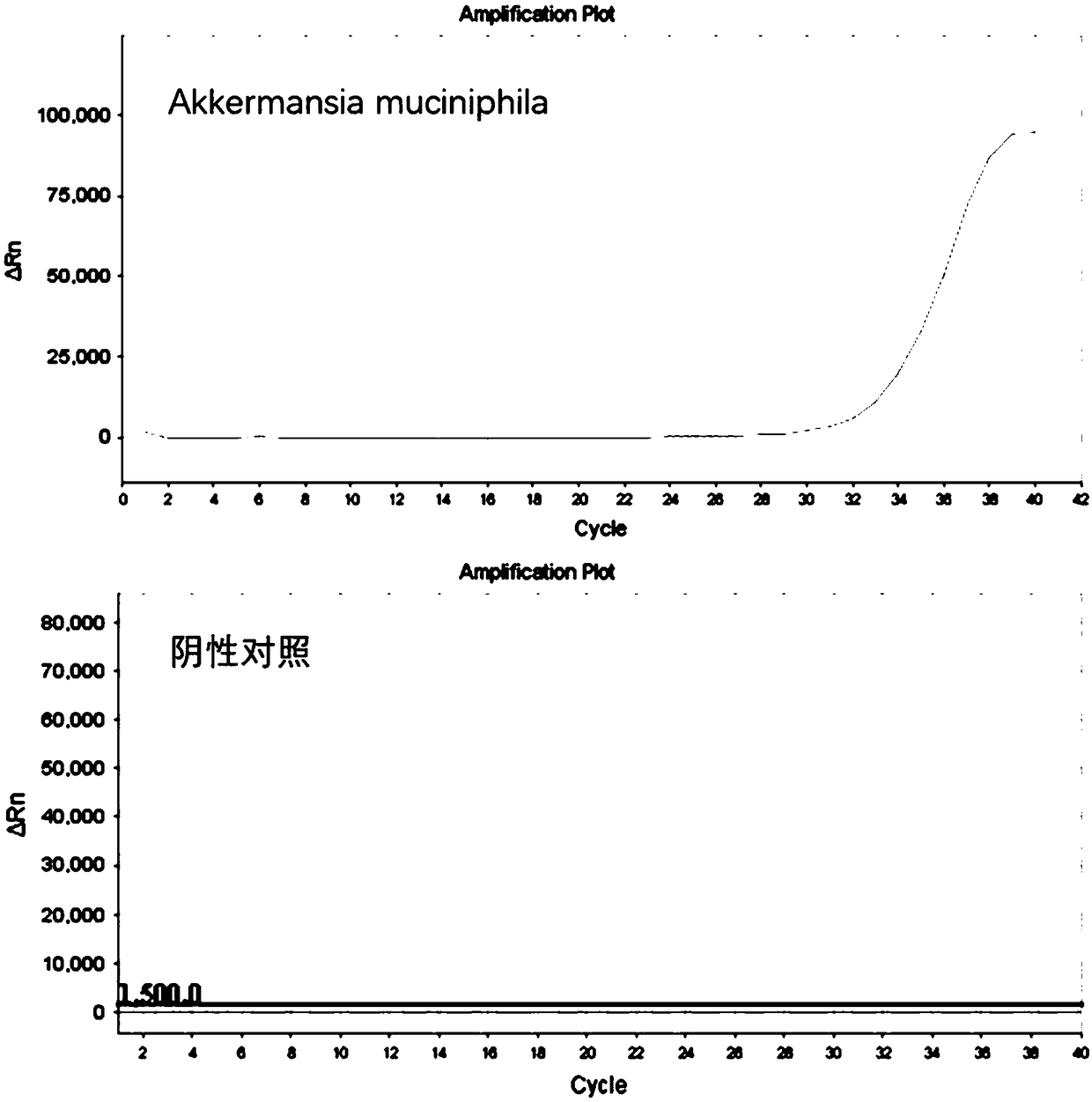
ActiveCN109266764AGood amplification specificityHigh amplification efficiencyMicrobiological testing/measurementMicroorganism based processesBifidobacteriumProbiotic
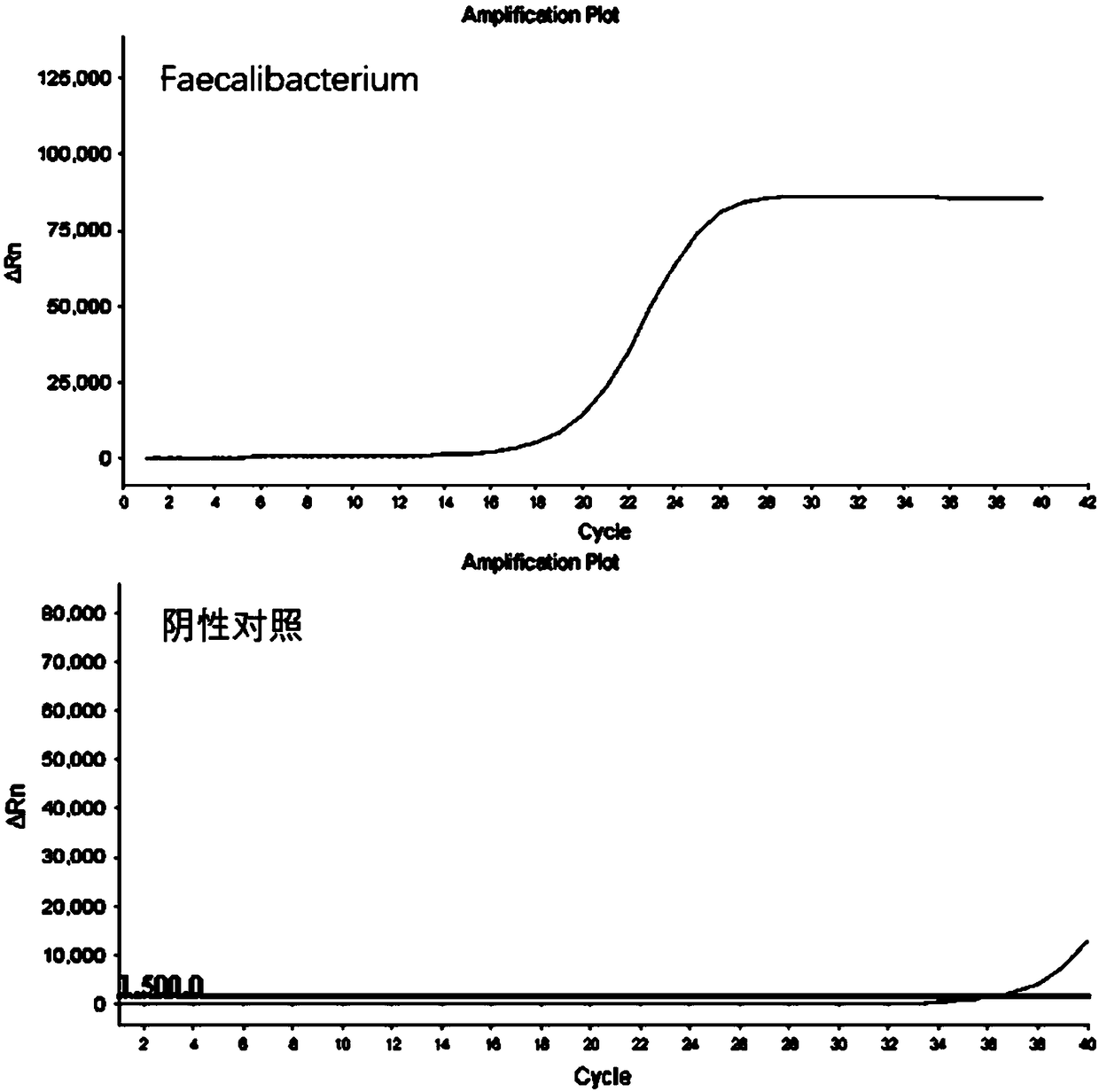
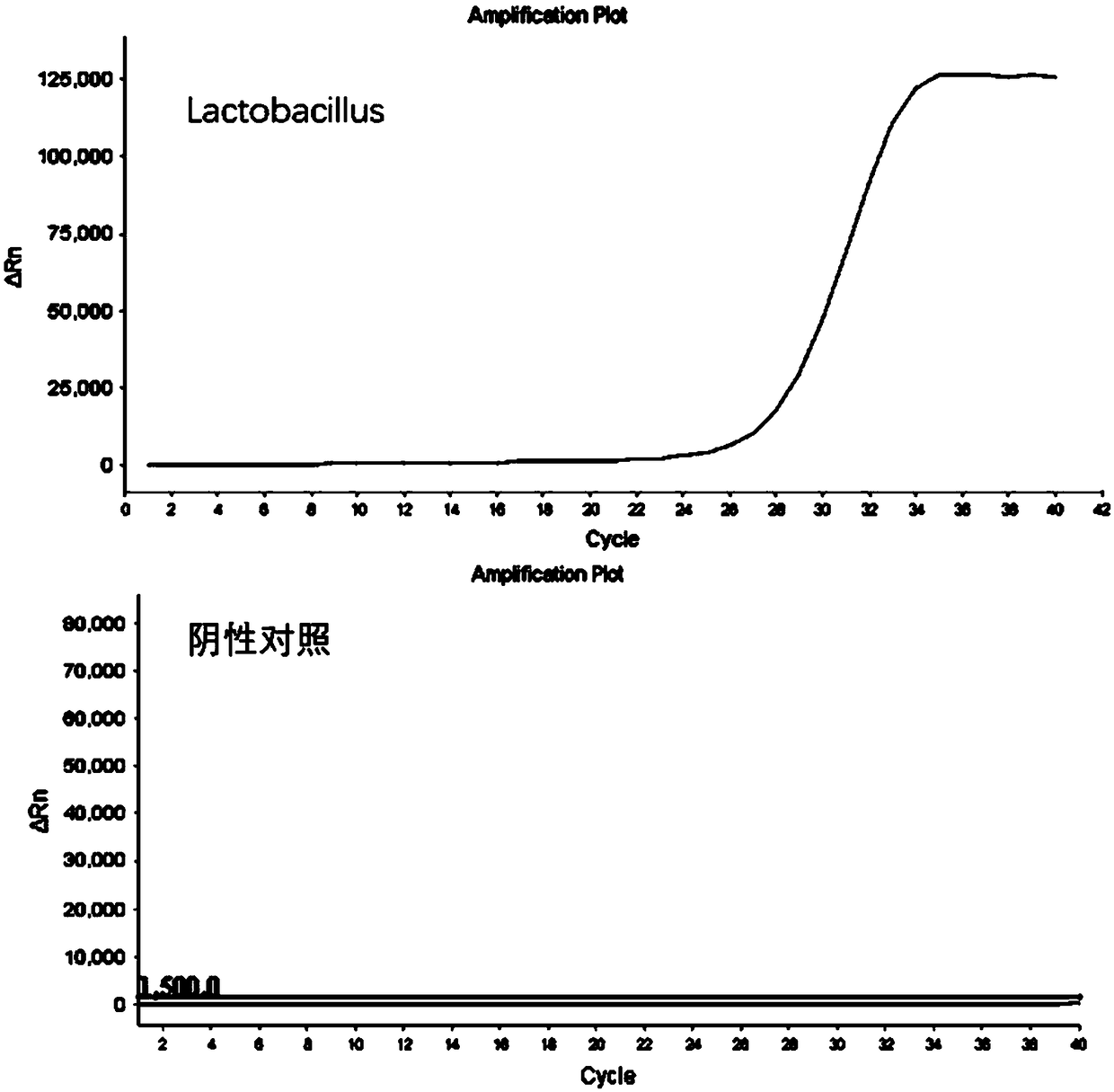
The invention discloses a kit for detecting the abundance of common probiotics, as a result of their own technology, analyzing and designing from a large amount of data, the primers with the best specificity and amplification efficiency were screened out, including Faecalibacterium, lactobacillus, Bifidobacterium, akkermansia muciniphil and total bacterial prim that primers of the invention have good amplification specificity and high amplification efficiency, and can specifically amplify the correspond probiotics from the total DNA of the complex intestinal microbial flora, and the result isthe same as that of the mature method. The reagent kit of the invention can realize the rapid and intuitive detection of common probiotics, shortens the detection period and reduces the detection cost.
Owner:GENETALKS BIO TECH CHANGSHA CO LTD
Polyase chain reaction optimization method based on arborized polymer
InactiveCN101343650AEasy to storeImprove amplification specificityFermentationBiotechnologyPolymer science
The invention discloses a polymerase chain reaction optimizing method based on dendritic polymer, which belongs to the technical field of the biology. A dendritic polymer material is added into a polymerase chain reaction system as an additive agent to optimize the polymerase chain reaction, the dendritic polymer comprises multivalent nucleus combined with at least two dendritic branching covalences and a branch structure which can ensure the polymerization degree to achieve at least 2.0 through at least two generations of expansion. The dendritic polymer is diluted or concentrated and added into the polymerase chain reaction system in a gradient way, so as to achieve the range of the optimization purpose through actual amplification, and the range is the effective use amount range of the required dendritic polymer. The invention has the advantages that the optimization and amplification effects are remarkable, the cost of the additive agent is low, the amplified product is easy to be preserved, the application of the amplified product is wide, the specificity of the amplified product is obviously improved, and the output of the product in certain reactions is obviously enhanced.
Owner:SHANGHAI JIAO TONG UNIV
Primer composition for detecting harmful gene of deficiency of uridine monophosphate synthase of cattle, kit with primer composition and application of kit
ActiveCN103627804AStrong amplification specificityHigh amplification efficiencyMicrobiological testing/measurementDNA/RNA fragmentationNucleotide sequencingUridine monophosphate
The invention discloses a primer composition for detecting a harmful gene of deficiency of uridine monophosphate synthase of cattle, a kit with the primer composition and an application of the kit. The primer composition disclosed by the invention is composed of a primer group A and a primer group B, wherein the primer group A is composed of a primer 1 and a primer 2, the primer group B is composed of a primer 3 and a primer 4, and the nucleotide sequences of the primer 1, the primer 2, the primer 3 and the primer 4 are respectively shown as SEQIDNO. 1-4. The invention also provides the kit with the primer composition. The method for applying the kit disclosed by the invention to the detection of the harmful gene of the deficiency of uridine monophosphate synthase of cattle comprises the steps of extracting the complete set of DNA (Deoxyribonucleic acid) in cattle blood as a template to carry out nested PCR (Polymerase Chain Reaction) amplification to obtain a PCR product, and sequencing the obtained PCR product so as to directly know about the basic group change on a mutation site according to a sequenced result, thereby ensuring the accuracy of the result and meeting the requirements of a detecting technology for characteristics such as high speed, precision, high throughput and the like.
Owner:SOUTH CHINA AGRI UNIV
Primer, detection kit and method for detecting TERC gene whole exome sequence mutation
InactiveCN108531575AStrong specificityImprove amplification specificityMicrobiological testing/measurementDNA/RNA fragmentationDyskeratosis congenitaSanger sequencing
The invention discloses a method, primer and detection kit for detecting TERC gene whole exome sequence mutation of a dyskeratosis congenita patient. The primer comprises forward and reverse primers amplifying a TERC whole exome sequence and a pair of sequencing primers. According to the method, primer and detection kit for detecting TERC gene whole exome sequence mutation of the dyskeratosis congenita patient, Touch-down PCR amplification and Sanger sequencing are combined so that the mutation situation of the TERC whole exome sequence in the body of the dyskeratosis congenita patient can befast detected, and a TERC gene has two mutation sites of C408G and GC107-108AG.
Owner:杭州艾迪康医学检验中心有限公司
Primer group and kit for simultaneously amplifying 34 human STR gene loci and application of primer group and kit
PendingCN112852972AImprove individual recognitionHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationCore geneY chromosome deletions
The invention provides a primer group and a kit for simultaneously amplifying 34 human STR gene loci and application of the primer group and the kit, and belongs to the technical field of molecular genetics. In the invention, the 34 STR gene loci comprise 29 autosomal STR gene loci, one Y chromosome STR gene locus and 4 sex identification STR gene loci. According to the invention, 34 STR gene loci can be simultaneously amplified in one reaction, 20 core gene loci and 10 preferable gene loci specified by Ministry of Public Security are included, all loci of mainstream kits in the current market are included, and meanwhile, 5 gender identification gene loci are also included; and the risk of gender identification errors caused by Y chromosome deletion can be effectively prevented. 34 STR gene loci are combined, so that the method has the characteristics of high individual recognition capability and high non-father exclusion rate.
Owner:百特元生物科技(北京)有限公司
Multiplex amplification kit for simultaneously detecting autosome and Y chromosome STR loci
PendingCN110157812ASolve the problem of micro inspection materialsAccelerated exclusionMicrobiological testing/measurementGeneticsY-STR
The invention discloses a multiplex amplification kit for simultaneously detecting autosome and Y chromosome STR loci. The kit contains 36 groups of specific amplification primer pairs corresponding to 38 loci, and comprises sequences SEQ ID NO. 1-74. A-STR data and Y-STR data can be simultaneously analyzed and used for rapid screening of cases, so that the detection time is shortened, and meanwhile the efficiency of excluding and determining criminal suspects is improved.
Owner:苏州市公安局刑事科学技术研究所 +1
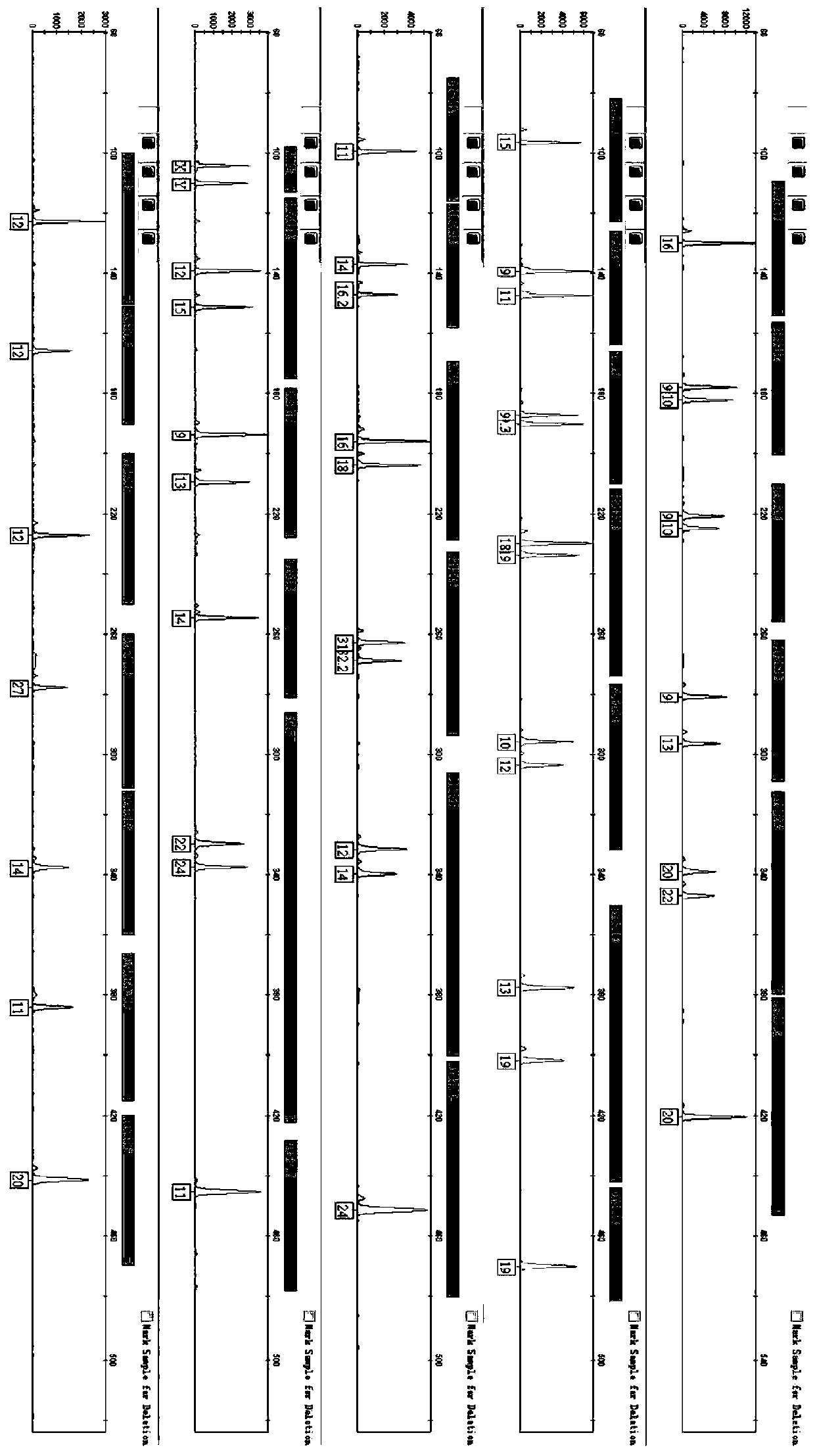
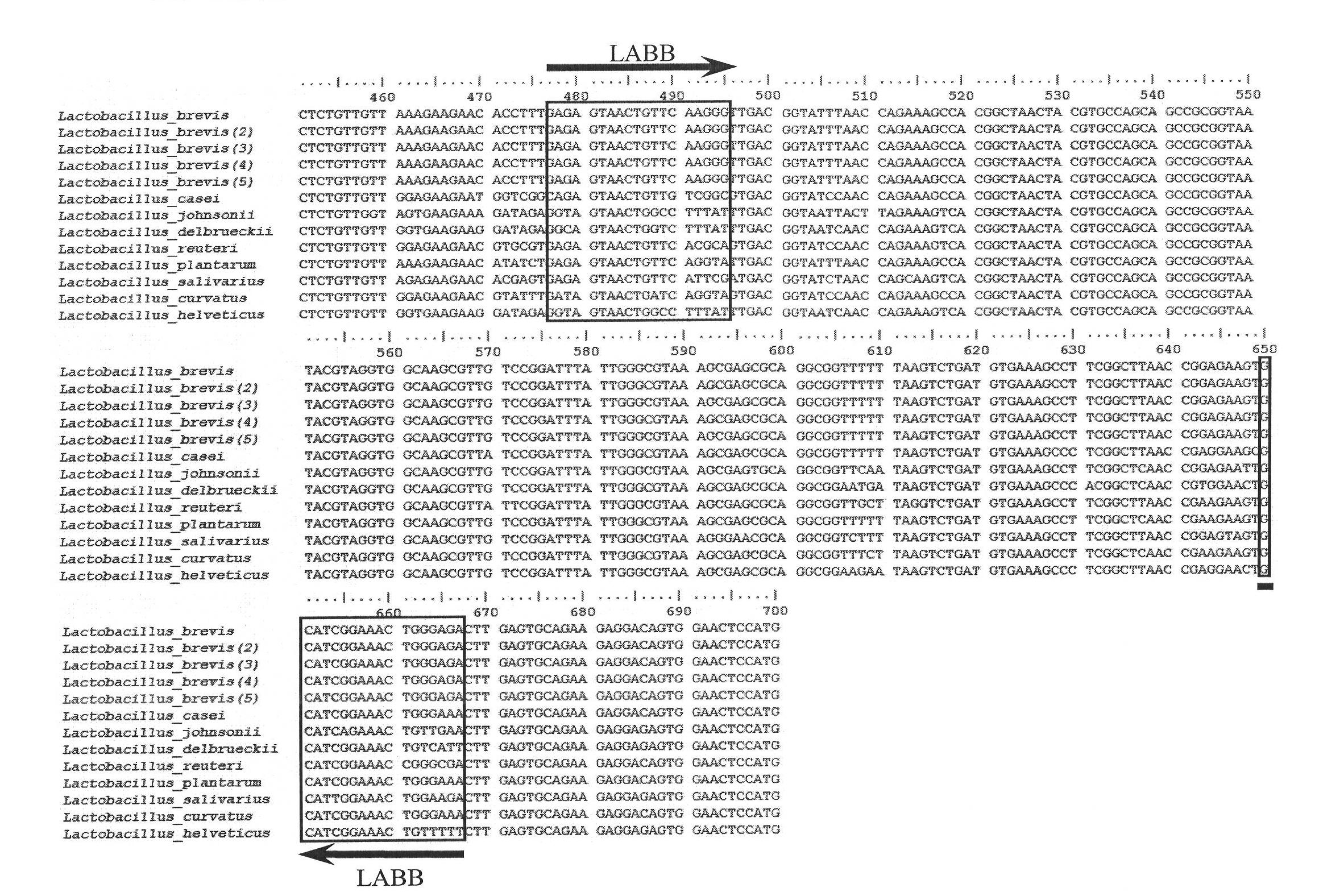
Primer sequence for identifying lactobacillus brevis and application thereof
ActiveCN102094089AImprove amplification specificitySimple processMicrobiological testing/measurementBase sequenceLactobacillales
The invention discloses a primer sequence for identifying lactobacillus brevis and an application thereof, belonging to the field of molecular biology. The base sequences of a primer disclosed by the invention are shown in SEQ ID NO:1 and SEQ ID NO:2. The primer sequence disclosed by the invention can be used for identifying lactobacillus brevis. The primer can specifically amplify the partial sequence of lactobacillus brevis 16SrDNA, has high amplification specificity, and can accurately and quickly identify lactobacillus brevis. The process of identifying lactobacillus brevis by using the primer is simple, the identification method is stable and efficient, the detection time is shortened, and the detection cost is low. The invention provides a detection method for identifying germ plasmresources of lactobacillus brevis, and lays a good foundation for screening fine strains of lactobacillus brevis.
Owner:福建大北农华有水产科技集团有限公司 +2
Method and primer as well as kit for detecting mutation sites of promoters C250T and C228T of TERT (Telomerase Reverse Transcriptase) gene
InactiveCN105695596AImprove amplification specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationSanger sequencingTelomerase reverse transcriptase
The invention discloses a method, a primer and a kit for detecting mutation sites of promoters C228T and C250T of a telomerase reverse transcriptase (TERT) gene. The primer comprises a forward primer and a reverse primer for amplifying the mutation sites of the promoters C228T and C250T of the TERT gene, and in addition, can also comprise one pair of sequencing primers. A Touch-down PCR (Polymerase Chain Reaction) amplification technique is combined with a Sanger sequencing method; the occurrence conditions of the mutation sites of the promoters C228T and C250T of the TERT gene in a body of a patient suffered from a brain glioma can be quickly detected.
Owner:南京艾迪康医学检验所有限公司
RT-RPA primer pair, probe, kit and detection method for detecting cat coronavirus
ActiveCN111471797AImprove amplification specificityIncreased sensitivityMicrobiological testing/measurementAgainst vector-borne diseasesFeline coronavirusSerotype
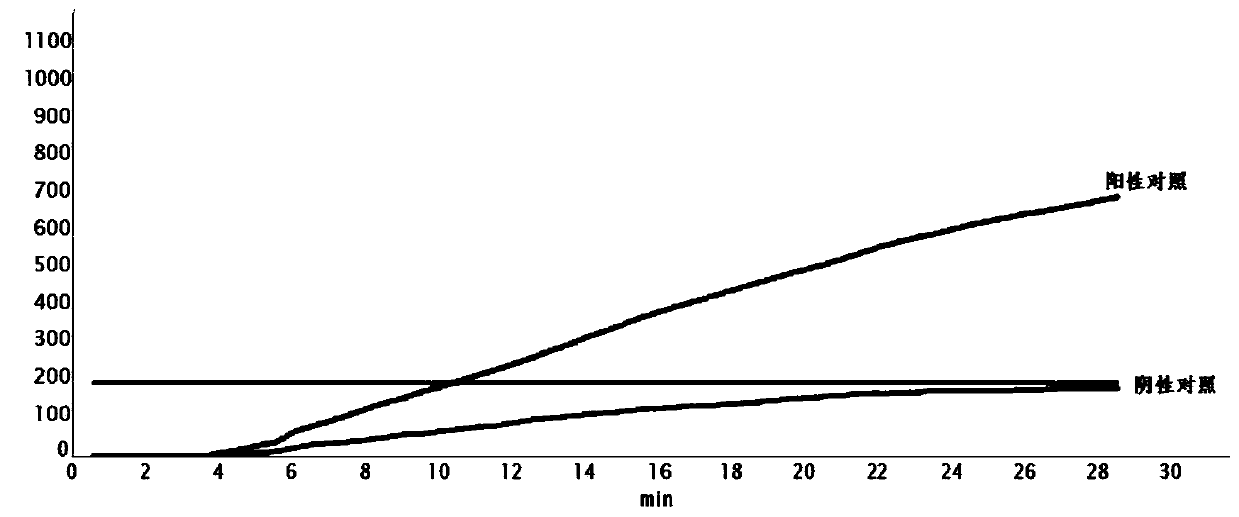
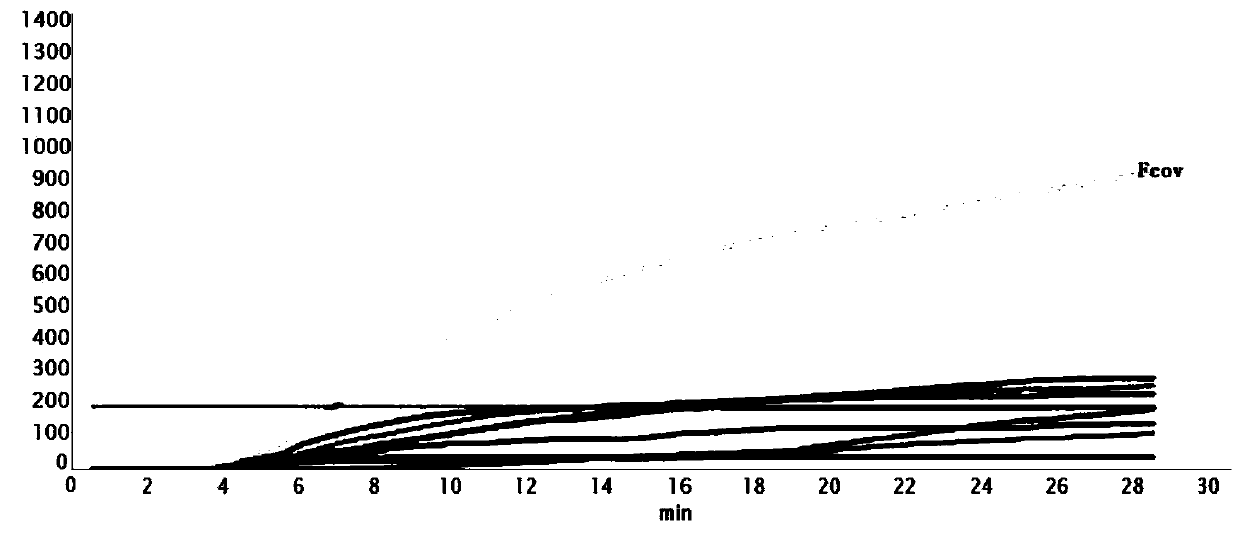
The invention provides an RT-RPA primer pair, a probe, a kit and a detection method for detecting cat coronavirus, and belongs to the technical field of molecular biological detection. Aiming at an MPgene sequence of a cat coronavirus feline infectious peritonitis viruses (Fcov), a specific primer pair and a probe are designed, and a real-time fluorescence RPA detection method is established through repeated tests. The method disclosed by the invention is strong in amplification specificity and high in sensitivity, and can be used for detecting two viruses of FCoV-I and FCoV-II serotypes which both can cause feline infectious peritonitis (FIP) in one detection test, so that the method is more efficient. Technical reference is provided for rapid diagnosis and prevention and control of Fcov.
Owner:GUANGDONG LAB ANIMALS MONITORING INST +1
RPA (recombinase polymerase amplification) primer pair, probe, kit and detection method for rapidly detecting Marek's disease virus (MDV)
ActiveCN110699485ARapid implementation of diagnostic testingEnables diagnostic testingMicrobiological testing/measurementMicroorganism based processesMarek's diseaseRecombinase Polymerase Amplification
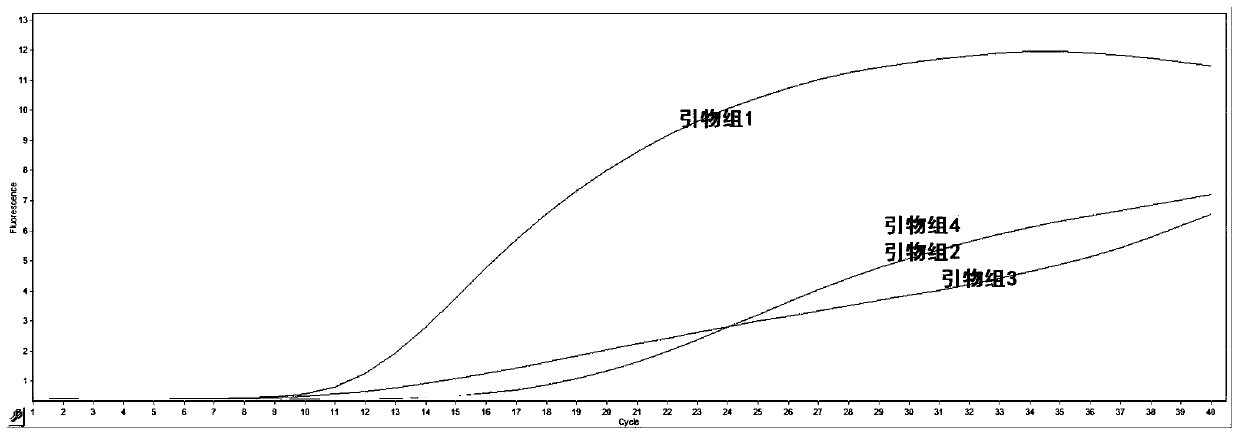
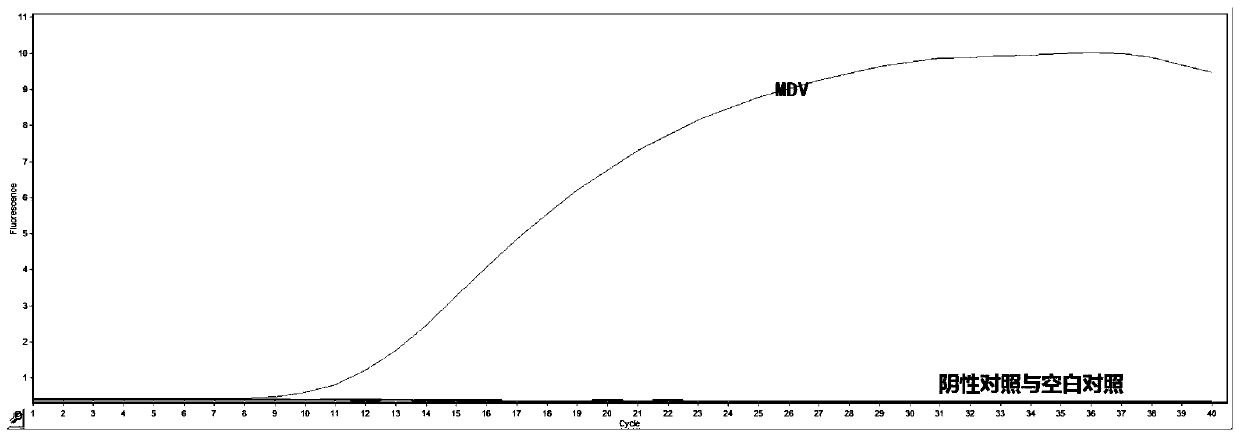
The invention provides an RPA (recombinase polymerase amplification) primer pair, probe, kit and detection method for rapidly detecting Marek's disease virus (MDV), and belongs to the technical fieldof molecular biological detection. Sequences of the RPA primer pair are shown in SEQ ID No.1 and SEQ ID No.2; and a sequence of the RPA probe is shown in SEQ ID No.3. The detection method can completedetection at 39 DEG C within 20 min, has the characteristics of being rapid, sensitive, convenient to operate and applicable to laboratories and field rapid detection, and provides a technical reference for rapid diagnosis, prevention and control of the MDV.
Owner:广州千寻生物技术有限公司 +2
PCR (polymerase chain reaction) primer and method for identifying authenticity of bulbus fritillariae cirrhosae or detecting degree of adulteration of bulbus fritillariae cirrhosae
ActiveCN109913570AEasy to operateImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceBulbus fritillariae cirrhosae
The invention discloses a PCR (polymerase chain reaction) primer and a method for identifying the authenticity of bulbus fritillariae cirrhosae or detecting the degree of adulteration of the bulbus fritillariae cirrhosae. The PCR primer comprises an identification primer for authentic bulbus fritillariae cirrhosae and / or an identification primer for counterfeit bulbus fritillariae cirrhosae. The 75th base 'C' of an ITS1 spacer of the bulbus fritillariae cirrhosae serves as the 3' end of the identification primer for the authentic bulbus fritillariae cirrhosae, and the 75th base 'T' of the ITS1spacer of the bulbus fritillariae cirrhosae serves as the 3' end of the identification primer for the counterfeit bulbus fritillariae cirrhosae. The base mismatch is introduced to third, ninth and tenth bases at the 3' ends of the two primers to obtain the identification primer for the authentic bulbus fritillariae cirrhosae and the identification primer for the counterfeit bulbus fritillariae cirrhosae, wherein the identification primer for the authentic bulbus fritillariae cirrhosae is represented by SEQ ID No. 1 or 2, and the identification primer for the counterfeit bulbus fritillariae cirrhosae is represented by SEQ ID No. 3 or 4. When the PCR primer is used for authenticating bulbus fritillariae cirrhosae samples, the authenticity of the bulbus fritillariae cirrhosae or the degree of adulteration of the bulbus fritillariae cirrhosae can be accurately and quickly identified.
Owner:GUIZHOU MEDICAL UNIV
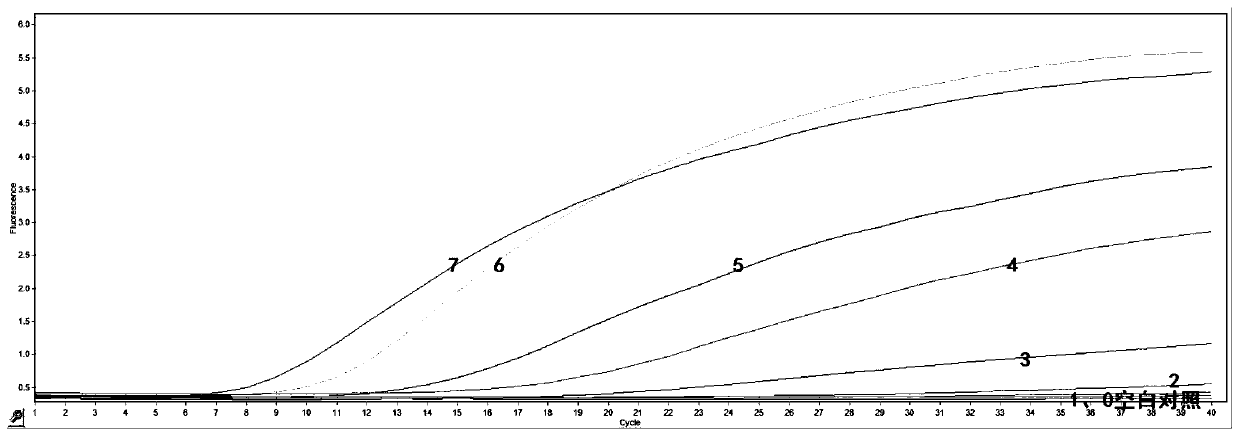
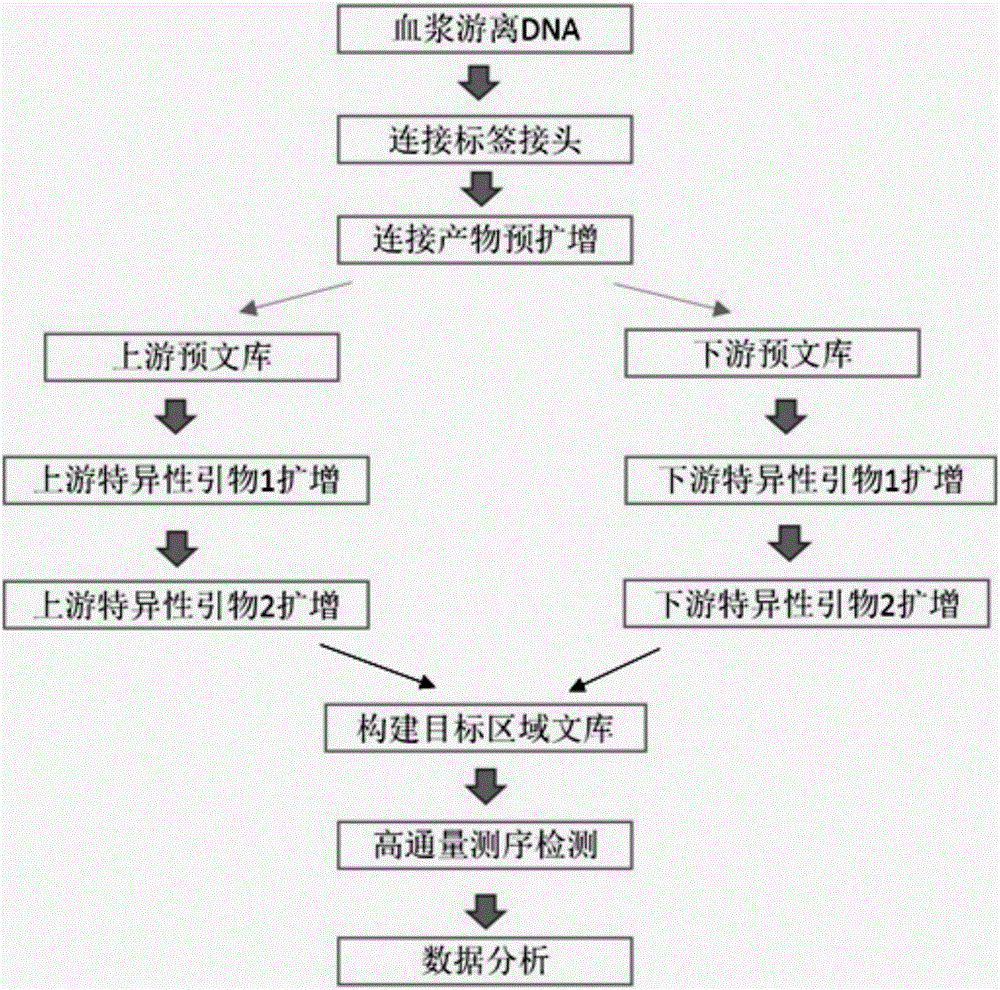
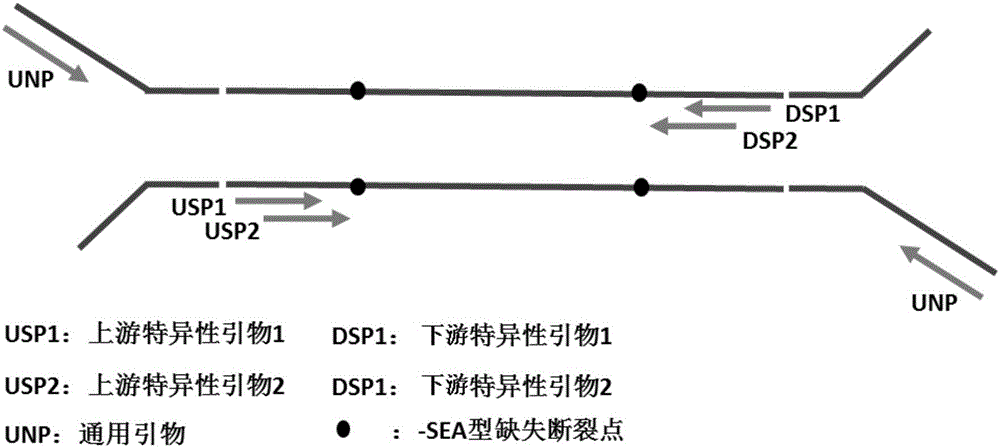
Noninvasive antepartum fetal alpha<-SEA> type thalassemia gene mutation detection library building method, detection method and kit
PendingCN106755484AHigh specificityComputationally efficientMicrobiological testing/measurementLibrary creationGene mutationPeripheral blood
The invention discloses a noninvasive antepartum fetal alpha<-SEA> type thalassemia gene mutation detection library building method, a detection method and a kit. In the library building method, a specific connector is connected onto a maternal peripheral blood free DNA (deoxyribonucleic acid) fragment; then, a connector connection product pre-amplification product is divided into two parts; two rounds of specific amplification are performed by respectively and independently using forward primers and reverse primers aiming at target sites; the target sites can be enriched at high specificity; the amplification specificity of the primers can be obviously improved. The forward and reverse primer groups of a plurality of SNP (single nucleotide polymorphism) sites used for calculating the fetal free DNA proportion are respectively used for performing two-round specific amplification; the fetal thalassemia genotype can be efficiently and accurately judged. The library is subjected to sequence testing; the alpha<-SEA> type thalassemia gene mutation can be accurately and effectively detected; the result is identical to the result of the amniocentesis detection and typing; the safety, the noninvasion and the high efficiency are obviously superior to those of the amniocentesis detection.
Owner:GENETALKS BIO TECH CHANGSHA CO LTD
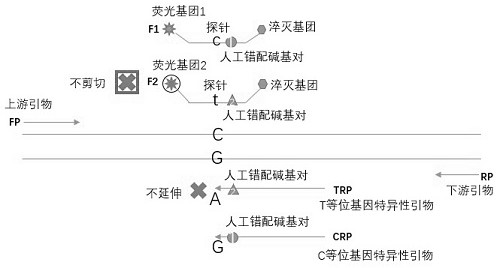
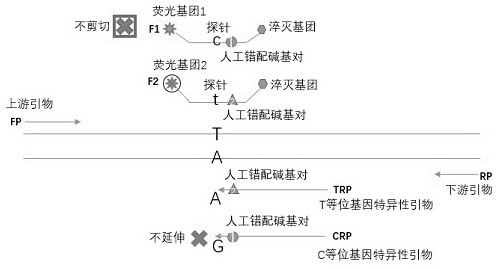
Composition for detecting single nucleotide polymorphism an application of composition
ActiveCN111996244AReduced probability of non-specific bindingReduce design difficultyMicrobiological testing/measurementDNA/RNA fragmentationMultiple allelesAllele specific
The invention belongs to the technical field of fluorescent probe detection of genes, and particularly relates to a composition for detecting single nucleotide polymorphism and an application of the composition. The composition for detecting single nucleotide polymorphism comprises an upstream primer and a downstream primer which are shared among different alleles, allele specific primers for eachspecific allele, and probes being respectively complementary and associated with various allele specific primers to use different fluorescence labelling, wherein the probes cover single nucleotide polymorphism sites. The composition for detecting single nucleotide polymorphism is adopted for gene detection, so that single-pipe multi- allele detection can be realized, the sensitivity is high, heterozygosis or homozygosis types of the genes can also be distinguished, repeated optimization of the probe is not needed, the cost is saved, the detection flux can be increased, the results are accurate and analysis is facilitated.
Owner:浙江绍兴鼎晶生物医药科技股份有限公司
Two-step detection method of PCR (Polymerase Chain Reaction) of emulsion
ActiveCN106148569AWith anti-jammingHigh sensitivityMicrobiological testing/measurementMicroorganism based processesInterference resistanceOil phase
The invention discloses a two-step detection method of PCR (Polymerase Chain Reaction) of an emulsion, which belongs to the technical fields of biomedicine and clinical medicine. The method specifically comprises the steps of preparing a water-in-oil emulsion first, and then loading a PCR tube, filled with the water-in-oil emulsion, into a PCR instrument for amplification for the first step; then centrifuging an amplified product, and discarding an upper oil phase, so as to prepare an oil-in-water emulsion, loading into the PCR tube for amplification for the second step, and performing agarose gel electrophoresis detection after amplification is ended. According to the two-step detection method, the yield is further increased, and the gene detection sensitivity is increased; the two-step detection method has the characteristics of interference resistance and high specificity of the PCR of the emulsion and has the characteristic of high yield of the PCR of the emulsion in a conventional PCR method, and additionally, the processes of diethyl ether demulsification, purification and the like are not applied, so that the effects of simplicity, convenience and economy are achieved.
Owner:江苏护理职业学院
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com