Primer sequence for identifying lactobacillus brevis and application thereof
A primer sequence and Lactobacillus technology, applied in the field of molecular biology, can solve the problems of long detection period, poor accuracy, and low degree of automation, and achieve the effect of low detection cost, simple process, and rapid identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Step 1 Design and synthesis of primers
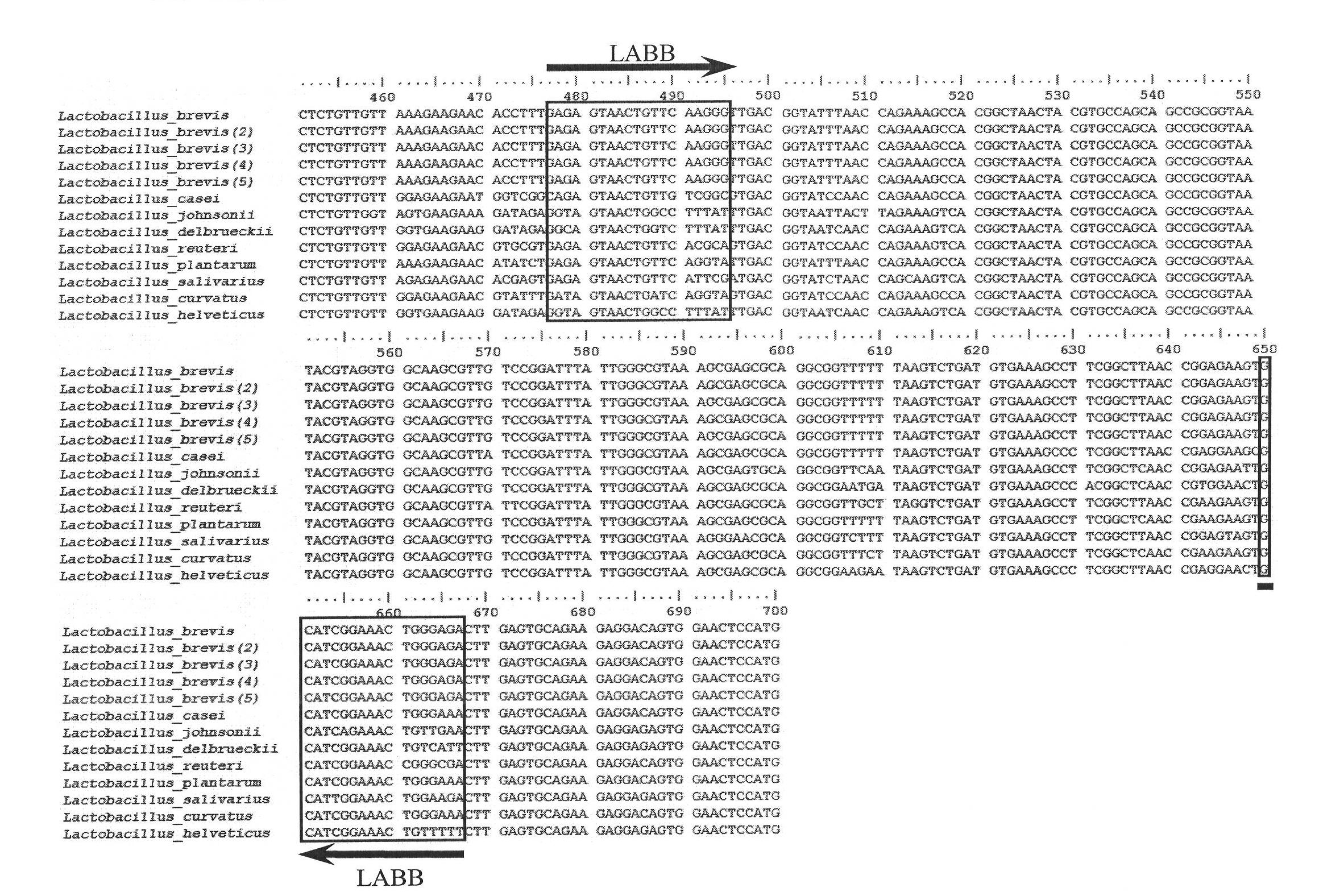
[0022] Obtain the 16S rDNA sequence of Lactobacillus pumilus and its homologous Lactobacillus from the Genebank database, search for specific regions that are conserved in Lactobacillus pumilus but not conserved in other Lactobacillus species, design primers, and screen and optimize specific sequences. Finally, a pair of primers LABBF and LABBR with better parameters were obtained, and the above two primers were synthesized.
[0023] The specific sequences of the above primers and the corresponding relationship in the sequence listing are shown in Table 2 below:
[0024] Table 2
[0025] Primer name
sequence
serial number
LABBF
5`-GAGAGTAACTGTTCAAGGG-3`
SEQ ID NO: 1
LABBR
5`-TCTCCCAGTTTCCGATGC-3`
SEQ ID NO: 2
[0026] The physical positions of the above primers on the 16S rDNA sequence are as follows: figure 1 shown.
[0027] Step 2 Preparation of PCR template
[0028] 1)...
Embodiment 2
[0056] The design and synthesis of step 1 primers are the same as the primer sequences shown in the primer list in Example 1
[0057] Step 2 Preparation of PCR template
[0058] 1) Pick a single colony of each strain on the petri dish to a 1.5mL sterilized centrifuge tube with 50μL sterile water, and mix well;
[0059] 2) Boil the centrifuge tube in boiling water at 100°C for 5 minutes;
[0060] 3) Place the centrifuge tube in an ultra-low temperature freezer at -80°C for 15 minutes;
[0061] 4) Boil the centrifuge tube in boiling water at 100°C for 5 minutes;
[0062] 5) Centrifuge at 4000 rpm for 10 minutes, and take the supernatant as a PCR template.
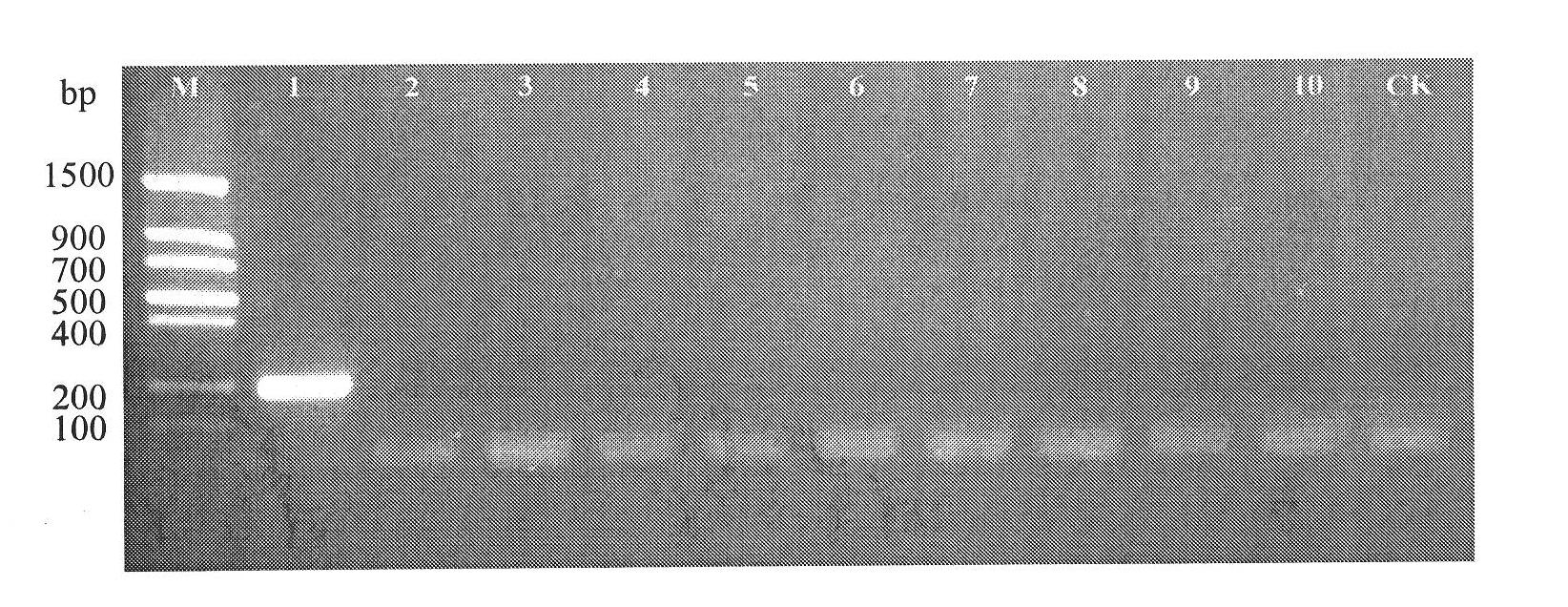
[0063] Step 3PCR amplification
[0064] 1. PCR reaction system
[0065] Use the DNA prepared in step 2 as a template, and use the base sequence synthesized in step 1 as a primer to carry out a PCR reaction. The reaction system is:
[0066]Template 1μL
[0067] 10×PCR Buffer 2.5μL
[0068] dNTP (10mM) 0.5μL
[0069] L...
Embodiment 3
[0084] The design and synthesis of step 1 primers are the same as the primer sequences shown in the primer list in Example 1
[0085] Step 2 Preparation of PCR template
[0086] 1) Pick a single colony of each strain on the petri dish to a 1.5mL sterilized centrifuge tube with 100μL sterile water, and mix well;
[0087] 2) Boil the centrifuge tube in boiling water at 100°C for 15 minutes;
[0088] 3) Place the centrifuge tube in an ultra-low temperature freezer at -80°C for 40 minutes;
[0089] 4) Boil the centrifuge tube in boiling water at 100°C for 15 minutes;
[0090] 5) Centrifuge at 8000 rpm for 5 minutes, and take the supernatant as a PCR template.
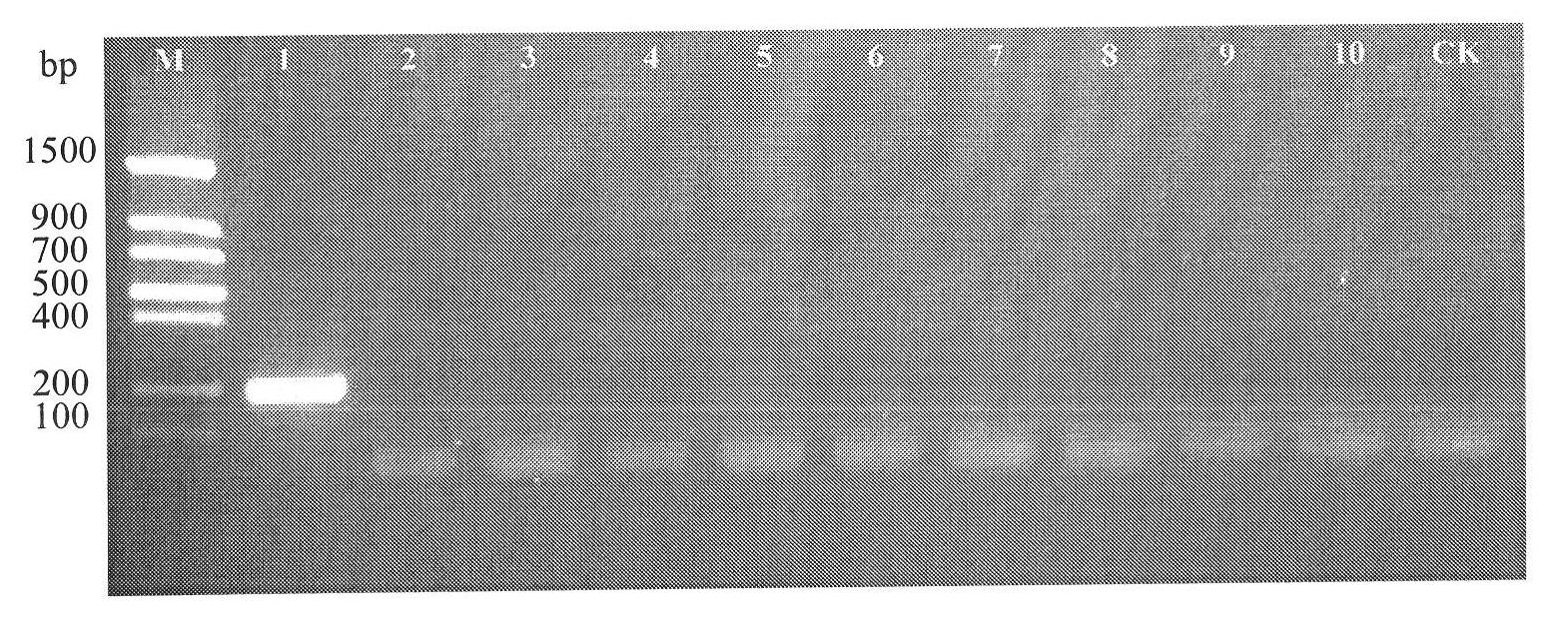
[0091] Step 3PCR amplification
[0092] 3. PCR reaction system
[0093] Use the DNA prepared in step 2 as a template, and use the base sequence synthesized in step 1 as a primer to carry out a PCR reaction. The reaction system is:
[0094] Template 1μL
[0095] 10×PCR Buffer 2.5μL
[0096] dNTP (10mM) 0.5μL
[0097...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com