Composition for detecting single nucleotide polymorphism an application of composition
A single nucleotide polymorphism and composition technology, which is applied in the field of gene fluorescent probe detection, can solve the problems of time-consuming, time-consuming, labor-intensive, and difficult operations, and achieves a reduction in the probability of non-specific binding, design difficulty, and development savings. cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
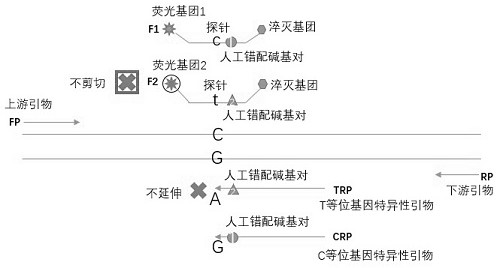
[0043] Embodiment 1: as figure 1 As shown, the allele-specific primers and their associated probes were designed and used for the detection of the rs429358 (T / C) SNP site of the ApoE gene; the PCR template was the genomic DNA of the cell line.
[0044] Reaction liquid system preparation: H 2 O, purified water, 16.7 μL; PCR buffer (Mg 2+ plus), 5X, 5μL; dNTPs (each 10mM dATP, dCTP, dGTP; 20mM dUTP), 10mM, 0.5μL; allele-specific primer 1, 100uM, 0.1μL; allele-specific primer 2, 100uM, 0.1μL; Allele-specific primer-associated probe 1, 100uM, 0.05μL; Allele-specific primer-associated probe 2, 100uM, 0.05μL; Upstream primer, 100uM, 0.1μL; Downstream primer, 100uM, 0.1μL; UDG enzyme, 1U / μL, 0.1μL; polymerase, 5U / μL, 0.2μL; total volume 23μL.
[0045] Operation steps: Prepare relevant liquids according to the above reaction liquid system preparation, shake and mix for 15 seconds, and centrifuge quickly for 15 seconds, dispense 23 μL per tube into PCR reaction tubes, add 2 μL of ...
Embodiment 2
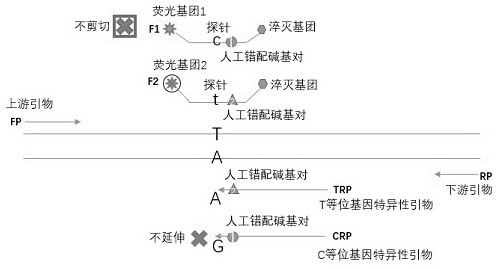
[0059] Embodiment 2: as figure 2 As shown, the allele-specific primer complementary probe PCR detection technology was used for the detection of the MTHFR gene 677 (C / T) SNP site; the PCR template was the genomic DNA of the cell line.
[0060] Reaction liquid system preparation: H 2 O, purified water, 16.7 μL; PCR buffer (Mg 2+ plus), 5X, 5μL; dNTPs (each 10mM dATP, dCTP, dGTP; 20mM dUTP), 10mM, 0.5μL; allele-specific primer 1, 100uM, 0.1μL; allele-specific primer 2, 100uM, 0.1μL; Allele-specific primer-associated probe 1, 100uM, 0.05μL; Allele-specific primer-associated probe 2, 100uM, 0.05μL; Upstream primer, 100uM, 0.1μL; Downstream primer, 100uM, 0.1μL; UDG, 1U / μL, 0.1 μL; polymerase, 5 U / μL, 0.2 μL; total volume 23 μL.
[0061] Operation steps: Prepare relevant liquids according to the above reaction liquid system preparation, shake and mix for 15 seconds, and centrifuge quickly for 15 seconds, dispense 23 μL per tube into PCR reaction tubes, add 2 μL of the samples...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com