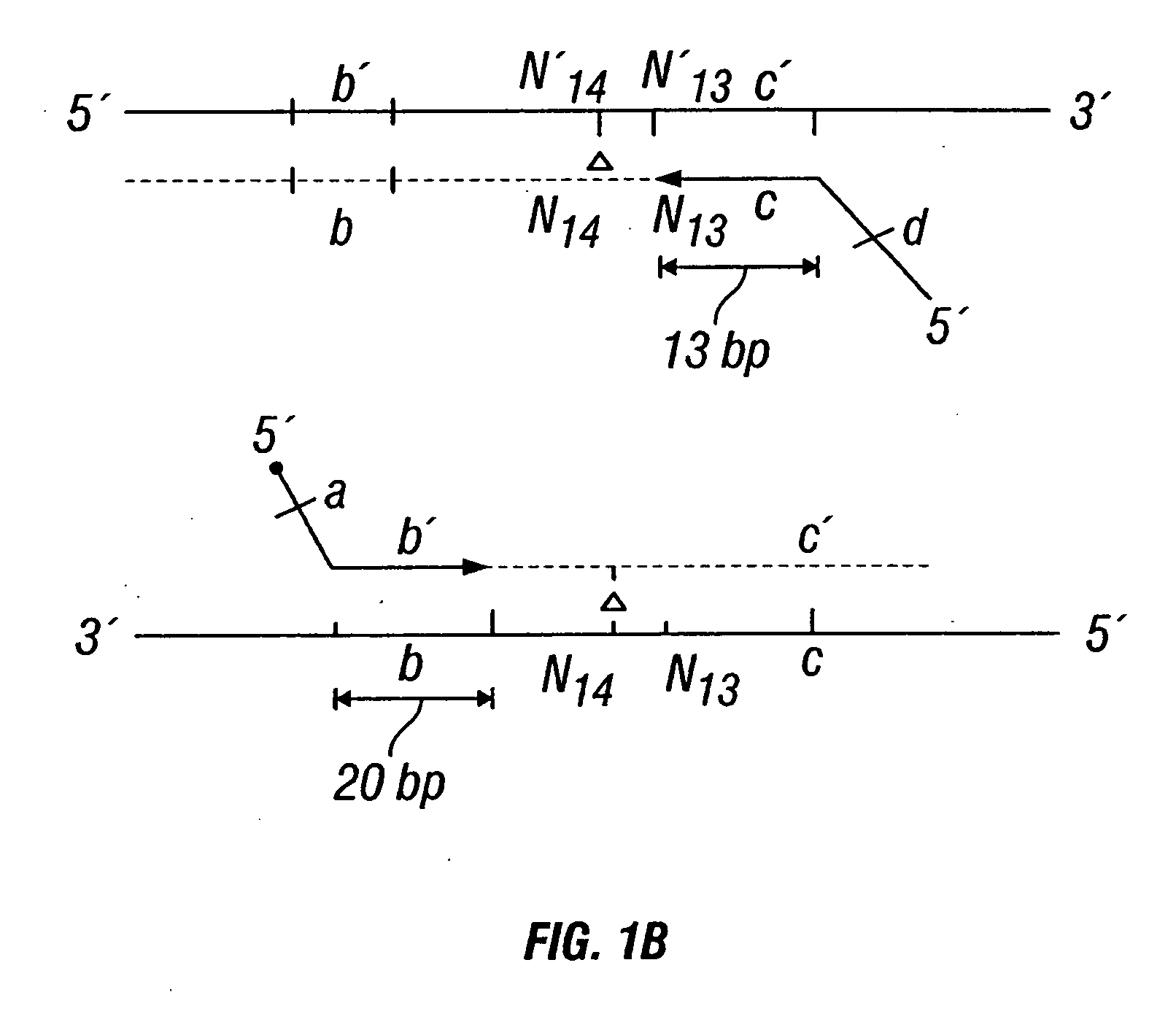
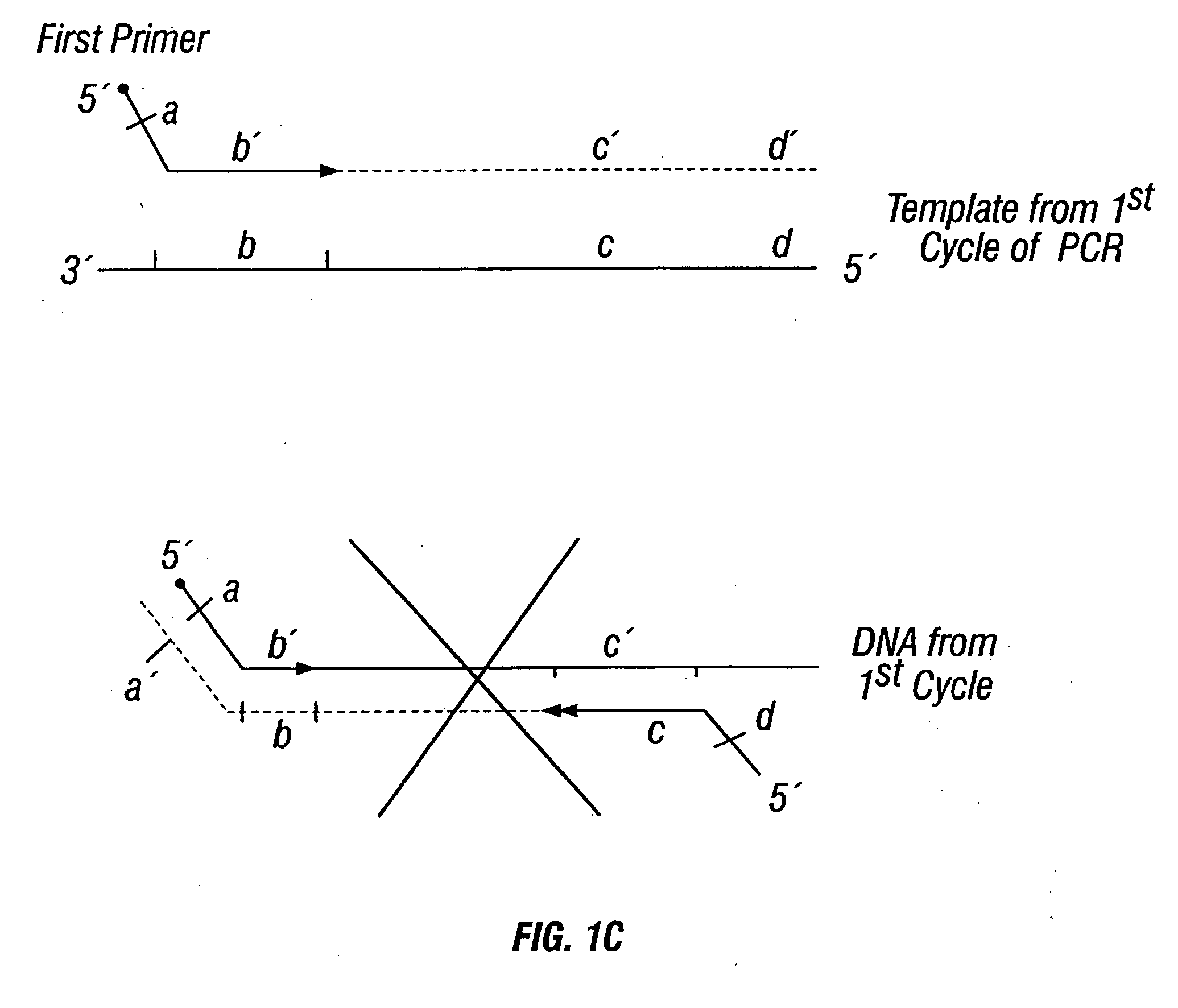
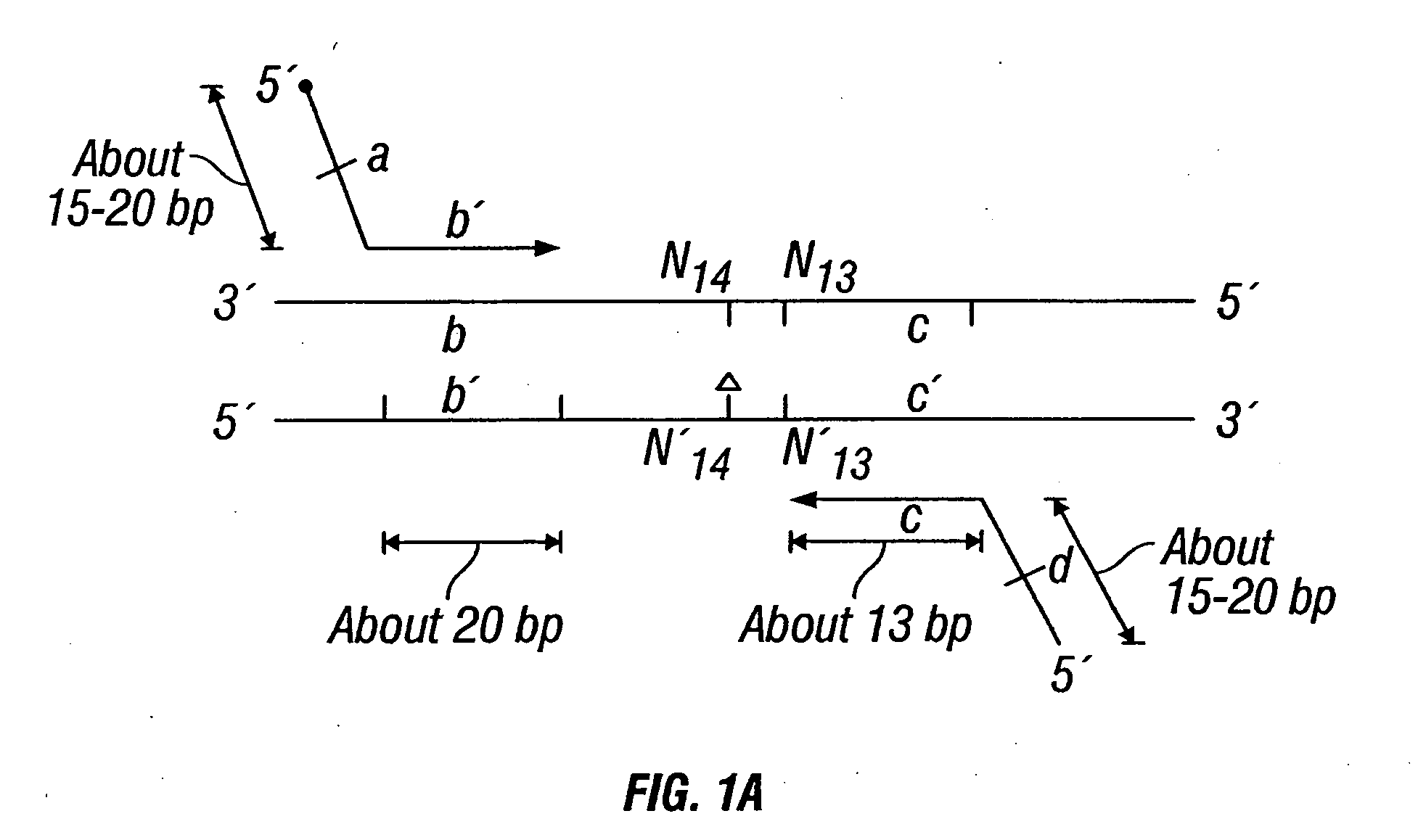
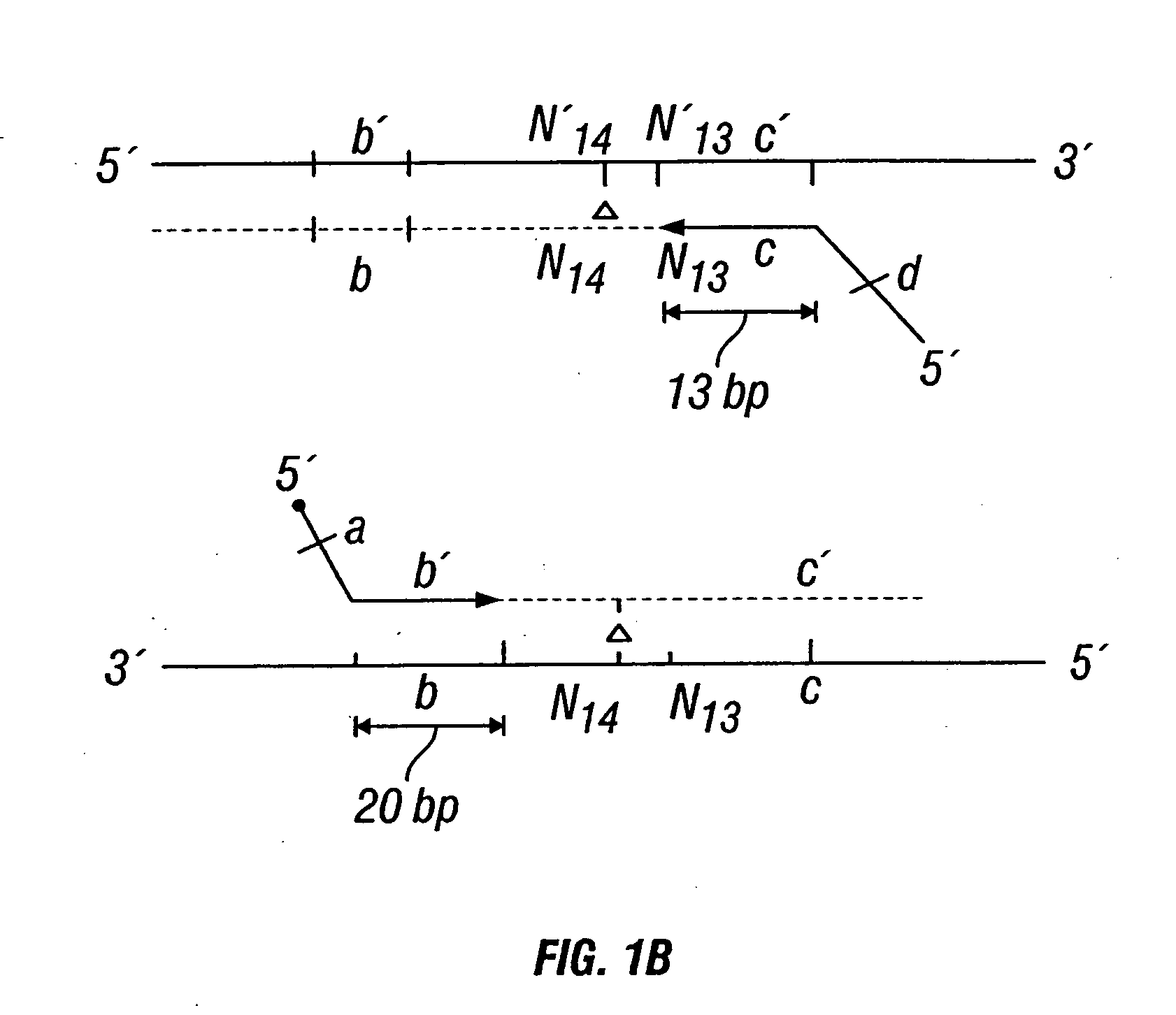
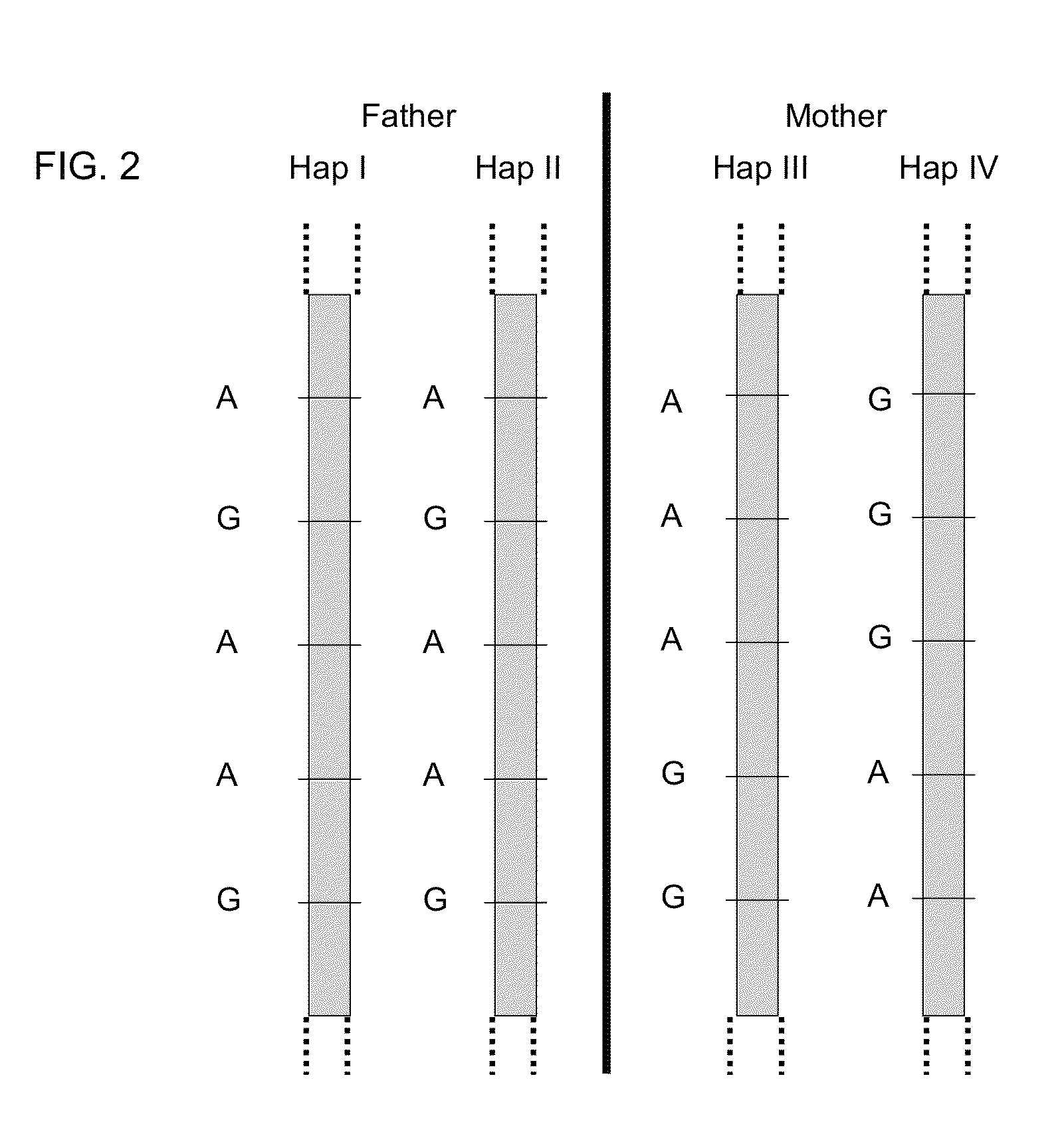
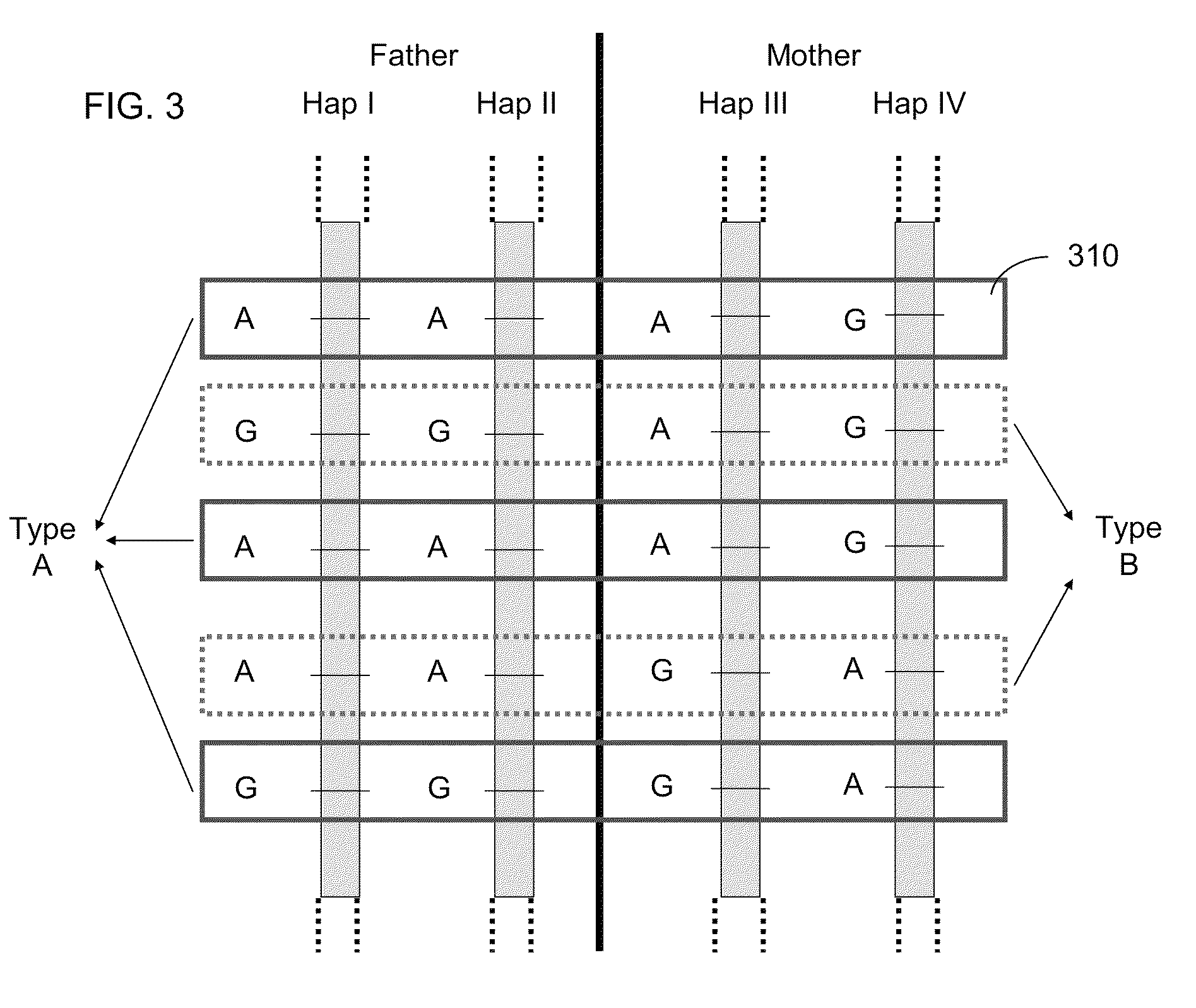
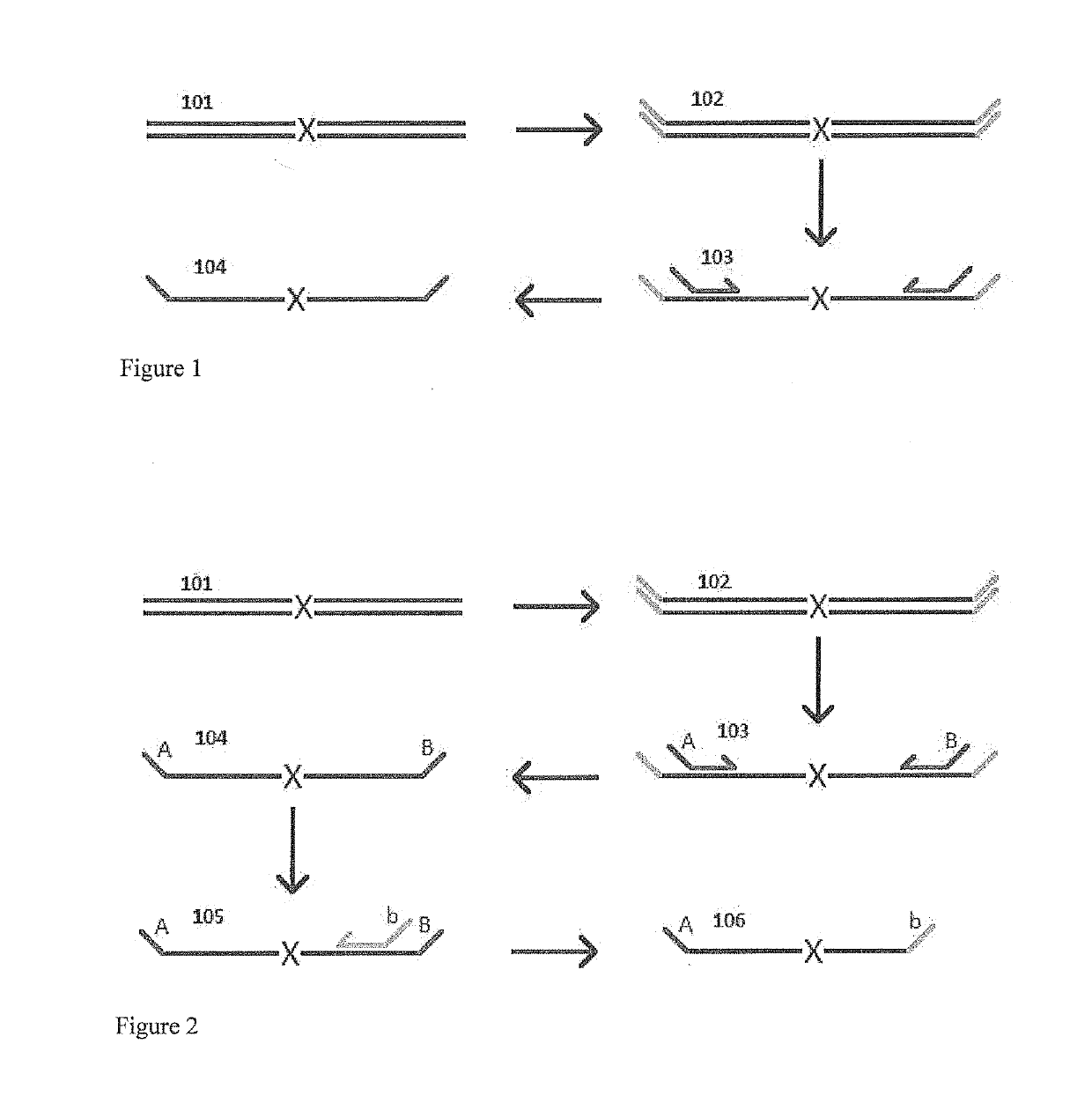
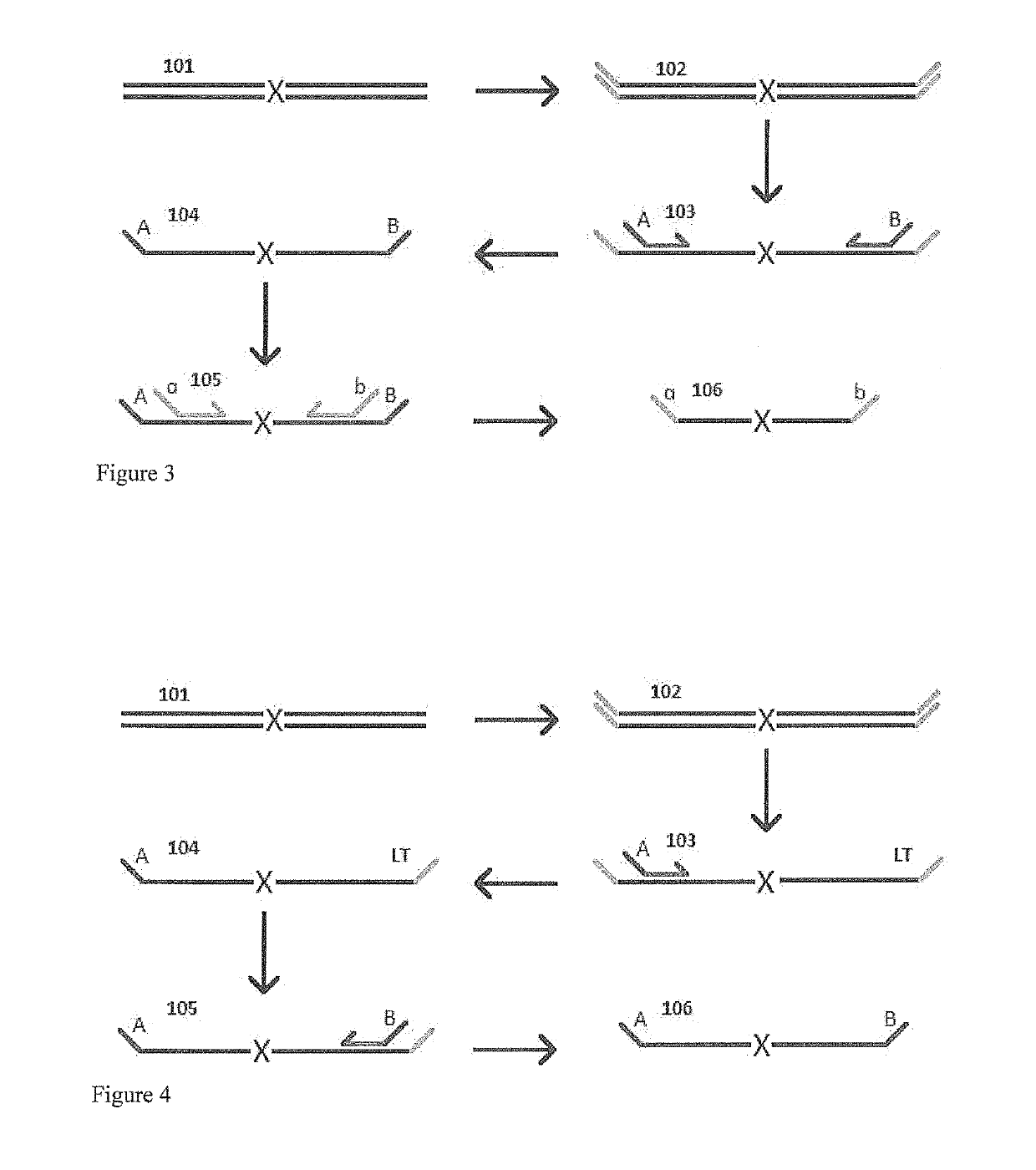
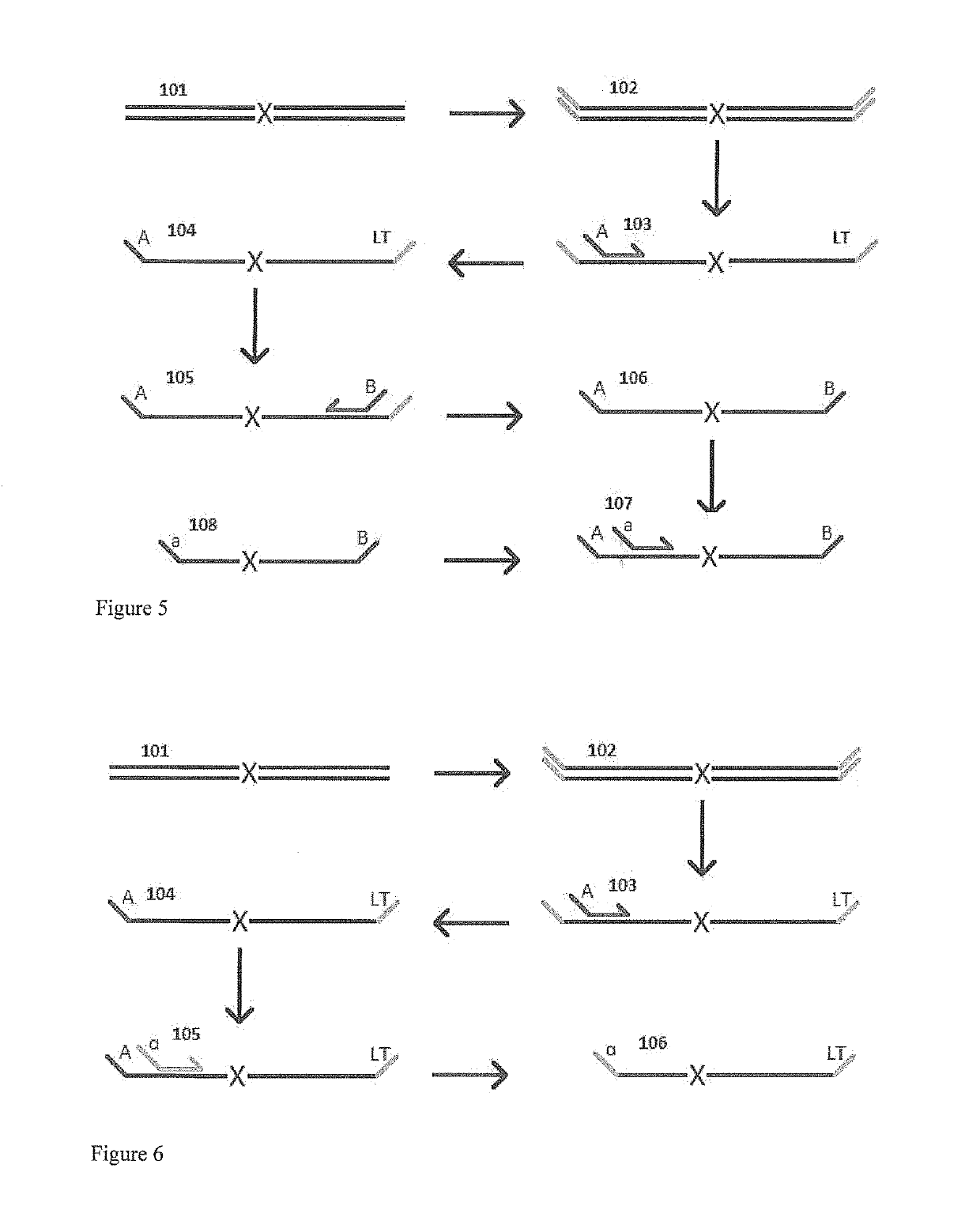
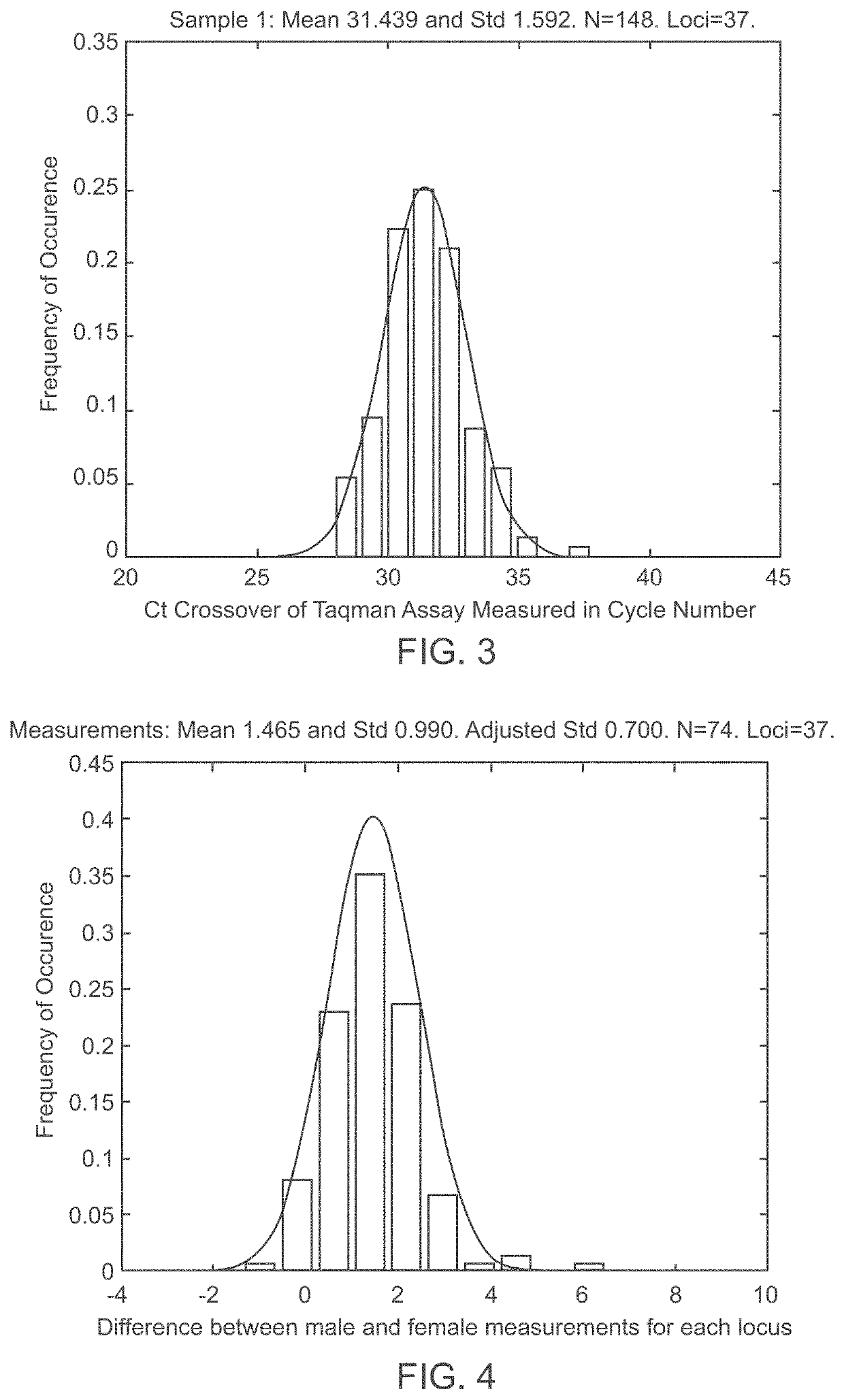
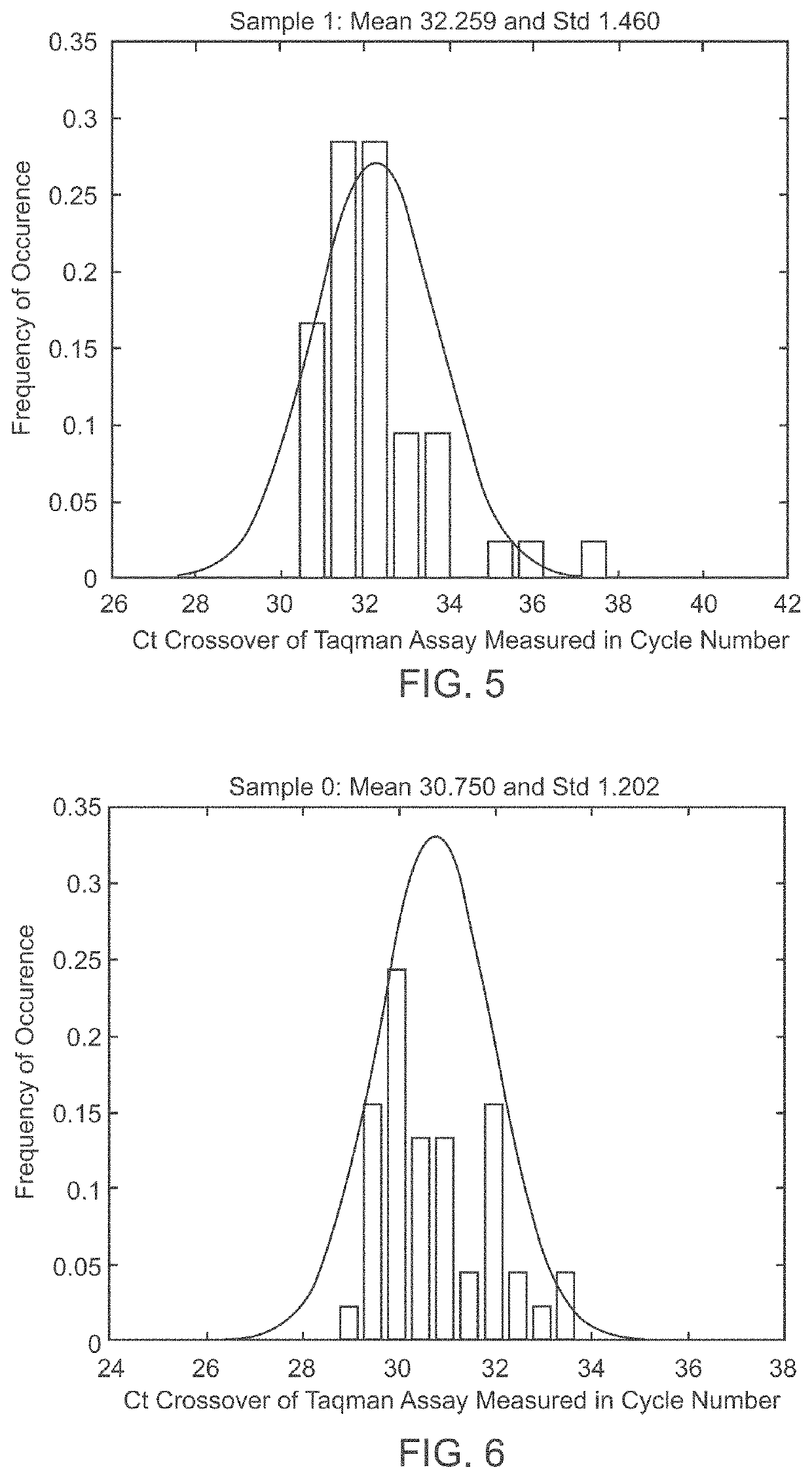
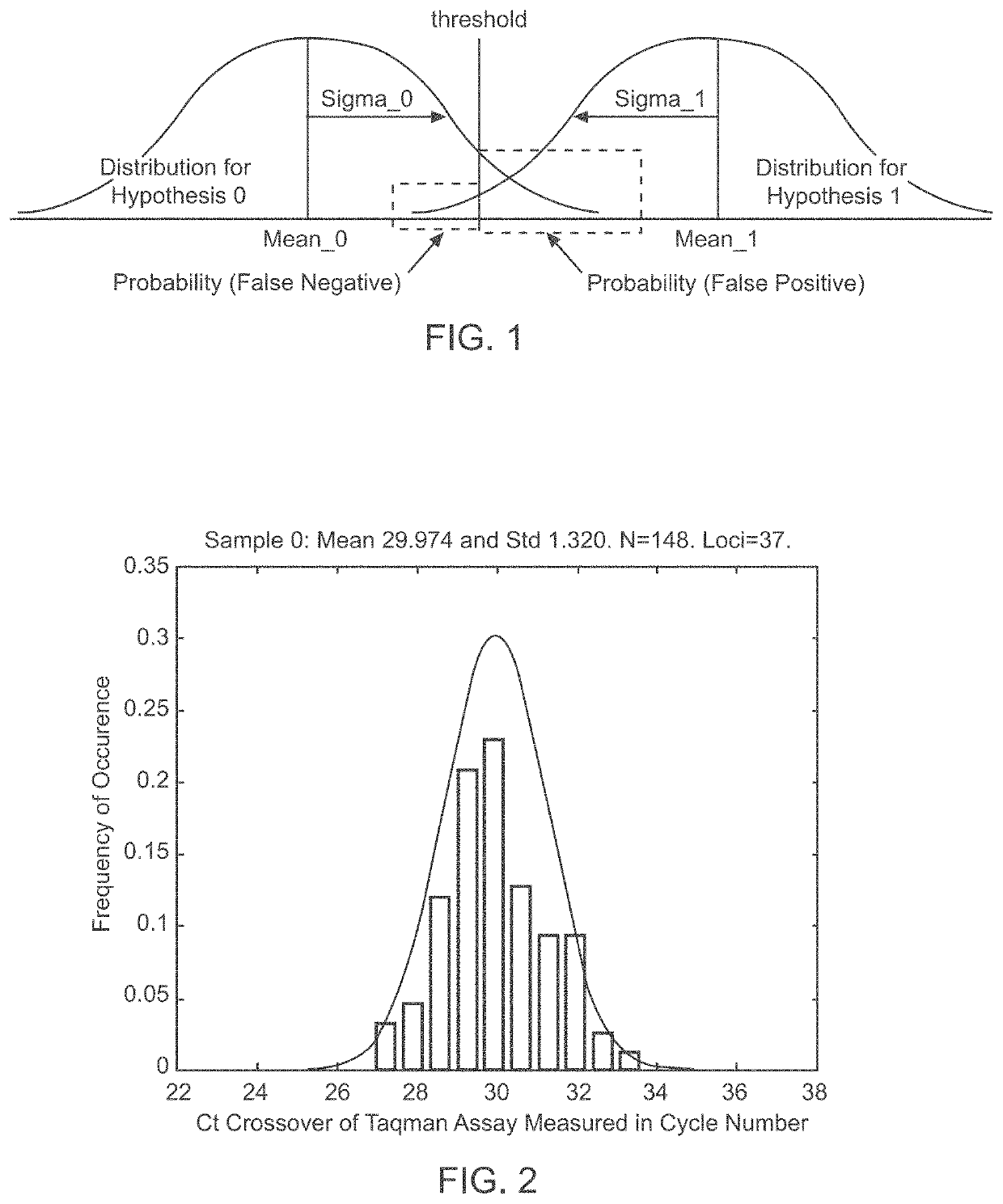
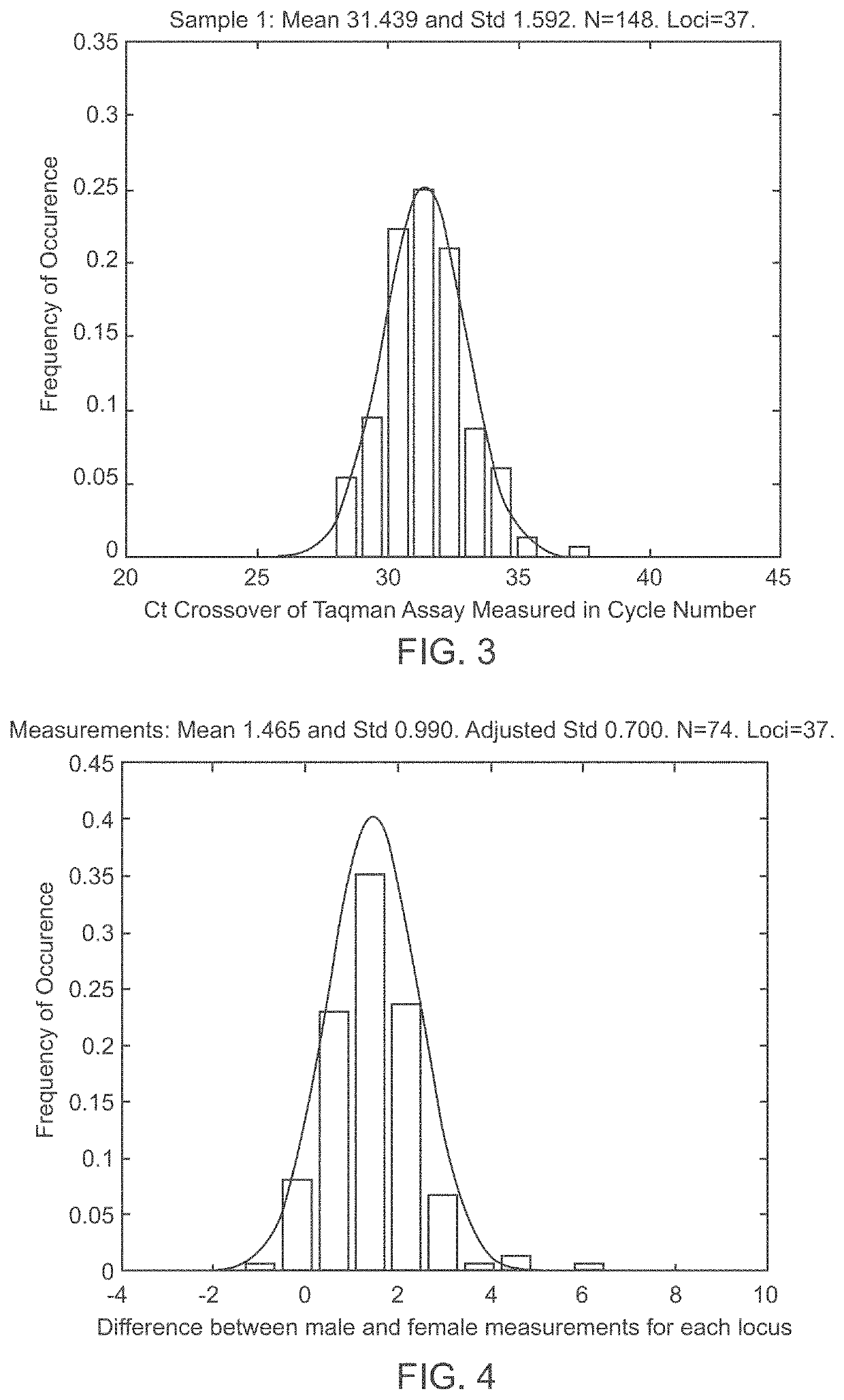
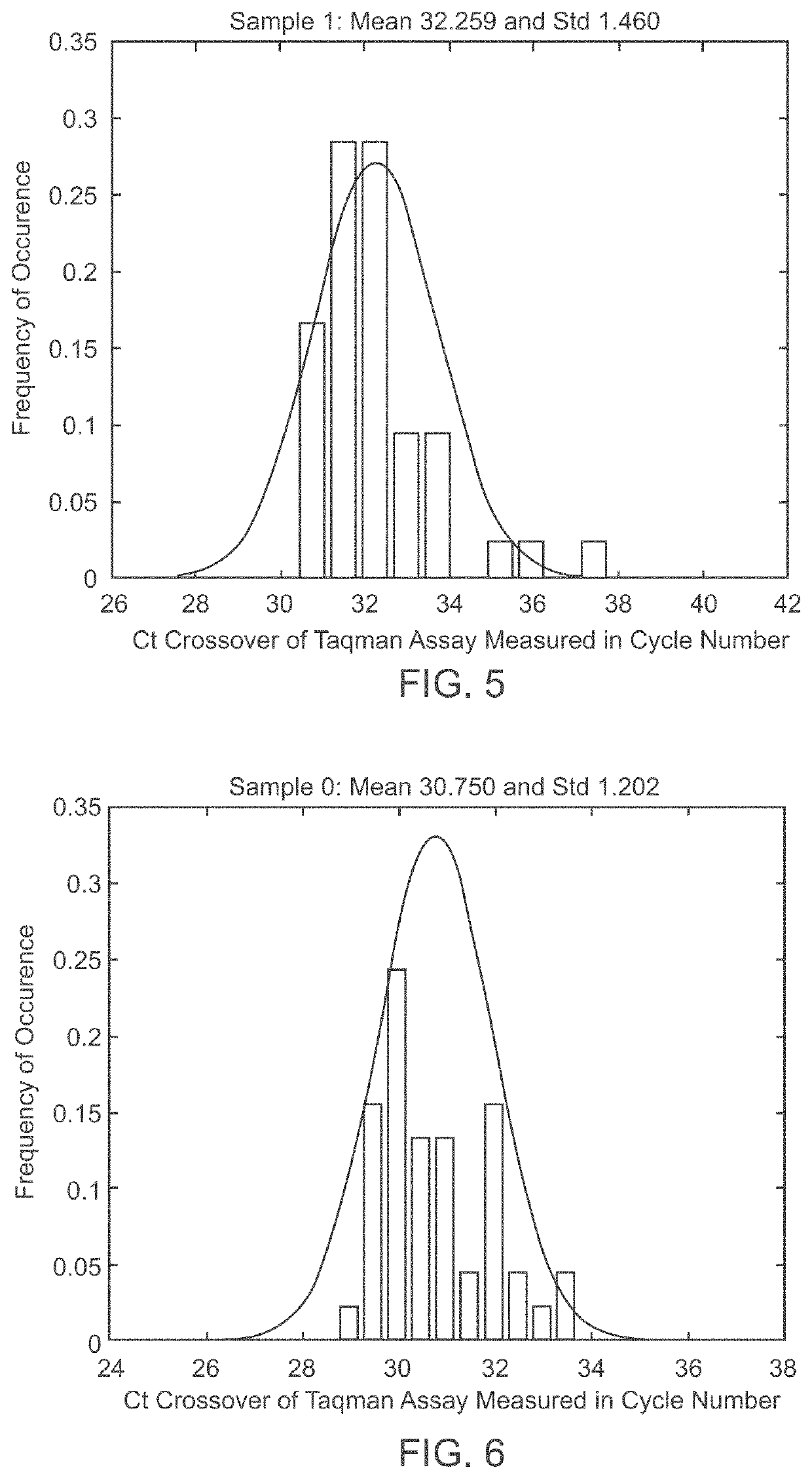
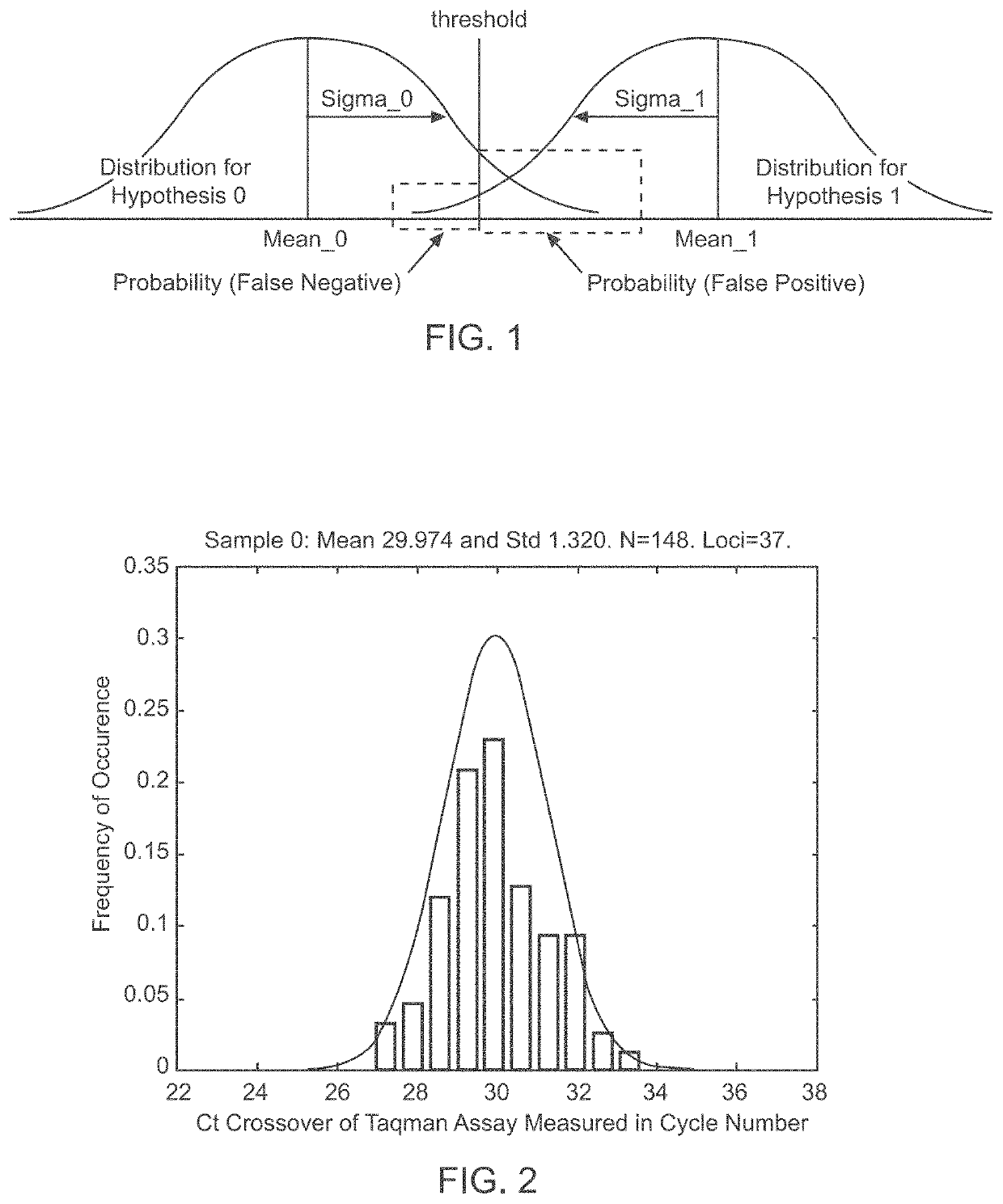
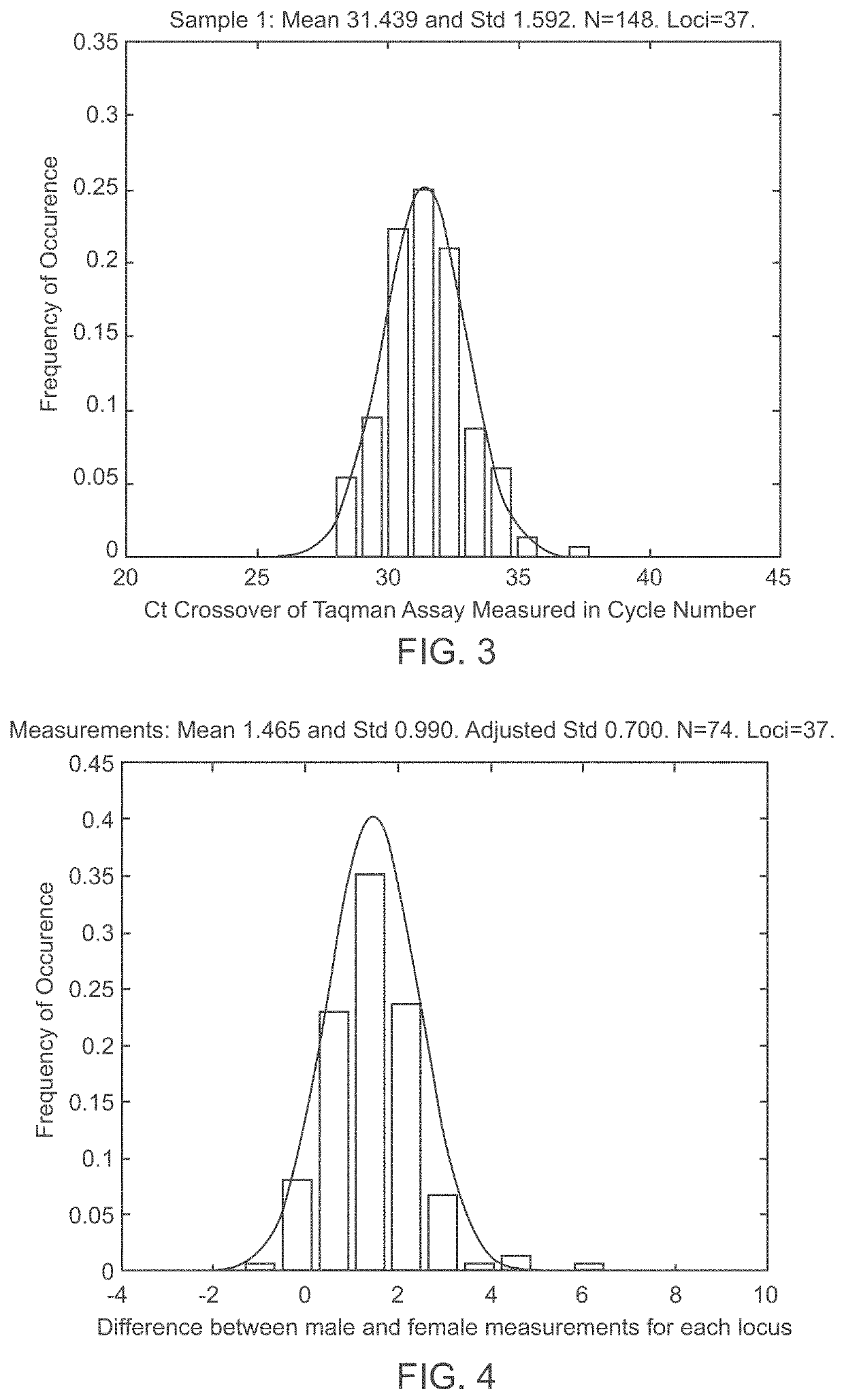
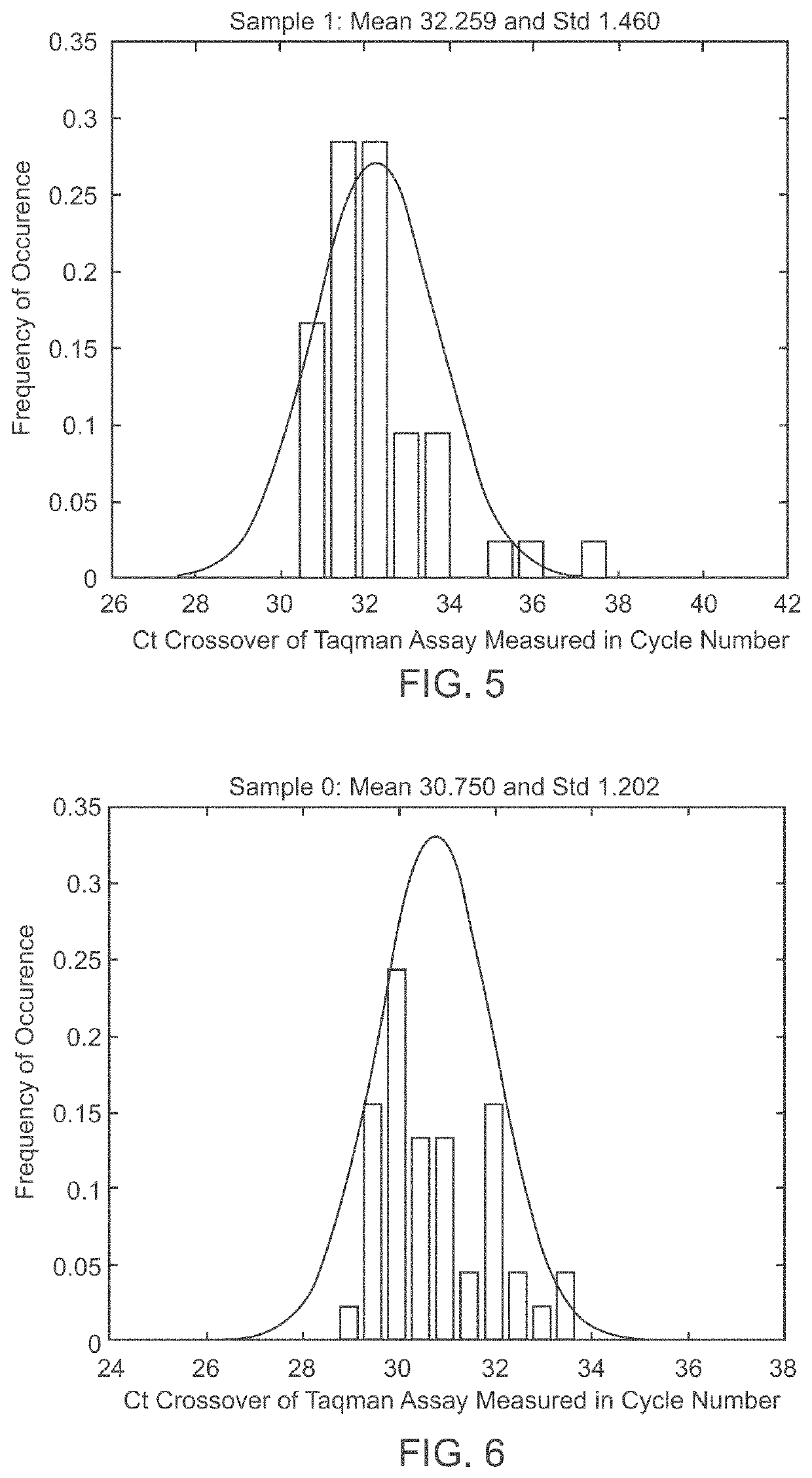
Patents
Literature
1635 results about "Allelic gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
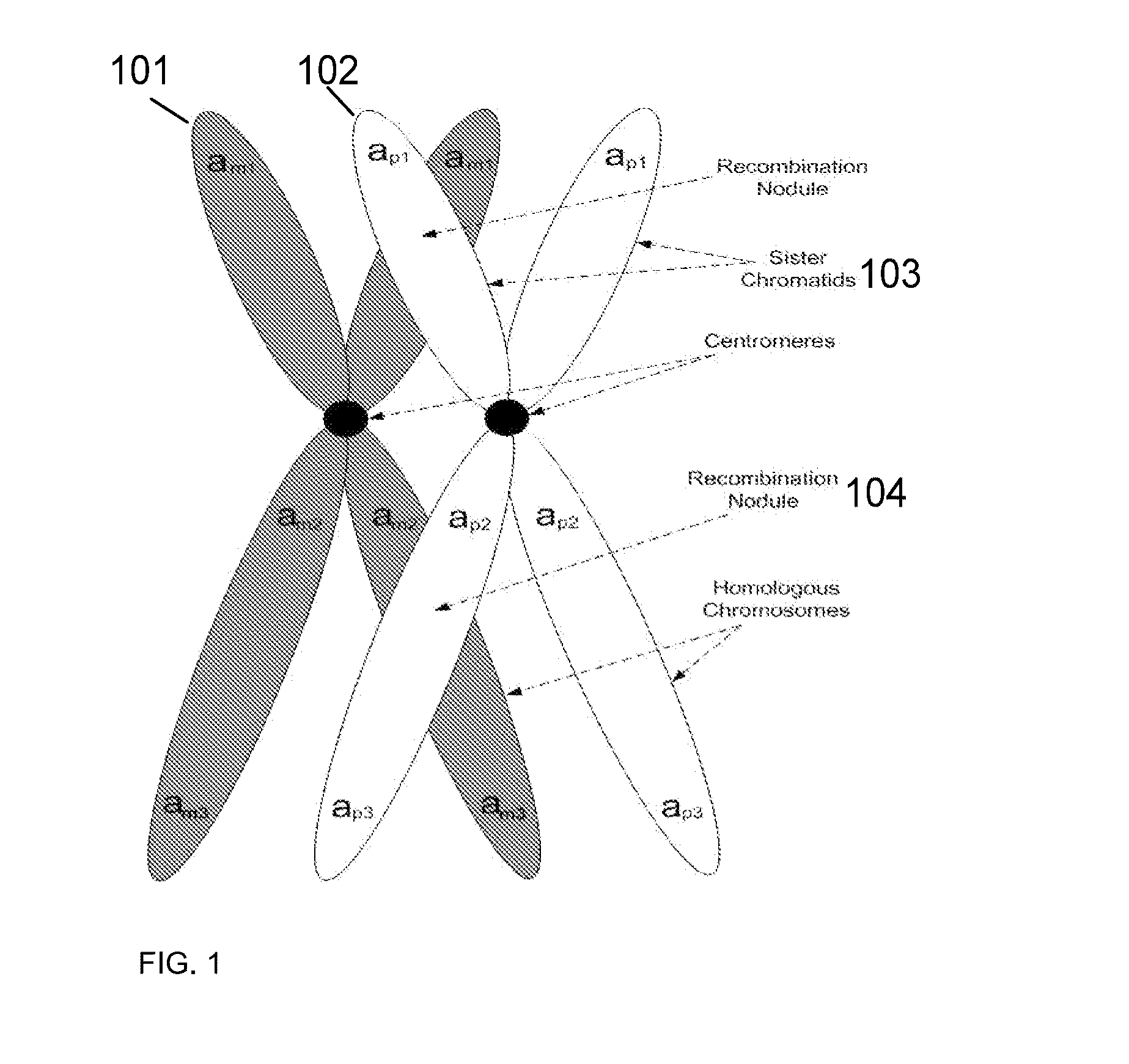
Such genes are allelic genes. Now if there is no gene aligned on similar locus for a particular character then it will be called non allelic gene. In other words one can say that the genes which do not show any major characters can be considered to be non allelic genes.
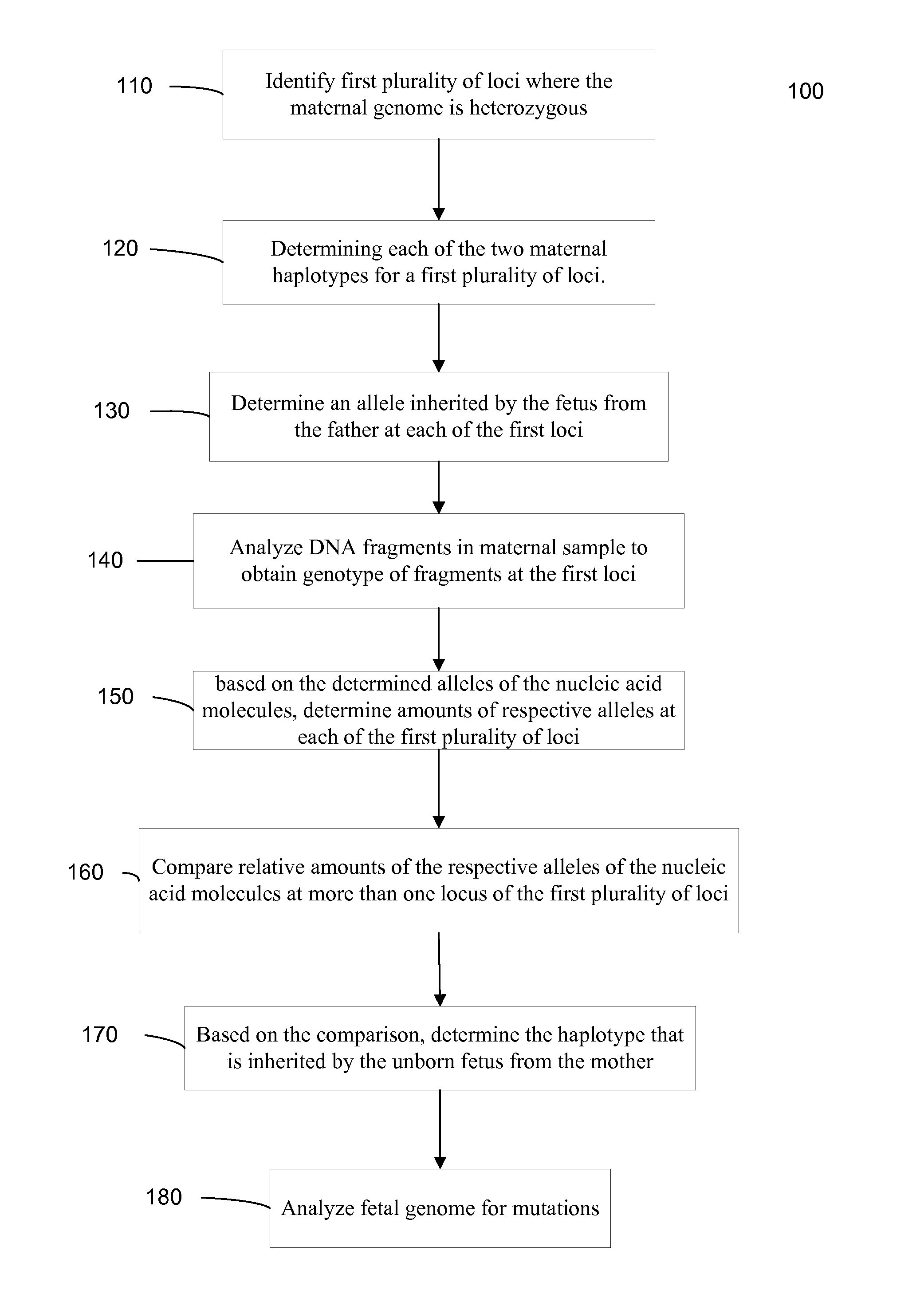
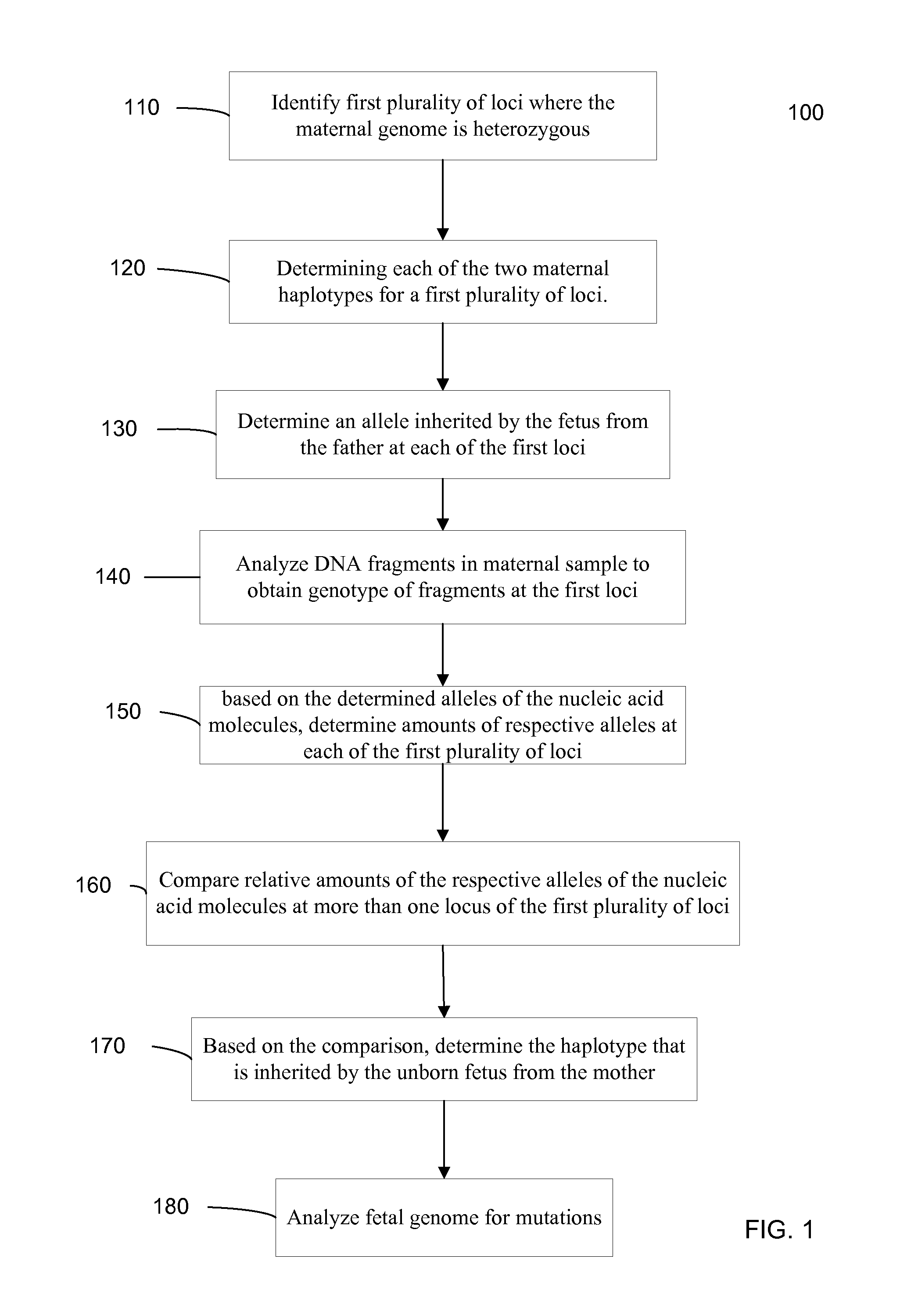
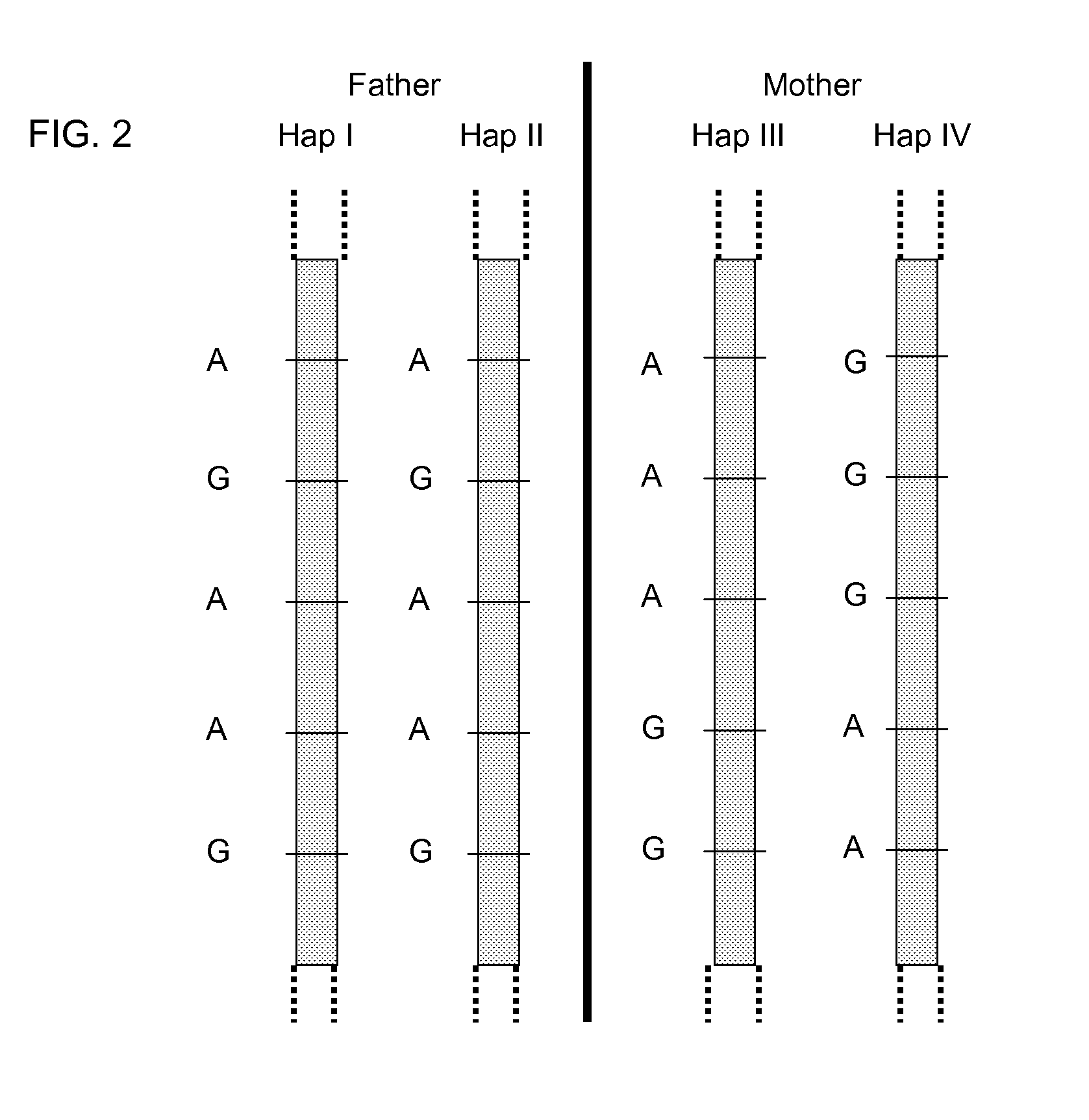
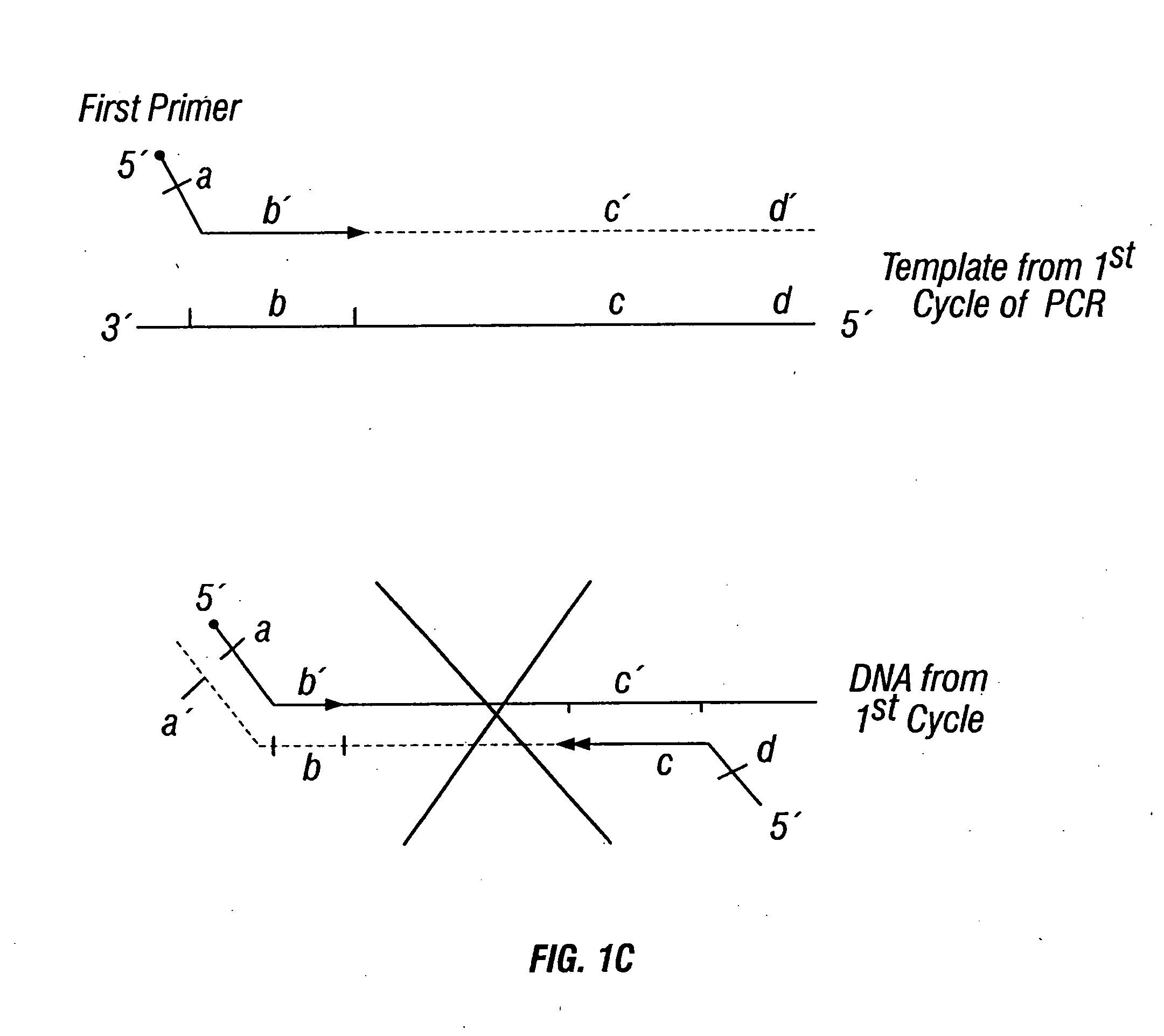
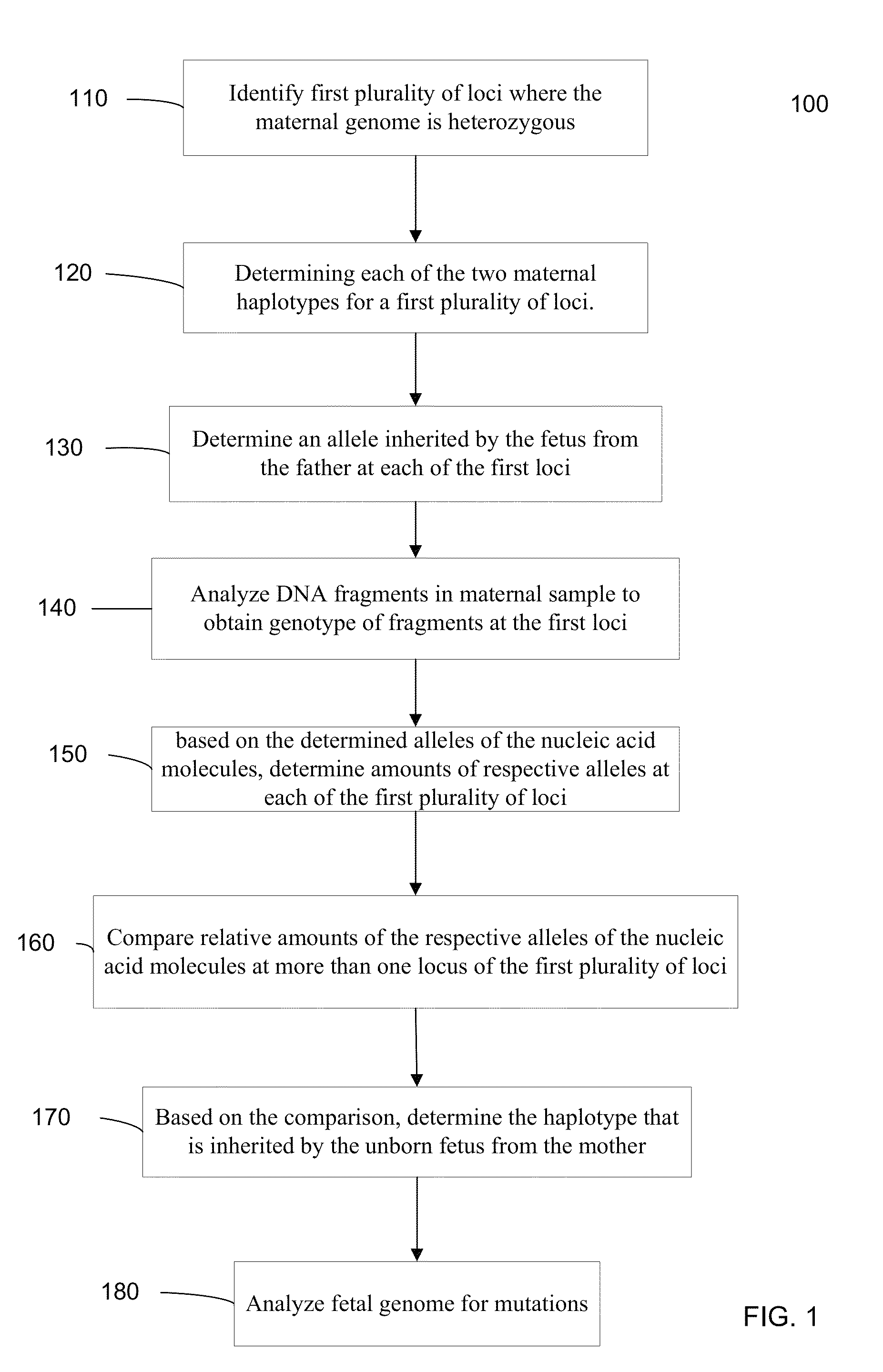
Fetal Genomic Analysis From A Maternal Biological Sample
ActiveUS20110105353A1Microbiological testing/measurementLibrary screeningDna concentrationPopulation
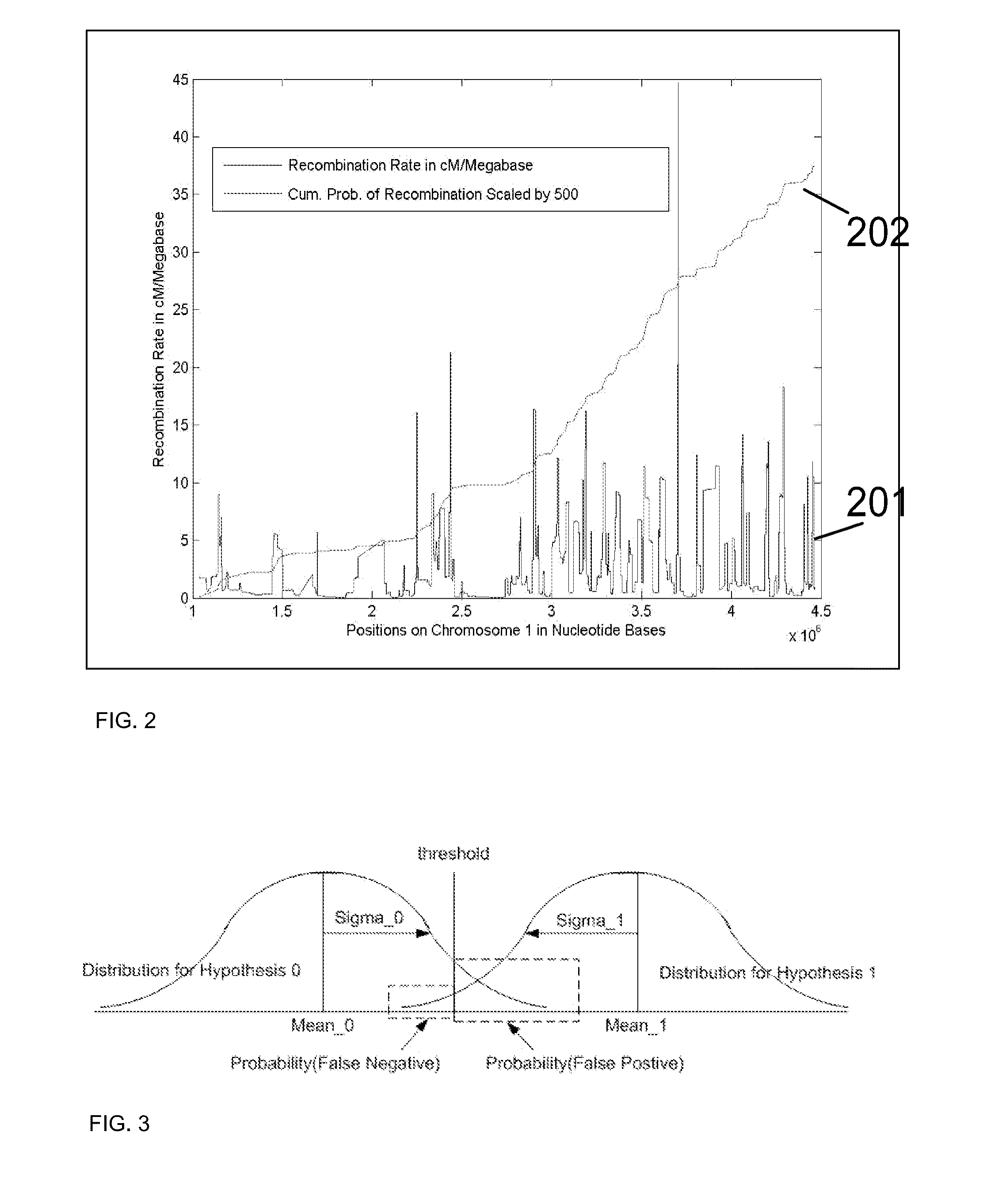
Systems, methods, and apparatus for determining at least a portion of fetal genome are provided. DNA fragments from a maternal sample (maternal and fetal DNA) can be analyzed to identify alleles at certain loci. The amounts of DNA fragments of the respective alleles at these loci can be analyzed together to determine relative amounts of the haplotypes for these loci and determine which haplotypes have been inherited from the parental genomes. Loci where the parents are a specific combination of homozygous and heterozygous can be analyzed to determine regions of the fetal genome. Reference haplotypes common in the population can be used along with the analysis of the DNA fragments of the maternal sample to determine the maternal and paternal genomes. Determination of mutations, a fractional fetal DNA concentration in a maternal sample, and a proportion of coverage of a sequencing of the maternal sample can also be provided.
Owner:SEQUENOM INC +1
Methods for detection of genetic disorders
InactiveUS20070178478A1Minimize disruptionMaximize efficiencyMicrobiological testing/measurementFermentationGeneticsAllele
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
Methods for detection of genetic disorders
ActiveUS20060121452A1Minimize disruptionMaximize efficiencySugar derivativesMicrobiological testing/measurementNon invasiveChromosomal Abnormality
The invention provides a method useful for detection of genetic disorders. The method comprises determining the sequence of alleles of a locus of interest, and quantitating a ratio for the alleles at the locus of interest, wherein the ratio indicates the presence or absence of a chromosomal abnormality. The present invention also provides a non-invasive method for the detection of chromosomal abnormalities in a fetus. The invention is especially useful as a non-invasive method for determining the sequence of fetal DNA. The invention further provides methods of isolation of free DNA from a sample.
Owner:RAVGEN INC
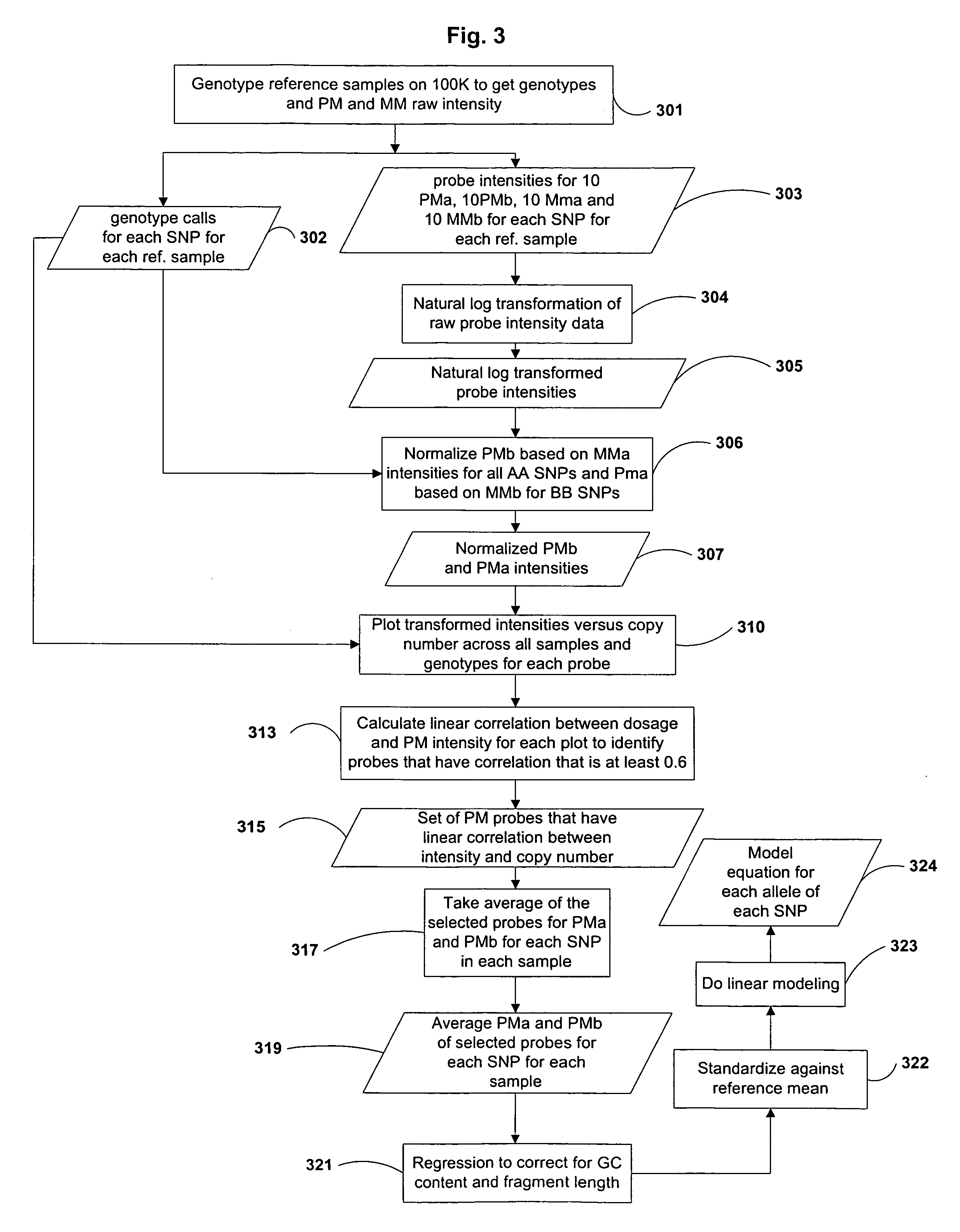
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS20080243398A1Significant resultImprove fidelityData processing applicationsMicrobiological testing/measurementGenetic MaterialsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic material from the target individual is acquired, amplified and the genetic data is measured using known methods. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment of the invention, the chromosome copy number can be determined from the measured genetic data of a single or small number of cells, with or without genetic information from one or both parents. In another embodiment of the invention, these determinations are made for the purpose of embryo selection in the context of in-vitro fertilization. In another embodiment of the invention, the genetic data can be reconstructed for the purposes of making phenotypic predictions.
Owner:NATERA
Fetal genomic analysis from a maternal biological sample
Systems, methods, and apparatus for determining at least a portion of fetal genome are provided. DNA fragments from a maternal sample (maternal and fetal DNA) can be analyzed to identify alleles at certain loci. The amounts of DNA fragments of the respective alleles at these loci can be analyzed together to determine relative amounts of the haplotypes for these loci and determine which haplotypes have been inherited from the parental genomes. Loci where the parents are a specific combination of homozygous and heterozygous can be analyzed to determine regions of the fetal genome. Reference haplotypes common in the population can be used along with the analysis of the DNA fragments of the maternal sample to determine the maternal and paternal genomes. Determination of mutations, a fractional fetal DNA concentration in a maternal sample, and a proportion of coverage of a sequencing of the maternal sample can also be provided.
Owner:SEQUENOM INC +1
Plant breeding method
InactiveUS20050144664A1Other foreign material introduction processesAgricultureNumeral systemPlant variety
Methods for using genetic marker genotype (e.g., gene sequence diversity information) to improve the process of developing plant varieties (e.g., single cross hybrids) with improved phenotypic performance are provided. Methods for predicting the value of a phenotypic trait in a plant are provided. The methods use genotypic, phenotypic, and optionally family relationship information for a first plant population to identify an association between at least one genetic marker and the phenotypic trait, and then use the association to predict the value of the phenotypic trait in one or more members of a second, target population of known marker genotype. Methods for identifying new allelic variants affecting the trait are also provided. Plants selected, provided, or produced by any of the methods herein, transgenic plants created by any of the methods herein, and digital systems for performing the methods herein are also provided.
Owner:EI DU PONT DE NEMOURS & CO +1
Probe-based analysis of heterozygous mutations using two-color labelling
InactiveUS6342355B1Bioreactor/fermenter combinationsBiological substance pretreatmentsVariant alleleGenomic clone
The invention provides methods of analyzing a nucleic acid in a target sample for variant alleles. In such methods, a first-labelled control sample and a second-labelled target sample are hybridized to at least one set of probes. The control sample comprises a homozygous reference allele. The target sample comprises the homozygous reference allele, or variant alleles differing from the reference allele at a locus, or one variant allele differing from the reference allele at the locus and one reference allele. The probes in the probe set span the locus and are complementary to the reference allele. After hybridization the intensity of first and second label bound to each probe in the set is measured. This information is then used to indicate the presence of one variant allele and one reference allele, or the presence of two variant alleles in the target sample.
Owner:UNITED STATES OF AMERICA +1
Methods for genetic analysis
Several methods are described for assessing an individual's likelihood of developing or exhibiting a multifactorial trait. The methods include determining a plurality of genotypes for the individual at a plurality of biallelic polymorphic loci, using the genotypes to compute a score for the individual, and comparing the score to at least one threshold value.
Owner:GENETIC TECHNOLOGIES LIMTIED
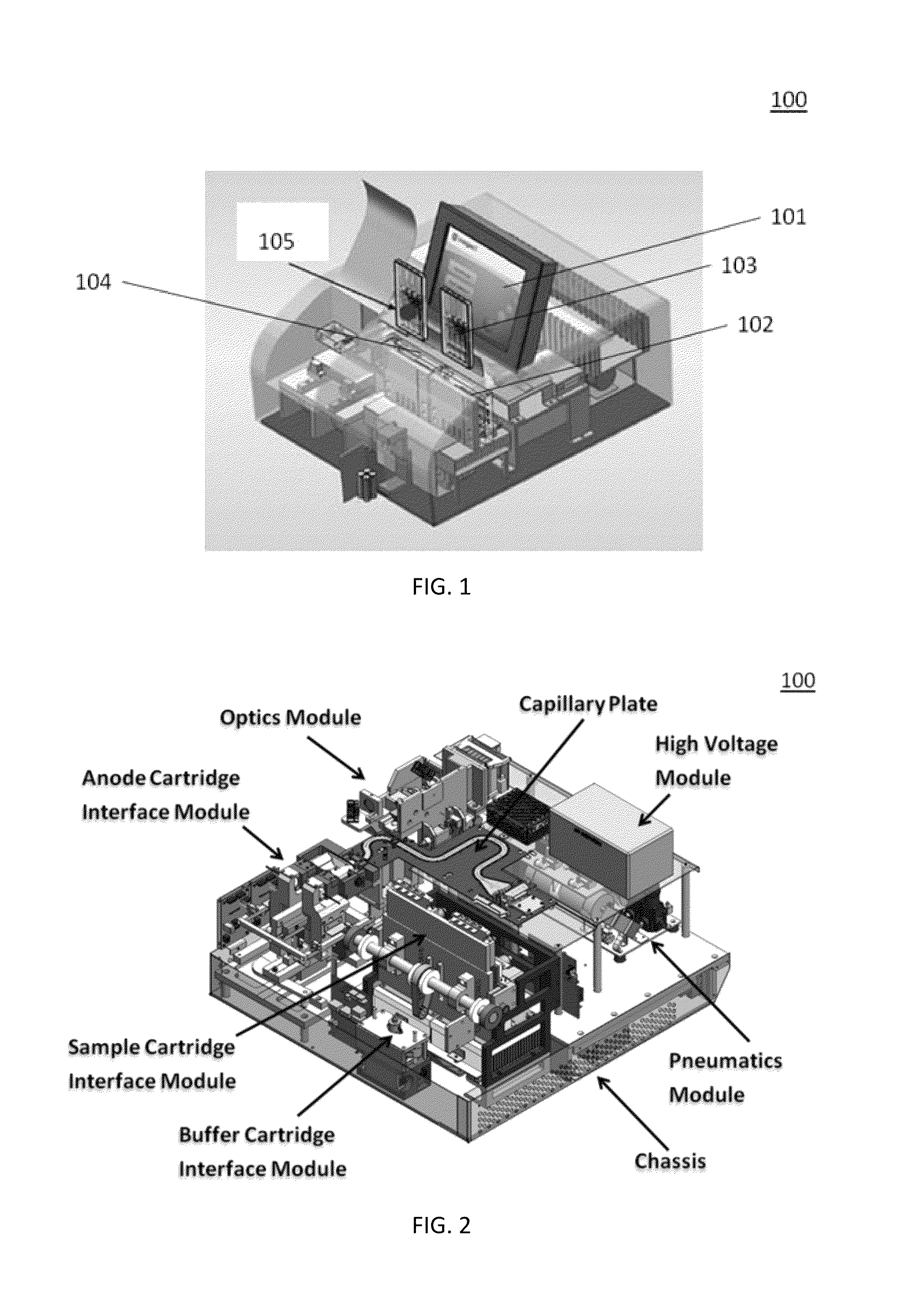
Sample Preparation, Processing and Analysis Systems
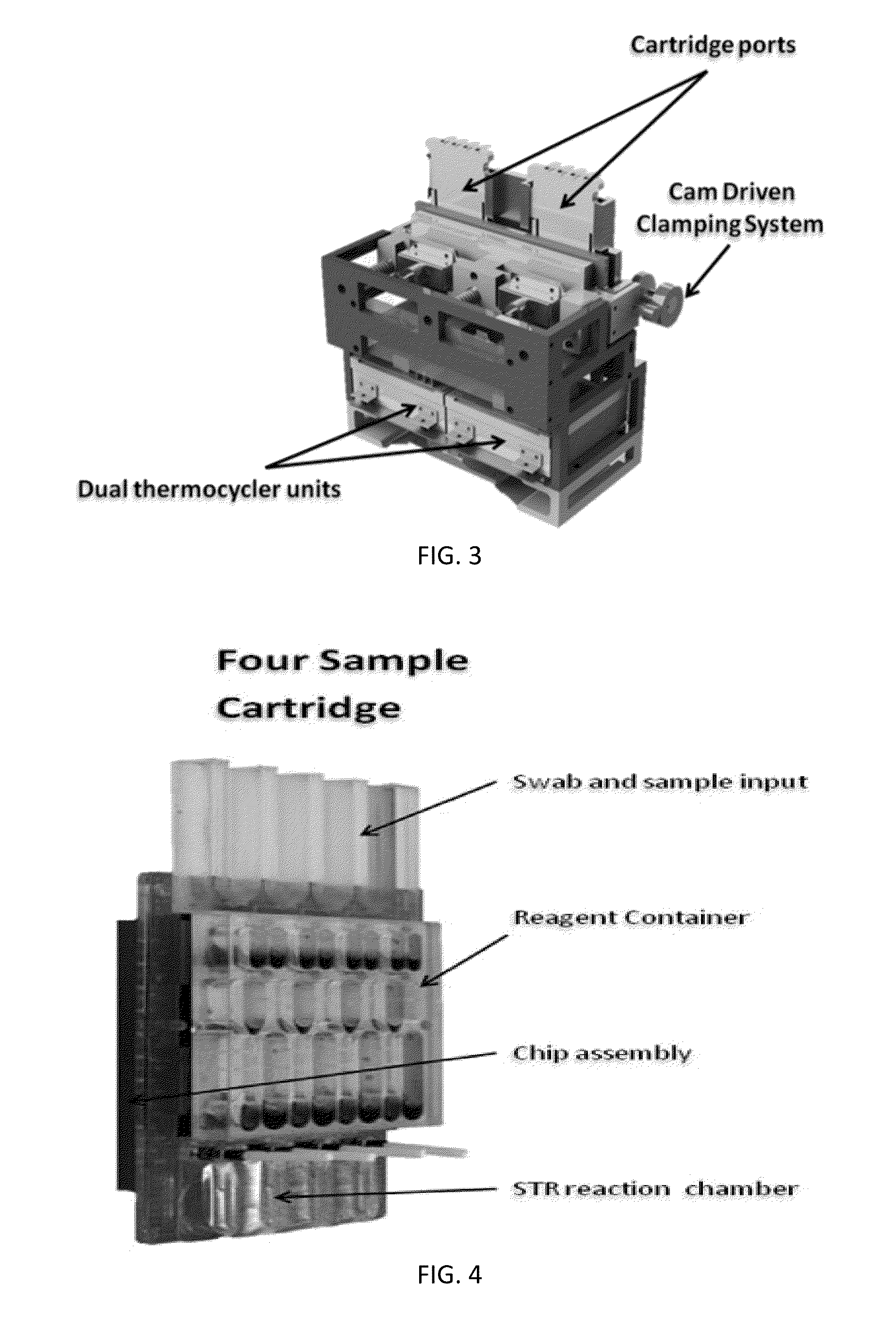
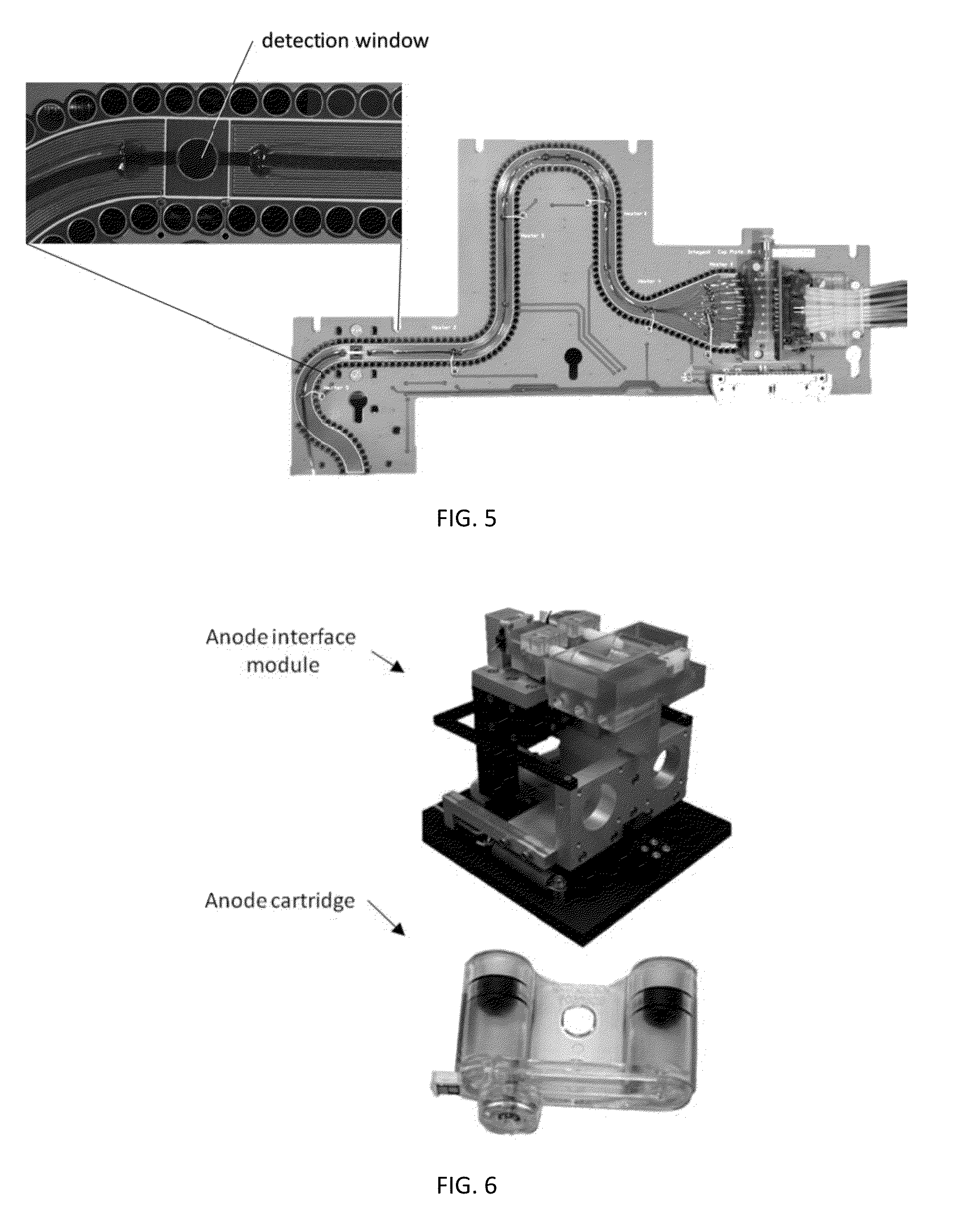
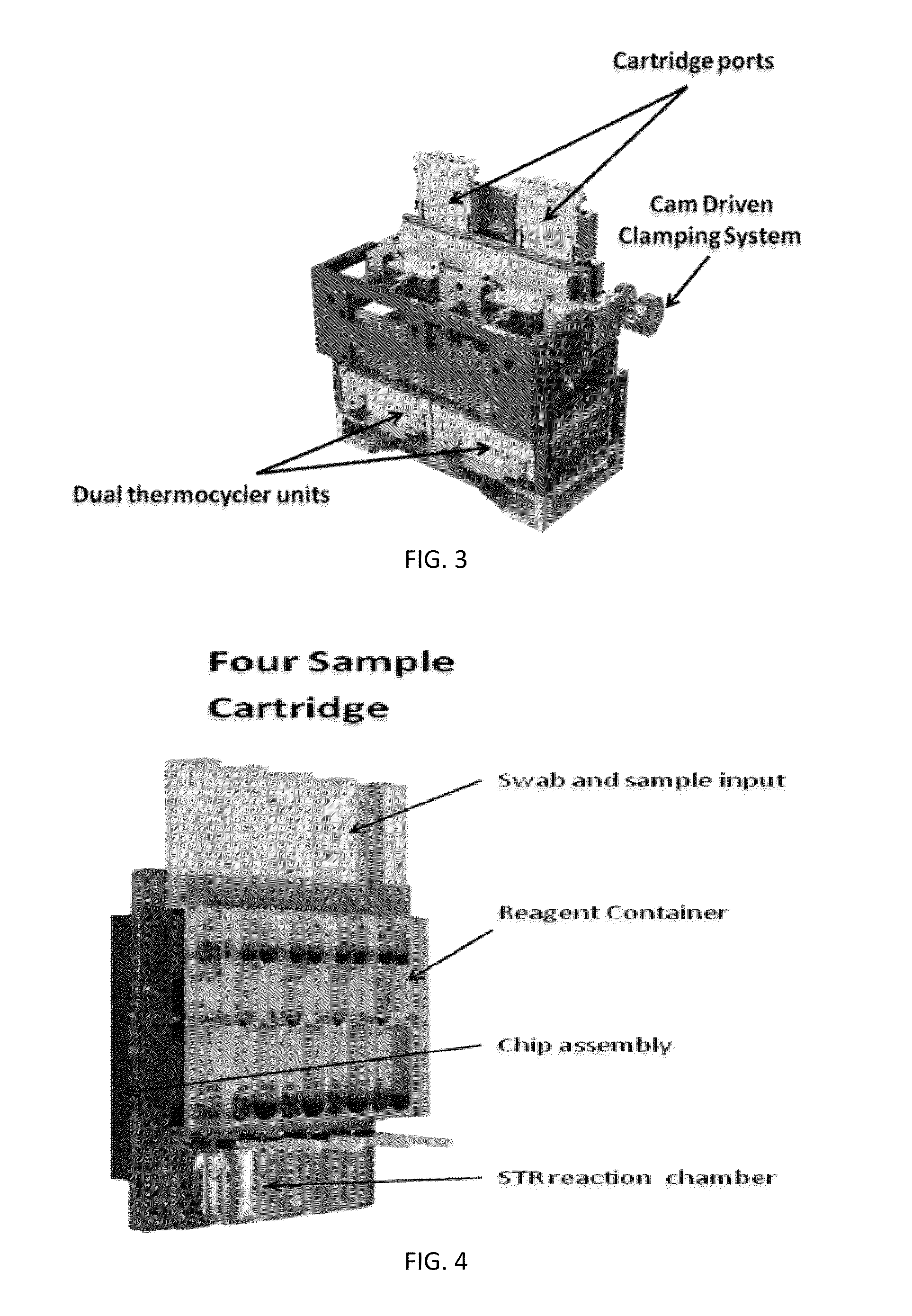
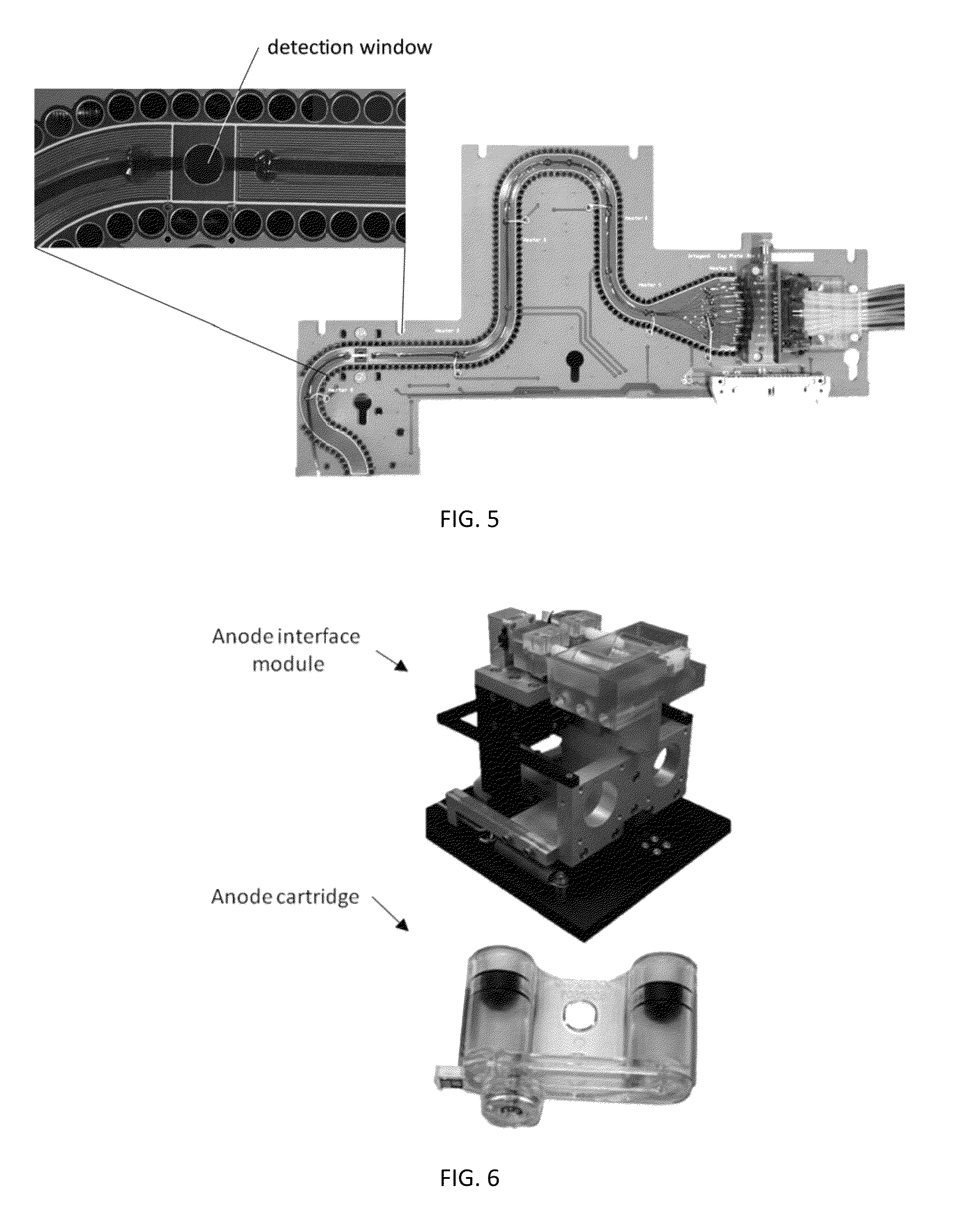
ActiveUS20130115607A1Small footprintFast timeBioreactor/fermenter combinationsBiological substance pretreatmentsGenetic profileBioinformatics
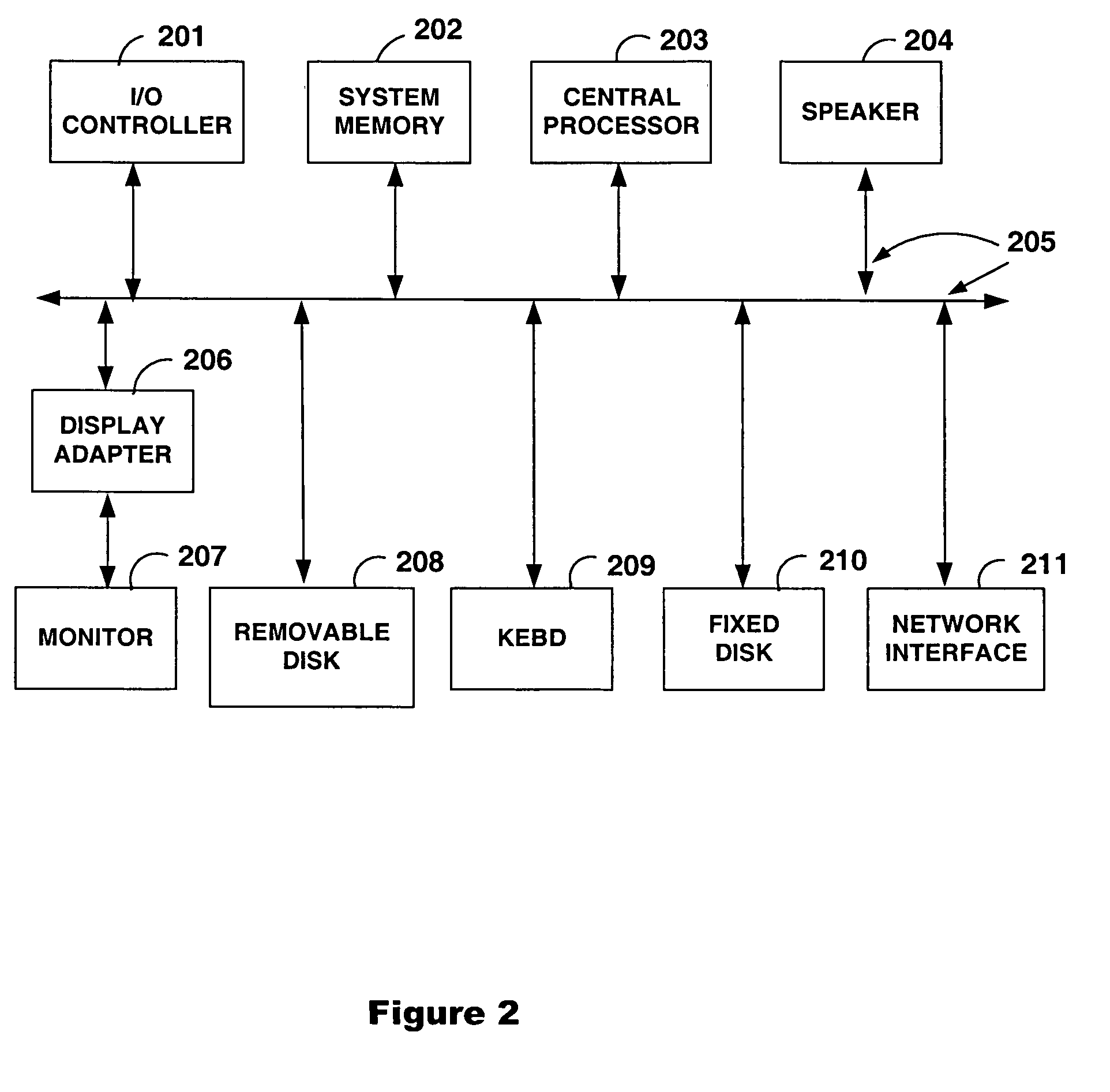
This disclosure provides an integrated and automated sample-to-answer system that, starting from a sample comprising biological material, generates a genetic profile in less than two hours. In certain embodiments, the biological material is DNA and the genetic profile involves determining alleles at one or a plurality of loci (e.g., genetic loci) of a subject, for example, an STR (short tandem repeat) profile, for example as used in the CODIS system. The system can perform several operations, including (a) extraction and isolation of nucleic acid; (b) amplification of nucleotide sequences at selected loci (e.g., genetic loci); and (c) detection and analysis of amplification product. These operations can be carried out in a system that comprises several integrated modules, including an analyte preparation module; a detection and analysis module and a control module.
Owner:INTEGENX
System, method and software arrangement for bi-allele haplotype phasing
ActiveUS20050255508A1Determine an individual's haplotypeMicrobiological testing/measurementProteomicsHaplotypeSystems approaches
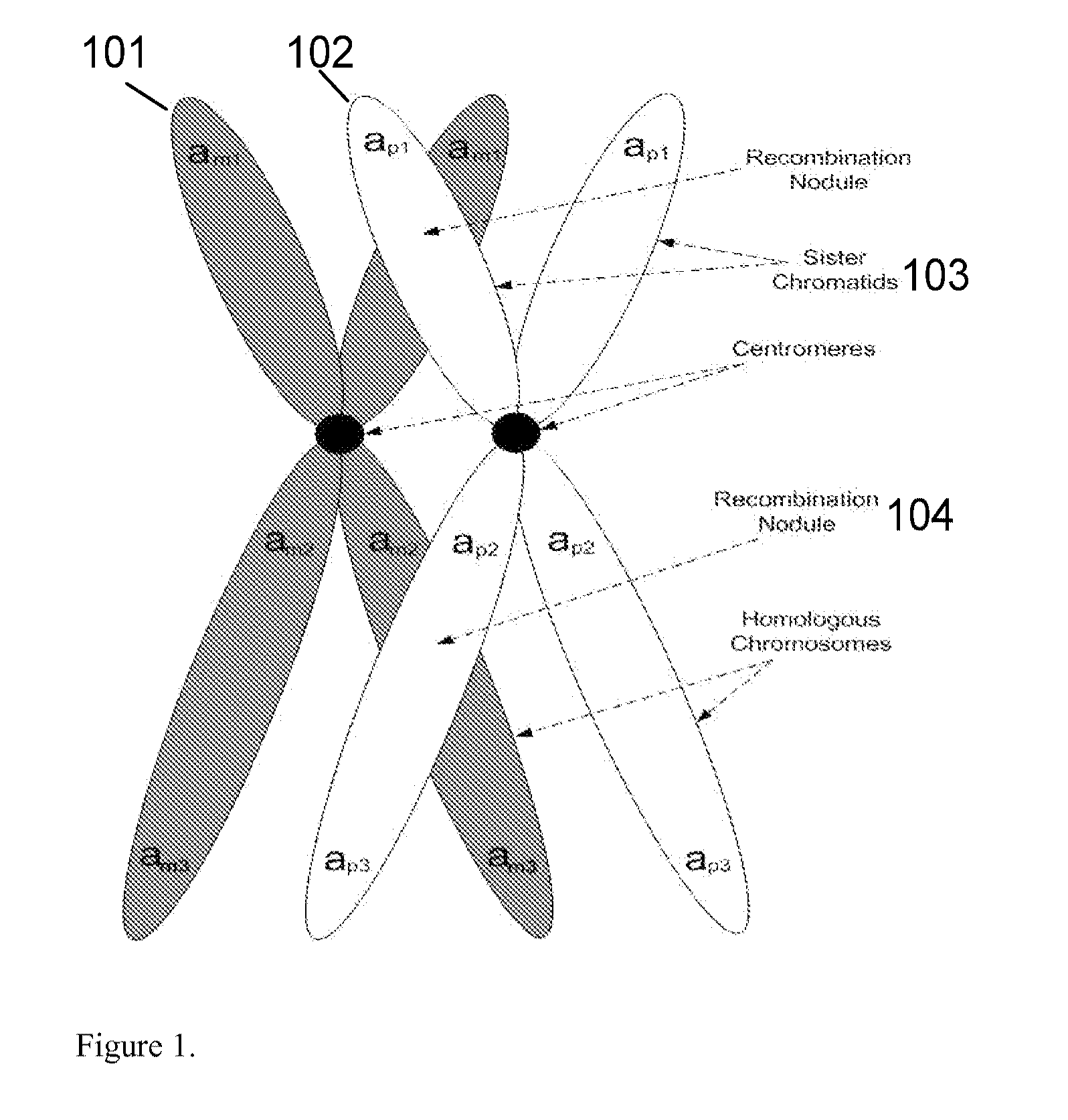
The present invention relates to a method, system and software arrangement for determining the co-associations of allele types across consecutive loci and hence for reconstructing two haplotypes of a diploid individual from genotype data generated by mapping experiments with single molecules, families or populations. The haplotype reconstruction system, method and software arrangement of the present invention can utilize a procedure that is nearly linear in the number of polymorphic markers examined, and is therefore quicker, more accurate, and more efficient than other population-based approaches. The system, method, and software arrangement of the present invention may be useful to assist with the diagnosis and treatment of any disease, which has a genetic component.
Owner:NEW YORK UNIV +1
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS20140032128A1Significant resultImprove fidelityMicrobiological testing/measurementBiostatisticsDiploid cellsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
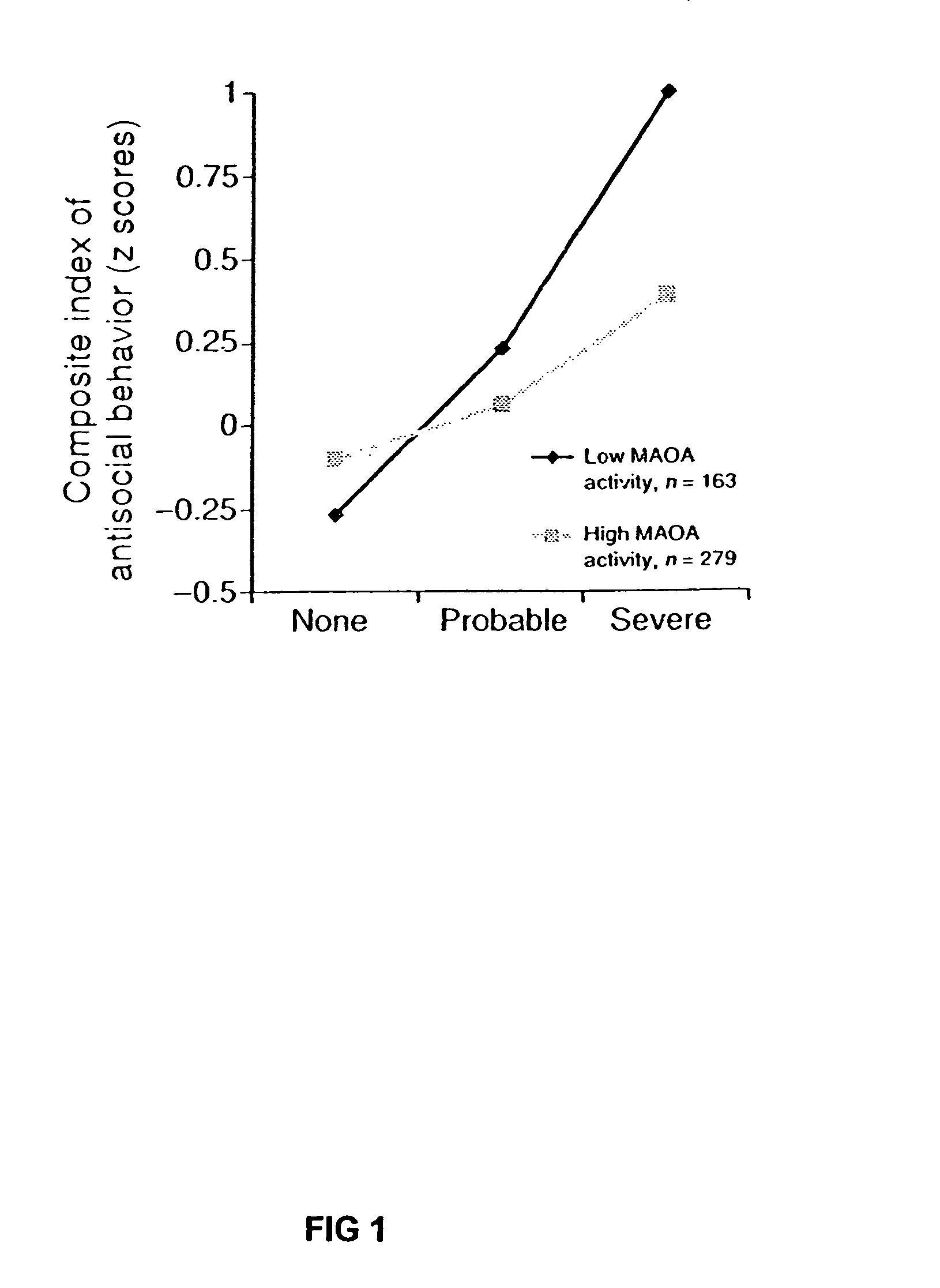
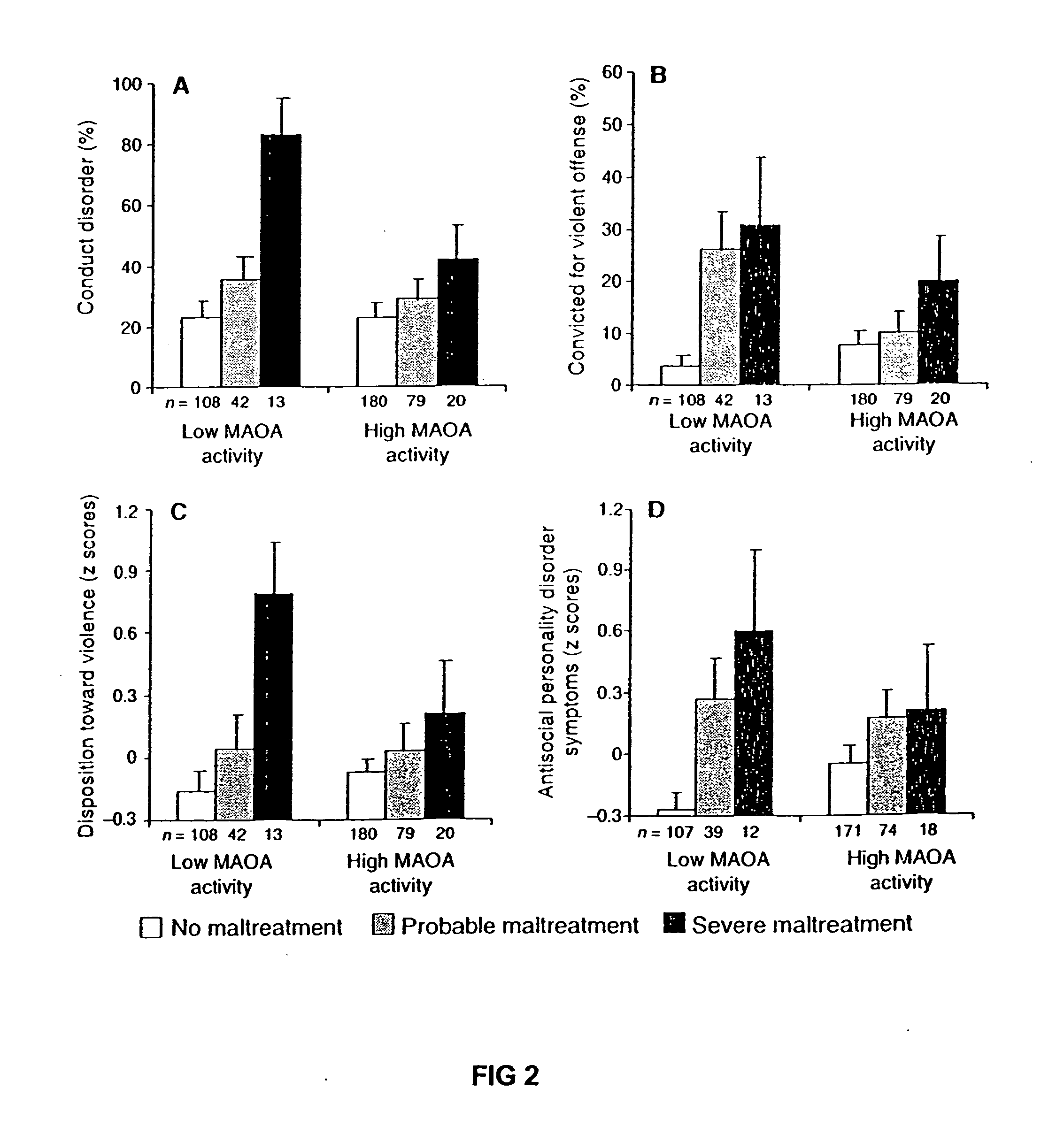
Method for assessing behavioral predisposition
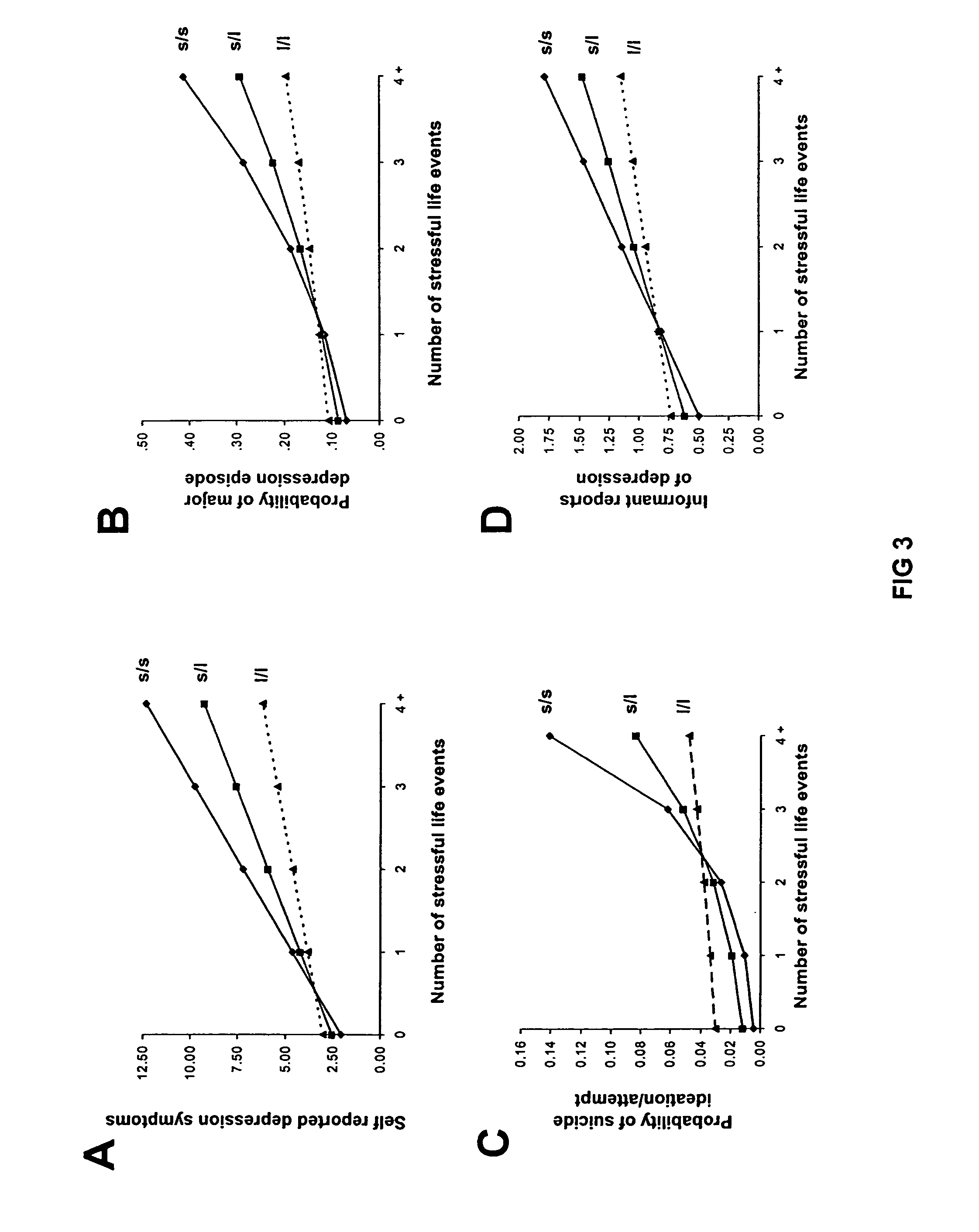
The present invention relates to diagnostic methods for assessing predisposition of a subject to a mental disorder phenotype having an association with an at-risk allele of a brain-functional gene having a plurality of alleles, the association being conditioned by a pathogenic environmental risk factor status condition. Additionally, the invention relates to methods for discovering a conditional association between a mental disorder phenotype and an at-risk allele of a brain-functional gene having a plurality of alleles, the association being conditioned by a pathogenic environmental risk factor status condition.
Owner:WISCONSIN ALUMNI RES FOUND
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
Methods for determining genetic haplotypes and DNA mapping
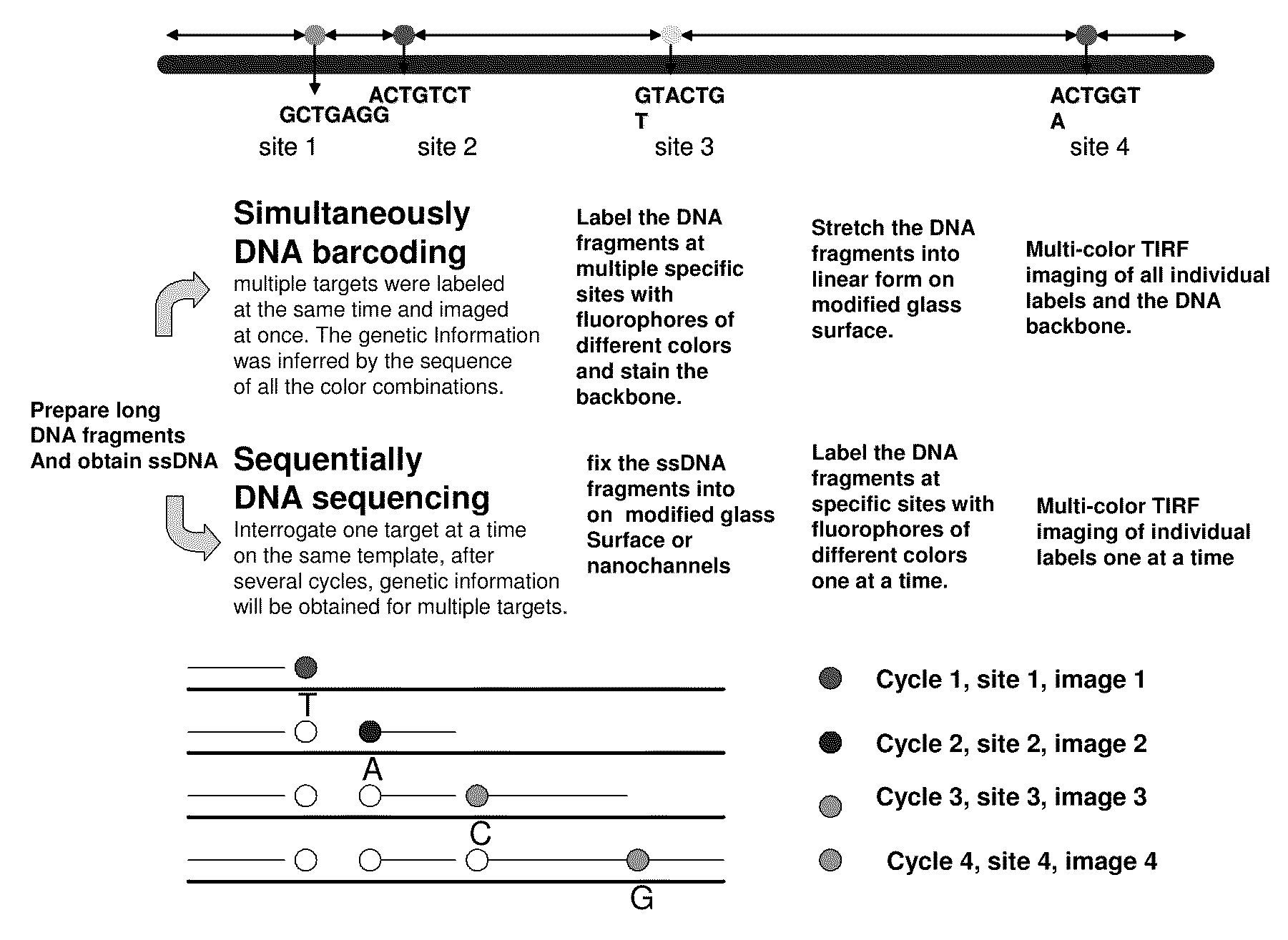
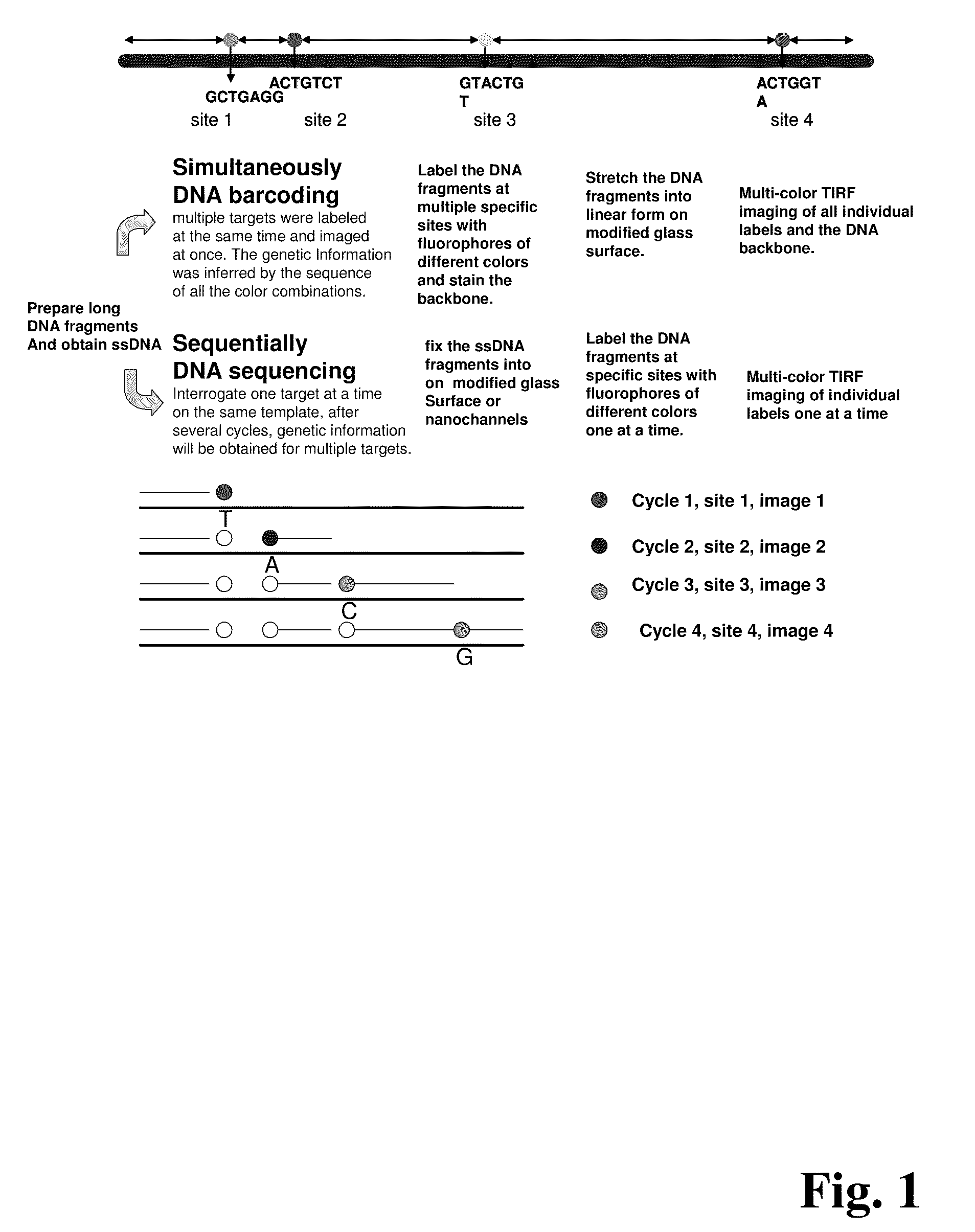
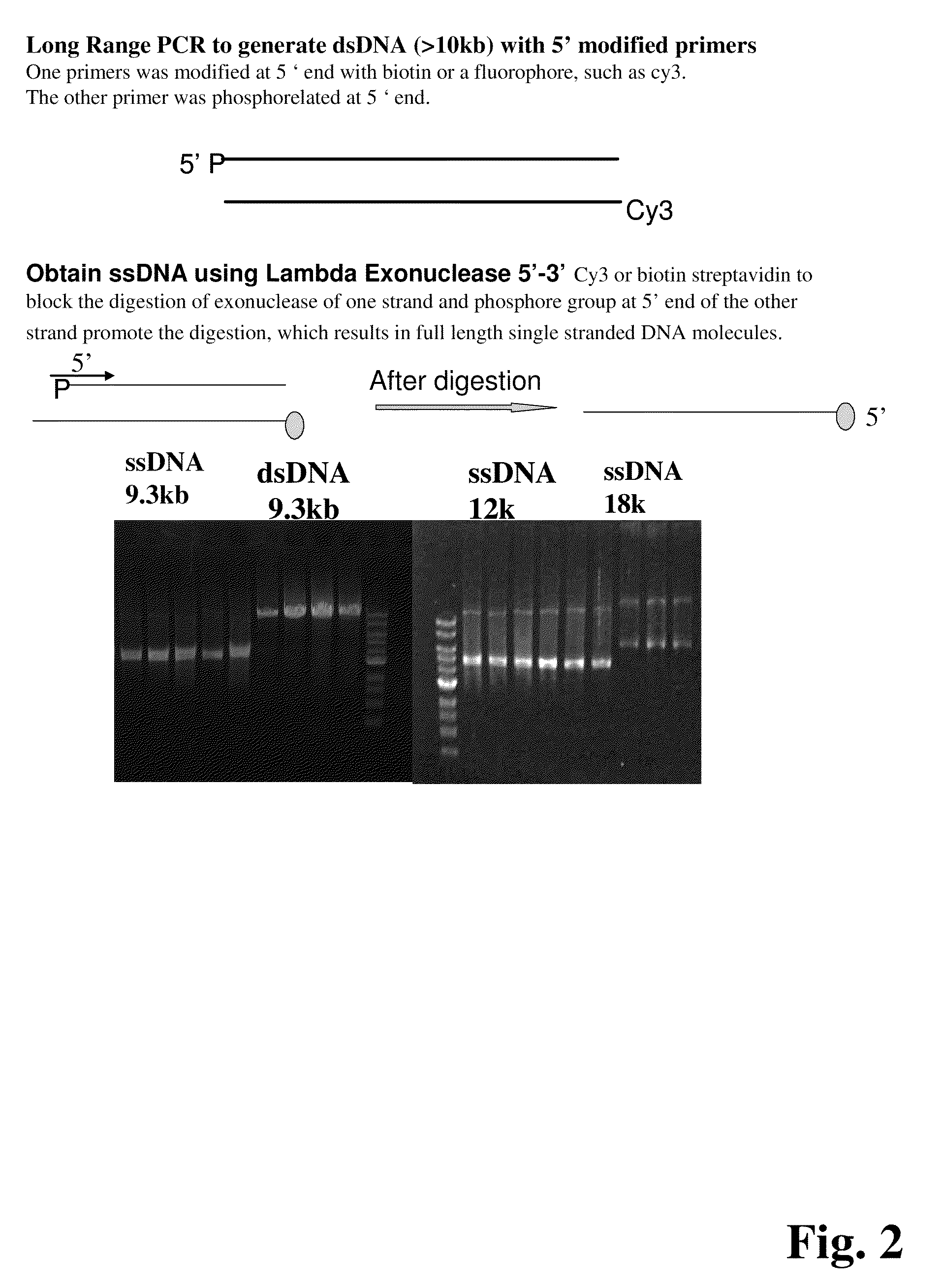
ActiveUS20090155780A1Improve labeling efficiencyStable labelingMicrobiological testing/measurementFermentationDirect imagingDNA barcoding
Improved methods of genetic haplotyping and DNA sequencing and mapping, including methods for making amplified ssDNA, methods for allele determination, and a DNA barcoding strategy based on direct imaging of individual DNA molecules and localization of multiple sequence motifs or polymorphic sites on a single DNA molecule.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
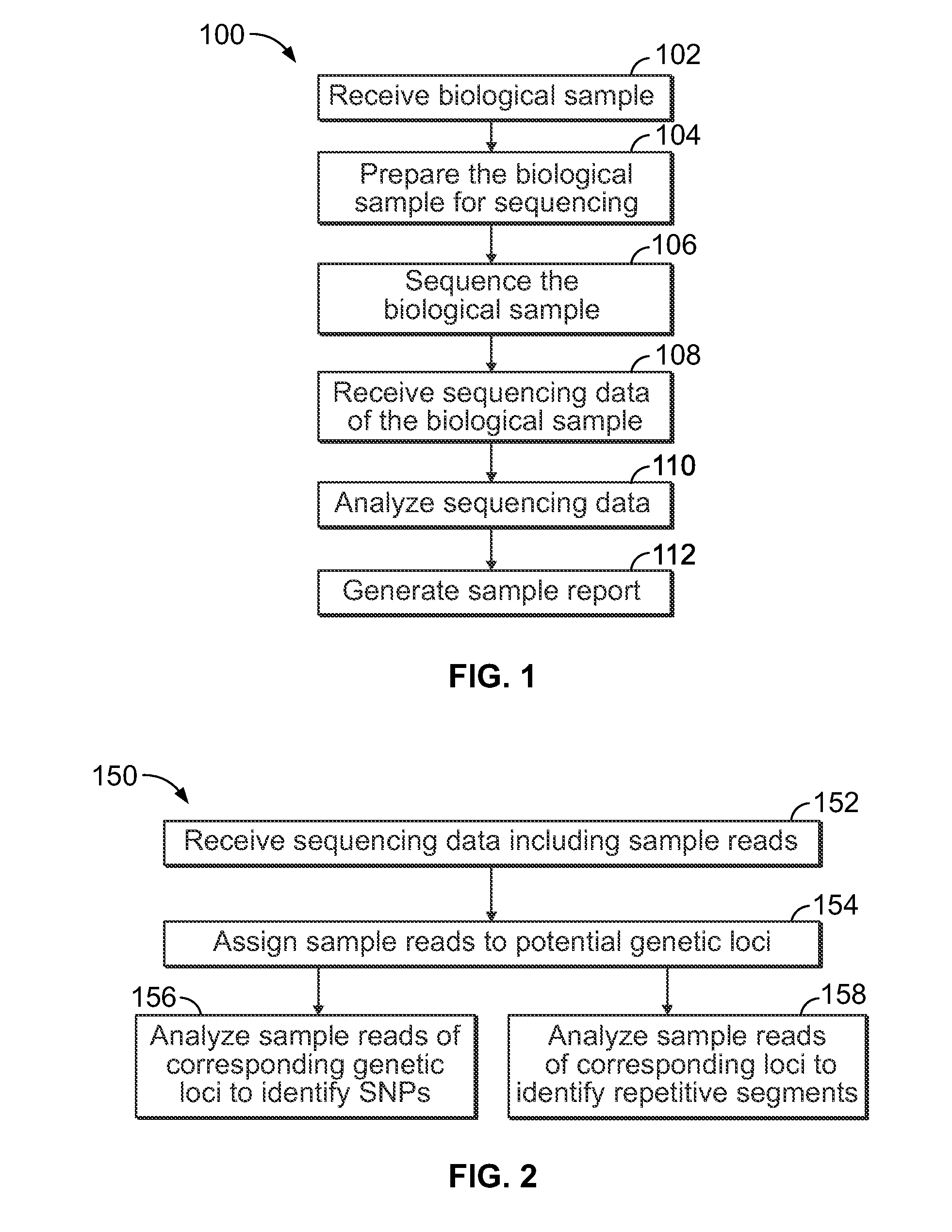
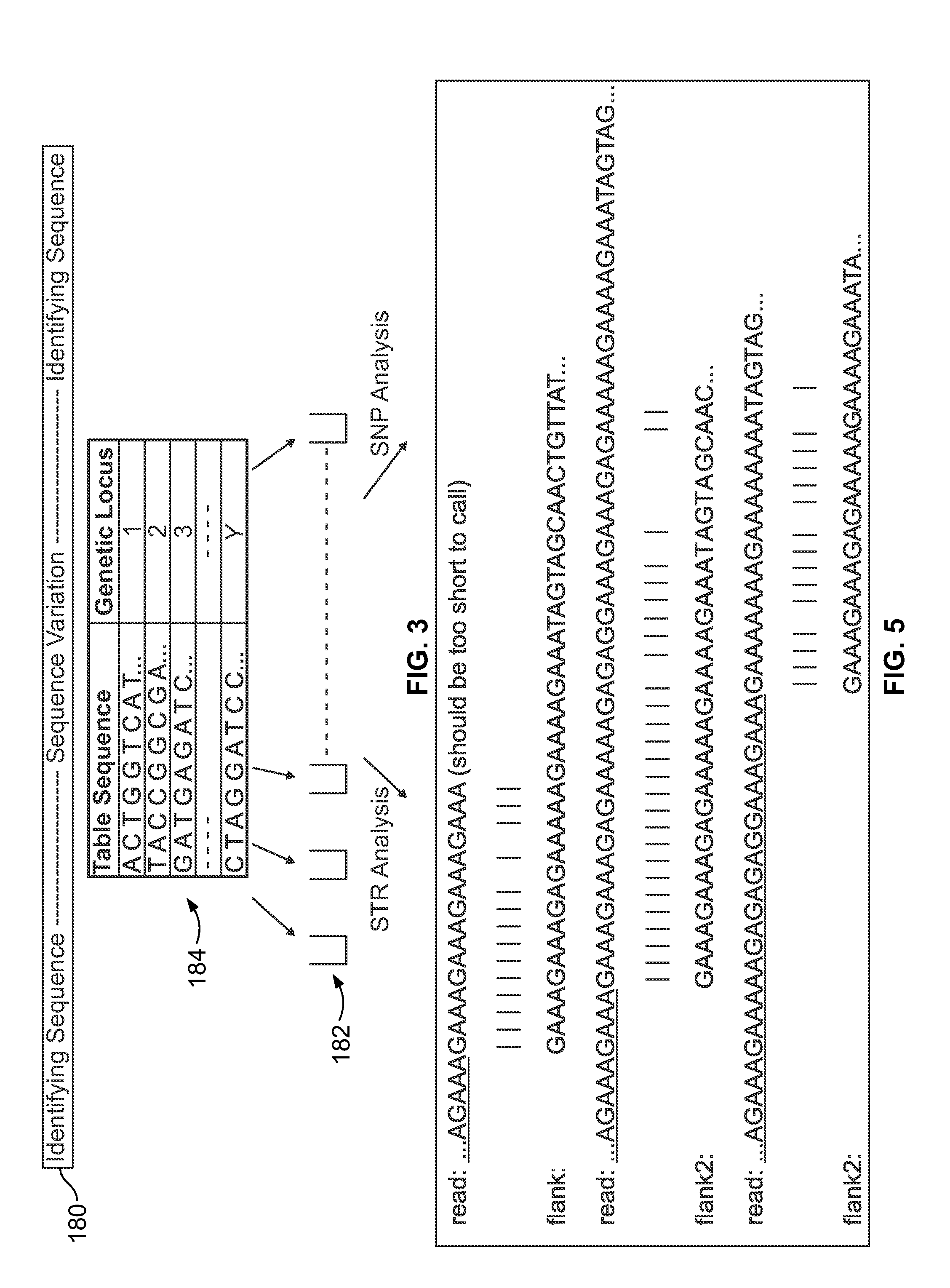
Methods and systems for analyzing nucleic acid sequencing data
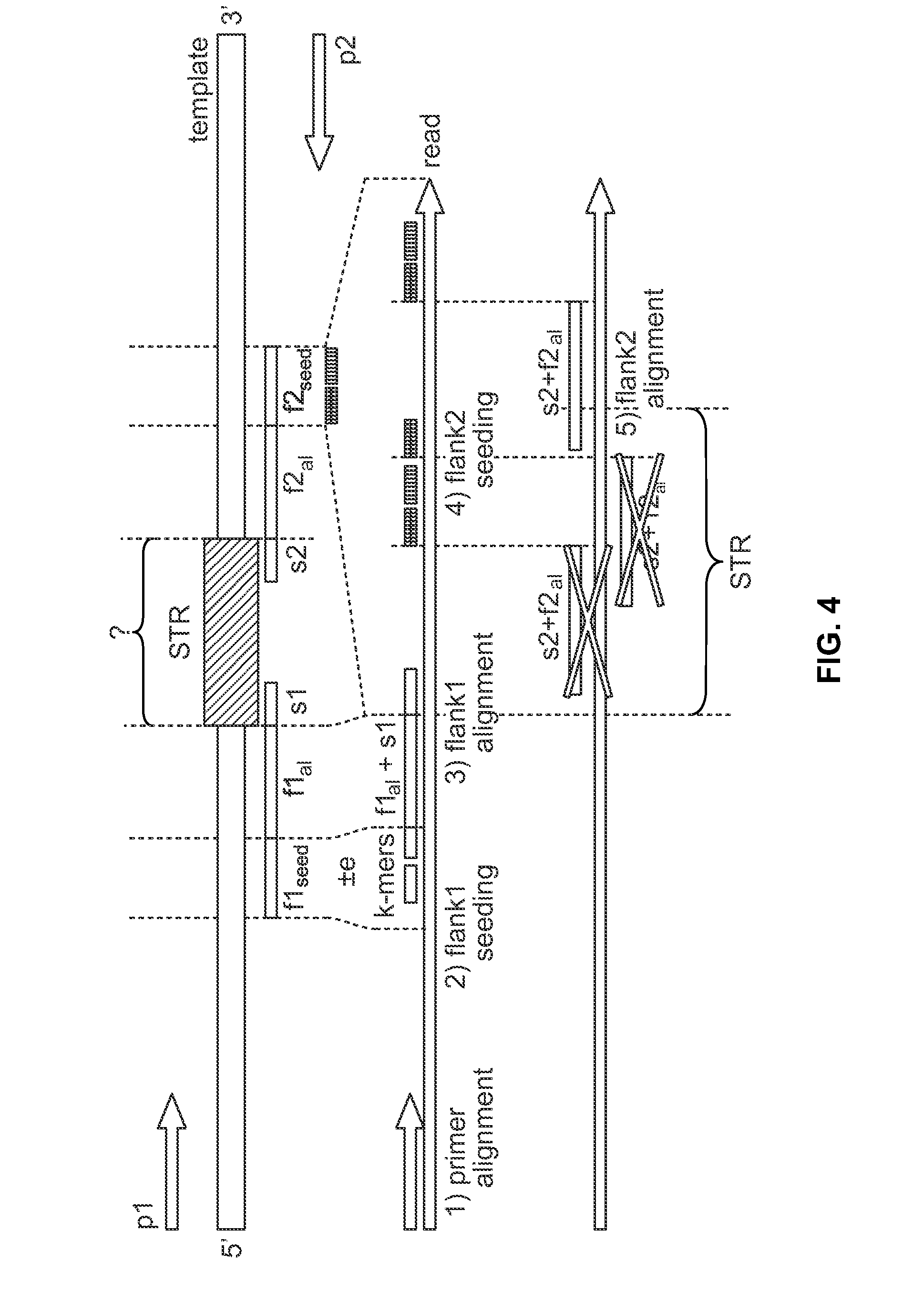
Method includes receiving sequencing data including a plurality of sample reads that have corresponding sequences of nucleotides and assigning the sample reads to designated loci. The method also includes analyzing the assigned reads for each designated locus to identify corresponding regions-of-interest (ROIs) within the assigned reads. Each of the ROIs has one or more series of repeat motifs. The method also includes sorting the assigned reads based on the sequences of the ROIs such that the ROIs with different sequences are assigned as different potential alleles. The method also includes analyzing, for designated loci having multiple potential alleles, the sequences of the potential alleles to determine whether a first allele of the potential alleles is suspected stutter product of a second allele of the potential alleles.
Owner:ILLUMINA INC
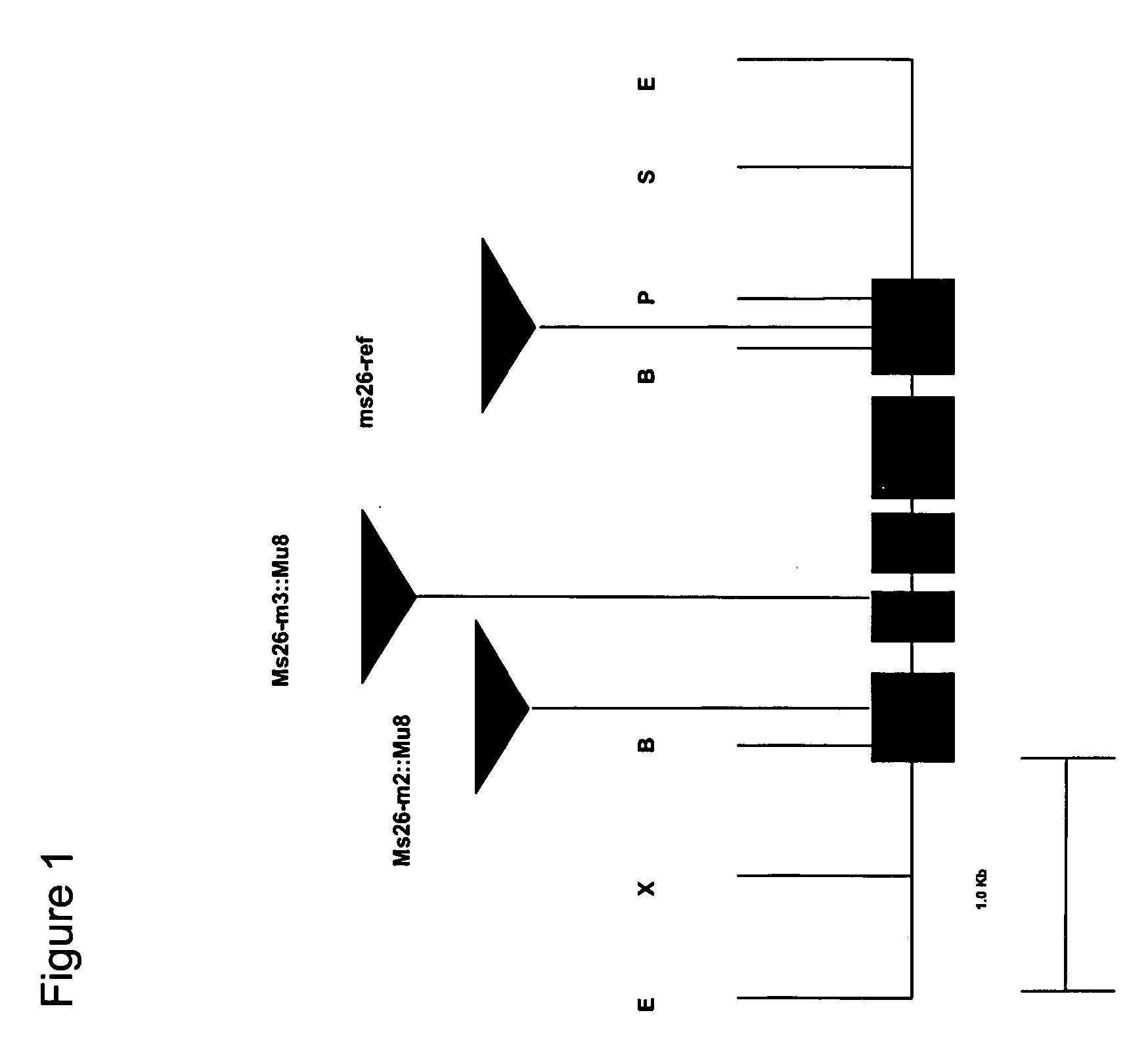
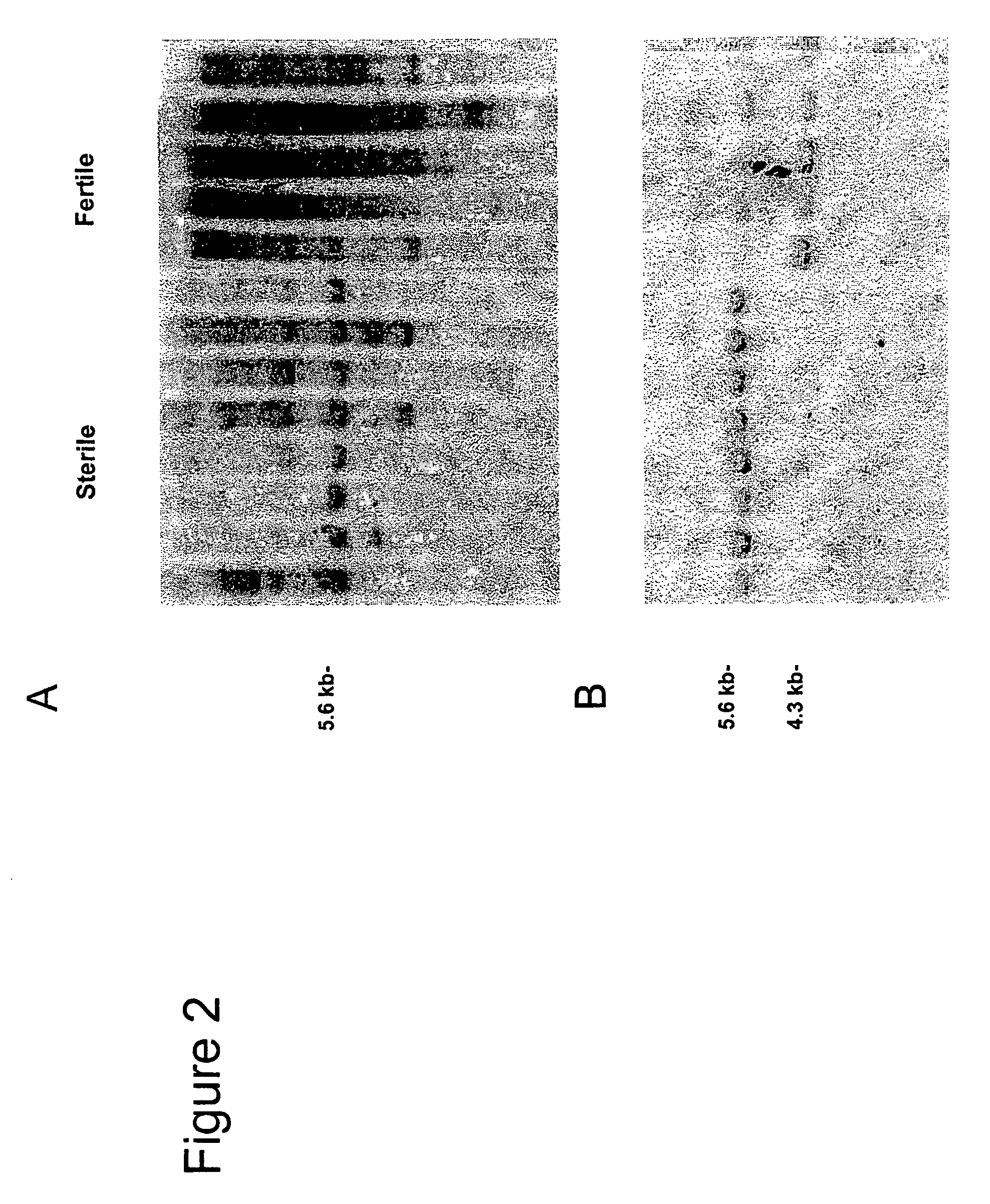
Nucleotide sequences mediating male fertility and method of using same
InactiveUS20060288440A1Microbiological testing/measurementOther foreign material introduction processesAdemetionineGamete
Nucleotide sequences mediating male fertility in plants are described, with DNA molecule and amino acid sequences set forth. Promoter sequences and their essential regions are also identified. The nucleotide sequences are useful in mediating male fertility in plants. In one such method, the homozygous recessive condition of male sterility causing alleles is maintained after crossing with a second plant, where the second plant contains a restoring transgene construct having a nucleotide sequence which reverses the homozygous condition. The restoring sequence is linked with a hemizygous sequence encoding a product inhibiting formation or function of male gametes. The maintainer plant produces only viable male gametes which do not contain the restoring transgene construct. Increase of the maintainer plant is also provided by self-fertilization, and selection for seed or plants which contain the construct.
Owner:PIONEER HI BRED INT INC
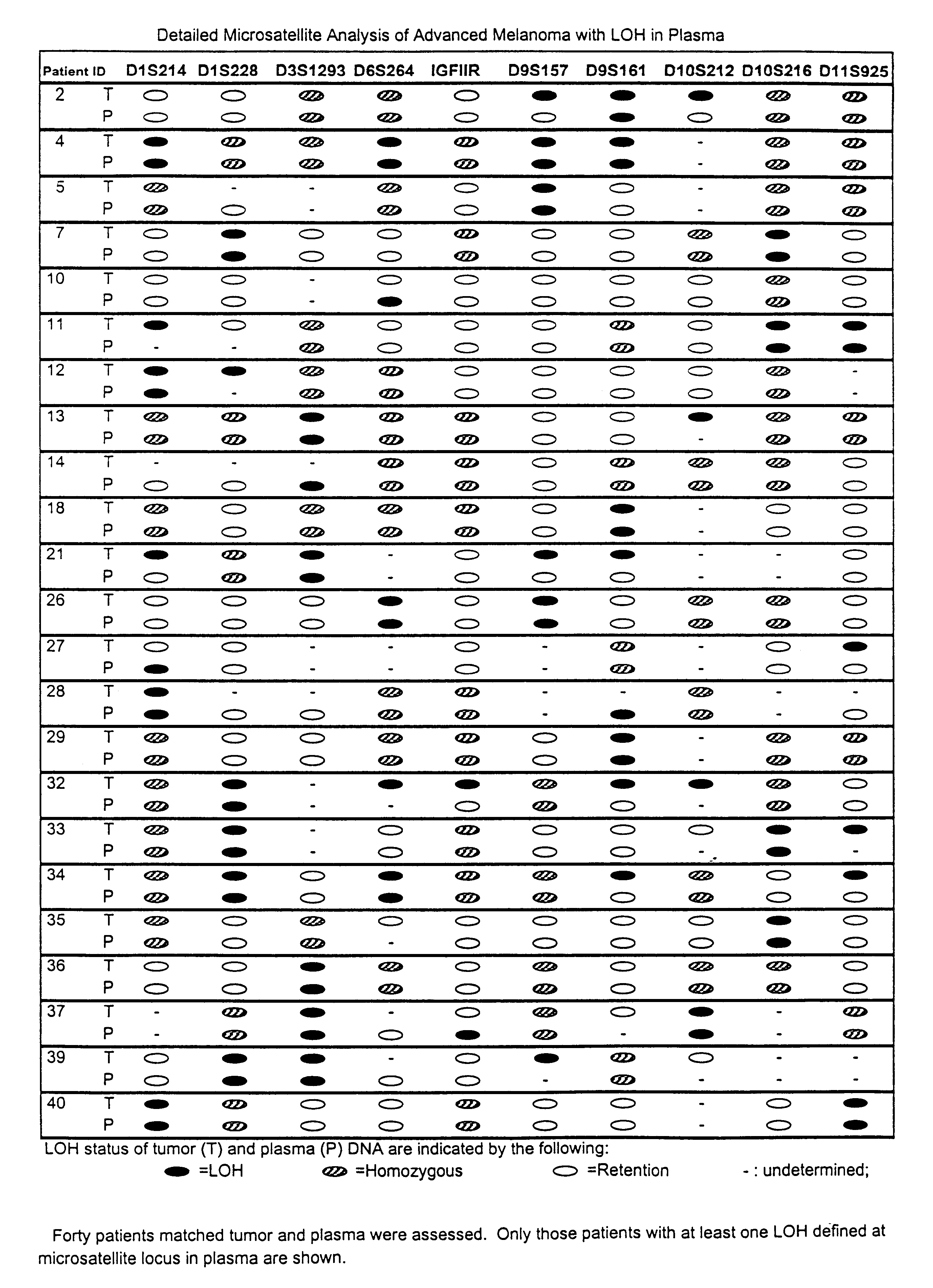
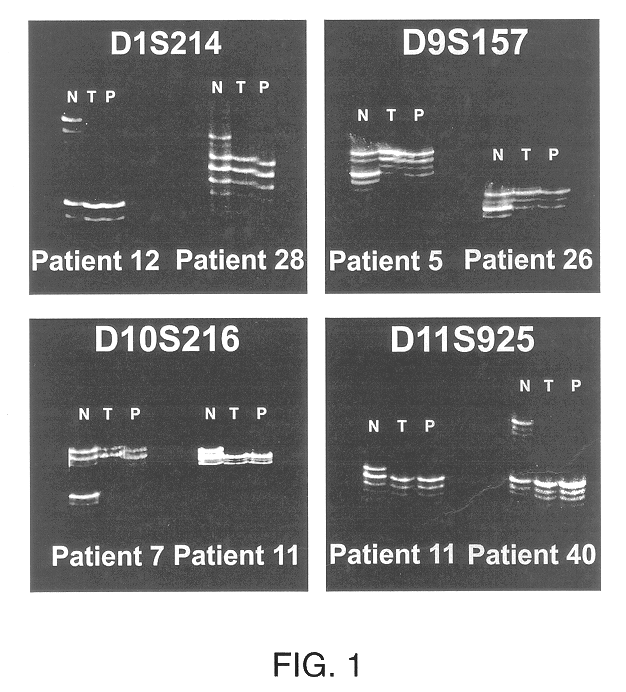
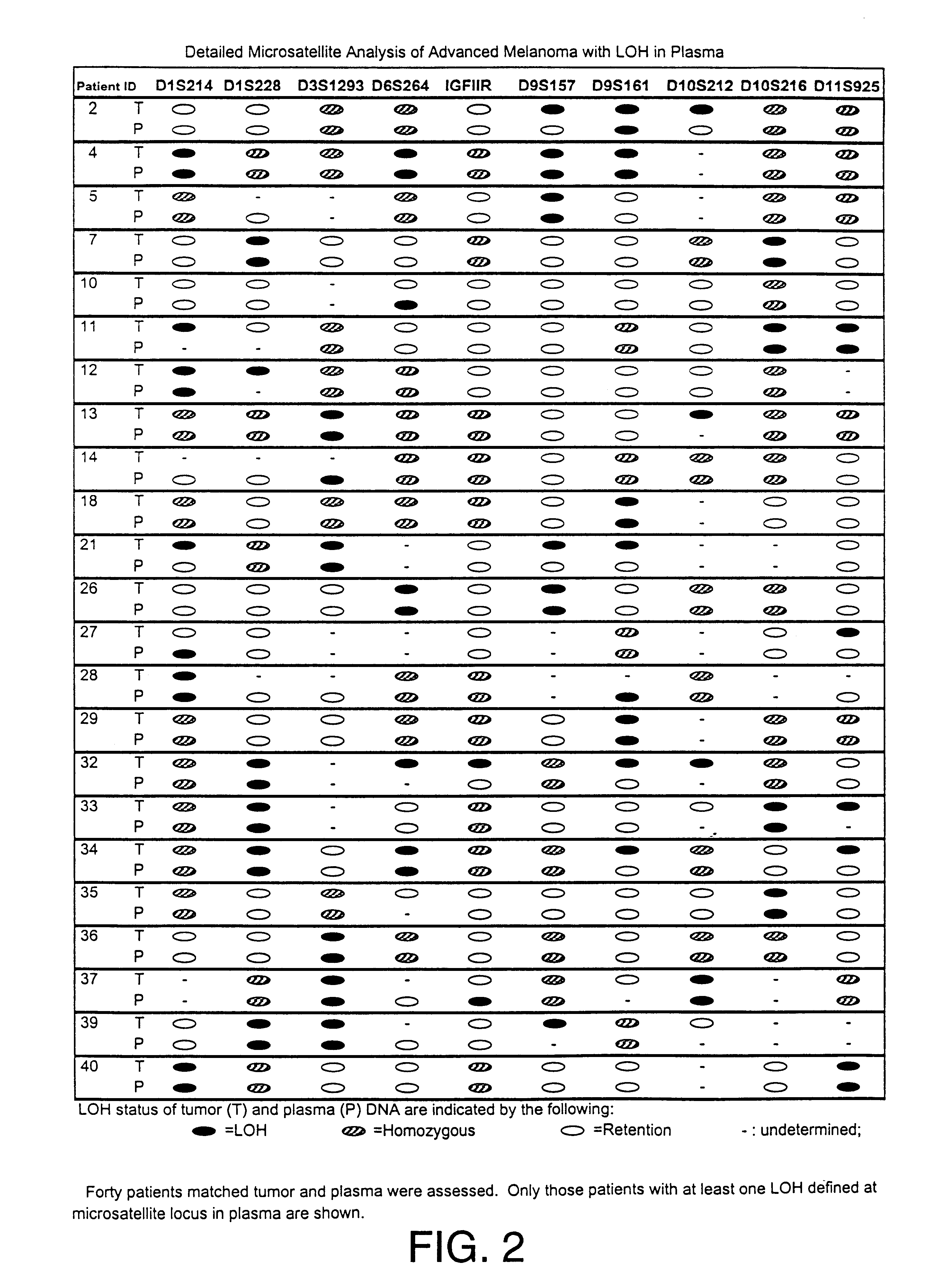
Detection of loss of heterozygosity in tumor and serum of melanoma patients
A method is provided for assessing allelic losses on specific chromosomal regions in melanoma patents. The method relies on the evidence that free DNA may be released in the plasma / serum of cancer patients allowing the detection of DNA with LOH in the plasma / serum of cancer patients by analysis for microsatellite markers. The amount of and specific allelic loss allows a prognosis to be made regarding tumor diagnosis and progression, tumor metastasis, tumor recurrence, and mortality.
Owner:JOHN WAYNE CANCER INST
Sample preparation, processing and analysis systems
ActiveUS8894946B2Small footprintFast timeBioreactor/fermenter combinationsHeating or cooling apparatusAnalyteAllele
This disclosure provides an integrated and automated sample-to-answer system that, starting from a sample comprising biological material, generates a genetic profile in less than two hours. In certain embodiments, the biological material is DNA and the genetic profile involves determining alleles at one or a plurality of loci (e.g., genetic loci) of a subject, for example, an STR (short tandem repeat) profile, for example as used in the CODIS system. The system can perform several operations, including (a) extraction and isolation of nucleic acid; (b) amplification of nucleotide sequences at selected loci (e.g., genetic loci); and (c) detection and analysis of amplification product. These operations can be carried out in a system that comprises several integrated modules, including an analyte preparation module; a detection and analysis module and a control module.
Owner:INTEGENX
Nucleic acid encoding reactions
ActiveUS20160208322A1Microbiological testing/measurementLibrary member identificationNucleotideBarcode
Described herein are methods useful for incorporating one or more adaptors and / or nucleotide tag(s) and / or barcode nucleotide sequence(s) one, or typically more, target nucleotide sequences. In particular embodiments, nucleic acid fragments having adaptors, e.g., suitable for use in high-throughput DNA sequencing are generated. In other embodiments, information about a reaction mixture is encoded into a reaction product. Also described herein are methods and kits useful for amplifying one or more target nucleic acids in preparation for applications such as bidirectional nucleic acid sequencing. In particular embodiments, methods of the invention entail additionally carrying out bidirectional DNA sequencing. Also described herein are methods for encoding and detecting and / or quantifying alleles by primer extension.
Owner:FLUIDIGM CORP
Methods for non-invasive prenatal ploidy calling
PendingUS20190211392A1Fill in the blanksMathematical modelsMicrobiological testing/measurementMedicinePloidy
The present disclosure provides methods for determining the ploidy status of a chromosome in a gestating fetus from genotypic data measured from a mixed sample of DNA comprising DNA from both the mother of the fetus and from the fetus, and optionally from genotypic data from the mother and father. The ploidy state is determined by using a joint distribution model to create a plurality of expected allele distributions for different possible fetal ploidy states given the parental genotypic data, and comparing the expected allelic distributions to the pattern of measured allelic distributions measured in the mixed sample, and choosing the ploidy state whose expected allelic distribution pattern most closely matches the observed allelic distribution pattern. The mixed sample of DNA may be preferentially enriched at a plurality of polymorphic loci in a way that minimizes the allelic bias, for example using massively multiplexed targeted PCR.
Owner:NATERA
Methods for identifying DNA copy number changes
Methods of identifying allele-specific changes in genomic DNA copy number are disclosed. Methods for identifying homozygous deletions and genetic amplifications are disclosed. An array of probes designed to detect presence or absence of a plurality of different sequences is also disclosed. The probes are designed to hybridize to sequences that are predicted to be present in a reduced complexity sample. The methods may be used to detect copy number changes in cancerous tissue compared to normal tissue. The methods may be used to diagnose cancer and other diseases associated with chromosomal anomalies.
Owner:AFFYMETRIX INC
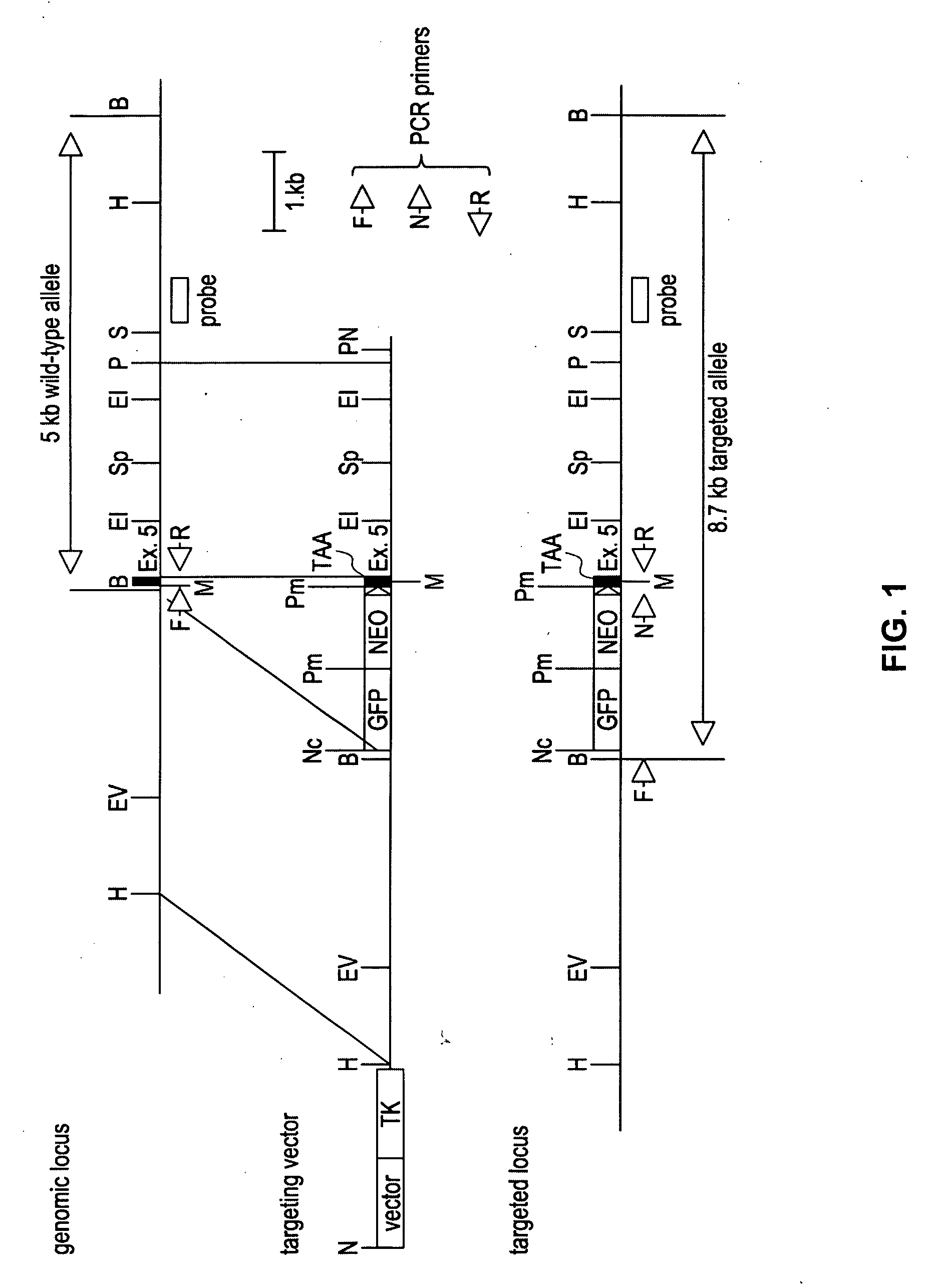
Universal stem cells
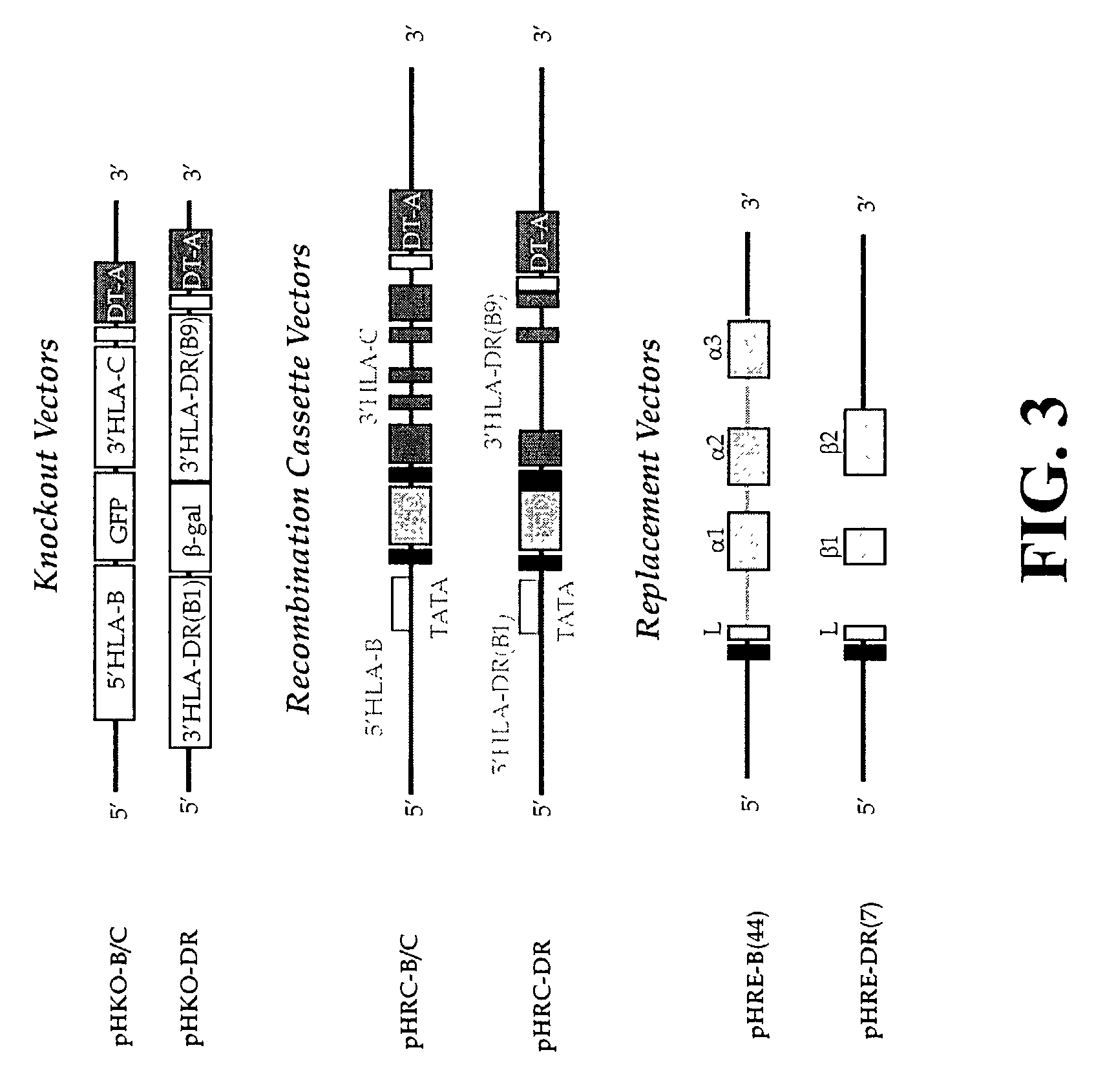
The subject invention pertains to materials and methods for preparing multi-potential stem cells having a pre-selected expression of MHC antigens. Stem cells of the subject invention can be used to generate histocompatible tissues / organs for transplantation. The process of the subject invention comprises the use of targeting vectors capable of gene knockout, insertion of site-specific recombination cassettes, and the replacement of histocompatibility alleles in the stem cell. Novel knockout vectors are used to delete designated regions of one chromosome. Recombination cassette vectors are then used to delete the same region on the second chromosome and deposit a site-specific recombination cassette which can be utilized by replacement vectors for inserting the new MHC genes on the chromosome of the engineered cell. The subject invention also pertains to cells, tissues, and transgenic mammal prepared using the methods and materials of the invention.
Owner:MORPHOGENESIS
Allele-specific expression patterns
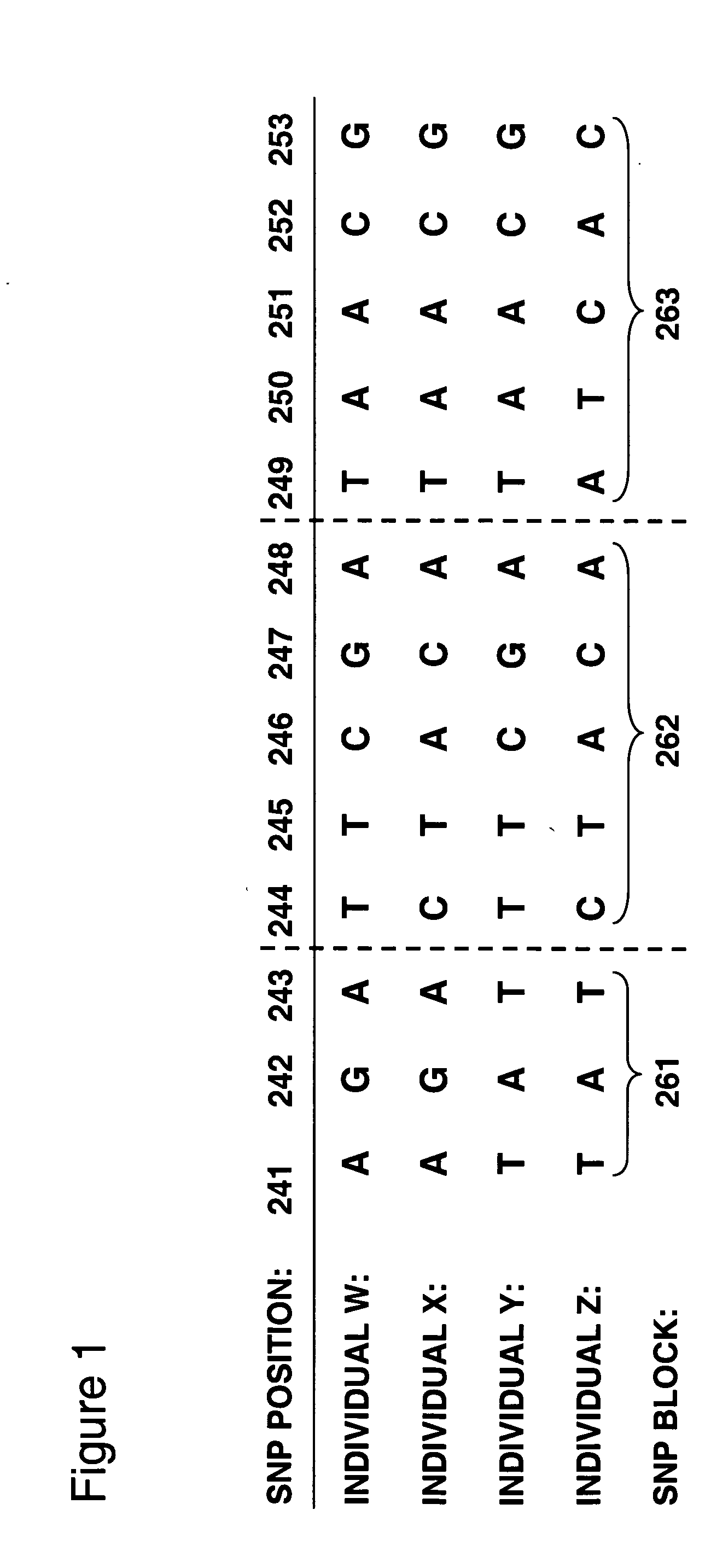
The invention provides methods of analyzing genes for differential relative allelic expression patterns. Haplotype blocks throughout the genomes of individuals are analyzed to identify haplotype patterns that are associated with specific differential relative allelic expression patterns. Haplotype blocks that contain associated haplotype patterns may be further investigated to identify genes or variants of genes involved in differential relative allelic expression patterns.
Owner:PERLEGEN SCIENCES INC
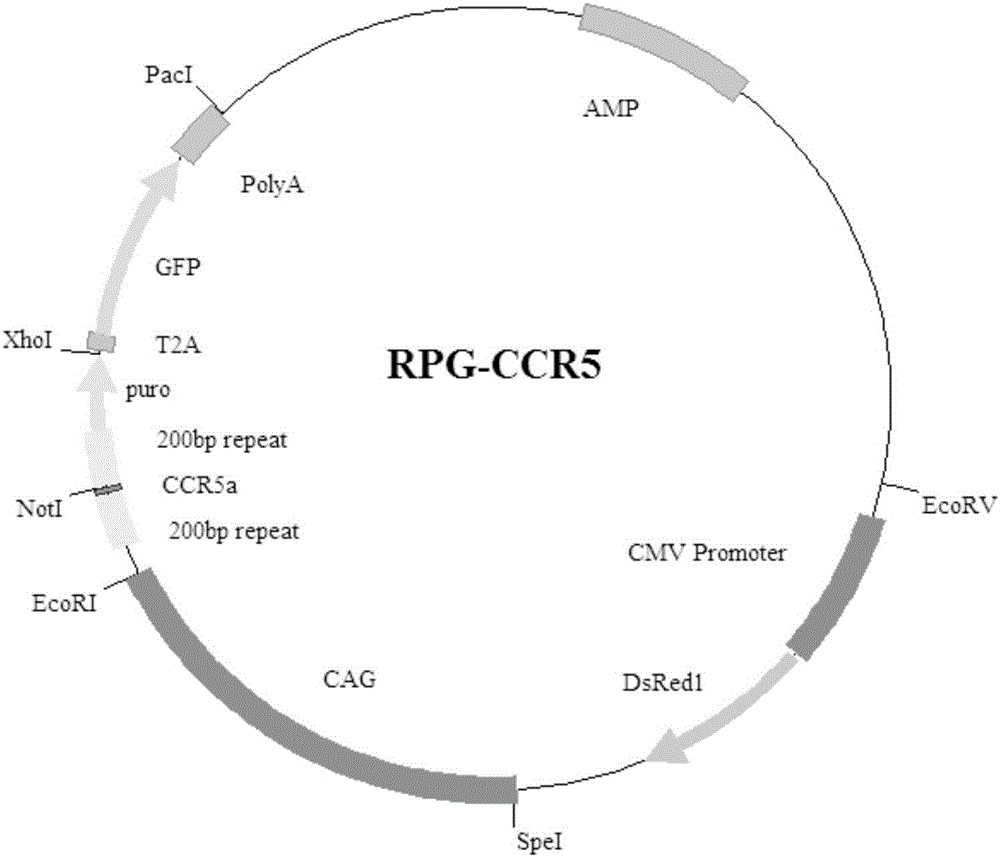
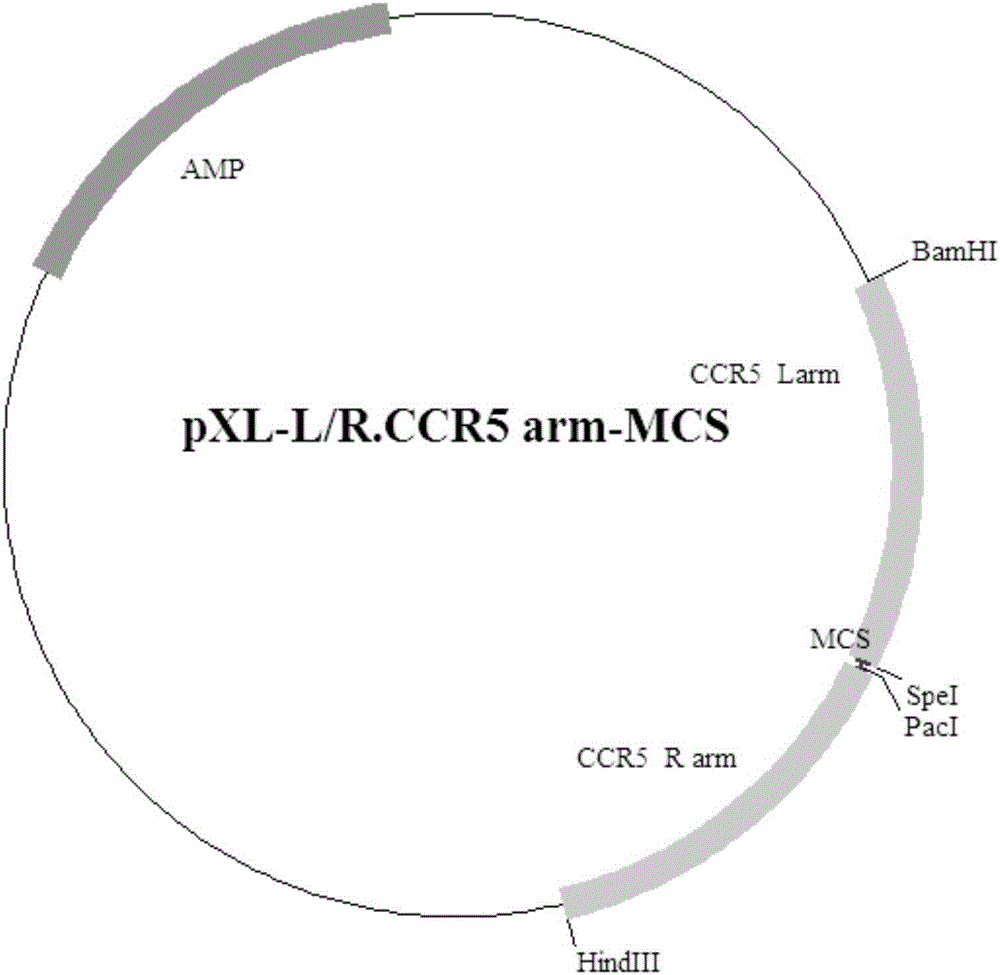
Exogenous gene knocking-in and integrating system on basis of CRISPR/Cas9, method for establishing exogenous gene knocking-in and integrating system and application thereof
ActiveCN106191116ANucleic acid vectorVector-based foreign material introductionTarget geneIntegrated systems
The invention provides an exogenous gene knocking-in and integrating system on the basis of CRISPR / Cas9, a method for establishing the exogenous gene knocking-in and integrating system and application thereof. The exogenous gene knocking-in and integrating system comprises vectors with report / donor functions and Cas9 expression vectors. Each report / donor vector comprises two target gent homologous arms and an exogenous sequence fragment positioned between the two target gene homologous arms; homologous sequences, which are positioned on a target gene, of the two target gene homologous arms of each report / donor vector are respectively positioned on two sides of a target sequence of the target gene and are connected with the target sequence of the target gene; the exogenous sequence fragments comprise promoters, resistant genes, shorn peptide sequences, report genes and polyA tails which are sequentially arrayed, two SSA repair homologous sequences of each resistant gene are inserted into the resistant gene, and the target sequence of each target gene is inserted in a space between the two corresponding SSA repair homologous sequences. The exogenous gene knocking-in and integrating system, the method and the application have the advantages that exogenous genes can be integrated with endogenous gene sequences in an efficient site-directed and accurately targeted manner, and double-chromosome allelic gene double-knocking-in can be efficiently carried out.
Owner:成都中科奥格生物科技有限公司
System and method for cleaning noisy genetic data and determining chromosome copy number
InactiveUS20200224273A1Improve fidelityMicrobiological testing/measurementBiostatisticsGenetic correlationDiploid cells
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
System and method for cleaning noisy genetic data and determining chromosome copy number
PendingUS20200248264A1Improve fidelityMicrobiological testing/measurementBiostatisticsGenetic correlationDiploid cells
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS20200232036A1Significant resultImprove fidelityMicrobiological testing/measurementBiostatisticsGenetic correlationDiploid cells
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
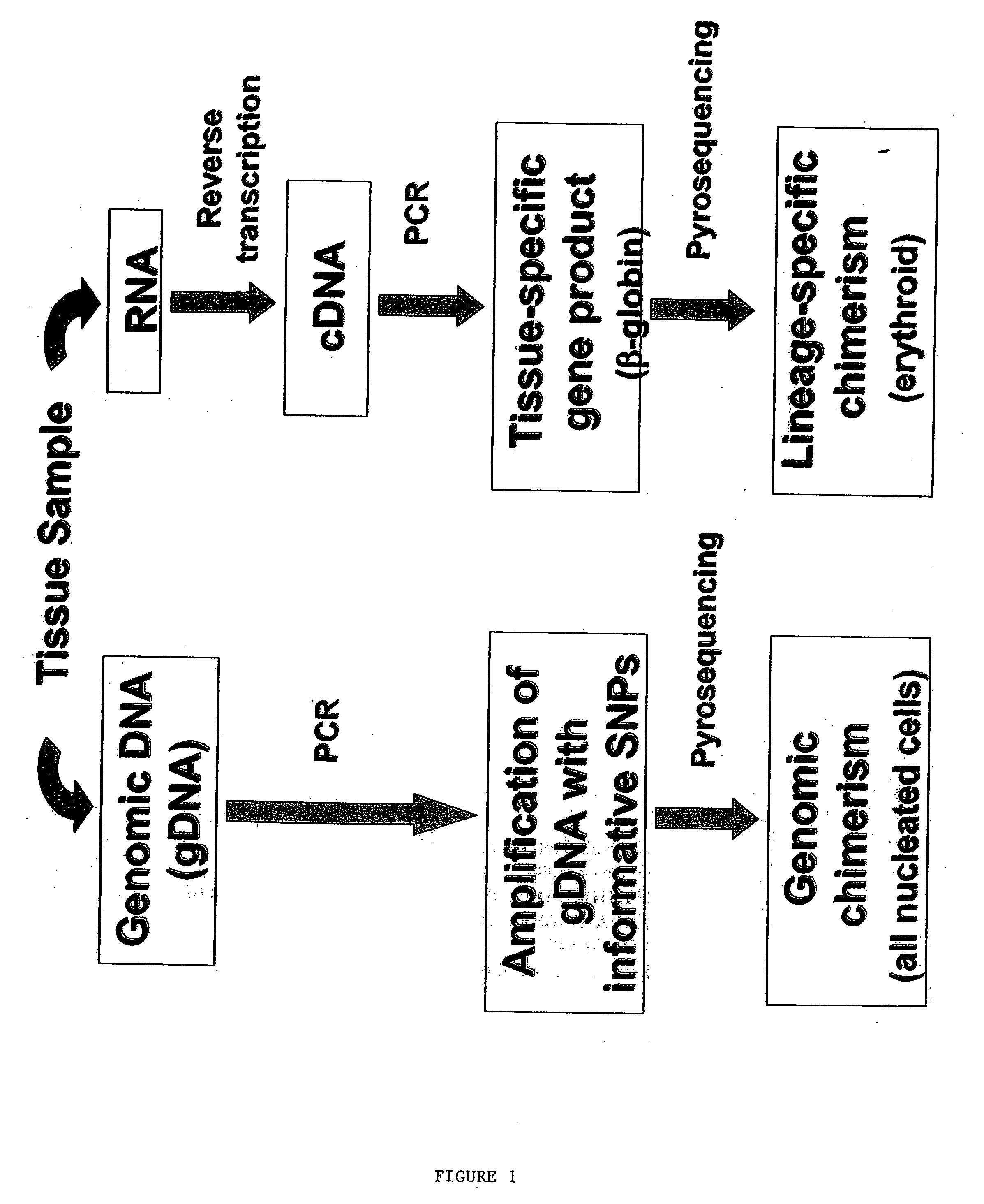
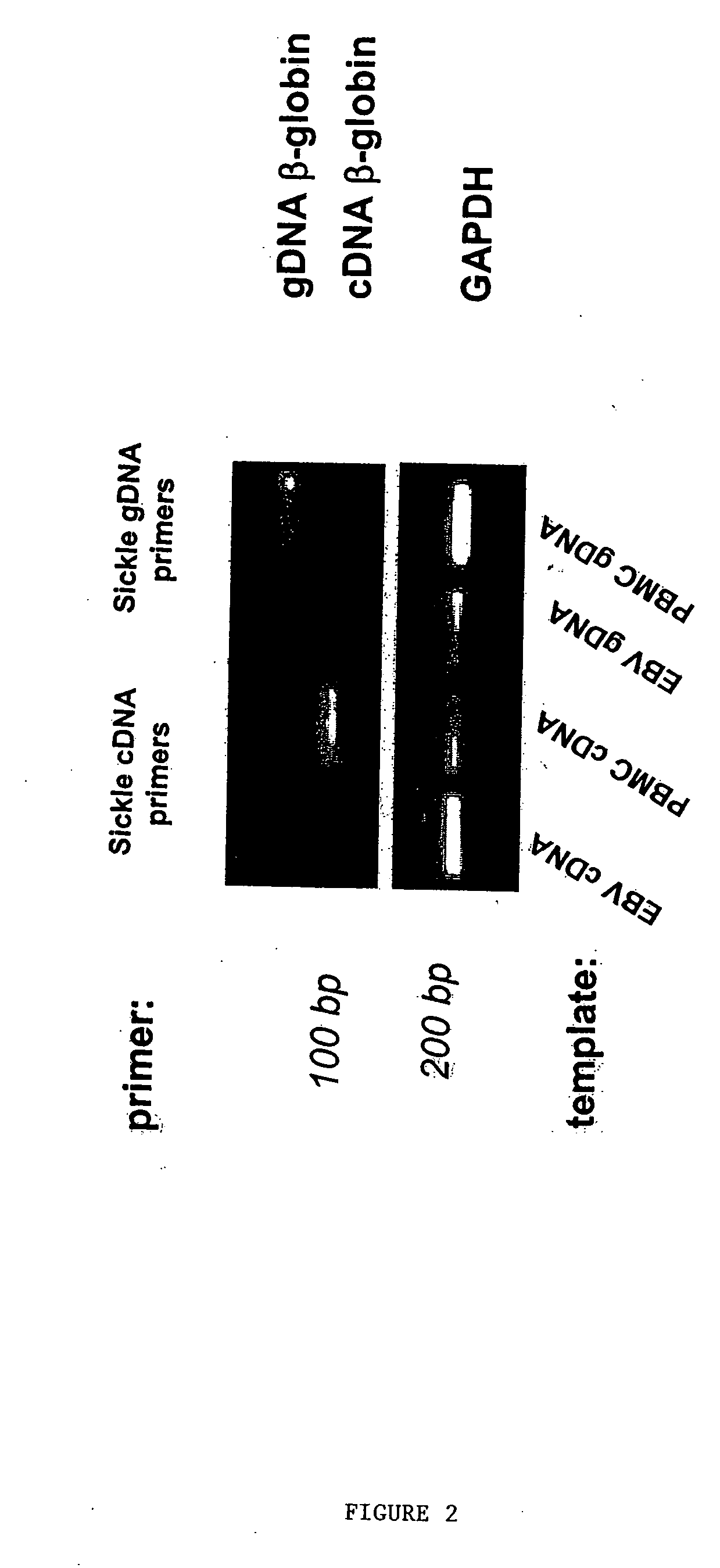
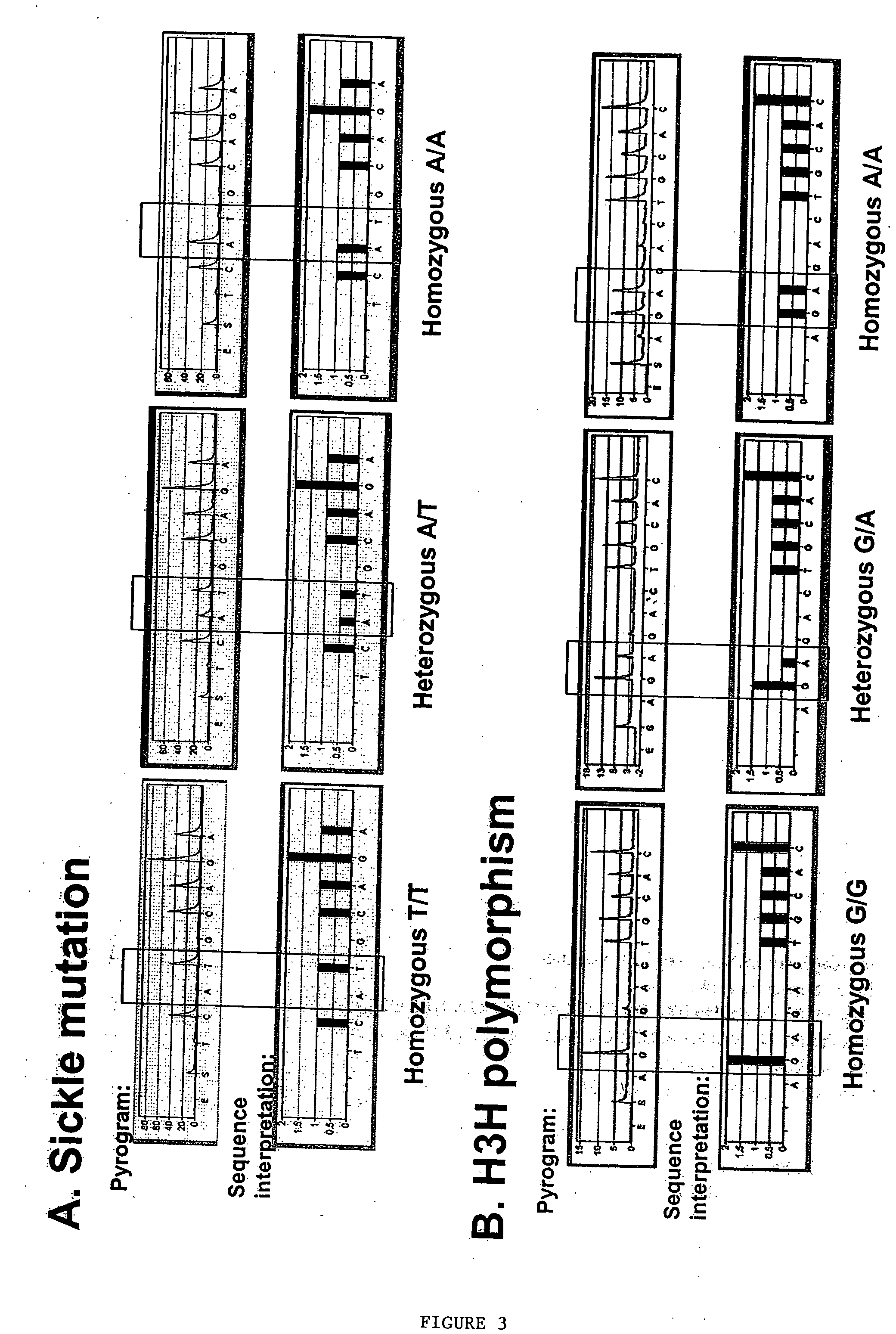
Methods to detect lineage-specific cells
The present invention is based, at least in part, on methods to identify and quantify lineage-specific cells. The present invention provides methods of detecting lineage-specific cells in a biological sample, and monitoring the effectiveness of progenitor cell transfer in a subject. The invention further provides methods of determining an effective dose of progenitor cell transfer in a subject and methods of quantifying progenitor cell transfer in a subject. Additionally, methods are provided to identify allelic variants in lineage-specific cells.
Owner:DANA FARBER CANCER INST INC
Muir-Torre-like syndrome in Fhit deficient mice
InactiveUS20060075511A1Drug screeningNucleic acid vectorParanasal Sinus CarcinomaGastrointestinal cancer
The invention provides nonhuman transgenic animals with a disrupted FHIT gene. The invention further provides transgenic mice in which one or both Fhit alleles have been inactivated. Preferably, the Fhit-deficient mice develop multiple tumors of both visceral and sebaceous origin, similar to those of Muir-Torre familial cancer syndrome. The present invention further relates to the generation of these transgenic mice and their use as model systems to study the effects of carcinogenic agents in promoting clonal expansion of neoplastic cells in cancers, preferably gastrointestinal cancers of which Muir-Torre syndrome is a subset. The invention further relates to testing therapeutic agents for their efficacy in the prevention and treatment of cancer, preferably gastrointestinal cancer.
Owner:CROCE CARLO +1
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com