PCR (Polymerase Chain Reaction) primer for detecting and identifying porcine circovirus 3 (PCV3) and detection method and detection kit
The technology of a porcine circovirus and a kit, which is applied in the field of animal disease sanitation inspection, can solve the problems that there is no method or kit for detecting and identifying porcine circovirus type 3, so as to facilitate clinical detection and carry out epidemiological investigation, Great application prospect and high specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Implementation example 1 The design of porcine circovirus type 3 PCR primers
[0047] 1. The present invention finds that porcine circovirus type 3 encodes two main open reading frames: rep and cap, both of which are in opposite directions on the DNA chain. By referring to and comparing all PCV3 cap gene sequences in GenBank, selecting a conserved nucleotide sequence as an amplification region, a pair of PCR primers for detecting porcine circovirus type 3 have been designed; the PCR primers are as follows:
[0048] Upstream primer PCV3-F: 5'-GGGCACACAGCCATAGAT-3';
[0049] Downstream primer PCV3-R: 5'-TTCCGGGACATAAATGCT-3'.
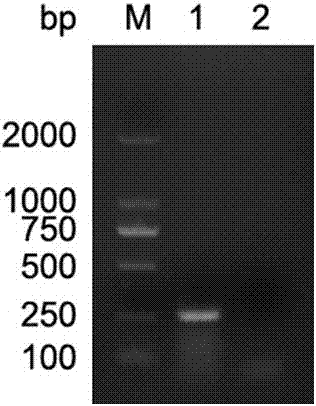
[0050] 2. After verification, the DNA of porcine circovirus type 3 was used as a template for amplification, and the results showed that the primer set could specifically amplify and detect porcine circovirus type 3.
Embodiment 2
[0051] Example 2 PCR primers for detection of porcine circovirus type 3 samples
[0052] 1. DNA extraction
[0053] (1) Take 30mg of swine disease tissue, cut the tissue into pieces as much as possible, and put them into 1.5ml centrifuge tubes.
[0054] (2) Add 450µL of lysis buffer (10mM Tris-HCl pH8.0; 100mM EDTA, pH8.0) to each tube.
[0055] (3) Add 50-100 µL of 10% SDS to each tube, and then add 2.5-5 µL of proteinase K (20 mg / ml) to make the final concentration reach 100 µg / mL, mix well, and incubate at 55°C for 3 hours to overnight (during this period, turn the tube upside down a few times) Second-rate).
[0056] (4) Add 150µL NaCl to the sample, centrifuge at 8000rpm for 20min at room temperature, remove the precipitate, then add an equal volume (500µL) of pre-cooled isopropanol to the supernatant, mix well, treat at 20°C for 30min, centrifuge at 14000rpm for 10min, remove the supernatant liquid. Wash the precipitate with 70% ALC and centrifuge to remove ALC, evapo...
Embodiment 3
[0072] Implementation example 3 The specific detection of porcine circovirus type 3 PCR primers
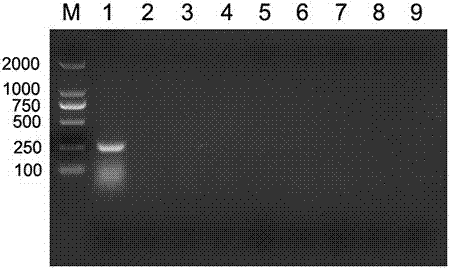
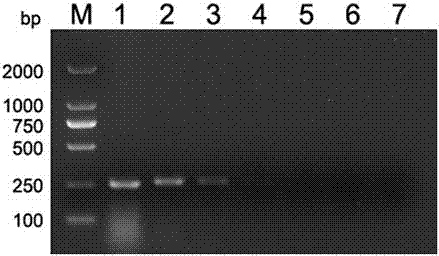
[0073] With reference to step 1 to step 3 in embodiment example 2, utilize the PCR primer described in embodiment 1 to porcine circovirus type 3, foot-and-mouth disease virus, porcine epidemic diarrhea virus, porcine senega valley virus, porcine deltacoronavirus, porcine Kub virus, porcine Boca virus and porcine Sapero virus were detected by PCR respectively. The virus strains were all donated by Guangdong Wen's Group.
[0074] Electrophoresis results such as figure 2 Shown, PCR primer of the present invention is positive reaction to porcine circovirus type 3, but to porcine vesicular virus, foot-and-mouth disease virus, porcine epidemic diarrhea virus, porcine senega valley virus, porcine deltacoronavirus, porcine Kubu Viruses, porcine Boca virus and porcine Sapero virus were negative, indicating that the primer has a strong specificity for porcine circovirus type 3.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com