RT-RPA primer pair, probe, kit and detection method for detecting cat coronavirus
A technology of RT-RPA and feline coronavirus, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., to achieve low temperature and machine requirements, high sensitivity, and short time-consuming effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
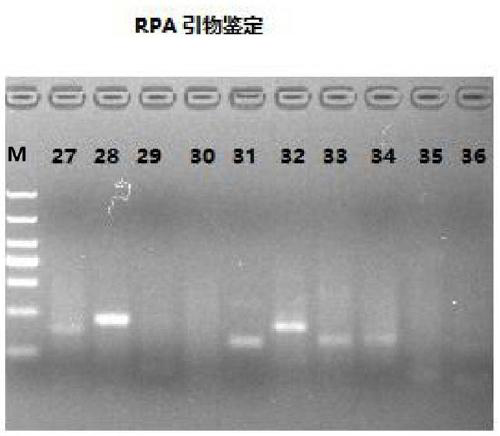
[0036] Embodiment 1. is used for detecting the design and screening of the RT-RPA primer pair of feline coronavirus, probe
[0037] 1. Virus Extraction
[0038] RNA was extracted from ascites samples of cats according to the instructions of the RNA extraction kit from TIANGEN Company, and stored at -80°C for later use.
[0039] 2. Design of RT-RPA primers and probes
[0040] According to the recommendation of the TwistDX design guide, the Fcov sequence (accession number: KY566209.1) was downloaded from NCBI and RNAstar was used to design RT-RPA-specific primers and probes based on the highly conserved MP gene sequence. The designed primers range in length from 30 to 35 bases, and the probes range in length from 48 to 52 bases. The RPA probe should have four groups, including a fluorophore, a quencher, tetrahydrofuran (THF) and a 3-terminal blocker (C3spacer). All primers and probes were synthesized by Shanghai Sangon Bioengineering Co., Ltd. after the design was completed. ...
Embodiment 2
[0045] Embodiment 2. is used to detect the establishment of the RT-RPA method of feline coronavirus
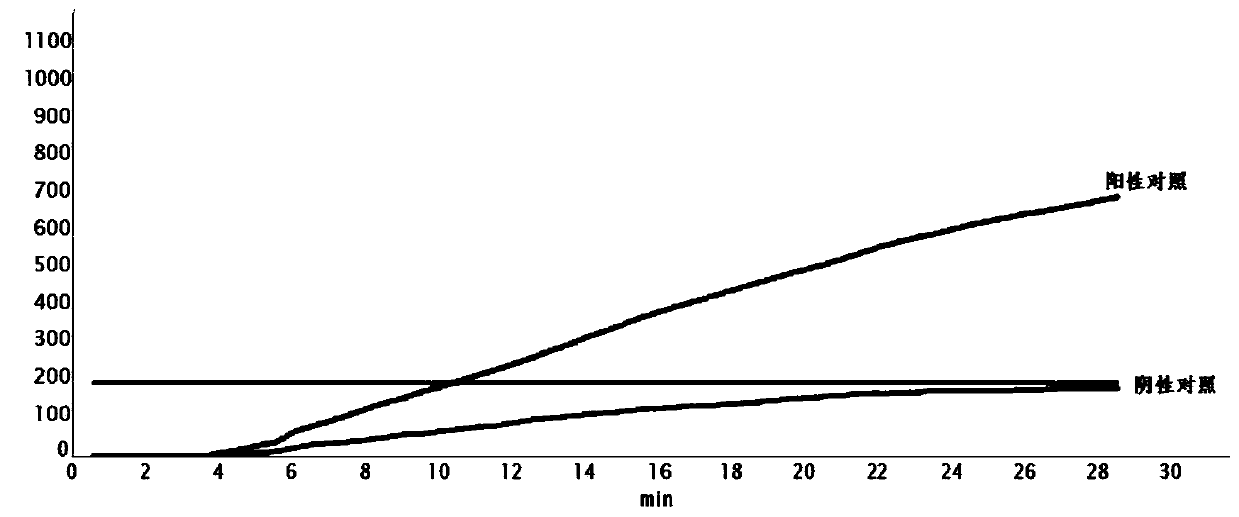
[0046] Use the fluorescent RT-RPA kit of Hangzhou Zongce Biotechnology Co., Ltd. to carry out the RPA reaction with a 50uL reaction system. The optimal reaction system and conditions are: forward primer (10um) 2ul, reverse primer (10um) 2ul, RNA template 2ul, ABuffer 40.9ul, B Buffer 2.5ul, probe (10um) 0.6ul. The RT-RPA reaction was amplified at 39°C for 30 minutes in a real-time constant temperature fluorescence detector (Guangzhou Double Helix Gene Technology Co., Ltd.). Select a positive control and a negative control (both identified by qPCR) respectively, and carry out fluorescent RT-RPA identification, and the identification results are consistent with qPCR (see figure 2 ).
Embodiment 3
[0047] Embodiment 3, the present invention detects the RT-RPA method specificity test and sensitivity test of feline coronavirus
[0048] 3.1 Method
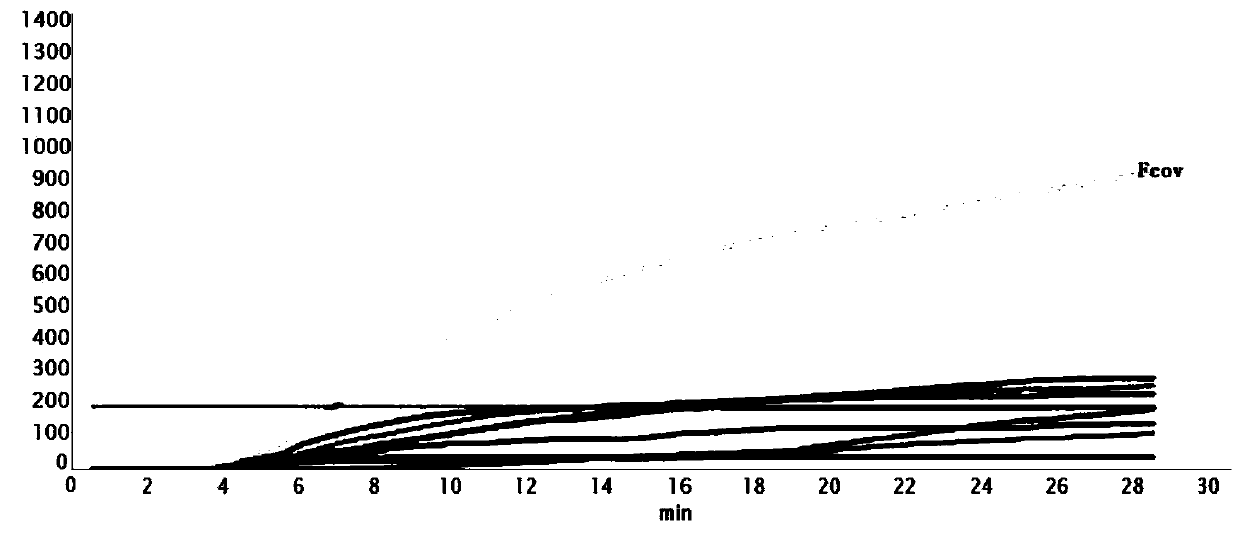
[0049] Specificity test: The specificity of the constructed real-time RPA method was determined by detecting nucleic acids containing Fcov-positive nucleic acids as well as nucleic acids from other pathogens. Nucleic acids from other pathogens include canine coronavirus (CCV), canine parvovirus (CPV), canine distemper (CDV), canine adenovirus (CAV), feline herpesvirus (FHV), feline parvovirus (FPV), cat cup Coronavirus (FCV). Sensitivity test: using plasmid diluted standards, with ddH 2 O was diluted to 7 concentrations at a ratio of 10 times, and the template copy numbers after dilution were 1*10 6 copies / μL to 1 copies / μL.
[0050] 3.2 Results
[0051] The specificity of the constructed real-time RPA method was determined by detecting nucleic acids containing Fcov-positive nucleic acids as well as nucleic acids from other...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com