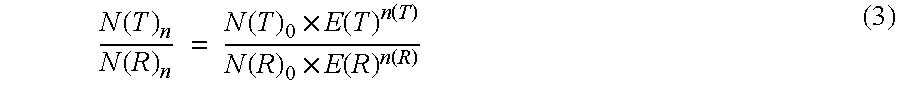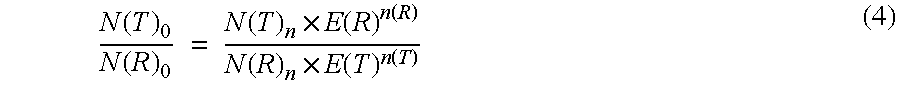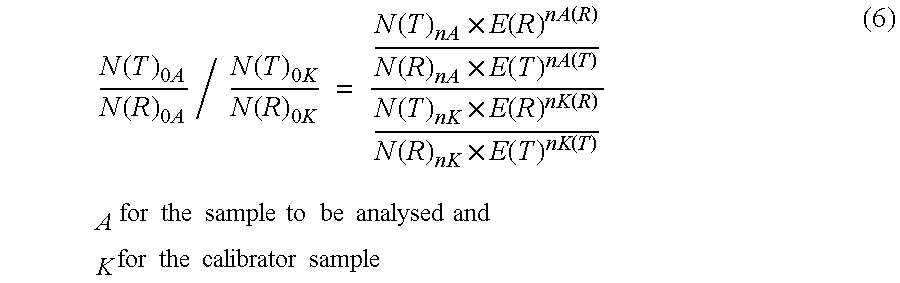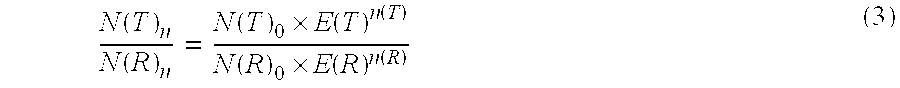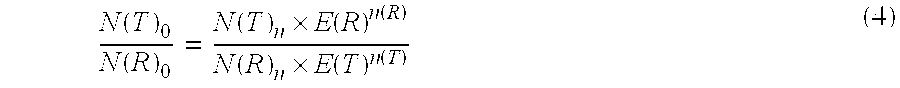Patents
Literature
348 results about "Absolute quantification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
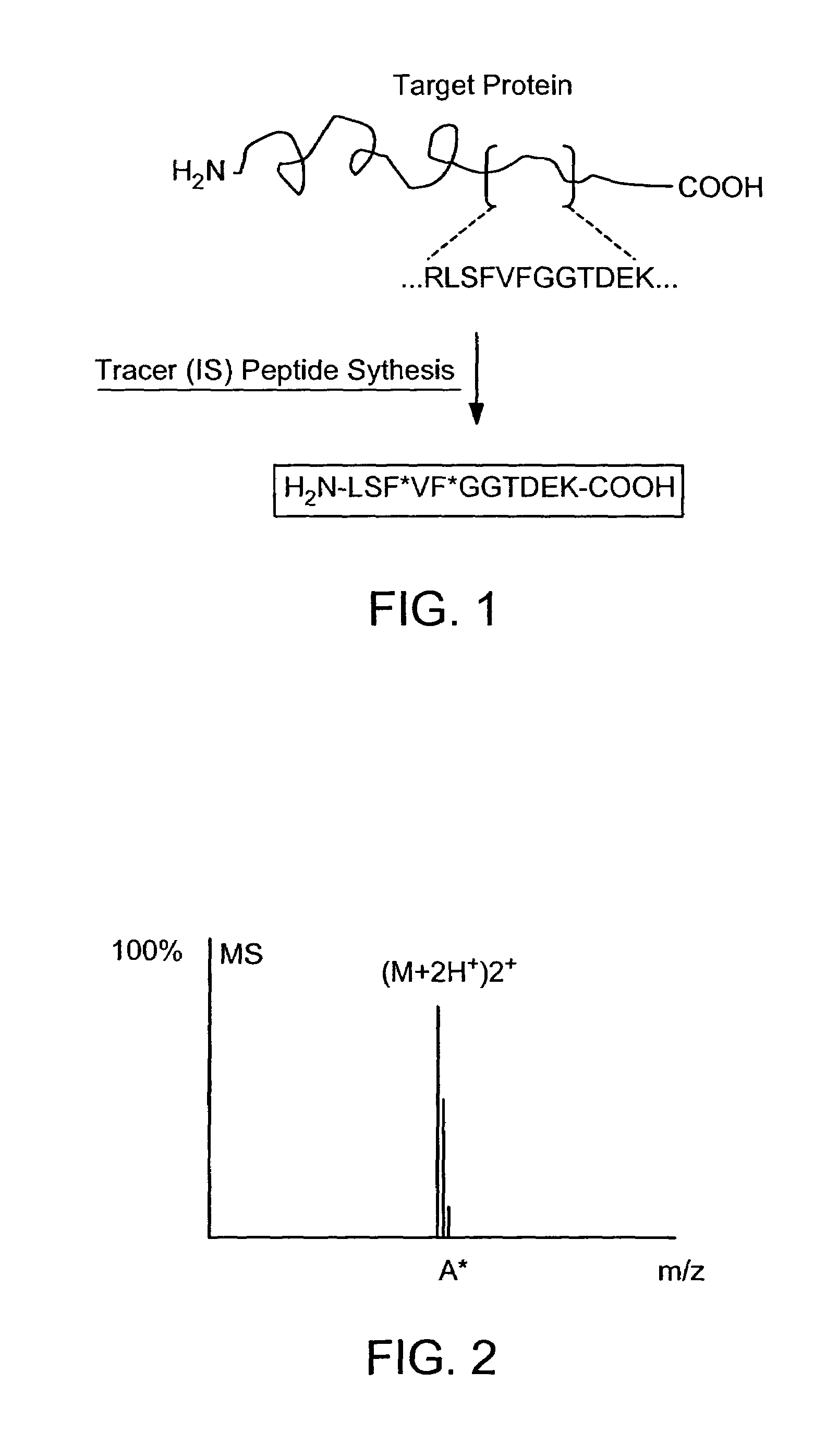
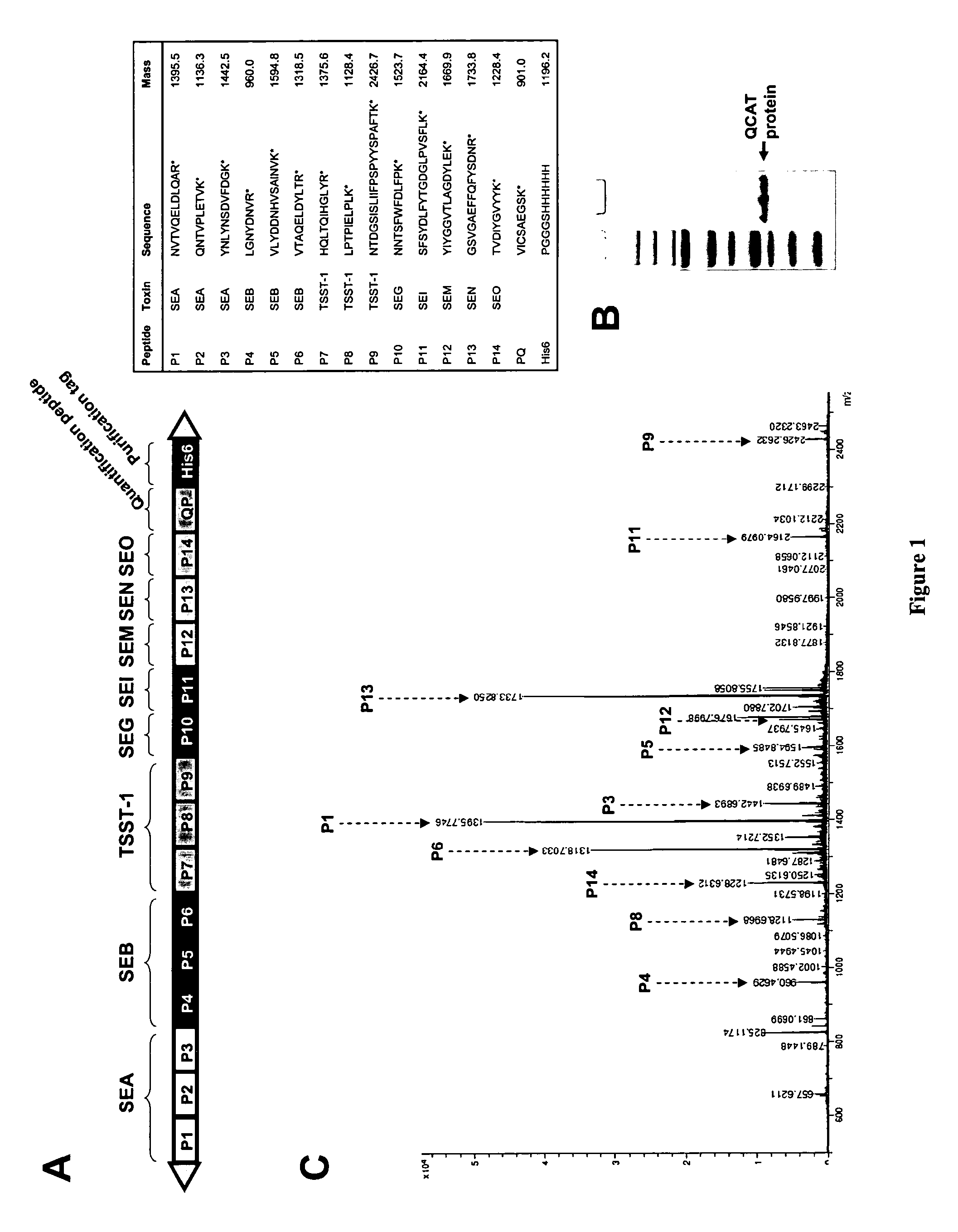
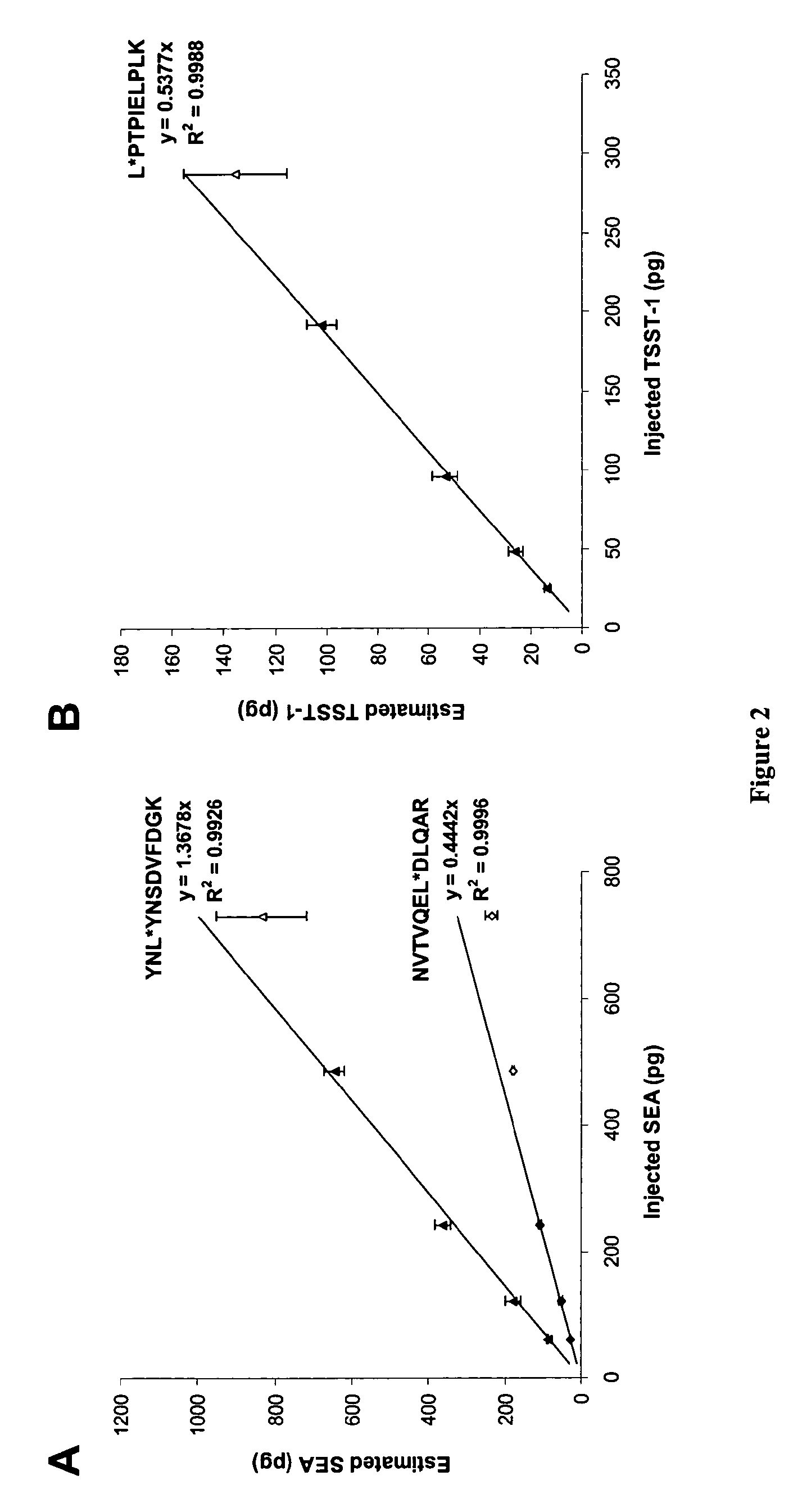
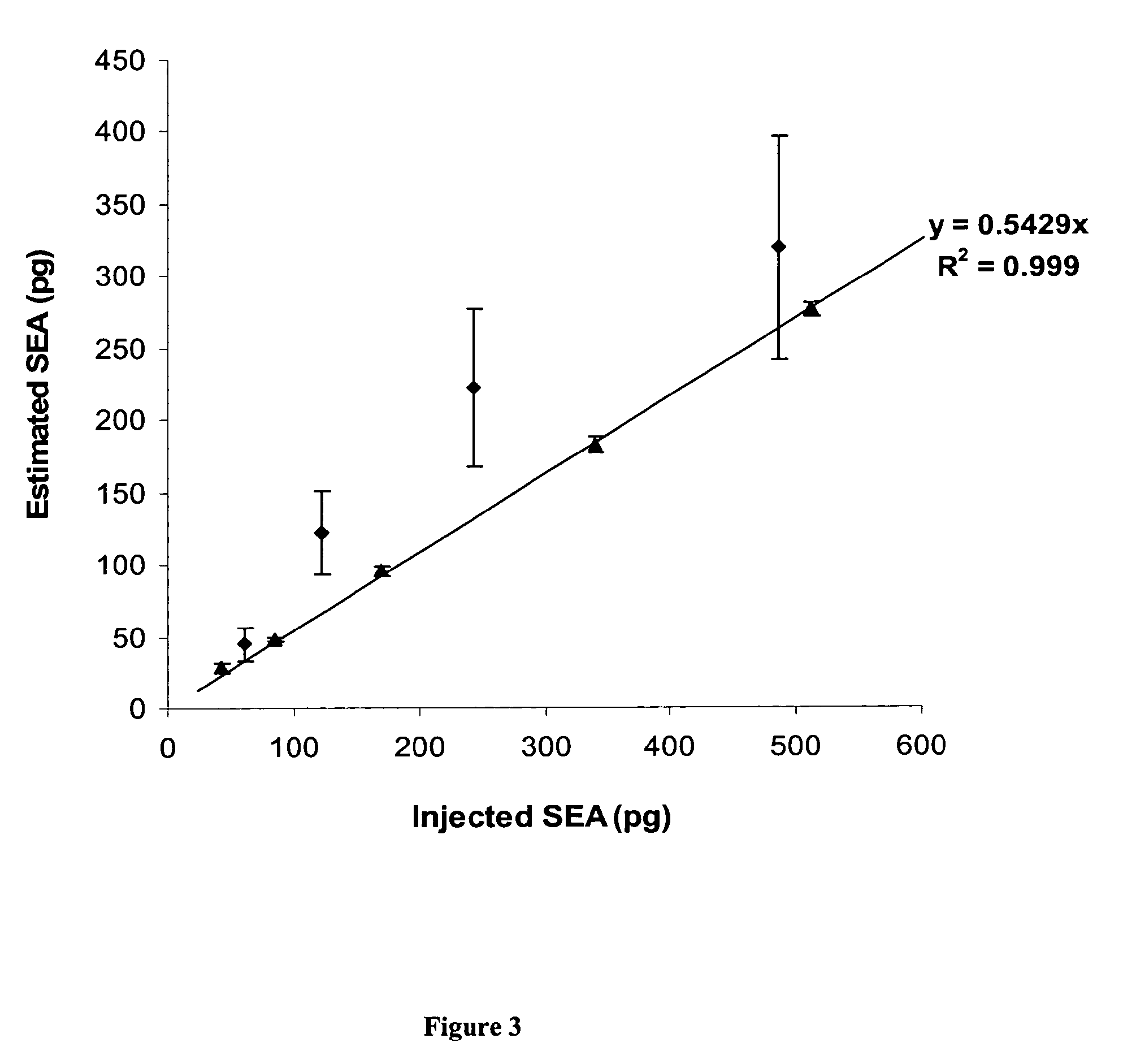
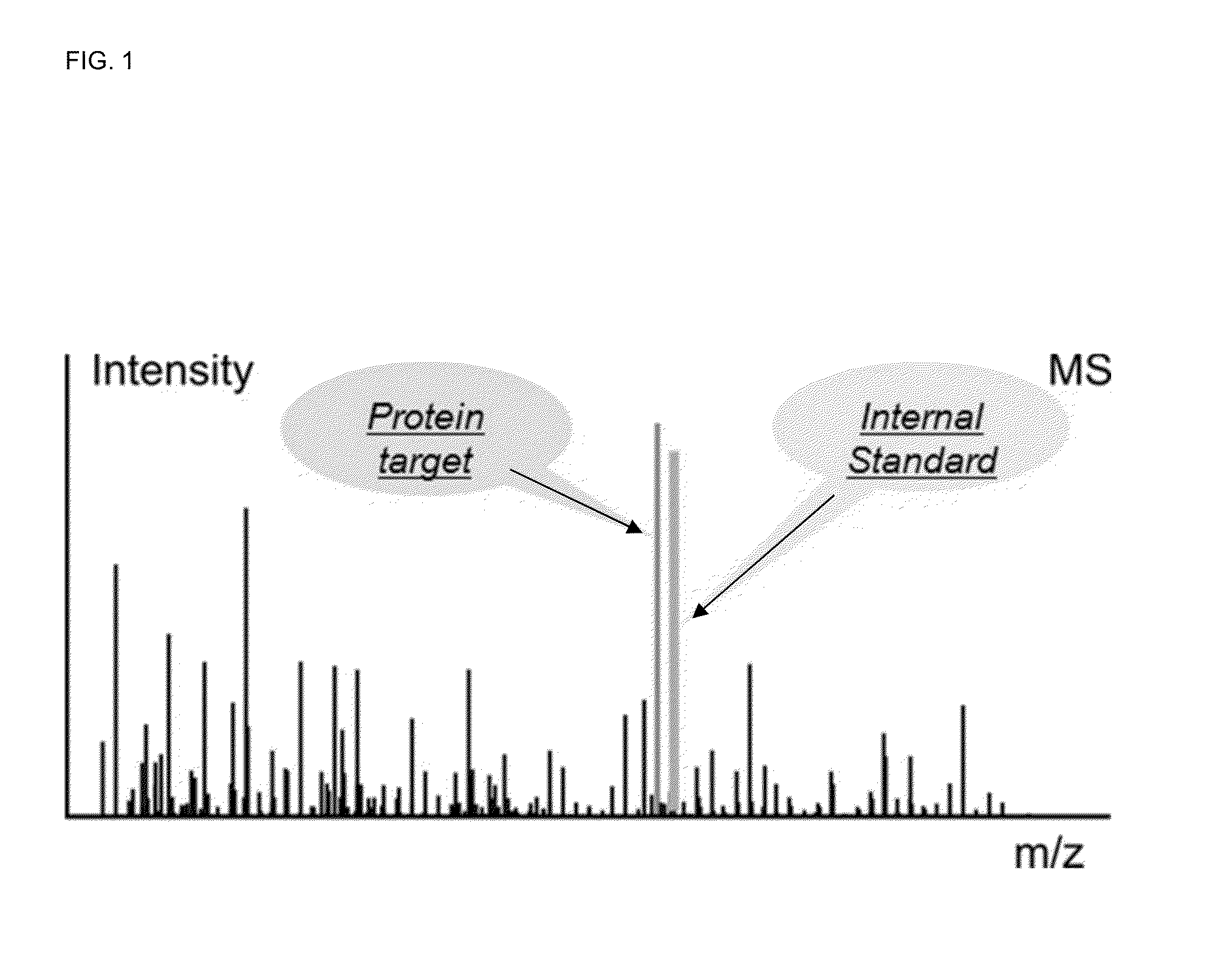
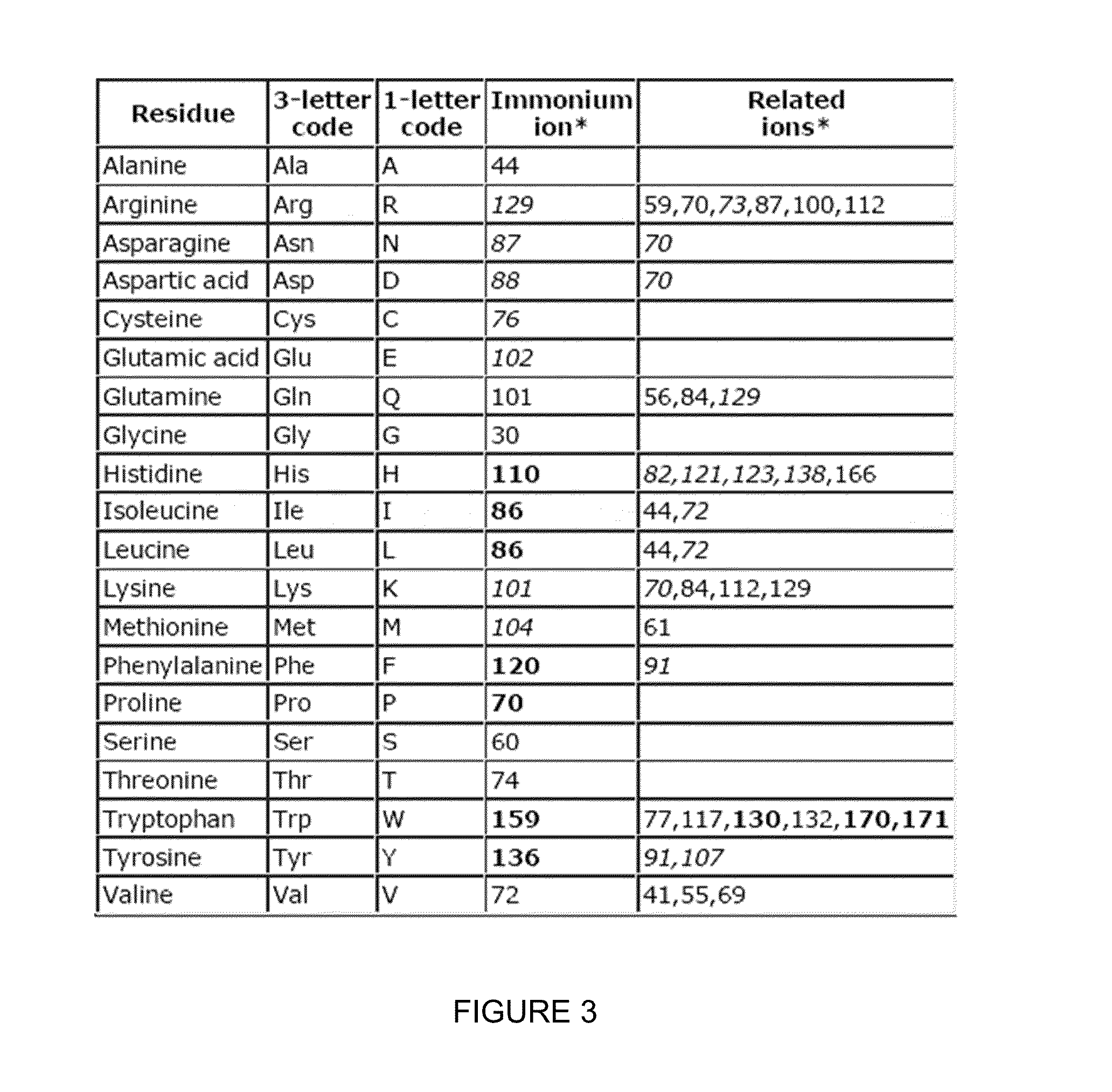
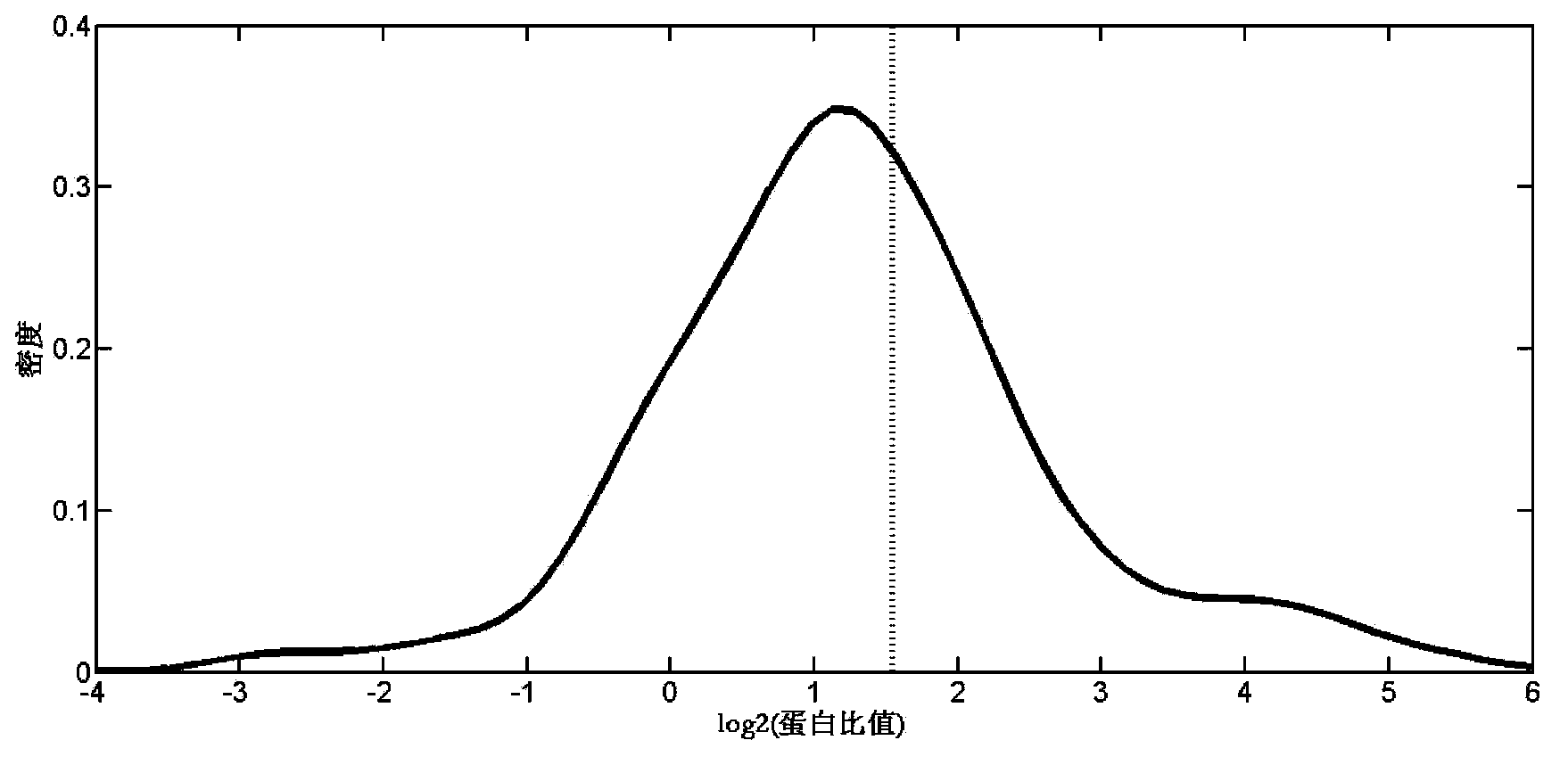
Absolute Quantification (AQUA) Absolute Quantification is a targeted quantitative proteomics technique that exhibits robust efficacy and is being increasingly utilized for a wide variety of quantitative proteomics studies. AQUA strategy is for the absolute quantification (AQUA) of proteins and their modification states.
Method for the efficiency-corrected real-time quantification of nucleic acids
InactiveUS6691041B2Microbiological testing/measurementRecombinant DNA-technologyHousekeeping geneInternal standard
The present invention concerns a method for the quantification of a target nucleic acid in a sample comprising the following steps: (i) determination of the amplification efficiency of the target nucleic acid under defined amplification conditions, (ii) amplification of the target nucleic acid contained in the sample under the same defined reaction conditions, (iii) measuring the amplification in real-time, (iv) quantification of the original amount of target nucleic acid in the sample by correction of the original amount derived from step (iii) with the aid of the determined amplification efficiency. The efficiency correction of PCR reactions according to the invention for the quantification of nucleic acids can be used for absolute quantification with the aid of an external or internal standard as well as for relative quantification compared to the expression of housekeeping genes.
Owner:ROCHE MOLECULAR SYST INC
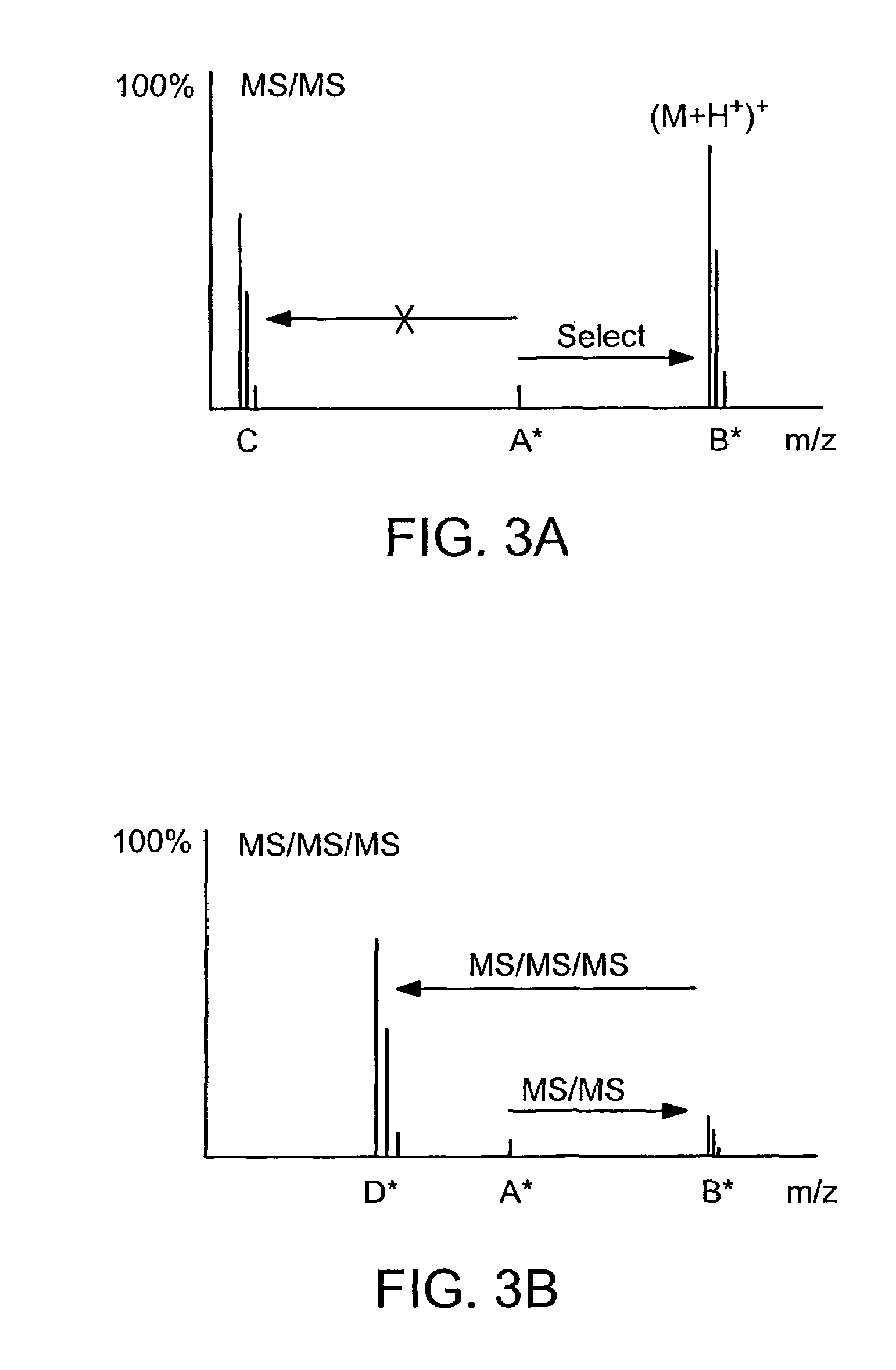
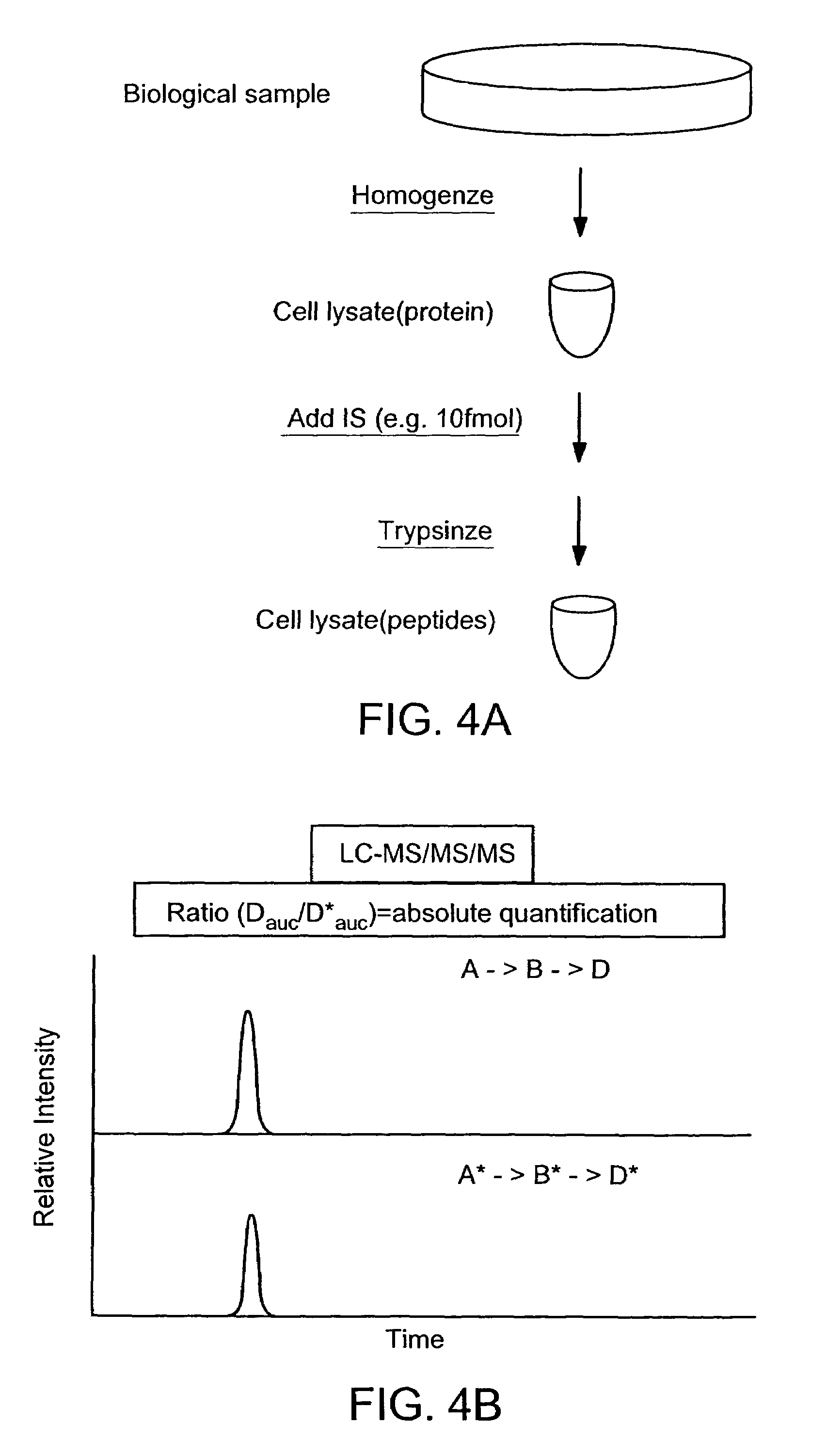
Absolute quantification of proteins and modified forms thereof by multistage mass spectrometry
ActiveUS7501286B2Rapid and high throughput analysisQuantitative precisionDepsipeptidesPeptide preparation methodsStable Isotope LabelingIsotope
The invention provides reagents, kits and methods for detecting and / or quantifying proteins in complex mixtures, such as a cell lysate. The methods can be used in high throughput assays to profile cellular proteomes. In one aspect, the invention provides a peptide internal standard labeled with a stable isotope and corresponding in amino acid sequence to the amino acid sequence of a subsequence of a target polypeptide. In another aspect, the peptide internal standard is labeled at a modified amino acid residue and is used to determine the presence of, and / or quantitate the amount of a particular modified form of a protein.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Method for the efficiency-corrected real-time quantification of nucleic acids
InactiveUS20030165832A1Minimize the differenceDifference in efficiencyMicrobiological testing/measurementRecombinant DNA-technologyHousekeeping geneInternal standard
The present invention concerns a method for the quantification of a target nucleic acid in a sample comprising the following steps: (i) determination of the amplification efficiency of the target nucleic acid under defined amplification conditions, (ii) amplification of the target nucleic acid contained in the sample under the same defined reaction conditions, (iii) measuring the amplification in real-time, (iv) quantification of the original amount of target nucleic acid in the sample by correction of the original amount derived from step (iii) with the aid of the determined amplification efficiency. The efficiency correction of PCR reactions according to the invention for the quantification of nucleic acids can be used for absolute quantification with the aid of an external or internal standard as well as for relative quantification compared to the expression of housekeeping genes.
Owner:ROCHE MOLECULAR SYST INC
Absolute quantification type digital nucleic acid analytic system based on efficient liquid drop microreactor
ActiveCN106434330AReduce usageSimplify complexityBioreactor/fermenter combinationsBiological substance pretreatmentsFluorescenceQuarantine
The invention relates to inspection and quarantine systems for biomedicine, agriculture, food hygiene and the like and particularly relates to an absolute quantification type digital nucleic acid analytic system based on an efficient liquid drop microreactor. The absolute quantification type digital nucleic acid analytic system is mainly used for detecting the molecular structure of nucleic acid so as to analyze attributes of organisms and belongs to the fields of biological and medical detection. The absolute quantification type digital nucleic acid analytic system comprises a confocal fluorescence image collecting system, a micro-fluidic chip, a temperature circulating heating system, an electric loading platform, an automatic sample feeding device, a controller and a computer. The formation and PCR amplification of micro-droplets are finished by virtue of the same micro-fluidic chip, namely the micro-fluidic chip has a droplet formation function and a PCR amplification (microreactor) function. According to the absolute quantification type digital nucleic acid analytic system, the complexity of the absolute quantification type nucleic acid analysis process is reduced, the nucleic acid analysis process is finished in a one-button manner, and culture and bacteria-proliferating processes are omitted, so that the rapid detection is realized, the sensitivity of the absolute quantification type nucleic acid detection is greatly improved, meanwhile, the sample consumption of the absolute quantification type nucleic acid analysis is substantially reduced, and the high throughput detection is realized.
Owner:SUZHOU HOTTOP INN INSTR TECH CO LTD CHINA +2
Method for Absolute Quantification of Polypeptides
InactiveUS20100173786A1Reduce yieldHigh yieldPeptide librariesMicrobiological testing/measurementAbsolute quantificationBiology
Owner:INST NAT DE LA SANTE & DE LA RECHERCHE MEDICALE (INSERM) +1
Absolute quantitation of proteins and protein modifications by mass spectrometry with multiplexed internal standards
ActiveUS20140364337A1Improve accuracyHigh sensitivityPeptide librariesLibrary screeningSpectroscopyMass spectrometry imaging
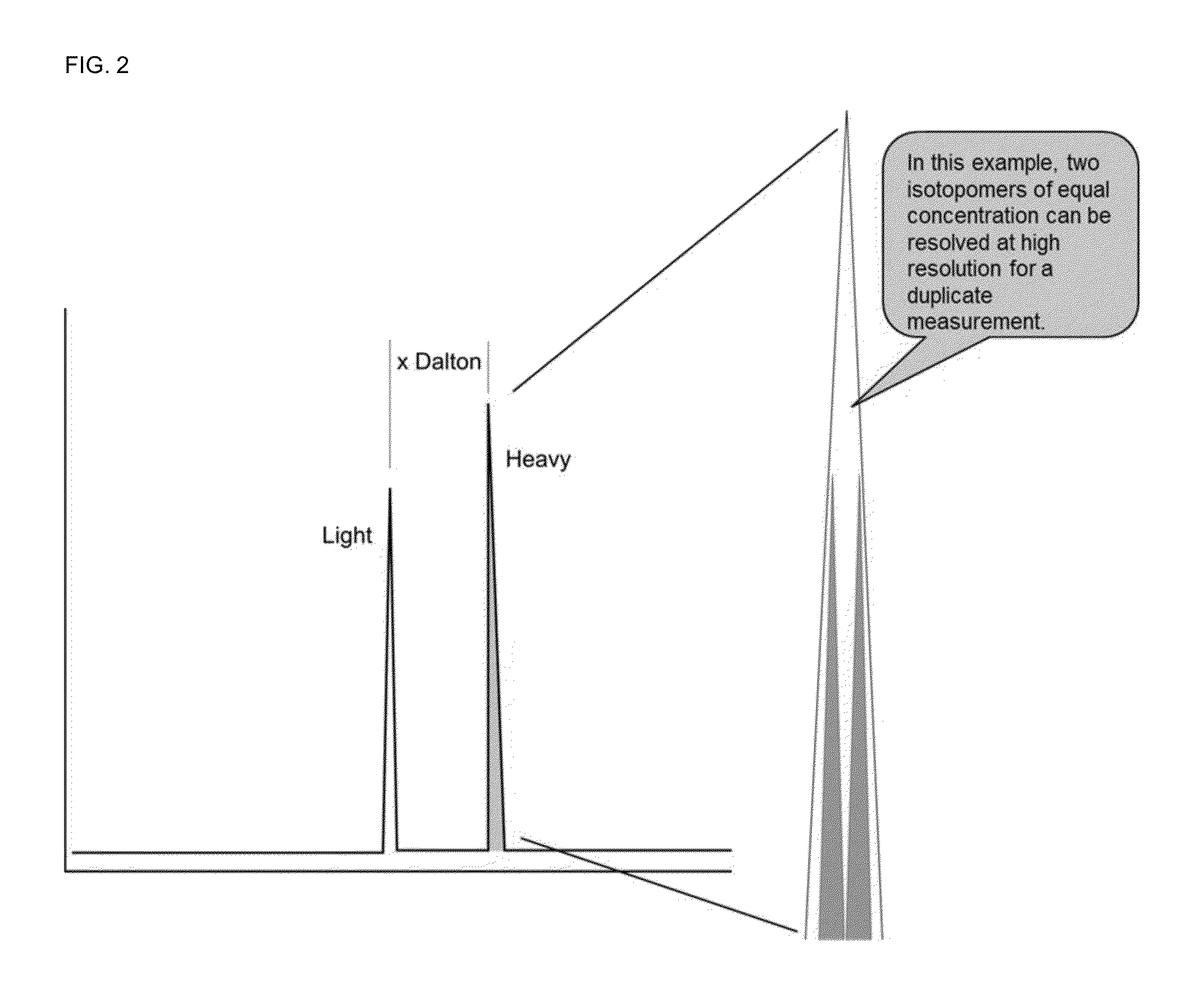
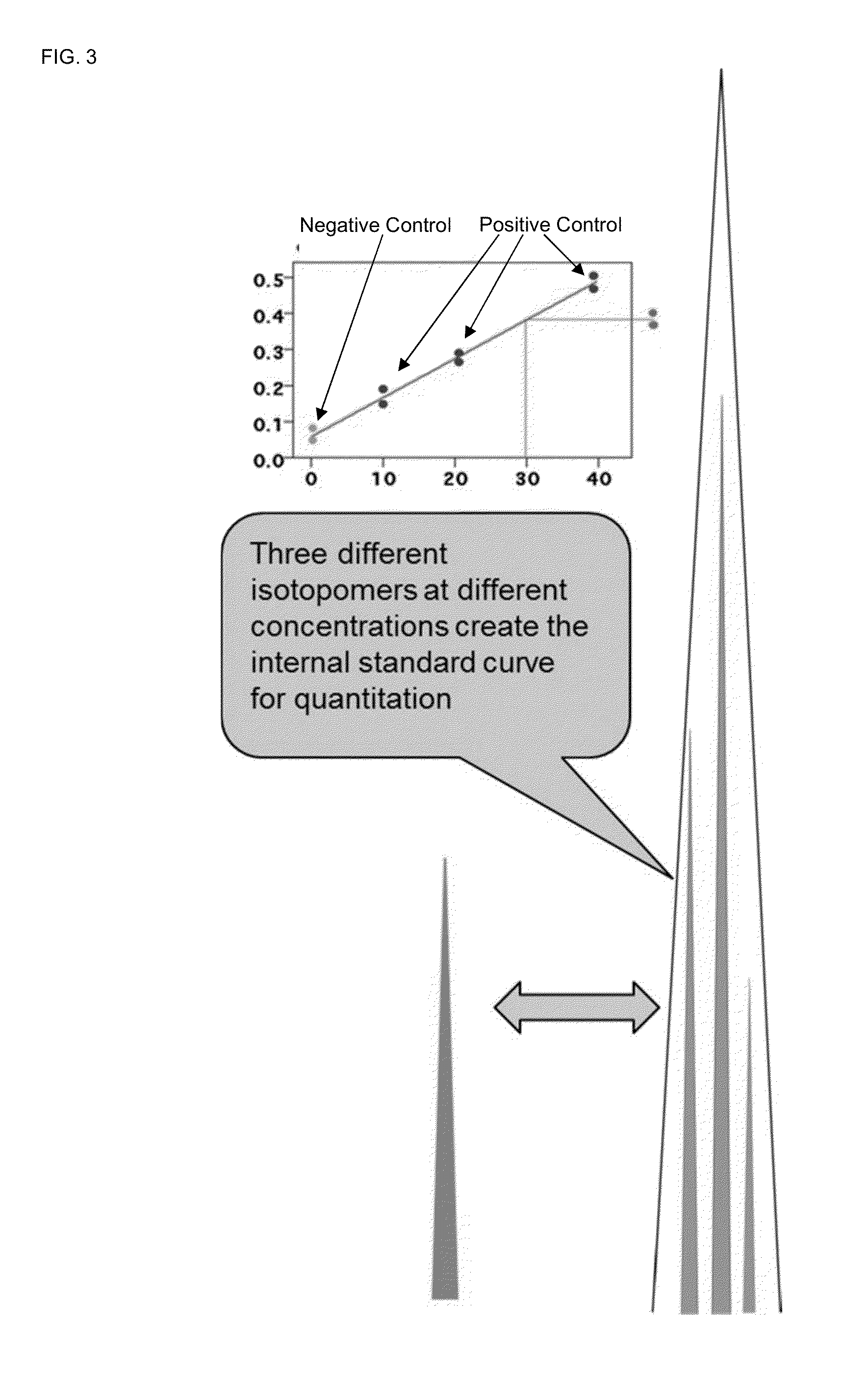
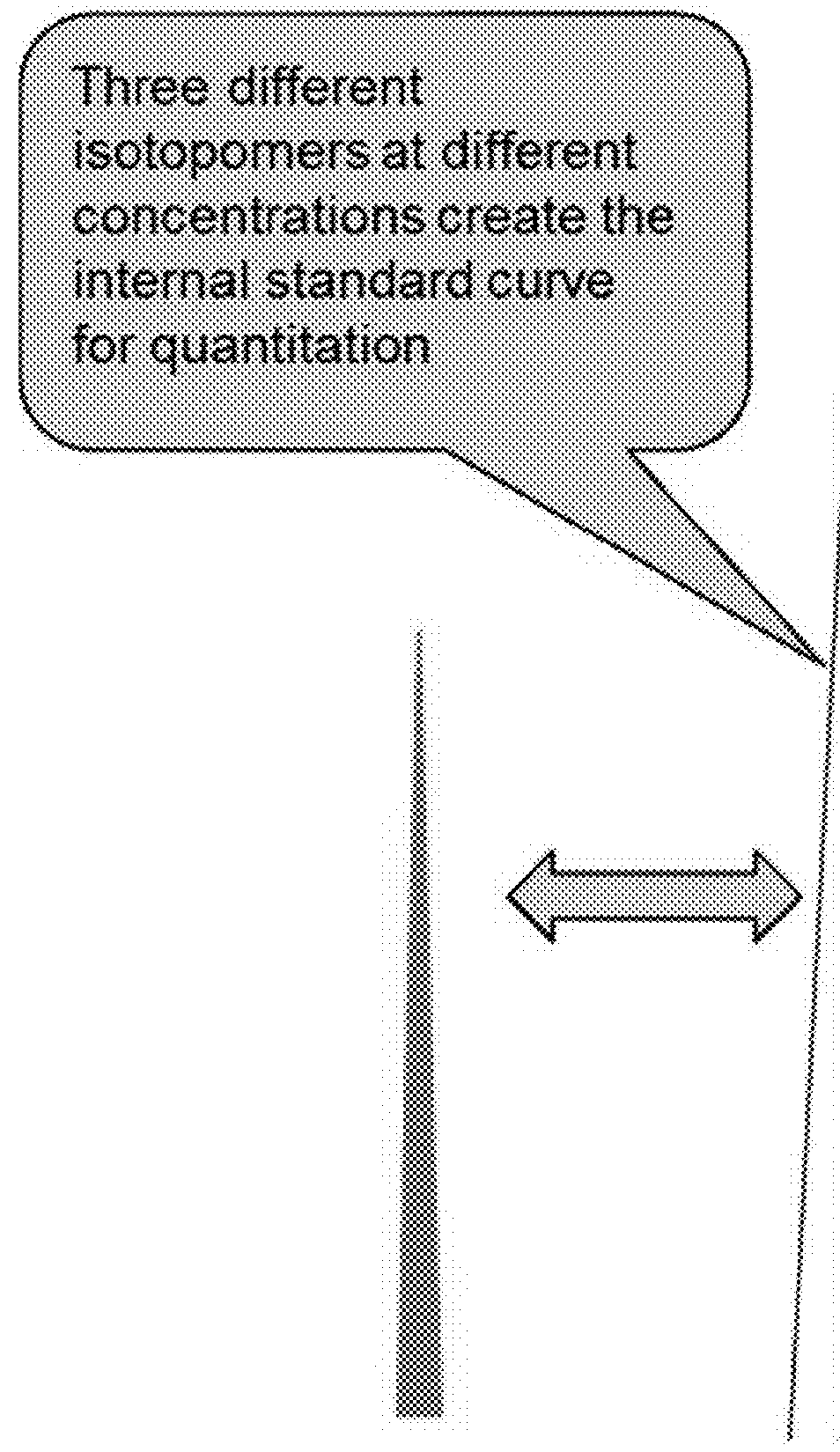
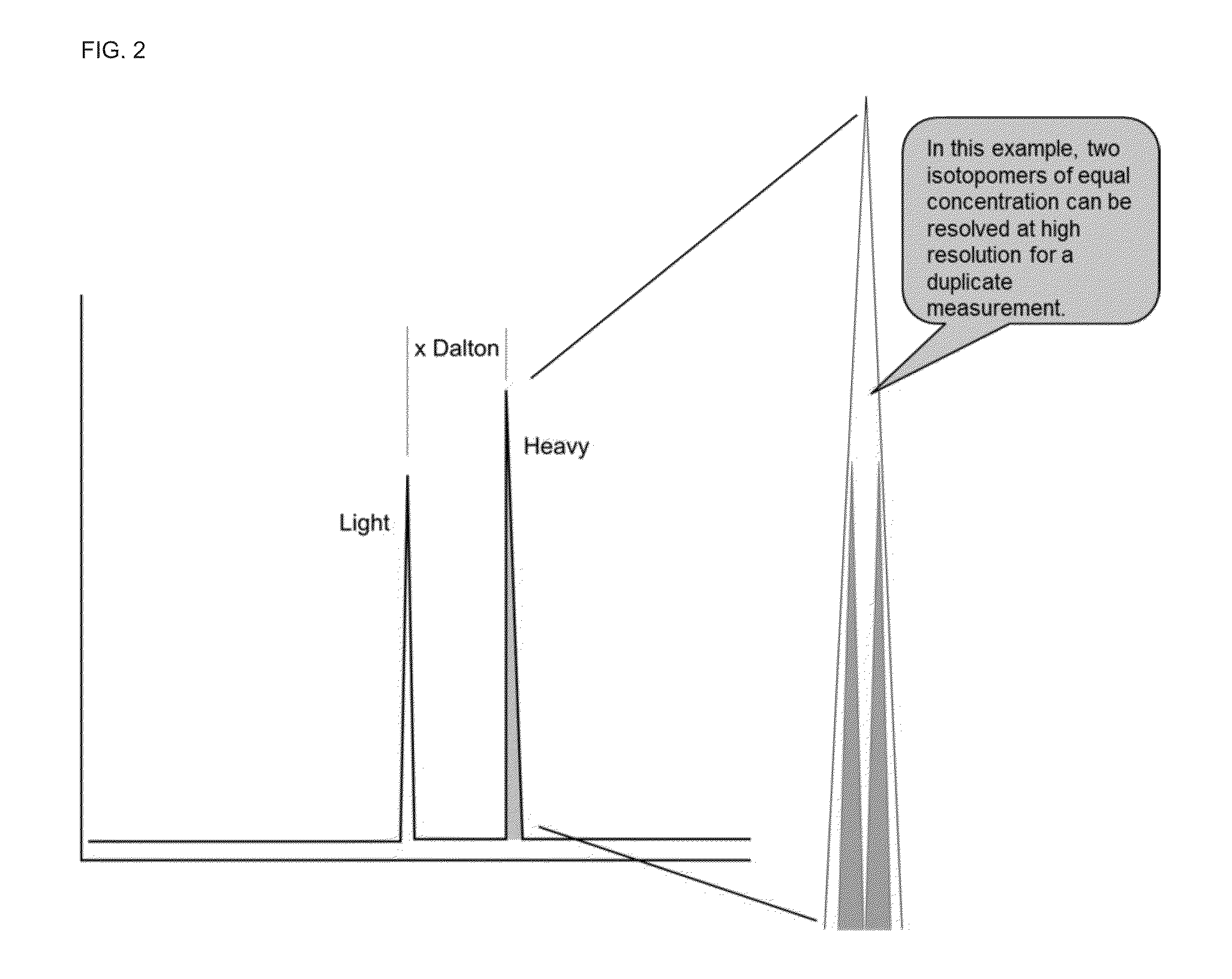
A method for absolute protein or peptide quantitation by mass spectroscopy. A sample containing a protein or peptide of interest is prepared for mass spectroscopy analysis. The sample is subjected to mass spectroscopy analysis at low resolution whereby a single additive mass spectroscopy peak is obtained, then is subjected to high resolution mass spectroscopy analysis whereby a plurality of mass spectroscopy peaks are obtained. The intensity of each of the plurality of mass spectroscopy peaks is quantitated either by comparison to an internal standard set, or by using a standard curve generated for each isotopologue set. Quantitation using a standard curve enhances quantitation across a dynamic range of analyte.
Owner:PIERCE BIOTECHNOLOGY
Analysis of protein isoforms using unique tryptic peptides by mass spectrometry and immunochemistry
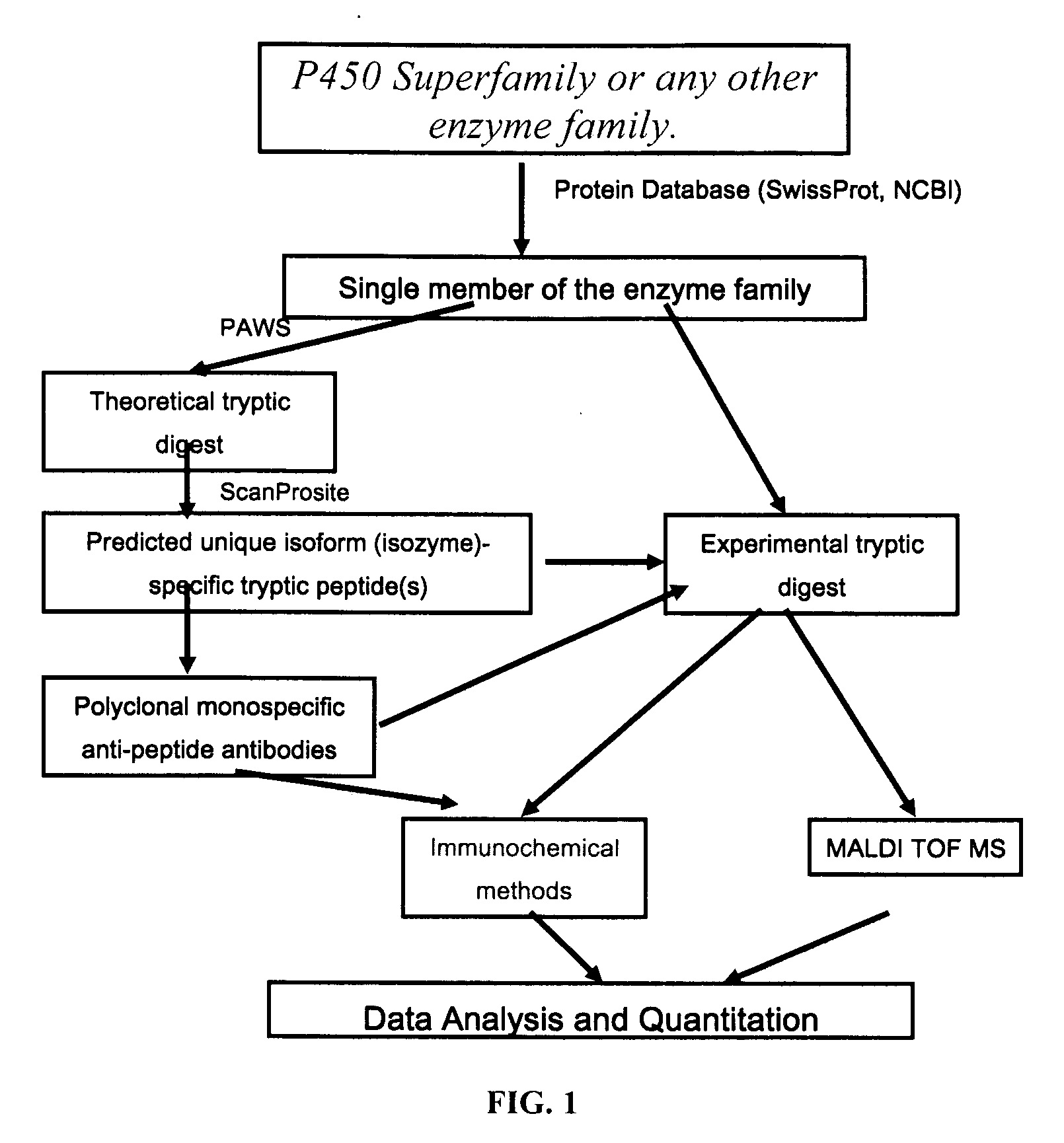
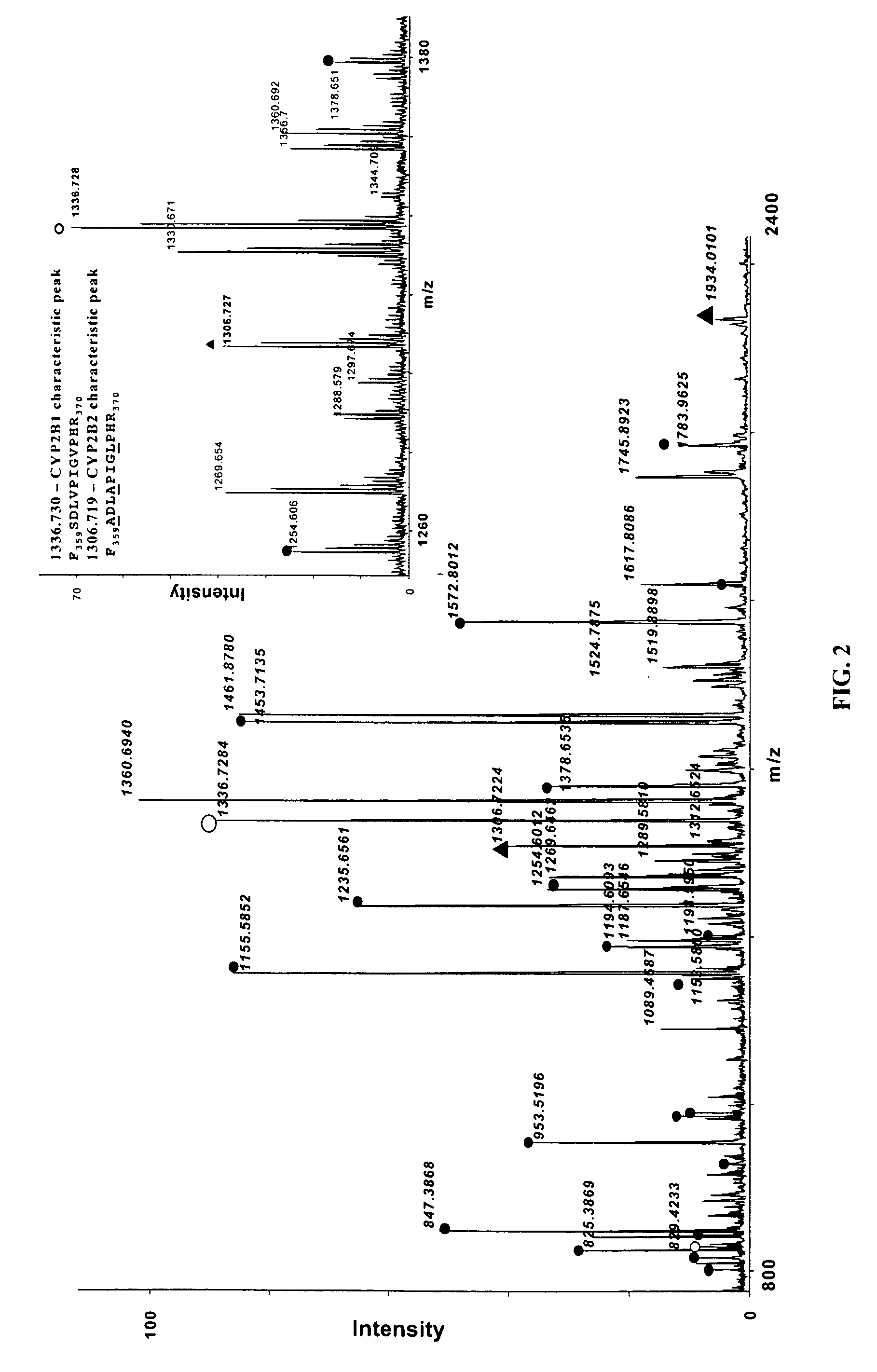
InactiveUS20070092926A1Accurate measurementReliable detectionMicrobiological testing/measurementBiological material analysisIsozymeMass Spectrometry-Mass Spectrometry
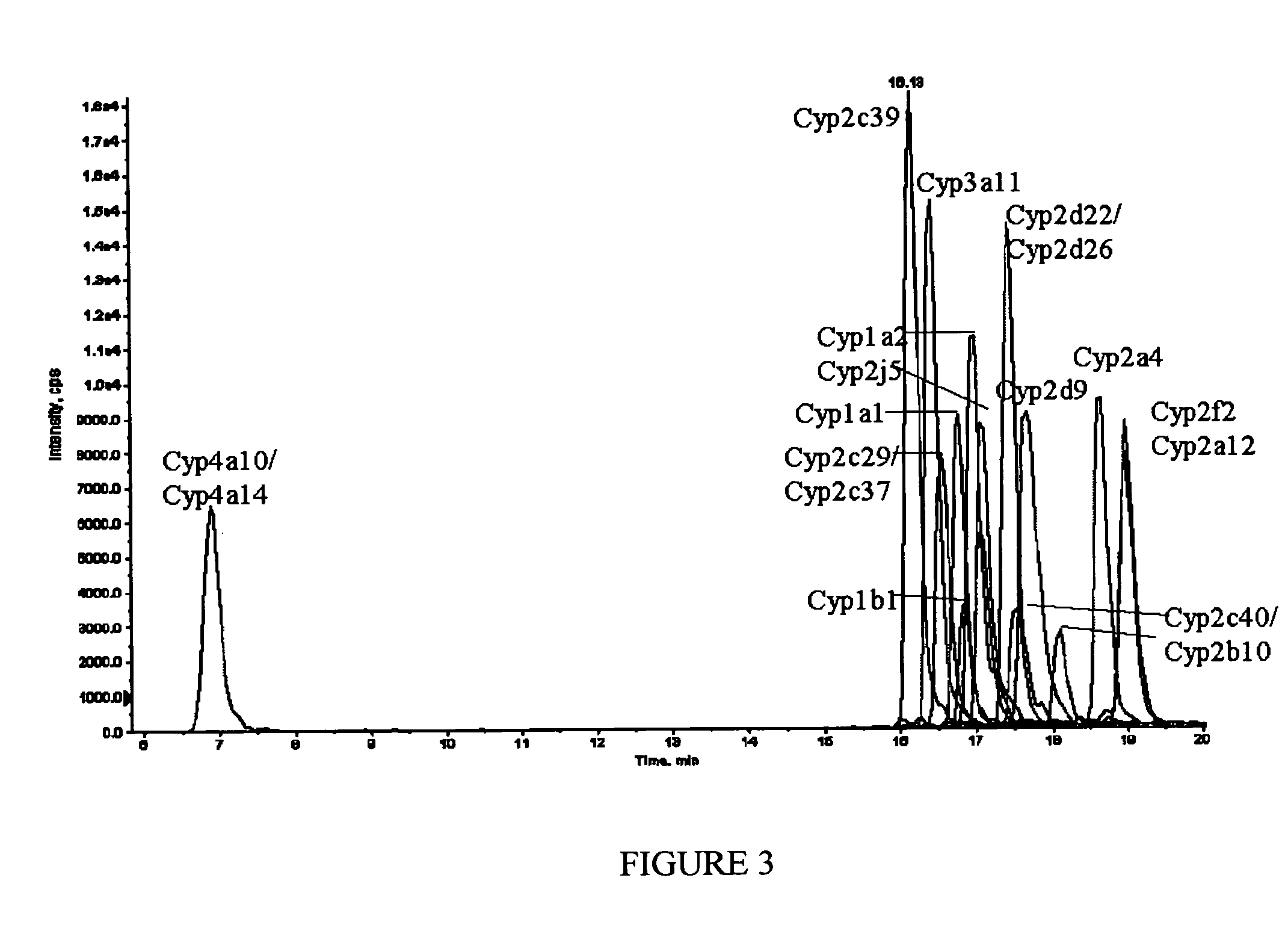
A method for qualitatively and quantitatively detecting a protein isoform (p450 isozyme) in a sample using MALDI-TOF mass spectrometry or immunochemistry using a unique proteolytic peptide for the isoform. Relative and absolute quantitation can be performed using calibration curves with P450 isozyme-specific peptide standards.
Owner:KANSAS UNIV OF
Protein characteristic spectrum of active tuberculosis in children and method for creating protein characteristic spectrum
The invention discloses a protein characteristic spectrum of active tuberculosis in children and a method for creating the protein characteristic spectrum. The protein characteristic spectrum of the active tuberculosis in the children is obtained by comparing proteomics difference of an active tuberculosis group and a healthy control group. In addition, the invention further relates to the method for creating the protein characteristic spectrum of the active tuberculosis in the children, protein in blood plasma can be effectively identified and relatively quantified by utilizing a non-marked tandem mass spectrum technology of relative and absolute quantification, and a protein expression difference mass spectrum in the blood plasma of a patient suffering from the active tuberculosis in the children can be obtained by adopting the technology. A series of discovered proteins provide a foundation and resources for searching new more ideal markers; compared with a conventional blood plasma detection method, the method has relatively high sensitivity and specificity, and can be used for screening drugs for the active tuberculosis in the children; at the same time, a new way is provided for a mechanism for exploring occurrence and development of diseases.
Owner:BEIJING CHILDRENS HOSPITAL AFFILIATED TO CAPITAL MEDICAL UNIV
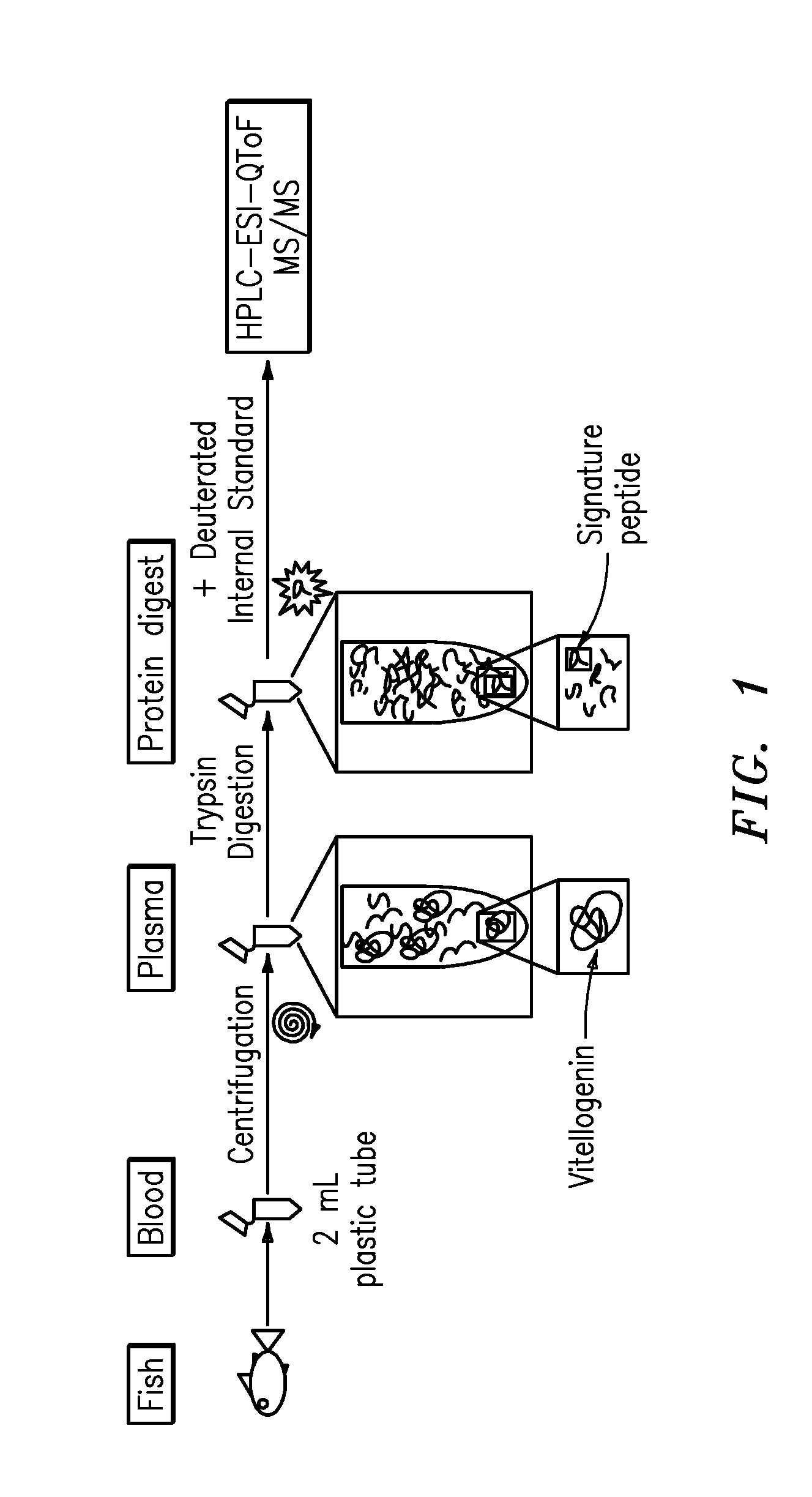
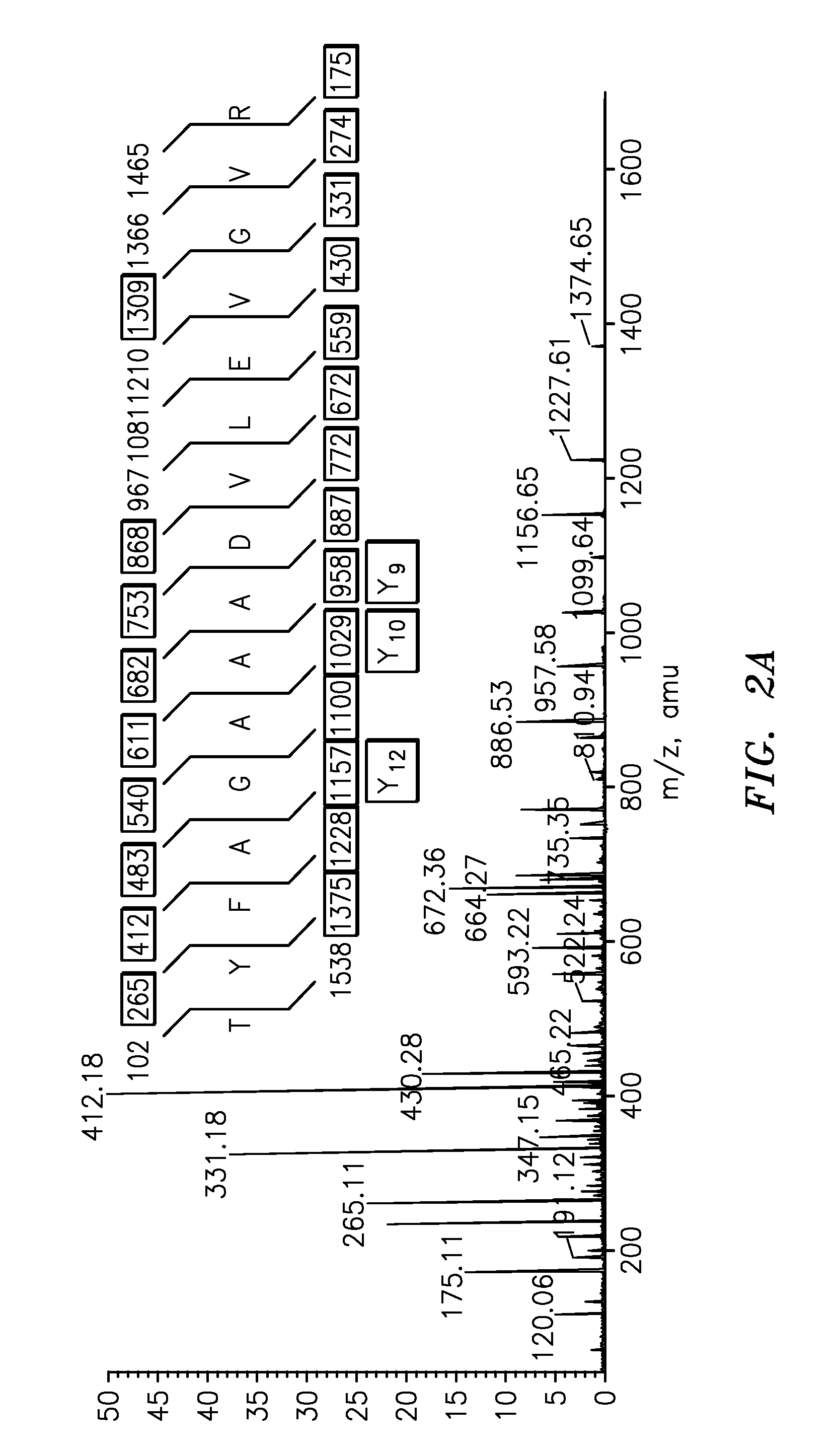
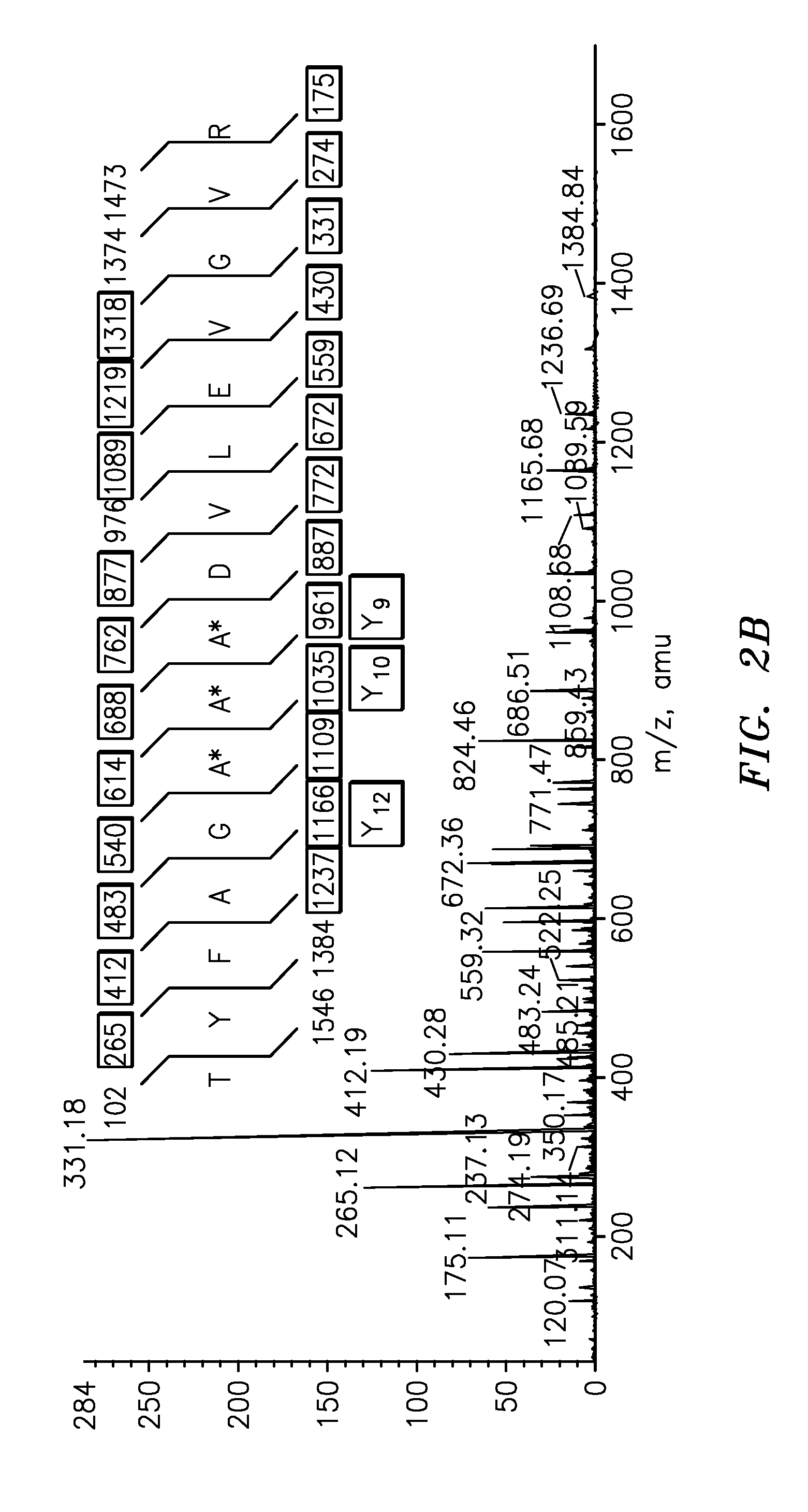
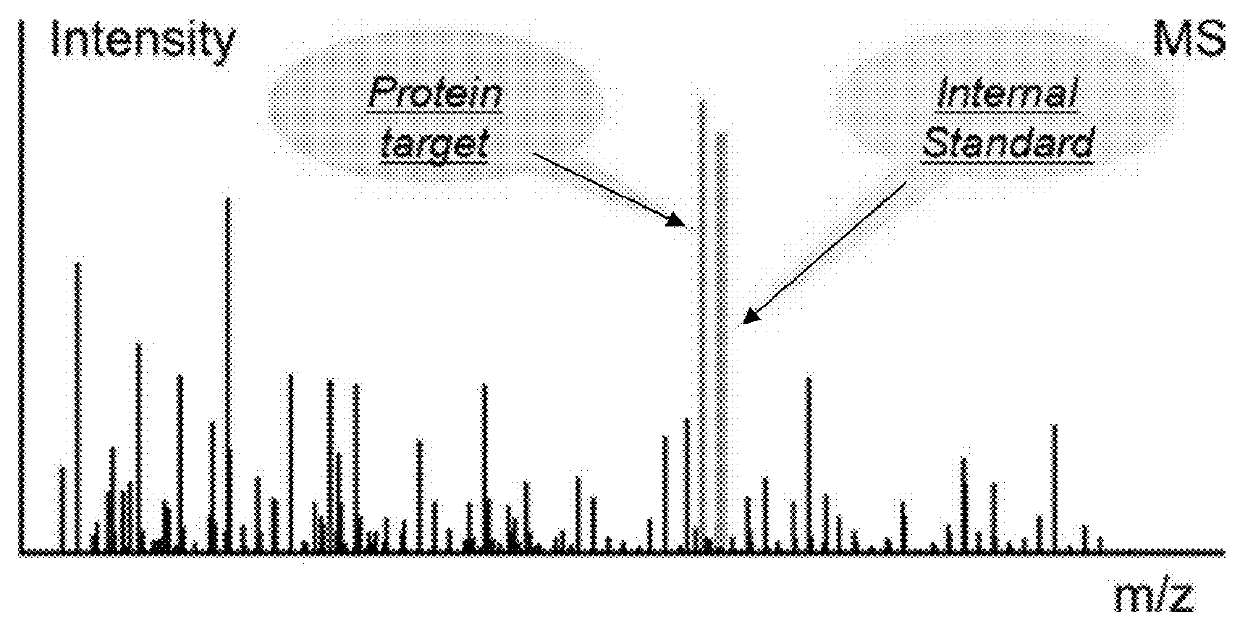
Quantification of vitellogenin
InactiveUS20090011447A1Simple methodMicrobiological testing/measurementPeptidesVitellogeninsGlu-Val-Gly
The present invention is directed to a simple method for absolute quantification of plasma vitellogenin from two or more different fish species such as Rainbow trout and Atlantic salmon, or Atlantic cod and haddock. In the case of Rainbow trout and Atlantic salmon, plasma samples obtained from control and β-estradiol induced fish were digested with trypsin. A characteristic ‘signature peptide’ was selected and analyzed by high performance liquid chromatography coupled to an electrospray quadrupole-time-of-flight tandem mass spectrometer, using a deuterated homologue peptide as an internal standard. The hybrid tandem mass spectrometer was operated in a ‘pseudo’ selected reaction monitoring mode by which three diagnostic product ions were monitored for identification and quantification purposes. The reproducibility (coefficient of variation ˜5%) and sensitivity (limit of quantification of 0.009 mg / mL) achieved by this simple assay allow it to be considered as an alternative to immunological assays. In the case of Atlantic cod and haddock, the amino acid sequence of the vitellogenin protein has not yet been determined, but, the Atlantic cod vitellogenin has been characterized using a ‘bottom-up’ mass spectrometric approach. Vitellogenin synthesis was induced ‘in vivo’ with β-Estradiol, and subjected to trypsin digestion for characterization by matrix-assisted laser desorption / ionization-Quadrupole-Time-of-flight tandem mass spectrometry. A peptide mass fingerprint was obtained and ‘de novo’ sequencing of the most abundant tryptic peptides was performed by low energy collision induced dissociation-tandem mass spectrometry. Thus, the sequences of various tryptic peptides have been elucidated. It has also been determined that Atlantic cod vitellogenin shares a series of common peptides with the two different known vitellogenin sequences of Haddock, a closely related species. There are also disclosed novel isolated signature peptides, namely Thr-Tyr-Phe-Ala-Gly-Ala-Ala-Ala-Asp-Val-Leu-Glu-Val-Gly-Val-Arg, Asp Leu Gly Leu Ala Tyr Thr Glu Lys, Phe Phe Gly Gln Glu Ile Ala Asn Ile Asp Lys, Glu Ile Val Leu Leu Gly Tyr Gly Thr Met Ile Ser Lys and Tyr Glu Ser Phe Ala Val Ala Arg.
Owner:BANOUB JOSEPH H +2
Absolute quantitation of proteins and protein modifications by mass spectrometry with multiplexed internal standards
ActiveUS9252003B2Fast and accurate and precise quantitationImprove accuracyParticle spectrometer methodsMass spectrometersSpectroscopyMass spectrometry imaging
A method for absolute protein or peptide quantitation by mass spectroscopy. A sample containing a protein or peptide of interest is prepared for mass spectroscopy analysis. The sample is subjected to mass spectroscopy analysis at low resolution whereby a single additive mass spectroscopy peak is obtained, then is subjected to high resolution mass spectroscopy analysis whereby a plurality of mass spectroscopy peaks are obtained. The intensity of each of the plurality of mass spectroscopy peaks is quantitated either by comparison to an internal standard set, or by using a standard curve generated for each isotopologue set. Quantitation using a standard curve enhances quantitation across a dynamic range of analyte.
Owner:PIERCE BIOTECHNOLOGY
Expression quantification using mass spectrometry
InactiveUS20060078960A1Easy to identifyHelp studyParticle separator tubesMicrobiological testing/measurementMass spectrometry imagingProtein mass spectrometry
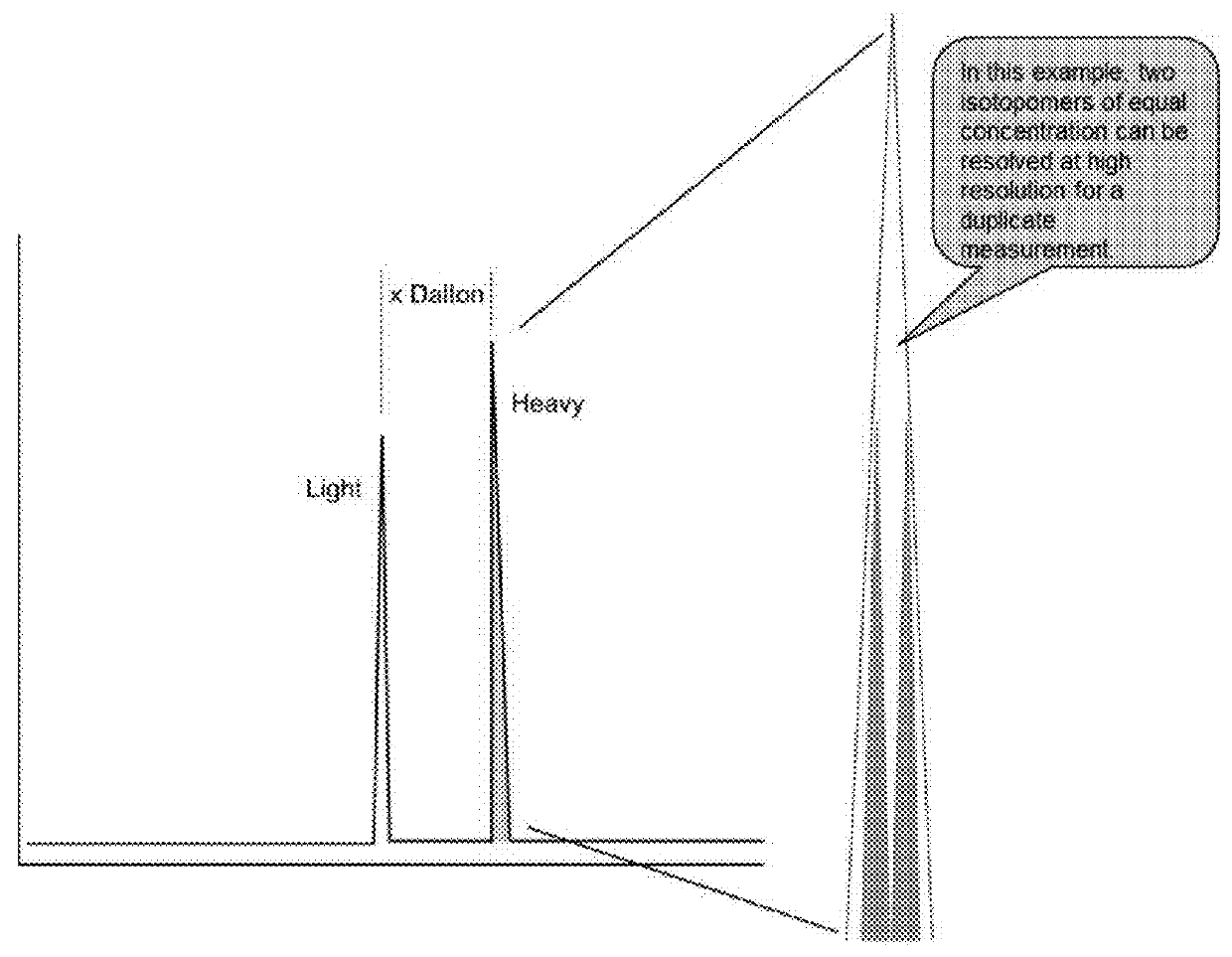
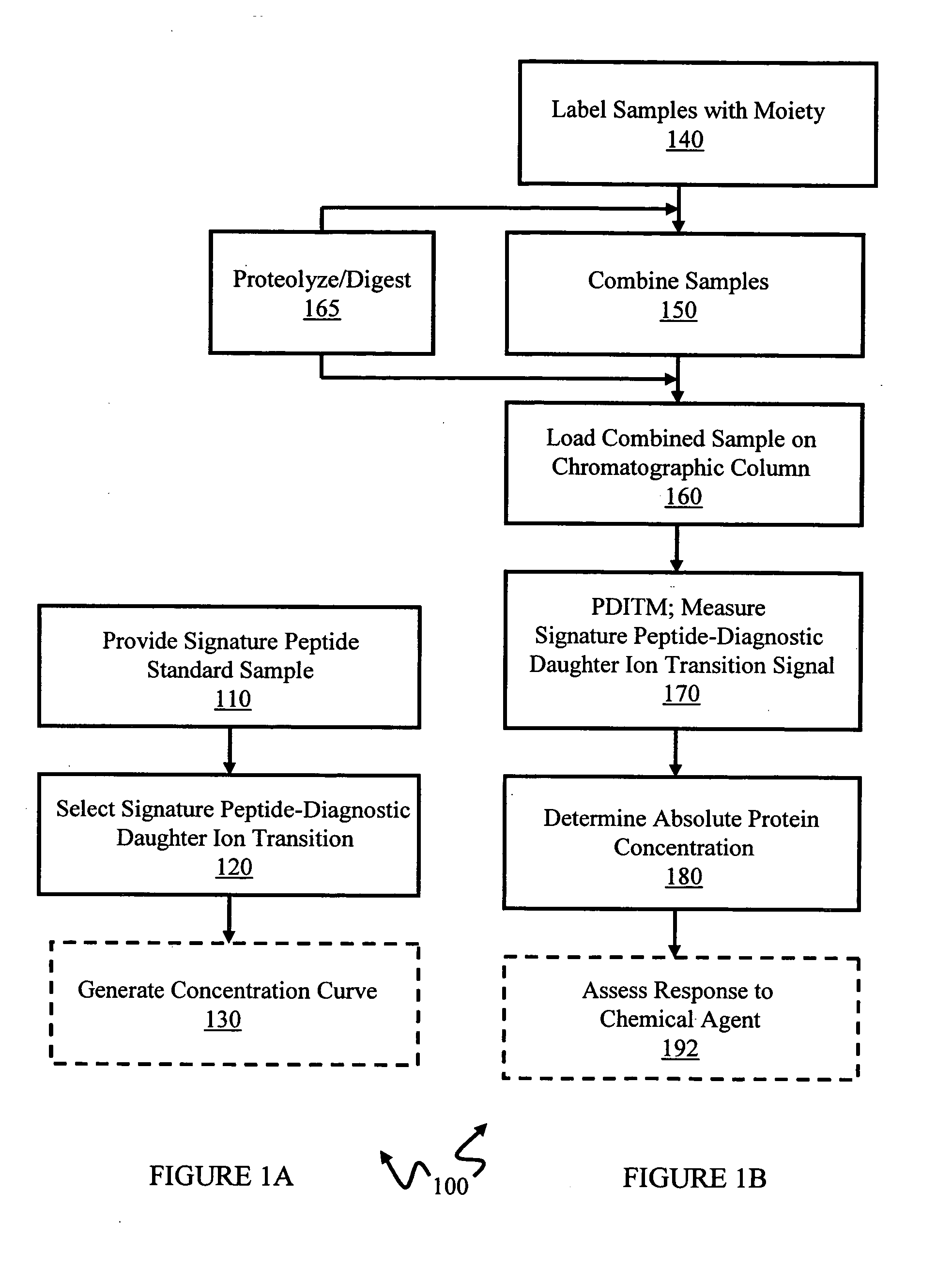
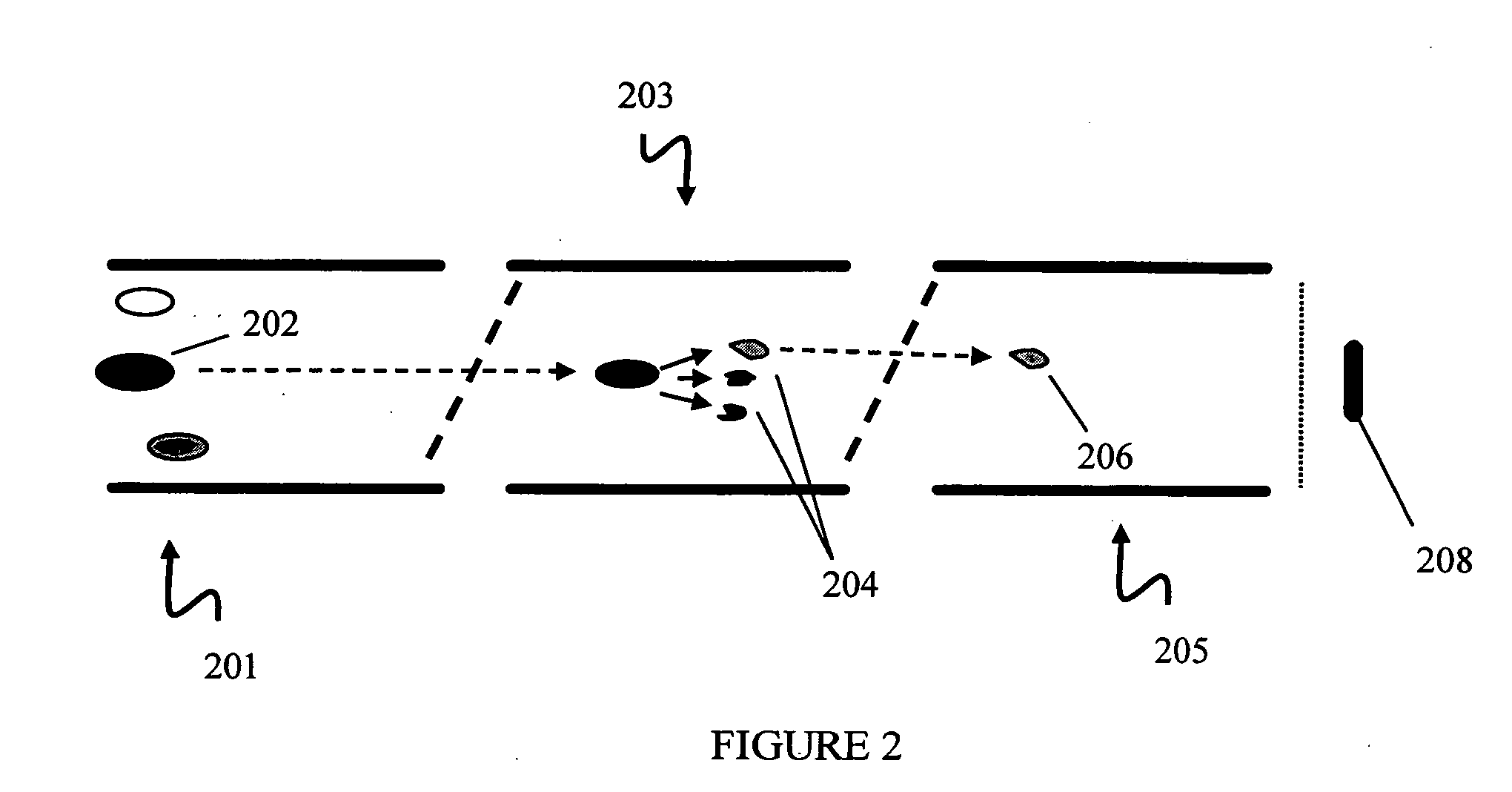
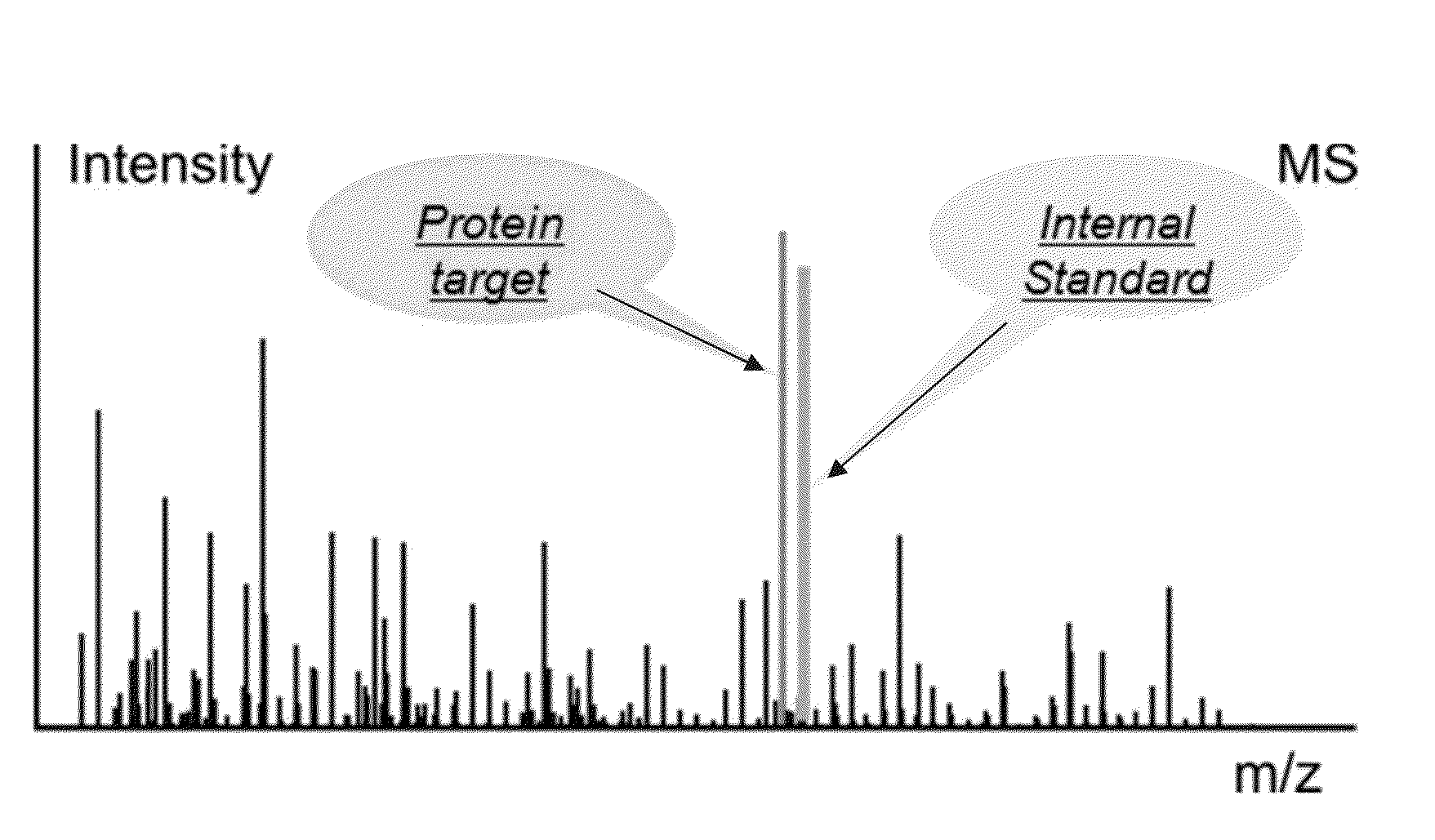
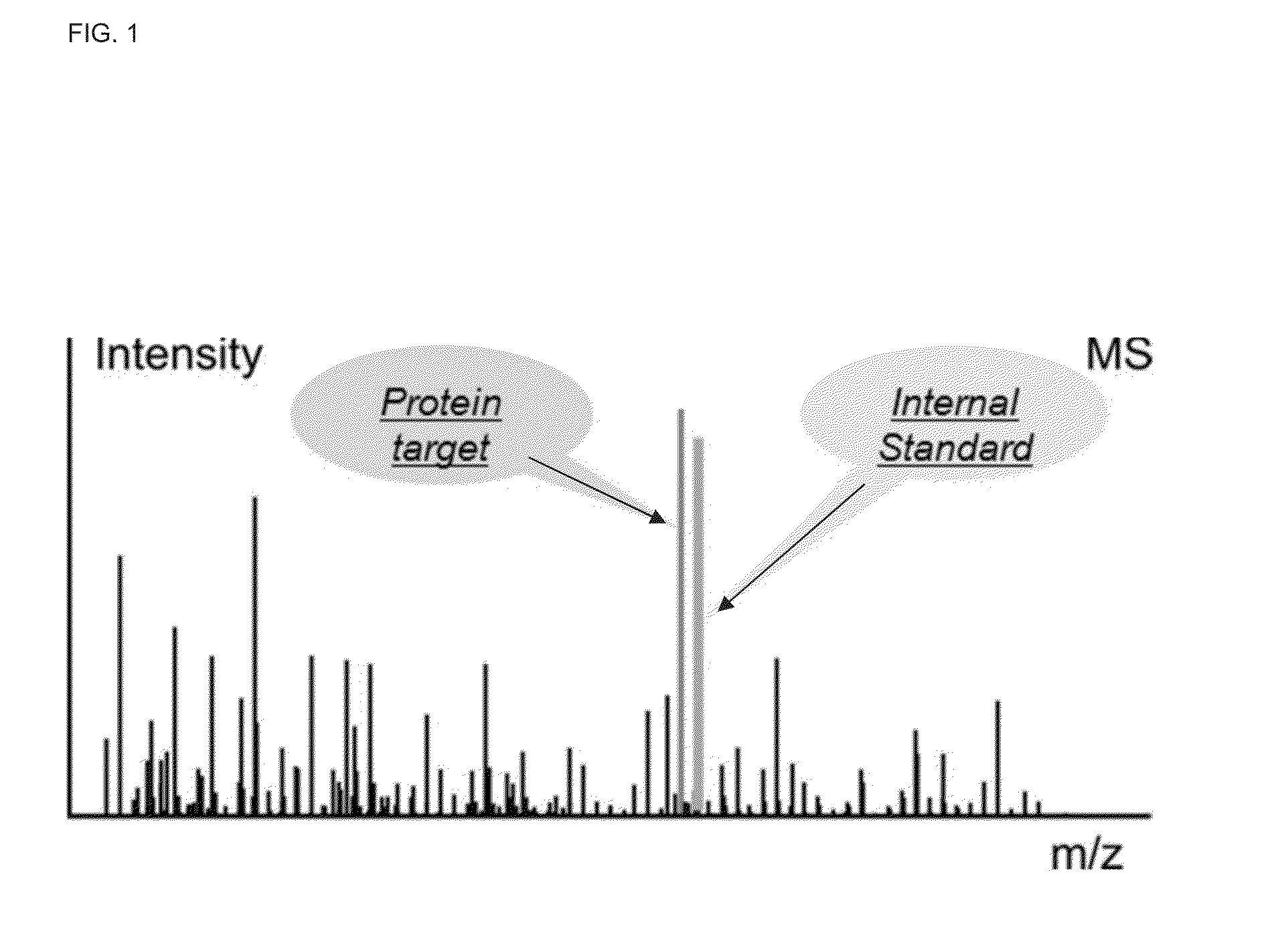
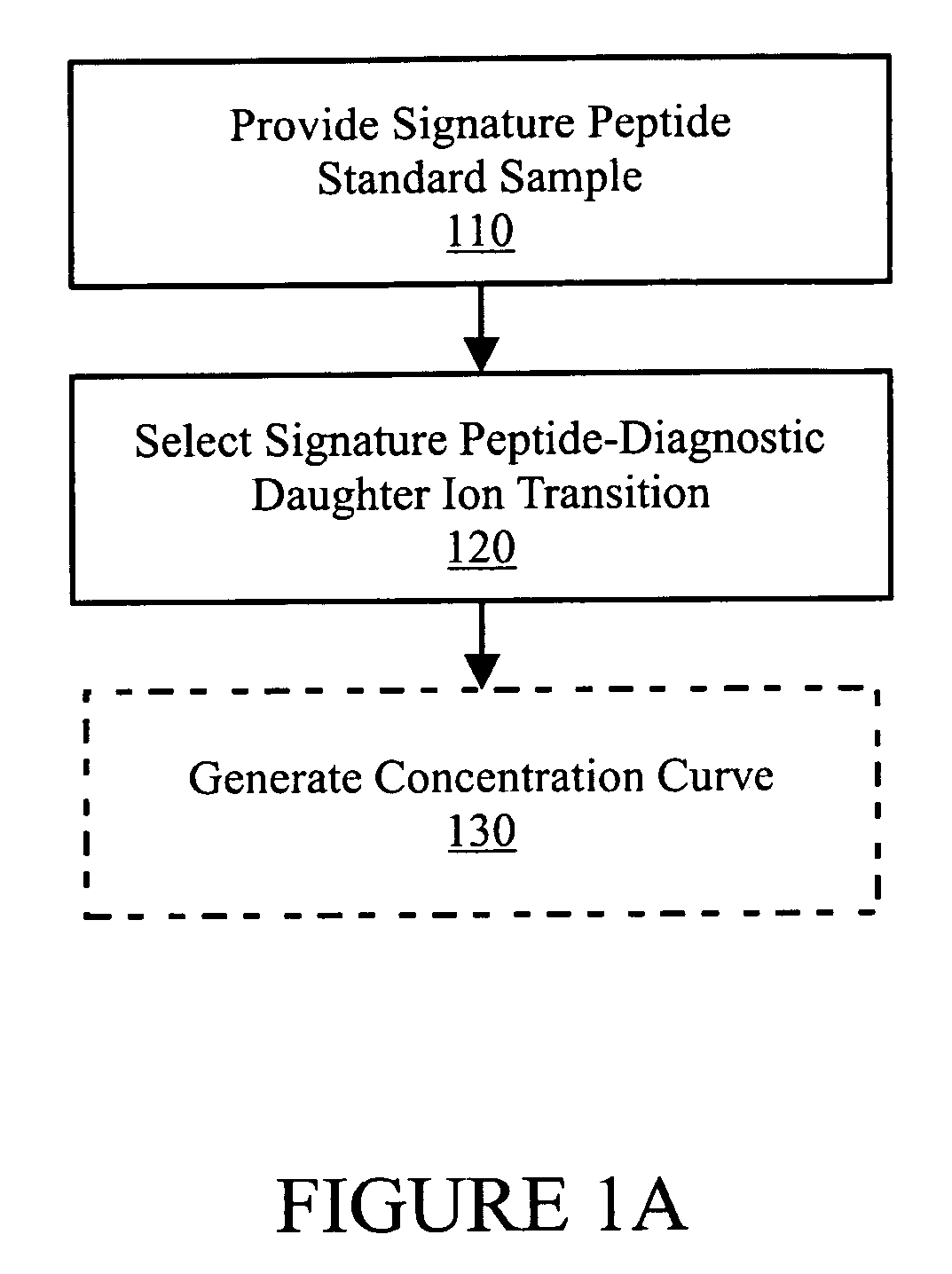
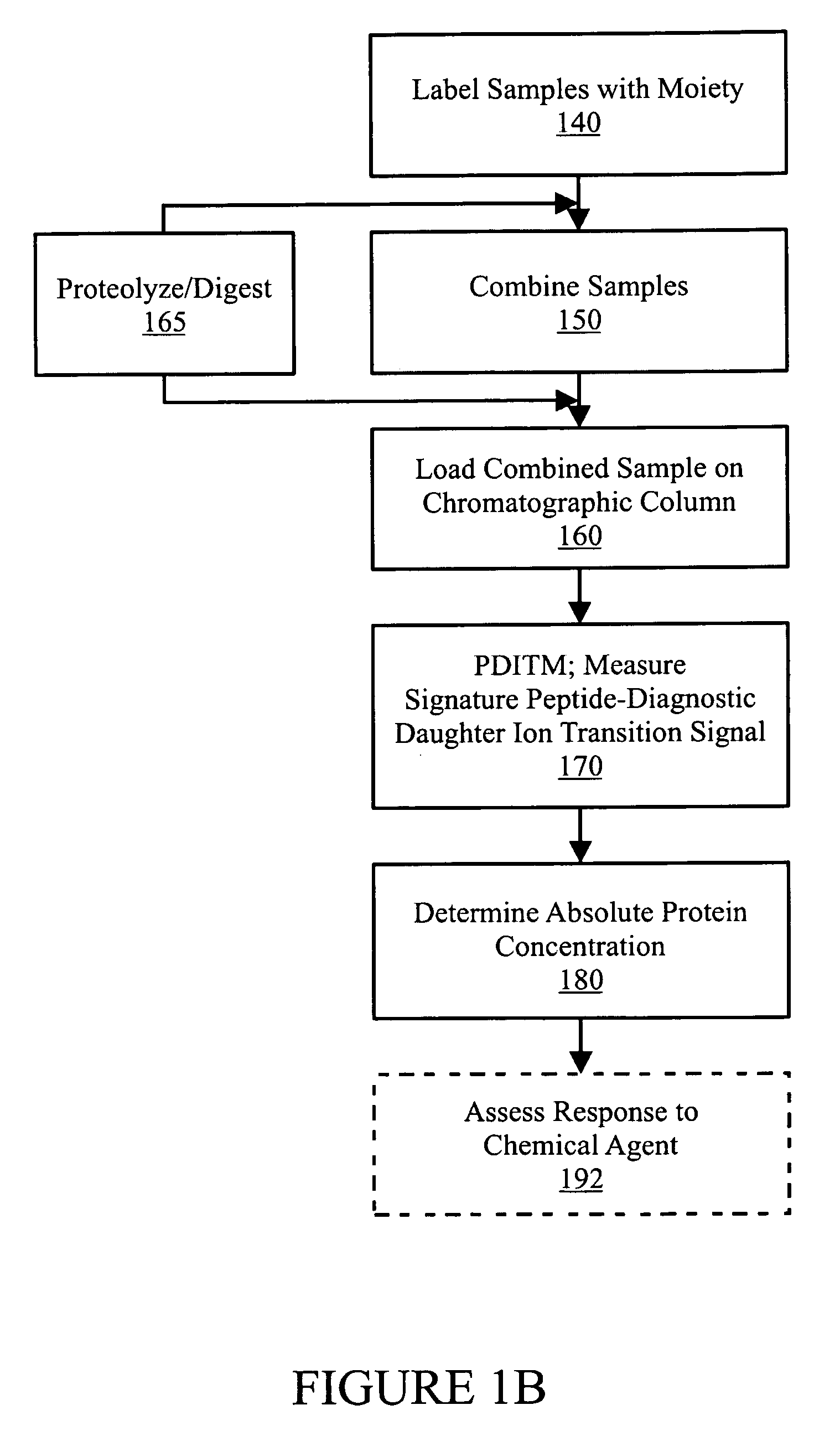
In various aspects, the present teachings provide systems, methods, assays and kits for the absolute quantitation of protein expression. In various aspects, the present teachings provide methods of determining the concentration of one or more proteins of interest in one or more samples of interest. In various aspects, the present teachings provide methods of determining the absolute concentration of one or more isoforms of a protein using standard samples of signature protein fragments and parent-daughter ion transition monitoring (PDITM). In various embodiments, the absolute concentration of multiple isoforms of a biomolecule in a sample, multiple proteins in a biological process, a combination of multiple samples, or combinations thereof, can be determined in a multiplex fashion using the present teachings. In various aspects, provided are methods of assessing the response of a biological system to a chemical agent.
Owner:APPLERA
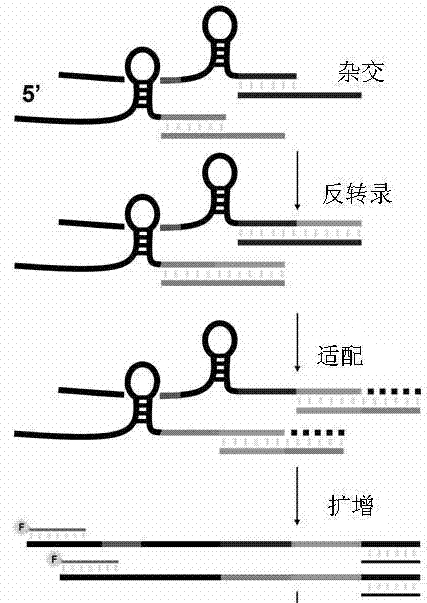
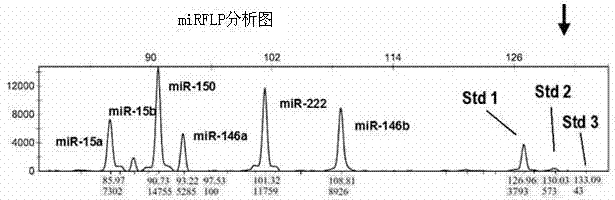
Method for measuring short chain RNA by amplifying length polymorphism of DNA fragment
ActiveCN104120184AEliminate distractionsMicrobiological testing/measurementFermentationA-DNAAmplified fragment length polymorphism
The invention discloses a method for measuring a short chain RNA by amplifying length polymorphism of a DNA fragment and belongs to the l field of molecular biological techniques. The method comprises the following steps: first, by using at least two synthesized small RNAs without natural homologous sequences as compared with the short chain RNA to be measured as an interior label of measurement, mixing the synthesized small RNAs in different numbers of molecules to form a dynamic small RNA standard molecular gradient; and then, mixing with the short chain RNA to be measured in equal amount according to the dynamic small RNA standard, and carrying out RNA inverse transcription, cDNA tailing, PCR synchronous amplification and fluorescent quantitative analysis on length polymorphism fragment of DNA of the PCR product to measure a relative proportion of the fluorescence intensity of the DNA fragment amplified by short chain RNA to be measured in the dynamic small RNA standard fluorescence intensity gradient. The method can absolutely quantify the short chain RNA, particularly miRNA, has high specificity, high sensitivity, objective result and good repeatability, can realize accurate quantification in a copy number range of 100-10<6> and is also suitable for RNA crude extracts.
Owner:CHENGDU NUOEN BIOLOGICAL TECH
PCR micro droplet kit for examining human EGFR gene mutation
InactiveCN106319042AWith absolute quantificationStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationEGFR Gene MutationMutant
The application relates to the examining field of gene mutation and particularly relates to a PCR micro droplet kit for examining human EGFR gene mutation. The kit includes a nucleic acid amplification reagent; a reference substance and micro droplet PCR generated oil. The nucleic acid amplification reagent includes a primer and a probe for examining the No. 18 extron mutant, a primer and a probe for examining the No. 19 extron mutant, a primer and a probe for examining the No. 20 extron mutant, and a primer and a probe for examining the No. 21 extron mutant. The present application can be used for examining the seven species of the No. 18, 19, 20, and 21 extron mutants for the EGFR genes in the DNAs extracted from the paraffin-embedded pathology tissue slices and the peripheral blood dissociate DNAs. The present application provides a reference for the patients to choose the tumor molecularly targeted drug remedy and monitor. The present invention has absolute ration, high specificity, accuracy and high sensitivity.
Owner:上海睿昂基因科技股份有限公司
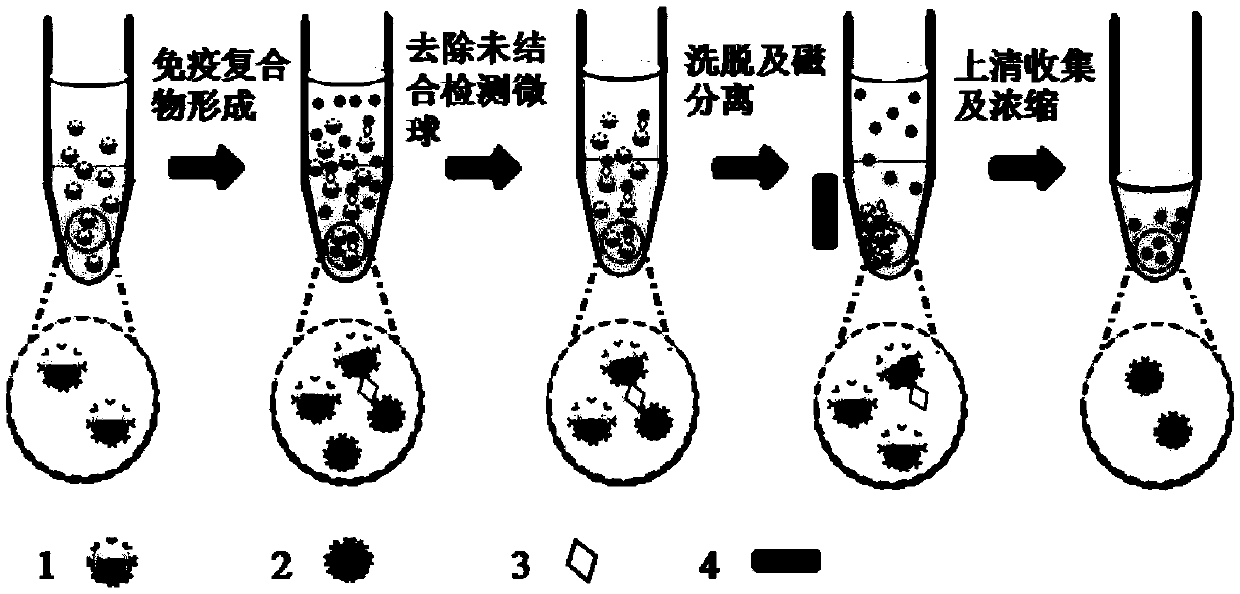
Low abundant protein absolute quantification method based on digital immunoassay technology
InactiveCN109521202AAchieve ultra-sensitivityAchieving Absolute Quantitative AnalysisBiological testingProtein targetImmune complex deposition
The invention relates to a low abundant protein absolute quantification method based on a digital immunoassay technology. The method comprises the following steps: carrying out an immunoreaction on captured beads, target antigens and detected particles so as to form an immune complex, eluting the detected particles in the immune complex, collecting and concentrating the eluant, transferring the concentrated solution to a micro-fluidic particle counting chip, depositing and fixing the detected particles on the micro-fluidic particle counting chip after a solvent in the concentrated solution iscompletely volatilized, performing imaging recording on all the detected particles fixed on the micro-fluidic particle counting chip, and counting the quantity of the detected particles on the micro-fluidic particle counting chip, thereby obtaining the molecular number of the target protein. The method disclosed by the invention has the advantages that ultra-sensitive and absolute quantification analysis of disease-related proteins in clinical samples can be realized, any standard substance is not needed, a standard curve does not need to be drawn, and the method has the advantages of being wide in application range, high in detection sensitivity, high in analytical result accuracy, excellent in precision degree and high in throughput.
Owner:XUZHOU NORMAL UNIVERSITY
Novel Isobaric Tandem Mass Tags for Quantitative Proteomics and Peptidomics
ActiveUS20130078728A1High quantitation efficacyLow costOrganic chemistryBiological testingCombinatorial chemistryIsotope
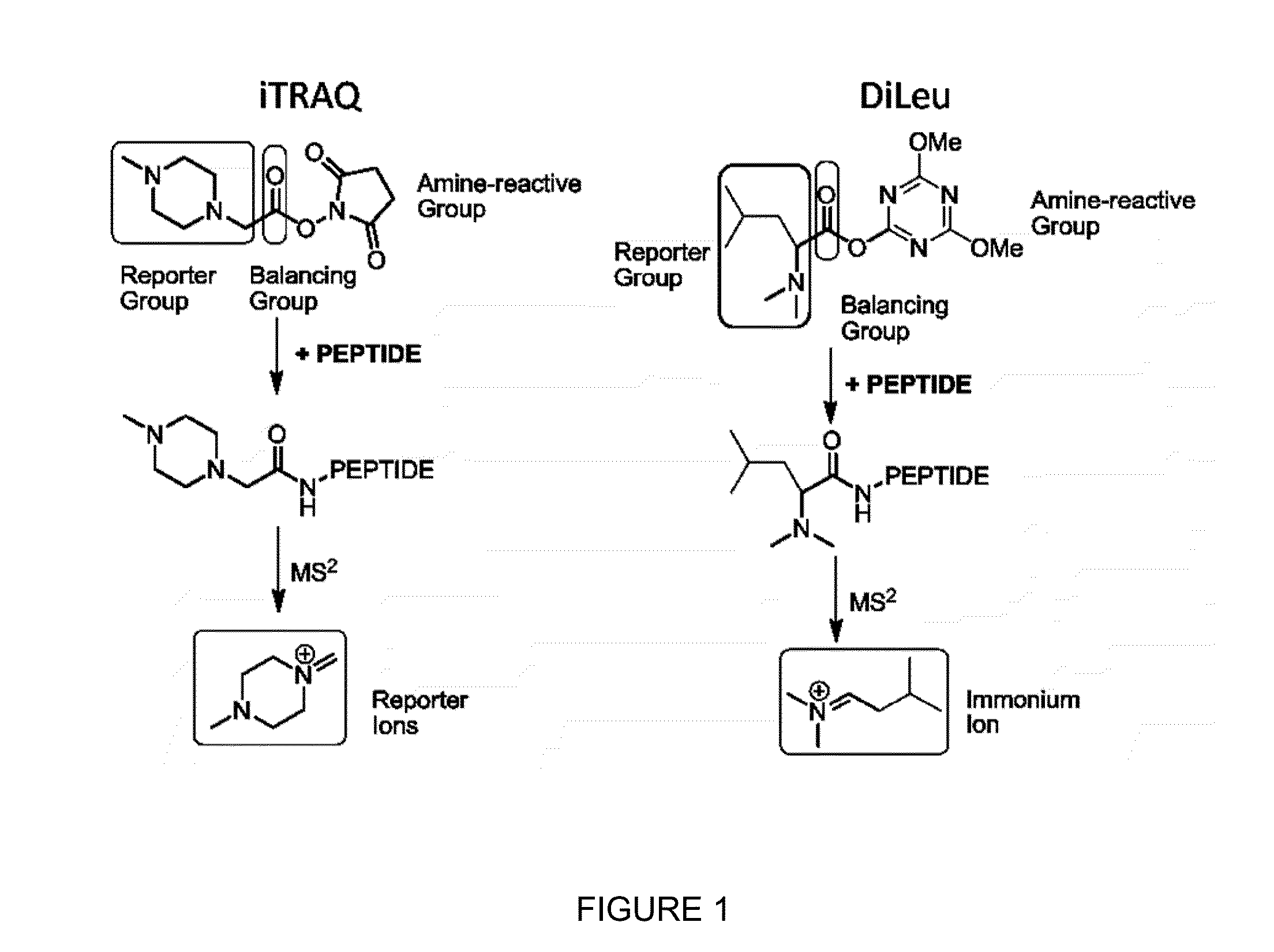
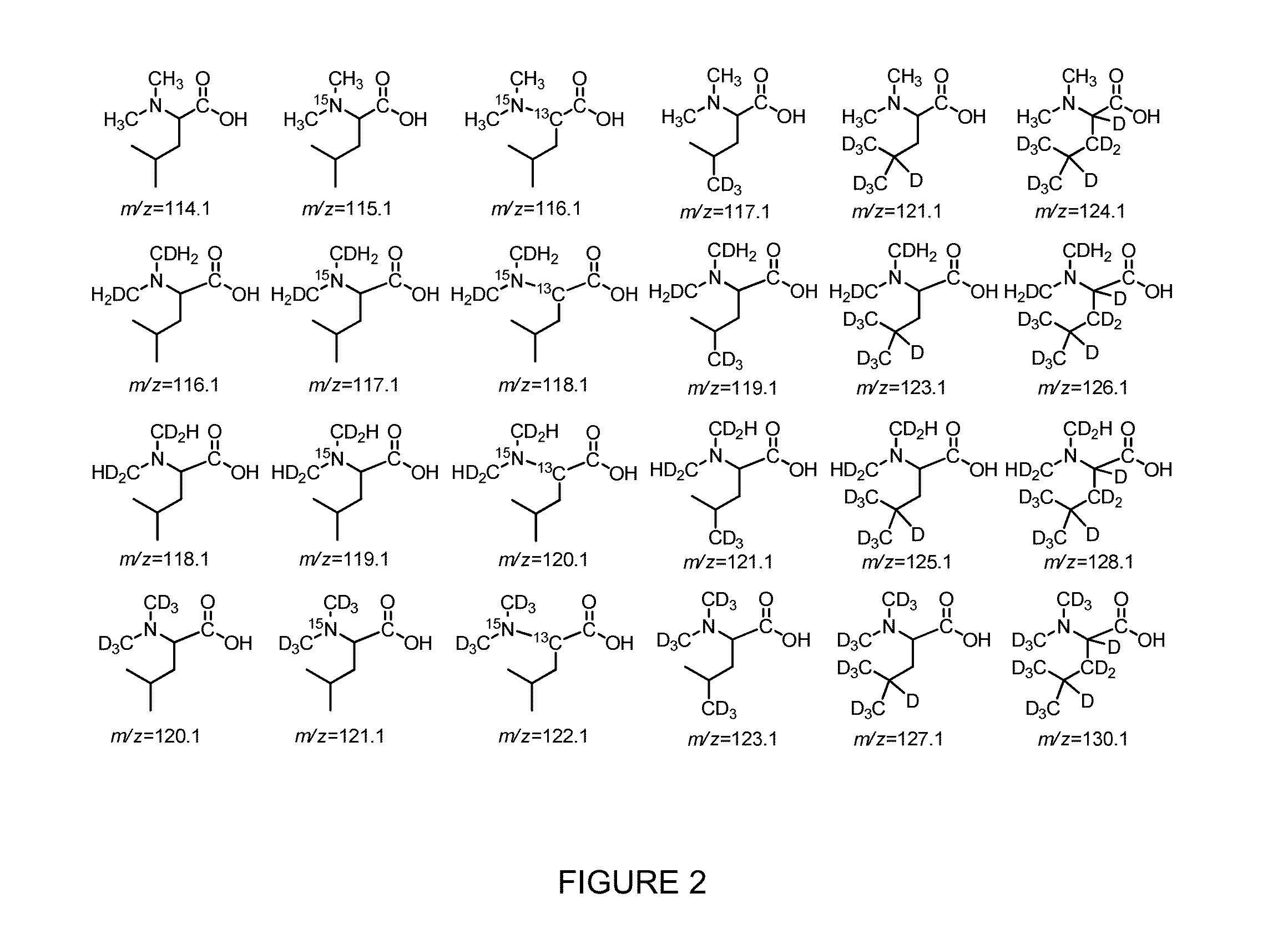
Compositions and methods of tagging peptides and other molecules using novel isobaric tandem mass tagging reagents, including novel N,N-dimethylated amino acid 8-plex and 16-plex isobaric tandem mass tagging reagents. The tagging reagents comprise: a) a reporter group having at least one atom that is optionally isotopically labeled; b) a balancing group, also having at least one atom that is optionally isotopically labeled, and c) an amine reactive group. The tagging reagents disclosed herein serve as attractive alternatives for isobaric tag for relative and absolute quantitation (iTRAQ) and tandem mass tags (TMTs) due to their synthetic simplicity, labeling efficiency and improved fragmentation efficiency.
Owner:WISCONSIN ALUMNI RES FOUND
Method for determining soybean seed vigor based on nuclear magnetic resonance technique
ActiveCN106680307AReduce dosageEasy to operateAnalysis using nuclear magnetic resonanceNMR - Nuclear magnetic resonanceMetabolite
The invention discloses a method for determining soybean seed vigor based on a nuclear magnetic resonance technique. The method is characterized by comprising the steps of carrying out absolute quantification analysis on metabolite in soybean seed extract by using a nuclear magnetic resonance wave spectrum technique to obtain a metabolite attribution map, screening specific metabolite based on an established partial least square method multiple regression model, then determining the vigor level by using an absolute content value of the specific metabolite in the soybean seeds, and providing reference for determining the soybean seed vigor and screening anti-aging soybean seed resources. The method is simple and rapid to operate and reliable in result, and can provide reference for determining and screening high-quality soybean seed resources with anti-aging potential.
Owner:SICHUAN AGRI UNIV
Absolute Quantitation of Proteins and Protein Modifications by Mass Spectrometry with Multiplexed Internal Standards
InactiveUS20160154006A1Improve accuracyHigh sensitivityPeptide librariesLibrary screeningAnalyteImage resolution
A method for absolute protein or peptide quantitation by mass spectroscopy. A sample containing a protein or peptide of interest is prepared for mass spectroscopy analysis. The sample is subjected to mass spectroscopy analysis at low resolution whereby a single additive mass spectroscopy peak is obtained, then is subjected to high resolution mass spectroscopy analysis whereby a plurality of mass spectroscopy peaks are obtained. The intensity of each of the plurality of mass spectroscopy peaks is quantitated either by comparison to an internal standard set, or by using a standard curve generated for each isotopologue set. Quantitation using a standard curve enhances quantitation across a dynamic range of analyte.
Owner:PIERCE BIOTECHNOLOGY INC
Expression quantification using mass spectrometry
InactiveUS20070054345A1Facilitate validationConvenient verificationMicrobiological testing/measurementBiological material analysisOrganismal ProcessMass Spectrometry-Mass Spectrometry
In various aspects, the present teachings provide systems, methods, assays and kits for the absolute quantitation of protein expression. In various aspects, the present teachings provide methods of determining the concentration of one or more proteins of interest in one or more samples of interest. In various aspects, the present teachings provide methods of determining the absolute concentration of one or more isoforms of a protein using standard samples of signature protein fragments and parent-daughter ion transition monitoring (PDITM). In various embodiments, the absolute concentration of multiple isoforms of a biomolecule in a sample, multiple proteins in a biological process, a combination of multiple samples, or combinations thereof, can be determined in a multiplex fashion using the present teachings. In various aspects, provided are methods of assessing the response of a biological system to a chemical agent.
Owner:APPLERA
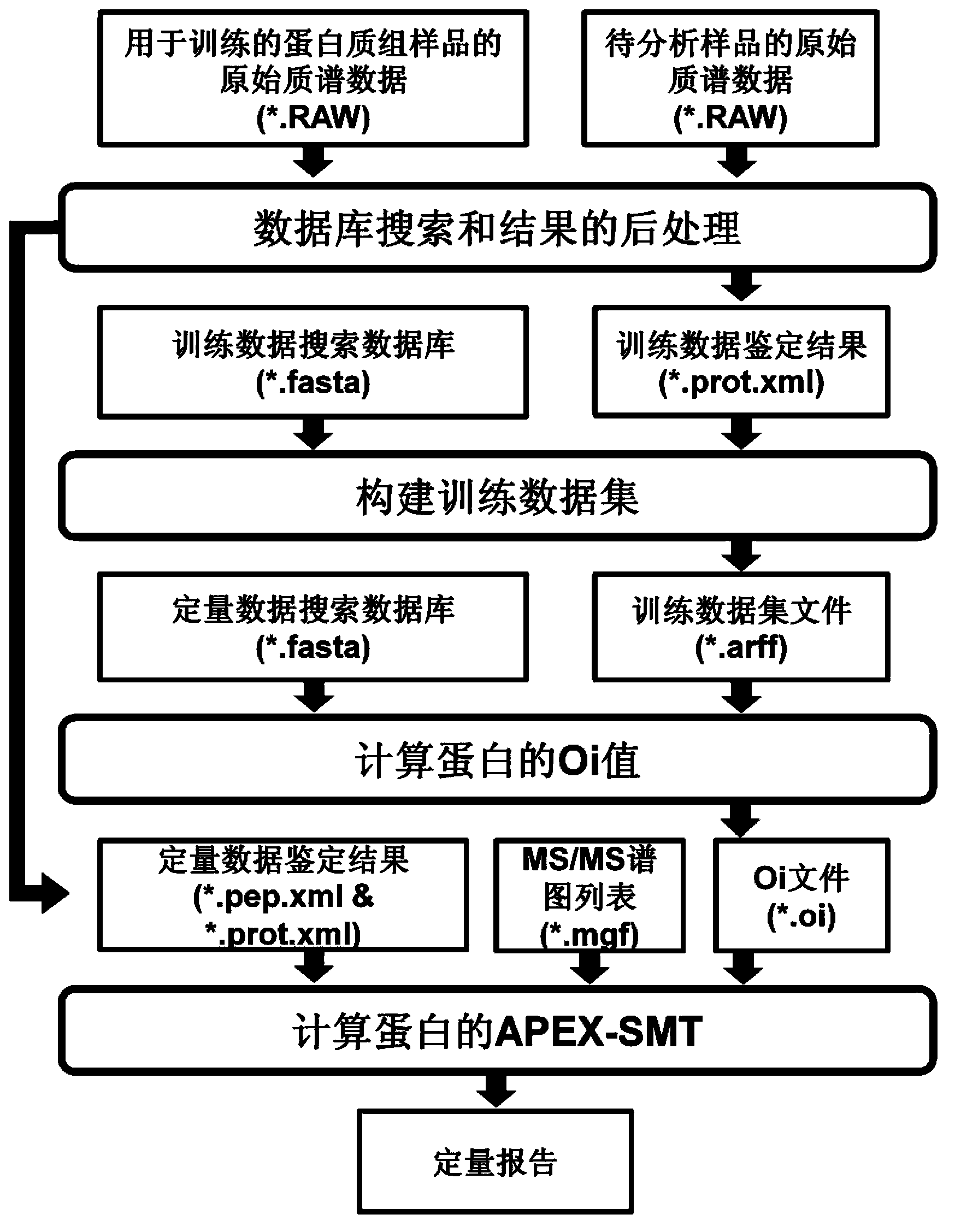
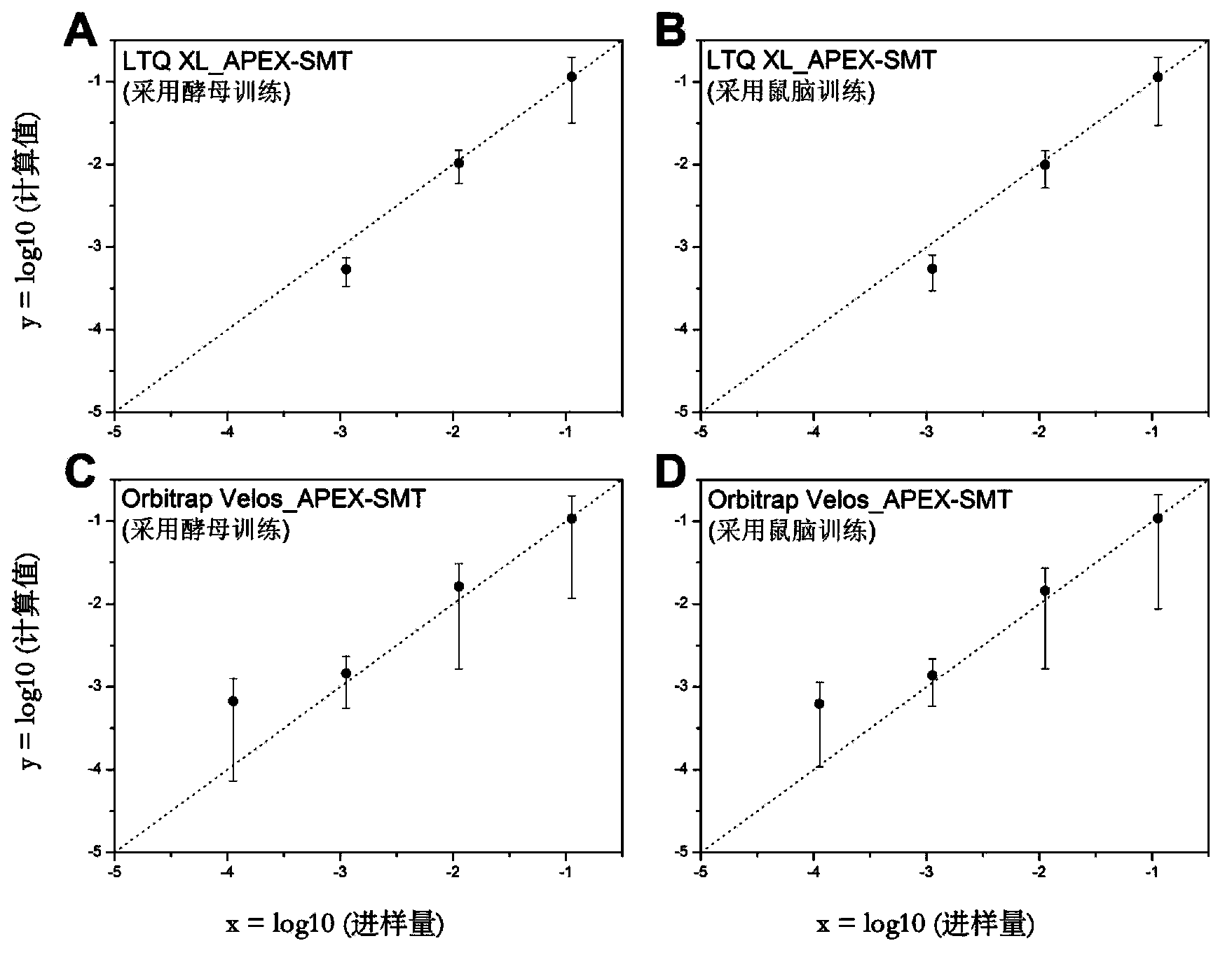
Proteome label-free quantification method combining tandem mass spectrometry with machine learning algorithm
ActiveCN103884806AMinimize response varianceQuantitative results are accurateComponent separationData setMass spectrometric
The invention relates to a proteome label-free quantification method combining tandem mass spectrometry with machine learning algorithm, which is used for absolute and relative quantitative analysis of a proteome level. The method firstly analyzes an enzymolytic peptide fragment mixture of a proteome actual sample for establishing a training data set and an enzymolytic peptide fragment mixture of a proteome sample to be analyzed by using a liquid chromatography-tandem mass spectrometry system. The total amount of the samples can be obtained by cell counting or protein concentration determination, and the absolute amount of each protein can be calculated according to the percents and sample total amount calculated in the last step. The absolute amounts of the same protein in different samples are compared to obtain relative quantitative information of the protein in different samples. The method has good accuracy in both absolute quantification and relative quantification.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
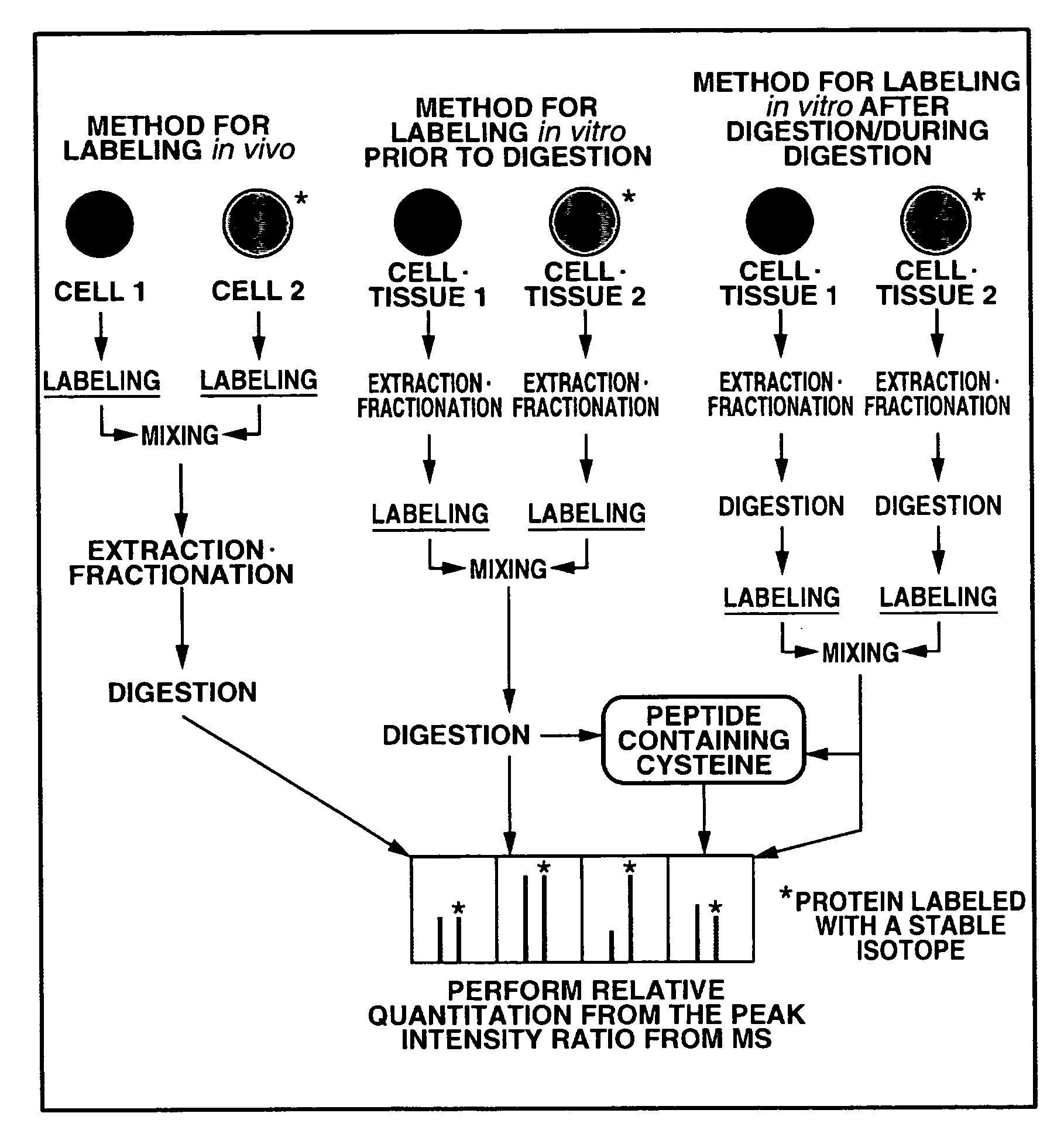
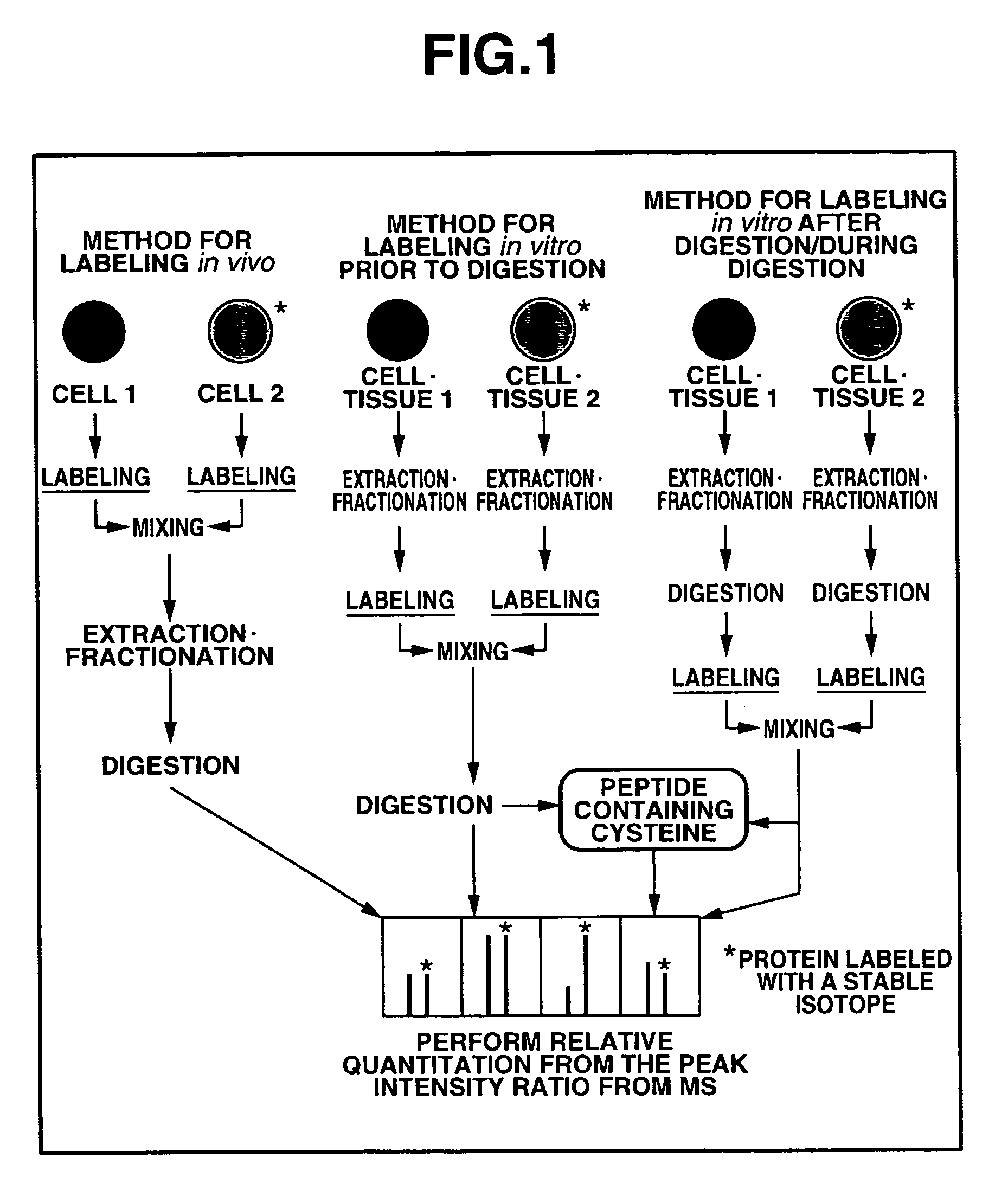
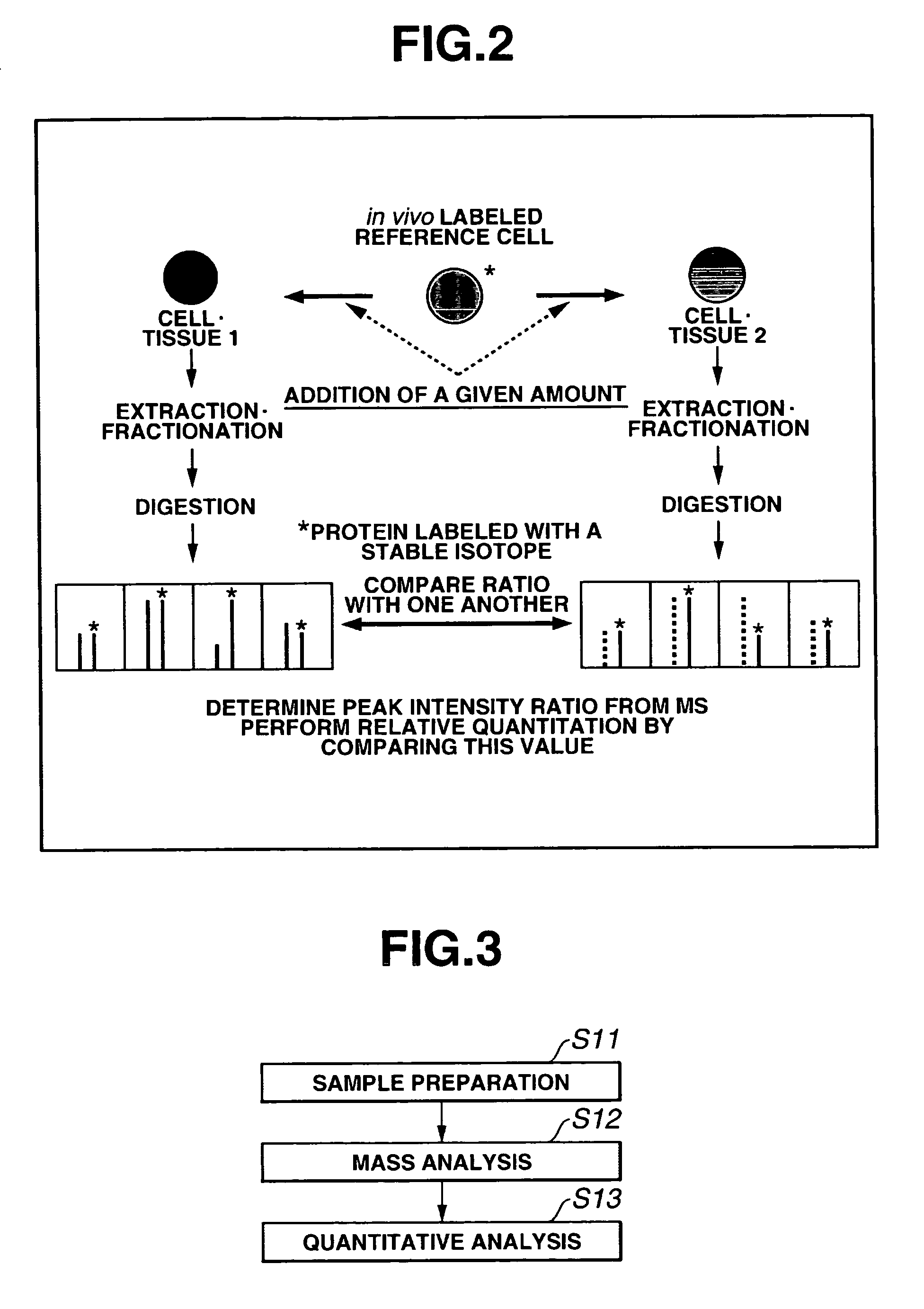
Mass spectrometric quantitation method for biomolecules based on metabolically labeled internal standards
InactiveUS7759130B2Improve accuracyEasy to compareMaterial analysis by electric/magnetic meansBiological testingIsotopeProtein-protein complex
An object of the present invention is to quantitate with good accuracy, furthermore, quantitate absolutely, one or a plurality of biological molecules in a sample such as a tissue, a biological fluid, a cell, a cell organ or protein complex.By adding a metabolically isotope labeled biological molecule as an internal standard substance and measuring with a mass spectrometer, quantitating with good accuracy one or a plurality of target molecules in a sample has become possible. In addition, by performing waveform separation processing during mass analysis, a highly accurate quantitative analysis method of mass analysis is provided.
Owner:EISIA R&D MANAGEMENT CO LTD
Digital PCR-based novel coronavirus nucleic acid quantitative detection kit and application
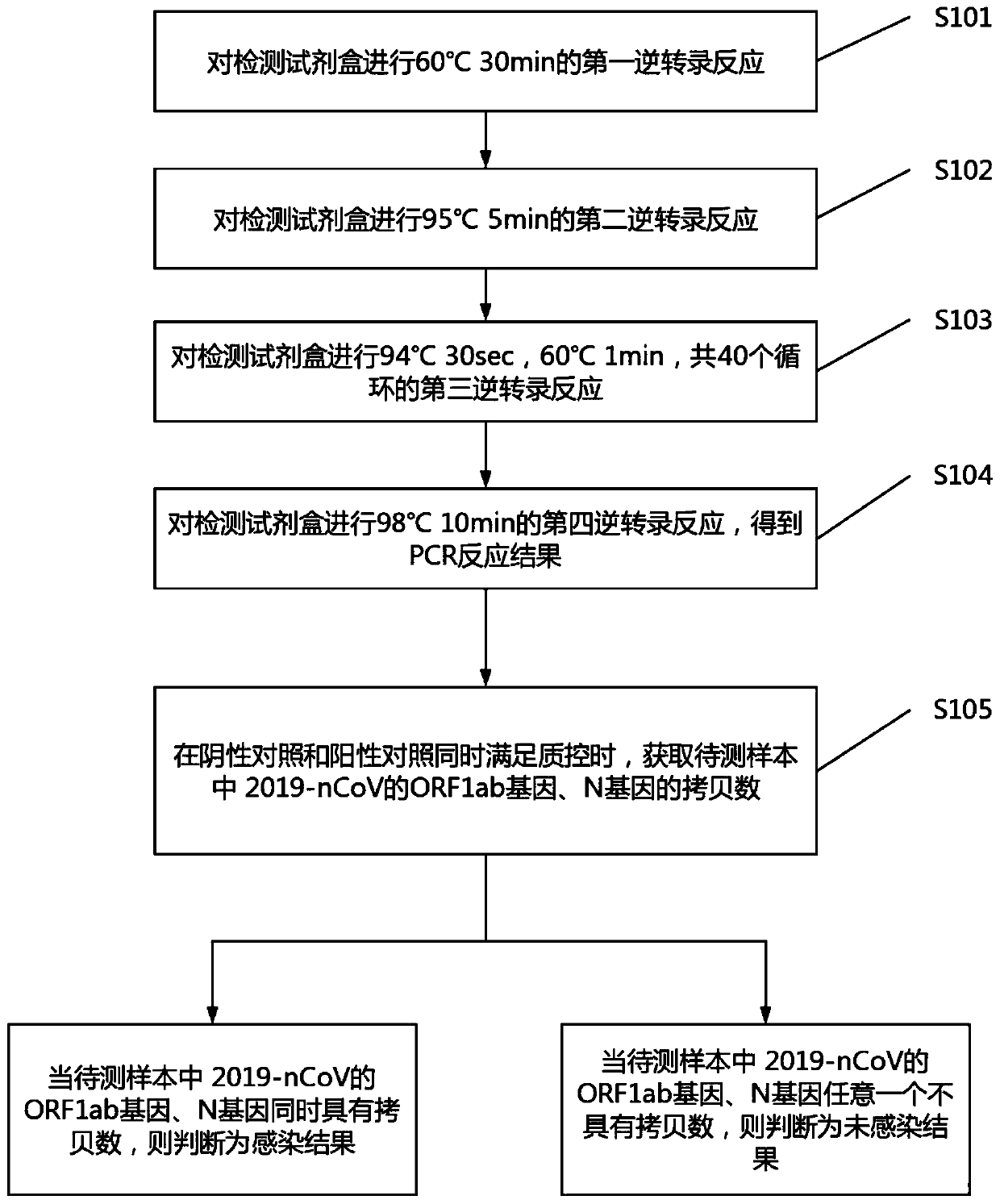
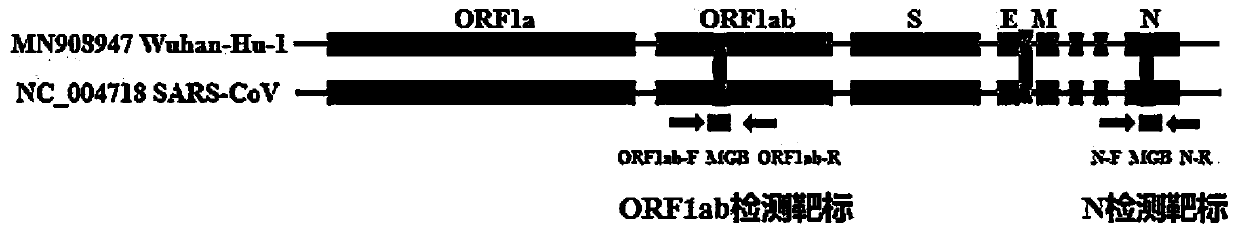
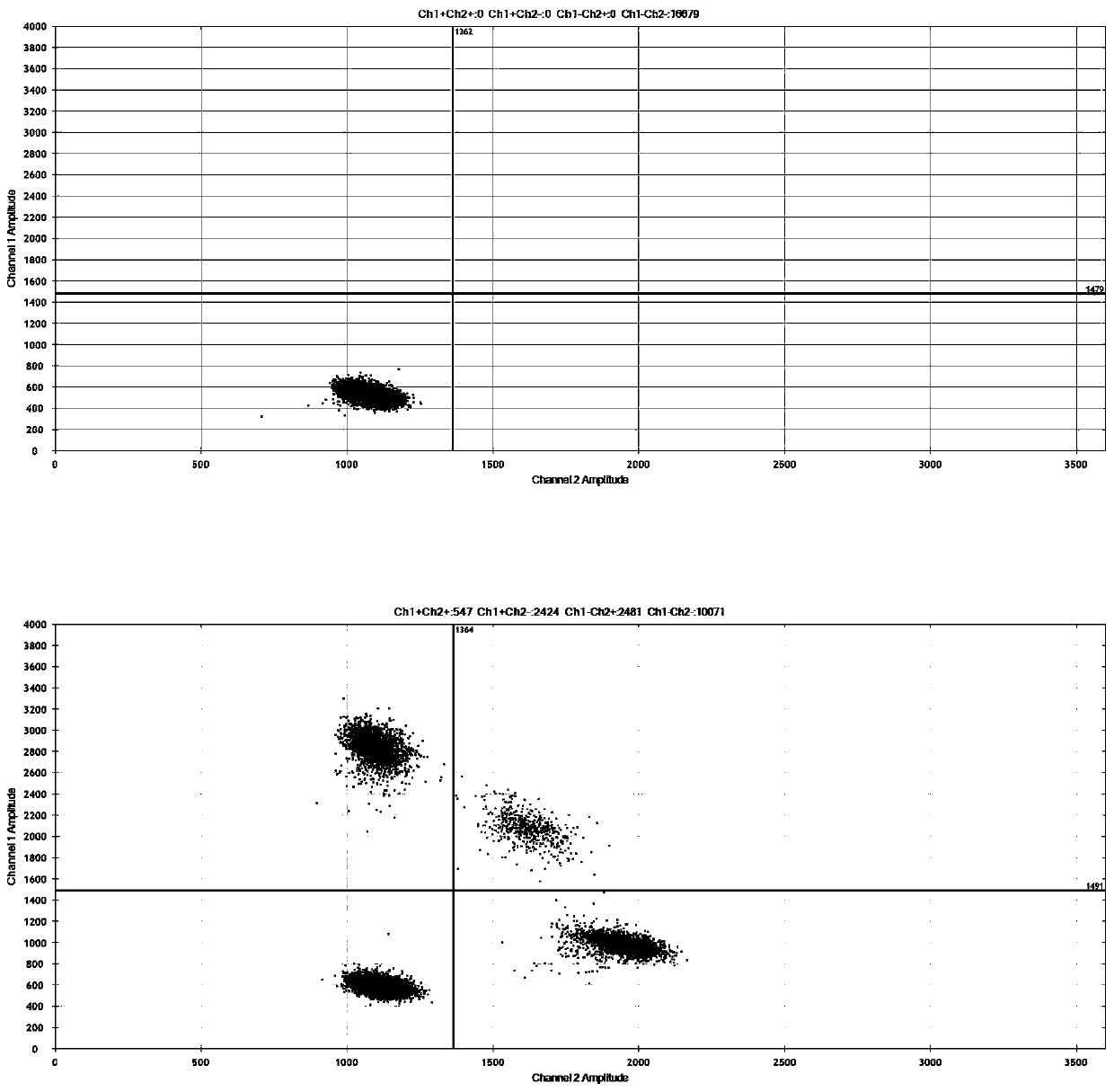
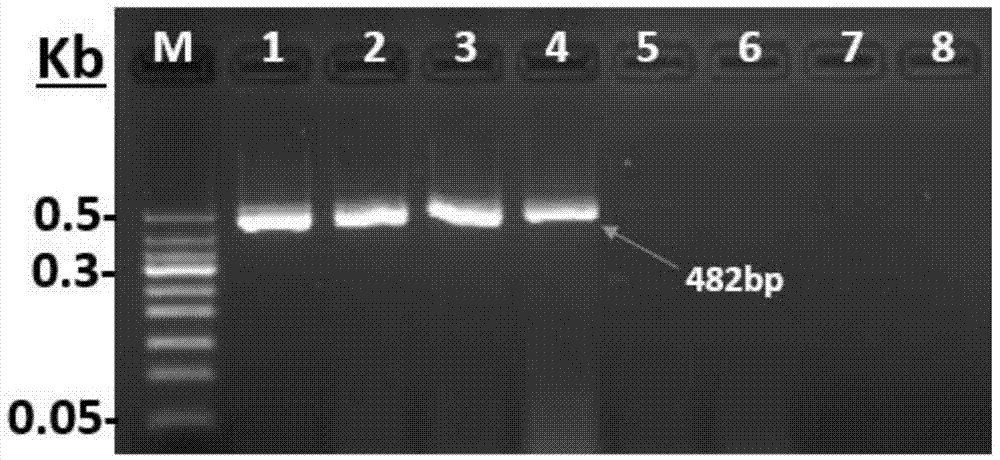
ActiveCN111394519AHigh sensitivityStrong specificityMicrobiological testing/measurementAgainst vector-borne diseasesRNA extractionAbsolute quantification
The invention discloses a digital PCR-based novel coronavirus nucleic acid quantitative detection kit and an application. The reaction total system of the digital PCR-based novel coronavirus nucleic acid quantitative detection kit has 20 ul, comprising 10ul of 2x One-Step RT-ddPCR Supermix, 0.8ul of 25mM manganese acetate solution, 5ul of to-be-detected sample RNA, 1ul of ORFlab gene primer probeworking solution, 1ul of N gene primer probe working solution, 1ul of RPP30 gene primer probe working solution and 1.2ul of Nuclease-Free Water, wherein 5ul of negative control RNA extraction solutionand 5ul of positive control RNA extraction solution are adopted to replace the to-be-detected sample RNA in a negative control reaction system and a positive control reaction system respectively. Based on the innovative RNA one-step reverse transcription microdroplet type digital PCR technology, nucleic acid absolute quantification is carried out specific to highly conservative ORFlab gene and Ngene in the novel coronavirus (2019-nCoV) genome, so that the detection accuracy is improved, and the kit can be used for clinical assisted diagnosis and viral load analysis of novel coronavirus (2019-nCoV) infection, and has a wide clinical application value.
Owner:南京实践医学检验有限公司
Primer and method for carrying out specific detection and absolute quantification on tomato spotted wilt virus
InactiveCN104120193AMicrobiological testing/measurementDNA/RNA fragmentationSpecific detectionFluorescence
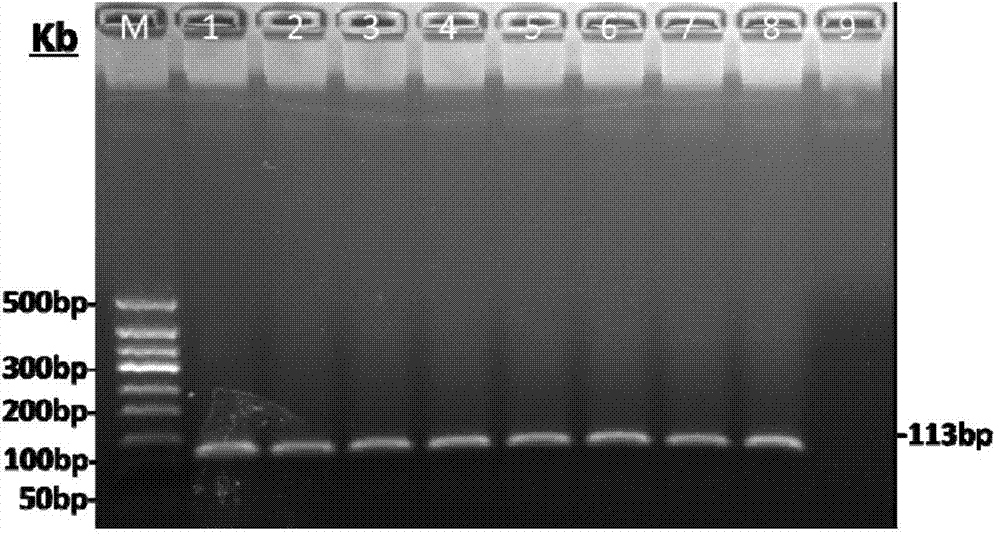
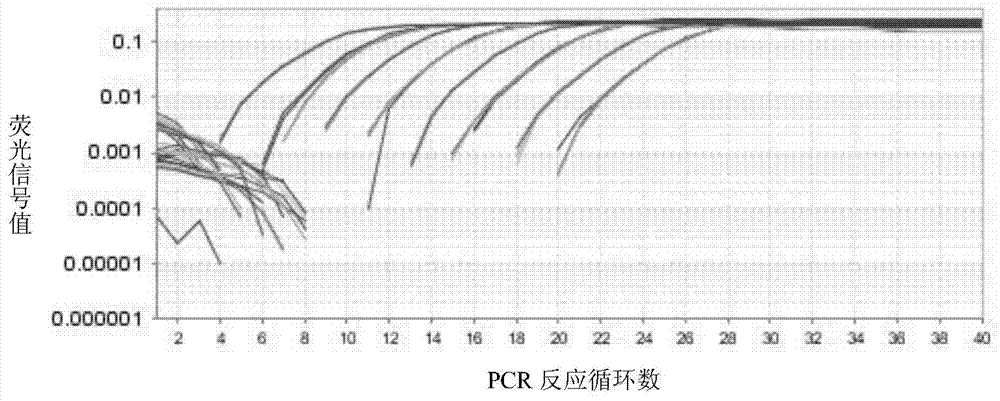
The invention relates to a primer and a method for carrying out specific detection and absolute quantification on a tomato spotted wilt virus and belongs to the field of plant virus detection. According to the primer and method disclosed by the invention, the tomato spotted wilt virus is taken as an object of study, a more rapid, simple, accurate and sensitive method for quantitatively detecting a plant virus is established, the method comprises the following steps of (1) establishing a standard curve according to the standard plasmid; (2) obtaining the cDNA of a sample to be detected; synthesizing cDNA by virtue of reverse transcription; and carrying out fluorescence quantitative PCR by virtue of using cDNA as a template and TSWV-113F / TSWV-113R as a primer to obtain the Ct value of the sample to be detected; and (3) calculating the copy number of the tomato spotted wilt virus in the sample to be detected by virtue of the standard curve. By virtue of the method, tomato spotted wilt virus within the sample can be accurately quantified in 4-5 hours, the time is saved and the experimental steps are simplified, the method is contributed to taking corresponding remedial measures before the onset of plant diseases to prevent huge economic losses brought by the widespread of the virus among crops.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Kit and method for detecting mutation of EGFR gene
ActiveCN108949990AEfficient amplificationHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationEGFR Gene MutationAbsolute quantification
The invention provides a kit and method for detecting the mutation of an EGFR gene and particularly discloses a method, a primer and a probe for detecting the mutation of the EGFR gene, a kit including prime probe mixing liquid, a primer and a probe for detecting the mutation of a C797S locus, a primer and a probe for detecting the mutation of a T790M locus, a primer and a probe for detecting themutation of a 19del locus and a primer and a probe for detecting the mutation of a L858 locus. According to the method provided by the invention, 0.1% mutation rate can be detected based on a digitalPCR platform; and the method has the advantages that the optimization process is simple, much detected mutation types can be detected, the absolute quantification is realized, the sensitivity is high,samples are easily acquired, and the like.
Owner:DAAN GENE CO LTD
Typing detecting kit and method for detecting digital PCR absolute quantification of HBV-B/C
ActiveCN105441595AStrong amplification specificityAccurate Quantification of Copy NumberMicrobiological testing/measurementFluorescenceA-DNA
The invention discloses a typing detecting kit and method for detecting the digital PCR absolute quantification of HBV-B / C. The kit comprises a DNA extraction solution, a digital PCR reaction buffer solution A, a digital PCR reaction buffer solution B, an HBV-B virus gene positive quality control, an HBV-C virus gene positive quality control, a negative quality control and an internal standard solution. The method includes the following steps of firstly, processing a to-be-detected sample; secondly, processing a quality control product; thirdly, preparing a digital PCR reaction mixed solution; fourthly, generating a micro-reaction liquid drop, and conducting digital PCR reaction amplification; fifthly, reading a fluorescent signal, analyzing an HBV type and calculating the number of copying times. By means of the digital PCR technology and the dual-color florescent probe, two subtypes of HBV are detected at the same time; without depending on an external standard object or a standard curve, the kit and method are easy and convenient to operate and can directly detect the HBV precise absolute quantification.
Owner:ANHUI UNIV OF SCI & TECH
System and method for absolute quantitation of proteins using LC/MS
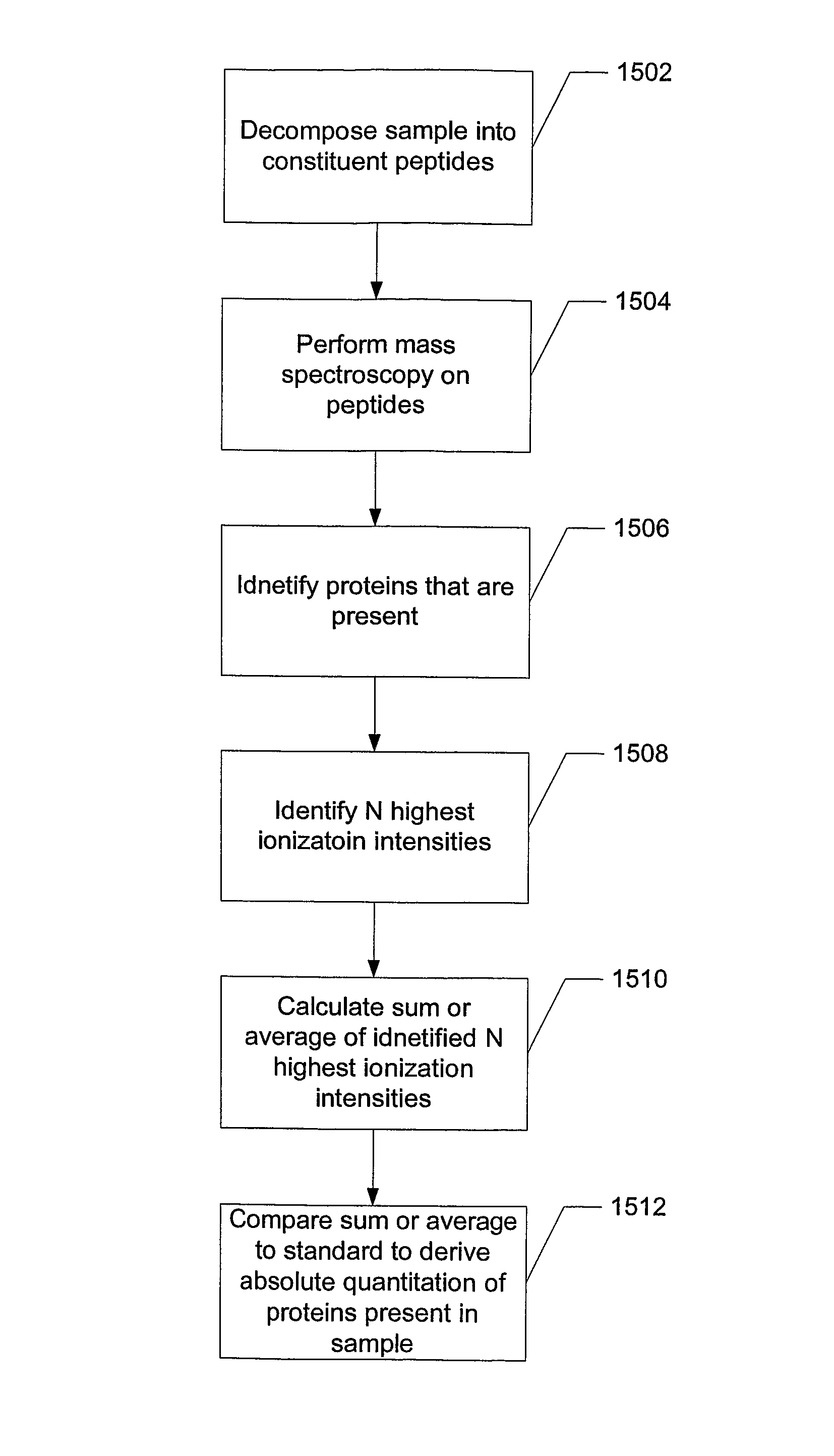
ActiveUS8271207B2Reduce errorsComponent separationBiological testingConversion factorLiquid chromatography mass spectroscopy
Absolute quantitation of protein in a sample is provided by comparing a sum or average of the N highest ionization intensities observed for peptides of a particular protein along with a calibration standard. The calibration standard can be in the form of a table generated by prior protein peptide analysis performed using one or more pre-determined proteins. The comparison is used to determine a corresponding absolute quantity of protein based on the observed sum or average of ionization intensities. A simple conversion factor can be applied to the calibration standard value to determine the absolute quantity of protein in the sample.
Owner:WATERS TECH CORP
Absolute PCR Quantification
ActiveUS20100137152A1Heating or cooling apparatusMicrobiological testing/measurementAbsolute quantificationBiology
The present application provides methods and devices for absolute quantification of polymerase chain reaction target nucleic acids. In particular, the methods and devices of the present application provide for splitting a nucleic acid sample to be analyzed into small, isolated volumes, conducting the method of polymerase chain reaction (PCR) on said volumes, detecting PCR amplification products, analyzing said detected PCR amplification products, performing absolute quantification of the PCR target and presenting said quantification results.
Owner:THE RES FOUND OF STATE UNIVOF NEW YORK
Quantitative detection method for transgene protein CP4-EPSPS in plant
ActiveCN103698418AAchieving absolute quantificationHigh sensitivityComponent separationInternal standardDigestion
The invention relates to a quantitative detection method for transgene protein CP4-EPSPS in a plant. The quantitative detection method comprises: extracting protein from a plant to obtain a crude protein extraction solution; and carrying out digestion on the crude protein extraction solution, adopting three 18O-labeled CP4-EPSPS specific peptide segments as internal standards, adopting UPLC / ESI-QQQ MS detection, and carrying out quantification on CP4-EPSPS in a peptide segment level, wherein the three specific peptide segment sequences are ITGLLEGEDVINTGK, SFMFGGLASGETR and LAGGEDVADLR. According to the present invention, the method has high sensitivity, and absolute CP4-EPSPS quantification can be achieved through combination of an isotope dilution method and an MRM strategy.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
HBV nucleic acid quantitative detection system based on micro-droplet digital PCR technology
InactiveCN108342507AAccurate Absolute QuantificationMicrobiological testing/measurementHepatitis B virus DNAPcr ctpp
The invention provides a HBV nucleic acid quantitative detection system based on a micro-droplet digital PCR technology, the HBV nucleic acid quantitative detection system comprises an upstream primerand a downstream primer for amplifying hepatitis B virus DNA, a specific probe for detecting the hepatitis B virus DNA; a non-infected linearizing double-stranded plasmid quality control standard product and a probe for detecting the quality control standard product; and a buffer system comprising deoxyribonucleoside triphosphates dATP, dCTP, dGTP, and dUTP, and UNG. The HBV nucleic acid quantitative detection system is based on the micro-droplet digital PCR technology to realize the accurate absolute quantification of HBV DNA, faces the clinical actual demand, and has an important application prospect.
Owner:TARGETINGONE CORP +1
Method for detecting number of arbuscular mycorrhizal fungi in wheat rhizosphere soil based on real-time fluorescence quantification PCR
InactiveCN106011256AStable and repeatable resultsStrong specificityMicrobiological testing/measurementMicroorganism based processesArbuscular mycorrhizal fungiFluorescence
The invention provides a method for detecting the number of arbuscular mycorrhizal fungi in wheat rhizosphere soil based on real-time fluorescence quantification PCR. The method includes the steps that firstly, sample total DNA of wheat rhizosphere soil is extracted and serves as a template of PCR amplification; then, amplification is conducted with SEQ ID No.1 and SEQ ID No. 2 as primers, and specific fragments of 129 bp are obtained; after bond and conversion are conducted, qPCR amplification is conducted. By means of the method, arbuscular mycorrhiza is authenticated, absolute quantification can be conducted on the arbuscular mycorrhiza accurately, the copy number of the arbuscular mycorrhizal fungi in all periods of duration of transgenic wheat is obtained, and in the growth and development stages of wheat, the arbuscular mycorrhiza copy number tends to be gradually increased as a whole. The rapid, convenient and precise authentication method is built, a theoretical and test basis is provided for subsequent tests, great significance is provided for further evaluation of safety of transgenic crops, and compared with an existing method, the method is high in sensitivity, high in specificity, good in repeatability, convenient to operate and visual in result.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
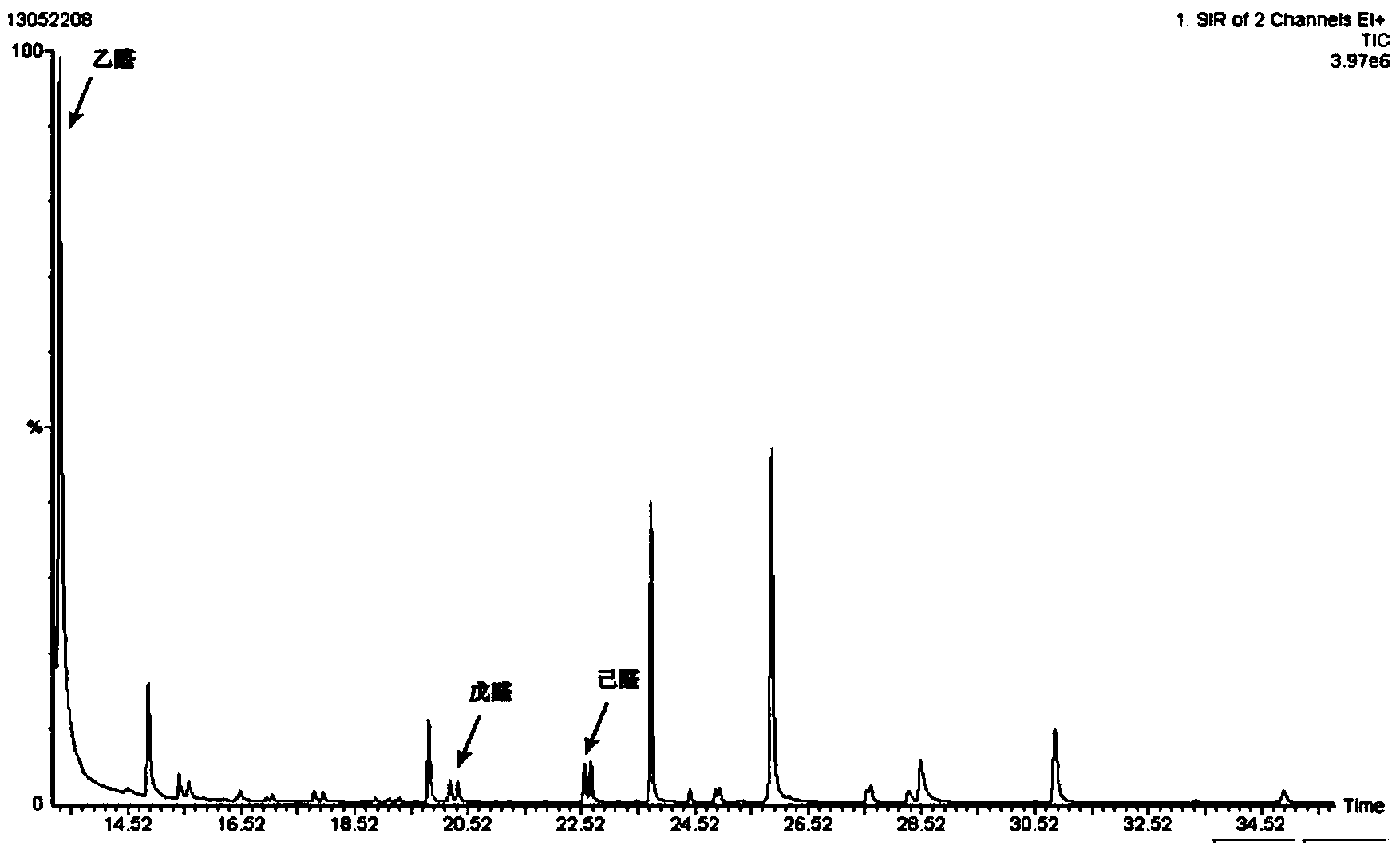
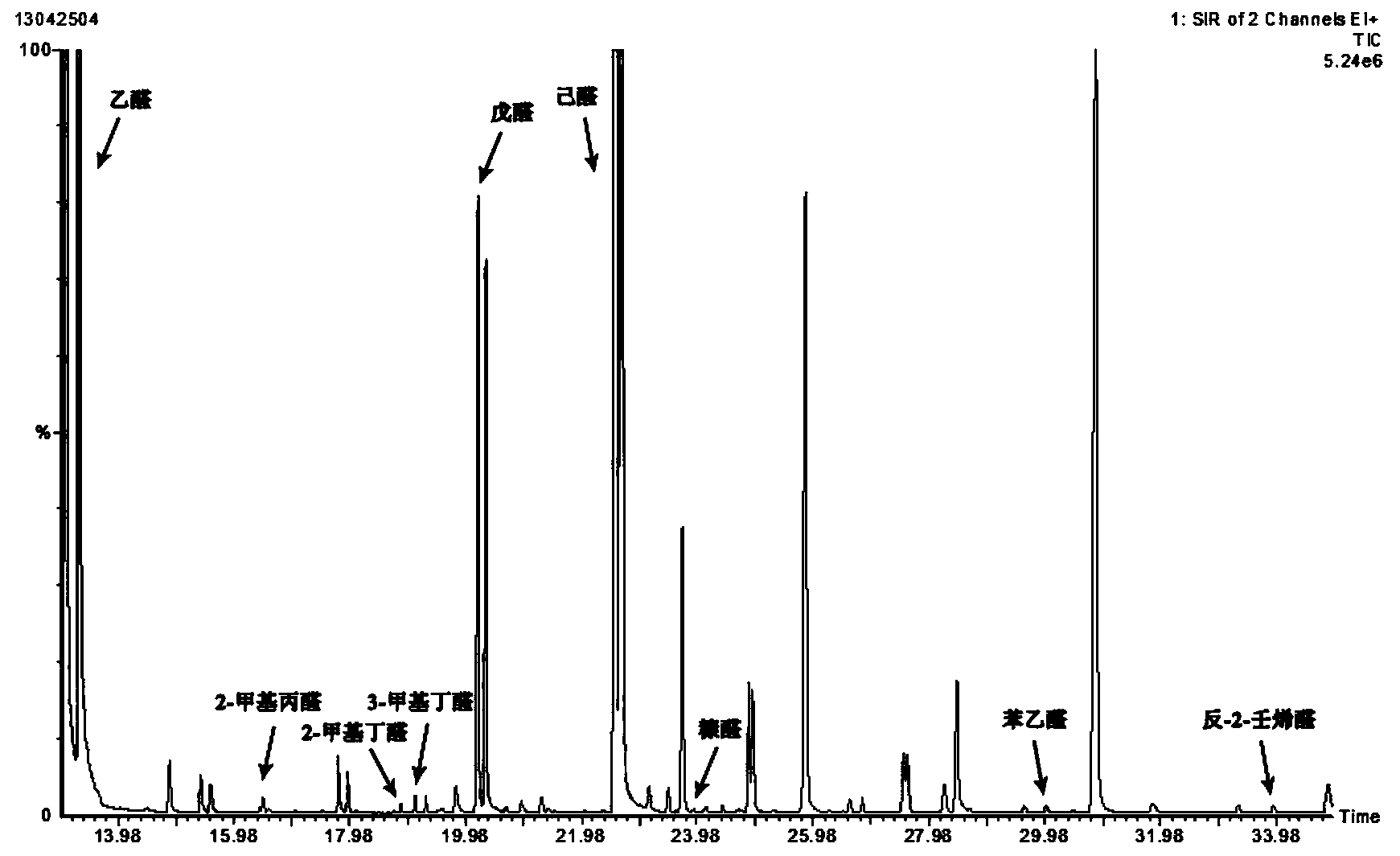
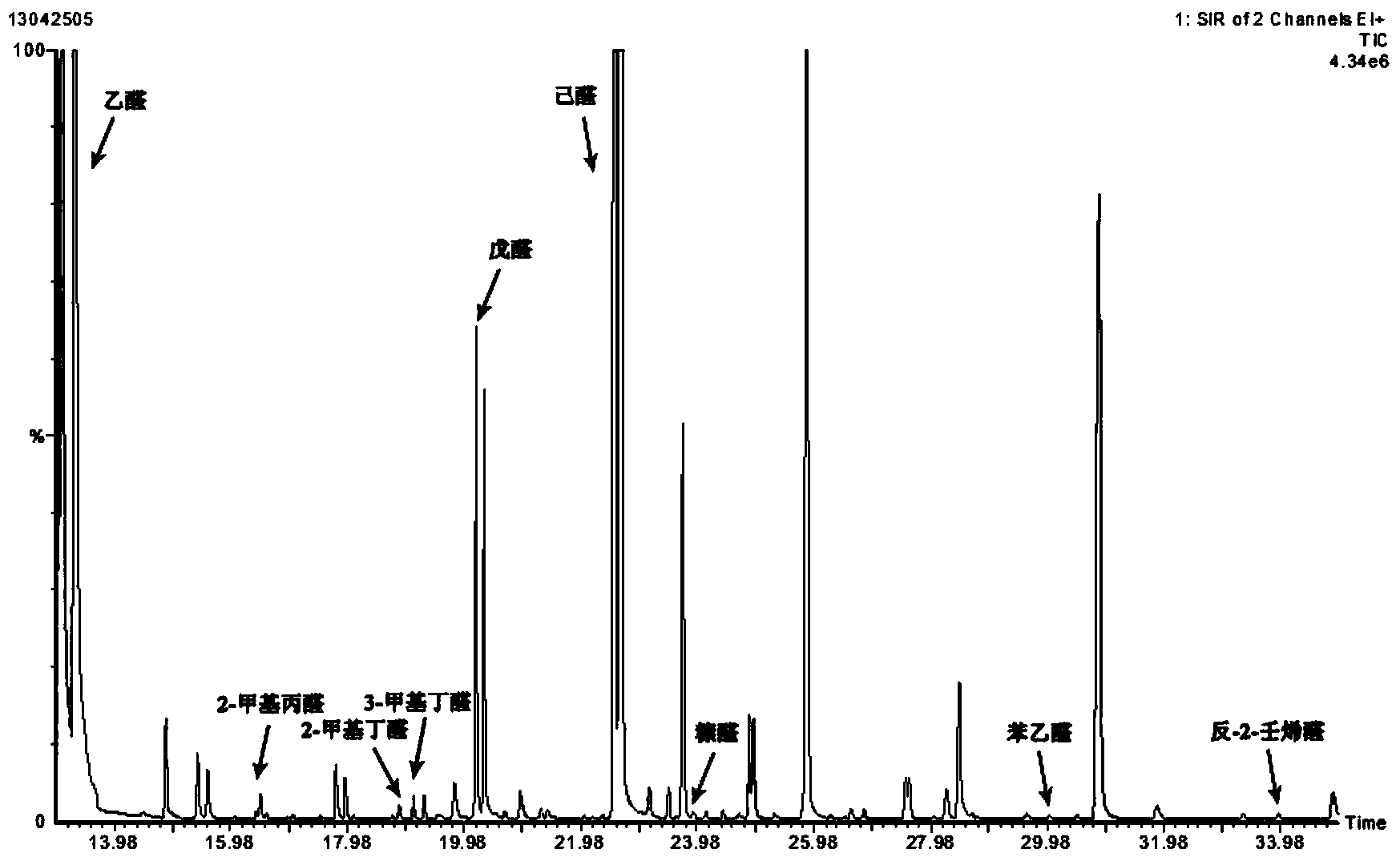
Method for detecting aldehydes in rice
ActiveCN103364515AAvoid distortionTruly reflect the actual contentComponent separationGas phaseMass chromatography
The invention discloses a method for detecting aldehydes in rice. According to the method, the method is realized by employing a headspace derived solid phase micro-extraction-gas chromatography-mass spectrometry method. The method comprises the following steps of: grinding a rice sample, adding the detected sample into a headspace bottle, introducing protective gas, inserting an extraction head into the headspace bottle filled with derivative liquid for deriving for 20 minutes, and inserting the extraction head into a headspace sample introduction bottle, wherein the extraction temperature is 40 DEG C, and the extraction time is 80 minutes; detecting in a gas chromatograph-mass spectrometer. The fineness and headspace derivatization conditions of the ground rice particles are optimized, so that newly generated aldehydes in the analysis process are avoided, and the accuracy of an experimental result is improved. According to the detection method, five steps of adsorbing derivative liquid of the extraction head, deriving and extracting the aldehydes, performing sample introduction, separating and measuring can be sequentially finished in a process, automation is realized, the operation is simple, the detection efficiency and accuracy are improved, and absolute quantification of nine aldehydes in rice is realized.
Owner:TSINGTAO BREWERY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com