Primer and method for carrying out specific detection and absolute quantification on tomato spotted wilt virus
A technology of tomato spotted wilt virus and spotted wilt virus, applied in biochemical equipment and methods, microbe measurement/testing, DNA/RNA fragments, etc., to achieve high sensitivity, prevent economic loss, and save time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Embodiment 1, design and synthesis of primers
[0060] Step 1: Primer Design
[0061] According to the TSWV gene sequence reported in the GENBANK database, after downloading as many TSWV sequences as possible (such as JN601525.1), use DNAMAN software for sequence comparison analysis; find the virus-specific and conserved regions of different strains and design primers (Table 1 ), the primers were synthesized by Shanghai Sangon Co., Ltd.
[0062] The target fragment primers are:
[0063] TSWV-482F: 5'-TCTGGTAGCATTCAACTTCA-3',
[0064] TSWV-482R: 5'-CTTCTCTGGTGTCATACTTCT-3'.
[0065] The fluorescent quantitative primers are: TSWV-113F: CTTGCCATAATGCTGGGAGGTAG, TSWV-113R: TCCCGAGGTCTTTGTATTTTGC.
[0066] Table 1 Primers used in this experiment
[0067]
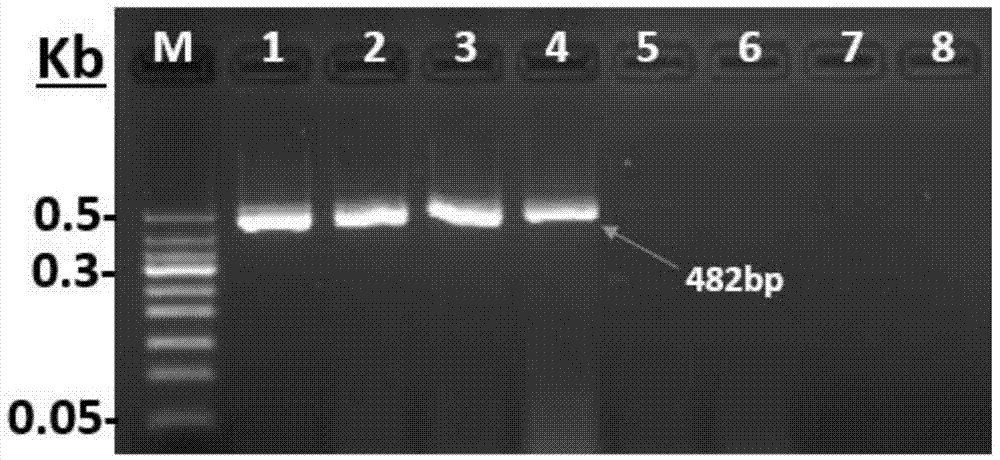
[0068] Step 2: Specificity verification of specific primers TSWV-482F / TSWV-482R:
[0069] Material:
[0070] Fresh datura leaves with TSWV,
[0071] Pepper leaves in the greenhouse of Institute of Vegetable and ...
Embodiment 2
[0081] Embodiment 2, the preparation of target fragment PCR amplification and plasmid standard
[0082] (1) Extraction of plant total RNA, synthesis of cDNA first strand and amplification of target fragments
[0083] Datura datura leaves with TSWV were used as the material, and the cDNA of Datura datura leaves with TSWV was prepared by the method in step 2 of Example 2, verified by PCR amplification, and the positive cDNA samples were quantified to 1ug / ul, and stored at -80°C for later use .
[0084] (2) Preparation of plasmid standard
[0085] The target fragment of the positive sample amplified in step (1) is recovered and purified, and cloned into pMD TM Place on 18-T carrier, place on ice for 30min, add 50ul transT1 competent cells, then add 500uL LB medium without ampicillin, culture with shaking at 37°C, recover for 1h, on solid LB agar containing X-Gal, IPTG and ampicillin Cultivate overnight on the plate; select white single colonies, add 3ml of LB liquid medium con...
Embodiment 3
[0086] Embodiment 3, the establishment of TSWV real-time fluorescent quantitative PCR method
[0087] (1) Gradient PCR to verify the specificity of fluorescent quantitative primers
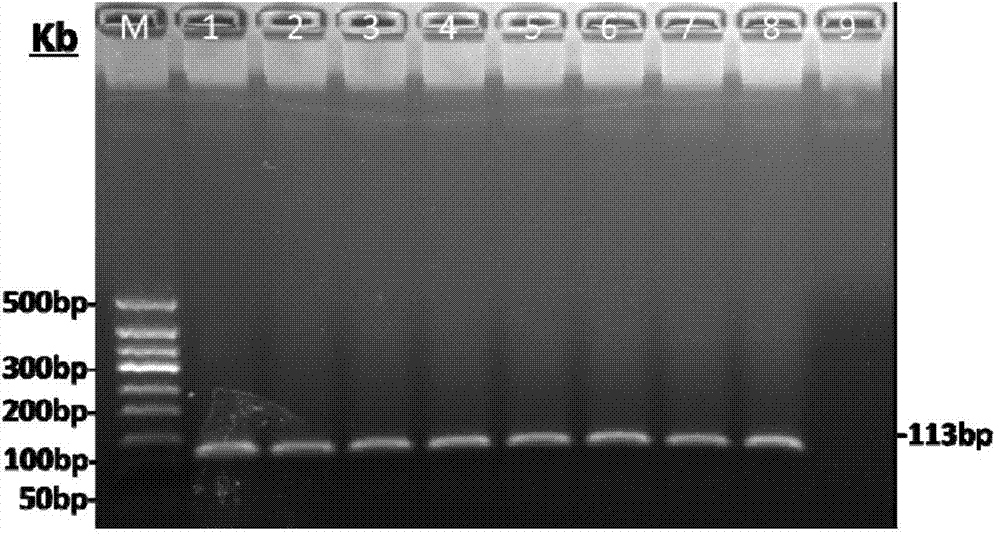
[0088] The fluorescent quantitative primer (113bp) used in this experiment was designed in the region of the target gene fragment (482bp).
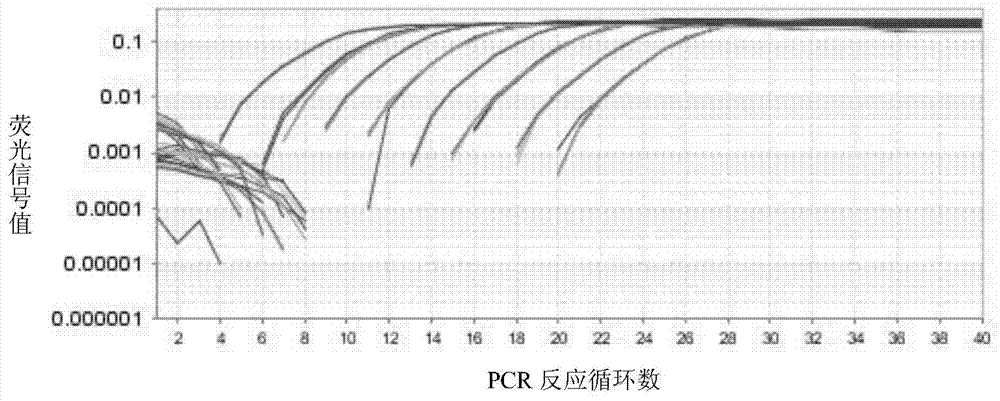
[0089] The extracted TSWV Datura leaf RNA was reverse-transcribed and RT-PCR amplified with fluorescent quantitative primers. Step is the same as step 2 of embodiment 1. The annealing temperature in the PCR program was set to 8 gradients: 65°C, 64.5°C, 63.5°C, 62°C, 60.2°C, 58.9°C, 57.8°C, 57.0°C.
[0090] Take 10uL of the amplification product and electrophoresis with 2% agarose gel in TBE buffer, the result is as follows figure 2 , PCR products at 8 annealing temperatures were all single bands of 113bp, indicating that the fluorescent quantitative primers had strong specificity. The PCR product was sent to Beijing Qingke Xinye Biotechnology Co., Ltd. f...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com