CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) based PCR (Polymerase Chain Reaction) typing method and application thereof
A reaction and reaction reagent technology, applied in the field of CRISPR typing PCR, can solve the problems of false positives, non-specific amplification, and weak primer specificity in PCR detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
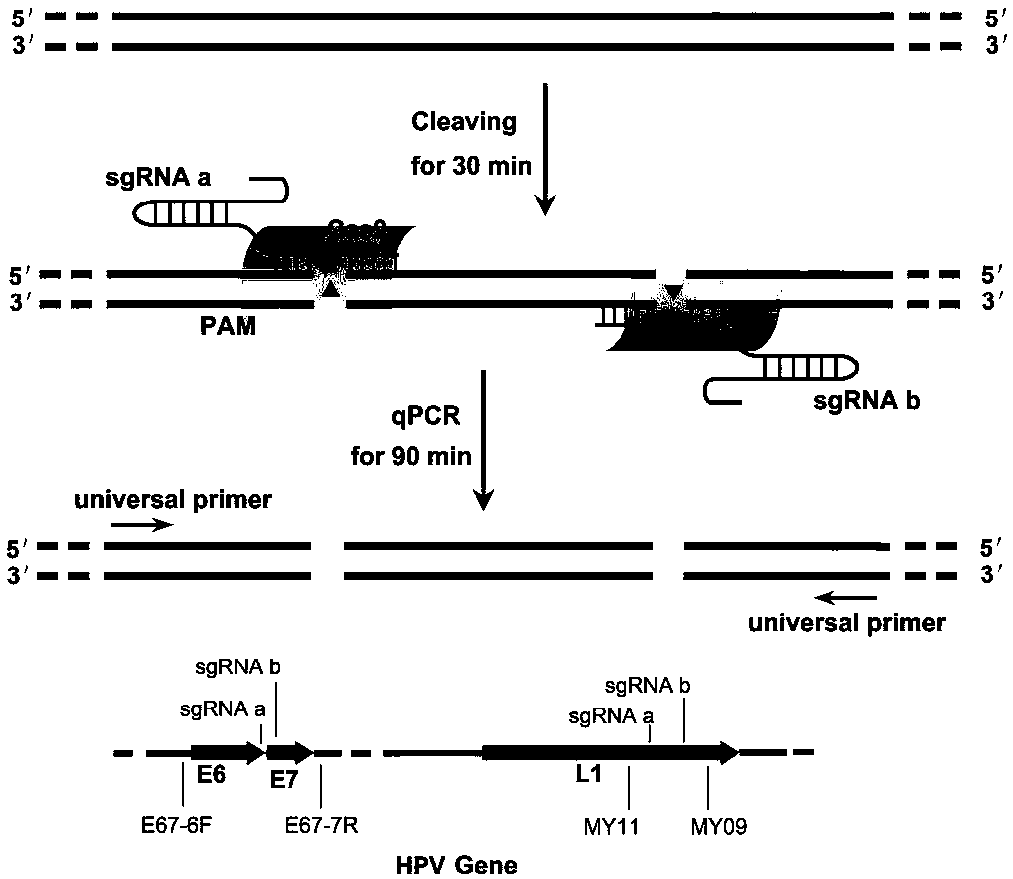
[0042] Schematic diagram of the principle and flow chart of ctPCR3.0 detection and typing of DNA molecules figure 1 shown. Schematic diagram of ctPCR3.0 detection of DNA molecules ( figure 1 ). The method detects target DNA homogeneously in one step: the detected DNA sample is first cleaved by a pair of Cas9 / sgRNA complexes, followed by qPCR with a pair of universal primers. All detection components (DNA to be detected, Cas9 protein, sgRNA and qPCR reagents) are pre-mixed in one PCR tube. The whole detection process is to add a period of constant temperature incubation time (37° C., 30 minutes) before the common PCR procedure. During the constant temperature incubation step, Cas9 nucleases complexed with a pair of sgRNAs (sgRNAa and b) respectively and cut DNA samples simultaneously. Specific cleavage of target DNA by Cas9 / sgRNA will result in increased Ct values in qPCR amplification.
Embodiment 2
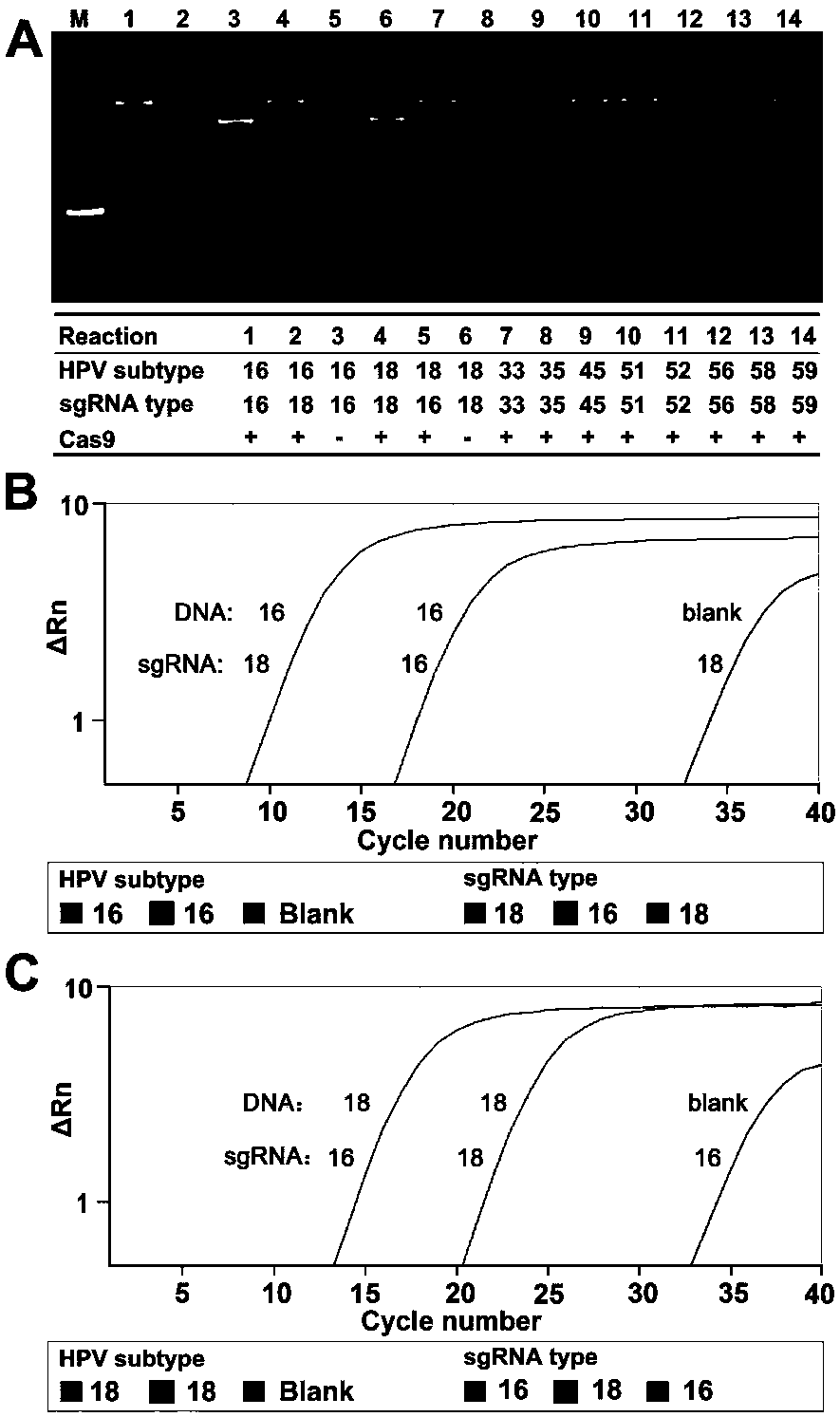
[0045] Cutting HPV plasmids with Cas9 / sgRNA and identifying HPV16 and 18 with ctPCR3.0
[0046] experimental method:
[0047] Preparation of sgRNA:
[0048] Preparation of sgRNA in vitro transcription template: PCR1: According to the backbone part of sgRNA, first design a pair of primers (F1 and R as shown in Table 1) for PCR. PCR reaction system (30 μL): 2 μL F1 (Table 1), 2 μL R (Table 1), 15 μL 2×primestar (TAKARA), with H 2 O Make up the volume to 30 μL. PCA reaction program: 95°C for 2 minutes; 7 cycles: 95°C for 15 seconds, 72°C for 1 minute. Then use 1.5% agarose gel 100V electrophoresis for 40 minutes, recover and purify with gel recovery kit (Axygen), dissolve in 25 μ L of eluent, and use Nanodrop2000 spectrophotometer to detect its DNA concentration and purity, and this fragment is named fragment 1. Store at -20°C for later use. PCR2: PCR amplification was carried out using fragment 1 as a template and F2 and Sg-R as primers. PCR reaction system (50 μL): 2 μL F...
Embodiment 3
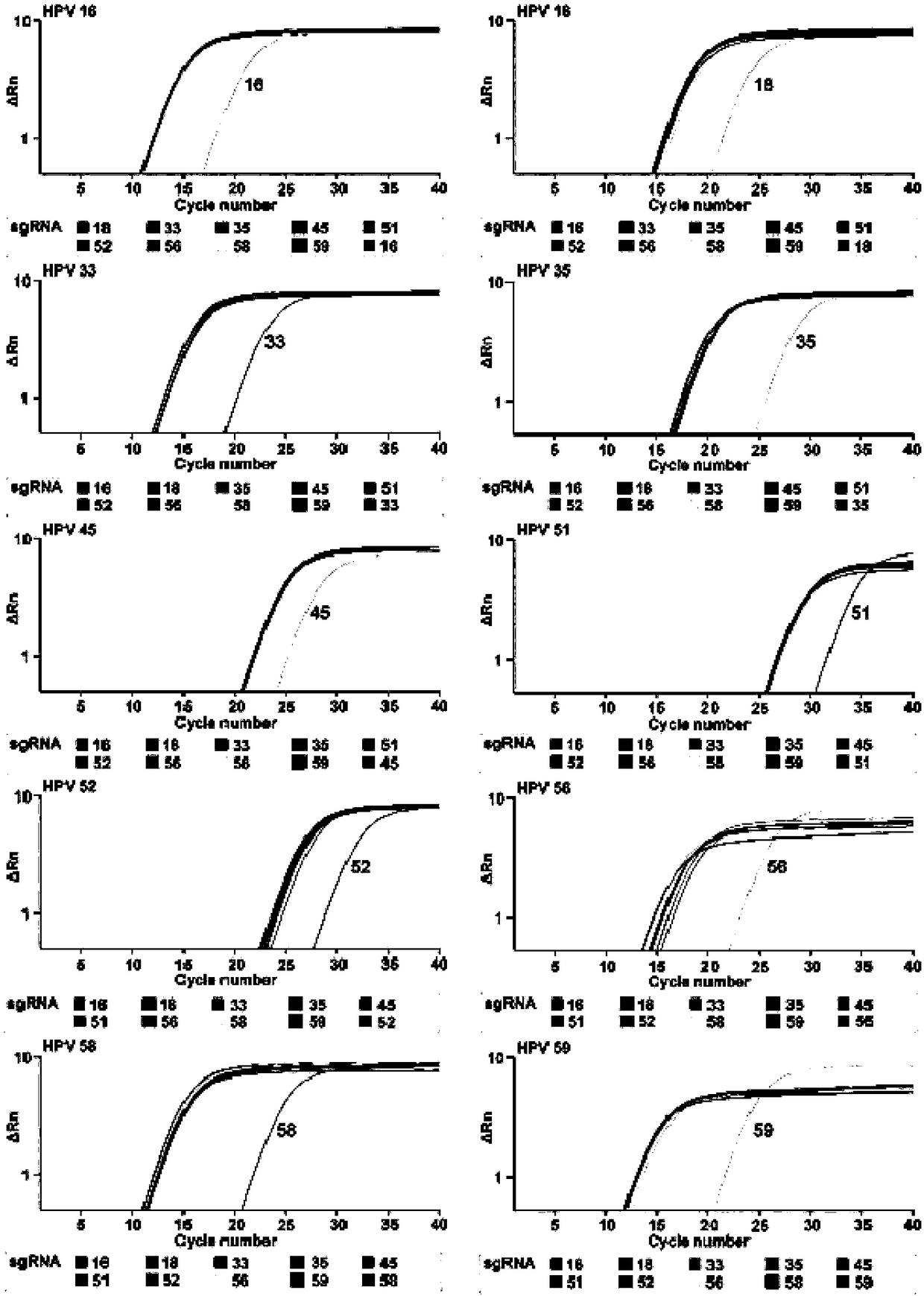
[0067] Detection of L1 gene in HPV subtype plasmid by ctPCR3.0
[0068] experimental method:
[0069] HPV plasmid DNA (2ng) was cleaved by sgRNAs / Cas9 nuclease and directly entered into qPCR reaction. ctPCR3.0 reaction (30 μL): 15 μL 2×SYBR Green Master Mix (Yeasen), 1 μM Cas9 nuclease (NEB), 300 nM sgRNAa (Table 2), 300 nM sgRNAb (Table 2), 500 nM L1-MY09 (Table 3), 500 nM L1-MY11 (Table 3) and 2ng HPV plasmid DNA. Run protocol: 37°C for 30 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, 58.5°C for 30 seconds and 72°C for 45 seconds. The reaction was carried out on the real-time PCR device StepOne plus (ABI).
[0070] Experimental results:
[0071] In order to further verify the specificity of ctPCR3.0. There are 10 subtypes of HPV (high risk: 16, 18, 33, 35, 45, 51, 52, 56, 58, and 59) L1 plasmid DNA, and 10 pairs of sgRNAs are combined with Cas9 to cut each subtype HPV plasmid DNA (37°C, 30 minutes). The Cas9 protein was then inactivated at 95°C while...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com