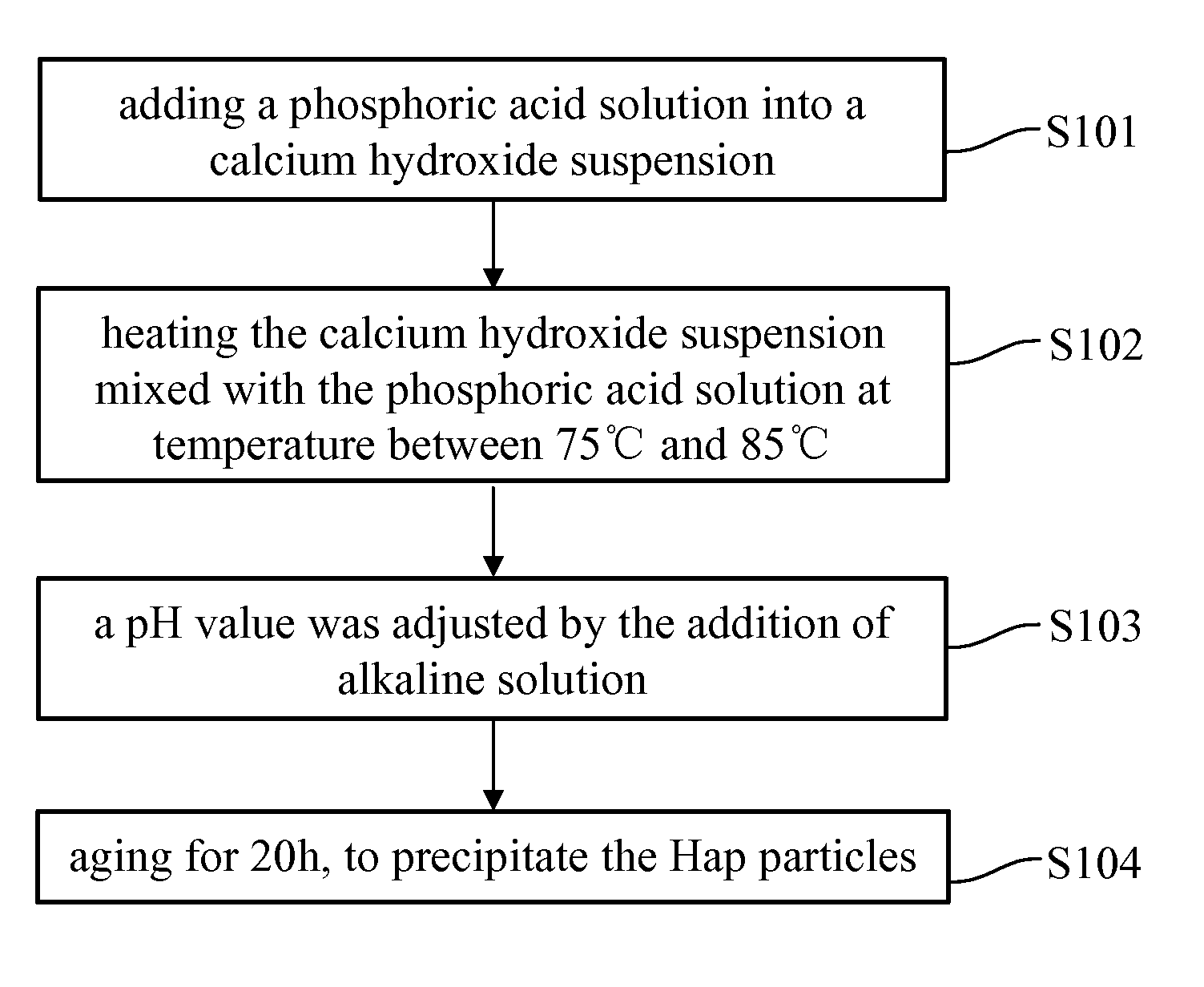
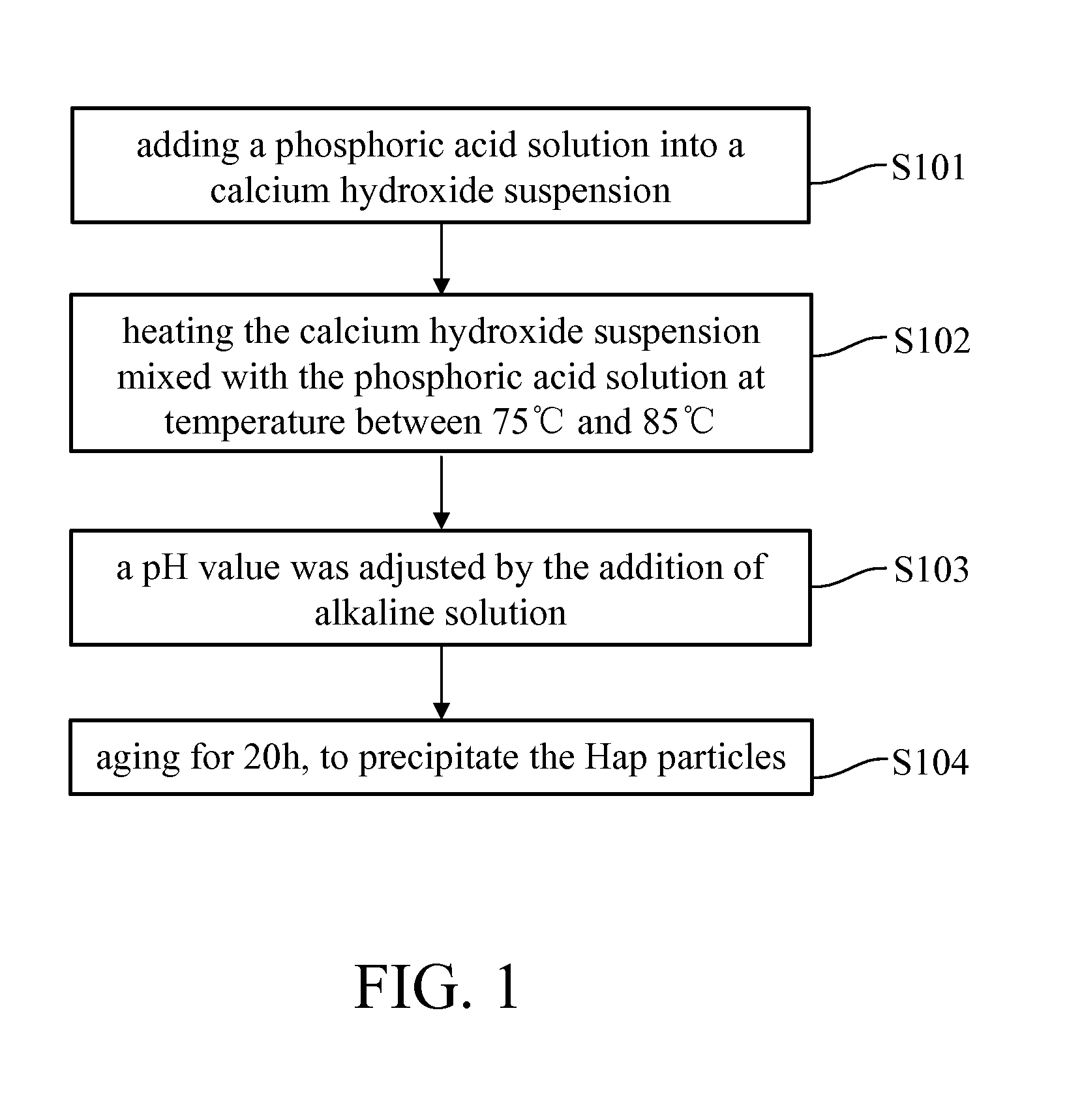
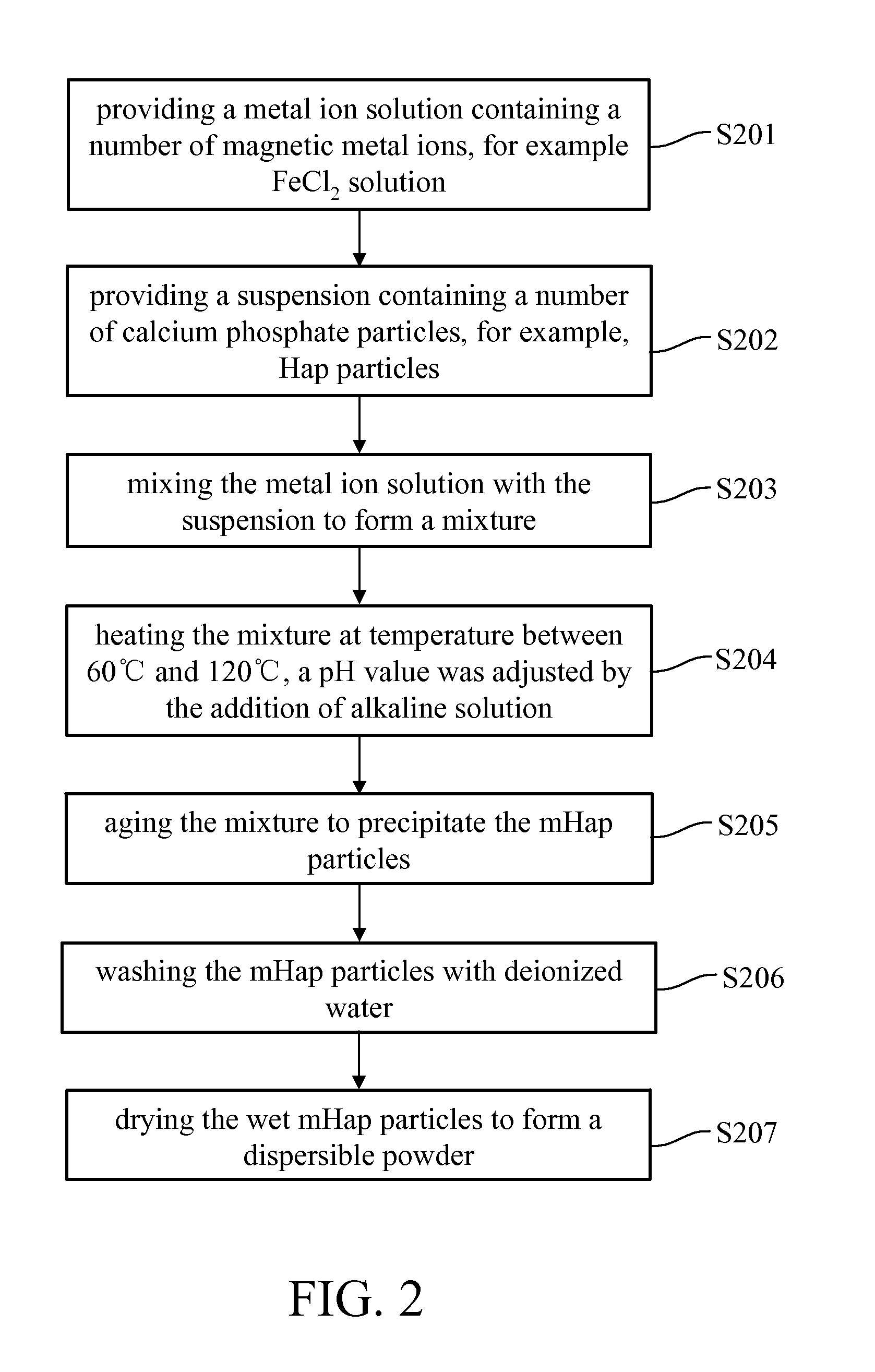
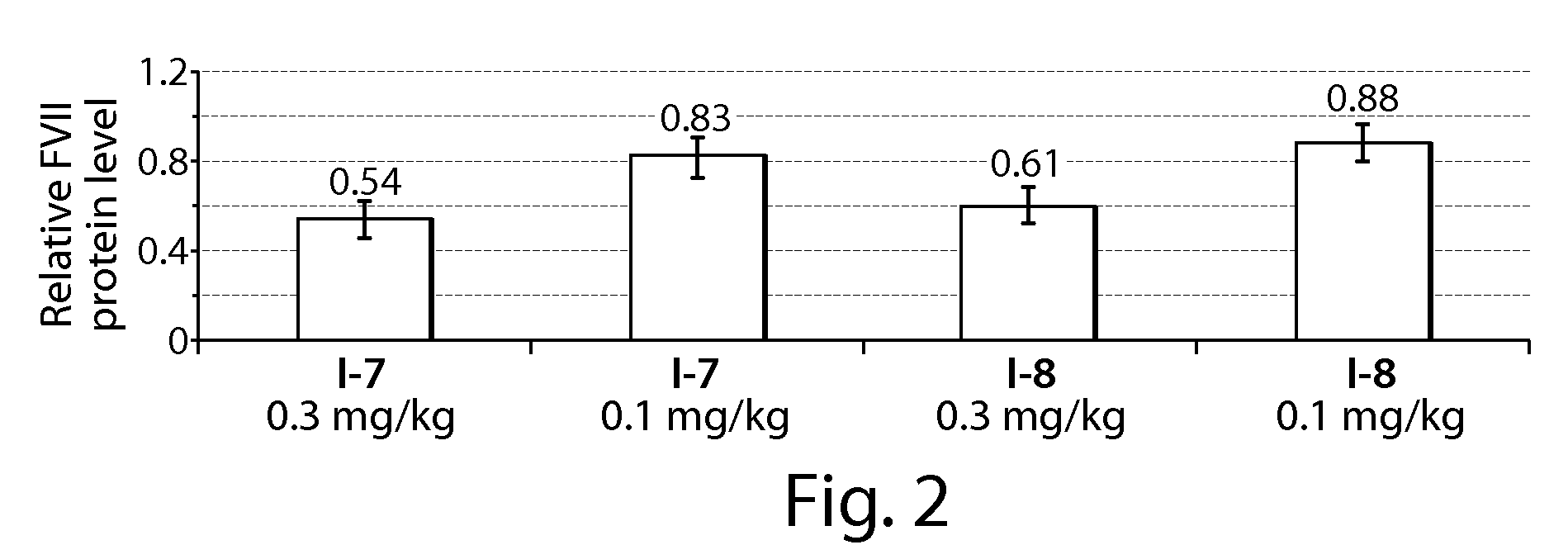
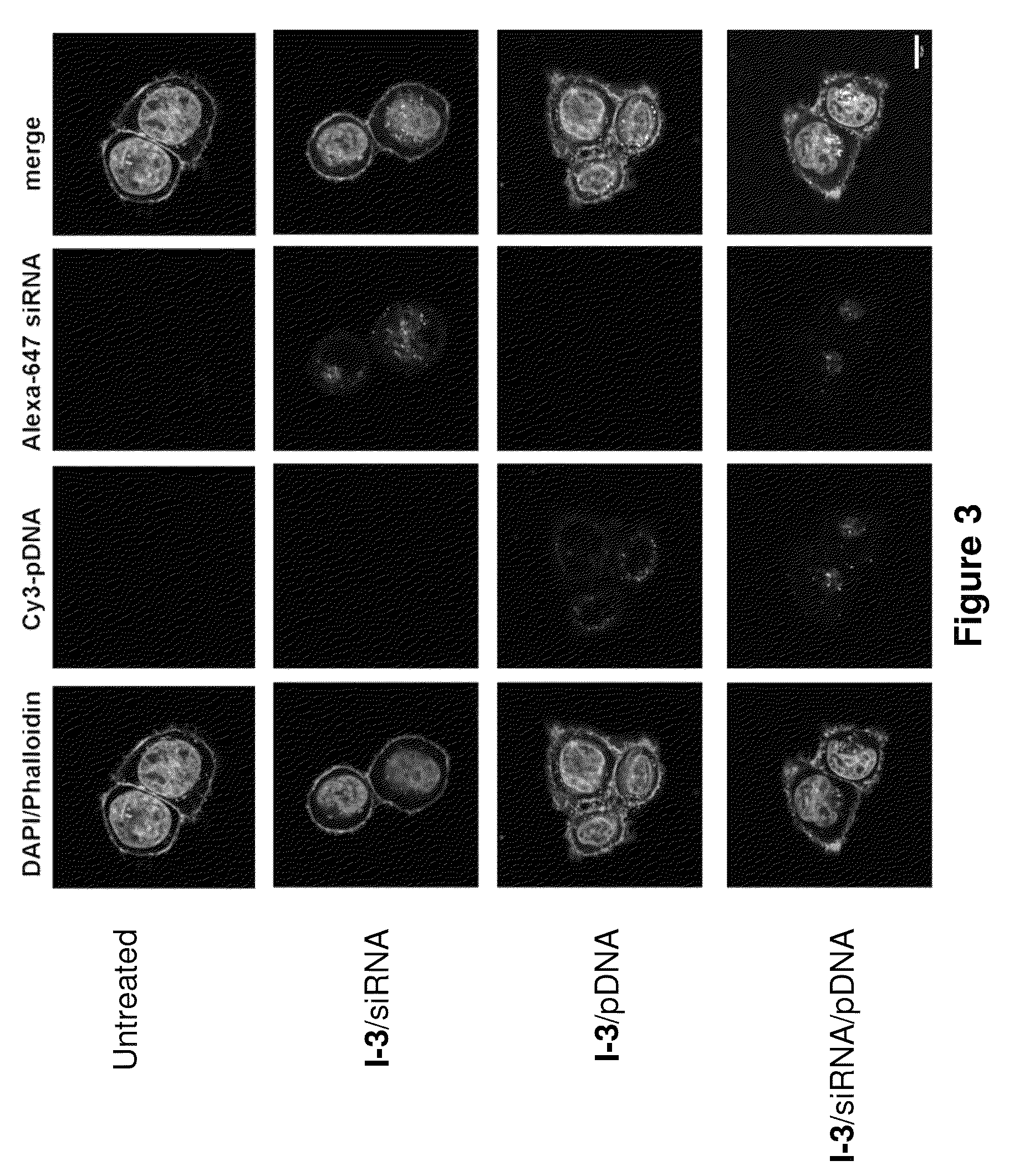
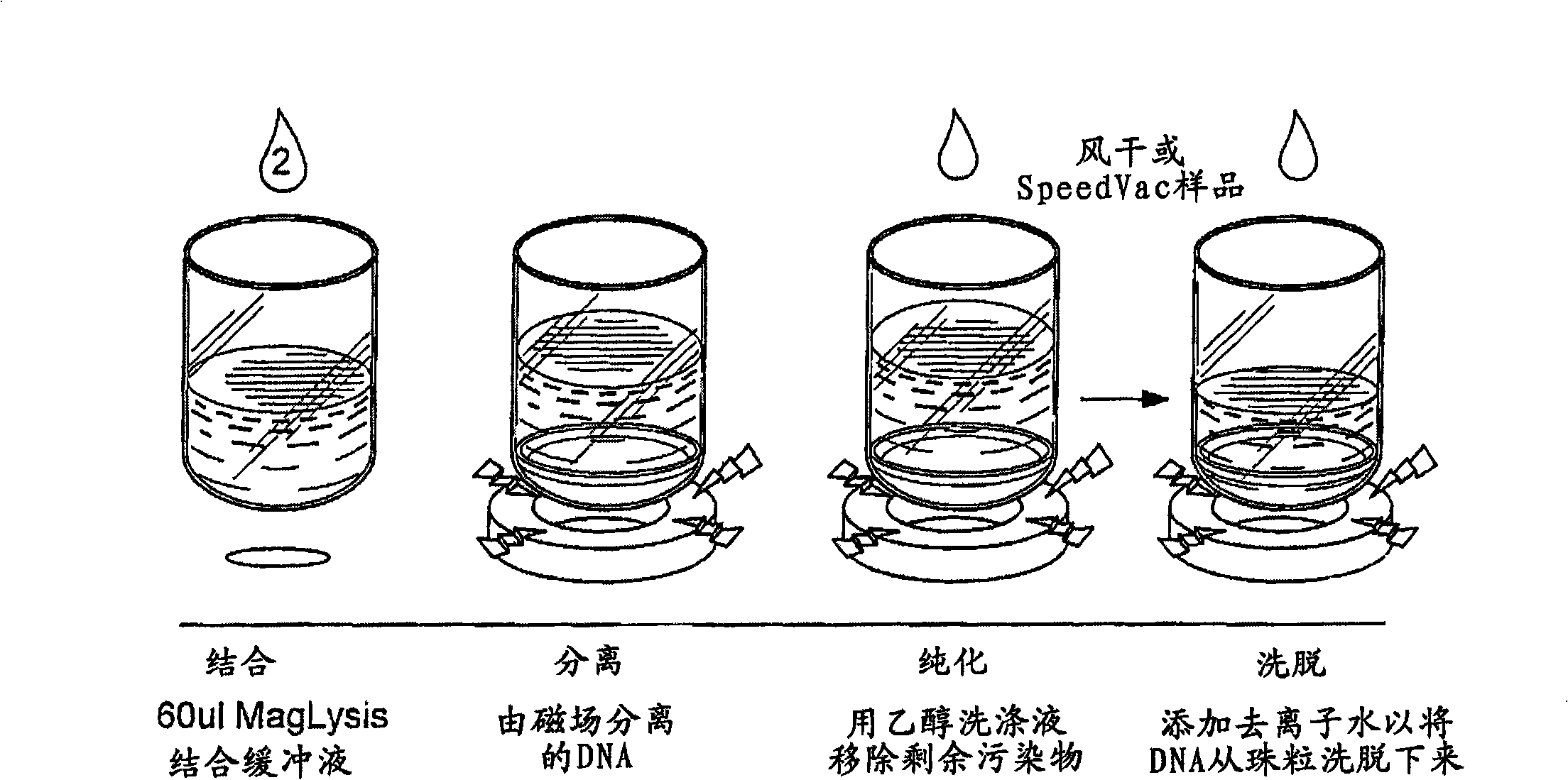
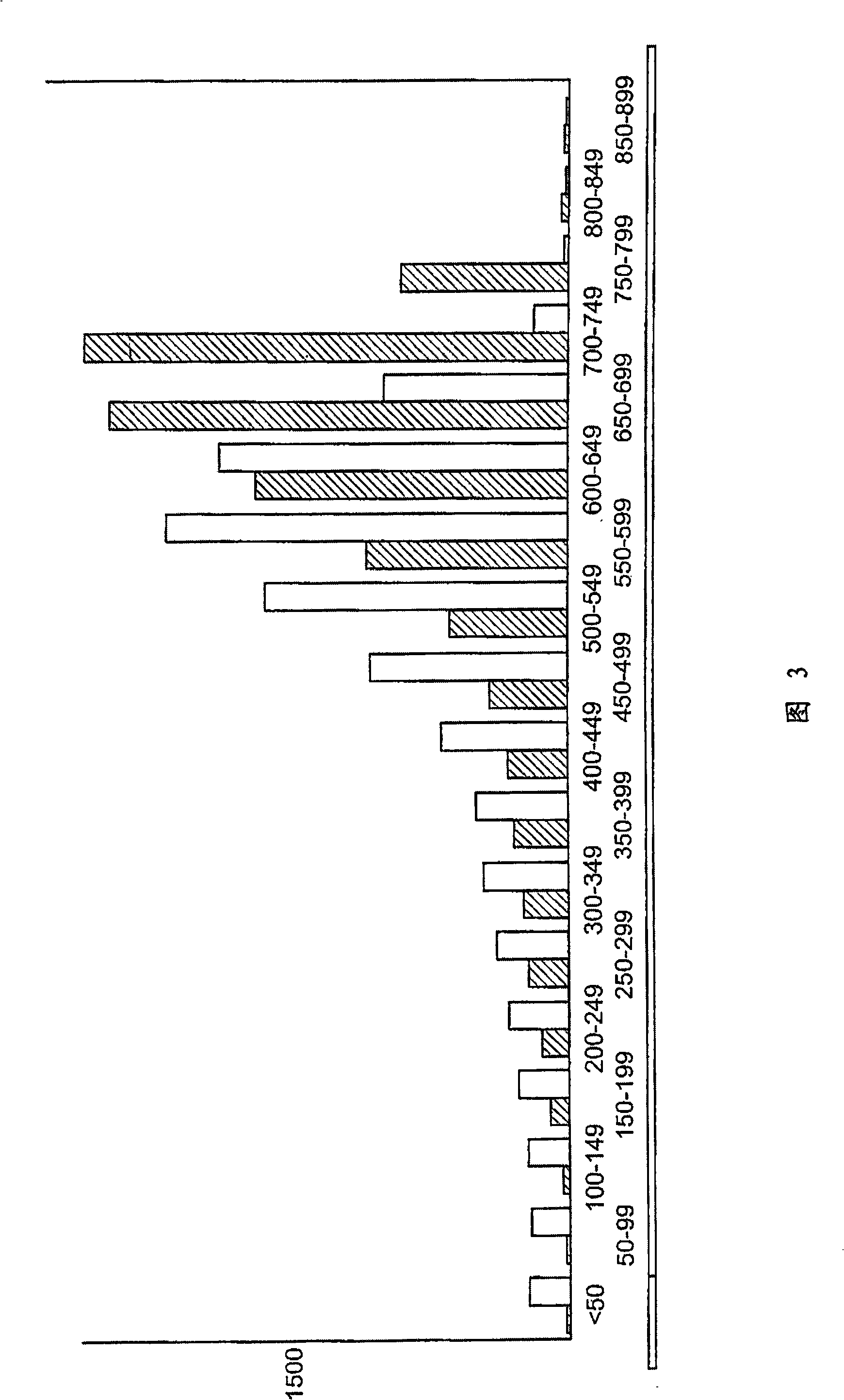
Patents
Literature
624 results about "Plasmid dna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
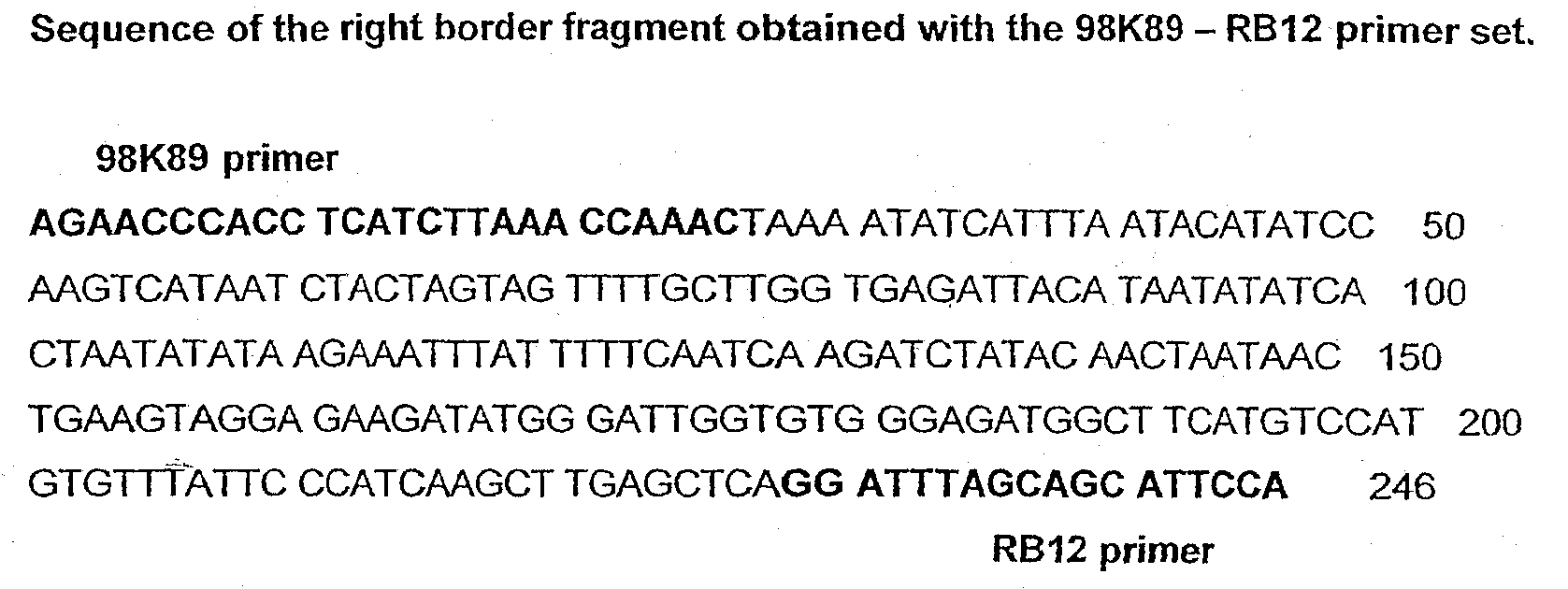
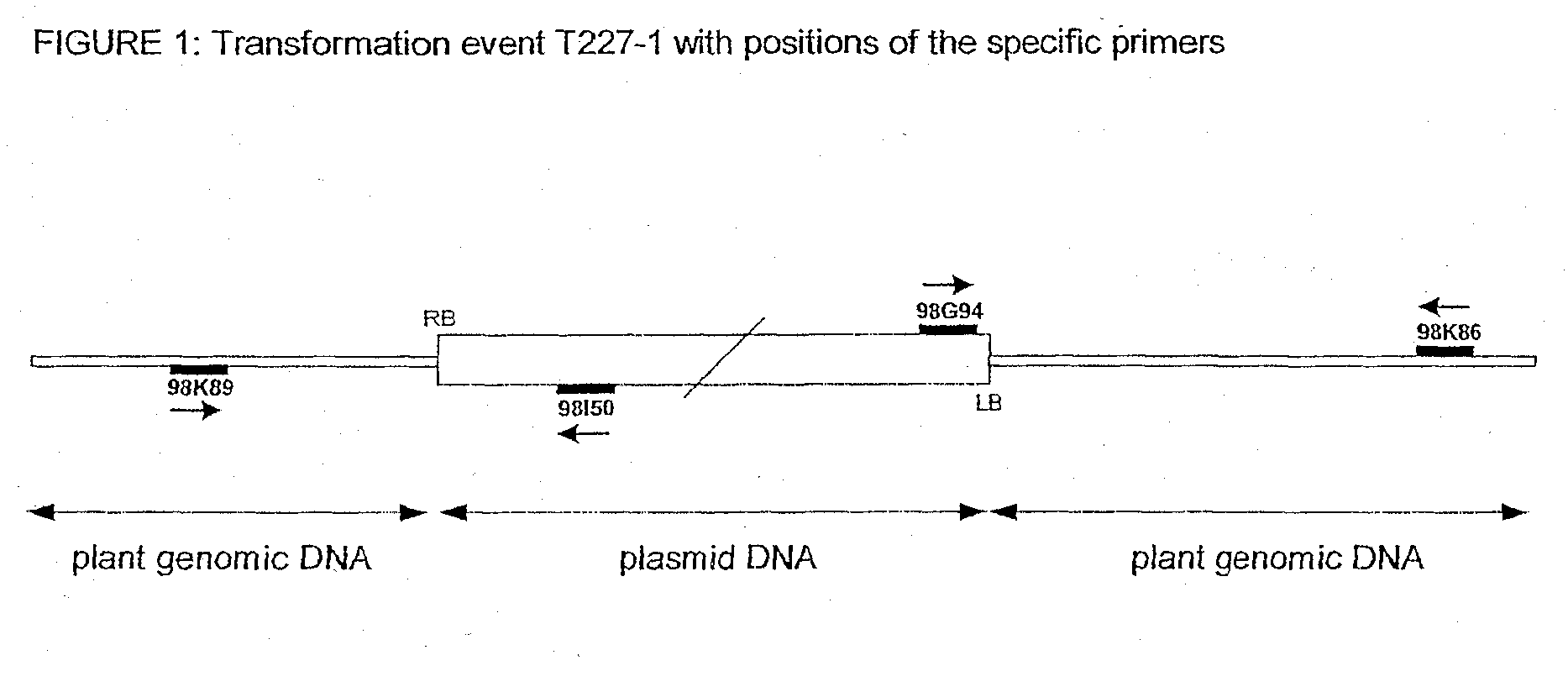
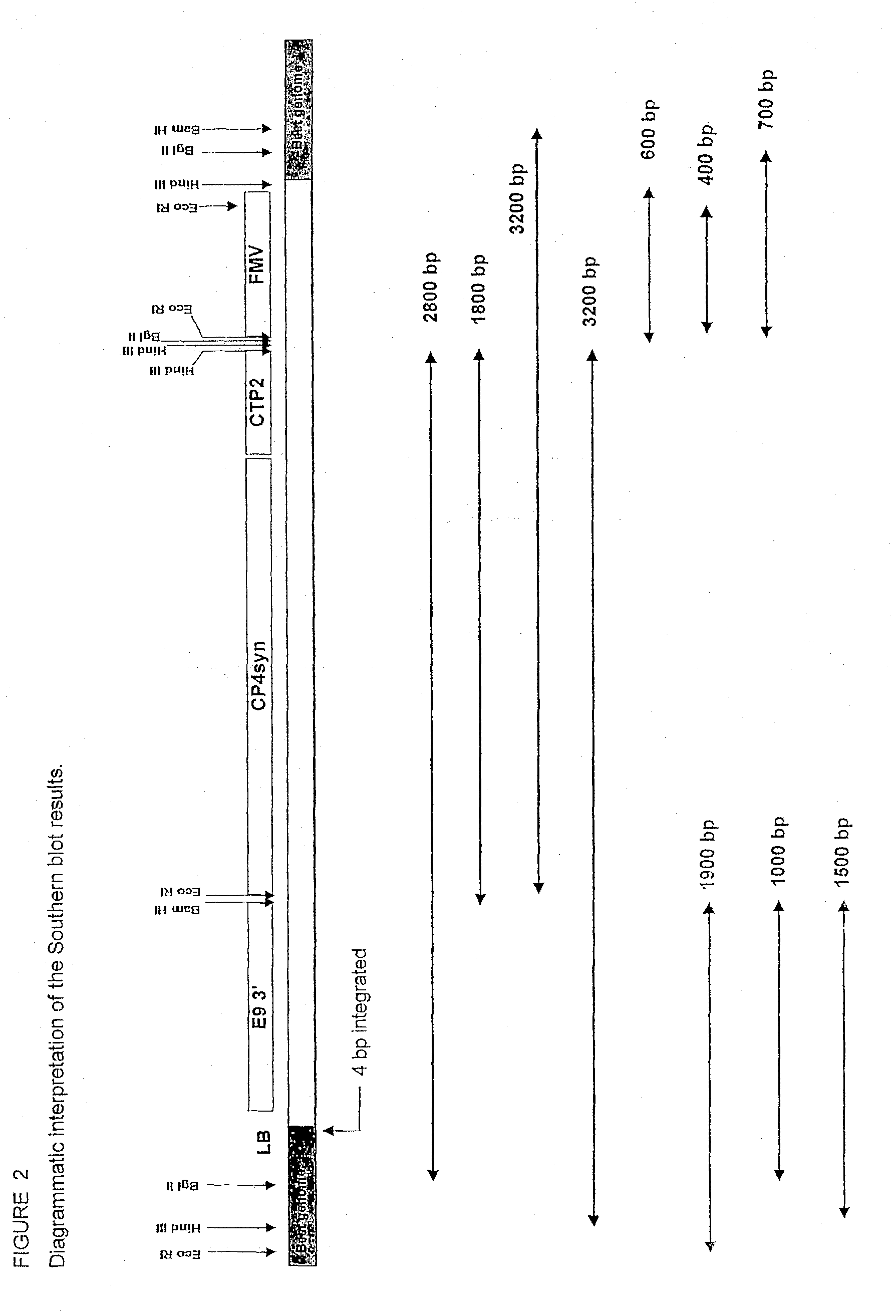
T227-1 flanking sequence
A herbicide resistant transformed sugar beet that is detectable by the specific primers developed to match the DNA sequences that flank the left and / or right border region of the inserted transgenic DNA and the method of identifying primer pairs containing plant genomic DNA / plasmid DNA. More specifically, the present invention covers a specific glyphosate resistant sugar beet plant having an insertion of the transgenic material identified as the T227-1 event. The present invention additionally covers primer pairs: plant genomic DNA / Plasmid DNA that are herein identified. Additionally, these primer pairs for either the left or the right flanking regions make an event specific test for the T227-1 insert of transgenic material.
Owner:SES EURO N V
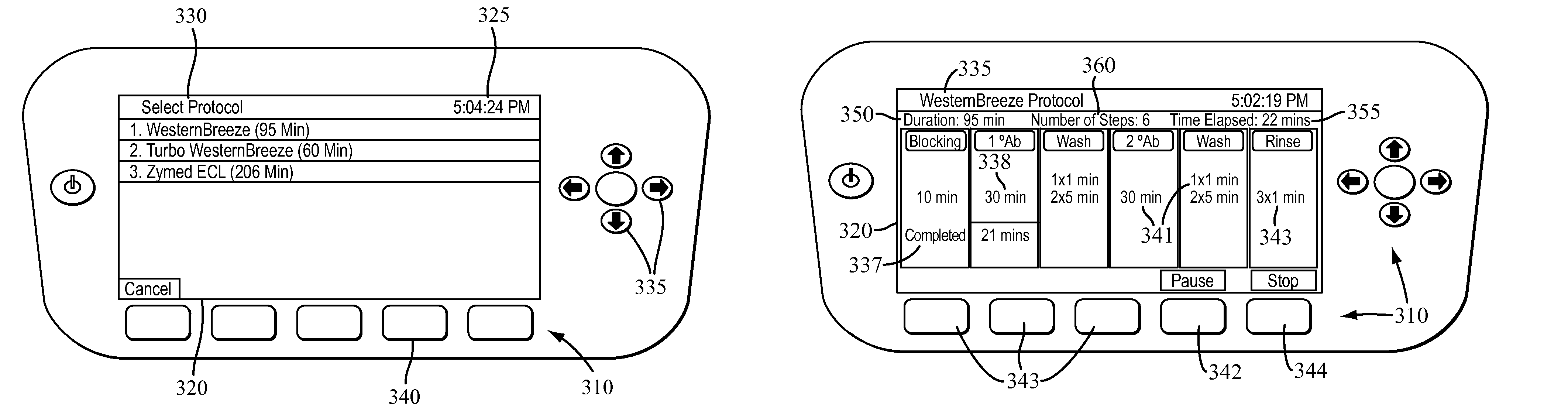
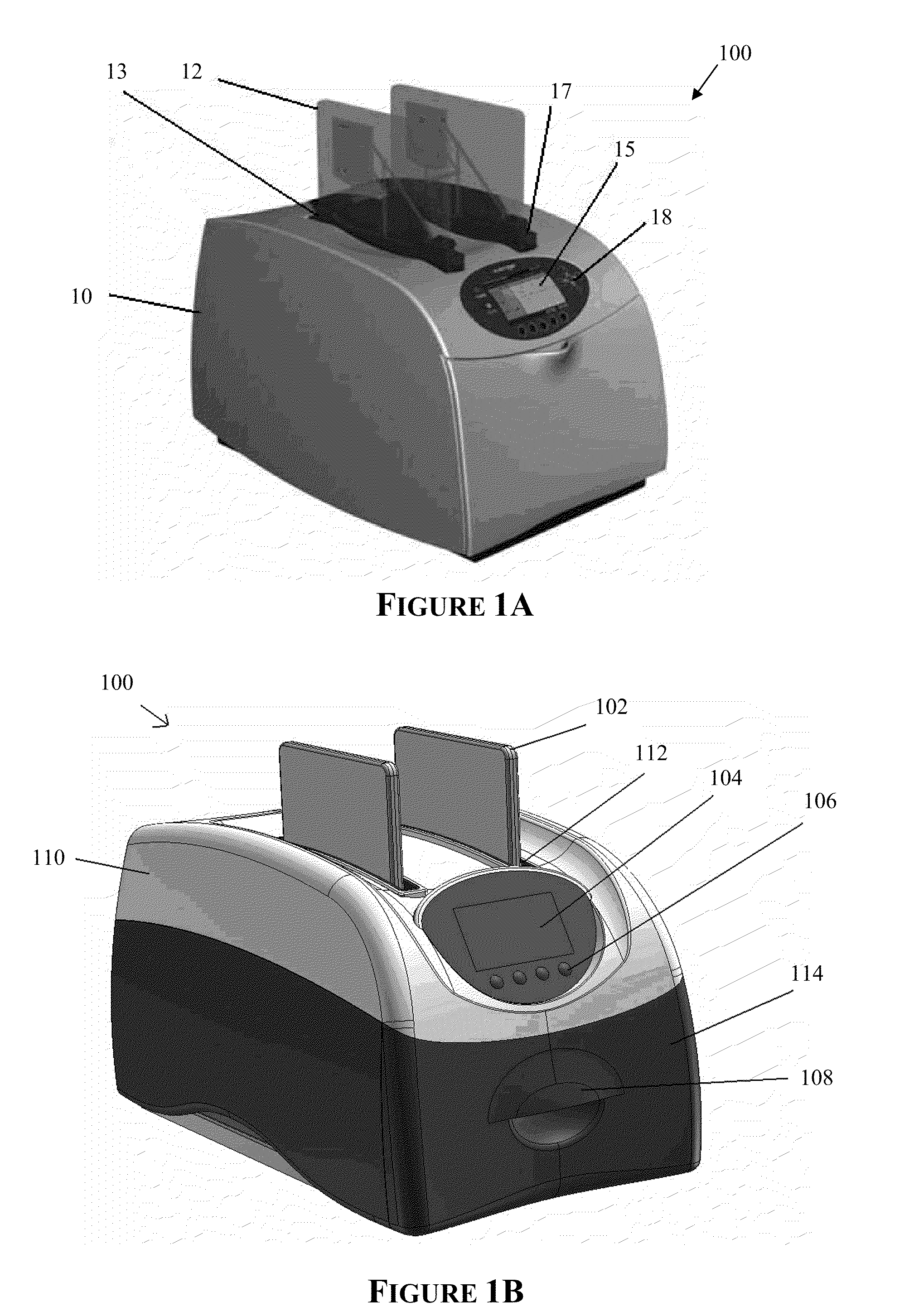
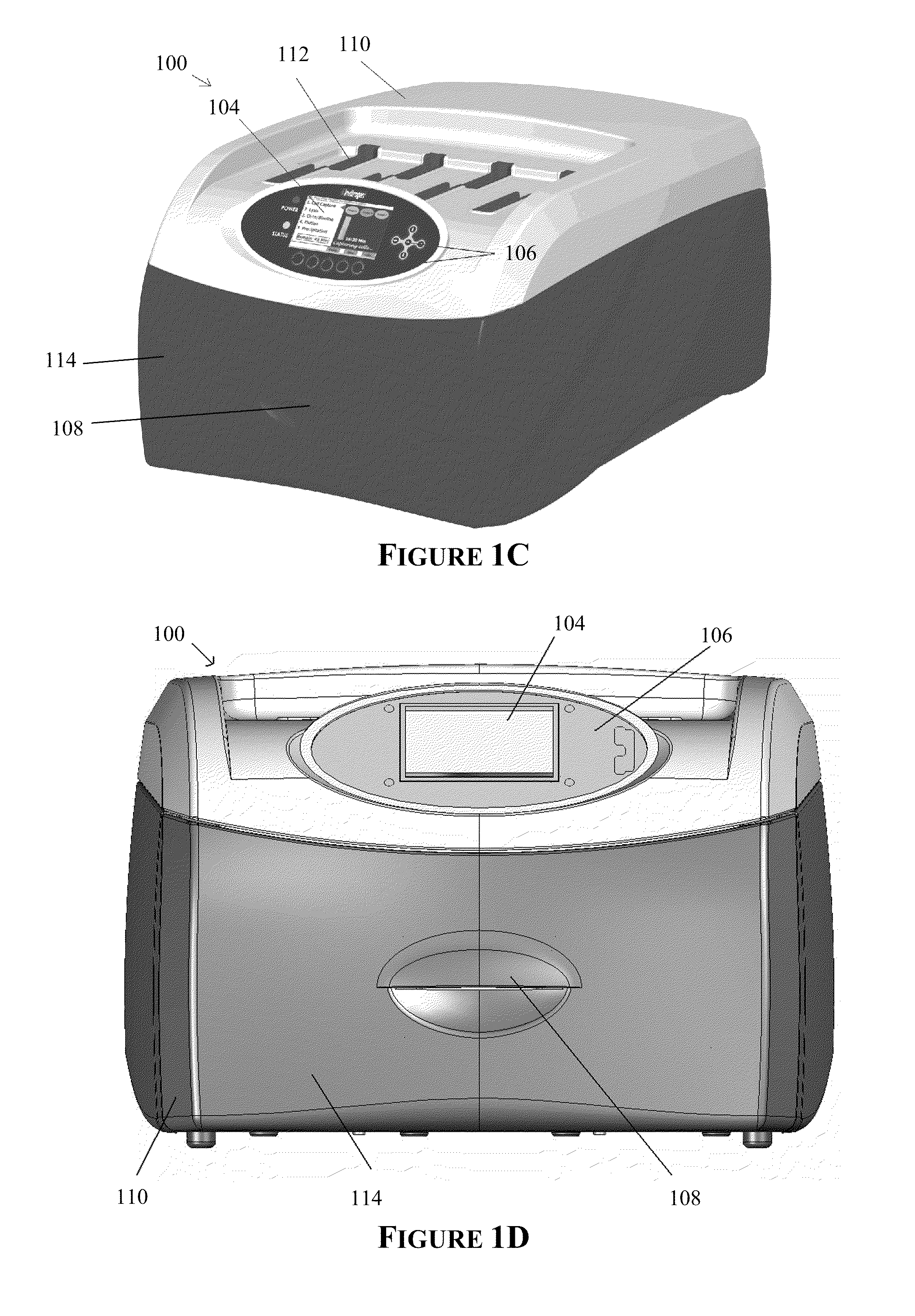
Apparatus for and method of processing biological samples
The present invention provides systems, devices, apparatuses and methods for automated bioprocessing. Examples of protocols and bioprocessing procedures suitable for the present invention include but are not limited to: immunoprecipitation, chromatin immunoprecipitation, recombinant protein isolation, nucleic acid separation and isolation, protein labeling, separation and isolation, cell separation and isolation, food safety analysis and automatic bead based separation. In some embodiments, the invention provides automated systems, automated devices, automated cartridges and automated methods of western blot processing. Other embodiments include automated systems, automated devices, automated cartridges and automated methods for separation, preparation and purification of nucleic acids, such as DNA or RNA or fragments thereof, including plasmid DNA, genomic DNA, bacterial DNA, viral DNA and any other DNA, and for automated systems, automated devices, automated cartridges and automated methods for processing, separation and purification of proteins, peptides and the like.
Owner:LIFE TECH CORP
Porous nanoparticle-supported lipid bilayers (protocells) for targeted delivery and methods of using same
ActiveUS20140079774A1Promoting death of cancer cellEfficient packagingBiocideSpecial deliveryLipid formationBinding peptide
The present invention is directed to protocells for specific targeting of hepatocellular and other cancer cells which comprise a nanoporous silica core with a supported lipid bilayer; at least one agent which facilitates cancer cell death (such as a traditional small molecule, a macromolecular cargo (e.g. siRNA or a protein toxin such as ricin toxin A-chain or diphtheria toxin A-chain) and / or a histone-packaged plasmid DNA disposed within the nanoporous silica core (preferably supercoiled in order to more efficiently package the DNA into protocells) which is optionally modified with a nuclear localization sequence to assist in localizing protocells within the nucleus of the cancer cell and the ability to express peptides involved in therapy (apoptosis / cell death) of the cancer cell or as a reporter, a targeting peptide which targets cancer cells in tissue to be treated such that binding of the protocell to the targeted cells is specific and enhanced and a fusogenic peptide that promotes endosomal escape of protocells and encapsulated DNA. Protocells according to the present invention may be used to treat cancer, especially including hepatocellular (liver) cancer using novel binding peptides (c-MET peptides) which selectively bind to hepatocellular tissue or to function in diagnosis of cancer, including cancer treatment and drug discovery.
Owner:NAT TECH & ENG SOLUTIONS OF SANDIA LLC +1
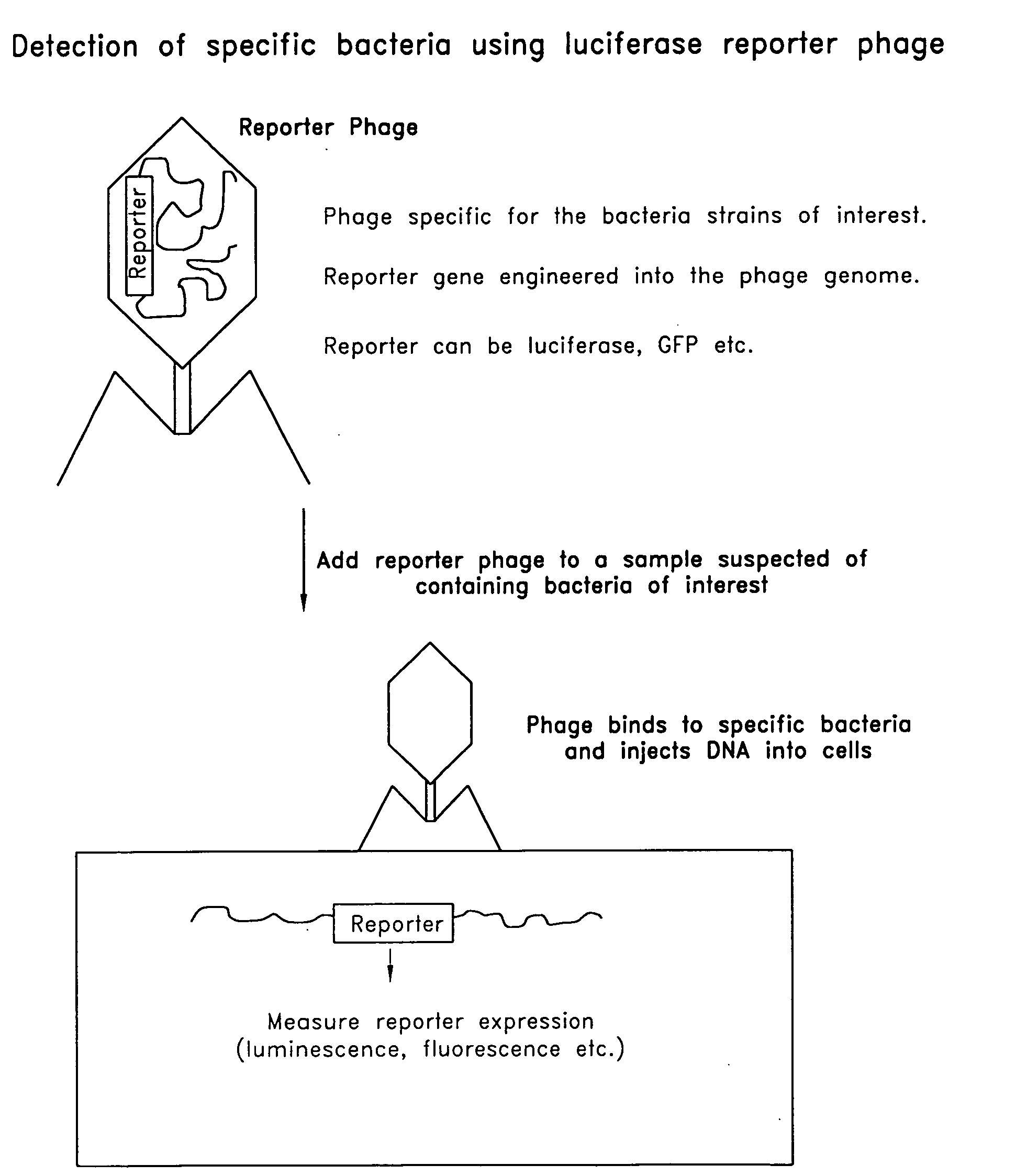
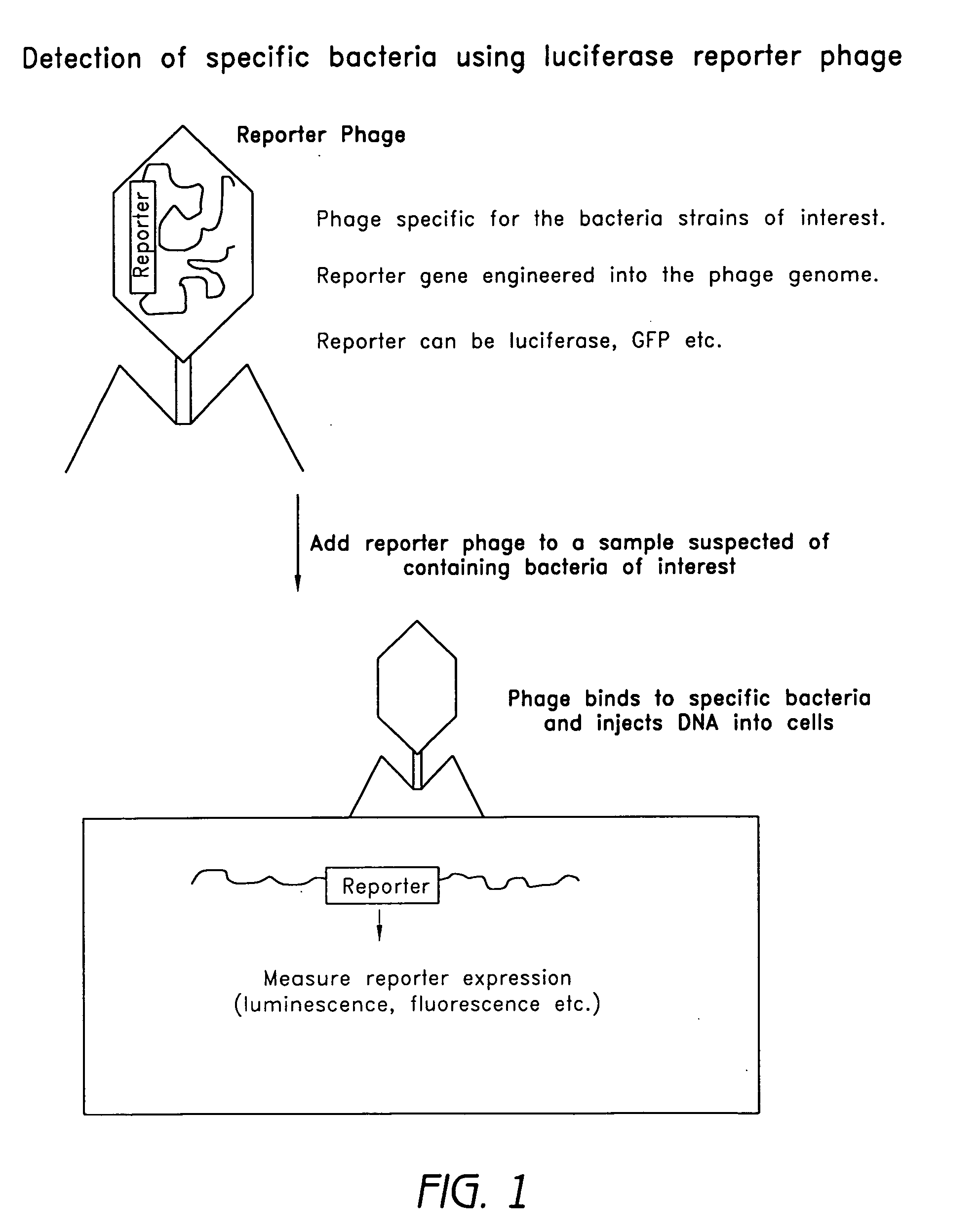
Reporter plasmid phage packaging system for detection of bacteria
InactiveUS20090155768A1Microbiological testing/measurementOther foreign material introduction processesBacteroidesOrigin of replication
The invention is related to a transducing particle that comprises a bacteriophage coat and a DNA core that comprises plasmid DNA comprising: a) a host-specific bacteriophage packaging site wherein the packaging site is substantially in isolation from sequences naturally occurring adjacent thereto in the bacteriophage genome, b) a reporter gene, c) a bacteria-specific promoter operably linked to said reporter gene, d) a bacteria-specific origin of replication, and optionally e) an antibiotic resistance gene. The invention includes phage transducing particles, methods of making transducing particles, and methods of using the transducing particles in bacterial detection.
Owner:UNITED STATES OF AMERICA
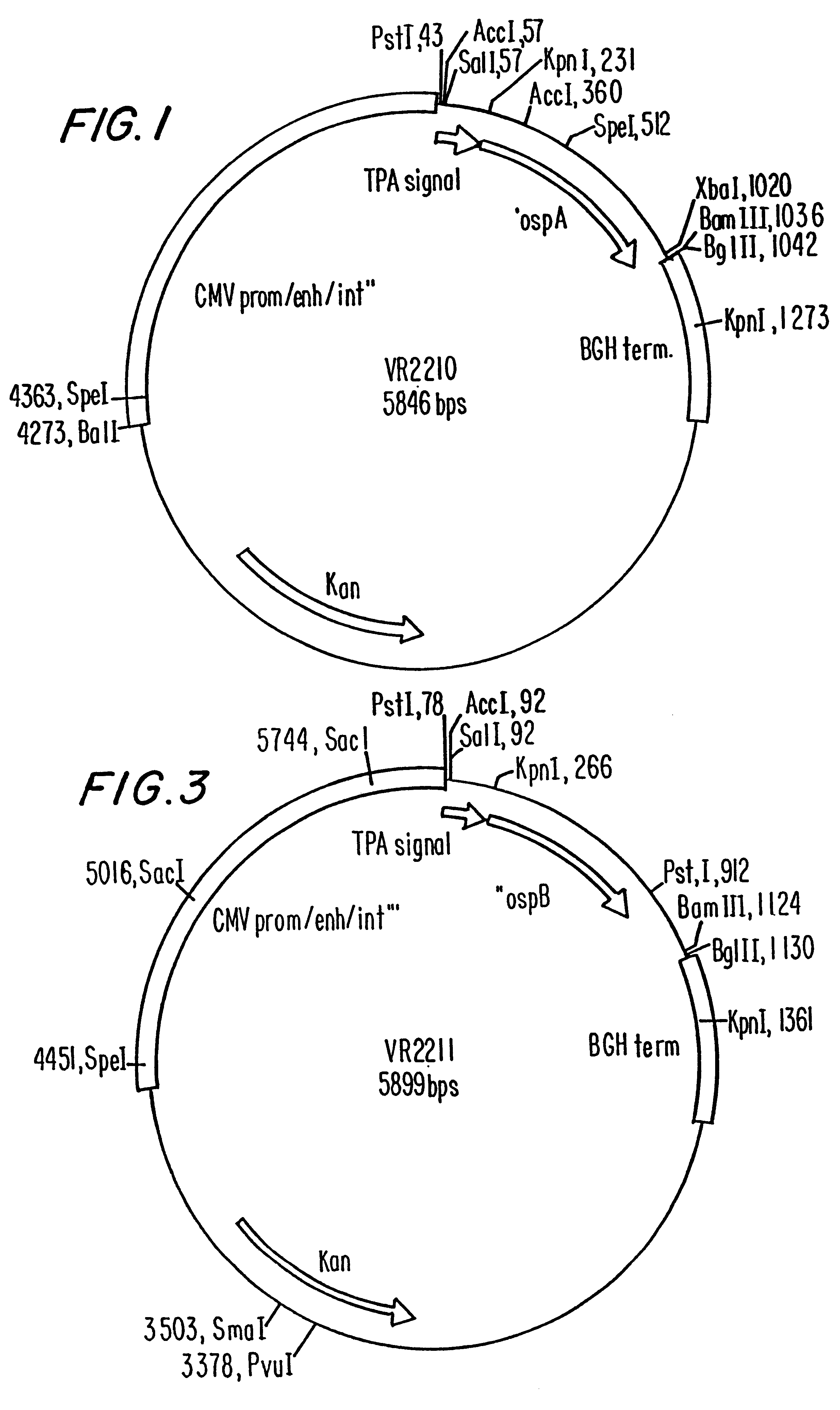
Compositions and methods for administering Borrelia DNA
Disclosed is a vaccine against Lyme Disease or its causative agent Borrelia burgdorferi (sensu stricto or sensu lato) containing a plasmid a DNA encoding a promoter for driving expression in a mammalian cell, DNA encoding a leader peptide for facilitating secretion / release of a prokaryotic protein sequence from a mammalian cell, a DNA encoding Borrelia OspA or OspB, and a DNA encoding a terminator. Disclosed too is an immunogenic composition against Lyme Disease or its causative agent Borrelia burgdorferi (sensu stricto or sensu lato) containing a plasmid comprising a DNA encoding a promoter for driving expression in a mammalian cell, DNA encoding a leader peptide for facilitating secretion / release of a prokaryotic protein sequence from a mammalian cell, a DNA encoding a Borrelia OspC, and a DNA encoding a terminator. And, methods for making and using such vaccines and the immunogenic composition are also disclosed.
Owner:PASTEUR MERIEUX SERUMS & VACCINS SA
Integrated system for high throughput capture of genetic diversity
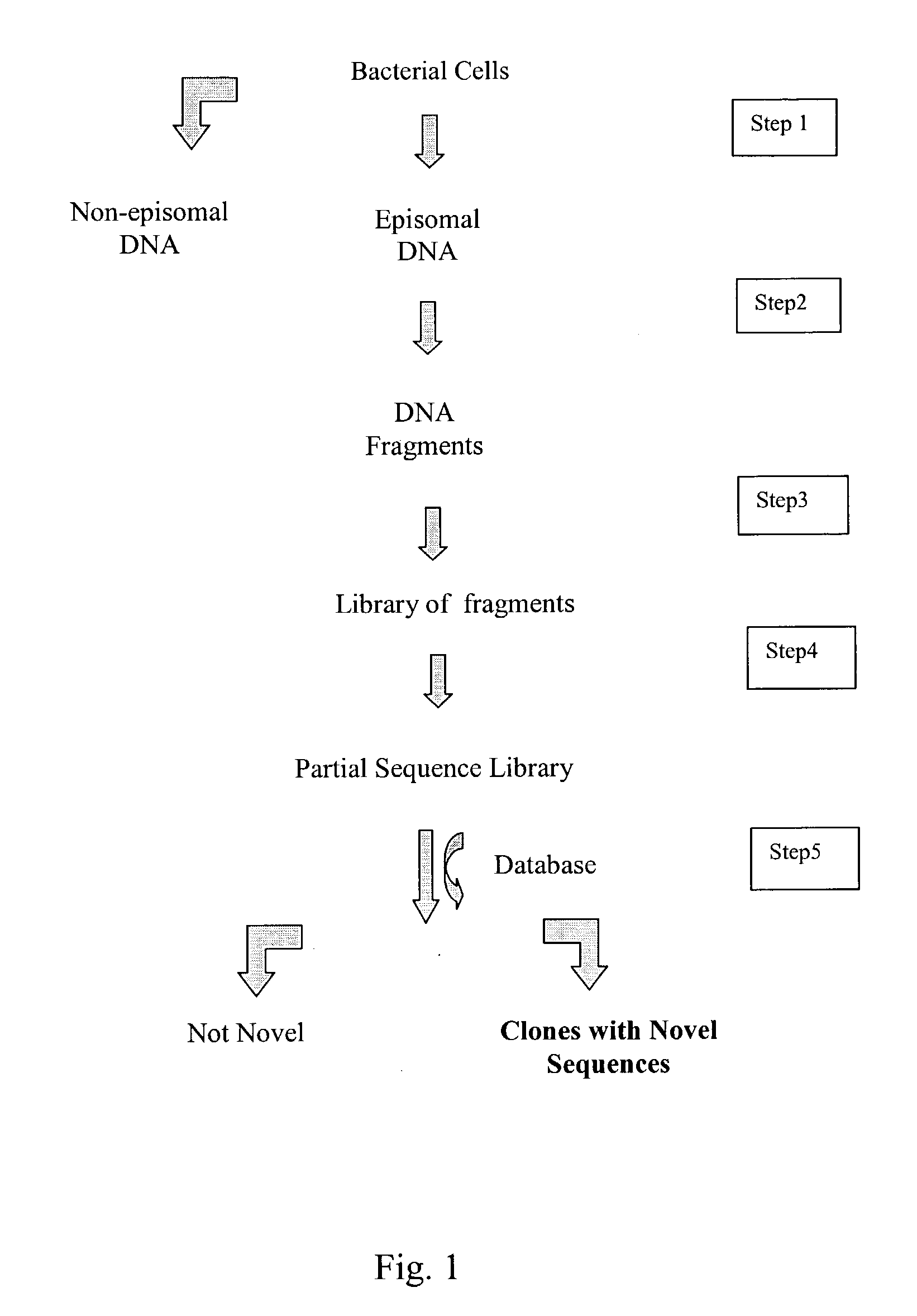
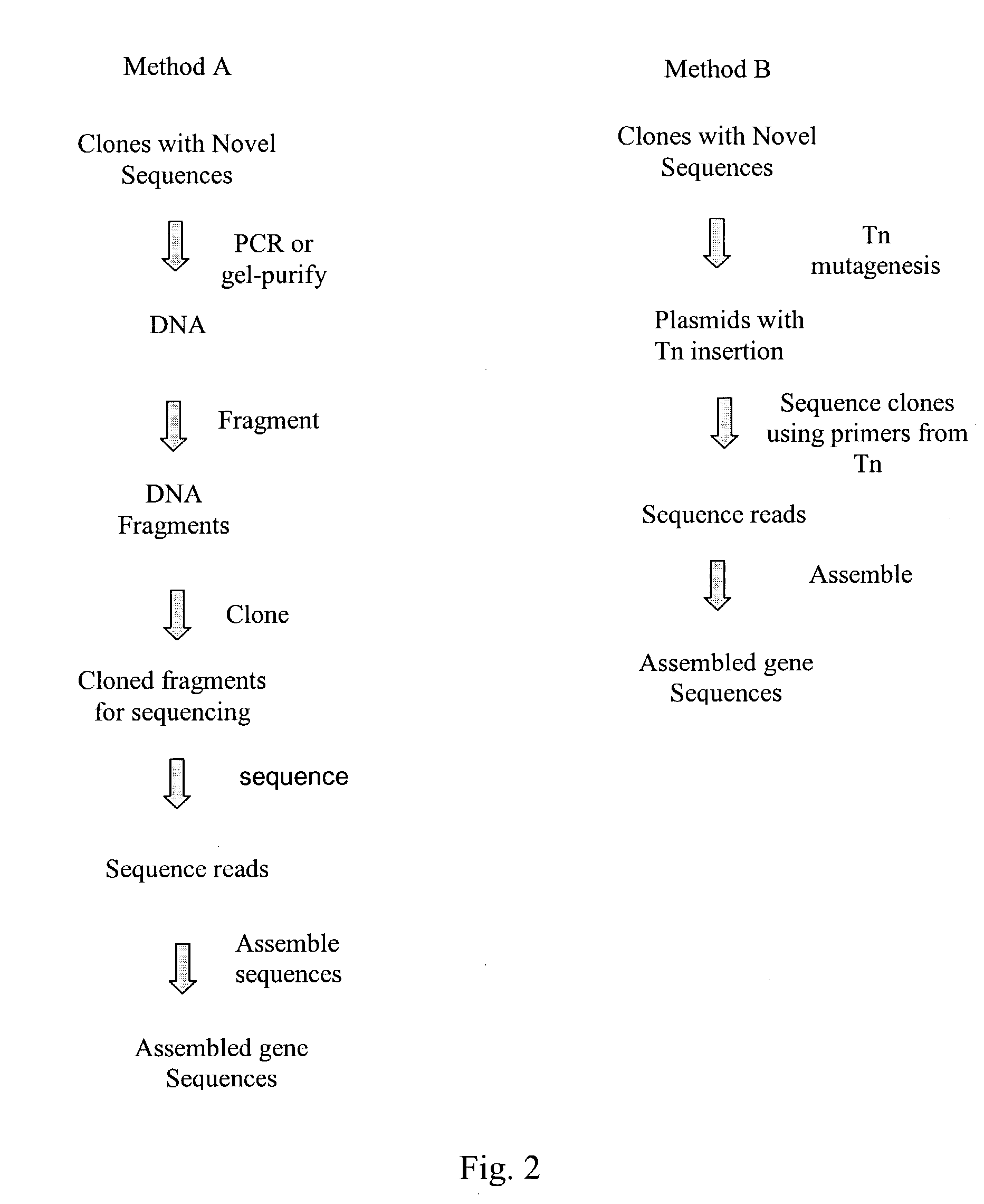
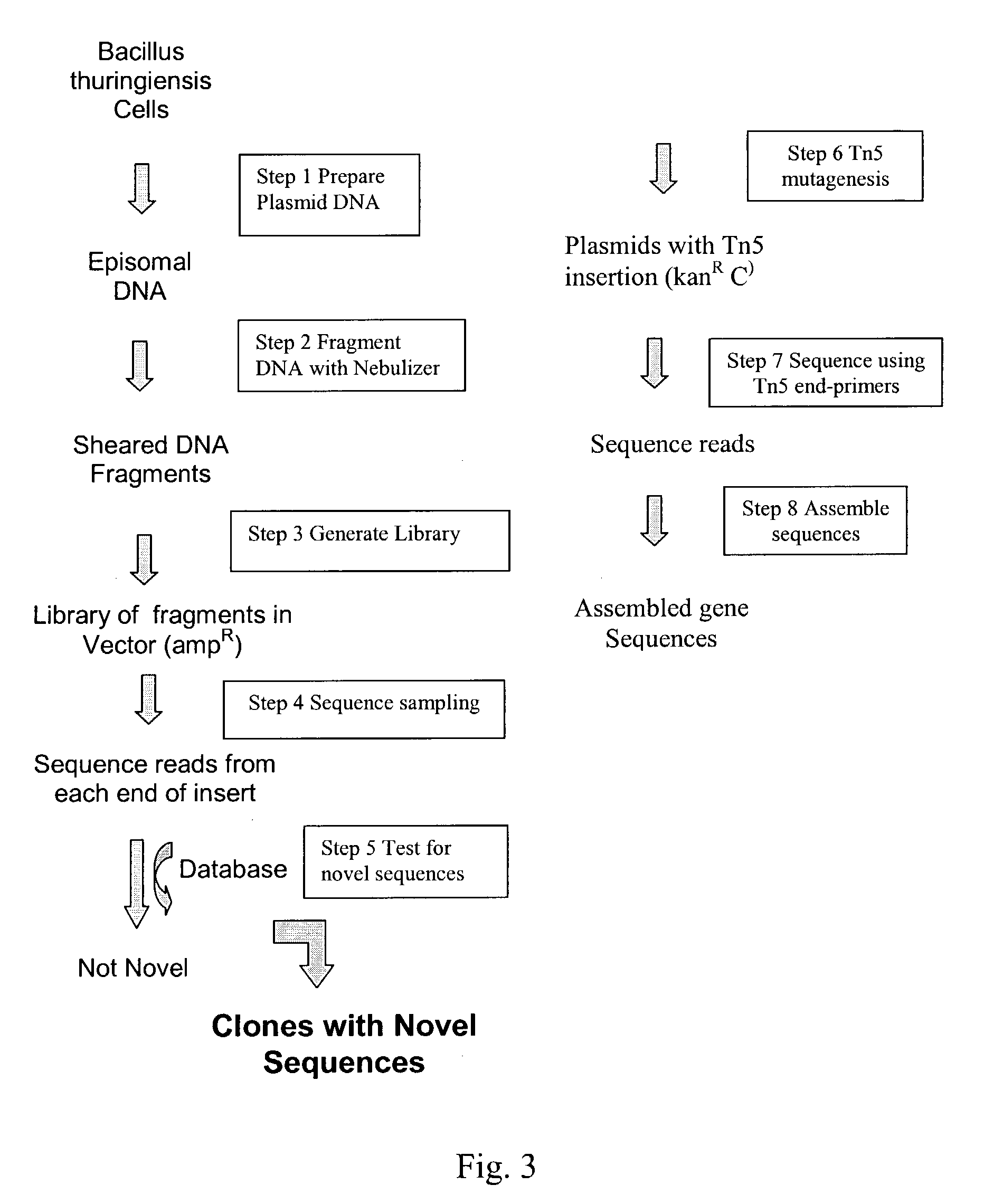
InactiveUS20040014091A1Rapid and highly efficient characterizationFast wayMicrobiological testing/measurementBiological testingGenetic diversitySequence database
Compositions and methods for rapid and highly efficient characterization of genetic diversity in organisms are provided. The methods involve rapid sequencing and characterization of extrachromosomal DNA, particularly plasmids, to identify and isolate useful nucleotide sequences. The method targets plasmid DNA and avoids repeated cloning and sequencing of the host chromosome, thus allowing one to focus on the genetic, elements carrying maximum genetic diversity. The method involves generating a library of extrachromosomal DNA clones, sequencing a portion of the clones, comparing the sequences against a database of existing DNA sequences, using an algorithm to select said novel nucleotide sequence based on the presence or absence of said portion in a database, and identification of at least one novel nucleotide sequence. The DNA sequence can also be translated in all six frames and the resulting amino acid sequences can be compared against a database of protein sequences. The integrated approach provides a rapid and efficient method to identify and isolate useful genes. Organisms of particular interest include, but are not limited to bacteria, fungi, algae, and the like. Compositions comprise a mini-cosmid vector comprising a stuffer fragment and at least one cos site.
Owner:BASF AGRICULTURAL SOLUTIONS SEED LLC
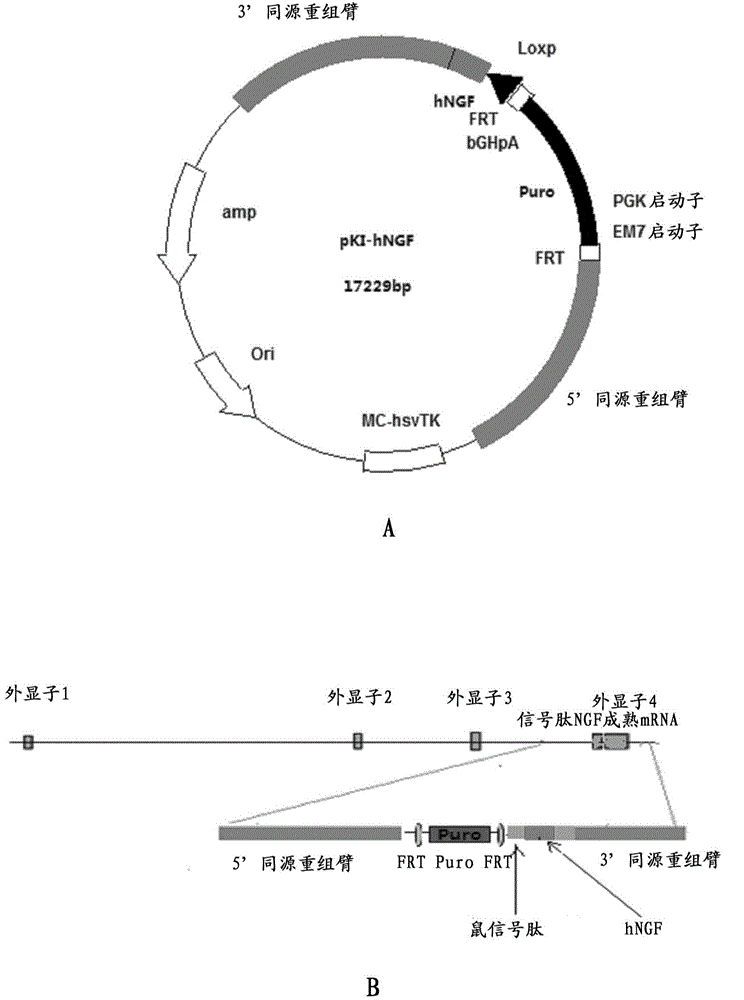
Preparation method for transgenic mice capable of producing human nerve growth factor
ActiveCN104561095AEasy to operateIncrease success rateAnimals/human peptidesVector-based foreign material introductionPlasmid dnaGenetically modified mouse
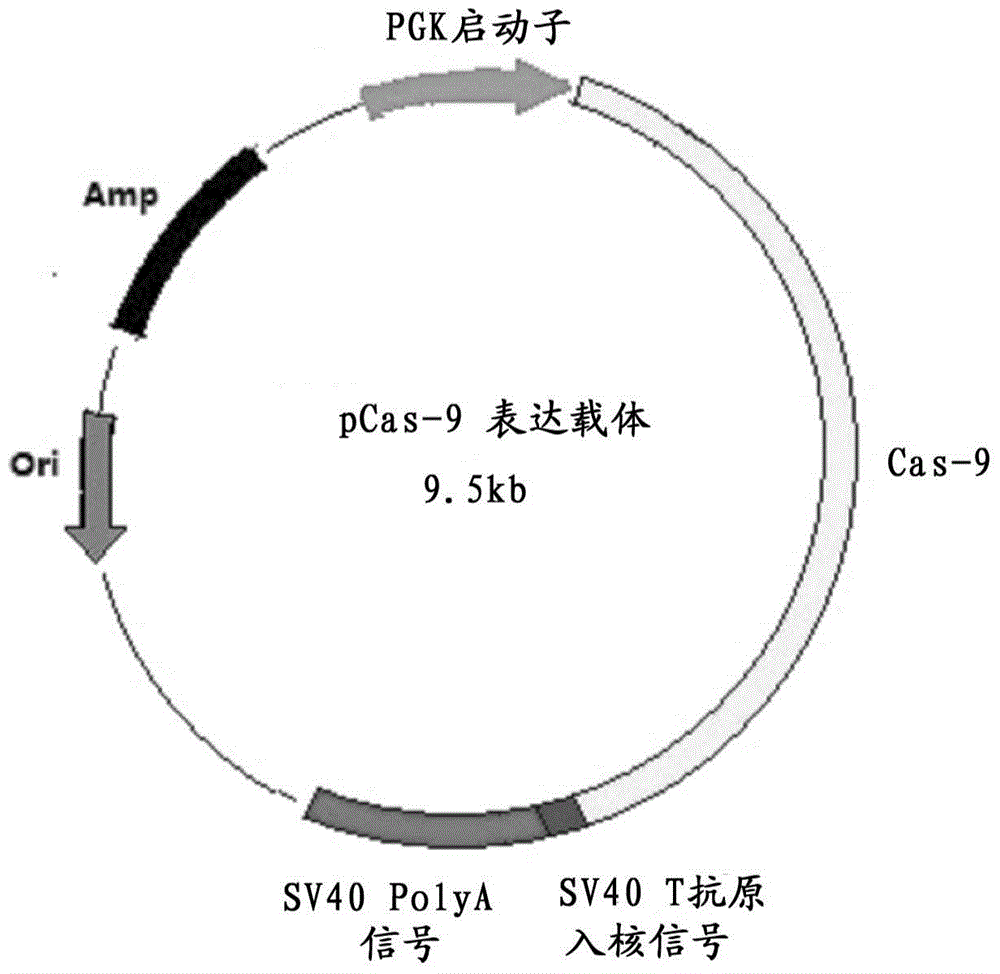
The invention discloses a method for producing transgenic mice through a homologous recombination technology. The method comprises the following steps: replacing mouse NGF genes on mouse chromosomes by human NGF genes through a Cas-9 / CRISPR gene knock-in technology, and obtaining the genes with homozygous human NGF genes through breeding and knocking the genes in mice, thus obtaining filial generation mice with salivary glands capable of secreting human NGF. Compared with the conventional targeting technology utilizing positive and negative double screening homologous recombination genes, the method disclosed by the invention is simple to operate, capable of being realized only by transfecting mouse embryonic stem cells by three plasmid DNAs or carrying out pronucleus injection on the zygotes of mice, and high in success rate which is up to 2-5% and remarkably higher than the mouse embryonic stem cell positive rate of 0.1% of the common gene targeting technology.
Owner:深圳市国创纳米抗体技术有限公司
Insect bioreactor expressing multiple exogenous genes and its construction method and application
The invention discloses an insect bioreactor capable of expressing multiple exogenous genes, and a construction method and application thereof. The construction method comprises the following steps: (1) introducing multicopy high-efficiency bacteria DNA (deoxyribonucleic acid) replication initiator into chitinase and cysteine proteinase genes of a baculovirus genome to obtain a baculovirus shuttle plasmid; (2) replacing virus duplicated essential gene downstream the polyhedrosis gene of the baculovirus shuttle plasmid with antibiotic gene to obtain a baculovirus plasmid DNA; and (3) replacingother virus duplicated and infected nonessential genes in the baculovirus shuttle plasmid with reverse selection marker gene to obtain the insect bioreactor. The antibiotic gene or reverse selection marker gene in the insect bioreactor which is constructed by replacing the exogenous target genes can express multiple exogenous genes in a host insect or insect cell. The insect bioreactor disclosed by the invention can efficiently expressing one or more exogenous genes in an insect body at the same time, and can produce massive recombinant proteins at low cost.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
Compositions comprising high concentration of biologically active molecules and processes for preparing the same
InactiveUS20090004716A1High purityMicroorganism lysisPeptide preparation methodsHigh concentrationLysis
Large scale processes for producing high purity samples of biologically active molecules of interest from bacterial cells are disclosed. The methods comprise the steps of producing a lysate solution by contacting a cell suspension of said plurality of cells with lysis solution; neutralizing said lysate solution with a neutralizing solution to produce a dispersion that comprises neutralized lysate solution and debris; filtering the dispersion through at least one filter; performing ion exchange separation on said neutralized lysate solution to produce an ion exchange eluate; and performing hydrophobic interaction separation on said ion exchange eluate to produce a hydrophobic interaction solution. Further, provided are compositions comprising large scale amounts of plasmid DNA produced by the disclosed large scale processes.
Owner:VGXI
1,3,5-triazinane-2,4,6-trione derivatives and uses thereof
The present invention provides novel 1,3,5-triazinane-2,4,6-trione derivatives, such as compounds of any one of Formulae (I) and (II), and salts thereof, and methods of preparing the compounds. Also provided are compositions including a compound of the invention and an agent (e.g., an siRNA, mRNA, or plasmid DNA). The present invention also provides methods and kits using the compositions for delivering an agent to a subject (e.g., to the liver, spleen, or lung of the subject) or cell and for treating and / or preventing a range of diseases, such as genetic diseases, proliferative diseases, hematological diseases, neurological diseases, liver diseases, and lung diseases.
Owner:MASSACHUSETTS INST OF TECH
Methods for purifying nucleic acids
InactiveUS6011148AEasy to purifyImprove efficacySugar derivativesPeptide/protein ingredientsUltrafiltrationPlasmid dna
Methods are provided for producing highly purified compositions of nucleic acids by using tangential flow ultrafiltration. A scaleable process for producing pharmaceutical grade plasmid DNA, useful for gene therapy, is provided, which is efficient and avoids the use of toxic organic chemicals.
Owner:URIGEN PHARMA INC
Method for purifying plasmid DNA and plasmid DNA substantially free of genomic DNA
InactiveUS6214586B1Efficient and cost-effectiveHigh puritySugar derivativesBacteriaGenomic DNAPlasmid dna
A method is described for purifying plasmid DNA from a mixture containing plasmid DNA and genomic DNA. A solution containing both plasmid DNA and genomic DNA is treated with at least 80% by weight saturation ammonium sulfate, thereby precipitating the genomic DNA and providing purified plasmid DNA in solution. Genomic DNA levels in the purified plasmid DNA product are less than 1% by weight based on the plasmid DNA. The purified plasmid DNA is suitable for use in humans.
Owner:GENZYME CORP
Method for isolating nucleic acids
InactiveUS20060078923A1Simple procedureImprove automationMicrobiological testing/measurementNucleic acid reductionBiological bodyPlasmid dna
Described herein is a method in which genomic nucleic acid of a cell can be separated from nucleic acid having a molecular weight that is lower than the molecular weight of the genomic nucleic acid (e.g., plasmid DNA) of the cell directly from a cell growth culture. Also described herein, a method in which genomic nucleic acid can be separated from nucleic acid having a molecular weight that is lower than the molecular weight of the genomic nucleic acid in a cell lysate without the need to prepare a cleared lysate. The present invention is also directed to a method of isolating genomic nucleic acid (e.g., RNA, DNA) of a cell or an organism.
Owner:AGENCOURT BIOSCI CORP
Apparatus, methods and compositions for biotechnical separations
InactiveUS20020010145A1Low costImprove performanceGenetic material ingredientsTyresFractional PrecipitationPlasmid dna
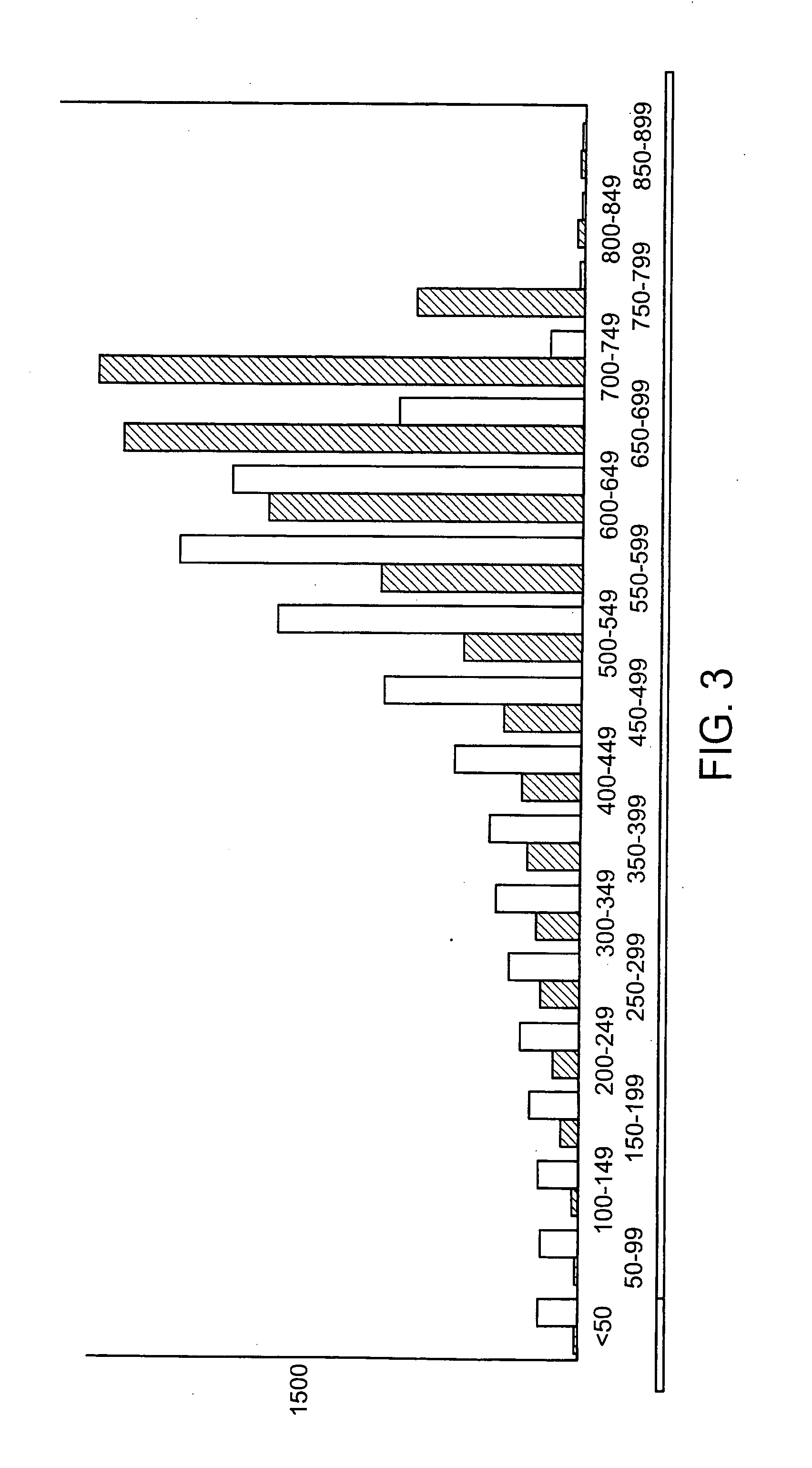
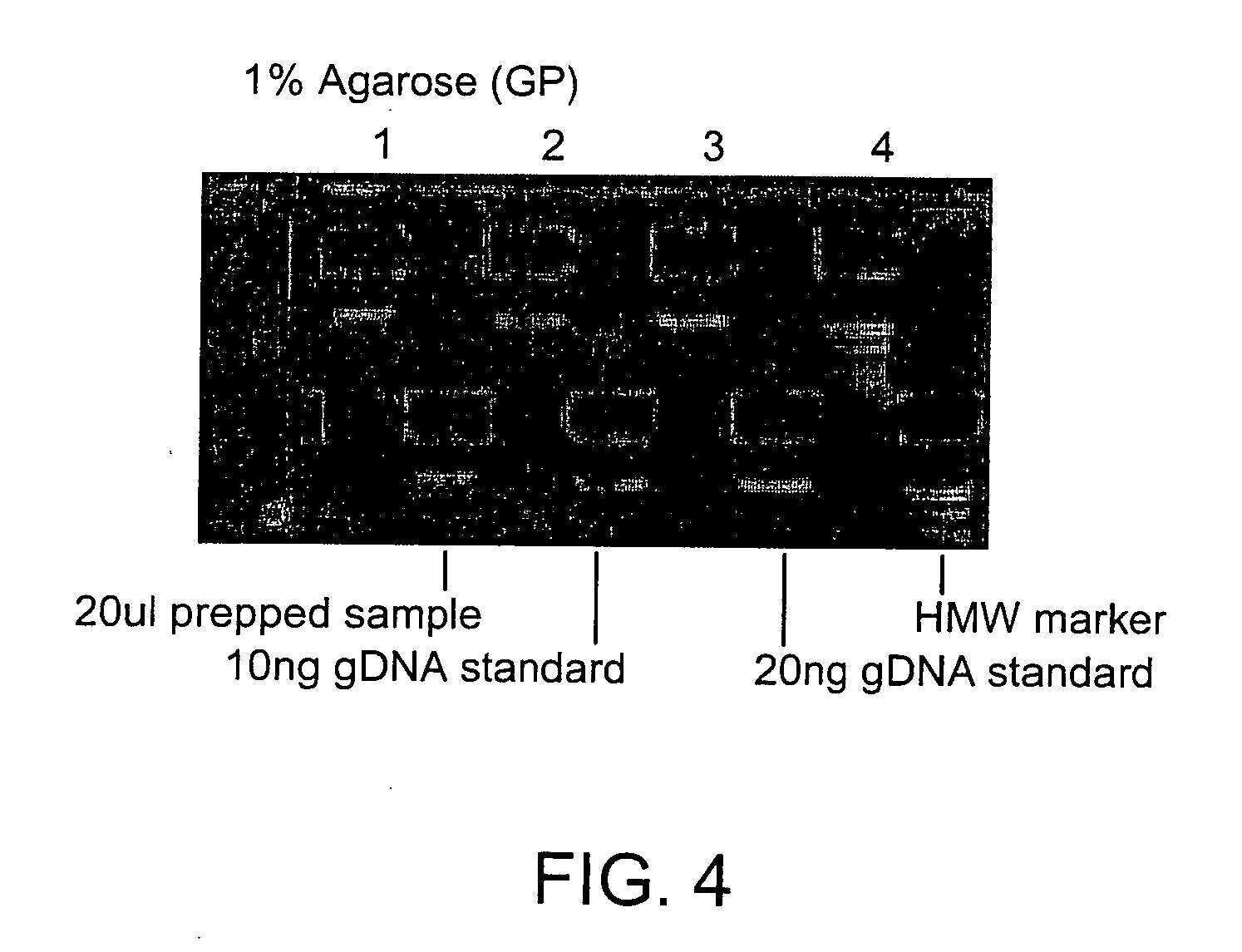
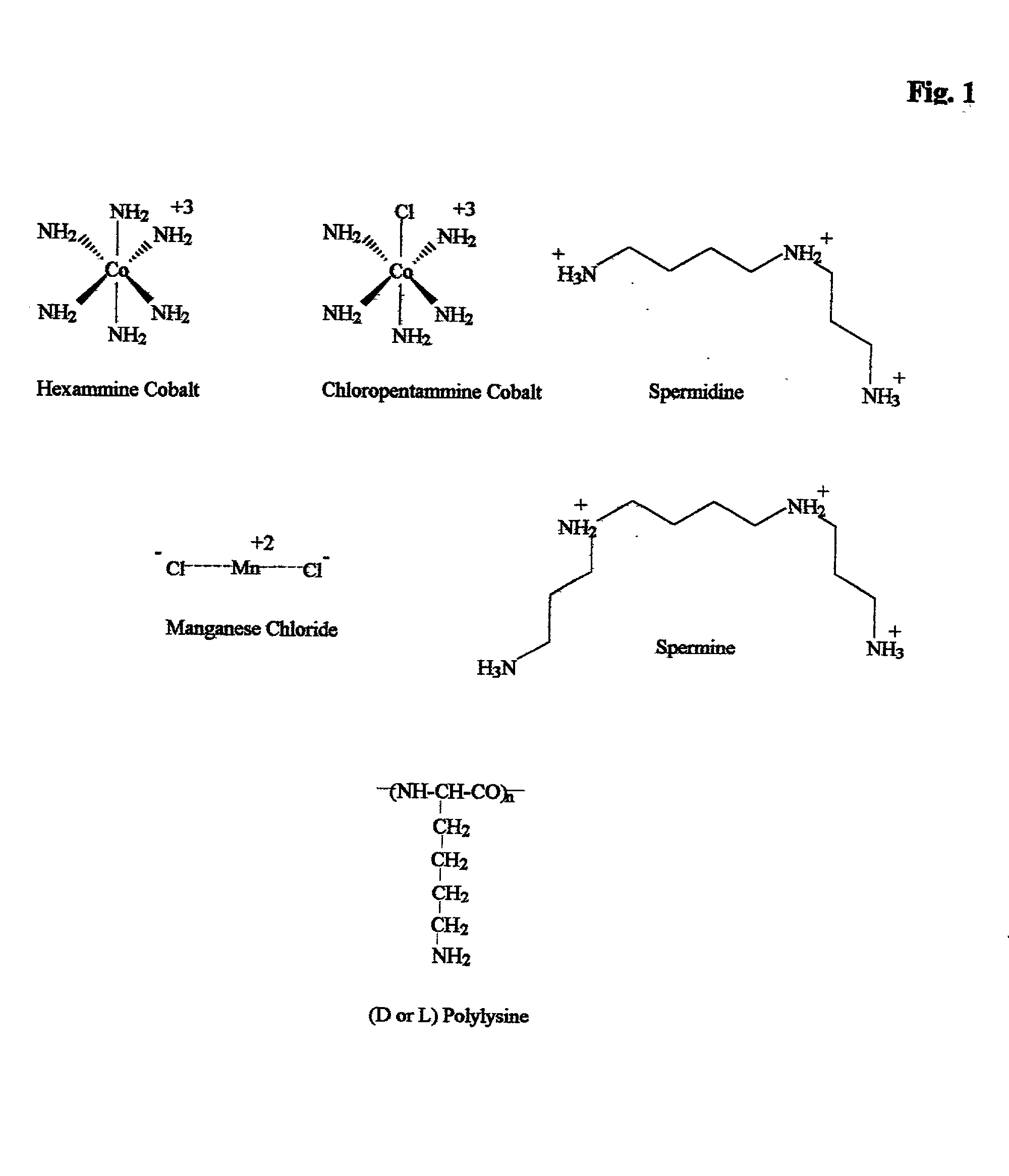
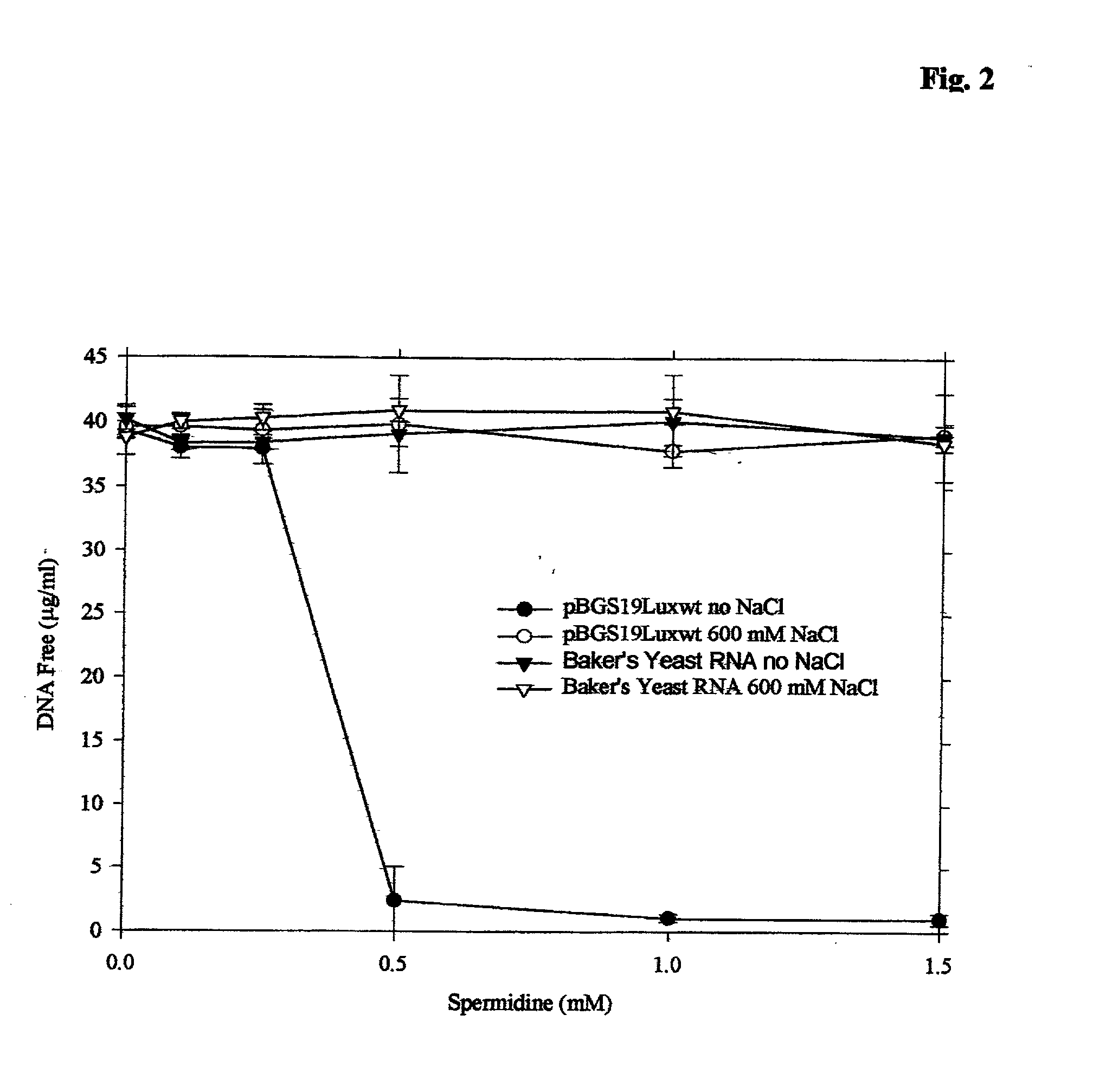
embodiments of the invention include purification of DNA, preferably plasmid DNA, by use of selective precipitation, preferably by addition of compaction agents Also included is a scaleable method for the liquid-phase separation of DNA from RNA. RNA may also be recovered by fractional precipitation according to the invention. RNA, commonly the major contaminant in DNA preparations, can be left in solution while valuable purified plasmid DNA is directly precipitated. Endotoxin can also be kept to very low levels. The invention includes mini-preps, preferably of plasmid and chromosomal DNA to obtain sequenceable and restriction digestible DNA in high yields in multiple simultaneous procedures. As a method of assay, a labeled probe is precipitated by hybridizing it to a target, (erg. chromosomal DNA, oligonucleotides, Ribosomal RNA, tRNA), and thereafter precipitating the probe / target complex with compaction agents and leaving in solution any unhybridized probe.
Owner:WILLSON RICHARD C III +1
Modified spin column for simple and rapid plasmid DNA extraction
InactiveUS20080300397A1Rapid isolationImprove system performanceSamplingSugar derivativesCellular DebrisSpins
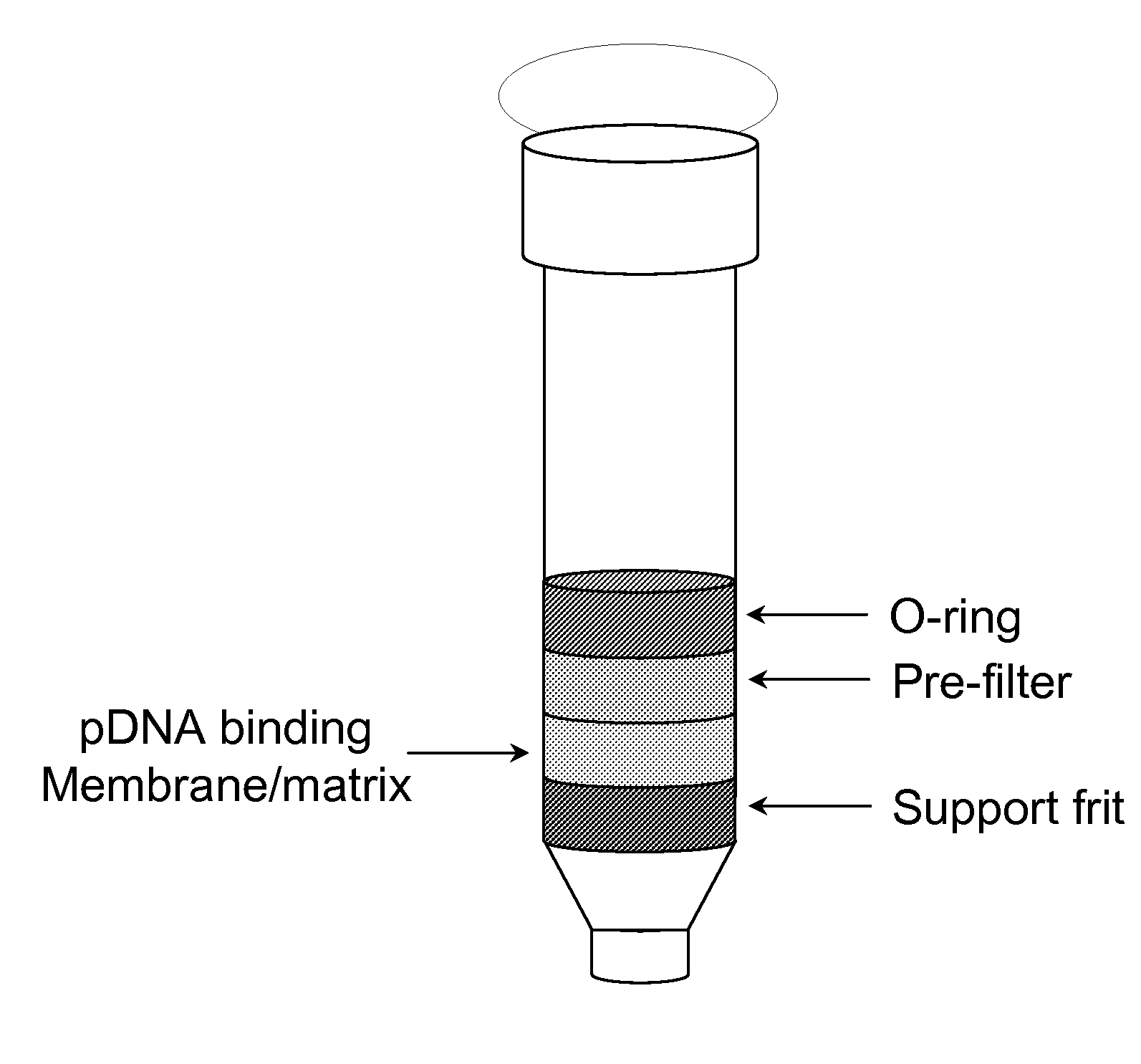
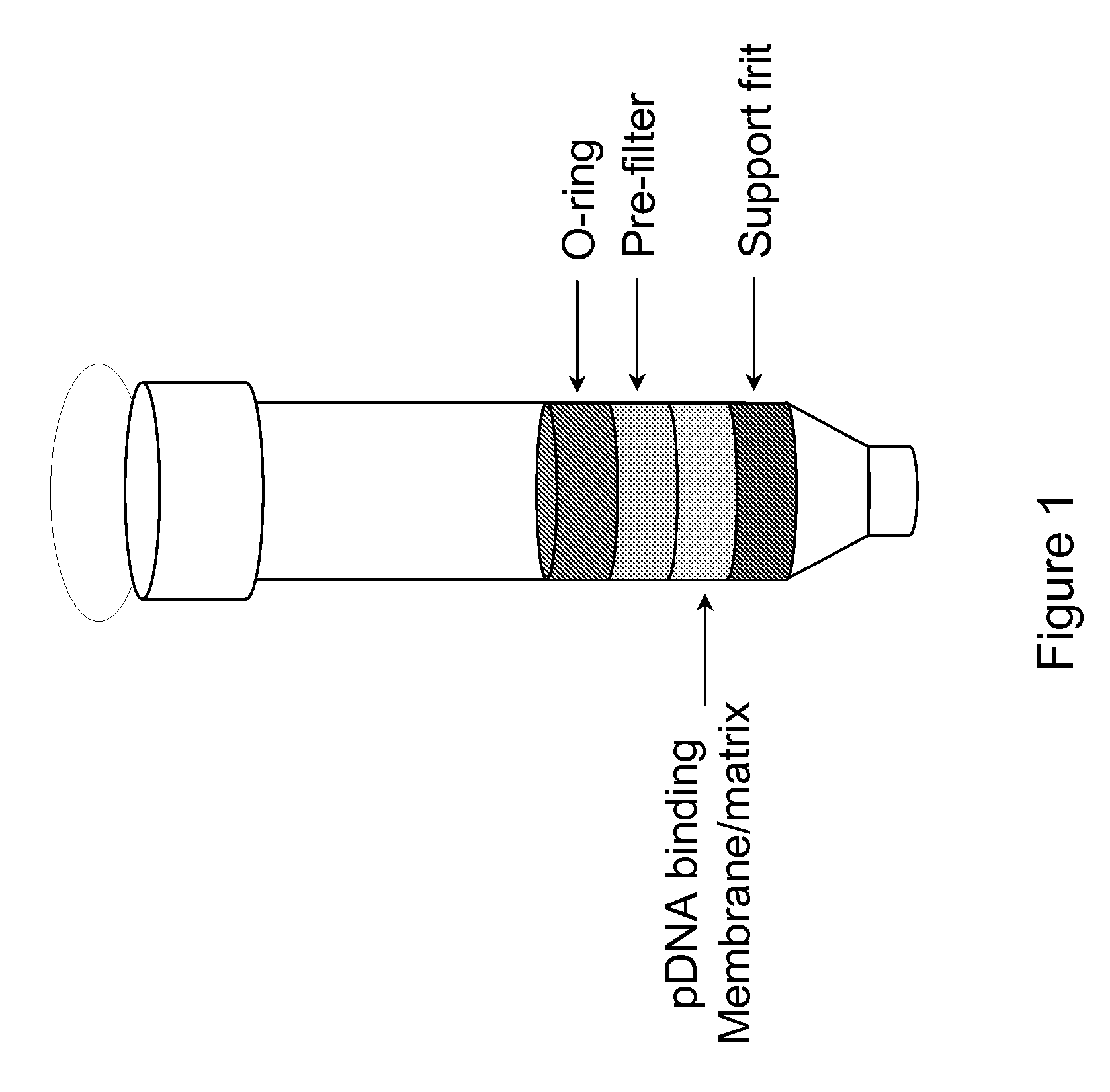
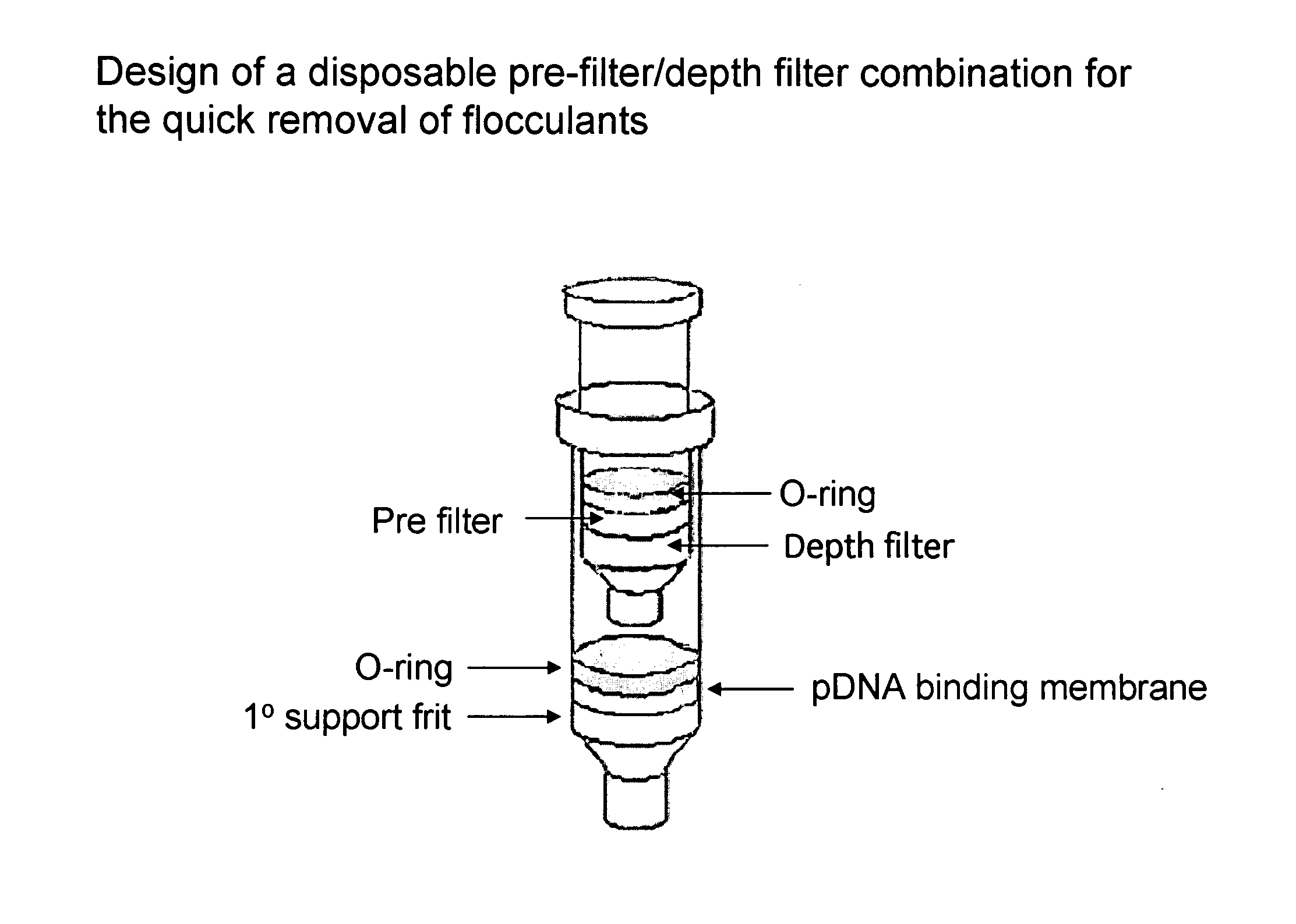
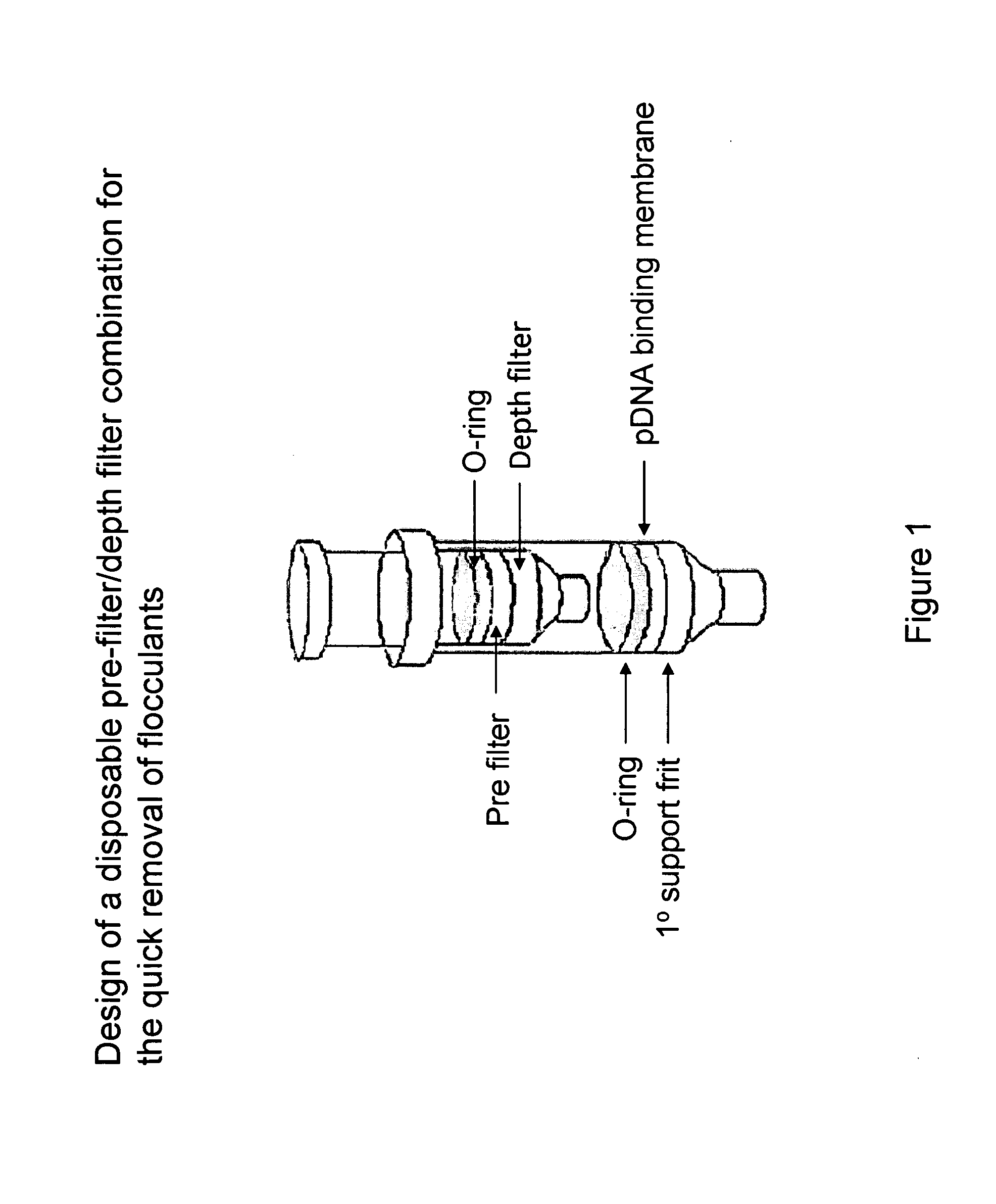
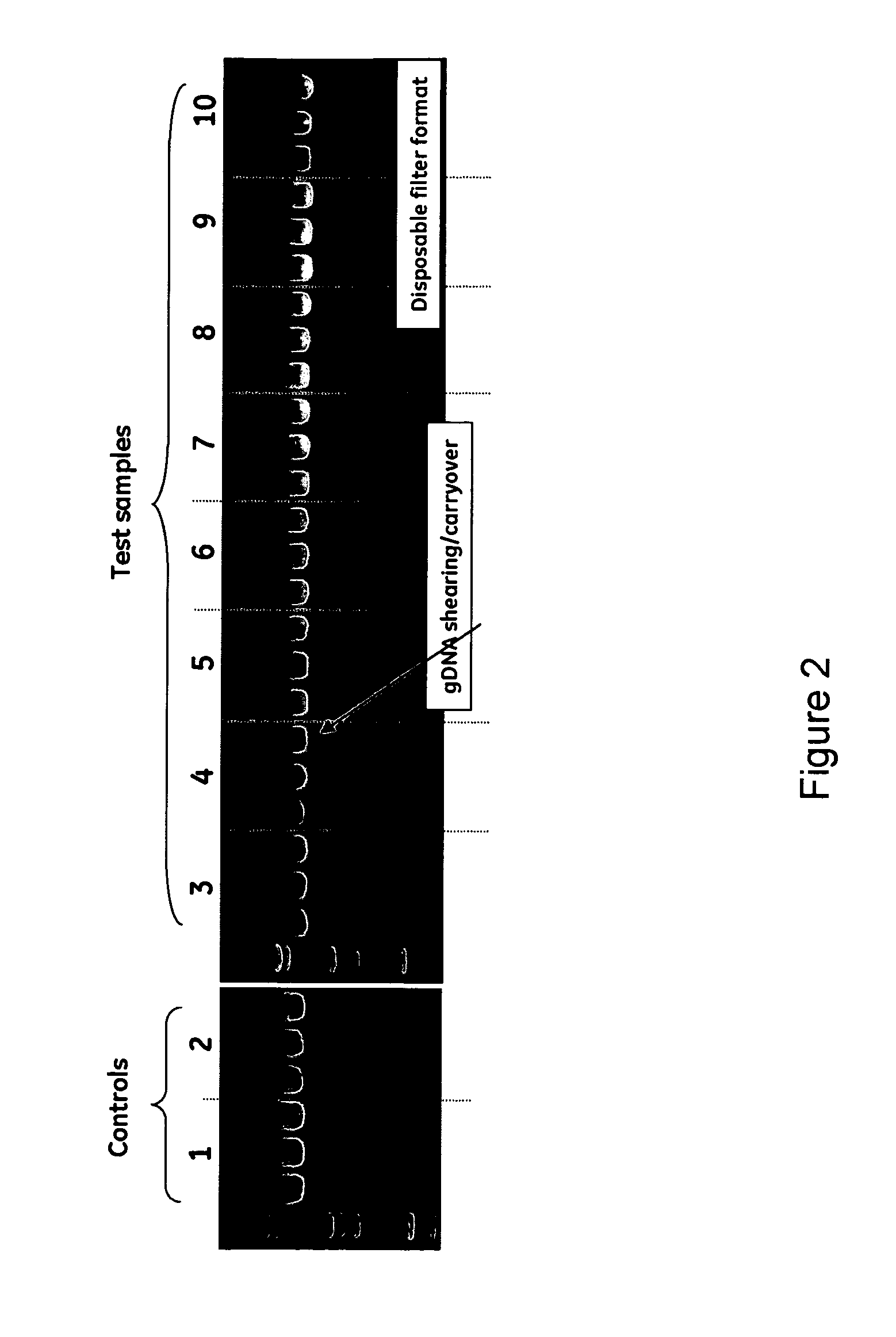
The invention relates to a modified spin column for the isolation and purification of plasmid DNA. A pre-filtration disc is included in a traditional spin column. During plasmid DNA isolation, the lysate can be loaded directly to the modified spin column, eliminates the need to first remove the flocculants containing cellular debris. This results in a much shortened process. Variation of the invention includes a depth filter in between the pre-filtration disc and the main separation matrix. Also provided are kits for isolation of plasmid DNA including the modified spin columns.
Owner:GE HEALTHCARE LTD
Miniprep system for simple and rapid plasmid DNA extraction
InactiveUS20080299621A1Rapid isolationBioreactor/fermenter combinationsBiological substance pretreatmentsCellular DebrisSpins
The invention relates to a modified microspin system for the isolation and purification of plasmid DNA. A disposable pre-filter column is used in combination with the traditional microspin column for increase speed and quality of plasmid DNA preparation. The disposable pre-filter column includes a pre-filter and a depth filter for optimal result. During plasmid DNA isolation, the lysate can be loaded directly to the assembly including the disposable pre-filter column and the microspin column. A quick spin causes the plasmid DNA to bind to the microspin column while the flocculents remain on top of the disposable pre-filter column, eliminates the need to first remove the flocculants containing cellular debris. This results in a much shortened process. Also provided are kits for isolation of plasmid DNA including the pre-filter column and the microspin columns.
Owner:GE HEALTHCARE LTD
Method for establishing Gadd45a knockout rabbit model by adopting knockout technology
InactiveCN107630043APredictive effectReduce the risk of research and developmentStable introduction of DNAAnimal husbandryRabbit modelPlasmid dna
The invention relates to a method for establishing a Gadd45a knockout rabbit model by adopting a knockout technology and belongs to the technical field of biotechnology. The invention aims to establish a rabbit model by knocking out GADD45a gene by utilizing a Grispr / cas9 technology and to provide the method for establishing the Gadd45a knockout rabbit model by adopting the knockout technology andfor researching the influence of the gene on animal liver. The method provided by the invention comprises the following steps: establishing sgRNA; synthesizing double-stranded DNA; linearizing p UC-57 carrier; linking p UC-57 carrier with double-stranded DNA; converting; performing monoclonal picking and plasmid DNA extraction; identifying plasmid sequence; expressing plasmid with CAS9; and performing digestion linearization for in vitro transcription. Through related detection, the invention successfully acquires the Gadd45a knockout rabbit model; the model is established for simulating theclinic pathological processes of the liver diseases, such as, fatty liver, liver cirrhosis and liver cancer after giving the corresponding alcohol stimulation, is capable of effectively forecasting the clinic application effects of new vaccine, new drugs and new diagnostic reagents, and meanwhile, and is capable of greatly reducing the risk in researching and developing the new drugs; and a basismodel is supplied for the clinical research.
Owner:JILIN UNIV
New yeast-bacteria shuttle vector
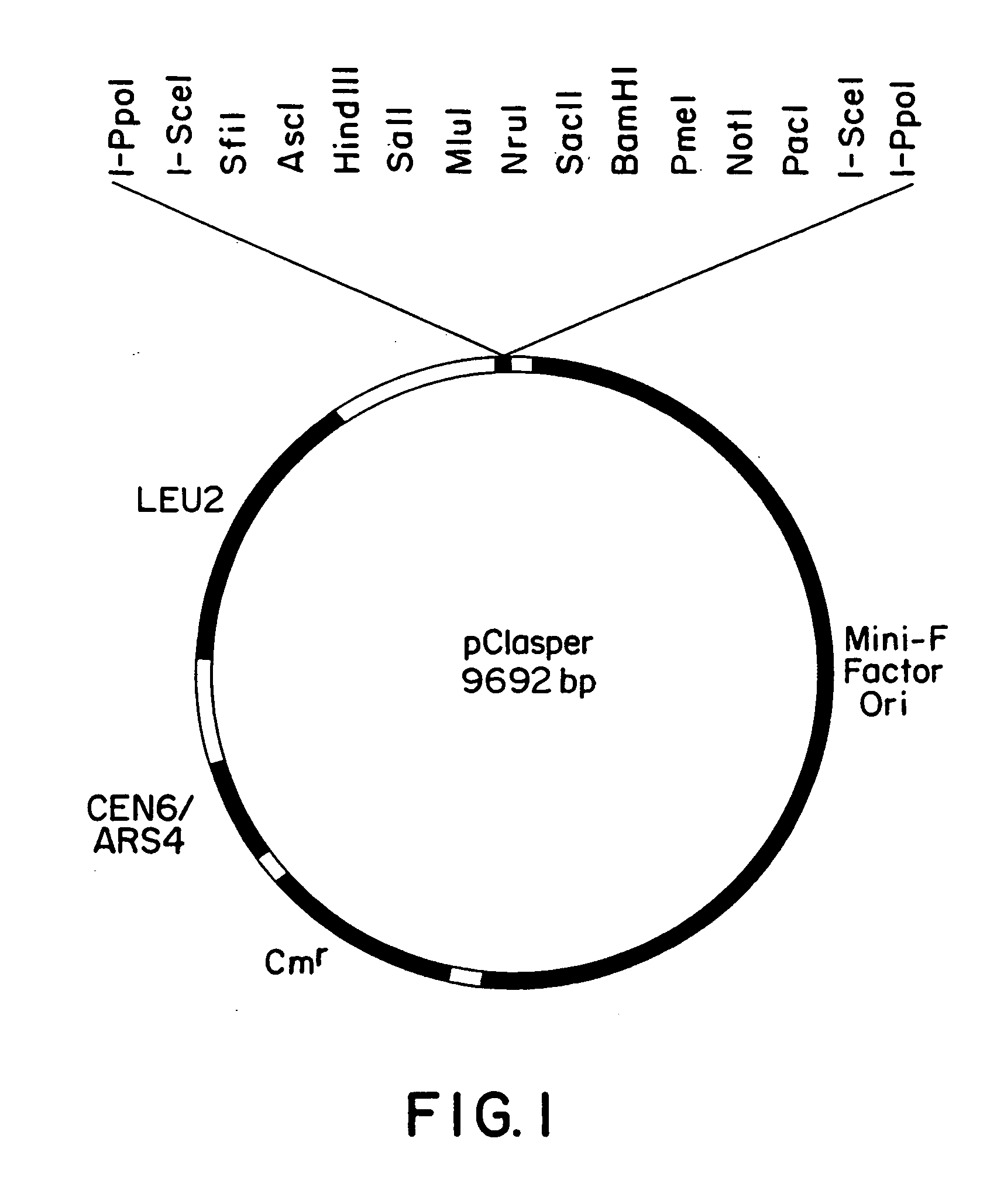
The functional analysis of genes frequently requires the manipulation of large genomic regions. A yeast-bacteria shuttle vector is described, that can be used to clone large regions of DNA by homologous recombination. The important feature of present invention is the presence of the a bacterial replication origin, which allows large DNA insert capacity. The utility of this vector lies in its ability to isolate, manipulate and maintain large fragments in bacteria and yeast, allowing for mutagenesis by yeast genetics and simplified preparation of plasmid DNA in bacteria.
Owner:BRADSHAW M SUZANNE +2
Nucleic acid separation using immobilized metal affinity chromatography
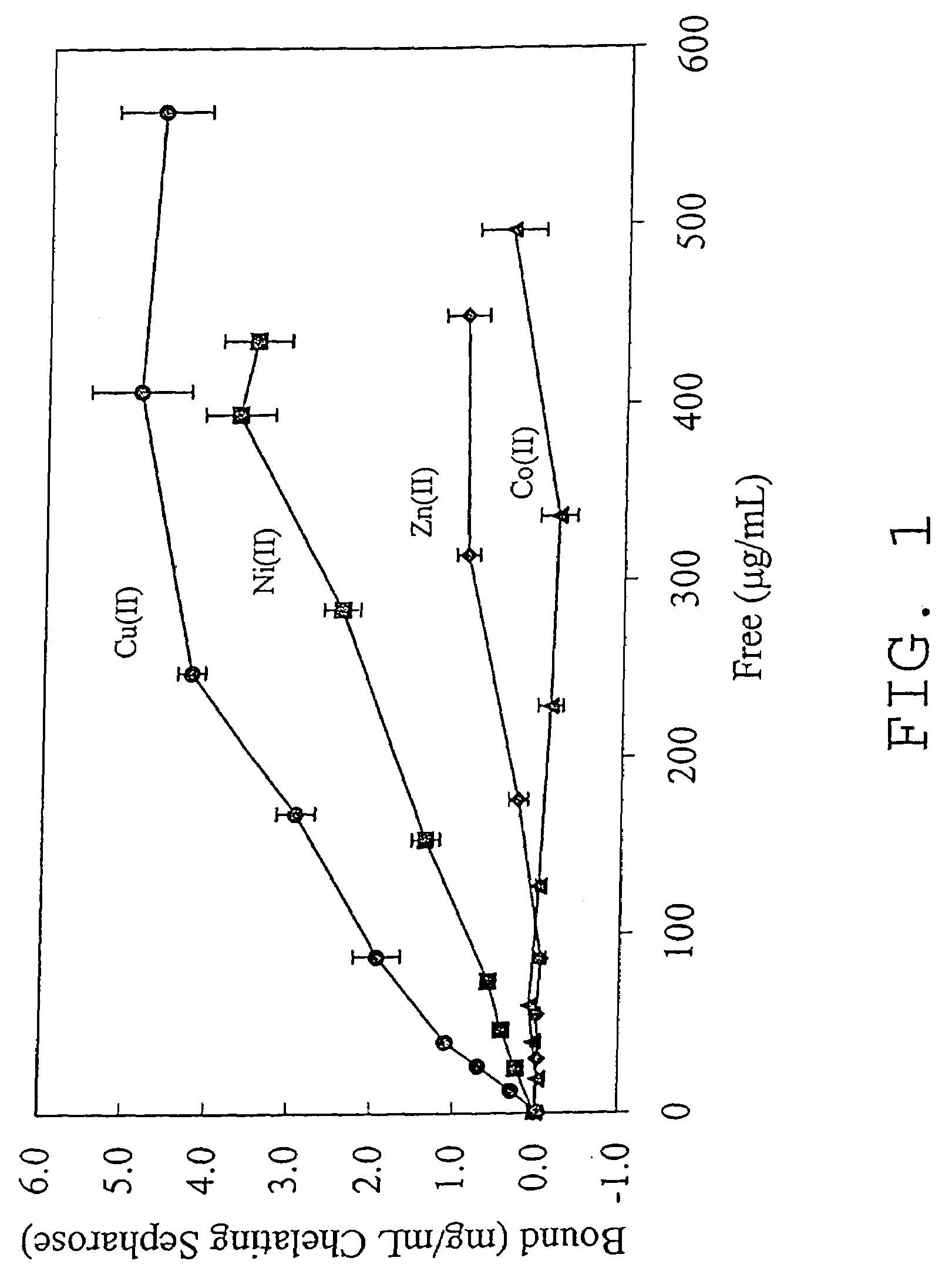
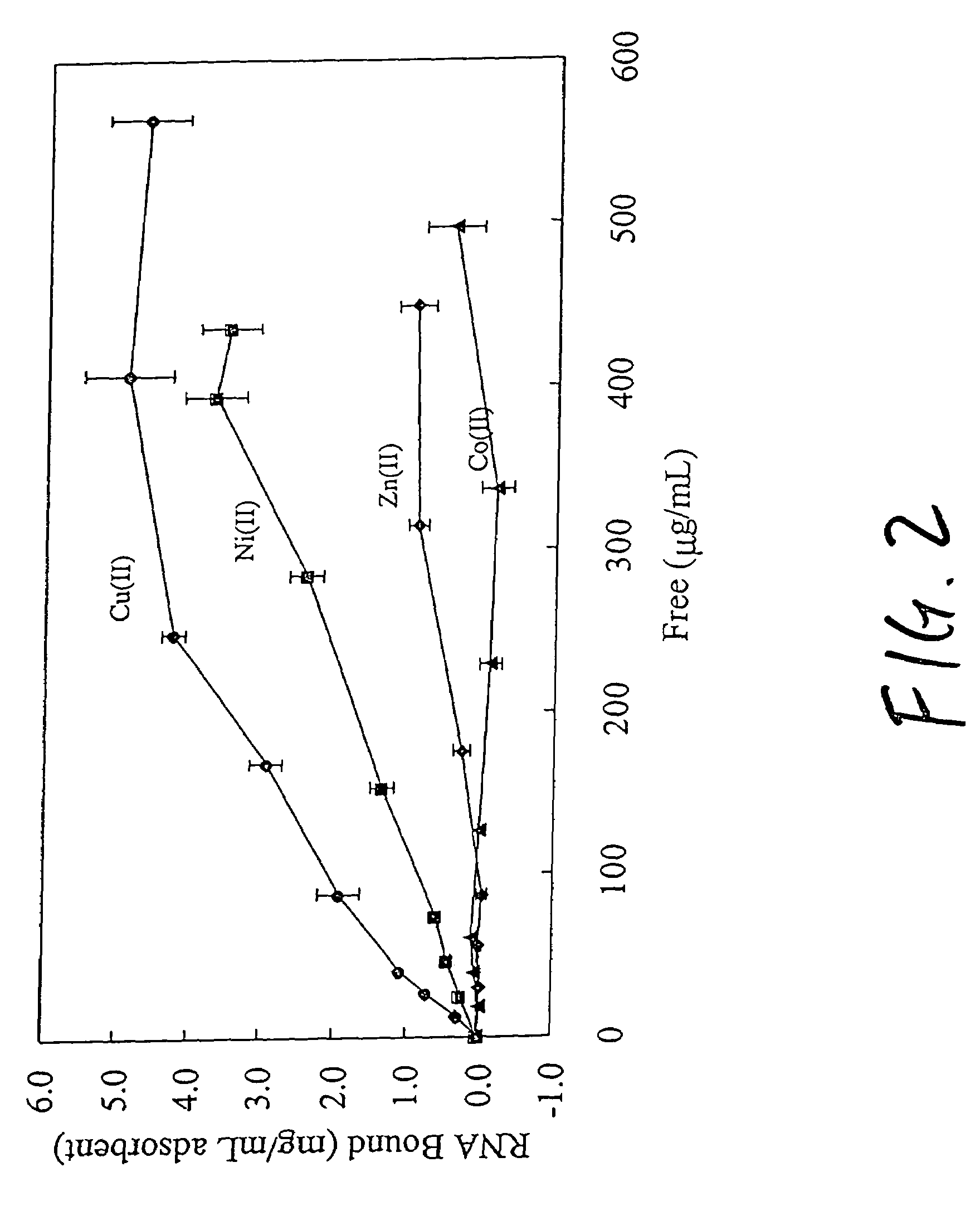
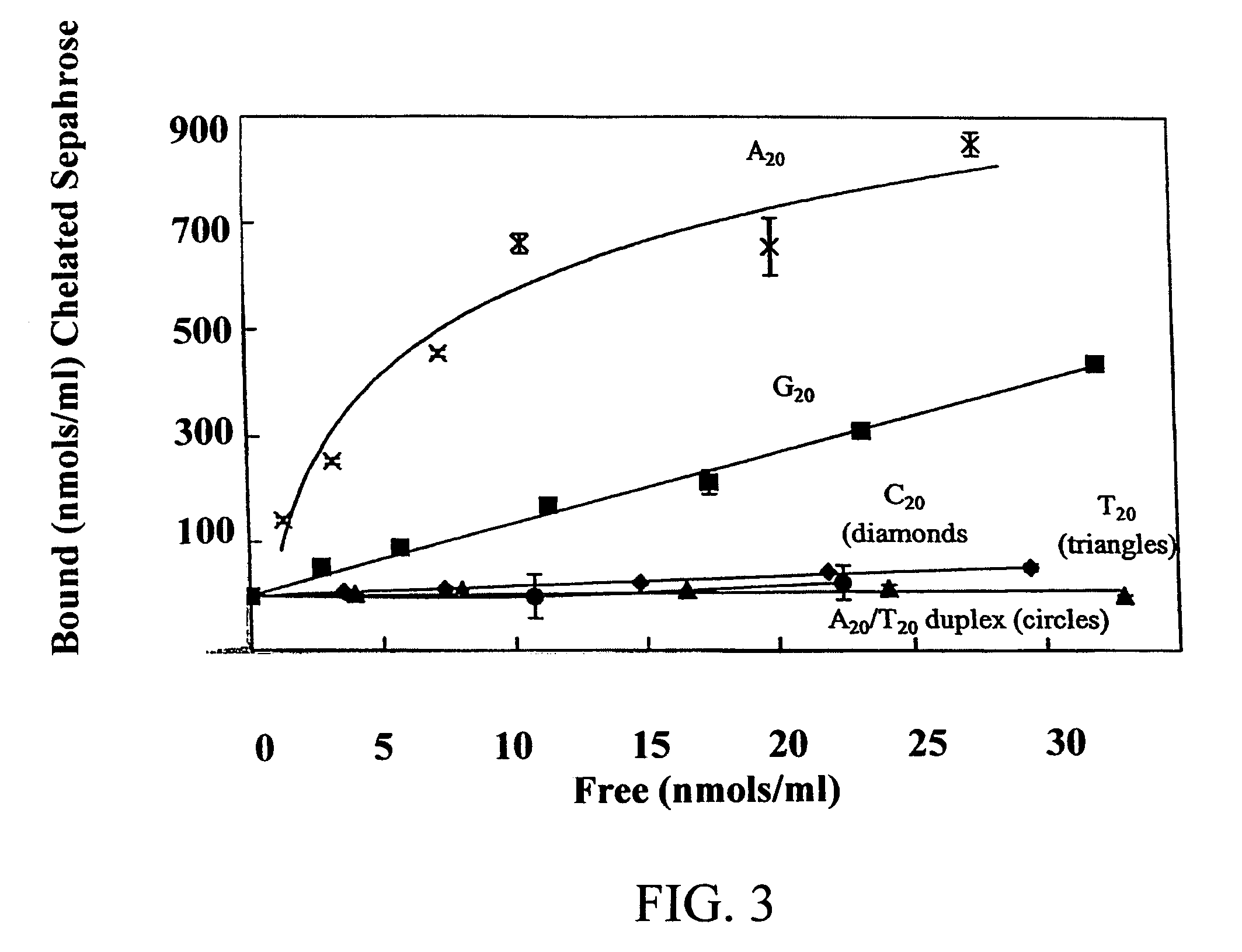
An immobilized metal affinity chromatography (IMAC) method for separating and / or purifying compounds containing a non-shielded purine or pyrimidine moiety or group such as nucleic acid, presumably through interaction with the abundant aromatic nitrogen atoms in the purine or pyrimidine moiety. The method can also be used to purify compounds containing purine or pyrimidine moieties where the purine and pyrimidine moieties are shielded from interaction with the column matrix from compounds containing a non-shielded purine or pyrimidine moiety or group. Thus, double-stranded plasmid and genomic DNA, which has no low binding affinity can be easily separated from RNA and / or oligonucleotides which bind strongly to metal-charged chelating matrices. IMAC columns clarify plasmid DNA from bacterial alkaline lysates, purify a ribozyme, and remove primers and other contaminants from PCR reactions. The metal ion affinity of yeast RNA decreases in the order: copper (II), nickel (II), zinc (II), and cobalt (II).
Owner:UNIV HOUSTON SYST
Host-vector system for antibiotic-free CoIE1 plasmid propagation
InactiveUS20060063232A1Inhibit transcriptionRegulate expressionBacteriaFermentationVector systemAntibiotic free
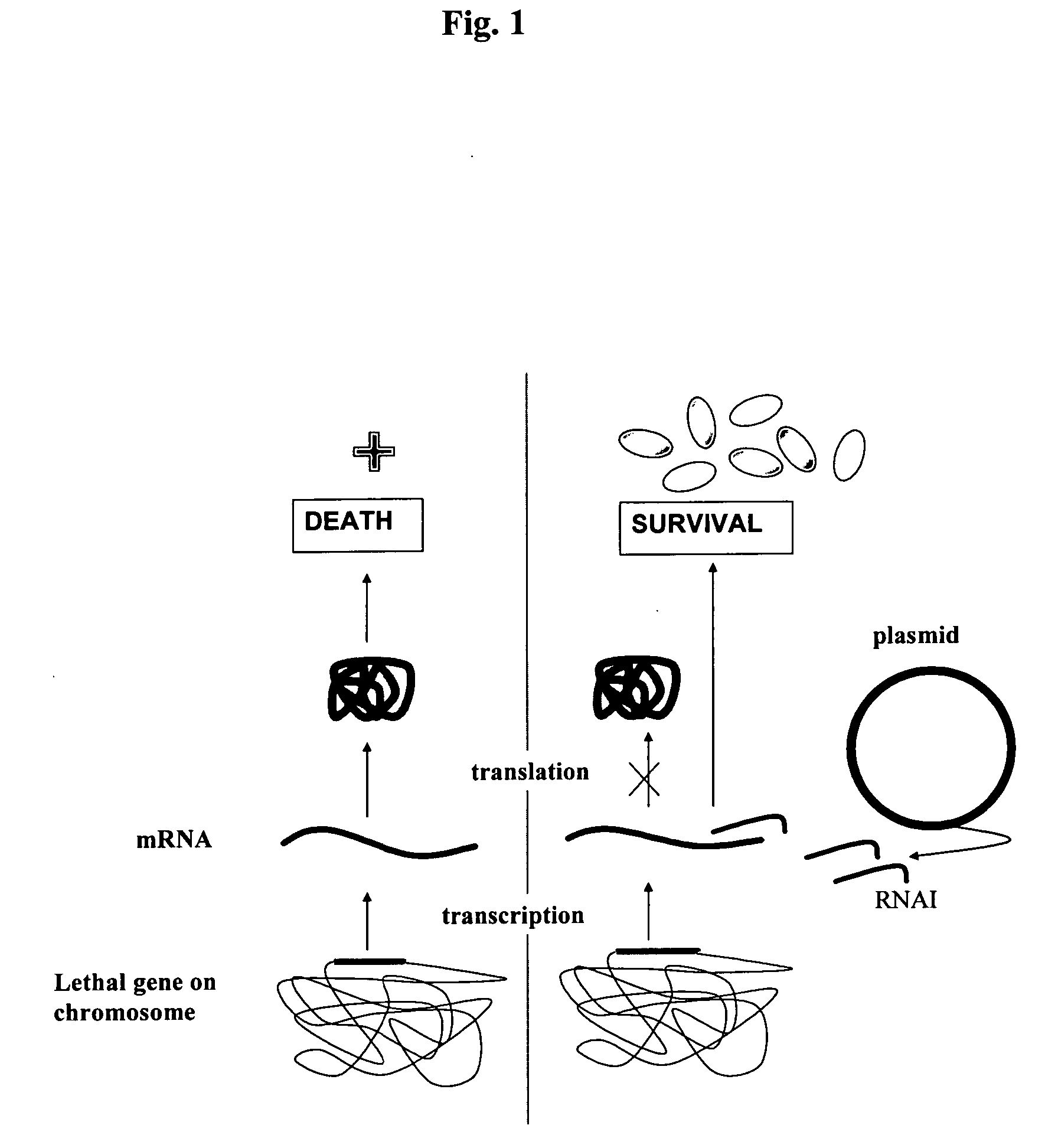
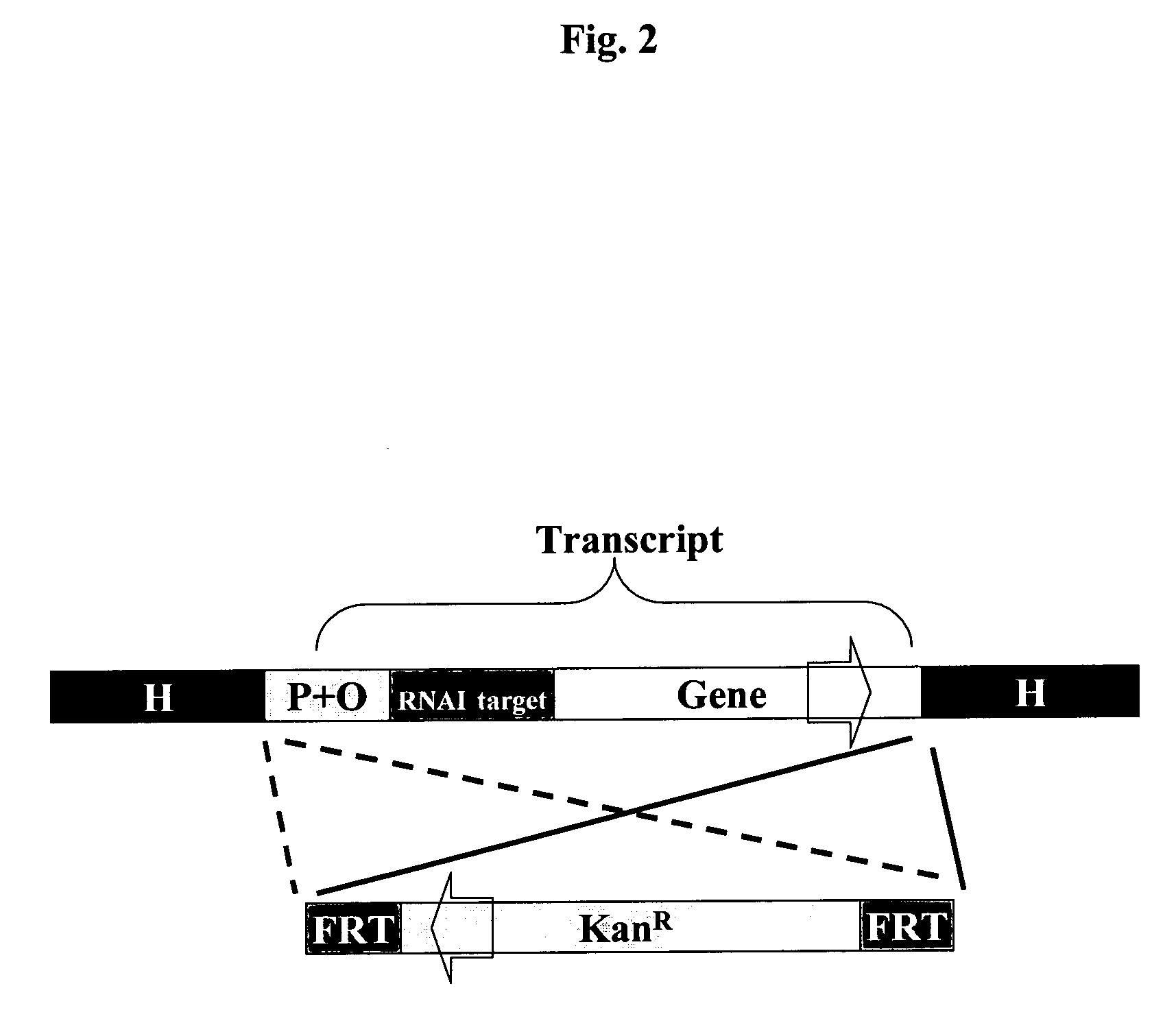
A host-vector system that uses the RNA-based copy number control mechanism of ColE1-type plasmids for regulating the expression of a marker gene allows for antibiotic-free selection of plasmids and is useful for production of plasmid DNA and recombinant proteins.
Owner:BOEHRINGER INGELHEIM RCV GMBH & CO KG
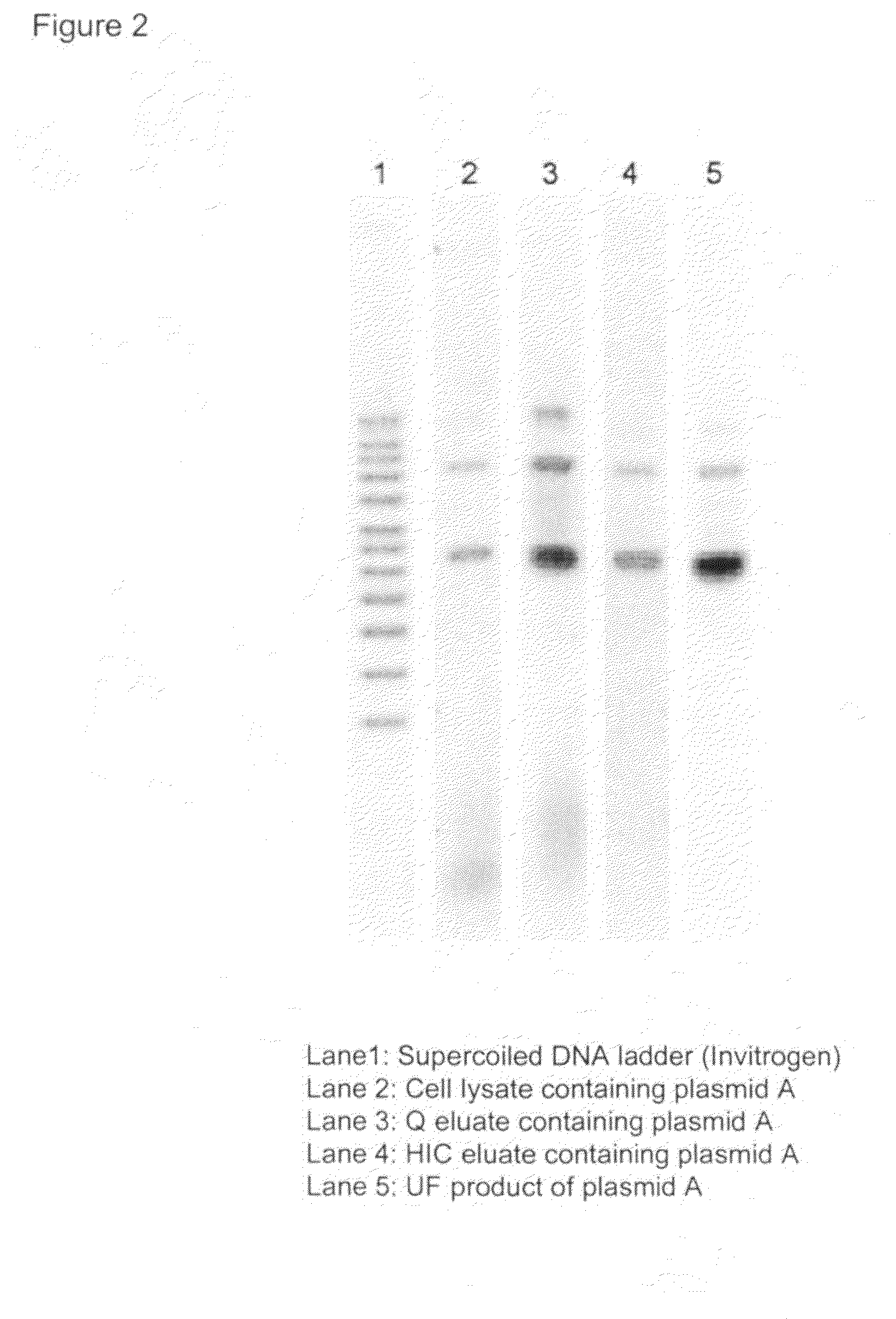
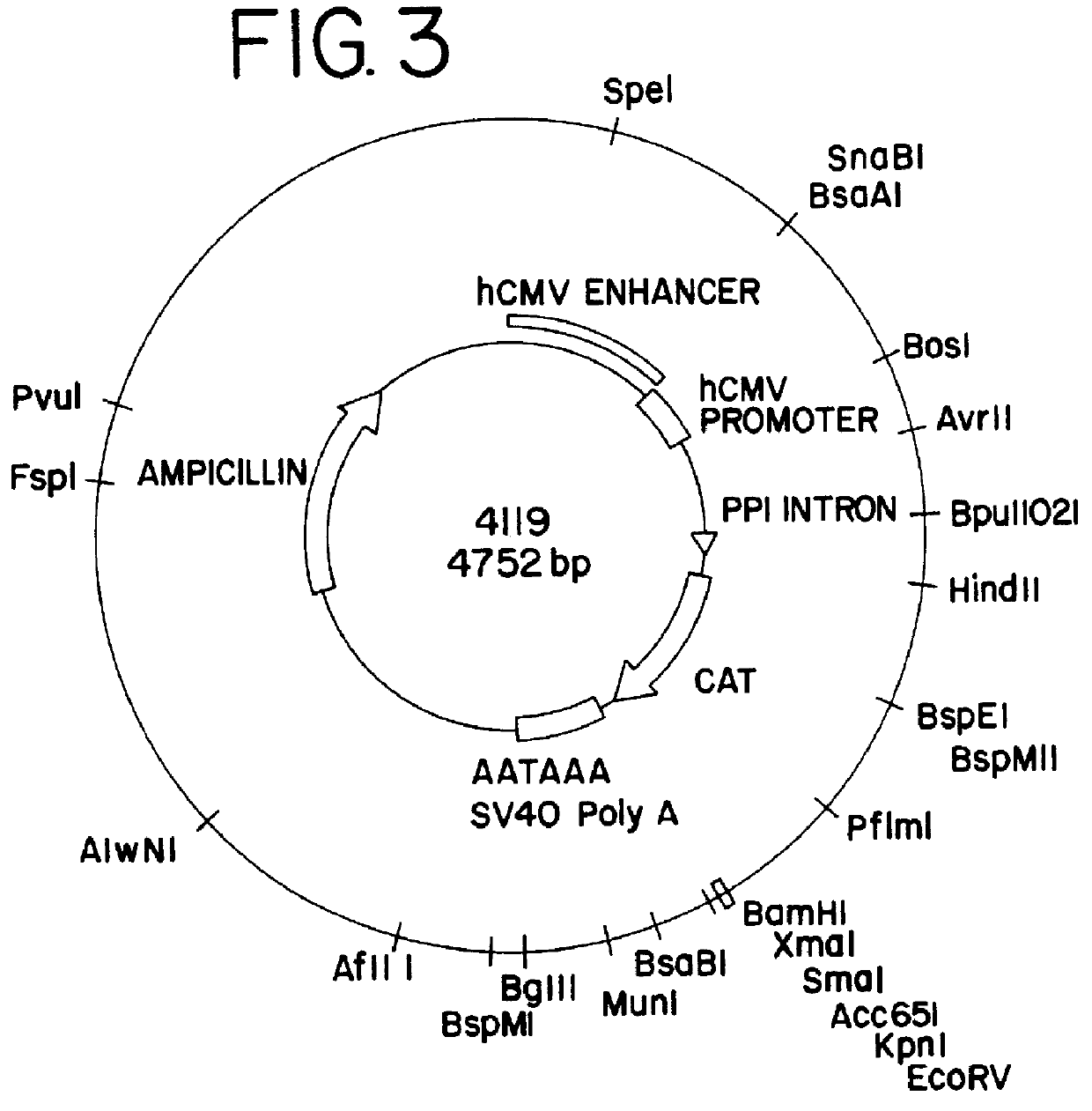
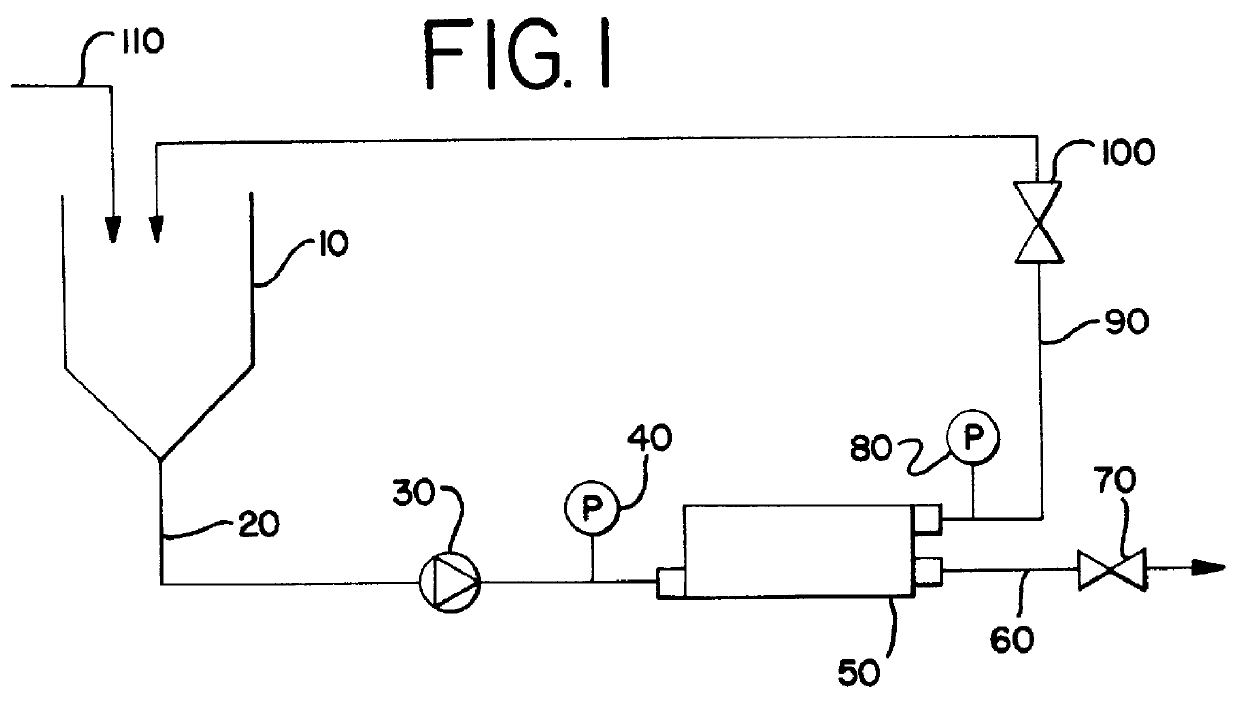
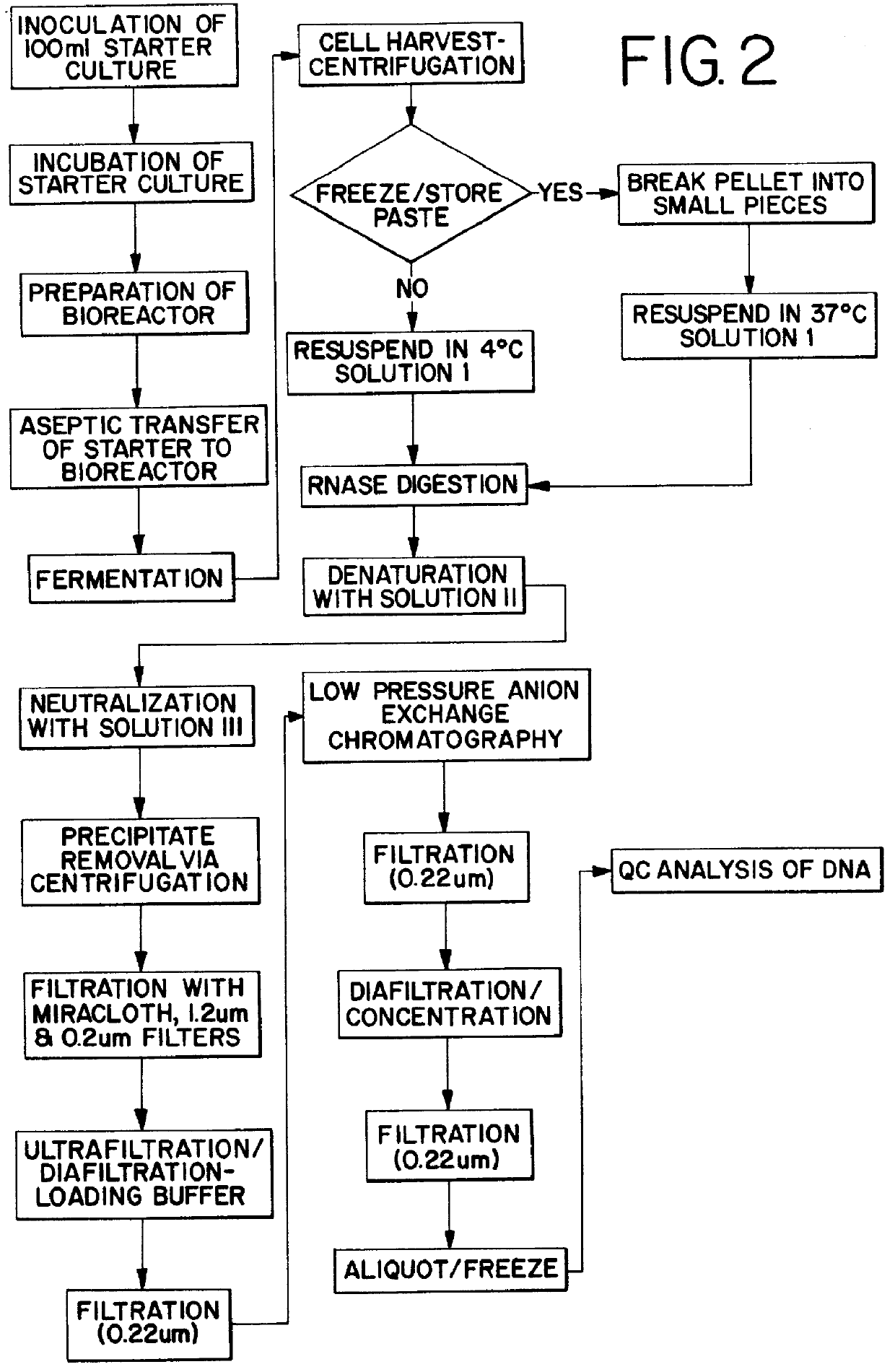
Purification process for plasmid DNA
ActiveUS7767399B2Low production costImprove robustnessMicrobiological testing/measurementMicroorganism lysisClinical gradePurification methods
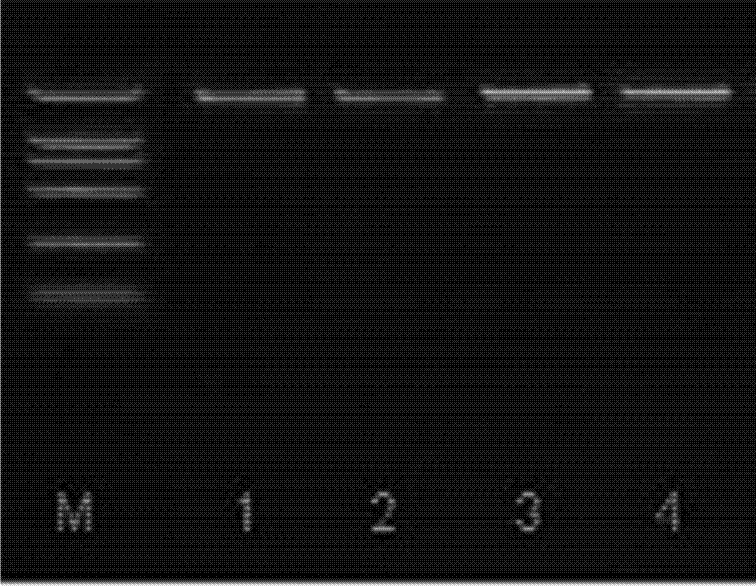
Methods of isolating clinical-grade plasmid DNA from manufacturing processes, including large-scale fermentation regimes, are disclosed which encompass alternatives to two core unit operations common to plasmid DNA purification processes. The novel upstream and downstream purification processes disclosed herein provide for reduced production costs and increase process robustness. Either or both of the purification processes disclosed herein may be used in combination with additional purification steps known in the art that are associated with DNA plasmid purification technology.
Owner:MERCK SHARP & DOHME LLC
Devices and methods for biomaterial production
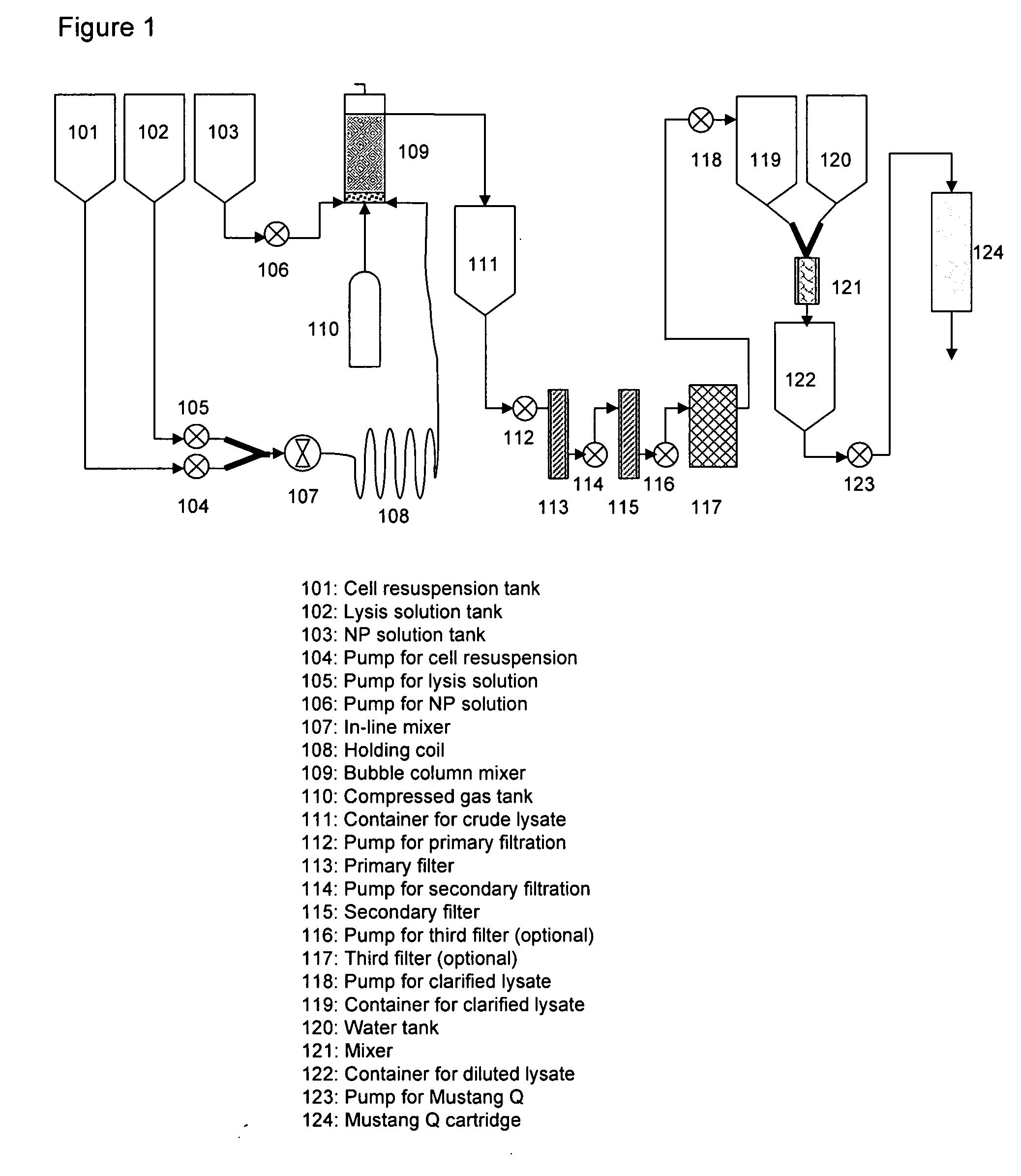
ActiveUS20050014245A1Avoid excessive fragmentationIndependent controlBioreactor/fermenter combinationsBiological substance pretreatmentsCellular componentUltrafiltration
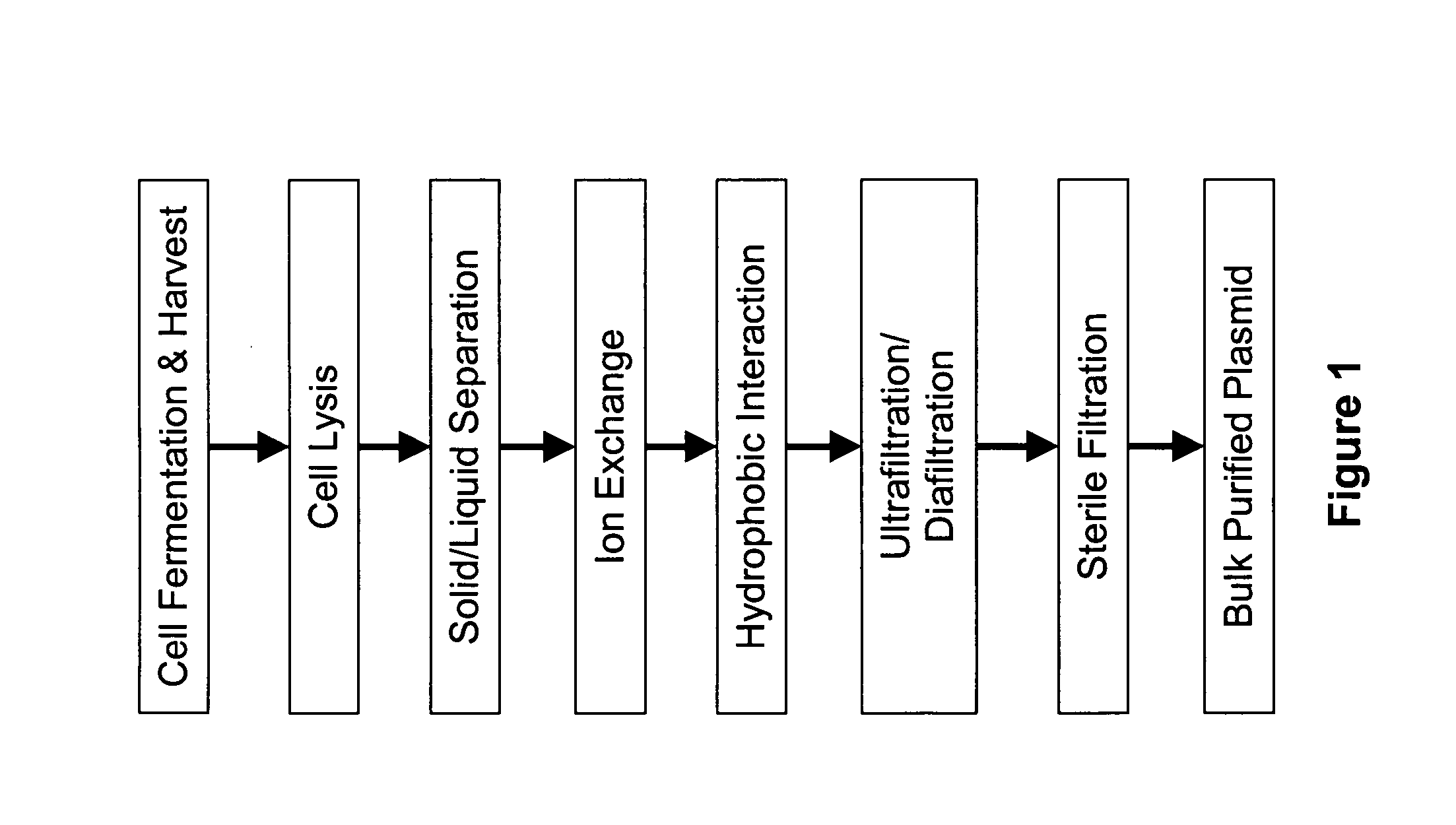
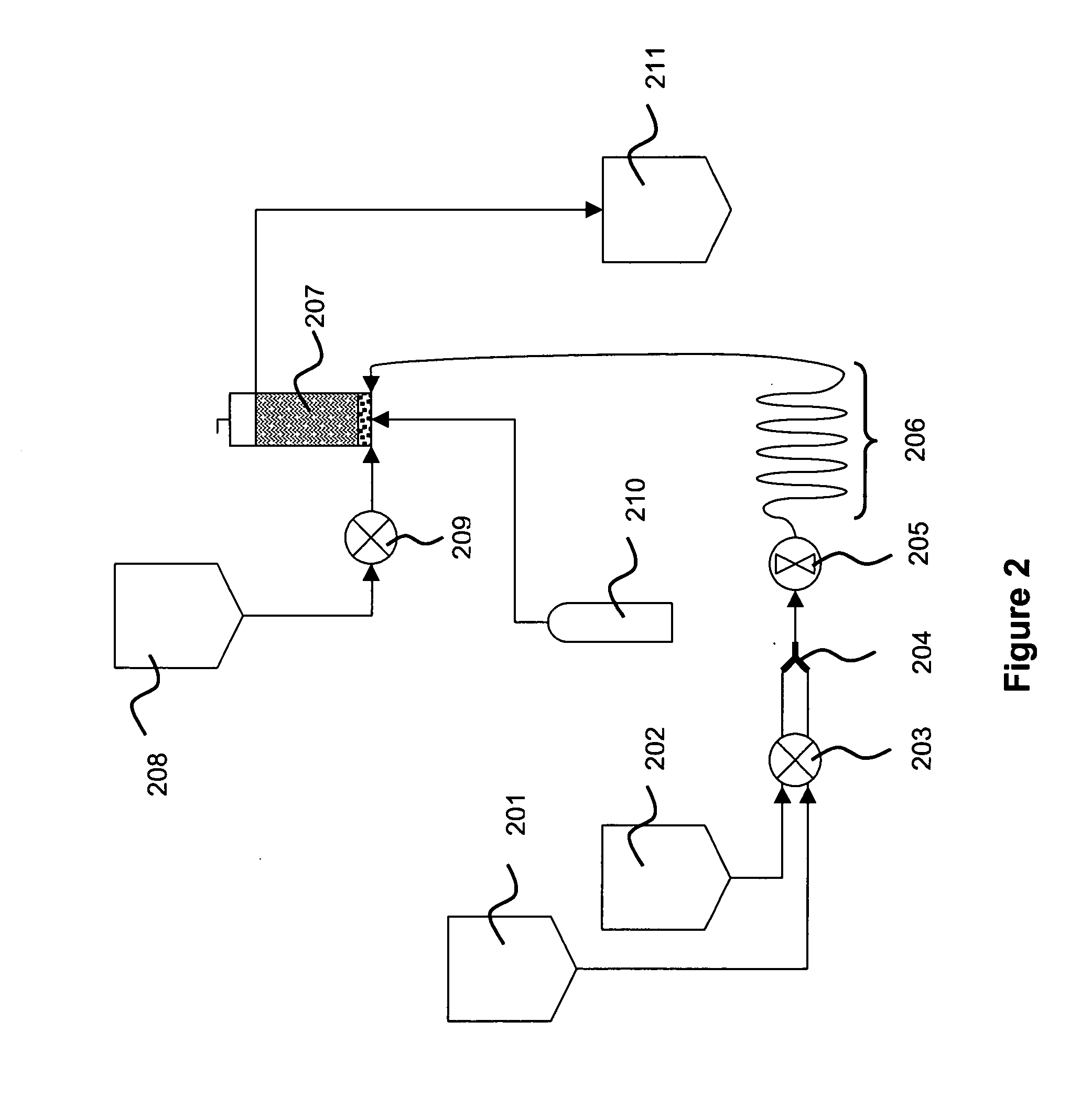
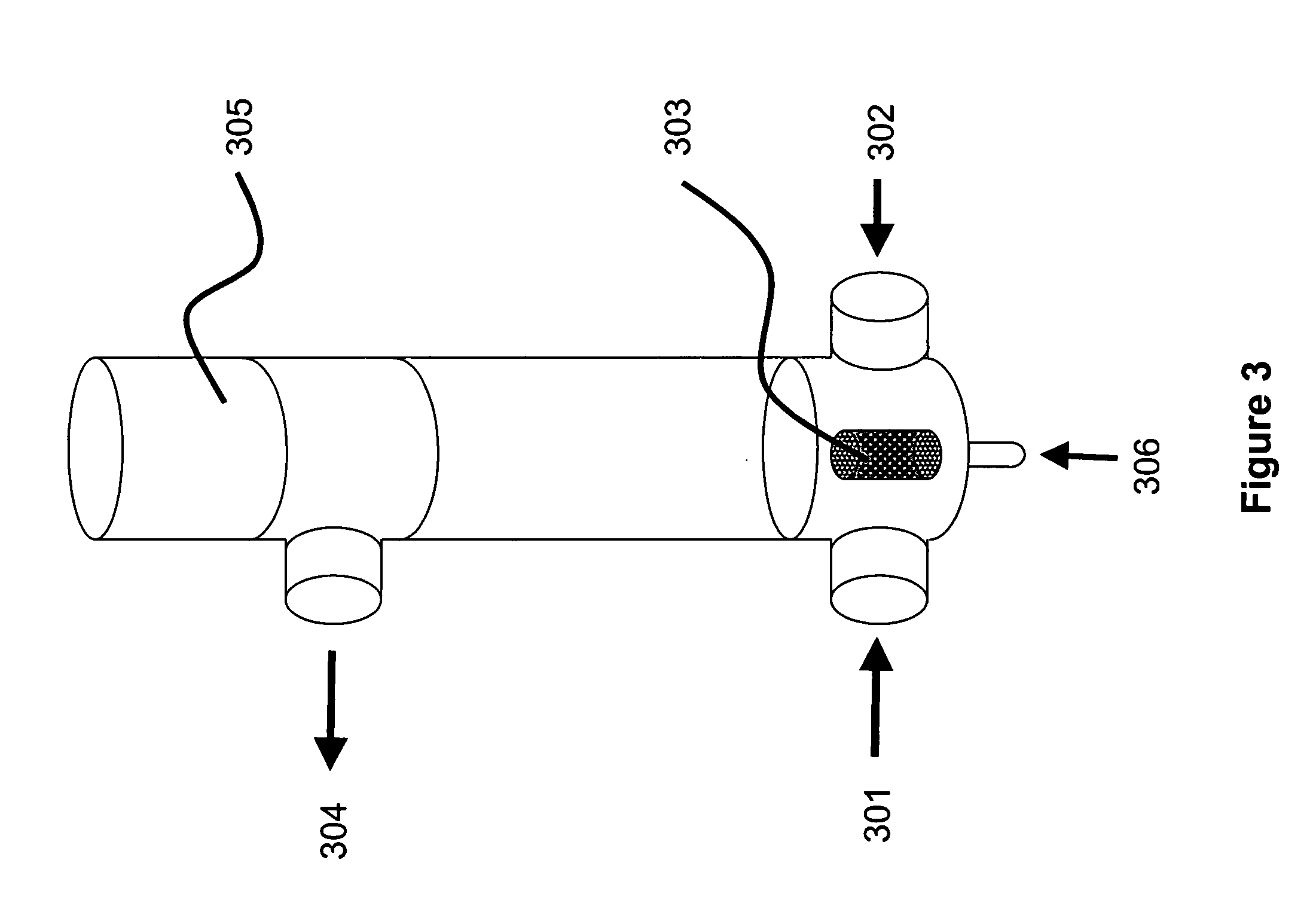
An apparatus and a method for isolating a biologic product, such as plasmid DNA, from cells. The method involves lysing cells in a controlled manner separate insoluble components from a fluid lysate containing cellular components of interest, followed by membrane chromatographic techniques to purify the cellular components of interest. The process utilizes a unique lysis apparatus, ion exchange and, optionally, hydrophobic interaction chromatography membranes in cartridge form, and ultrafiltration. The process can be applied to any biologic product extracted from a cellular source. The process uses a lysis apparatus, including a high shear, low residence-time mixer for advantageously mixing a cell suspension with a lysis solution, a hold time that denatures impurities, and an air-sparging bubble mixer that gently yet thoroughly mixes lysed cells with a neutralization / precipitation buffer and floats compacted precipitated cellular material.
Owner:VGXI
Vectors comprising stuffer/filler polynucleotide sequences and methods of use
Owner:COLGATE PALMOLIVE CO
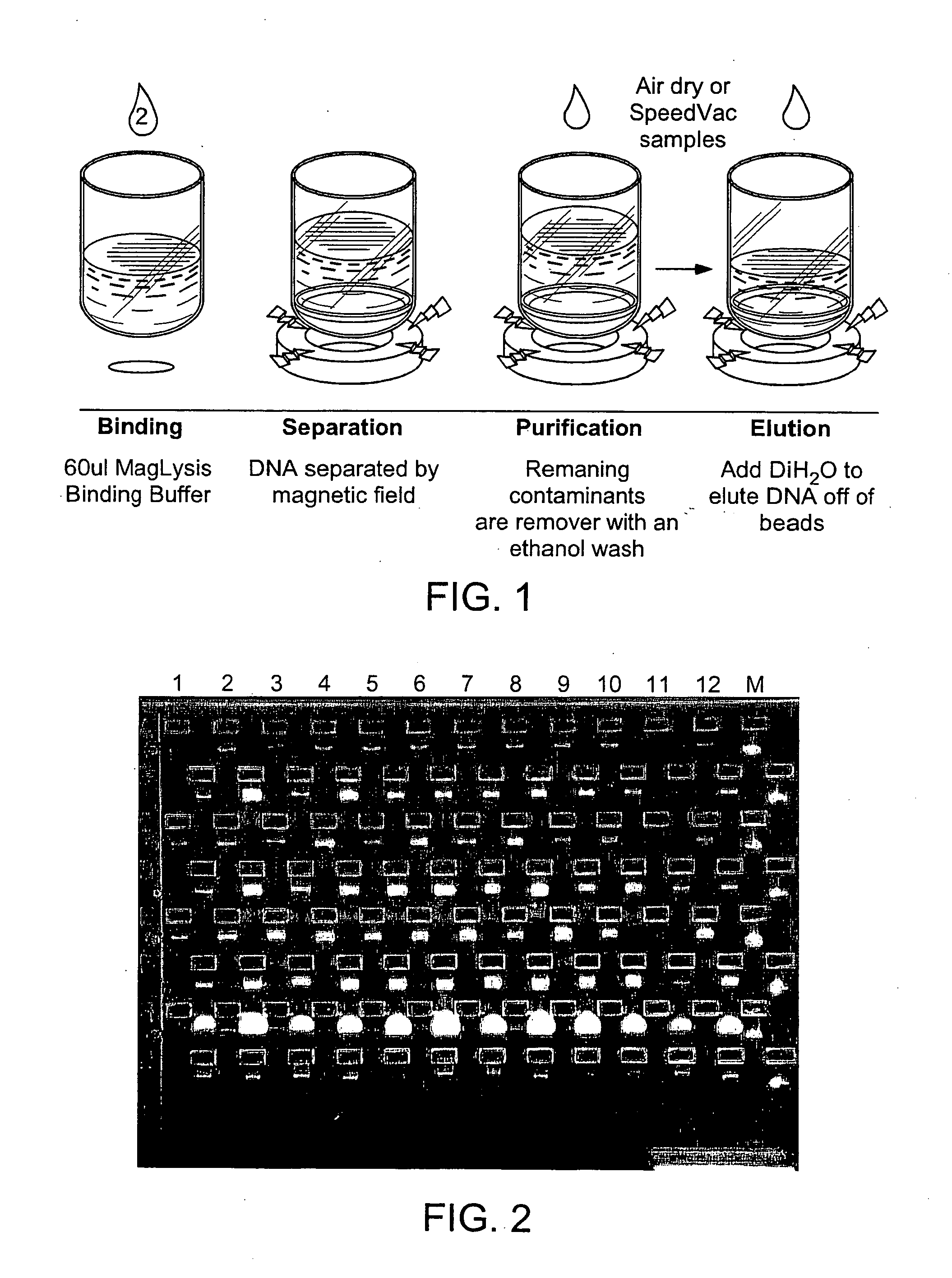
Kit for extracting bacterial plasmid DNA (deoxyribonucleic acid) by magnetic beads, and extraction method thereof
The invention discloses a kit for extracting bacterial plasmid DNA (deoxyribonucleic acid) by magnetic beads, and an extraction method thereof. The kit comprises six components: a bacterial resuspension solution, a cracking solution, a neutralizing solution, a magnetic bead combining solution, a washing solution and an eluting solution. The extraction method of the kit comprises the following steps of: resuspending bacteria, cracking, neutralizing, combining with magnetic beads, washing and eluting. The plasmid DNA extracted by the invention has high purity and complete fragments, and the extraction process is convenient, quick, efficient and safe.
Owner:HENAN HUIER NANO TECH
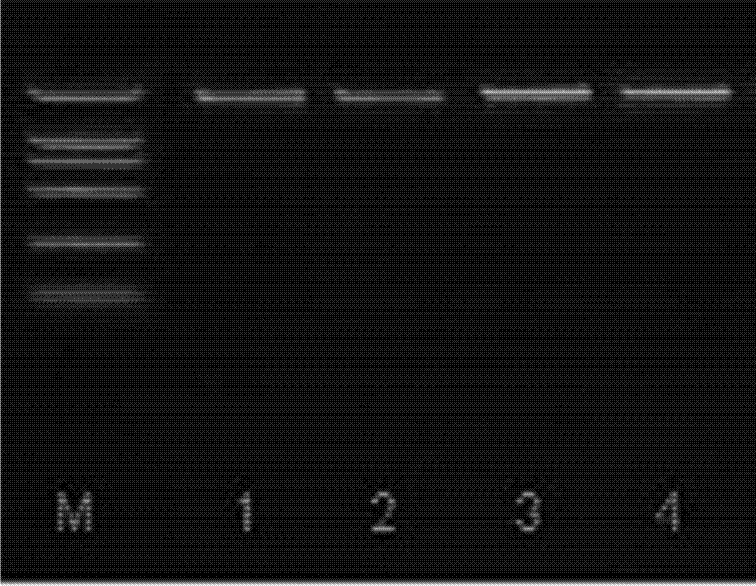
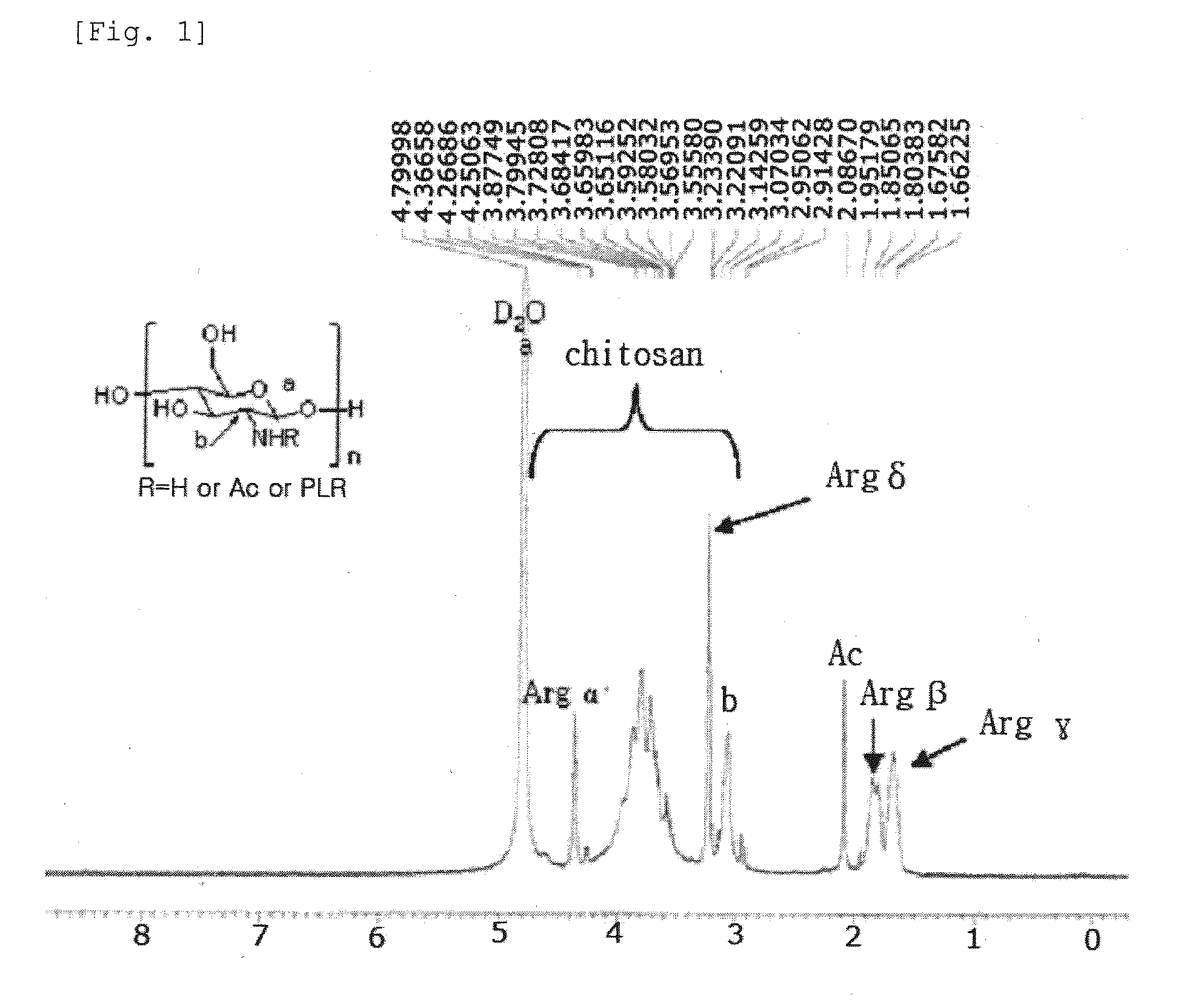
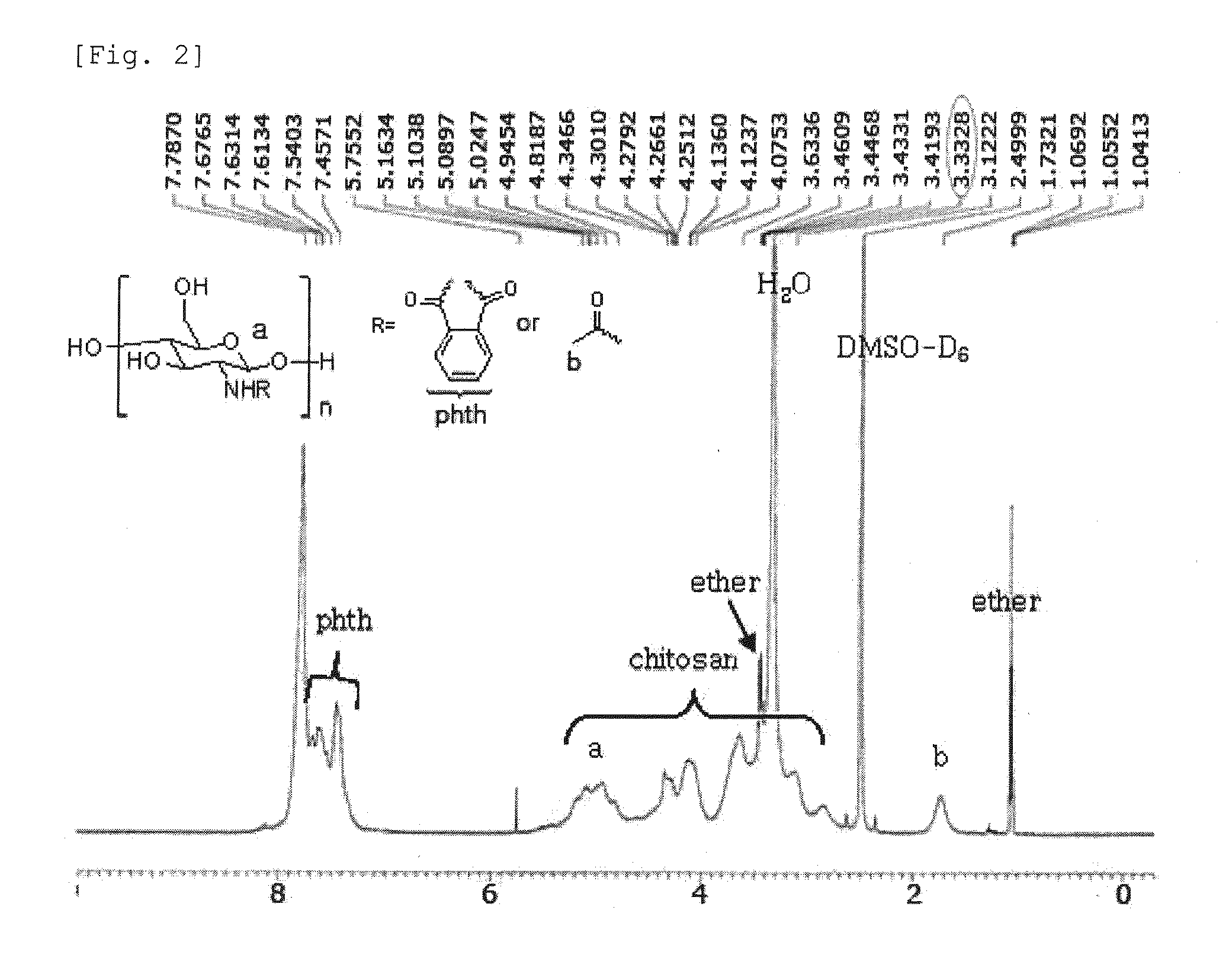
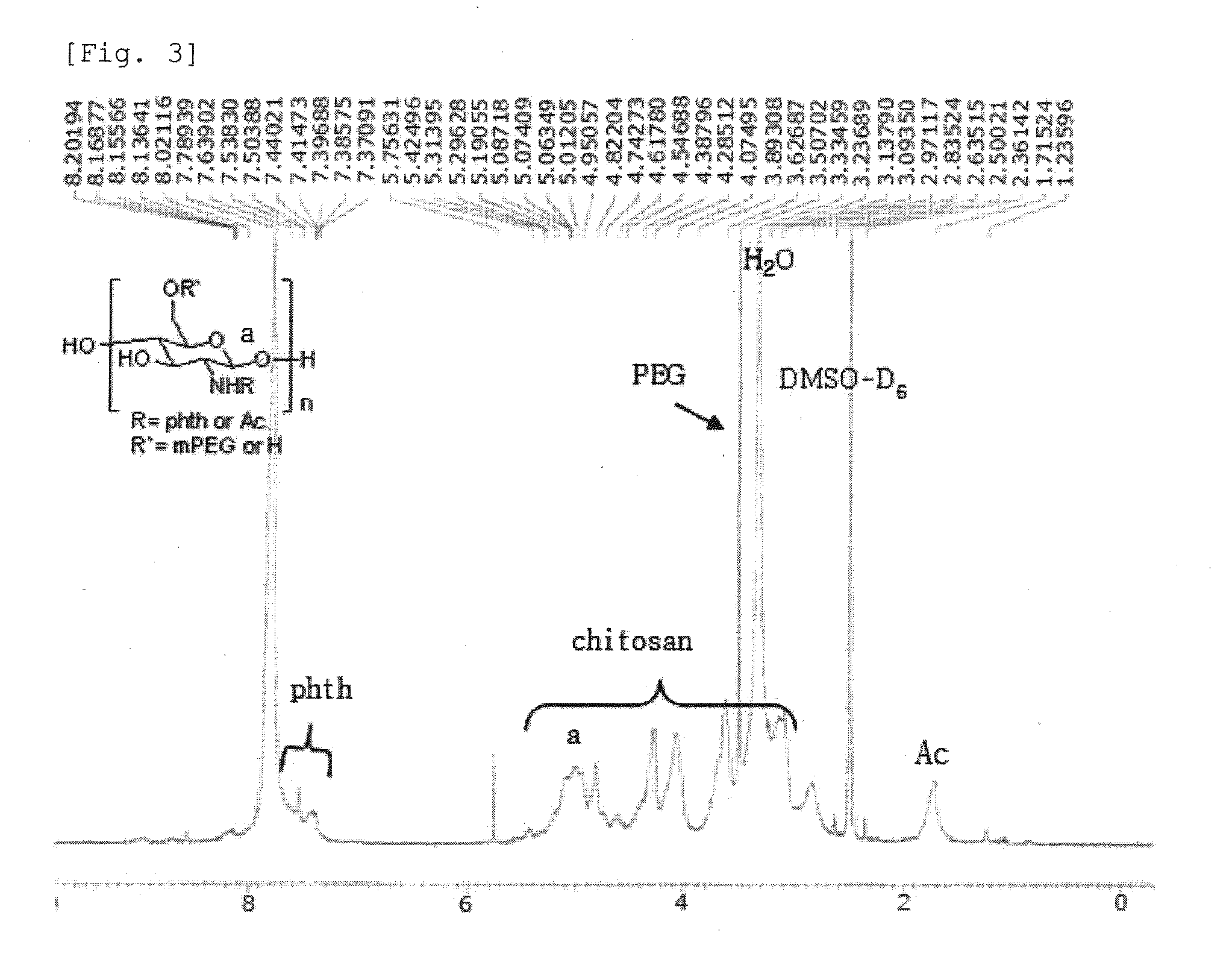
Chitosan Based Polymer Conjugate and a Method for Producing the Same
InactiveUS20100113559A1Improve in vivo transfer efficiencyLow cytotoxicityPeptide/protein ingredientsGenetic material ingredientsPolyethylene glycolCytotoxicity
The present invention relates to a conjugate of chitosan and polyamine polymer that is useful for transferring a desired gene medicine into cells, and a method for preparing the same. In particular, the present invention relates to a double conjugate that is prepared by 1 in king poly-L-arginine to low molecular weight chitosan or triple conjugate that is prepared by additionally linking polyethylene glycol (PEG) to the double conjugate, and a method for preparing the same. The chitosan based cationic polymer conjugate of the present invention forms a complex with negatively charged gene medicine such as plasmid DNA and small interfering RNA to efficiently transfer the desired gene medicine into cells with low cytotoxicity. Accordingly, the conjugate can be used as an effective delivery system for in vivo administration of gene medicine.
Owner:ENGENE INC
Superparamagnetic nanoparticles IN MEDICAL THERAPEUTICS and manufacturing method THEREOF
InactiveUS20110135577A1Safe and effectiveAvoid problemsBiocidePeptide/protein ingredientsDelivery vehicleMagnetite
The successful transfer of therapeutic agents such as genetic materials (e.g. nucleic acid) or drug into living cells is the most important issue depending on the development of the delivery carrier. A method for manufacturing superparamagnetic nanoparticles in medical therapeutics is described to develop nano-sized calcium phosphate (CaP) mineral was rendered magnetic as delivery vehicle. The CaP-based magnetized nanoparticles (NPs) were possessed superparamagnetic property by hetero-epitaxial growth of magnetite on the CaP crystallites and also showed no harm to the cultured cells and elicited no cytotoxicity. The magnetized CaP was demonstrated to have good plasmid DNA binding affinity or drug carrying capacity. It significantly increased the expression of gene transfection and efficiency in delivery to mesenchymal stem cells (MSCs) under exogenous magnetic field. According to the above facts, this newly-synthesized magnetized CaP NPs has great potential as a novel non-viral targeted delivery vehicle to be applied for medical applications.
Owner:NAT TAIWAN UNIV
Preparation method of double-targeting cationic ultrasound microbubbles carrying cell-penetrating peptide iRGD
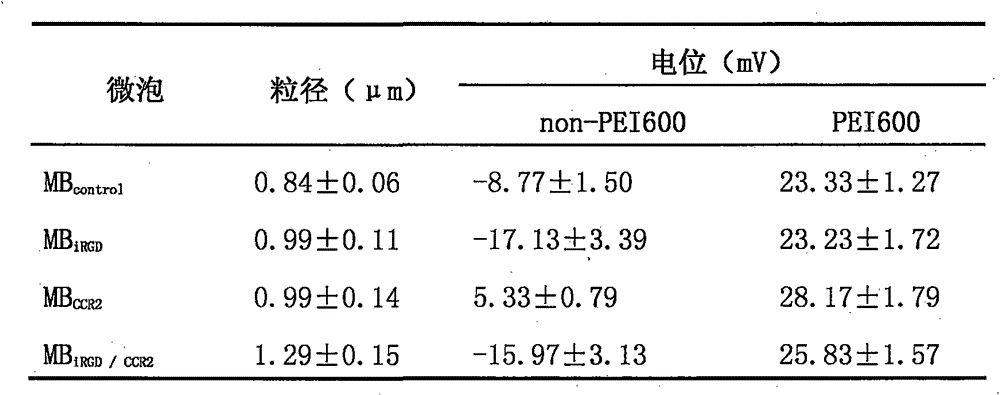
InactiveCN105106977AUniform particle size distributionSpeed up entryEnergy modified materialsGenetic material ingredientsPolyetherimidePlasmid dna
The invention discloses a preparation method of double-targeting cationic ultrasound microbubbles carrying cell-penetrating peptide iRGD and belongs to the field of fundamental research. The preparation method uses materials, including DSPC, DSPE-PEG2000-maleimide, DSPE-PEG2000-Biotin, iRGD peptide, CCR2 antibody, stearic acid, branched-chain polyetherimide-600, N, N'-carbonyldiimidazole, and avidin; by bonding integrin Alpha v Beta3 targeting cell-penetrating peptide iRGD and the CCR2 antibody to surfaces of the microbubbles, double-targeting cationic ultrasonic contrast agent is constructed and has the double functions of carrying plasmid DNA and targeting tumor cells; combining ultrasound-mediated biological effect and iRGD cell-penetrating action, the agent is capable of promoting entry of genes into the tumor cells and improving gene transfection efficiency and is expected to be developed and applied to visual ultrasound-controlled-release genetic treatment of tumors.
Owner:SHENZHEN PEOPLES HOSPITAL
Paired-end library construction method and method for sequencing genome by using library
InactiveCN102181943AReduce workloadLow costMicrobiological testing/measurementVector-based foreign material introductionGenomic sequencingDNA fragmentation
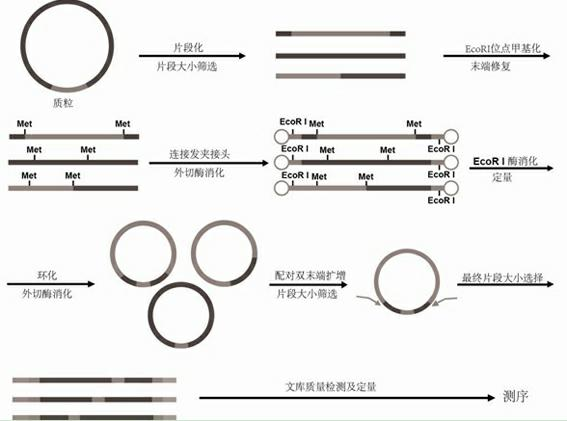
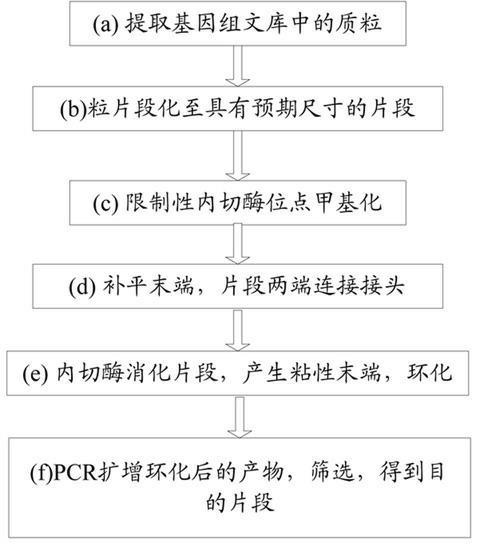
The invention relates to a paired-end library construction method and a method for sequencing a genome by using the library. The paired-end library construction method comprises the following steps of: performing plasmid extraction on clone in a constructed genome DNA library, fragmenting the extracted plasmid DNA, performing methyl protection on the restriction endonuclease site on the fragmented DNA, adding a hairclip type joint, performing enzyme cutting and cyclization to obtain circular DNA, then amplifying the genome library paired-end fragment in the circular DNA through a pair of composite primers to obtain a paired-end sequence of ultra-long span, and finally performing high flux sequencing. The paired-end sequence obtained by using the method can be applied to splicing of a new species genome sequence so as to further improve the splicing quality.
Owner:SUN YAT SEN UNIV
1,3,5-triazinane-2,4,6-trione derivatives and uses thereof
ActiveUS9315472B2Good curative effectDelay moreOrganic chemistryHeterocyclic compound active ingredientsDiseaseHepatic Diseases
The present invention provides novel 1,3,5-triazinane-2,4,6-trione derivatives, such as compounds of any one of Formulae (I) and (II), and salts thereof, and methods of preparing the compounds. Also provided are compositions including a compound of the invention and an agent (e.g., an siRNA, mRNA, or plasmid DNA). The present invention also provides methods and kits using the compositions for delivering an agent to a subject (e.g., to the liver, spleen, or lung of the subject) or cell and for treating and / or preventing a range of diseases, such as genetic diseases, proliferative diseases, hematological diseases, neurological diseases, liver diseases, and lung diseases.
Owner:MASSACHUSETTS INST OF TECH
Method for isolating nucleic acids
InactiveCN101268189AAchieve separationSimplify automationDNA preparationNucleic acid detectionPlasmid dna
Described herein is a method in which genomic nucleic acid of a cell can be separated from nucleic acid having a molecular weight that is lower than the molecular weight of the genomic nucleic acid (e.g., plasmid DNA) of the cell directly from a cell growth culture. Also described herein, a method in which genomic nucleic acid can be separated from nucleic acid having a molecular weight that is lower than the molecular weight of the genomic nucleic acid in a cell lysate without the need to prepare a cleared lysate. The present invention is also directed to a method of isolating genomic nucleic acid (e.g., RNA, DNA) of a cell or an organism.
Owner:亚钧阔特生命科学公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com