New yeast-bacteria shuttle vector
a shuttle vector and yeastbacteria technology, applied in the field of new yeastbacteria shuttle vectors, can solve the problems of cumbersome isolating sufficient quantities of yac dna from agarose gel for microinjection or electroporation, and affecting the efficiency of agarose gel agarose synthesis, and achieves the effect of high versatility of the cloning system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of Shuttle Vector
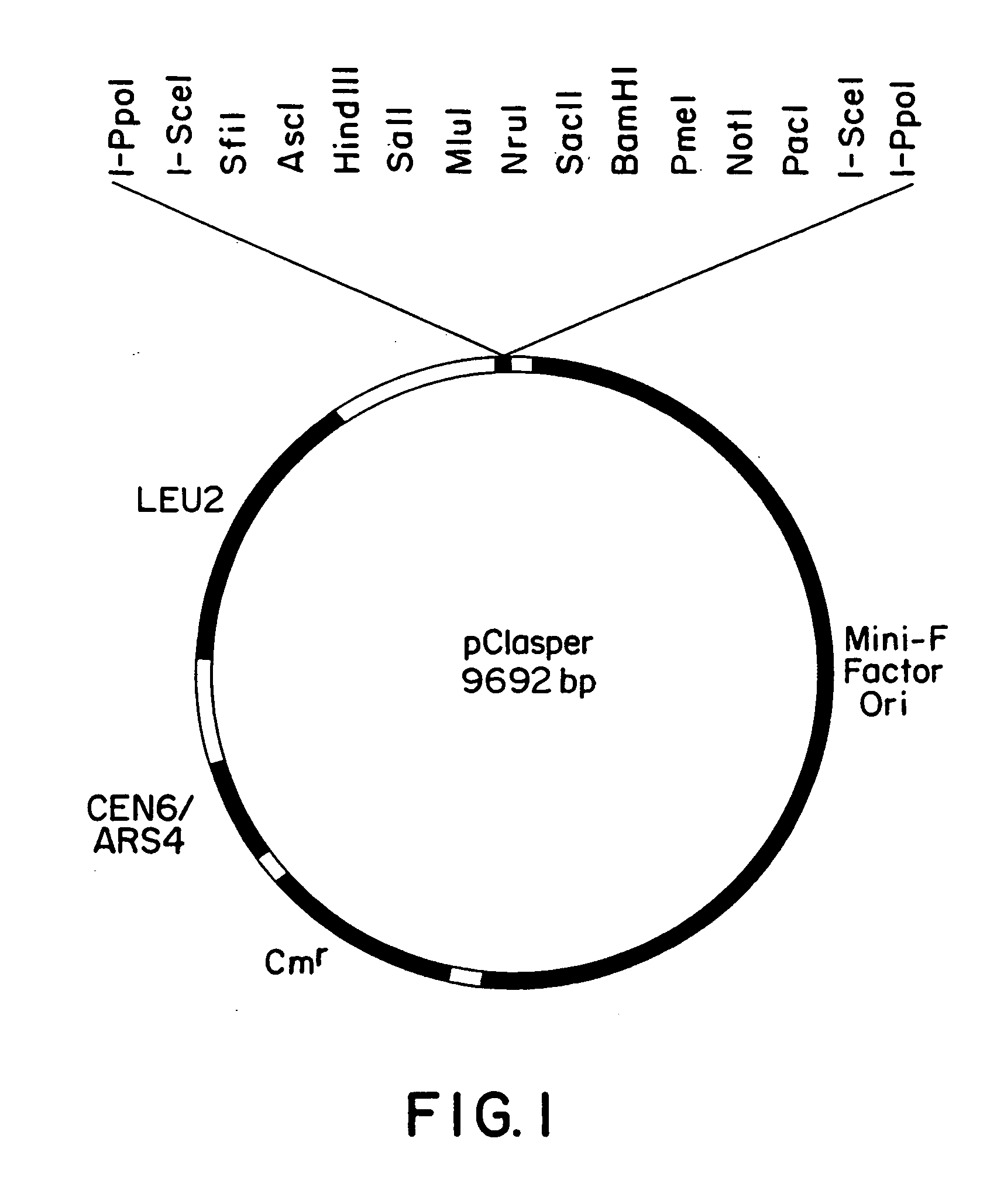
[0040] The vector, pCLASPER. (FIG. 1), was constructed as follows. The PmlI site in pRS415 (Stratagene) was cleaved to insert a linker with a SacI site flanked on one side with a 10 bp reduplication of centromeric DNA from chromosome VI (CEN6) and with a random sequence on the other side to ascertain the orientation by PCR after cloning. The resultant plasmid was cleaved at the BamHI site in the polylinker of pRS415 and at a single DraIII site between the polylinker and the beta-isopropyl malate dehydrogenase gene (LEU2) to insert a 100 bp synthetic DNA fragment containing part of a newly designed polylinker. The new plasmid was digested with Sacl and BamHI to isolate the CEN6 / ARS4 origin of replication, the LEU2 gene, and one half of the new polylinker. Plasmid pBCSK (Stratagene) was used as a substrate to generate by PCR a 1103 bp fragment encompassing, the chloramphenicol acetyl transferase (Cmr) gene from Tn9. The PCR primer for the 5′ end of the g...
example 2
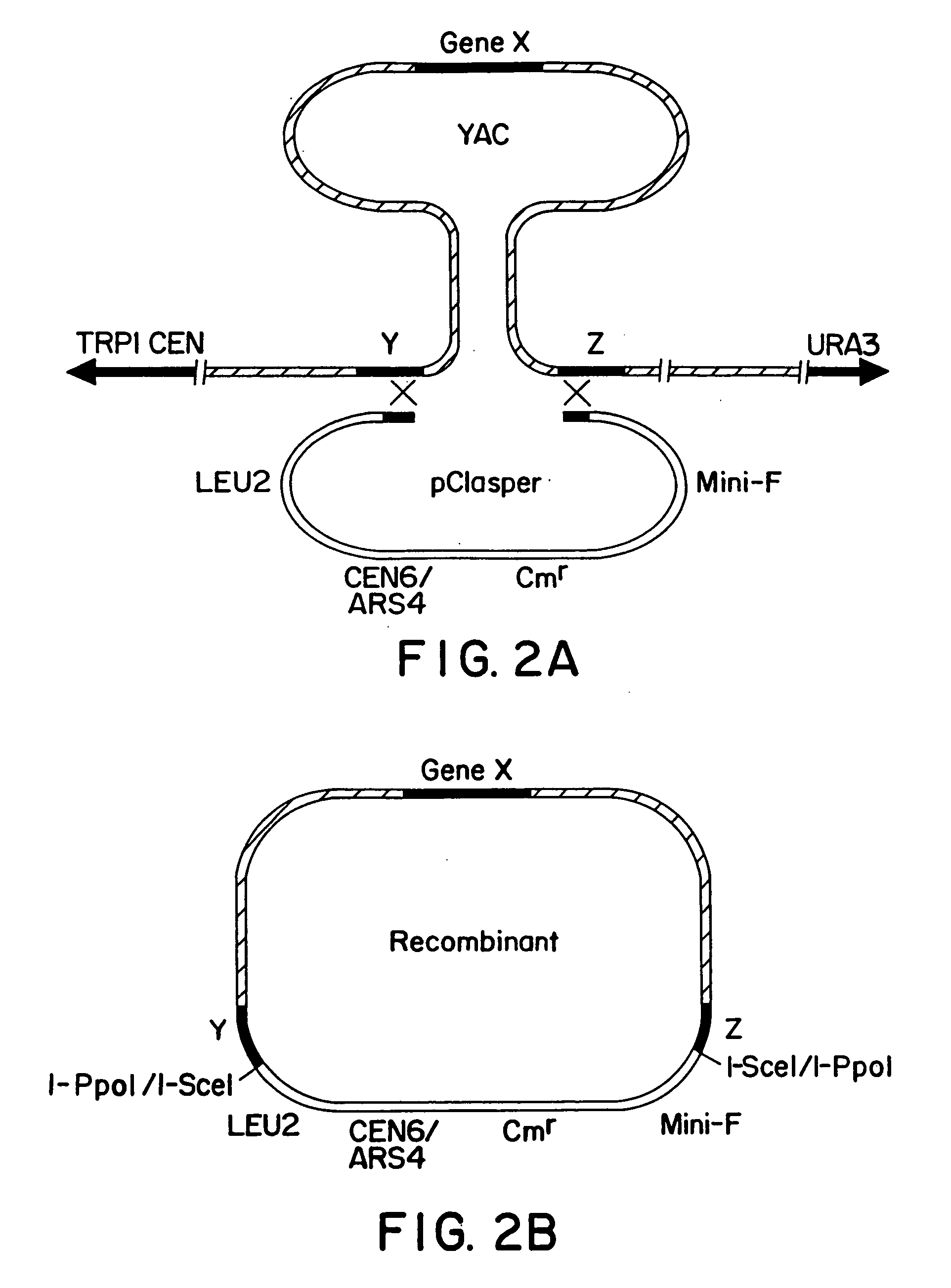
Construction of Targeting Vectors
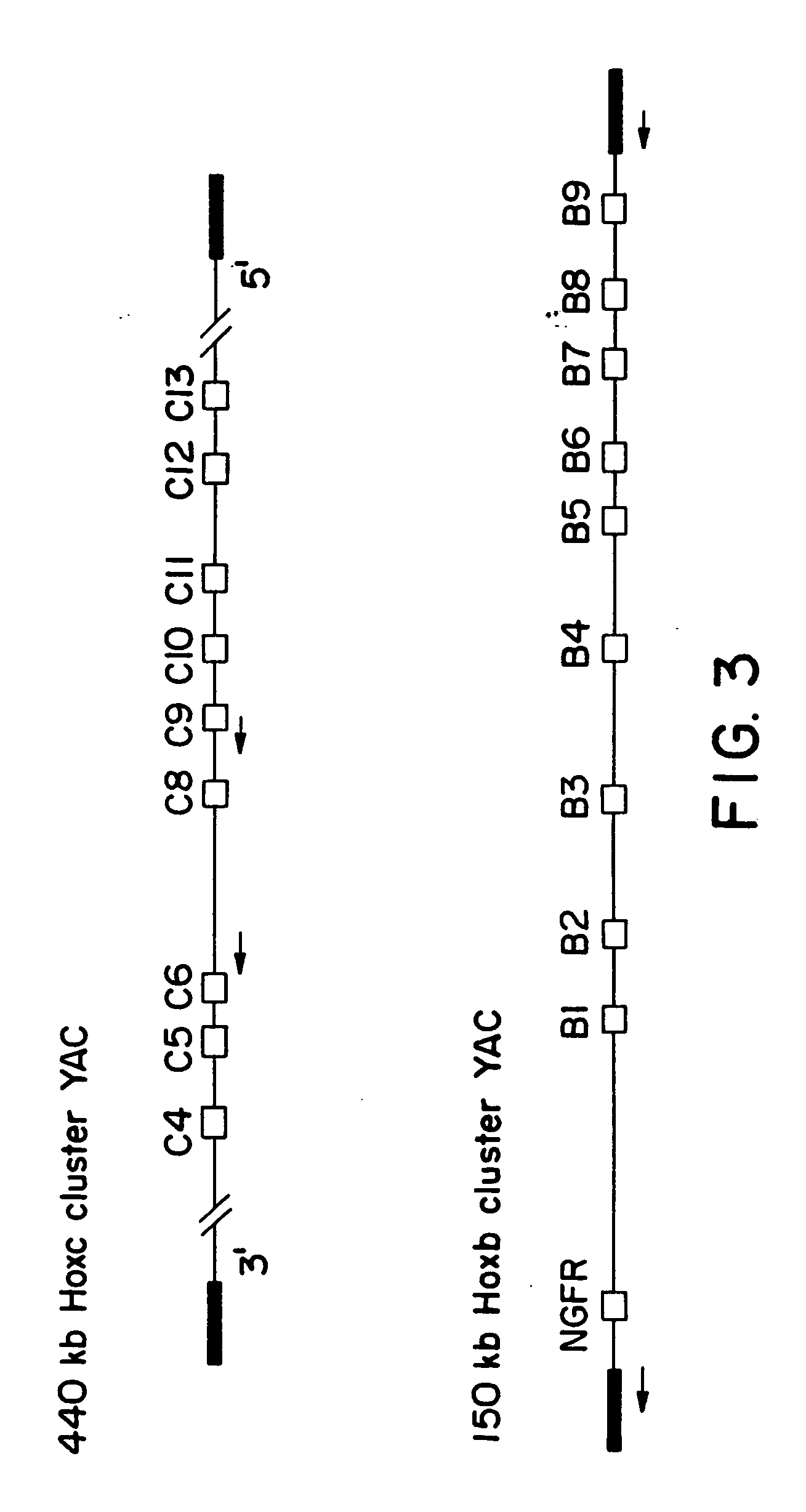
[0041] The YACs to be targeted were obtained from the Princeton mouse genomic YAC library (6). Isolation and characterization of the Hoxb and Hoxc cluster YACs have been previously described (7, 8). The targeting vector designed to clone the Hoxc-8 gene, pClC9C6, was made as follows. PCR primers for the Hoxc-6 gene (9) were designed to the 5′ untranslated region of the gene. The 5′ primer was: 5′-TAGATCTGTTTGTCTCCCACATGCC-3′ and contained a Nrul linker. The 3′ primer was: 5′-AGGTGGCAGGATAAGGAAGGGTTAG-3′ and contained a HindIII linker. The product was 706 bp. The PCR primers for the Hoxc-9 gene (10) were designed to amplify a 624 bp fragment of the 3′ untranslated region. The 5′ primer was: 5′-CGACAAGGAACAAATCCTAAGCCC-3′ and contained a BamHI linker. The 3′ primer was: 5′-TGCATTTGCAGCCTGATCCAGCCA-3′ and contained a NruI linker. All polymerase chain reactions (PCR) was carried out in a Hybard Omnigene machine. Reactions contained 1× Boehringer Mannhei...
example 3
Preparation of DNA for Yeast Transformations
[0045] Since pCLASPER is maintained at single copy, the following procedures facilitate the preparation of large quantities of plasmid DNA. E. coli strain DH1OB cells containing the targeting vectors were grown overnight in 1-2 liters TB media containing 20 μg / ml chloramphenicol. Each one liter culture was divided into 3 centrifuge bottles to pellet. The Plasmid Maxi Protocol for Qiagen-tip 500 (Qiagen) was followed for each pellet with the following changes. After the addition of 10 ml each of buffers P1, P2 and P3 to each pellet, the suspension was centrifuged at 4° C. for 30 minutes at 15,000×g. The supernatant was removed, then recentrifuged under the same conditions. The supernatants were pooled, passed through a 0.45 micron filter and loaded on a single Qiagen-tip 500 following the suppliers directions. Generally, at least 20 μg vector DNA per liter of cells was obtained as determined by comparing both optical density and visual est...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electrical resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com