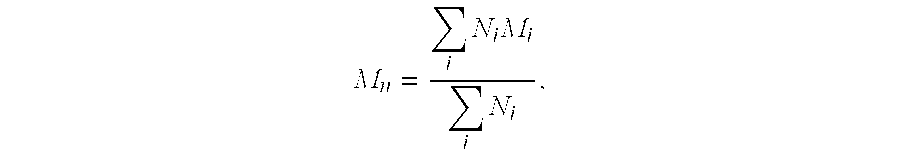Patents
Literature
324results about How to "Highly versatile" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
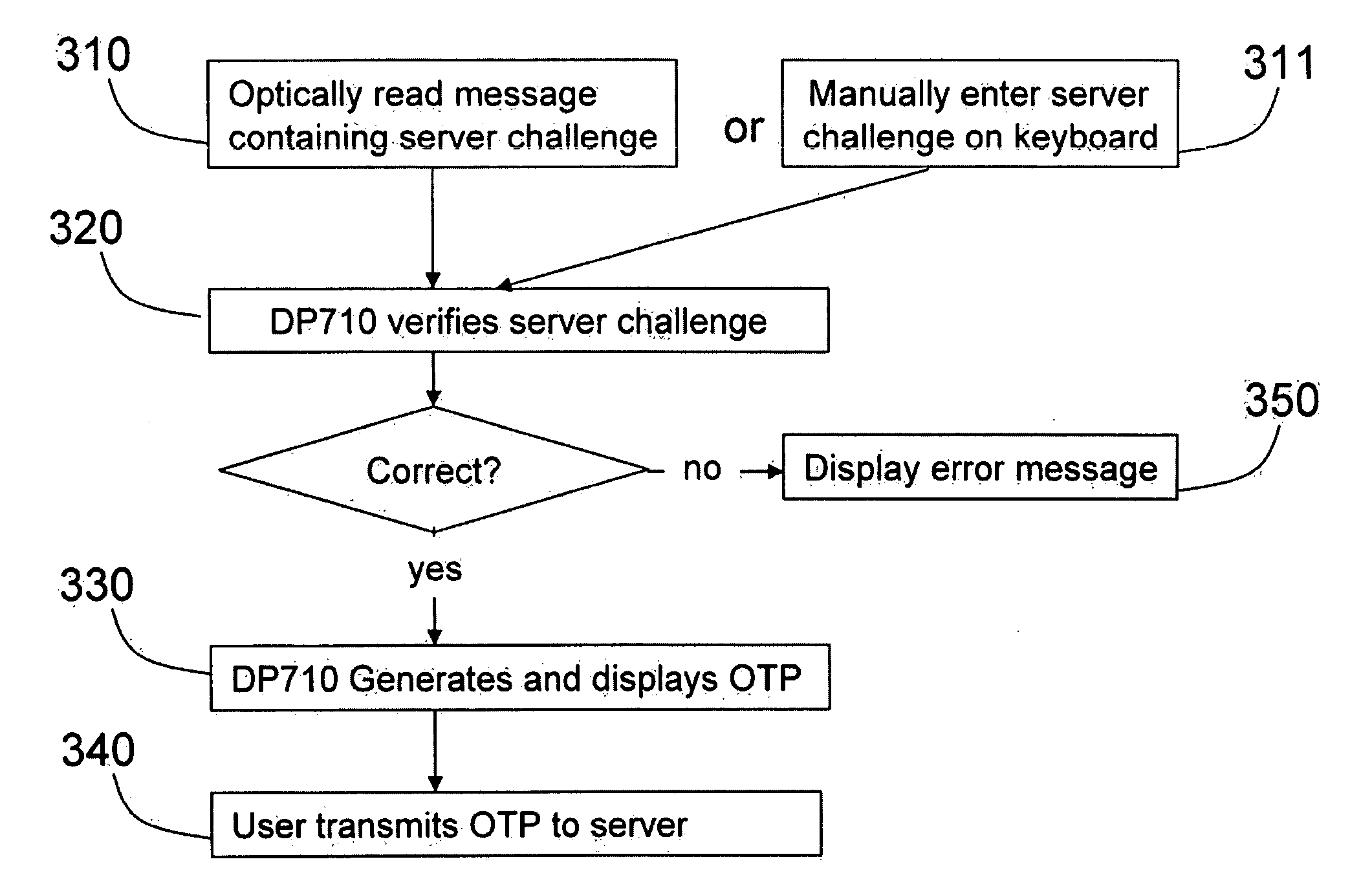
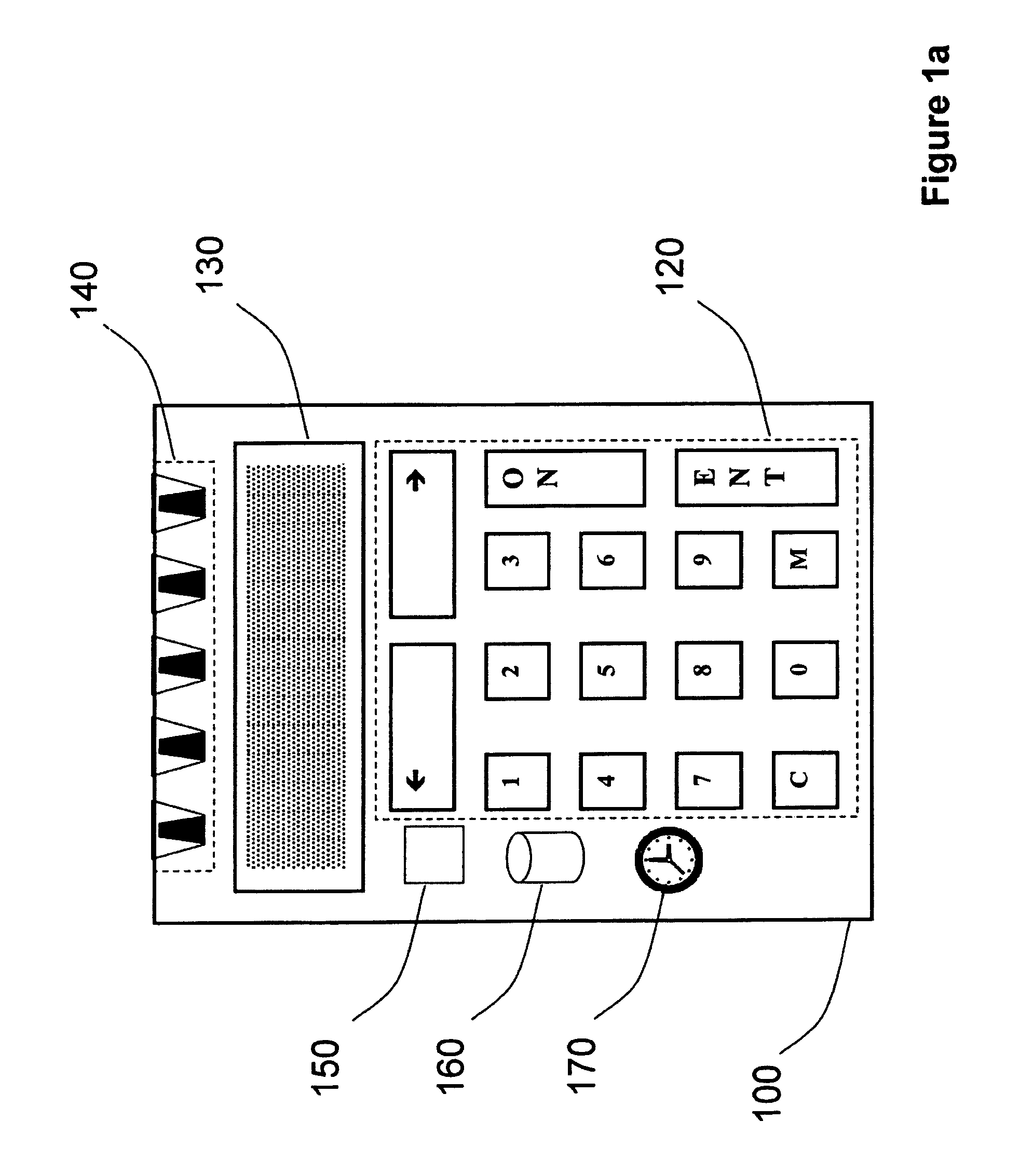
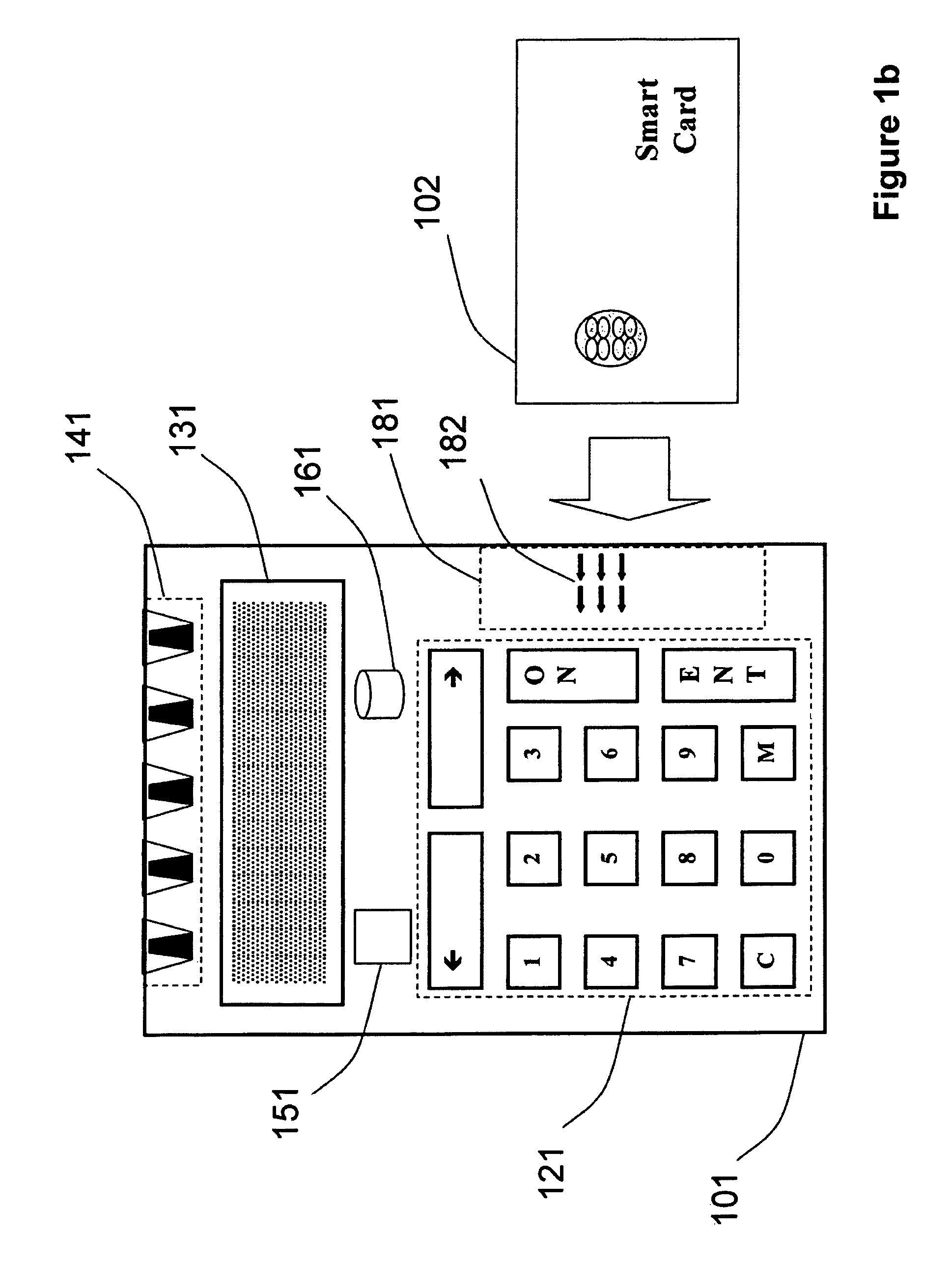
Strong authentication token generating one-time passwords and signatures upon server credential verification
ActiveUS20090235339A1Transfer size reductionHighly secureDigital data processing detailsMultiple digital computer combinationsInternet privacyTransaction data
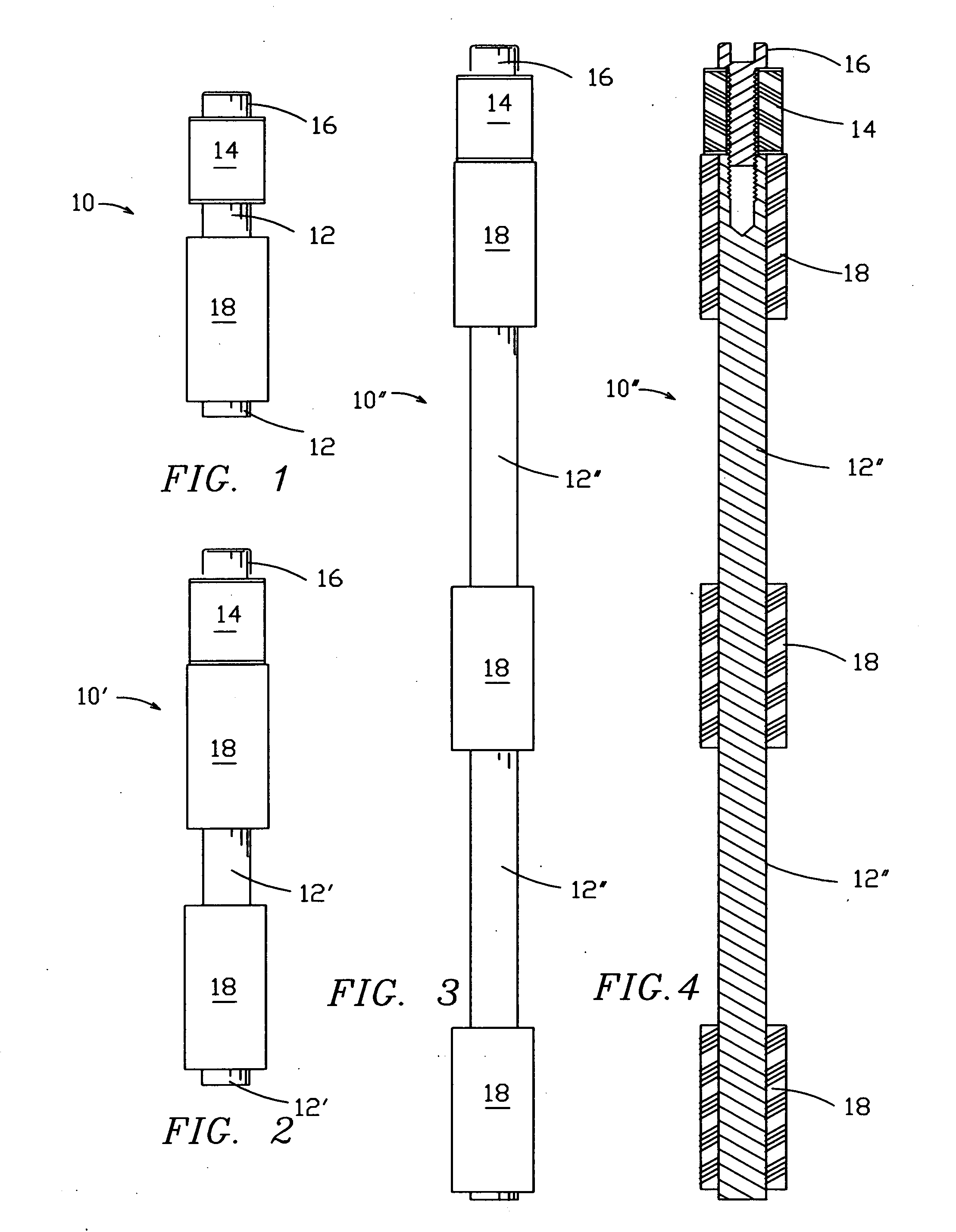
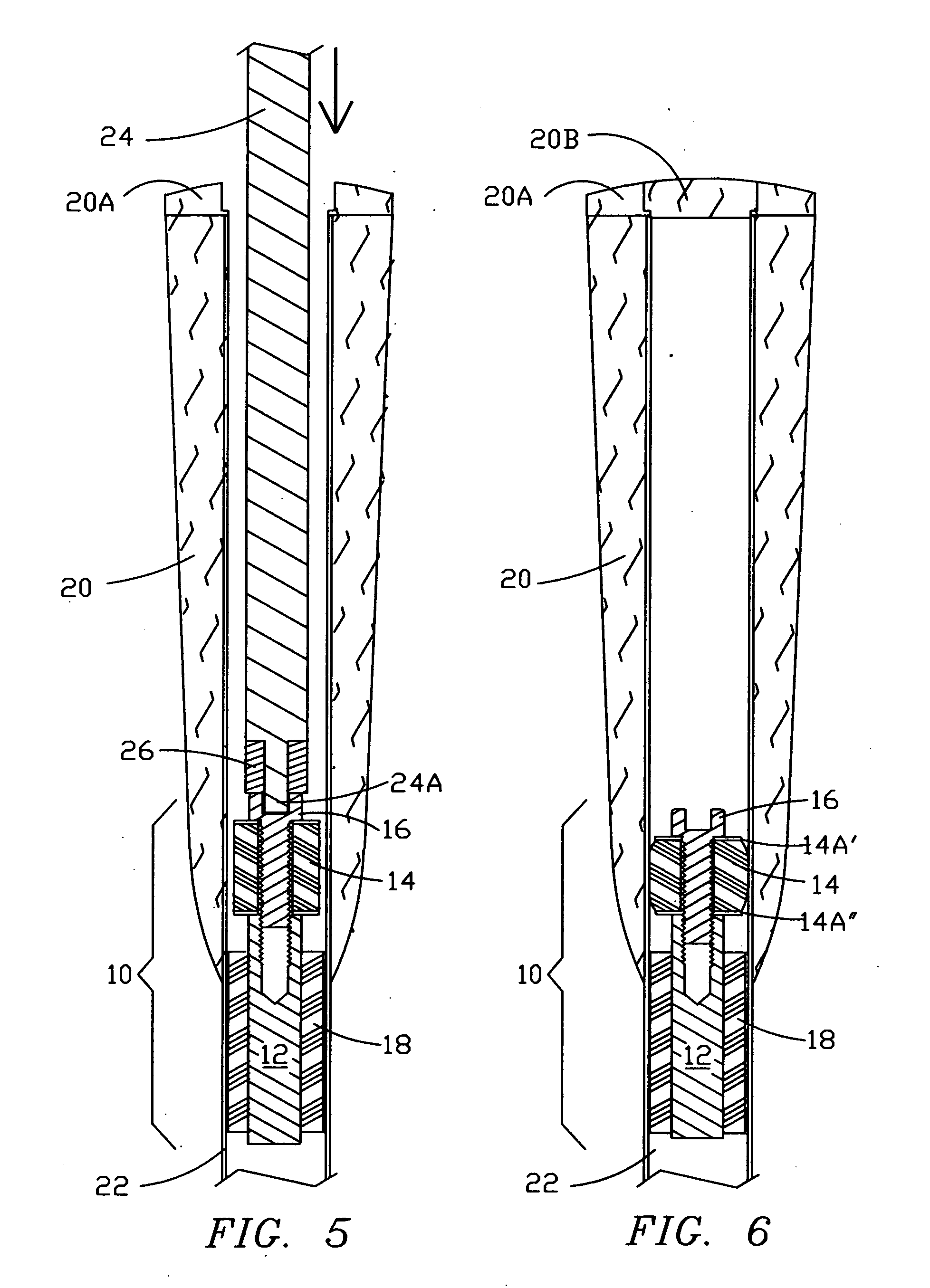
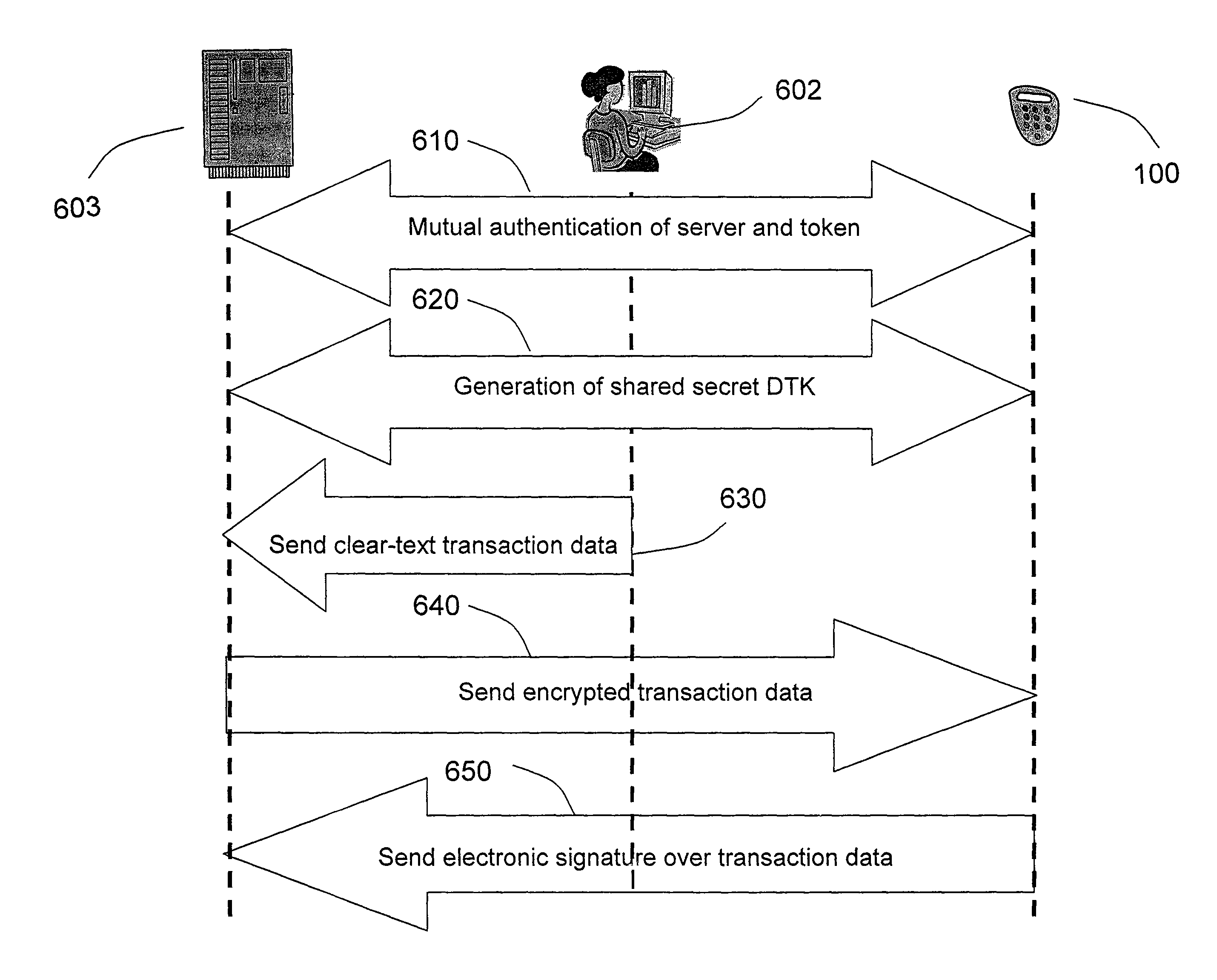
The invention defines a strong authentication token that remedies a vulnerability to a certain type of social engineering attacks, by authenticating the server or messages purporting to come from the server prior to generating a one-time password or transaction signature; and, in the case of the generation of a transaction signature, signing not only transaction values but also transaction context information and, prior to generating said transaction signature, presenting said transaction values and transaction context information to the user for the user to review and approve using trustworthy output and input means. It furthermore offers this authentication and review functionality without sacrificing user convenience or cost efficiency, by judiciously coding the transaction data to be signed, thus reducing the transmission size of information that has to be exchanged over the token's trustworthy interfaces
Owner:ONESPAN NORTH AMERICA INC
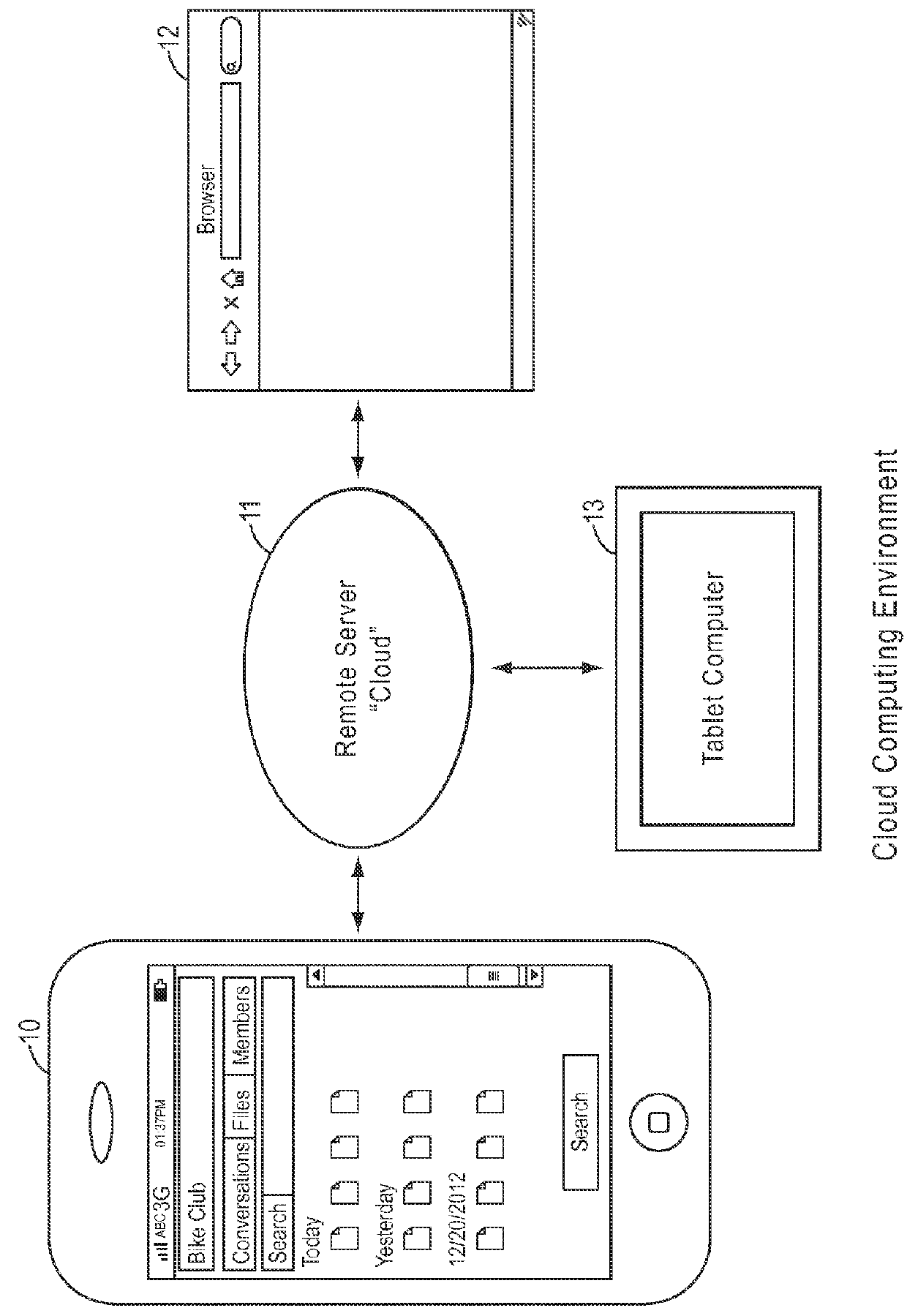
Online systems and methods for advancing information organization sharing and collective action
ActiveUS9253609B2Easily scaledHighly versatileInput/output for user-computer interactionData processing applicationsKnowledge organizationMobile device
Methods and systems and mobile device interfaces for creating, joining, organizing and managing via mobile devices affinity groups in a cloud computing environment for social and business purposes.
Owner:HOSIER DOUG
Programmable AC power switch
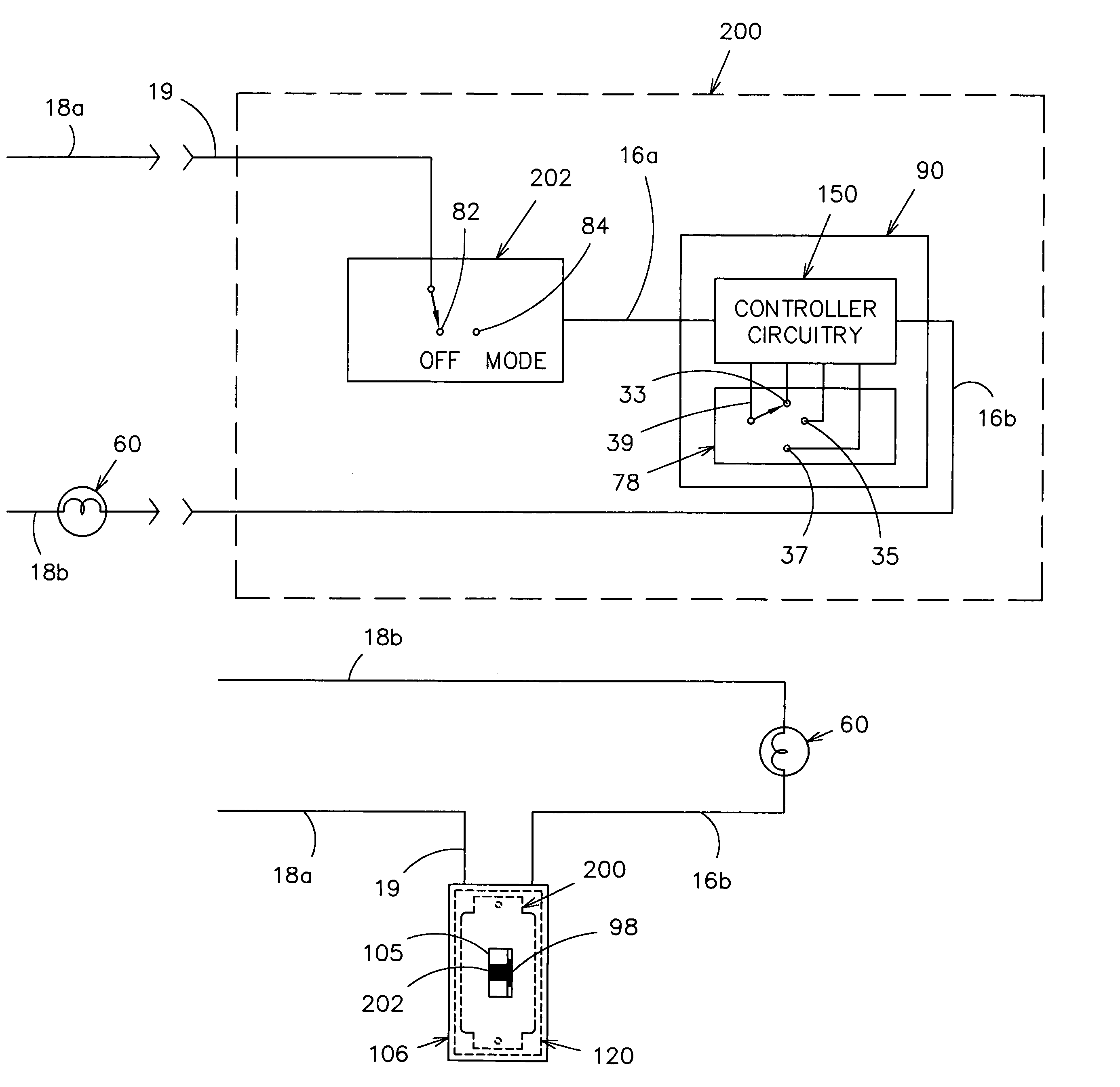
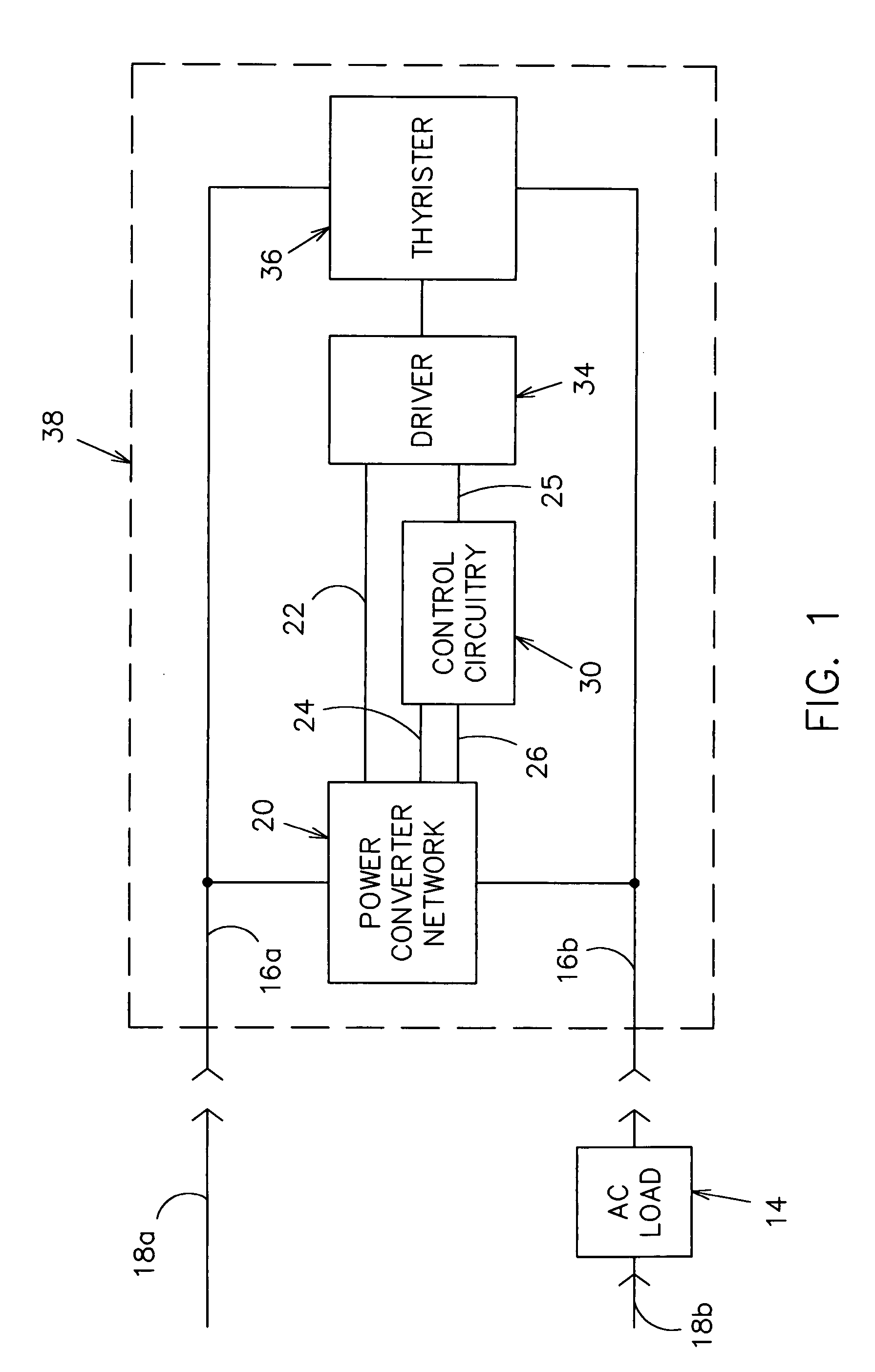
InactiveUS6933686B1Low costInexpensive and effectiveTime indicationBoards/switchyards circuit arrangementsMicrocontrollerTime delays
An AC controller provides programmable switching of AC power flow, together with producing a source of DC power for the AC controller. The AC controller is connected in series with only one side of the AC power source and the AC load, and uses a microcontroller or a Programmable Logic Device (PLD) for the programmable capability. The AC controller can be programmed to provide a light flasher function, a time delay off function, an automatic fade function, a dimming function, a burglar deterrent function, and a time delay dim function. With a multiple-position switch replacing a current wall switch, the operator can easily select many pre-programmed functions.
Owner:BISHEL RICHARD A
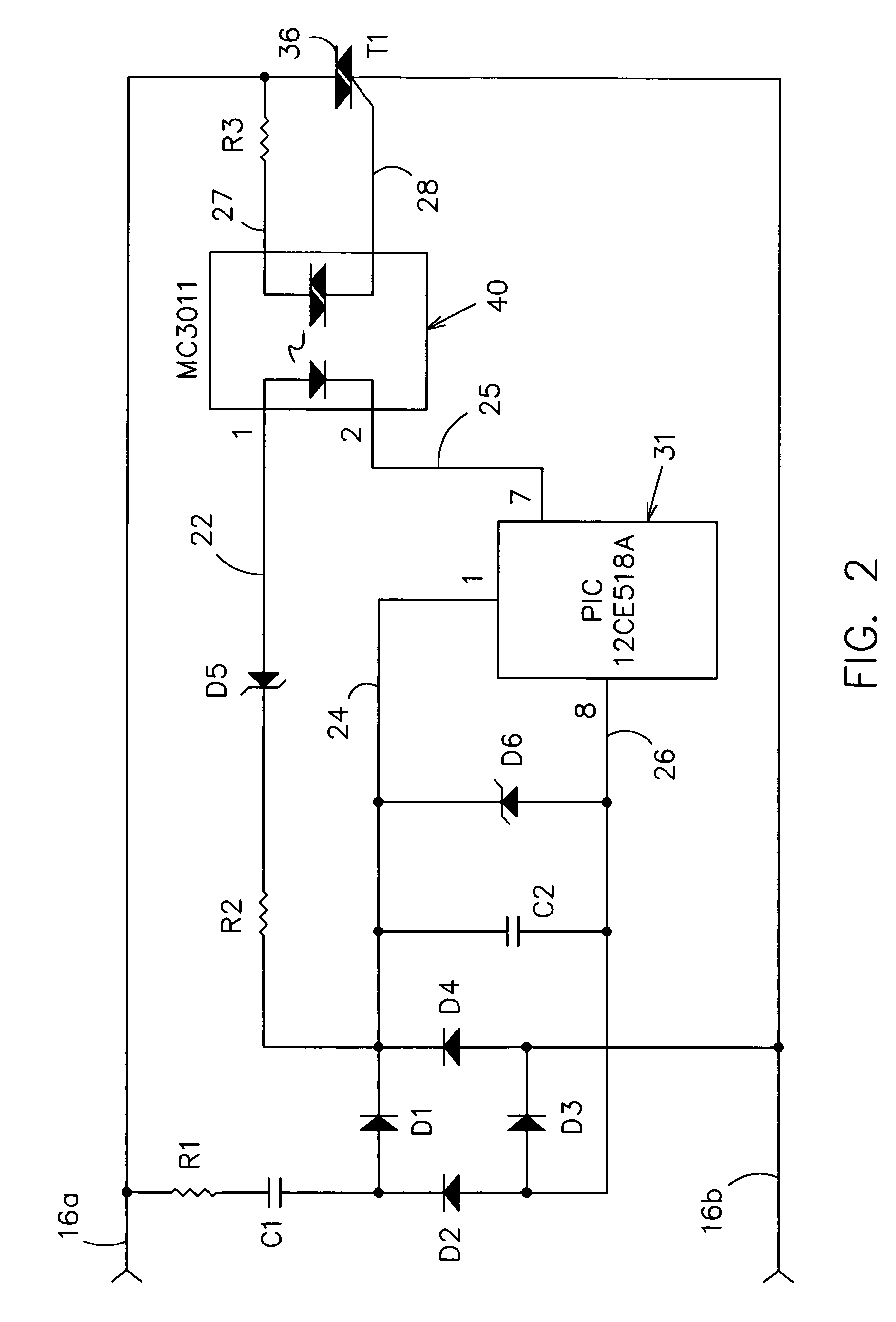
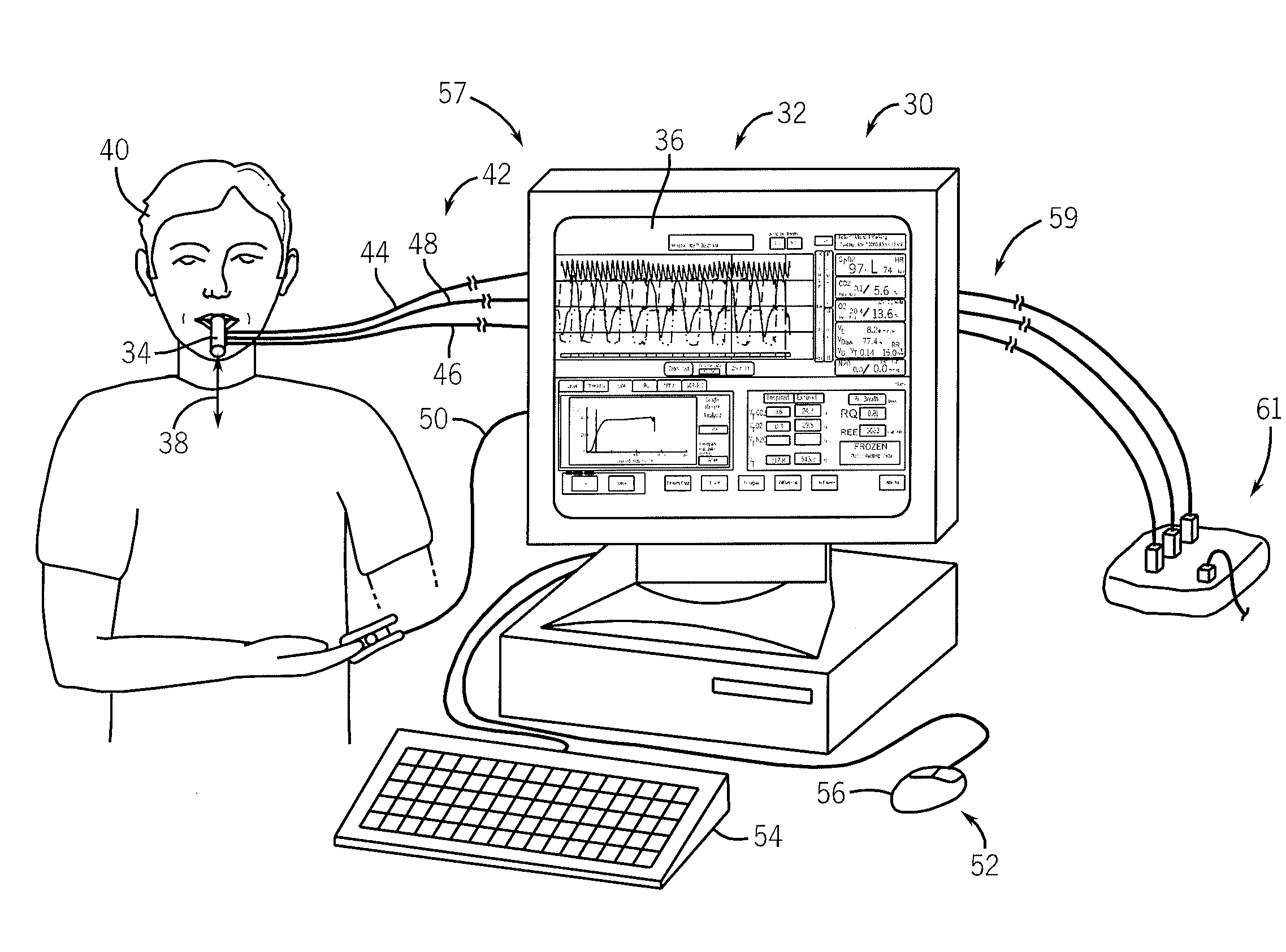
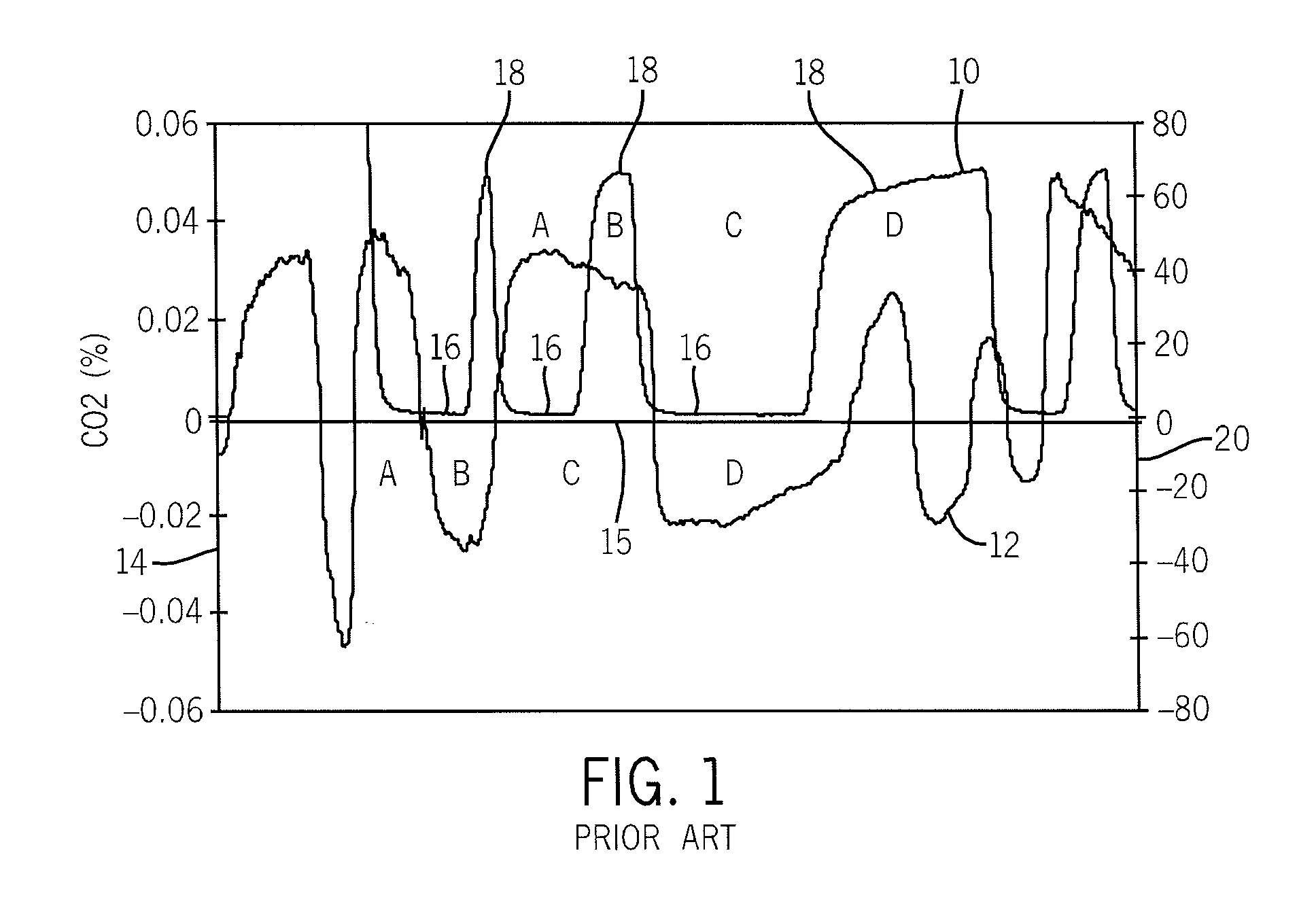
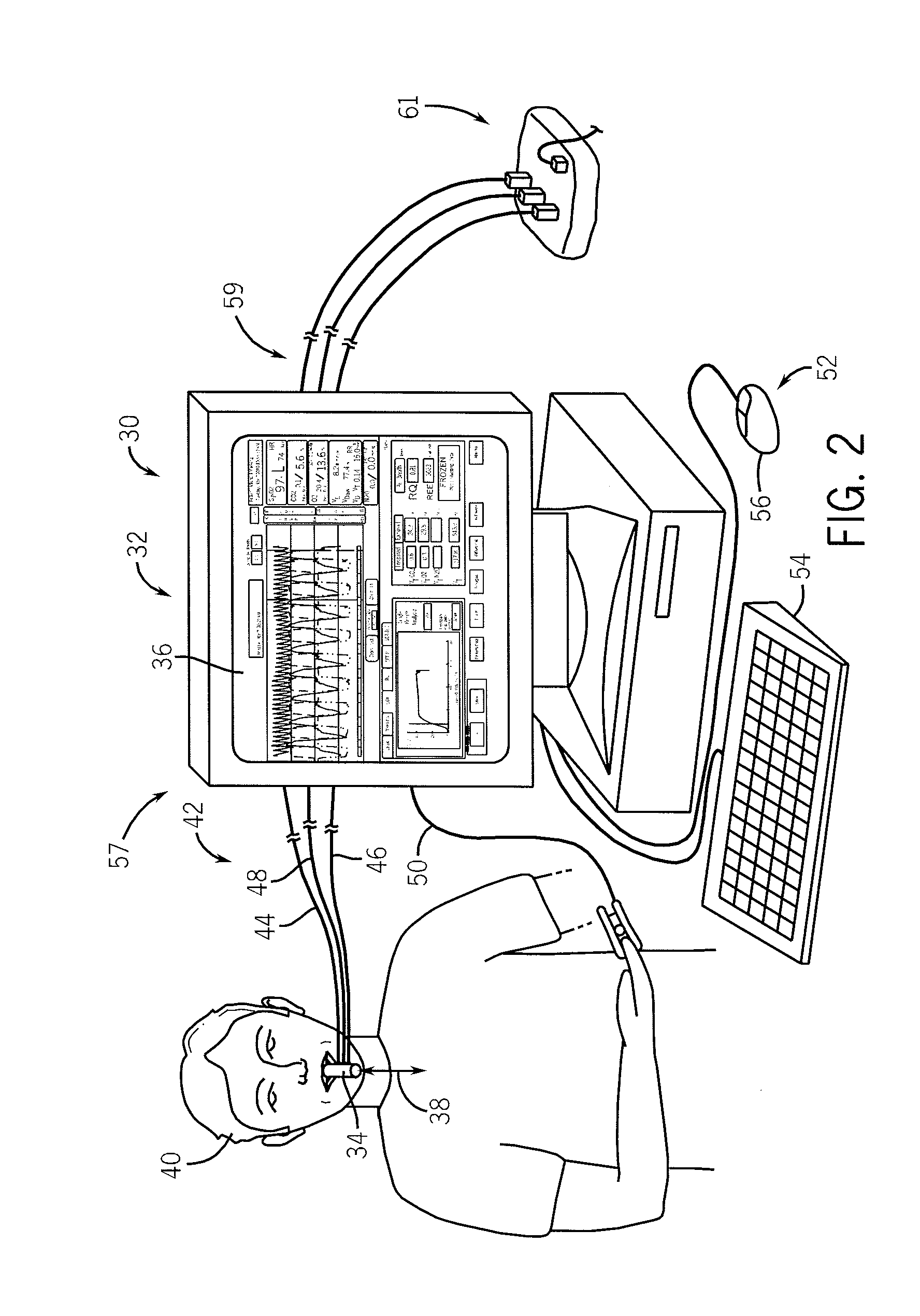
Side-stream respiratory gas monitoring system and method
ActiveUS20070107728A1Simple and efficient to manufacture and operateQuick diagnosis and analysisRespiratorsOperating means/releasing devices for valvesTraffic volumeCarbon dioxide
A side-stream respiration monitoring system includes a sensor, a monitor, and a display. The sensor is constructed to engage respiration flows having a variable flow amounts and acquire a respiration sample from the flow. The monitor is connected to the sensor and configured to determine the amount of respiration flow as well as the amount of several constituents of the respiration flow such as oxygen, carbon dioxide, water, and nitrous oxide. The monitor is also configured to adjust the determined values for ambient environment variations and is self-calibrating with respect to the ambient conditions. The monitor temporally aligns the acquired data to account for dead-space respiration events and physiological respiration modifications such as cardiac events. Information generated by the monitor is communicated to an operator via a display configured to display the real-time respiration performance.
Owner:TREYMED
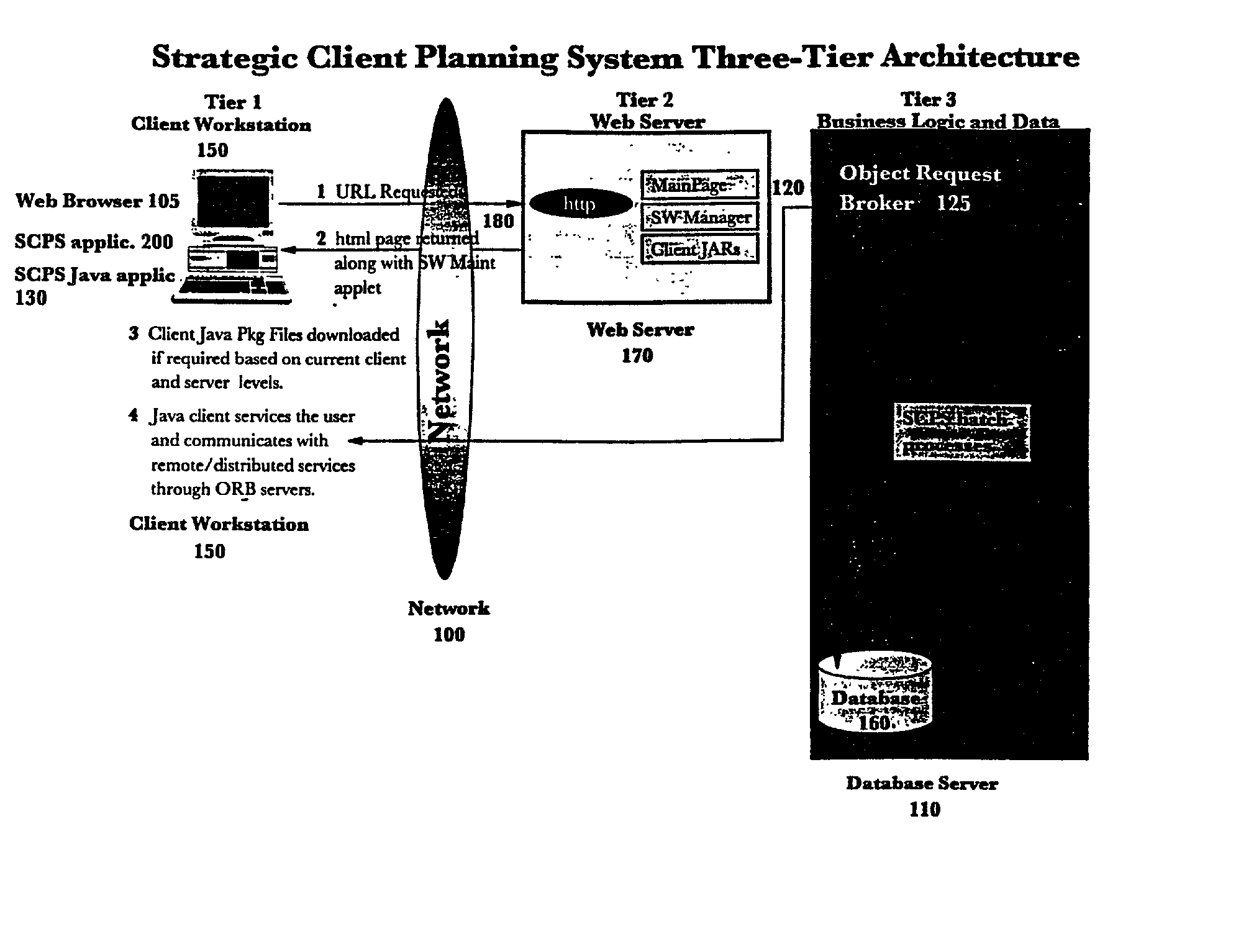
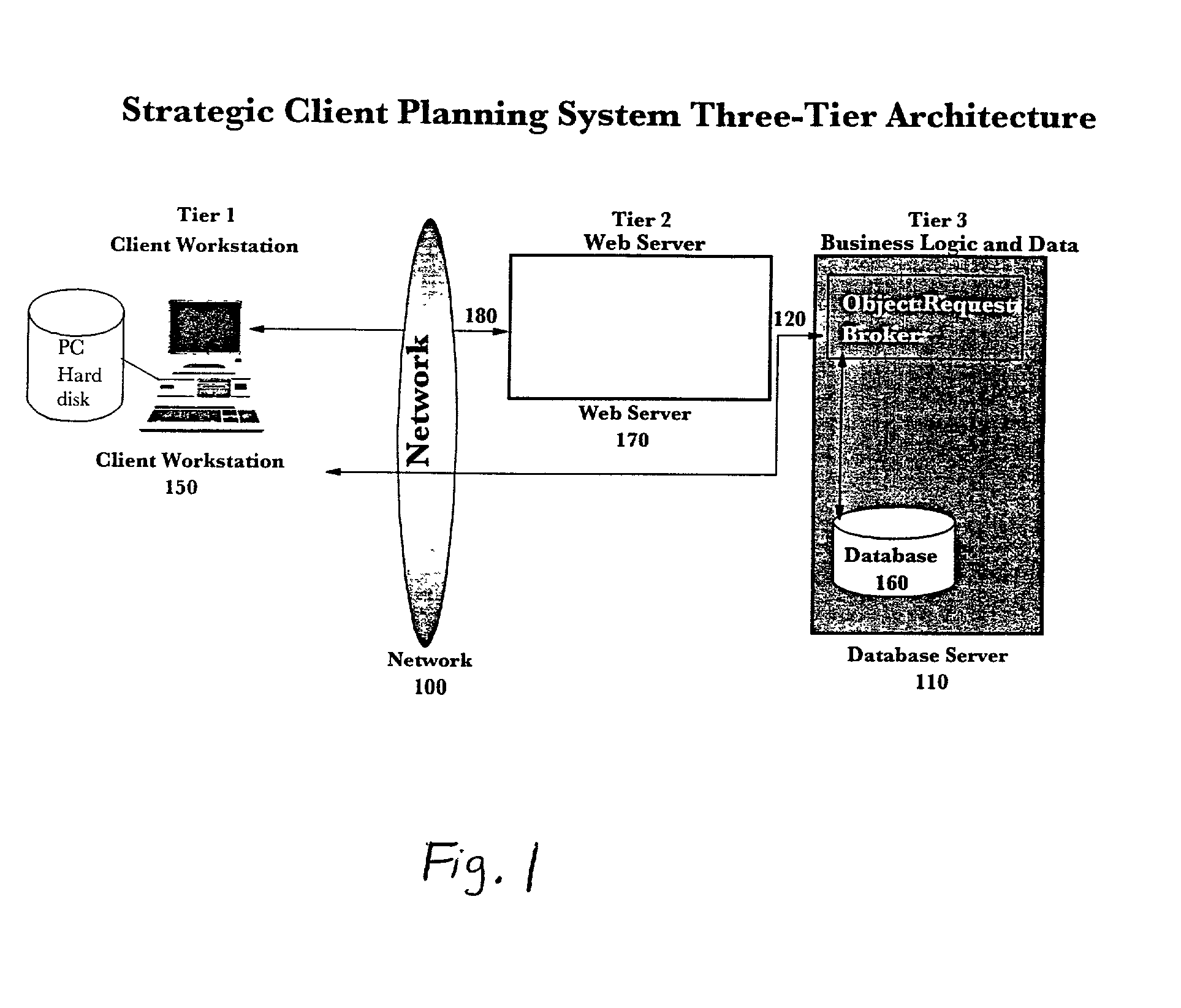
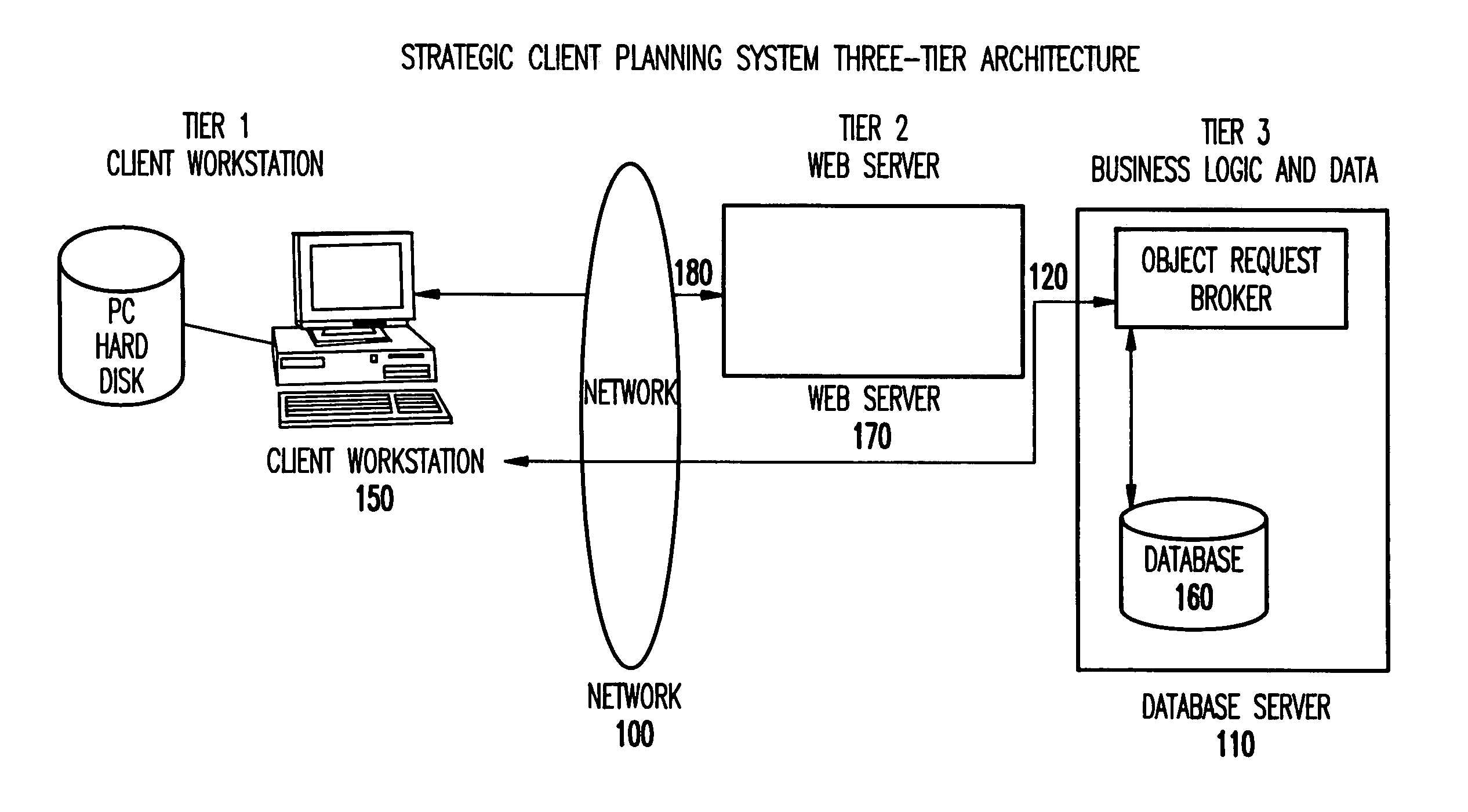
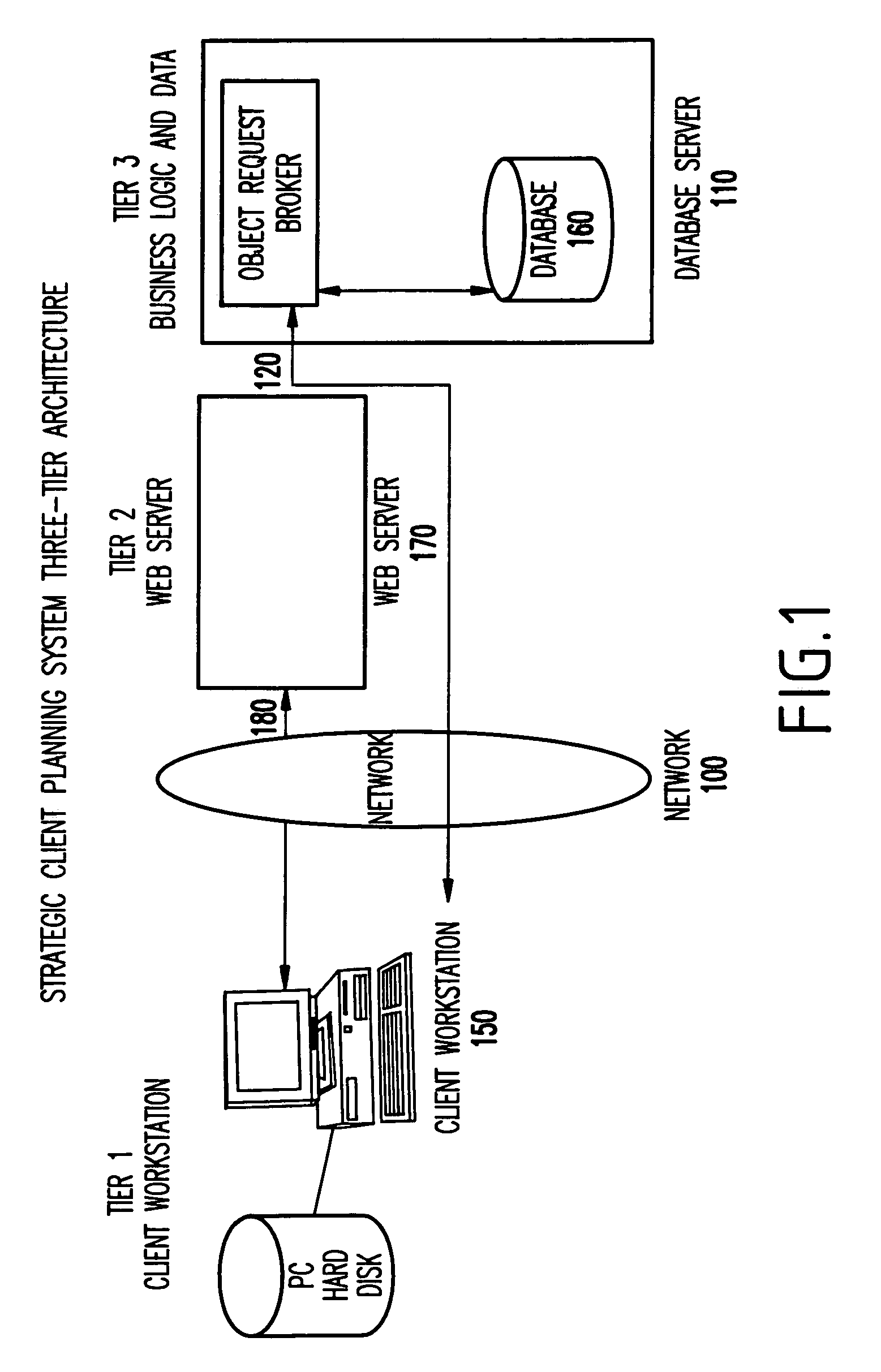
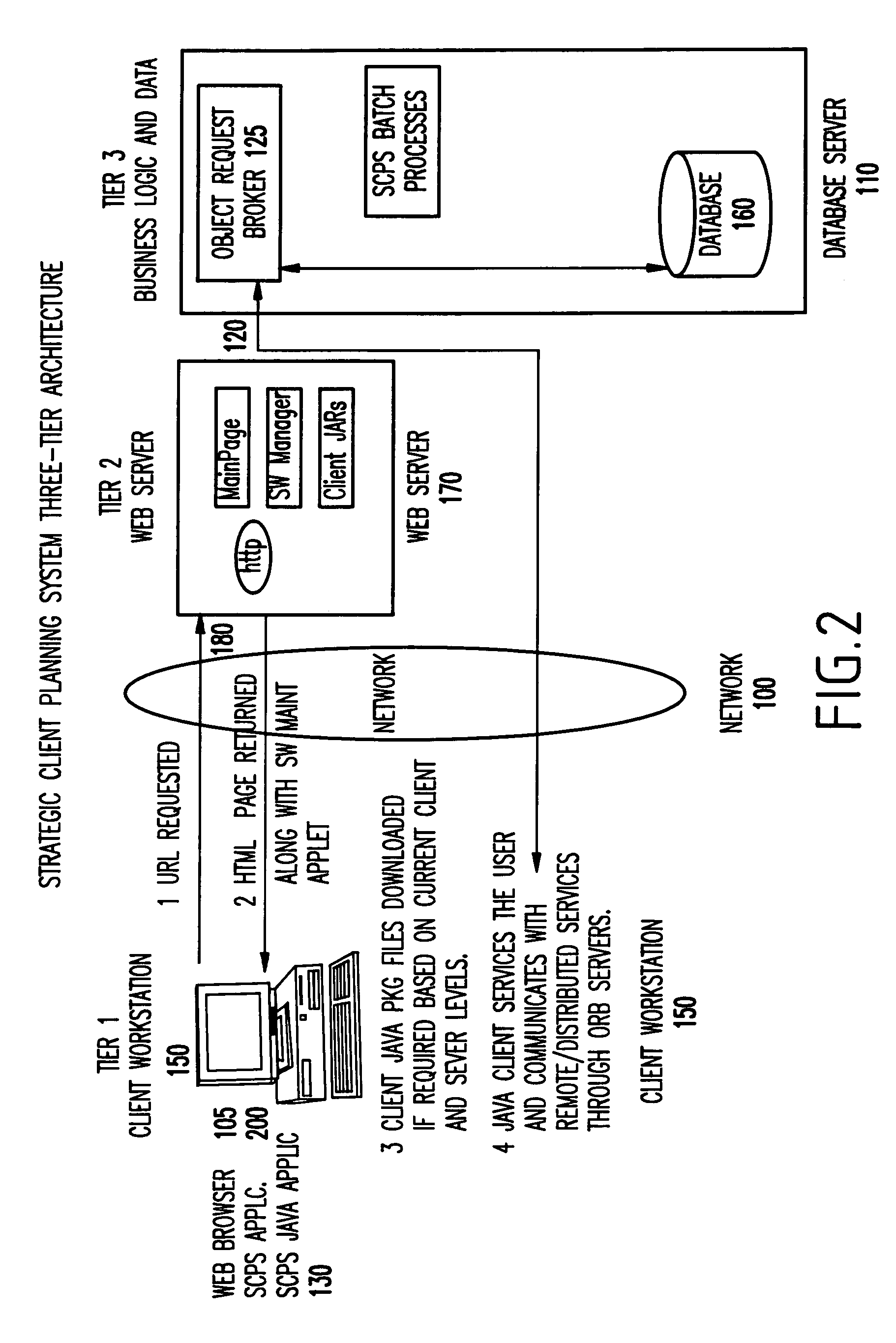
Web-based strategic client planning system for end-user creation of queries, reports and database updates
InactiveUS20030018644A1Easy to useEasy to openData processing applicationsDigital data information retrievalProgram planningClient-side
A method and program storage device for creating tabular data stream flow for sending rows of secure data between a client workstation and a server computer over a network using a common object request broker architecture (CORBA). This method includes receiving a request to create a query form at the client workstation, receiving a worksheet grid form defining selected tabular data, and packaging the worksheet grid form representing an updated status of the data for the tabular data stream flow.
Owner:IBM CORP
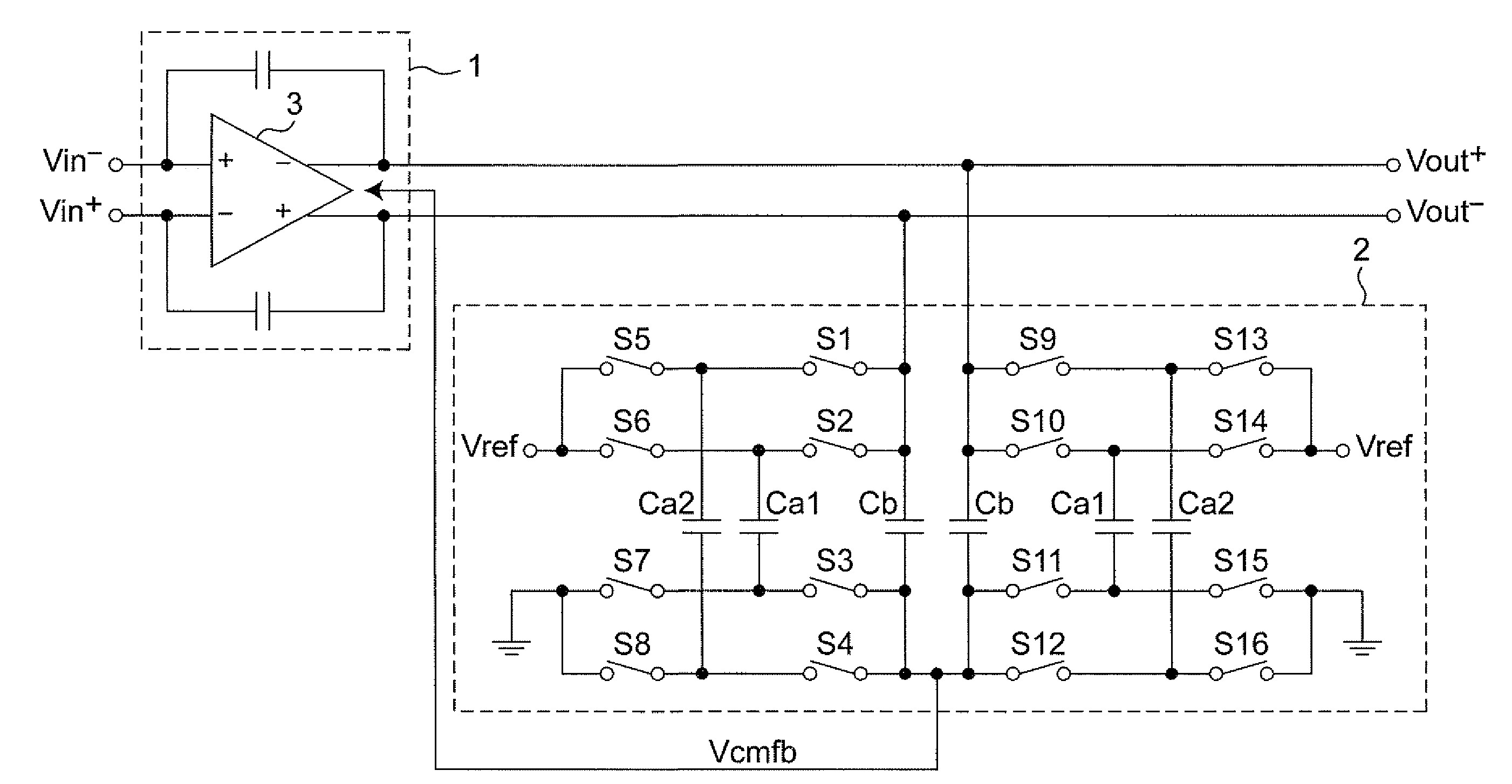
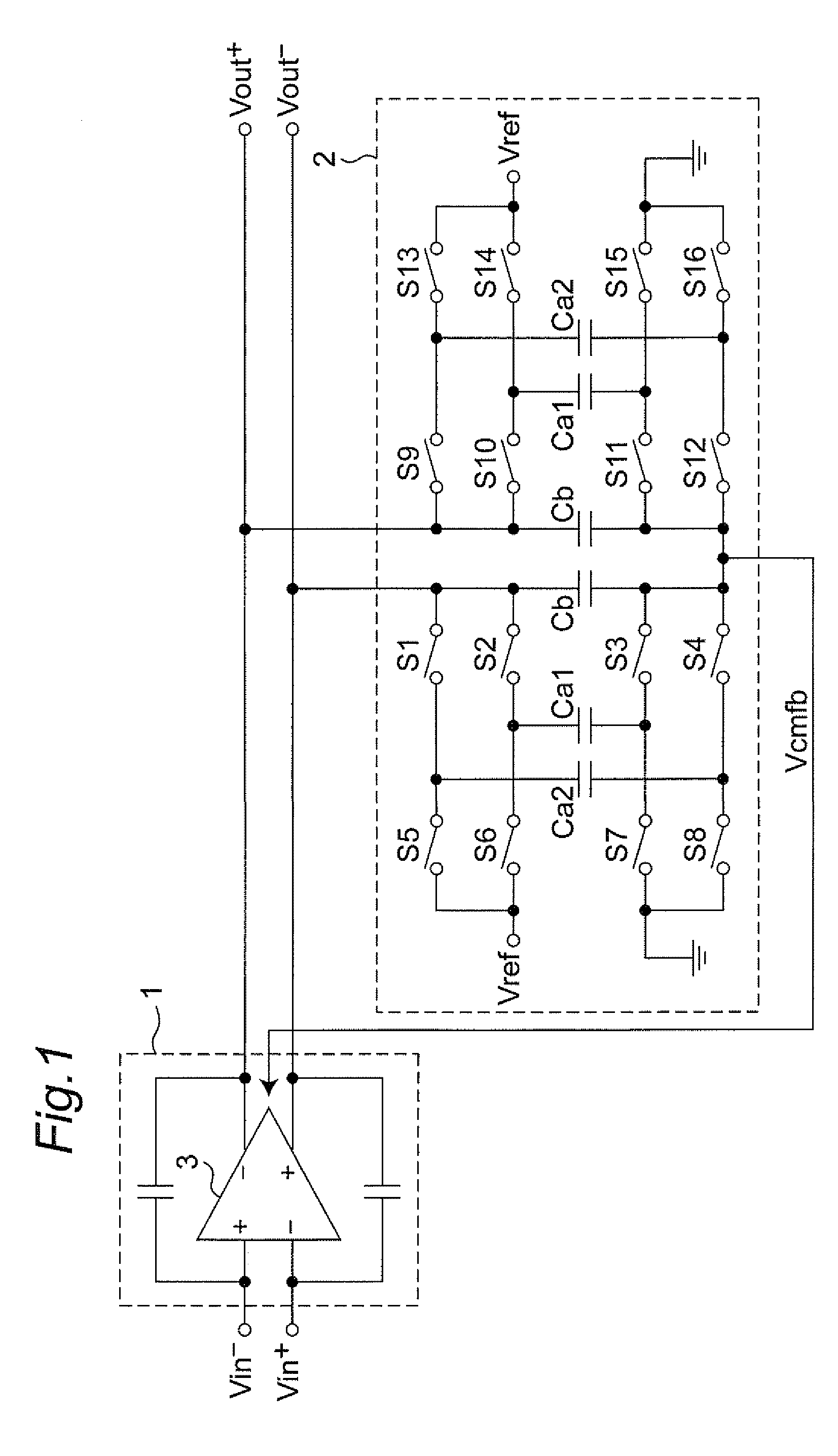
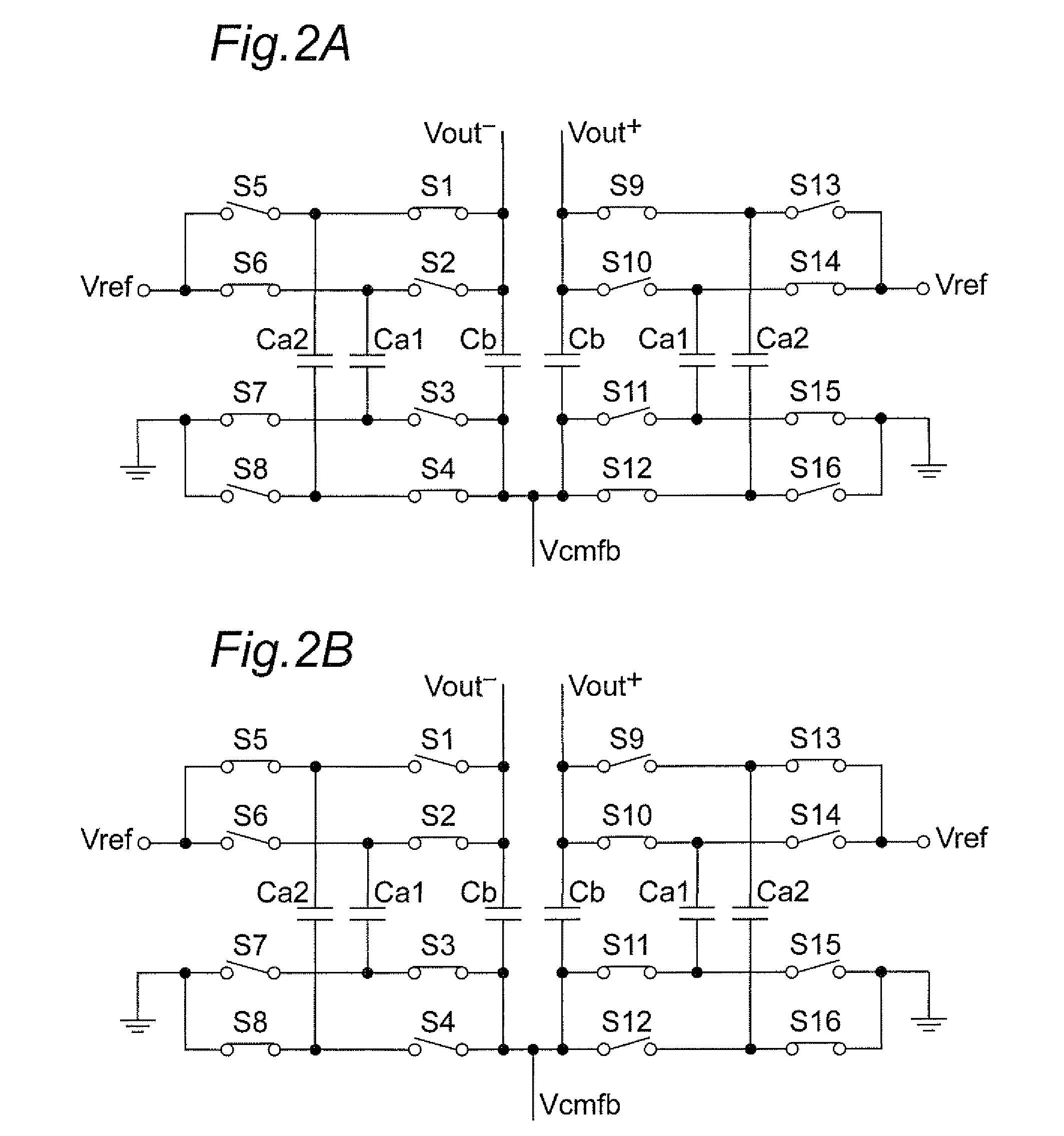
Discrete time amplifier circuit and analog-digital converter
ActiveUS20090115523A1Simple circuit configurationShorten convergence timeElectric signal transmission systemsAnalogue-digital convertersAudio power amplifierA d converter
The present invention is intended to attain simplified circuit configuration and low current consumption in a discrete time amplifier circuit and an AD converter, to improve the convergence from the transient response state to the steady state of the amplifier circuit and to reduce noise and distortion owing to the variation in the output common-mode voltage. The discrete time amplifier circuit and the AD converter are provided with a switched-capacitor common-mode feedback (CMFB) circuit capable of detecting and feeding back the output common-mode voltage at every sampling timing in the case that the circuit operates at double sampling timing (every ½ cycle).
Owner:SOCIONEXT INC
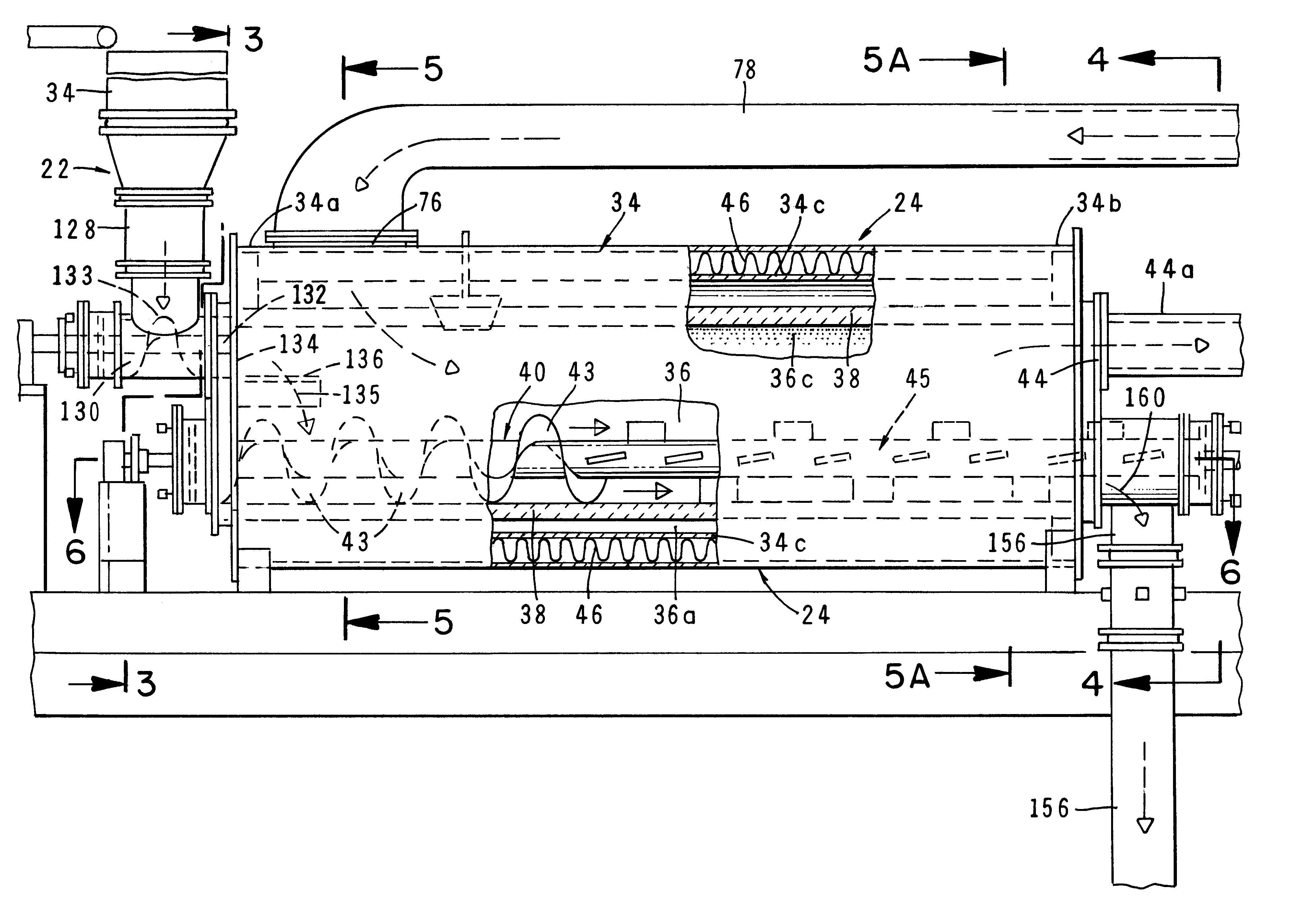
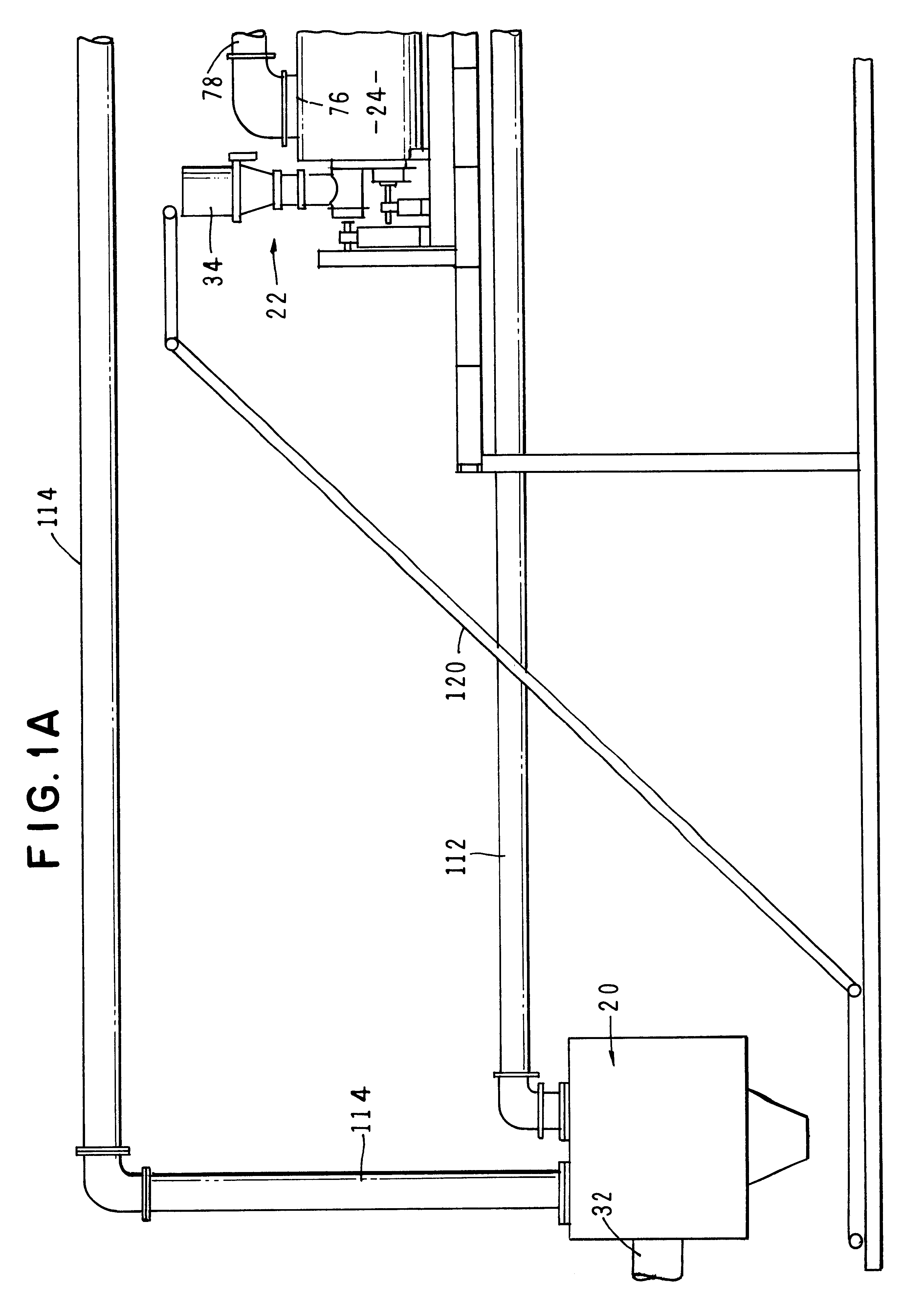
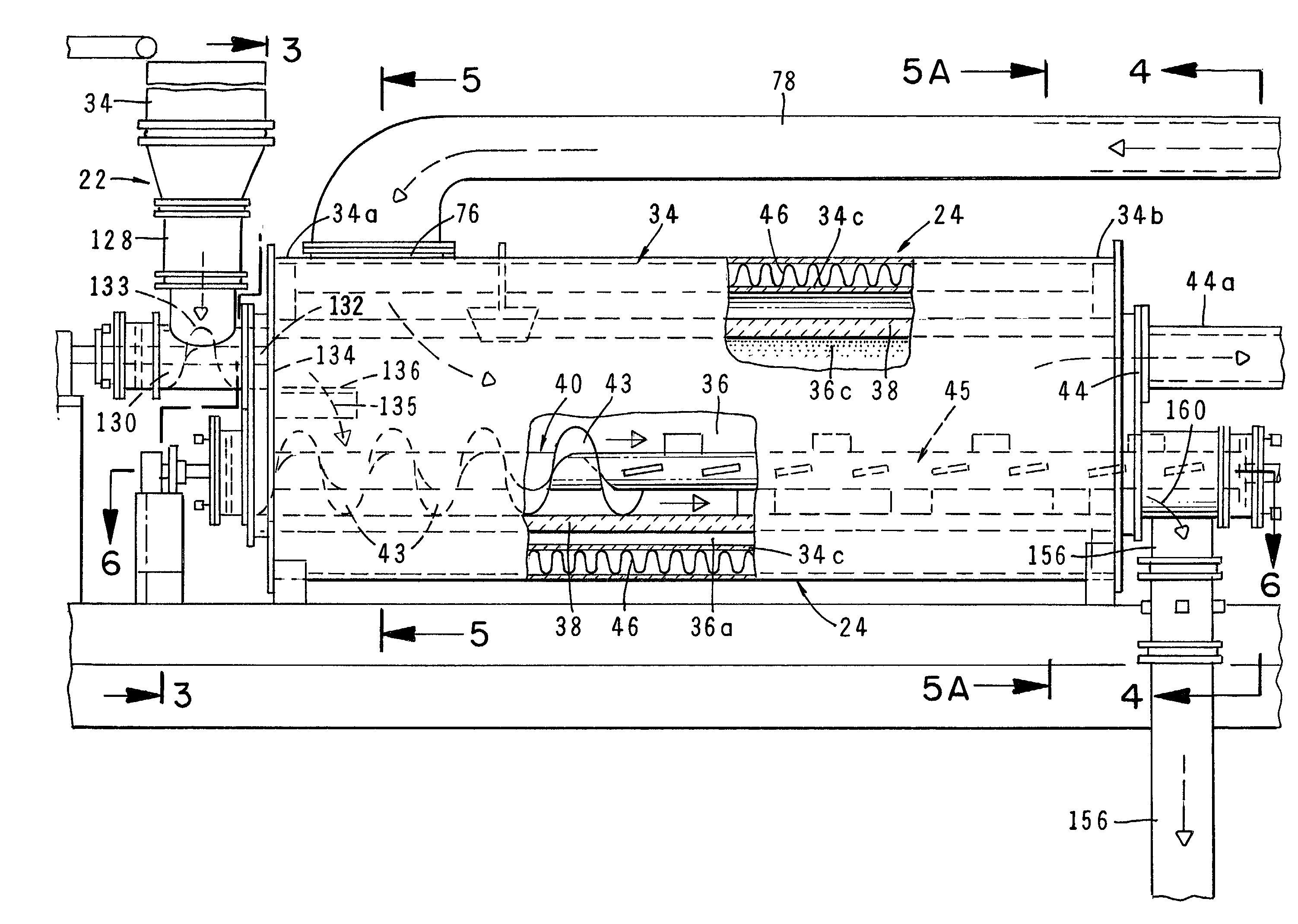
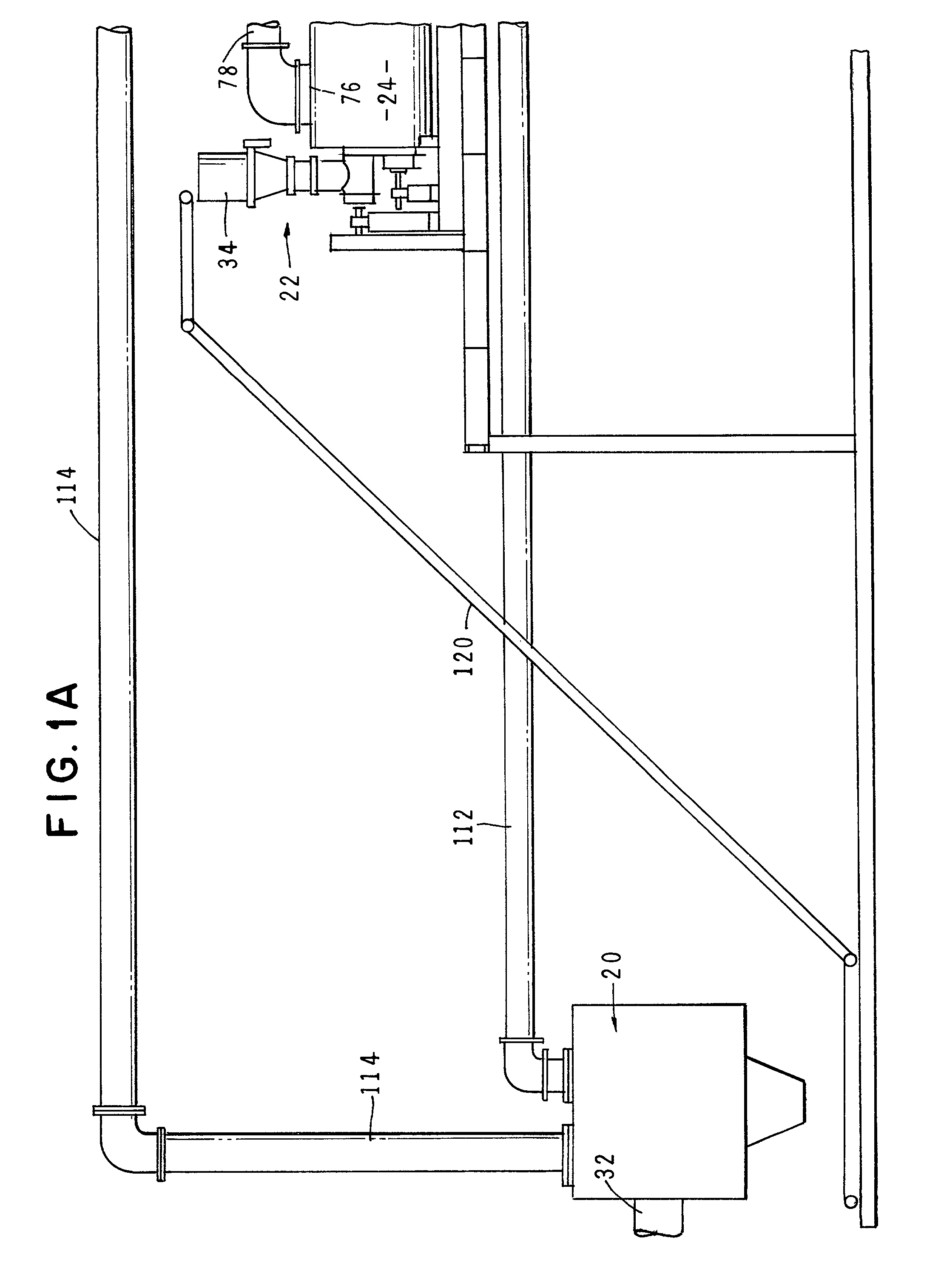
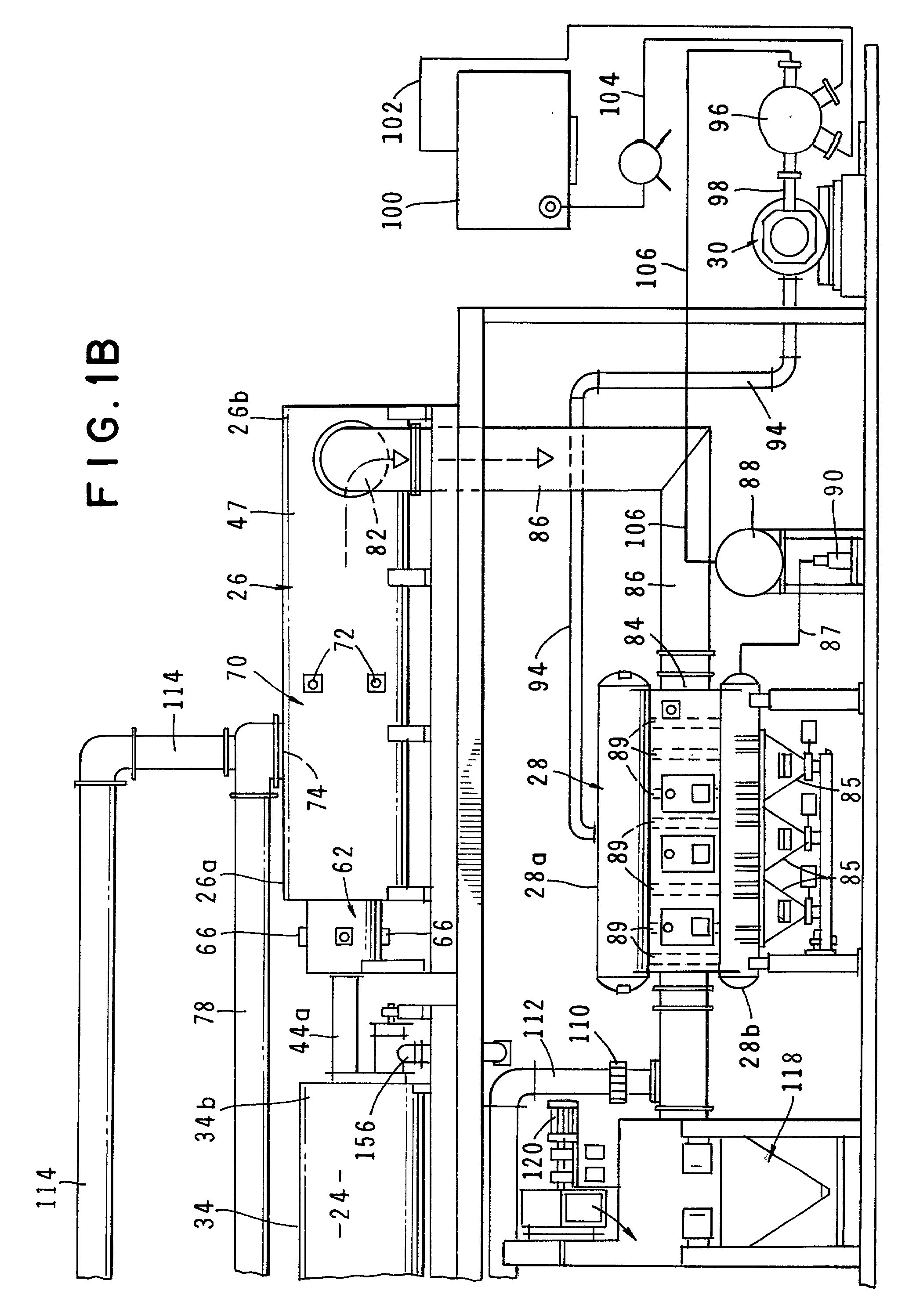
Method and apparatus for treatment of waste
InactiveUS6619214B2Efficient and reliable in operationEasy maintenanceSolid fuel combustionIncinerator apparatusEngineeringTransfer mechanism
An apparatus for treating waste material that comprises four major cooperating subsystems, namely a pyrolytic converter, a two-stage thermal oxidizer, a steam generator and a steam turbine driven by steam generated by the steam generator. In operation, the pyrolytic converter is uniquely heated without any flame impinging on the reactor component and the waste material to be pyrolyzed is transported through the reaction chamber of the pyrolytic converter by a pair of longitudinally extending, side-by-side material transfer mechanisms. Each of the transfer mechanisms includes a first screw conveyor section made up of a plurality of helical flights for conveying the heavier waste and a second paddle conveyor section interconnected with the first section for conveying the partially pyrolyzed waste, the second section comprising a plurality of paddle flights. Once operating, the apparatus is substantially self-sustaining and requires a minimum use of outside energy sources for pyrolyzing the waste materials.
Owner:APS IP HLDG
Calibratable analog-to-digital converter system
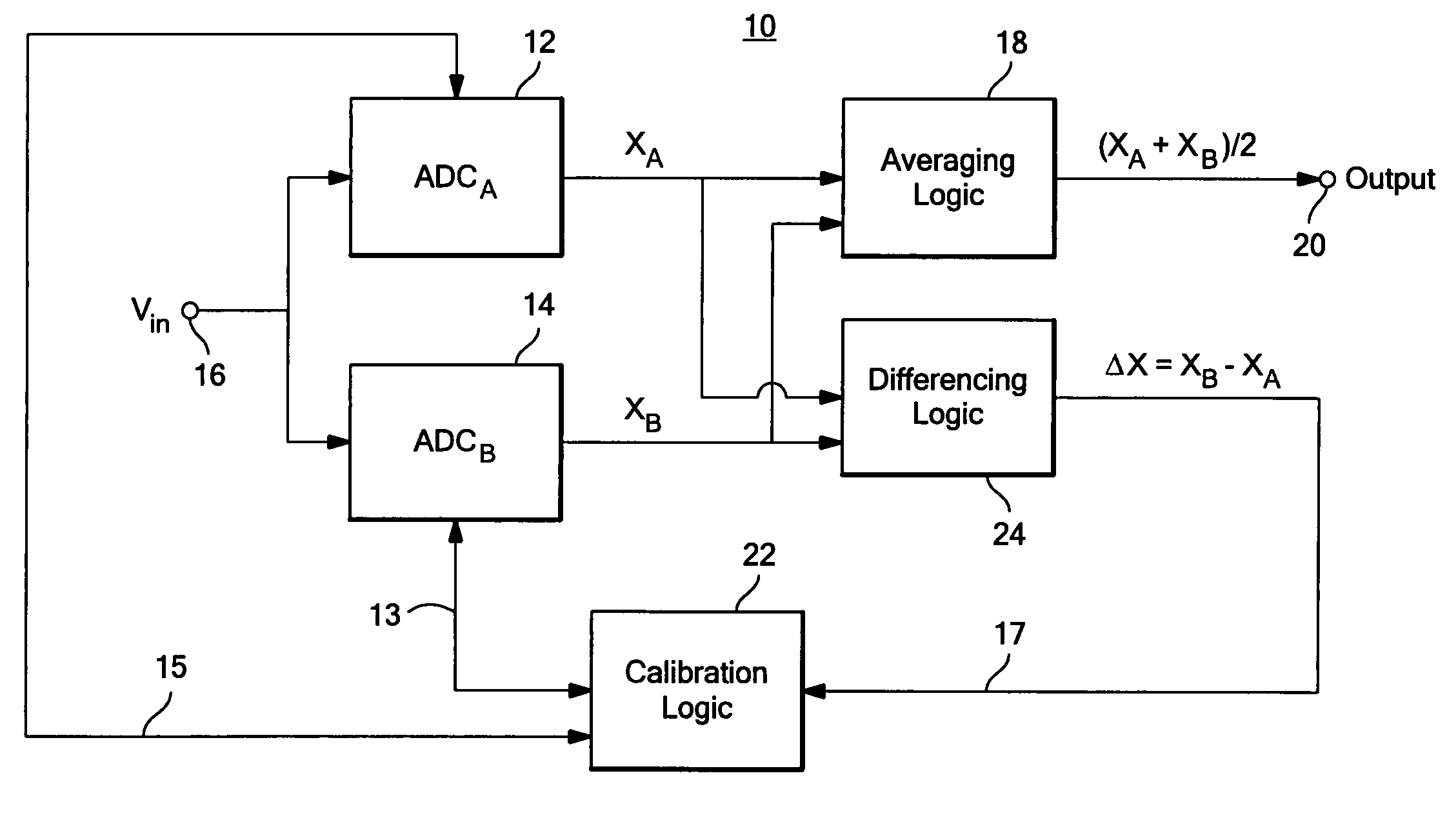
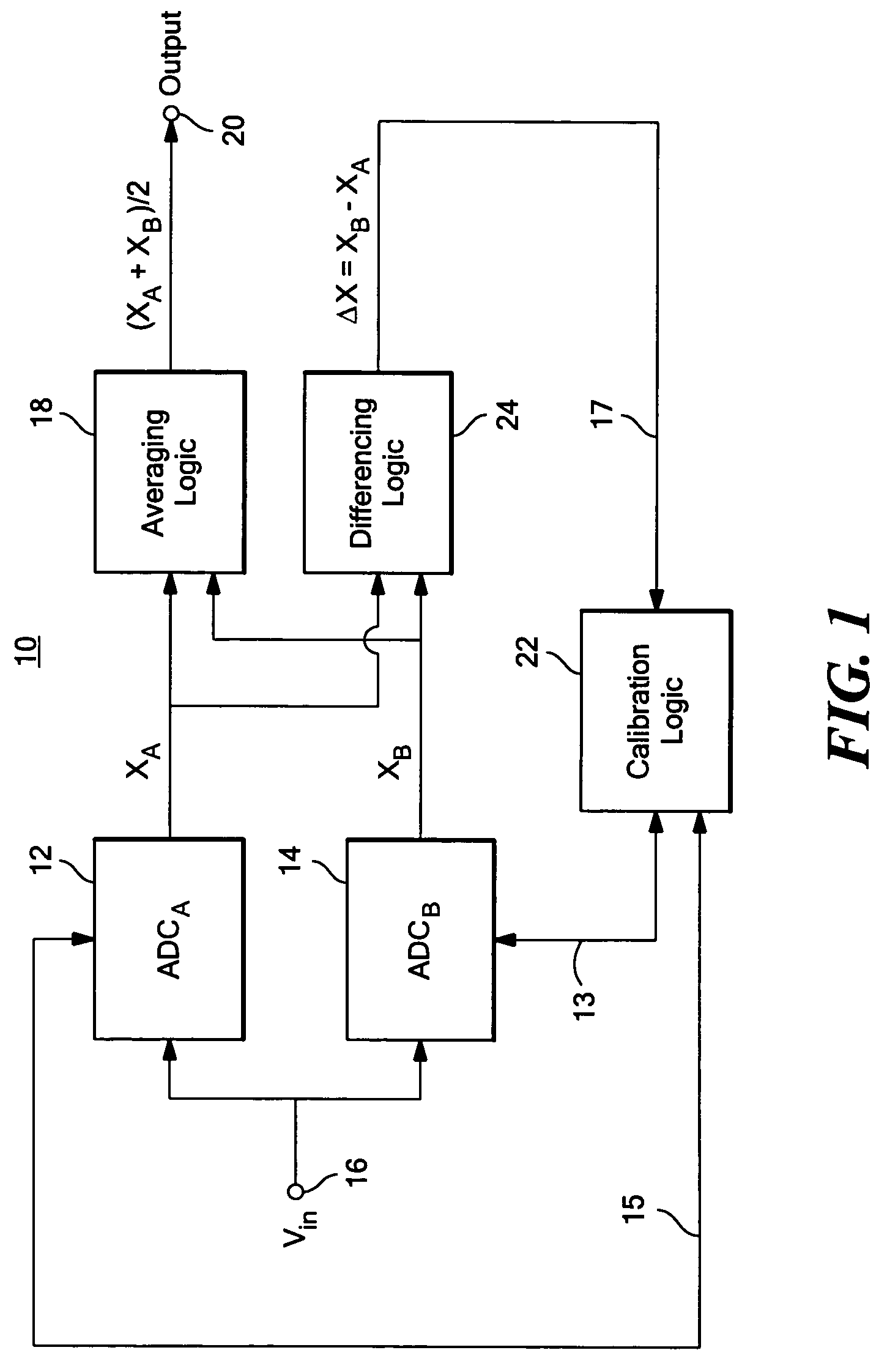
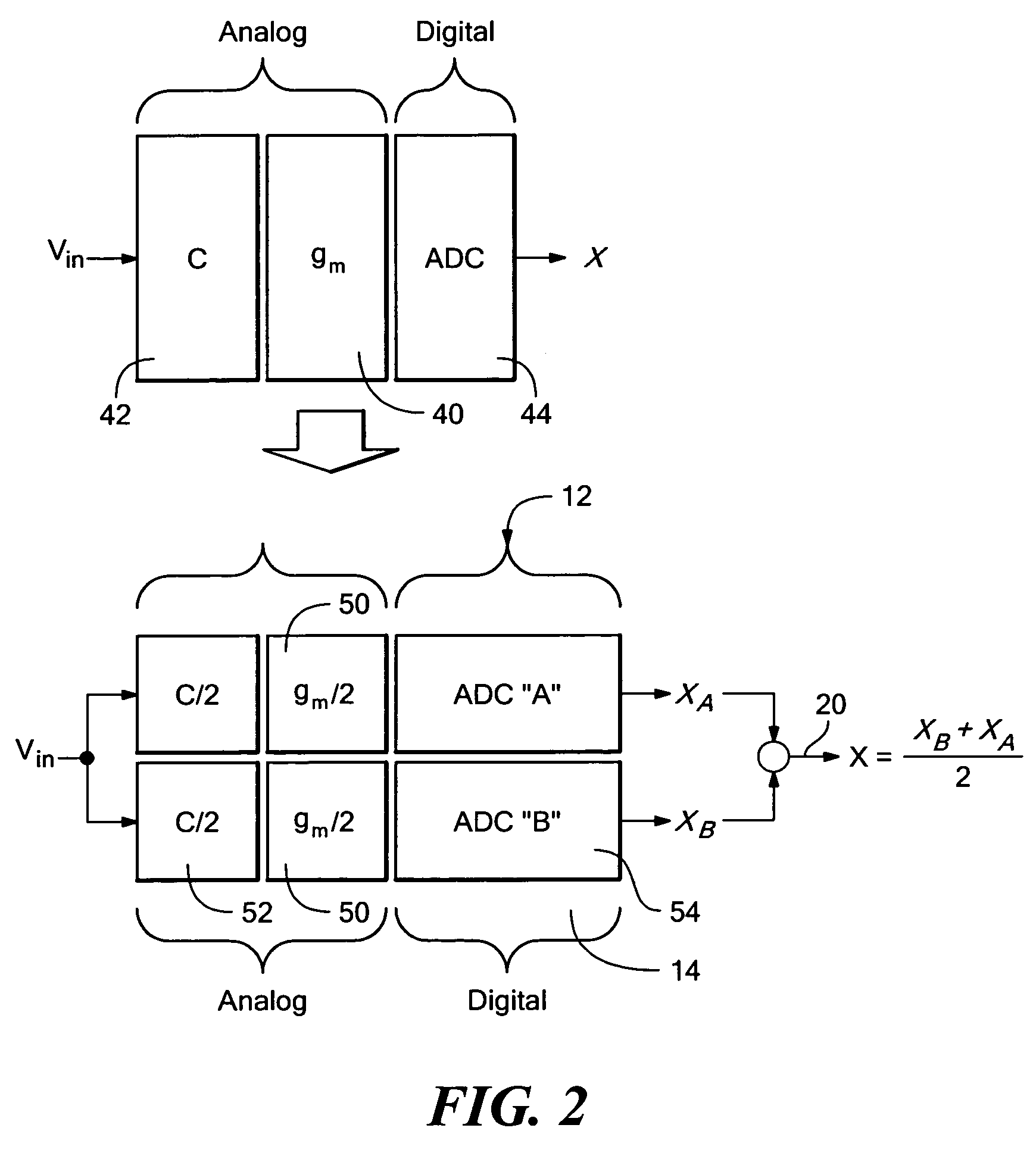
ActiveUS7312734B2Control areaLimited powerElectric signal transmission systemsAnalogue-digital convertersAnalog signalEngineering
A calibratable analog-to-digital converter system with a split analog-to-digital converter architecture including N Analog-to-Digital Converters (ADCs) each configured to convert the same analog input signal into a digital signal. Calibration logic is responsive to the digital signals output by the N ADCs and is configured to calibrate each of the ADCs based on the digital signals output by each ADC.
Owner:ANALOG DEVICES INC
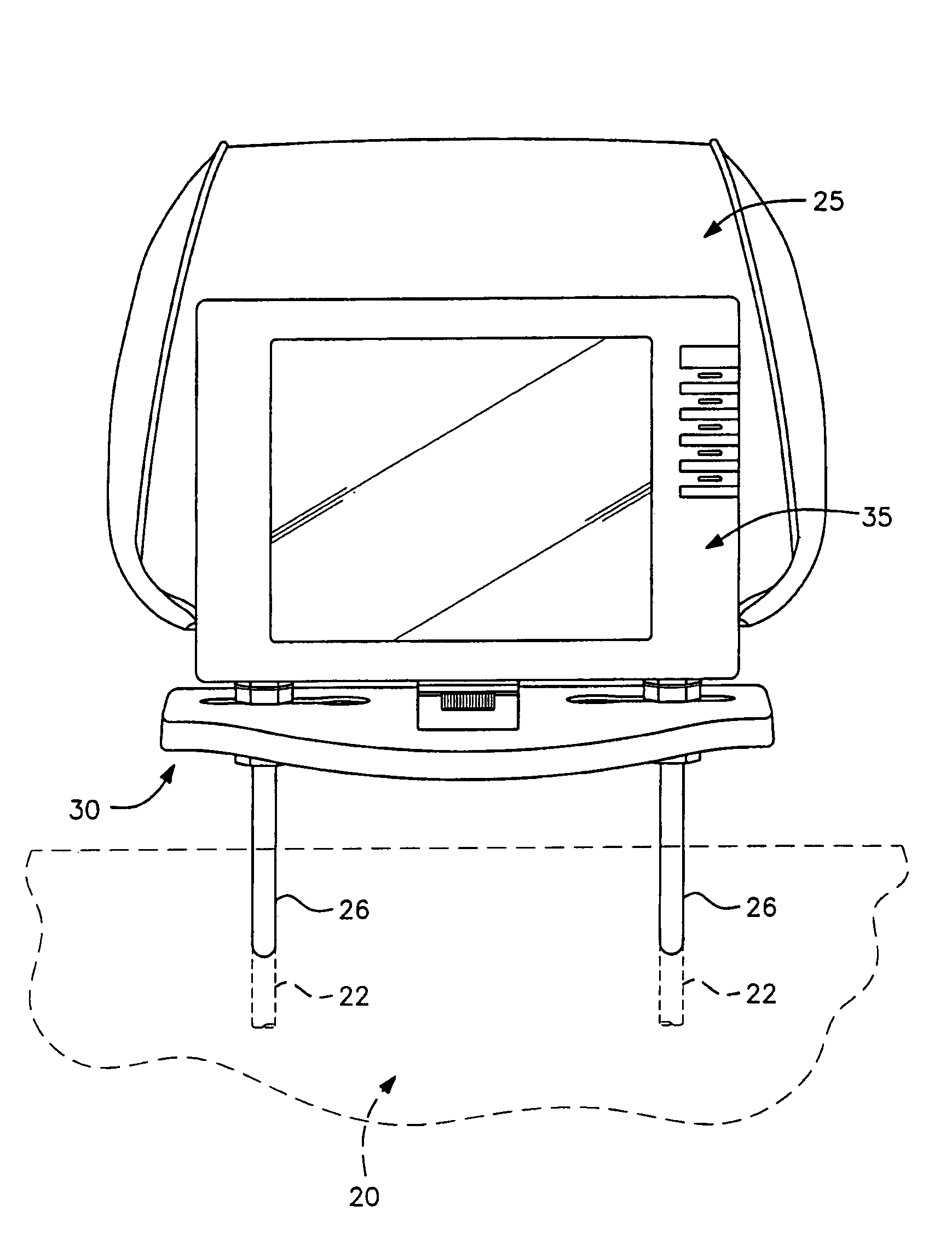
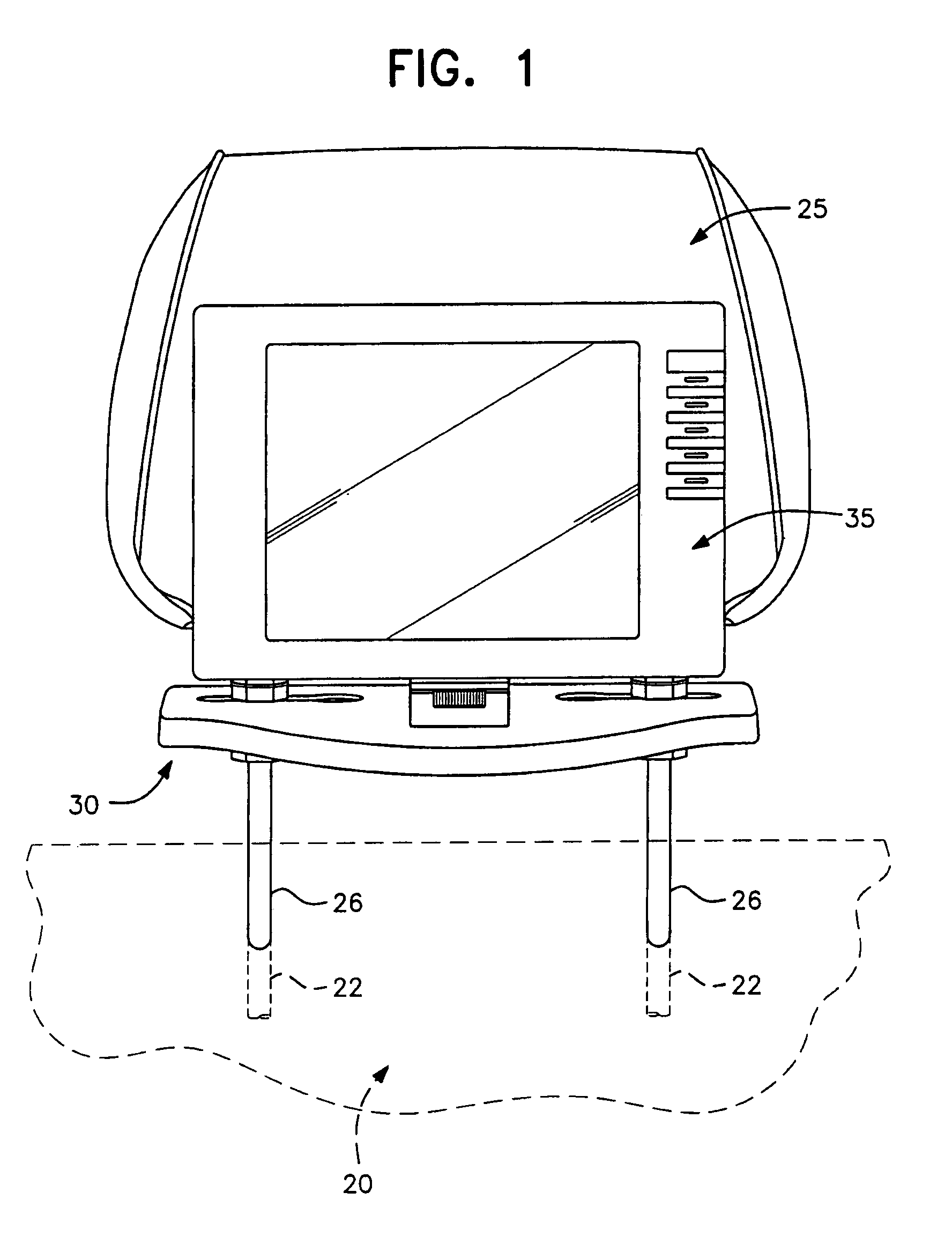
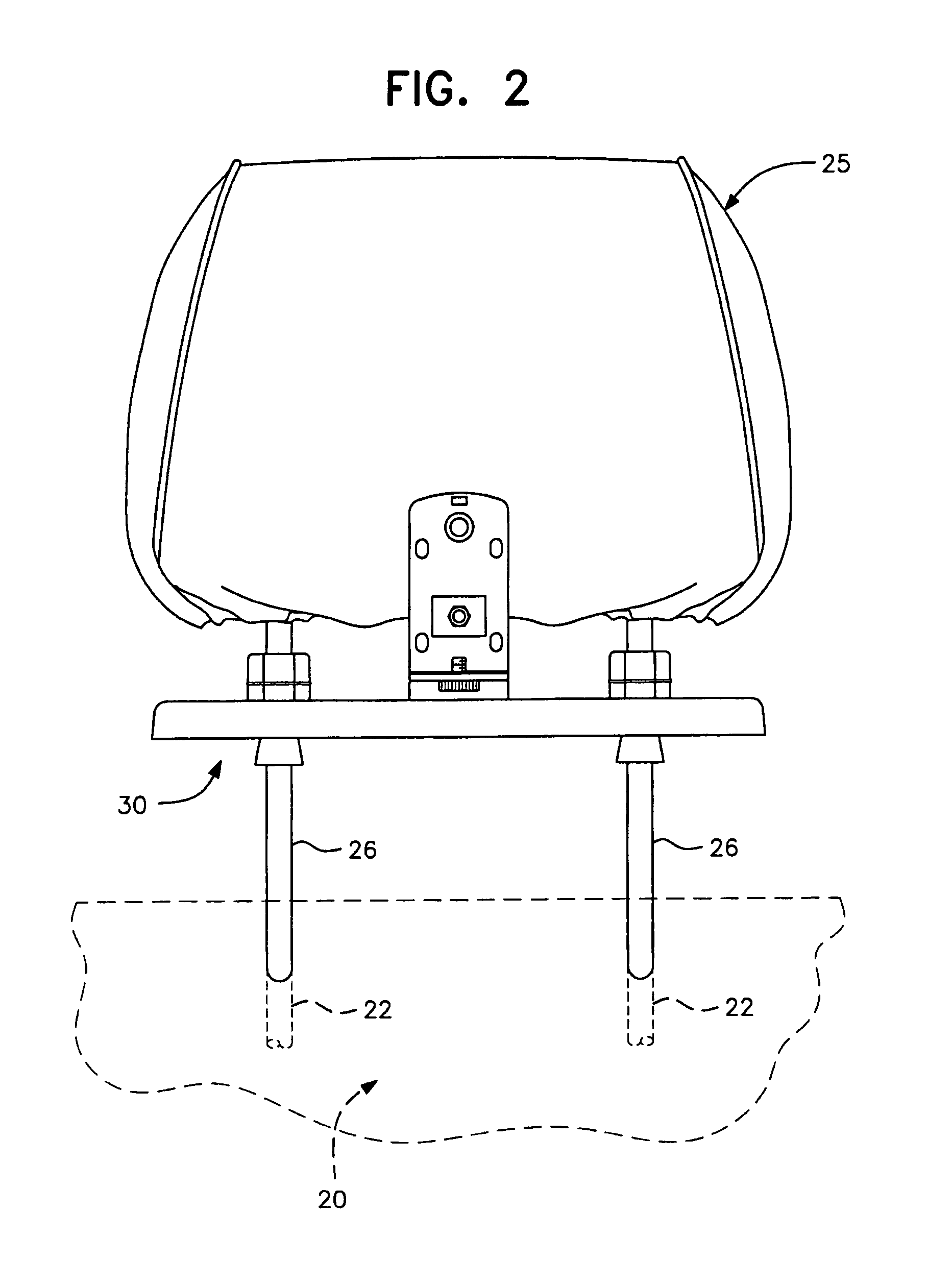
Universal vehicle headrest monitor supporting bracket assembly
InactiveUS7070237B2Easy to installEasy to removeTelevision system detailsOperating chairsMechanical engineeringTroffer
A universal headrest mounting collar which is engaged with the rods of a headrest on a vehicle seat back and supports a video monitor thereon for viewing by a passenger in a rear seat. The bracket can be secured at a fixed vertical position on the headrest rods and collar assemblies surrounding the rods are horizontally slidable in grooves formed in the base of the bracket assembly to accommodate different spacing between headrest rods by different vehicle manufacturers. The monitor mounting element is universally adjustable to modify the viewing angle of the monitor screen.
Owner:EPSILON ELECTRONICS
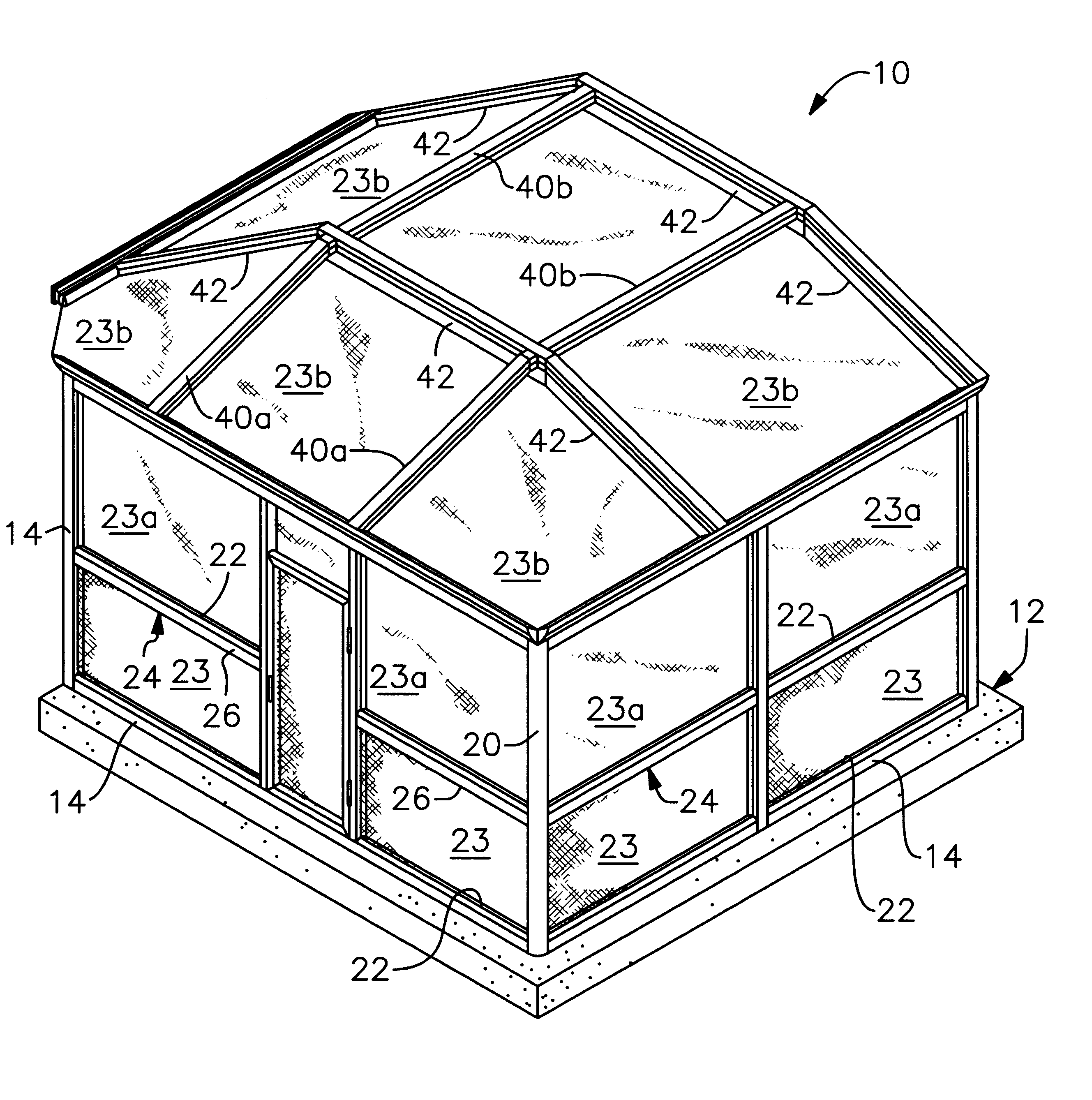
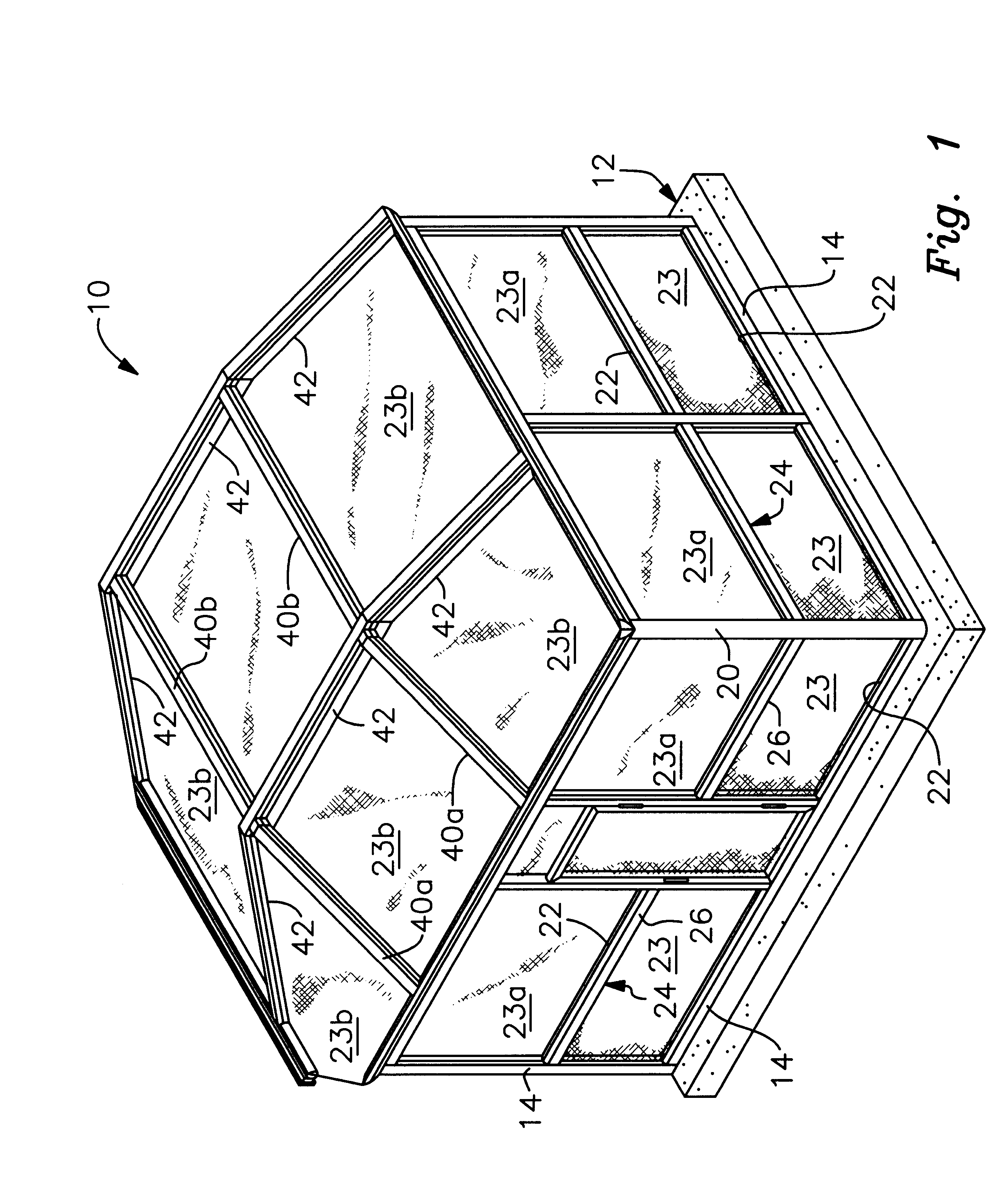
Modular pool enclosure system having aesthetic appeal
InactiveUS6192643B1Easy to assembleHighly versatileRoof covering using slabs/sheetsWallsModularityFastener
An aesthetic pool enclosure of modular design includes cover members that hide from view various utilitarian structural components such as bolts, screws, rubber splines for holding screens, and the like. The structure is made of extruded posts and beams and interlocking members for interlocking the posts and beams to one another to reduce the number of fasteners needed and to provide enhanced strength of the finished assembly. The extruded pieces include corner posts for ninety and sixty degree corners, doorjambs, bottom door sweeps, and other specialized parts for completing the assembly. Many of the parts are snap fit to one another to further reduce the number of fasteners required to assemble the enclosure and to facilitate the construction process.
Owner:ZADOK YIGEL
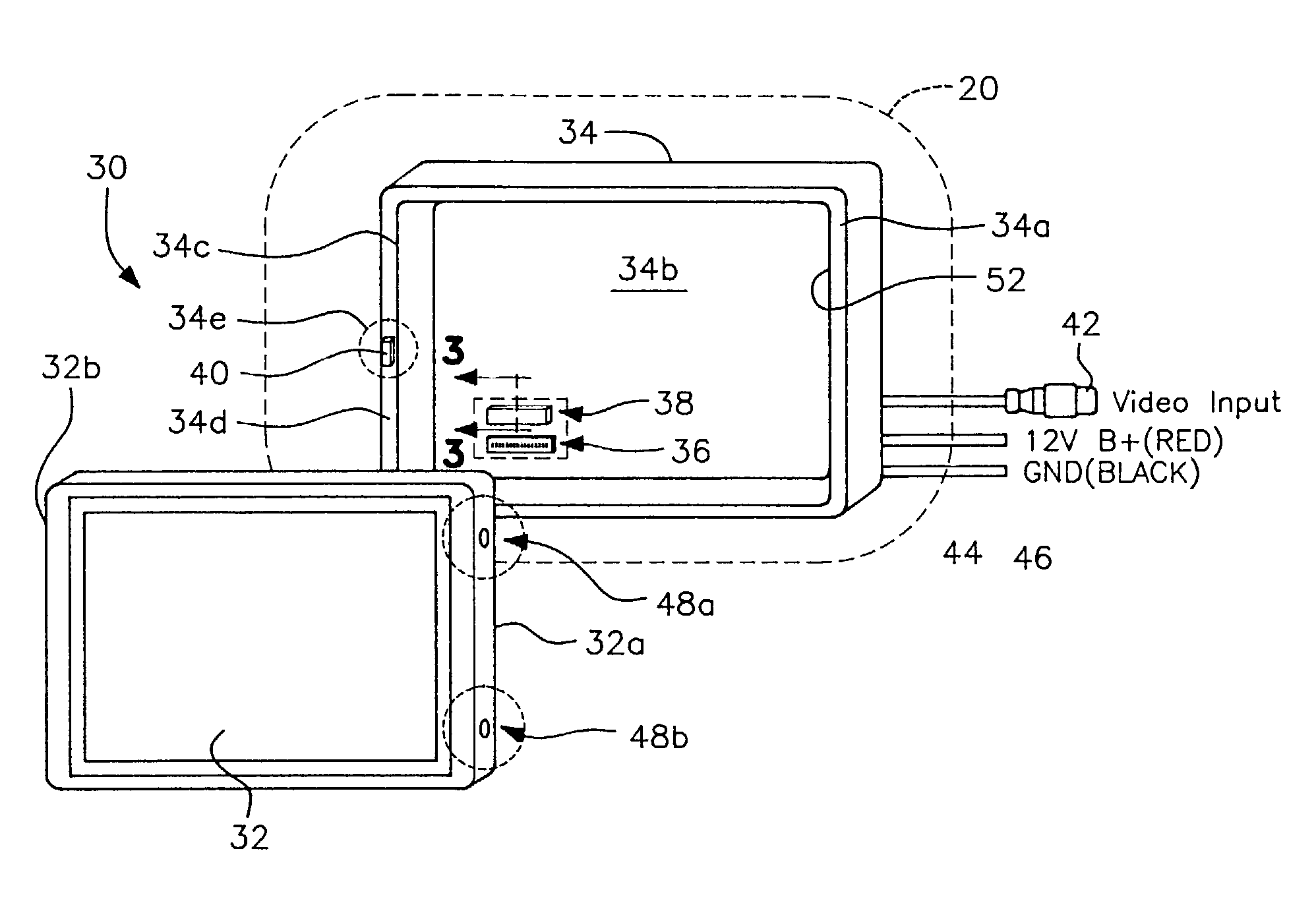
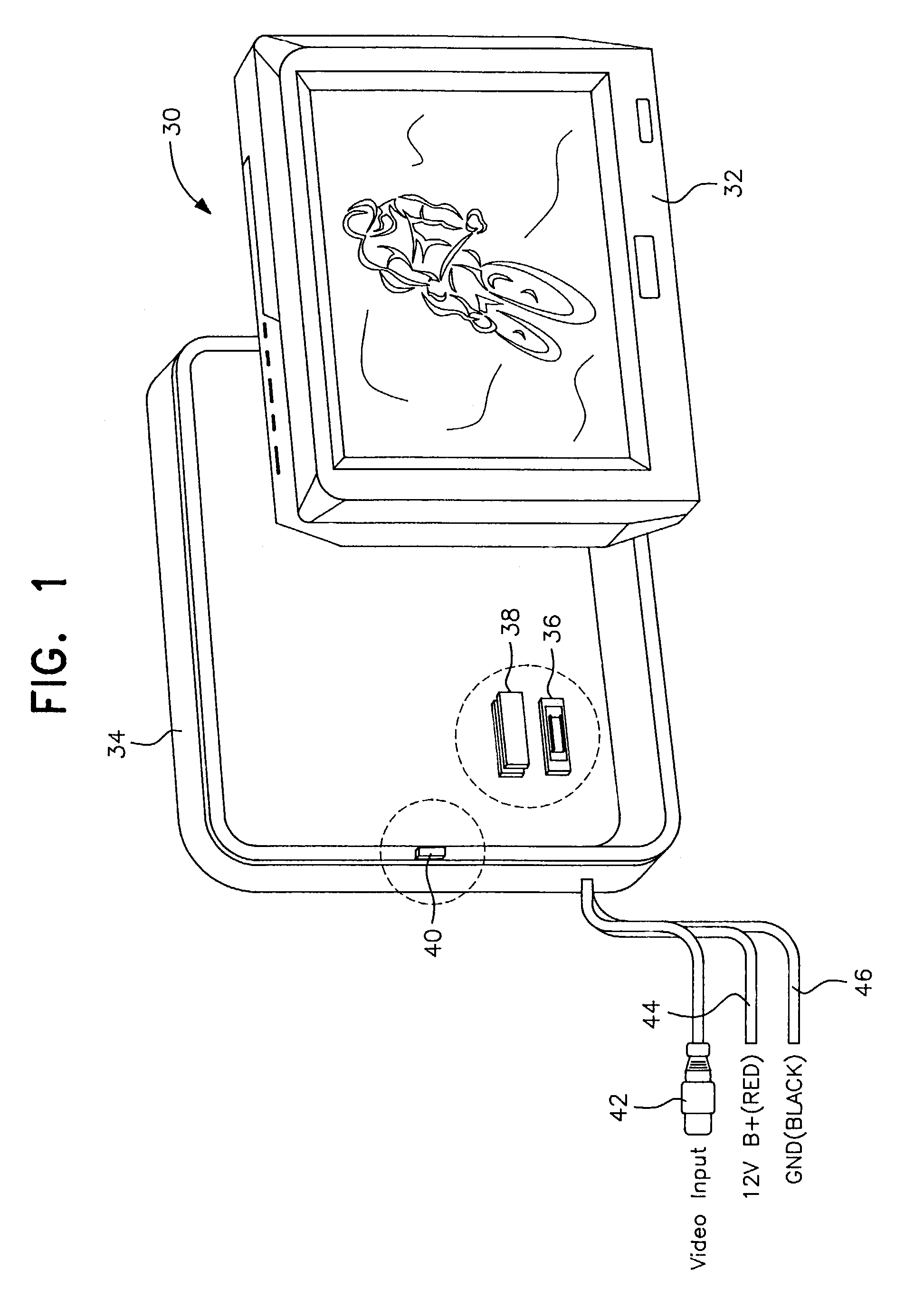
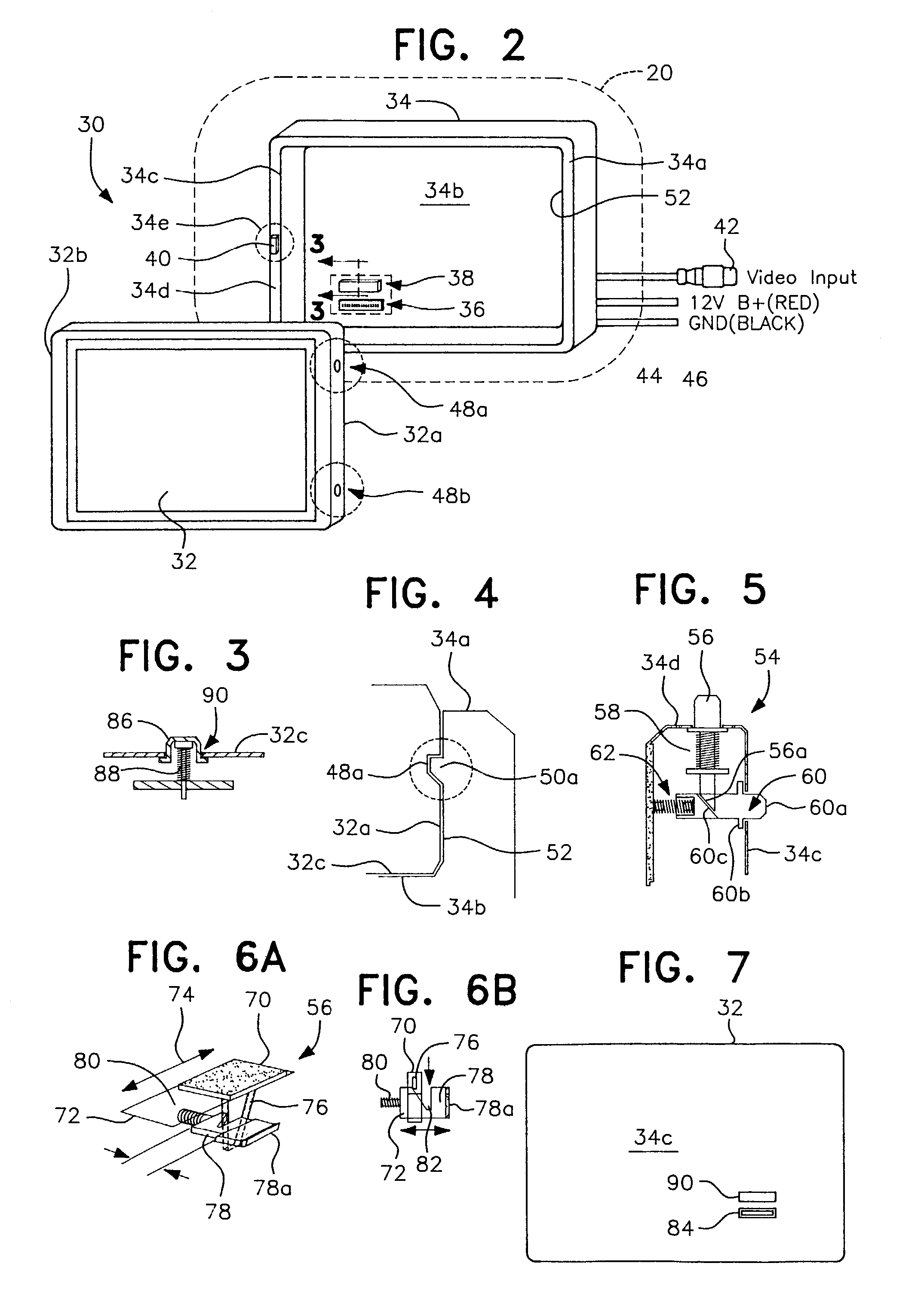
Detachable vehicle monitor
InactiveUS7019794B2Facilitate quick disconnectionEasy reconnectionTelevision system detailsMachine supportsEngineeringAssembly design
Owner:EPSILON ELECTRONICS
Method and apparatus for treatment of waste
InactiveUS20020195031A1Efficient and reliable in operationEasy maintenanceSolid fuel combustionIncinerator apparatusEngineeringTransfer mechanism
An apparatus for treating waste material that comprises four major cooperating subsystems, namely a pyrolytic converter, a two-stage thermal oxidizer, a steam generator and a steam turbine driven by steam generated by the steam generator. In operation, the pyrolytic converter is uniquely heated without any flame impinging on the reactor component and the waste material to be pyrolyzed is transported through the reaction chamber of the pyrolytic converter by a pair of longitudinally extending, side-by-side material transfer mechanisms. Each of the transfer mechanisms includes a first screw conveyor section made up of a plurality of helical flights for conveying the heavier waste and a second paddle conveyor section interconnected with the first section for conveying the partially pyrolyzed waste, the second section comprising a plurality of paddle flights. Once operating, the apparatus is substantially self-sustaining and requires a minimum use of outside energy sources for pyrolyzing the waste materials.
Owner:APS IP HLDG
Light-Emitting Element, Light-Emitting Device, Electronic Device, and Lighting Device
InactiveUS20100123152A1Manufacturing cost be reduceEmission efficiency be improveElectroluminescent light sourcesSolid-state devicesTitaniumTitanium oxide
Provided is a light-emitting element including an anode over a substrate, a layer containing a composite material in which a metal oxide is added to an organic compound, a light-emitting layer, and a cathode having a light-transmitting property. The anode is a stack of a film of an aluminum alloy and a film containing titanium or titanium oxide. The film containing titanium or titanium oxide is in contact with the layer containing a composite material.
Owner:SEMICON ENERGY LAB CO LTD
Systems and methods for testing the performance of and simulating a wireless communication device
InactiveUS20070127559A1Exact reproductionMaintain optimum performancePolarisation/directional diversityTransmission monitoringControl signalData file
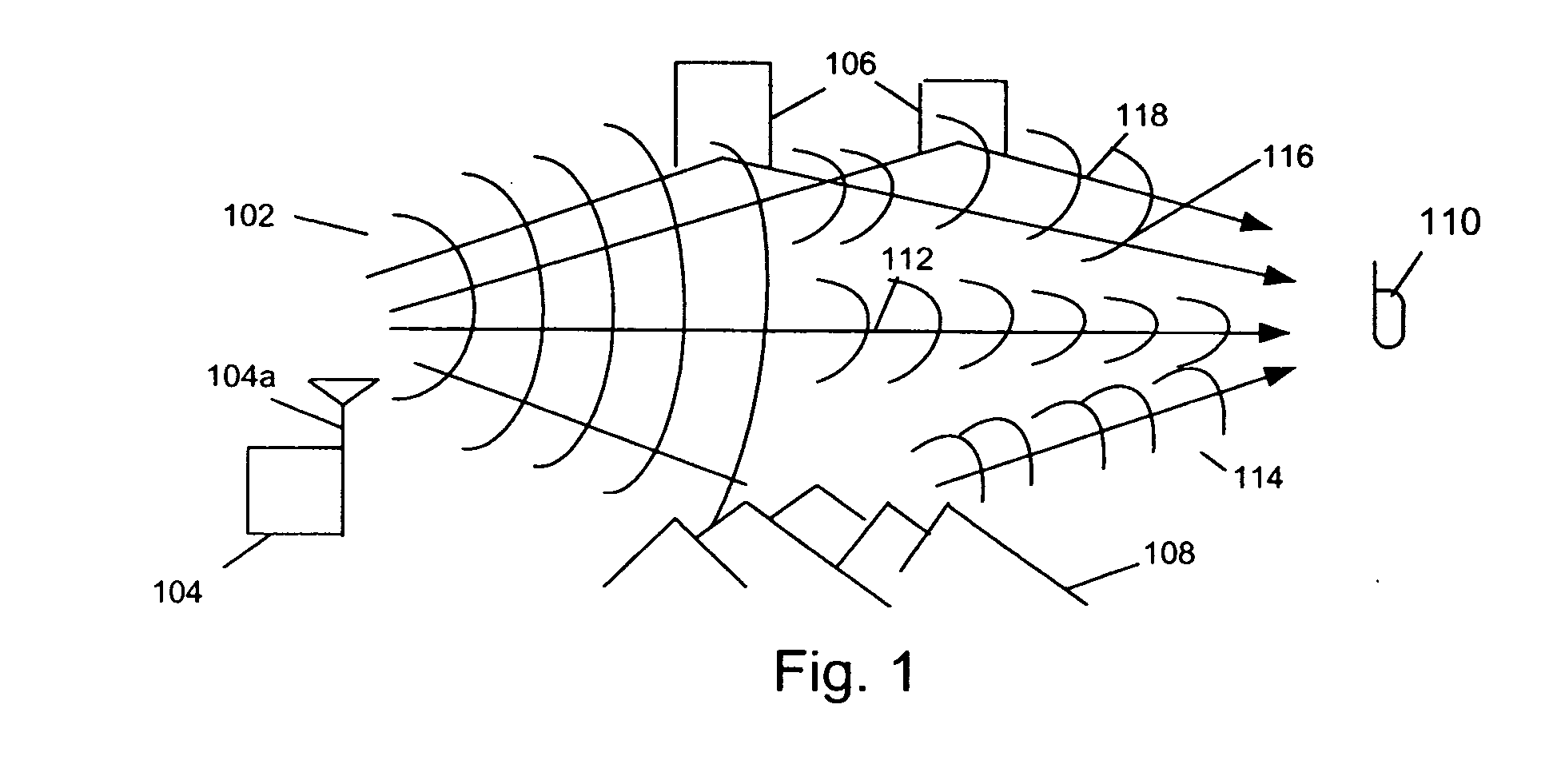
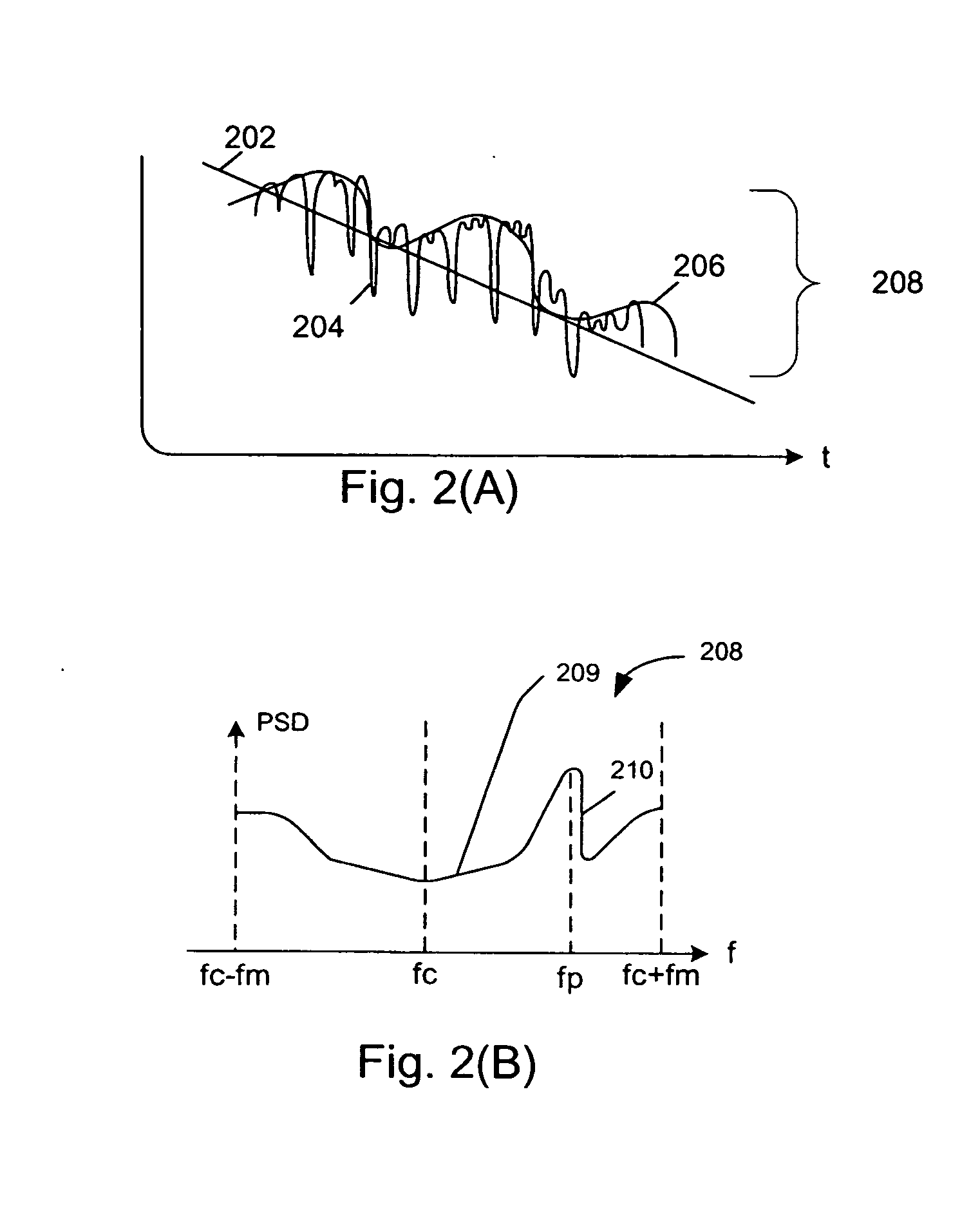
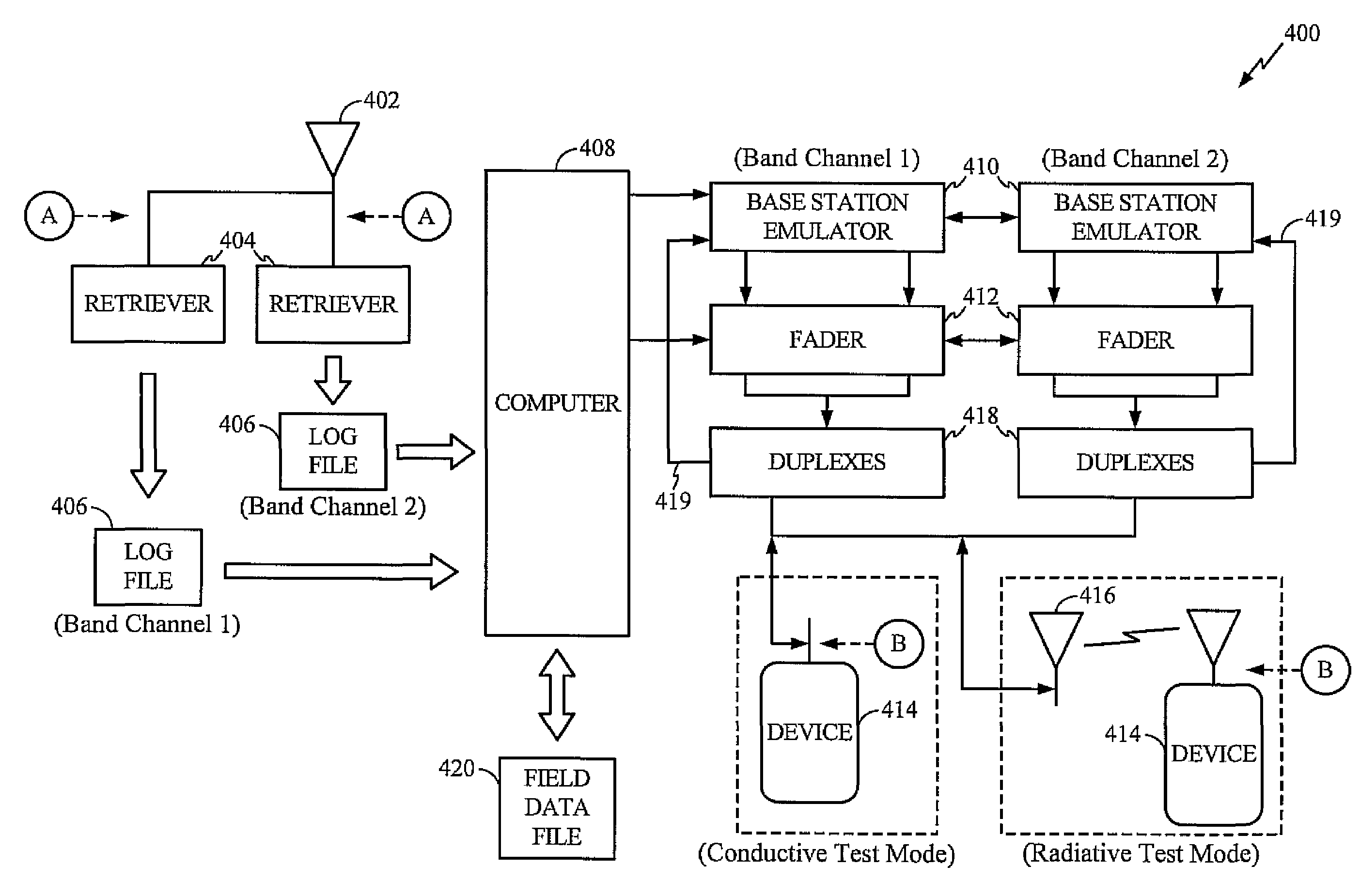
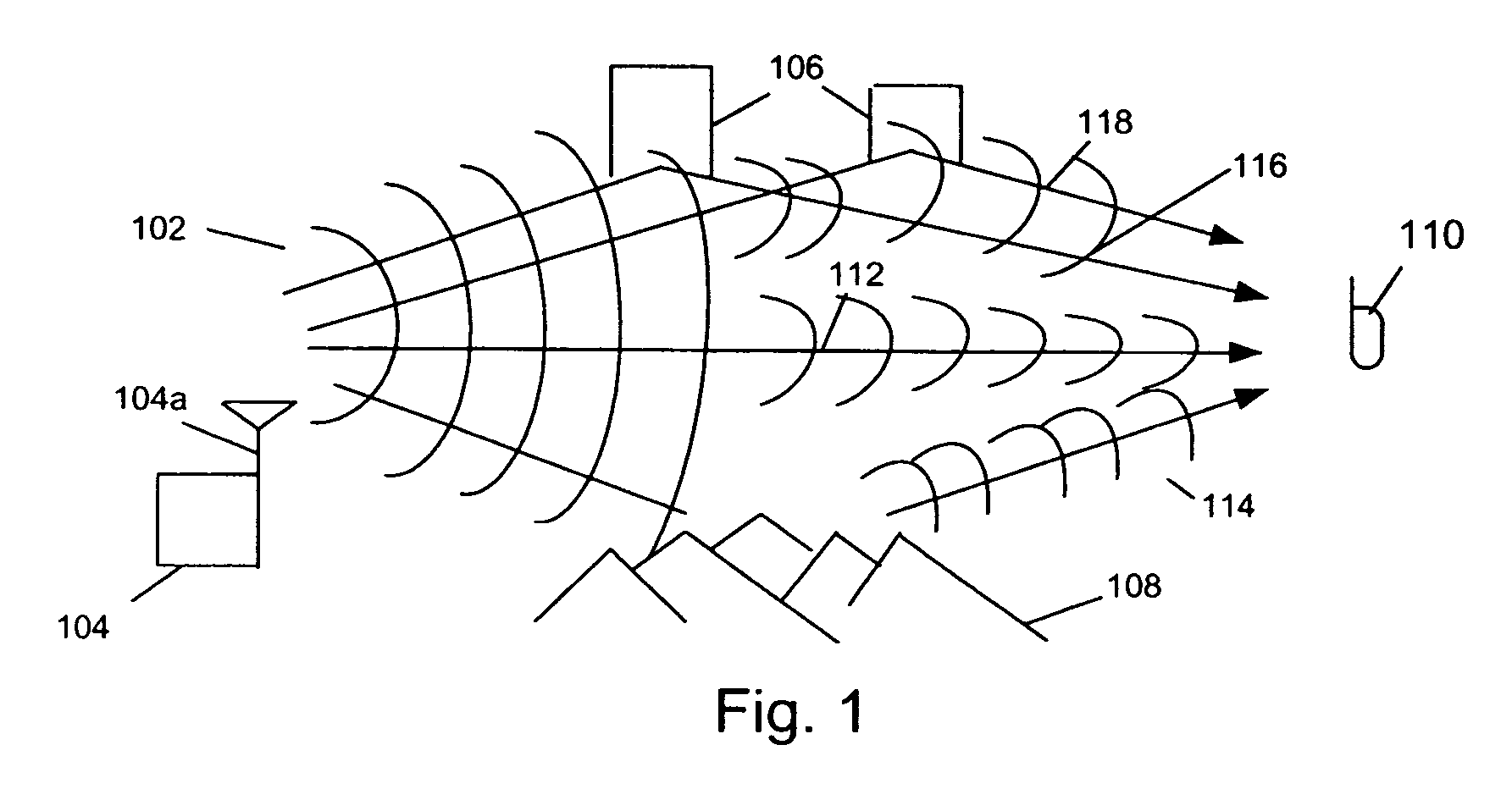
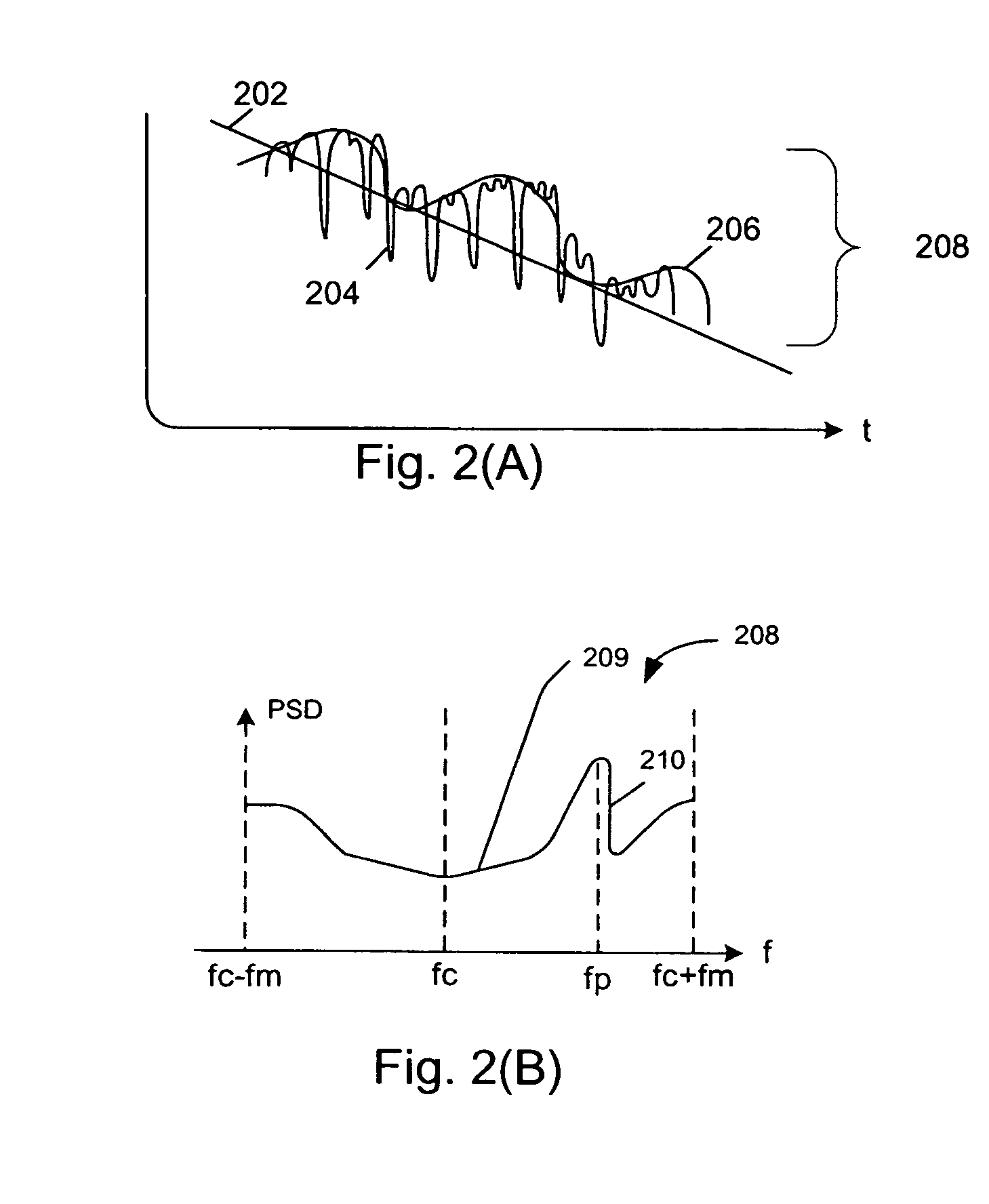
A system abstracts channel information from field data gathered in actual wireless communication system environments. The abstracted data is then transformed into control signals or programming codes that can be used to control channel simulators so as to recreate the field conditions, including path loss, slow fading, fast fading, path delay, fading power spectral density with and without line-of-sight (LOS), and different kind of handoff scenarios, such as soft, softer, intra-band hard, inter-band hard handoffs. The system thus can accurately simulate a realistic wireless communication link originated from multiple signal sources in different band channels and formed by multipath signal propagation. The simulated realistic wireless communication link can be condensed by selecting the most useful scenarios from its original field data files or modified by tuning its parameters as desired.
Owner:INTERDIGITAL PATENT HLDG INC
Information processing device, control device, communication device, communication equipment, electronic device, information processing system, power management method, power management program, and recording medium
InactiveUS20050048960A1Low power operationSave powerPower managementEnergy efficient ICTInformation processingControl communications
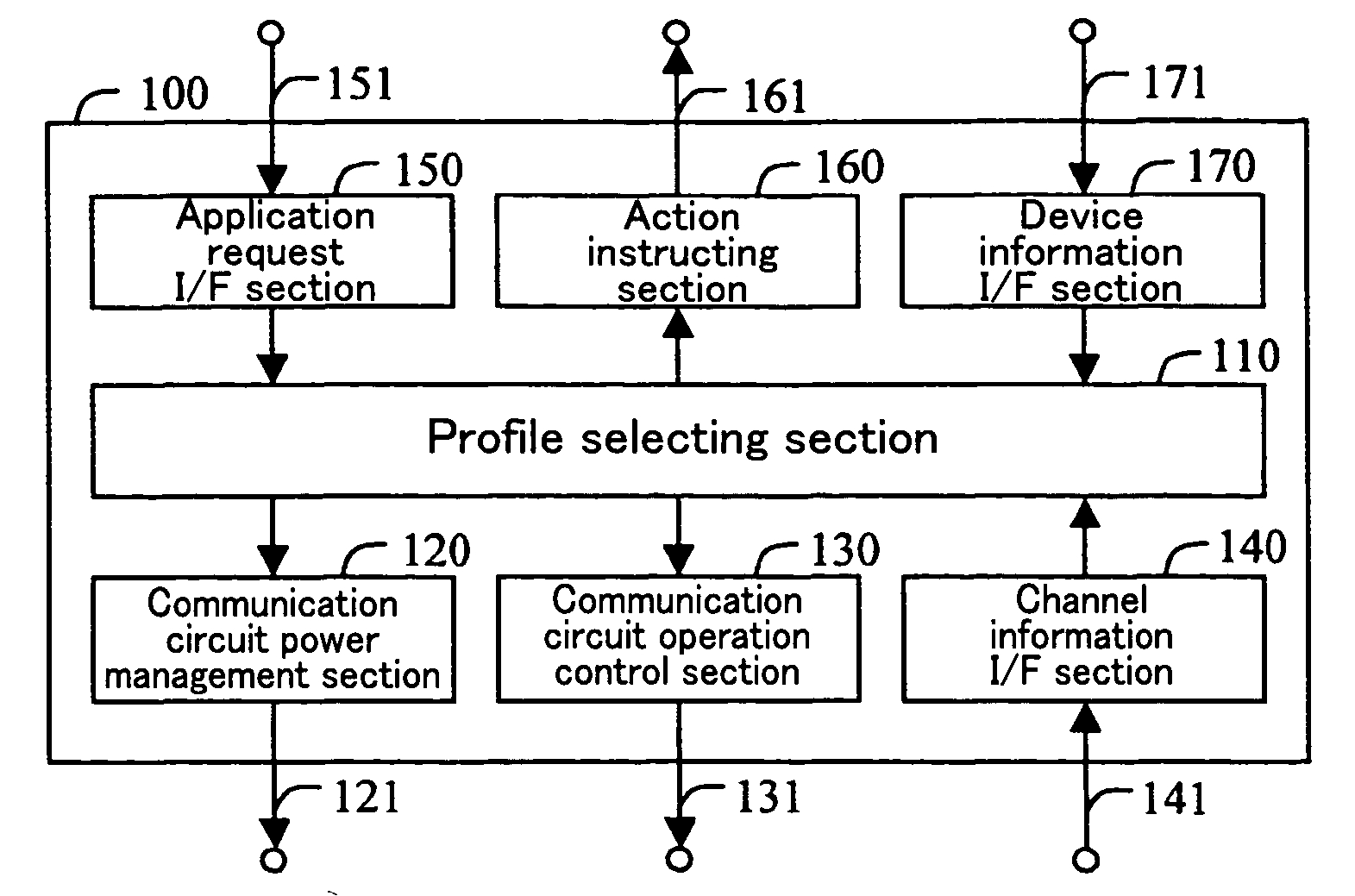
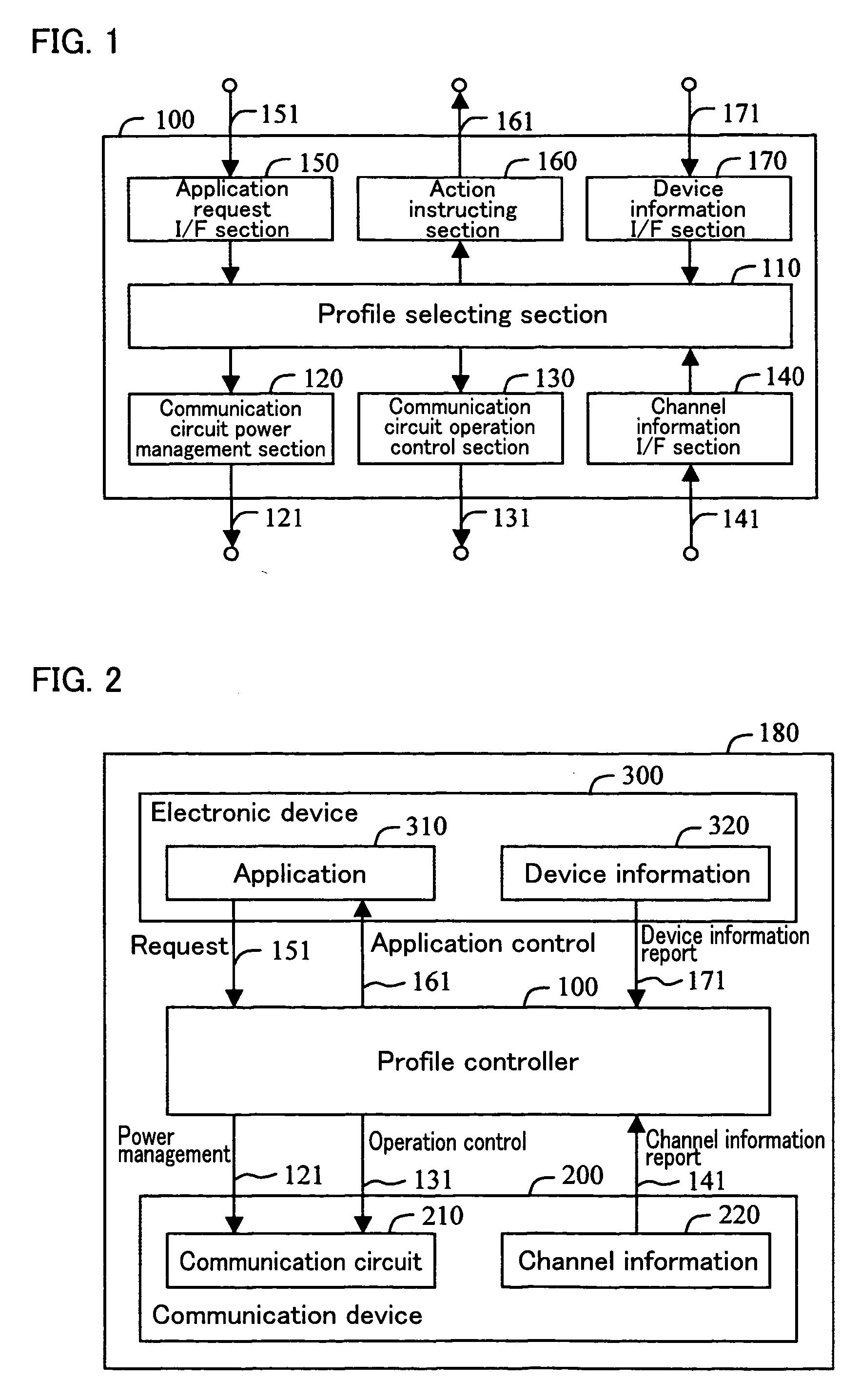
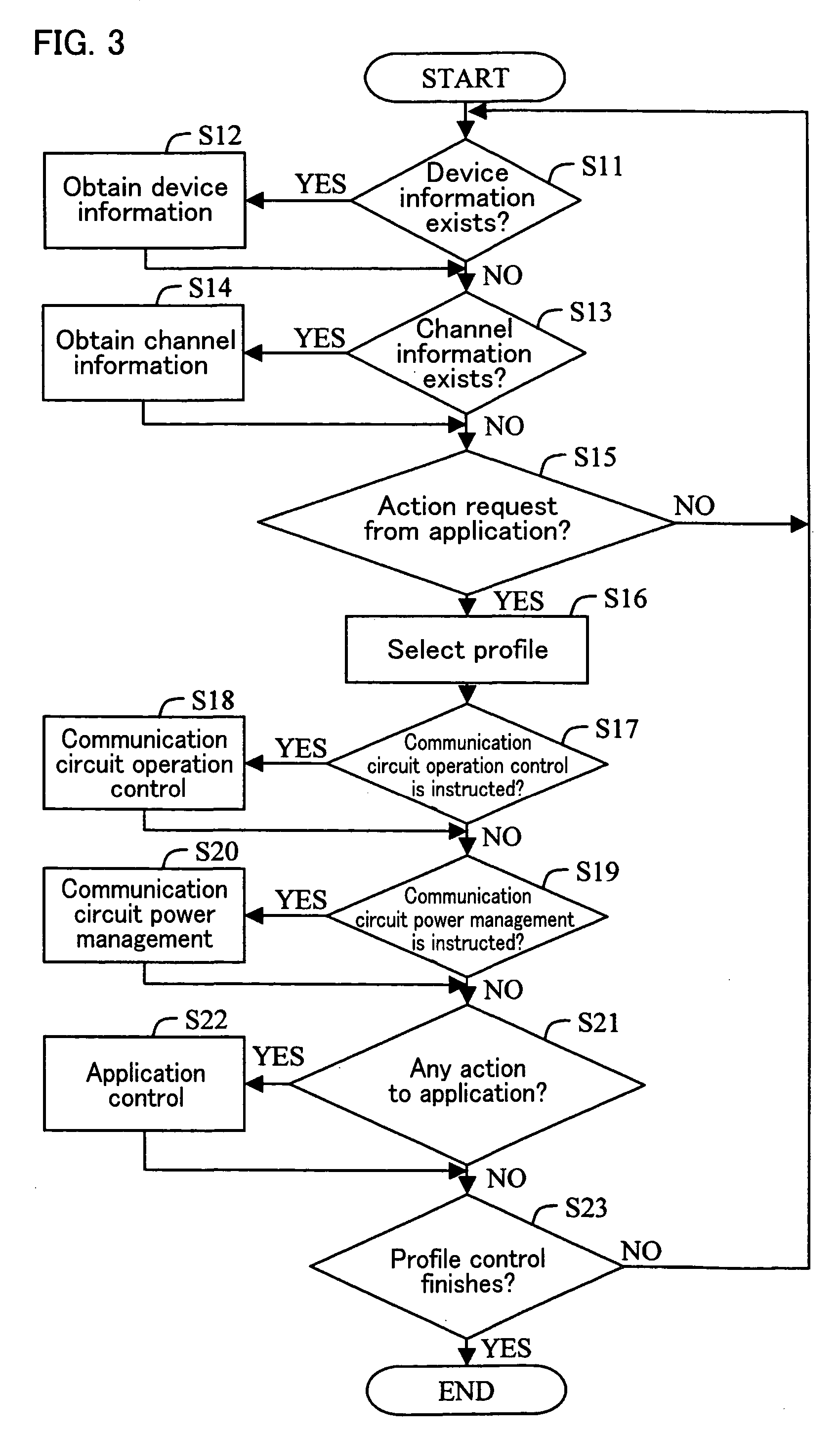
A system controller is provided between a communication device and an electronic device which performs communication using the communication device. The system controller controls the communication device, in accordance with device information from the electronic device, a request from the application, and channel information from the communication device. With this arrangement, the communication device can realize an effective low-power-consumption operation, while having a high degree of versatility.
Owner:SHARP KK
Systems and methods for testing the performance of and simulating a wireless communication device
InactiveUS7508868B2Exact reproductionTest accuratePolarisation/directional diversitySupervisory/monitoring/testing arrangementsControl signalData file
A system abstracts channel information from field data gathered in actual wireless communication system environments. The abstracted data is then transformed into control signals or programming codes that can be used to control channel simulators so as to recreate the field conditions, including path loss, slow fading, fast fading, path delay, fading power spectral density with and without line-of-sight (LOS), and different kind of handoff scenarios, such as soft, softer, intra-band hard, inter-band hard handoffs. The system thus can accurately simulate a realistic wireless communication link originated from multiple signal sources in different band channels and formed by multipath signal propagation. The simulated realistic wireless communication link can be condensed by selecting the most useful scenarios from its original field data files or modified by tuning its parameters as desired.
Owner:INTERDIGITAL PATENT HLDG INC
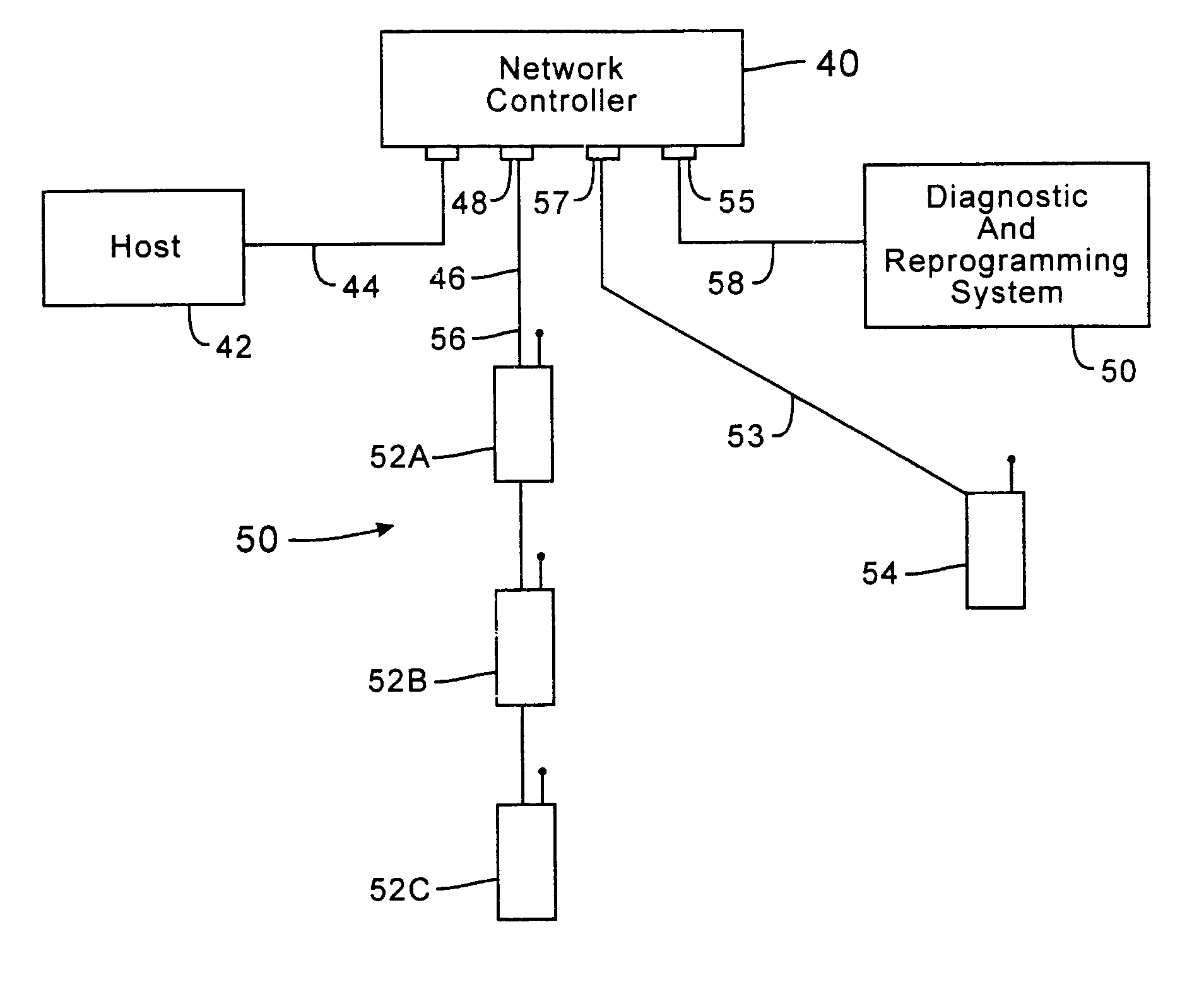
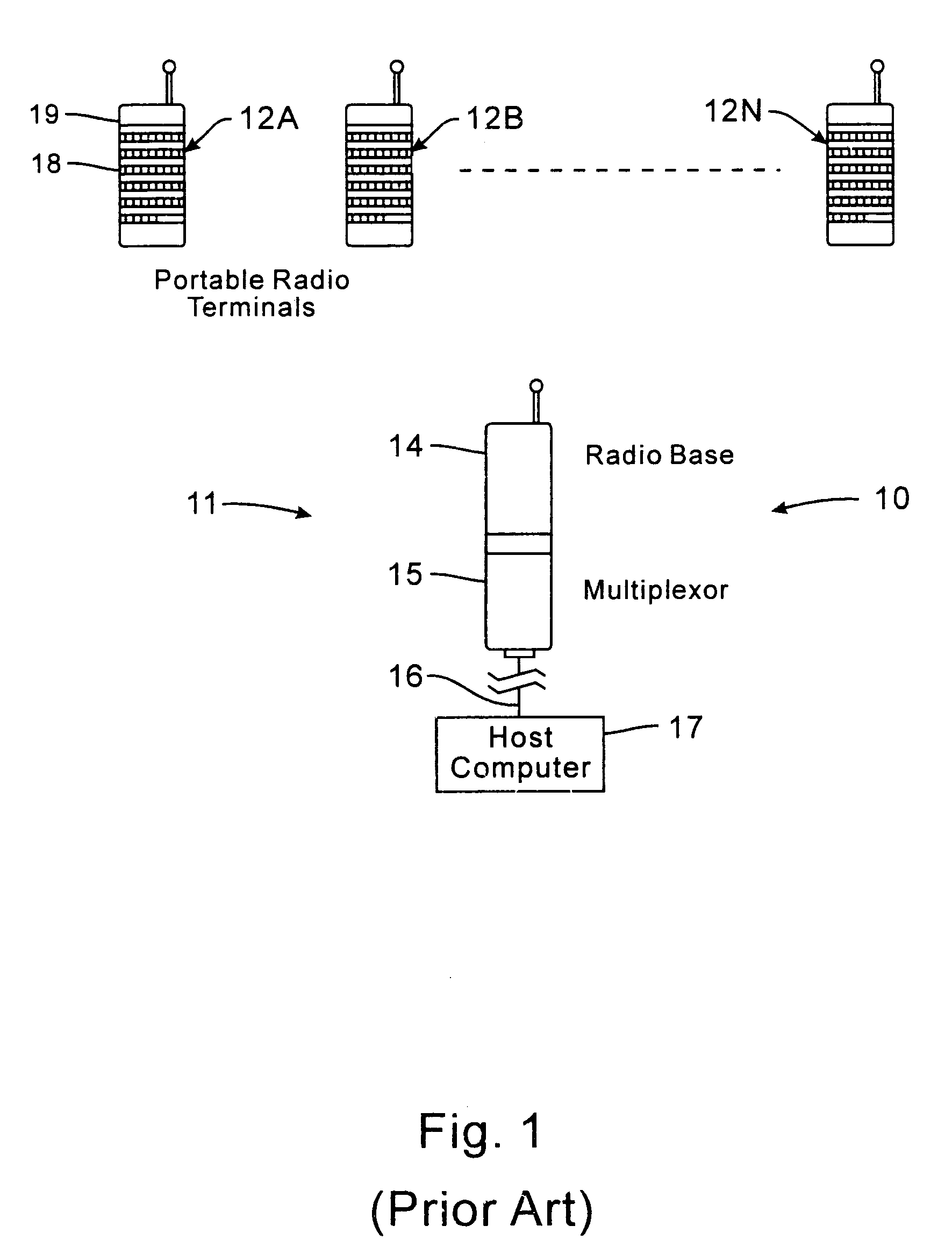
Network supporting roaming, sleeping terminals
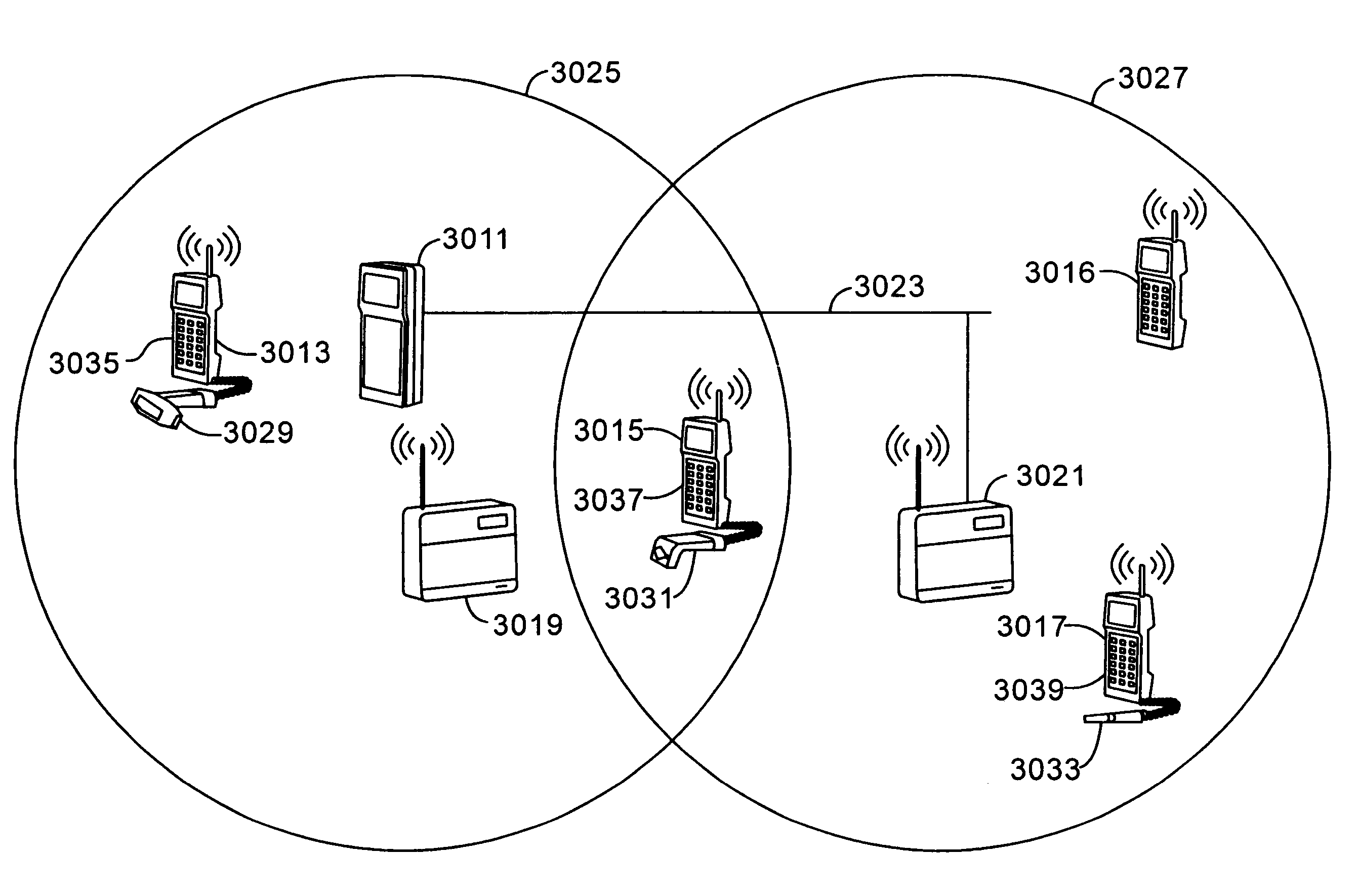
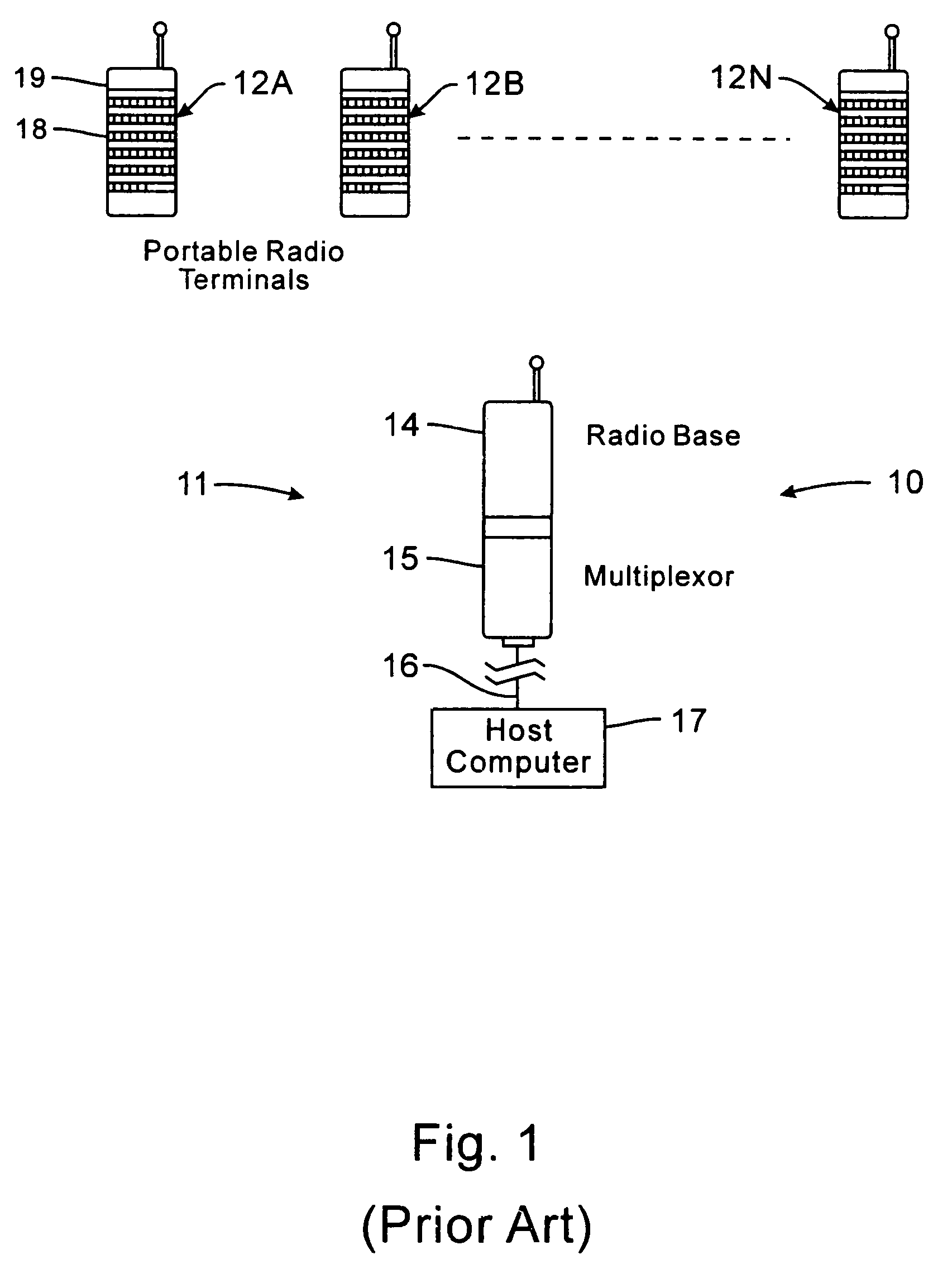
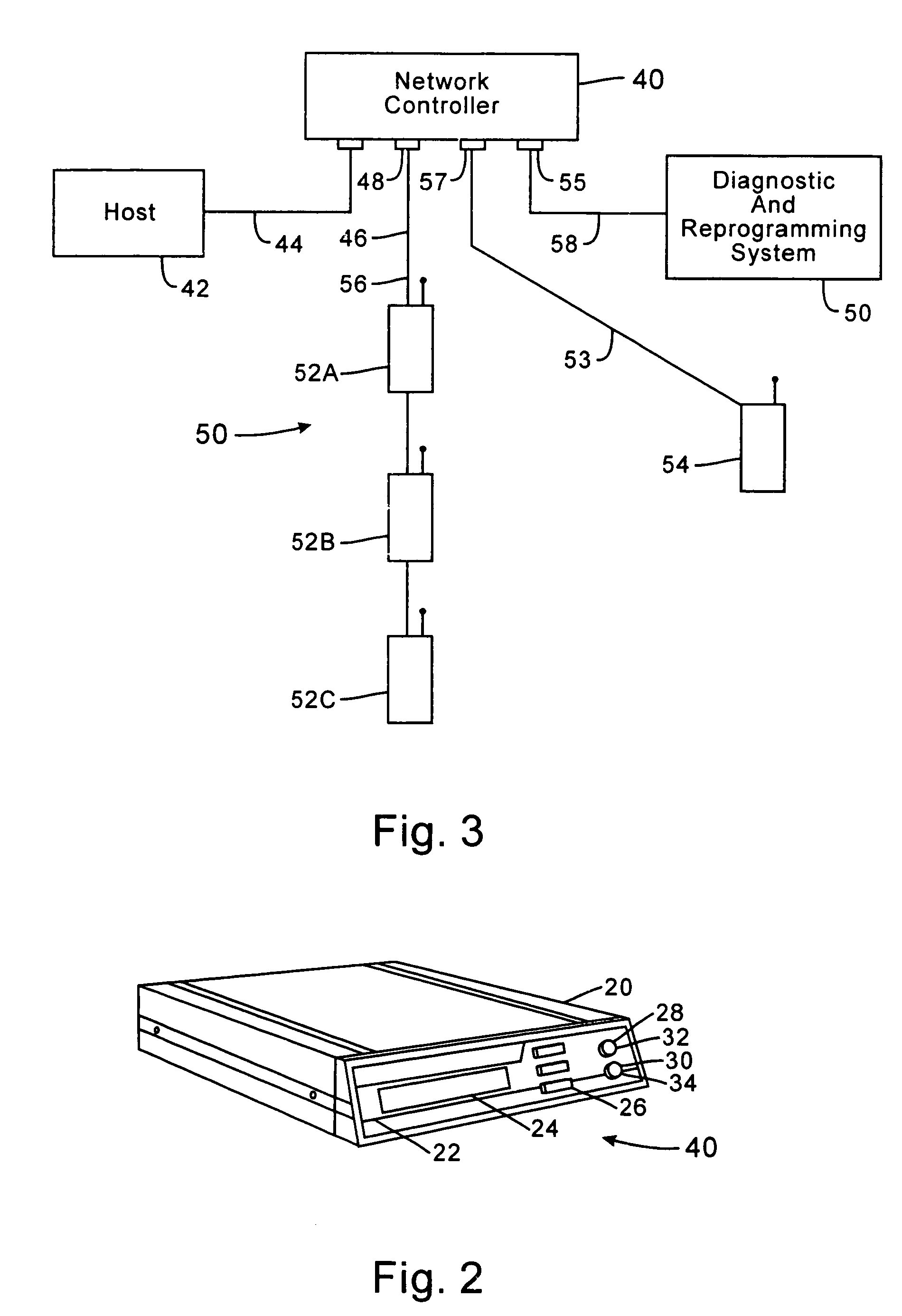
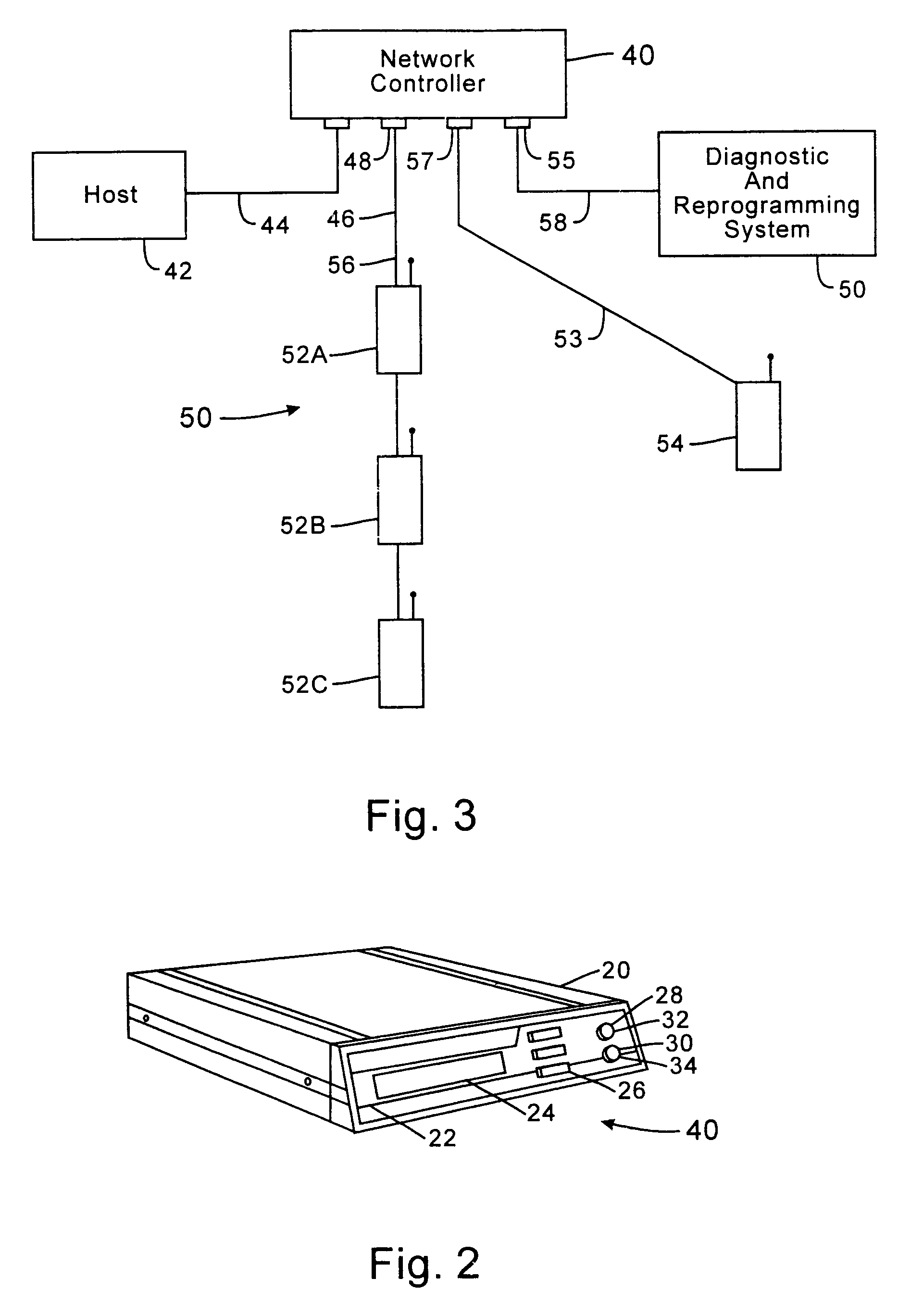
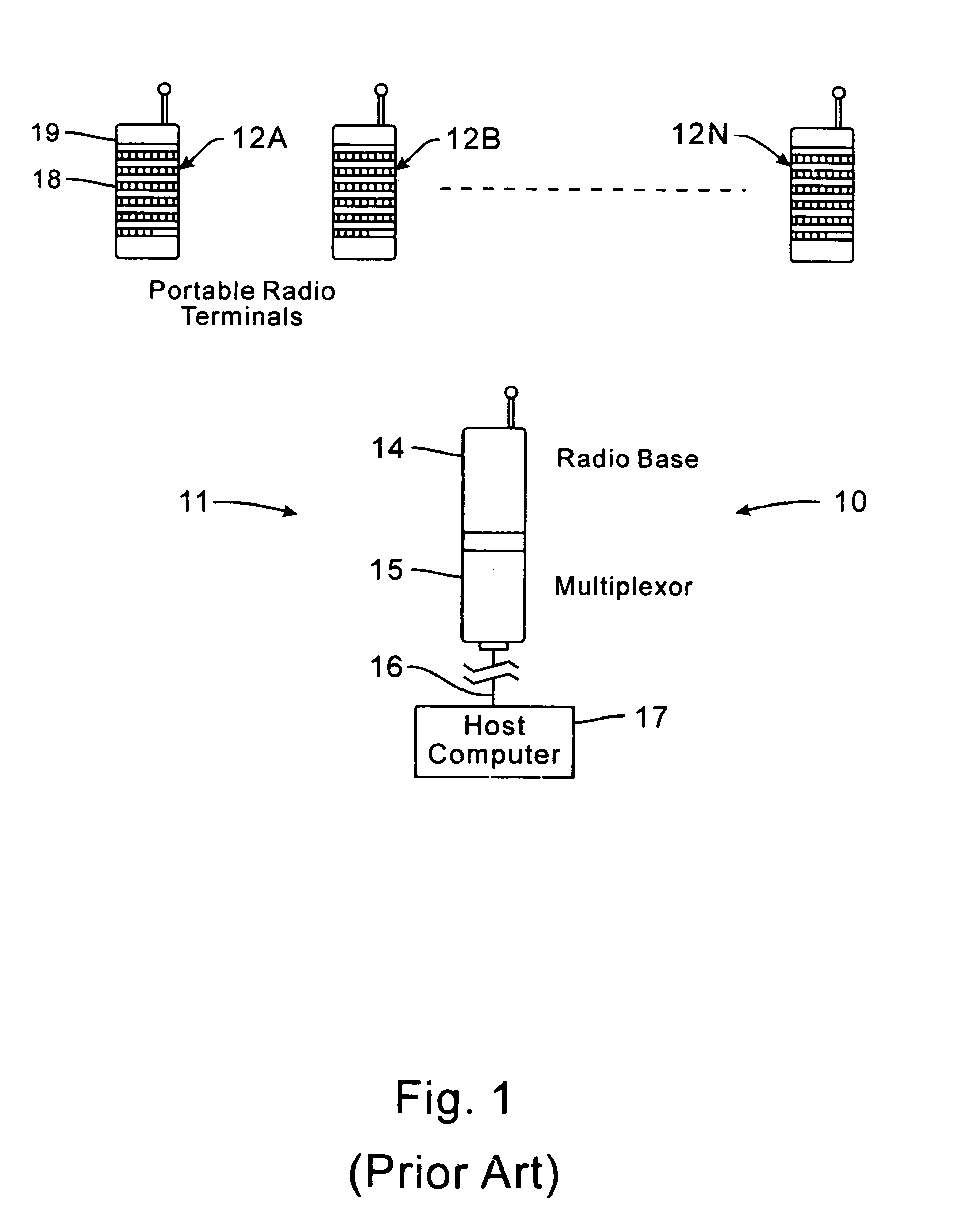
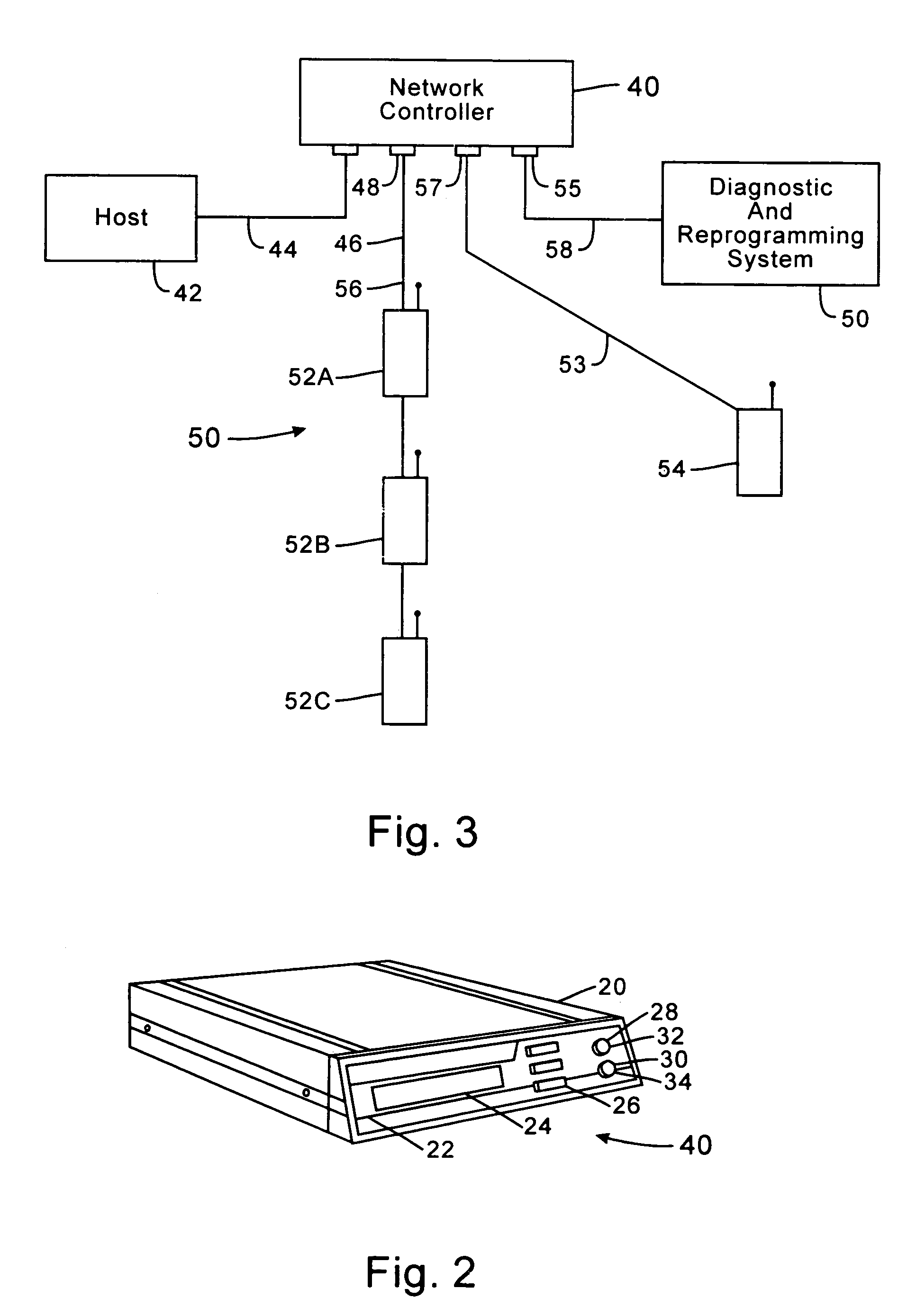
InactiveUS7536167B2Minimize collisionHighly versatileEnergy efficient ICTPower managementTransceiverCommunications system
Improved apparatus for a radio communication system having a multiplicity of mobile transceiver units selectively in communication with a plurality of base transceiver units which, in turn, communicate with one or more host computers for storage and manipulation of data collected by bar code scanners or other collection means associated with the mobile transceiver units. A network controller and an adapter which has a simulcast and sequential mode provide selective interface between host computers and base transceivers. A scheme for routing data through the communication system is also disclosed wherein the intermediate base stations are organized into an optimal spanning-tree network to control the routing of data to and from the RF terminals and the host computer efficiently and dynamically. Additionally, redundant network and communication protocol is disclosed wherein the network utilizes a polling communication protocol which, under heavy loaded conditions, requires that a roaming terminal wishing to initiate communication must first determine that the channel is truly clear by listing for an entire interpoll gap time. In a further embodiment, a criterion used by the roaming terminals for attaching to a given base station reduces conflicts in the overlapping RF regions of adjacent base stations.
Owner:AVAGO TECH WIRELESS IP SINGAPORE PTE +1
Versatile vibration-damped golf swing-weight system
ActiveUS20100105498A1Reliably securedConveniently releasedMetal working apparatusGolf clubsAxial pressureEngineering
A highly versatile damper-weight system enables the installation of adjustable swing-weight in a vibration-damped manner inside a golf club shaft. A plug assembly of selectable weight is inserted through a circular opening in the golf grip cap with a special tool, moved to any desired location within the shaft and securely fastened in place in a vibration-damped manner by radial expandable of a cylindrical resilient expandable element. A weight rod, made available in different materials, lengths and weights, is spaced from the shaft by one or more resilient damper sleeves to minimize shaft vibration. The expandable element is secured by a machine screw threaded into the upper end of the weight rod, and is dimensioned (unexpanded) to enable easy insertion and location adjustment of said plug assembly. The tool provides dual functions: as a screw head driver to expand / contract the expandable element radially by axial pressure / release as required, and as a removable coupler capable of pulling the plug assembly (with the expandable element unexpanded) upwardly, as well as pressing it downwardly within the shaft for adjustment to any desired location.
Owner:JOHNSON JOHN
Robot control device
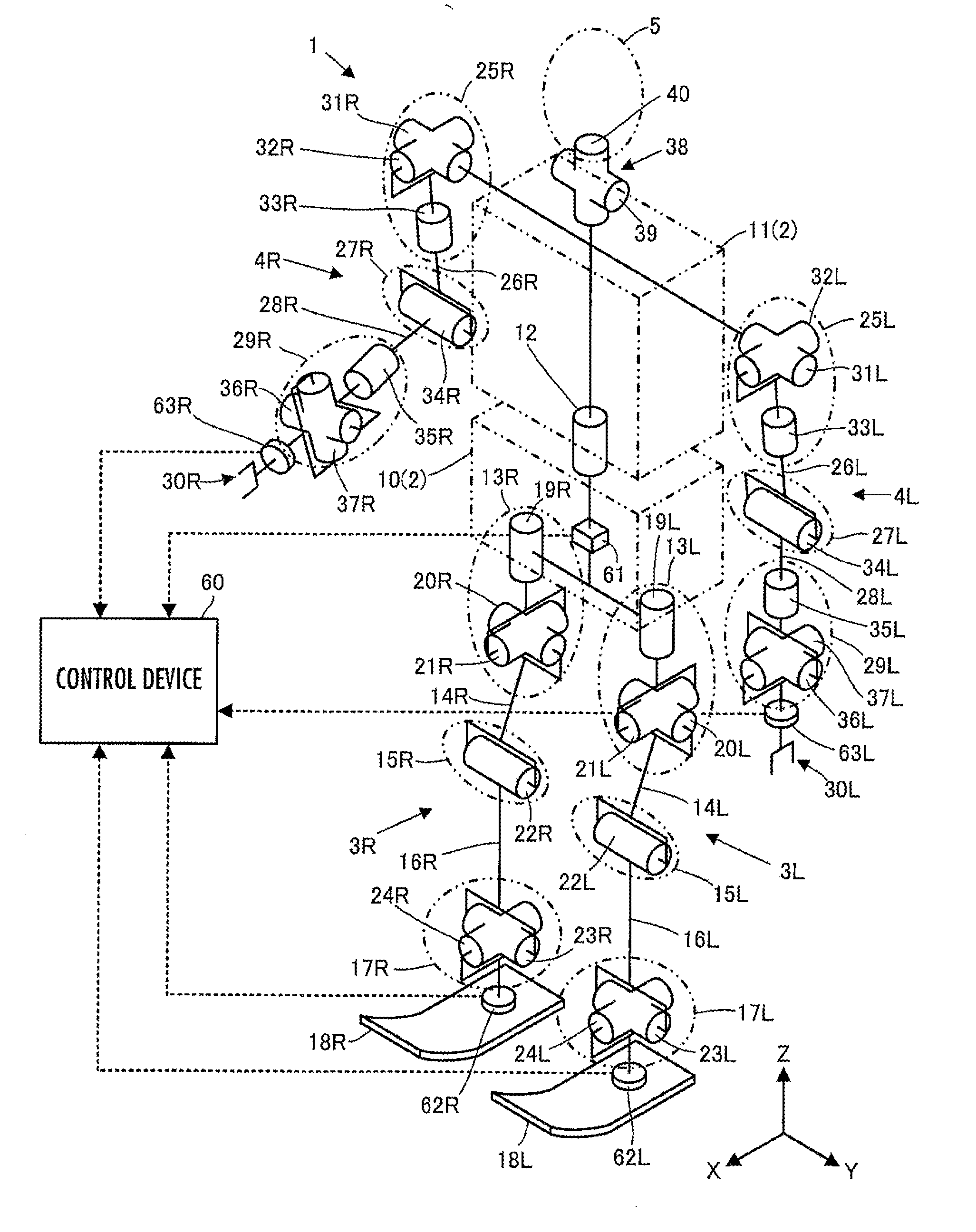
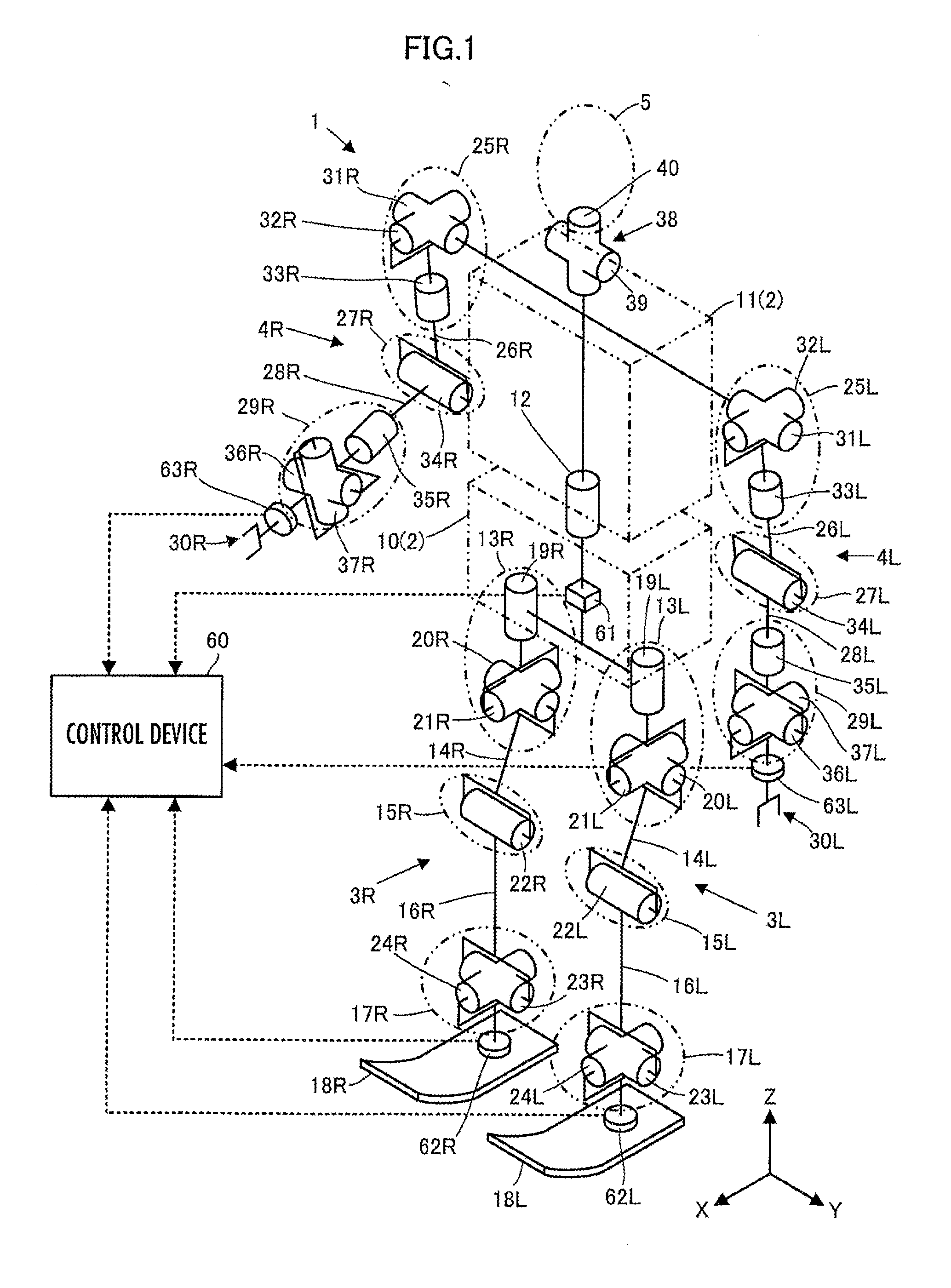
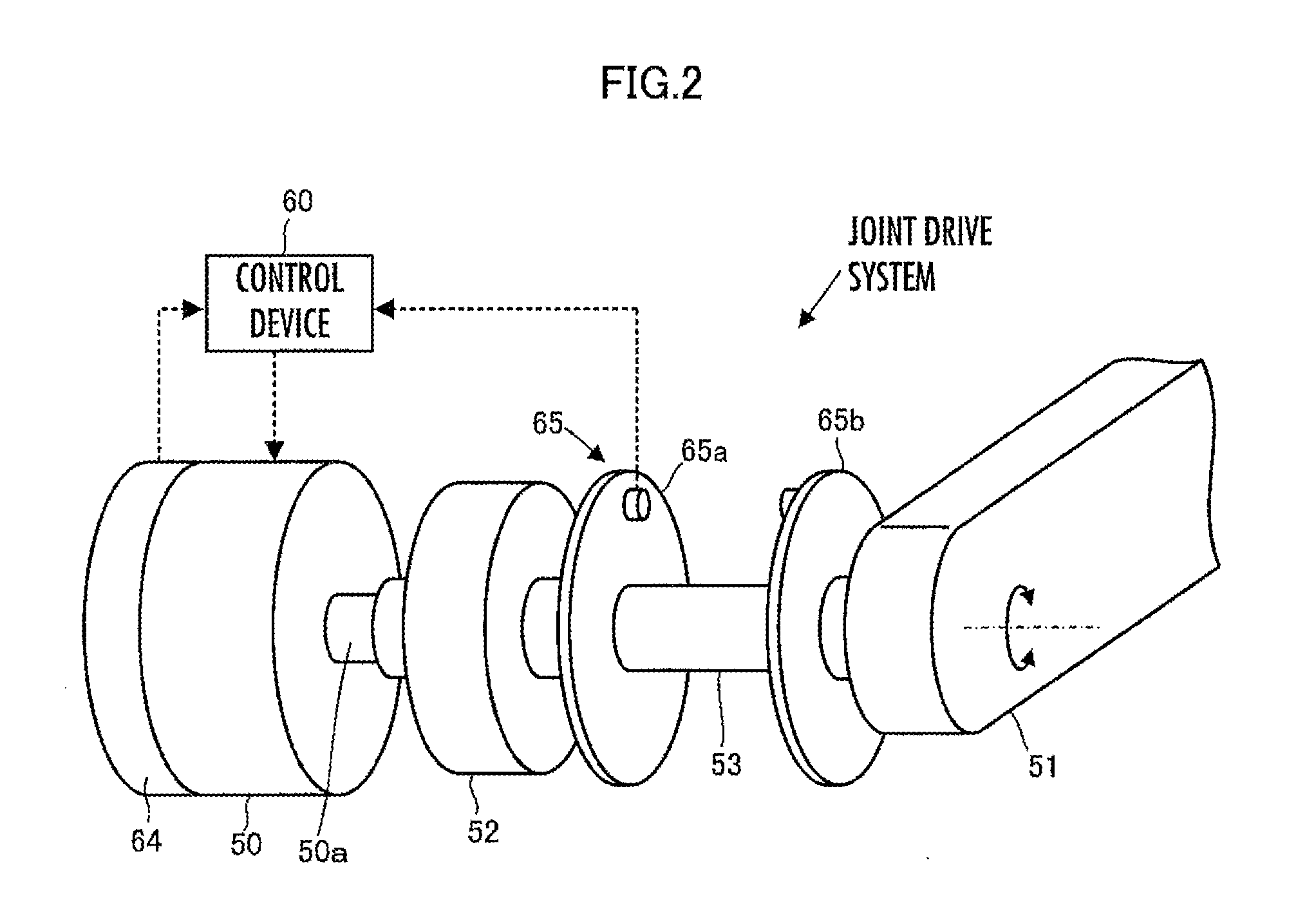
ActiveUS20120259463A1Satisfy the demandHighly versatileProgramme-controlled manipulatorRobotRobot controlControl theory
Disclosed is a robot control device including a means for determining control inputs classified by state quantity for achieving respective target values of a plurality of types of state quantities of a robot, and a means for determining a synthesized control input by synthesizing control inputs classified by frequency region while determining control inputs classified by frequency region in a plurality of respective frequency regions, according to control inputs classified by state quantity. The means determines a control input classified by frequency region corresponding to any one of the frequency regions by synthesizing the plurality of control inputs classified by state quantity in a mutually non-interfering manner. The operation of the robot is controlled so that, under a variety of operating conditions of the robot, a plurality of types of state quantities are efficiently controlled to target values which correspond to the respective types of state quantities.
Owner:HONDA MOTOR CO LTD
Watercraft docking system and propulsion assembly
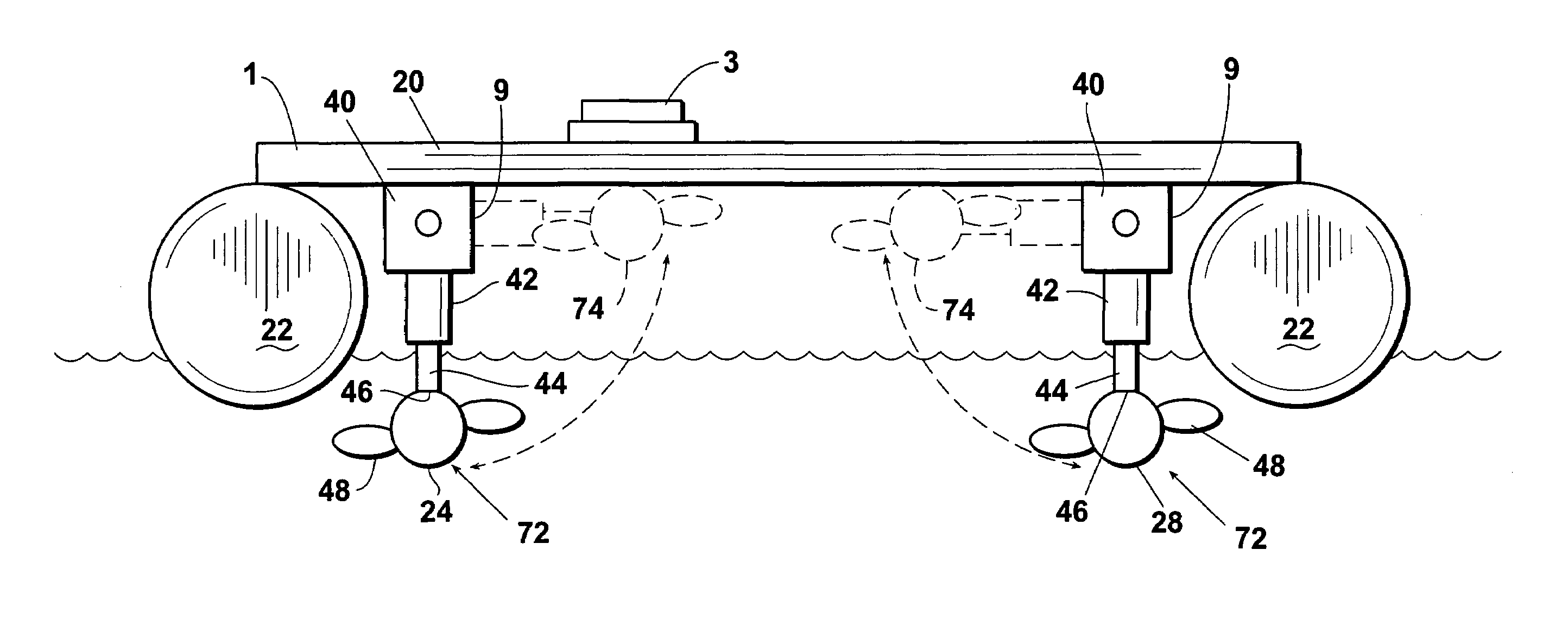
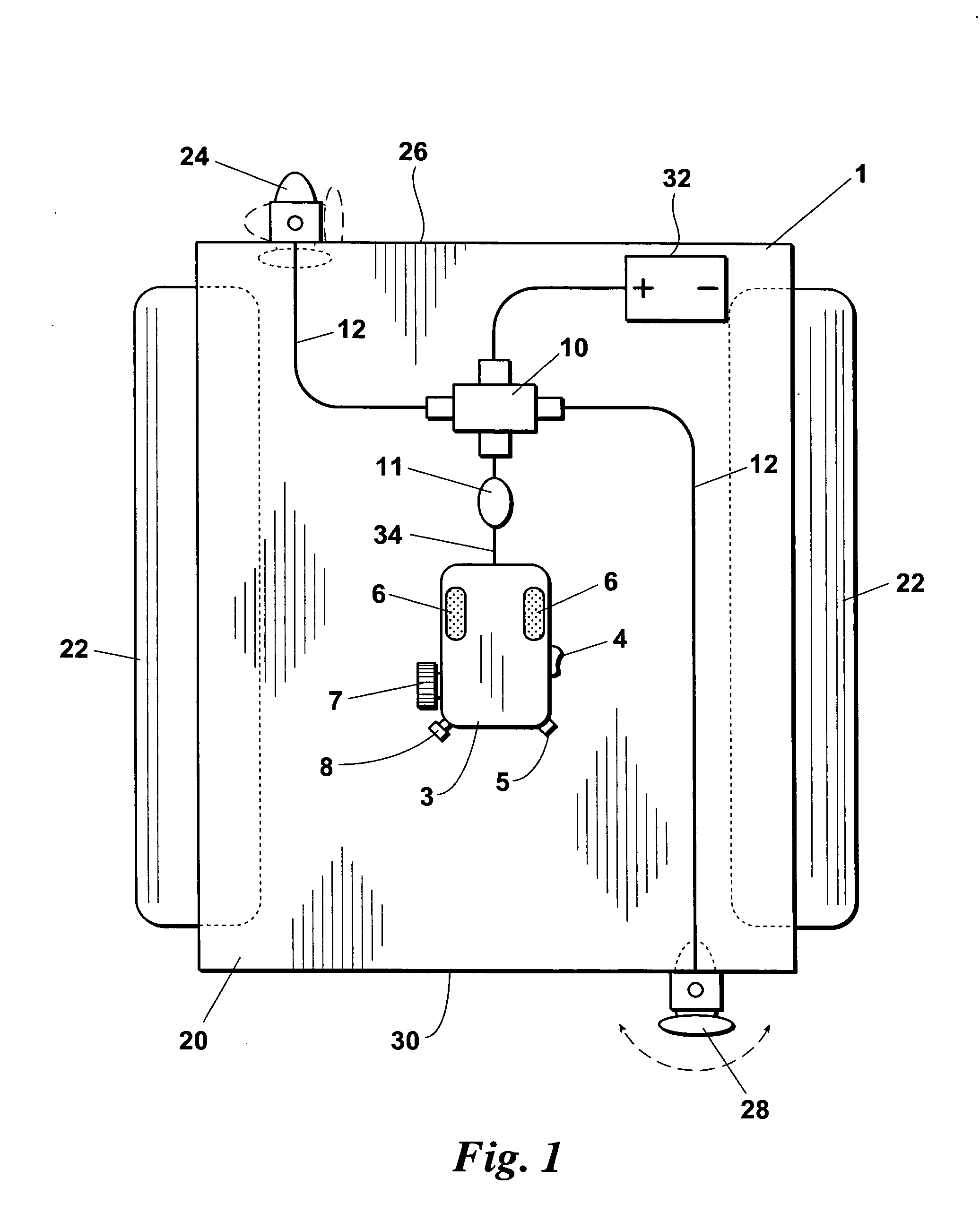
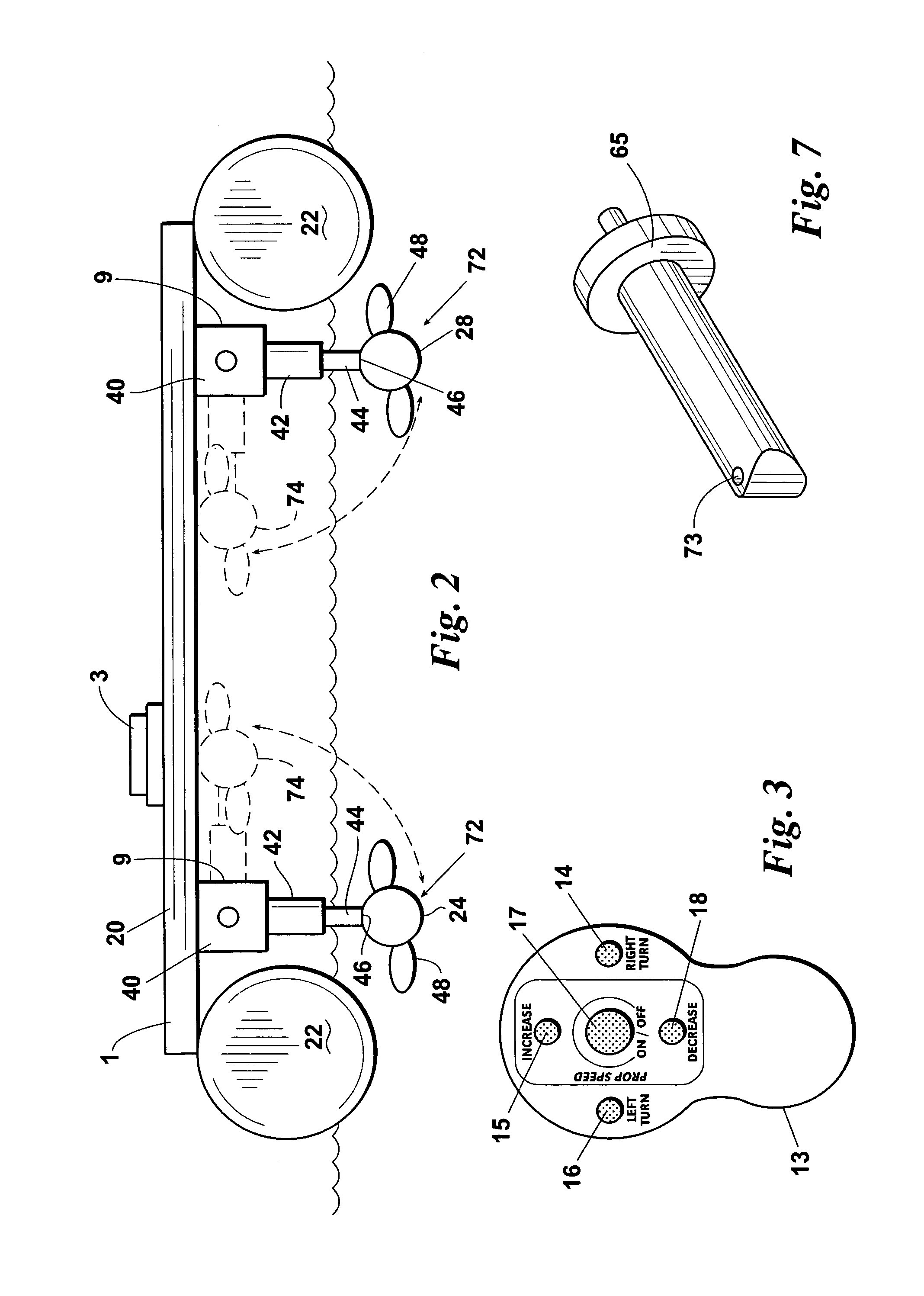
ActiveUS7150662B1Highly maneuverableHighly versatileSpeed controllerPropulsion power plantsSteering columnOperating point
An improved docking system for a watercraft and a propulsion assembly therefor wherein the docking system comprises a plurality of the propulsion assemblies and wherein each propulsion assembly includes a motor and propeller assembly provided on the distal end of a steering column and each of the propulsion assemblies is attachable in an operating position such that the motor and propeller assembly thereof will extend into the water and can be turned for steering the watercraft.
Owner:BRUNSWICK CORPORATION
Processing apparatus which performs predetermined processing while supplying a processing liquid to a substrate
InactiveUS20050212837A1Efficiently formedLower capability requirementsBioreactor/fermenter combinationsBurnersEngineeringLaser
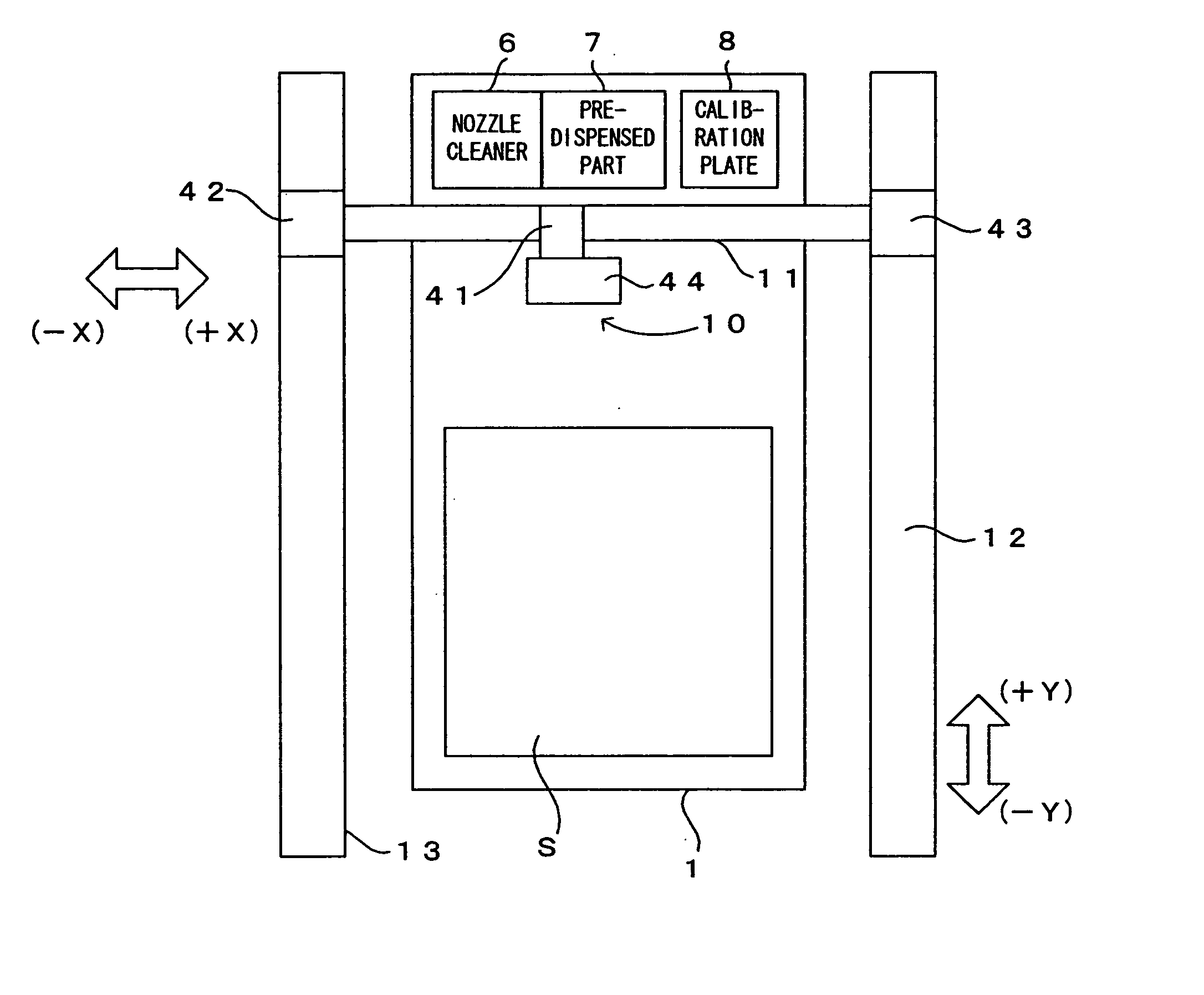
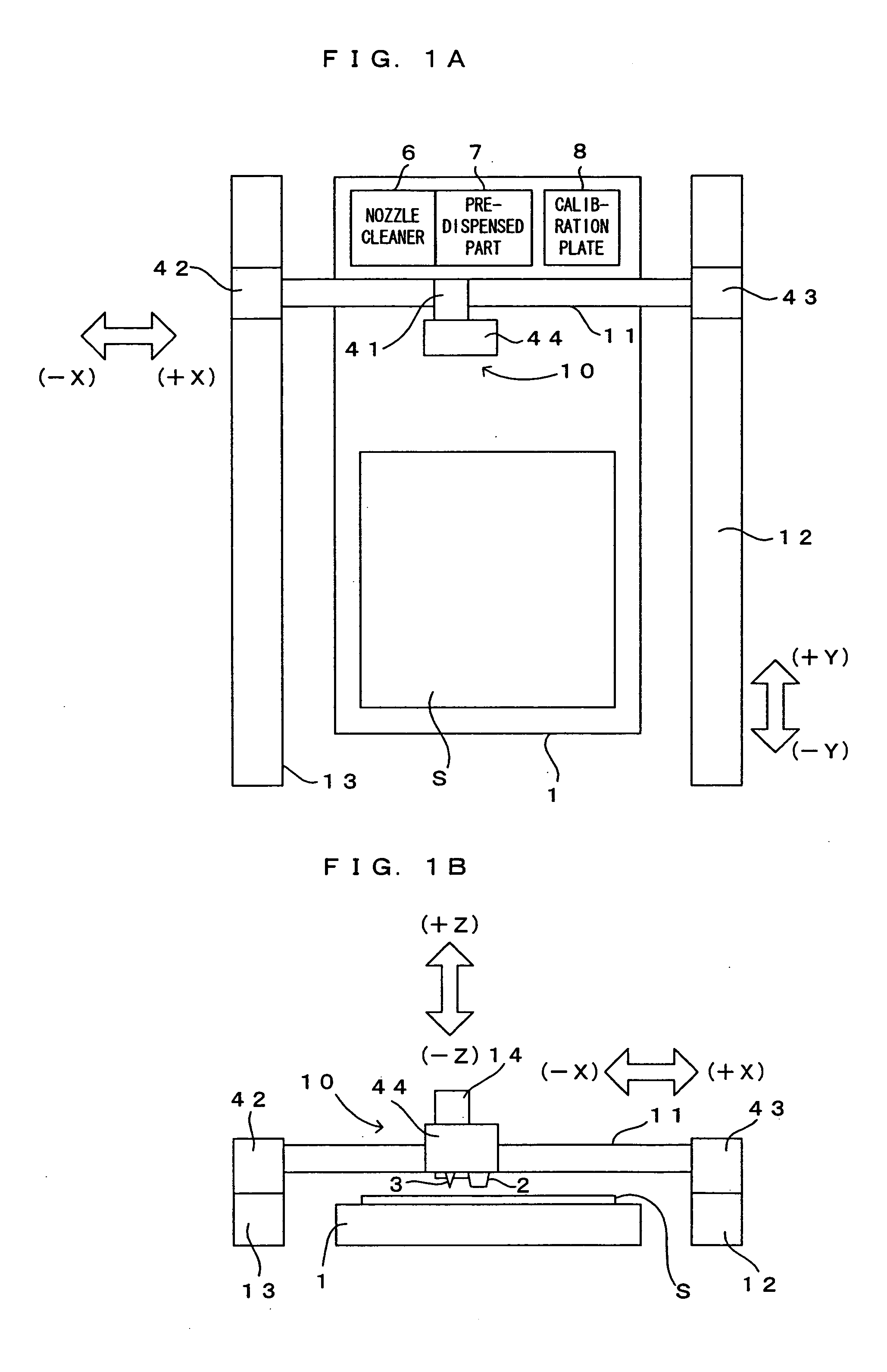
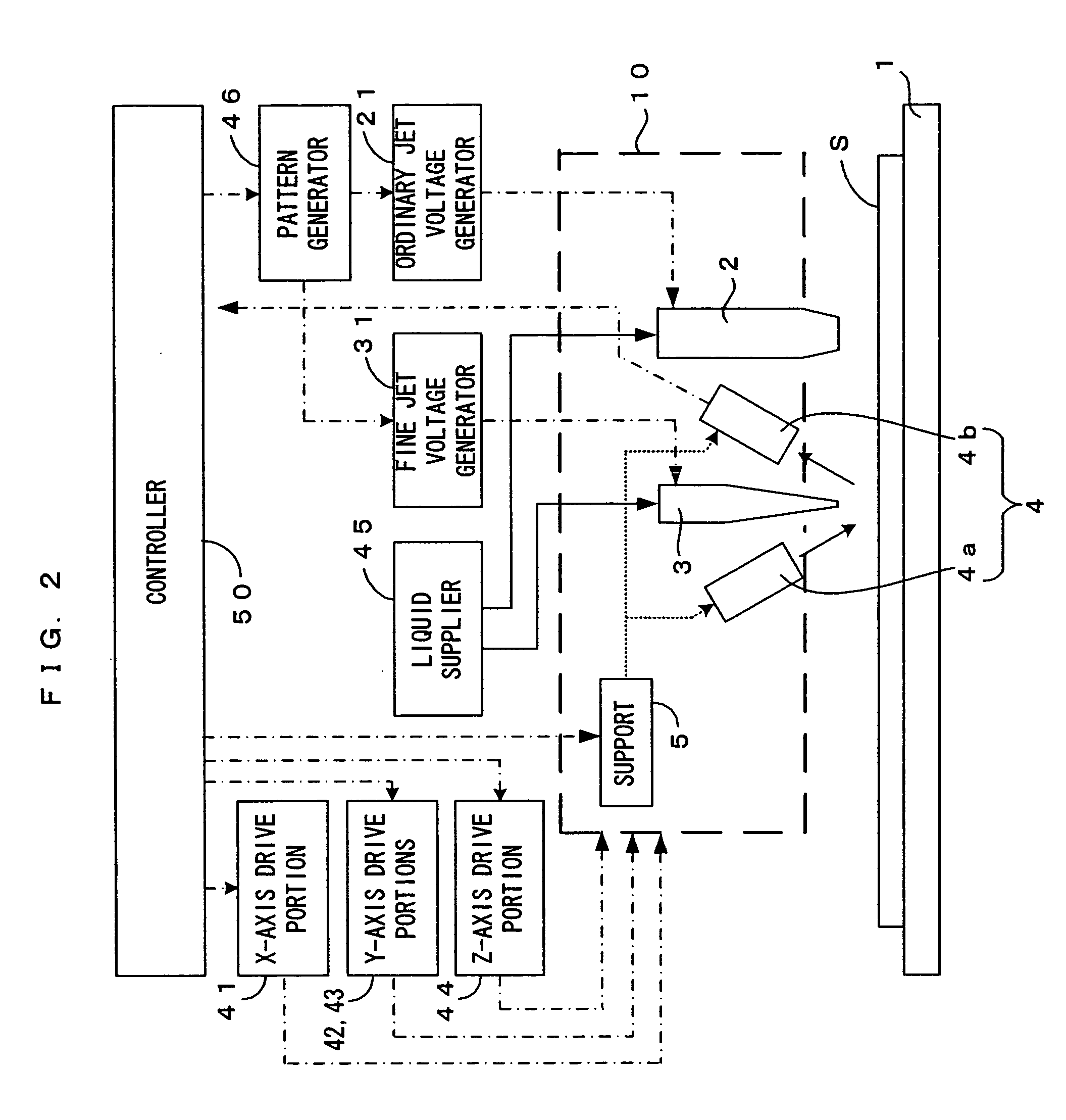
Above a substrate holder 1, an ordinary fluid jet nozzle 2 which is capable of injecting drops of a solution having ordinary particle diameters, a fine fluid jet nozzle 3 which is capable of injecting drops of a solution having fine particle diameters which are smaller than the ordinary particle diameters, and a regular reflection laser displacement gauge 4 are disposed. These are attached on the same plate, thereby forming a head 10. The head 10 freely moves in directions X, Y and Z, as an X-axis drive portion 41, Y-axis drive portions 42 and 43 and a Z-axis drive portion 44 operate in accordance with operation commands received from a controller 50 which controls the apparatus as a whole. It is therefore possible to set the head 10 to a predetermined position on a substrate S which is held by the substrate holder 1. Thus, the respective nozzles supply the solutions to any desired positions the substrate S.
Owner:NAT INST OF ADVANCED IND SCI & TECH
Strong authentication token generating one-time passwords and signatures upon server credential verification
ActiveUS8302167B2Transfer size reductionHighly secureDigital data processing detailsMultiple digital computer combinationsInternet privacyTransaction data
The invention defines a strong authentication token that remedies a vulnerability to a certain type of social engineering attacks, by authenticating the server or messages purporting to come from the server prior to generating a one-time password or transaction signature; and, in the case of the generation of a transaction signature, signing not only transaction values but also transaction context information and, prior to generating said transaction signature, presenting said transaction values and transaction context information to the user for the user to review and approve using trustworthy output and input means. It furthermore offers this authentication and review functionality without sacrificing user convenience or cost efficiency, by judiciously coding the transaction data to be signed, thus reducing the transmission size of information that has to be exchanged over the token's trustworthy interfaces.
Owner:ONESPAN NORTH AMERICA INC
Amine-aldehyde resins and uses thereof in separation processes
InactiveUS20080029460A1Highly versatileHigh selectivitySpecific water treatment objectivesWater contaminantsParticulatesKaolin clay
Amine-aldehyde resins are disclosed for removing a wide variety of solids and / or ionic species from the liquids in which they are suspended and / or dissolved. These resins are especially useful as froth flotation depressants, for example in the beneficiation of value materials (e.g., bitumen, coal, or kaolin clay) to remove impurities such as sand. The resins are also useful for treating aqueous liquid suspensions to remove solid particulates, as well as for removing metallic ions in the purification of water.
Owner:INGEVITY SOUTH CAROLINA
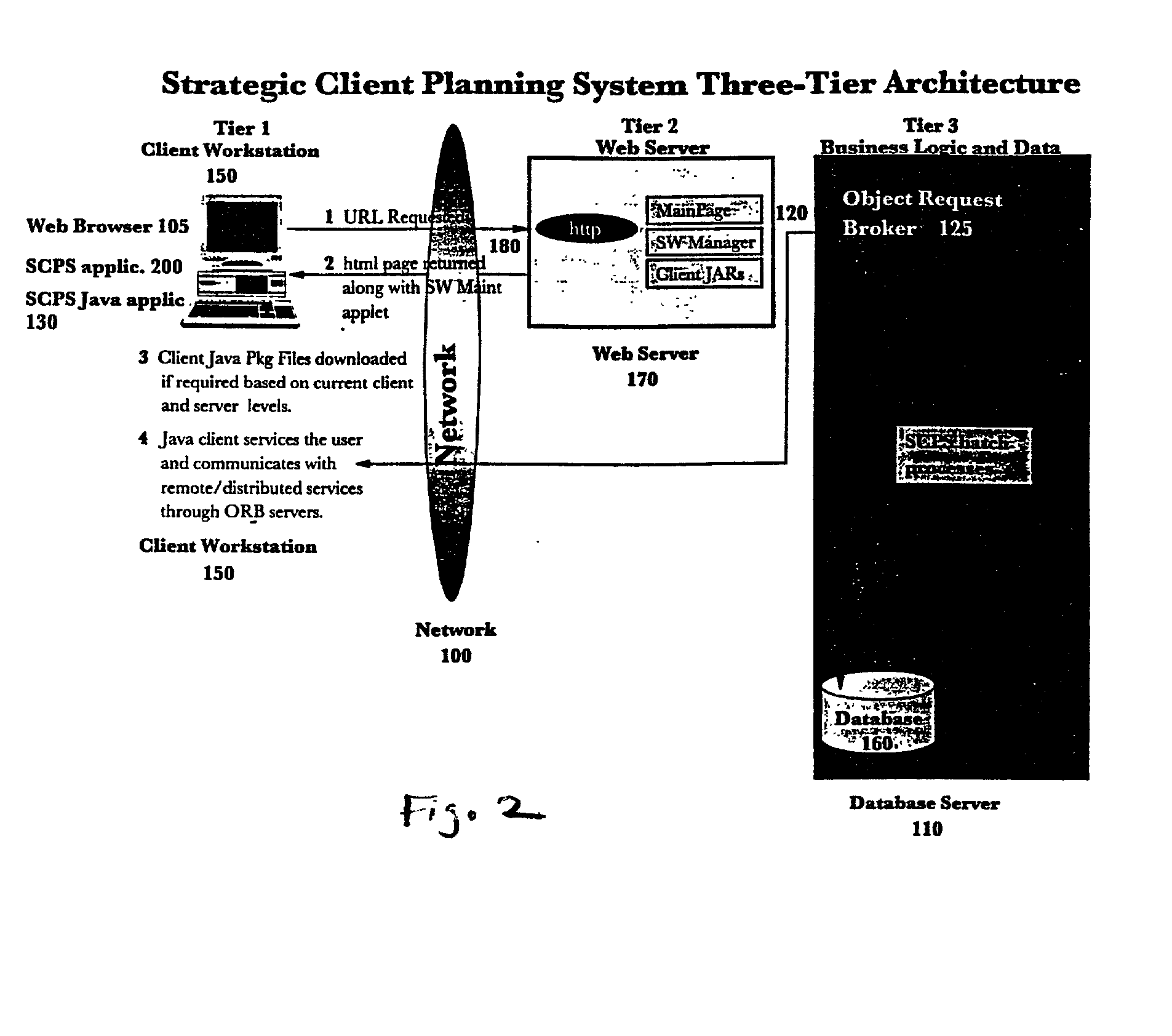
Web-based strategic client planning system for end-user creation of queries, reports and database updates
InactiveUS7013312B2Easy to useEasy to openData processing applicationsDigital data information retrievalProgram planningClient-side
A method and program storage device for creating tabular data stream flow for sending rows of secure data between a client workstation and a server computer over a network using a common object request broker architecture (CORBA). This method includes receiving a request to create a query form at the client workstation, receiving a worksheet grid form defining selected tabular data, and packaging the worksheet grid form representing an updated status of the data for the tabular data stream flow.
Owner:IBM CORP
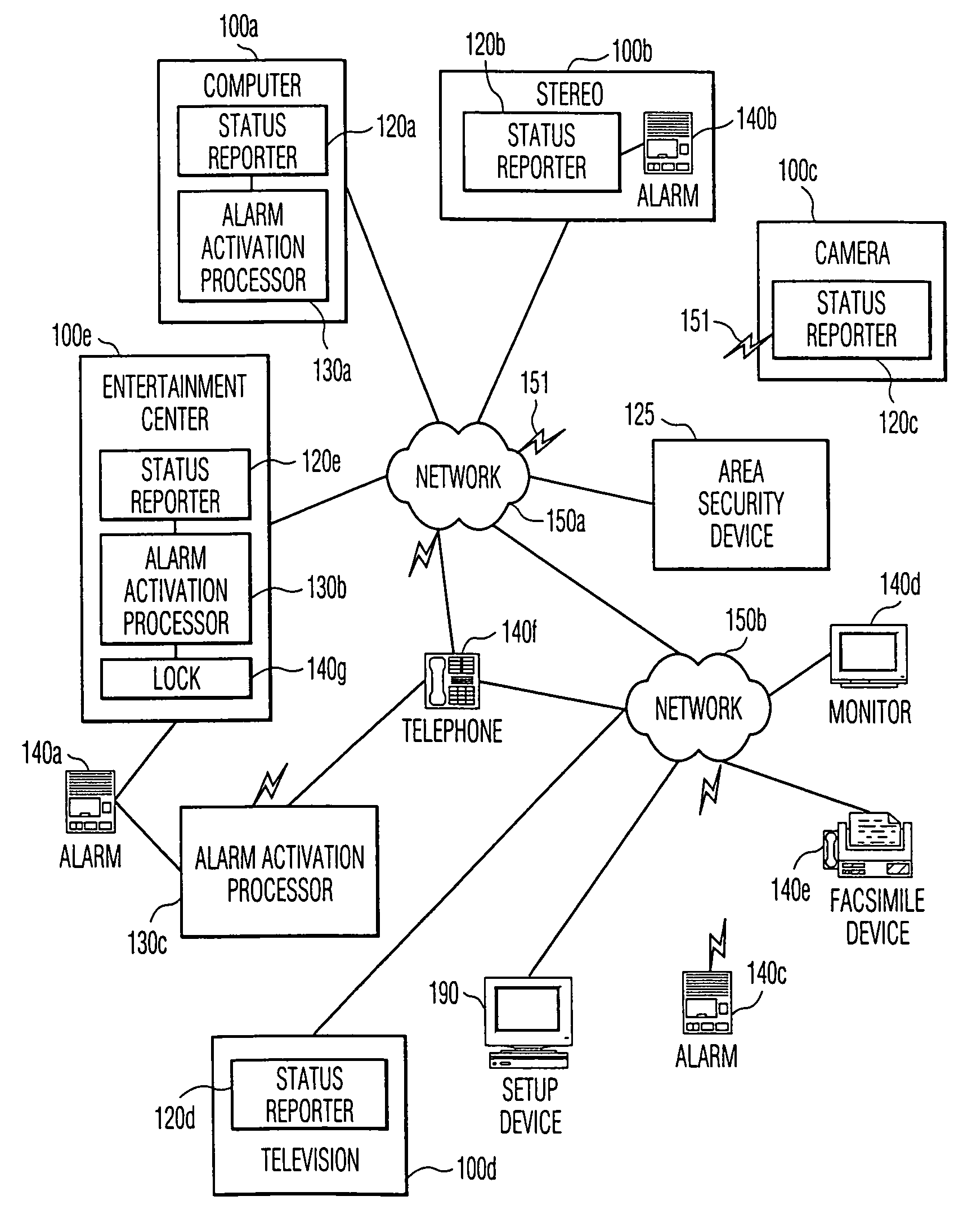
Distributed software controlled theft detection
InactiveUS6970081B1Highly flexibleHighly versatileElectric signal transmission systemsFrequency-division multiplex detailsBiological activationSecurity system
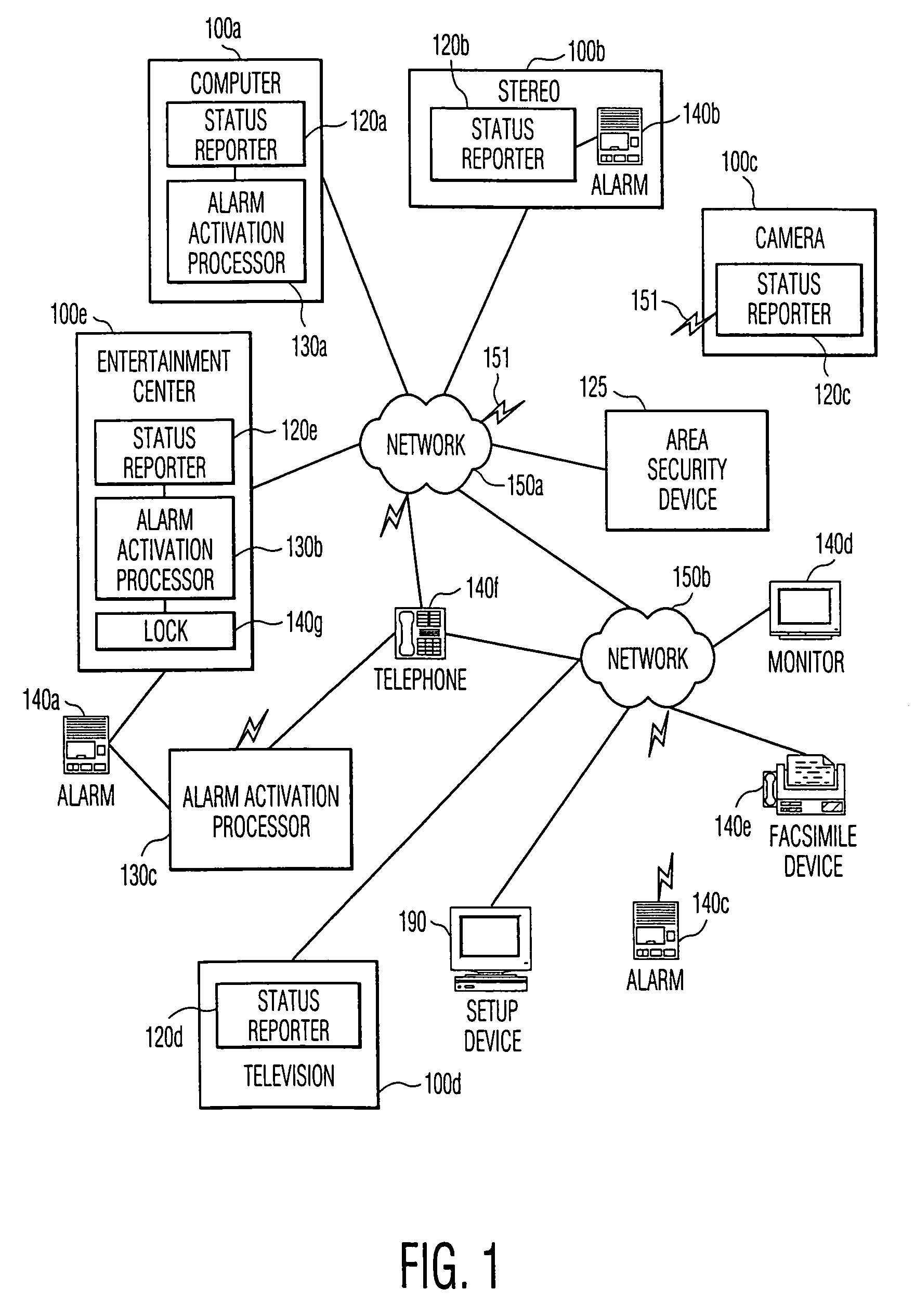
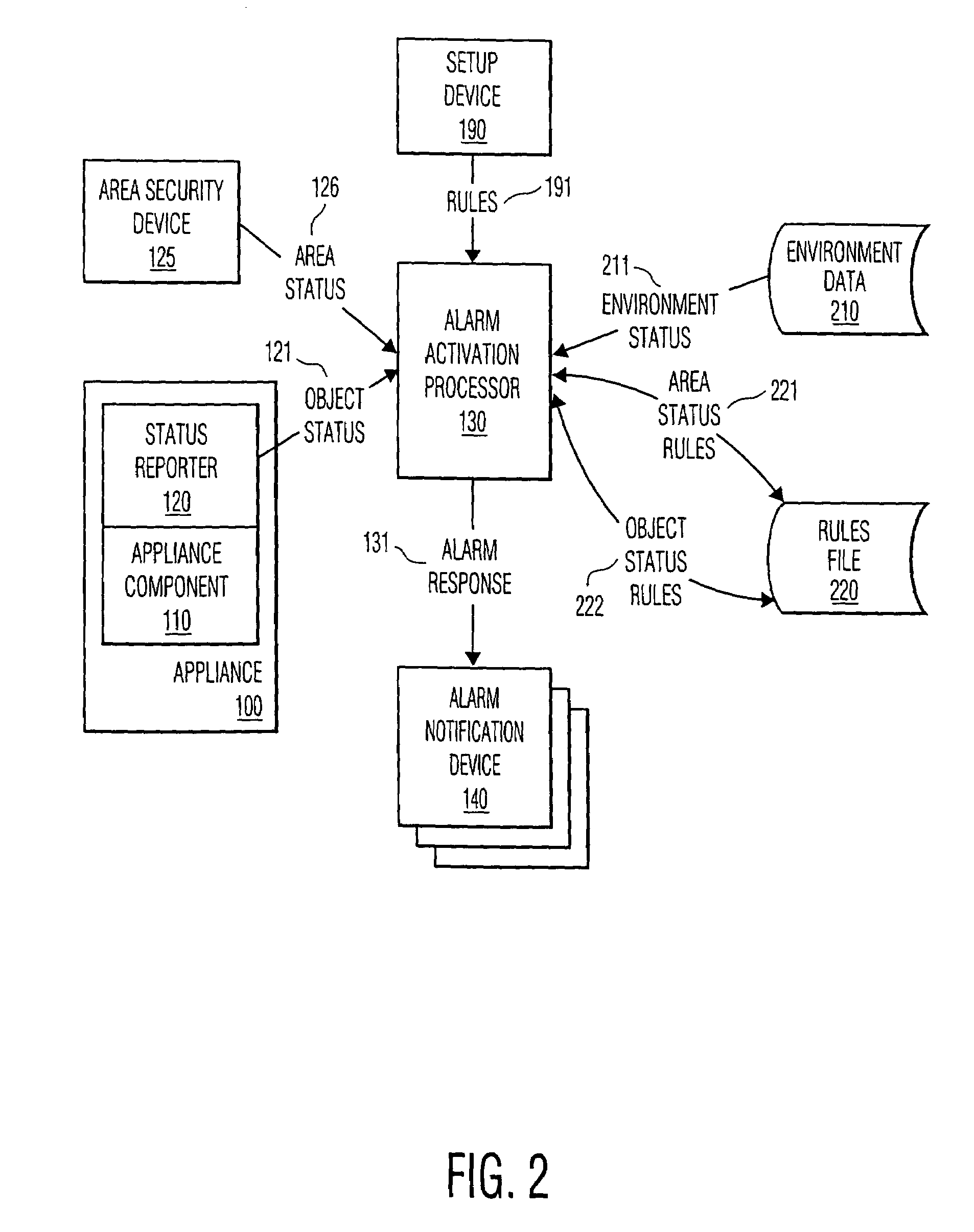
This invention provides the framework of a security system that is property specific as well as area specific. The invention includes a distributed software controlled security system that monitors and assesses the status of individual property items and conventional area security devices within a secured area. The activation of an alarm and the action taken in response to the alarm are determined by the state of the secured area, the state of the individual property item, and rules associated with each state or combination of states. Depending upon the capabilities of the individual property items, or appliances, and the area security devices, the security capabilities of this invention are distributed amongst a variety of appliances. In a preferred embodiment, the communication of messages and signals among the components of the system is in accordance with existing or proposed standards, such as HAVi and Home API, thereby allowing interoperability among components of varying vendors.
Owner:KONINKLIJKE PHILIPS ELECTRONICS NV
Method and kit for the detection and/or quantification of homologous nucleotide sequences on arrays
InactiveUS20050272044A1Easy to identifyHighly versatileSugar derivativesMicrobiological testing/measurementBiological bodyHomologous sequence
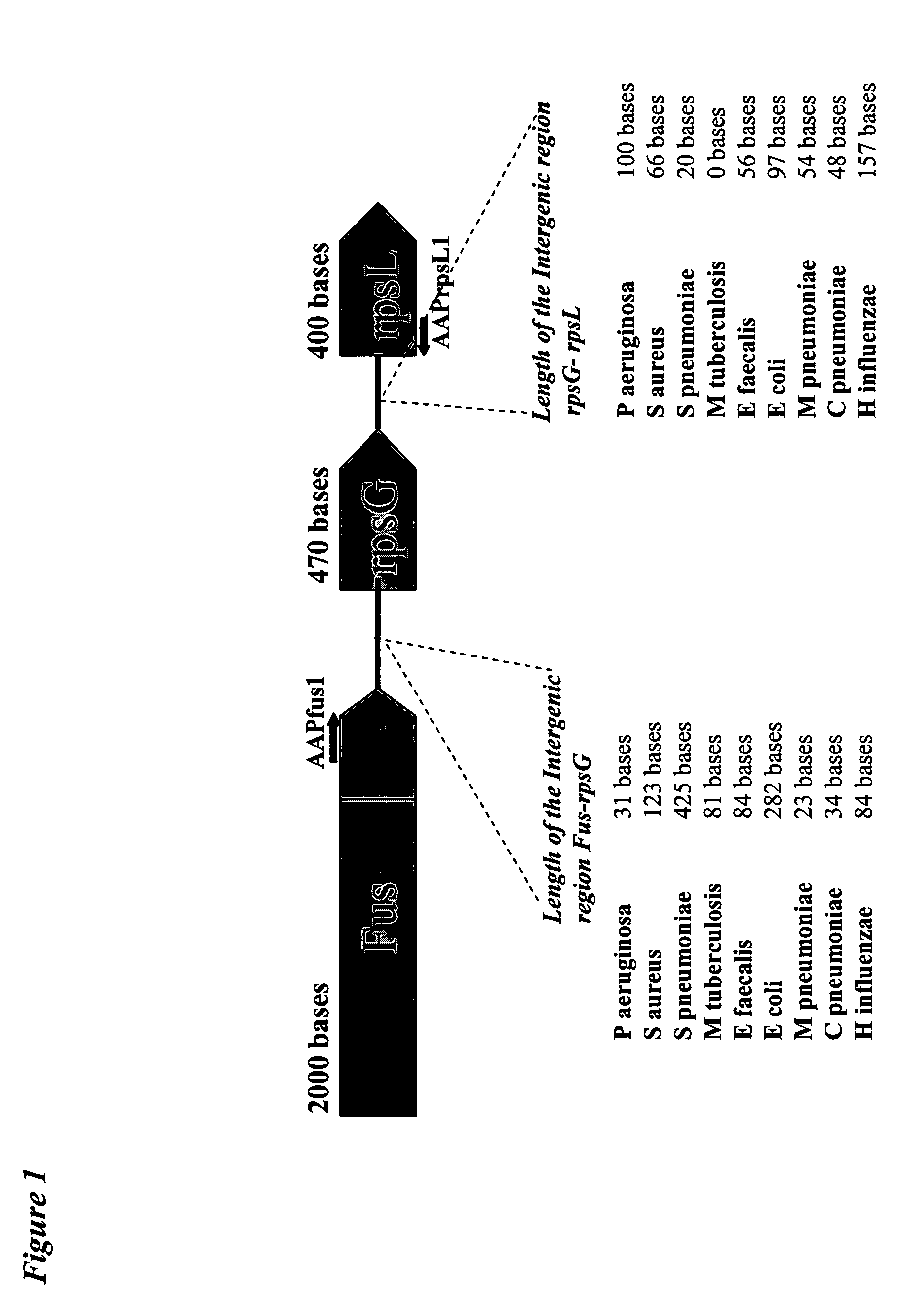
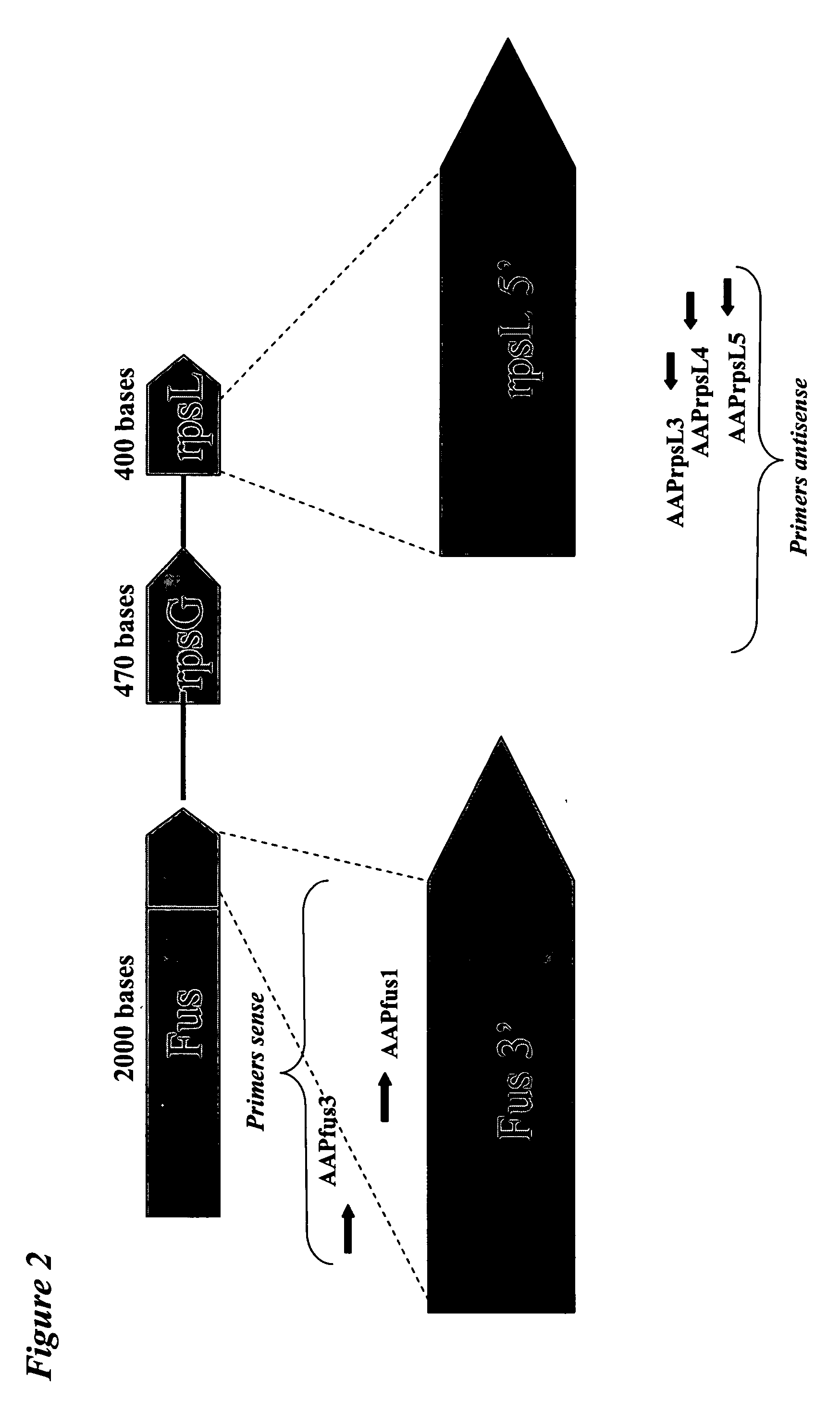
The invention relates to a method and a kit for the specific identification and / or quantification of one or several among at least 7 organisms or parts thereof, in a biological sample being possibly contaminated by at least 4 other organisms, by detecting at least one nucleotide sequence specific of each of the organisms possibly present in said biological sample, wherein said nucleotide sequence is homologous with at least 4 other nucleotide sequences. The method comprises the steps of: amplifying the nucleotide sequences specific of said organisms into target amplified nucleotide sequences using at least 2 different primer pairs, each one being capable of amplifying at least 4 of said homologous nucleotide sequences from other organisms and having an homology higher than 85% with each of the said amplified homologous nucleotide sequences to be amplified; providing an array onto which single-stranded capture nucleotide sequences are arranged at pre-determined locations, said single-stranded capture nucleotide sequences being covalently bound to an insoluble support, via a spacer which is at least 6.8 nm in length, and wherein said capture nucleotide sequences comprise a nucleotide sequence of about 10 to 50 bases which is able to specifically bind to one target amplified sequence without binding to said other amplified homologous nucleotide sequences and presenting an homology lower than 85% with the other capture nucleotide sequences of the said other amplified homologous sequences, contacting said target amplified sequences with the array in one solution under conditions allowing hybridization of the target amplified sequences to complementary capture nucleotide sequences present on the array; and detecting and quantifying signals present on specific locations on the array; wherein the intensities of the signals in specific locations allows identification of the organisms.
Owner:EPPENDORF ARRAY TECH SA
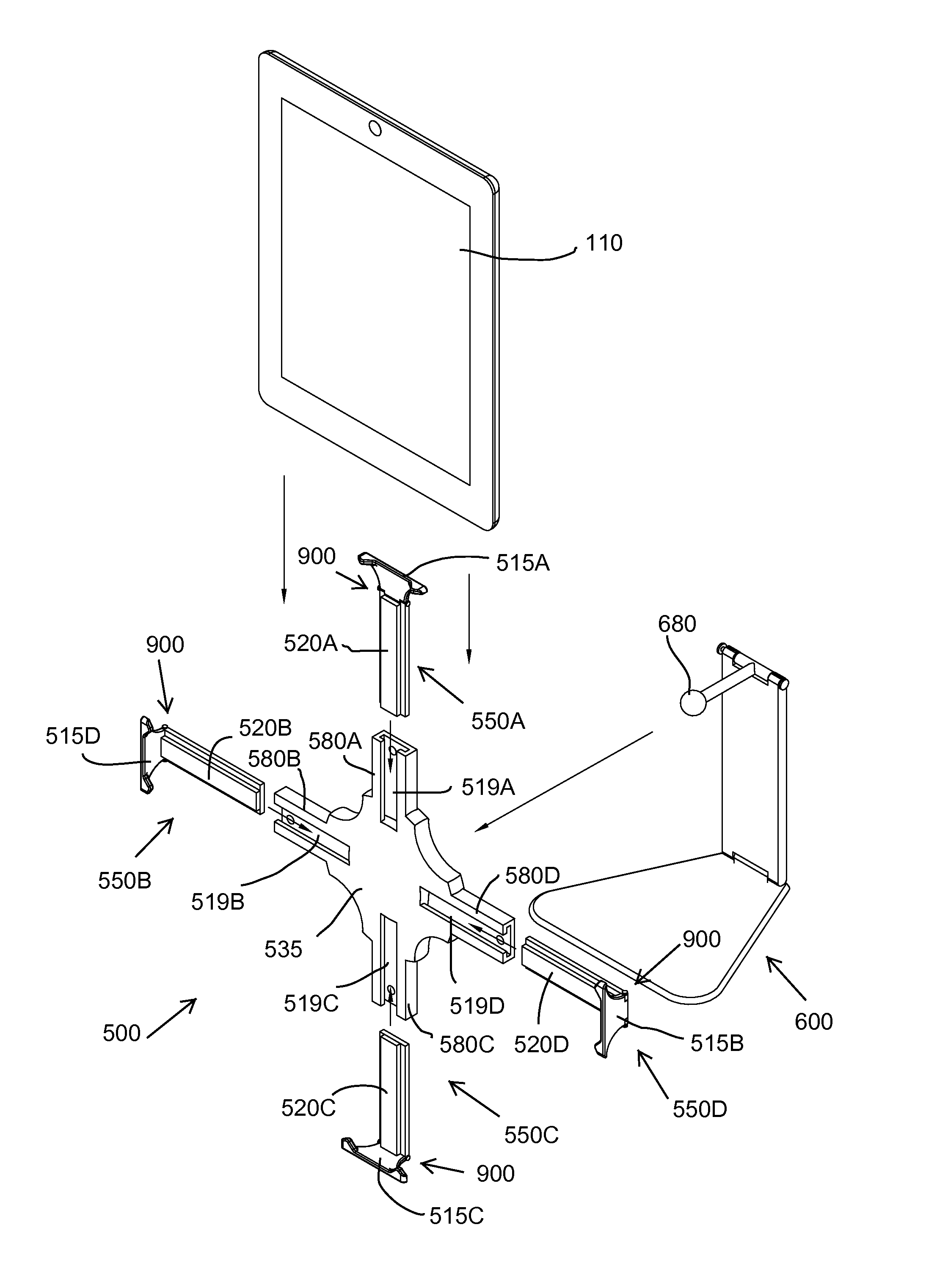
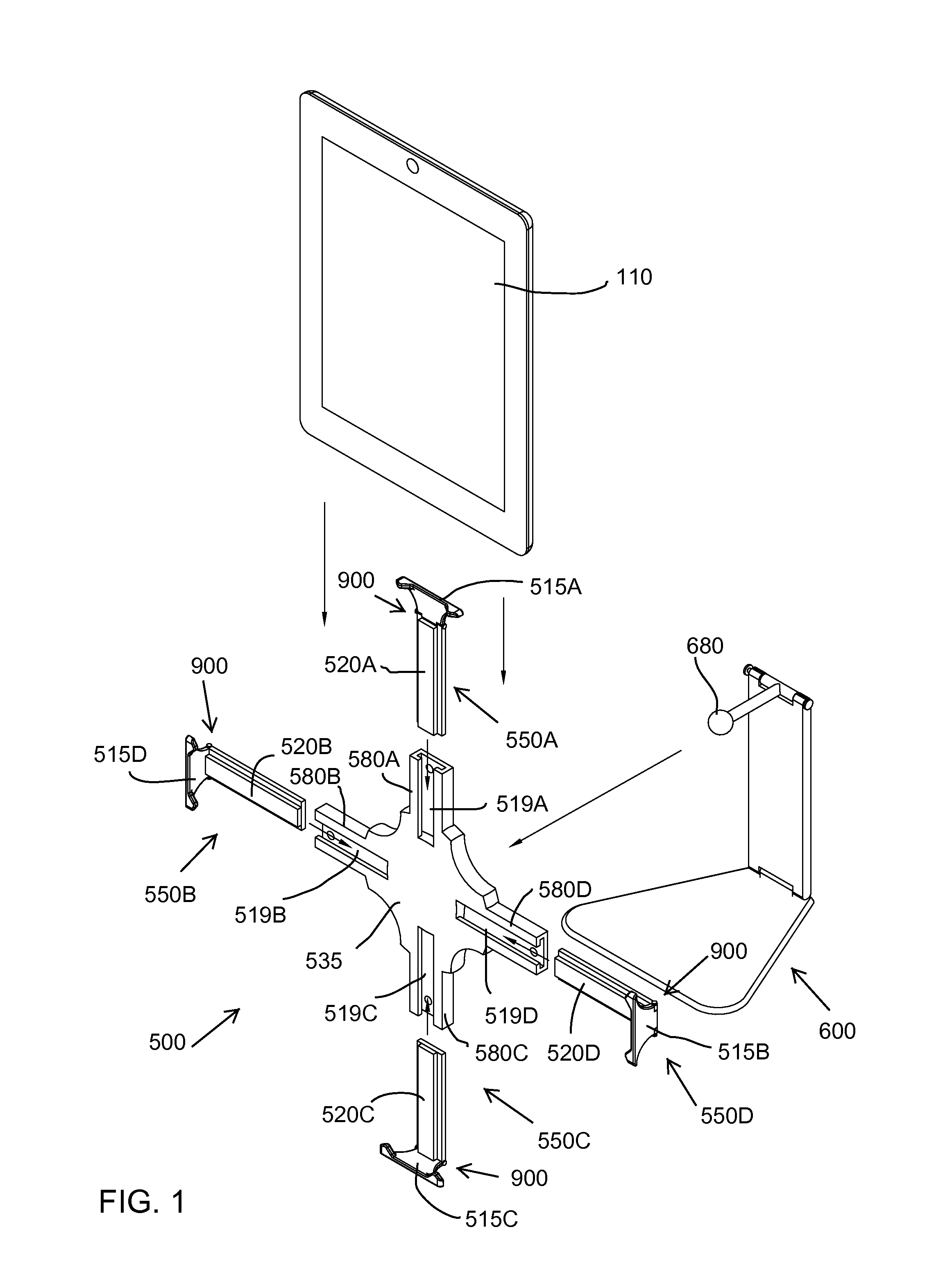
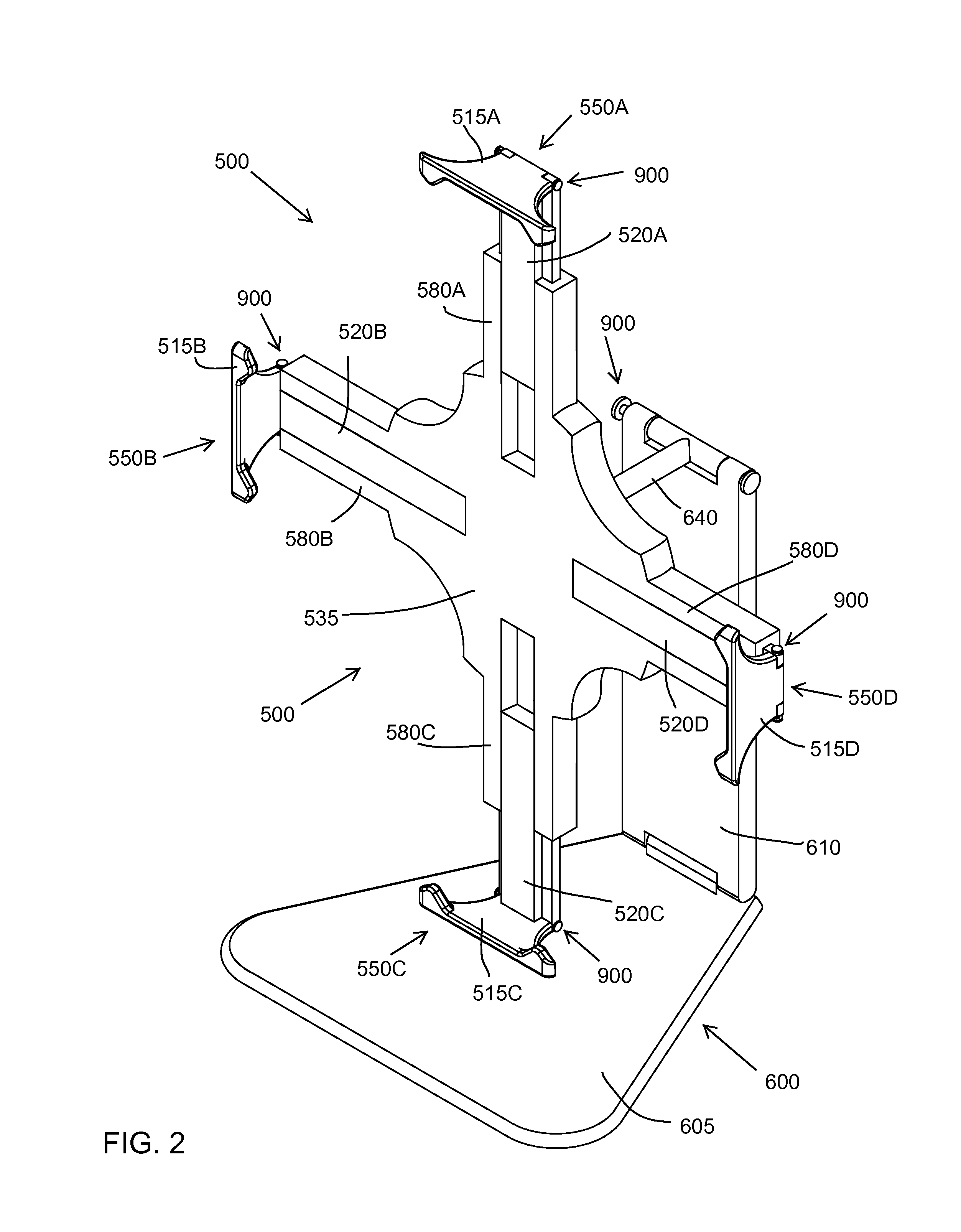
Quick-release universal tablet holder for desktops and floor stands
InactiveUS9568141B1Quick and efficientImprove experienceBed-tablesStands/trestlesCouplingLocking mechanism
A tablet holder including: a mounting plate including a back surface with a coupling for a support device and a front surface with slots configured to receive arms; the arms are slidably mounted in the slots and each include an end extending outward from the mounting plate; a locking mechanism releasably securing each of the arms to the mounting plate; and for each arm, a swiveling ledge connected by a locking hinge to the end of the arm, wherein the hinge includes a releasable locking mechanism.
Owner:INTEGRATED TECH
Ocular therapeutic agent delivery devices and methods for making and using such devices
ActiveUS7658364B2Increase release rateReached efficientlyOrganic active ingredientsSenses disorderDual modeOcular implant
Owner:UNITED STATES OF AMERICA
Network supporting roaming, sleeping terminals
InactiveUS7826818B2Highly versatileConfiguration choice is simplifiedEnergy efficient ICTCurrent supply arrangementsCommunications systemTransceiver
Improved apparatus for a radio communication system having a multiplicity of mobile transceiver units selectively in communication with a plurality of base transceiver units which, in turn, communicate with one or more host computers for storage and manipulation of data collected by bar code scanners or other collection means associated with the mobile transceiver units. A network controller and an adapter which has a simulcast and sequential mode provide selective interface between host computers and base transceivers. A scheme for routing data through the communication system is also disclosed wherein the intermediate base stations are organized into an optimal spanning-tree network to control the routing of data to and from the RF terminals and the host computer efficiently and dynamically. Additionally, redundant network and communication protocol is disclosed wherein the network utilizes a polling communication protocol which, under heavy loaded conditions, requires that a roaming terminal wishing to initiate communication must first determine that the channel is truly clear by listing for an entire interpoll gap time. In a further embodiment, a criterion used by the roaming terminals for attaching to a given base station reduces conflicts in the overlapping RF regions of adjacent base stations.
Owner:INNOVATIO IP VENTURES
Low-power messaging in a network supporting roaming terminals
InactiveUS7558557B1Highly versatileConfiguration choice is simplifiedEnergy efficient ICTCurrent supply arrangementsTransceiverSleep state
A data collection network supporting roaming terminals which may enter and exit a sleep mode to conserve power. A plurality of base stations each periodically transmit pending message indications to those roaming data collection terminals within range. Instead of requiring the transceiver within each terminal to always be active, the receiver is activated in synchrony with the transmissions of pending message indications from at least one of the base stations. Otherwise, the terminal may deactivate the transceiver (i.e., place it in a sleep state). The terminal may also sleep through pluralities of transmissions of pending message indications, and, thereafter, awake in synchrony to receive subsequent pending message indications. To minimize noise, the terminal may operate using a lower frequency system clock when not collecting data. When collecting data, especially where processing time proves critical, the system clock frequency is increased.
Owner:INNOVATIO IP VENTURES
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com