Patents
Literature
154 results about "Cellular Debris" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Cellular debris (uncountable) Organic waste left over after a cell dies by undergoing apoptosis or lysis. It is a natural waste product in animals, and in some cases is cleared away by the immune system.
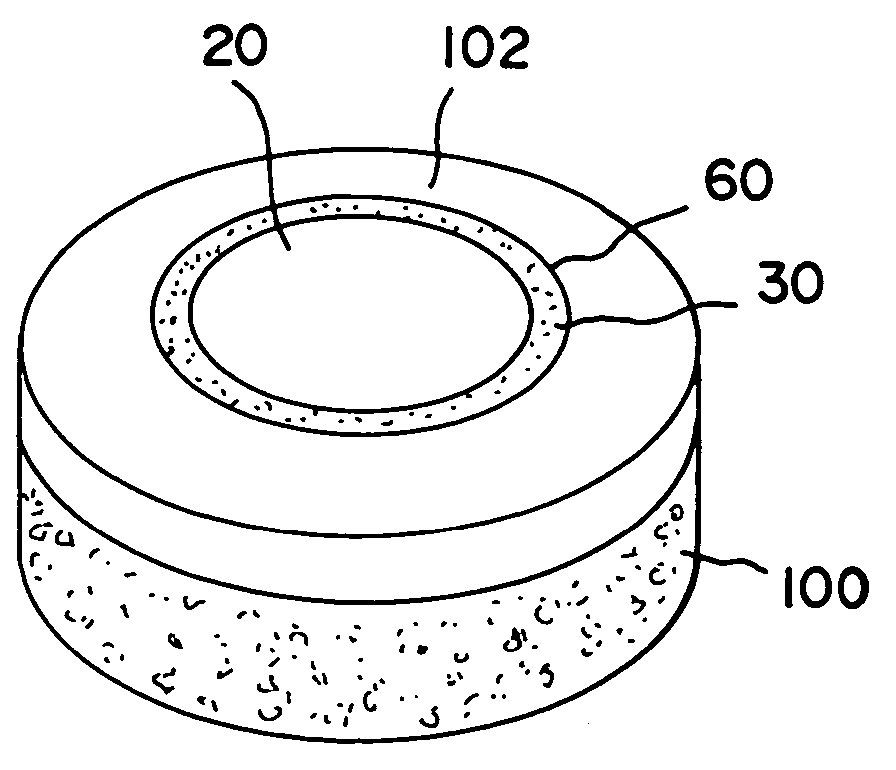
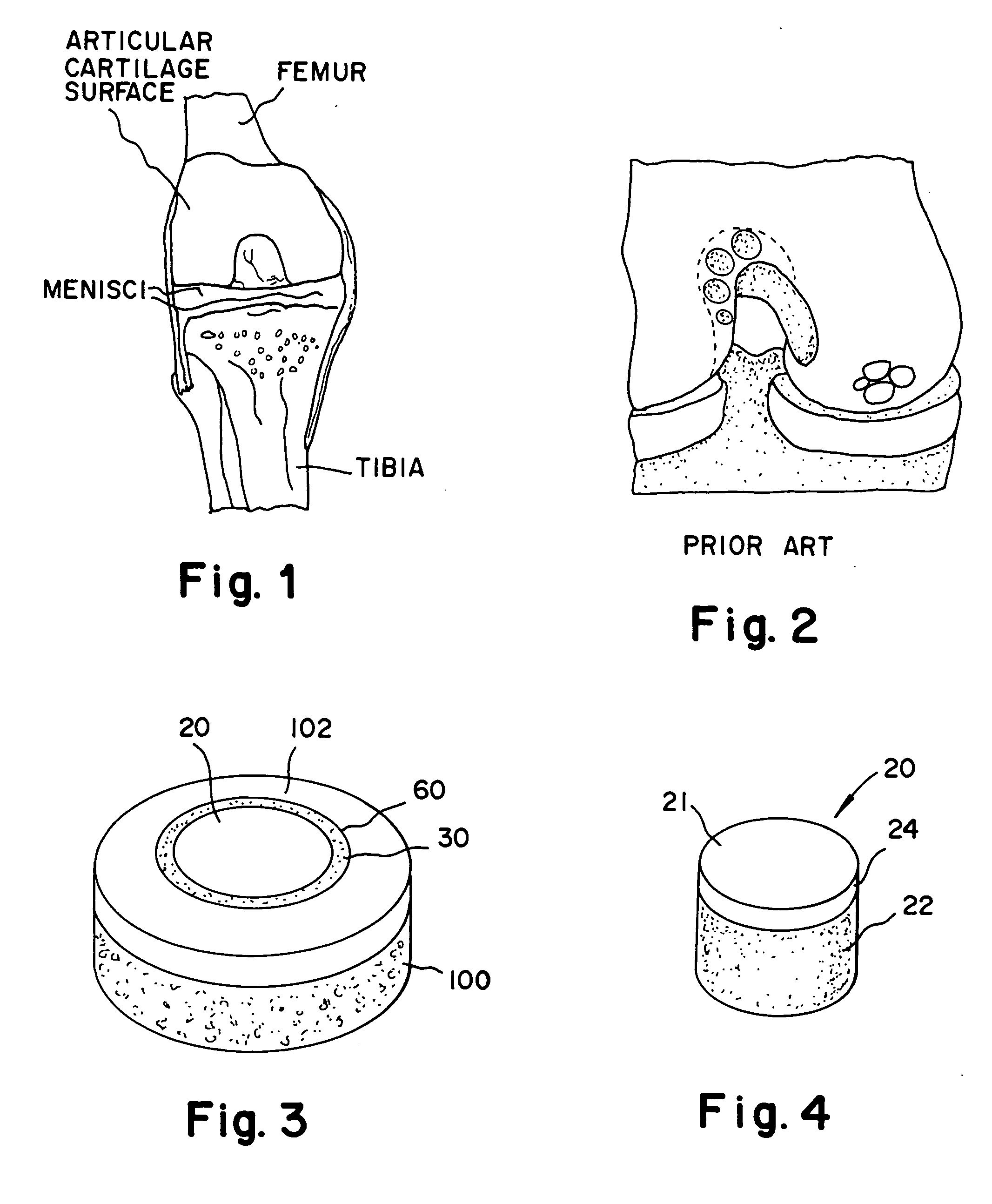
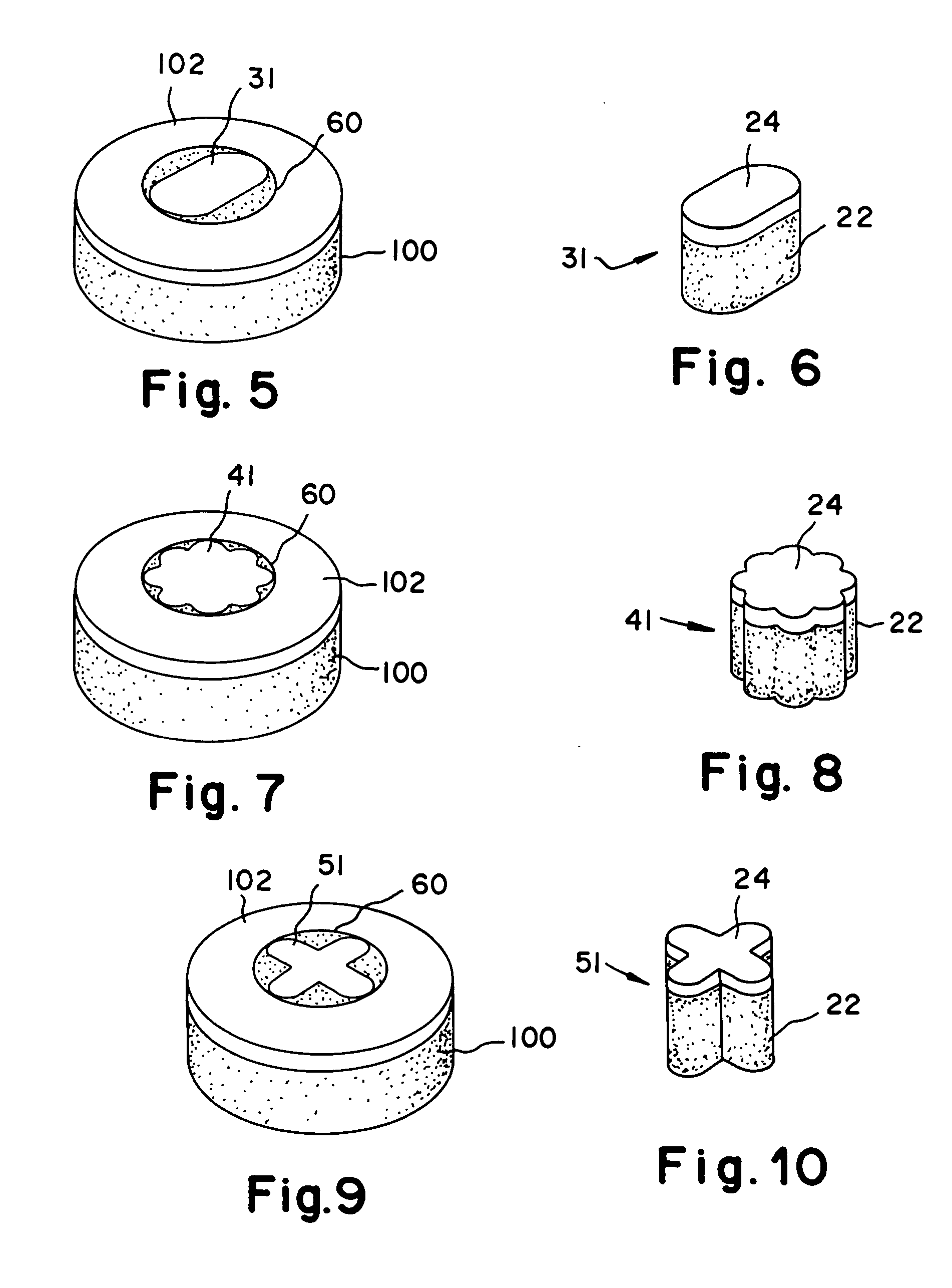
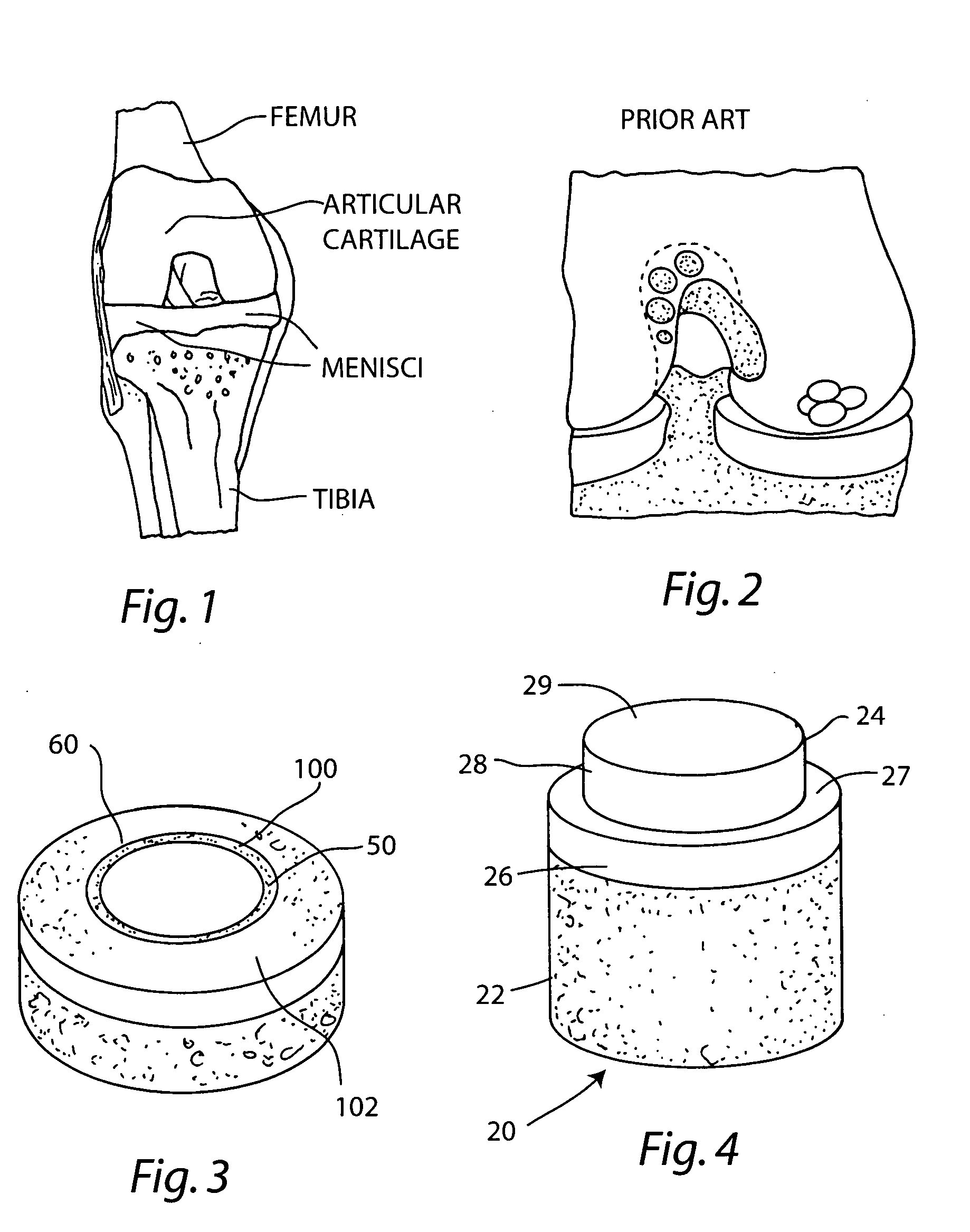
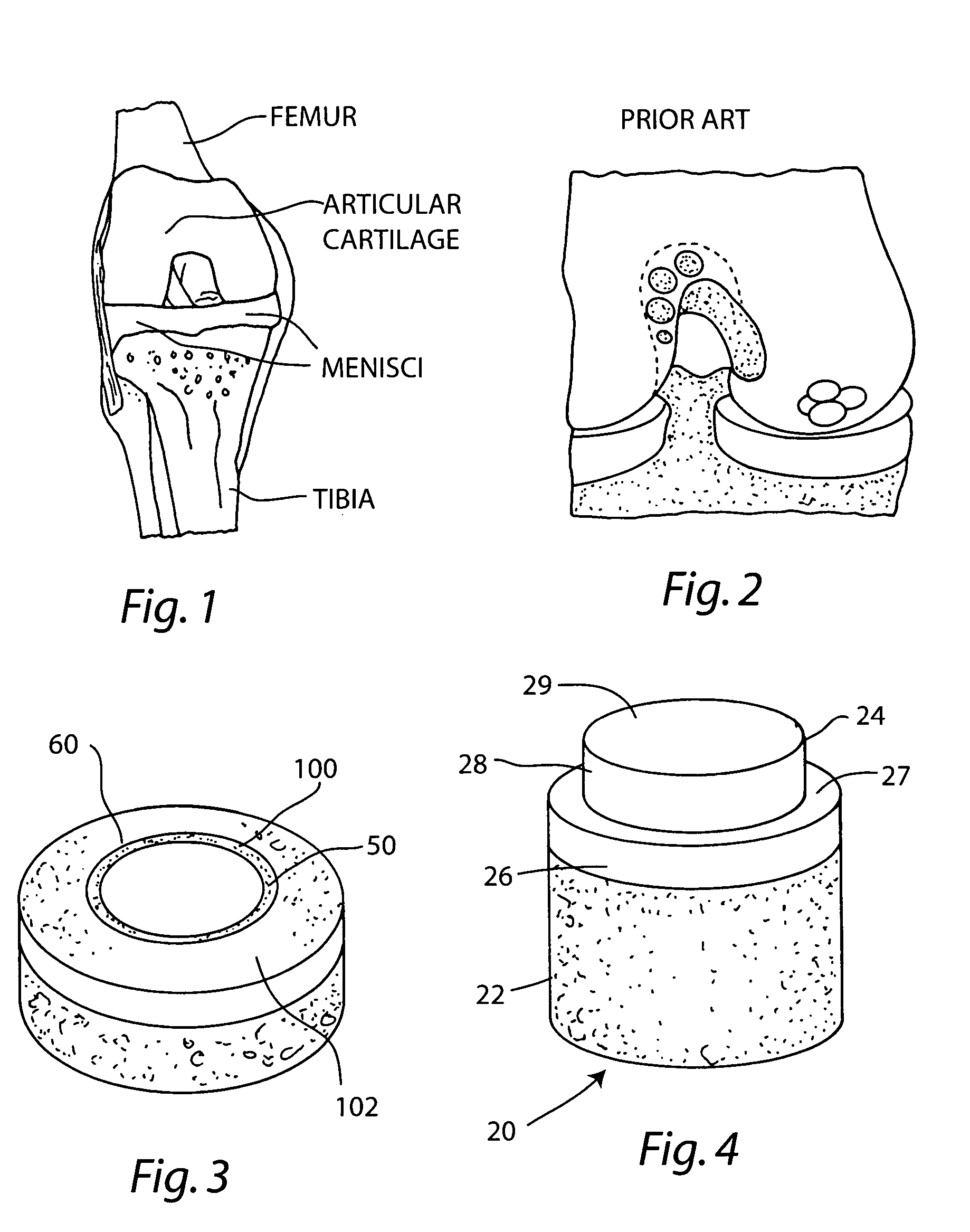
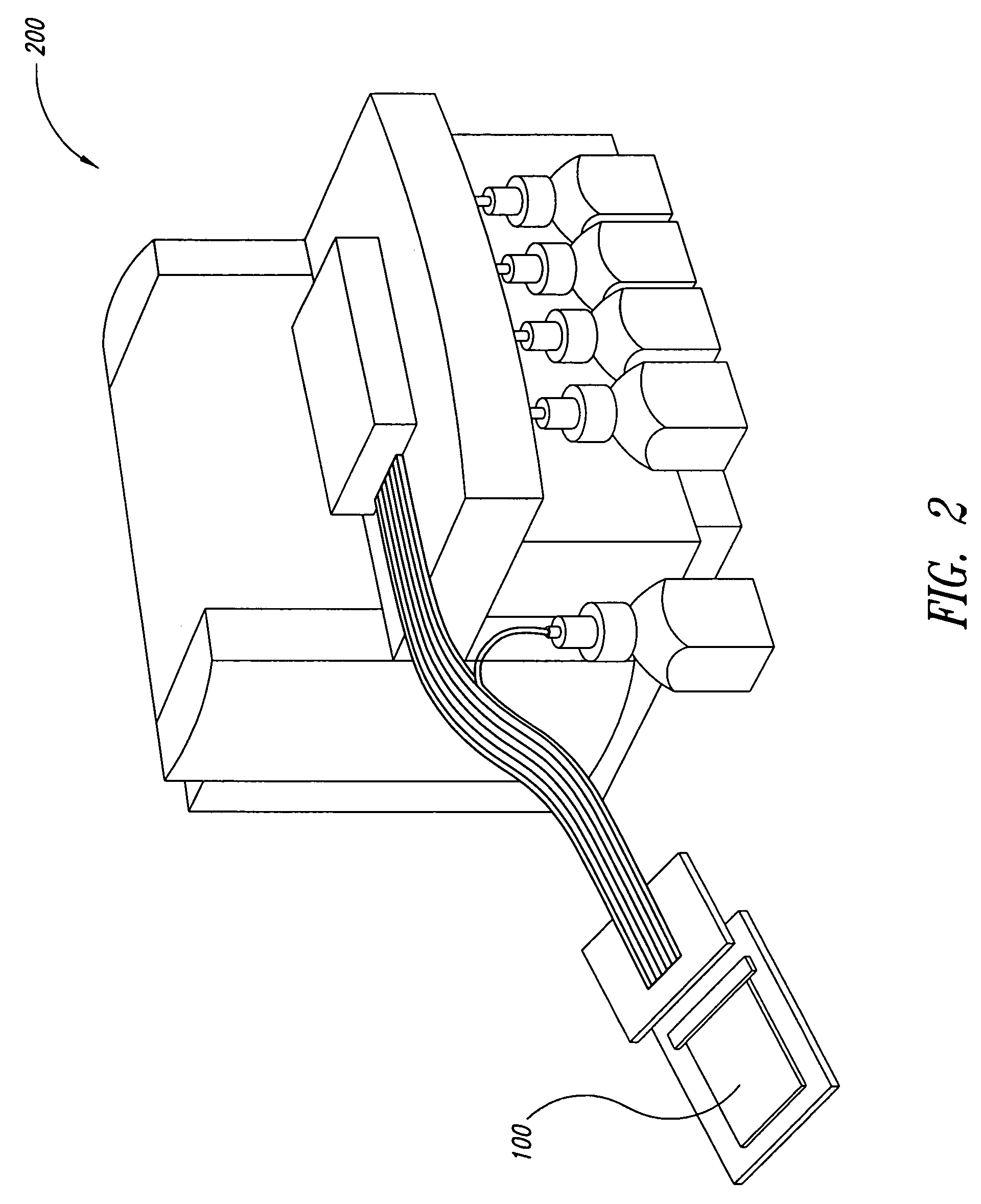
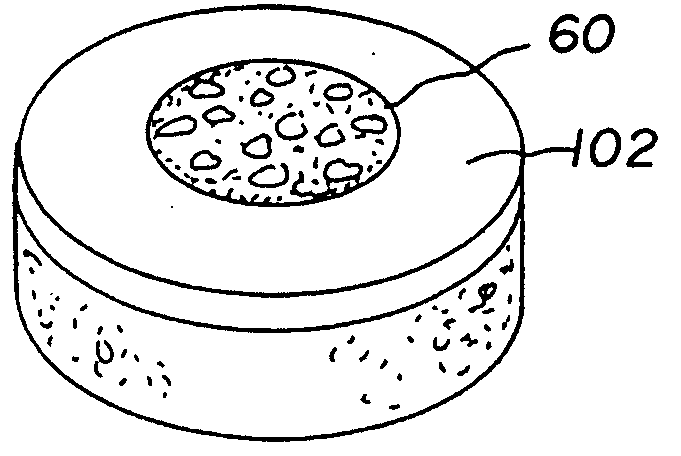
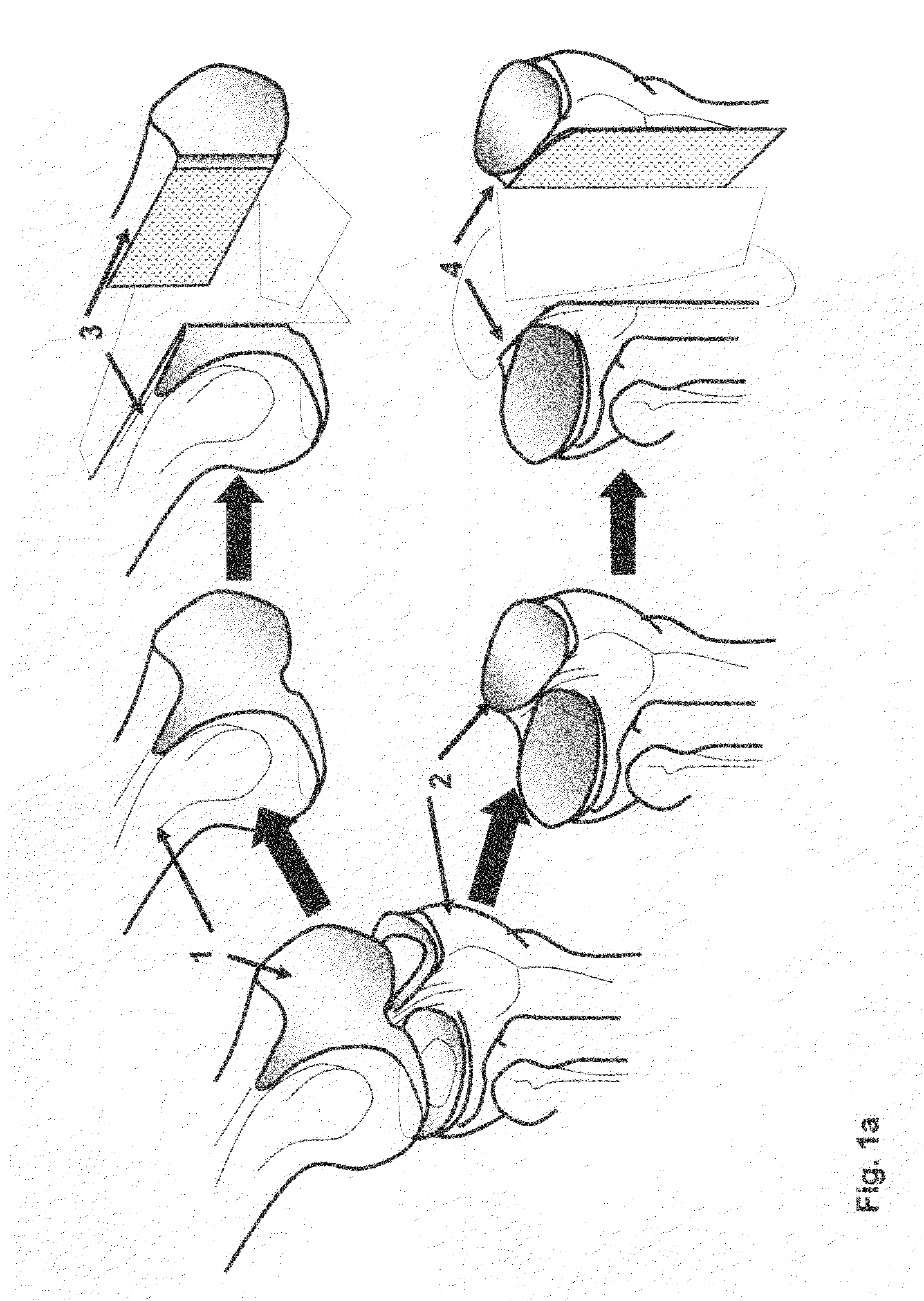
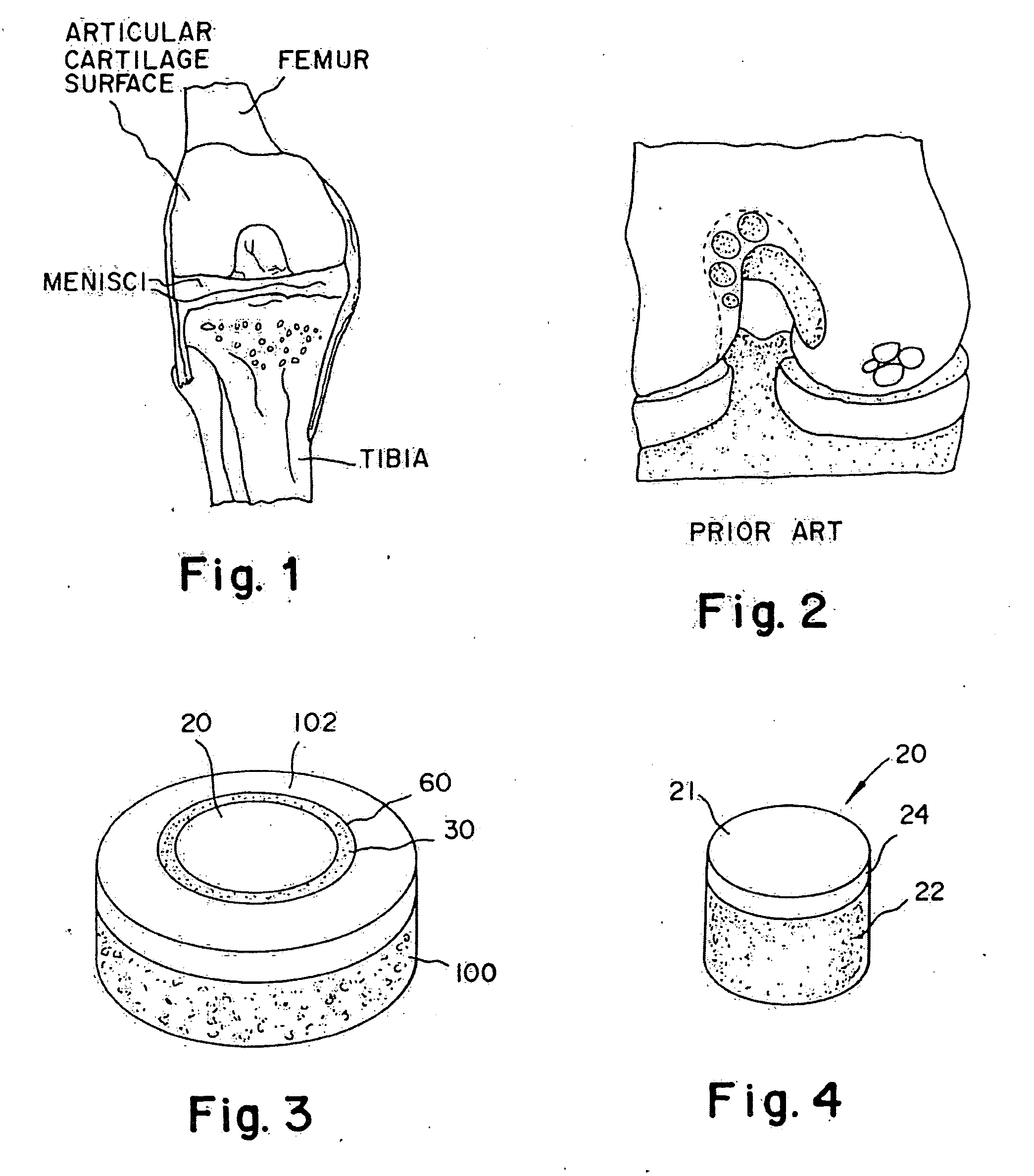
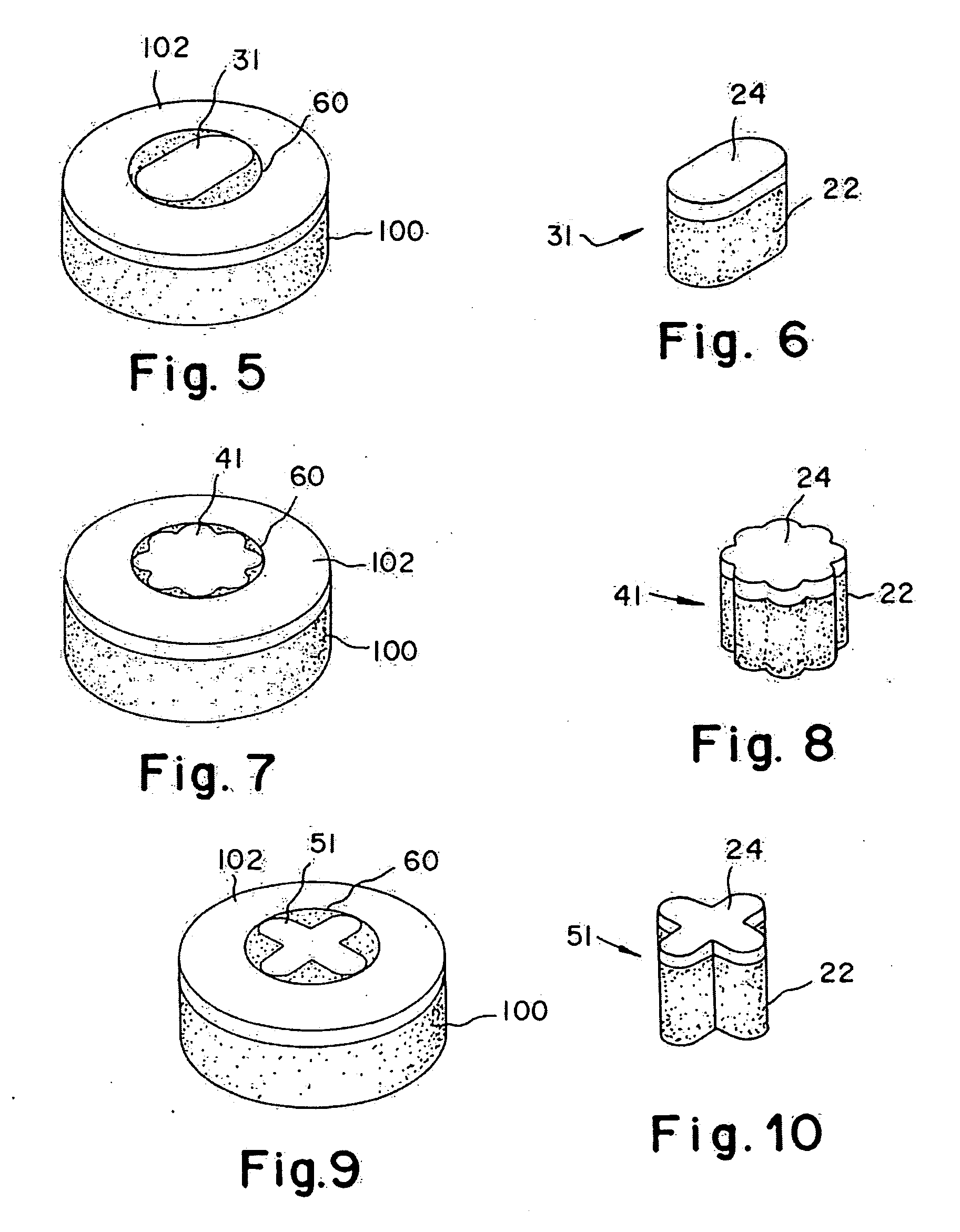
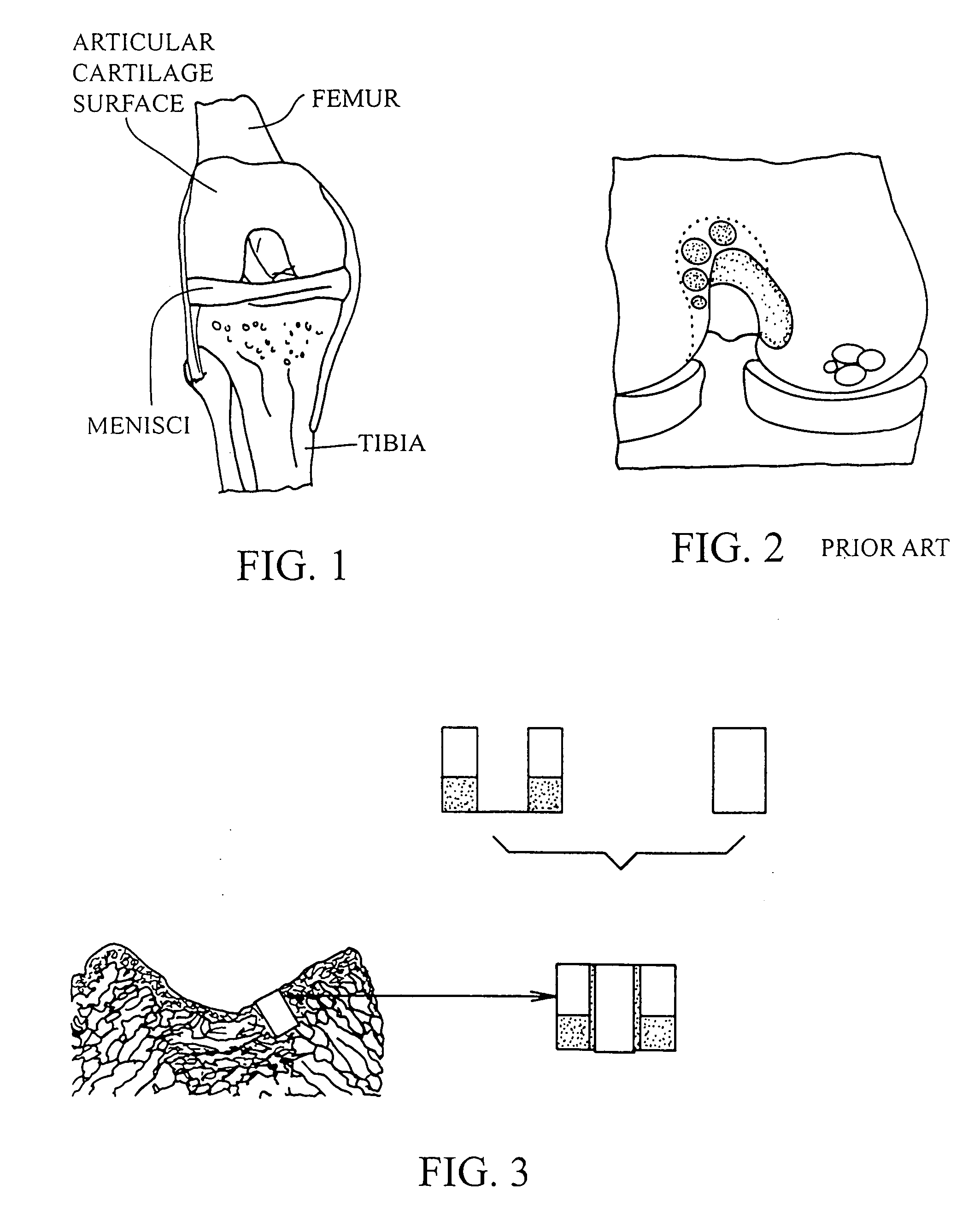
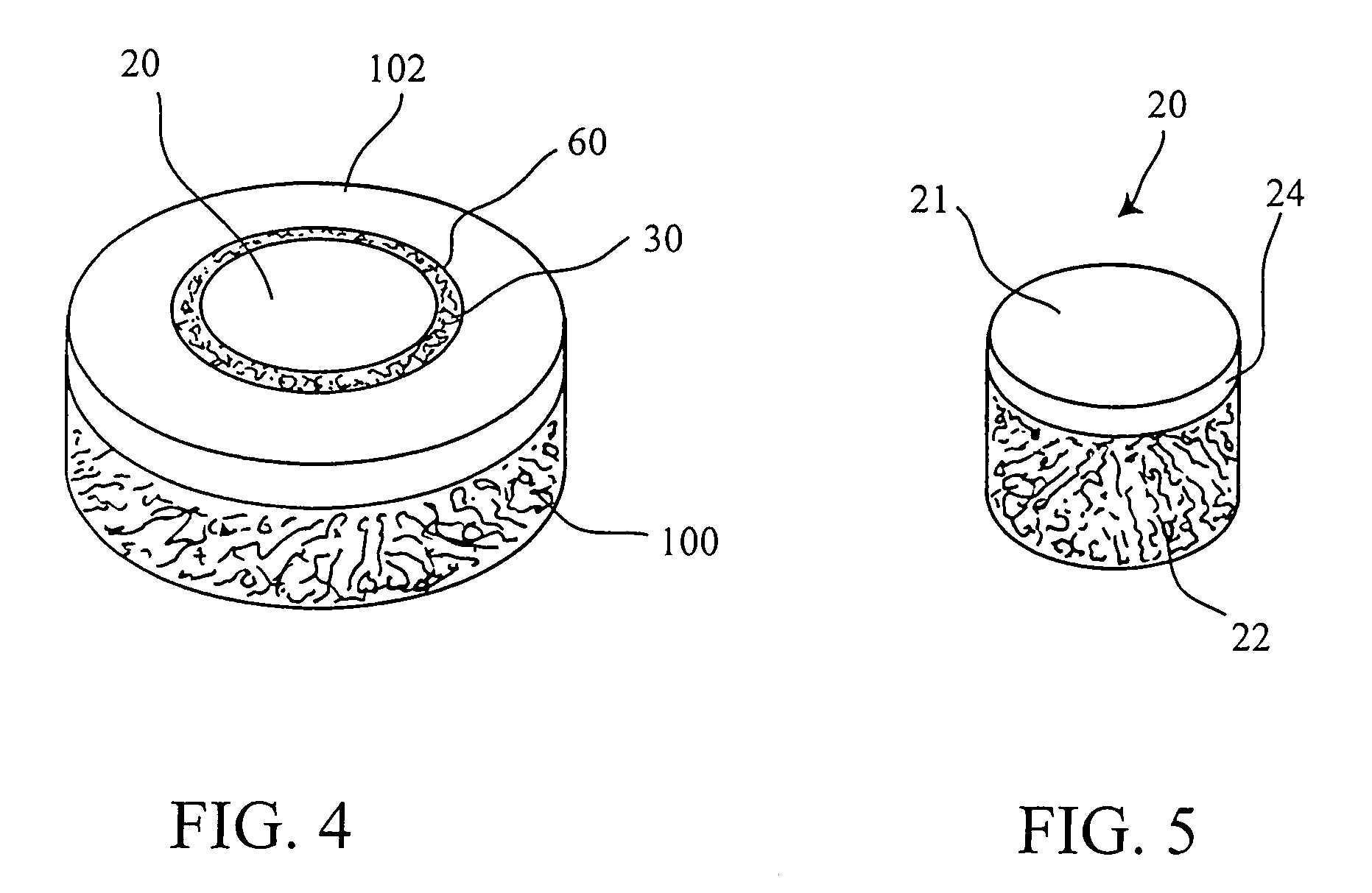
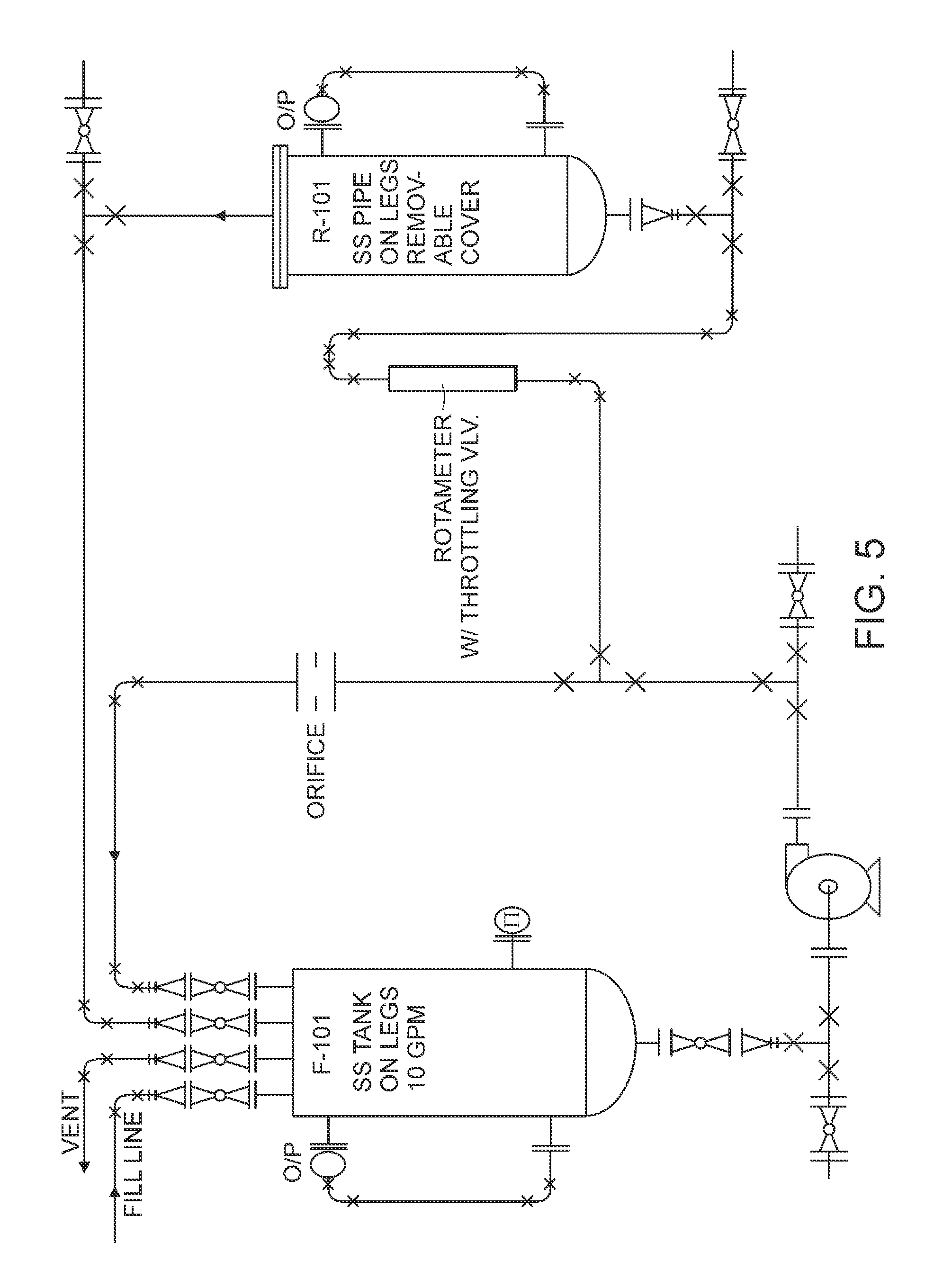
Cartilage implant assembly and method for implantation
InactiveUS20050222687A1Recovery functionRelief the painBone implantSurgerySubchondral boneInterference fit
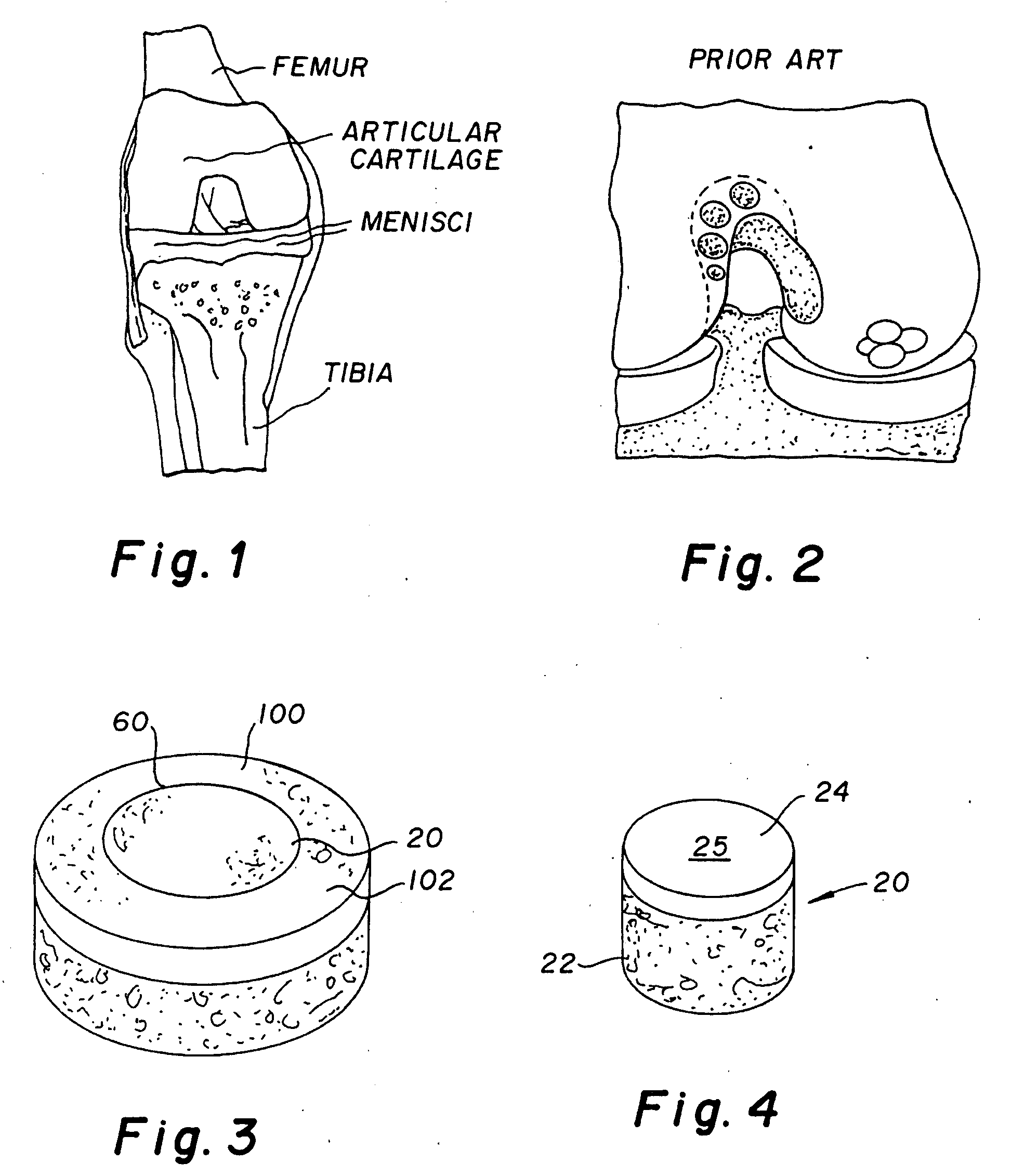
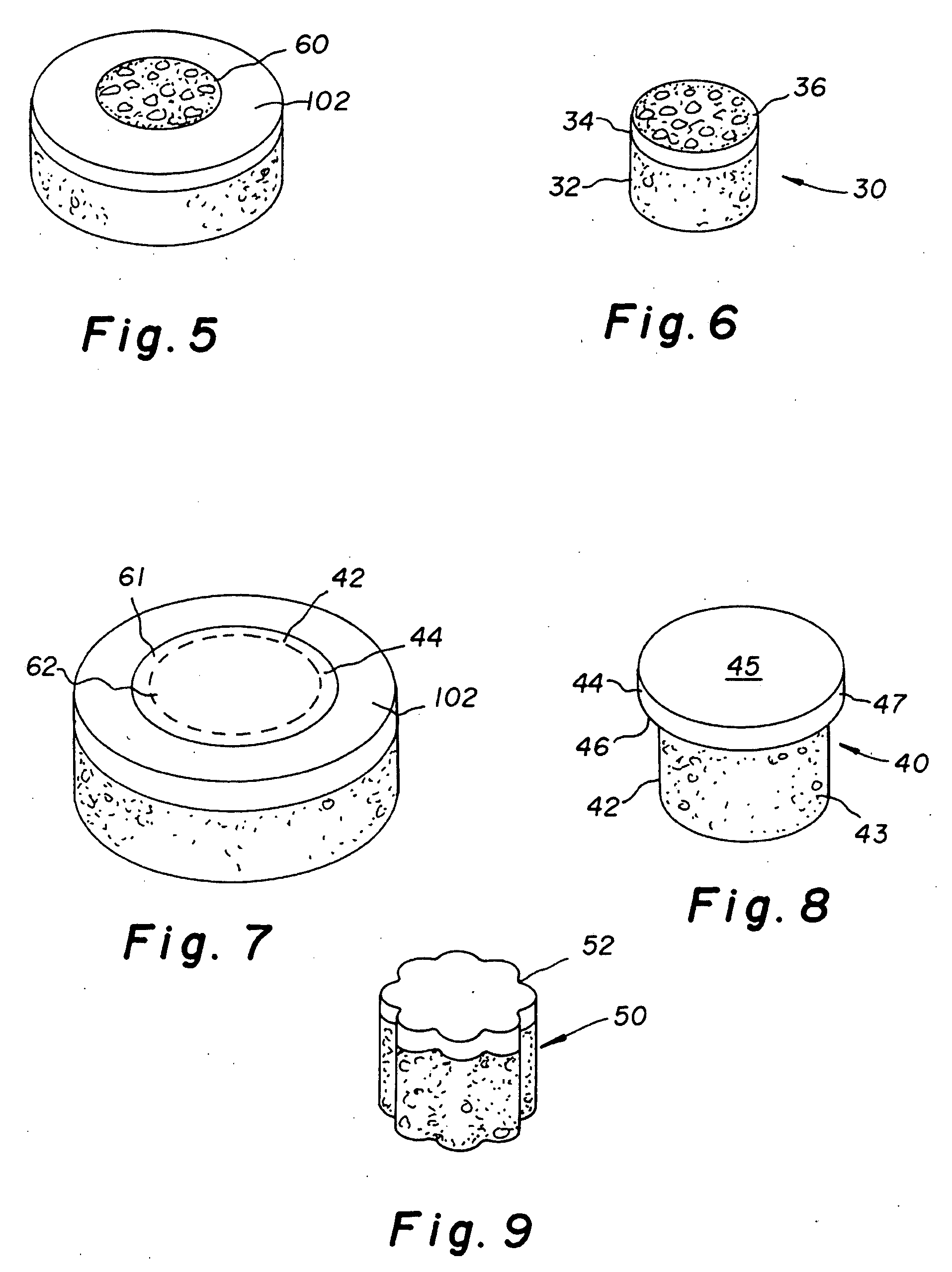
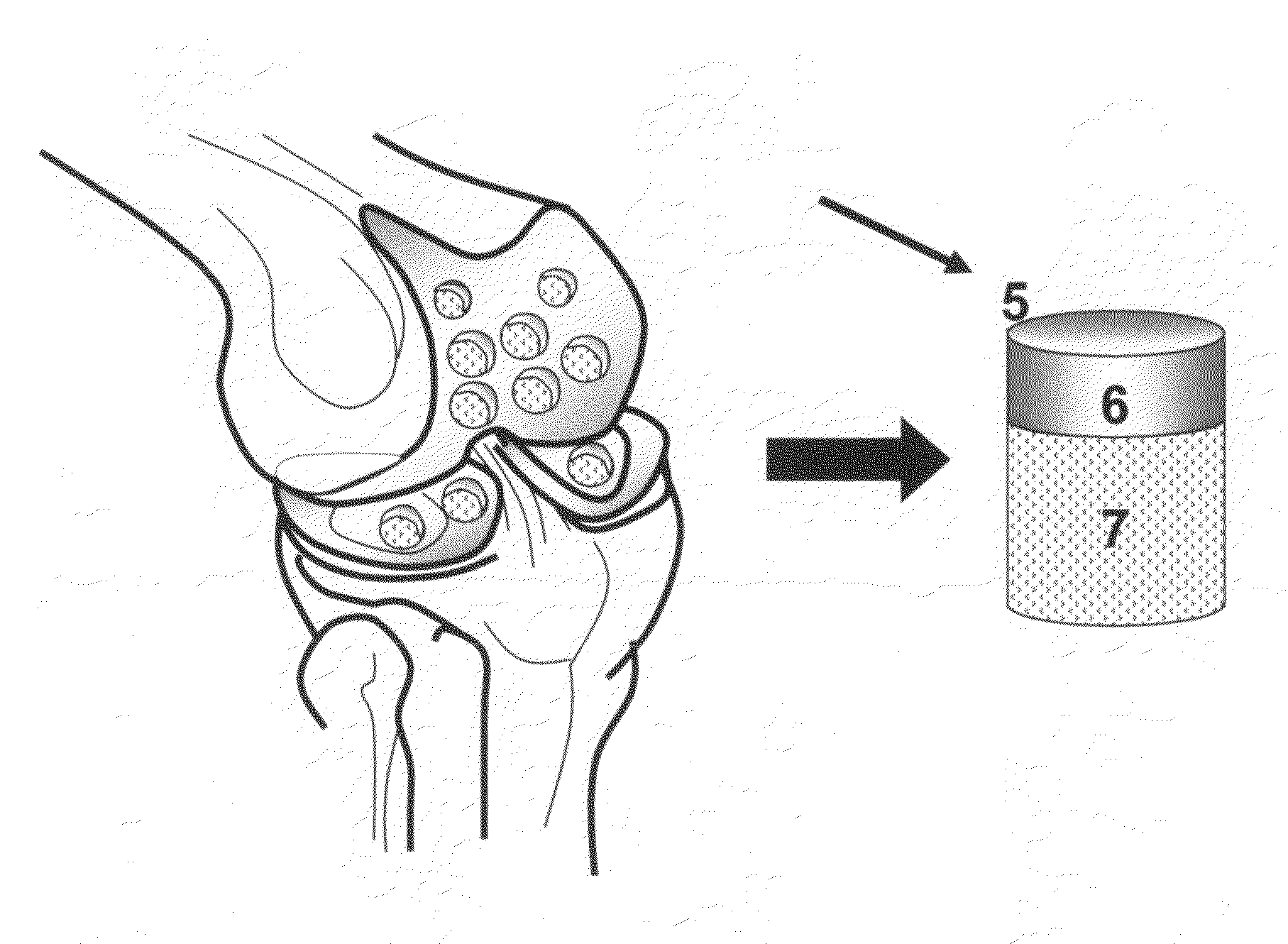
The invention is directed toward a cartilage repair assembly comprising a shaped structure of subchondral bone with an integral overlying cartilage cap which is treated to remove cellular debris and proteoglycans and milled cartilage in a bioabsorbable carrier. The shaped structure is dimensioned to fit in a drilled bore in a cartilage defect area so that said shaped bone and cartilage cap when centered in the bore does not engage the side wall of the bore in an interference fit and is surrounded by milled cartilage and carrier. A method for inserting the assembly into a cartilage defect area is disclosed.
Owner:MUSCULOSKELETAL TRANSPLANT FOUND INC +1
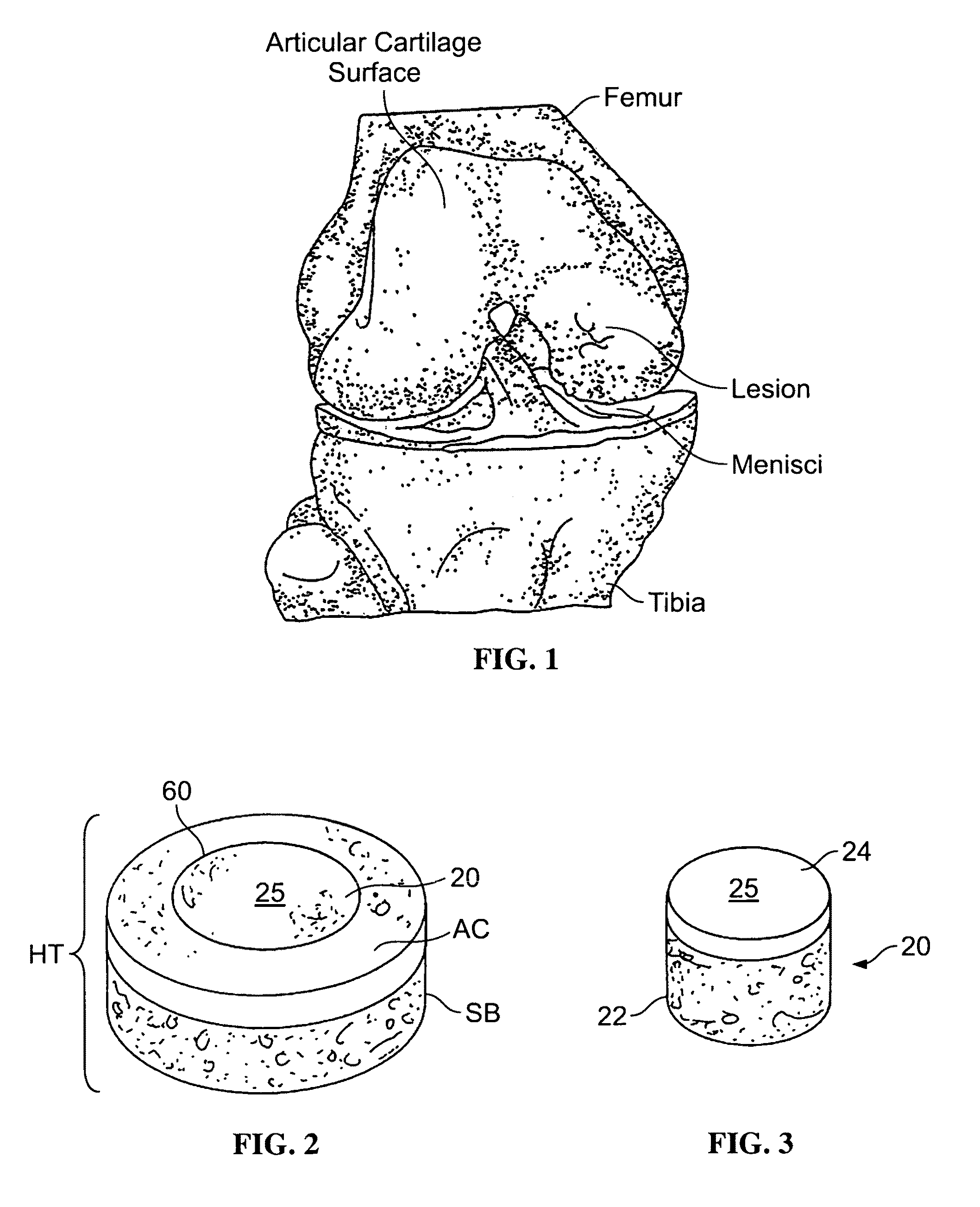
Cartilage allograft plug
InactiveUS20050251268A1Increase chondrocyte migrationIncreased proliferationBone implantLigamentsInterference fitSubchondral bone
The invention is directed toward a cartilage repair assembly comprising a cylindrically shaped allograft structure of subchondral bone with an integral overlying smaller diameter cartilage cap which is treated to remove cellular debris and proteoglycans. The shaped structure is dimensioned to fit in a drilled bore in a cartilage defect area so that the subchondral bone of the structure engages the side wall of the bone portion of the drilled bore in an interference fit while the cartilage cap is spaced from cartilage portion of the side wall of the drilled bore forming a gap in which a milled cartilage and biocompatible carrier mixture is placed allowing cell transfer throughout the defect area. A method for inserting the shaped allograft structure into a cartilage defect area is also disclosed.
Owner:MUSCULOSKELETAL TRANSPLANT FOUND INC
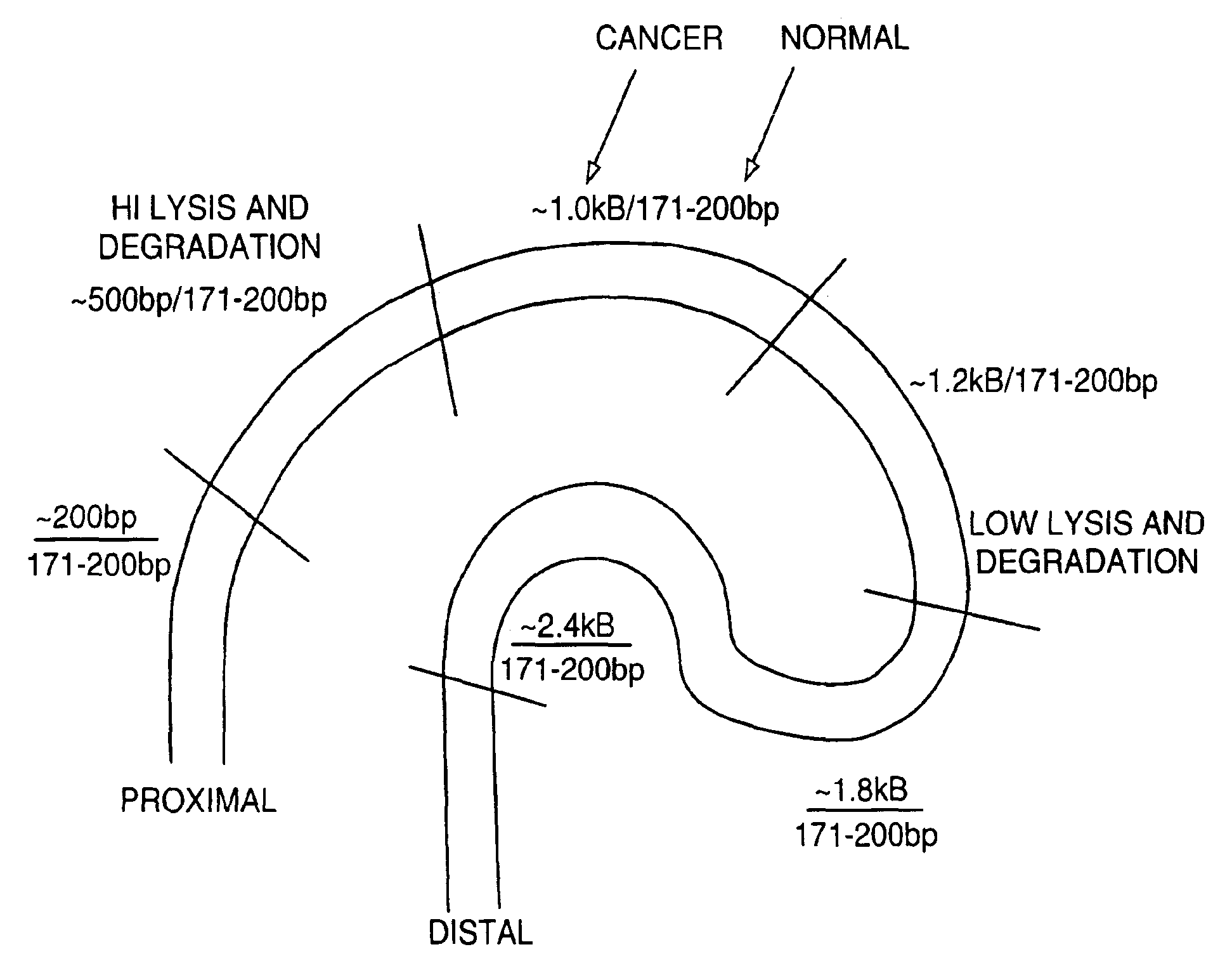
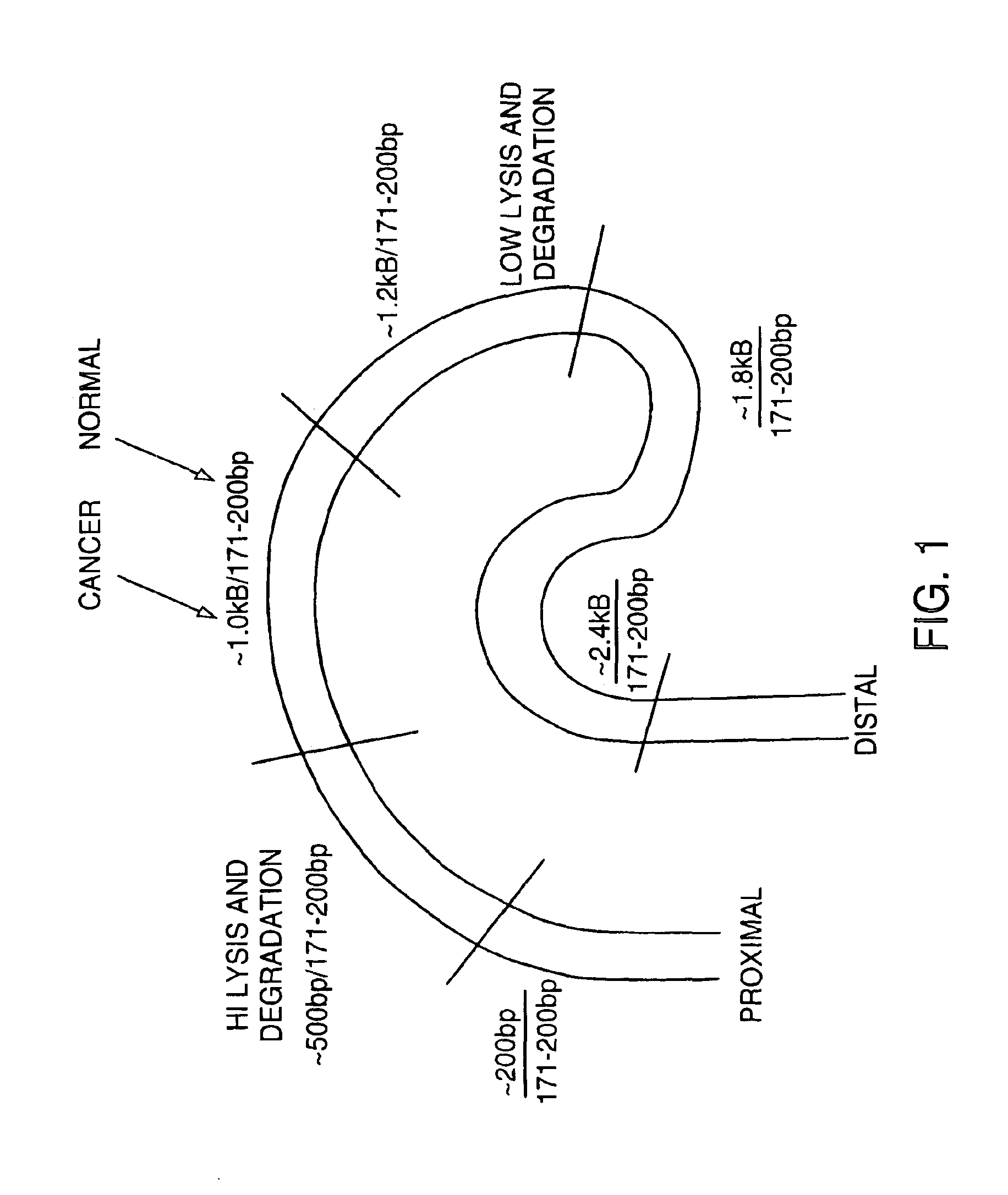
Methods for detecting nucleic acids indicative of cancer
InactiveUS6964846B1High molecular weightMicrobiological testing/measurementRecombinant DNA-technologyCellular DebrisBody fluid sample
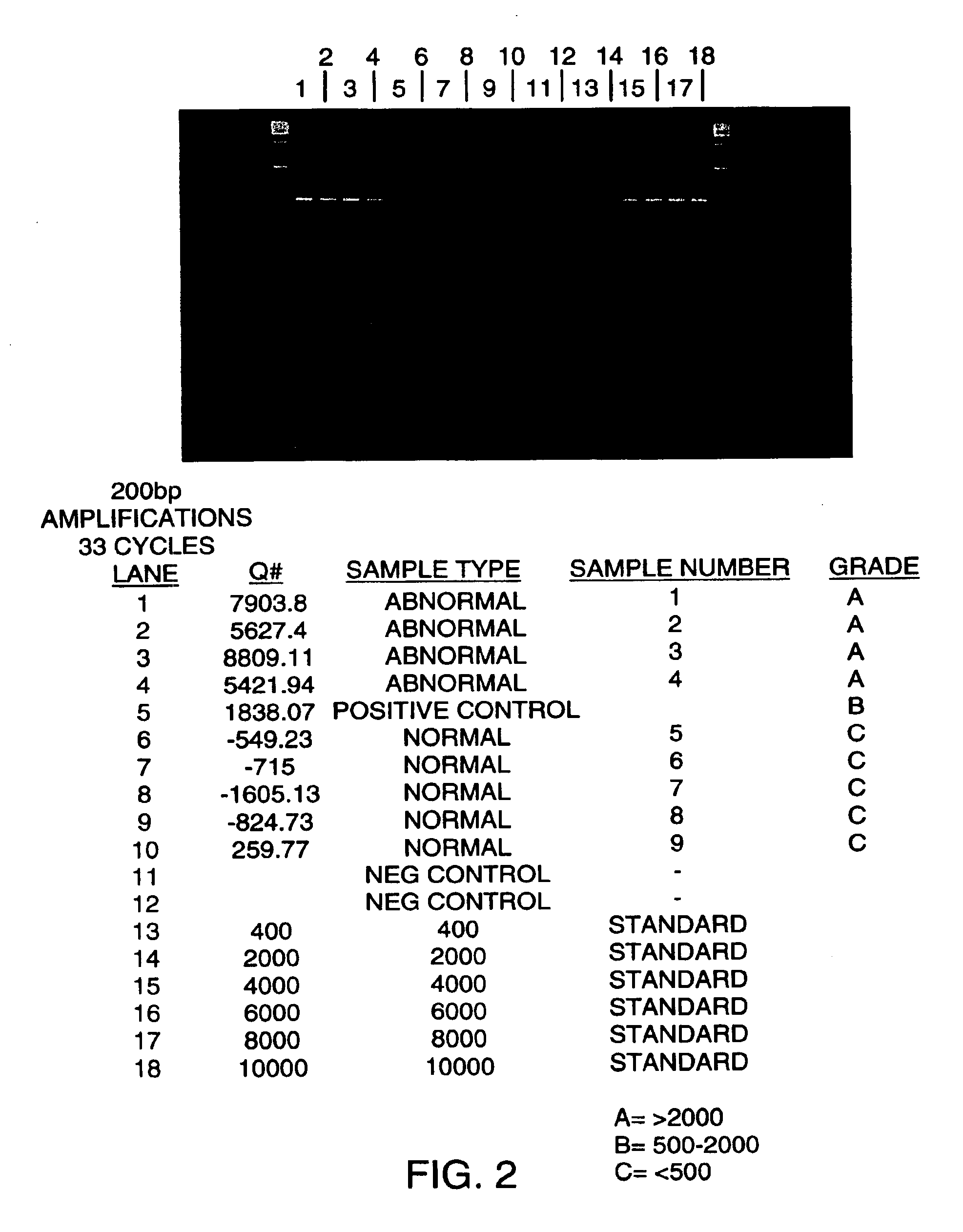
The invention provides methods for screening tissue or body fluid samples for nucleic acid indicia of cancer or precancer.Method are provided for screening a patient for cancer or precancer by detecting the presence of nucleic acid fragments that are longer than nucleic acid fragments expected to be present in a sample obtained from a healthy individual. In one embodiment, a positive screen for cancer or precancer is identified when a patient tissue or body fluid sample comprising exfoliated cells or cellular debris contains an amount of nucleic acid of a length greater than about 200 base pairs that exceeds a predetermined amount.
Owner:EXACT SCI CORP
Transdermal device for administration through de-epithelialized skin
InactiveUS6264979B1Prevent proliferationImprove adhesionAdhesive dressingsAbsorbent padsCellular DebrisActive agent
A transdermal device is described suitable for delivery of a pharmaceutical to the systemic circulation through de-epithelialized skin. In its various embodiments the device includes means to prevent the attack of any protein or polypeptide active agent included therein by proteolytic enzymes which exude from the lesion, means to prevent the ingress of bacteria and other cellular debris from the lesion and means to ensure that substantially all said active agent is directed to the de-epithelialized lesion.
Owner:SVEDMAN PAL
Method and system for microfluidic manipulation, amplification and analysis of fluids, for example, bacteria assays and antiglobulin testing
A microfluidic system for isolation and amplification of DNA or RNA from aqueous solutions and detection of the DNA or RNA on a lateral flow detection strip, including a disposable microfluidic card for use in analysis of bacteria in platelets and an analysis of sexually transmitted diseases (STD) in urine. The card will include an embedded membrane that filters out cells and cellular debris. Any biological debris on the membrane will be lysed and the DNA or RNA amplified via PCR amplification protocol, including appropriate reagents and thermal cycling conditions. The amplified DNA or RNA are transferred to a lateral flow detection strip for a visual diagnostic read out. An alternate embodiment includes a microfluidic card for use in typing antiglobulin assays.
Owner:PERKINELMER HEALTH SCIENCES INC
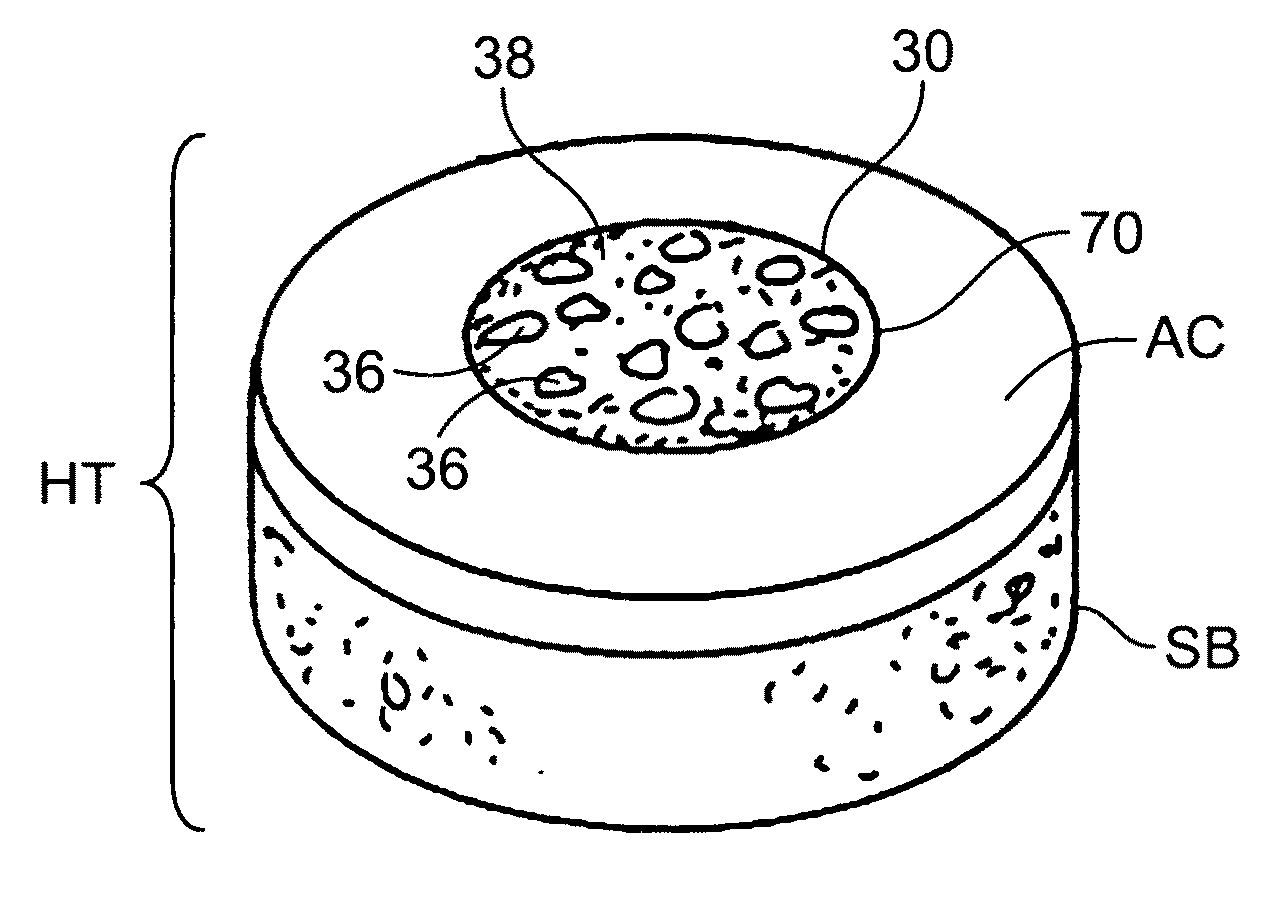
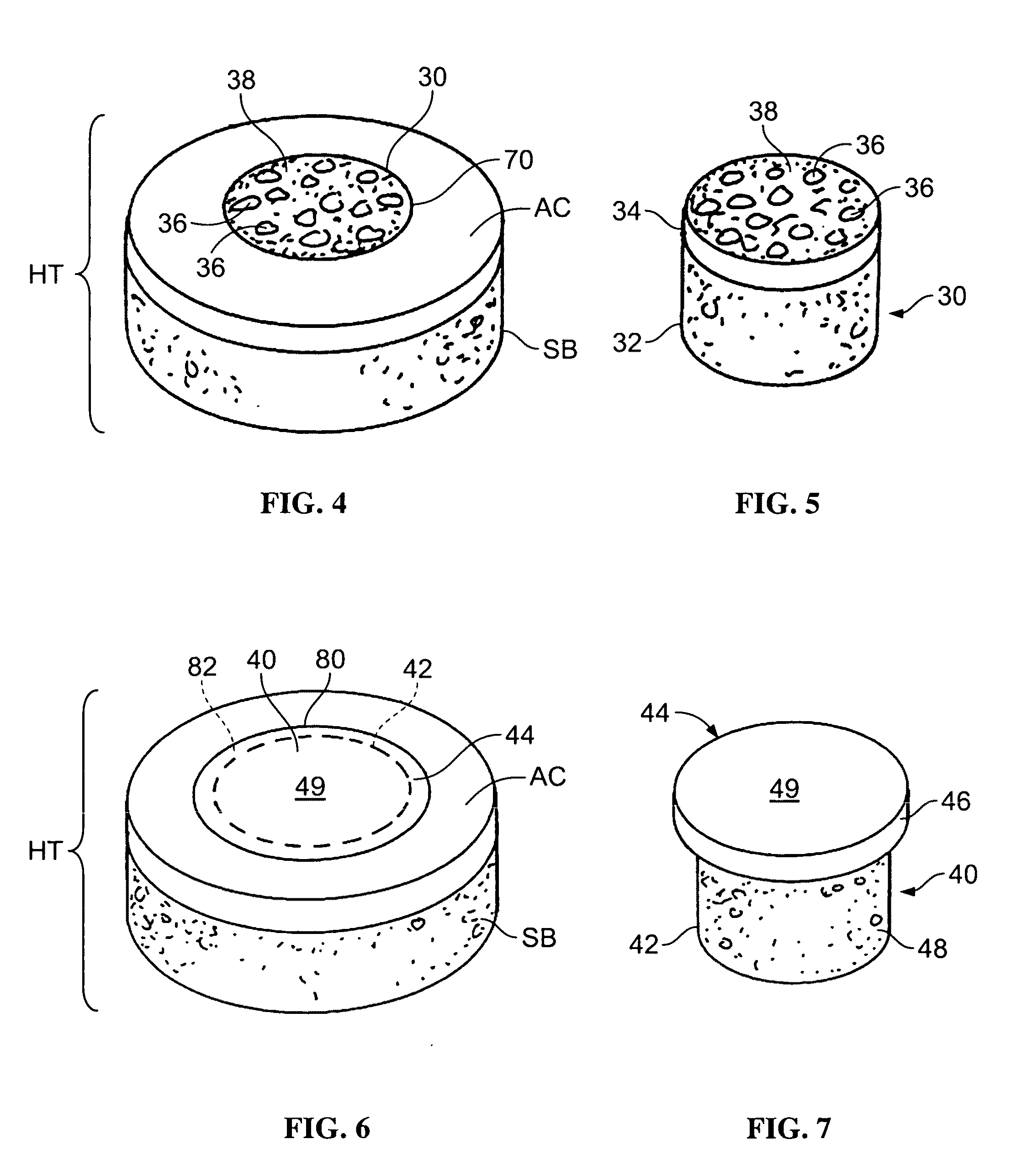
Cleaning and devitalization of cartilage
InactiveUS20080077251A1Improve recellularizationBone implantDead animal preservationCellular DebrisMedicine
The invention is further directed to producing a cleaned, disinfected, and devitalized cartilage graft by optionally cleaning and disinfecting the cartilage graft; treating the cartilage graft in a pretreatment solution; treating the cartilage graft in an extracting solution; washing the extracted cartilage graft with a rinsing solution; and subsequently soaking the devitalized cartilage graft in a storage solution. The devitalized cartilage graft is essentially free from metabolically viable and / or reproductively viable cells and the rinsing solution is hypotonic solution or isotonic solution. The present invention is further directed to a cleaned, disinfected, and devitalized cartilage graft and a process for cleaning, disinfecting, and devitalizing cartilage grafts. The invention also relates to a process for repairing a cartilage defect and implantation of a cartilage graft into a human or animal by crafting the cartilage matrix into individual grafts, disinfecting and cleaning the cartilage graft, applying a pretreatment solution to the cartilage graft, removing cellular debris using an extracting solution to produce a devitalized cartilage graft, implanting the cartilage graft into the cartilage defect with or without an insertion device, and sealing the implanted cartilage graft with recipient tissue. The devitalized cartilage graft is optionally recellularized in vitro, in vivo, or in situ with viable cells to render the tissue vital before or after the implantation. The devitalized cartilage graft is also optionally stored between the removing cellular debris and the recellularizing steps.
Owner:LIFENET HEALTH
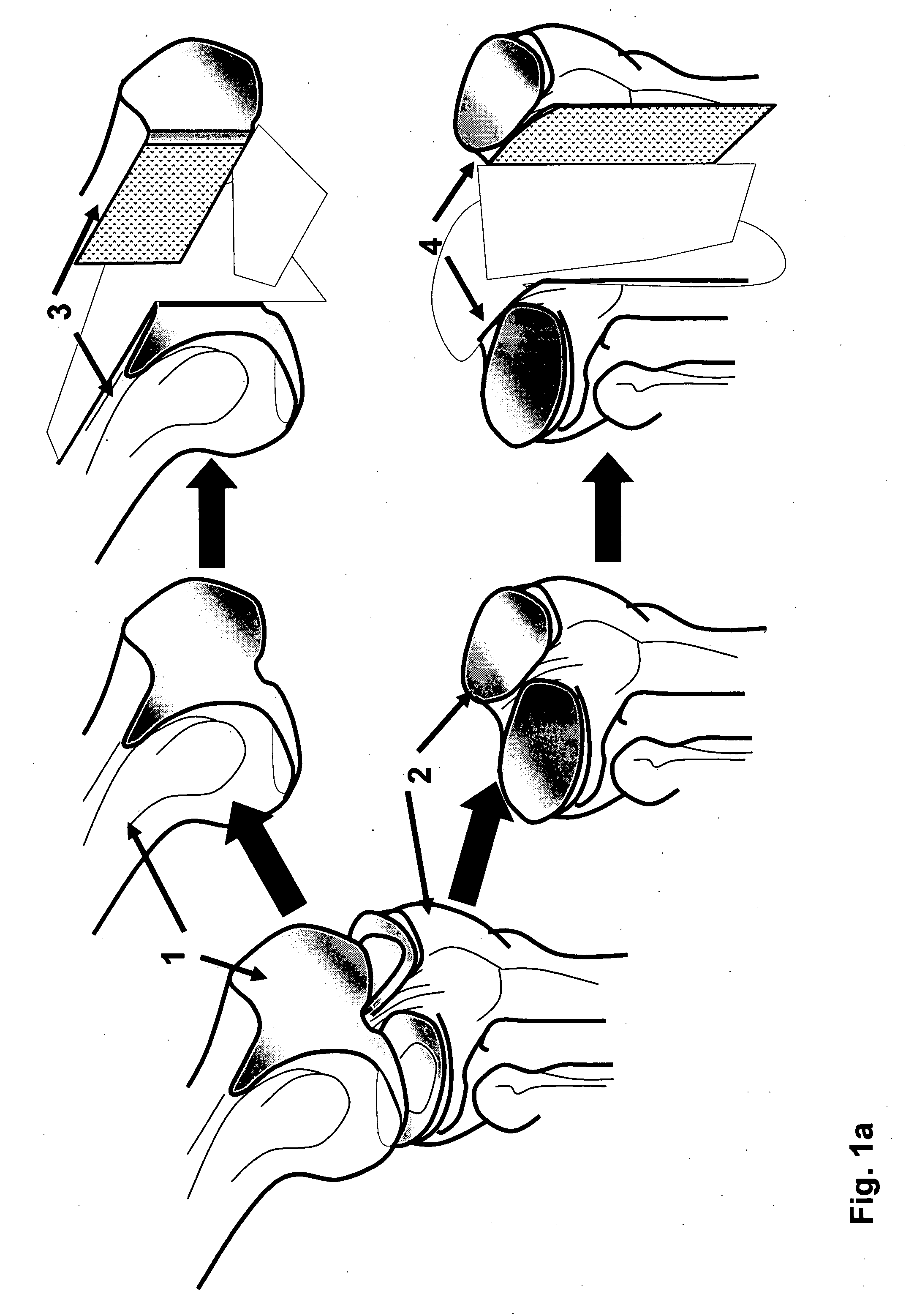
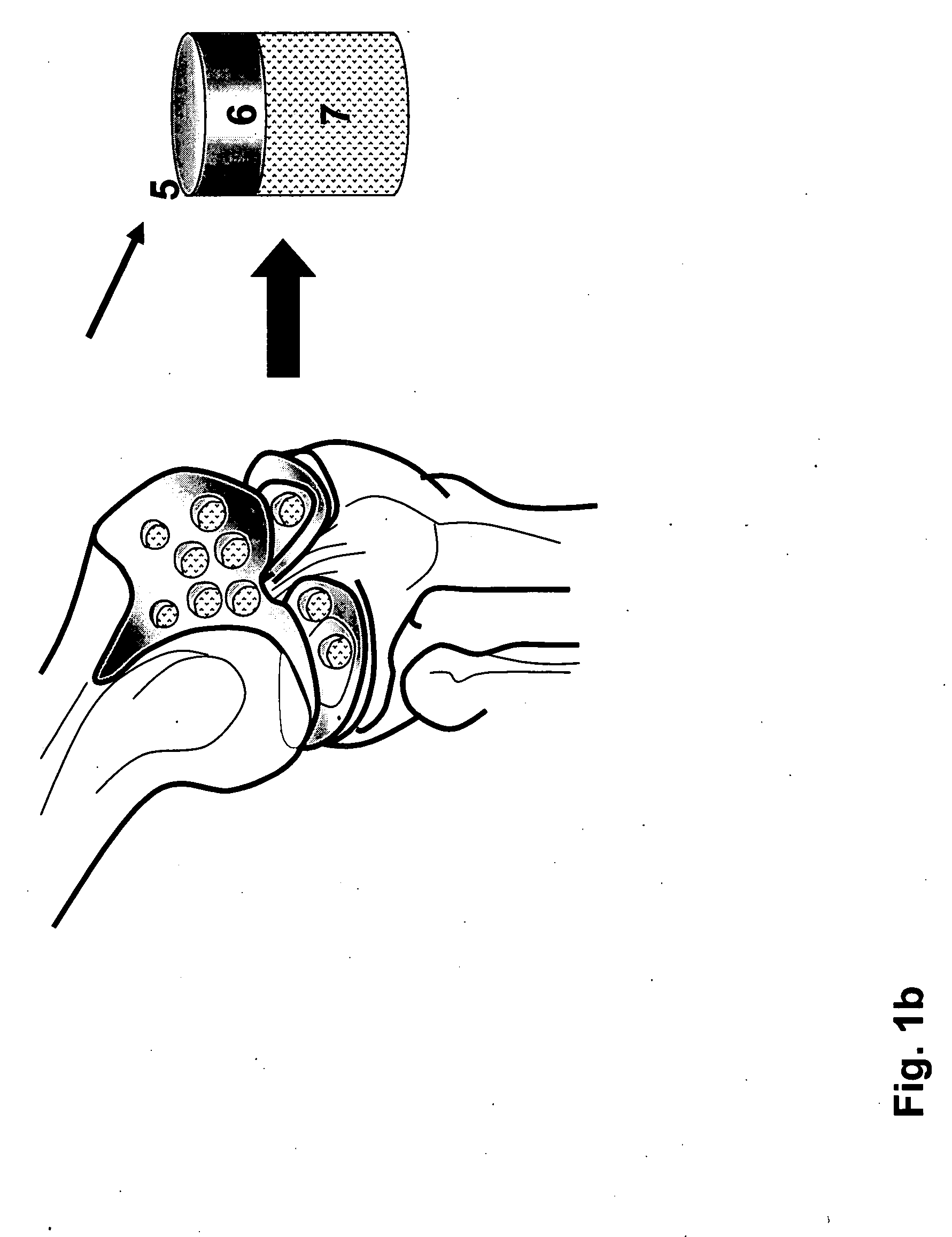
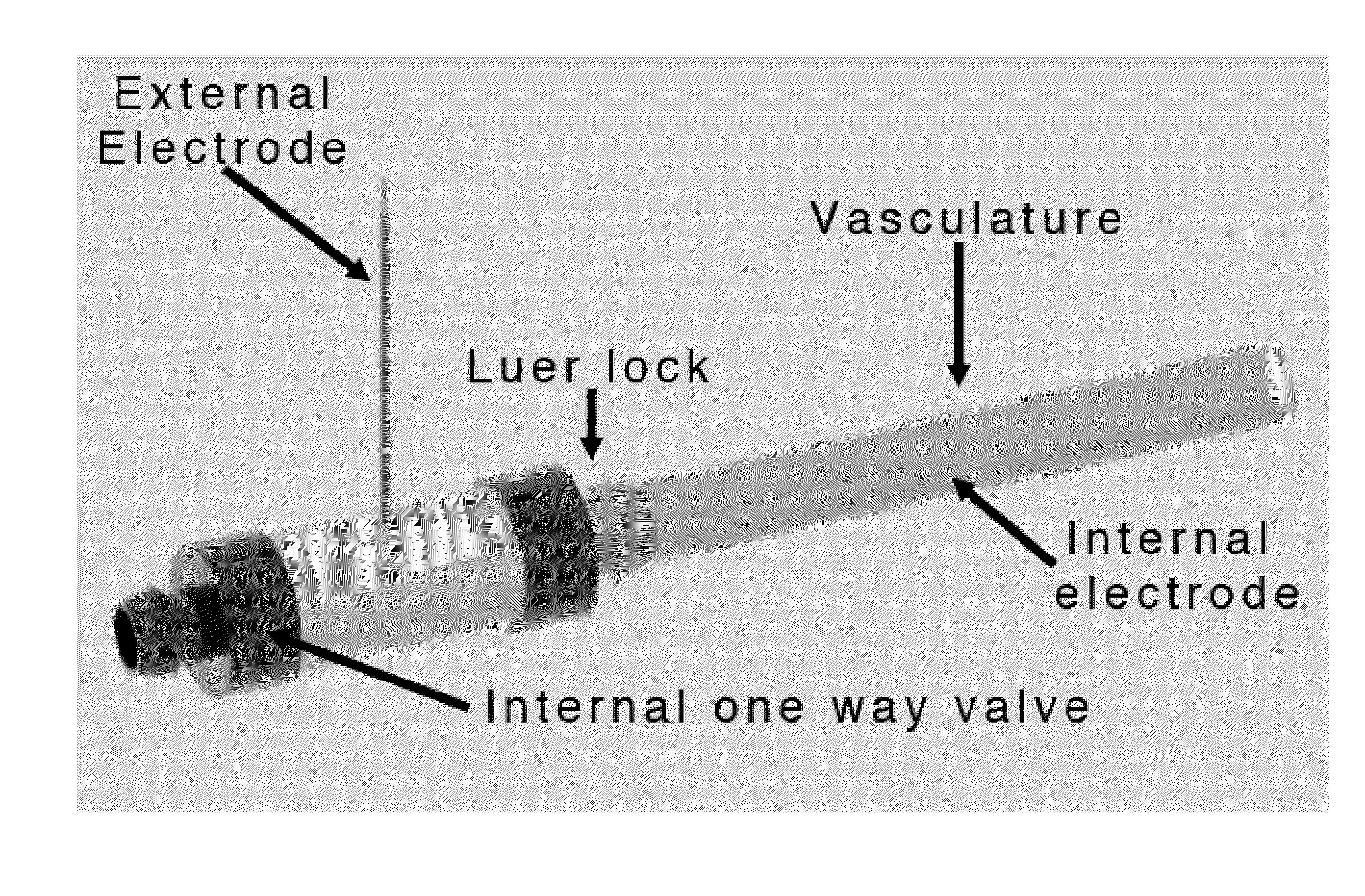
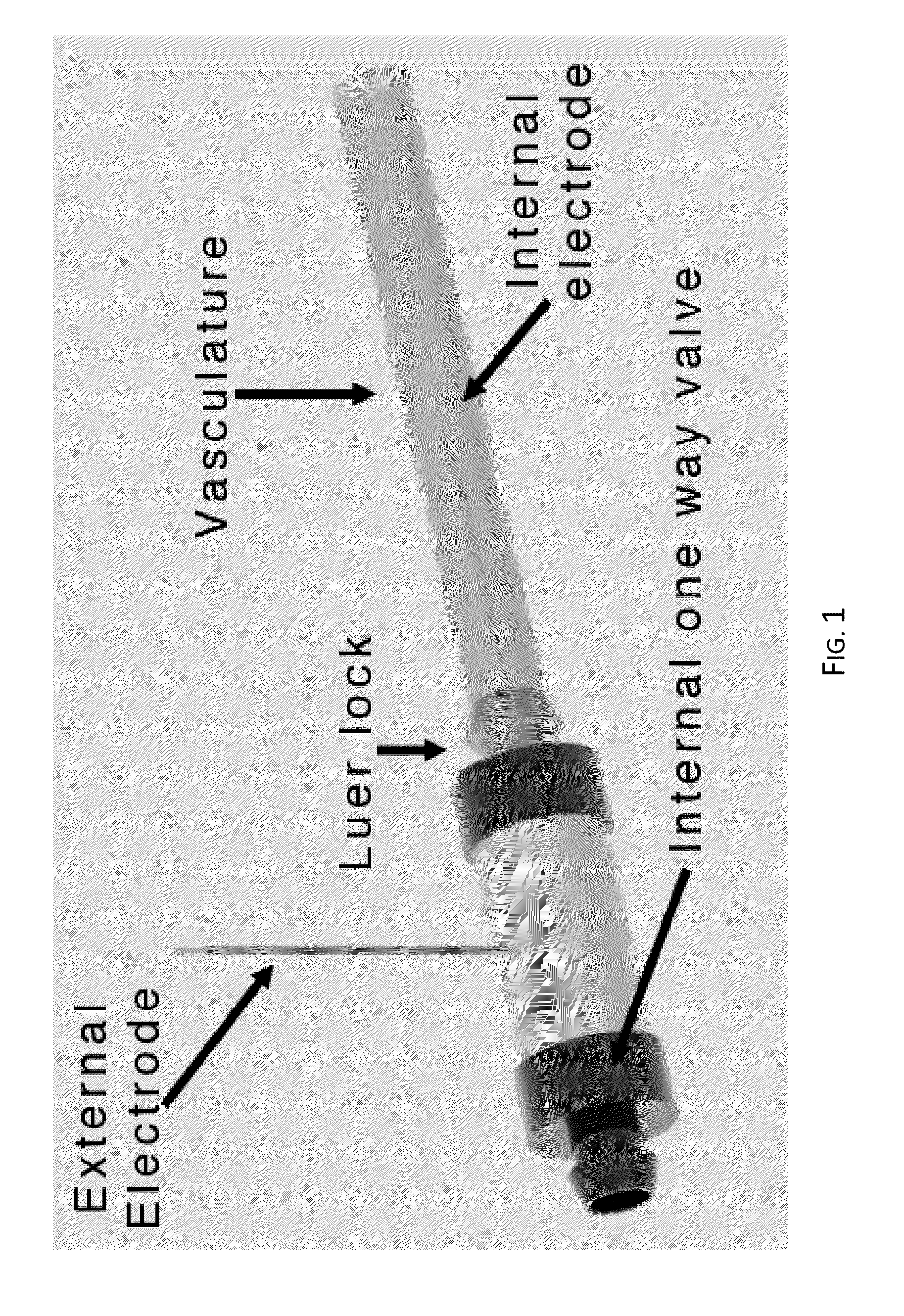
Irreversible electroporation using tissue vasculature to treat aberrant cell masses or create tissue scaffolds
ActiveUS20130253415A1Easy to storeImprove breathabilityElectrotherapyIntravenous devicesDiseaseNatural source
The present invention relates to the field of medical treatment of diseases and disorders, as well as the field of biomedical engineering. Embodiments of the invention relate to the delivery of Irreversible Electroporation (IRE) through the vasculature of organs to treat tumors embedded deep within the tissue or organ, or to decellularize organs to produce a scaffold from existing animal tissue with the existing vasculature intact. In particular, methods of administering non-thermal irreversible electroporation (IRE) in vivo are provided for the treatment of tumors located in vascularized tissues and organs. Embodiments of the invention further provide scaffolds and tissues from natural sources created using IRE ex vivo to remove cellular debris, maximize recellularization potential, and minimize foreign body immune response. The engineered tissues can be used in methods of treating subjects, such as those in need of tissue replacement or augmentation.
Owner:VIRGINIA TECH INTPROP INC
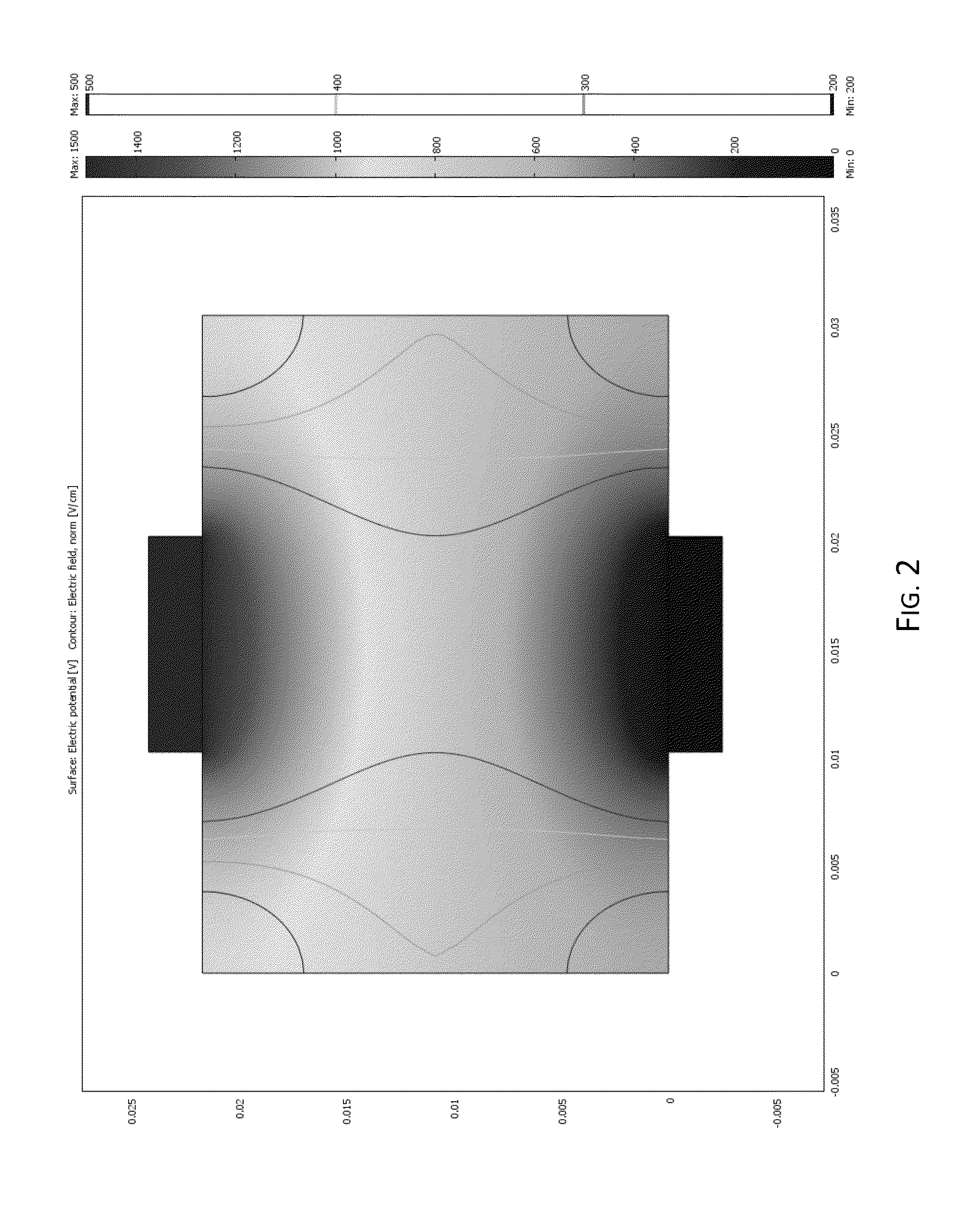
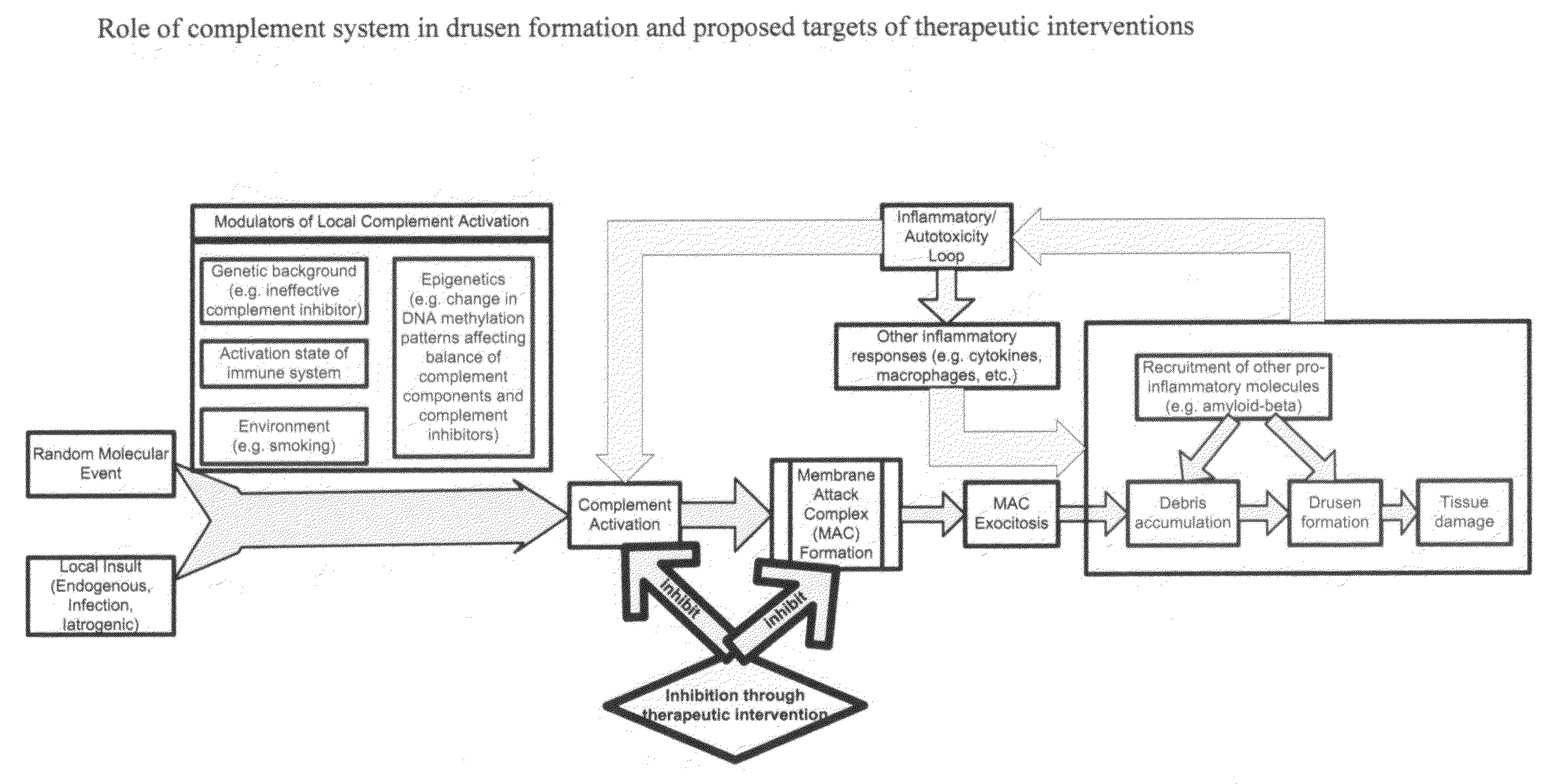
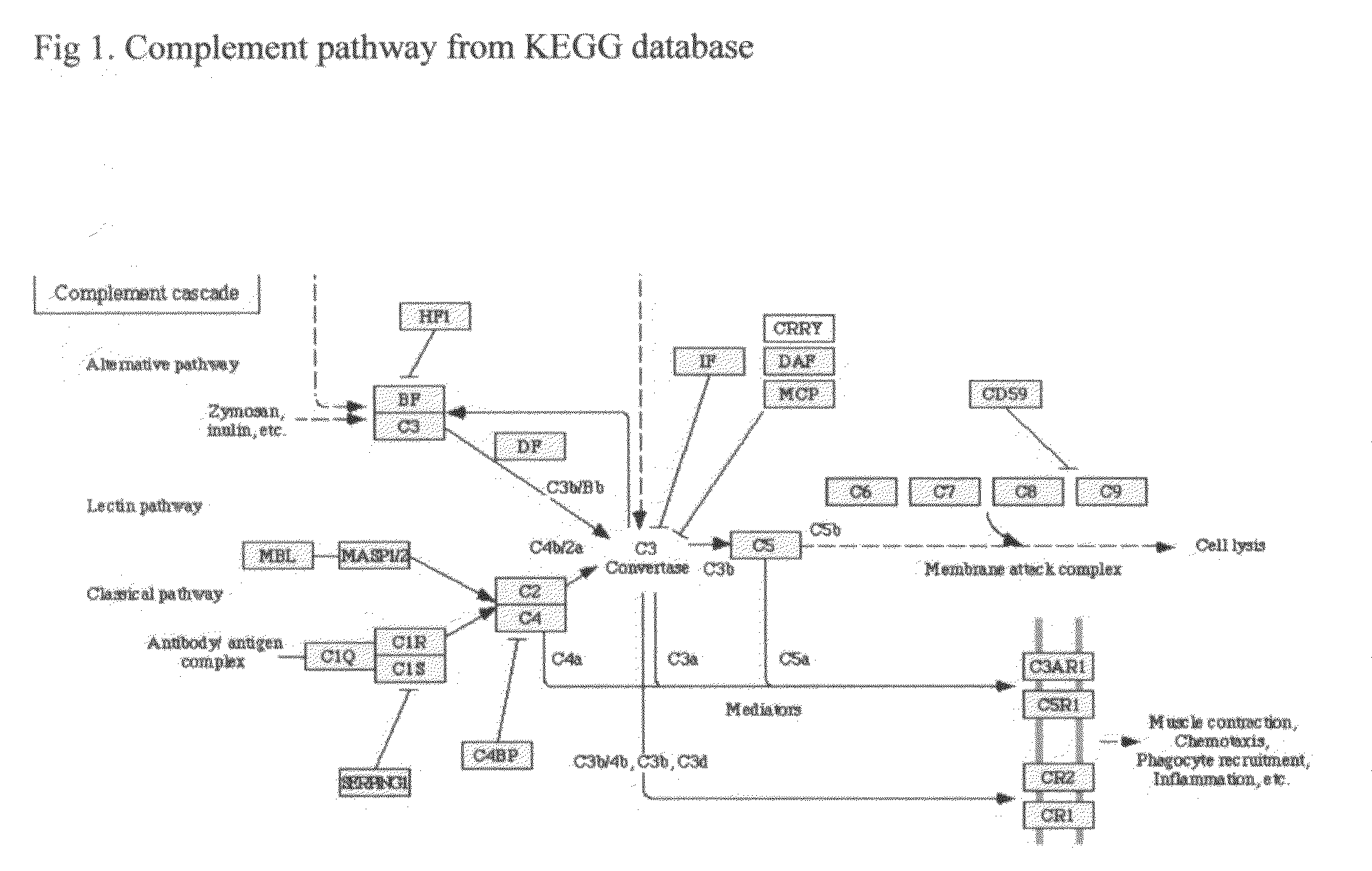
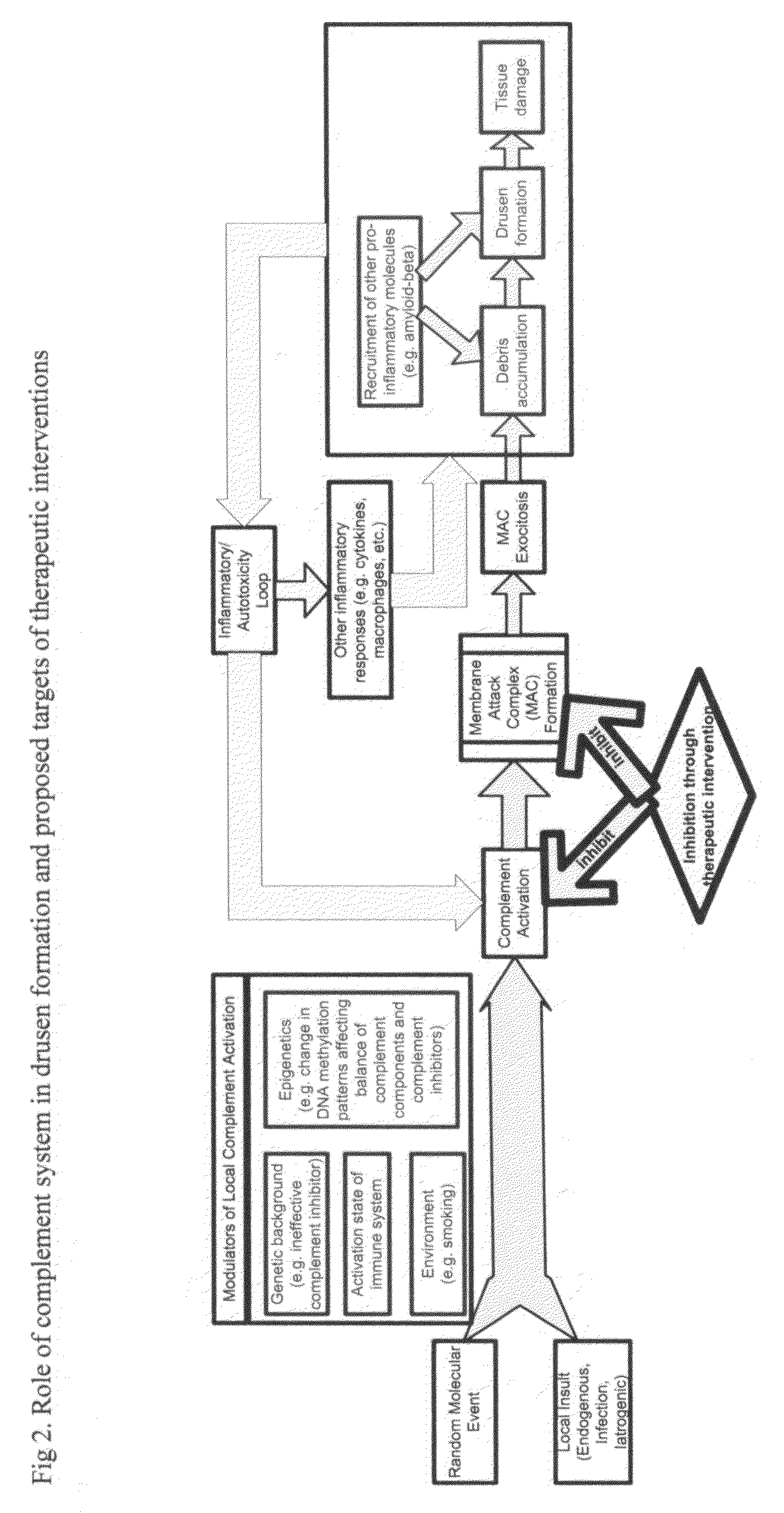
Methods of preventing and treating Alzheimer's disease, age related macular degeneration and other diseases involving extra-cellular debris through the inhibition of the complement system
This invention proposes that the best therapeutic strategy for treating and / or preventing Alzheimer's disease (AD), age related macular degeneration (AMD) and other diseases that exhibit extra-cellular debris deposits, such as atherosclerosis, is the inhibition of the complement pathway. A model for the accumulation of extra-cellular debris through the activation of the complement pathway is presented, and the primary pathogenic role of the debris in the etiology of the disorders is explained. Previously identified complement inhibitors are identified as therapeutic agents for the treatment and / or prevention of AD, AMD and other diseases that exhibit extra-cellular debris deposits.
Owner:DINU VALENTIN
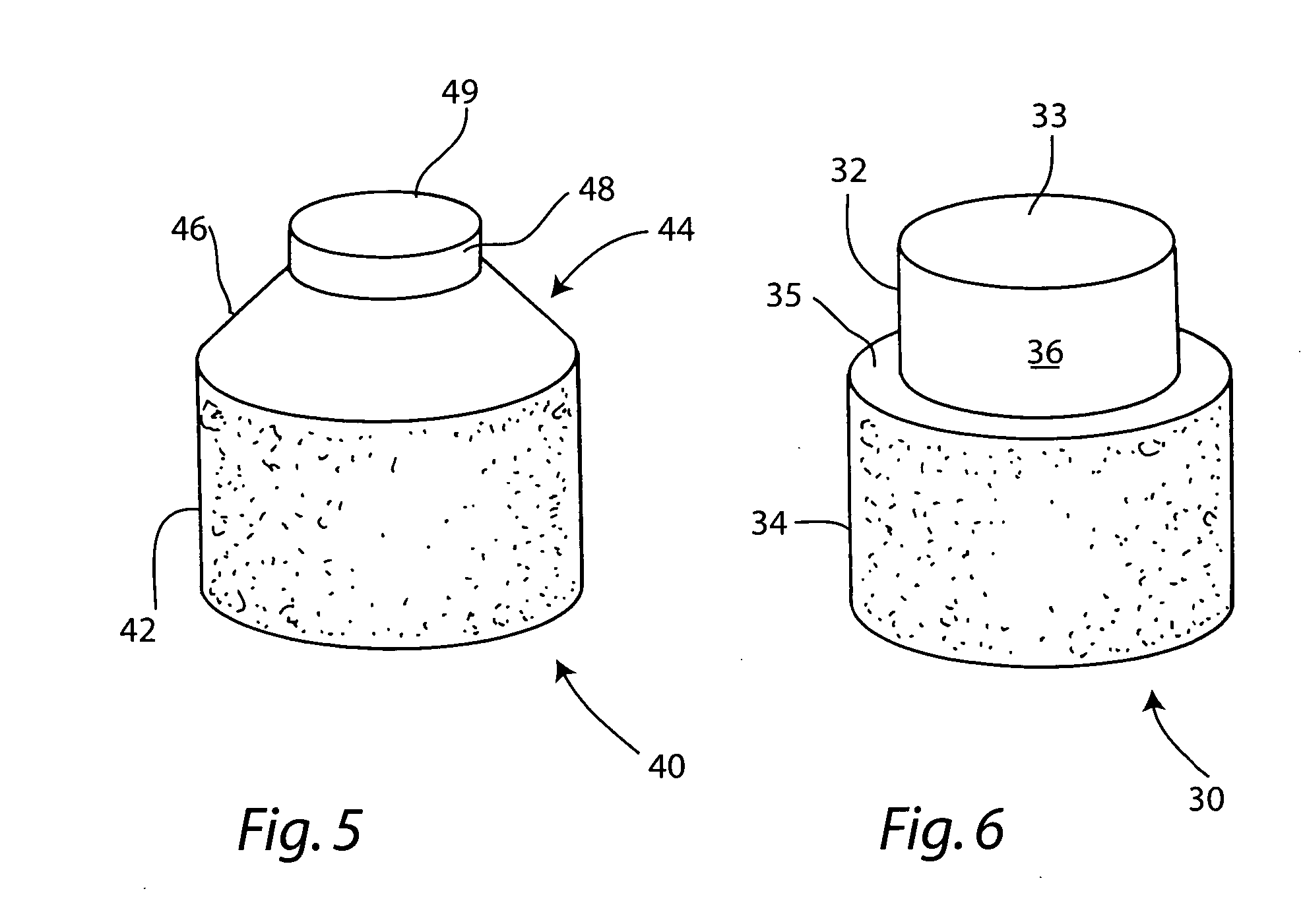
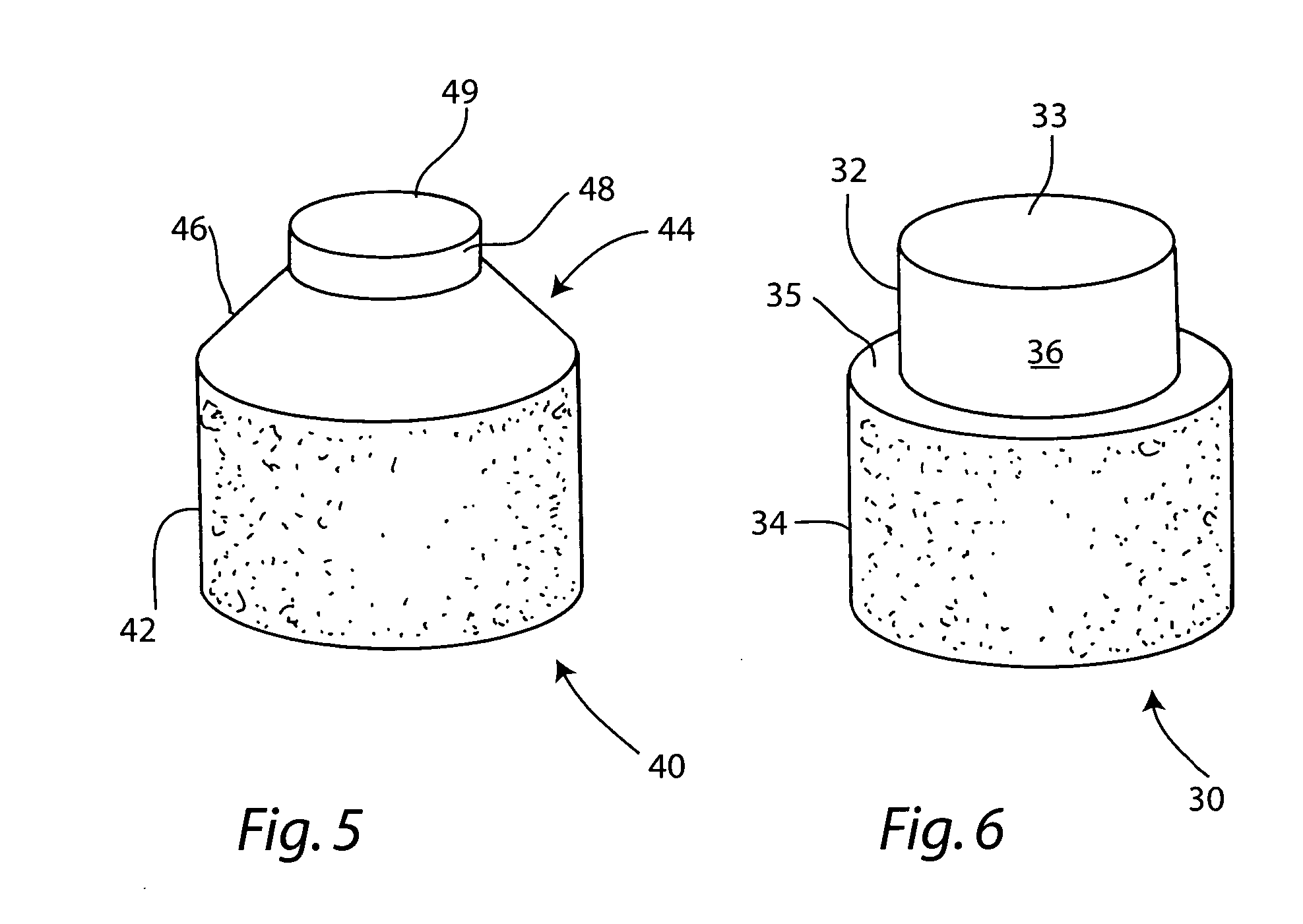
Cartilage allograft plug
InactiveUS7901457B2Promote migrationIncreased proliferationBone implantJoint implantsSubchondral boneInterference fit
The invention is directed toward a cartilage repair assembly comprising a shaped allograft structure of subchondral bone with an integral overlying cartilage cap which is treated to remove cellular debris and proteoglycans and milled allograft cartilage in a bioabsorbable carrier. The shaped structure is dimensioned to fit in a drilled bore in a cartilage defect area so that either the shaped bone or the cartilage cap engage the side wall of the drilled bore in an interference fit and is in contact with a milled cartilage and biocompatible carrier mixture allowing cell transfer throughout the defect area. A method for inserting the shaped allograft structure into a cartilage defect area is also disclosed.
Owner:MUSCULOSKELETAL TRANSPLANT FOUND INC
Cartilage allograft plug
InactiveUS7488348B2Guaranteed functionEasily placed in defect areaBone implantLigamentsInterference fitSubchondral bone
The invention is directed toward a cartilage repair assembly comprising a cylindrically shaped allograft structure of subchondral bone with an integral overlying smaller diameter cartilage cap which is treated to remove cellular debris and proteoglycans. The shaped structure is dimensioned to fit in a drilled bore in a cartilage defect area so that the subchondral bone of the structure engages the side wall of the bone portion of the drilled bore in an interference fit while the cartilage cap is spaced from cartilage portion of the side wall of the drilled bore forming a gap in which a milled cartilage and biocompatible carrier mixture is placed allowing cell transfer throughout the defect area. A method for inserting the shaped allograft structure into a cartilage defect area is also disclosed.
Owner:MUSCULOSKELETAL TRANSPLANT FOUND INC
Method and system for microfluidic manipulation, amplification and analysis of fluids, for example, bacteria assays and antiglobulin testing
Owner:PERKINELMER HEALTH SCIENCES INC
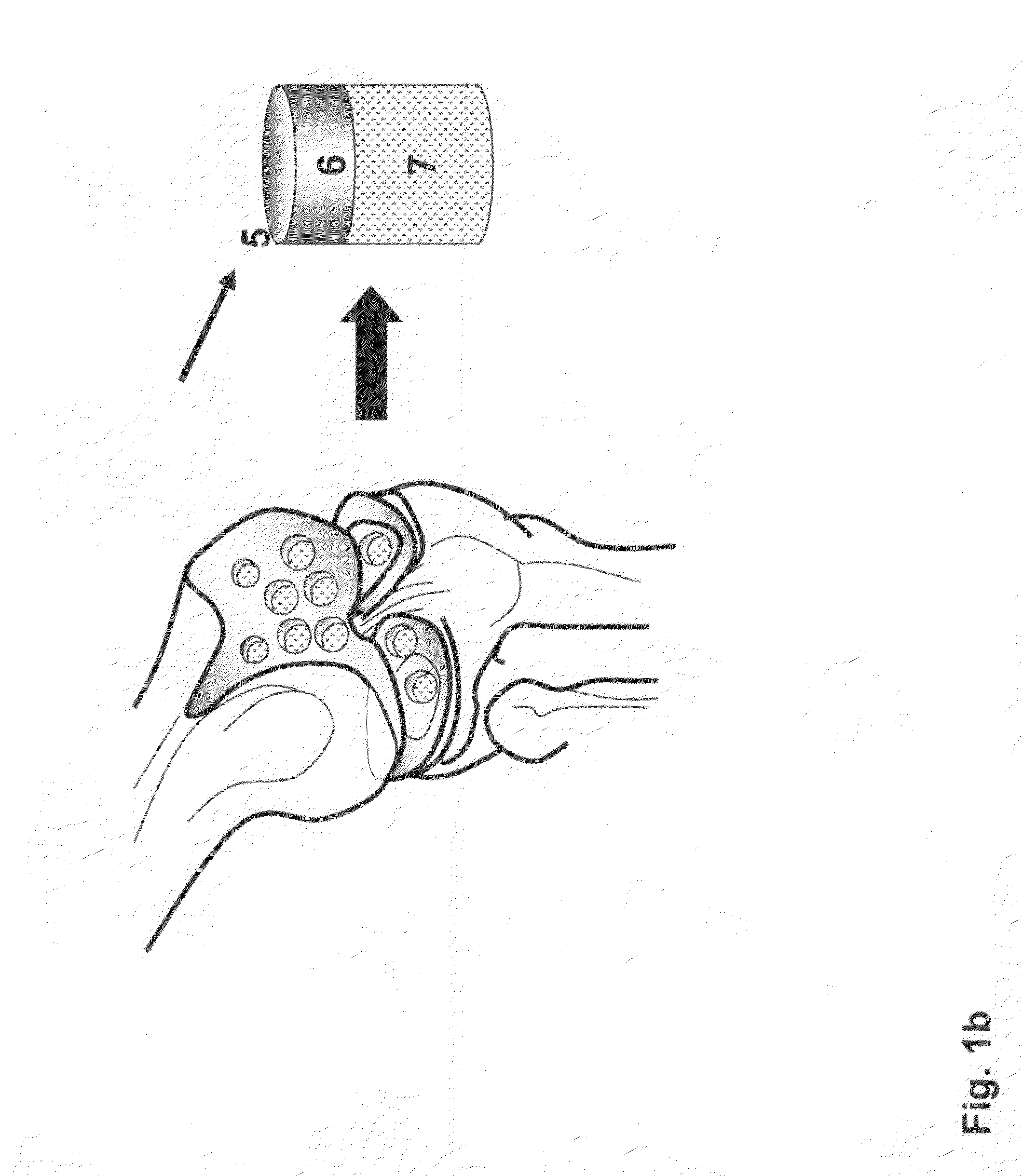
Allograft osteochondral plug combined with cartilage particle mixture
InactiveUS20090291112A1Promoting cartilage cell migration intoPromoting proliferationBiocidePeptide/protein ingredientsInterference fitSubchondral bone
An allograft osteochondral plug is combined with a mixture that includes freeze-milled cartilage particles, and such combination is used to repair defects in articular cartilage. The plug includes an subchondral bone portion and an integral overlying cartilage cap which is treated to remove cellular debris and proteoglycans. At least a portion of the plug has a lateral dimension selected to form an interference fit against a tissue layer exposed as a result of a bore formed in a defect area in articular cartilage of a host. The cartilage particle mixture is placed adjacent at least a portion of the plug for promoting cartilage cell migration into (i.e., from the adjacent host cartilage) and proliferation in the bore, and for enhancing tissue integration between the plug and patient (i.e., host) tissue when the plug is inserted into the bore. Methods for surgical implantation of the plug into a patient are also disclosed.
Owner:MUSCULOSKELETAL TRANSPLANT FOUND INC
Cartilage allograft plug
ActiveUS20090069901A1Increase chondrocyte migrationIncreased proliferationBone implantJoint implantsSubchondral boneInterference fit
The invention is directed toward a cartilage repair assembly comprising a shaped allograft structure of subchondral bone with an integral overlying cartilage cap which is treated to remove cellular debris and proteoglycans and milled allograft cartilage in a bioabsorbable carrier. The shaped structure is dimensioned to fit in a drilled bore in a cartilage defect area so that either the shaped bone or the cartilage cap engage the side wall of the drilled bore in an interference fit and is in contact with a milled cartilage and biocompatible carrier mixture allowing cell transfer throughout the defect area. A method for inserting the shaped allograft structure into a cartilage defect area is also disclosed.
Owner:MUSCULOSKELETAL TRANSPLANT FOUND INC
Crafting of cartilage
The invention is directed to producing a shaped cartilage matrix isolated from a human or animal where the cartilage has been crafted to facilitate disinfection, cleaning, devitalization, recellularization, and / or integration after implantation. The invention relates to a process for repairing a cartilage defect and implantation of a cartilage graft into a human or animal by crafting the cartilage matrix into individual grafts, disinfecting and cleaning the cartilage graft, applying a pretreatment solution to the cartilage graft, removing cellular debris using an extracting solution to produce a devitalized cartilage graft, implanting the cartilage graft into the cartilage defect with or without an insertion device, and sealing the implanted cartilage graft with recipient tissue. The devitalized cartilage graft is optionally recellularized in vitro, in vivo, or in situ with viable cells to render the tissue vital before or after the implantation. The devitalized cartilage graft is also optionally stored between the removing cellular debris and the recellularizing steps.
Owner:LIFENET HEALTH
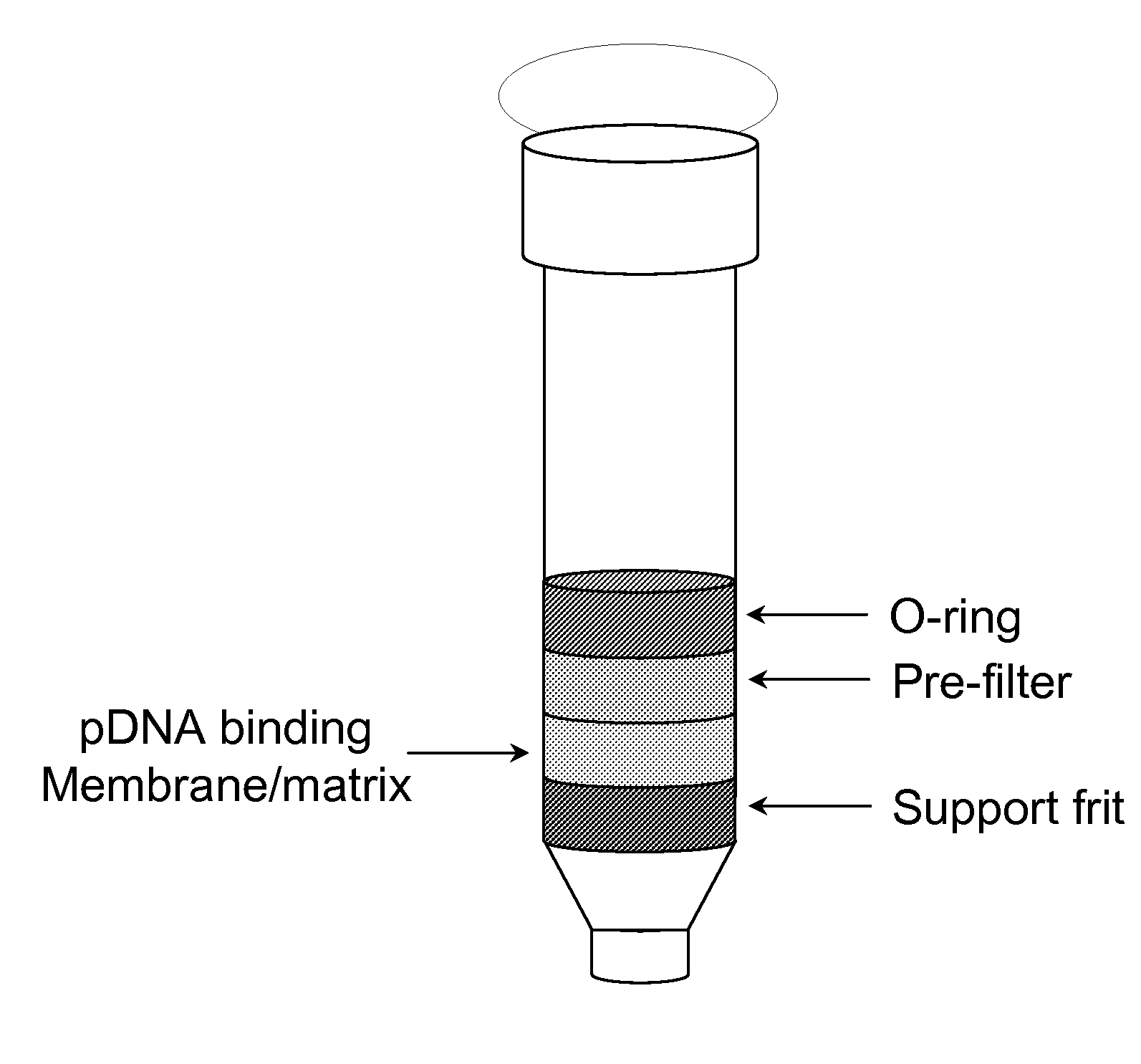
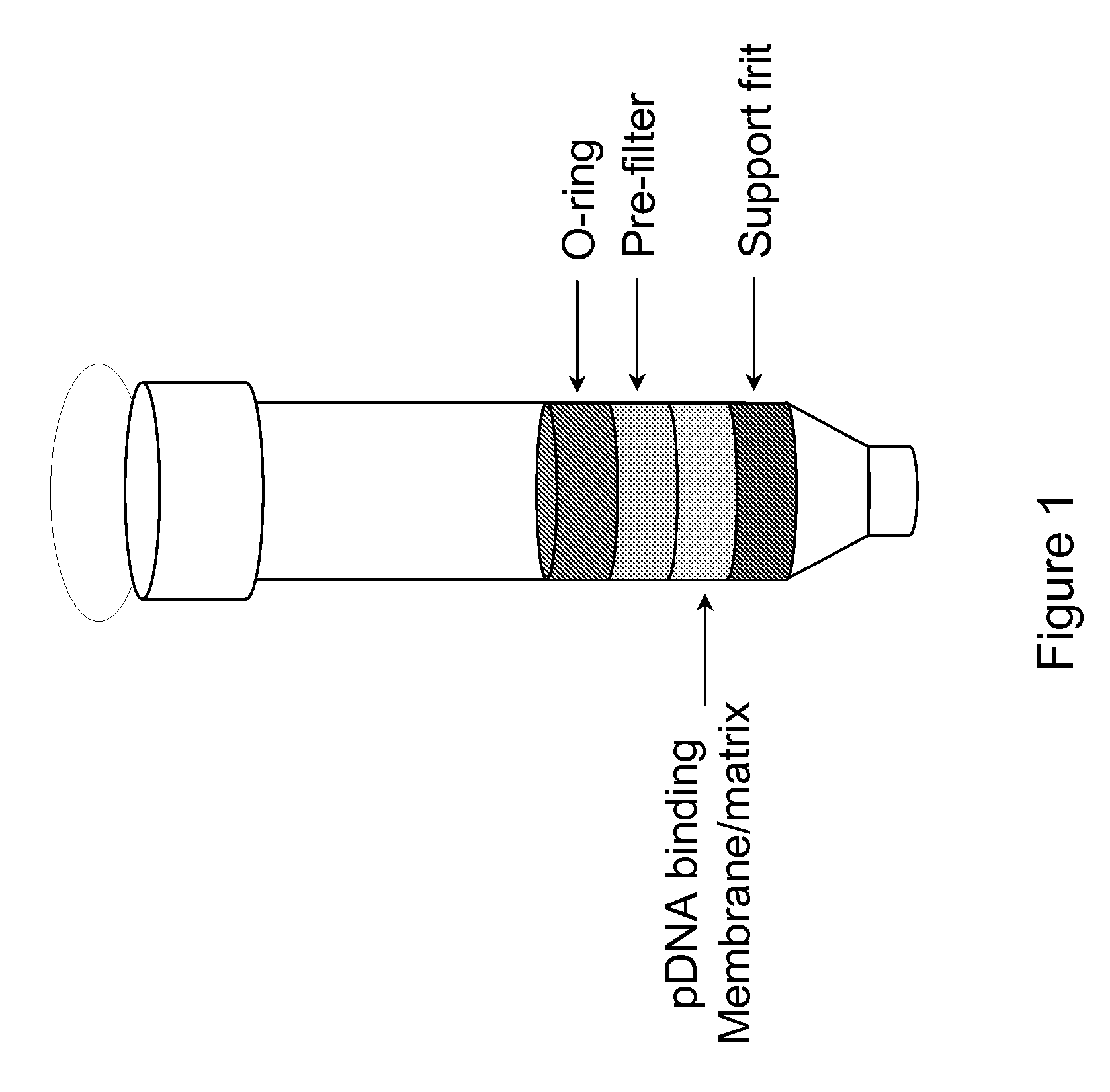
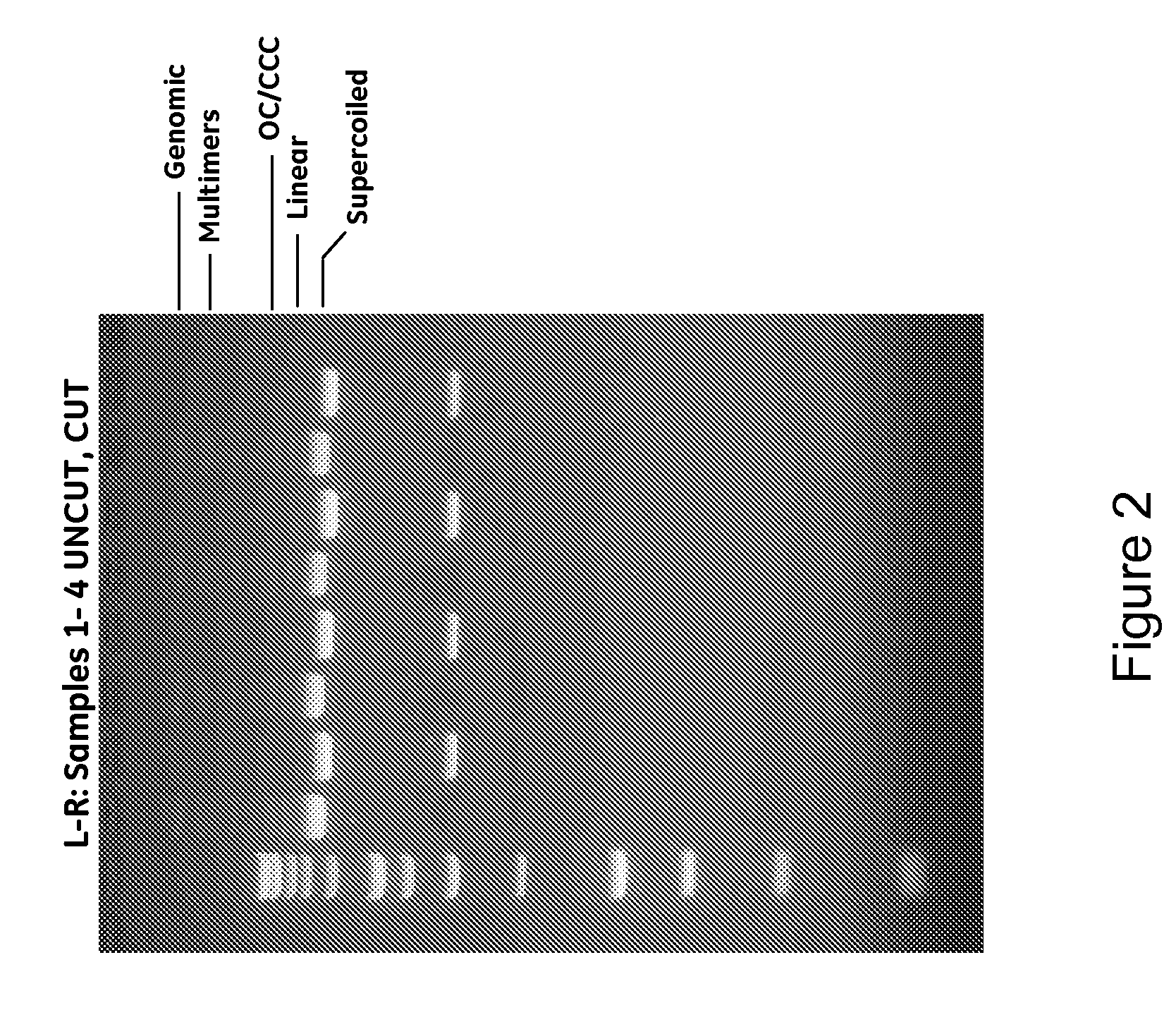
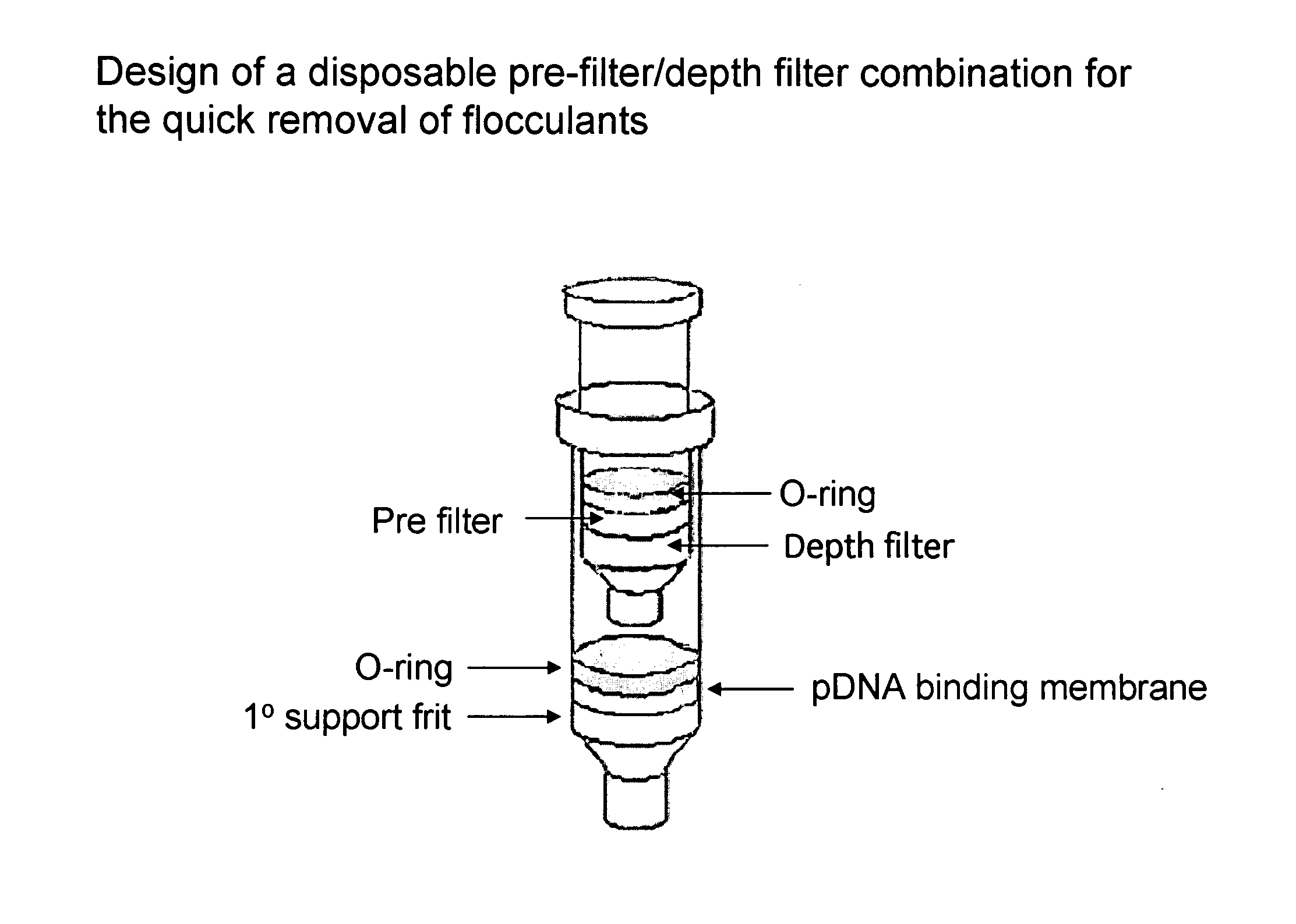
Modified spin column for simple and rapid plasmid DNA extraction
InactiveUS20080300397A1Rapid isolationImprove system performanceSamplingSugar derivativesCellular DebrisSpins
The invention relates to a modified spin column for the isolation and purification of plasmid DNA. A pre-filtration disc is included in a traditional spin column. During plasmid DNA isolation, the lysate can be loaded directly to the modified spin column, eliminates the need to first remove the flocculants containing cellular debris. This results in a much shortened process. Variation of the invention includes a depth filter in between the pre-filtration disc and the main separation matrix. Also provided are kits for isolation of plasmid DNA including the modified spin columns.
Owner:GE HEALTHCARE LTD
Irreversible electroporation using tissue vasculature to treat aberrant cell masses or create tissue scaffolds
ActiveUS9867652B2Easy to storeImprove breathabilityElectrotherapyIntravenous devicesAbnormal tissue growthNatural source
The present invention relates to the field of medical treatment of diseases and disorders, as well as the field of biomedical engineering. Embodiments of the invention relate to the delivery of Irreversible Electroporation (IRE) through the vasculature of organs to treat tumors embedded deep within the tissue or organ, or to decellularize organs to produce a scaffold from existing animal tissue with the existing vasculature intact. In particular, methods of administering non-thermal irreversible electroporation (IRE) in vivo are provided for the treatment of tumors located in vascularized tissues and organs. Embodiments of the invention further provide scaffolds and tissues from natural sources created using IRE ex vivo to remove cellular debris, maximize recellularization potential, and minimize foreign body immune response. The engineered tissues can be used in methods of treating subjects, such as those in need of tissue replacement or augmentation.
Owner:VIRGINIA TECH INTPROP INC
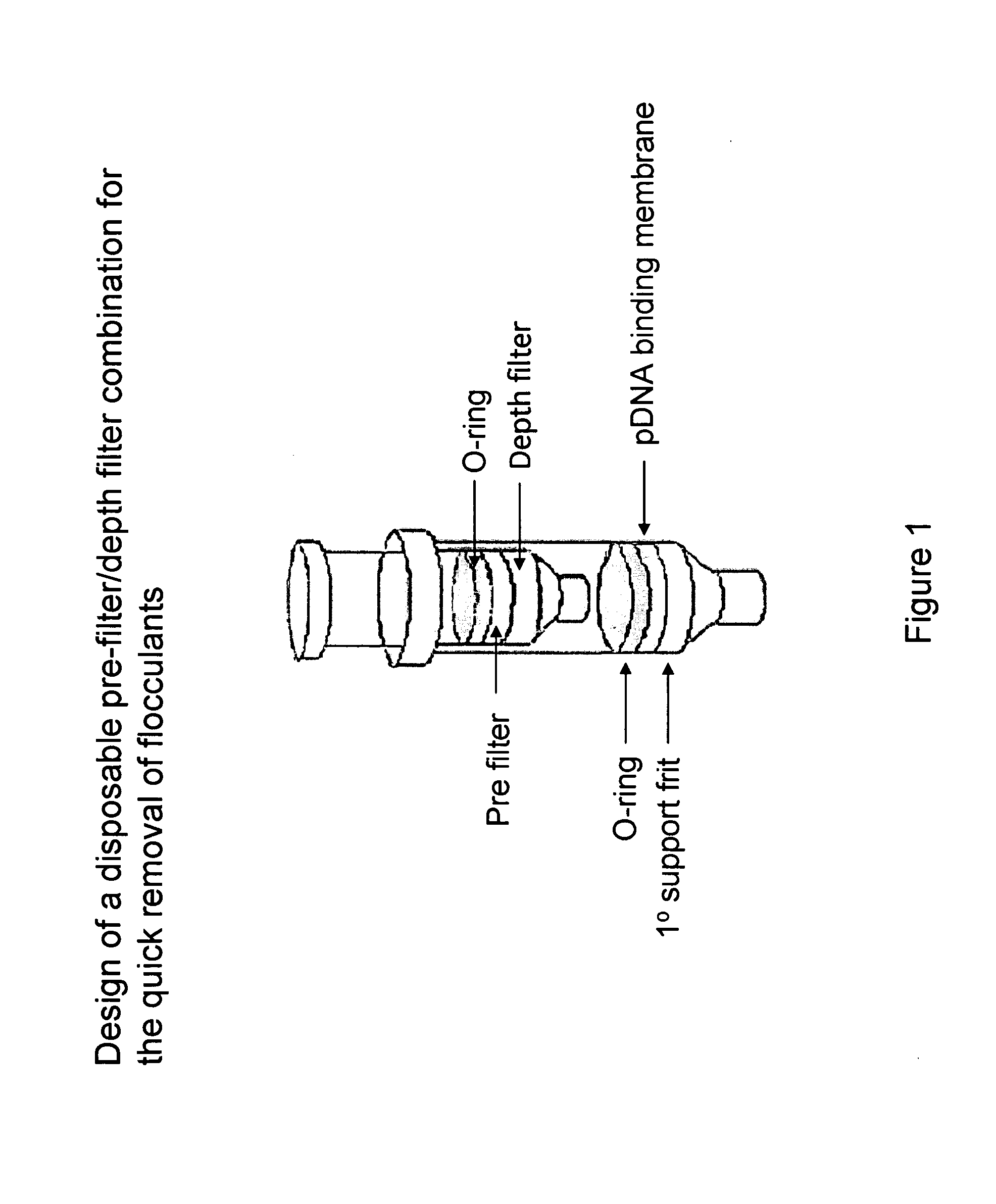
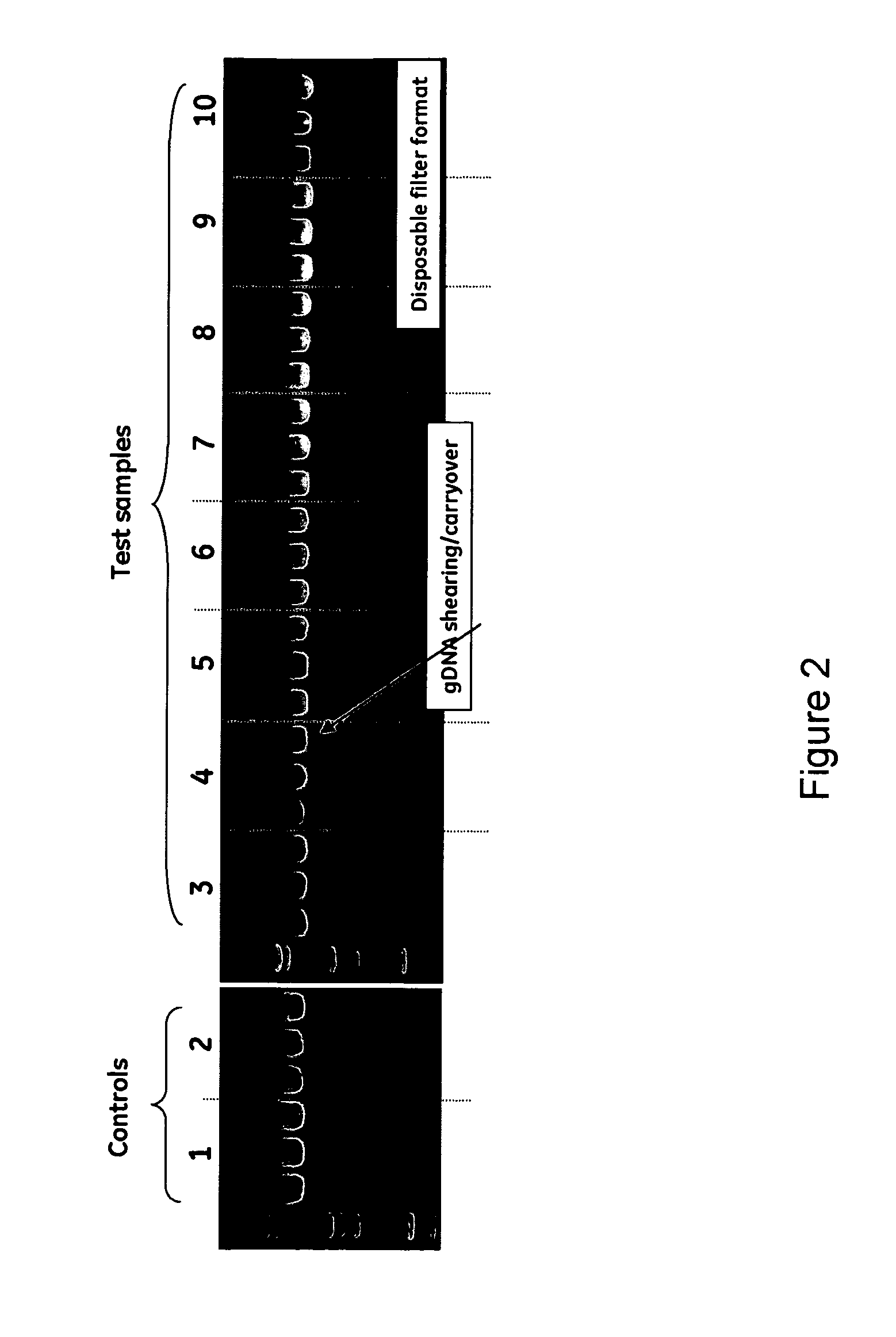
Miniprep system for simple and rapid plasmid DNA extraction
InactiveUS20080299621A1Rapid isolationBioreactor/fermenter combinationsBiological substance pretreatmentsCellular DebrisSpins
The invention relates to a modified microspin system for the isolation and purification of plasmid DNA. A disposable pre-filter column is used in combination with the traditional microspin column for increase speed and quality of plasmid DNA preparation. The disposable pre-filter column includes a pre-filter and a depth filter for optimal result. During plasmid DNA isolation, the lysate can be loaded directly to the assembly including the disposable pre-filter column and the microspin column. A quick spin causes the plasmid DNA to bind to the microspin column while the flocculents remain on top of the disposable pre-filter column, eliminates the need to first remove the flocculants containing cellular debris. This results in a much shortened process. Also provided are kits for isolation of plasmid DNA including the pre-filter column and the microspin columns.
Owner:GE HEALTHCARE LTD
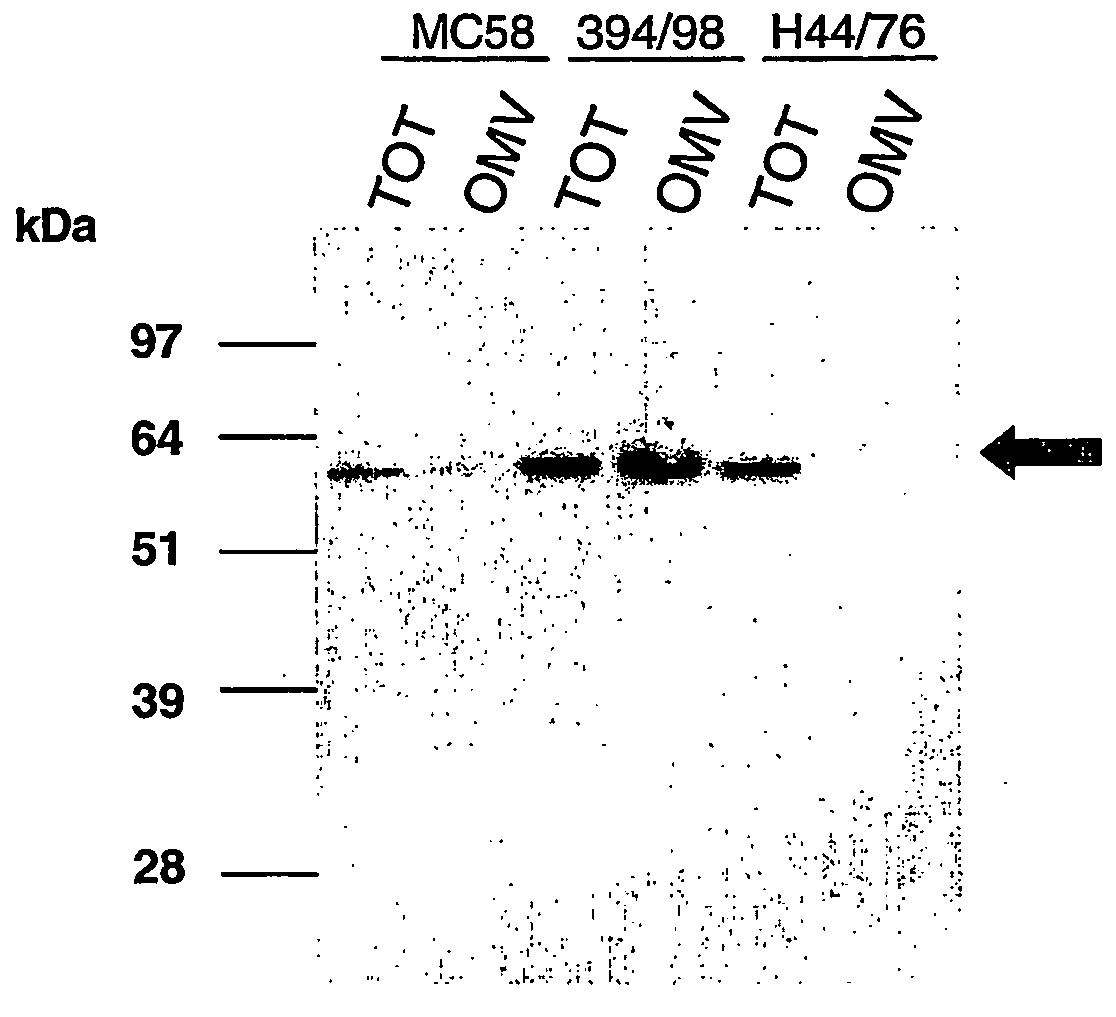
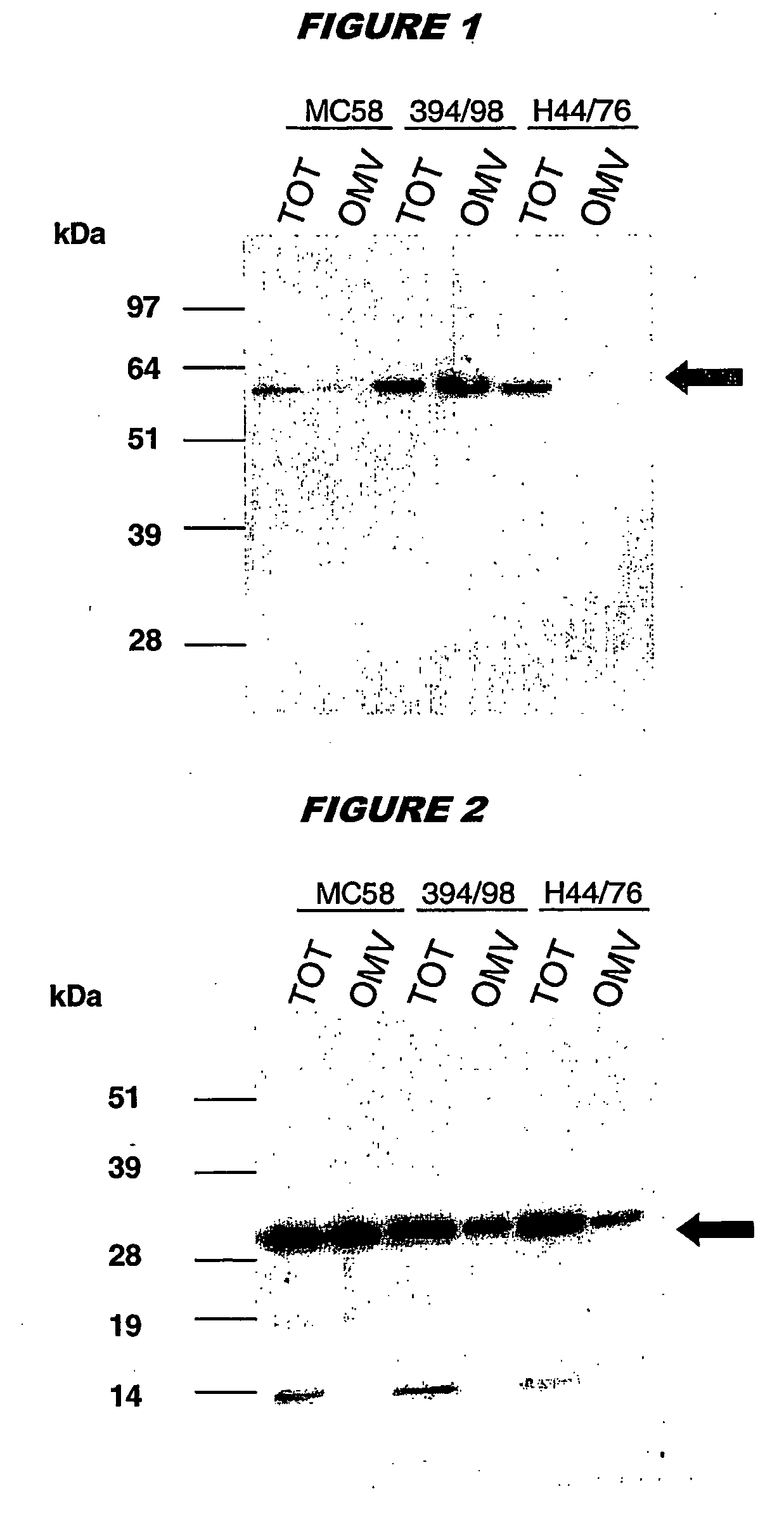
Bacterial outer memberane vesicles
InactiveUS20060166344A1Good curative effectAvoid problemsAntibacterial agentsBacteriaImmunogenicityTreated cell
Existing methods of meningococcal OMV preparation involve the use of detergent during disruption of the bacterial membrane. According to the invention, membrane disruption is performed substantially in the absence of detergent. The resulting OMVs which retain important bacterial immunogenic components, particularly (i) the protective NspA surface protein, (ii) protein NMB2132 and (iii) protein NMB 1870. A Typical process involves the following steps: (a) treating bacterial cells in the substantial absence of detergent; (b) centrifuging the composition from step (a) to separate the outer membrane vesicles from treated cells and cell debris, and collecting the supernatant; (c) performing a high speed centrifugation of the supernatant from step (b) and collecting the outer membrane vesicles in a pellet; (d) re-dispersing the pellet from step (c) in a buffer; (e) performing a second high speed centrifugation in accordance with step (c), collecting the outer membrane vesicles in a pellet; (f) re-dispersing the pellet from step (e) in an aqueous medium.
Owner:NOVARTIS AG
Cartilage implant plug with fibrin glue and method for implantation
Owner:MASSACHUSETTS INST OF TECH +1
Depth Filters For Disposable Biotechnological Processes
InactiveUS20130012689A1Avoid concentrationEfficient separationIon-exchanger regenerationUltrafiltrationChemical treatmentColloidal particle
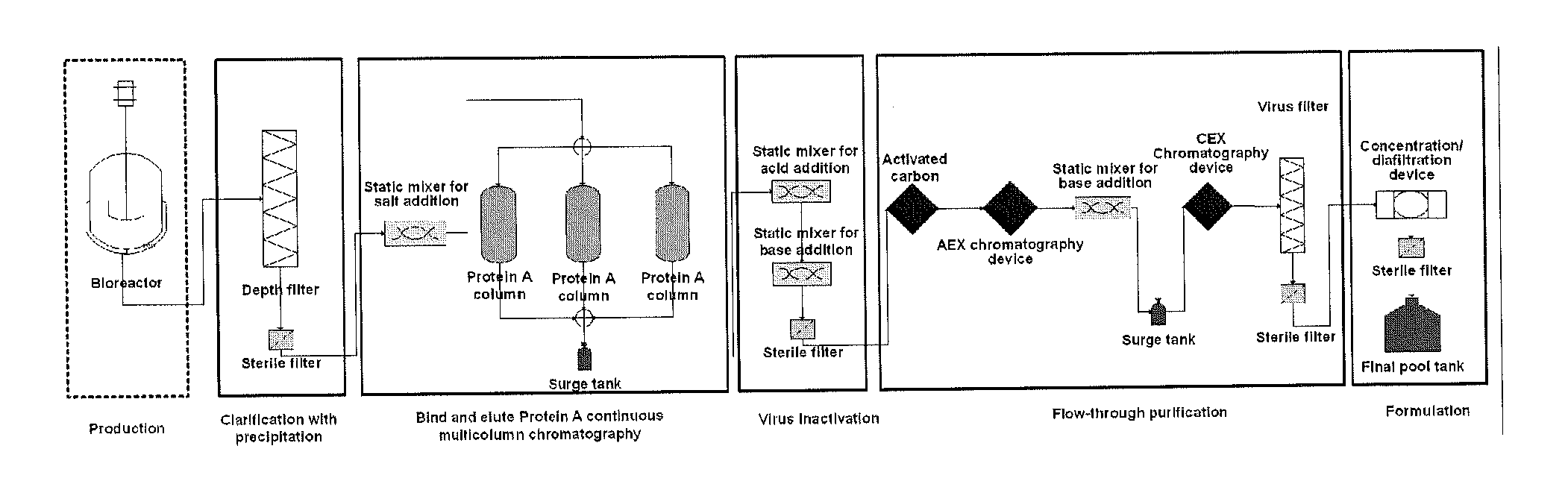
A process for the primary clarification of feeds, including chemically treated flocculated feeds, containing the target biomolecules of interest such as mAbs, mammalian cell cultures, or bacterial cell cultures, using a primary clarification depth filtration device without the use of a primary clarification centrifugation step or a primary clarification tangential flow microfiltration step. The primary clarification depth filtration device contains a porous depth filter having graded porous layers of varying pore ratings. The primary clarification depth filtration device filters fluid feeds, including chemically treated flocculated feeds containing flocculated cellular debris and colloidal particulates having a particle size distribution of approximately about 0.5 μm to 200 μm, at a flow rate of about 10 litres / m2 / hr to about 100 litres / m2 / hr. Kits and methods of using and making the same are also provided.
Owner:MILLIPORE CORP
Cartilage implant plug with fibrin glue and method for implantation
InactiveUS20080274157A1Guaranteed functionEasy to placeSurgical adhesivesPeptide/protein ingredientsSubchondral boneFibrin glue
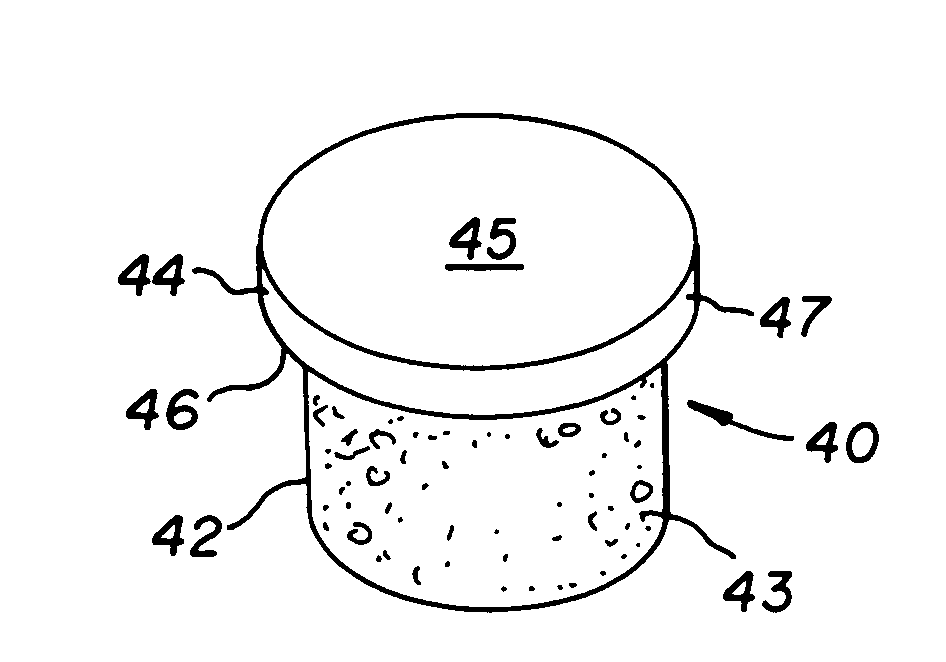
The invention is directed toward a cartilage repair assembly comprising a shaped structure of subchondral bone with an integral overlying cartilage cap which is treated to remove cellular debris and proteoglycans and milled cartilage in a bioabsorbable carrier. The shaped structure is dimensioned to fit in a drilled bore in a cartilage defect area so that said shaped bone and cartilage cap when centered in the bore does not engage the side wall of the bore and is positioned from the side wall of the bone a distance ranging from 10 microns to 1000 microns and is surrounded by milled cartilage and a fibrin thrombin glue. A method for inserting the assembly into a cartilage defect area is disclosed.
Owner:MASSACHUSETTS INST OF TECH +1
Process for treating sewage to generate active mud and extract protein
InactiveCN1369445AReduce pollutionSimple processSludge treatment by de-watering/drying/thickeningSludge treatment by thermal conditioningCellular DebrisSludge
A process for extracting protein from the active mud generated by treating sewage includes dewatering said mud, stirring at 15-90 deg.C for 5-60 min, regulating pH value to 7-12, stirring at constanttemp for 5-30 min, filter to obtain aqueous solution of protein, stirring until the pH value of 3-7, laying aside, depositing, removing supernatant, and drying. Its filtered dregs can be used as fuelor feed.
Owner:邵胜学
Method and apparatus for the treatment of fluid waste streams
InactiveUS20060286006A1Combination devicesShaking/oscillating/vibrating mixersCyclosarinWaste stream
This invention relates generally to methods and apparatus for the detoxification of fluid streams, for example, wastewater contaminated with neurotoxins, particularly organophosphorous compounds, and comprises contacting the fluid stream with a bioactive coating. More particularly, the invention relates to chemical reactors for detoxifying fluid streams and also, bioactive coated support components comprising rigid, semi-rigid, or flexible support materials coated with a bioactive coating compriseing dessicated whole cells, whole cell fragments, enzymes, and combinations thereof that are capable of hydrolizing neurotoxic organophosphorous chemical compounds. Organophosphorus hydrolases that are capable of detoxifying organophosphorus compounds that are: chemical weapons agents, in particular, tabun (“GA”), sarin (“GB”), soman (“GD”), cyclosarin, VX, and its isometric analog Russian VX (“VR” or “R-VX”); chemical weapons agent analogs, chemical weapons surrogates; and pesticides are most preferred. The process and apparatus embodiments of the present invention are designed to detoxify organophosphorus compounds continuously, semi-continuously and and in batch operation.
Owner:MCDANIEL C STEVEN +2
Flow-cytometry-based method for rapidly measuring heterotrophic bacteria in eutrophic lake
InactiveCN103926189AShorten the timeRapid detection quantityIndividual particle analysisEutrophicationStaining
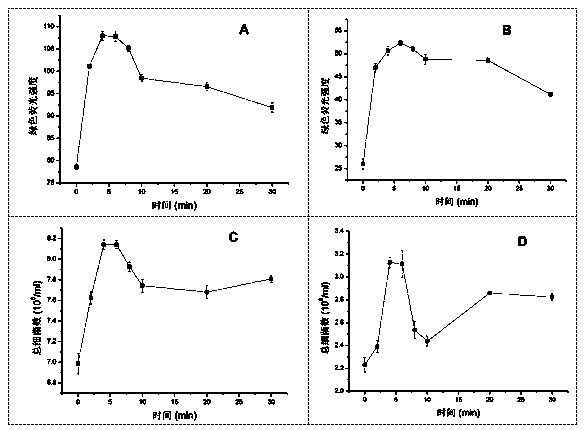
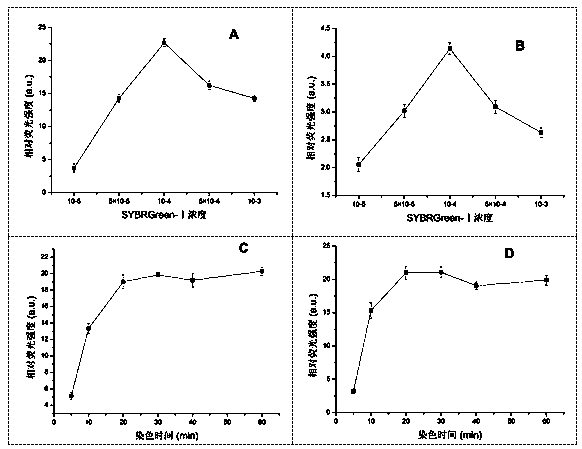
The invention aims to develop a flow-cytometry-based method for rapidly measuring heterotrophic bacteria in a eutrophic lake. The method is a flow-cytometry-based bacterium counting method. The method comprises the following steps of fixing a sample, performing ultrasonic processing, filtering, performing SYBR Green I dyeing, detecting the heterotrophic bacteria by utilizing a flow cytometer, and acquiring forward and lateral scattering light, an SYBR Green I green fluorescence signal and a chlorophyll a red fluorescence signal of autotrophic plankton, wherein the SYBR Green I green fluorescence signal is used for setting a threshold value; and eliminating the influence of the nannoplankton on bacterium detection by utilizing the forward and lateral scattering light, separating the plankton from abiological particles and cellular debris according to the intensity of the SYBR Green I green fluorescence signal, and separating the bacteria from phytoplankton according to the intensity of the chlorophyll a red fluorescence signal of the autotrophic plankton, thereby obtaining the accurate number of the heterotrophic bacteria. The method has the time-saving and labor-saving effects, the large-scale ecological survey can be developed, and the counting precision and accuracy of the bacteria are improved.
Owner:NANJING INST OF GEOGRAPHY & LIMNOLOGY
Nucleic acid extraction solution and use thereof
InactiveUS6939672B2Quick and reliableSugar derivativesMicrobiological testing/measurementHydrogenCellular Debris
Disclosed are methods and compositions for extracting nucleic acids from a biological sample. In particular, disclosed is a nucleic acid extraction solution together with methods using such a solution for extracting nucleic acid sequences from biological samples containing cells, cellular debris or both. The nucleic acid extraction solution contains a molecule having the formula R1O—CH2—CH2—OR2, wherein R1 and R2 independently are selected from the group consisting of hydrogen and an alkyl group.
Owner:CYTYC CORP
Process for protein extraction
ActiveUS20060106205A1High viscosityReduced pHImmunoglobulins against growth factorsDepsipeptidesSolubilityEscherichia coli
The invention includes a process for extracting a target protein from E. coli cells that includes lowering the pH of a whole E. coli cell solution to form an acidic solution, disrupting the cells to release the protein into the acidic solution, and separating the cellular debris from the released protein to obtain a protein product enriched in the heterologous target protein. The invention also includes addition of a solubility enhancer.
Owner:GENENTECH INC
Devices and methods for processing a biological sample
InactiveUS20150177111A1Bioreactor/fermenter combinationsBiological substance pretreatmentsCellular DebrisMagnetic separation
Aspects of the present disclosure include methods for processing a biological sample. Methods according to certain embodiments include disrupting a biological sample to produce a disrupted biological sample and acoustically separating larger components from smaller components in the disrupted biological sample. In certain embodiments, methods may include monitoring aggregation while the biological sample is being processed. Methods, in certain instances, also include acoustically separating cells from cellular debris and non-cellular macromolecules as well as magnetically separating magnetically labelled moieties from unlabeled moieties. Systems, including a disrupter, one or more acoustic concentrator devices, feedback monitors and magnetic separation devices suitable for practicing the subject methods are also described.
Owner:BECTON DICKINSON & CO
Preparation method of anti-infectious lower biomaterial of small intestinal mucosa
InactiveCN109331228AImprove solubilityPromote digestionProsthesisFreeze-dryingScanning electron microscope
The invention relates to a preparation method of an anti-infectious lower biomaterial of small intestinal mucosa. The method comprises the following steps: primarily treating; degreasing; inactivatingvirus; decellularizing; loading antibacterial components; forming; freezing drying or soaking in a storing solution; packing; sterilizing. The lower biomaterial prepared according to the method is good in decellularizing effect; the damage to the structure of a raw material is reduced; an SIS material subjected to decellularizing is complete and uniform in a collagenous fiber structure by observing through a light microscope; the collagenous fiber beam is complete; extremely few cells are residual; complete cells cannot be observed; the collagenous fibers of the material are of three-dimensional net shaped structures by being observed under a scanning electron microscope, and cells are not observed, and few broken cell pieces are residual; after the antibacterial components are loaded tothe SIS, the antibacterial effect is obvious; the clinical demand can be met.
Owner:陈德夫
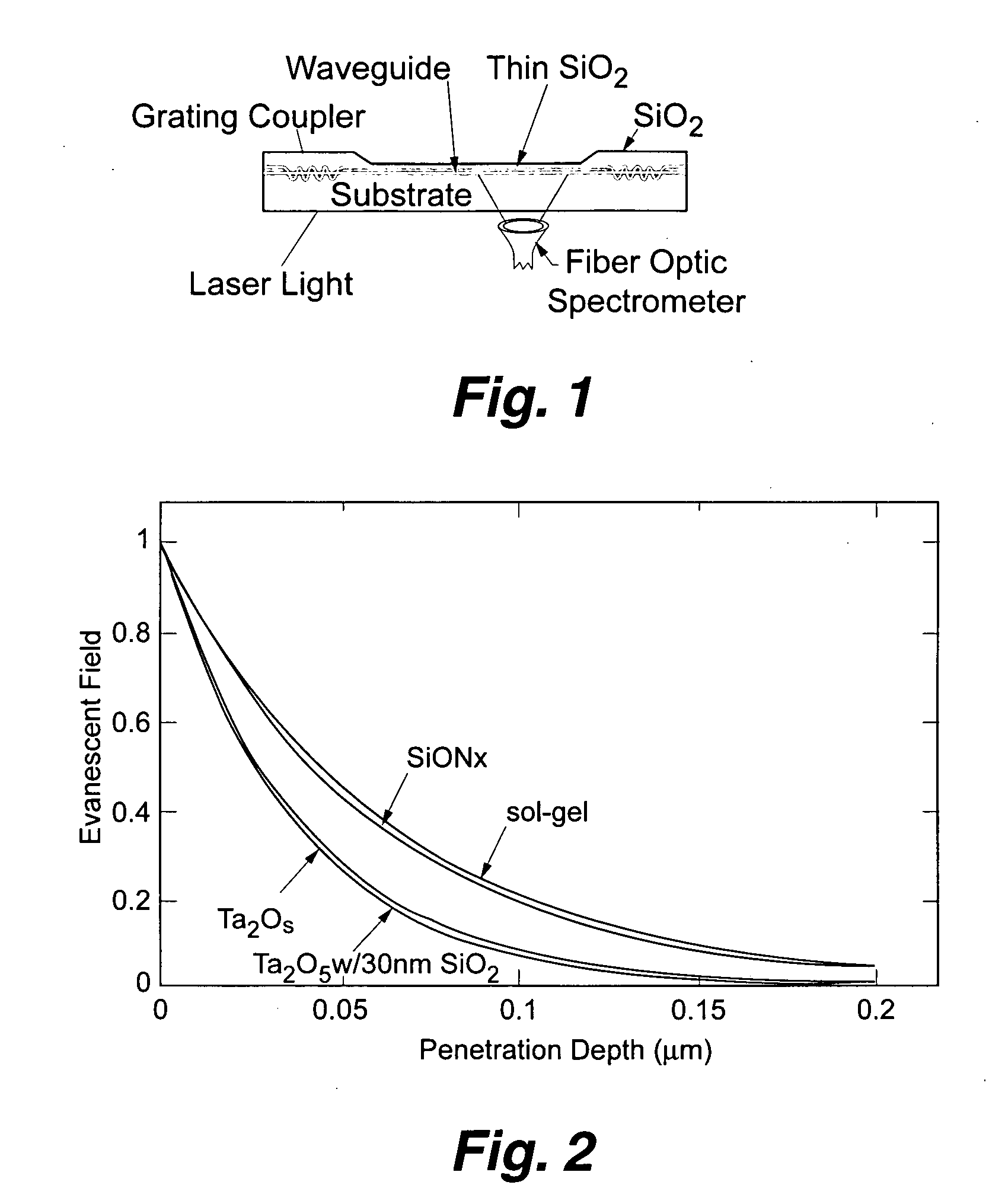
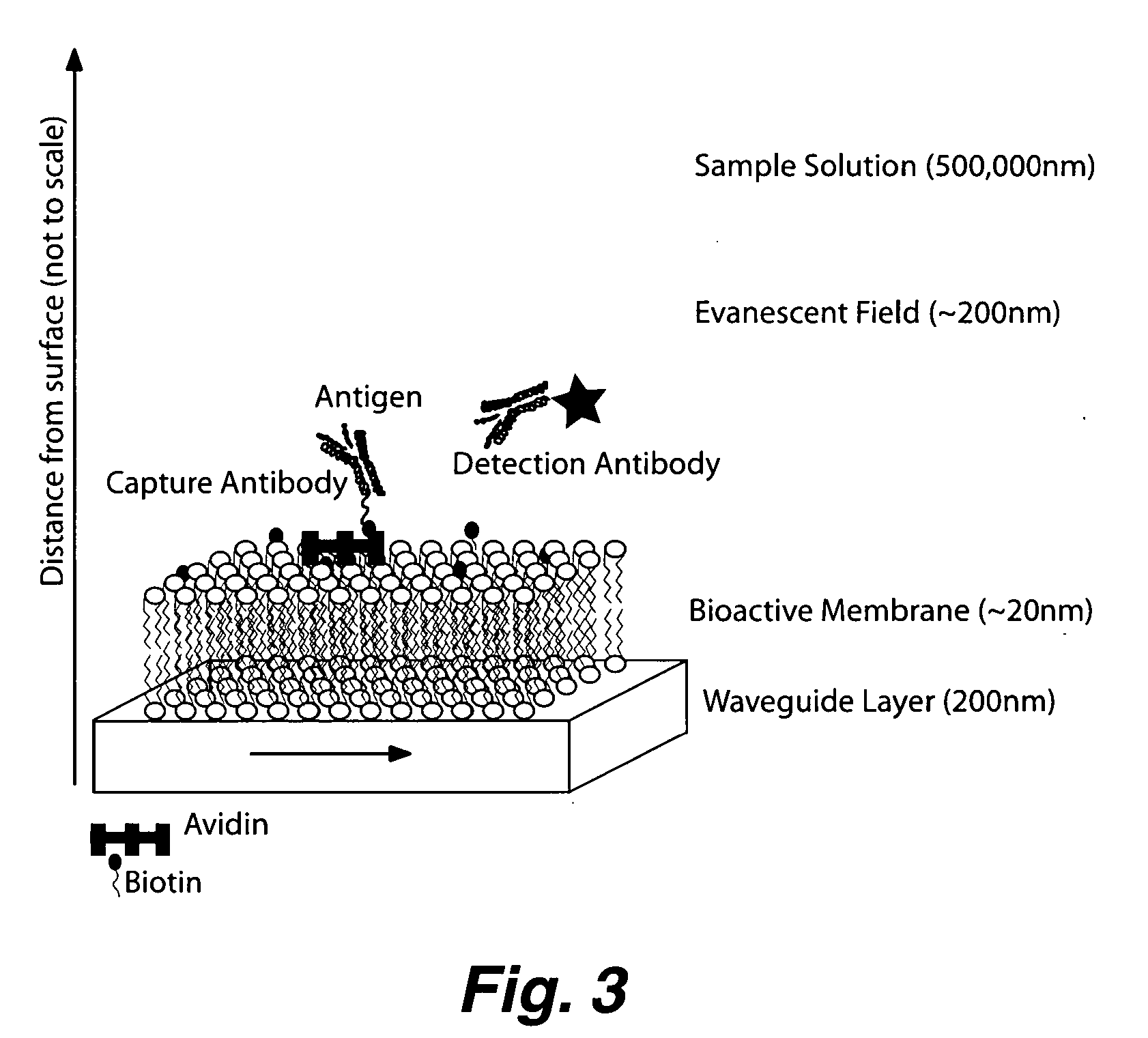
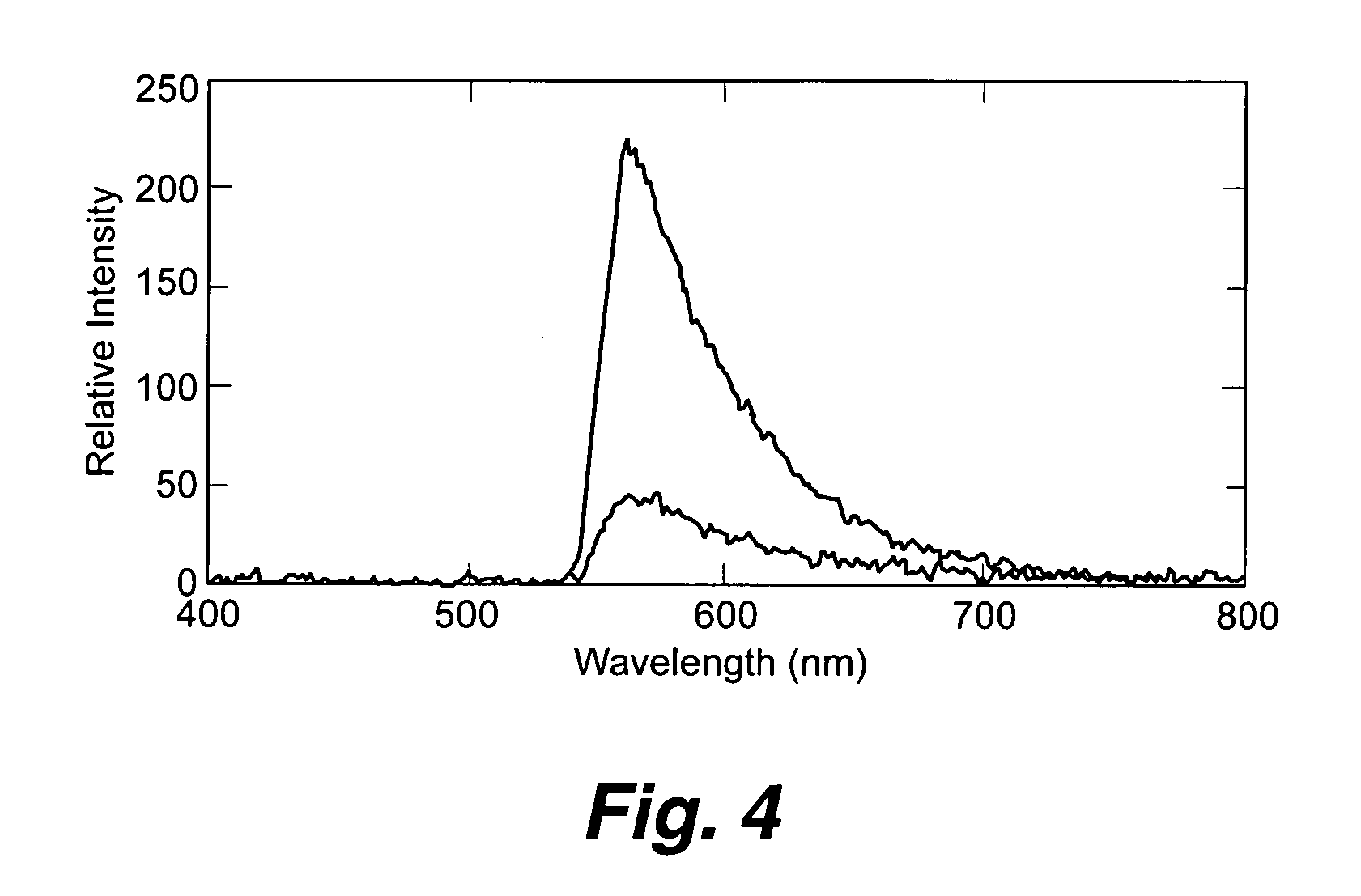
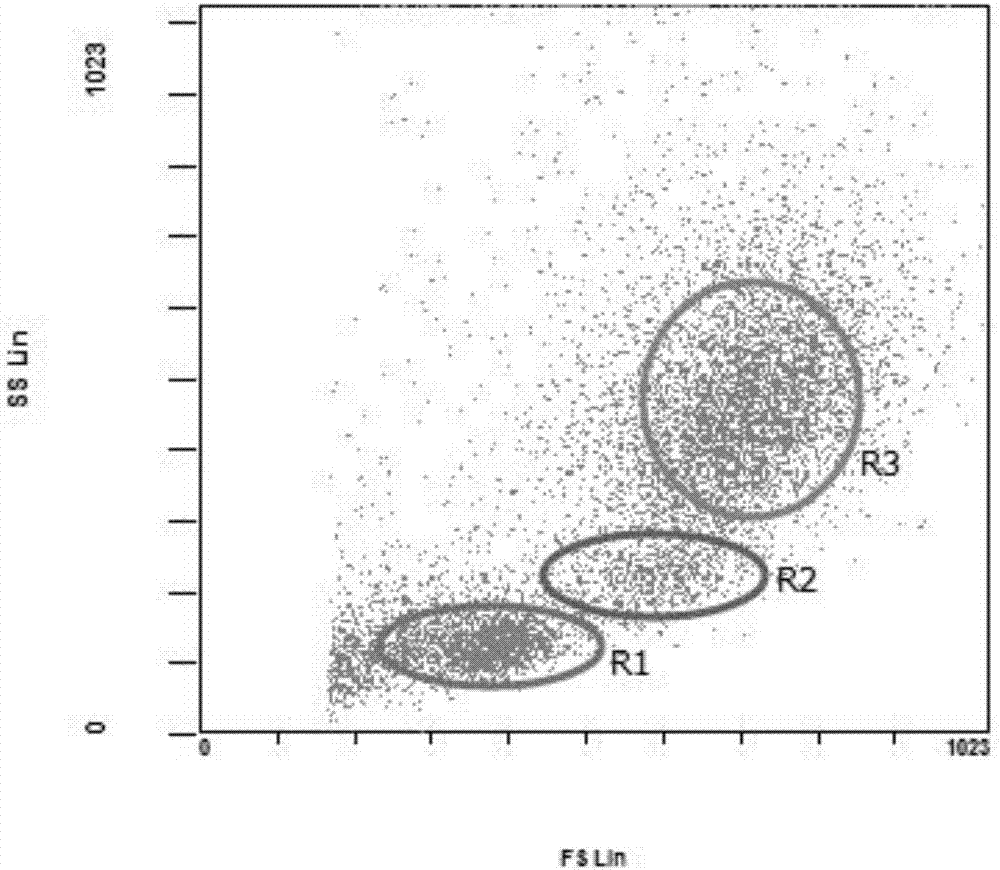
Planar optical waveguide based sandwich assay sensors and processes for the detection of biological targets including protein markers, pathogens and cellular debris
InactiveUS20060019244A1Enough timeBioreactor/fermenter combinationsMaterial nanotechnologyProtein markersSelf-assembled monolayer
An assay element is described including recognition ligands bound to a film on a single mode planar optical waveguide, the film from the group of a membrane, a polymerized bilayer membrane, and a self-assembled monolayer containing polyethylene glycol or polypropylene glycol groups therein and an assay process for detecting the presence of a biological target is described including injecting a biological target-containing sample into a sensor cell including the assay element, with the recognition ligands adapted for binding to selected biological targets, maintaining the sample within the sensor cell for time sufficient for binding to occur between selected biological targets within the sample and the recognition ligands, injecting a solution including a reporter ligand into the sensor cell; and, interrogating the sample within the sensor cell with excitation light from the waveguide, the excitation light provided by an evanescent field of the single mode penetrating into the biological target-containing sample to a distance of less than about 200 nanometers from the waveguide thereby exciting the fluorescent-label in any bound reporter ligand within a distance of less than about 200 nanometers from the waveguide and resulting in a detectable signal.
Owner:TRIAD NAT SECURITY LLC
Preparation method of fish single-cell suspension
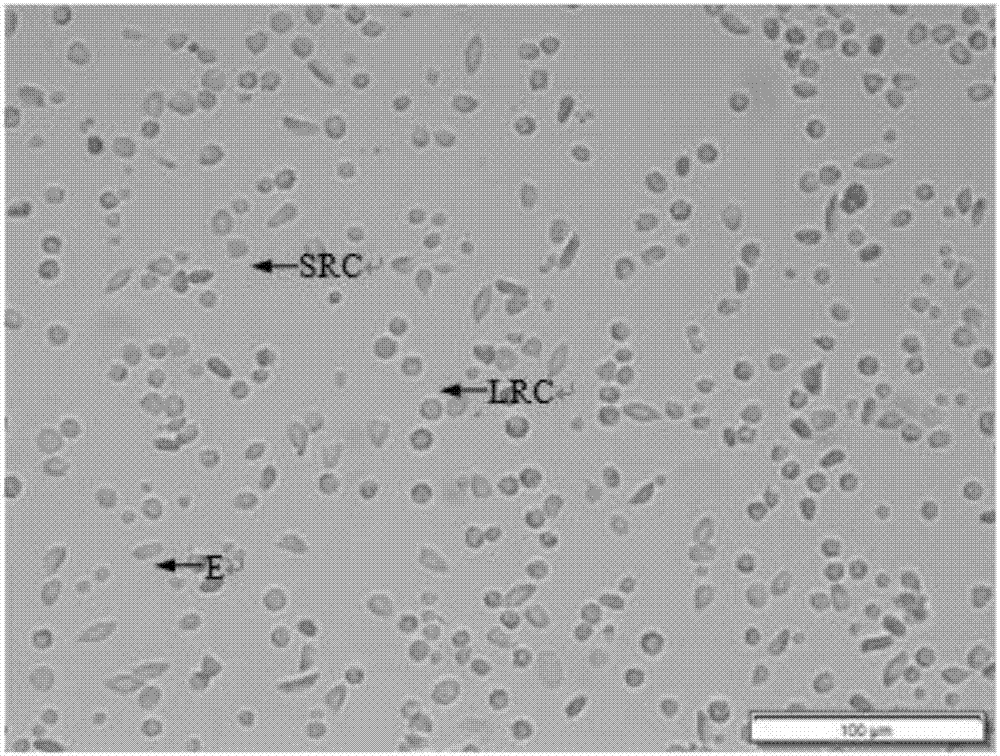
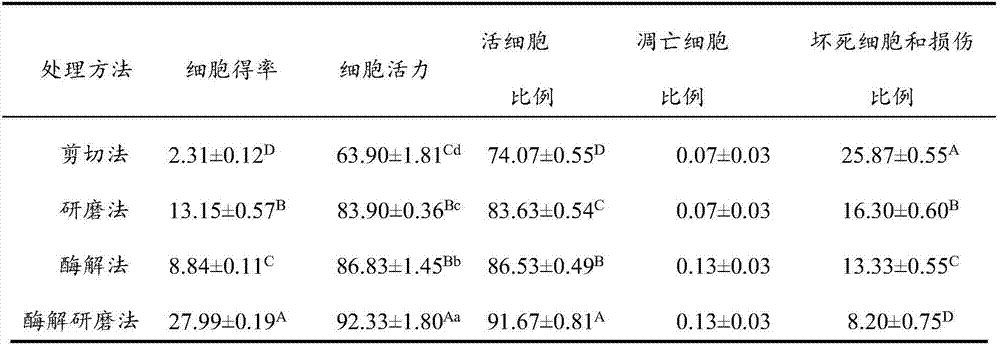
InactiveCN106929468ABest preparation methodGood dispersionCell dissociation methodsArtificial cell constructsEnzymatic digestionFiltration
The application discloses a preparation method of a fish single-cell suspension. The preparation method comprises the following steps: preparing a PBS reagent, a D-Han liquid and an IV type collagenase liquid, preparing a kidney tissue block of a fish, and washing the kidney tissue for more than three times with PBS; putting the kidney tissue block in a flat vessel of an ice bath, adding PBS, shearing the kidney tissue block into small pieces, and putting the tissue blocks in a high throughput tissue centrifuge tube of a burnisher; adding the IV type collagenase liquid into the centrifuge tube and meanwhile, adding PBS to fix the volume to 4ml, putting a zirconium oxide wall to grind, and performing enzymatic digestion on a tissue homogenate; and performing filtration, centrifugalization, washing with PBS, performing centrifugalization to abandon supernate and removing cell debris, filtering the cell suspension and fixing the volume to 3ml with PBS, and storing the cell suspension for later use at 4 DEG C. By adopting an enzymatic hydrolysis grinding method, advantages of a griding method and an enzymatic hydrolysis method are combined. The tissue is more fully treated by enzymatic hydrolysis grinding, so that the problem that the cell yield is low and the cell viability is low for preparing the fish single-cell suspension can be solved.
Owner:SOUTHWEST UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com