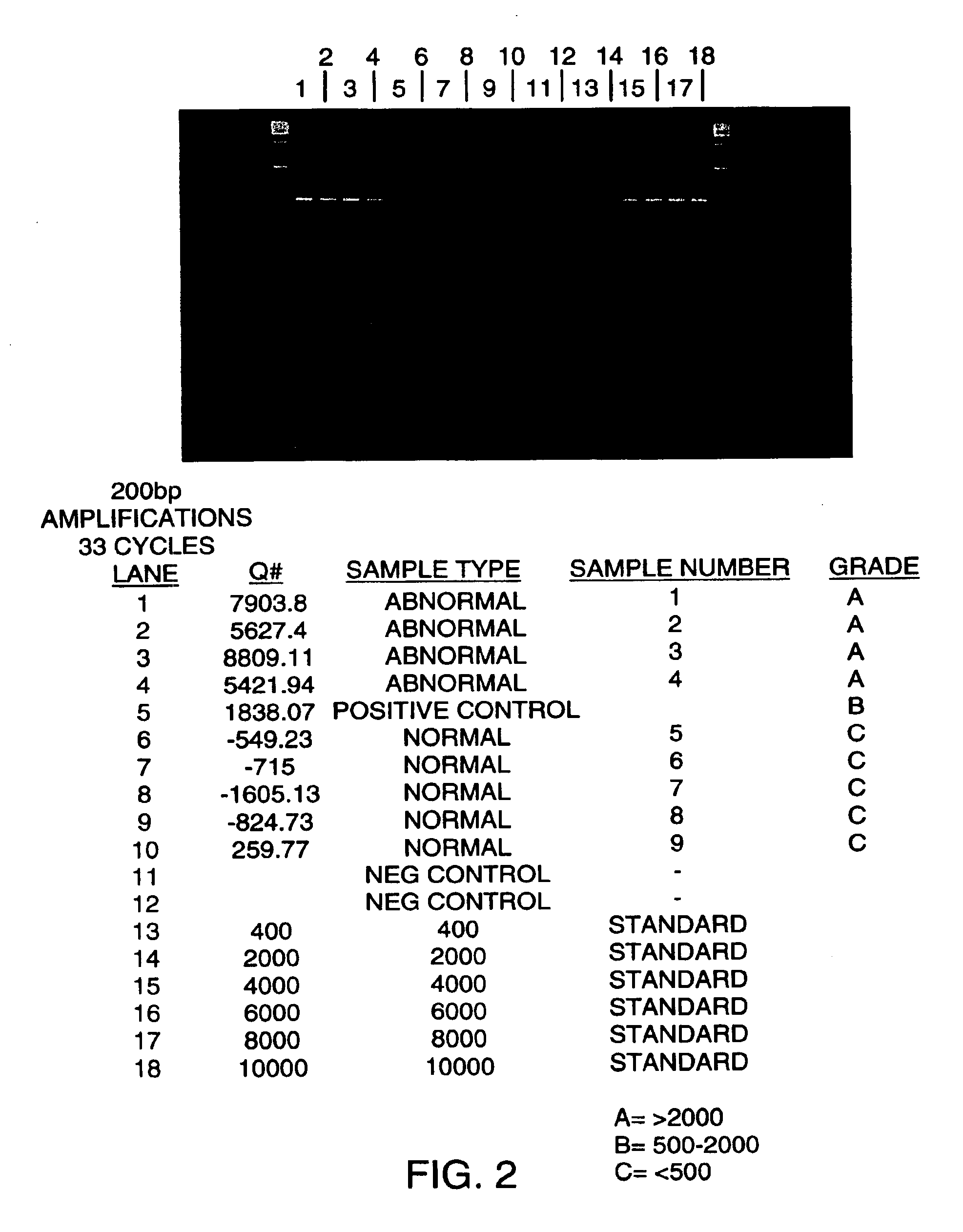
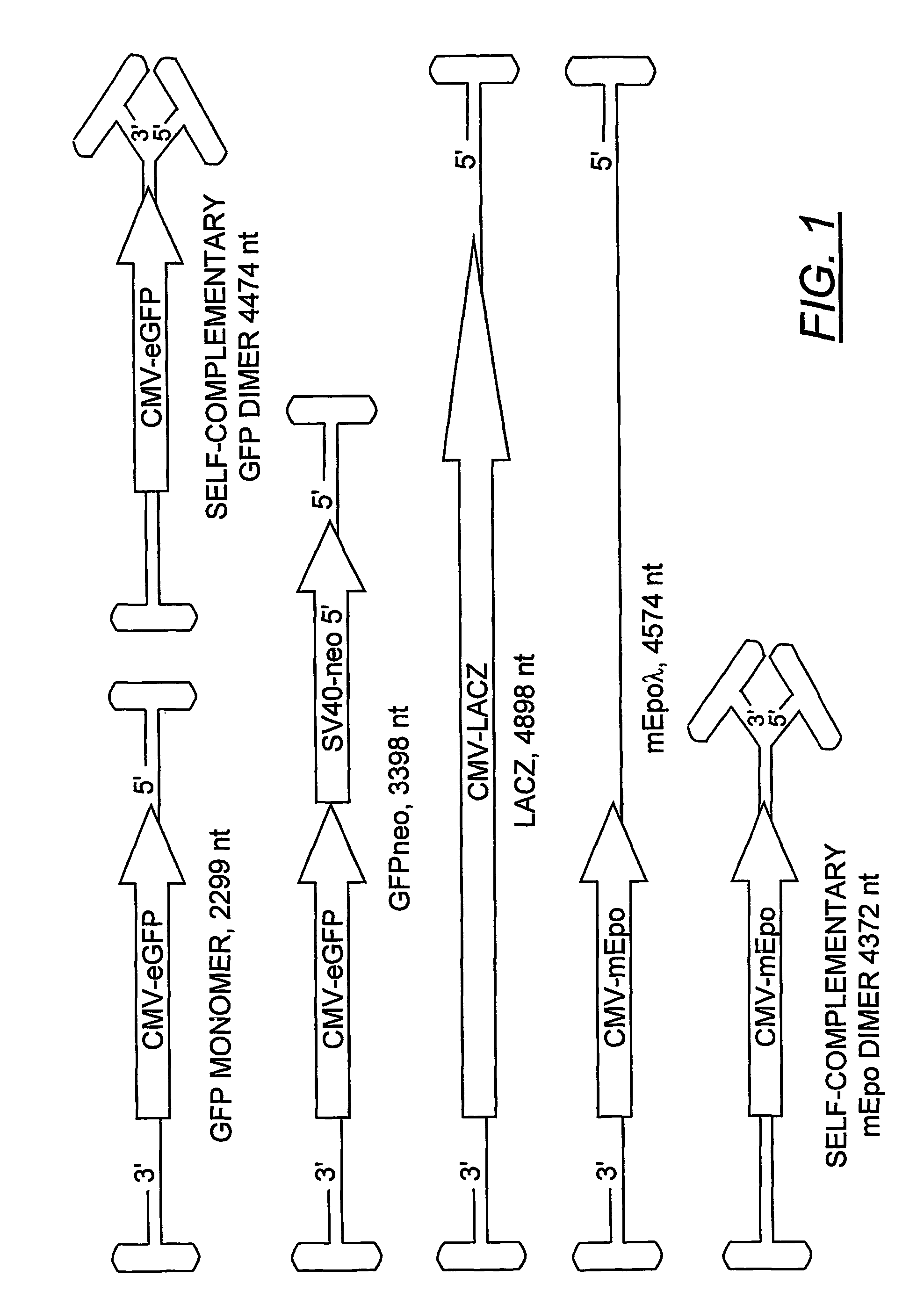
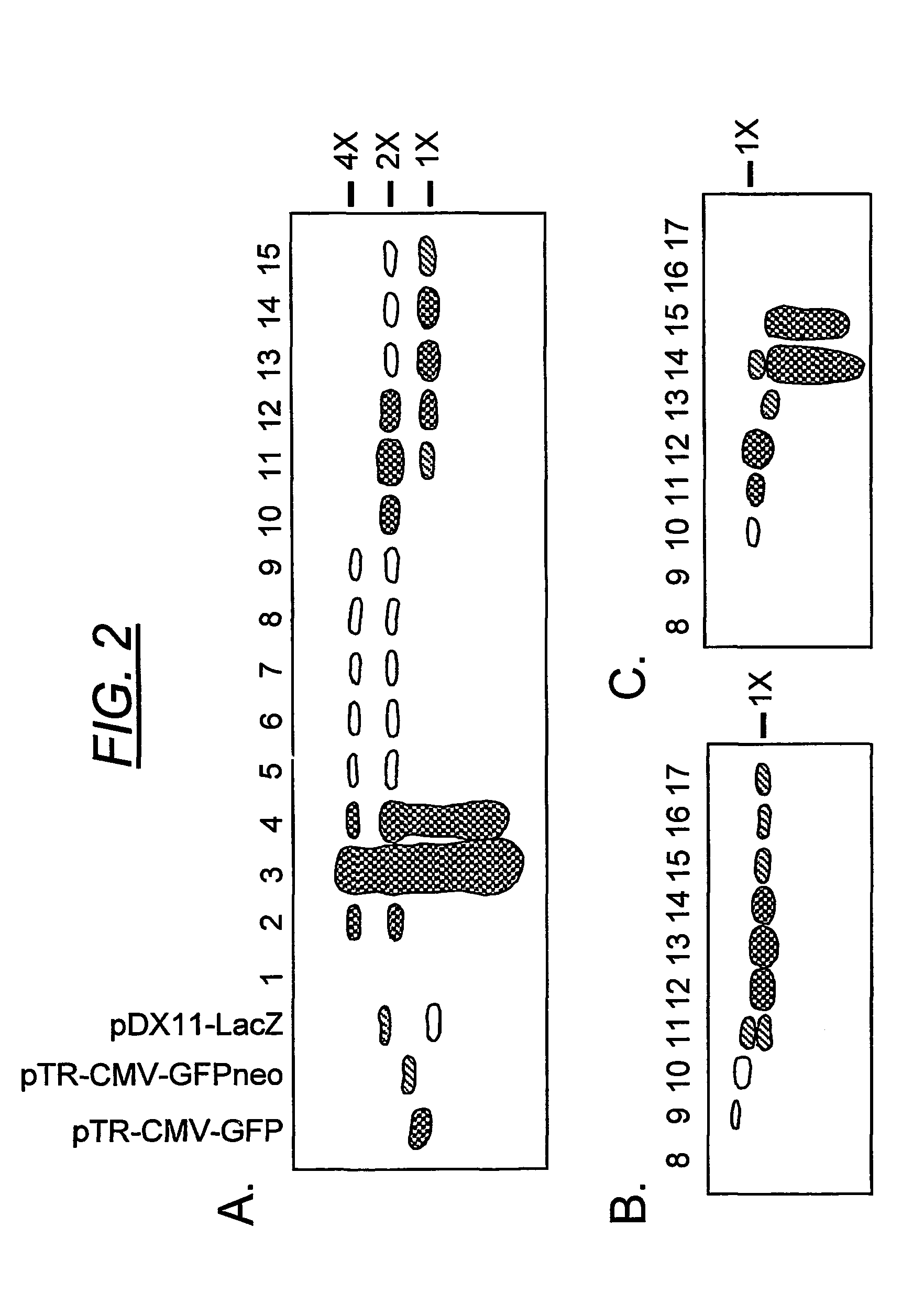
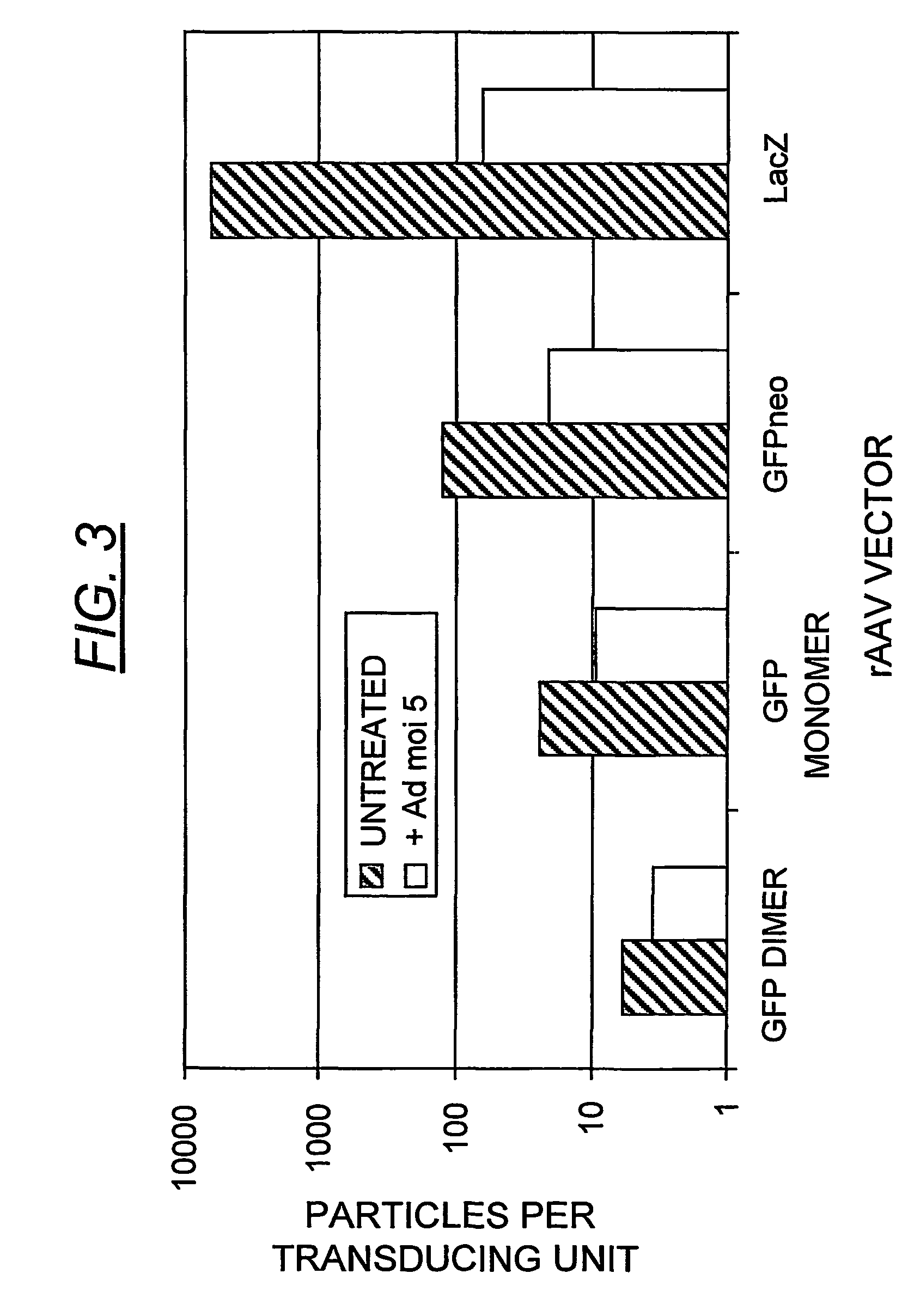
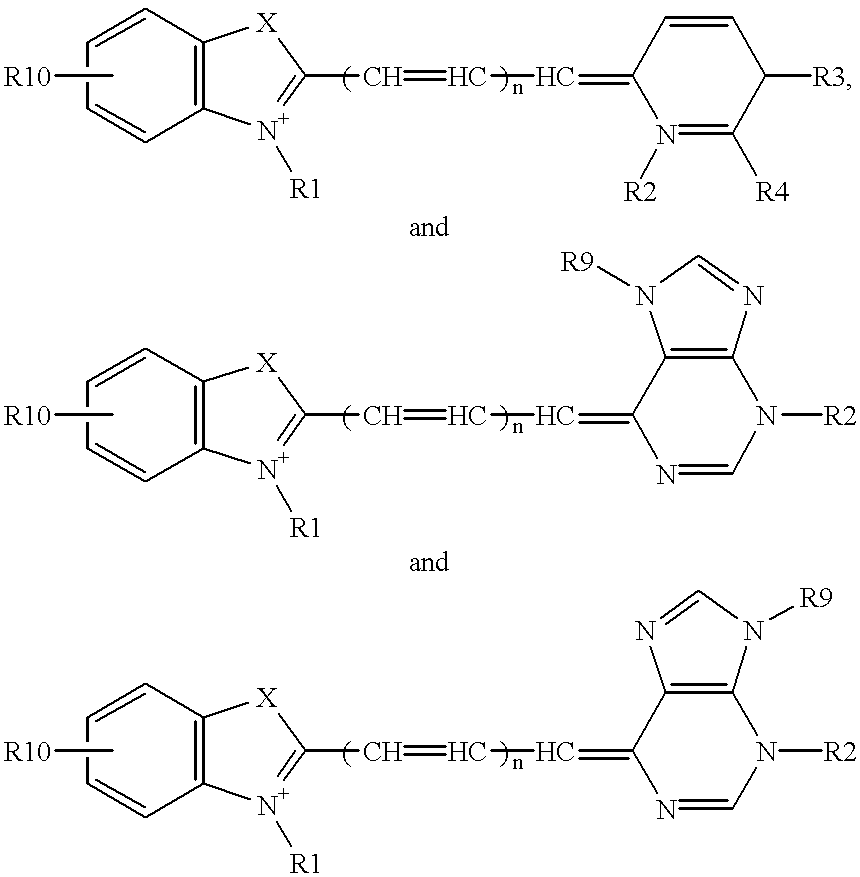
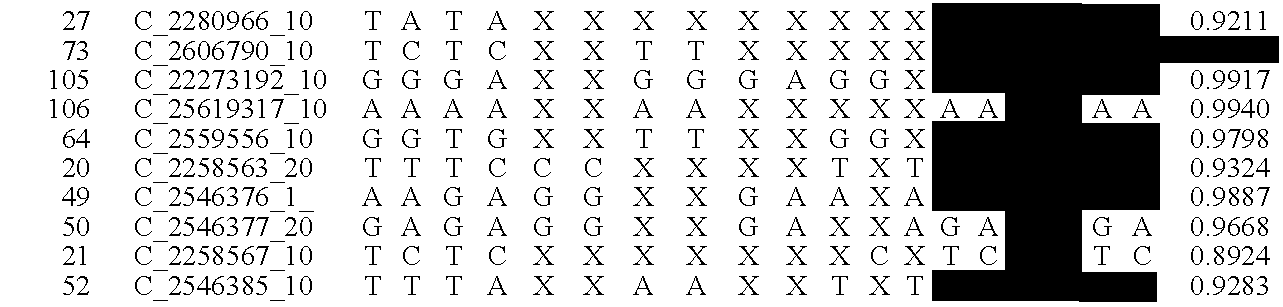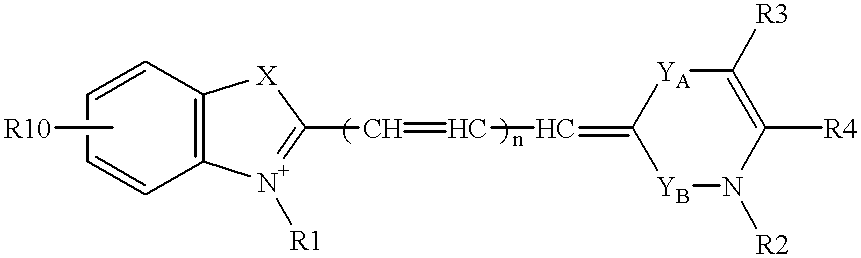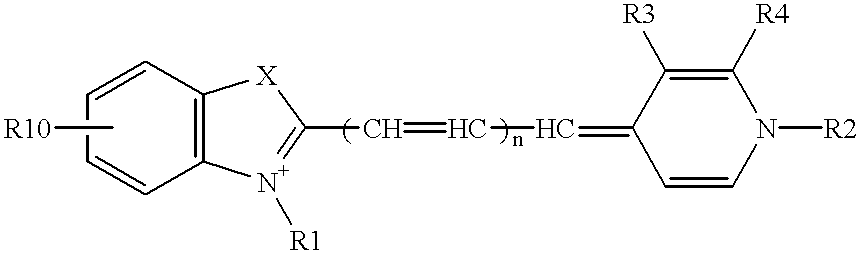Patents
Literature
743 results about "Base pair" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
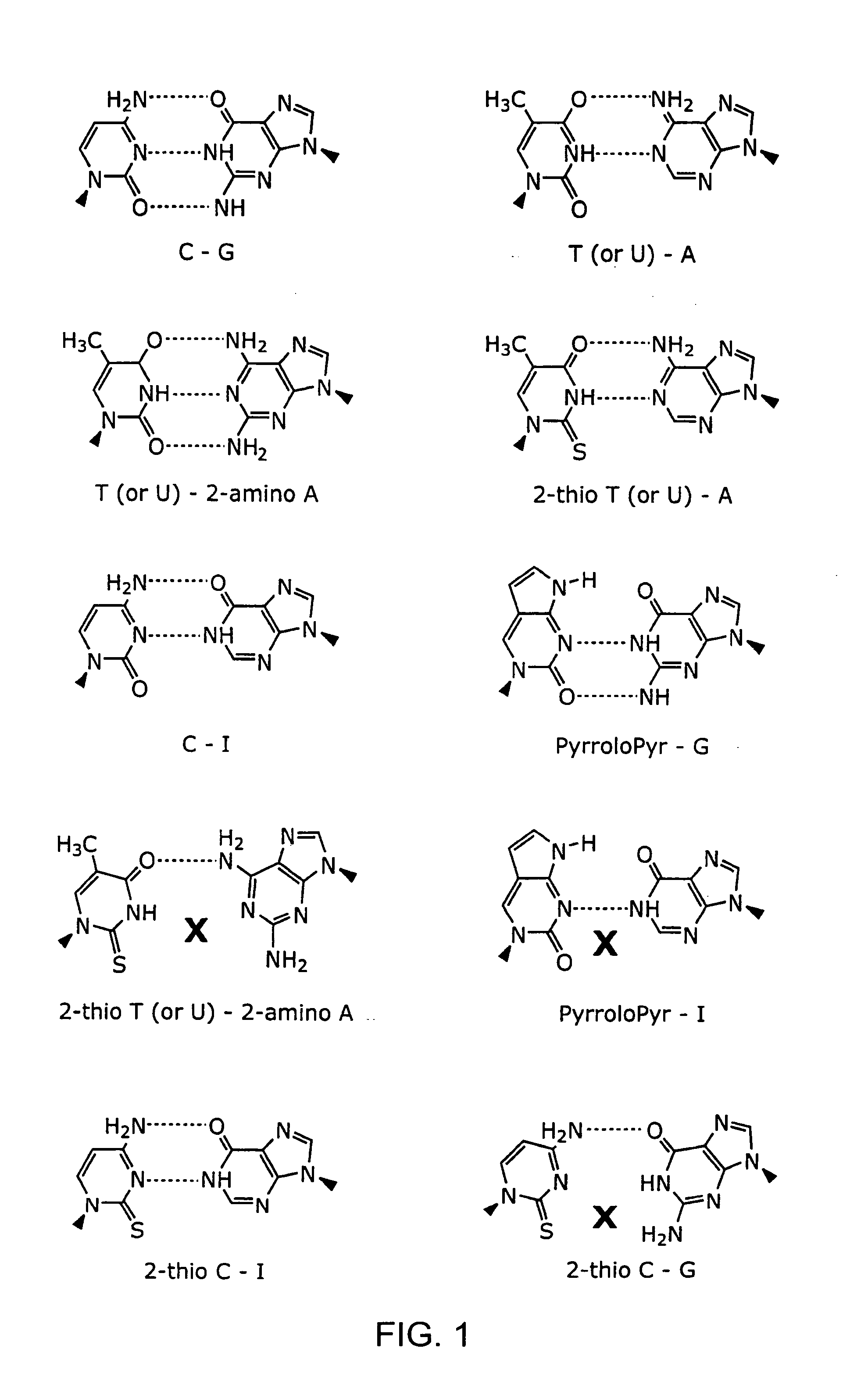
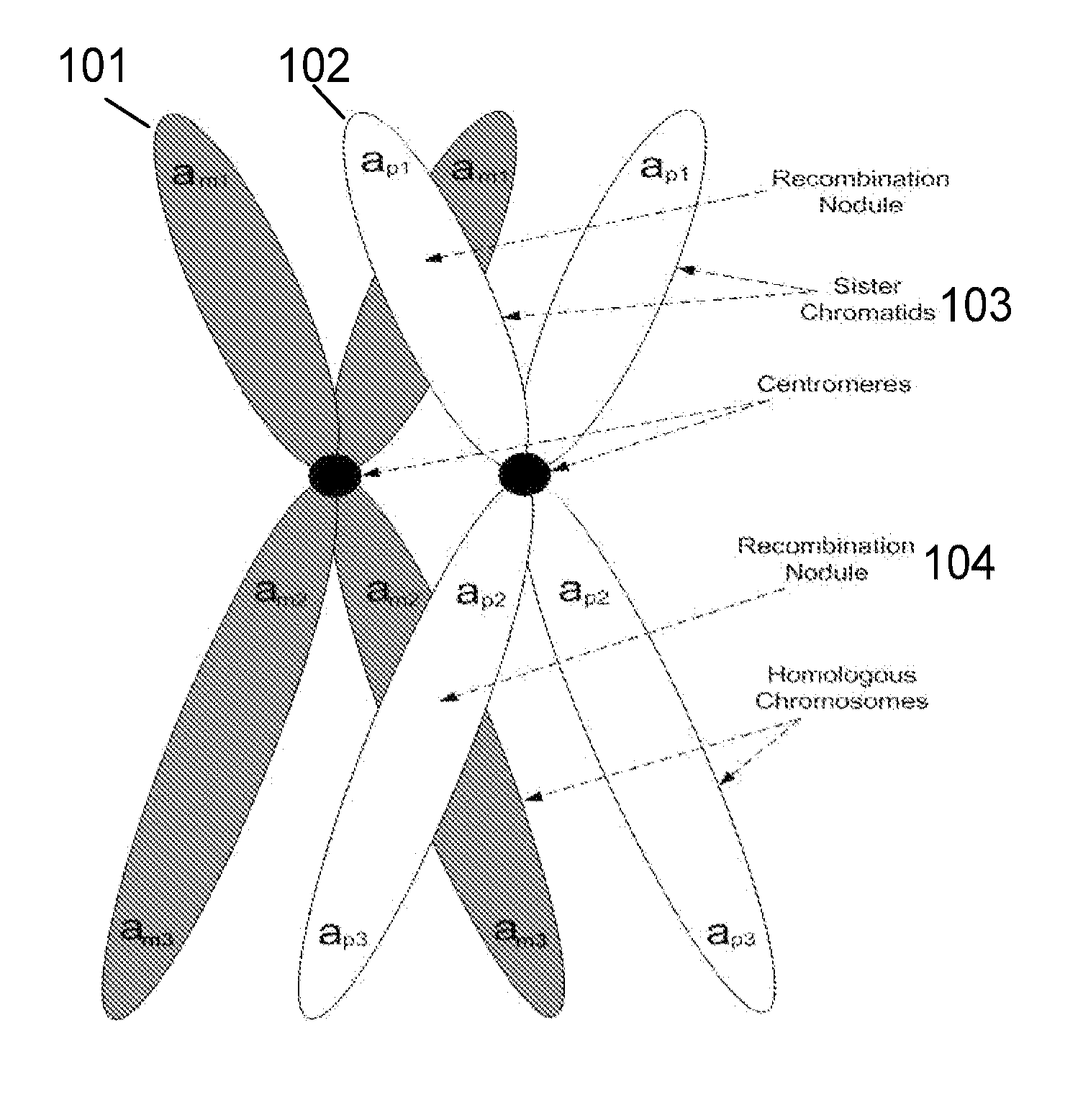
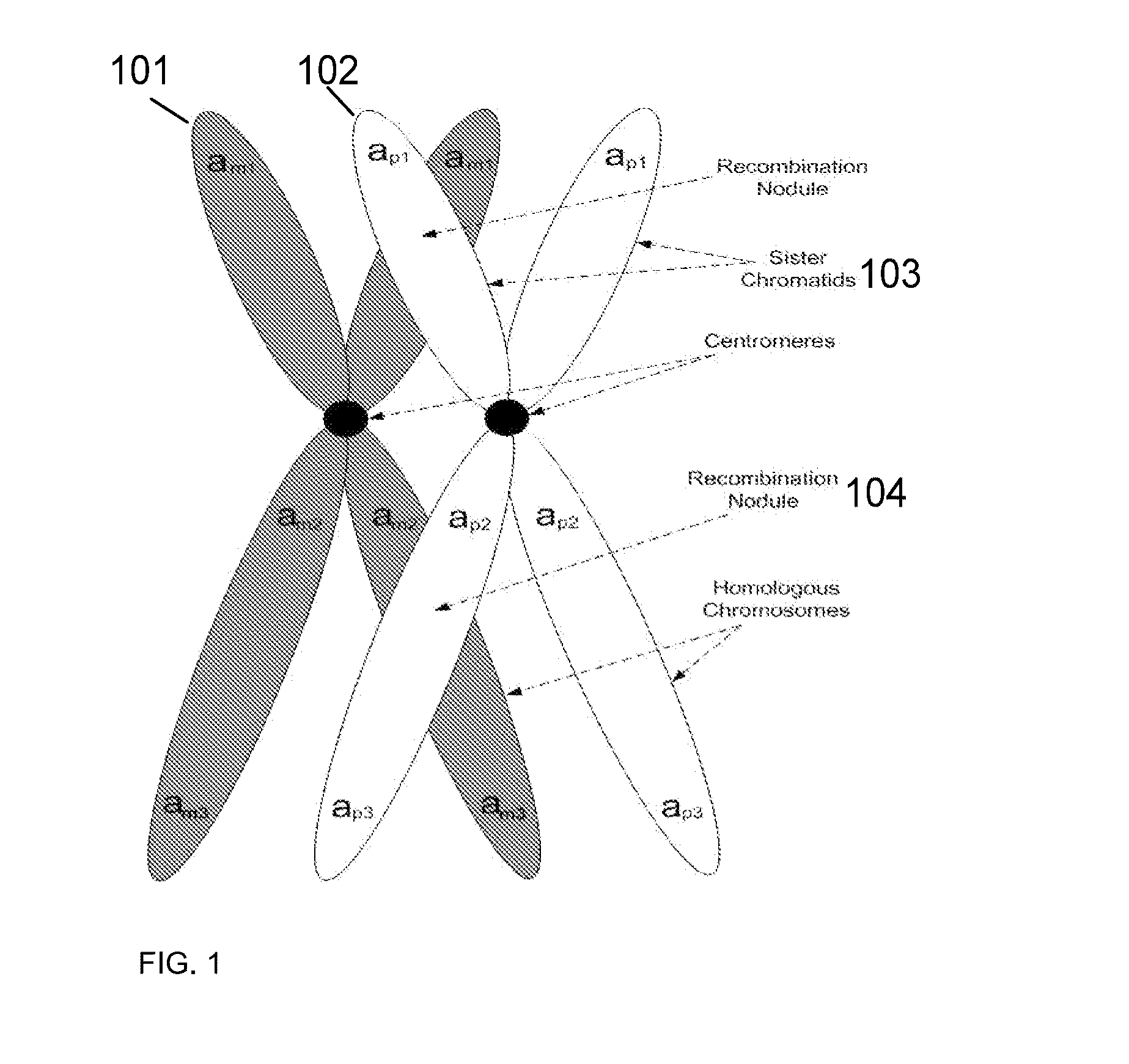
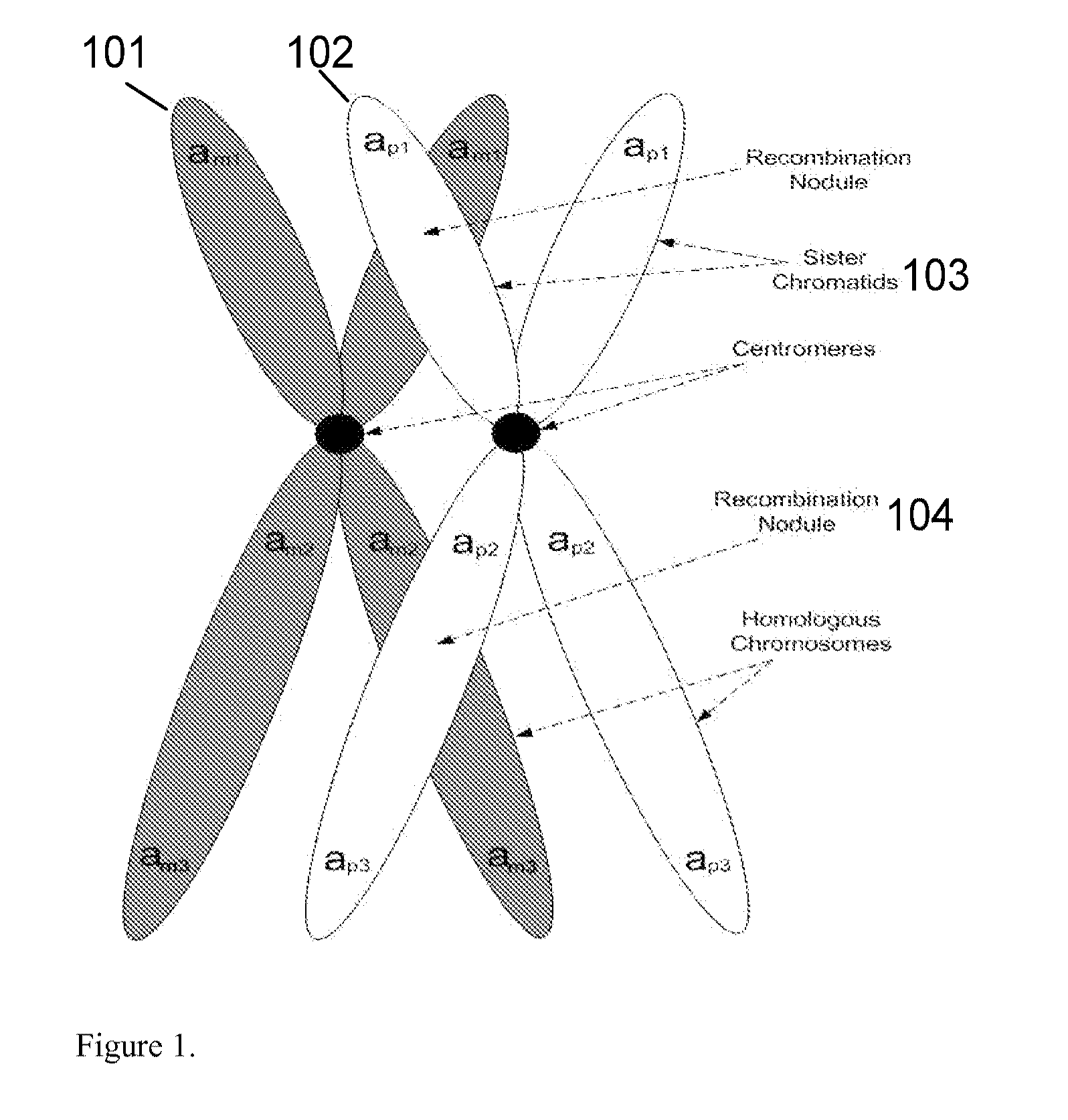
A base pair (bp) is a unit consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, Watson–Crick base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins can recognize specific base-pairing patterns that identify particular regulatory regions of genes.
Non-invasive detection of fetal genetic traits
InactiveUS20050164241A1Facilitates non-invasive detectionComponent separationOther chemical processesPregnancyNon invasive
Blood plasma of pregnant women contains fetal and (generally>90%) maternal circulatory extracellular DNA. Most of said fetal DNA contains ≦500 base pairs, said maternal DNA having a greater size. Separation of circulatory extracellular DNA of <500 base pairs results in separation of fetal from maternal DNA. A fraction of a blood plasma or serum sample of a pregnant woman containing, due to size separation (e.g. by chromatography, density gradient centrifugation or nanotechnological methods), extracellular DNA substantially comprising ≦500 base pairs is useful for non-invasive detection of fetal genetic traits (including the fetal RhD gene in pregnancies at risk for HDN; fetal Y chromosome-specific sequences in pregnancies at risk for X chromosome-linked disorders; chromosomal aberrations; hereditary Mendelian genetic disorders and corresponding genetic markers; and traits decisive for paternity determination) by e.g. PCR, ligand chain reaction or probe hybridization techniques, or nucleic acid arrays.
Owner:SEQUENOM INC
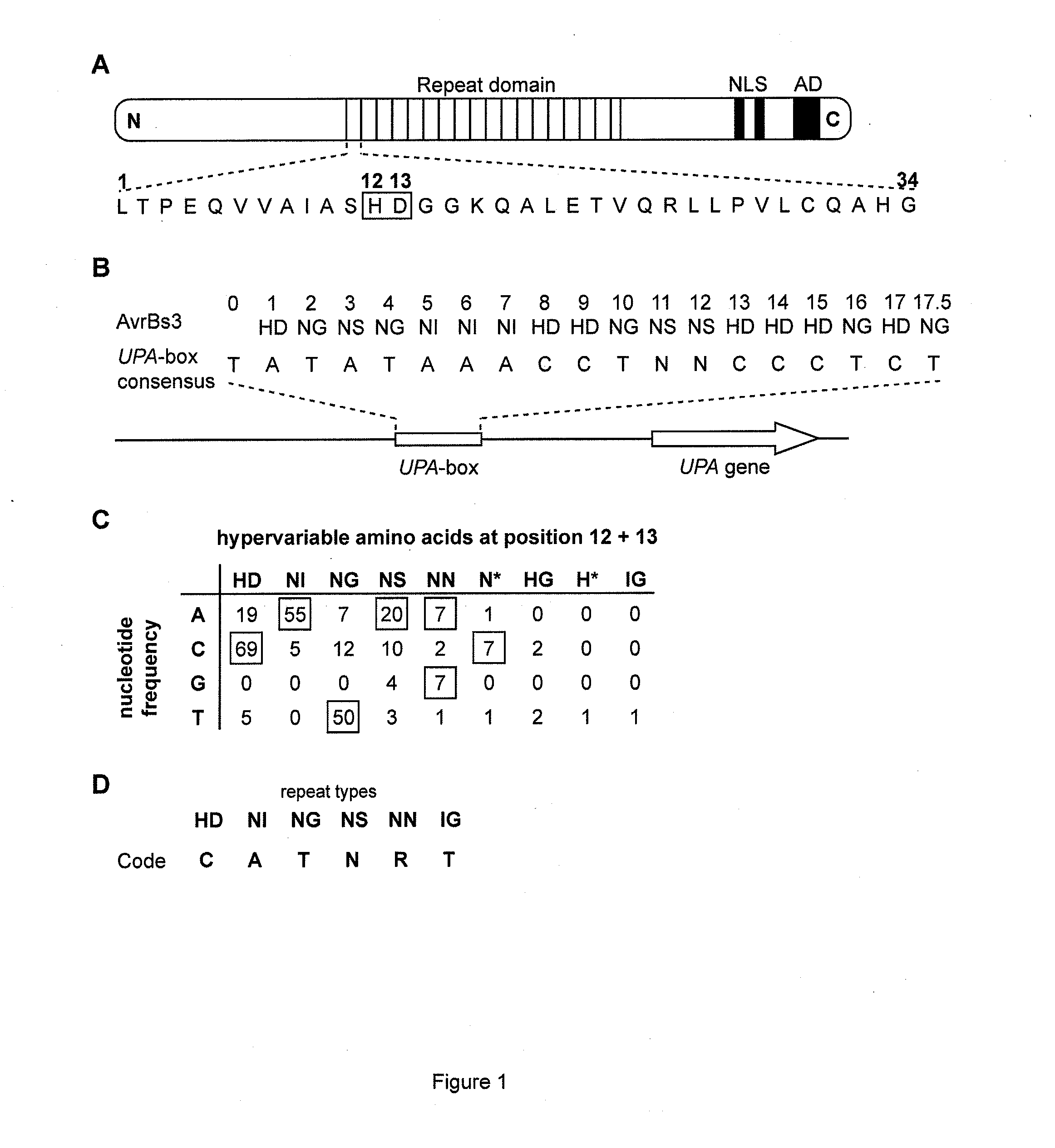
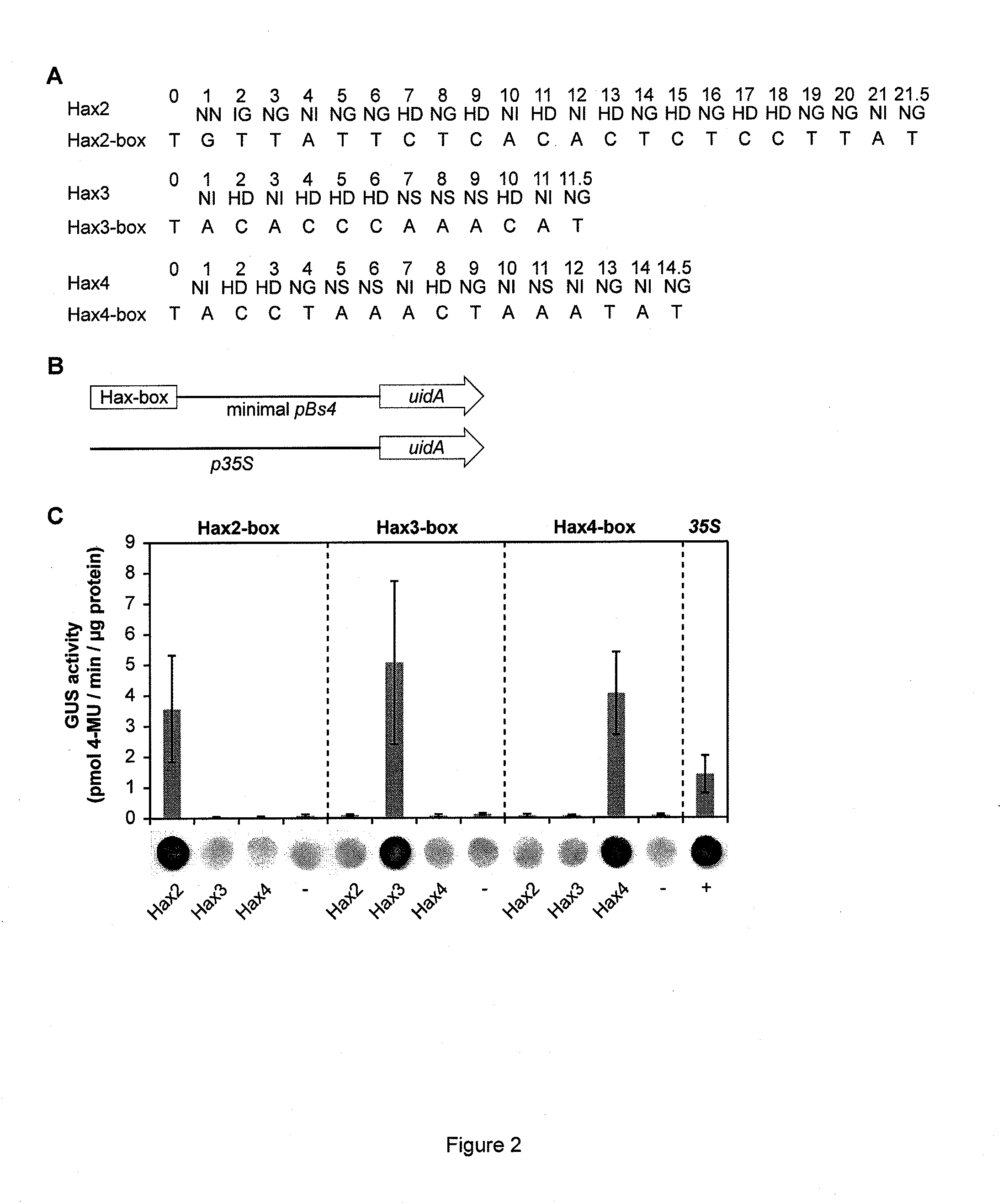
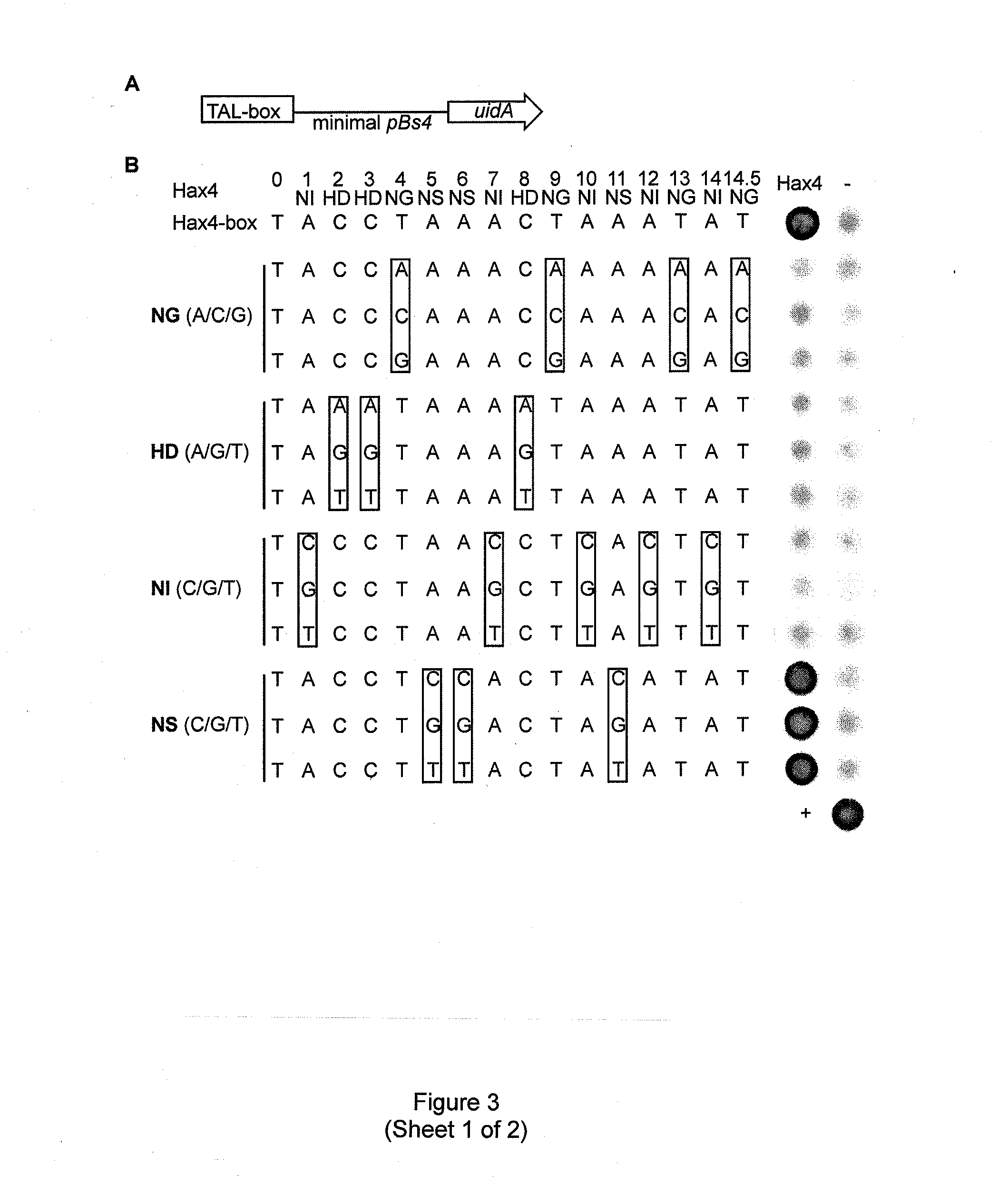
Modular dna-binding domains and methods of use
InactiveUS20110239315A1Enabling targeted DNA modificationFungiBacteriaDNA-binding domainDna targeting
The present invention refers to methods for selectively recognizing a base pair in a DNA sequence by a polypeptide, to modified polypeptides which specifically recognize one or more base pairs in a DNA sequence and, to DNA which is modified so that it can be specifically recognized by a polypeptide and to uses of the polypeptide and DNA in specific DNA targeting as well as to methods of modulating expression of target genes in a cell.
Owner:BONAS ULLA +2
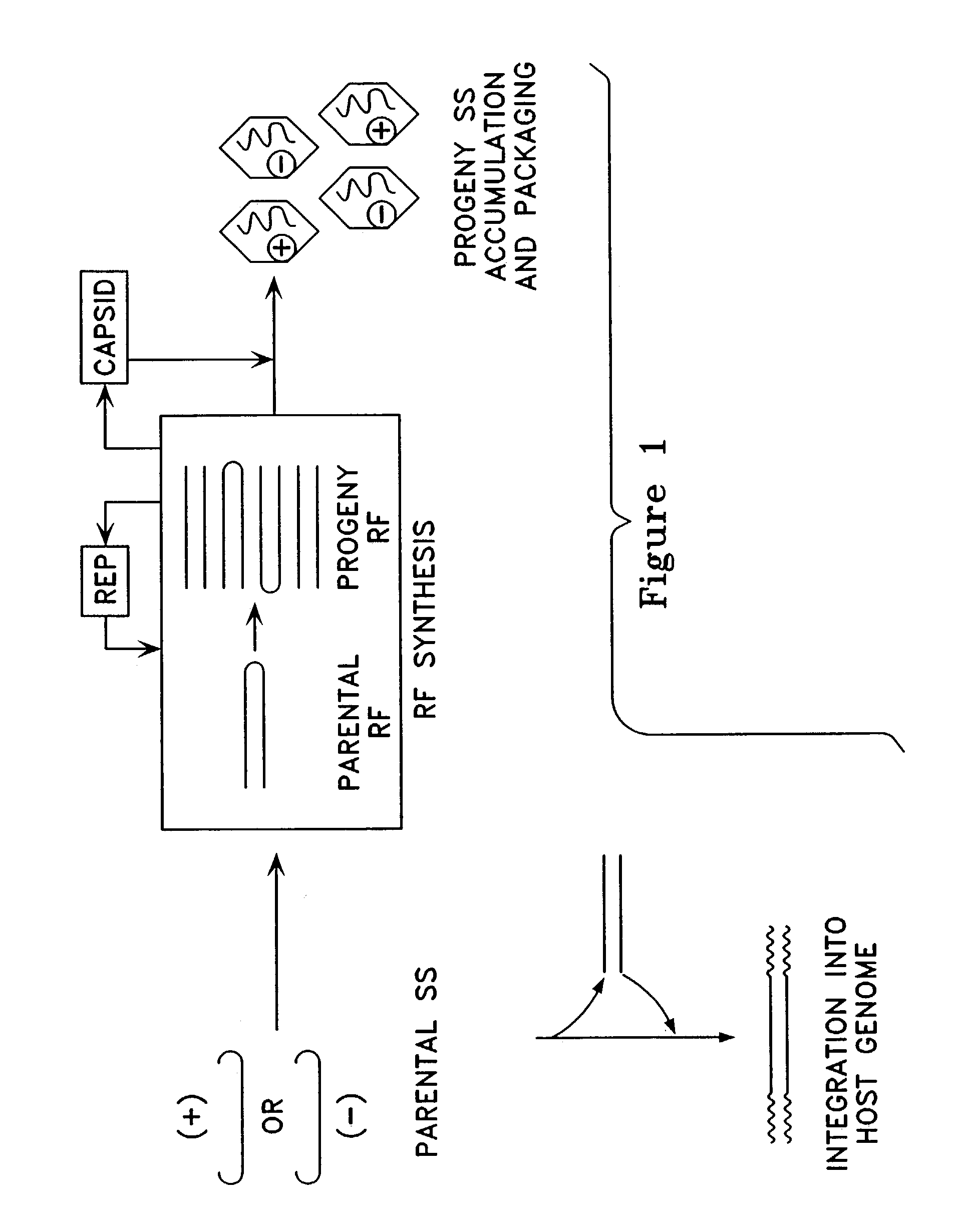
Metabolically activated recombinant viral vectors and methods for their preparation and use
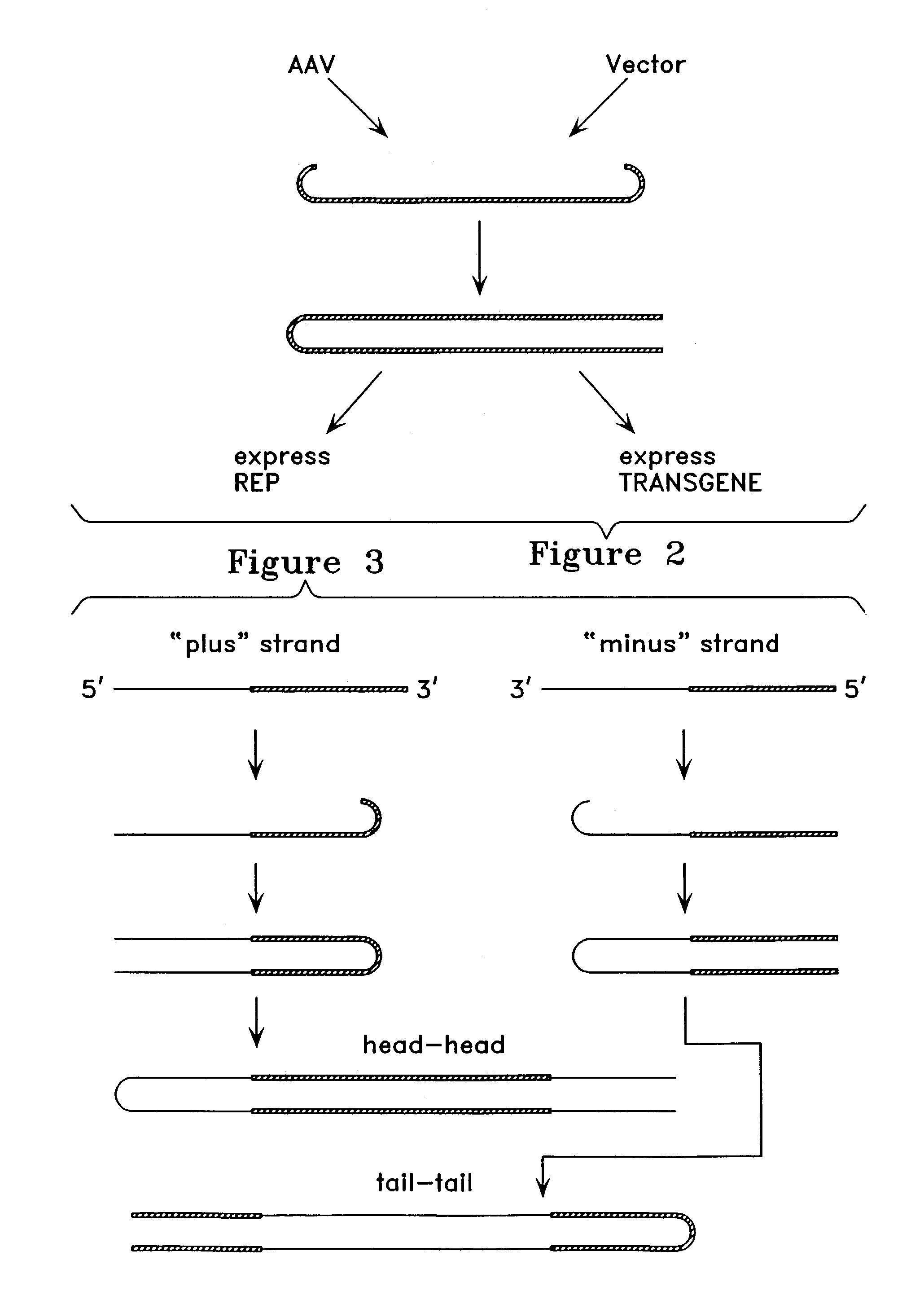
Recombinant viral vectors, especially parvovirus vectors such as adeno-associated virus (AAV) vectors, capable of enhanced expression of heterologous sequences, and methods for their construction and use, are provided. The vectors have a structure, or are capable of rapidly adopting a structure, which involves intrastrand base pairing of at least one region in a heterologous sequence.
Owner:GENZYME CORP
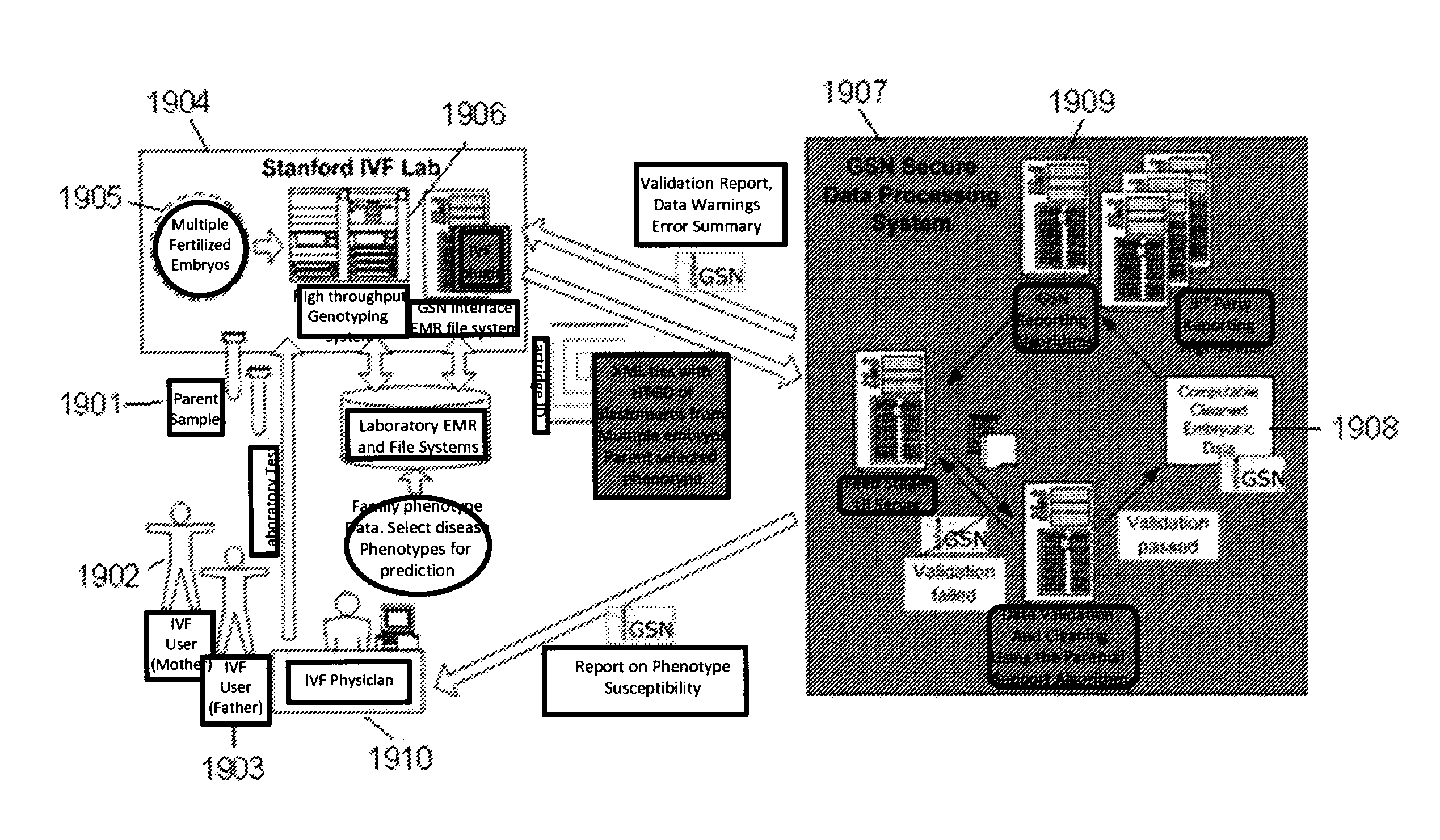
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
ActiveUS20070184467A1Significant resultMicrobiological testing/measurementProteomicsUniparental disomyEmbryo
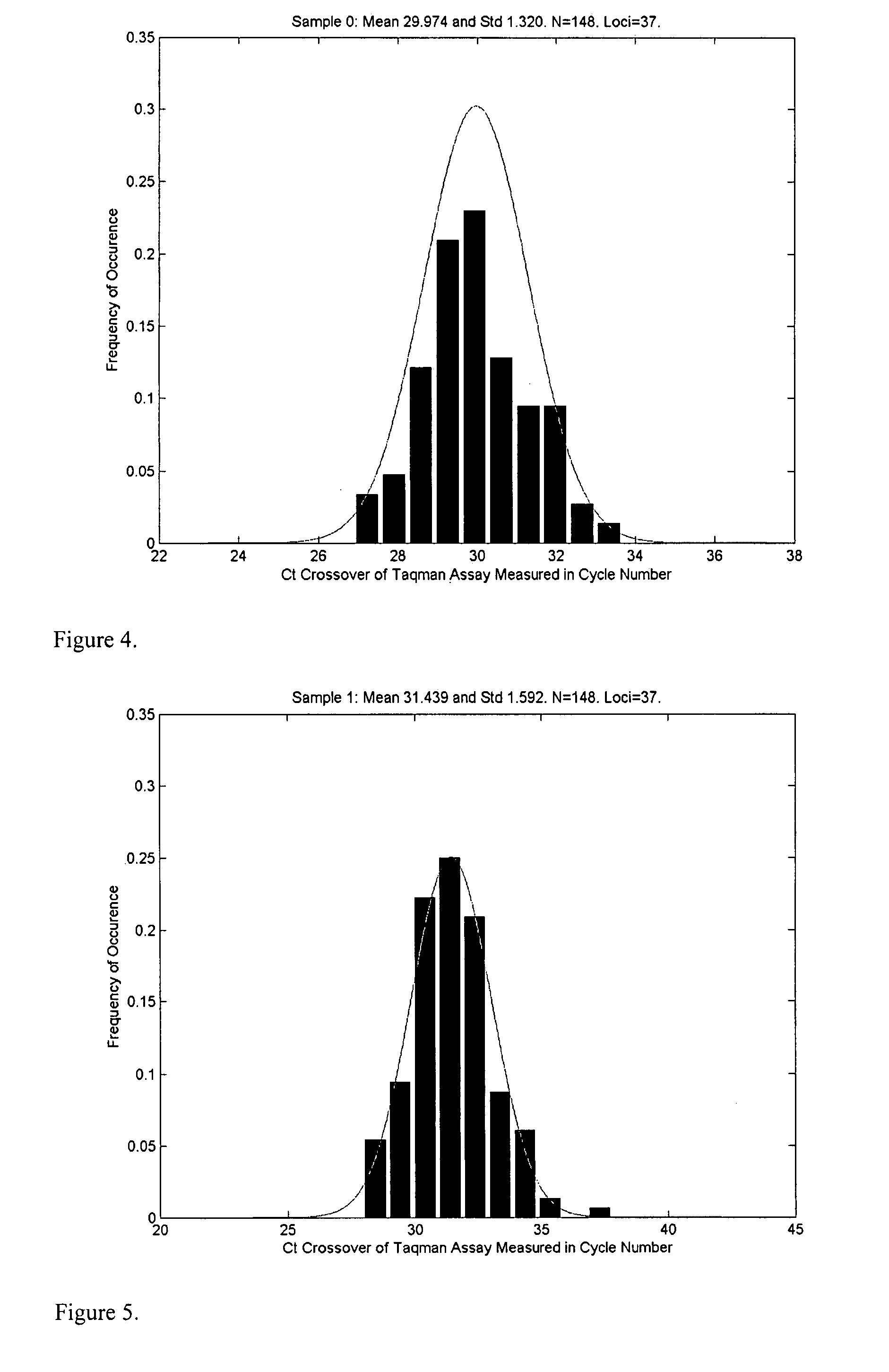
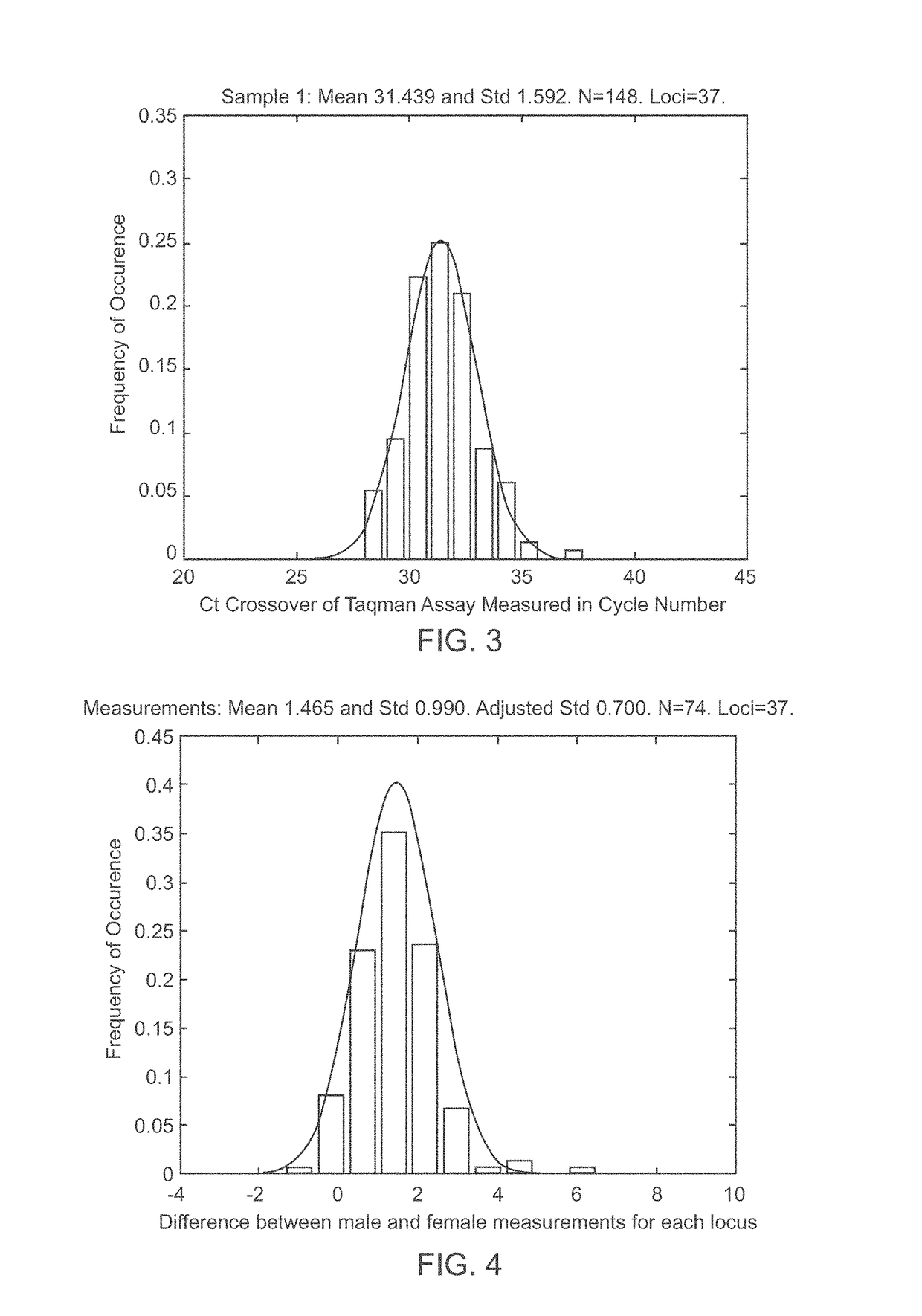
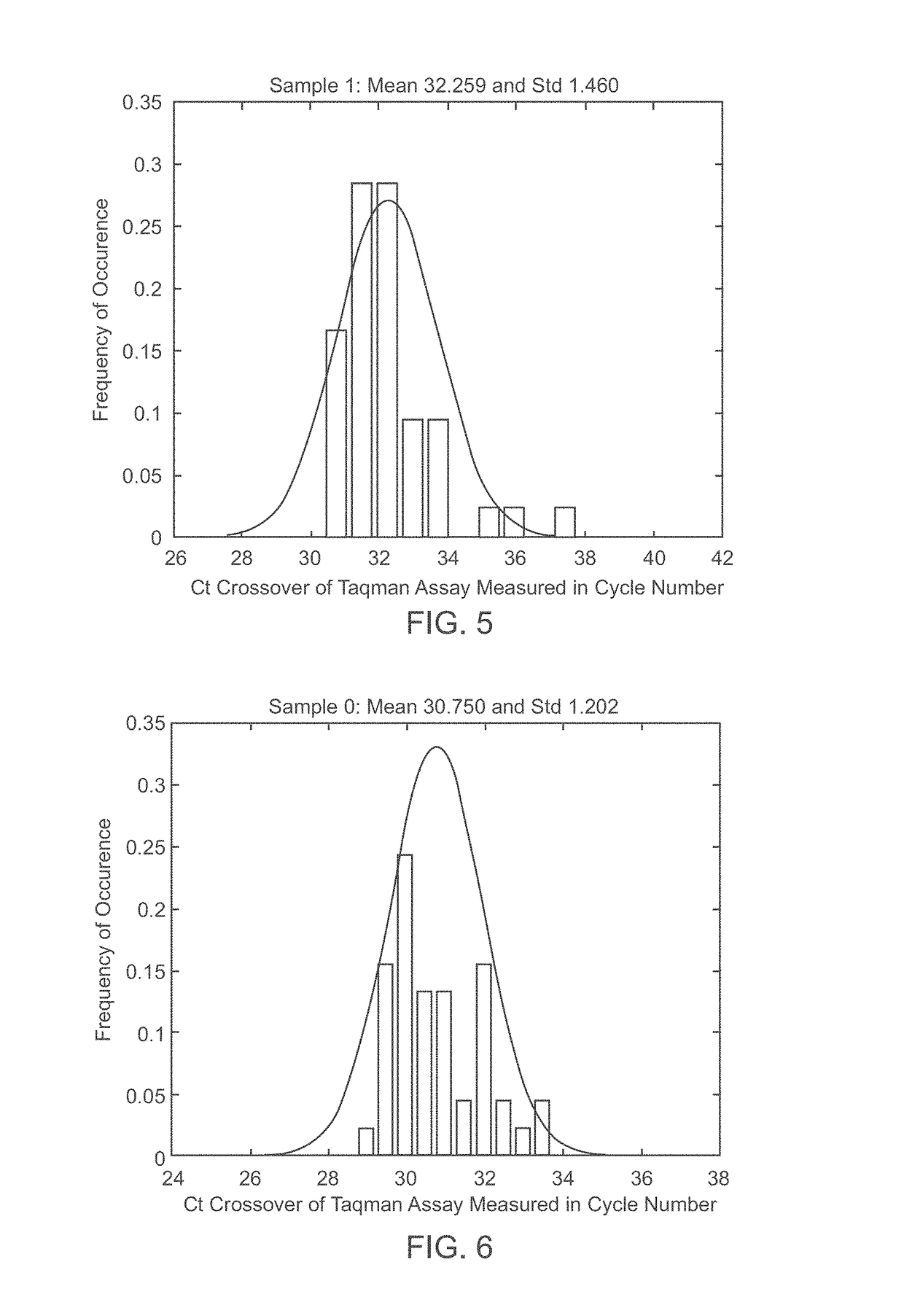
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell is reconstructed using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In accordance with another embodiment of the invention, incomplete genetic data from a fetus is acquired from fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In one embodiment, the genetic data can be reconstructed for the purposes of making phenotypic predictions. In another embodiment, the genetic data can be used to detect for aneuploides and uniparental disomy.
Owner:NATERA
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS20080243398A1Significant resultImprove fidelityData processing applicationsMicrobiological testing/measurementGenetic MaterialsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic material from the target individual is acquired, amplified and the genetic data is measured using known methods. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment of the invention, the chromosome copy number can be determined from the measured genetic data of a single or small number of cells, with or without genetic information from one or both parents. In another embodiment of the invention, these determinations are made for the purpose of embryo selection in the context of in-vitro fertilization. In another embodiment of the invention, the genetic data can be reconstructed for the purposes of making phenotypic predictions.
Owner:NATERA
Oligonucleotide analogs having cationic intersubunit linkages
ActiveUS7943762B2High expressionHigh activitySugar derivativesActivity regulationNeutral Amino AcidsOligomer
Owner:AVI BIOPHARMA
P element derived vector and methods for its use
Novel P element derived vectors and methods for their use to insert an exogenous nucleic acid into the genome of a target cell are provided. The subject vectors have a pair of P element transposase recognized insertion sites, e.g. 31 base pair inverted repeats, flanking at least two transcriptionally active genes. In practicing the subject methods, a vector of the subject invention is introduced into the target cell under conditions sufficient for transposition to occur. The subject methods find use in a variety of applications in which the insertion of an exogenous nucleic acid into the genome of a target cell is desired, e.g. include research, synthesis and therapeutic applications.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Non-invasive detection of fetal genetic traits
ActiveUS20080071076A1Facilitates non-invasive detectionSugar derivativesOther chemical processesPregnancyNon invasive
Blood plasma of pregnant women contains fetal and (generally>90%) maternal circulatory extracellular DNA. Most of said fetal DNA contains .Itoreq.500 base pairs, said maternal DNA having a greater size. Separation of circulatory extracellular DNA of .Itoreq.500 base pairs results in separation of fetal from maternal DNA. A fraction of a blood plasma or serum sample of a pregnant woman containing, due to size separation (e.g. by chromatography, density gradient centrifugation or nanotechnological methods), extracellular DNA substantially comprising .Itoreq.500 base pairs is useful for non-invasive detection of fetal genetic traits (including the fetal RhD gene in pregnancies at risk for HDN; fetal Y chromosome-specific sequences in pregnancies at risk for X chromosome-linked disorders; chromosomal aberrations; hereditary Mendelian genetic disorders and corresponding genetic markers; and traits decisive for paternity determination) by e.g. PCR, ligand chain reaction or probe hybridization techniques, or nucleic acid arrays.
Owner:SEQUENOM INC
Modulating ph-sensitive binding using non-natural amino acids
InactiveUS20090035836A1High affinitySugar derivativesPeptide/protein ingredientsAbnormal tissue growthSide chain
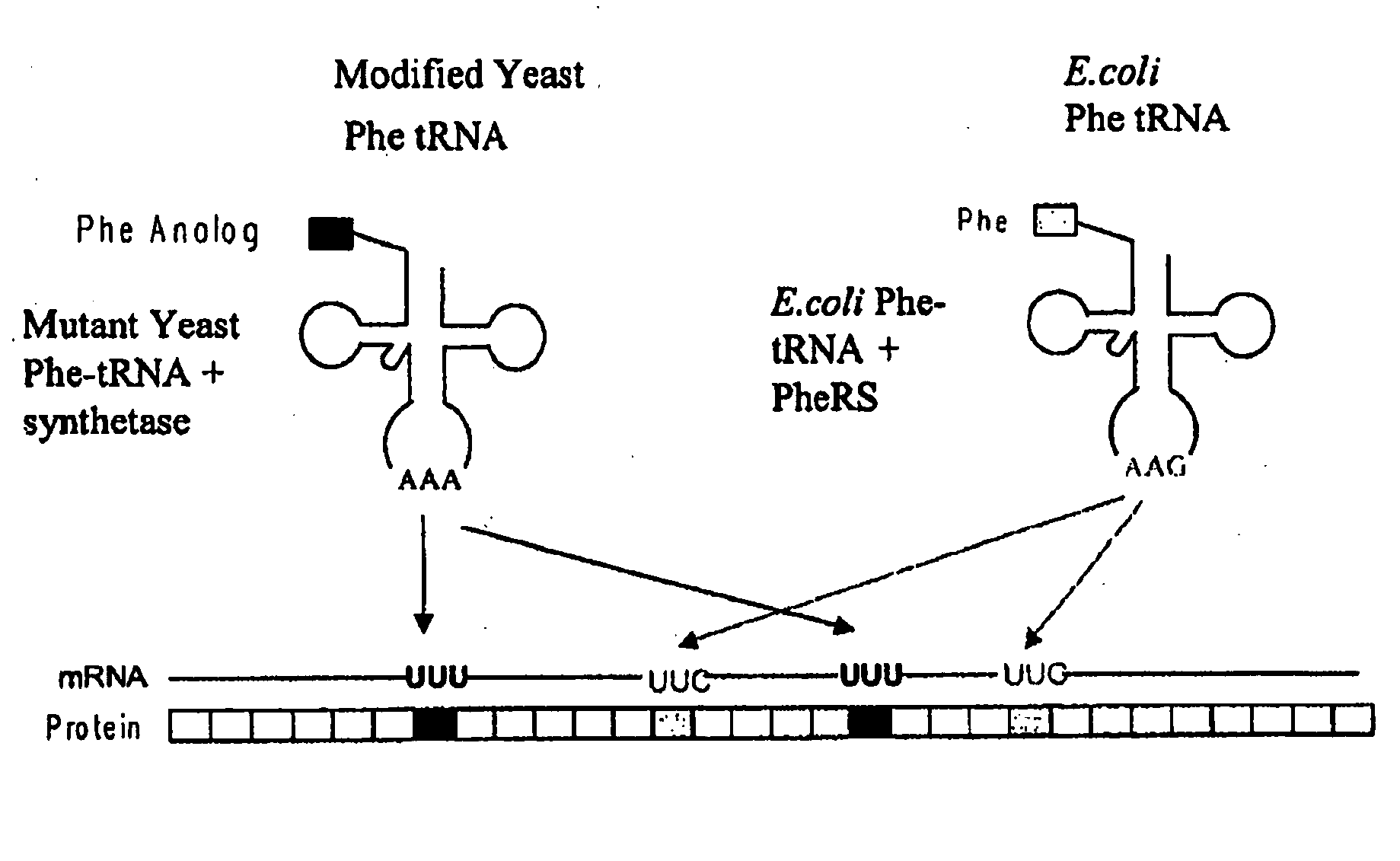
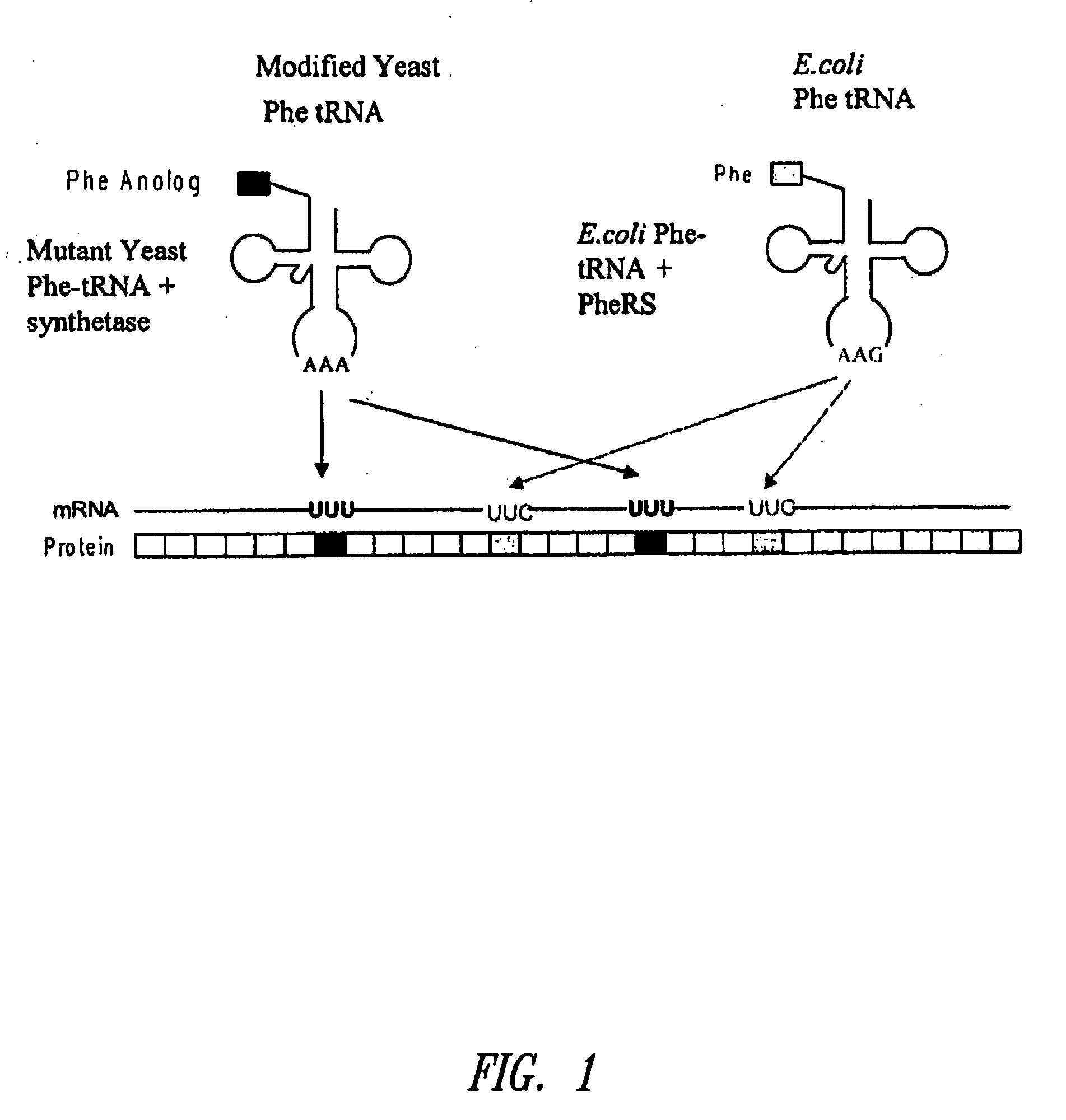
The invention provides methods, systems and reagents for regulating pH-sensitive protein interaction by incorporating non-natural amino acids into the protein (e.g. an antibody, or its functional fragment, derivative, etc.). The invention also relates to specific uses in regulating pH-sensitive binding of antibodies to tumor site, by conferring enhanced tumor-specificity / selectivity. In that embodiment, the non-natural amino acids preferably have desirable side-chain pKa's, such that at below physiological pH (e.g. about pH 6.3-6.5) the non-natural amino acid confer enhanced binding to tumor antigens in acidic environments. Such non-natural amino acids can be incorporated by any suitable means, such as by utilizing a modified aminoacyl-tRNA synthetase to charge the nonstandard amino acid to a modified tRNA, which forms strict Watson-Crick base-pairing with a codon that normally forms wobble base-pairing with natural tRNAs (e.g. the degenerate codon orthogonal system.
Owner:CALIFORNIA INST OF TECH
Oligonucleotide analogs having cationic intersubunit linkages
ActiveUS20090088562A1Avoid infectionReduce viral infectionSugar derivativesActivity regulationNeutral Amino AcidsOligomer
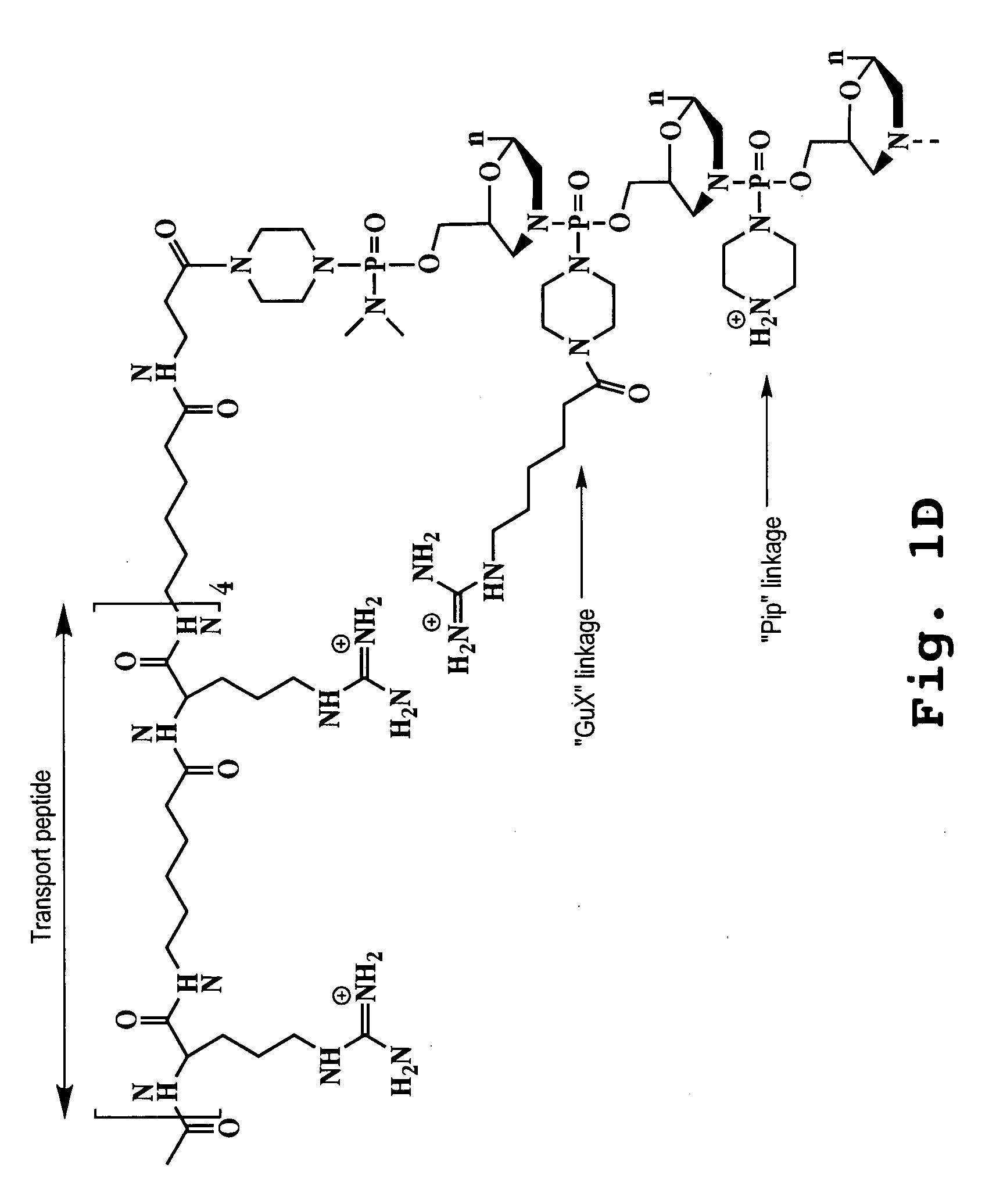
Morpholino oligomers containing both uncharged and cationic intersubunit linkages are provided. The oligomers are oligonucleotide analogs containing predetermined sequences of base-pairing moieties. The presence of the cationic intersubunit linkages in the oligomers, typically at a level of about 10-50% of total linkages, provides enhanced antisense activity, in various antisense applications, relative to the corresponding uncharged oligomers. Also provided are such oligomers conjugated to peptide transporter moieties, where the transporters are preferably composed of arginine subunits, or arginine dimers, alternating with neutral amino acid subunits.
Owner:AVI BIOPHARMA
Methods for detecting nucleic acids indicative of cancer
InactiveUS6964846B1High molecular weightMicrobiological testing/measurementRecombinant DNA-technologyCellular DebrisBody fluid sample
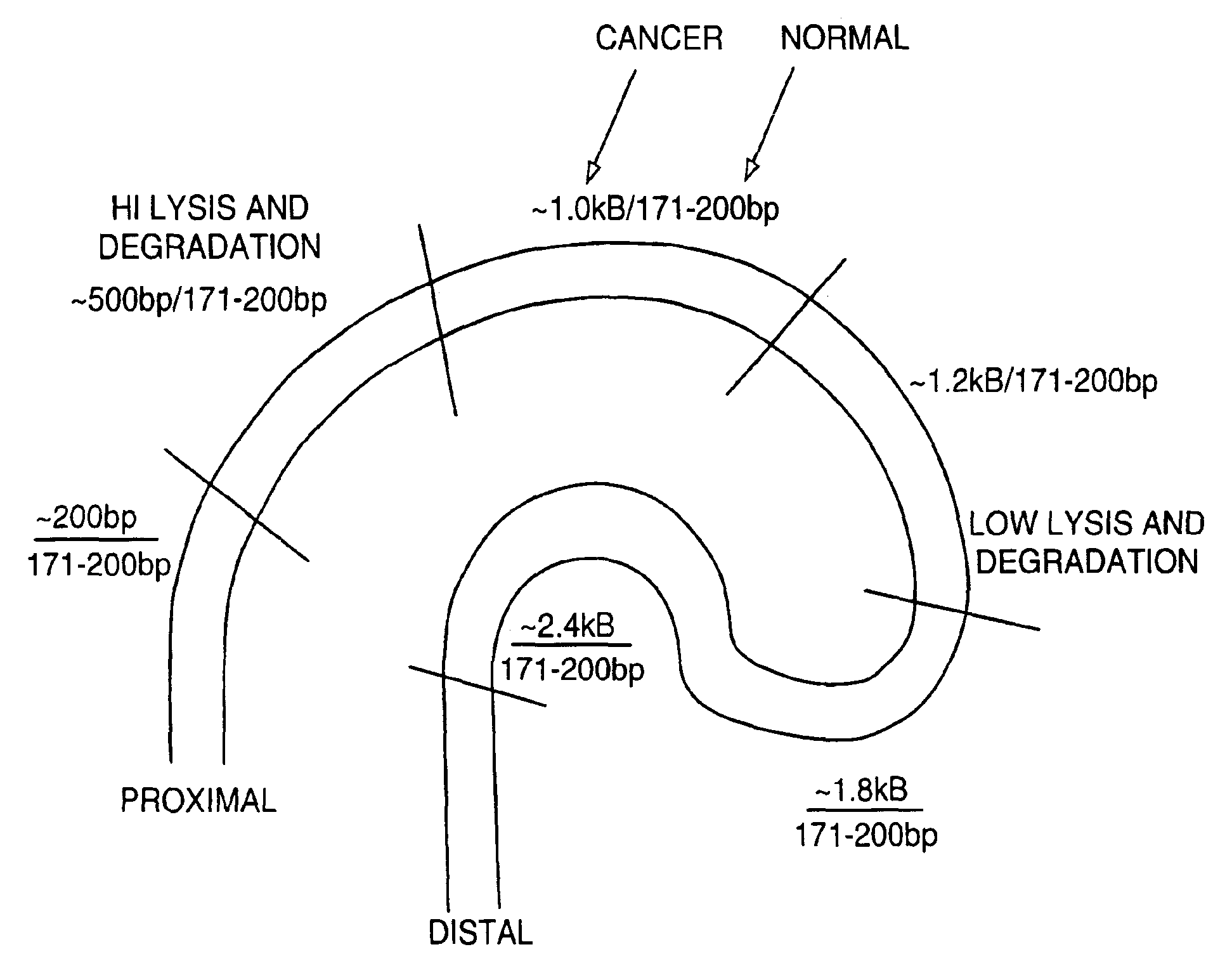
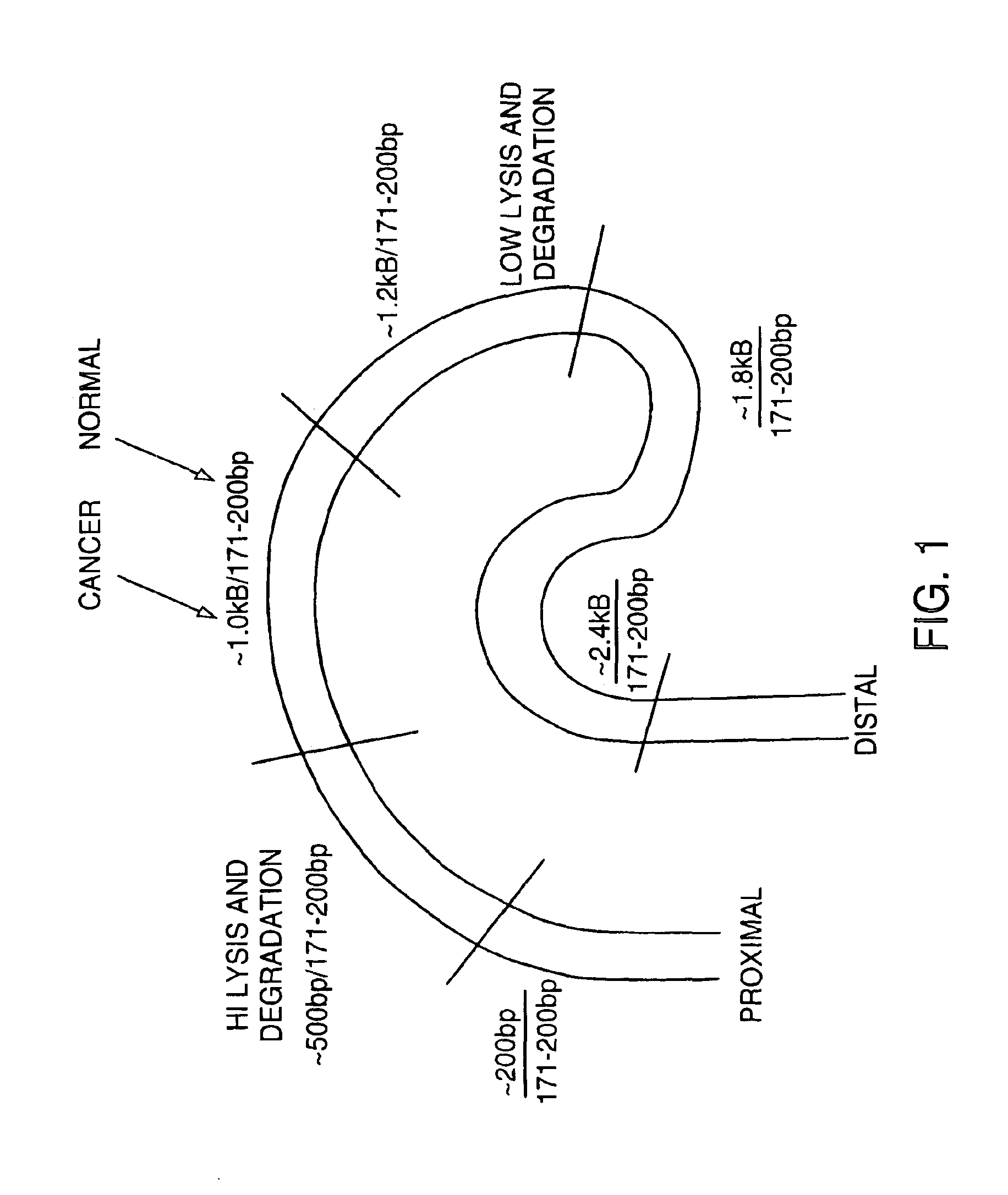
The invention provides methods for screening tissue or body fluid samples for nucleic acid indicia of cancer or precancer.Method are provided for screening a patient for cancer or precancer by detecting the presence of nucleic acid fragments that are longer than nucleic acid fragments expected to be present in a sample obtained from a healthy individual. In one embodiment, a positive screen for cancer or precancer is identified when a patient tissue or body fluid sample comprising exfoliated cells or cellular debris contains an amount of nucleic acid of a length greater than about 200 base pairs that exceeds a predetermined amount.
Owner:EXACT SCI CORP
Duplexed parvovirus vectors
InactiveUS7465583B2High transduction efficiencyRapid onsetBiocidePeptide/protein ingredientsGeneticsViral vector
The present invention provides duplexed parvovirus vector genomes that are capable under appropriate conditions of forming a double-stranded molecule by intrastrand base-pairing. Also provided are duplexed parvovirus particles comprising the vector genome. Further disclosed are templates and methods for producing the duplexed vector genomes and duplexed parvovirus particles of the invention. Methods of administering these reagents to a cell or subject are also described. Preferably, the parvovirus capsid is an AAV capsid. It is further preferred that the vector genome comprises AAV terminal repeat sequences.
Owner:THE UNIV OF NORTH CAROLINA AT CHAPEL HILL
Probe for analysis of target nucleic acids
The invention is a probe for detecting nucleic acids having a particular sequence. It is composed of two joint units. One unit is chemically different from natural nucleic acids, but has the ability to recognize a particular sequence of bases or base pairs in single or double-stranded DNA of RNA. The other unit is a compound whose detectable properties are altered upon binding to nucleic acids.
Owner:LIGHTUP TECH
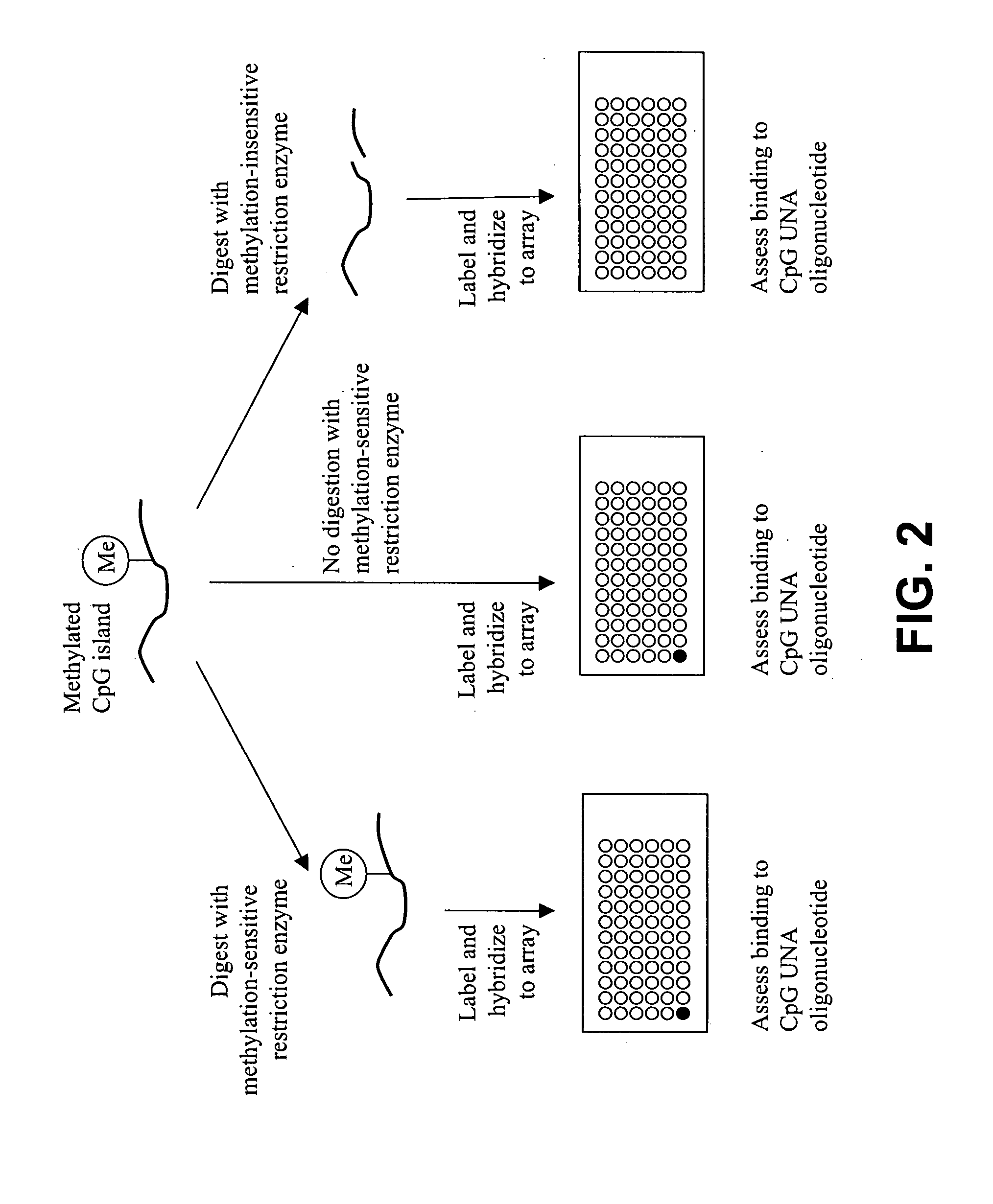
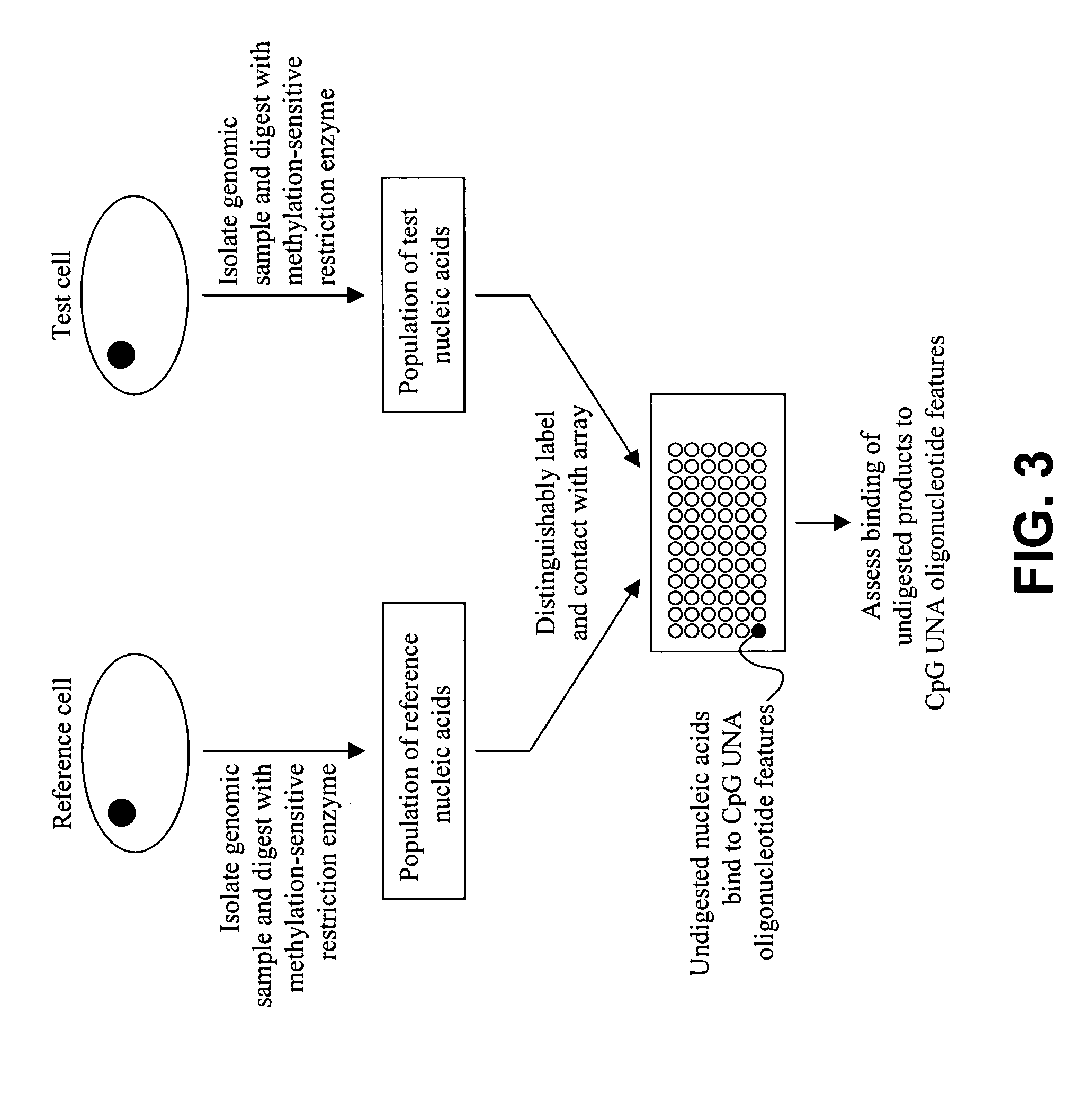
Methods and compositions for assessing CpG methylation
InactiveUS20050233340A1Bioreactor/fermenter combinationsBiological substance pretreatmentsDNA methylationCpg island methylation
Methods and compositions for assessing CpG island methylation are provided. Specifically, the invention provides an unstructured nucleic acid (UNA) oligonucleotide that base pairs with, i.e., hybridizes to, CpG islands. The subject oligonucleotide may be present in an array, and find use in methods for evaluating methylation of CpG islands in cells. Kits and computer programming for use in practicing the subject methods are also provided.
Owner:AGILENT TECH INC
Method for determining the number of copies of a chromosome in the genome of a target individual using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell is reconstructed using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In accordance with another embodiment of the invention, incomplete genetic data from a fetus is acquired from fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals. In one embodiment, the genetic data can be reconstructed for the purposes of making phenotypic predictions. In another embodiment, the genetic data can be used to detect for aneuploides and uniparental disomy.
Owner:NATERA
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS8515679B2Improve fidelityData processing applicationsMicrobiological testing/measurementGenetic MaterialsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Genetic material from the target individual is acquired, amplified and the genetic data is measured using known methods. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment of the invention, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment of the invention, the chromosome copy number can be determined from the measured genetic data of a single or small number of cells, with or without genetic information from one or both parents. In another embodiment of the invention, these determinations are made for the purpose of embryo selection in the context of in-vitro fertilization. In another embodiment of the invention, the genetic data can be reconstructed for the purposes of making phenotypic predictions.
Owner:NATERA
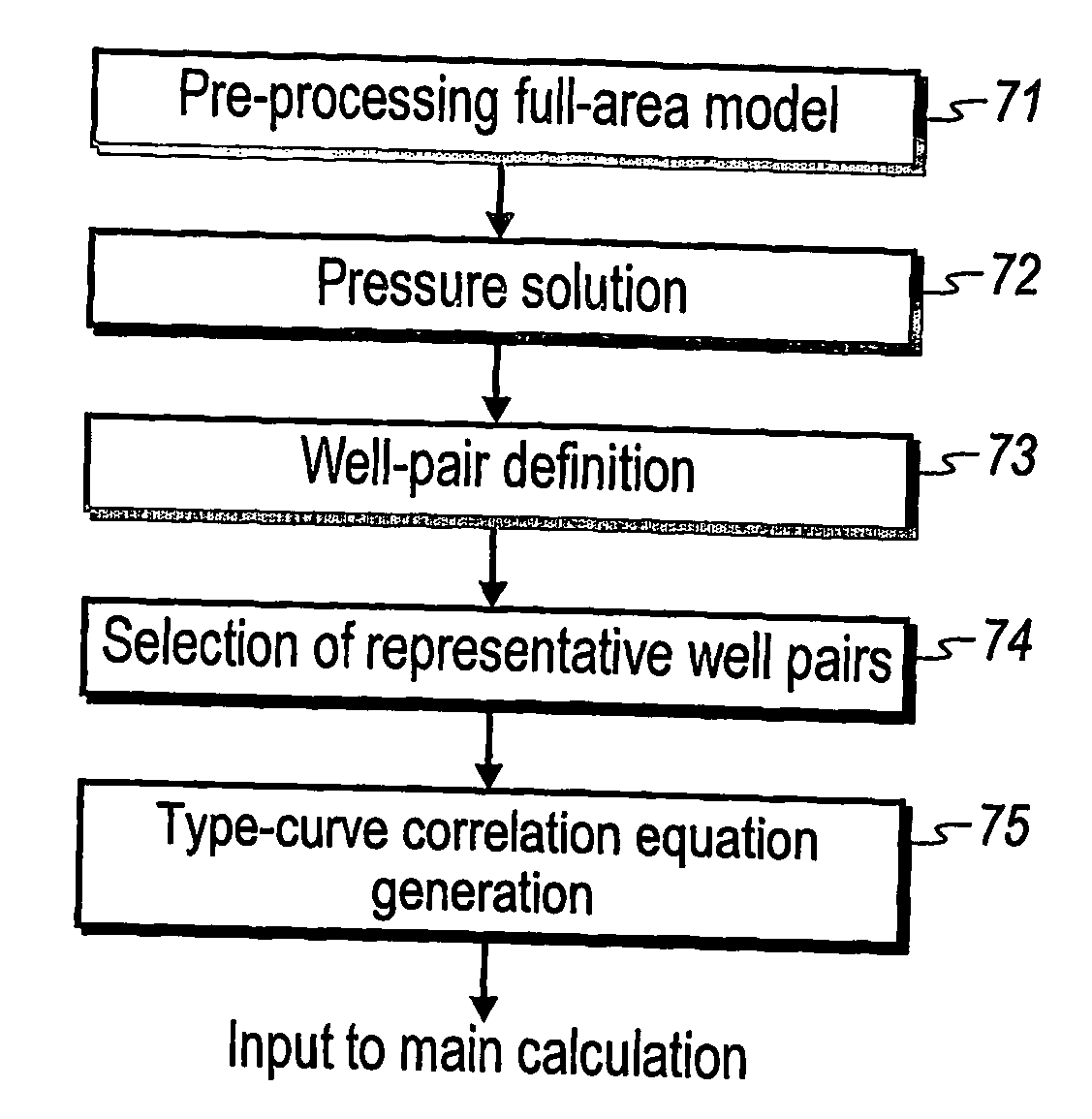
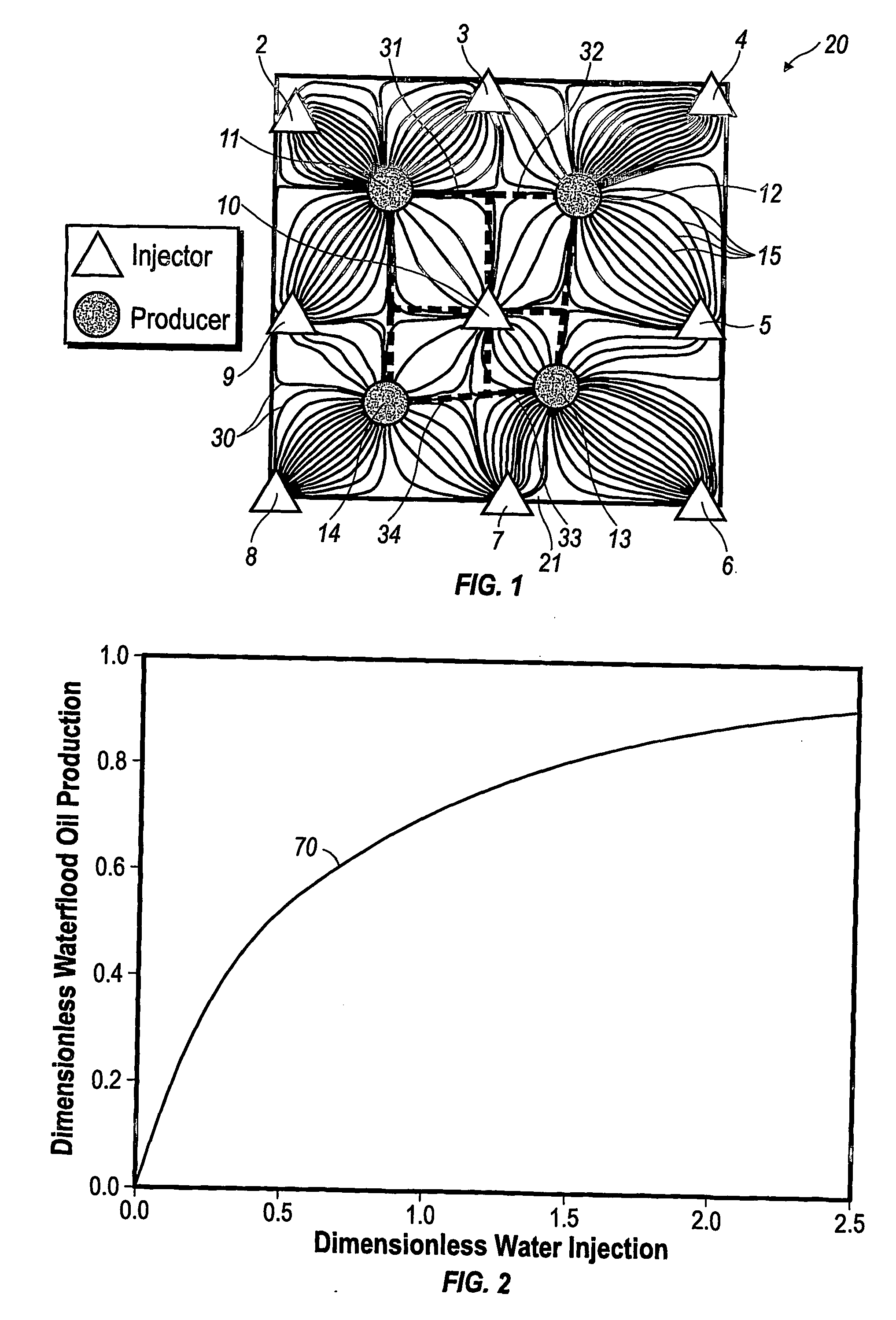
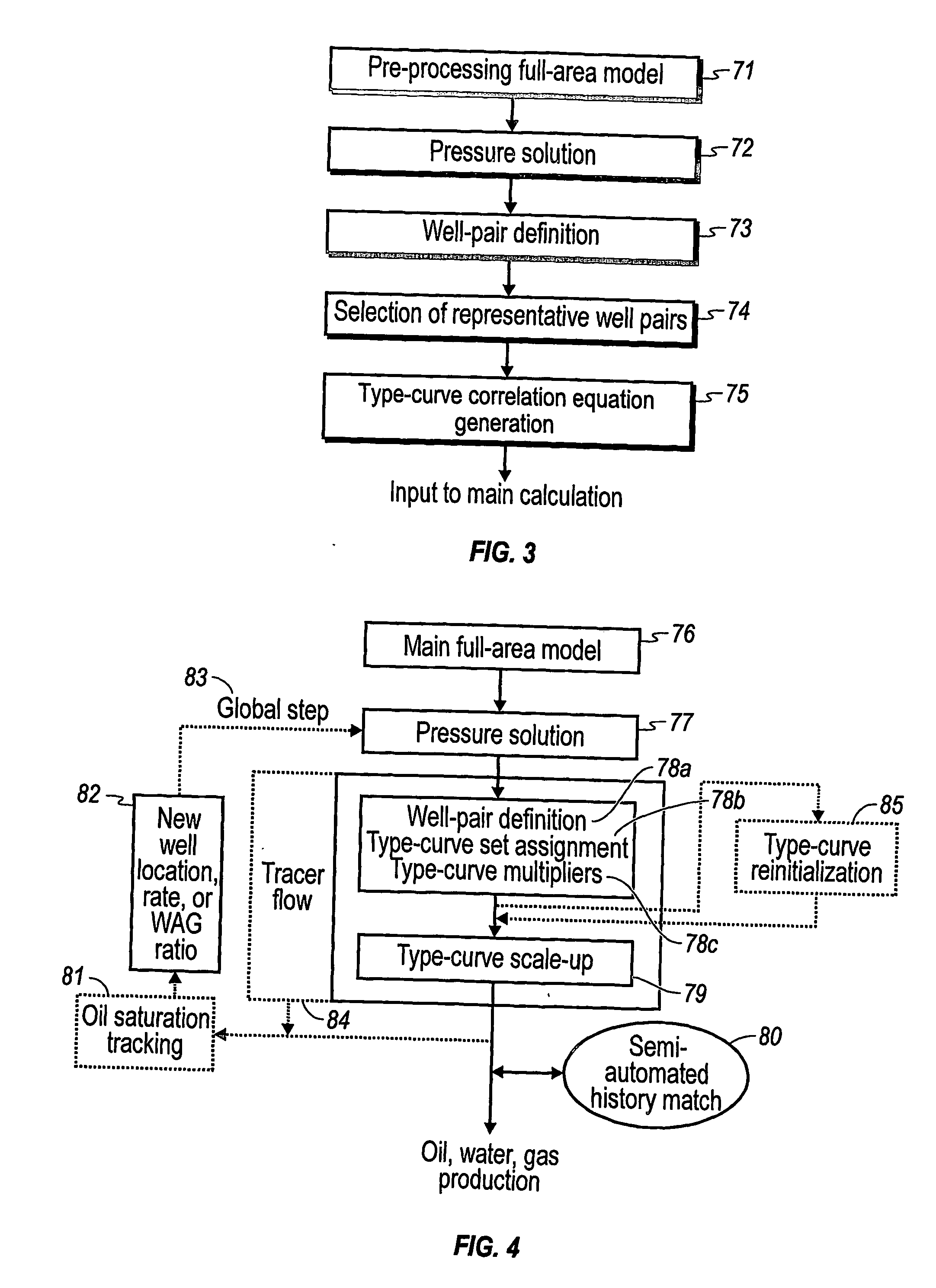
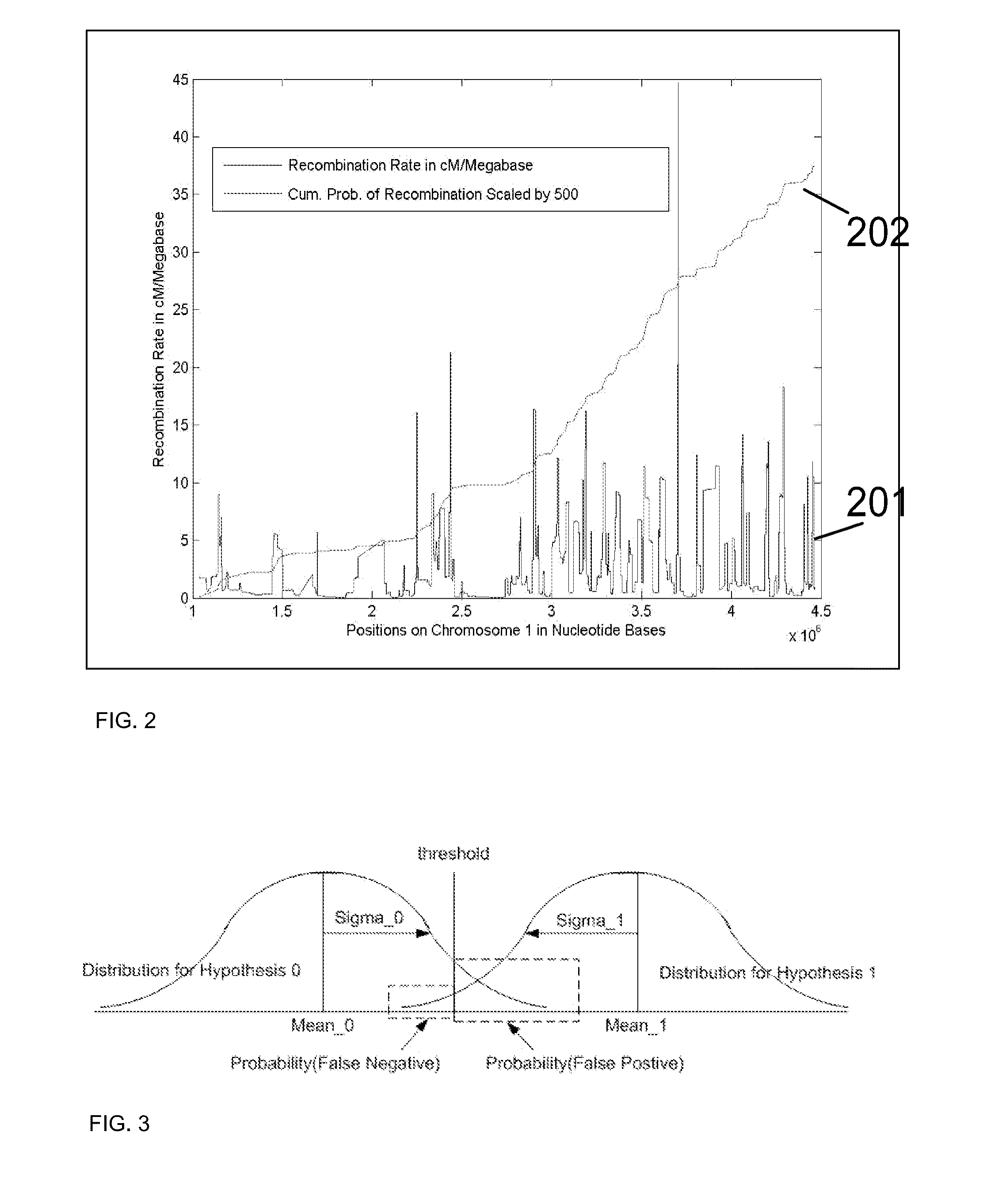
Performance prediction method for hydrocarbon recovery processes
The invention relates to a method for predicting the performance of large-scale hydrocarbon-bearing reservoir floods. One embodiment of the invention includes a method for predicting performance of a patterned flooding process in a subterranean hydrocarbon-bearing formation, said formation being penetrated by a plurality of injector wells and producer wells, comprising the steps of: determining flow-based pairs of injector to producer wells [FIG. 4, item 78a] (first well pairs) using a geological model [item 76]; developing a connective pore volume distribution curve for each first well pair item [78b]; selecting at least two first well pairs (selected well pairs) that reflect narrow and wide connective pore volume distributions that correspond to high and lower oil recovery levels; developing a 3-D simulation model for each selected well pair, performing a reservoir simulation for each selected well pair for the corresponding flooding process; and generating prototype performance curves for each selected well pair. An alternate embodiment of the invention includes a method for predicting the performance of large-scale hydrocarbon-bearing reservoir floods where injection well location, production well location, a process parameter, or a well processing rate is modified.
Owner:EXXONMOBIL UPSTREAM RES CO
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS20140032128A1Significant resultImprove fidelityMicrobiological testing/measurementBiostatisticsDiploid cellsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals
A system and method for determining the genetic data for one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available, are disclosed. Genetic data for the target individual is acquired and amplified using known methods, and poorly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related subjects. In accordance with one embodiment of the invention, incomplete genetic data is acquired from embryonic cells, fetal cells, or cell-free fetal DNA isolated from the mother's blood, and the incomplete genetic data is reconstructed using the more complete genetic data from a larger sample diploid cells from one or both parents, with or without genetic data from haploid cells from one or both parents, and / or genetic data taken from other related individuals.
Owner:NATERA
Modular dna-binding domains and methods of use
ActiveUS20120064620A1Enabling targeted DNA modificationFusion with DNA-binding domainSugar derivativesDNA-binding domainDna targeting
The present invention refers to methods for selectively recognizing a base pair in a DNA sequence by a polypeptide, to modified polypeptides which specifically recognize one or more base pairs in a DNA sequence and, to DNA which is modified so that it can be specifically recognized by a polypeptide and to uses of the polypeptide and DNA in specific DNA targeting as well as to methods of modulating expression of target genes in a cell.
Owner:BONAS ULLA +3
CATIONIC PEPTIDES FOR siRNA INTRACELLULAR DELIVERY
What is described is a composition for delivery of a RNA molecule to a cell, comprising: a double stranded RNA (dsRNA) molecule of about 15 to about 40 base pairs; and a polynucleotide delivery-enhancing peptide, comprising a region of alternating lysine and histidine residues, or of alternating D and L forms of arginine.
Owner:NASTECH PHARMA
Site-specific incorporation of amino acids into molecules
The invention provides certain embodiments relating to methods and compositions for incorporating non-natural amino acids into a polypeptide or protein by utilizing a mutant or modified aminoacyl-tRNA synthetase to charge the non-natural amino acid to a the corresponding tRNA. In certain embodiments, the tRNA is also modified such that the complex forms strict Watson-Crick base-pairing with a codon that normally forms wobble base-pairing with unmodified tRNA / aminoacyl-tRNA synthetase pairs.
Owner:CALIFORNIA INST OF TECH
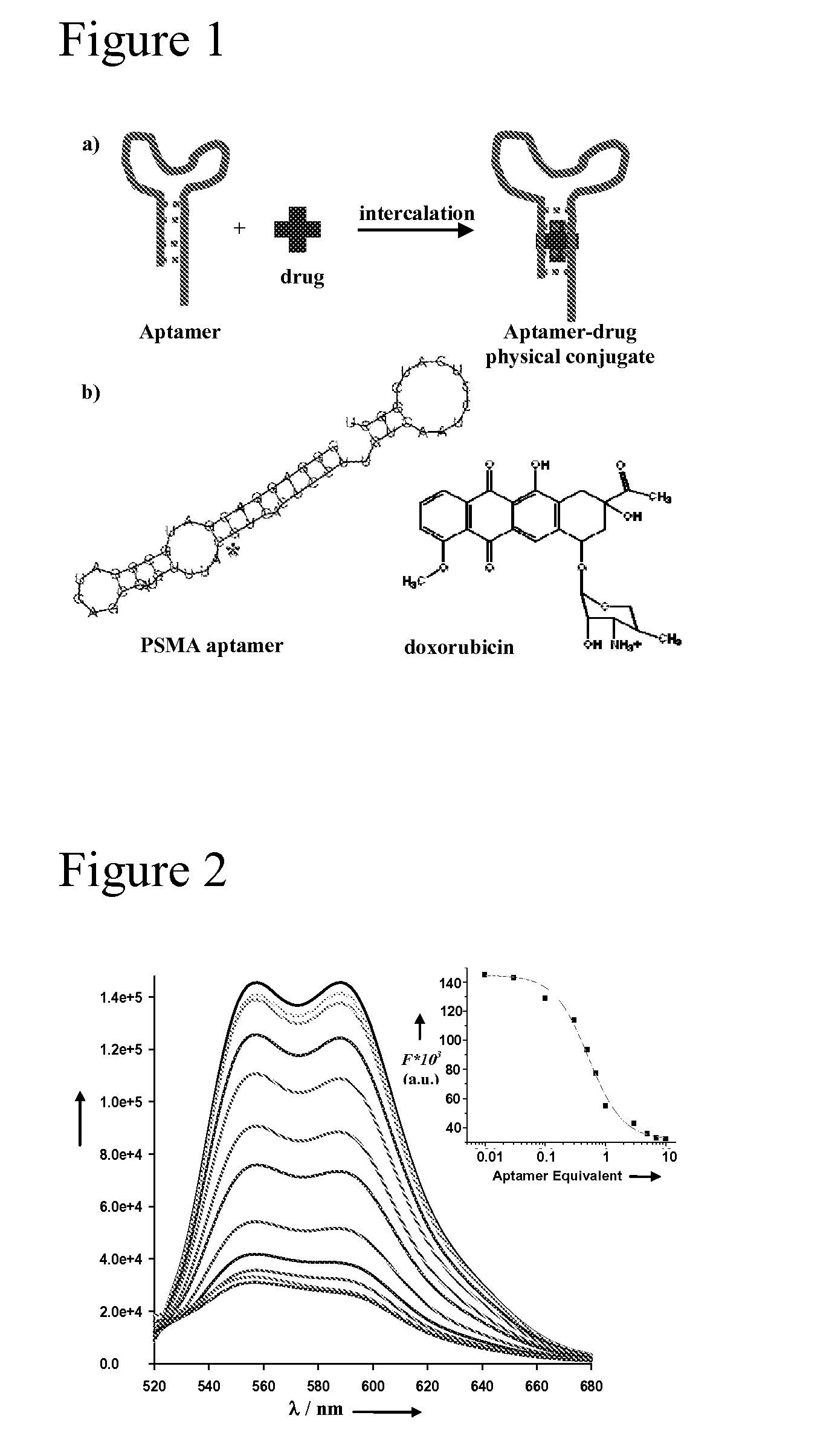
Aptamer-Directed Drug Delivery
InactiveUS20110052697A1Reduce severityReduce morbidityOrganic active ingredientsPowder deliveryCell-Extracellular MatrixDiagnostic agent
The present invention provides systems, methods, and compositions for targeted delivery of a therapeutic agent organs, tissues, cells, extracellular matrix components, and intracellular compartments. The present invention provides a complex comprising a therapeutic or diagnostic agent and a nucleic acid targeting moiety, wherein the agent non-covalently associates with base pairs of the nucleic acid targeting moiety. The invention provides targeted particles comprising a particle and an inventive complex. The present invention provides methods of designing, manufacturing, and using inventive complexes and targeted particles.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC +2
Characterizing Stretched Polynucleotides in a Synthetic Nanopassage
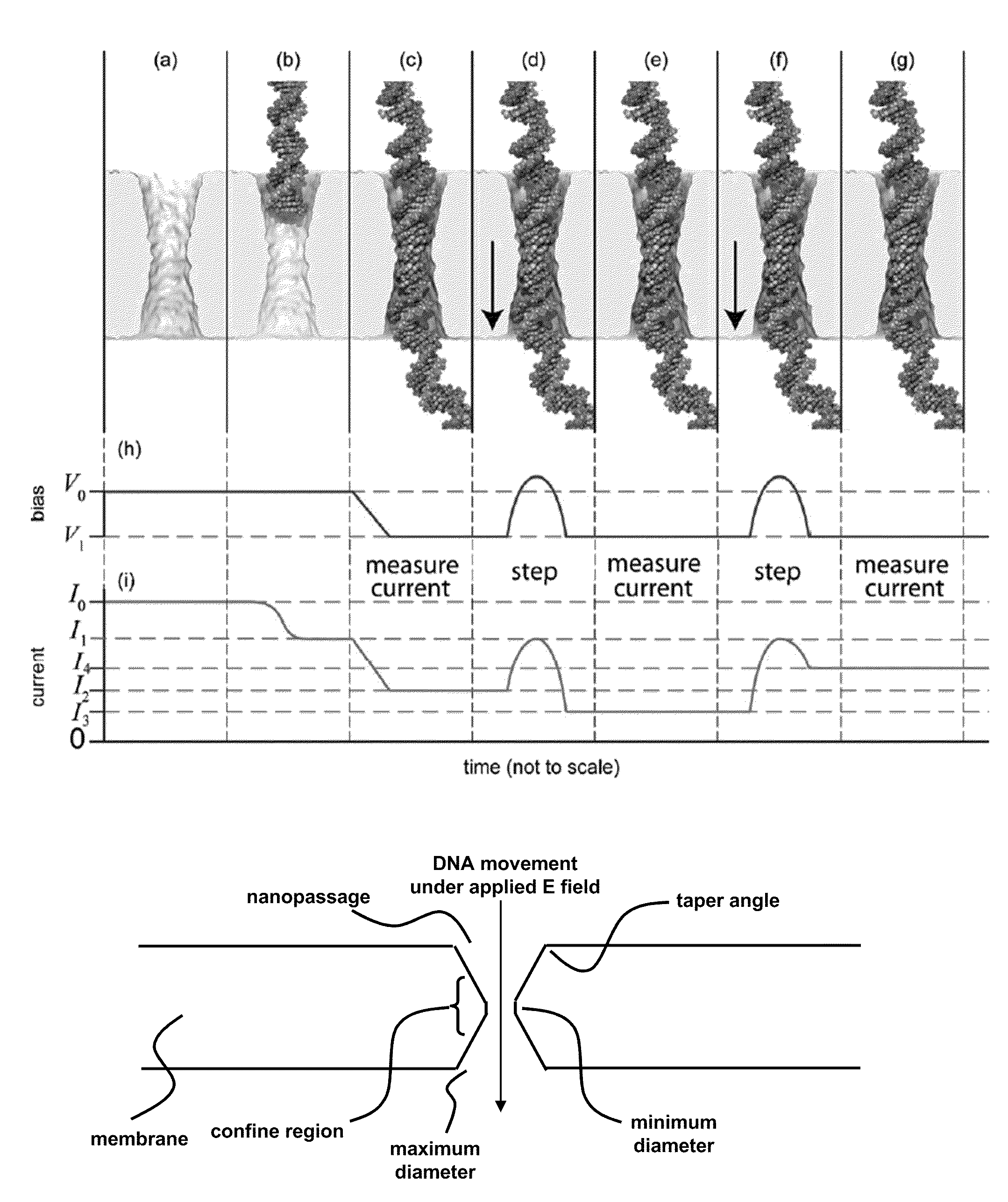
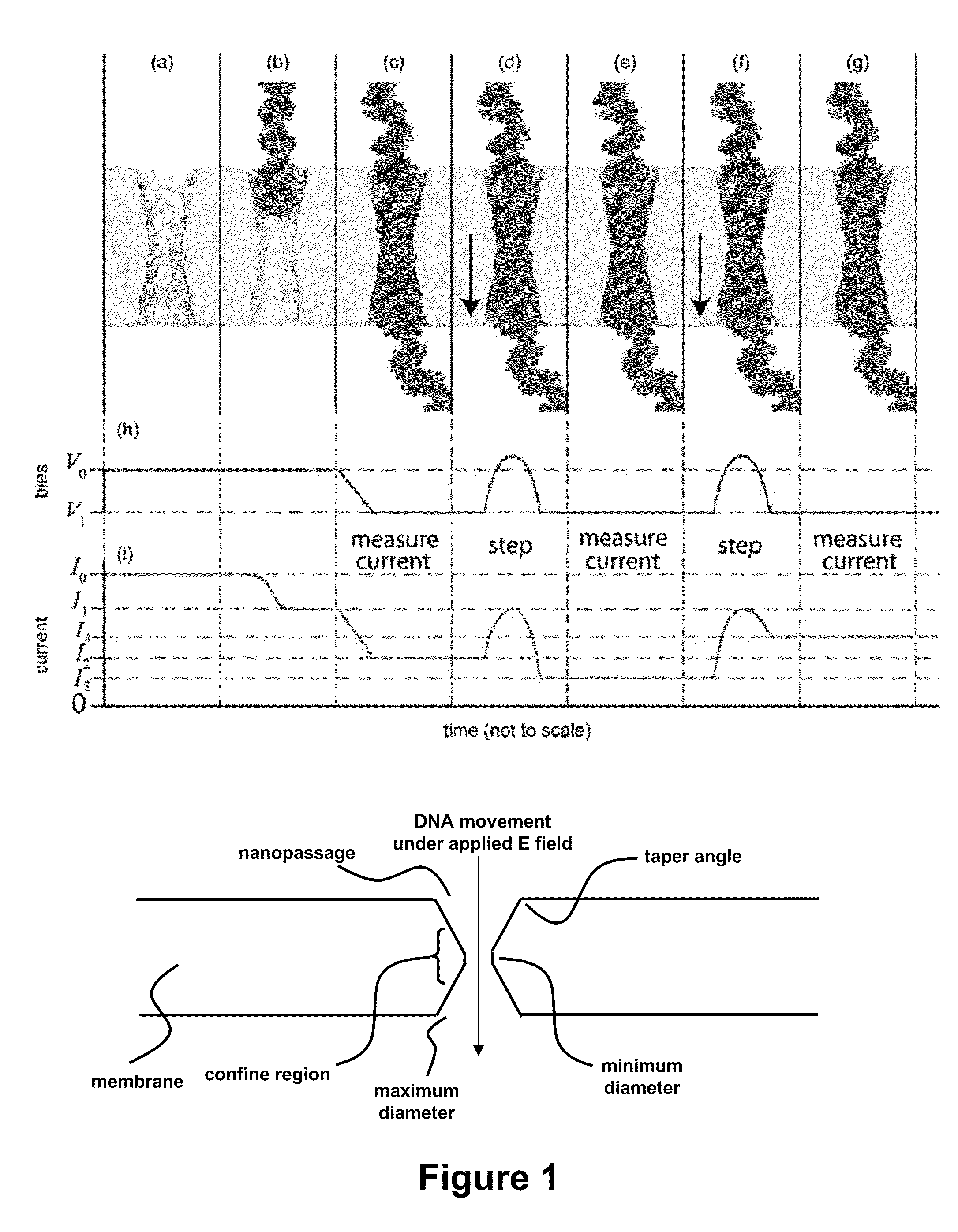
ActiveUS20110226623A1Longer read lengthIncrease speedElectrolysis componentsVolume/mass flow measurementFluid compartmentsTrapping
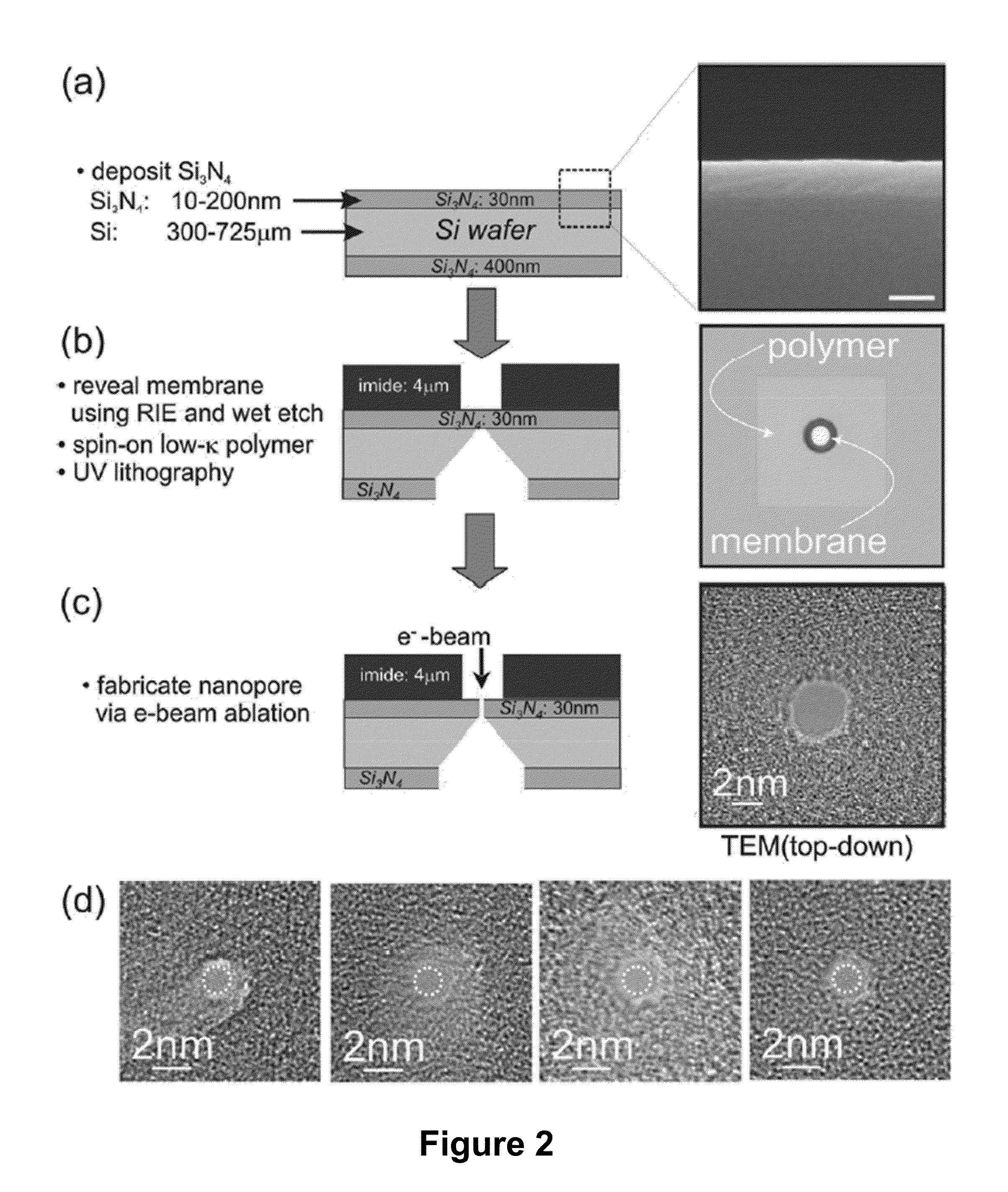
Methods of trapping a deformed portion of a double-stranded polynucleotide in a membrane nanopassage are provided. In an aspect, the membrane has a nanopassage that defines a confine region, wherein the membrane separates a first fluid compartment from a second fluid compartment, and the nanopassage is in fluid communication with the first and second compartments. A polynucleotide is provided to the first fluid compartment and optionally a threshold voltage for the membrane and the polynucleotide is determined. A driving voltage across the membrane that is greater than the threshold voltage is applied to force a portion of the polynucleotide sequence into the nanopassage confine region, and decreased to a holding voltage bias to trap the polynucleotide portion in the nanopassage confine region. In particular, at least one nucleotide base-pair is fixably positioned in the nanopassage confine volume. In further embodiments, any of the trapping methods are used to characterize or sequence double stranded DNA.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
Methods of PCR-Based Detection of "Ultra Short" Nucleic Acid Sequences
InactiveUS20100068711A1Reducing nucleic acid degradationInhibiting nuclease activityMicrobiological testing/measurementNucleic acid sequencingBody fluid
The present invention provides highly sensitive methods used for diagnosing and monitoring various diseases and disorders by detecting and analyzing “ultra short” (20-50 base pair) nucleic acids obtained from bodily fluids.
Owner:TROVAGENE
Method and apparatus for identifying classifying or quantifying DNA sequences in a sample without sequencing
InactiveUS6141657ARapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsDigital data processing detailsSequence databaseDNA Sequence Databases
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
System and method for cleaning noisy genetic data and determining chromosome copy number
ActiveUS10083273B2Improve fidelityData processing applicationsMicrobiological testing/measurementDiploid cellsEmbryo
Disclosed herein is a system and method for increasing the fidelity of measured genetic data, for making allele calls, and for determining the state of aneuploidy, in one or a small set of cells, or from fragmentary DNA, where a limited quantity of genetic data is available. Poorly or incorrectly measured base pairs, missing alleles and missing regions are reconstructed using expected similarities between the target genome and the genome of genetically related individuals. In accordance with one embodiment, incomplete genetic data from an embryonic cell are reconstructed at a plurality of loci using the more complete genetic data from a larger sample of diploid cells from one or both parents, with or without haploid genetic data from one or both parents. In another embodiment, the chromosome copy number can be determined from the measured genetic data, with or without genetic information from one or both parents.
Owner:NATERA
Method and apparatus for identifying, classifying, or quantifying DNA sequences in a sample without sequencing
InactiveUS6418382B2Rapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsMicrobiological testing/measurementSample sequenceSingle sequence
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com