Paired-end library construction method and method for sequencing genome by using library
A library construction, paired-end technology, applied in the field of paired paired-end library construction, can solve problems such as low recovery rate, difficult cyclization, and influence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
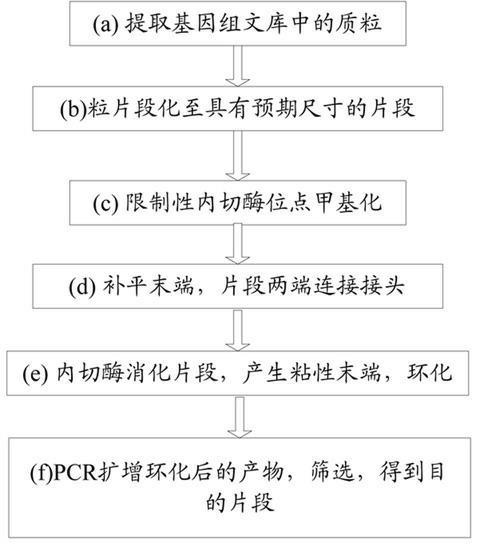
[0027] Such as figure 2 As shown, in another embodiment of the present invention, a flowchart of paired paired-end library construction, the specific process is:
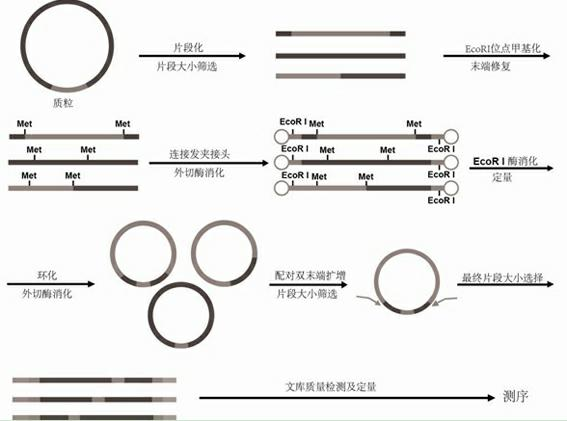
[0028] (1) Plasmid DNA extraction: extract large fragments of plasmid DNA;
[0029] (2) Plasmid fragmentation: take no less than 20ug of the plasmid in step (1), shear the sample with an instrument, and obtain a fragment that is 100-1500bp larger than the vector used in the genome library by the method of gel cutting and recovery;
[0030] (3) EcoR I restriction site methylation: under the action of EcoR I methylase, the EcoR I restriction site on the fragment is methylated and protected;
[0031] (4) Fill in the ends of fragments: Fill in the ends of the fragments under the action of T4 DNA polymerase and T4 polynucleotide kinase; connect Hairpin Adaptor: under the action of ligase, add at both ends of the methylated fragments On the Hairpin Adaptor linker, the linker has a stem-loop structure and contains an EcoR I restr...
Embodiment 2
[0039] A paired-end (Paired-End) high-throughput sequencing of a Bacterial Artificial Chromosome (BAC) library with an average insert length of ~80kb.
[0040] 1) Bacchus leucocephalus BAC library: The BAC library used in this experiment is the BAC library of leucocephalus Chinese leucocephalus. The vector of the library is CopyControl? pCC1BAC? Vector (Epicentre), with a total length of 8128bp. The library consists of 44,706 clones, and the average length of the insert is about 80kb.
[0041] 2) Reagents and instruments: The HydroShear genomic DNA shearing instrument from Digilab Genomic Solutions is used for DNA fragmentation. SAM, EcoR I methylase, λ-exonuclease, T7 exonuclease, and exonuclease I used in the experiment were purchased from NEB; bovine serum albumin (BSA), ATP, PCR nucleic acid mixture, T4 DNA polymerase , T4 Polynucleotide Kinase (PNK), Hairpin Adaptor, Rapid Ligase, EcoRI (high concentration), GC-RICH PCR system, GS FLX Titanium Amplicon emPCR kit purchased fro...
Embodiment 3
[0113] A paired-end (Paired-End) high-throughput sequencing of a Bacterial Artificial Chromosome (BAC) library with an average insert length of ~160kb
[0114] 1) Bacchus leucocephalus BAC library: The BAC library used in this experiment is the Bacillus leucocephalus BAC library of China. The vector of the library is CopyControl® pCC1BAC™ vector (Epicentre), with a total length of 8128bp. The library consists of 15,302 clones, with an average insert length of ~160kb.
[0115] 2) Reagents and instruments: The HydroShear Genomic DNA Shearing Machine from Digilab Genomic Solutions is used for DNA fragmentation. SAM, EcoR I methylase, λ-exonuclease, T7 exonuclease, and exonuclease I used in the experiment were all purchased from NEB; bovine serum albumin (BSA), ATP, PCR nucleic acid mixture, T4 DNA polymerase , T4 Polynucleotide Kinase (PNK), Hairpin Adaptor, Rapid Ligase, EcoRI (high concentration), GC-RICH PCR system, GS FLX Titanium Amplicon emPCR kit purchased from Roche; Advantag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com