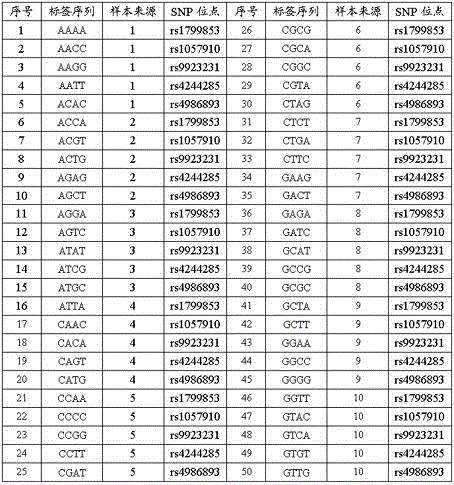
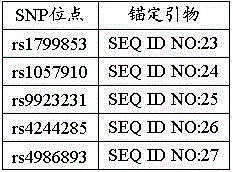
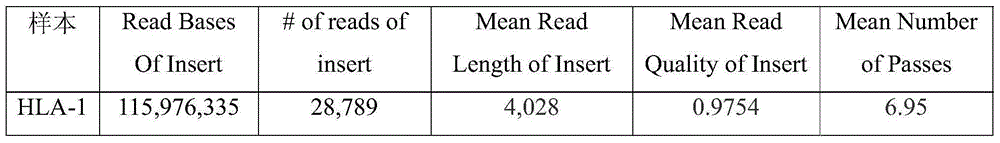
Patents
Literature
208 results about "Typing methods" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Drug related gene type database, gene typing and drug action detection method
ActiveCN103198238AGive quickly and accuratelyFast and accurate auxiliary basisMicrobiological testing/measurementProteomicsTyping methodsGenotype
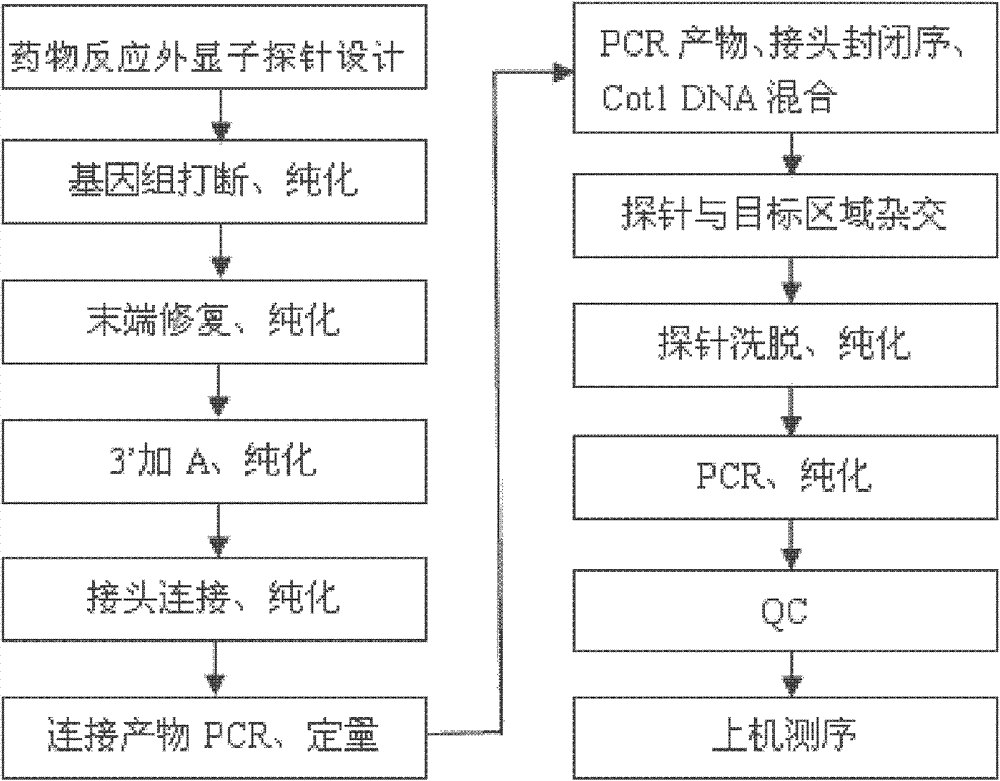
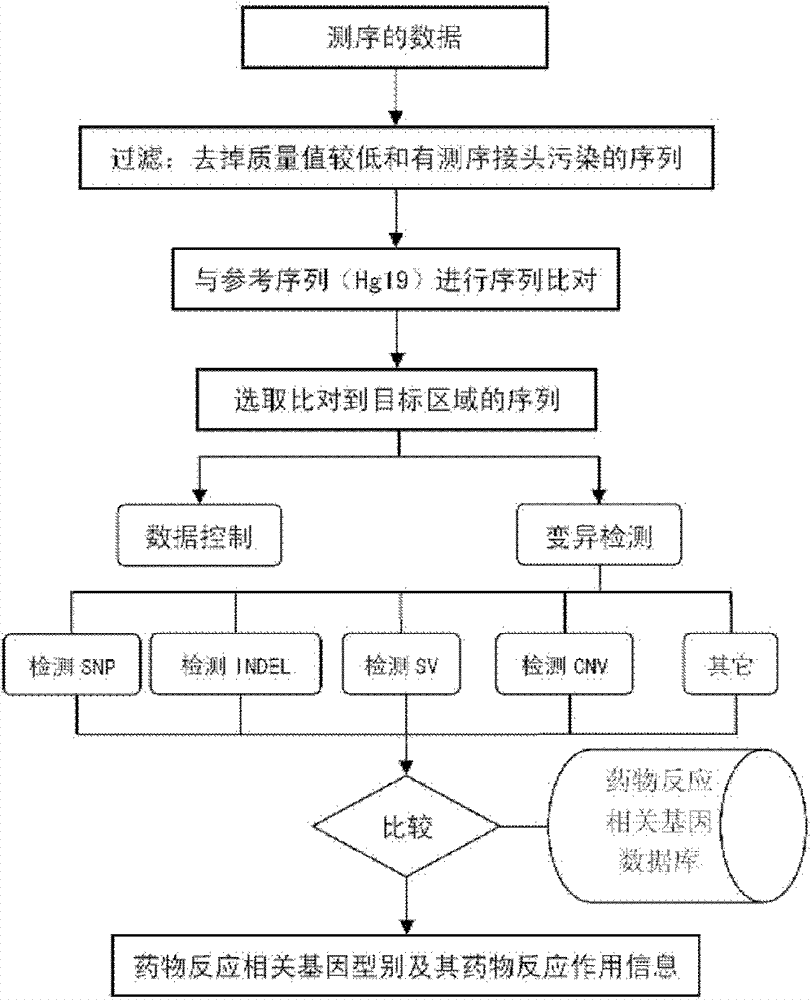
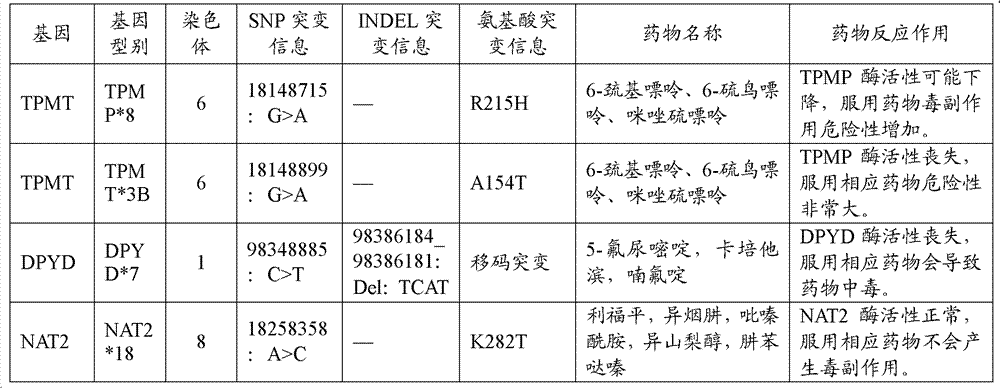
The invention discloses a standard gene type database of drug action related genes and a construction method of the standard gene type database. The construction method comprises the following steps: comparing the corresponding special sequence of mutation information with the human whole genome group standard sequence to obtain the corresponding relation between the special sequence and the human whole genome group standard sequence; and according to the corresponding relation, converting the gene type into the standard gene type corresponding to the human whole genome group standard sequence. On the basis, the invention discloses a drug action related gene typing method and a drug action detection method. The database provides a unified standard for drug action related gene typing and a more accurate basis for clinical medication. The gene typing and drug action detection method covers 48 human drug action related genes and can carry out multi-sample detection on all the known mutation sites and the corresponding drug information at the same time. The gene typing method can detect unknown polymorphic sites, and lays the foundation of researching and finding new polymorphic sites affecting the drug action.
Owner:BGI GENOMICS CO LTD
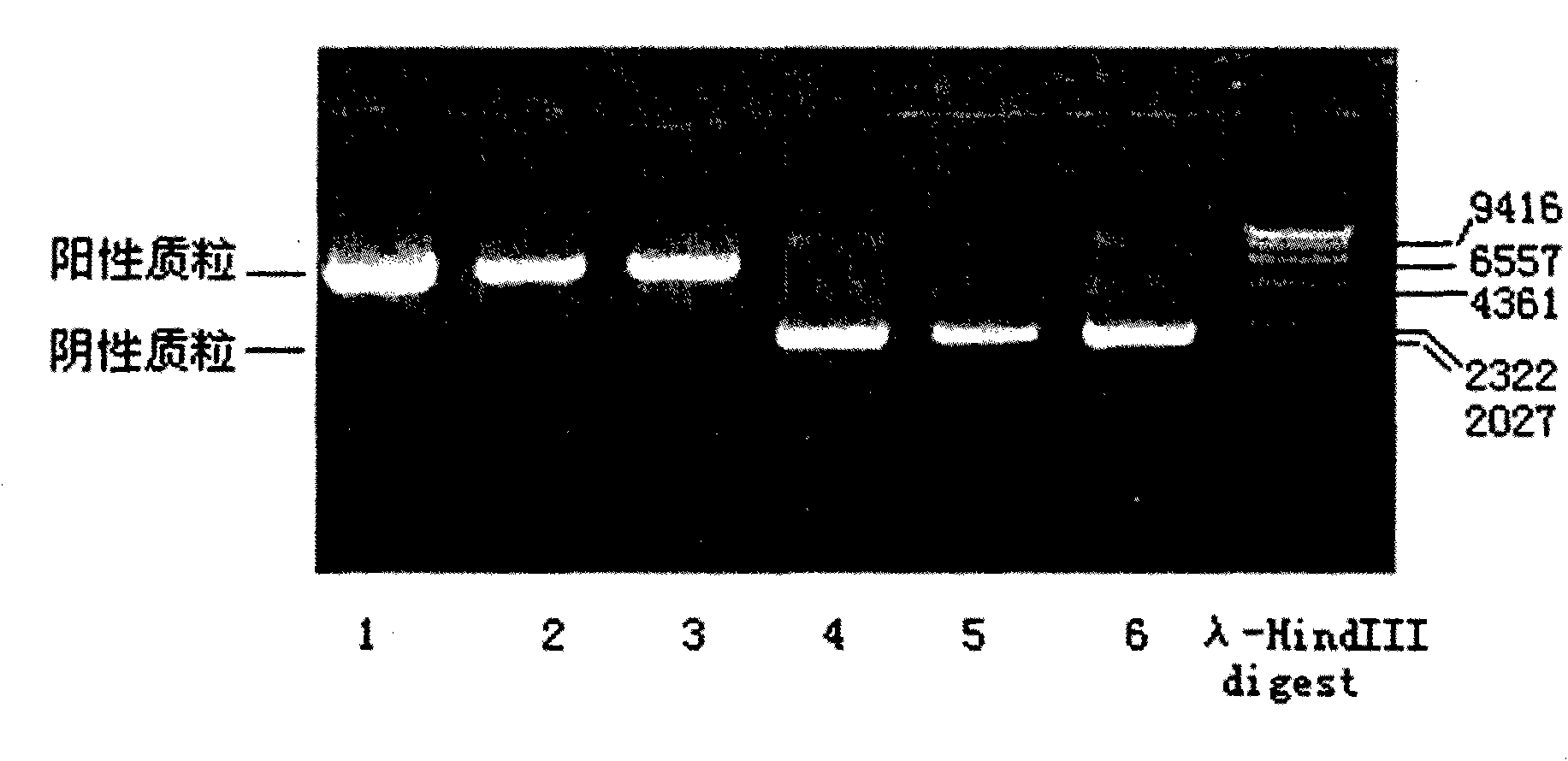
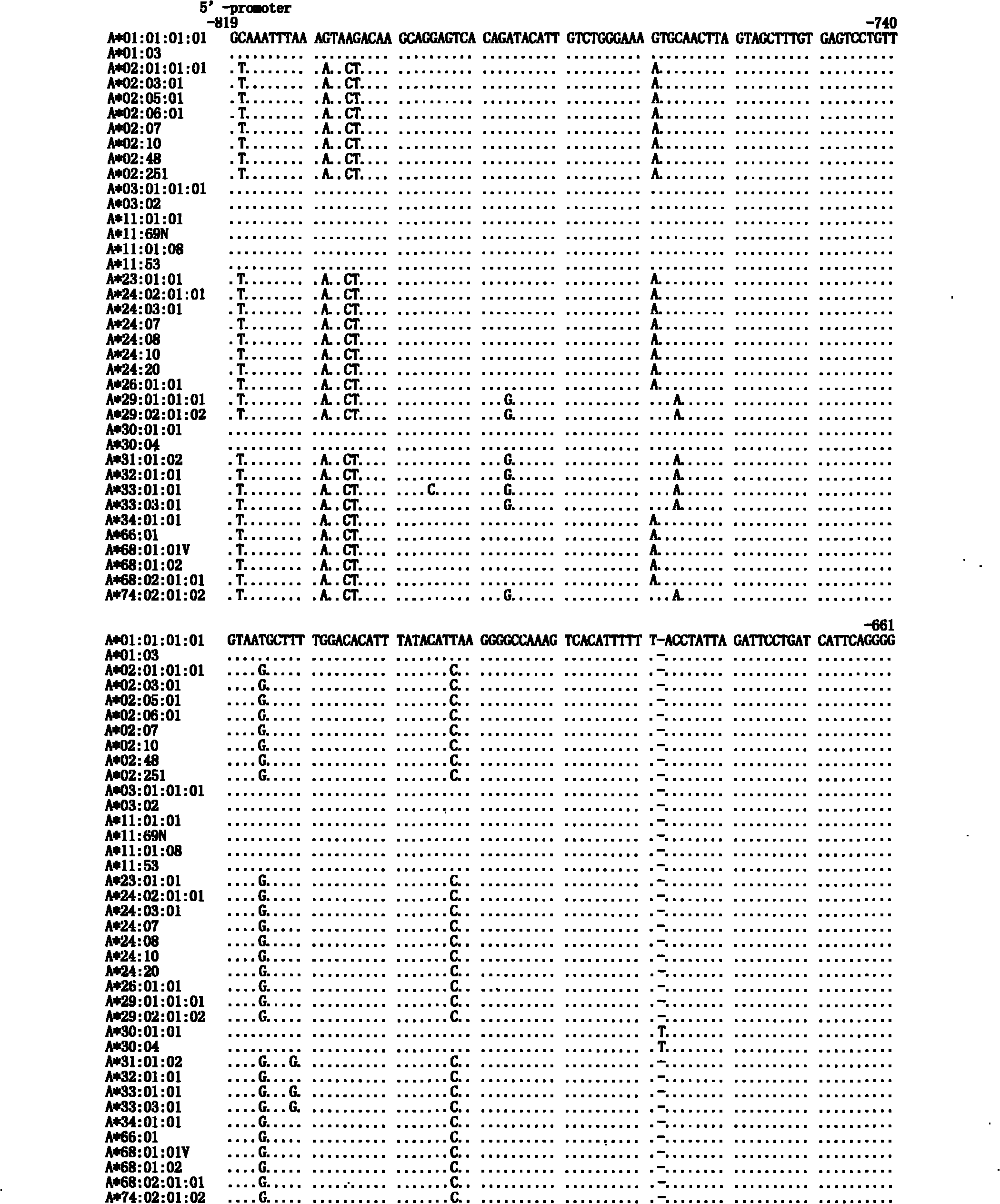
Human leukocyte antigen HLA-A and HLA-B gene full-length sequencing method and HLA gene sequencing and typing method
InactiveCN101962676AWide detection coverageResolving Ambiguous Results IssuesMicrobiological testing/measurementDNA/RNA fragmentationTyping methodsHLA-B gene
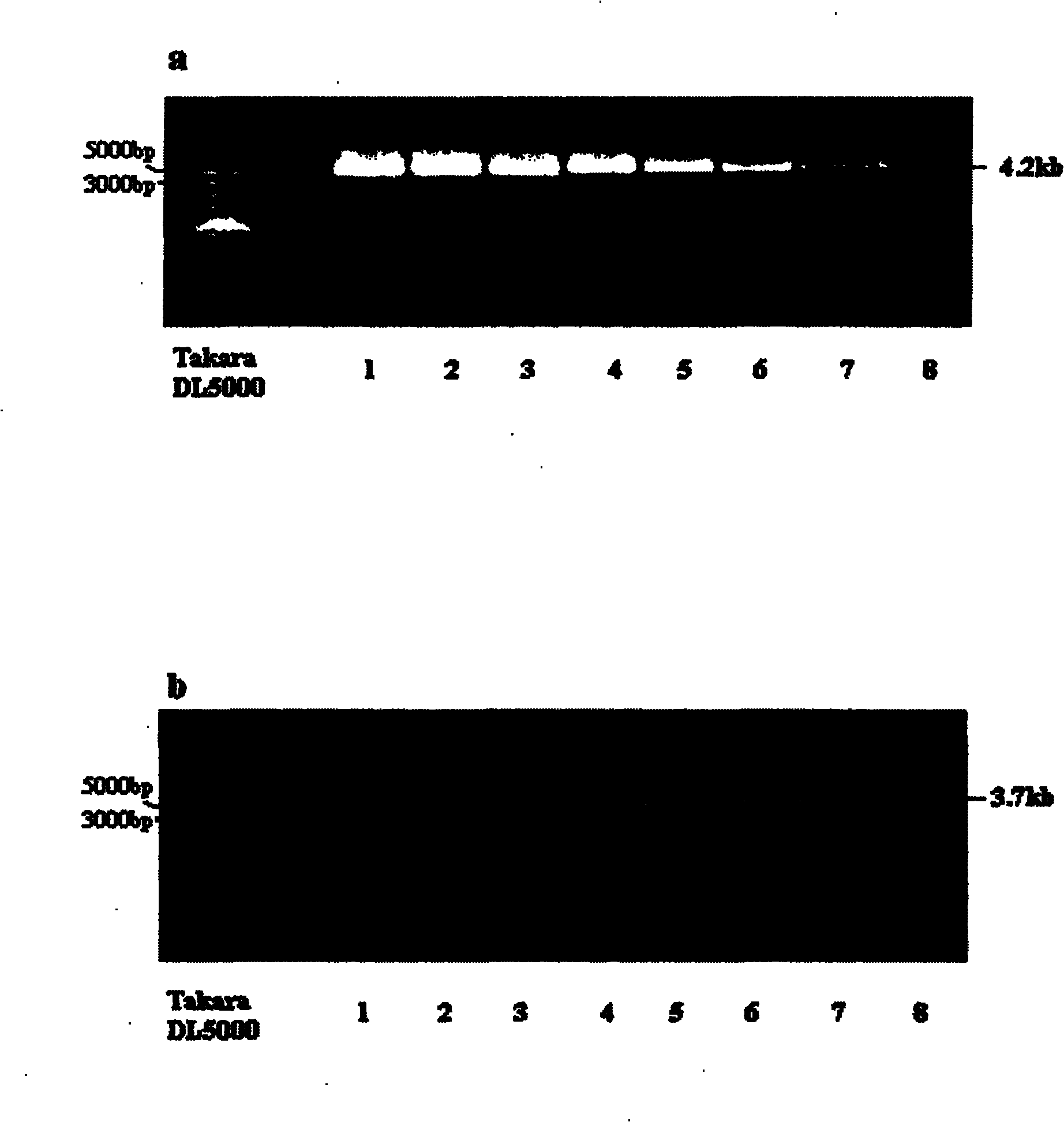
The invention discloses a human leukocyte antigen HLA-A and HLA-B gene full-length sequencing method and an HLA gene sequencing and typing method. The HLA-A and HLA-B gene full-length sequencing method comprises the following steps of: a, performing PCR amplification on about 4kb full-length sequences of HLA-A and HLA-B genes by using a pair of primers respectively; and b, cloning the amplification products to a pGEM-Tea sy vector, sequencing the full-length sequences by using ten walking sequencing primers in positive and negative directions respectively, and totally obtaining 38 allele 4.3kb full-length sequences of the HLA-A and 30 allele 3.7kb full-length sequences of the HLA-B. The HLA-A and HLA-B sequencing and typing method comprises the following steps of: performing PCR amplification on typing target areas of the HLA-A and HLA-B by using two pairs of primers respectively; and performing two directional sequencing on products by using fourteen sequencing primers respectively, wherein the HLA-DRB1 and HLA-DQB1 sequencing and typing method comprises the following steps of: amplifying sequences of second and third exons of DRB1 and DQB1 by adopting group specificity primers respectively; performing two directional sequencing on the second and third exons of the DRB1 by adopting eight group specificity primers and three sequencing primers; and performing two directional sequencing on the second and third exons of the DQB1 by adopting four sequencing primers respectively.
Owner:SHENZHEN BLOOD CENT
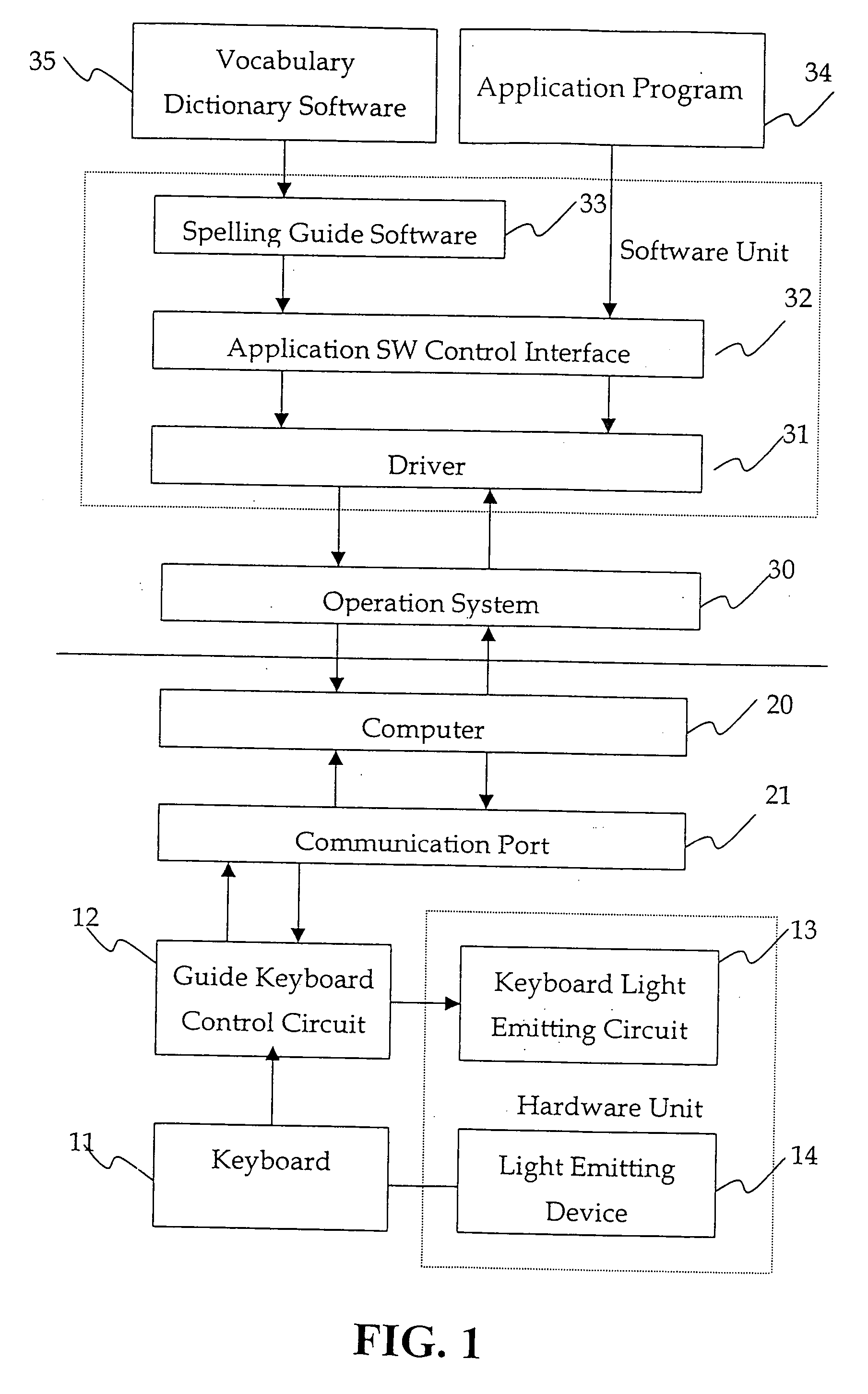
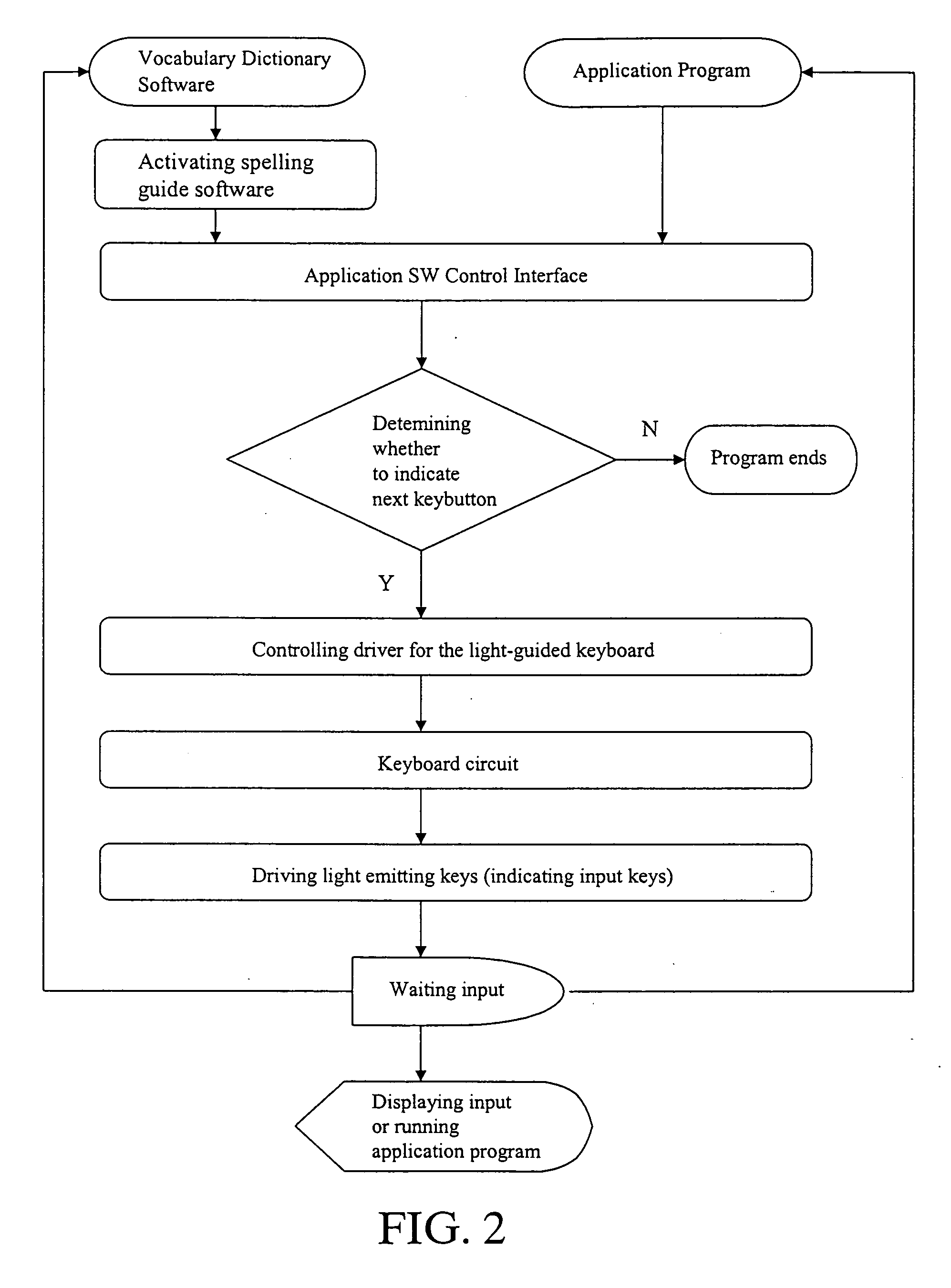
Light guided keyboard system
InactiveUS20060158353A1Faster and easy keyboard operationMinimize operation errorOperation facilitationLegendsLight guideTyping methods
A light guided keyboard system essentially comprised of a light emission circuit and a guide control circuit built in the keyboard; an EL lamp or other light emitting device being connected to the light emission circuit; and guide software such as spelling guide or typing method guide, application software control interface and driver being included in the computer software unit to guide typing of spelling, characters, applications, web pages, game software, and pre-schooling education programs in faster and easier operation of the keyboard.
Owner:IDEAL DESIGN TECH CORP
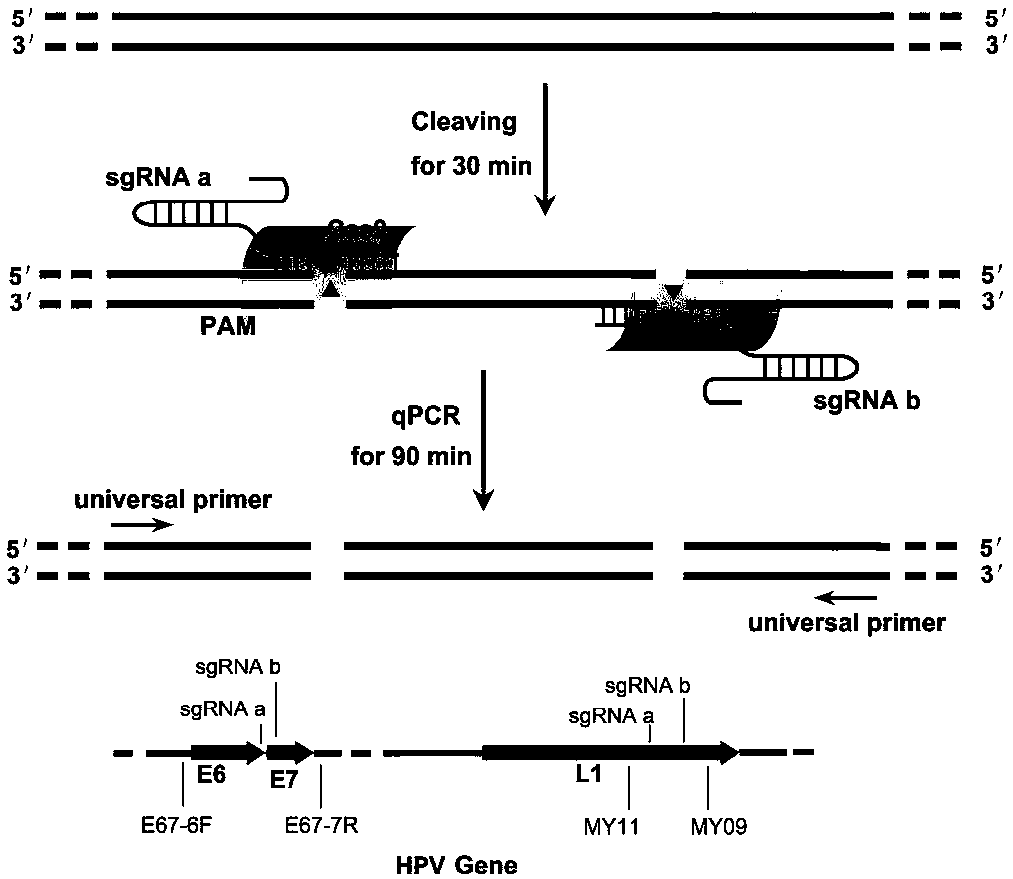
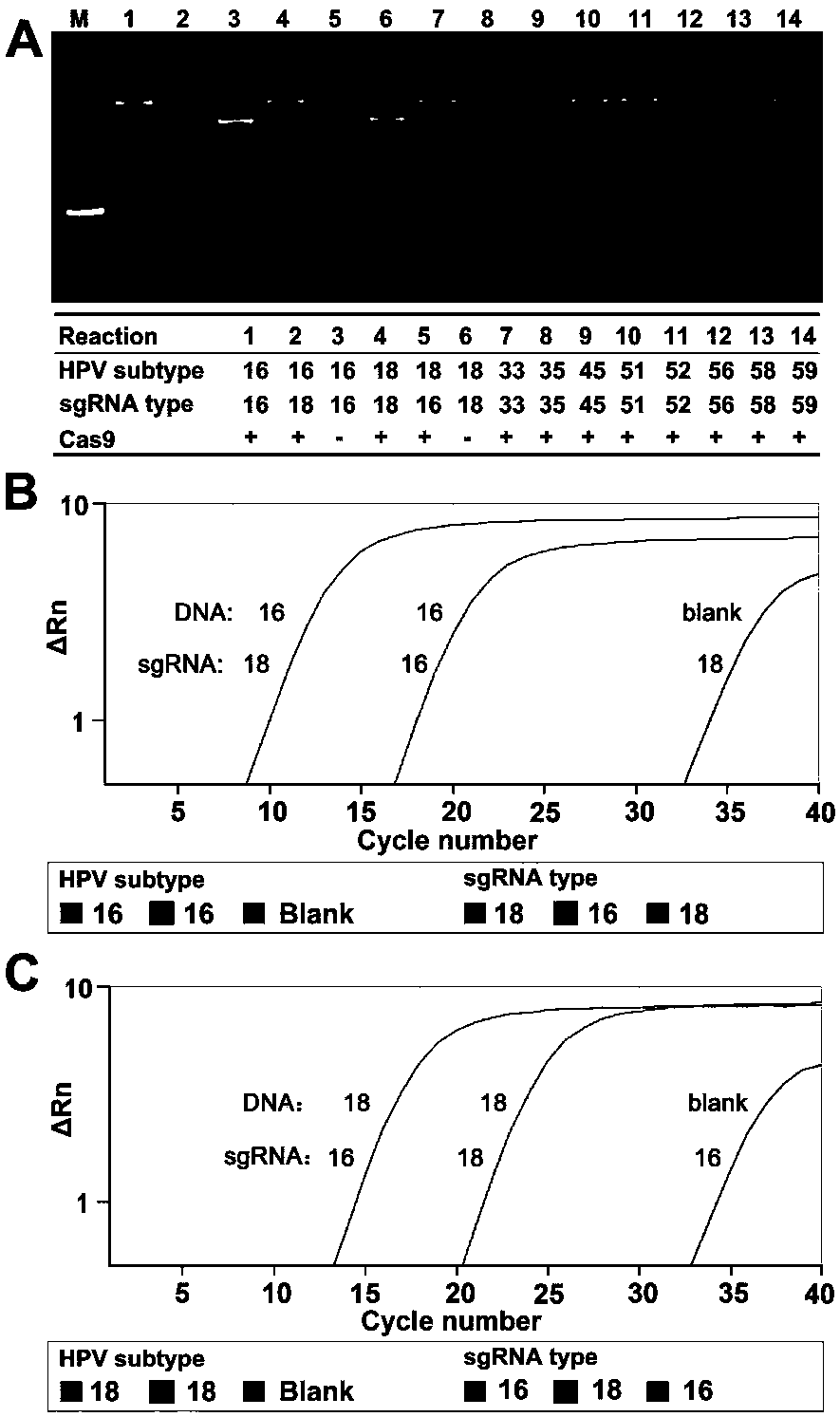
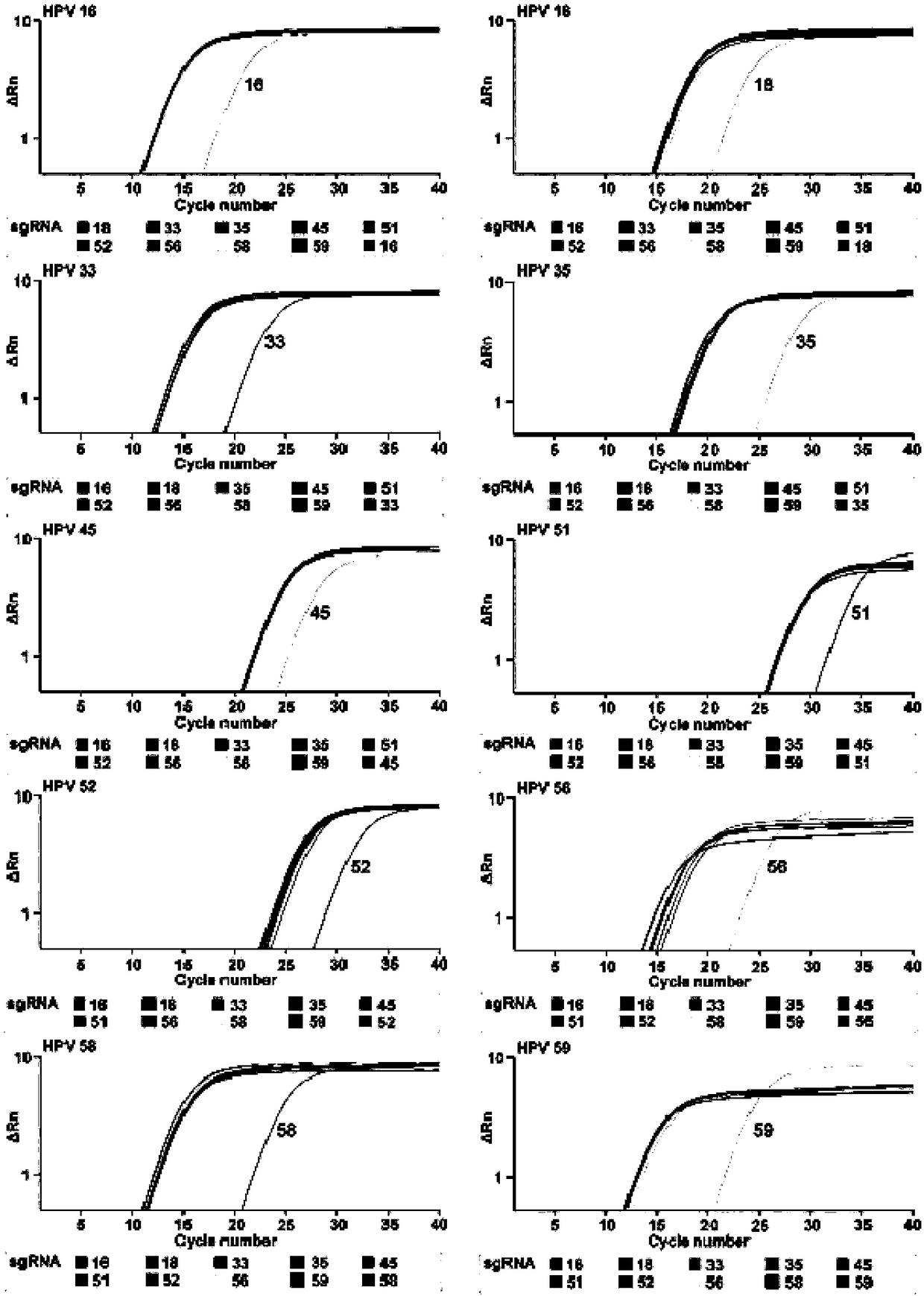
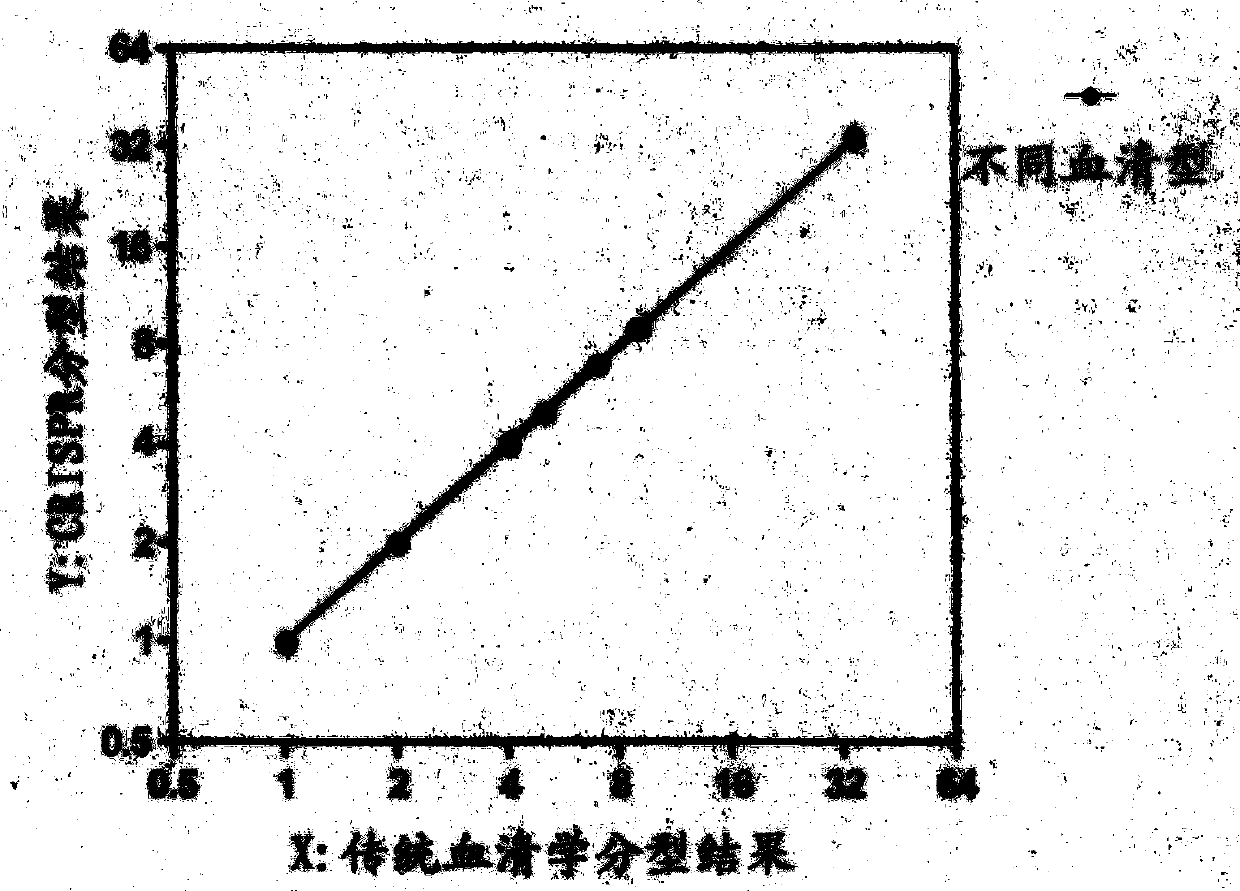
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) based PCR (Polymerase Chain Reaction) typing method and application thereof
ActiveCN108486234ASimplified typesQuick checkMicrobiological testing/measurementSpecific detectionTyping methods
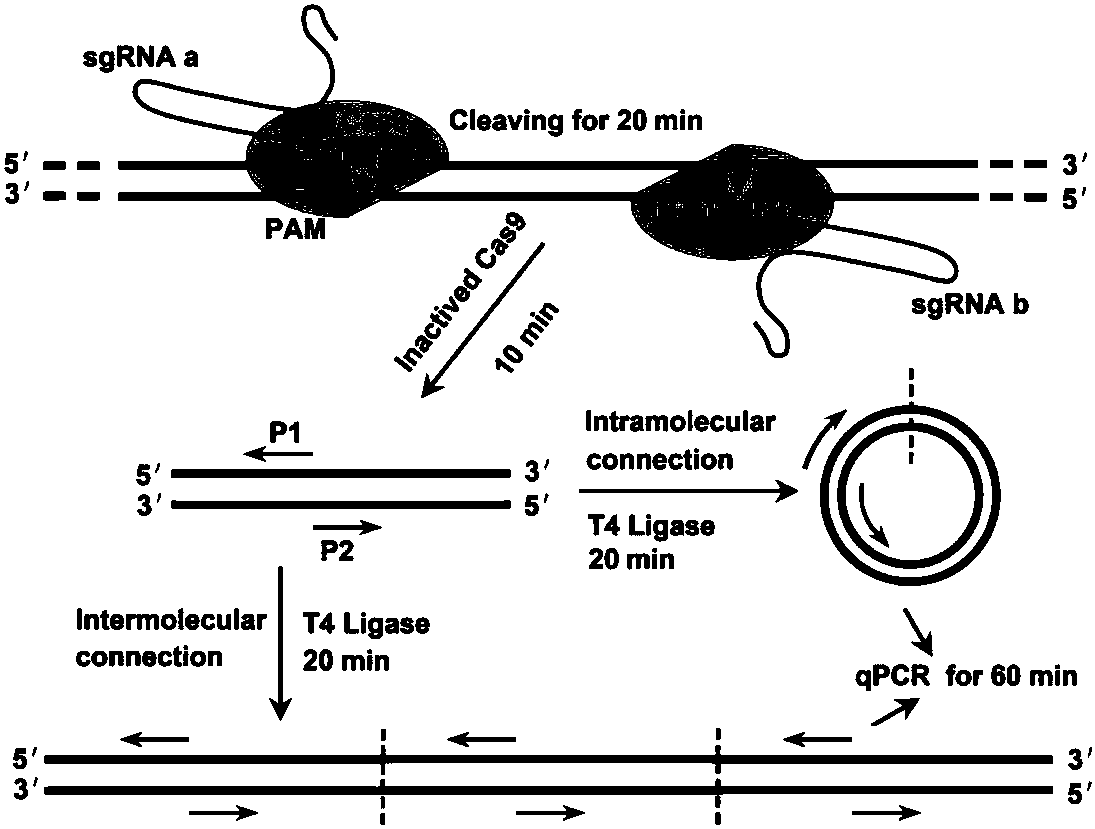
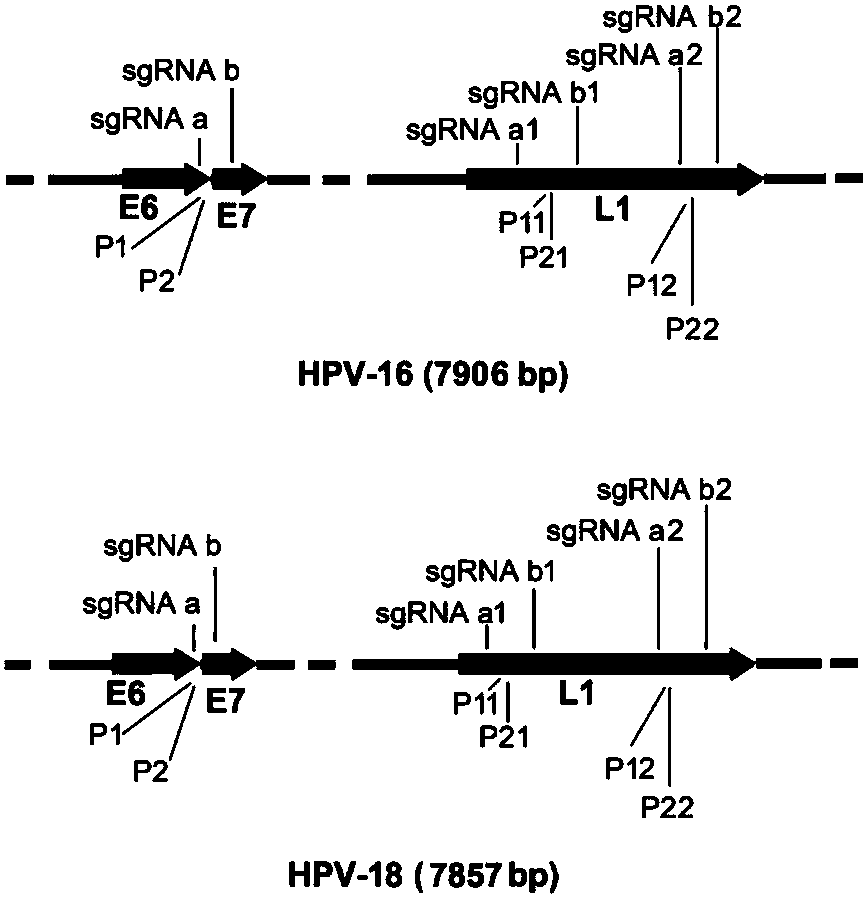
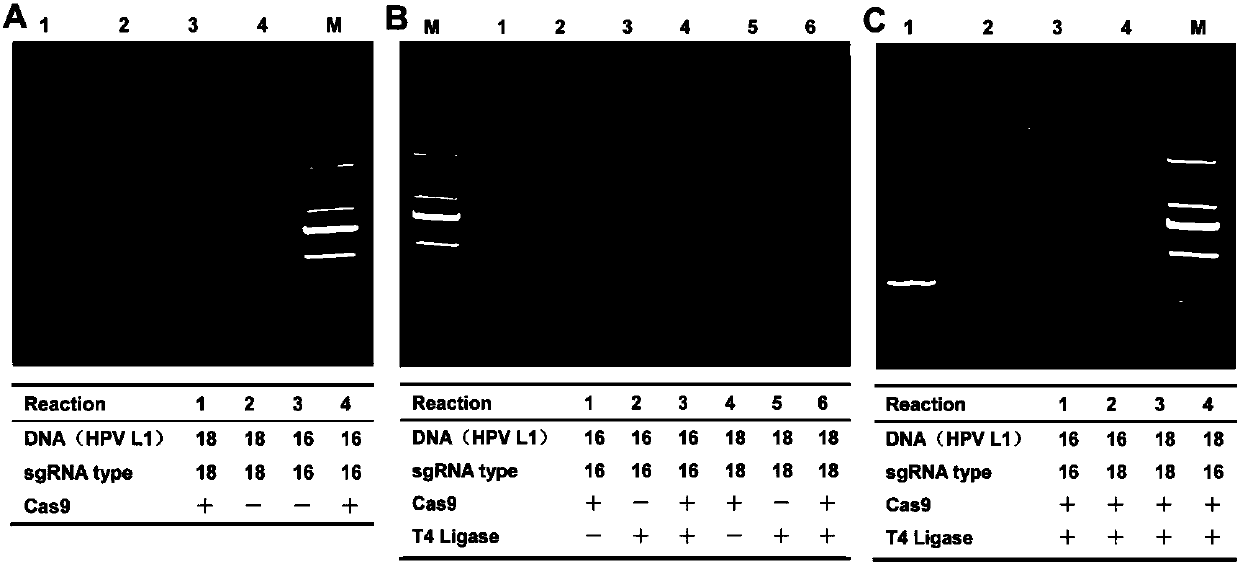
The invention discloses a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) based PCR (Polymerase Chain Reaction) typing method and the application thereof. The method is a method forperforming DNA detection and typing. The method comprises the following steps: adding a Cas9 protein and sgRNA for targeting target DNA into a conventional PCR reaction system, adding a short-term thermostatic incubation program before the PCR reaction program, and starting a conventional PCR amplification program. The CRISPR based PCR typing method disclosed by the invention adopts a homogeneousdetection technology, and detection can be completed by a PCR amplification step only. By utilizing a specific recognition cutting characteristic of the CRISPR technology on the DNA, the target DNA can be simply, homogeneously, rapidly and sensitively subjected to specific detection and typing, and the method is a novel DNA detection method with high specificity and sensitivity. According to themethod in the invention, human HPV DNA in clinical samples can be successfully detected.
Owner:SOUTHEAST UNIV
CRISPR (clustered regularly interspaced short palindromic repeat) based DNA detecting and typing method and application thereof
ActiveCN107828874ASimple detectabilitySimplified typesMicrobiological testing/measurementNucleic acid detectionSpecific detection
The invention discloses a CRISPR (clustered regularly interspaced short palindromic repeat) based DNA detecting and typing method and an application thereof. The method comprises following steps: (1),target DNA is subjected to PCR (polymerase chain reaction) amplification by a pair of universal primers; (2), the amplified target DNA is cut by Cas9 / sgRNA; (3), the cut target DNA is linked with DNAligase; (4), the linked target DNA is subjected to PCR amplification. According to the method, the target DNA can be subjected to specific detection and typing simply, rapidly and sensitively according to the specific identification cutting characteristics of the CRISPR technology on DNA, the method is one novel DNA detection method with high specificity and sensitivity, and the critical bottleneck problems about nucleic acid hybridization, specific PCR primer design and the like in current nucleic acid detection and typing fields are solved successfully. With the application of the method, L1 and E6 / E7 genes of HPV16 and HPV18 in human cervical cancer cells are detected successfully.
Owner:SOUTHEAST UNIV
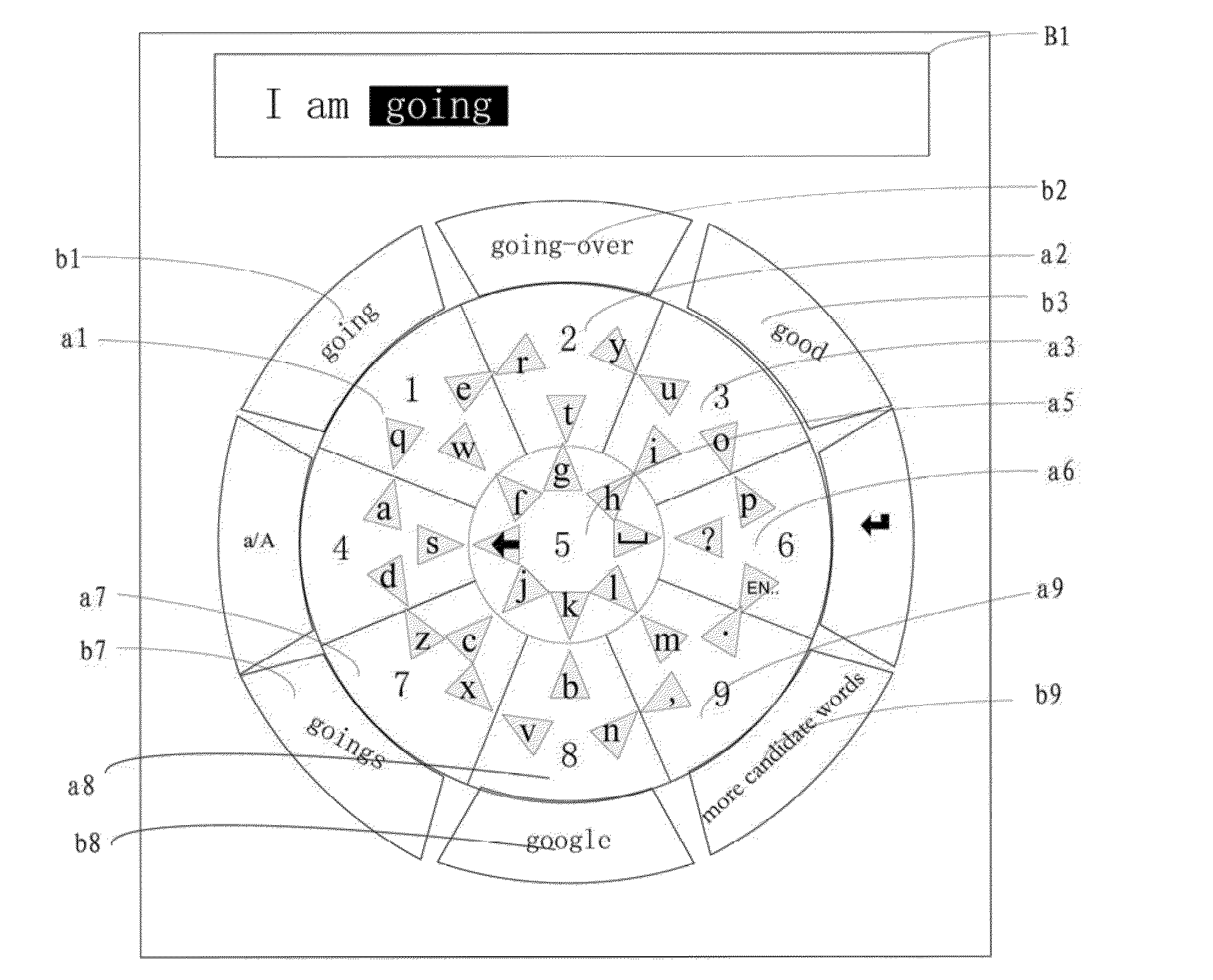
Input method and apparatus of circular touch keyboard
InactiveUS20150355727A1Typing fast and intuitiveInput textInput/output for user-computer interactionCathode-ray tube indicatorsTyping methodsHuman–computer interaction
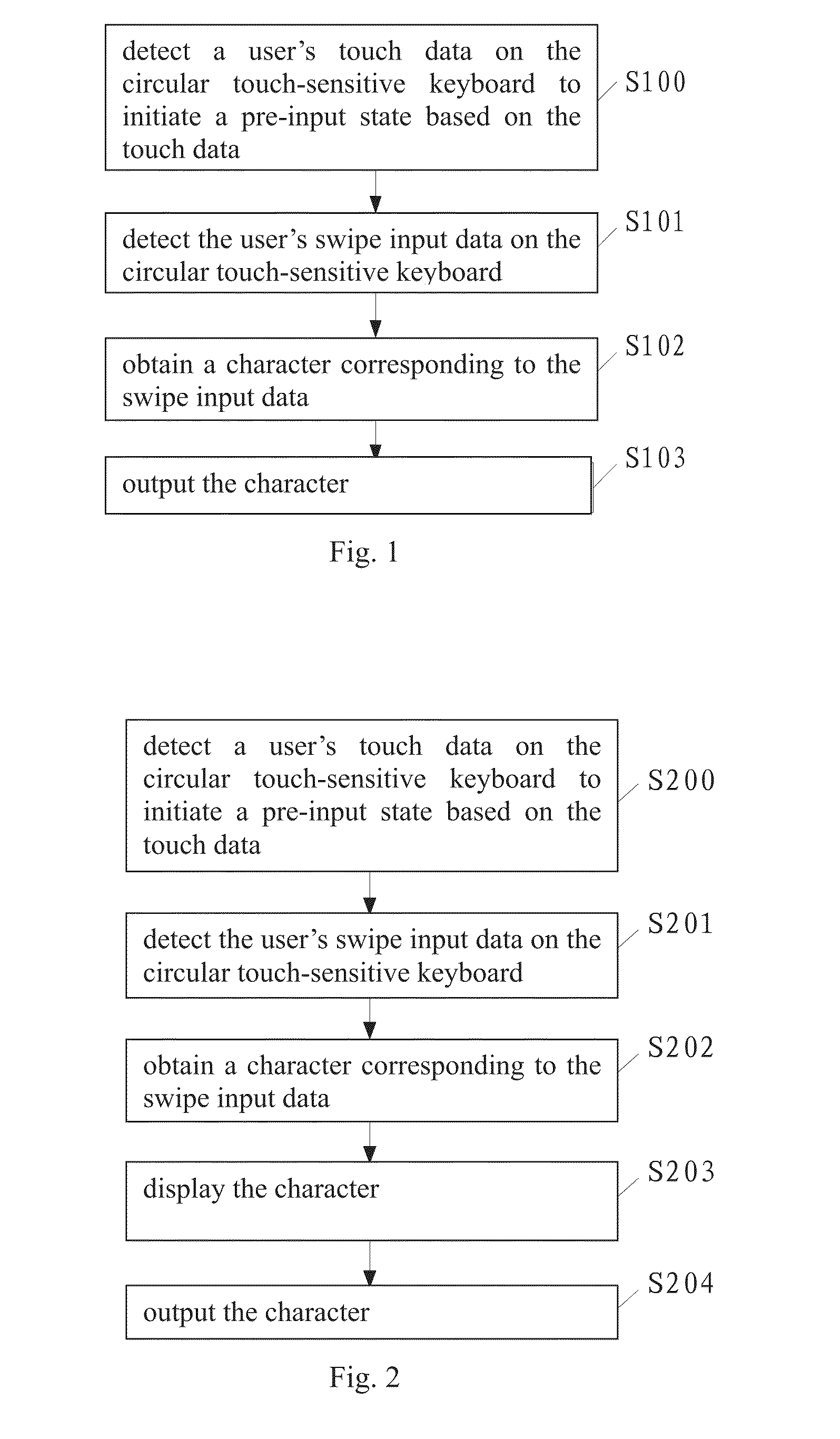
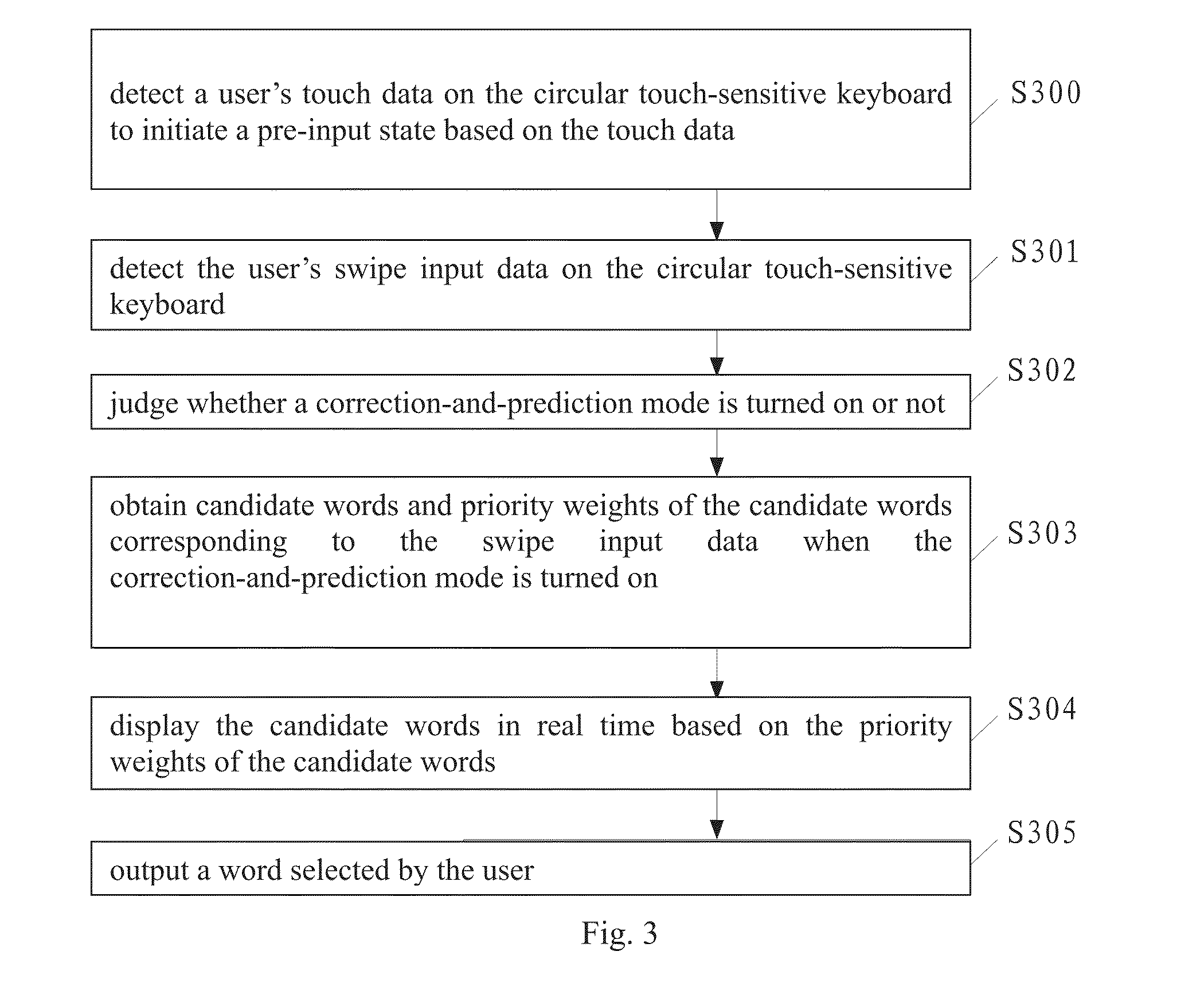
Disclosed is a typing method for a circular touch-sensitive keyboard, comprising detecting a user's touch data on the circular touch-sensitive keyboard to initiate a pre-input state based on the touch data; detecting the user's swipe input data on the circular touch-sensitive keyboard; obtaining a character corresponding to the swipe input data; and outputting the character; wherein the circular touch-sensitive keyboard comprises an inner key zone and an outer key area, the outer key area comprises eight outer key zones, the inner key zone comprises eight sectors, each outer key zone comprises four sectors, each sector is assigned with a particular character, the circular touch-sensitive keyboard is provided with an input module for detecting the swipe input data. Also disclosed is a typing device for a circular touch-sensitive keyboard. The typing method and typing device of the present invention can increase typing speed.
Owner:HU JINGTAO
CYP450 gene type database and gene typing and enzymatic activity identification method
ActiveCN103198236AGive quickly and accuratelyAccurate judgment basisMicrobiological testing/measurementProteomicsDiseaseGene type
The invention discloses a method for building a standard CYP450 (cytochrome p450) gene type database. The method comprises the steps as follows: comparing a specific sequence corresponding to mutation information of a CYP450 gene type with a standard human whole genome sequence to obtain a corresponding relation between the specific sequence and the standard human whole genome sequence; and converting the CYP450 gene type into a gene type taking the standard human whole genome sequence as a reference sequence according to the corresponding relation. The invention further discloses a database built by the method and a CYP450 gene typing and enzymatic activity identification method based on the database. The database is convenient for research of CYP450; the gene typing method provides unified standards for CYP450 gene typing and provides more accurate judgment criterions for diseases, drugs or the like; unknown mutation sites in CYP450 genes can be effectively detected; hundreds of samples can be detected at the same time; and the coverage range is comprehensive and wide.
Owner:北京六合华大基因科技有限公司
HLA gene specific PCR amplification primer, HLA typing method and kit
InactiveCN103045591ANo StrapReasonable and accurateMicrobiological testing/measurementDNA/RNA fragmentationNucleotideTyping methods
The invention discloses an HLA (human leukocyte antigen) gene specific PCR (polymerase chain reaction) amplification primer, an HLA typing method and an HLA typing kit. A sequence of the PCR amplification primer comprises a primer sequences shown as SEQ ID No. (sequence identification number) 1-2, 3-4, 5-6, 7-8 and 9-10. The HLA gene typing method comprises the steps that the HLA gene specific PCR amplification primer is used for conducting PCR amplification and purification on sample DNA (deoxyribonucleic acid); a gene exon sequencing primer is used for conducting sequencing PCR amplification, purification and sequencing on an HLA gene amplification product, the sequence of the HLA gene amplification product is compared with a standard sequence; and the type of the HLA gene is determined. The HLA gene typing kit comprises the PCR amplification primer and the sequencing primer. A qualitative leap in aspects of detection flux, data quality, cost control and the like is achieved; data is more reliable and realer; and the problem that typing cannot be conducted when nucleotide of certain alleles is positioned outside an amplification area is solved.
Owner:SHANGHAI TISSUEBANK BIOTECH +3
Detection and parting method of human papillomavirus and reagent box
ActiveCN1948503AHigh sensitivityImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationHuman papillomavirusInfection rate
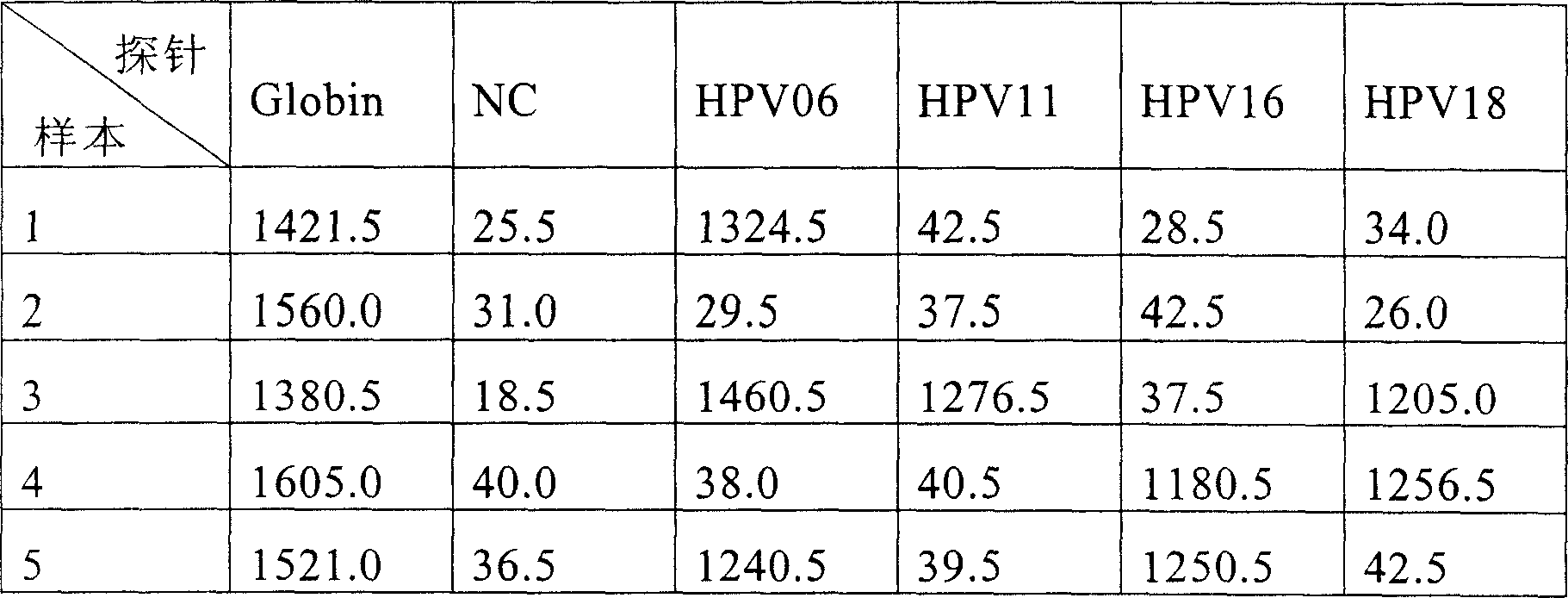
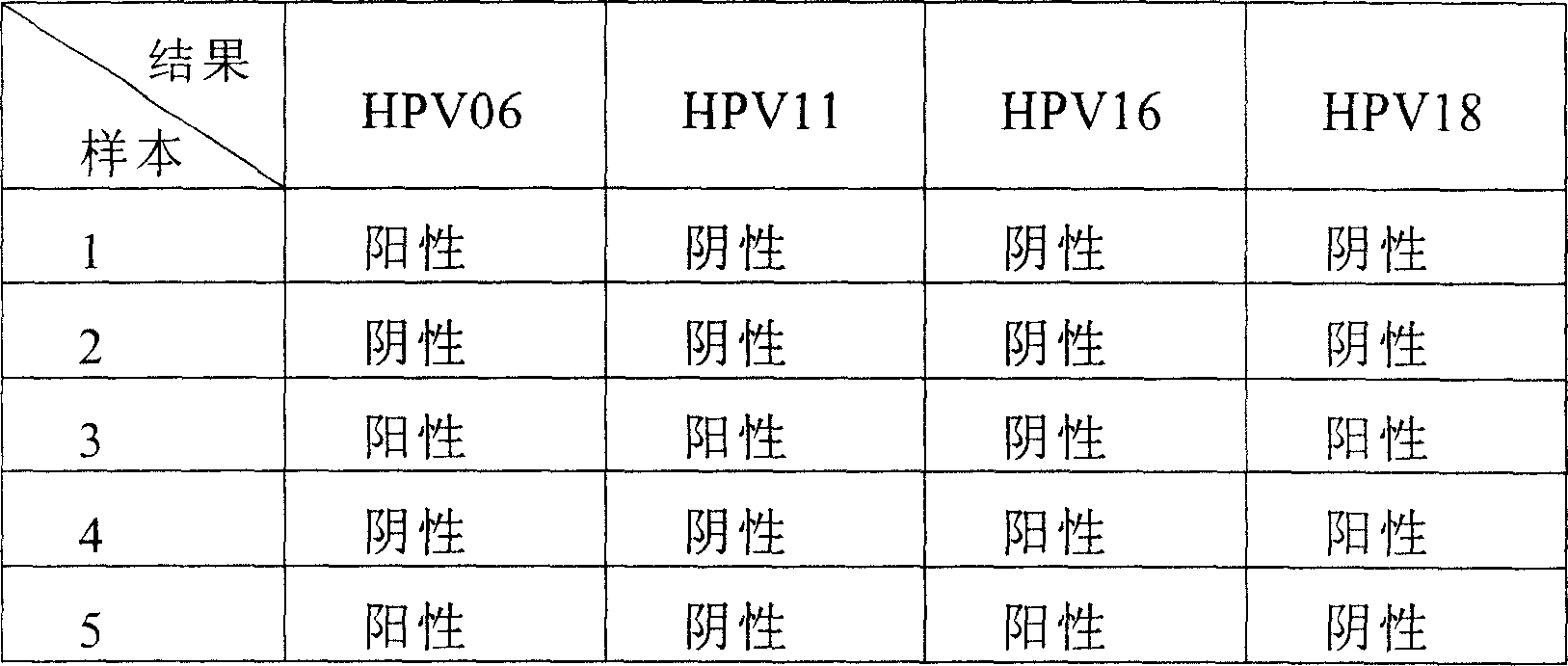
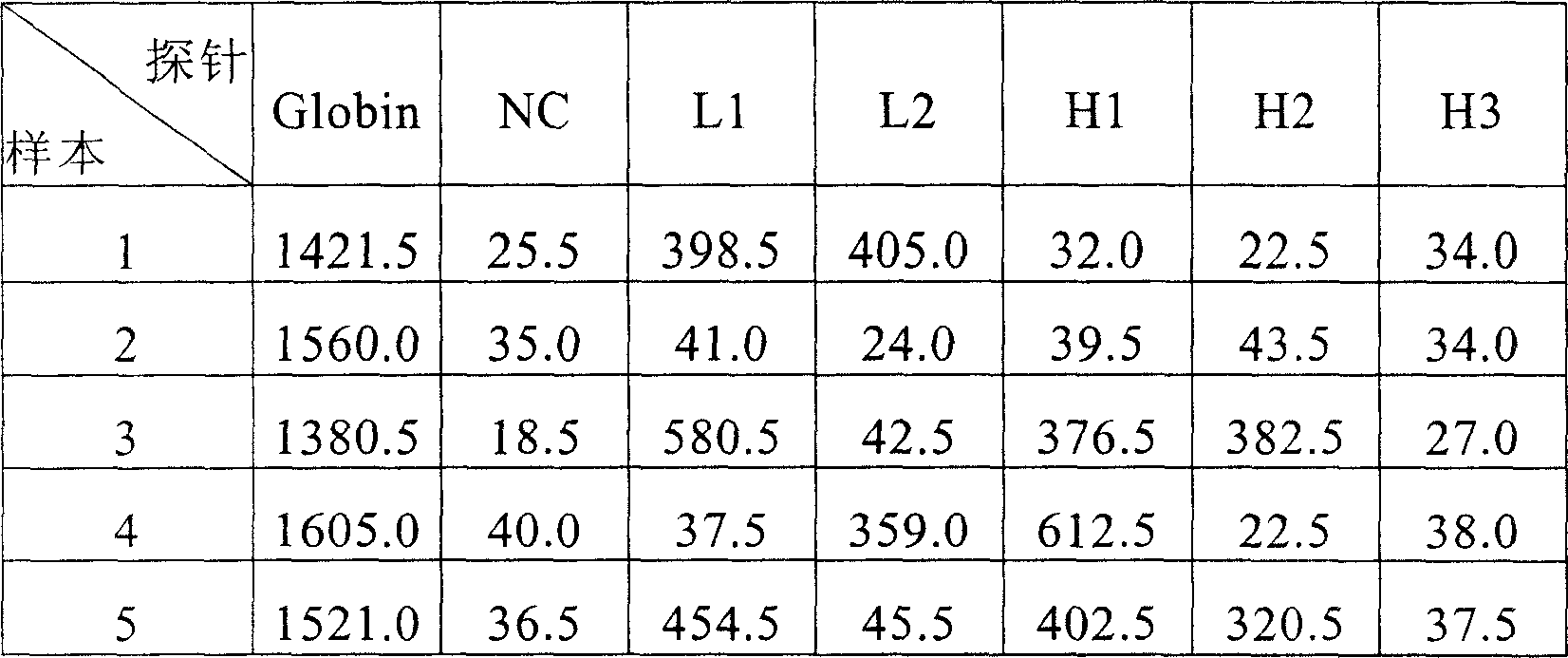
A testing and typing method and its reagent boxes for Human Papillomavirus(HPV), relating to exciters and nucleic acid probes which are especially for testing and typing of HPV. The invention fixes three types of special probes including different subgroups of low-hazard HPV and high-hazard HPV, subgroups with extra-low infection rate of low-hazard or high-hazard HPV, onto the same position or mediator. The invention also fixes oligomeric nucleoside probes of special HPV onto microballoons based on Luminex xMAP technical platform, which forms microballoons probes for testing HPV subgroups and typing slugs of HPV. A testing and typing method with simplicity of operator and its reagent boxes are invented. The method has much strongpoint including high-sensitivity, high-flux, stable result and good repeatability.
Owner:SHANGHAI TELLGEN LIFE SCI CO LTD
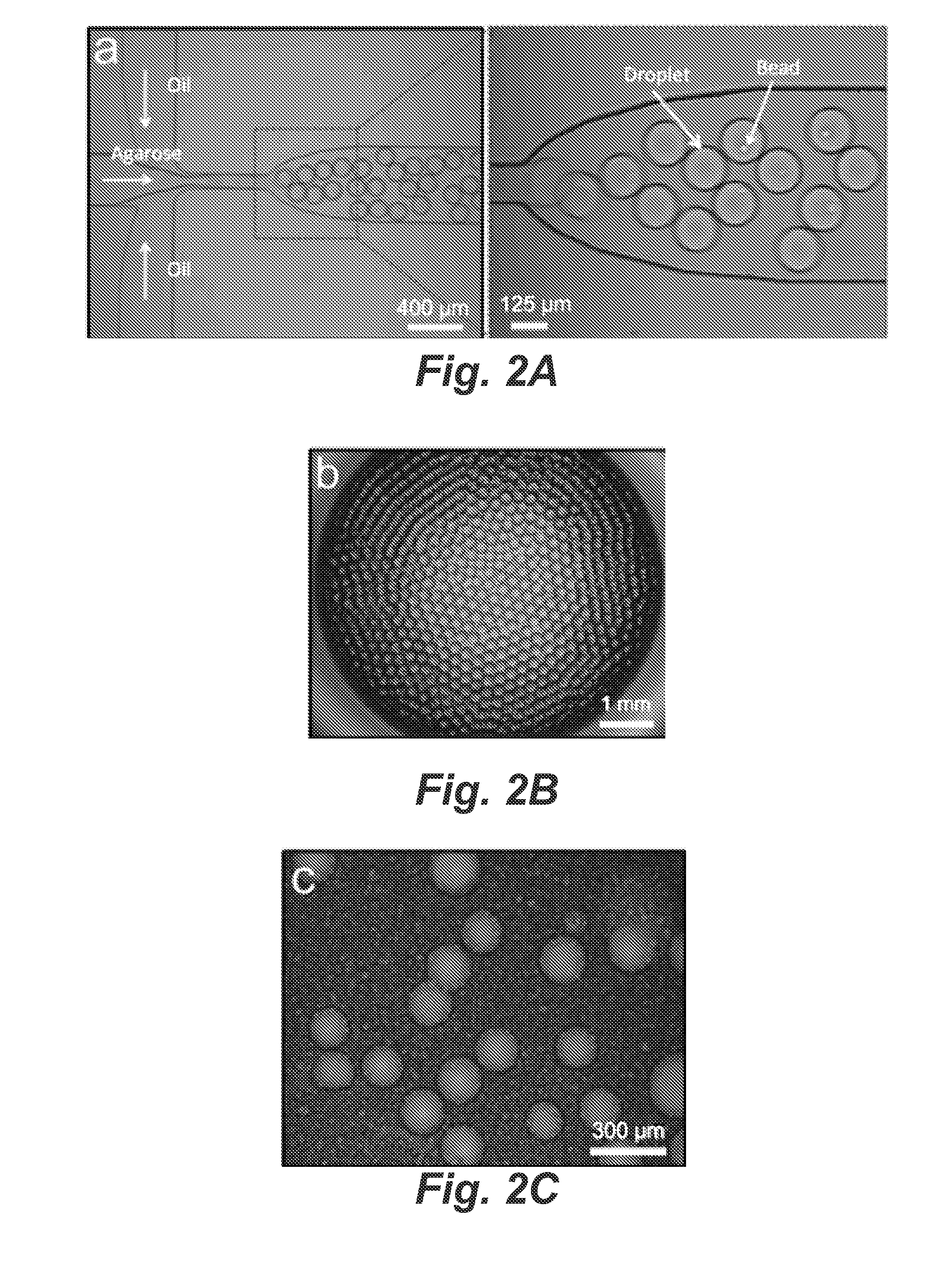
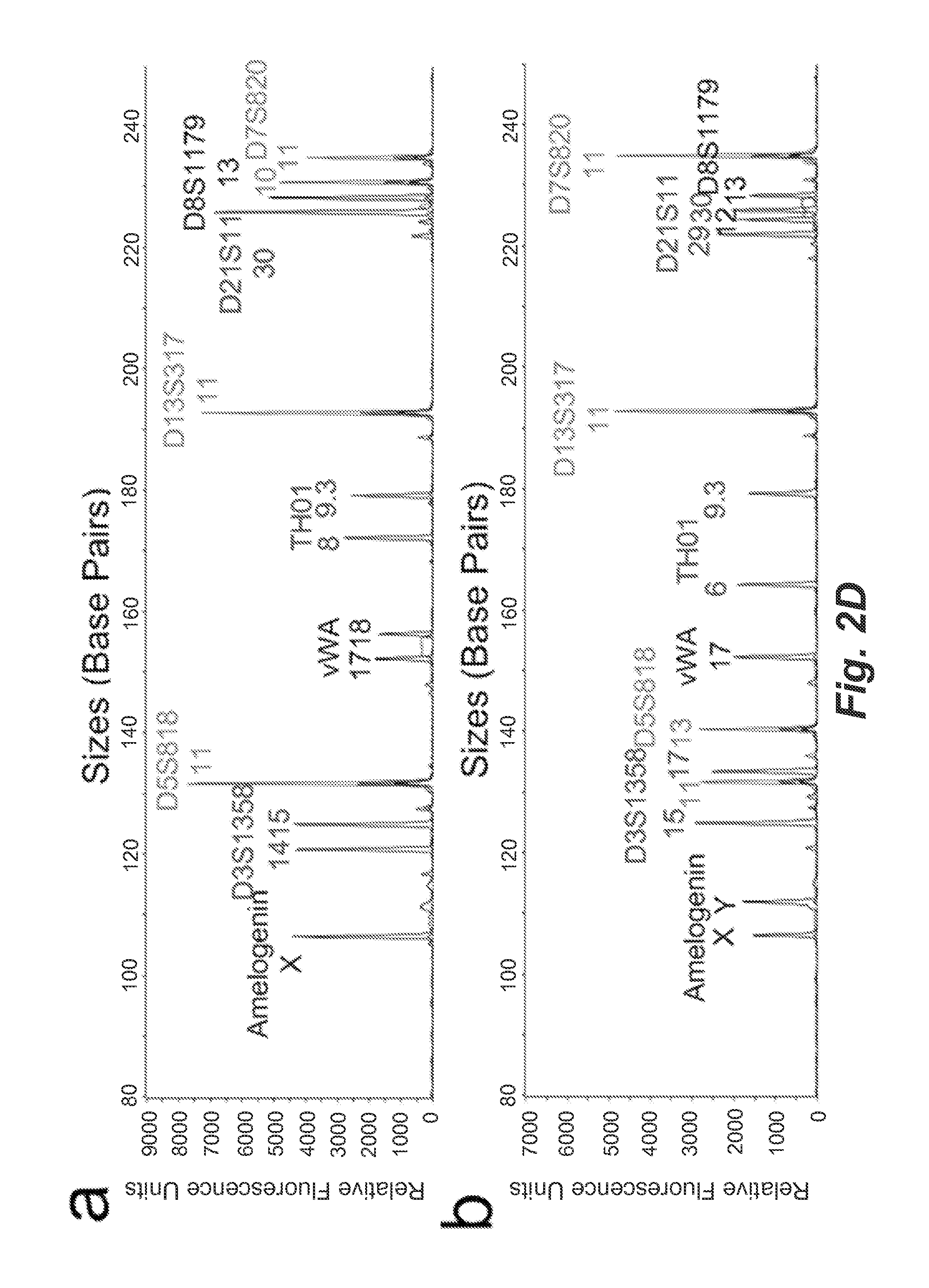
Single-cell forensic short tandem repeat typing within microfluidic droplets
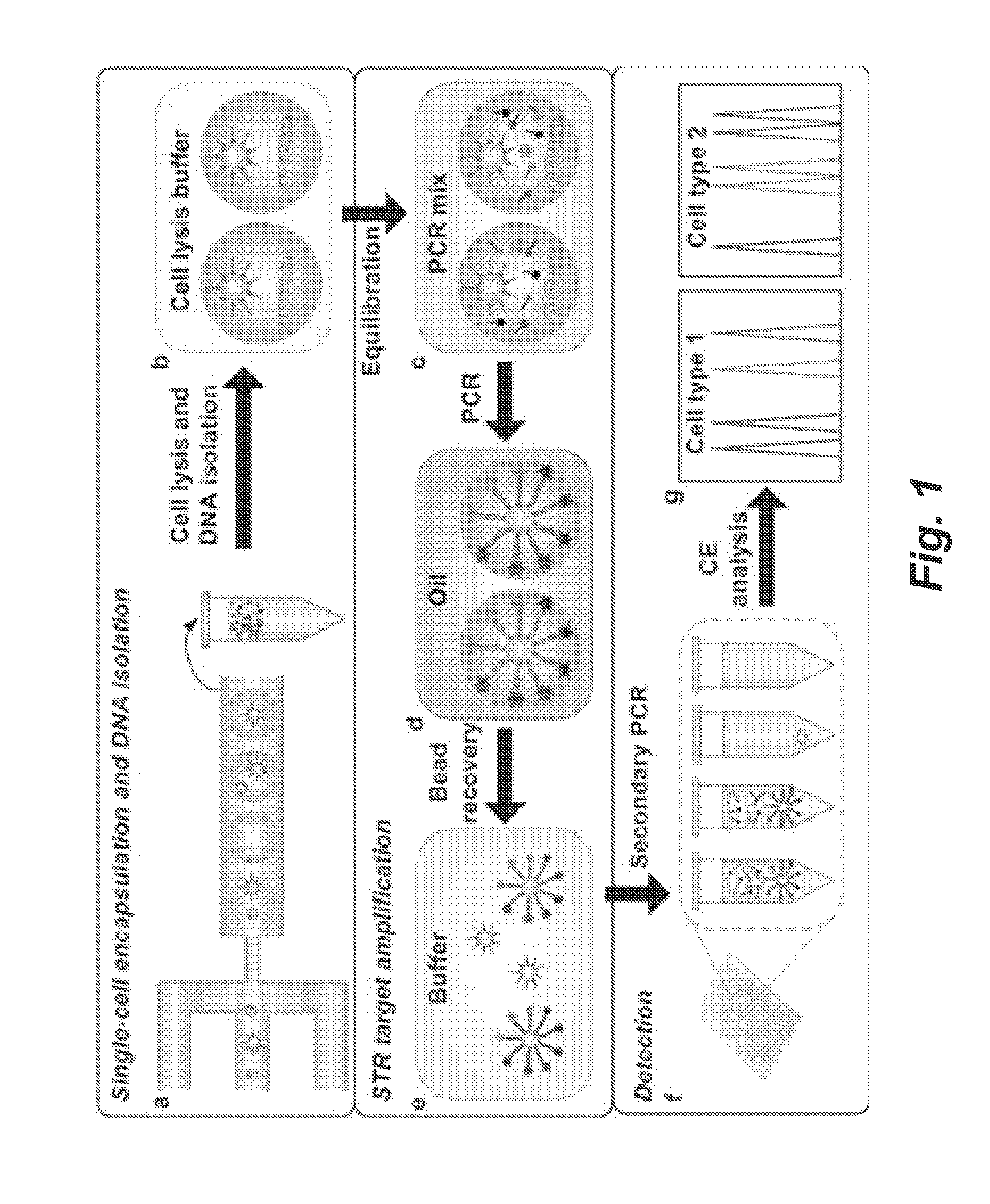
A short tandem repeat (STR) typing method and system are developed for forensic identification of individual cells. Agarose-in-oil droplets are produced with a high frequency using a microfluidic droplet generator. Statistically dilute single cells, along with primer-functionalized microbeads, are randomly compartmentalized in the droplets. Massively parallel single-cell droplet PCR is performed to transfer replicas of desired STR targets from the single-cell genomic DNA onto a coencapsulated microbead. These DNA-conjugated beads are subsequently harvested and reamplified under statistically dilute conditions for conventional capillary electrophoresis STR fragment size analysis. The methods and systems described herein are valuable for the STR analysis of samples containing mixtures of cells / DNA from multiple contributors and for low concentration samples.
Owner:RGT UNIV OF CALIFORNIA
SNP markers related to grass carp growth speed and application thereof
ActiveCN106381331AEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationStart codonNucleotide
Through amplification of a part of fragments of an NPY gene, and through an SNaPshot SNP typing method, provided is SNP markers related to the grass carp growth speed, wherein S1 is located at a 639th locus from an initiation codon (ATG, with an ATG first nucleotide as the first) in an intron 1 sequence of the NPY gene, and a base at the position is T or A; S2 is located at an 892nd locus from the initiation codon in the intron 1 sequence of the NPY gene, and a base at the position is C or A. The two SNP loci can be used for identification or breeding of grass carp with faster growth speed, and lay the foundation for screening and establishment of grass carp fast-growth new varieties.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
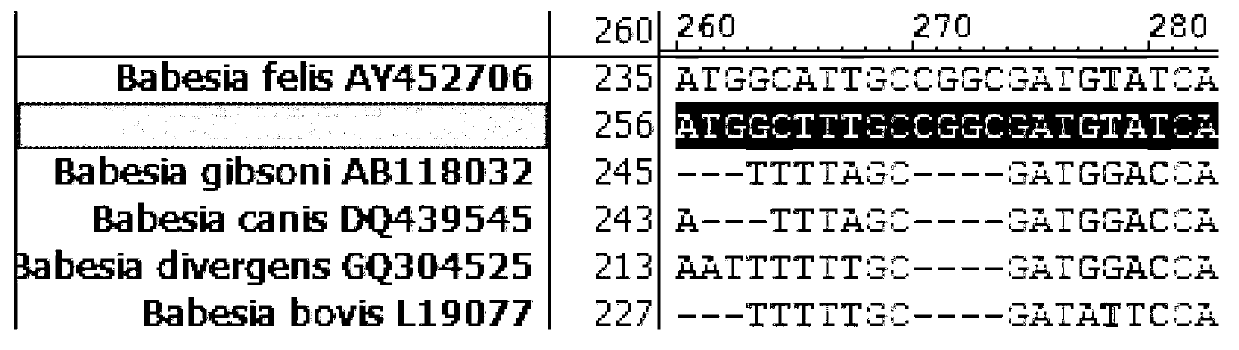
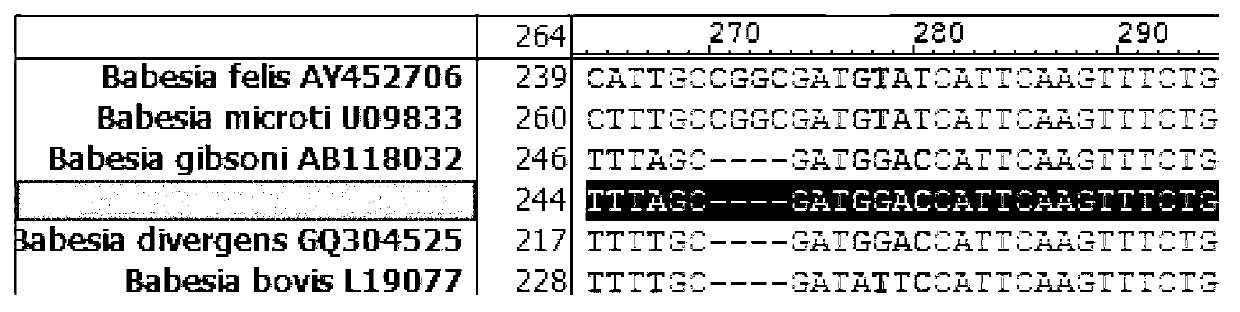
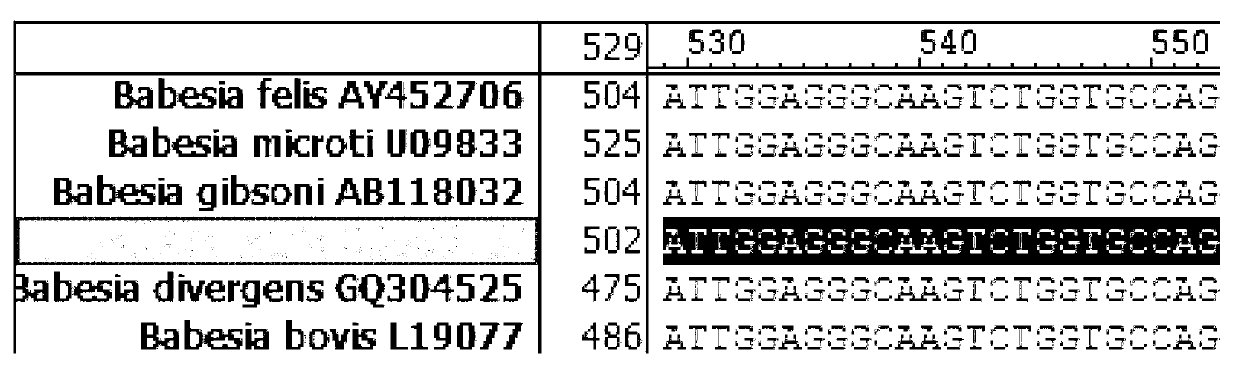
Primer, probe and kit for fluorescence quantitative PCR (Polymerase Chain Reaction) detecting and typing of babesia
InactiveCN103103286AQuick checkSensitive detectionMicrobiological testing/measurementDNA/RNA fragmentationPositive sampleFluorescence
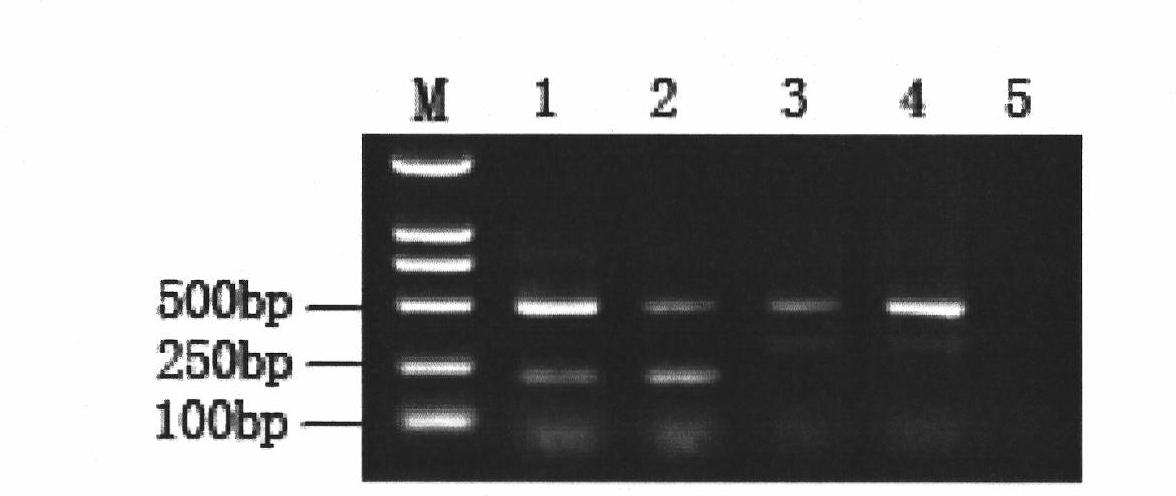
The invention discloses a primer, a probe and a kit for fluorescence quantitative PCR (Polymerase Chain Reaction) detecting and typing of babesia. According to the invention, the primer sequence is as shown in SEQID (SEQuence IDentifier) NO.1, 2 and 3 and the probe is as shown in SEQIDNO.4 and 5. A typing method of the babesia comprises the following steps of: carrying out PCR amplification on the to-be-detected bacterium by using the primer and the probe to obtain Erich bacterium positive samples of 290bp (base pair) bands, wherein the fluorescence of the samples is enhanced at the wavelength of 640nm in fluorescence detection; and typing the babesia according to the melting temperature. The primer, the probe and the kit disclosed by the invention have obvious technical advantages, have very high specificity and sensitivity and can be used for conveniently detecting the babesia of important species.
Owner:YANGZHOU UNIV
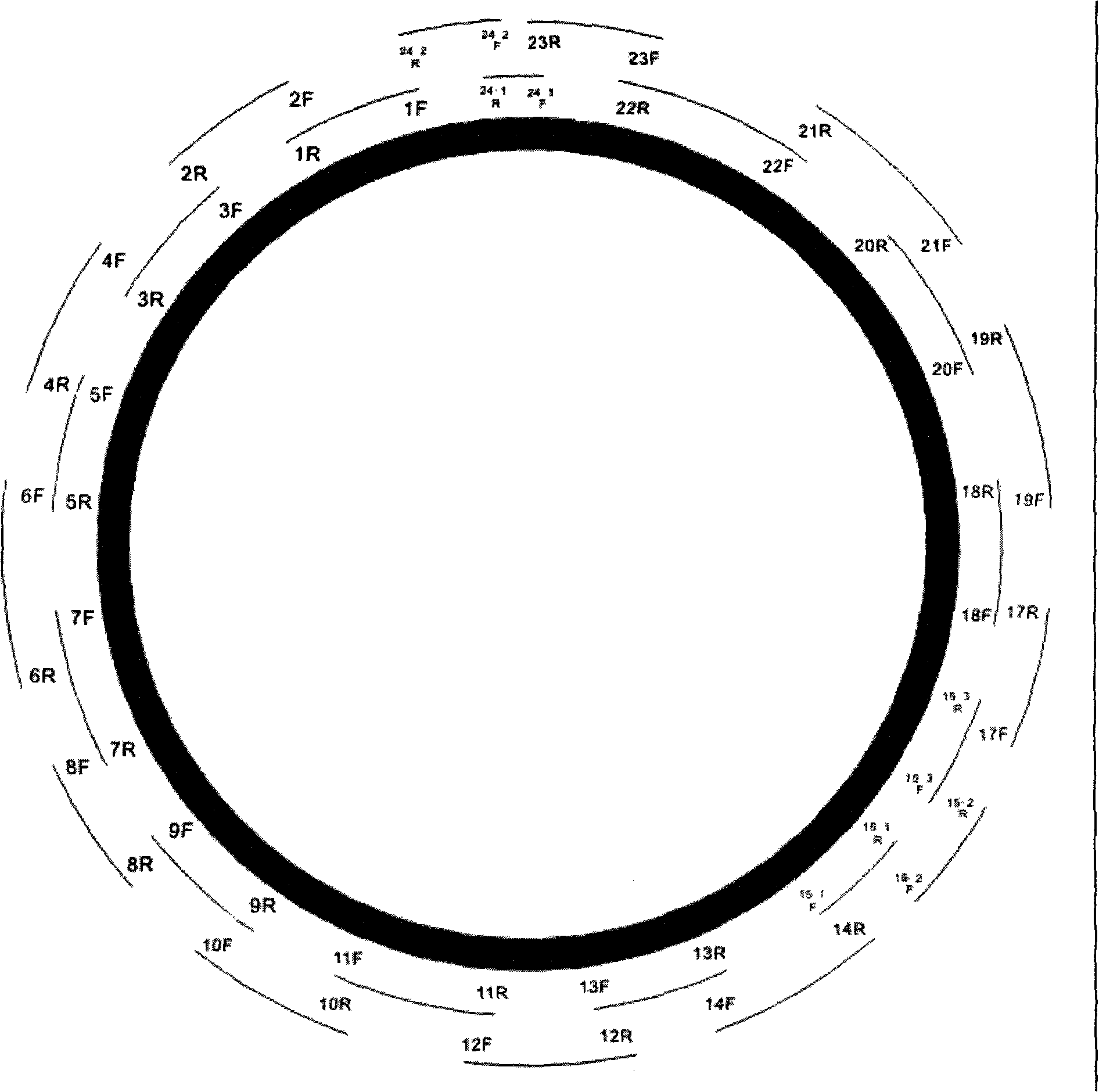
26-pair PCR primer for mitochondrion sequencing and parting method based on the primer
InactiveCN101270390AOvercoming demandsMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceTyping methods
The invention relates to a detection method of nucleic acid, particularly discloses a whole base sequence typing of mitochondria and a determination method thereof, specifically 26 pairs of PCR primer for mitochondria sequencing and a typing method based on the primers. The 26 pairs of PCR primer which are adopted by the invention cover the total length of mitochondria genome, wherein, primer 15-1, 15-2, 15-3, 24-1 and 24-2 are designed renewedly based on Chinese population and has the pertinence of the typing of Chinese population. The amplified fragment which corresponds to the primer 24-1 is the minimum and equals to 420bp. The amplified fragment which corresponds to the primer 22 is the maximum and equals to 1162bp. The dimensions of all PCR fragments are moderate and favorable for PCR amplification. The 26 pairs of PCR primer of the invention is utilized in the laboratory of applicant for investigating the community information of Chinese nation, and the whole sequence information of Chinese population is submitted to GENEBANK databases. The result shows that the invention can be definitely applied in the field of individual recognitions, paternity testing and gene diagnosis of the field of forensic medicine, anthropology, genetics and disease.
Owner:XI AN JIAOTONG UNIV
Apparatus and method for finger to finger typing
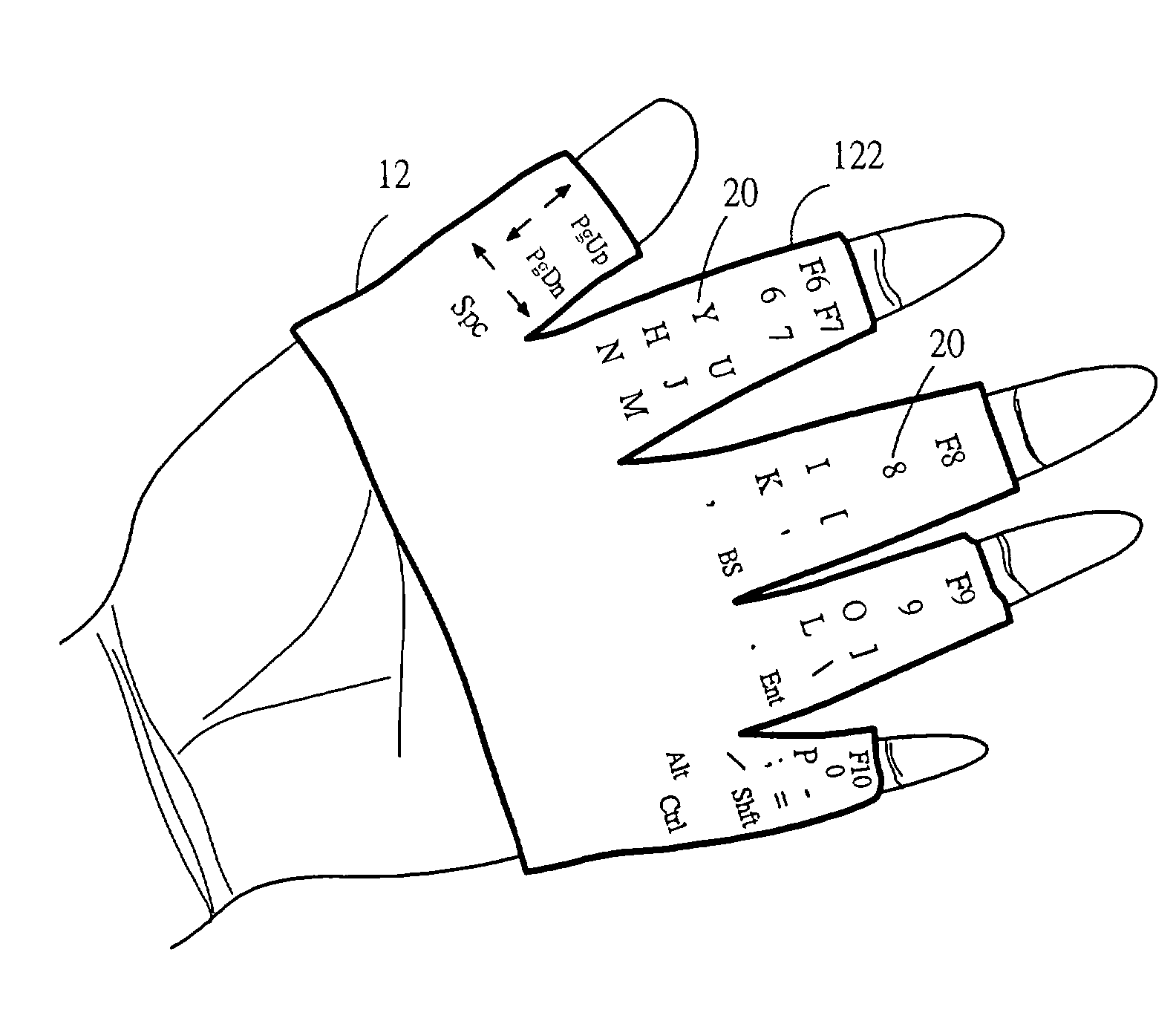
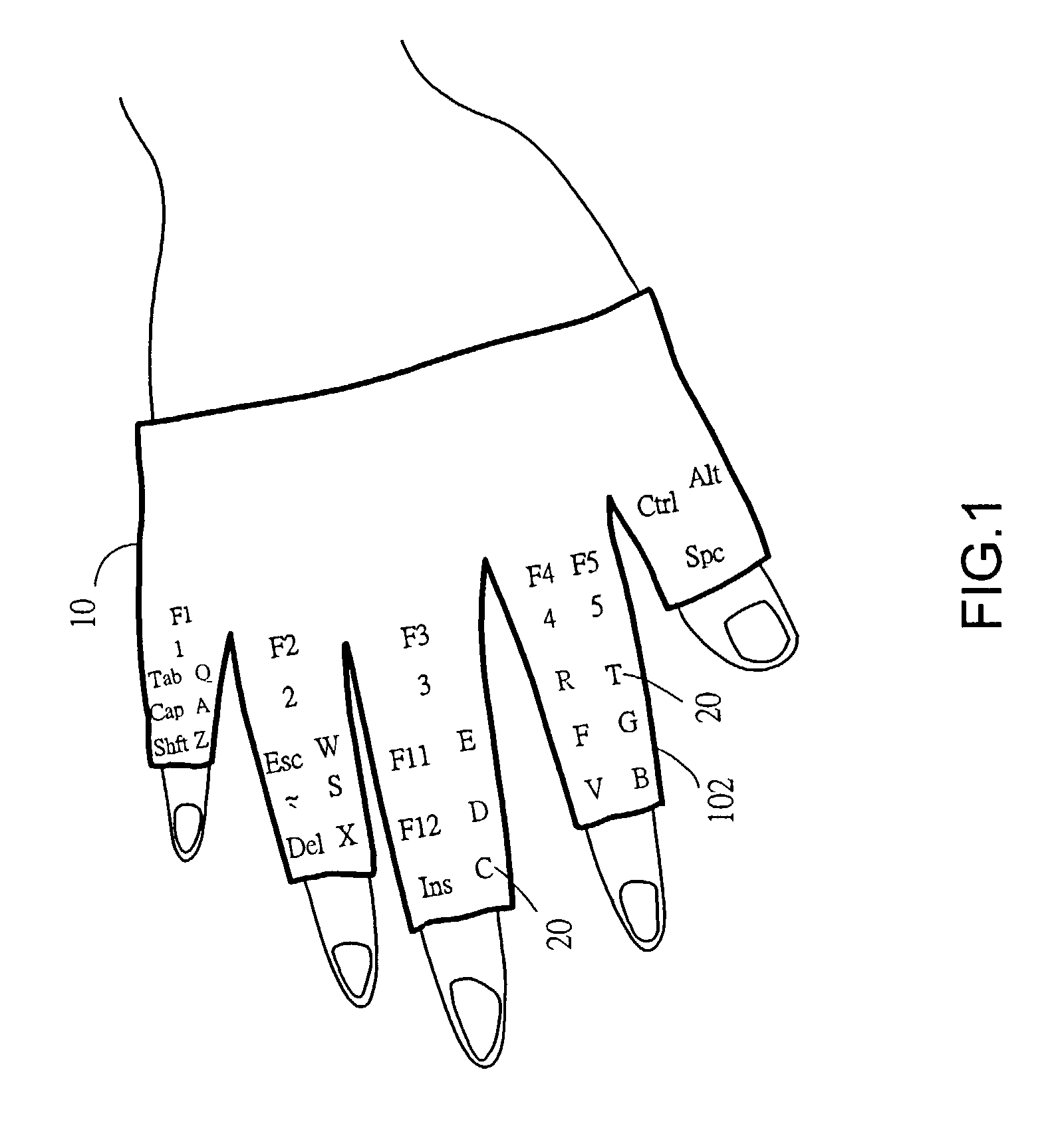
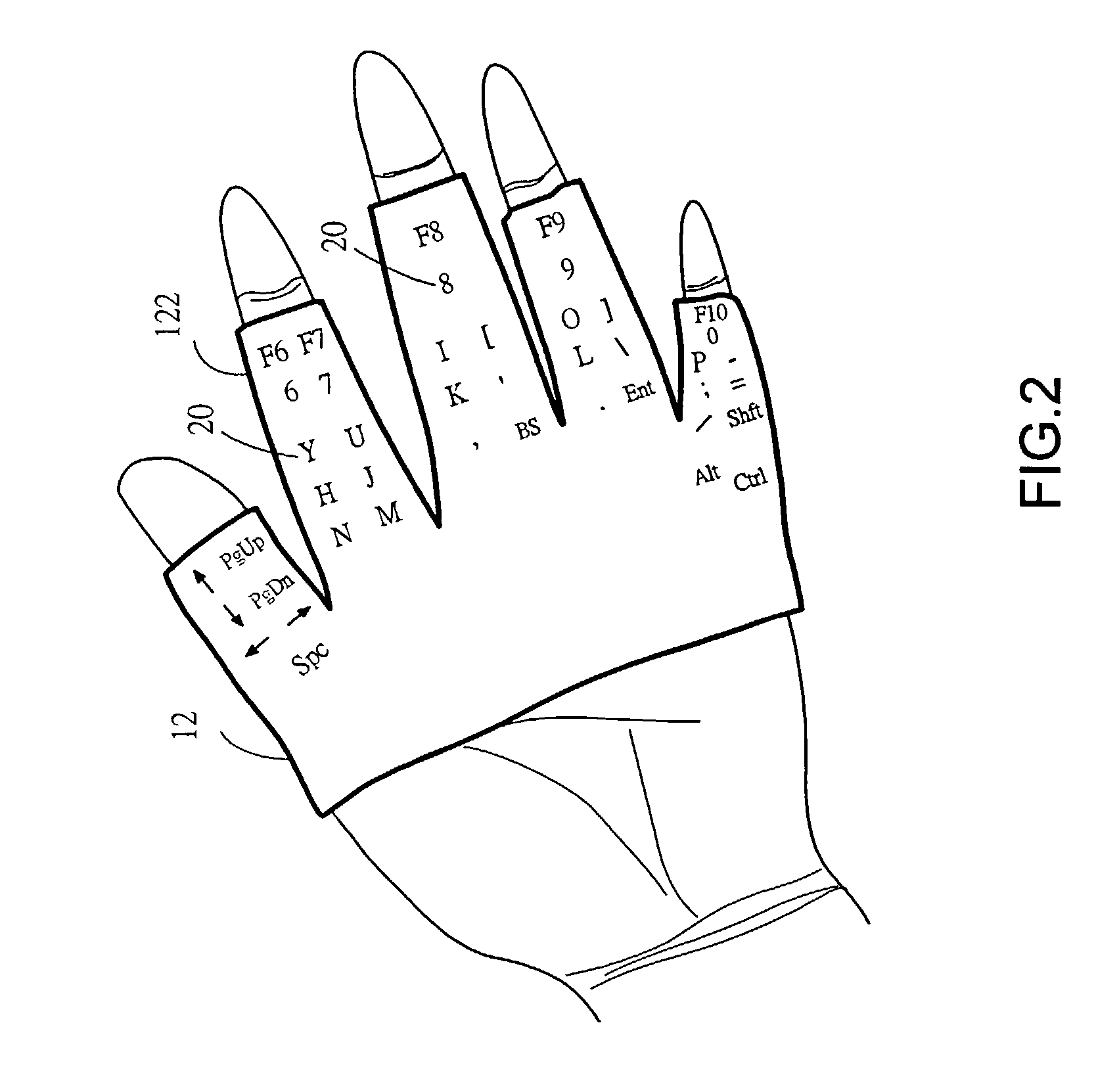
InactiveUS20040036678A1Input/output for user-computer interactionCathode-ray tube indicatorsFinger tappingTyping methods
An input apparatus comprises two glove bodies with glove fingers, each of which is mounted with key buttons as a keyboard for an information system. And a corresponding input method, with the keyboard apparel worn on both hands of a user, is to use a finger of one hand striking on a particular key mounted on a glove finger of the other hand. A half number of keys are mounted on the palm face of the glove fingers of one hand while other keys on the back face of the other hand. With such a keyboard and typing method, a user is able to type with ten fingers away-from-desk by mutually using fingers of each hand to type on the fingers of the other hand.
Owner:ZNGF FRANK
Monomolecular gene typing method based on DNA paper folding probe specificity marking and application thereof
InactiveCN106893781AImprove accuracyOvercoming limited quantitiesMicrobiological testing/measurementOptical diffractionFluorescence
The invention provides a monomolecular gene typing method based on DNA paper folding probe specificity marking and application thereof. The method comprises the following steps: 1) preparing a DNA paper folding probe; 2) carrying out specificity marking on a target DNA through the DNA paper folding probe; 3) carrying out imaging observation on the marked target DNA under an atomic force microscope. Moreover, the monomolecular gene typing method can also be applied to haplotype typing of patient samples and haplotype typing of practical samples with unknown haplotype information. All in all, according to the method provided by the invention, single nucleotide polymorphism can be effectively detected, and the position of a single basic group is observed at high resolution under the atomic force microscope, so that the problem that fluorescent molecular marking is limited to optical diffraction limit under an electron microscope is successfully avoided, and the method can be widely applied to the research fields such as in situ test of chromogene variation and clinic gene diagnosis.
Owner:SHANGHAI INST OF APPLIED PHYSICS - CHINESE ACAD OF SCI
Genotyping detection method of drug resistance to Sanmate of Fusarium graminearum
The invention belongs to a Sanmate resistance genotype molecular detection and SNP typing method of Fusarium graminearum, which can be used for drug resistance monitoring and drug resistance genotype diagnosis to Sanmate and other benzimidazole type bactericides of the Fusarium graminearum which can cause wheat scab. The detection method comprises the following three major steps: (1) extracting DNA of a nuclear genome of a strain to be detected; (2) applying three groups of primer pairs to carry out PCR reaction; and (3) identifying the genotype of the strain with the resistance to the Sanmate according to a PCR product electrophoretogram. By adopting the Tetra-primer ARMS PCR (Tetra-primer amplification refractorymutation system-PCR) technology, the strain with the drug resistance of theFusarium graminearum and the genotype thereof can be fast and accurately detected, and the level of the drug resistance can be further judged. The detection accuracy is above 95%.
Owner:NANJING AGRICULTURAL UNIVERSITY
Microsatellite loci marker combination and application thereof
InactiveCN101705299AAppropriate size intervalEasy to detect simultaneouslyMicrobiological testing/measurementMaterial analysis by electric/magnetic meansFluorescenceHigh flux
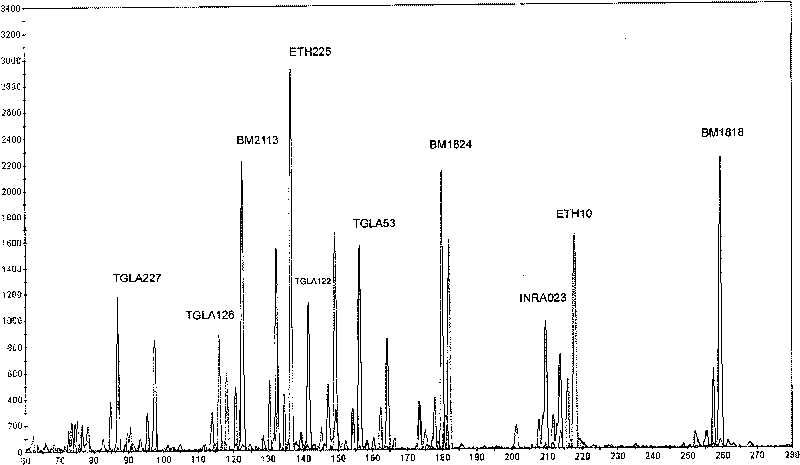
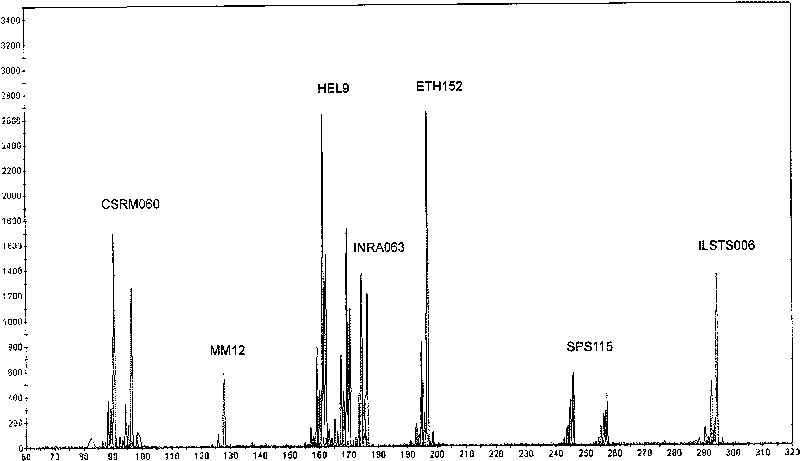
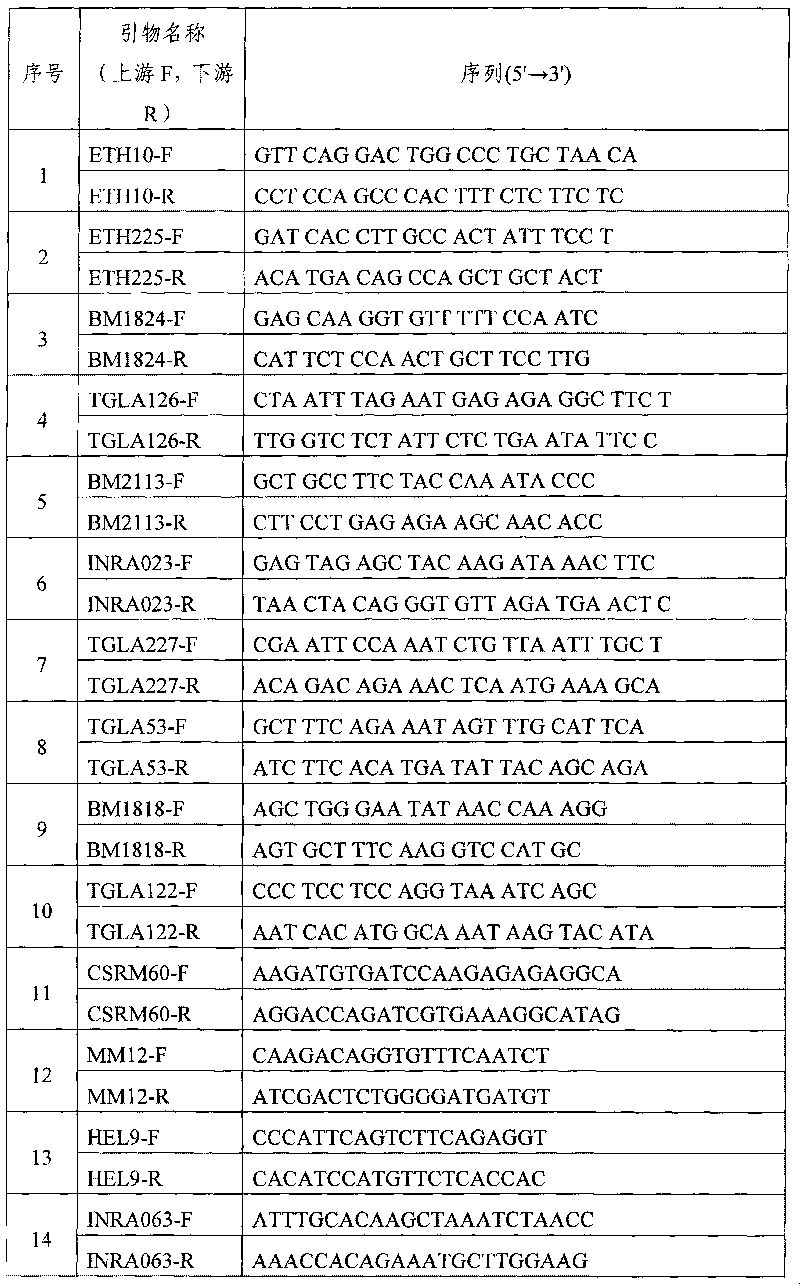
The invention discloses a microsatellite loci marker combination which comprises 17 microsatellite loci markers, and simultaneously provides an efficient detection method based on the 17 microsatellite loci markers; the markers have high polymorphism, are not linked with each other and have proper fragments interval, and are easy to be detected simultaneously. The invention is improved in a detection technology, optimizes an experimental system by utilizing a multi-marker amplification technology and a fluorescent semi-automatic microsatellite typing method, thus establishing high flux, accurate and effective 17 cattle microsatellite marker compound amplification and a gene typing method.
Owner:CHINA AGRI UNIV
Salmonella CRISPR (clustered regularlay interspaced short palindromic repeats) sequencing typing method
ActiveCN103981256ALower requirementMethod saves timeMicrobiological testing/measurementMicroorganism based processesFluorescenceTyping methods
The invention provides a salmonella CRISPR (clustered regularlay interspaced short palindromic repeats) sequencing typing method; according to the method, by DNA sequencing of a latest interval sequence between Salmonella CRISPR1 site and CRISPR2 site, CRISPR type of a strain to be detected can be determined, and the Serotype can be predicted. The method is simple, rapid, low-cost and high-resolution, is high in consistency in salmonella serotyping results, and low in requirements on laboratory equipment and software, a new CRISPR type comprises bacterial evolution and regional and other various information, and by combination of a fluorescence probe and other methods, real time and rapid detection can be realized, and the method has popularization and application value.
Owner:INST OF PLA FOR DISEASE CONTROL & PREVENTION
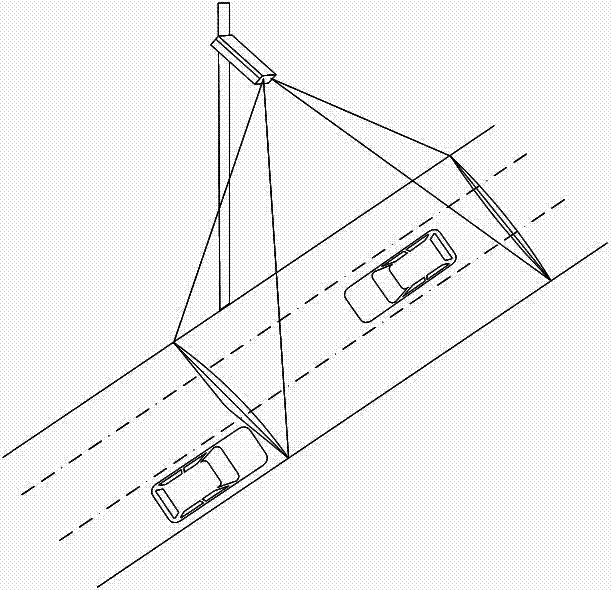
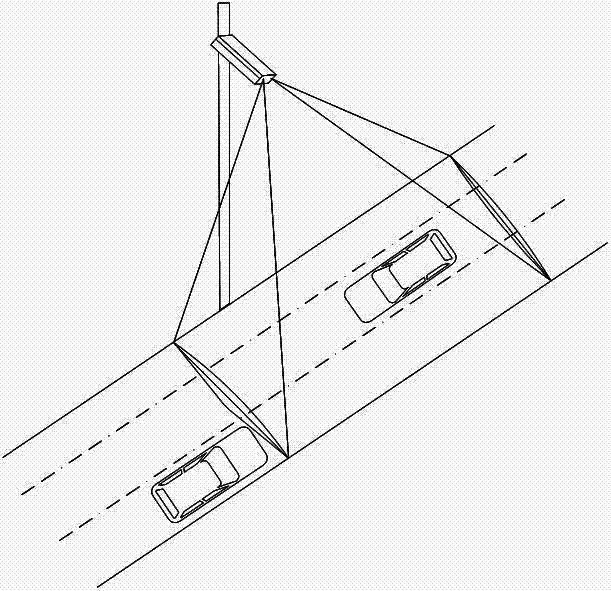
Vehicle Classification Method Based on Two-column Vertical Radar Waves
InactiveCN102289938ASolve the vehicle classification functionReduce the impact of detection accuracyRoad vehicles traffic controlRadio wave reradiation/reflectionVehicle detectorTyping methods
The invention discloses a vehicle classification method for double-row vertical radar waves. A microwave vehicle detector is installed on the side of the road. The microwave vehicle detector adopts an array type double microwave radar. The microwave vehicle detector is composed of a vector decomposition module and a frequency analysis module. and a microprocessor. The method of the present invention is a comprehensive identification and analysis of the echo characteristics, analyzing the characteristic value of the echo to obtain a group of typing results, analyzing the frequency of the echo, and then obtaining a group of typing results; finally, the The two classification results are weighted to obtain the final vehicle classification result. Because this product adopts the two-level small network classification method in the neural network, it can greatly improve the classification speed, solve the real-time problem of vehicle classification, and improve the accuracy and stability of classification.
Owner:LIAONING JINYANG GROUP INFORMATION TECH
Genome cytosine site epigenome typing method
ActiveCN106845152AMature technologyLow costMicrobiological testing/measurementSequence analysisCytosineTyping methods
The invention provides a genome cytosine site epigenome typing method which includes the steps: 1) performing bisulfite whole genome methylation sequencing on male and female parents of samples to be tested and offspring samples to obtain male and female parent and offspring sample genome sequences; 2) comparing the male and female parent and offspring sample genome sequences with a reference genome to obtain comparison results to determine a cytosine site to be tested; 3) performing chromosome coordinate sequencing and reads replication removal on the comparison results, performing Call SNPs on 5-10bp upstream and downstream sequences of a known cytosine site through GATK2-V3.2 and accordingly distinguishing offspring allelic gene sequences; 4) comparing the obtained distinguished offspring allelic gene sequences with the male and female parent genome sequences, and finishing cytosine epigenome typing. The method finishes genome cytosine site epigenome typing for the first time and is mature in technology, low in cost and easy to operate, popularize and apply.
Owner:BEIJING FORESTRY UNIVERSITY
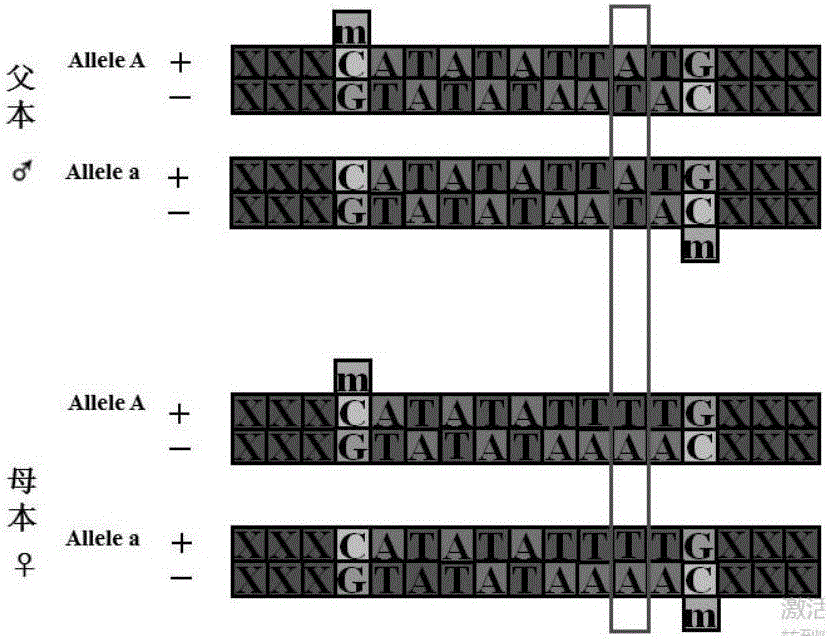
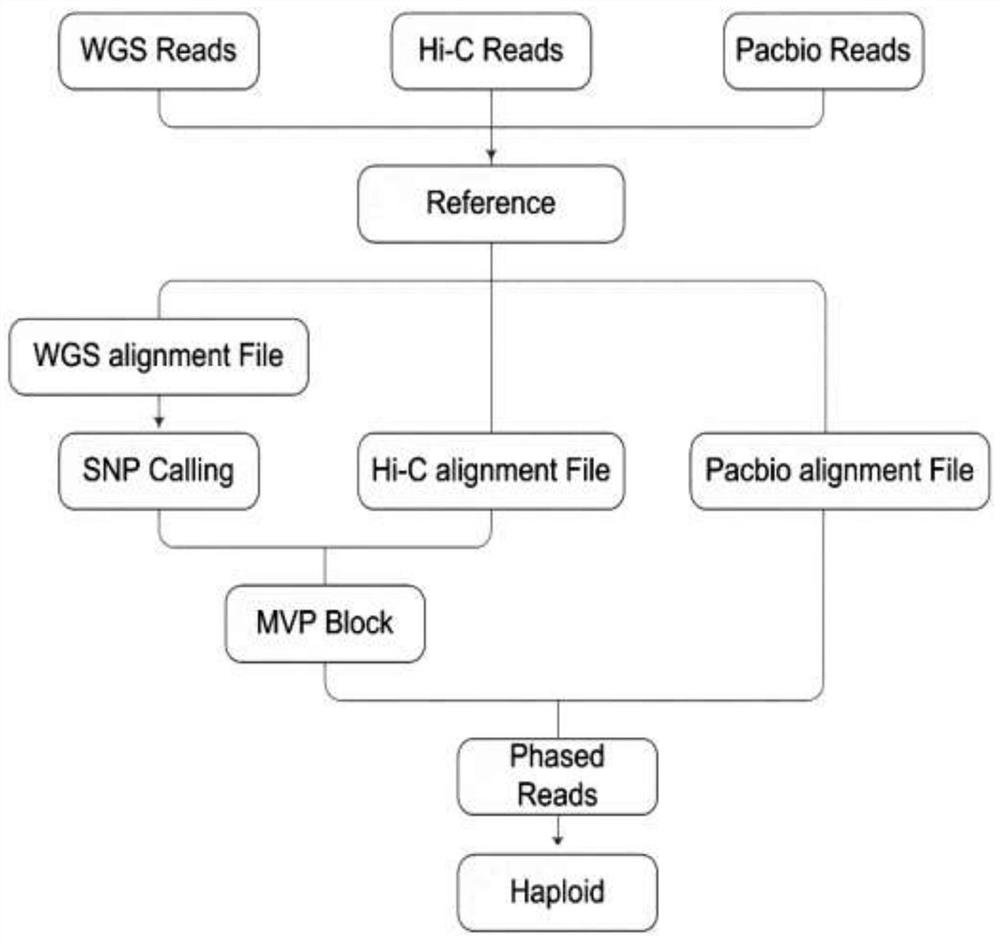
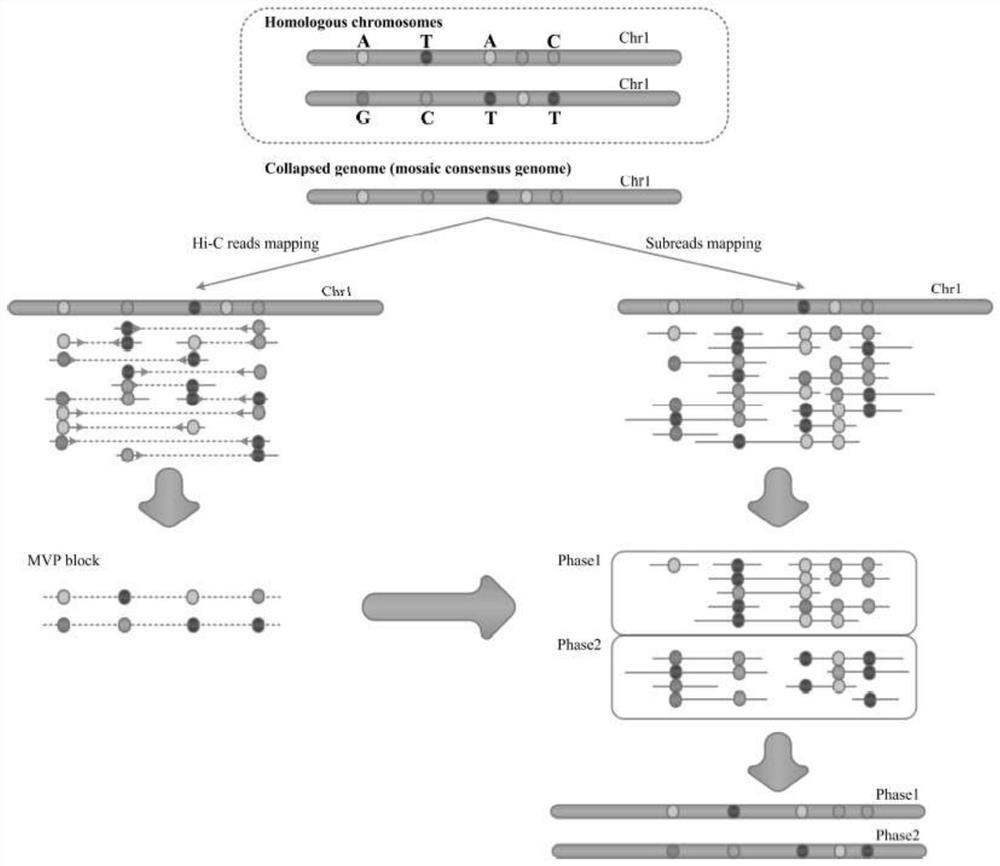
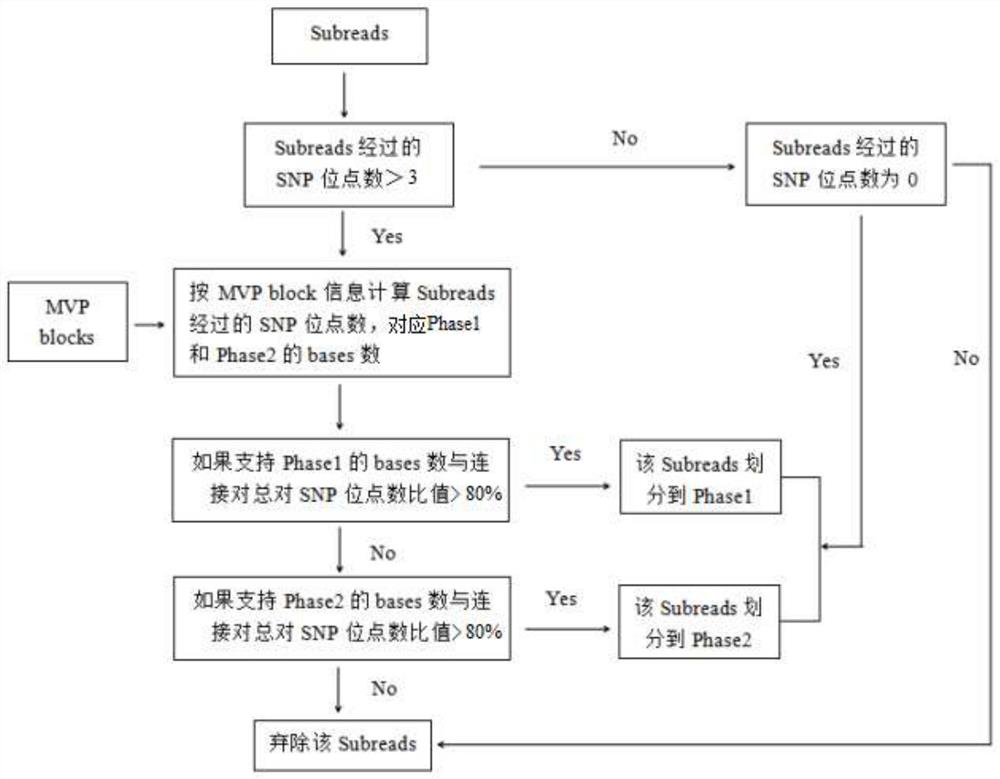
Whole genome typing method based on Pacio subreads and Hi-C reads
PendingCN111816248AGuaranteed accuracyReduced risk of errorProteomicsGenomicsRestriction Enzyme Cut SiteWhole genome sequencing
The invention relates to a whole genome typing method based on Pacbio subreads and Hi-C reads. The whole genome typing method comprises the following steps: 1) preparing a reference genome; 2) comparing second-generation sequencing data with the reference genome, and detecting all the SNP sites of each chromosome; 3) comparing Hi-C library building sequencing data with a reference genome, and building a linkage SNP group by adopting a combination of HapCUT2 with the SNP sites; 4) grouping the Pacbio subreads based on the MVP Block, and then respectively assembling the Pacbio subreads to finally obtain the sequence of each chromatid; and 5) performing whole genome sequencing on the parent genome, comparing a sequencing result to a chromatid sequence separated in the previous step, and dividing the chromatids into two groups according to a comparison result to correspond to male and female parent genomes. According to the method, the defect that contigs with too few restriction enzyme cutting sites cannot be assembled in the process of Hi-C data assembling is avoided, a linked SNP group is constructed from the aspect of whole genome, and then the Pacbio long reads are combined, so the error risk of typing is greatly reduced.
Owner:WUHAN FRASERGEN CO LTD
Typing method for human papilloma virus gene
InactiveCN101397590ALow costEasy to operateMicrobiological testing/measurementSpecial data processing applicationsDesign softwareTyping methods
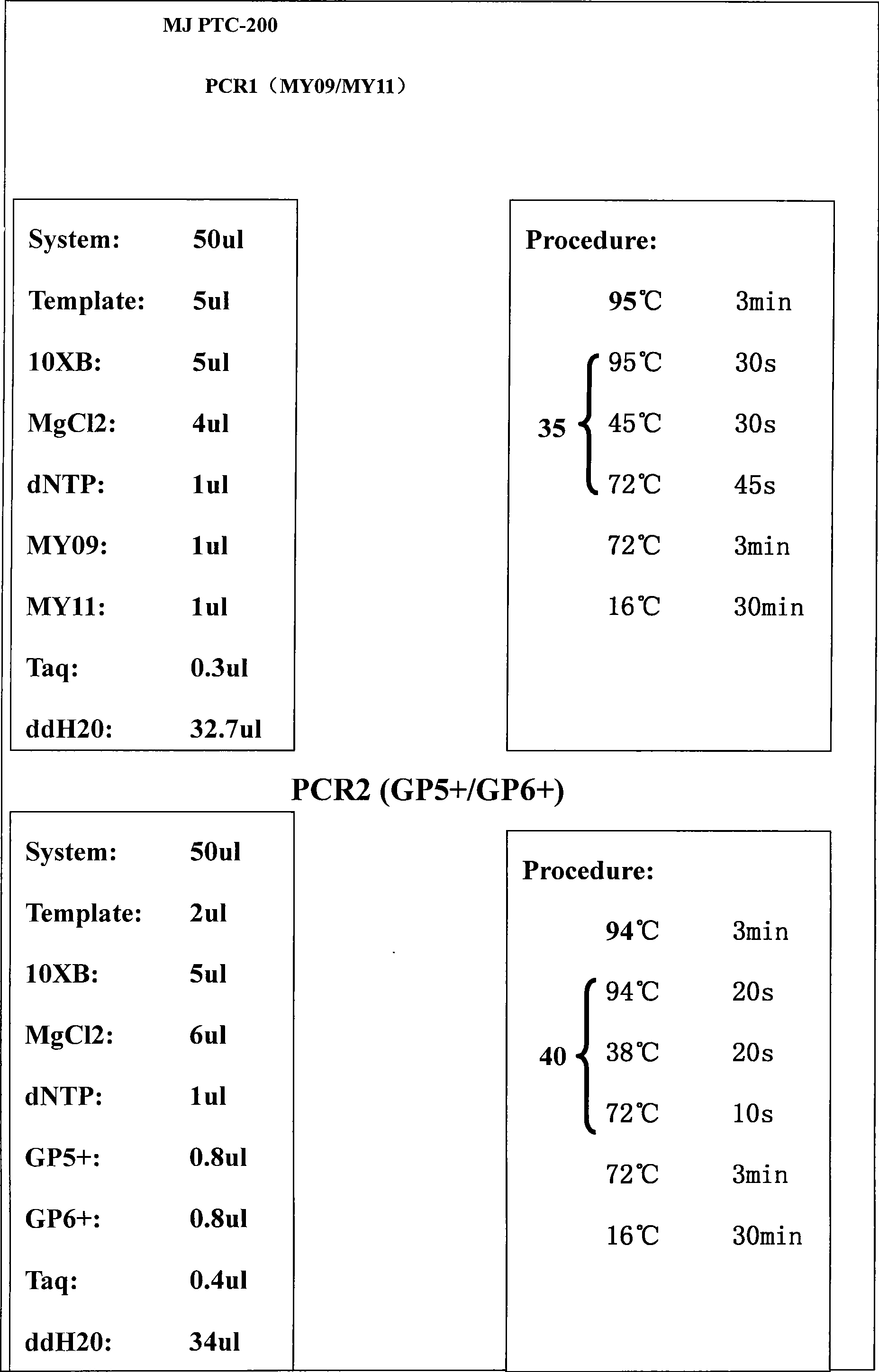
The invention discloses a genotyping method of human papilloma virus, which carries out permutation and combination on the sequences of 25bp behind GP5 of 40 common HPV subtypes by designed software, and the detailed steps are as follows: 1. the sequences of 25bp behind GP5 of a single type of the 40 common HPV subtypes; 2. mixed sequence mode of 25bp behind GP5 after arbitrary two types of the 40 common HPV subtypes are combined; 3. mixed sequence mode of 25bp behind GP5 after arbitrary three types of the 40 common HPV subtypes are combined; 4. mixed sequence mode of 25bp behind GP5 after arbitrary four types of the 40 common HPV subtypes are combined; 5. mixed sequence mode of 25bp behind GP5 after arbitrary five types of the 40 common HPV subtypes are combined; and 6. mixed sequence mode of 25bp behind GP5 after arbitrary six types of the 40 common HPV subtypes are combined. All the modes carry out pairing with the bases following GP5 by the ATCG sequence, and the pairing number is recorded, circulation is carried out in this way until the sequences of 25bp behind GP5 are all paired, then a pyrosequencing mode database infected by 1 to 6 types of different subtypes is created, the sample is carried out DNA extraction and MY09 / 11 amplification, the amplification product is taken as a template and then carried out amplification by GP5+ / 6+, then GP5 is taken as a sequencing primer, and the sequencing result is compared with the pyrosequencing mode database to determine the type. The genotyping method has the advantages of low cost, convenient operation, strong specificity and synchronously detecting a plurality of types of inflection.
Owner:HANGZHOU D A GENETIC ENG
Method for measuring pyrophosphoric acid and SNP typing method
InactiveUS20130189687A1Bioreactor/fermenter combinationsBiological substance pretreatmentsTyping methodsTonicity
One general aspect provides a method for detecting pyrophosphoric acid in a sample solution with high sensitivity and high accuracy by a small sensor element, and an SNP typing method. In the general aspect, a sample solution having a volume of more than that of a measurement cavity is supplied to the measurement cavity through a flow path, so as to expose a droplet from the opening. The droplet has a shape of sphere. The shape of the sphere is maintained by surface tension generated on a surface of the droplet. At least part of the sample solution contained in the droplet is evaporated so as to increase a concentration of pyrophosphoric acid in the sample solution included in the measurement cavity.
Owner:PANASONIC CORP
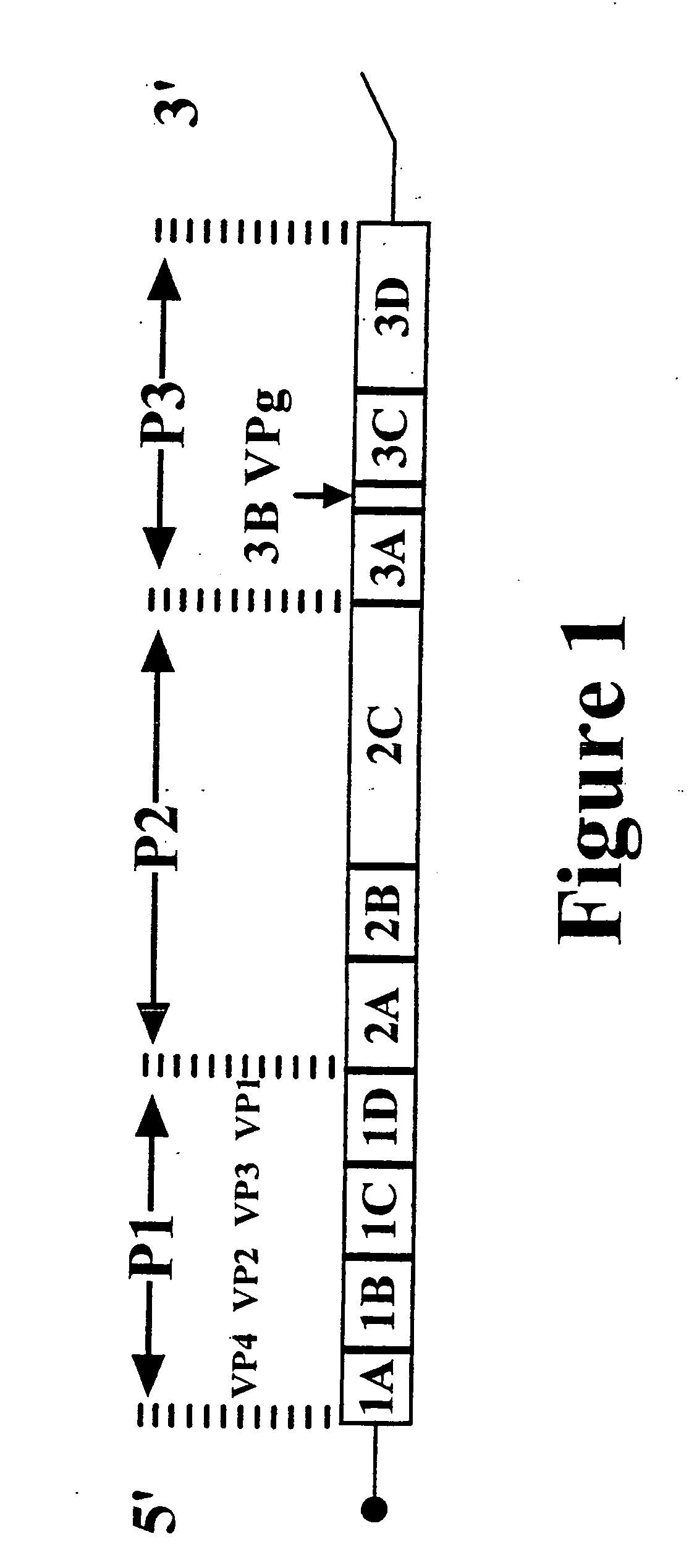
Typing of human enteroviruses
The present invention discloses a method for detecting the presence of an enterovirus in a clinical sample. The invention additionally discloses a method for typing an enterovirus in a clinical sample. Both methods employ a set of primer oligonucleotides for reverse transcription and amplification that hybridize to conserved regions of the enterovirus genome, and that provide amplicons that include significant portions of the VP1 region that are characteristic of the various serotypes. In the typing method, the invention further provides a database consisting of nucleotide sequences from prototypical enteroviral serotypes, which is used to type the clinical sample by comparing the sequence of its amplicon with each prototypical sequence in the database. The invention additionally provides mixtures of primer oligonucleotides, and a kit for use in conducting the typing method that includes a mixture of the primer oligonucleotides,
Owner:US DEPT OF HEALTH & HUMAN SERVICES
SNP typing method and kit
InactiveCN106834427AImprove accuracyReduce false signalsMicrobiological testing/measurementBinding siteTyping methods
The invention relates to the technical field of DNA detection and provides an SNP typing method and kit. A to-be-tested SNP site comprises SNP sites in CYP2C9, VKORC1 and CYP2C19 genes. The SNP typing method comprises the following steps: replacing a fluorescently-labeled oligonucleotide probe with a non-fluorescently-labeled oligonucleotide probe, performing one-time connection sequencing reaction on to-be-sequenced library molecules and then obtaining sequence information of the to-be-tested SNP site through connection sequencing. The SNP typing kit comprises a CYP2C9 specific primer pair, a VKORC1 specific primer pair, a CYP2C19 specific primer pair and the non-fluorescently-labeled oligonucleotide probe. The SNP typing method and kit provided by the invention have the benefit that through sealing non-specific binding sites in the to-be-sequenced library molecules, a false signal in a result obtained by the connection sequencing is reduced, so that the accuracy of SNP site detection is improved.
Owner:GUANGZHOU KANGXINRUI GENE HEALTH TECH CO LTD
HLA (human leucocyte antigen) typing method based on PacBio RS II sequencing platform
The invention discloses an HLA (human leucocyte antigen) typing method based on a PacBio RS II sequencing platform. A collected sample is subjected to DNA extraction and then to PCR (polymerase chain reaction) amplification, PCR products are mixed to establish a 10k library, and PacBio RS II sequencing is performed; then, original data obtained by sequencing are corrected, and HLA typing is performed with software programs. Compared with existing HLA typing methods, the HLA typing method has super-high resolution and is of important value to applications such as clinical graft tissue matching, population genetics, anthropology and evolutiology, as well as basic research work.
Owner:BEIJING GRANDOMICS BIOTECH
Constructing method of maternal plasma free DNA library and typing method of parental alleles
ActiveCN107119046AIncrease success rateGuaranteed coverageMicrobiological testing/measurementLibrary creationLibrary preparationGenomic DNA
The invention discloses a constructing method of a maternal plasma free DNA library and a typing method of parental alleles. Firstly, a group of specific primers for amplifying SNP in maternal plasma free DNA is designed, and comprises 720 pairs of primers, and the sequences of upstream and downstream primers are shown as SEQ ID NO:1-1440. Then, peripheral blood of pregnant women of 9-20 weeks is acquired for the extraction of the plasma free DNA, and a multiple composite PCR amplification technology is applied to the construction of the free DNA library; after the library is purified and quantified, computer sequencing is performed on an Ion TorrentTM platform. The same method is used for performing library preparation and sequencing on corresponding maternal cells and fetal tissue genomic DNA. According to sequencing results, the maternal cells are selected as homozygous SNP sites, the percentage concentrations of non-parental alleles of the free DNA in maternal plasma at the corresponding sites are analyzed, and the purpose of determining a paternal allele in the free DNA is achieved.
Owner:SUN YAT SEN UNIV
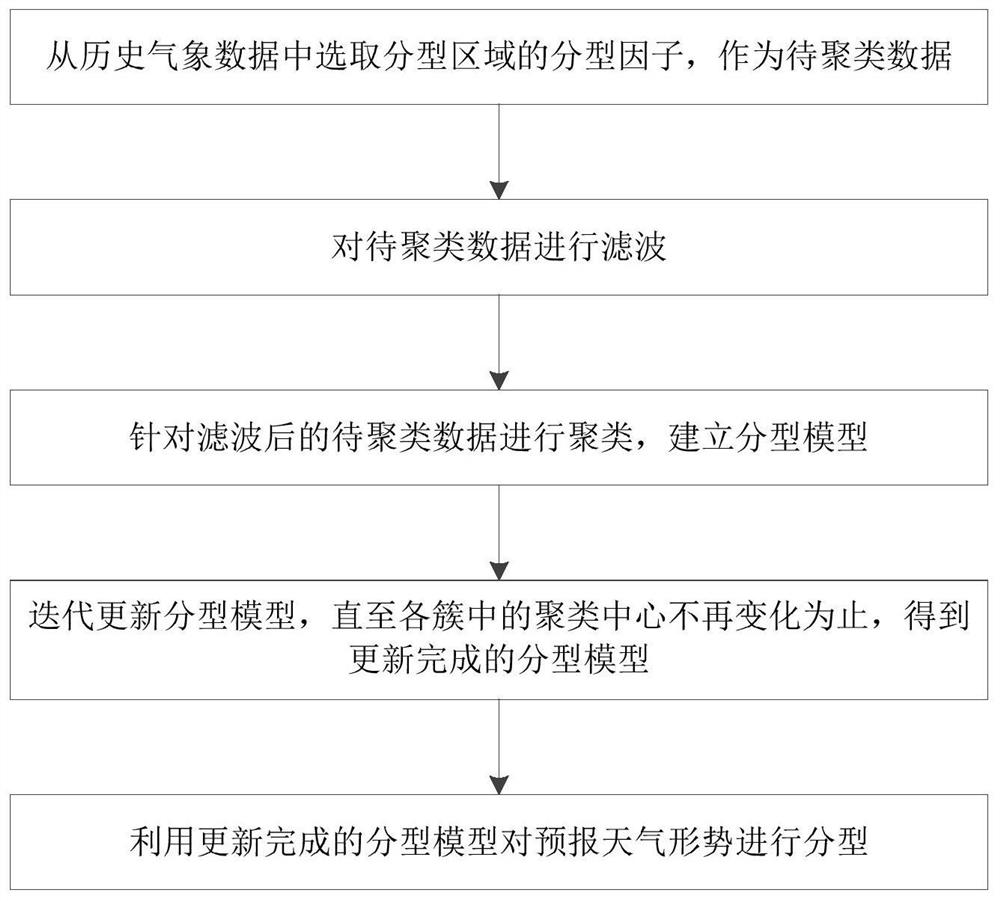
Weather situation typing method and air pollution condition prediction method and device
ActiveCN111612055AImprove practicalityReduce clustering errorForecastingCharacter and pattern recognitionClustered dataTyping methods
The invention discloses a weather situation typing method and an air pollution condition prediction method and device. The weather situation typing method comprises the steps of selecting a typing factor of a typing region from historical meteorological data as to-be-clustered data; filtering the to-be-clustered data; clustering the to-be-clustered filtered data and establishing a parting model; iteratively updating the parting model; and parting the forecast weather situation by using the parting model. According to the weather situation typing method provided by the invention, the initial cluster center is determined in a more reasonable manner, so that clustering errors can be effectively reduced, the similarity between classes is increased, and the difference between different classesis obvious; the similarity measurement index is improved, the similarity of each row and each column is calculated, the local difference condition is fully considered, the meteorological field distribution characteristics are fitted, the parting result practicability is enhanced, and the parting result is accurate.
Owner:北京中科三清环境技术有限公司 +1
Multiplex amplification system, and next generation sequencing typing kit and typing method for 21 micro-haplotype sites
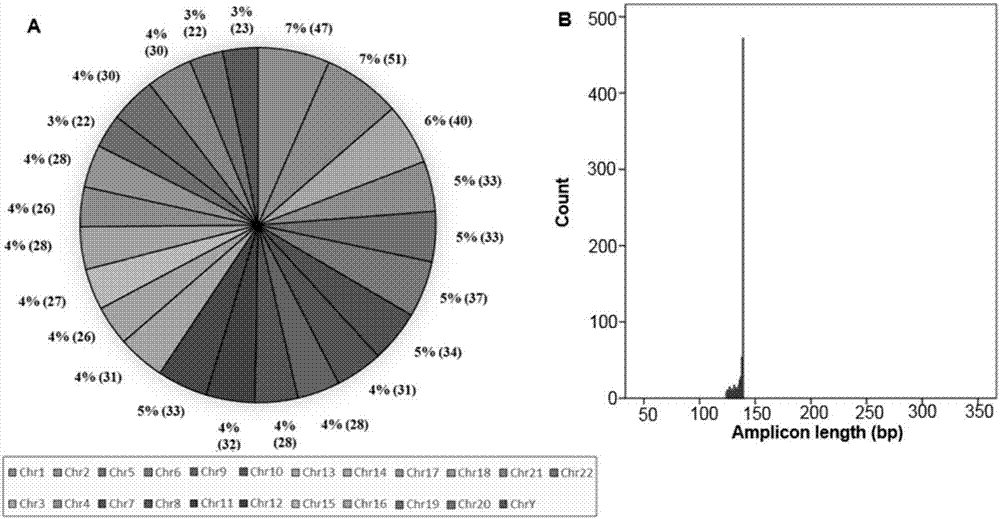
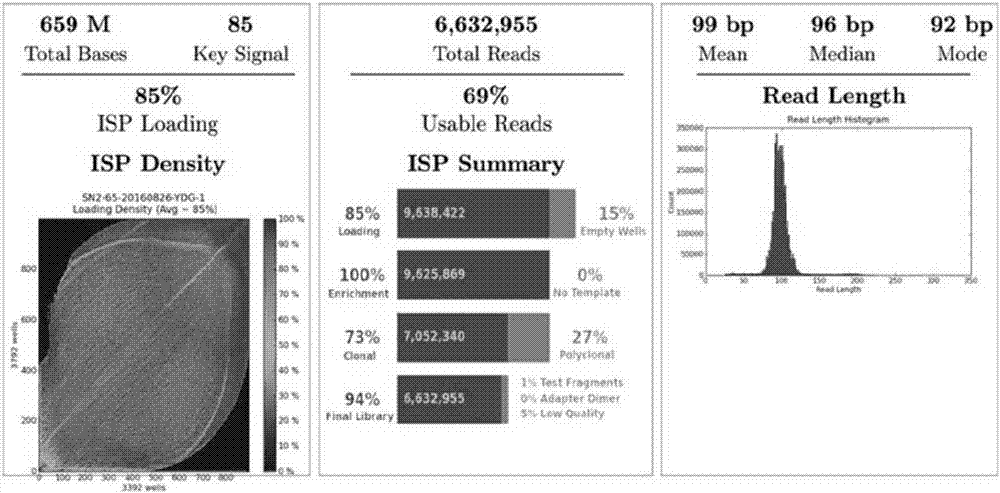
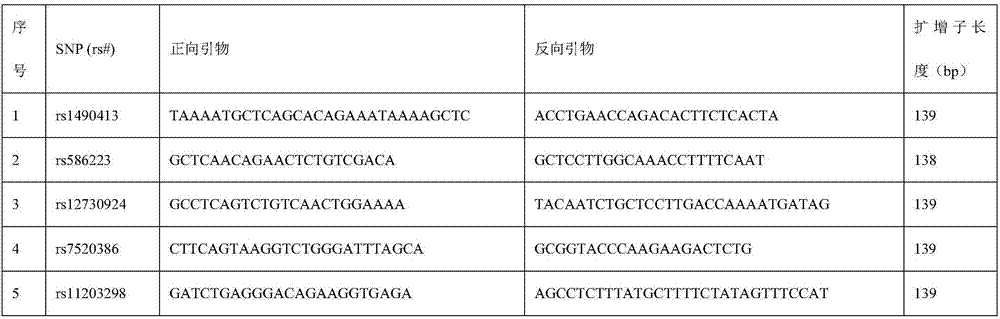
ActiveCN110218781AShorten the lengthLow mutation rateMicrobiological testing/measurementSequence analysisStr typingAllele frequency
The invention relates to the technical field of molecular biology, and especially relates to a multiplex amplification system, and a next generation sequencing typing kit and typing method for 21 micro-haplotype sites. The 21 micro-haplotype sites in the multiplex amplification system are derived from 21 autosomes, are balanced in allele frequency, are mutually independent between the sites, and have the advantages of polymorphism and low mutation rate. The kit contains 53 PCR-amplified single-end specific primers shown in SEQ ID NO. 1-SEQ ID NO.53, and can detect the 21 microhaplotype sites in a same reaction system. The multiplex amplification system, the kit and the typing method provided by the invention can solve the technical problem that a current STR typing technology cannot obtainan ideal typing result, and allele loss may occur due to excessive number of SNP sites.
Owner:HEBEI MEDICAL UNIVERSITY
New 10 Y-chromosome short tandem repeat locus parting method therefor
InactiveCN101225386AOvercome the defect of not being able to reflect the genetic characteristics of China's multi-ethnic groupsSimple instrumentMicrobiological testing/measurementFermentationTyping methodsForensic science
The invention discloses a new kind of ten STR gene loci for Y chromosome and the typing method, which is characterized in that polyacrylamide gel is utilized for electrophoresis typing; the ten STR gene loci for Y chromosome, which can be applied in forensic medicine field, are screened out with silver staining coloration method; furthermore, the allelic ladder of each locus is prepared; a PCR primer and the expansion condition of the Y chromosome STR locus are optimized; wherein the optimization to the PCR primer and the expansion condition and the preparation of allelic ladder can be standardized and simplified, which is suitable for popularization of base unit. The ten STR gene loci for Y chromosome can be applied for individual identification, paternity testing and gene diagnosis in forensic medicine, anthropology, genetics, disease and other field. The new kind of ten STR gene loci for Y chromosome and the typing method has the advantages of wide application prospect, which is particularly suitable for paternity testing in the condition of patrilateral loss such as death or missing in forensic medicine practice and individual identification of mixed stain in rape and gang-rape cases, in particular to the identification to suspect having azoospermia or oligospermia disease.
Owner:XI AN JIAOTONG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com