CRISPR (clustered regularly interspaced short palindromic repeat) based DNA detecting and typing method and application thereof
A typing method, DNA ligase technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., to achieve the effect of avoiding key bottleneck problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
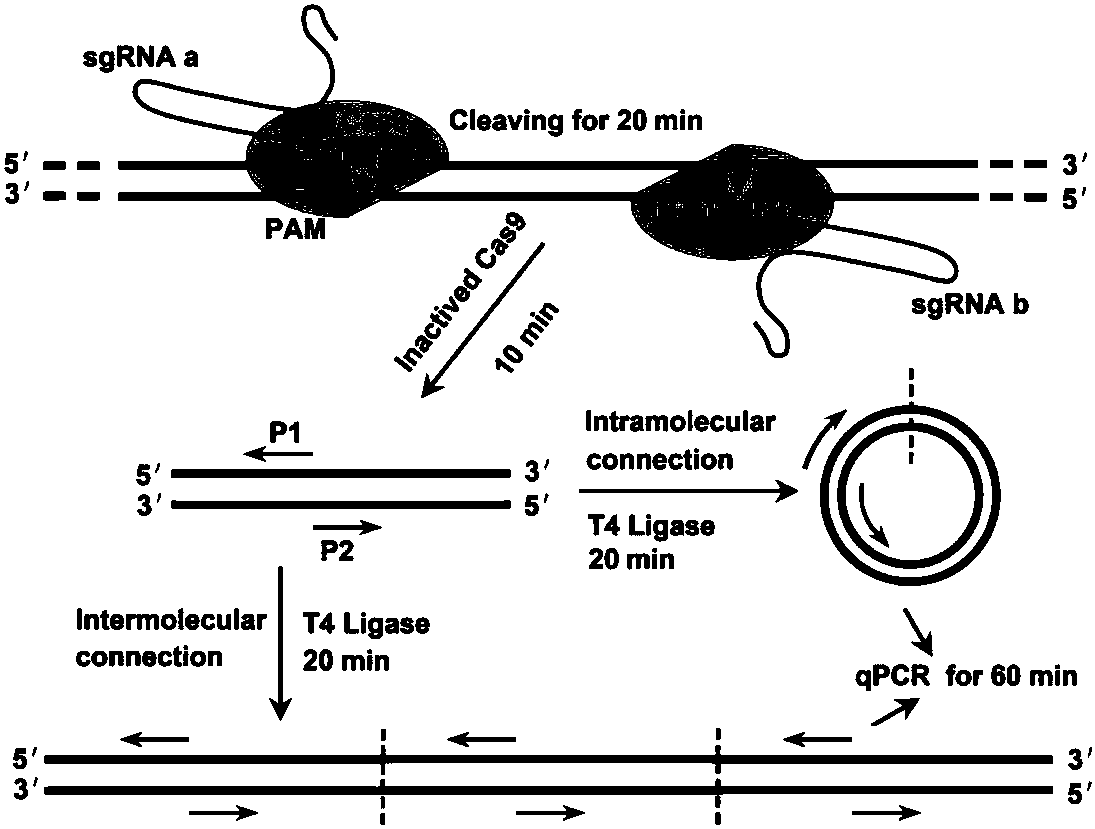
[0039] The schematic diagram of the principle and flow chart of CARP detection and typing of DNA molecules is as follows: figure 1 shown. CARP detection consists of three steps: (1) Cas9 endonuclease cleaves the target DNA. A pair of sgRNAs (sgRNA a and b) cleave the target DNA after binding to Cas9 nuclease; (2) the cleaved DNA is ligated by T4 DNA ligase (intermolecular and intramolecular ligation); (3) the ligated DNA is ligated by PCR Amplify. In the present invention, CARP is used to detect L1 and E6 / E7 genes of HPV16 and HPV18 to demonstrate the feasibility of CARP detection technology. In the PCR amplification step, conventional PCR (tPCR) and quantitative PCR (qPCR) were used, respectively.
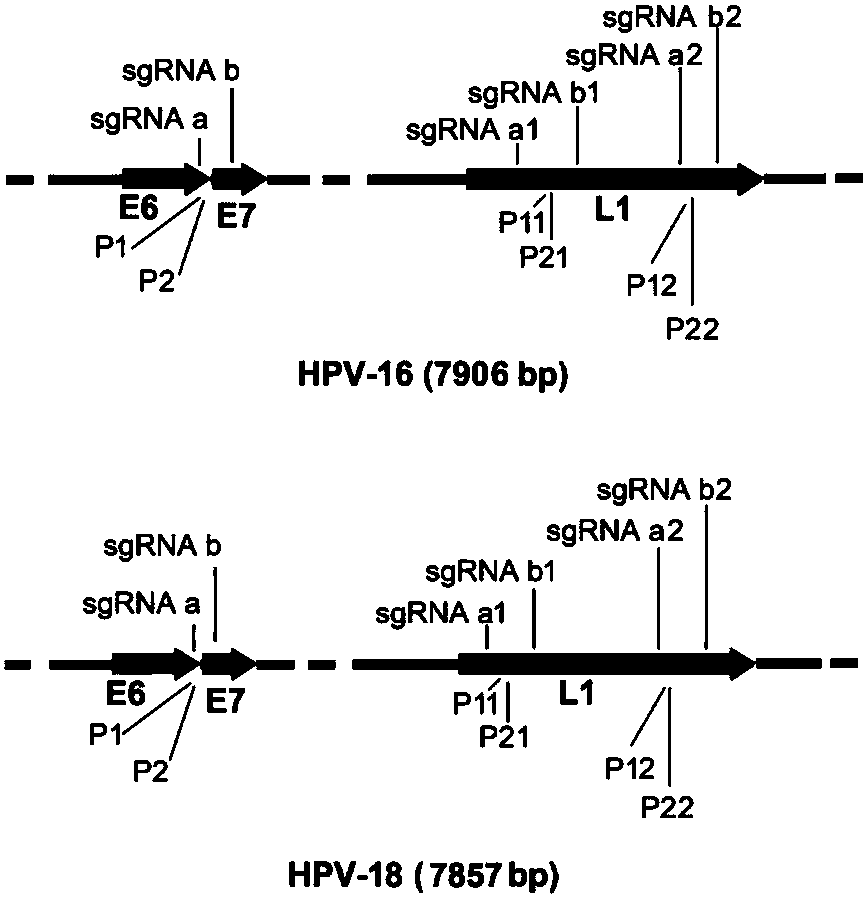
[0040] The L1 and E6 / E7 genes of the target DNA HPV16 and HPV18 detected in each embodiment of the present invention, and the positions of reverse PCR primers and sgRNA designed for these genes are as follows: figure 2 shown. The indication figure 2 It is helpful to unders...
Embodiment 2
[0041] Example 2 Cutting HPV 16 and HPV 18 L1 genes with Cas9 / sgRNA
[0042] experimental method:
[0043]Preparation of sgRNA: According to the instructions of T7 polymerase (New England Biolabs), sgRNA was synthesized by in vitro transcription with T7 polymerase. The DNA template of the sgRNA was amplified by PCR three times using the oligonucleotides listed in Table 1 (SEQ ID NO.1-27). The first PCR was performed with F1 and R (7 cycles). Use the product of the first PCR as a template, and use F2 and sgR as primers for the second PCR (30 cycles); use the product of the second PCR as a template, and use F3 and sgR as primers for the third PCR (30 cycles). cycle). The purified product of the third PCR was used as a template for in vitro transcription. The purified sgRNA template was then incubated overnight at 37°C with T7 RNA polymerase (New England Biolabs) for in vitro transcription. The in vitro transcribed RNA was mixed with Trizol solution, extracted sequentially w...
Embodiment 3
[0053] Embodiment 3 detects HPV16 and 18 L1 gene with CARP
[0054] experimental method:
[0055] The preparation of sgRNA and HPV L1 gene fragment was the same as that in Example 1.
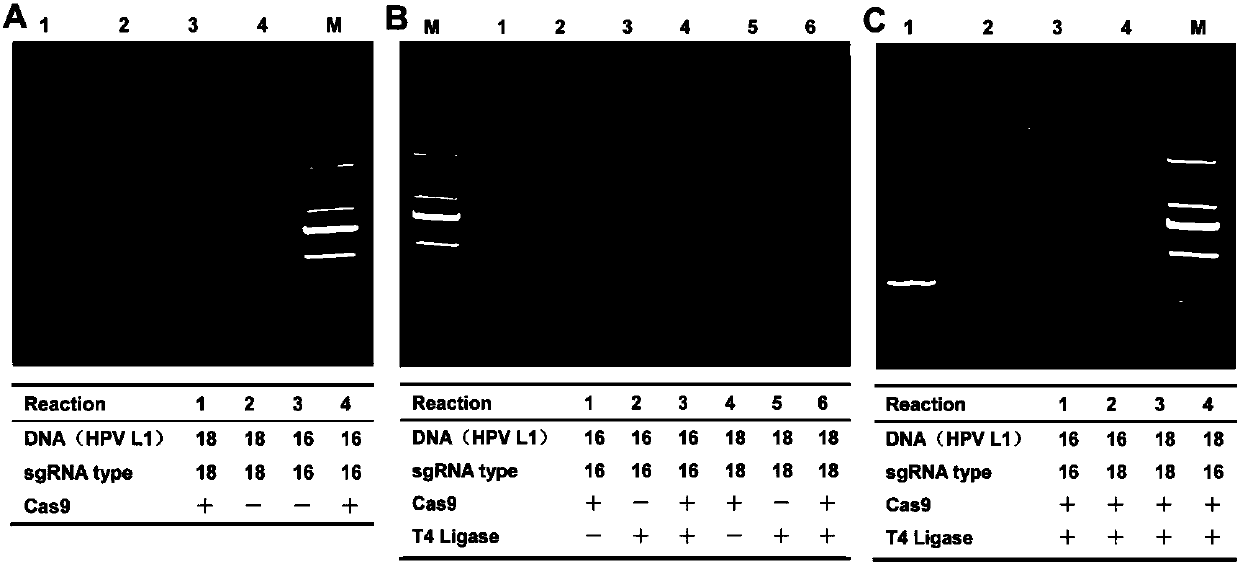
[0056] Cas9 / sgRNA cleavage of HPV16and 18 L1 gene: Recombinant Cas9 protein was purchased from New England Biolabs (NEB). Cas9 cleavage reaction (30 μL): 1× Cas9 nuclease reaction buffer, 1 μM Cas9 nuclease (NEB), 300 nM sgRNA a (16L1a1 or 18L1a1; Table 1) and 300 nM sgRNA b (16L1b1 or 18L1b1; Table 1). First, the Cas9 reaction solution was incubated at 25°C for 10 minutes. Then 200ng of HPV L1 gene fragment (substrate DNA) was added to the Cas9 reaction solution, and incubated at 37°C for 20 minutes. Finally, Cas9 was inactivated at 65 °C for 10 min. Electrophoresis was performed on 2% agarose gel.
[0057] Ligation reaction system (15 μL): 1×T4 ligase buffer, 5 U T4 DNA ligase (Thermo) and 5 μL Cas9 digested products of HPV16 and HPV18 L1 genes (as described above). Incubate at 22°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com