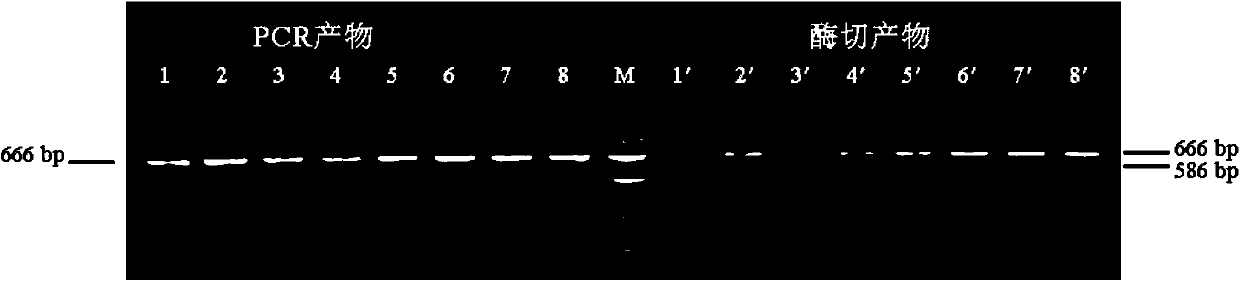
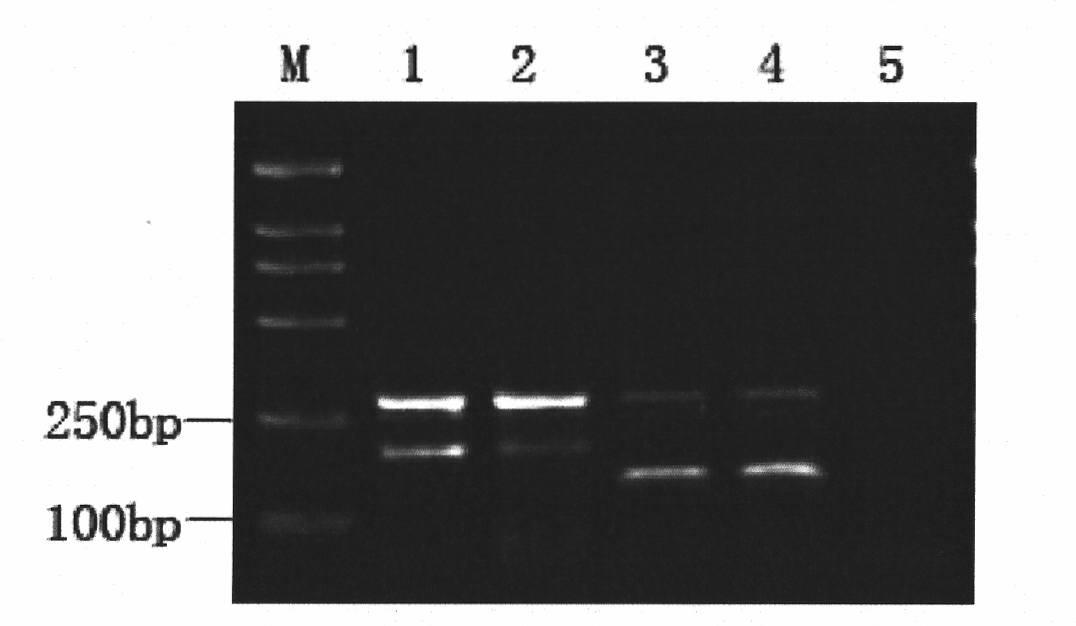
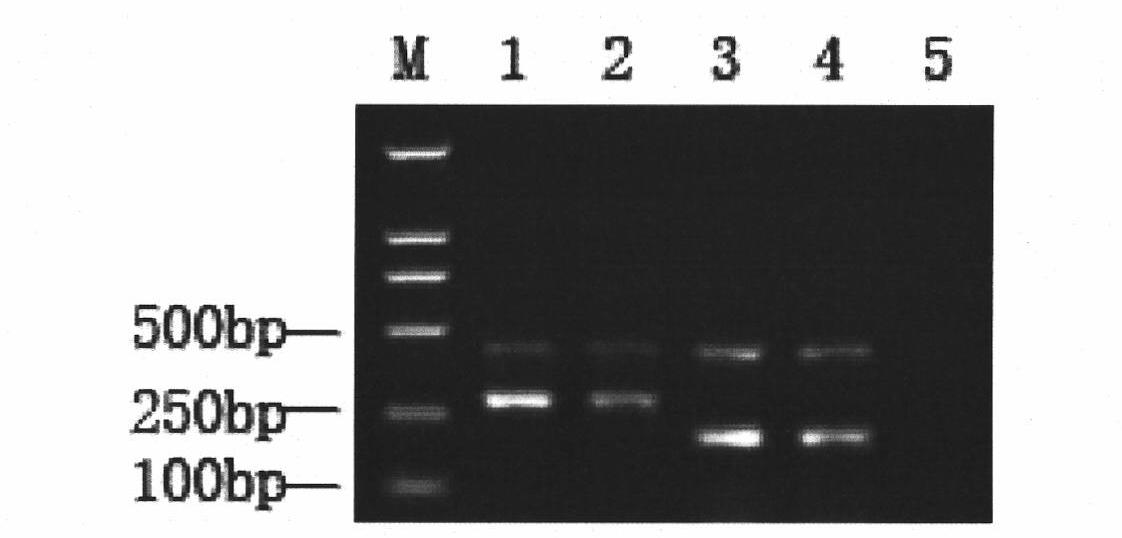
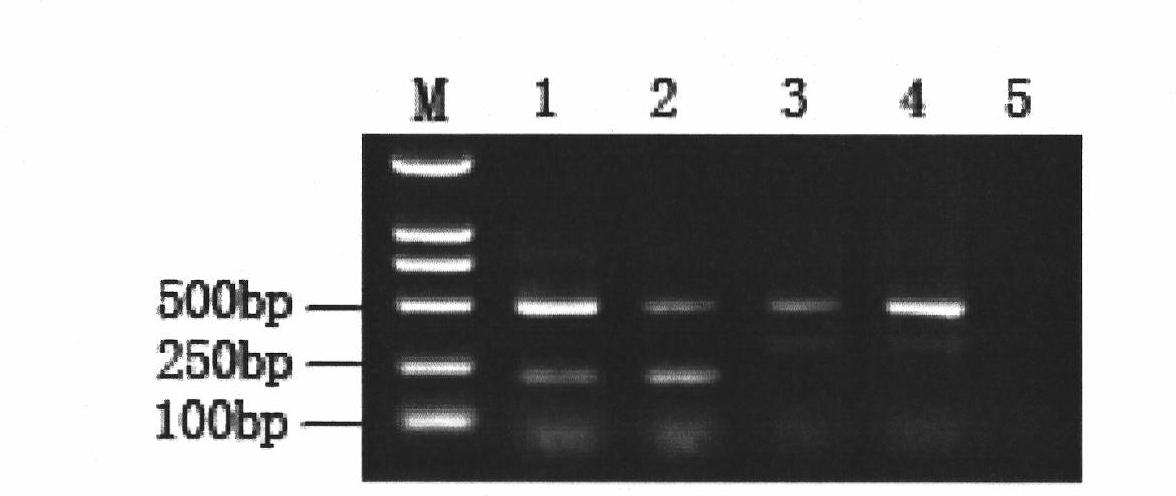
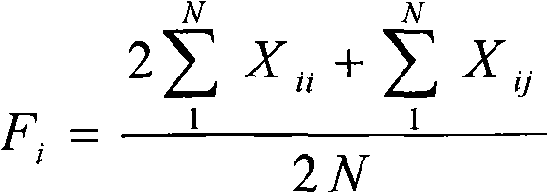Patents
Literature
180 results about "Electrophoretogram" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
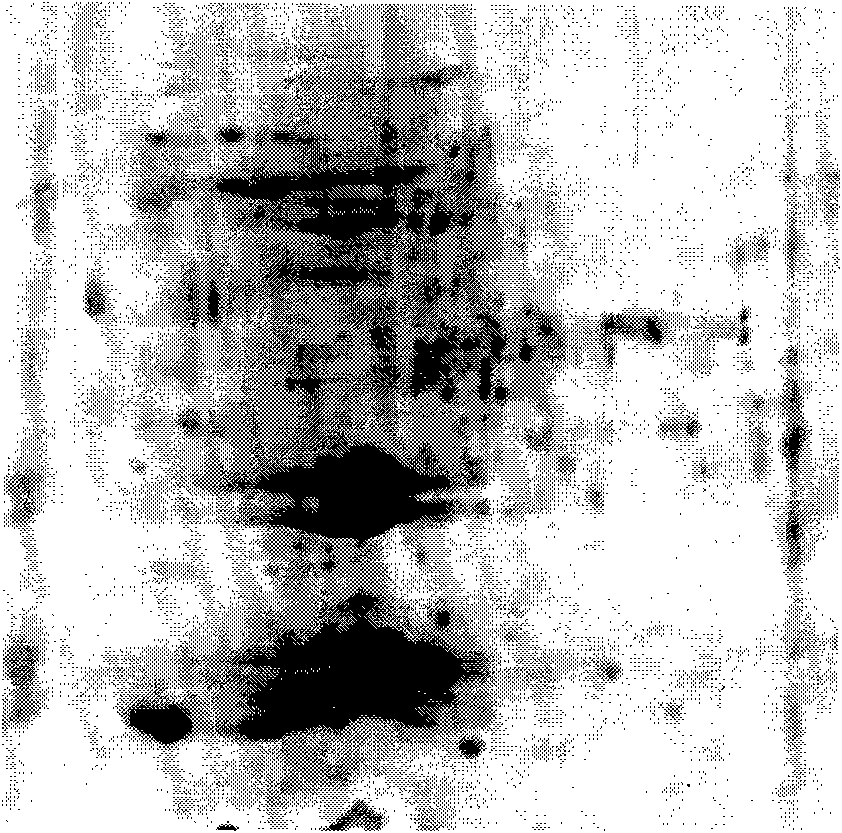
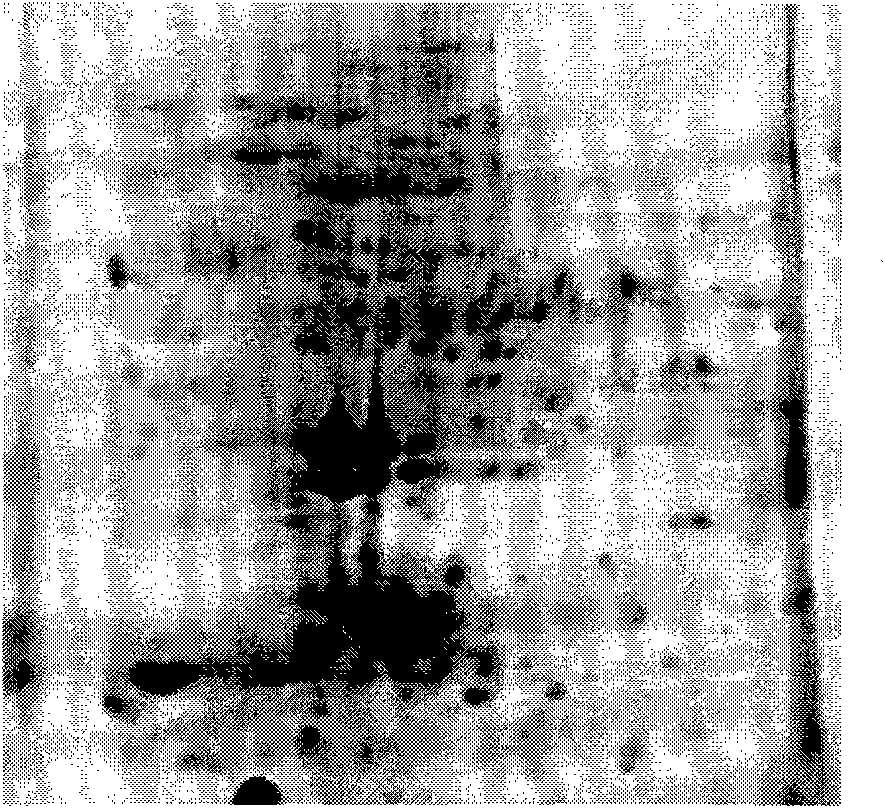
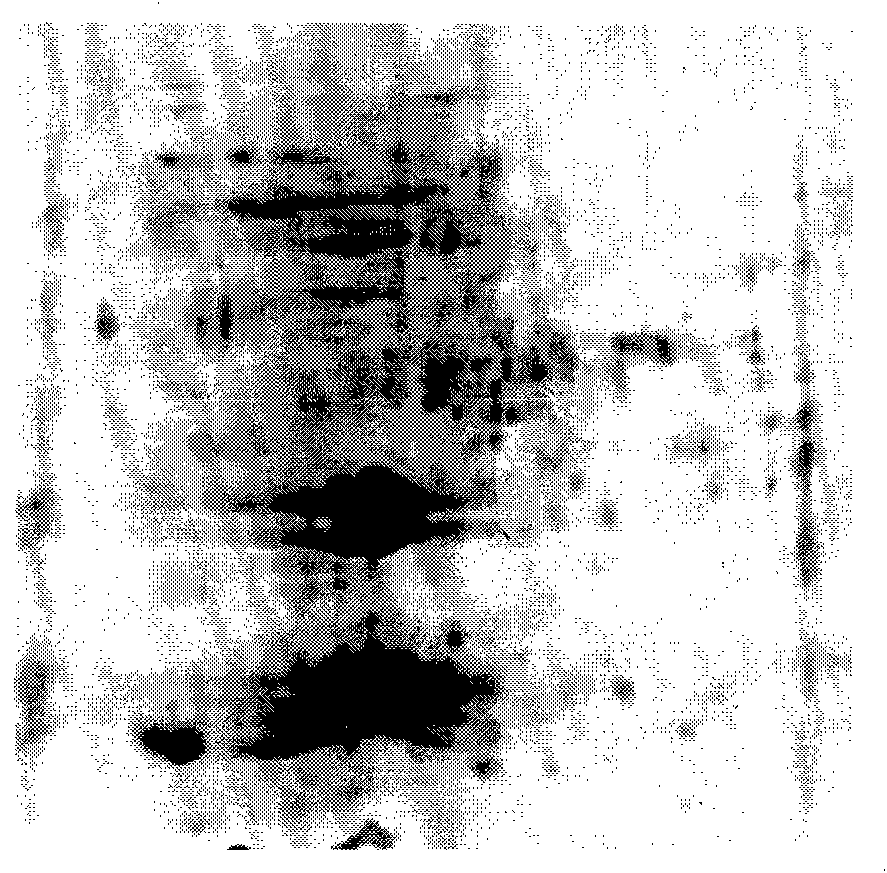
An electrophoretogram is the result of an electrophoresis, which gives the movement of charged particles over time in a gel, paper or another medium.
Detection method for Y chromosome micro-deleted gene
InactiveCN101575647AMeet the experimental conditionsSimple adjustment of primer concentrationMicrobiological testing/measurementMultiplex pcrsY-Chromosome Genes
The invention discloses a detection method for Y chromosome micro-deleted gene. In the method, after human peripheral blood genome DNA is extracted, multiple PCR primer design is used, wherein a multiple PCR reaction system comprises N pairs of chimeric primers for specifically amplifying the STSs site in an AZF segment of a Y chromosome and one pair of universal primers. N+1 pairs of primers are participated in the multiple PCR reaction, and the reactions of DNA template such as denaturalization, anneal and extending in circulating amplification target are carried out in a same reaction system; 10ulPCR products are taken and dye by EB, and electrophoresis is carried out through 3% agarose. The electrophoresis voltage is 4 V / cm, and the electrophoresis time is 20 minutes; and then the electrophoretogram is observed under an ultraviolet transilluminator so that the result is judged. Therefore, a detection method with simplified PCR reaction conditions, high amplification efficiency and easy operation for clinic experiments is established.
Owner:成都军区昆明总医院
Method for identifying indica rice and japonica rice by inserting or deleting molecular marker in rice DNA
InactiveCN101323878ASimple methodImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationDNA fragmentationAllele
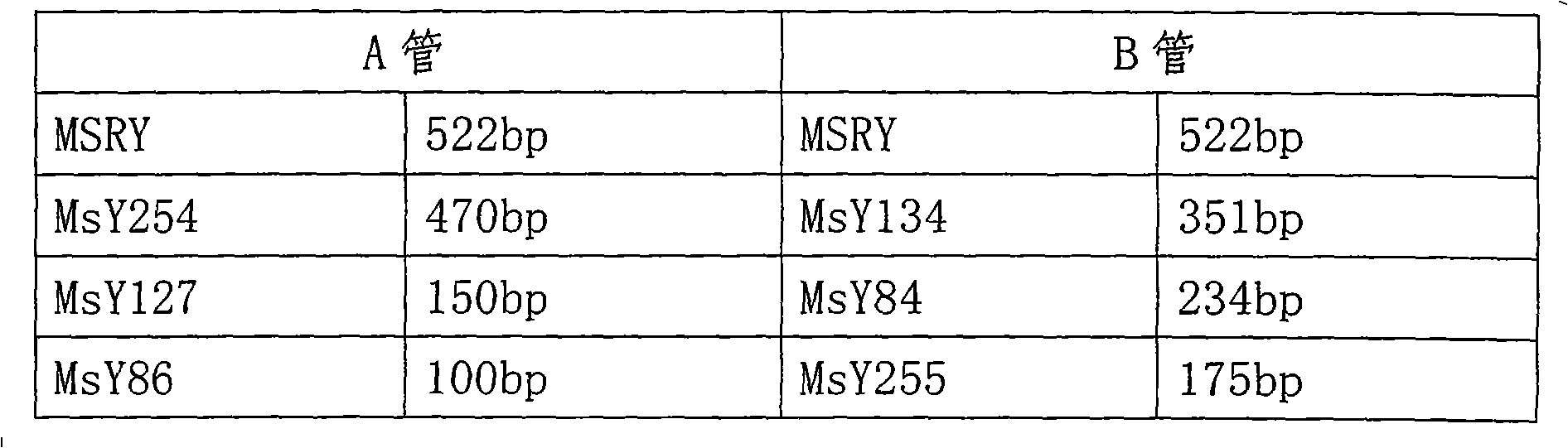
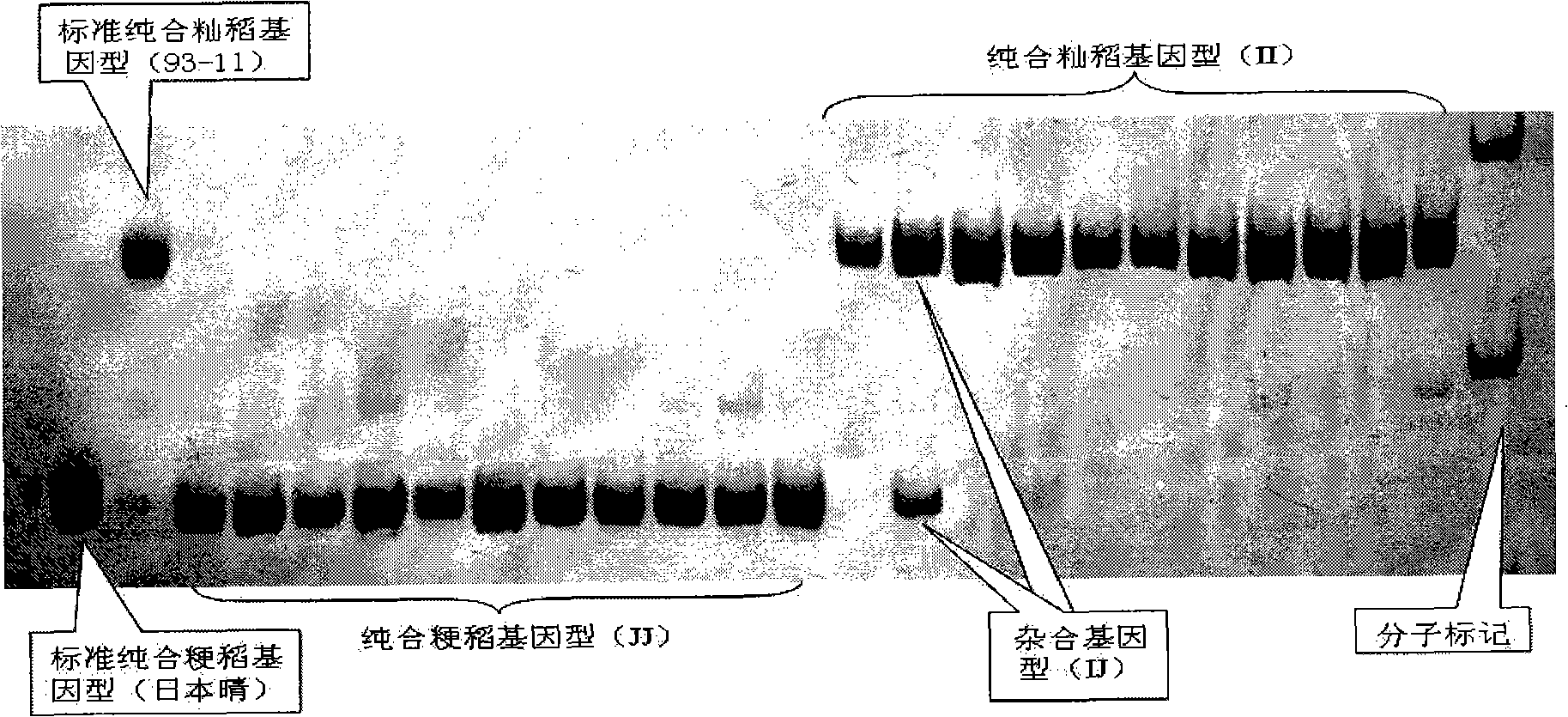
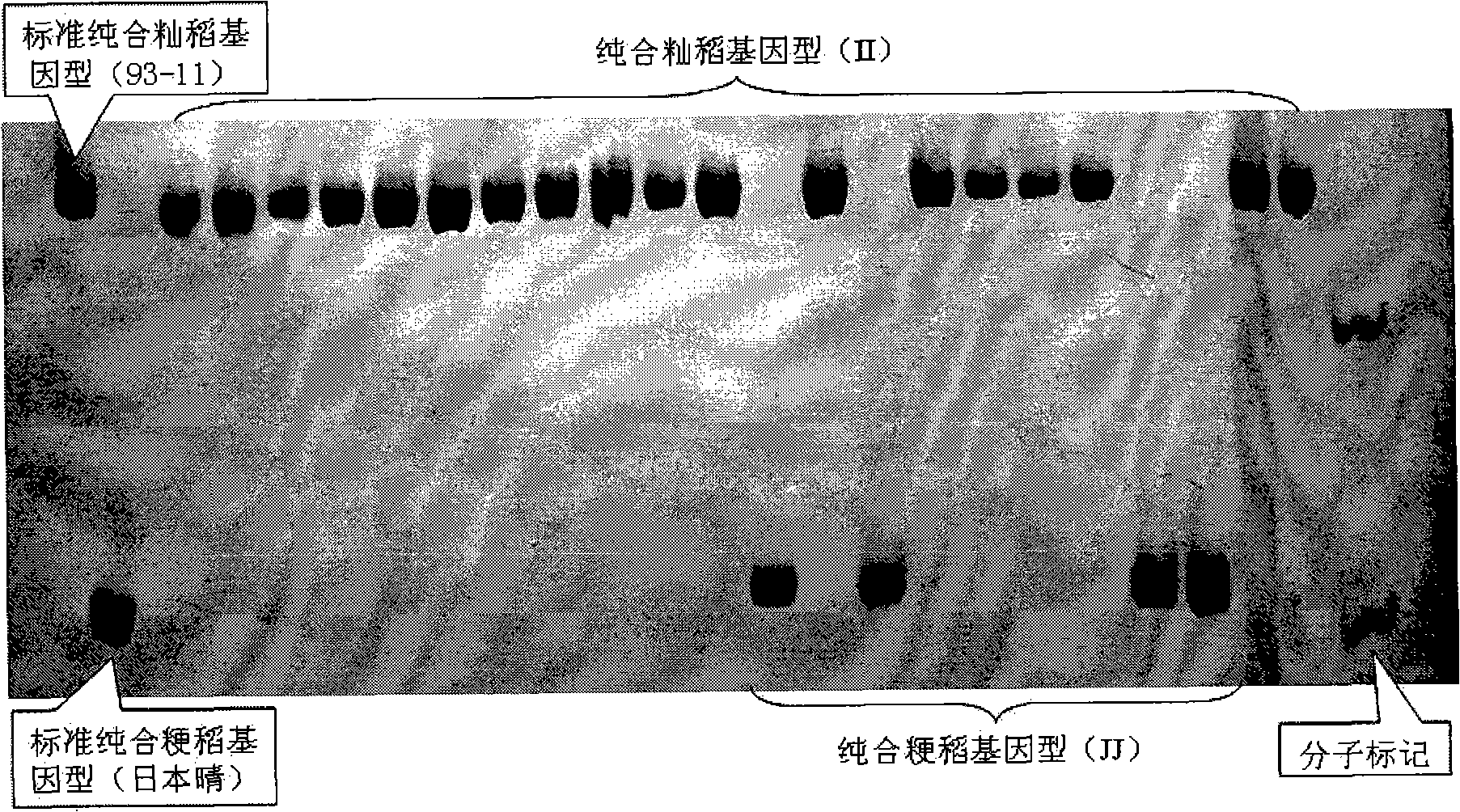
The invention pertains to the technical field of biological discrimination, in particular to a method for discriminating indica rice from japonica rice by utilizing molecular markers of rice DNA insertion or deletion. The method obtains 34 pairs of differential DNA primers designed by InDel differential fragment by making a comparison of whole genome sequence between indica rice cultivar 93-11 and japonica rice cultivar (Nipponbare). The DNA extraction, the amplification and ionophortic separation of DNA fragments and the analysis and calculation of an electrophoretogram of a target rice cultivar are carried out to discriminate indica rice from japonica rice. Precisely speaking, the 34 pairs of InDel primer groups are utilized and analysis and calculation are carried out according to a finger-print obtained on the basis of a polymerase chain reaction and vertical slab gel electrophoresis; the characteristics of indica rice or japonica rice of rice samples detected are worked out according to the average frequency (Fi or Fj) showing on indica rice alleles and japonica rice alleles on 34 InDel locuses. The invention has the advantages of simple, fast and convenient method, few samples for detection and correct discrimination results, thus having good prospect of promotion and application.
Owner:FUDAN UNIV
Wild ginseng protein extracting method suitable for dielectrophoresis
InactiveCN101906130AImprove solubilityReduce Denaturing PrecipitationPeptide preparation methodsElectrophoresisCentrifugation
The invention discloses a wild ginseng protein extracting method suitable for dielectrophoresis. The method comprises the following steps of: taking wild ginseng as an experimental material, fully grinding the wild ginseng in liquid nitrogen and transferring the obtained product to a centrifuge tube; adding acetone solution precooled at the temperature of -20 DEG C into the centrifuge tube, washing sample powder for 3 times, collecting sediments by centrifugation and drying the obtained product at room temperature for 20min to obtain the crude extract of the wild ginseng protein; dissolving the crude extract of the wild ginseng protein into lysis buffer, and performing ultrasonic cell disintegration and extracting the supernate by centrifugation to obtain the high-quality solution of the wild ginseng protein. The wild ginseng protein extracting method for the dielectrophoresis is simple and fast, can obtain an electrophoresis pattern having a high repeatability and resolution, is favorable for subsequent protein identification by mass spectrographic analysis and provides a technical support for the proteomic research of the wild ginseng.
Owner:BEIHUA UNIV
PCR-RFLP rapid detection method for common sturgeons
InactiveCN103436610AAccurate identificationImprove accuracyMicrobiological testing/measurement3-deoxyriboseSturgeon
The invention relates to the field of molecular biology and taxonomy and discloses a PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) rapid detection method for common sturgeons as well as a primer and a kit for the rapid detection method. The PCR-RFLP rapid detection method for the common sturgeons comprises the following steps: firstly, extracting a genome DNA (Deoxyribose Nucleic Acid) of a sample to be detected as a template; secondly, carrying out PCR amplification reaction to obtain a PCR product by taking the genome DNA as the template; thirdly, digesting the PCR product by using restriction endonuclease to obtain a digested product; fourthly, carrying out agarose gel electrophoresis detection on the PCR product and the digested product and identifying species according to electrophoretogram. According to the PCR-RFLP rapid detection method disclosed by the invention, the simplicity in operation is realized; a small amount of fin rays or muscle is needed to be sheared on the basis of ensuring the survival of fingerling; the identification of fingerlings of the common sturgeons is quickly and accurately carried out; the accuracy and the reliability of the identification result are improved; the working efficiency is greatly increased.
Owner:徐鹏
Genotyping detection method of drug resistance to Sanmate of Fusarium graminearum
The invention belongs to a Sanmate resistance genotype molecular detection and SNP typing method of Fusarium graminearum, which can be used for drug resistance monitoring and drug resistance genotype diagnosis to Sanmate and other benzimidazole type bactericides of the Fusarium graminearum which can cause wheat scab. The detection method comprises the following three major steps: (1) extracting DNA of a nuclear genome of a strain to be detected; (2) applying three groups of primer pairs to carry out PCR reaction; and (3) identifying the genotype of the strain with the resistance to the Sanmate according to a PCR product electrophoretogram. By adopting the Tetra-primer ARMS PCR (Tetra-primer amplification refractorymutation system-PCR) technology, the strain with the drug resistance of theFusarium graminearum and the genotype thereof can be fast and accurately detected, and the level of the drug resistance can be further judged. The detection accuracy is above 95%.
Owner:NANJING AGRICULTURAL UNIVERSITY
Mycoplasma bovis immunity-related protein P22, nucleotide sequence for coding same and application thereof
The invention discloses a mycoplasma bovis immunity-related protein P22, a nucleotide sequence for coding the same and an application thereof. A mycoplasma bovis Hubei strain is taken as a sample, and an efficient two-dimensional electrophoresis system of a mycoplasma bovis whole protein is established, so that a two-dimensional electrophoretogram with high resolution and high repeatability is obtained; an immunity-related protein is screened in combination with Western blot, and is named P22 protein; and as proved by a prokaryotic expression result of the protein, a Western blot rest result of a recombinant protein and a mycoplasma bovis positive serum is negative, and a Dot blot test result is positive. The protein is proved to be possibly a conformation-dependent immunity-related protein of mycoplasma bovis, and the mycoplasma bovis immunity-related protein P22 plays a guidance role in diagnosing, preventing and treating a mycoplasma bovis disease.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
Specific primer used for rapid detection of kiwi fruit soft rot pathogenic bacterium Botryosphaeria dothidea, and detection method of Botryosphaeria dothidea
InactiveCN103509856ASimple processThe detection method is reasonableMicrobiological testing/measurementMicroorganism based processesActinidiaKiwi fruit
The invention discloses a specific primer used for rapid detection of a kiwi fruit soft rot pathogenic bacterium, and a detection method of the kiwi fruit soft rot pathogenic bacteriaum. A problem that existing technology is incapable of realizing accurate and rapid detection of the kiwi fruit soft rot pathogenic bacterium is solved. The specific primer comprises BZY-1 and BZY-2, the sequence of BZY-1 is 5'-CCATCAAACTCCAGTCAGTAAACG-3', and the sequence of BZY-2 is 5'-CTGCGCTCCGAAGCGAGATGTATG-3. The detection method of the kiwi fruit soft rot pathogenic bacterium comprises following steps: (1) DNA of samples are extracted; (2) the sample DNA is taken as a template, and primer BZY-1, primer BZY-2, 2*TaqPCRMasterMix and sterile ultrapure water are added for uniform mixing, and then PCR amplification is performed; and (3) an amplification product is subjected to agarose gel electrophoresis analysis, and the existence of the kiwi fruit soft rot pathogenic bacterium is determined based on the existence of amplification bands. Specificity of the primer is extremely high, only the kiwi fruit soft rot pathogenic bacterium band can be obtained by amplification, so that it is just needed to extract genome DNA from kiwi fruit tissue to be detected for PCR amplification, and the existence of the kiwi fruit soft rot pathogenic bacterium is determined based on the existence of amplification bands in electrophoretograms.
Owner:SICHUAN AGRI UNIV
PCR-RFLP rapid detection method for common sturgeons
InactiveCN103436611AAccurate identificationImprove accuracyMicrobiological testing/measurement3-deoxyriboseSturgeon
The invention relates to the field of molecular biology and taxonomy and discloses a PCR-RFLP (Polymerase Chain Reaction-Restriction Fragment Length Polymorphism) rapid detection method for common sturgeons as well as a primer and a kit for the rapid detection method. The PCR-RFLP rapid detection method for the common sturgeons comprises the following steps: firstly, extracting a genome DNA (Deoxyribose Nucleic Acid) of a sample to be detected as a template; secondly, carrying out PCR amplification reaction to obtain a PCR product by taking the genome DNA as the template; thirdly, digesting the PCR product by using restriction endonuclease to obtain a digested product; fourthly, carrying out agarose gel electrophoresis detection on the PCR product and the digested product and identifying species according to electrophoretogram. According to the PCR-RFLP rapid detection method disclosed by the invention, the simplicity in operation is realized; small amount of fin rays or muscle is needed to be sheared on the basis of ensuring the survival of fingerlings; the identification of fingerlings of the common sturgeons is quickly and accurately carried out; the accuracy and the reliability of the identification result are improved; the working efficiency is greatly increased.
Owner:徐鹏
Method and kit for quality appraisal for amplification products after single cell genome amplification
InactiveCN104762405AReasonable designGuaranteed success rateMicrobiological testing/measurementA-DNAQuality appraisal
The invention relates to a method and a kit for quality appraisal for amplification products after single cell genome amplification. The method comprises the following steps: separating and splitting single cells to obtain the whole-genome DNA of the single cells; carrying out single cell whole-genome amplification on the whole-genome DNA to obtain whole-genome amplification products; placing specific primers in conserved segments on 5 pairs of genomes in an amplification system, carrying out PCR amplification by taking the whole-genome amplification products as a template, carrying out electrophoresis detection on 5 amplification products, judging the quality of the whole-genome amplification products according to a detection result, and showing a result that 3-5 amplification products of uniformly distributed strips on an electrophoretogram are amplification product samples meeting sequencing requirements; constructing a DNA sequencing library on the amplification products meeting the sequencing requirements; carrying out high-throughput sequencing on the DNA sequencing library.
Owner:PEKING JABREHOO MED TECH CO LTD +1
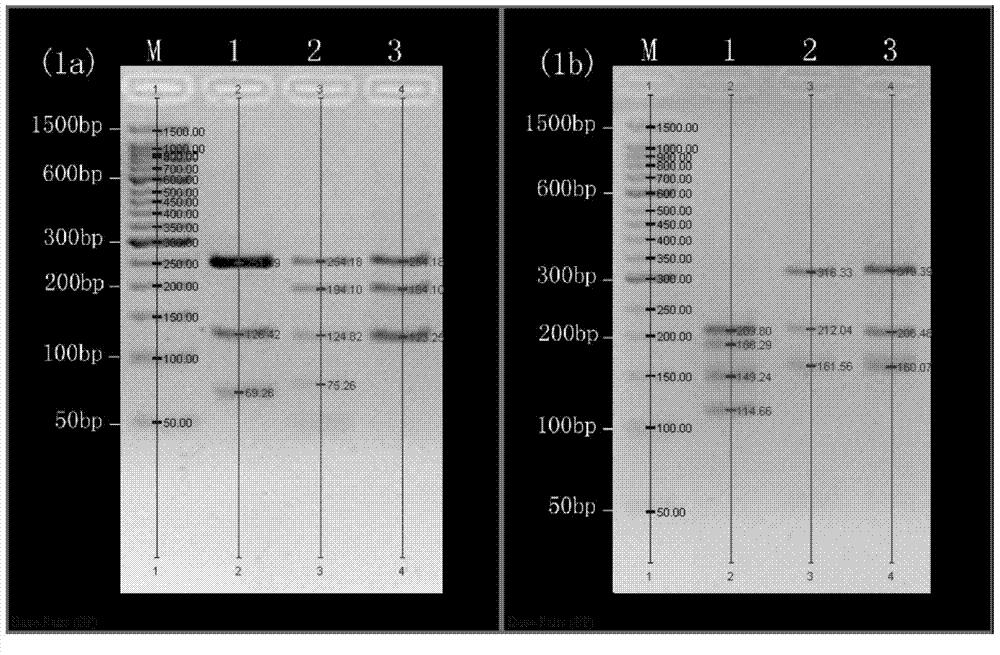
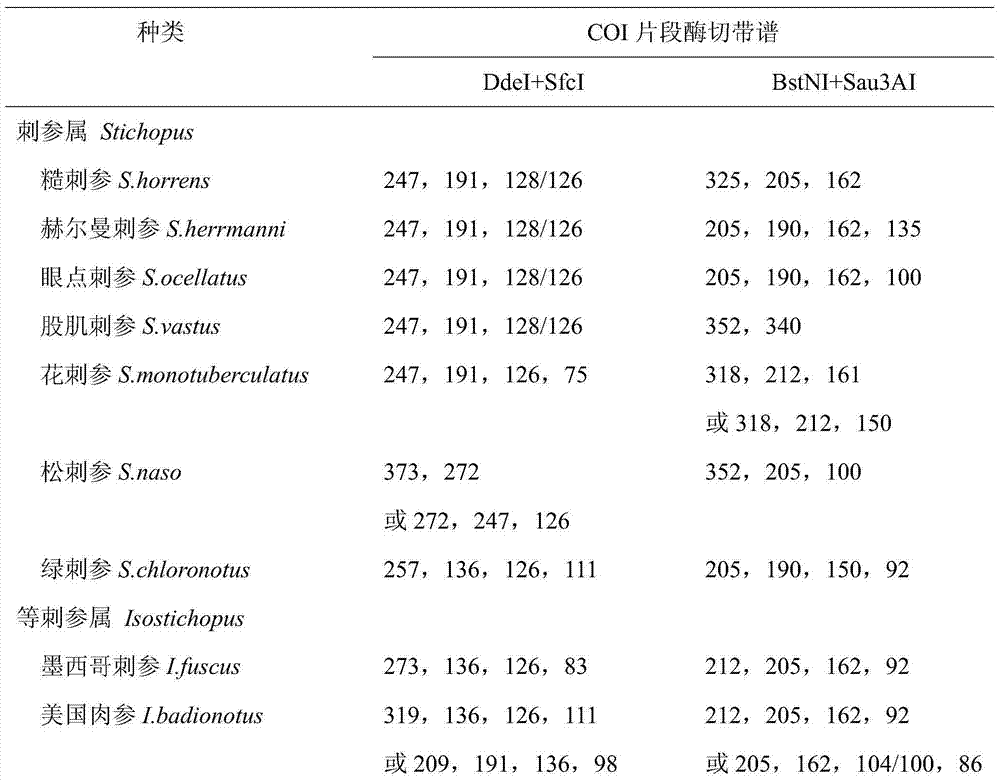
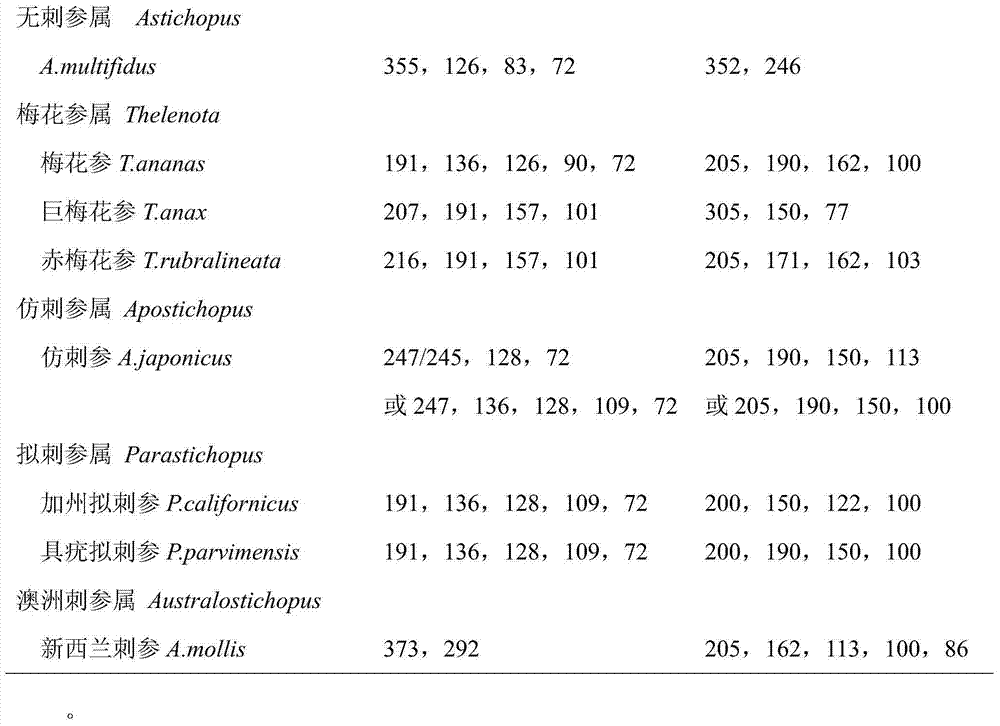
COI gene based PCR-RFLP discrimination method of seventeen economic sea cucumbers of Stichopodidae
ActiveCN103923972ALower requirementEasy to operateMicrobiological testing/measurementEnzyme digestionApplicability domain
The invention discloses a COI gene based PCR-RFLP discrimination method of seventeen economic sea cucumbers of Stichopodidae. The method comprises the following steps: 1, extracting DNA of a Stichopodidae sea cucumber to be detected as a detected sample, using the DNA as a template, and using primers COIe-F:5'-ATAATGATAGGAGGRTTTGG-3', COIe-R:5-GCTCGTGTRTCTACRTCCAT-3'PCR' to amplify a CO I fragment, wherein A is A / G; and 2, carrying out enzyme digestion on the CO I fragment by using a restrictive endonuclease group DdeI / SfcI or BstNI / Sau3AI, carrying out electrophoresis on the above obtained enzyme digestion product, estimating the size of each of the electrophoretic bands of samples to be detected, contrasting the number and the size of the electrophoretic bands in an electrophoretogram and the estimated value of the size with the reference band spectra of the seventeen sea cucumbers, and finding the standard spectral bands coupled with the number and the size of the electrophoretic bands, wherein the types represented by the standard spectral bands are the type of the detected sample. The method has the advantages of wide application range, simple operation, and accurate and fast detection, is especially suitable for the identification of a large batch of commodity sea cucumbers, so the method is a scientific method for identifying the sea cucumbers of Stichopodidae, and provides technologic supports for the standardization of the commodity market of the sea cucumbers and the protection of the sea cucumber resources.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Method for extracting protein from cotton leaves
InactiveCN103665121AGood repeatabilityHigh purityPeptide preparation methodsPlant peptidesWater bathsTrichloroacetic acid
The invention provides a method for extracting protein from cotton leaves. The method comprises the following steps of 1) grinding the cotton leaves; 2) transferring the ground leaves powder to a centrifugal pipe, adding trichloroacetic acid extraction liquid, uniformly mixing, and storing the mixture in a refrigerator with the temperature of -20 DEG C for more than 2h; 3) centrifuging in the centrifugal pipe, removing the supernatant, retaining the precipitate, adding acetone solution pre-cooled at the temperature of -20DEG C into the precipitate, uniformly mixing, and standing for 2h to 4h at the temperature of -20DEG C; 4) centrifuging in the centrifugal pipe, removing the supernatant, retaining the precipitate, washing the precipitate, and retaining the precipitate; 5) vacuumizing the precipitate, and volatizing acetone to obtain dry protein powder; 6) adding protein lysate into the dry protein powder, disintegrating for 1h in the water bath with temperature being 35DEG C, centrifuging, and taking the supernatant which is the cotton leave protein dissolved liquid. By adopting the method for extracting the protein from the cotton leaves, the extracted protein is stable, the repeatability is good, the purity of the protein is high, and the electrophoretogram show that no grain phenomena such as the cross stripes and streaking appear.
Owner:XINJIANG AGRI UNIV
Rapid detection method of fish parvalbumin by capillary electrophoresis
InactiveCN105911127AQuick checkAccurate detectionElectrophoretic profilingMaterial analysis by electric/magnetic meansCapillary electrophoresisAquatic product
The invention discloses a method for detecting fish parvalbumin by capillary electrophoresis. The method comprises the following steps: (1) extracting holoprotein from fish-muscle and dissolving in an equilibrium buffer with pH being 7-8, loading into an anion exchange chromatographic column, eluting with an eluant and collecting a solution corresponding to a flow-through peak so as to obtain a sample; and (2) carrying out capillary zone electrophoresis on the sample to obtain substituting peak area obtained in electrophoretogram into a standard curve regression equation of fish parvalbumin and calculating to obtain concentration of fish parvalbumin in the sample solution. The method of the invention is a method for rapid, qualitative and quantitative detection of aquatic product allergen fish parvalbumin by capillary electrophoresis. The method for qualitative and quantitative analysis of fish parvalbumin has been reported in the literature, and provides new basis for rapid and accurate detection of fish parvalbumin.
Owner:ZHEJIANG GONGSHANG UNIVERSITY
Primer and method for identifying germplasm of large yellow croakers and small yellow croakers
InactiveCN101942500AMicrobiological testing/measurementDNA/RNA fragmentationGermplasmDistilled water
The invention provides a primer and method for identifying germplasm of large yellow croakers and small yellow croakers. The invention is characterized in that the primer sequences are as follows: F1: 5'-TCGGCCTCCTGGGATTTATTGTC-3'; F2: 5'-ATTATCCTGACGGGTACGCTCTAT-3', R1: 5'-ACACGCGGGGGTCTAAC-3'; and R2: 5'-ATATGCATCGGGGTAATCTGAGTA-3'. The method comprises the following steps: adopting a phenol-chloroform extraction method to extract the DNA in the samples of the large yellow croakers or small yellow croakers; carrying out polymerase chain reaction (PCR): the PCR system is 25mu L, various components include Buffer, dNTP, Mg2+, four primers, Taq and suspected DNA in the samples of the large yellow croakers or small yellow croakers, and double distilled water is used for filling to 25mu L; and detecting the product by agarose gel electrophoresis after PCR, observing, photographing and recording the result and comparing the electrophoresis result with the provided standard electrophoretogram to identify the germplasm. The method is quick and accurate.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Primer for amplifying molecules of microbes in pit mud of Luzhou-flavor liquor and detection method
ActiveCN101974519AReflect the characteristics of the structureAbundant stripsMicrobiological testing/measurementDNA preparationMicroorganismCulture independent
The invention belongs to the technical field of wine making, which particularly relates to a primer for detecting molecules of microbes in pit mud of Luzhou-flavor liquor and a molecule detection method for microbes in pit mud of Luzhou-flavor liquor. The method is characterized in that protogenome of prokaryotic microbes in the microbes in pit mud is extracted by a culture-independent approach; the hypervariable regions V6-V8 of a microbe 16S rDNA are amplified through PCR (Polymerase Chain Reaction), and are processed by denaturing gradient gel electrophoresis (DGGE); and the diversity of the microbes and the variation of microbe structures can be analyzed through an electrophoretogram. By using the primer and the method, the 16S rDNA of the microbes in different kinds of pit mud in the process of making Luzhou-flavor liquor can be amplified quickly and accurately, the difference of microbes in different kinds of pit mud in the process of making Luzhou-flavor liquor can be further detected by DGGE, and thus realizing in-time monitoring on variations in the fermentation process of microbes in pit mud for making wine.
Owner:LUZHOU PINCHUANG TECH CO LTD
Method for labeling and identifying Szechwan white geese by utilizing ISSR (inter simple sequence repeat)
InactiveCN103276085AEasy to operateImprove stabilityMicrobiological testing/measurementBiotechnologyDNA
The invention belongs to the field of livestock or poultry genetic resource identification, and in particular relates to a method for identifying true or false of Szechwan white geese by utilizing an ISSR (inter simple sequence repeat) molecular marker technique, wherein three ISSR labels are used totally, and standard electrophoretograms after PCR (Polymerase Chain Reaction) amplification respectively of 11 goose variety (system) DNA by three genetic labels are formed for identifying whether the identified goose groups are true Szechwan white geese. According to the invention, the technical blank for labeling and identifying outstanding local goose variety of Szechwan white geese by utilizing molecular labeling is filled, and a reliable, rapid and accurate molecular biology method is provided for identifying the Szechwan white geese; unreliable factors for identifying Szechwan white geese by utilizing a traditional morphological identification method are eliminated; and technical guarantee of wide application in production of true high-fecundity Szechwan white geese can be provided.
Owner:SICHUAN AGRI UNIV
Molecular marker method for identifying tomato yellow leaf curl virus resistance gene Ty-1
ActiveCN104726602ADemanding conditionsHarsh omitMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMarker-assisted selection
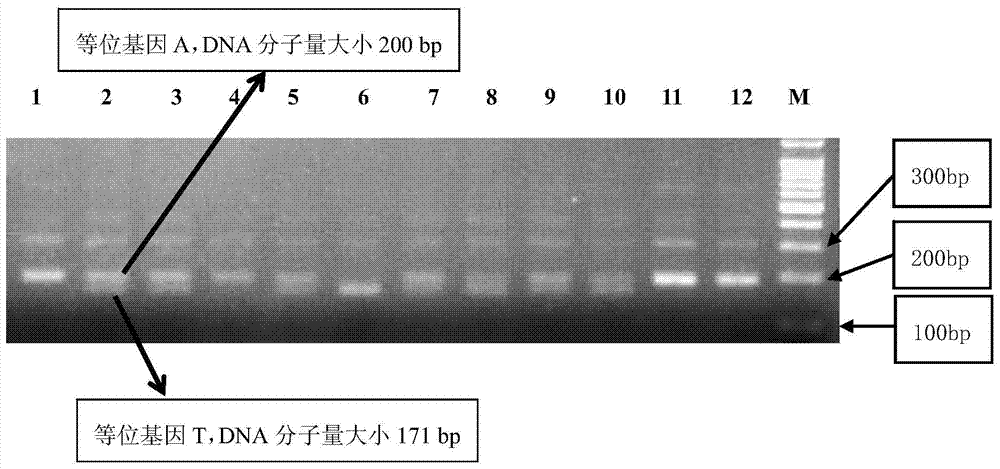
The invention discloses a molecular marker method for identifying a tomato yellow leaf curl virus resistance gene Ty-1. The method comprises the following steps: extracting a tomato gene group DNA; performing PCR amplification on the extracted tomato gene group DNA by using a molecular marker primer and performing electrophoretic analysis on the obtained PCR amplification product, representing that the yellow leaf curl virus resistance gene Ty-1 in a plant is a disease-resistant plant if a stripe with the length of 690bp is generated in an electrophoretogram, and representing that the plant is a plant system containing ty-1 gene site and is not a disease-resistant plant if the stripe with the length of 409bp is generated in the electrophoretogram. The electrophoretogram in the marker method is simple in stripe form, easy to identify, can be directly applied to marker assistant selection and can improve the identification accuracy; and moreover, the time is saving, and the material resources and the labor force are reduced.
Owner:上海孙桥现代农业联合发展有限公司 +1
SNP411871 marker associated to shell mould and weight of pinctada martensii, primer and application thereof
ActiveCN104711343AMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceHinge line
The invention discloses a SNP411871 marker associated to shell mould and weight of pinctada martensii, a primer and an application thereof. The primer comprises 411871FI: GGAAACGGAATTCACATTCATTATCTTTA; 411871RI: AATGTCAAGCTGGATATAATTTAGCTCTA; 411871FO: AAATTATTGAAACTATTTCACCGATTCG; and 411871RO: TTGTGATTTCATTTTCGTATCAATATTCTT. According to the invention, a specific SNP genotype site can be amplified by the primer, size of individual shell length and hinge line length as well as weight of soft part and adductor muscle can be visually compared according to a PCR electrophoretogram, nd the primer can be directly used for subsequent molecular mark assisted breeding.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Type H5 and H9 avian influenza virus and multiple detection method for newcastle disease virus
The invention discloses a method of detecting whether the birds and the related products are infected with H5 type or H9 type bird-flu virus or Newcastle disease virus. In the method of the invention, the trachea, cloacal swabs, blood, dung, or organs and tissues of dead birds are sterilely taken, and then milled into milk suspension by sterile physiological saline; the genomic RNA are extracted, and the RNA is used as template; the H5A, H9A and F are used as the primer for the targeted gene to undergo RT-PCR; the product obtained is electrophoresed; whether the birds detected are infected with H5, H9 type bird-flu virus or Newcastle disease virus are analyzed according to the electrophoretogram.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
Method for identifying immunofixation electrophoresis M protein components by using computer
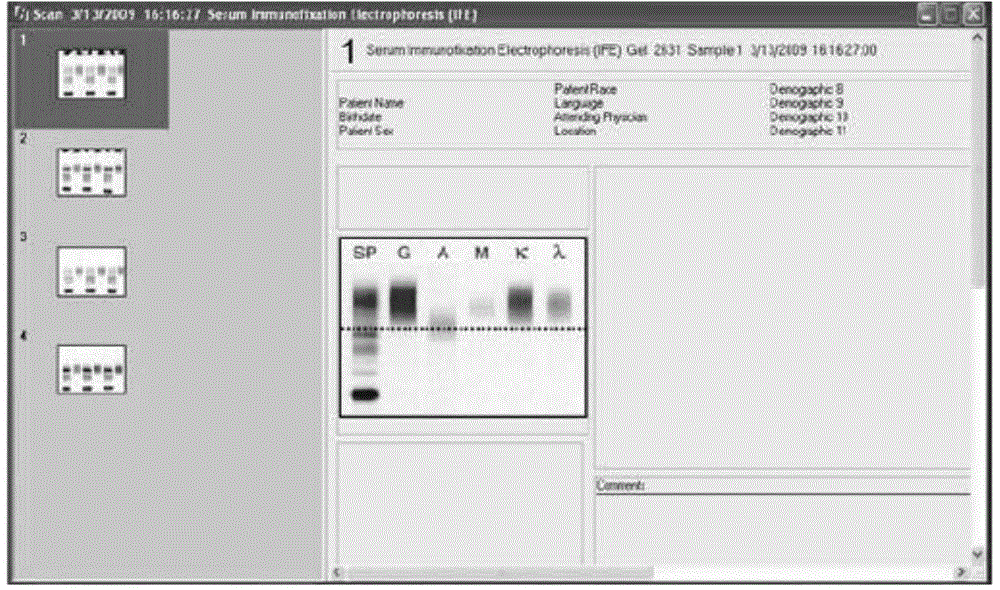
InactiveCN104598767ASpecial data processing applicationsImmunofixationImmunofixation electrophoresis
The invention belongs to the field of clinical differential diagnosis and particularly relates to a method and a system for identifying immunofixation electrophoresis M protein components by using a computer. A sample picture subjected to immunofixation electrophoresis is collected and then is scanned by the computer to be converted into six peak-shaped electrophoretograms; the six electrophoretograms are combined by using different colors to form one electrophoretogram; an M protein zone is found firstly, and then comprehensive matching treatment is carried out according to image information of other five different components. Automatic identification, classification and comprehensive analysis of serum protein electrophoresis and all components in the immunofixation electrophoretograms can be realized by using computer programming to obtain an accurate identification result. The clinical application accuracy of the method for identifying the immunofixation electrophoresis M protein components by using the computer is 96%; compared with a traditional method, the method has no remarkable difference in the aspect of statistics; the subjective errors caused by the fact that the current traditional method is used for judging the immunofixation electrophoresis M protein components by naked eyes are greatly avoided, and extreme convenience is provided for a clinical laboratory.
Owner:SHANGHAI FIRST PEOPLES HOSPITAL
DNA (Deoxyribonucleic Acid) molecular identification method for Luchuan pigs and meat products thereof
ActiveCN103215365AEasy to operatePolymorphism richMicrobiological testing/measurementBiotechnologyMolecular identification
The invention relates to a DNA (Deoxyribonucleic Acid) molecular identification method for Luchuan pigs and meat products thereof. According to the method, primers used in ISSR-PCR (Inter-Simple Sequence Repeat-Polymerase Chain Reaction) are WM14, WM12 and WM29, and nucleotide sequences of the primers are 5'-CTCCTCCTCGC-3', 5'-CACCACCACGC-3' and 5'-CTCCTCCTCAT-3'; and finally, seven electrophoretograms of the Luchuan pigs and the meat products thereof are drawn, a DNA fingerprint atlas of the Luchuan pigs and the meat products thereof is established, and the electrophoretograms and the DNA fingerprint atlas can serve as a basis for the distinction and identification of the Luchuan pigs and the meat products thereof. The method has the advantages that a reliable DNA molecular identification technique is adopted, so that the differences among the Luchuan pigs, the meat products of the Luchuan pigs, Luchuan pig hybrids, meat products of the Luchuan pig hybrids, Bama miniature pigs, meat products of the Bama miniature pigs, Huanjiang miniature pigs and meat products of the Huanjiang miniature pigs are identified quickly and accurately, and the development of advantaged brand pigs, namely the Luchuan pigs, is promoted.
Owner:GUANGXI UNIV
PCR detection method for identifying germplasms of four populations of Pelodiscus sinensis, primer group and kit
ActiveCN103276097ATemplate requirements are not highImprove reaction stabilityMicrobiological testing/measurementDNA/RNA fragmentationGermplasmMicrobiology
The invention discloses a PCR detection method for identifying germplasms of four populations of Pelodiscus sinensis, a primer group and a kit. The primer group comprises primers of SEQ No.1 / SEQ No.4, SEQ No.2 / SEQ No.5 and SEQ No.3 / SEQ No.6. The kit comprises a 2*TaqPCR premixed liquid, a 5U / mul enzyme BglI and 10*digestion buffer liquid A, a 5U / mul HpaII and 10*digestion buffer liquid B, a 5U / mul ClaI and 10*digestion buffer liquid C, 10mg / ml BSA, a contrast DNA template and a 10pM amplimer. The method comprises the following steps: carrying out PCR amplification of extracted DNA through using the primer group, digesting to obtain a digestion product, carrying out agarose gel electrophoresis, and comparing the obtained electrophoresis result with a standard electrophoretogram to obtain a result. The detection method has the advantages of simplicity, good reaction stability and repeatability, and cost saving.
Owner:ZHEJIANG FISHERIES TECH EXTENSION STATION
Method for identifying self-sterile S-gene type SSR molecular marker of sweet cherry variety
InactiveCN101838701AShorten the breeding processImprove detection efficiencyMicrobiological testing/measurementAgricultural scienceAgarose electrophoresis
The invention provides a method for identifying a self-sterile S-gene type SSR molecular marker of a sweet cherry variety. The method uses PTCR4SSR to mark, wherein the SSR primer sequence is upstream primer 5'-CATAGGTTCAAACCATACCCGTG-3' and downstream primer 5'-CTCATCTTTGTAGGGTATAATACC-3' to amplify the DNA of sweet cherry of different varieties (systems), if the amplified fragment of 255bp can be amplified, pollen S-gene exists, and a electrophoretogram can be obtained by agar gel electrophoresis; according to the agarose elelctrophoretogram, the self-sterile gene type of each variety (system) can be determined, wherein the same bands are the same S-gene type, otherwise being different S-gene types. The SAM method is used to develop SSR marked PTCR4 in sweet cherry for identifying the S-gene type of different sweet cherry self-sterile gene type varieties (systems) and guiding selection of breeding parents and configuration of pollination varieties thereof, so that the detection efficiency of the self-sterile gene type can be improved, the breeding process of the sweet cherry is shortened, and the production cost is saved.
Owner:SHANDONG INST OF POMOLOGY
Special primer for double PCR of contagious pustular dermatitis virus and mycoplasma ovipneumoniae
ActiveCN106636470AQuick checkAccurate diagnosisMicrobiological testing/measurementMicroorganism based processesImpetigo contagiosaEpidemiologic survey
The invention provides a special primer for double PCR of a contagious pustular dermatitis virus and mycoplasma ovipneumoniae. According to the special primer, two pairs of specific primers capable of amplifying a B2L gene and a P80 gene are designed according to the B2L gene of the contagious pustular dermatitis virus and the P80 gene of the mycoplasma ovipneumoniae. Through optimization of the conditions of the primer concentration, the annealing temperature and the like, a double PCR method for fast detecting the contagious pustular dermatitis virus and the mycoplasma ovipneumoniae is built. A specific segment of 401bp appearing in an electrophoretogram has positive contagious pustular dermatitis virus, and the specific segment of 700bp has positive mycoplasma ovipneumoniae. Detection of the contagious pustular dermatitis virus and the mycoplasma ovipneumoniae in the same reaction system can be achieved, the special primer has the advantage that the pathogens are fast, specifically and accurately diagnosed, and a novel method is provided for differential diagnosis of infection of the two pathogens in production and epidemiological investigation.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
PCR-RFLP method for distinguishing metacercaria of clonorchis sinensis from metacercaria of metorchis orientalis
InactiveCN104372084AImprove stabilityThe identification method is simpleMicrobiological testing/measurementMetorchis orientalisEnzyme digestion
The invention discloses a PCR-RFLP method for distinguishing metacercaria of clonorchis sinensis from metacercaria of metorchis orientalis, and relates to a molecular method for identifying metacercaria of clonorchis sinensis and metacercaria of metorchis orientalis by using a PCR-RFLP technology. A ribosome transcribed spacer sequence (ITS) of which the length is about 1200bp can be amplified from the metacercaria of clonorchis sinensis and the metacercaria of metorchis orientalis by using the same primers through PCR amplification; the PCR product is subjected to enzyme digestion by virtue of restriction endonuclease XhoI; the sample with two stripes displayed in an electrophoretogram is metacercaria of clonorchis sinensis; and the sample with one stripe displayed in the electrophoretogram is metacercaria of metorchis orientalis. Therefore, accurate and fast identification can be achieved according to the stripe change after enzyme digestion of the PCR product. The identification method disclosed by the invention is simple, relatively high in efficiency and accuracy rate, and fast in identification, only needs a working day, and simultaneously is relatively low in cost.
Owner:HEILONGJIANG BAYI AGRICULTURAL UNIVERSITY
Molecular biological method for rapidly identifying trichosanthin medicinal material
The invention relates to a new molecular identification method for identifying trichosanthin and its similar products; adventurous innovation is made in two key links of primer design and annealing temperature. Primer design is performed on a known ITS of trichosanthin medicinal materials; the length is about 20 bp; and general primer design principles are followed. Meanwhile, the annealing temperature is screened and determined according to the number of polymorphism strips. The characteristic identification strips of trichosanthin are determined to be 560, 960 bp strips, and 1930, 1400, 839, 715 bp strips are considered as auxiliary identification strips of trichosanthin. The invention has the following advantages of: (1) simplicity and practicality, wherein although the difficulty for primer design is increased slightly, the specificity is greatly improved, an ideal identification primer can be obtained after design of 2-3 primers, and thus the problem of large screening of random primers is avoided; (2) stability and repeatability, wherein since the specificity and annealing temperature of the primer are improved, the amplification strips are generally only 1-5 strips, and certified products with different producing areas and different storage time have no effect on PCR results; (3) large provided information content, wherein both certified products and most fake products can be amplified simultaneously, and thus standard identification electrophoretograms of the certified products and fake products can be established to realize accurate identification of the certified products and fake products.
Owner:QINGDAO VLAND BIOTECH INC
PCR (Polymerase Chain Reaction) detection method of mycoplasma
InactiveCN102242213AStrong specificityShort detection cycleMicrobiological testing/measurementPcr methodMycoplasma contamination
The invention relates to a PCR (Polymerase Chain Reaction) method for detecting mycoplasma, belonging to the technical field of biological detection. In the invention, PCR amplification and electrophoresis are carried out by extracting mycoplasma nucleic acids and applying mycoplasma specific primers to judge whether mycoplasma contamination exists in a detection sample or not, wherein if 435bp specific strips appear in an electrophoretogram, the result shows that the mycoplasma exists in the sample, and if 435bp specific strips do not appear, the result shows that the mycoplasma does not exist in the sample. By the detection and verification, the detection system provided by the invention has the characteristics of high specificity, good stability, short checking period and the like.
Owner:JINYUBAOLING BIO PHARM CO LTD
Molecular detection method for rapidly identifying carbendazim-resistant genotype-F200Y botrytis cinerea strain
ActiveCN104313177ARapid identificationRich technology system for resistance detectionMicrobiological testing/measurementMicroorganism based processesResistant genotypeSky
The invention discloses a molecular detection method for rapidly identifying a carbendazim-resistant genotype-F200Y botrytis cinerea strain. The molecular detection method is a molecular detection technology, which is established based on a LAMP (loop-mediated isothermal amplification) technology and is rapid, convenient, high in specificity and sensitivity and low in cost. The detection method comprises the following steps of designing two pairs of specific primers on a beta tubulin of a detected sample, performing LAMP, and judging whether the detected sample is a carbendazim-resistant genotype-F200Y botrytis cinerea strain or not according to the color of a reaction product; if the LAMP product is sky-blue and an electrophoretogram is a ladder band, determining that the product amplification exists and that the detected sample is the carbendazim-resistant genotype-F200Y botrytis cinerea strain; if the LAMP product is purple and the electrophoretogram is free of band, determining that the product amplification does not exist and that the detected sample is a carbendazim-nonresistant genotype-F200Y botrytis cinerea strain. The method is convenient, rapid, low in cost and significant for theoretically guiding the resistance risk evaluation and reasonable medication of botrytis of crops.
Owner:NANJING AGRICULTURAL UNIVERSITY
Rapid detection method of molecular variant of citrus tatter leaf virus
InactiveCN101560577AHigh speedSimple and fast operationMicrobiological testing/measurementMicroorganism based processesOperabilityBiology
The invention relates to a rapid detection method for diagnosing molecular variant of citrus tatter leaf virus (CTLV), comprising the steps of: designing a specific primer to perform a reverse transcription polymerase chain reaction augmentation (RT-PCR) on the nucleic acid sequences in a special area of CTLV genome, and using Taq I, Mse I, Hinf I or other restriction enzyme to digest the RT-PCR product, and then diagnosing the molecular variant of CTLV according to an electrophoretogram. The method of the invention is characterized by high speed, low cost, good repeatability, high stability and strong operability and the like, and is suitable for large-scale and high-flux sample analysis, and is applicable to various uses such as field sample detection, CTLV epidemic monitoring and detoxication plantlet identification.
Owner:SOUTHWEST UNIVERSITY +1
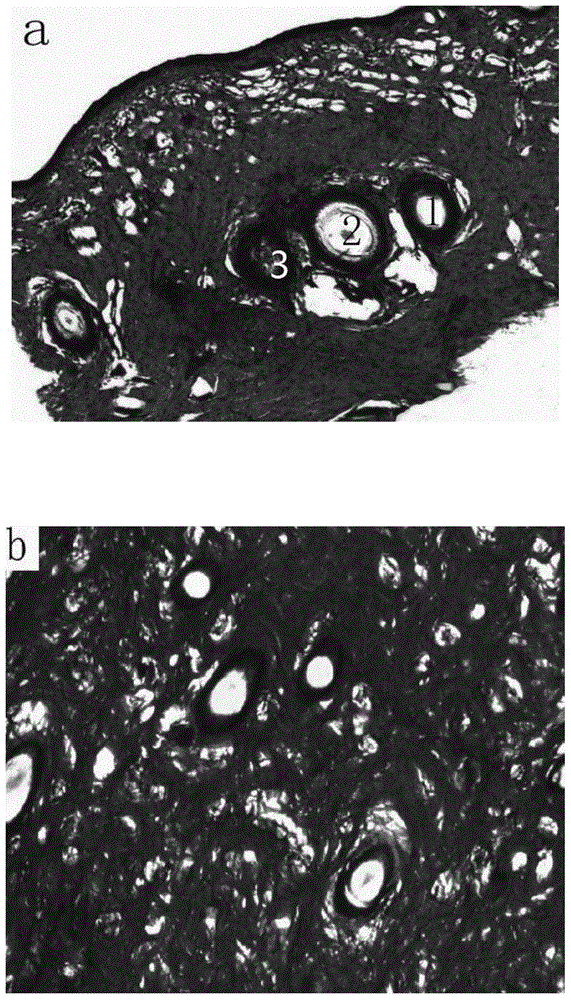
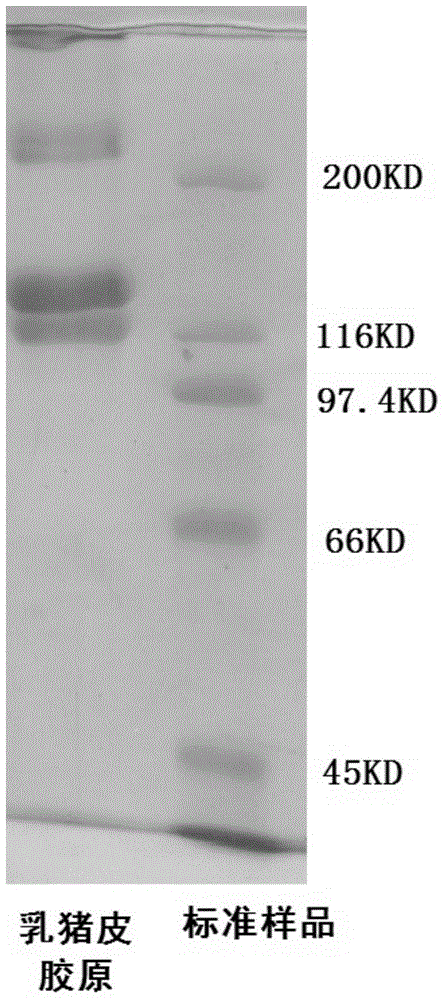
Method for preparing undenatured natural collagen from sucking pig skin
ActiveCN103601803AEffective hair removalEfficient removalConnective tissue peptidesPeptide preparation methodsHair rootsFreeze-drying
The invention discloses a method for extracting undenatured natural collagen from sucking pig skin. The method is characterized in that treatment is carried out by sub-step repeated different combinations in a manner that a degreasing agent is assisted by intermittent ultrasonic waves, an unhairing agent and a non-collagen removing agent are respectively used for unhairing and removing non-collagen, and no hair root exists in the skin after unhairing, wherein shown by determination, the fat content is lower than 0.5% and the ash content is lower than 0.3%; enzyme-soluble collagen is obtained through leaching by acetic acid-protease and is purified by high-speed centrifugation, sodium chloride salting-out and dialysis, and spongy sucking pig skin collagen is obtained after freeze-drying, wherein the collagen extraction ratio exceeds 50% (dry weight), and shown by electrophoretogram, the sucking pig skin collagen contains a beta chain and an alpha chain, of which the molecular weights are 200000 and 100000 respectively, and is free from other hybrid protein bands, so that the collagen reserves three helical structures and has relatively high purity.
Owner:SICHUAN UNIV
Method sutiable for two-dimensional electrophoresis extraction and separation of total protein of rat and mouse testicular tissue sample
InactiveCN103278370AReduce degradationImprove solubilityPreparing sample for investigationStainingTotal protein
The invention discloses a method for two-dimensional electrophoresis extraction and separation of total protein of a rat and mouse testicular tissue sample. The method comprises rat and mouse testicular tissue treatment, a preparation method for a testicular tissue total protein sample, a two-dimensional electrophoresis technology system, gel dyeing and spectrum analysis. The method optimizes a tedious universal two-dimensional electrophoresis technology system with relatively poor stability, greatly reduces time and reagent consumption for finding out conditions, saves experiment cost, and obtains a satisfactory two-dimensional electrophoresis spectrum that is convenient for subsequent analysis. The method is more suitable for the extraction of the rat and mouse testicular tissue protein sample than a universal homogenate extraction method, a lysate extraction method, a lysate extraction / acetone precipitation method at present. The method not only reduces the degradation of the protein but also increases solubility of the protein without increasing cost of the reagent, so that the two-dimensional electrophoresis spectrum with relatively many protein sites, clear horizontal stripes and relatively good repeatability can be obtained.
Owner:SHANXI AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com