COI gene based PCR-RFLP discrimination method of seventeen economic sea cucumbers of Stichopodidae
A technology of PCR-RFLP and sea cucumber, applied in biochemical equipment and methods, microbe measurement/inspection, etc., can solve the problems of lack of systematic research and few applications, and achieve accurate detection, low requirements for instruments and equipment, and wide application range wide effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Example 1: PCR-RFLP identification of Apostichopus japonicus samples
[0017] a. PCR amplification of the COI fragment of the Apostichopus japonicus sample:
[0018] The DNA of the Apostichopus japonicus samples was extracted by conventional methods, and then the COI fragment was amplified by PCR using it as a template. The PCR amplification reaction volume was 50 μL, and 200 μL PCR tubes were placed in an ice bath, followed by adding 37.6 μL sterile water, 5 μL 10 × PCRBuffer (including 15mMMg 2+ ), 1μL 10uMCOIe-F primer, 1μL 10uMCOIe-R primer, 4μL 2.5mM dNT Pmixture, 0.4μL 5U / μL TaKaRaTaqTM enzyme, 1μL Apostichopus japonicus sample DNA template; ABIVeriti 96-well thermal cycler was used for amplification, and the reaction conditions were pre-denaturation at 94°C for 2 minutes , followed by 35 cycles of denaturation at 94°C for 30 seconds, renaturation at 48°C for 30 seconds, and extension at 72°C for 45 seconds, with a final extension at 72°C for 7 minutes. The COIe...
Embodiment 2
[0028] Example 2: Identification of Apostichopus japonicus samples by PCR-RFLP
[0029] Steps a-c are the same as those in Example 1, except that Apostichopus japonicus is used as a sample.
[0030] d. Judgment of test results
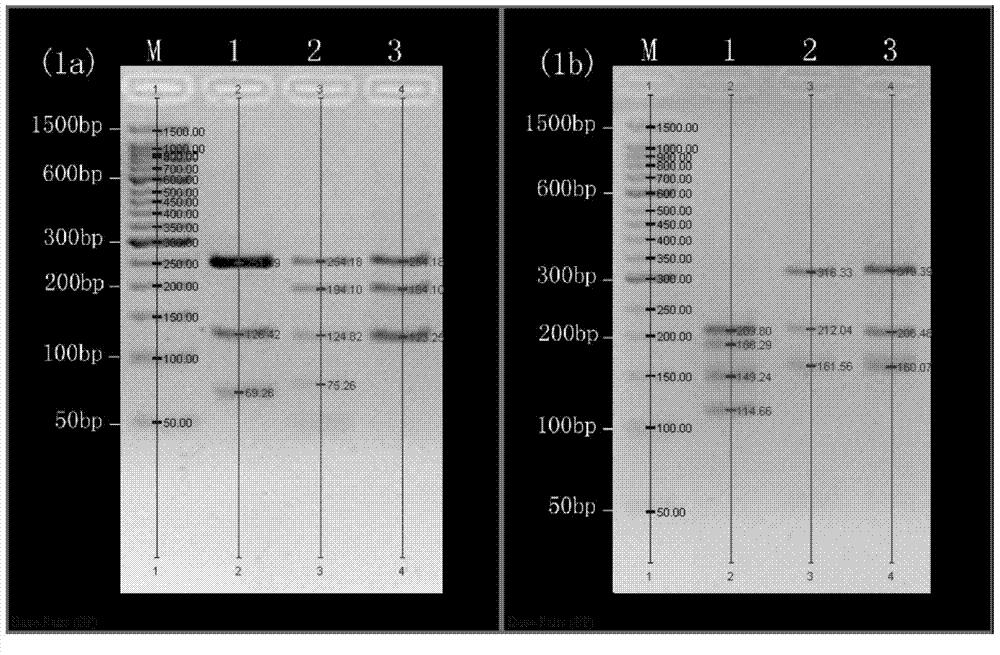
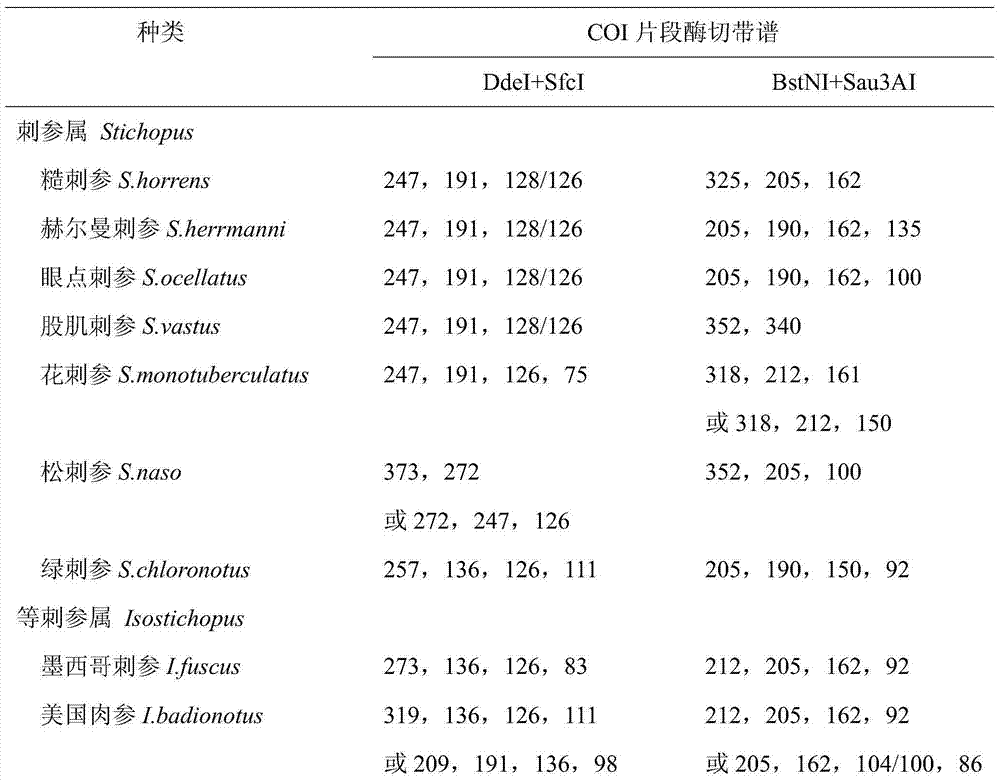
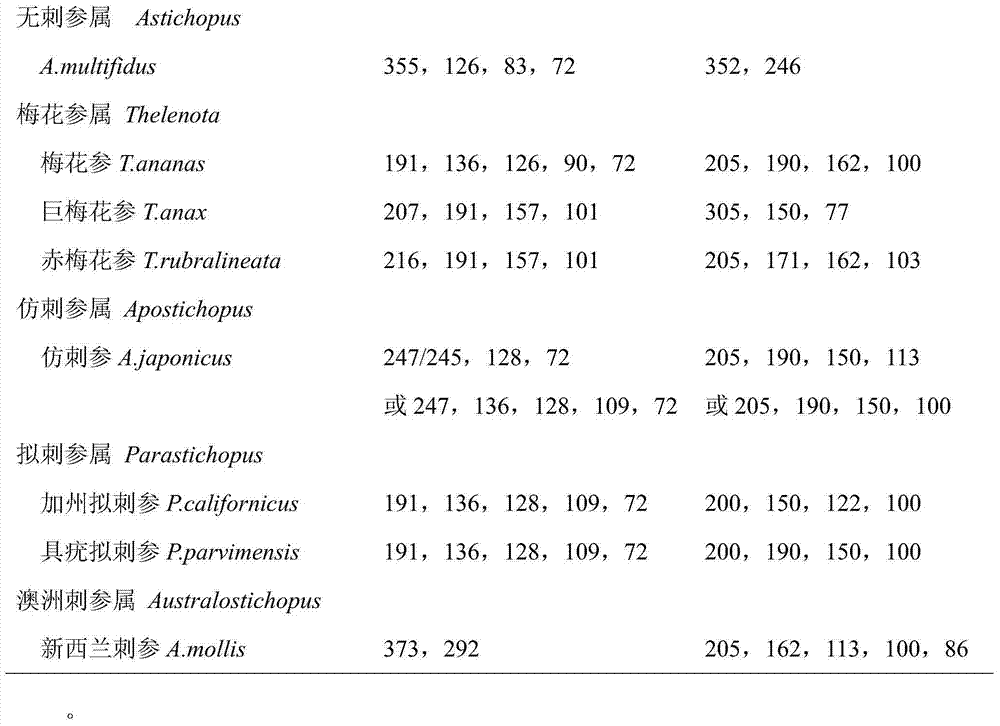
[0031] After electrophoresis, take pictures under the gel imaging system (Bio-Rad Company) to obtain the restriction electrophoresis diagrams of the two groups of endonucleases DdeI / SfcI and BstNI / Sau3AI in the COI fragment of the sea cucumber sample (see figure 1); the added 50bp DNALadder was set as the molecular weight standard, and the Quantityone software estimated that: ① The estimated value of the four bands cut by DdeI / SfcI double enzyme was 254.18bp, 194.10bp, 124.82bp and 75.26bp; ②BstNI / Sau3AI double enzyme cut The estimated values of the three bands are 316.33bp, 212.04bp and 161.56bp. Compared with Table 1, the number of bands is the same as that of A. japonicus, and the estimated value of each electrophoresis band matches the correspo...
Embodiment 3
[0032] Example 3: Identification of Apostichopus japonicus samples by PCR-RFLP
[0033] Steps a-c are the same as in Example 1, but Apostichopus japonicus is used as a sample.
[0034] d. Judgment of test results
[0035] After electrophoresis, take pictures under the gel imaging system (Bio-Rad Company) to obtain the restriction electrophoresis images of two groups of endonucleases DdeI / SfcI and BstNI / Sau3AI in the COI fragment of A. japonicus samples (see figure 1 ); the added 50bp DNALadder was set as the molecular weight standard, and Quantityone software estimated that: ① The estimated values of the three bands cut by DdeI / SfcI double enzymes were 254.18bp, 194.10bp and 123.25bp; ② The three bands cut by BstNI / Sau3AI double enzymes Band estimates are 319.39bp, 206.48bp and 160.07bp. Compared with Table 1, the number of bands is the same as that of A. japonicus, and the estimated value of each electrophoresis band matches the corresponding fragment size in the standard...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com