Rapid detection method of molecular variant of citrus tatter leaf virus
A technology for leaf-shattering virus and detection methods, which is applied in the directions of microorganism-based methods, microbial determination/inspection, biochemical equipment and methods, etc., which can solve the problems of complex staining procedures, difficulty in defining the type of variation, and long electrophoresis time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
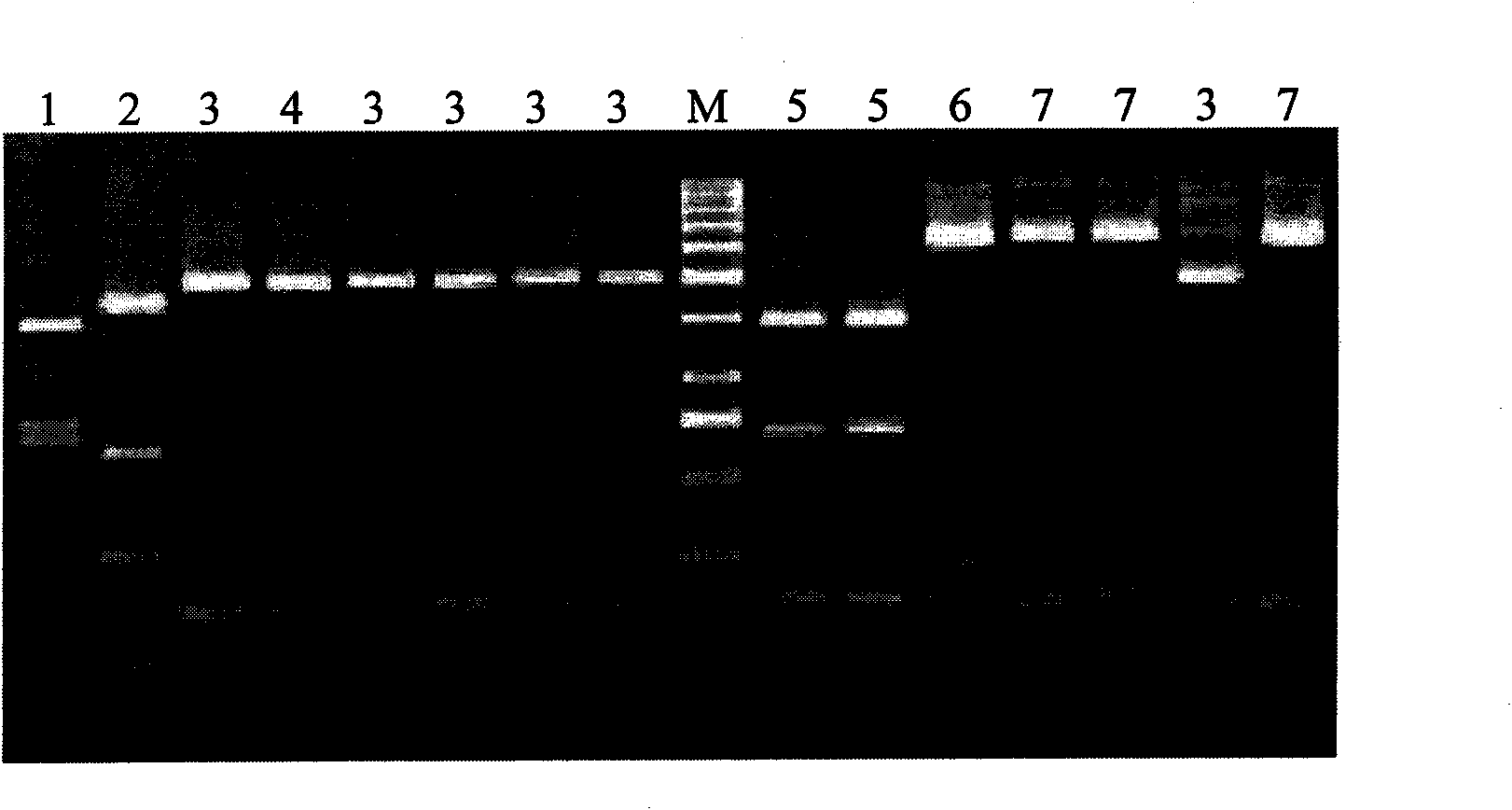
[0026] Embodiment 1: Nucleic acid extraction
[0027] Weigh 5 mg of virus host skin or leaf into a sterile centrifuge tube, grind in liquid nitrogen, add 60 μL of TES buffer solution and 60 μL of saturated phenol:chloroform:isoamyl alcohol (25:24:1), and mix well; Water bath at 70°C for 5 to 10 minutes; centrifuge at 13,000 rpm for 5 minutes at room temperature, absorb 40 μL of the supernatant and add it to a microcolumn made of Sephadex G-50-80, place in a new sterile centrifuge tube, centrifuge at 5,000 rpm for 4 minutes, and collect eluent.
Embodiment 2
[0028] Example 2: Reverse transcription polymerase chain amplification
[0029] Design specific primers CTLVR: 5′-AGAGTGGACA AACTCTAGAC-3′, CTLVF: 5′-CCCTCTCAGC TAGAATTGAA-3′ to amplify the 889bp nucleic acid sequence including the coat protein gene of citrus leaf scraper virus, reverse transcription polymerase chain Amplification was carried out in a PCR machine, and the total volume of the reaction system used was 10 μL, including: 5 μL of 2γPCR buffer, 1.25 μL of 10 μmol / μL primers, 0.3 μL of 50 mmol / L MgSO4, 0.2 μL of 5 U / μL RT / Taq enzyme (Invitrogen), 1 μL of nucleic acid mixture extracted according to the above method, 1.5 μL of ultrapure water; the reaction conditions are: 42°C, 30 minutes; 94°C, 2 minutes; 94°C for 20s, 54.5°C for 20s, 72°C for 45s, 40 cycles; 72 °C for 10 minutes.
Embodiment 3
[0030] Example 3: Restriction enzyme digestion
[0031] After reverse transcription polymerase chain amplification, directly add 10 μL of the following mixture for Hinf I digestion reaction: ultrapure water 6.25 μL; buffer B 2.5 μL; BSA (10 μg / μL) 0.25 μL; Hinf I (10U / μL ) 1 μL; after mixing thoroughly, bathe in water at 37°C for 1 hour.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com