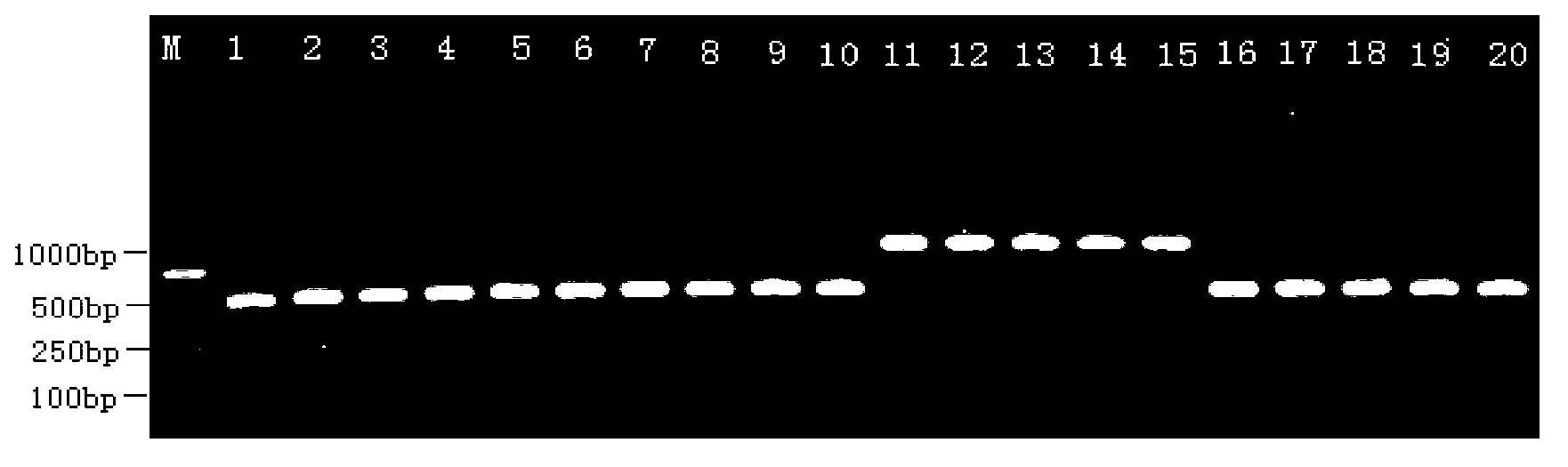PCR detection method for identifying germplasms of four populations of Pelodiscus sinensis, primer group and kit
A technology of Chinese soft-shelled turtle and detection method, which is applied in the field of aquatic germplasm detection, can solve the problems of limited reliability of identification results, high price, and great influence, and achieves saving detection time and cost, good reaction stability and repeatability , the result is intuitive
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
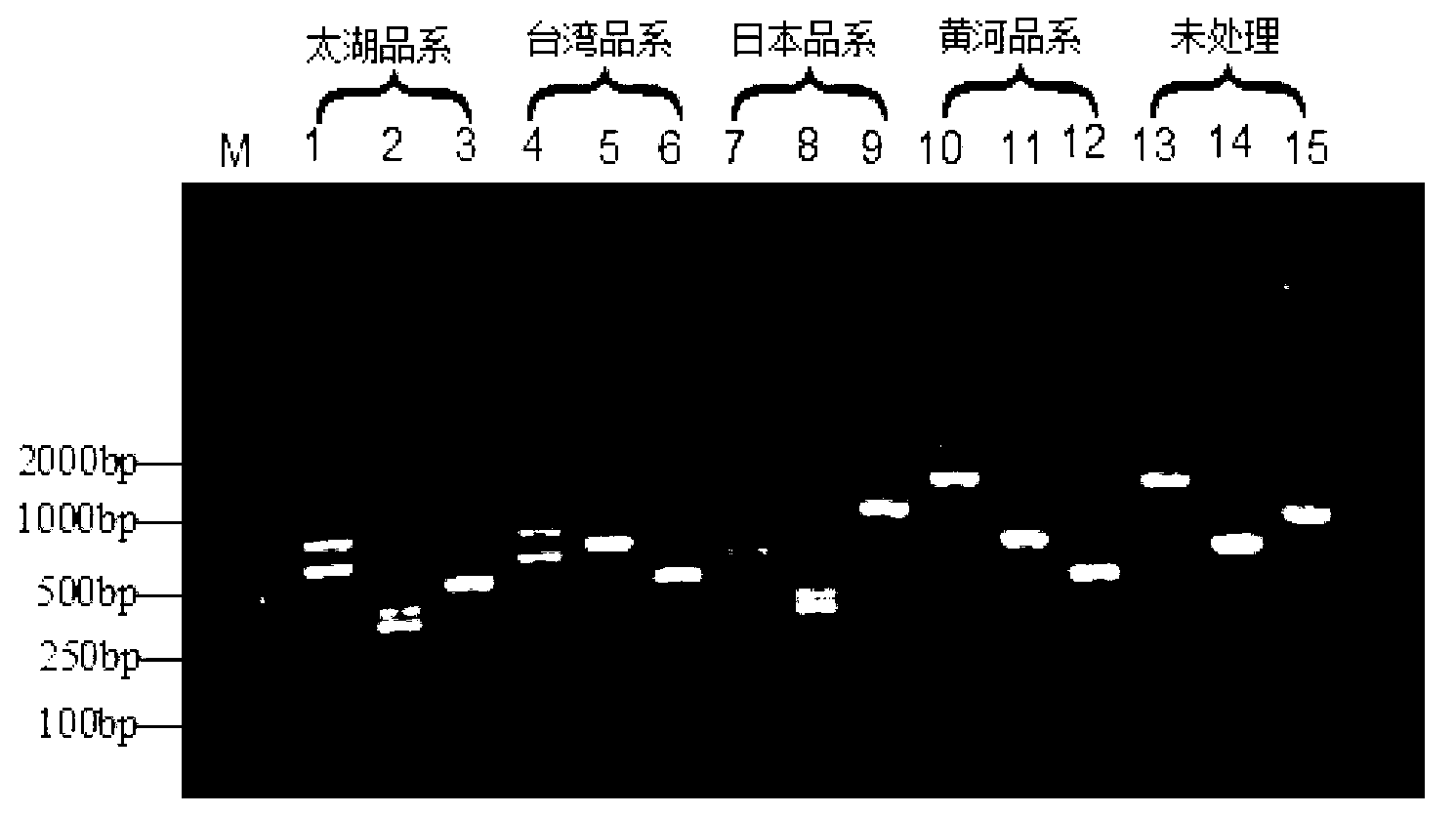
[0047] Example 1: Construction of standard PCR-RFLP profiles of four different populations of soft-shelled turtles:
[0048] (1) Extraction of total DNA: take the soft-shelled soft-shelled turtle tissue sample to be tested, refer to the Tiangen Total DNA Extraction Kit to extract the total DNA (other kits can also be used), and dilute the extracted DNA to 50-150ng / ul. Store at -20°C;
[0049] (2) PCR amplification: using the extracted total DNA as a template, use 3 pairs of amplification primers SEQ No.1 / SEQ No.4, SEQ No.2 / SEQ No.5 and SEQ No.3 / SEQ No. 6 Carry out PCR amplification, and also carry out PCR amplification to control template DNA at the same time,
[0050] The PCR amplification reaction system used is 50ul consisting of 25μl 2×Taq PCR master mix, 1μl upstream primer, 1μl corresponding downstream primer, 1μl template and the rest ddH 2 O composition.
[0051] The PCR amplification reaction conditions used were: pre-denaturation at 95°C for 3 min, denaturation at...
Embodiment 2
[0055] Embodiment 2: Germplasm identification of soft-shelled turtle Yellow River population:
[0056] (1) Extraction of total DNA: take the soft-shelled soft-shelled turtle tissue to be tested, and extract the total DNA according to the method in Example 1;
[0057] PCR amplification: use the extracted total DNA as a template, and use primers to perform PCR amplification of SEQ No.1 / SEQ No.4;
[0058] The PCR amplification reaction system used is 50ul consisting of 25μl 2×Taq PCR master mix, 1μl upstream primer SEQ No.1, 1μl corresponding downstream primer SEQ No.4, 1μl template and the rest of ddH 2 O composition, PCR amplification reaction condition is identical with embodiment example 1;
[0059] (2) Enzyme digestion reaction: use restriction endonuclease Bgl 1 enzyme digestion is carried out to the amplified product obtained in step (2);
[0060] The enzyme digestion reaction system used is 25 μl of the amplified product obtained from 15 μl of step (2), 2.5 μl of 10...
Embodiment 3
[0063] Embodiment 3: Germplasm identification of new varieties of Japanese strains:
[0064] (1) Extraction of total DNA: the extraction method of total DNA is the same as in Example 1;
[0065] (2) PCR amplification: use the extracted total DNA as a template, and use primers to perform PCR amplification of SEQ No.3 / SEQ No.6;
[0066] The PCR amplification reaction system used is 50ul consisting of 25μl 2×Taq PCR master mix, 1μl upstream primer SEQ No.3, 1μl corresponding downstream primer SEQ No.6, 1μl template and the rest of ddH 2 O composition, PCR amplification reaction condition is identical with embodiment example 1;
[0067] (3) Enzyme digestion reaction: use restriction endonuclease cla I digest the amplification product obtained in step (2),
[0068] The enzyme digestion reaction system used is 25 μl of the amplified product obtained from 15 μl of step (2), 2.5 μl of 10× digestion buffer C, 0.25 μl of BSA, and 5 U of restriction endonuclease cla I and ddH of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com