Detection method for Y chromosome micro-deleted gene
A technology of Y chromosome and detection method, applied in the field of genetic engineering, can solve the problems of increasing the cost of reagents, unable to completely detect AZF fragments, etc., and achieve the effects of reducing the formation of primer-dimers, facilitating clinical experiment operations, and simplifying PCR reaction conditions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Human Genomic DNA Extraction—Extract DNA from 250ul of anticoagulated whole blood by conventional boiling lysis method or column elution method, and the expected DNA yield is about 4-12ug, which can be used as template DNA.
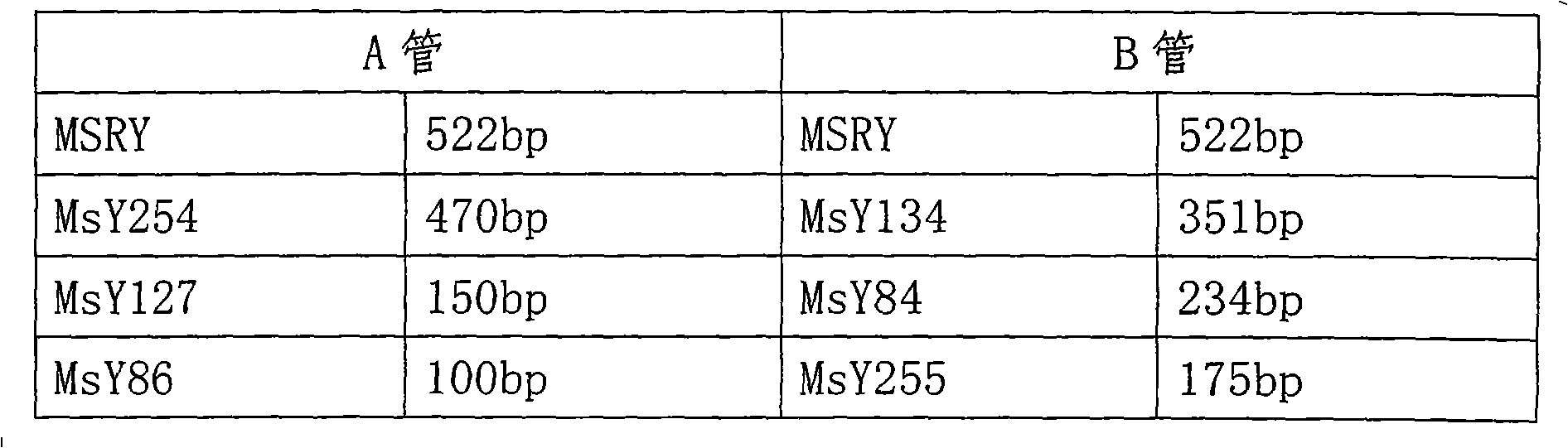
[0058] PCR amplification - multiplex PCR amplification is divided into tube A and tube B. The 30ul PCR reaction system is: 1×PCR buffer (50mmol / L KCl, 10mmol / L Tris-HCl, pH 9.0), 1.5mMMgCl 2 , 200uM dNTP, 5~10ng template DNA, and 1U Taq DNA polymerase (Promega); the concentrations of 5 pairs of primers added to tube A are: MSRY 0.12uM, MsY254 0.1uM, MsY127 0.05uM, MsY86 0.1uM, YUP 0.4uM; The concentrations of 5 pairs of primers added to tube B are: MSRY 0.1uM, MsY1340.12uM, MsY84 0.12uM, MsY255 0.05uM, YUP 0.4uM, and the reaction was carried out in a 9700 PCR reaction instrument of ABI Company. A positive control (genome DNA of normal fertile male), a negative control (genome DNA of female) and a water blank control were set up for each test.
[...
Embodiment 2
[0068] The selection of the STS site of the AZF fragment was based on the standards recommended by the "European Y Chromosome Microdeletion Molecular Diagnosis Guidelines 2004 Edition" issued by the European Association of Andrology EAA (clinical level) and the European Molecular Genetic Laboratory Quality Control Association EMQN (specific experimental operation level) performed (SIMONI M., et al. 2004, International Journal of Andrology, 27, 240-249). The AZFa region selects the sY84 and sY86 sites:
[0069] The base sequence of the primer for specifically amplifying AZFa region sY84 is:
[0070] sY84-F: 5’-AGA AGG GTC TGA AAG CAG GT-3’
[0071] sY84-R: 5'-GCC TAC TAC CTG GAG GCT TC-3'
[0072] The base sequence of the primer for specifically amplifying sY86 in the AZFa region is:
[0073] sY86-F: 5'-AGA CTA TGC TTC AGC AGG TC-3'
[0074] sY86-R: 5’-GAA CCG TAT CTA CCA AAG CAG C-3’
[0075] AZFb region selects sY127 and sY134 sites:
[0076] The base sequence of the pr...
Embodiment 3
[0101] Genomic DNA was extracted from human peripheral blood. For PCR reaction, multiplex PCR takes a 15ul PCR reaction system as an example. The reaction system (pH 8.5) includes the following reagents: PCR reaction solution, Taq enzyme, dATP, dGTP, dCTP, dTTP, MgCl 2 , primers, template DNA. The final concentration of dNTP reaction is 200uM; MgCl 2 The final concentration of the reaction was 1.5 mM. The PCR amplification was divided into two tubes: tube A with 5 pairs of primers for composite amplification; tube B with 5 pairs of primers for multiplex amplification, and both tubes were designed with MSRY primers as internal controls. Primer pairs are grouped as follows:
[0102] Group A: MSRY, MsY254(AZFc), MsY127(AZFb), MsY86(AZFa), YUP
[0103] Group B: MSRY, MsY134(AZFb), MsY84(AZFa), MsY255(AZFc), YUP
[0104] PCR cycle conditions: pre-denaturation at 95°C for 5 min, then denaturation at 95°C for 40 s, annealing at 54°C for 40 s, extension at 72°C for 40 s, after 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com