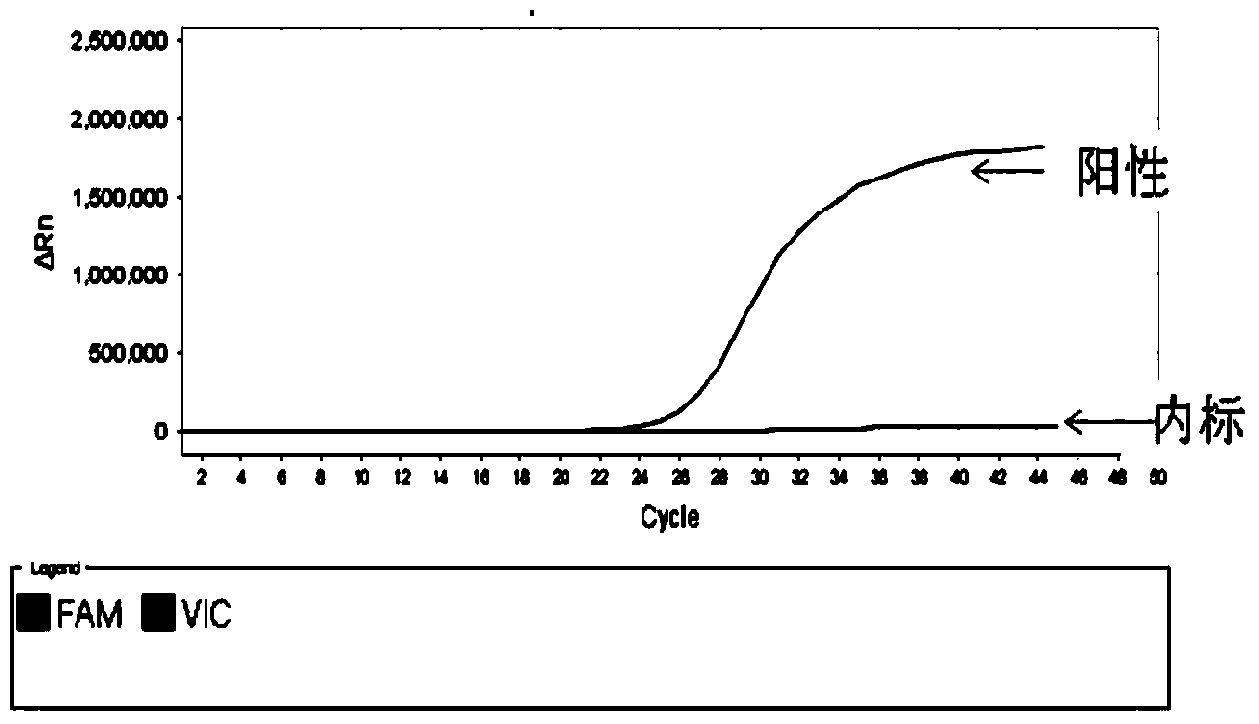
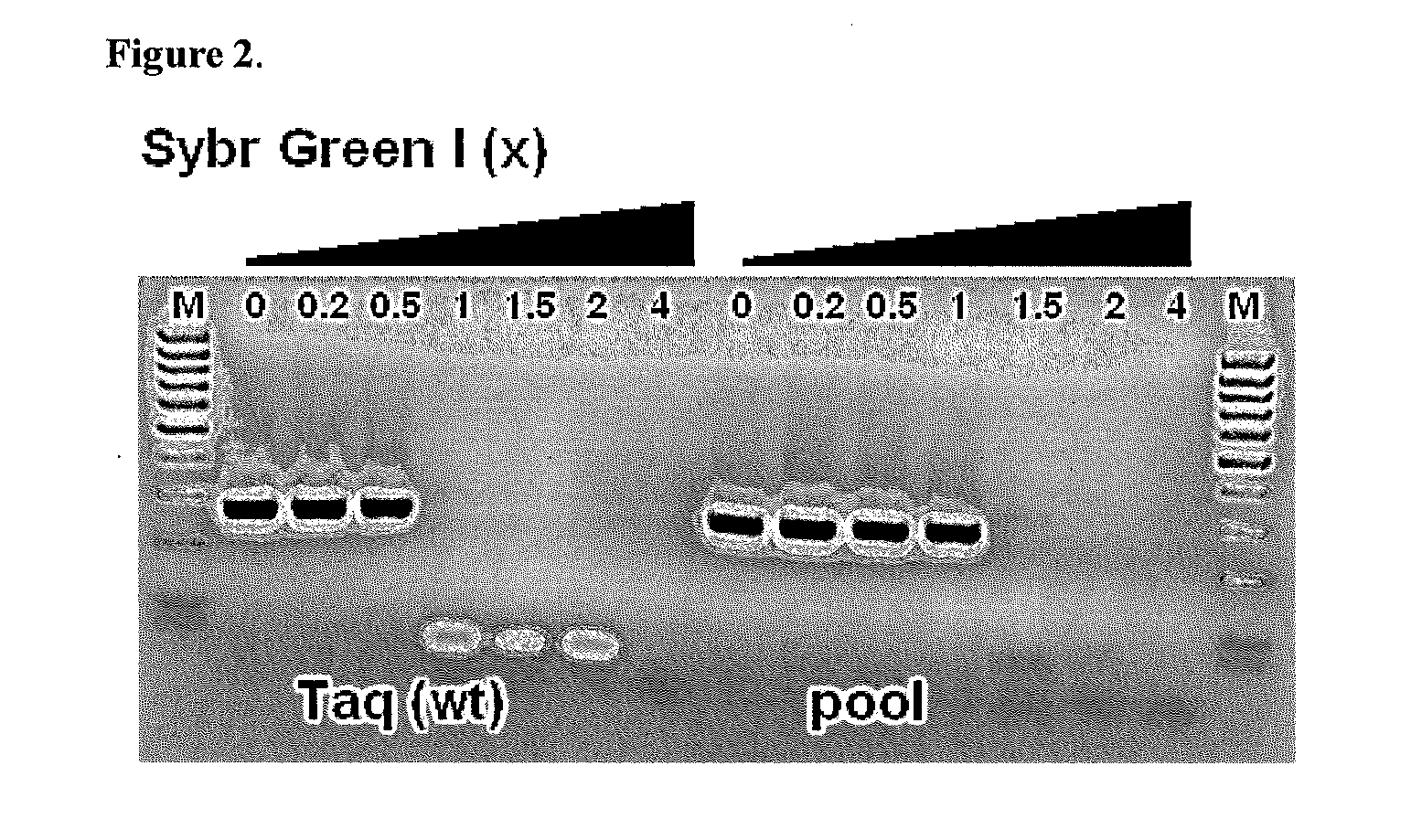
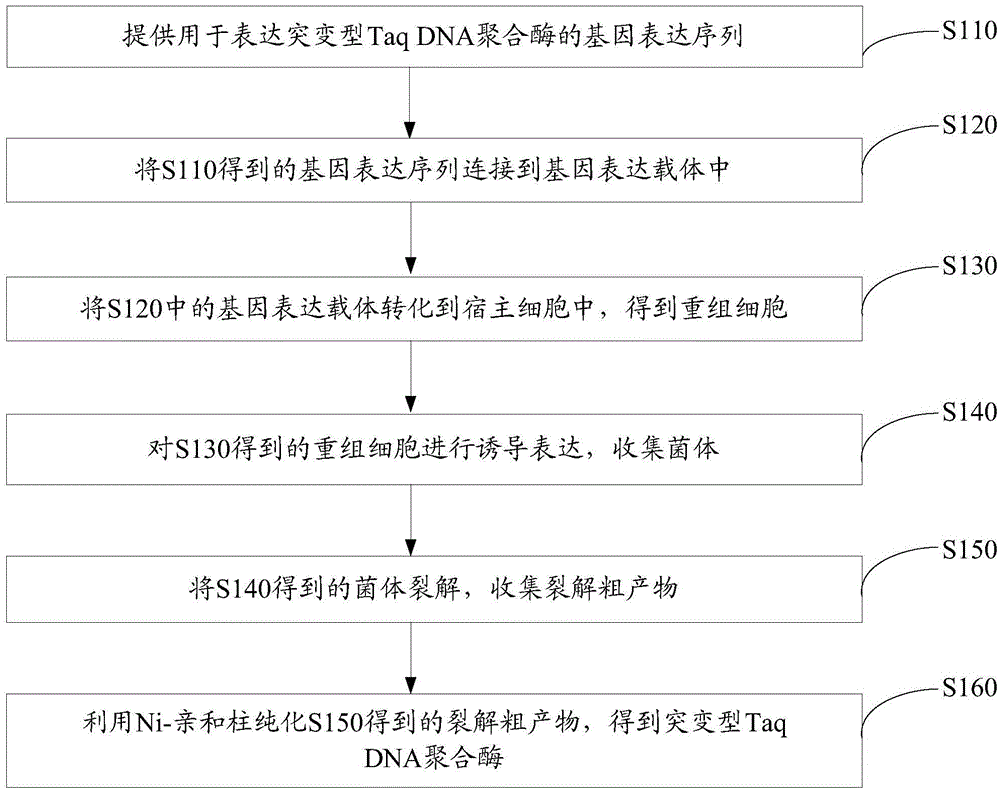
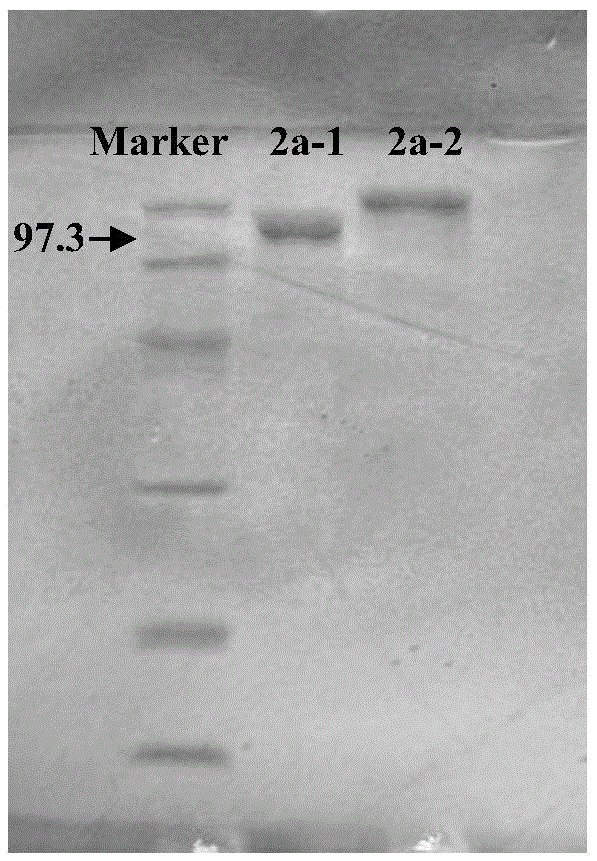
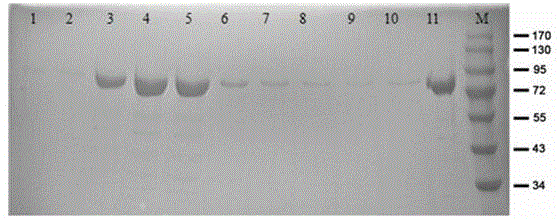
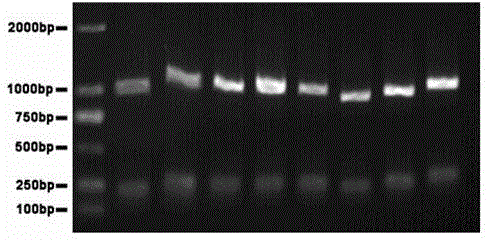
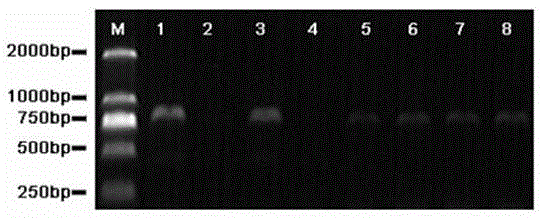
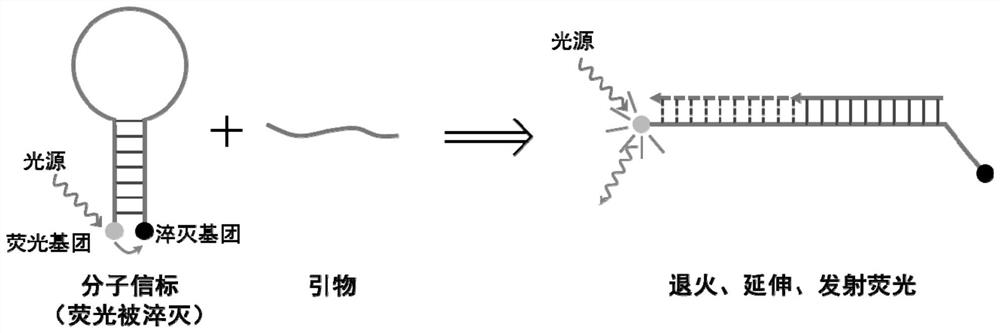
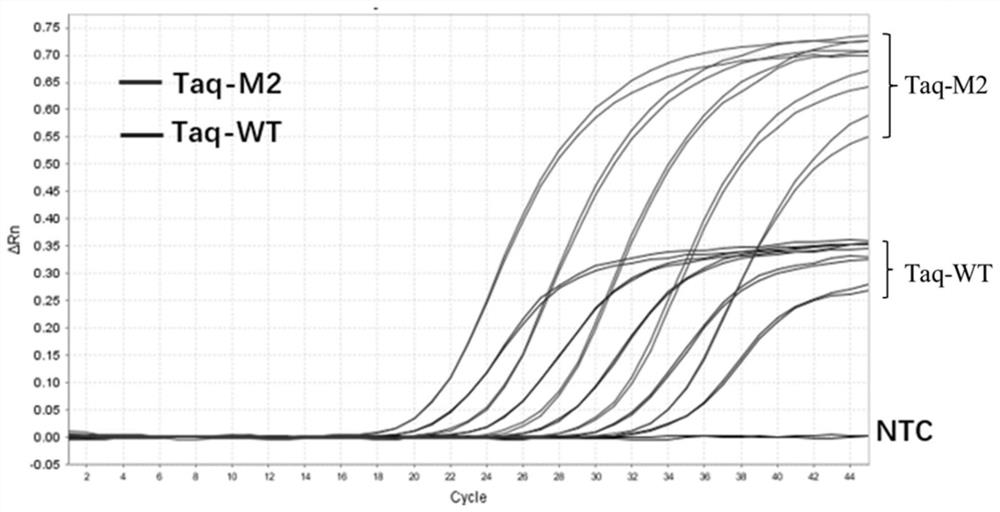
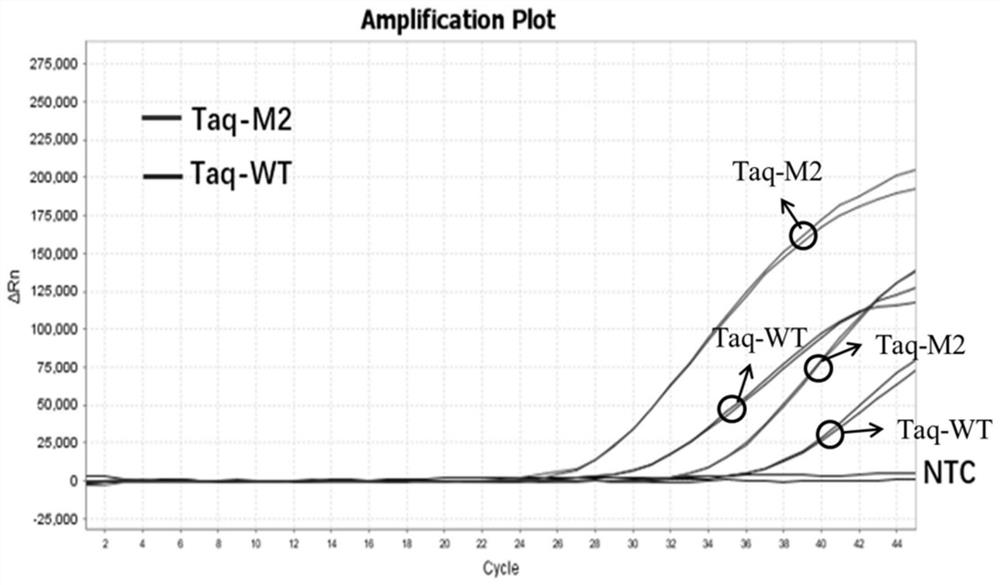
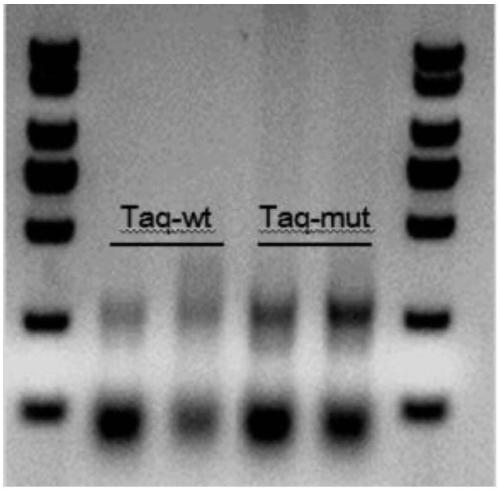
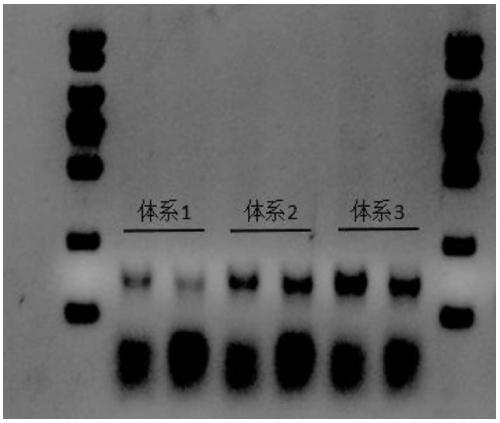
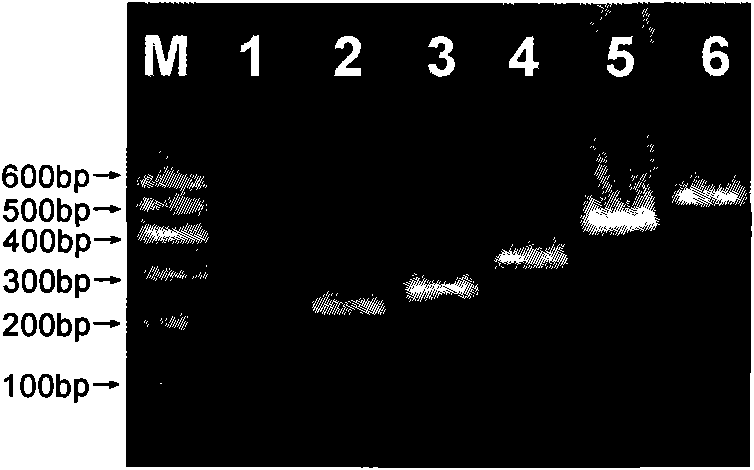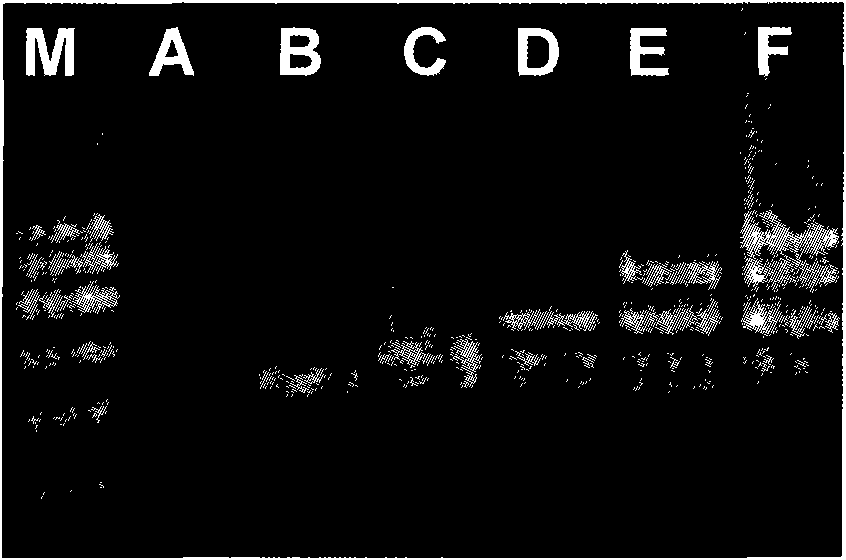Patents
Literature
370 results about "Taq DNA Polymerase" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Eventually, rather than isolating Taq polymerase from Thermus aquaticus cells, the pol gene from that bacteria was isolated and cloned to produce its genome in Escherichia coli (E. coli) cells. While newer thermostable DNA polymerases have been discovered, Taq polymerase remains the standard for PCR.
Reagents for reversibly terminating primer extension
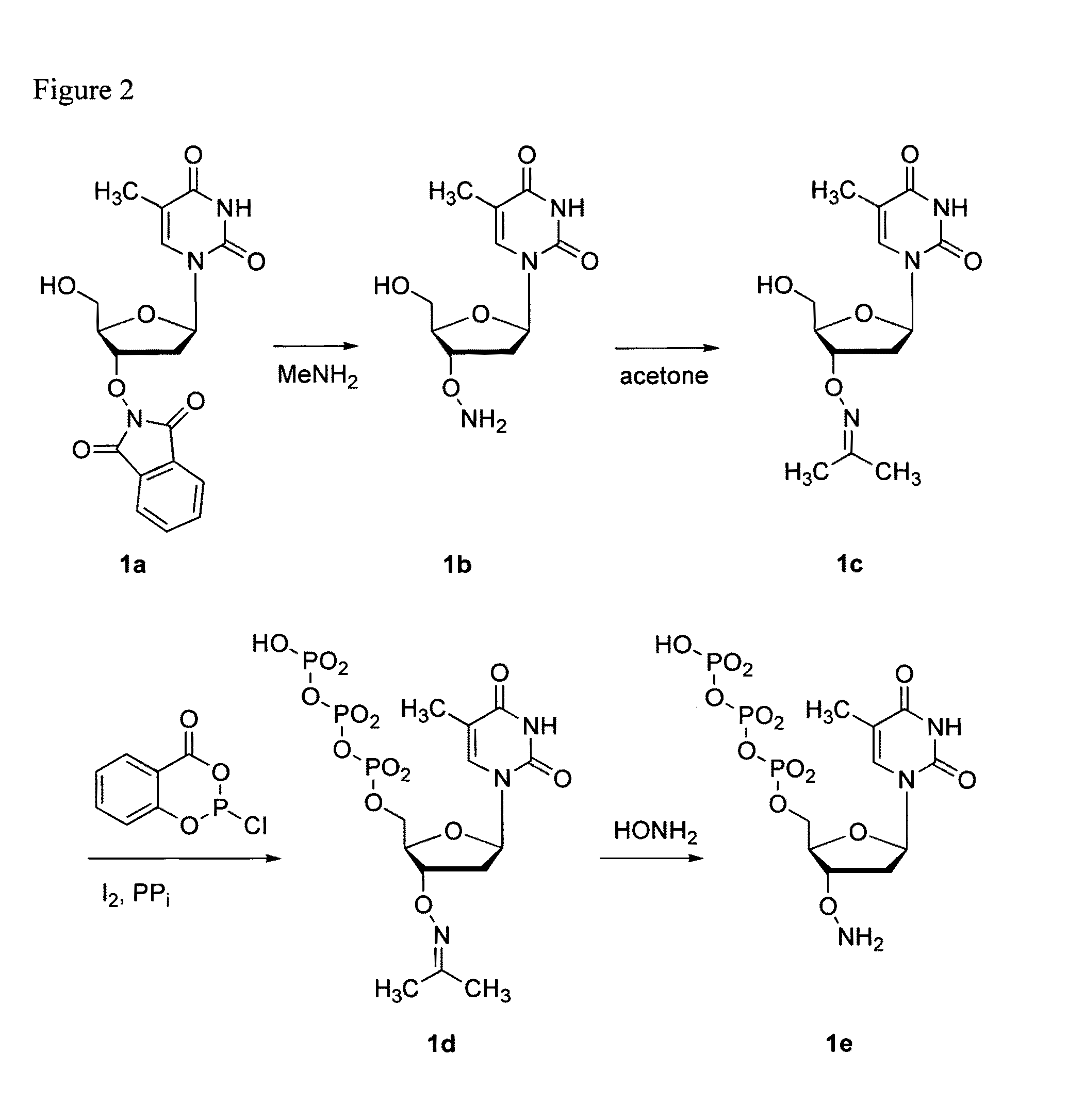
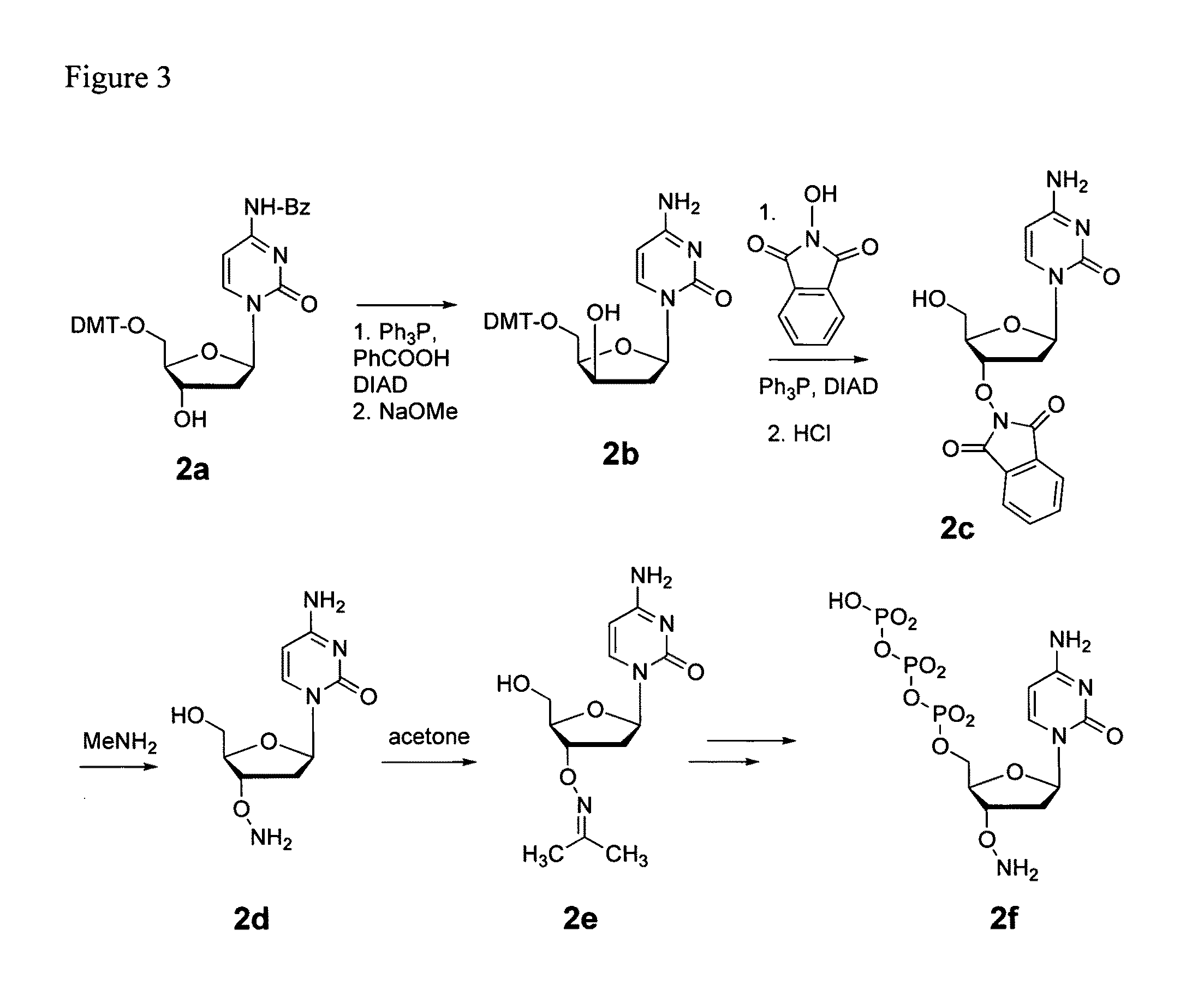
Processes are disclosed that use 3′-reversibly terminated nucleoside triphosphates to analyze DNA for purposes other than sequencing using cyclic reversible termination. These processes are based on the unexpected ability of terminal transferase to accept these triphosphates as substrates, the unexpected ability of polymerases to add reversibly and irreversibly terminated triphosphates in competition with each other, the development of cleavage conditions to remove the terminating group rapidly, in high yield, and without substantial damage to the terminated oligonucleotide product, and the ability of reversibly terminated primer extension products to capture groups. The presently preferred embodiments of the disclosed processes use a triphosphate having its 3′-OH group blocked as a 3′-ONH2 group, which can be removed in buffered NaNO2 and use variants of Taq DNA polymerase, including one that has a replacement (L616A).
Owner:BENNER STEVEN ALBERT +3
Thermostable type-a DNA polymerase mutants with increased polymerization rate and resistance to inhibitors
The present invention provides mutants of DNA polymerases having an increased rate of incorporation of nucleotides into nucleic acids undergoing polymerization and having an enhanced resistance to inhibitors of DNA polymerase activity. The mutant polymerases are well suited for fast PCR applications, for PCR amplification of targets in samples that contain inhibitors of wild-type polymerases, and for fast PCR amplification of samples containing DNA polymerase inhibitors. In exemplary embodiments, the mutants are mutants of Taq DNA polymerase.
Owner:AGILENT TECH INC
Fluorescent quantitative PCR (Polymerase Chain Reaction) detection reagent, kit and detection method for African swine fever virus (ASFV)
ActiveCN103757134AImprove throughputNo biological safety hazardMicrobiological testing/measurementDimethyl pyrocarbonateBiology
The invention discloses a fluorescent quantitative PCR (Polymerase Chain Reaction) detection reagent, a kit and a detection method for an African swine fever virus (ASFV). The detection reagent is targeted to a conserved segment of an ASFV VP72 gene, and comprises a pair of specific primers, a specific probe and an internal labeling probe. The kit comprises a PCR mixed solution, Taq DNA polymerase, DEPC (diethylpyrocarbonate)-H2O, an ASFV positive quality product, an ASFV negative quality product, a quantification standard substance and a pseudovirus internal labeling solution, wherein the PCR mixed solution comprises a pair of specific primers, a specific probe and an internal labeling probe. The detection method comprises the steps of extraction of total DNAs, preparation of reaction components, making of a standard curve by diluting the standard substance, amplifying and result judgment. The reagent and the kit have the advantages of high specificity, high sensitivity, no pollution and high flux, and can be used for accurately detecting infection, inapparent infection or continuous carrying of a toxic host of low-content ASFV. The detection method is quick, specific and sensitive, and can be used for quantitatively detecting a large quantity of samples at the same time.
Owner:深圳澳东检验检测科技有限公司
DNA Polymerases
ActiveUS20130034879A1Increase speedHigh affinityTransferasesDirected macromolecular evolutionPolymerase LDNA Polymerase Inhibitor
A DNA polymerase mutant comprising a Taq DNA polymerase amino acid sequence with a mutation at one or more of the following selected amino acid positions: E189K, E230K, E507K, H28R, L30R, G38R, F73V, H75R, E76A, E76G, E76K, E90K, K206R, E315K, A348V, L351F, A439T, D452N, G504S, E507A, D551N, L552R, I553V, D578N, H676R, Q680R, D732G, E734G, E734K, F749V; wherein the polymerase mutant exhibits relative to wild-type DNA polymerase increased polymerase speed, increased affinity to DNA substrate and / or increased resistance to a DNA polymerase inhibitor; and wherein, when the mutation is E507K in combination with two or more further mutations or the mutation is Q680R in combination with four or more further mutations, at least one of the further mutations is at one of the selected amino acid positions; and when the mutation is I553V, this is not in combination with D551S.
Owner:THERMO FISHER SCI BALTICS UAB
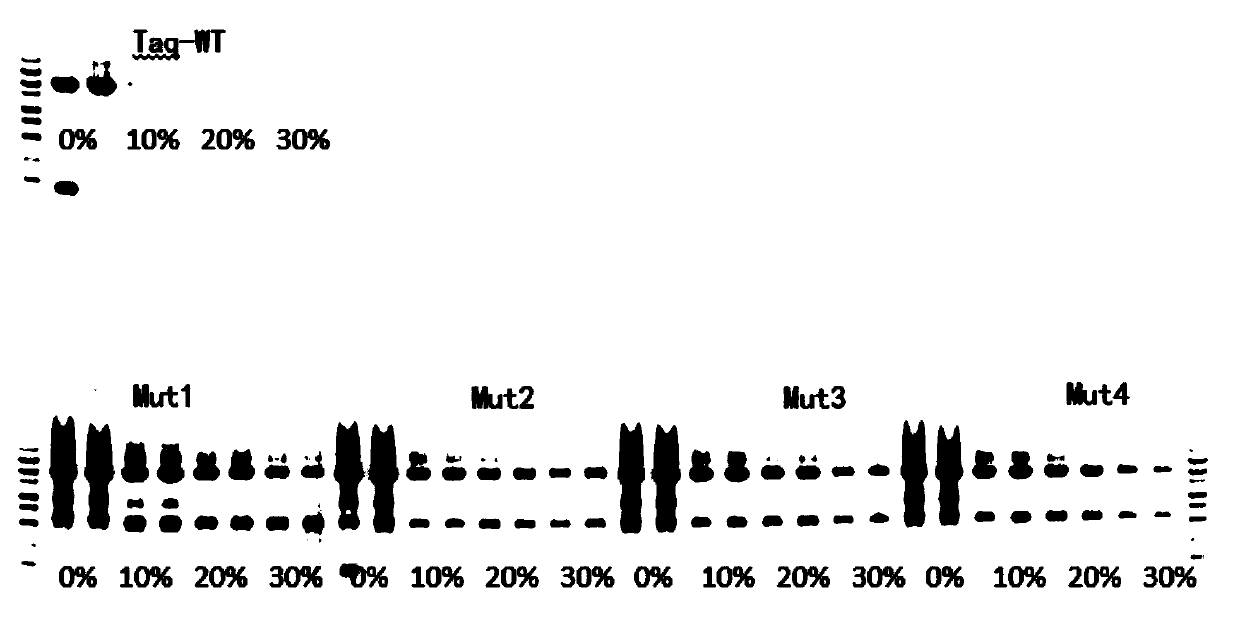
Mutant Taq DNA (deoxyribonucleic acid) polymerase, method for preparing same and application of mutant Taq DNA polymerase
The invention discloses a mutant Taq DNA (deoxyribonucleic acid) polymerase. Amino acid sequences of a wild Taq DNA polymerase are mutated, particularly, 605th leucine of the wild Taq DNA polymerase is mutated to obtain arginine, 617th arginine of the wild Taq DNA polymerase is mutated to obtain phenylalanine, 667th phenylalanine of the wild Taq DNA polymerase is mutated to obtain leucine, and 480th-485th amino acid of thumb zones of the Taq DNA polymerase is replaced by TBD zones, namely, 262nd-337th amino acid of T3 DNA polymerase. The mutant Taq DNA polymerase has the advantages that the activity, the tolerance and the like of the mutant Taq DNA polymerase can be obviously improved owing to site-directed mutation; the mutant Taq DNA polymerase with the high enzyme activity and the high tolerance can be used in direct amplification PCR (polymerase chain reaction) systems without nucleic acid extraction. The invention further discloses a method for preparing the mutant Taq DNA polymerase and application thereof.
Owner:FAPON BIOTECH INC +1
Human STRtyper PCR amplification fluorescence detection reagent kit
ActiveCN101144774AOptimize detection resultsImprove applicabilityMicrobiological testing/measurementMaterial analysis by optical meansFluorescenceDNA database
The invention discloses a human STRtyper PCR enlarged fluoroscopic examination reagent kit with quick speed detecting, accuracy, good repeatability, good versatility, low price and practicality. The applicability of the reagent kit for Chinese is remarkable in particular. The serial products of the invention comprise the three following kits: (1) a STRtyper-20G kit with the largest STR gene locus of the global and most powerful recognizing ability; (2) a STRtyper-16GC kit which is more suitably used to build a criminal DNA database of Chinese population and identify the identity; (3) a STRtyper-10F / G kit with brand new STR gene locus. Each kit product comprises the component of 1.1 ml * 1 tube of STR PCR reaction mixture; 0.55 ml * 1 tube of STRtyper primer mixture; 120 microlitre * 1 tube of HS-Taq DNA polymerase; 0.3ml * 1 tube of STR control DNA; 50 microlitre * 1 tube of STRtyper allelomerphic gene mixture; 0.5 ml * 1 tube of internal standard; 30 microlitre* 1 tube of 310 spectrum correcting standard; 30 microlitre * 1 tube of 3100 spectrum correcting standard.
Owner:NINGBO HEALTH GENE TECHNOLOGIES CO LTD
High-specificity kit for detecting deafness predisposing genes
InactiveCN102534031AAvoid false positivesMicrobiological testing/measurementTotal DeafnessFluorescence
The invention discloses a fluorescent detection kit for detecting 12 deafness predisposing genes simultaneously. The kit can detect 12 mutational hotspots in the most common deafness associated genes of the Chinese in 3 hours. The kit comprises reagents before amplification and reagents after amplification, wherein the reagents before amplification comprise a polymerase chain reaction (PCR) buffer solution, a reaction mixture of MgCl2 and deoxyribonucleoside triphosphates (DNTPs), Taq DNA polymerase, ultrapure water, and a primer mixture for amplifying loci of detection sites at high specificity; and the reagents after amplification comprise a genotyping standard and an internal standard. Deafness gene loci are simultaneously detected at high sensitivity and high specificity by combining a fluorescent labeling technology, a linolenic acid (LNA) nucleoside monomer doping-primer modification technology and a capillary electrophoresis technology for the first time, manpower and material resources and time are greatly saved, and pollution due to multi-step operation is prevented.
Owner:万戈江
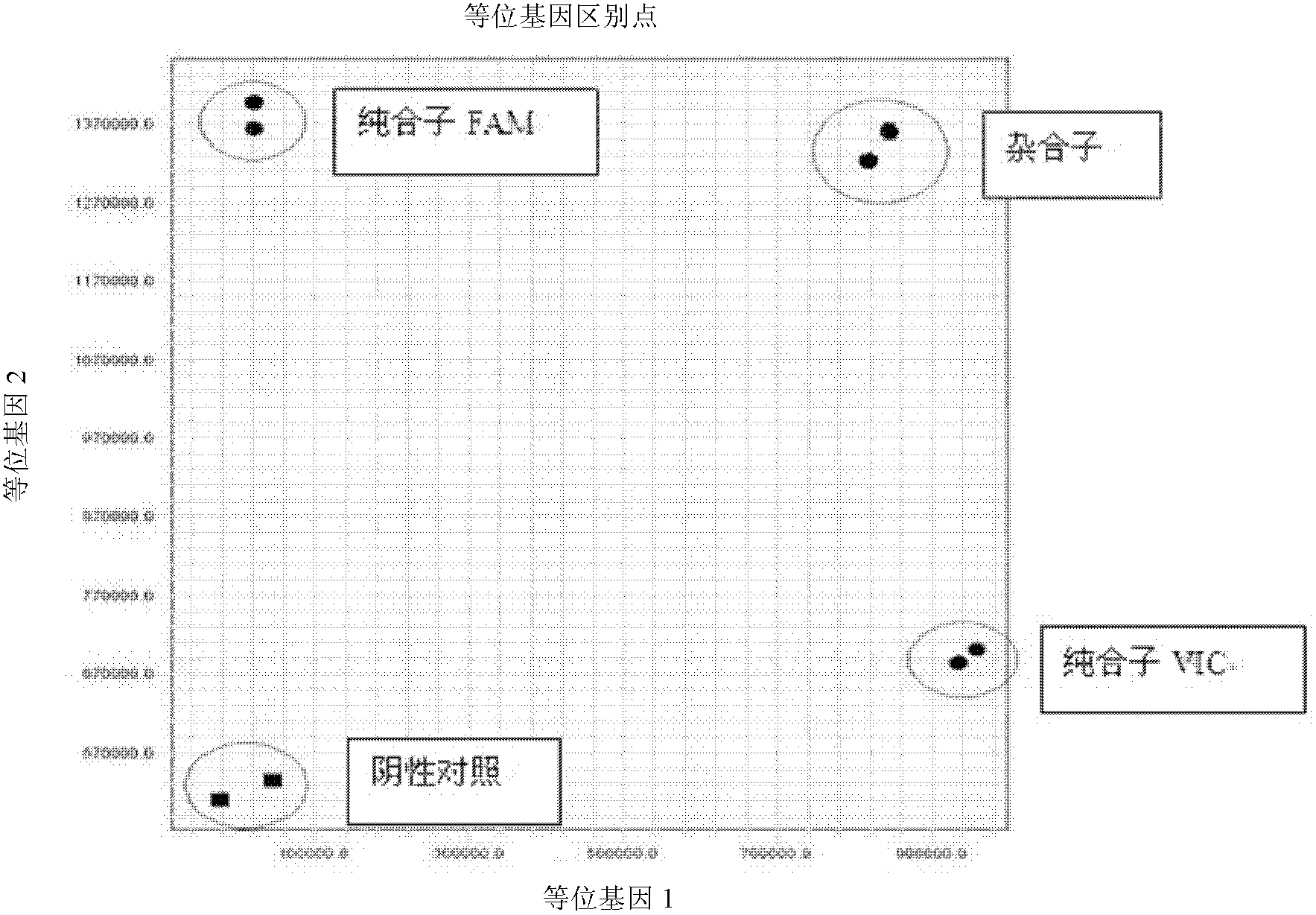
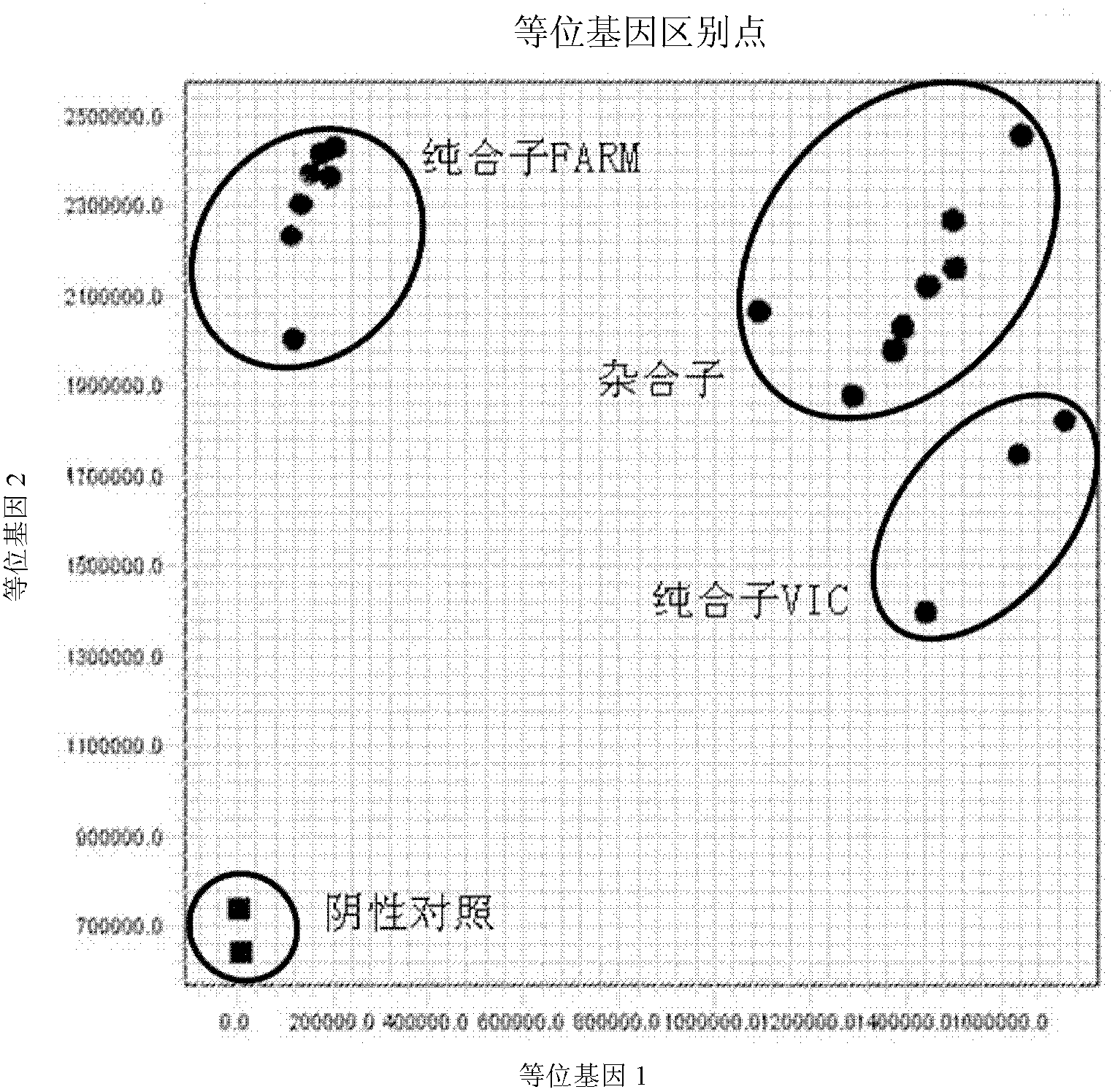
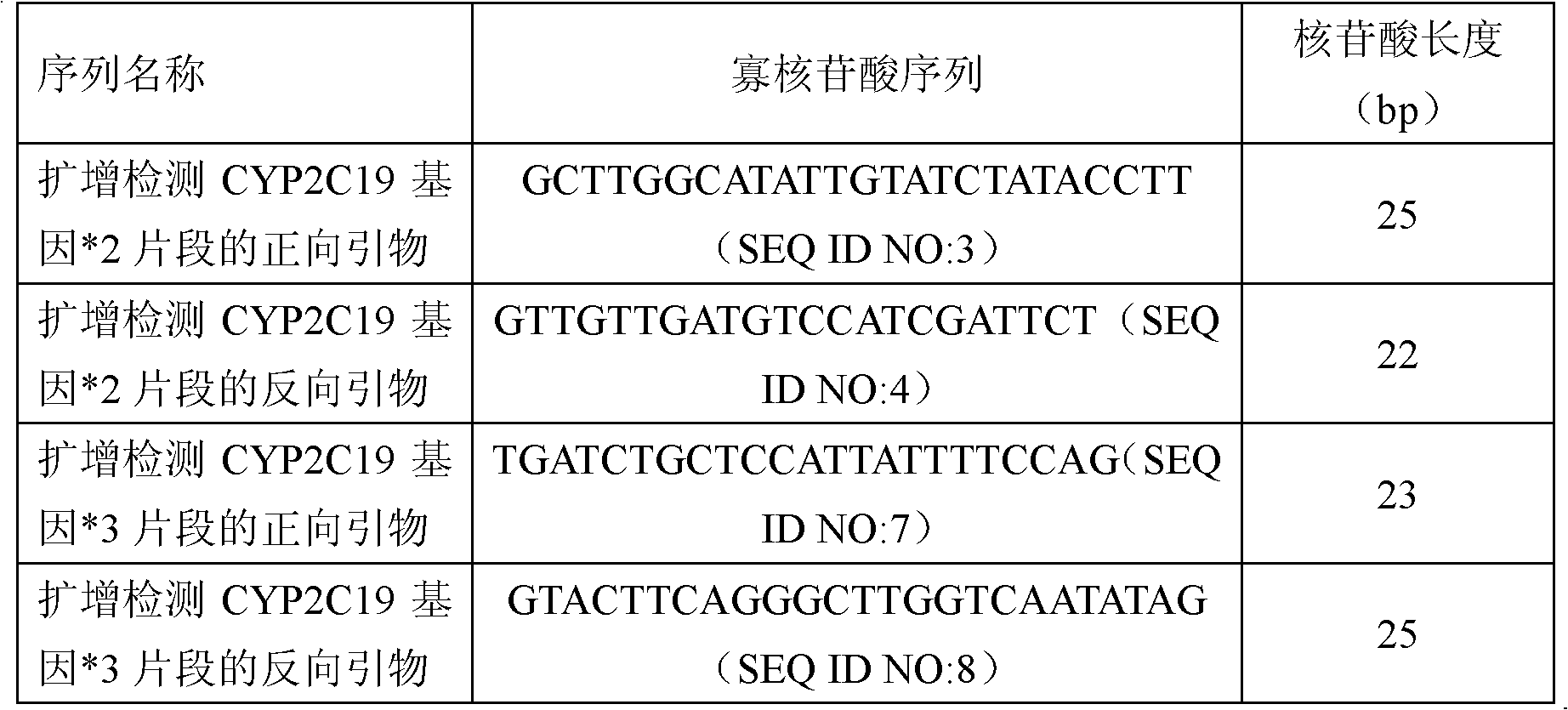
Fluorescent polymerase chain reaction (PCR) kit for detecting CYP2C19 genotypes
InactiveCN102534005AIncreased sensitivityImprove featuresMicrobiological testing/measurementProtein detectionNucleotide
The invention provides a fluorescent polymerase chain reaction (PCR) kit for detecting CYP2C19 genotypes, and belongs to the field of in-vitro nucleic acid testing. The fluorescent PCR kit comprises PCR reaction liquids for detecting CYP2C19*2 and CYP2C19*3 genotypes, Taq DNA polymerase, and uracil-DNA glycosylase, wherein the PCR reaction liquids for detecting the CYP2C19*2 and CYP2C19*3 genotypes respectively comprise PCR amplification primers, minor groove binder (MGB) probes and the like; and nucleotide sequences for detecting the CYP2C19*2 and CYP2C19*3 genotypes are shown as SEQ ID NO:3-4 and SEQ ID NO:5-6 respectively. The kit has high sensitivity and specificity, can monitor the reaction progress in real time, ensures short reaction time, avoids subsequent treatment, can avoid reaction product pollution to the greatest extent, and can replace the traditional protein detection or the common PCR detection to diagnose the CYP2C19 genotypes.
Owner:CHANGSHA 3G BIOTECH
Mutant TaqDNA polymerase and purification method thereof
ActiveCN108265039AImprove thermal stabilityStrong anti-inhibitory effectBacteriaMicrobiological testing/measurementEscherichia coliPurification methods
The invention discloses a mutant TaqDNA polymerase and a purification method thereof in escherichia coli. By adoption of a modern gene engineering technology, a natural TaqDNA polymerase is mutated toremove 5'-3' DNA exonuclease activity, and E507R, E742A and A743H mutants are introduced, wherein amino acid positions are determined according to a wild type TaqDNA polymerase. Compared with the wild type Taq DNA polymerase, the mutant Taq DNA polymerase has advantages of high thermal stability and resistance to inhibiting effects caused by impure components in biological samples to a polymerase, thereby meeting scientific research and clinical application requirements. In addition, in a purification process, by adoption of a specific affinity column which is simple, convenient, quick and high in yield, the obtained mutant TaqDNA polymerase is high in quality, low in cost and quite suitable for industrial large-scale production.
Owner:天津强微特生物科技有限公司
Quantitative PCR detection method based on gold nanoparticles
ActiveCN108342459ARealize highly sensitive quantitative analysisEfficient detectionMicrobiological testing/measurementDNA/RNA fragmentationModified dnaDynamic light scattering
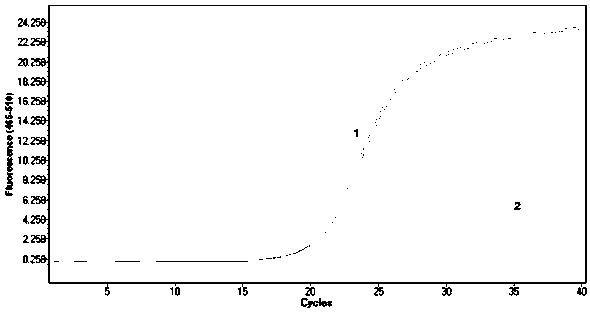
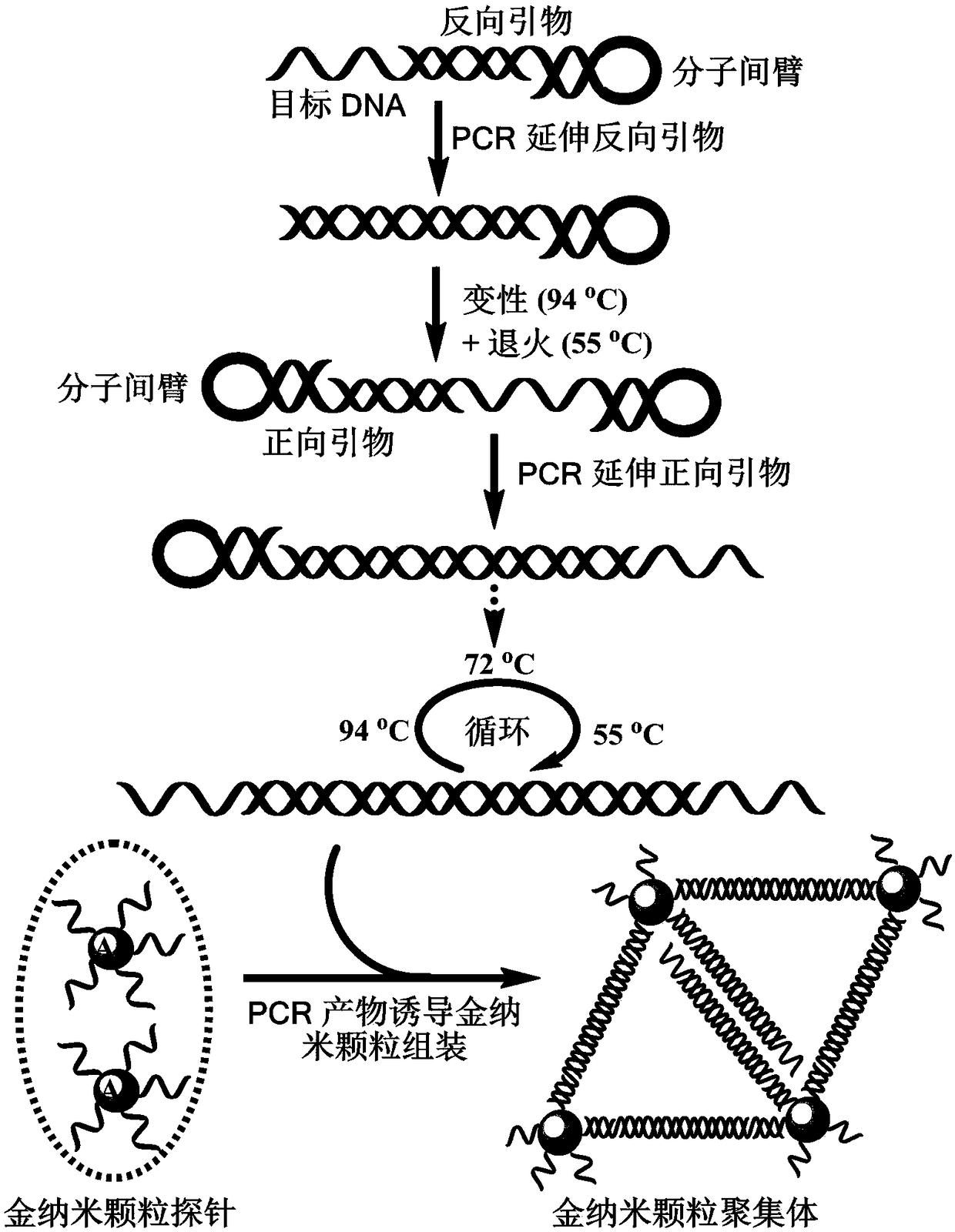
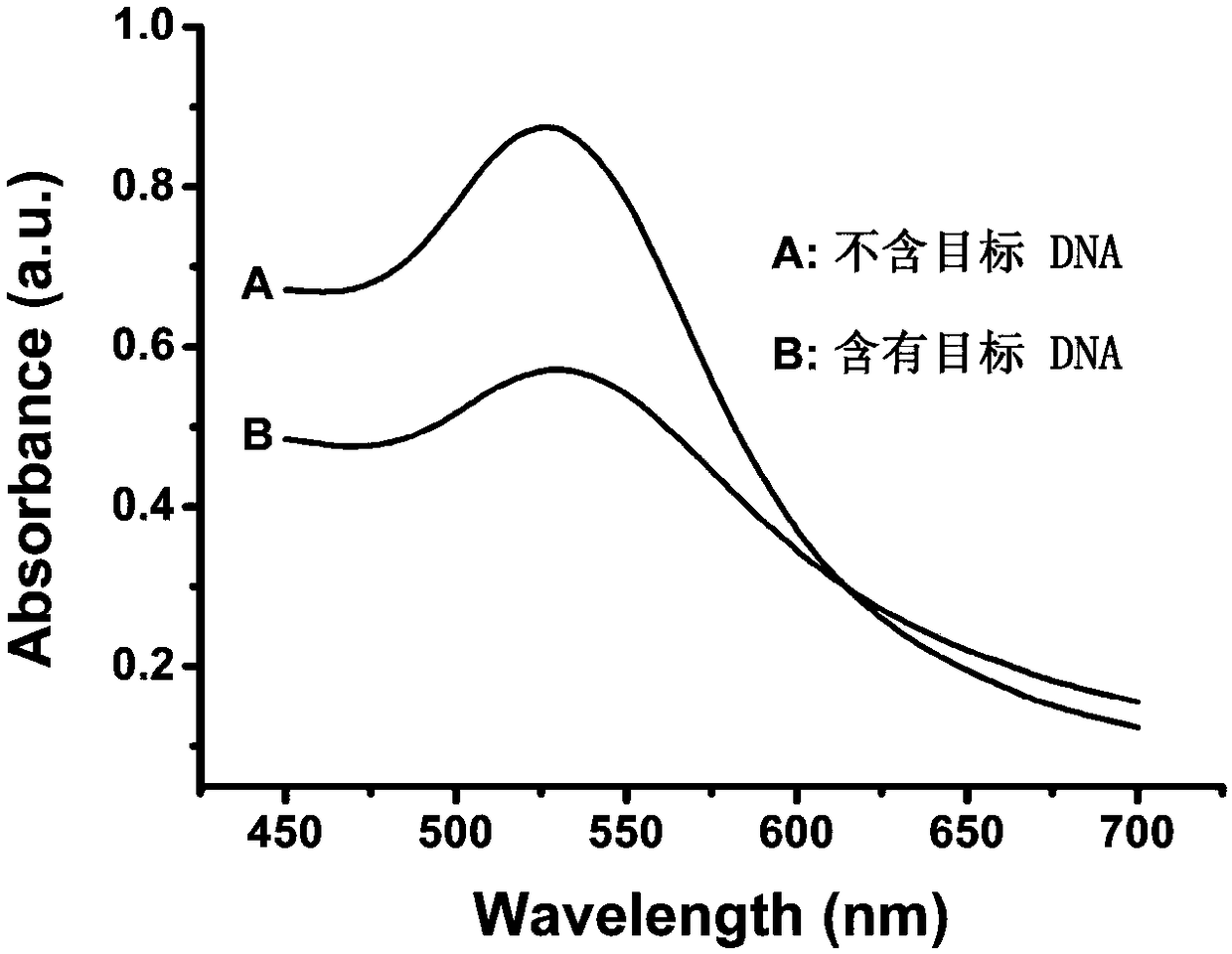
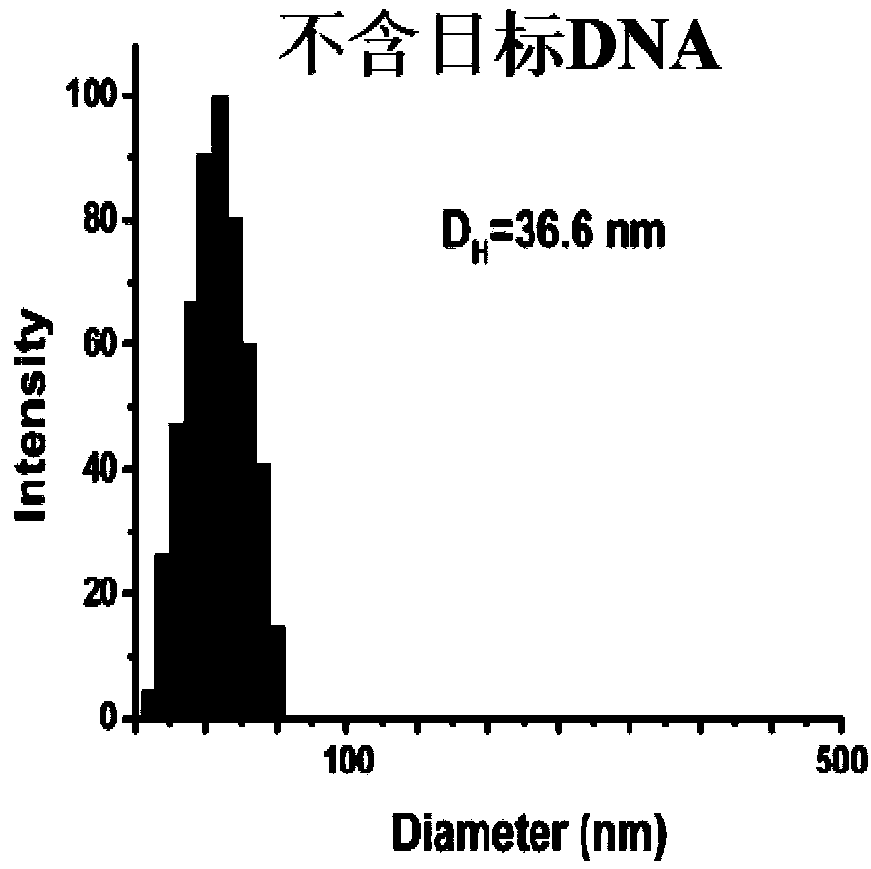
The invention provides a design method of amplification primers. The design method is characterized in that the amplification primers adopting hairpin structures are designed and single-stranded DNA fragments are formed at the two ends of an amplification product. The invention provides two gold nanoparticle probes; DNA sequences modified by the probes are complementary to the single-stranded DNAfragments at the two ends of a PCR product of DNA to be detected. The invention provides a quantitative PCR detection method based on gold nanoparticles. The quantitative PCR detection method comprises the following steps: (1), selecting a DNA sequence to be detected, and designing a pair of specially-structured amplification primers, wherein the pair of specially-structured amplification primersare designed to be the hairpin structures; (2), performing a PCR reaction in a system containing a DNA template to be detected, the pair of specially-structured amplification primers, Taq DNA polymerase and dNTP to obtain the PCR product; (3), preparing DNA-modified gold nanoparticle probes; (4), adding the probes into the PCR product, and hybridizing with the PCR product; (5), measuring the change of the absorbance or a dynamic light scattering signal of the gold nanoparticles. With the quantitative PCR detection method, the operation is simple, the PCR product can be rapidly detected, and the sensitivity is high.
Owner:SUN YAT SEN UNIV
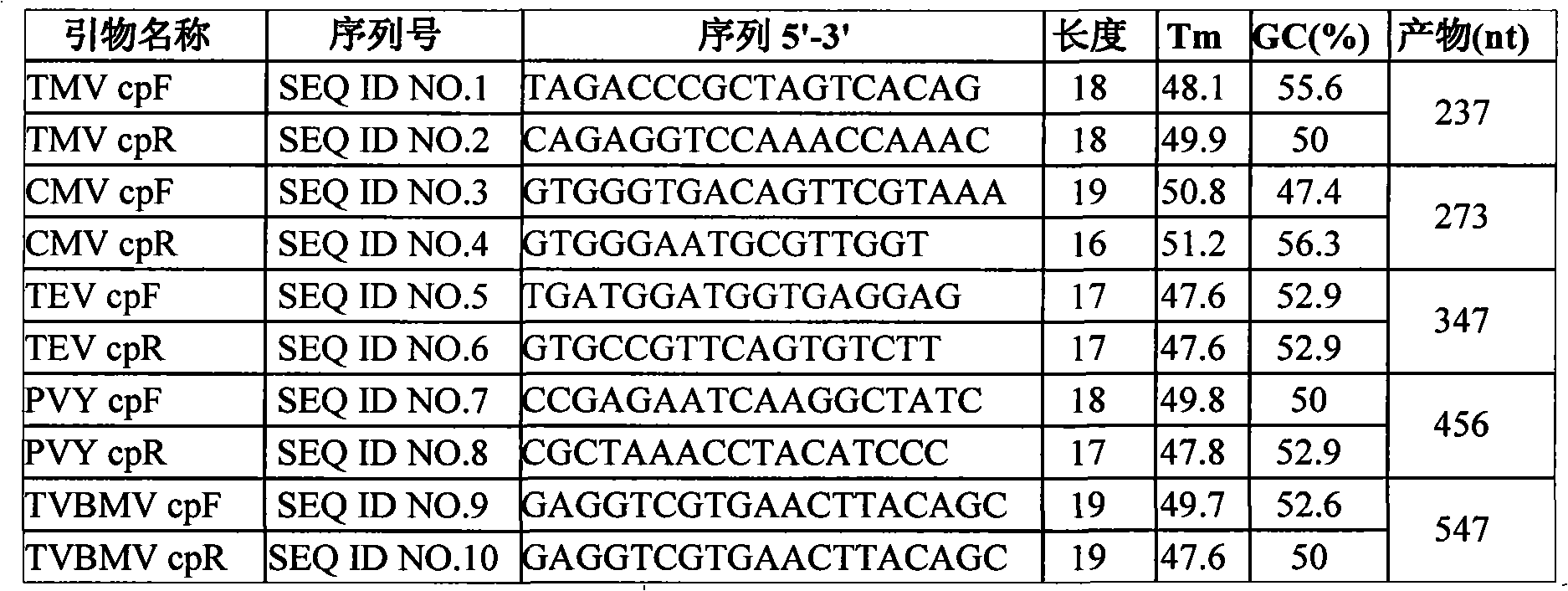
Multiple RT-PCR method for fast detecting four kinds of pepper viruses
ActiveCN105779652AReduce usageEasy to operateMicrobiological testing/measurementMicroorganism based processesDisease epidemiologyConserved sequence
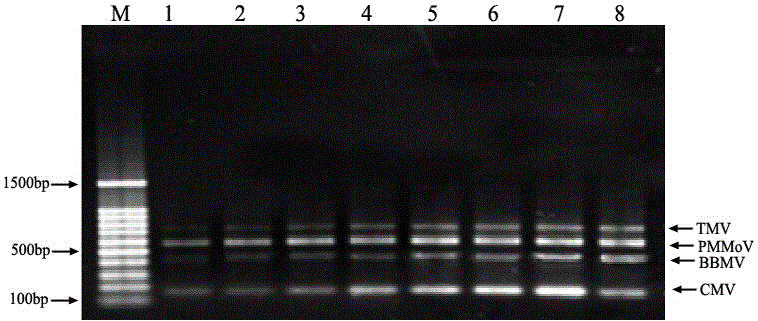
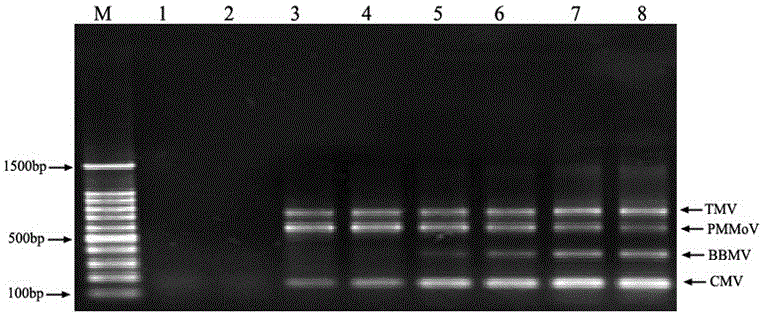
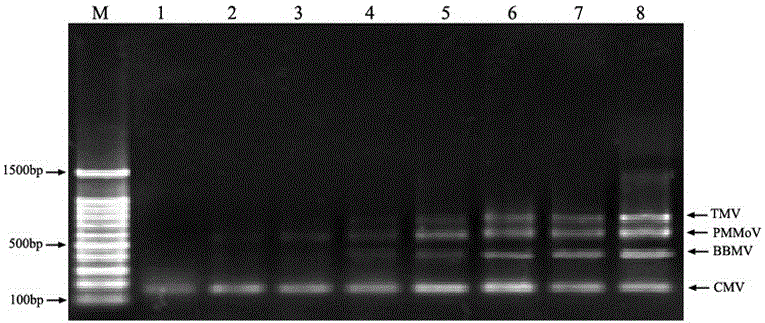
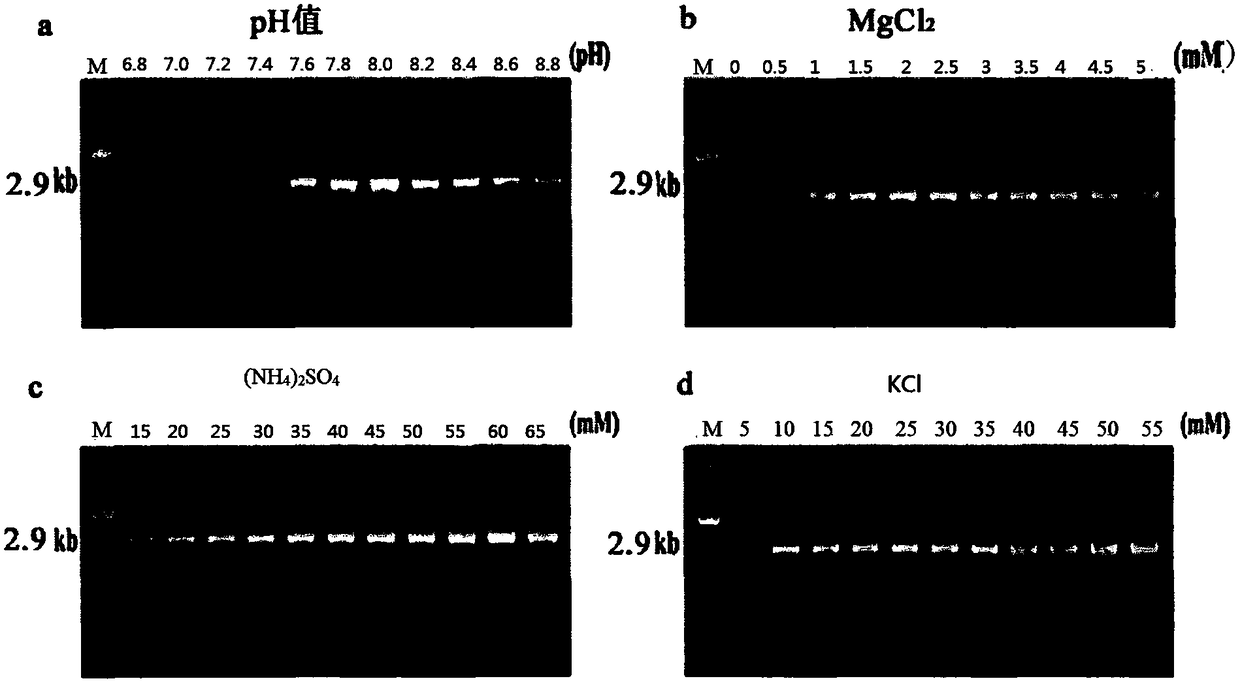
The invention relates to a multiple RT-PCR method for fast detecting four kinds of pepper viruses, and belongs to the field of virus detection.According to the method, specific primer pairs, namely, CMV-F / R, PMMoV-F / R, BBWV-F / R and TMV-F / R are designed and synthesized aiming at the nucleotide conserved sequences of the four kinds of viruses, multiple RT-PCR system optimization is carried out on the primer concentration, Mg2+ concentration, Taq DNA polymerase concentration, dNTPs concentration, annealing temperature and other aspects influencing multiple RT-PCR amplification, and the detecting method capable of detecting field samples compositely infected by the viruses of CMV, PMMoV, BBWV and TMV in pepper tissue at the same time is built.According to the multiple RT-PCR detecting method, the pepper poisoned condition can be detected fast, accurately, conveniently and economically so that effective prevention measures can be taken as early as possible, and very important significance is achieved on control of diseases in the field, reduction of economic losses, screening of disease resistance and predication of disease epidemiology.
Owner:XINJIANG AGRI UNIV
Mixed type hot-start DNA polymerase composition, PCR amplification kit, and applications thereof
ActiveCN108546749AImprove heat resistanceLow mismatch rateMicrobiological testing/measurementBiotechnologyWild type
The invention discloses a mixed type hot-start DNA polymerase composition, a PCR amplification kit, and applications thereof. The composition comprises Taq DNA polymerase, an anti-Taq antibody, and mutant type KOD DNA polymerase. The anti-Taq antibody is capable of specifically being combined with Taq DNA polymerase. The 147th site of wild type KOD DNA polymerase is histidine, and the 147th site of mutant type KOD DNA polymerase is lysine. The experiment results show that the mismatch rate is low when the mixed type hot-start DNA polymerase composition is used for PCR amplification, the composition has the dual characteristics of high efficient amplification and high fidelity and is heat resistant in amplification, and the amplification effect is good no matter for a long DNA segment, a template with a low amount, or a template with a high GC content.
Owner:深圳市艾伟迪生物科技有限公司
Recombinant Taq DNA polymerase and preparation method thereof
ActiveCN103602643AImprove expression efficiencyImprove purification efficiencyTransferasesMicroorganism based processesActive proteinGenetic engineering
The invention relates to the technical field of genetic engineering, and particularly relates to a method for preparing a gene recombinant expression enzyme of Thermusaquaticus TaqDNA polymerase. The nucleotide sequence of a gene and the amino acid sequence of an encoded protein are respectively shown as SEQIDNO:1 and SEQIDNO:2. According to the invention, the method comprises the following steps: cloning a gene sequence by using a gene cloning technique, and building a prokaryotic expression vector; by using a recombinant strain of a built TaqDNA polymerase gene, carrying out suspension by using an LB culture solution; after carrying out culturing for 4.5 hours at a temperature of 37 DEG C, adding IPTG (isopropyl-beta-d-thiogalactoside) with a final concentration of 0.5 mM into the obtained substance to induce for 16-20 hours; after thalli are collected in a centrifugal mode, dissociating the thalli by using 2-4 mg / ml lysozyme, so that protein fluid is obtained; and filtering the protein fluid by using a chromatographic column and a dialysis bag so as to obtain high-purity active protein TaqDNA polymerase. Compared with the prior art, stability of a production process is good, activity of obtained TaqDNA polymerase is high, the yield and purity of products are effectively ensured, and amplification effect when being applied to a gene PCR (polymerase chain reaction) is good.
Owner:哲尔基因科技(上海)有限公司
Taq DNA polymerase monoclonal antibody combination as well as reaction system and application thereof
ActiveCN111808197AHigh activityGuaranteed thermal stabilityMicrobiological testing/measurementTransferasesAntiendomysial antibodiesEnzyme
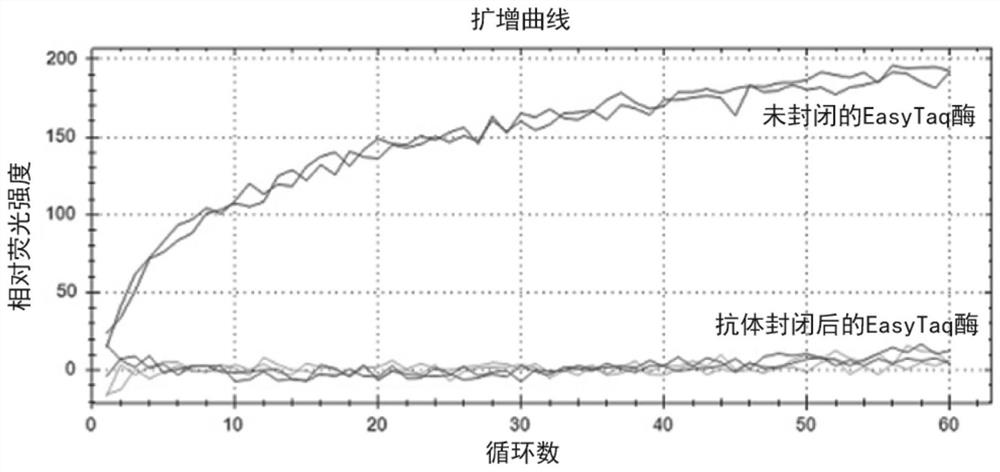
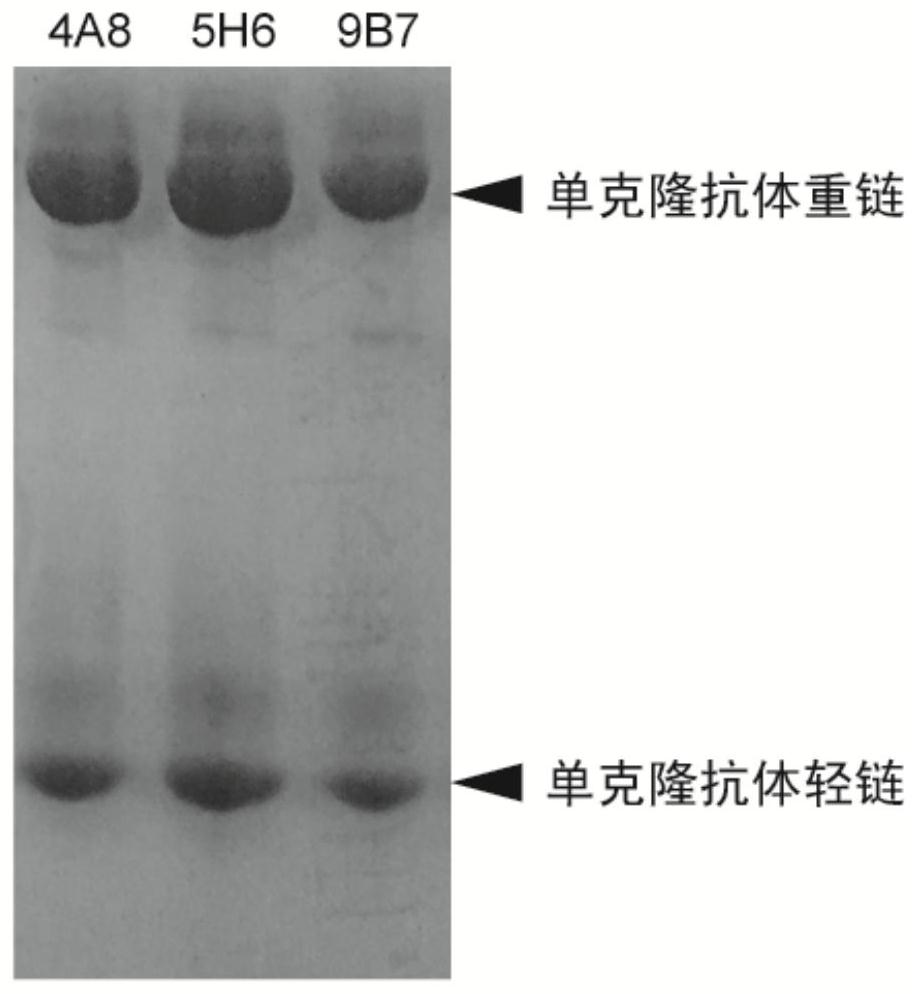
The invention discloses a Taq DNA polymerase monoclonal antibody combination as well as a reaction system and application thereof. The invention firstly discloses the Taq DNA polymerase monoclonal antibody combination. The combination comprises a monoclonal antibody 4A8 and a monoclonal antibody 9B7. The invention further discloses application of the antibody combination in reducing non-specific amplification of Taq DNA polymerase and / or ensuring thermal stability of Taq DNA polymerase, and a reaction system and a kit containing the antibody combination. The antibody combination and the optimized reaction solution can fully seal the active region of Taq DNA polymerase under the condition of low temperature, effectively reduce non-specific amplification and ensure the thermal stability of the enzyme, and can still effectively maintain the durability of the enzyme activity under the condition of higher amplification cycle number.
Owner:BEIJING TRANSGEN BIOTECH CO LTD
Full premix MTHFR and MTRR multiple PCR gene polymorphism detection kit and method thereof
InactiveCN107841537AActivity is not affectedStability is not affectedMicrobiological testing/measurementDNA/RNA fragmentationNucleotidePcr ctpp
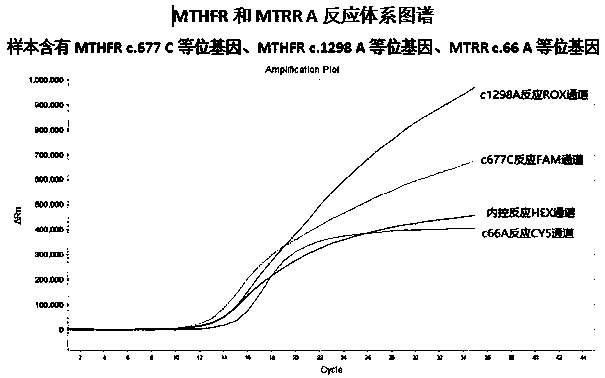
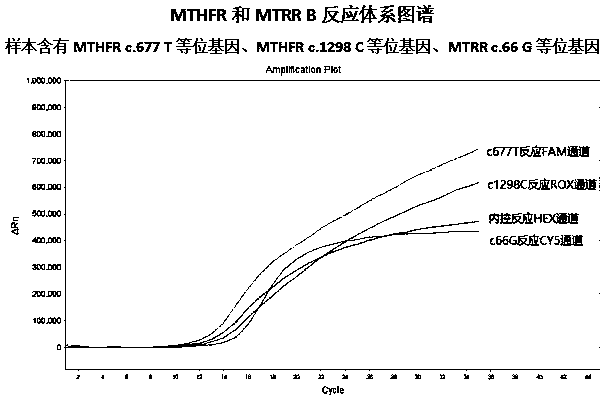
The invention provides a complete premixed multiple primer-specific PCR detection kit and method for detecting nucleotide polymorphisms at three sites of c.677 and c.1298 of the folic acid metabolic pathway and c.1298 of the MTRR gene. "Allele-specific PCR" principle, design multiple primer-specific PCR, realize two-tube reaction on the real-time fluorescent quantitative PCR technology platform, and simultaneously detect three folic acid metabolic pathway MTHFR genes c.677 and c.1298 and MTRR gene c.66 The nucleotide type of the site. In addition, modified Taq DNA polymerase and disaccharide PCR protective agent are added to the reaction system to increase the stability of the reaction system and realize the long-term stability of the pre-mixed detection system.
Owner:SUZHOU KUANGYUAN MOLECULAR BIOTECH
Nanometer PCR detection kit for rapidly identifying and diagnosing virulent virus and attenuated virus of porcine pseudorabies virus, and applications thereof
InactiveCN103224995AMicrobiological testing/measurementMicroorganism based processesPig farmsTime-Consuming
The invention discloses a nanometer PCR detection kit for rapidly identifying and diagnosing virulent virus and attenuated virus of porcine pseudorabies virus, and applications thereof. The kit comprises a 2*Nano PCR buffer, Taq DNA polymerase, a primer mixture, DNA polymerase, and nucleic acid-free ddH2O. In order to achieve a good detection effect, the kit further can contain a template mixture and a stabilizer. With the kit of the present invention, disadvantages of long time consuming, tedious property and low sensitivity of the existing kit are solved, sensitivity is improved, and early stage and rapid virus detection is achieved. The kit of the present invention has characteristics of strong specificity, high sensitivity and the like, is suitable for early stage and rapid diagnosis, and can be provided for identifying gene deletion strains and wild strain infection. In addition, the kit can be widely used in grassroots veterinary detection institutions, and can provide important significance for PRV detection, diagnosis and identifying diagnosis in large-scale pig farms.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
Multi-PCR detection kit for poultry salmonella and non-diagnostic detection method of poultry salmonella
ActiveCN105648055AHigh sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesBiologyMultiplex pcrs
The invention discloses a multi-PCR detection kit for poultry salmonella and a non-diagnostic detection method of the poultry salmonella. The kit comprises 10*PCR buffering liquid, 2.5 U / mu l Taq DNA polymerase, 10 Mm dNTPs, a multi-PCR detection primer group, a positive contrast material and a negative contrast material. The positive contrast material comprises salmonella enteritidis ATCC13076 genome DNA, salmonella typhimurium ATCC14028 genome DNA and salmonella gallinarum ATCC 9184 genome DNA; the negative contrast material is sterilized double distilled water. The invention further discloses a multi-PCR method for detecting the poultry salmonella by applying the kit. The method has the advantages of being rapid, simple, high in specificity and high in sensitivity, three types of salmonella can be detected and classified rapidly through a one-time PCR reaction, compared with traditional serological typing and ordinary PCR detection, great advantages are achieved on the aspect of detection time and detection cost, and the multi-PCR detection kit and the detection method are suitable for batch detection.
Owner:JIANGSU INST OF POULTRY SCI
Mutant type Taq DNA polymerase, nucleic acid extraction-free direct PCR amplification kit and application of mutant type Taq DNA polymerase
ActiveCN108130318AHigh activityImprove toleranceMicrobiological testing/measurementTransferasesBiotechnologyWild type
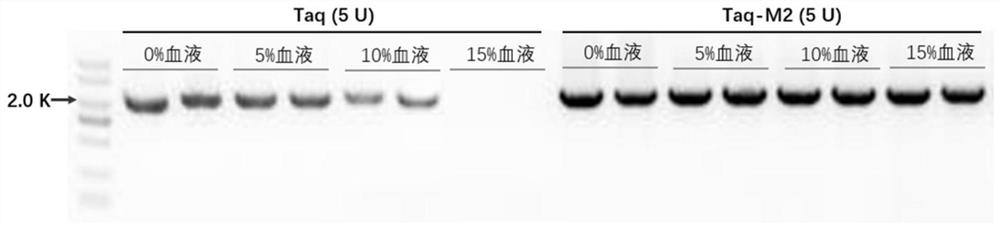
The invention discloses mutant type Taq DNA polymerase, a nucleic acid extraction-free direct PCR amplification kit and an application of the mutant type Taq DNA polymerase. By mutating 709-site glutamic acid of wild Taq DNA polymerase into glutamine, the activity, tolerance and the like of the obtained mutant type Taq DNA polymerase are obviously improved. The mutant type Taq DNA polymerase onlyhas a site, so that the mutation sites are few, and the preparation is easy. Experiment results show that the mutant type Taq DNA polymerase can be applied to a nucleic acid extraction-free direct PCRamplification reaction system, and the nucleic acid extraction-free direct PCR amplification of samples such as blood, foods and soil which contain a large number of inhibiting factors can be realized.
Owner:深圳市艾伟迪生物科技有限公司
Mink heart DNA detection kit and identification method
InactiveCN101434990ASpecificity Accurate Simultaneous DistinctionThe identification method is simpleMicrobiological testing/measurementMinkPositive control
The invention relates to the Chinese medicine detection technology field and belongs to a mink heart DNA detection kit, comprising buffer solution, 12.5mM dNTP, one type of 0.1mM primer 1 or 2, Taq DNA polymerase, sample DNA to be detected, and the identification reaction system of double-distilled water; containing buffer solution, 12.5mM dNTP, one type of 0.1mM primer 1 or 2, Taq DNA polymerase, mink heart DNA, and the positive control reaction system of the double-distilled water; including buffer solution, 12.5mM dNTP, one type of 0.1mM primer 1 or 2, Taq DNA polymerase, mixture of chicken heart, duck heart, goose heart and rabbit heart DNA according to 1:1, and the negative control reaction system of the double-distilled water. The mink heart DNA identification method comprises steps such as designing mink heart mitochondrial DNA two-pair specific oligonucleotide primer, artificially synthetizing mink heart mitochondrial DNA two-pair specific oligonucleotide primer, fixing reaction procedures and result judgment, and the like; the method can simultaneously and accurately differ the specificity of various animal hearts which are easily mixed with the mink heat; and the identification method has the advantages of simplicity, rapidness, and reliable detection result, etc.
Owner:BEIHUA UNIV
Narcissus latent virus detection kit and detection method thereof
ActiveCN102732642AQuick checkAccurate detectionMicrobiological testing/measurementMicroorganism based processesPositive controlReverse transcriptase
The invention relates to a narcissus latent virus detection kit and its detecting method that are special for narcissus latent virus detection. The kit comprises: an upstream primer, a downstream primer, an RT Buffer, an RNA enzyme inhibitor, a reverse transcriptase, dNTPs, a PCR Buffer, Mgcl2, Taq DNA polymerase, a positive control, a negative control and RNase-free ddH2O. In the invention, specific primers are designed according to a coat protein CP gene sequence of the narcissus latent virus, which is detected by a reverse transcription-polymerase chain reaction (RT-PCR) technology. With the obvious advantages of rapidity, accuracy, sensitivity, strong operability and simple kit preparation method, the kit and method of the invention are not only suitable for ports of China to conduct rapid detection on the narcissus latent virus of entry and exit narcissus, and also can be used for narcissus latent virus detection and diagnosis, epidemic surveillance as well as early warning and prediction, thus having broad application prospects.
Owner:INSPECTION & QUARANTINE TECH CENT OF FUJIAN ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Fusion type Taq DNA polymerase as well as preparation method and application thereof
ActiveCN111690626AHigh affinityImprove bindingMicrobiological testing/measurementTransferasesReverse transcriptaseWild type
The invention provides fusion type Taq DNA polymerase as well as a preparation method and application thereof. The fusion type Taq DNA polymerase comprises a Taq DNA polymerase mutant and DNA bindingprotein, wherein the Taq DNA polymerase mutant is protein obtained by carrying out substitution mutation on one or more sites of a helix-hairpin-helix DNA binding region of wild type Taq DNA polymerase. The fusion type Taq DNA polymerase has enhanced nucleic acid binding capacity, amplification speed and impurity tolerance, has good resistance to inhibition of reverse transcriptase, and has good amplification performance for trace templates, and a biological sample can be directly detected without a nucleic acid extraction step; and a constructed one-step RT-qPCR kit is high in detection speed, high in sensitivity and good in accuracy, and has a wide application prospect in the field of RNA virus detection.
Owner:VAZYME BIOTECH NANJING
Taq DNA polymerase mutant and application thereof
InactiveCN109402082AIncrease productionHigh sensitivityTransferasesFermentationBiotechnologyTaq polymerase
The invention provides a Taq DNA polymerase mutant and application thereof. The Taq DNA polymerase mutant is an amino acid sequence formed by insertion, substitution or depletion of one or more aminoacids in an amino acid sequence of Taq DNA polymerase shown as SEQ ID NO. 1, or by addition or deletion of one or more amino acids in the sequence of SEQ ID NO. 1. Compared with Taq DNA polymerase shown as SEQ ID NO. 1, the amino acid sequence has enhanced amplification sensitivity and increased yield. The Taq DNA polymerase mutant has higher sensitivity and yield than original Taq DNA polymeraseand is also suitable for multiple amplification of low-quality samples.
Owner:VAZYME BIOTECH NANJING
Method for analyzing genetic diversity of Amomum tsao-ko by using inter simple sequence repeat (ISSR) reaction system
ActiveCN107164476AOptimizing ISSR reaction conditionsHigh resolutionMicrobiological testing/measurementBiostatisticsAmomum tsao-koGenetic diversity
The invention belongs to the field of molecular biology DNA marker technique and application. The invention discloses a method for analyzing genetic diversity of Amomum tsao-ko by using inter simple sequence repeat (ISSR) reaction system. The ISSR reaction system comprises (per 25 muL) Mg<2+>-free 10*PCR buffer 3.0 muL, Taq DNA polymerase 1.5 U, Mg<2+> 1.5 mmol / L, dNTP 0.25 mmol / L, primers 0.3 mumol / , and template DNA 50 ng. The reaction mixture is amplified by the following procedures: pre-degeneration at 95 DEG C for 5 min, degeneration at 95 DEG for 1 min, annealing at 50-58 DEG C for 1 min, extension at 72 DEG C for 1 min, after 35 cycles, extension at 72 DEG C for 10 min, and storage at 4 DEG C. The method of the invention provides technical guide and theoretical support for Amomum tsao-ko resource identification, genetic diversity analysis and other scientific researches.
Owner:HONGHE COLLEGE
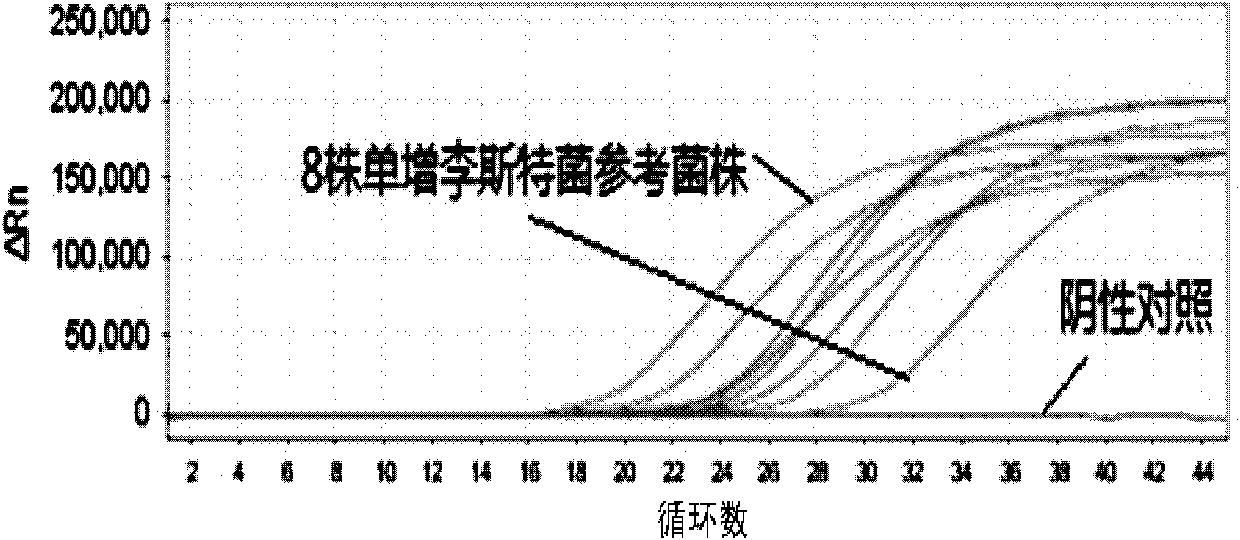
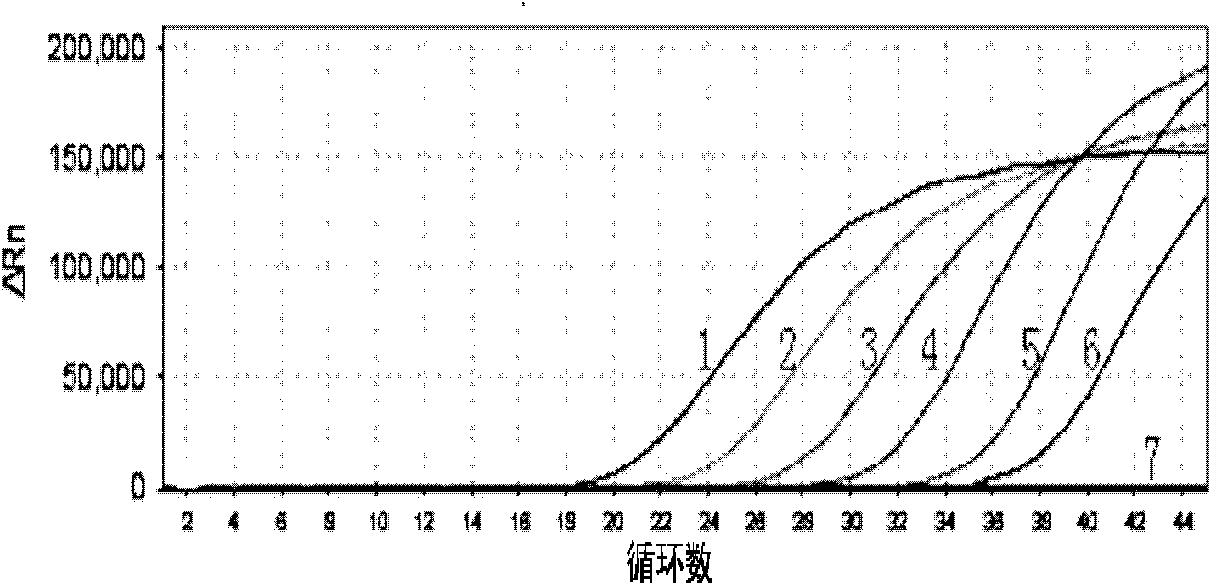
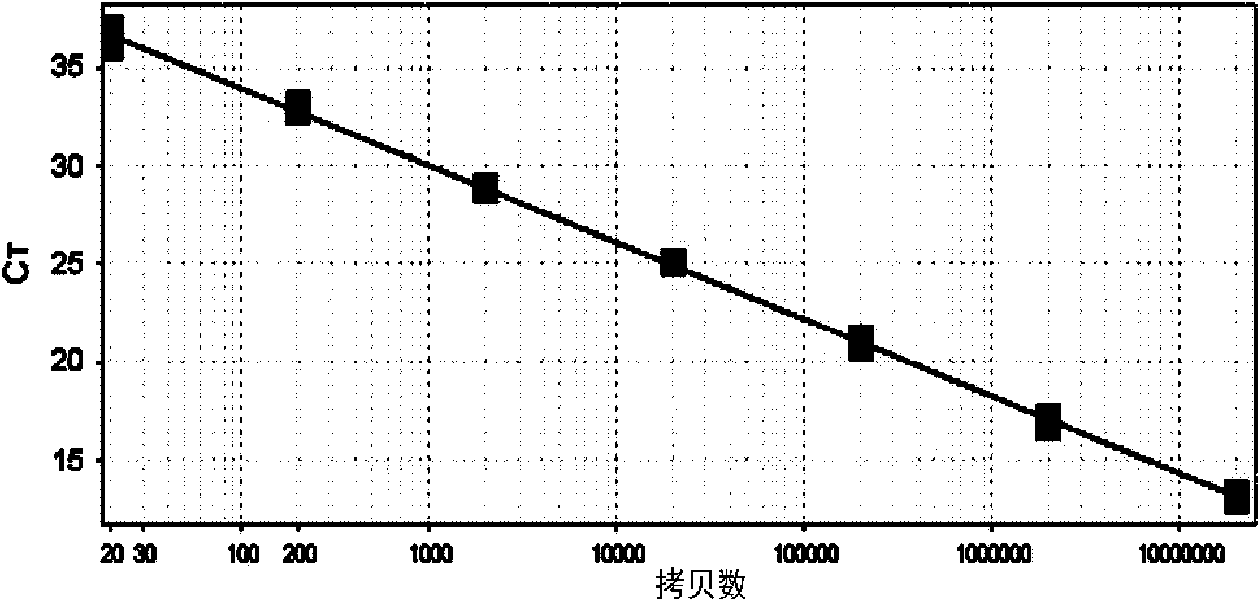
Listeria monocytogenes fluorescence quantitative PCR (Polymerase Chain Reaction) test kit and test method
InactiveCN101805799AHigh detection specificityHigh detection sensitivityMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceA-DNA
The invention relates to a Listeria monocytogenes fluorescence quantitative PCR (Polymerase Chain Reaction) test kit and a test method, belonging to the technical field of food safety. The test kit comprises a fluorescence quantitative PCR reaction liquid, a fluorescent probe, a warm start Taq DNA polymerase, a standard positive templet and a negative quality control standard product. The method comprises the following steps of: extracting a DNA of a sample to be tested with a common method; using the obtained DNA as a template; carrying out amplification to PCR by using the Listeria monocytogenes fluorescence quantitative PCR test kit to obtain reaction products; then placing the reaction products in a quantitative PCR instrument for fluorescence test; and carrying out a qualitative test or a quantitative test on the Listeria monocytogenes in the sample. The test kit of the invention has the advantages of high test specificity, high test sensitivity, accurate quantification and high test speed.
Owner:SHANGHAI JIAO TONG UNIV
Primers and probes for detecting genes associated with schizophrenia, bipolar affective disorders and major depression and kits and preparation methods thereof
InactiveCN102146479AMicrobiological testing/measurementDNA/RNA fragmentationMicrobiologySchizophrenia
The invention discloses a kit for detecting genes associated with schizophrenia, bipolar affective disorders and major depression, which belongs to the field of biotechnology, and consists of a polymerase chain reaction (PCR) reagent group and a ligase detection reaction (LDR) reagent group, wherein the PCR reagent group comprises buffer solution, deoxyribonucleotide triphosphate (dNTP) mixed solution, Taq DNA polymerase, pure water, an upstream primer represented by SEQ ID No.1 and a downstream primer represented by SEQ ID No.2; and the LDR reagent group comprises buffer solution, dNTP mixed solution Taq DNA polymerase, pure water, a probe represented by SEQ ID No.3, a probe represented by SEQ ID No.4 and a probe represented by SEQ ID No.5. In the invention, the operation is simple and convenient, the cost is low, and the kit is developed for bipolar affective disorders. The result is reliable, the stability is high and the sensitivity is high.
Owner:SHANGHAI JIAO TONG UNIV +1
Primer group and kit for synchronously detecting multiple tobacco viruses
InactiveCN101875981AEfficient detectionImprove detection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationReverse transcriptaseMicrobiology
The invention relates to a primer group and a kit for synchronously detecting multiple tobacco viruses. The kit consists of a component I and a component II, wherein the all the reagents in the component I and the component II are separately packed; the component I comprises reverse transcriptase, an RNA enzyme inhibitor, a dNTP mixture, a reverse primer sequence group, and reverse transcription reaction buffer; and the component II comprises PCR reaction buffer, a dNTP mixture, MgCl2, Taq DNA polymerase and a PCR primer group. The primer group is characterized in that: the reverse primer sequence group comprises SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 10; and the PCR primer group comprises SEQ ID NO: 1 to 10.
Owner:SHAANXI TOBACCO RES INST +1
Taq DNA polymerase mutant and application thereof
ActiveCN110747183AImprove tolerancePromote amplificationMicrobiological testing/measurementTransferasesAmino acid substitutionBlood specimen
The invention discloses a Taq DNA polymerase mutant and application thereof. The Taq DNA polymerase mutant has amino acid substitutions at one or more of the following amino acid positions in the sequence shown as SEQ ID NO. 1; and the various amino acid substitutions are represented in triplets as 'letters-numbers-letters', wherein the numbers reflect position of an amino acid mutation, the letters before the numbers are corresponding to an amino acid involved in the mutation, and the letters after the numbers reflect an amino acids used to replace the amino acid corresponding to the lettersbefore the numbers. The Taq DNA polymerase mutant disclosed by the invention is capable of changing configuration of an enzyme, thereby improving tolerance of the enzyme; and thus, the mutant can be very well applied in blood sample amplification.
Owner:VAZYME BIOTECH NANJING
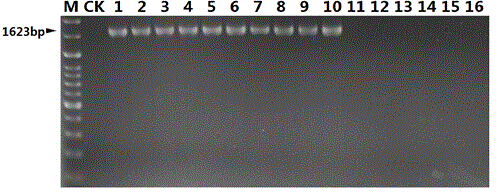
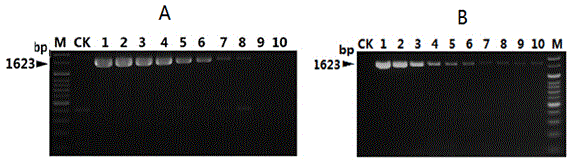
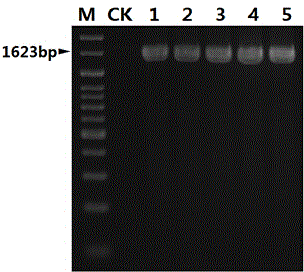
Nucleotide sequence for detecting listeria monocytogenes and detection method and detection kit
InactiveCN104450940AStrong specificityHigh sensitivityMicrobiological testing/measurementMicroorganism based processesMicrobiologyNucleotide sequencing
The invention discloses a nucleotide sequence for detecting listeria monocytogenes and a detection method and a detection kit, wherein the nucleotide sequence is expressed as SEQ ID NO. 1, the detection method is as follows: firstly, extracting the gene group DNA of the sample to be detected; secondly, taking the gene group DNA as the template, mixing with the specific primer Lm16, PCR buffer solution, MgCl2 solution, deoxidation nucleoside triphosphate mixture and Taq-DNA polymerase for preparing the PCR reaction system, carrying out the PCR reaction; finally, detecting whether the PCR product is the single amplification product, the size of which is 1623bp. The kit comprises the specific primer, PCR buffer solution, MgCl2 solution, deoxidation nucleoside triphosphate mixture and Taq-DNA polymerase. The kit can effectively detect the listeria monocytogenes in the food, and the detection sensitivity for the bacteria is high, the specificity is good and the antijamming capability is strong.
Owner:NANJING AGRICULTURAL UNIVERSITY
Human enterovirus four-color fluorescence RT-PCR detection kit and detection method
InactiveCN102851391AEffective monitoringStrong specificityMicrobiological testing/measurementMicroorganism based processesForward primerReverse transcriptase
The invention discloses a human enterovirus four-color fluorescence RT-PCR detection kit and a detection method; the detection kit comprises multiple fluorescence RT-PCR reaction mother liquor, AMV reverse transcriptase liquid, Taq DNA polymerase liquid, a positive control, and a negative control; the detection kit is characterized by further comprising an internal control, a multiple 10*PCR reaction buffer, bovine serum albumin with a concentration of 20 mg / ml, various forward primers and reverse primers with a concentration of 100 muM, and various specific probes with a concentration of 50 muM, wherein sequences of the forward primers, reverse primers, specific probes and the internal control are as shown in SEQIDNO. 1, NO. 2, NO.3, NO.4, NO.5, NO.6, NO.7. NO.8, NO.9, NO.10., NO.11., NO.12., and NO.13. The detection kit of the invention has the advantages of high detection sensitivity, good specificity, accuracy, high speed, and no false positivity.
Owner:NINGBO ACAD OF SCI & TECH FOR INSPECTION & QUARANTINE
Method for purifying recombinant escherichia coli Taq DNA polymerase
InactiveCN104560908AEconomical to useReduce sheddingTransferasesMicroorganism based processesEscherichia coliPurification methods
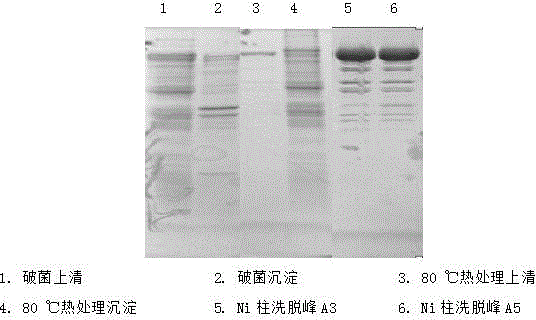
The invention discloses a method for purifying recombinant Escherichia coli Taq DNA polymerase. The method for purifying the recombinant Escherichia coli Taq DNA polymerase comprises the following steps: (1) breaking bacteria; (2) carrying out thermal treatment; (3) salting out; (4) carrying out Ni ion metal-chelated affinity chromatography; (5) dialyzing; and (6) obtaining preparation and carrying out split charging. The method for purifying the recombinant Escherichia coli Taq DNA polymerase has the advantages that a preparation technique is simple, easy and quick, quality of the obtained recombinant Escherichia coli Taq enzyme is high, cost is low, and the method for purifying the recombinant Escherichia coli Taq DNA polymerase is applicable to industrial large-scale production.
Owner:天津强微特生物科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com