Quantitative PCR detection method based on gold nanoparticles
A technology of gold nanoparticles and detection methods, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc. The success rate is not ideal and other problems, to achieve the effect of rapid detection, good versatility, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
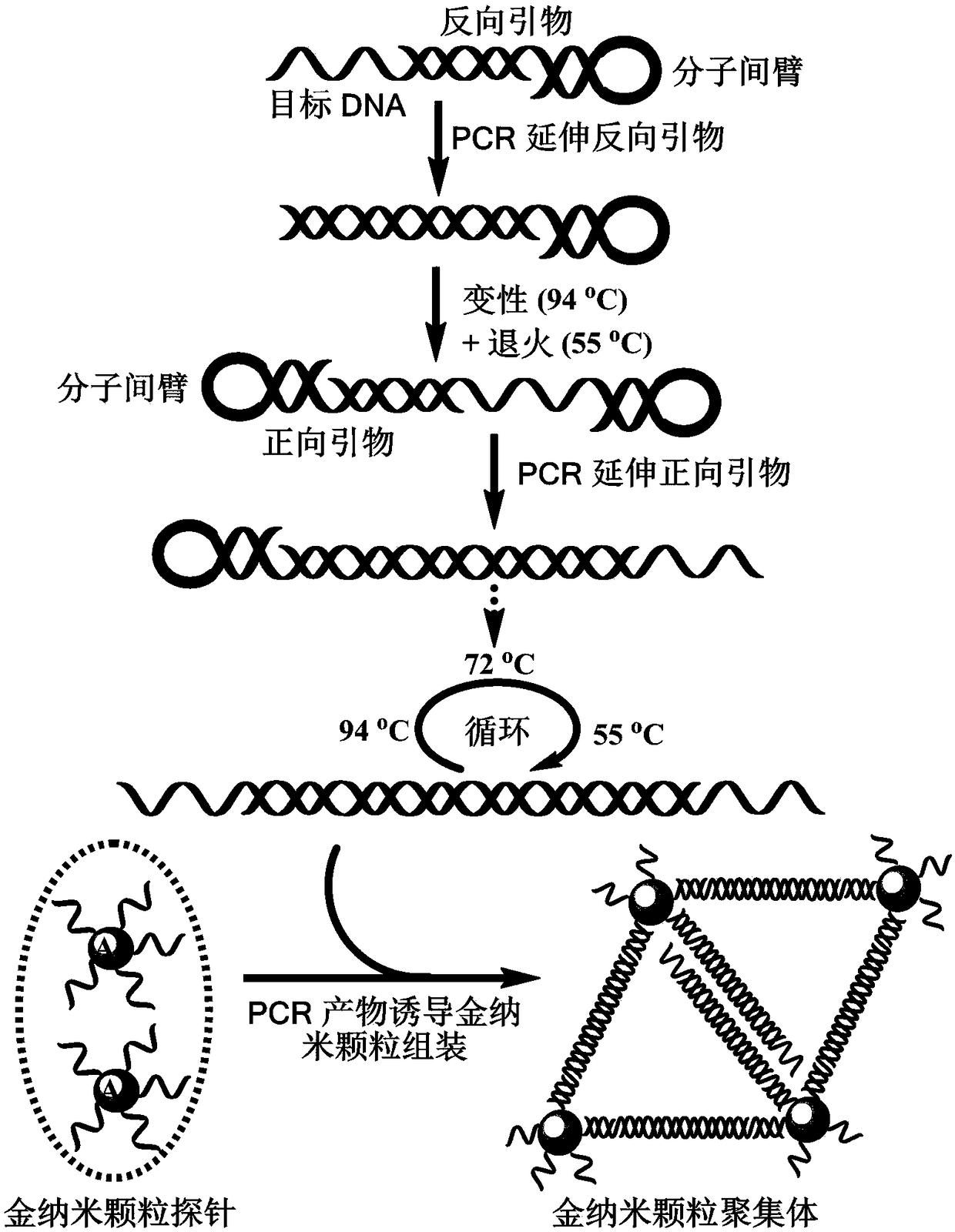
[0065] 1. Polymerase chain reaction based on special structural primers:
[0066] Design amplification primers with a hairpin structure, and introduce a hexaethylene glycol interarm (Spacer 18) into the loop. The sequences of the upstream and downstream primers are:
[0067] Upstream primer: 5’-GGG AGA GAA GAACT spacer18AG TTC TTC TCT CCC GAC AGG CCCGAA GGA ATA GA-3’
[0068] Downstream primer: 5'-GAG GAAGGAAAG CT spacer18AG CTT TCC TTC CTC CTC TCT CTCCAC CTT CTT CT-3';
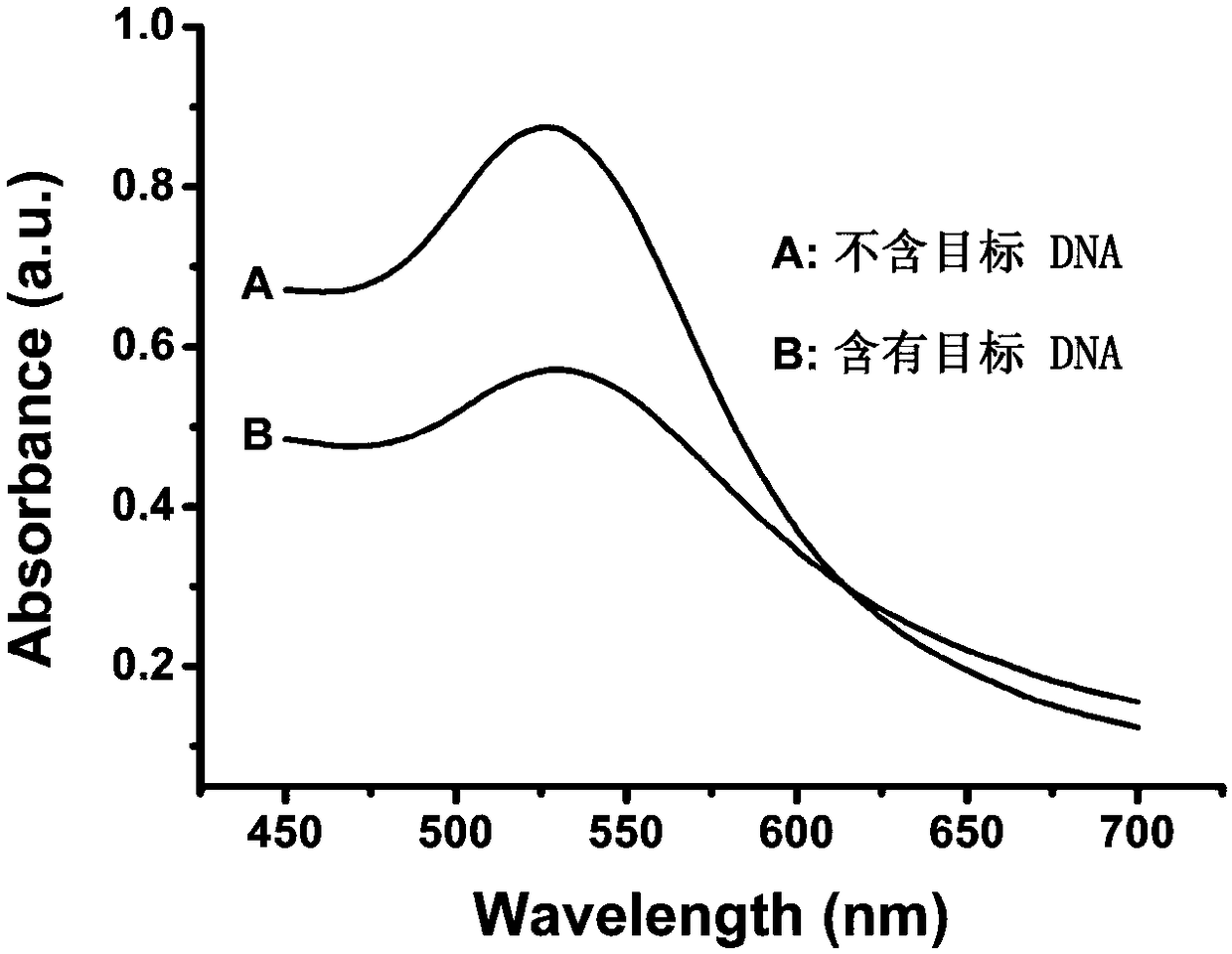
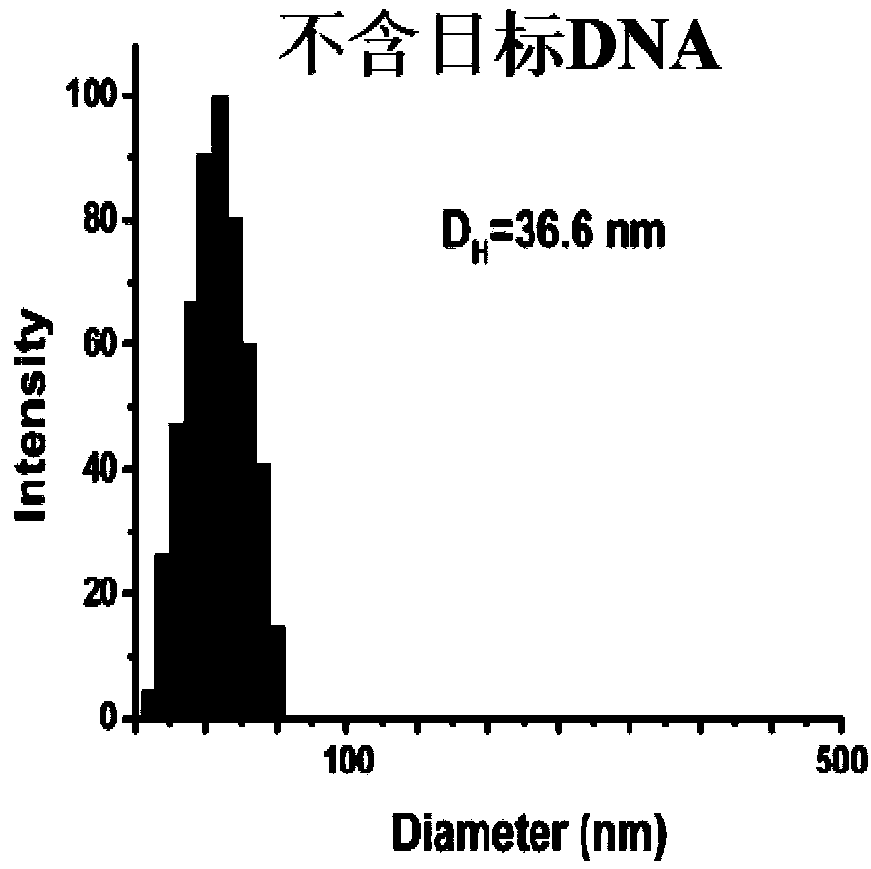
[0069] Carry out PCR reaction in a system containing template DNA (sequence is 5'-GAC AGG CCC GAA GGA ATA GAA GAA GAA GGT GGAGAG AGAG-3'), a pair of amplification primers, Taq DNA polymerase and dNTP to obtain bands at both ends PCR products with single-stranded DNA fragments.
[0070] The total volume of the PCR reaction solution is 50 μL, including 5 μL of 10× PCR buffer, 1 μL of 10 mM dNTP, 3 μL of 25 mM MgCl 2 , 0.5 μL 5U / μL Taq DNA polymerase, 10 μM / L upstream and downstream primers 2 μL each, 2 μL te...
Embodiment 2
[0087] 1. Polymerase chain reaction based on special structural primers:
[0088] Amplification primers with hairpin structure were designed, and a three-polyethylene glycol interarm (Spacer 9) was introduced into the loop, and the sequences of the upstream and downstream primers were:
[0089] Upstream primer: 5’-GGG AGA GAA GAA CT spacer9AG TTC TTC TCT CCC CTT CTC TTTGAT GTC ACG CA-3’
[0090] Downstream primer: 5’-GAG GAA GGA AAG CT spacer9AG CTT TCC TTC CTC GAT GCC ACAGGATTC CATA-3’
[0091] PCR reaction was carried out in a system containing template DNA (sequence 5'-CTT CTC TTT GAT GTC ACG CAT ATG GAATCC TGT GGCATC-3', a pair of amplification primers, Taq DNA polymerase and dNTP to obtain PCR products of stranded DNA fragments.
[0092] The total volume of the PCR reaction solution is 50 μL, including 5 μL of 10× PCR buffer, 1 μL of 10 mM dNTP, 3 μL of 25 mM MgCl 2, 0.5 μL 5U / μL Taq DNA polymerase, 10 μM / L upstream and downstream primers 2 μL each, 2 μL template DNA o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com