Taq DNA polymerase mutant and application thereof
A polymerase and mutant technology, applied in the biological field, can solve problems such as poor amplification of the target region, poor results in one degree and yield, and insufficient sensitivity of reagents, so as to improve sensitivity and compatibility, and library uniformity. Once good, the effects of increased sensitivity and yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
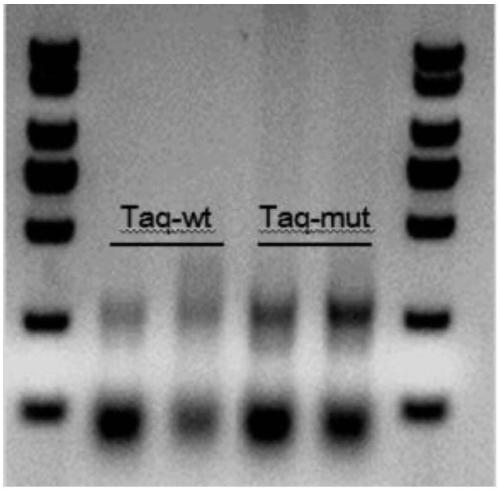
[0036] Use 2× of Nanjing Nuoweizan Biotechnology Co., Ltd. Max Master Mix (P515) and Ultra One Step Cloning Kit (C115) performs site-directed mutation on Taq DNA polymerase (sequence shown in SEQID NO.1), and the primers used for point mutation are as follows to obtain a mutant, called Taq-Mut, whose mutation site The points are: K53N, G364D, R636H (the sequence is shown in SEQ ID NO.2), the nucleic acid sequence before mutation is shown in SEQ ID NO.3, and the nucleic acid sequence after mutation is shown in SEQ ID NO.4.
[0037] The primer sequences (5'-3') used for point mutations are as follows:
[0038] AB-1F: GGTATATCTCTTCTTAAAGATGAGGGGGATGCTGCCC
[0039] AB-1R: TCCTTGAGGGCGTTGAGGAGGCTCTTGGCGAAGC
[0040] BC-1F:CTCCTCAACGCCCTCAAGGAG
[0041] BC-1R: AGGCCAAGGTCTTCCCTCAGGGCCAGAACG
[0042] CD-1F: CTGAGGGAAGACCTTGGCCTC
[0043] CD-1R: CGTGTGGATGTCGTGCCCCTCCTGGAAG
[0044] DE-1F: AGGGGCACGACATCCACACGGAGACCGCCAGCTGGATGT
[0045] DE-1R: GATAACAATTCCCCTCTAGATCCTTGGCGG...
Embodiment 2
[0050] Example 2 Application of Taq-Mut in Amplicon Library Construction
[0051] Use Taq-Mut to prepare the following different reaction systems for PCR:
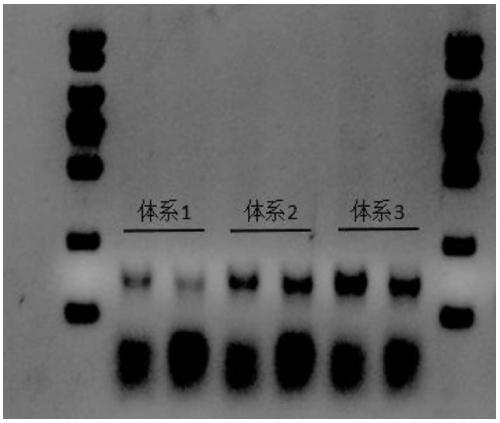
[0052] Multiplex amplification reaction system 1: Taq-Mut 2U, Tris 25mM, KCl 20mM, MgCl 2 1mM, dNTP 0.2mM, primer 10nM each, DNA input 1ng, pH7.6;
[0053] Multiplex amplification reaction system 2: Taq-Mut 2U, Tris 25mM, KCl 20mM, DMSO 2%, MgCl 2 1mM, dNTP 0.2mM, primer 10nM each, DNA input 1ng, pH7.6;
[0054] Multiplex amplification reaction system 3: Taq-Mut 2U, Tris 25mM, KCl 50mM, DMSO 2%, MgCl 2 1mM, dNTP 0.2mM, primer 10nM each, DNA input 1ng, pH7.6;
[0055] The above three systems were subjected to multiple amplification, and the input amount of 293T cell nucleic acid DNA was 1ng. The reaction procedure was as follows: 99°C for 2min; (99°C for 15sec; 60°C for 4min); 22 cycles; 72°C for 10min; 4°C hold. Run nucleic acid electrophoresis on a 3% agarose gel to obtain figure 2 The results shown, system 1 is...
Embodiment 3
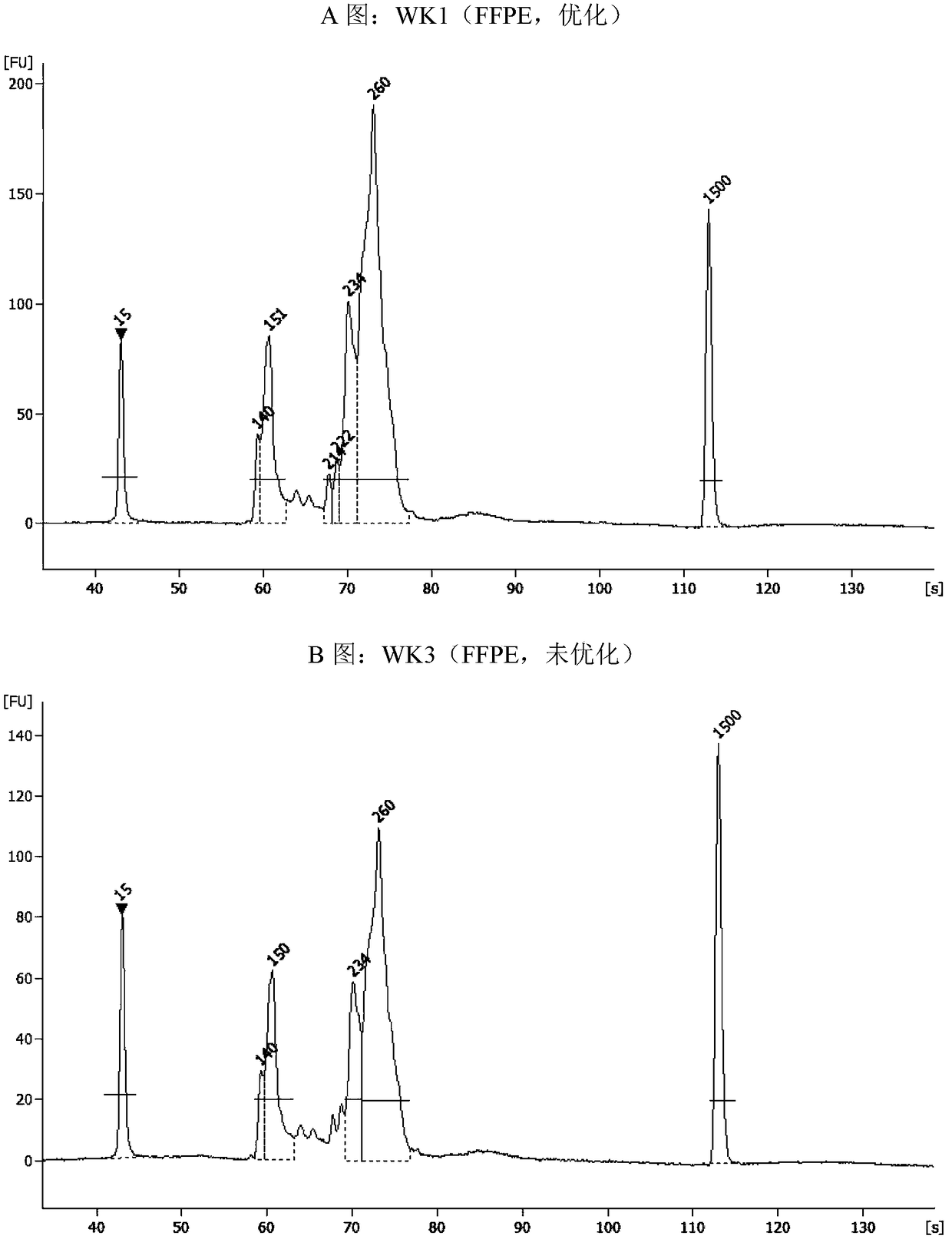
[0057] The reagents required to prepare for multiplex amplification include the amplification reagent Multi-PCR Mix, which is 2× concentration, and the concentration of key components is: Taq-Mut 4U, Tris 50mM, KCl 100mM, DMSO 4%, MgCl 2 2mM, dNTP0.4mM, pH7.6. Combined with the Adapters and Ligation Enzyme Mix required for conventional ligation, 207 primer pairs (Nanjing Nuoweizan Biotechnology Co., Ltd., VAHTS AmpSeqCancer HotSpot Panel NA102) for amplifying hot spot mutation regions of human tumor-related genes were used. The samples were human FFPE samples and One human cfDNA sample, after extracting DNA in a conventional way, the reaction system (1×concentration): Taq-Mut 2U, Tris 25mM, KCl 50mM, MgCl 2 1mM, DMSO2%, dNTP 0.2mM, primer 10nM each, pH7.6, FFPE DNA input amount 10ng / cfDNA input amount 1ng, primer digestion, adapter ligation, purification, library amplification, library purification, and amplicon library WK1, WK2, the amplicon libraries WK3 and WK4 obtained...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com