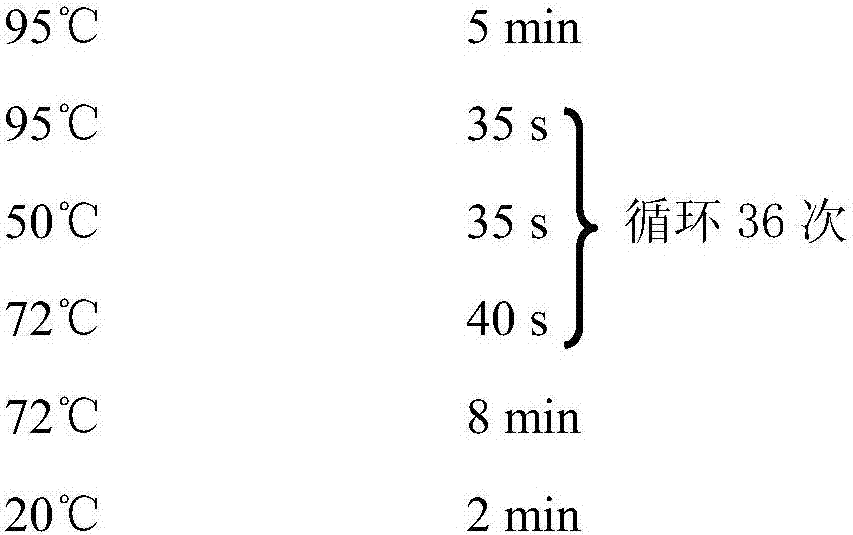
Patents
Literature
1395 results about "Pcr ctpp" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method of detecting gene mutation based on Blocker primers and ARMS primers, and kit
ActiveCN103255201AAchieve enrichmentReduce stepsMicrobiological testing/measurementPcr ctppReaction system
The invention discloses a method of detecting gene mutation based on Blocker primers and ARMS primers, and a kit. According to the invention, corresponding Blocker primers are designed according to an annealing temperature, the Blocker primers are combined with a wild-type template under the annealing temperature of the Blocker to block amplification of the wild type, and simultaneously an ARMS technology is used to reduce amplification of the wild-type background. The method integrates a Blocker enrichment technology and an ARMS fluorescent quantification PCR technology in one reaction system, and thus realizes enrichment and identification for the mutated genes for one step, reduces operation steps, and improves detection sensitivity and singularity.
Owner:北京宏微特斯生物科技有限公司
Method for carrying out high-throughput sequencing on TCR (T cell receptor) or BCR (B cell receptor) and method for correcting multiplex PCR (polymerase chain reaction) primer deviation by utilizing tag sequences
InactiveCN103710454AFully functionalReduce sequencing errorsMicrobiological testing/measurementDNA/RNA fragmentationV regionPcr ctpp
The invention provides a method for carrying out high-throughput sequencing on a TCR (T cell receptor) or a BCR (B cell receptor). The method is characterized by designing upstream primers according to gene features of a V region of the TCR or the BCR and designing downstream primers according to gene features of a C region or a J region of the TCR or the BCR and obtaining sequences of the of the TCR or the BCR in combination with the multiplex PCR (polymerase chain reaction) technology and high-throughput sequencing, thus analyzing the rearrangement information of the TCR or the BCR. Compared with 25-30 cycles of existing multiplex PCR, two cycles of the multiplex PCR technology provided by the invention can conduce to greatly reducing the sequencing errors caused by primer amplification preference. Besides, the invention also provides a method for correcting multiplex PCR (polymerase chain reaction) primer deviation by utilizing DNA (deoxyribonucleic acid) tag sequences, thus further reducing the sequencing errors caused by primer amplification preference and intrinsic sequencing errors of high-throughput sequencing.
Owner:SOUTH UNIVERSITY OF SCIENCE AND TECHNOLOGY OF CHINA +1
Process for high throughput DNA methylation analysis
InactiveUS7112404B2Sugar derivativesMaterial analysis by observing effect on chemical indicatorDNA methylationHybridization probe
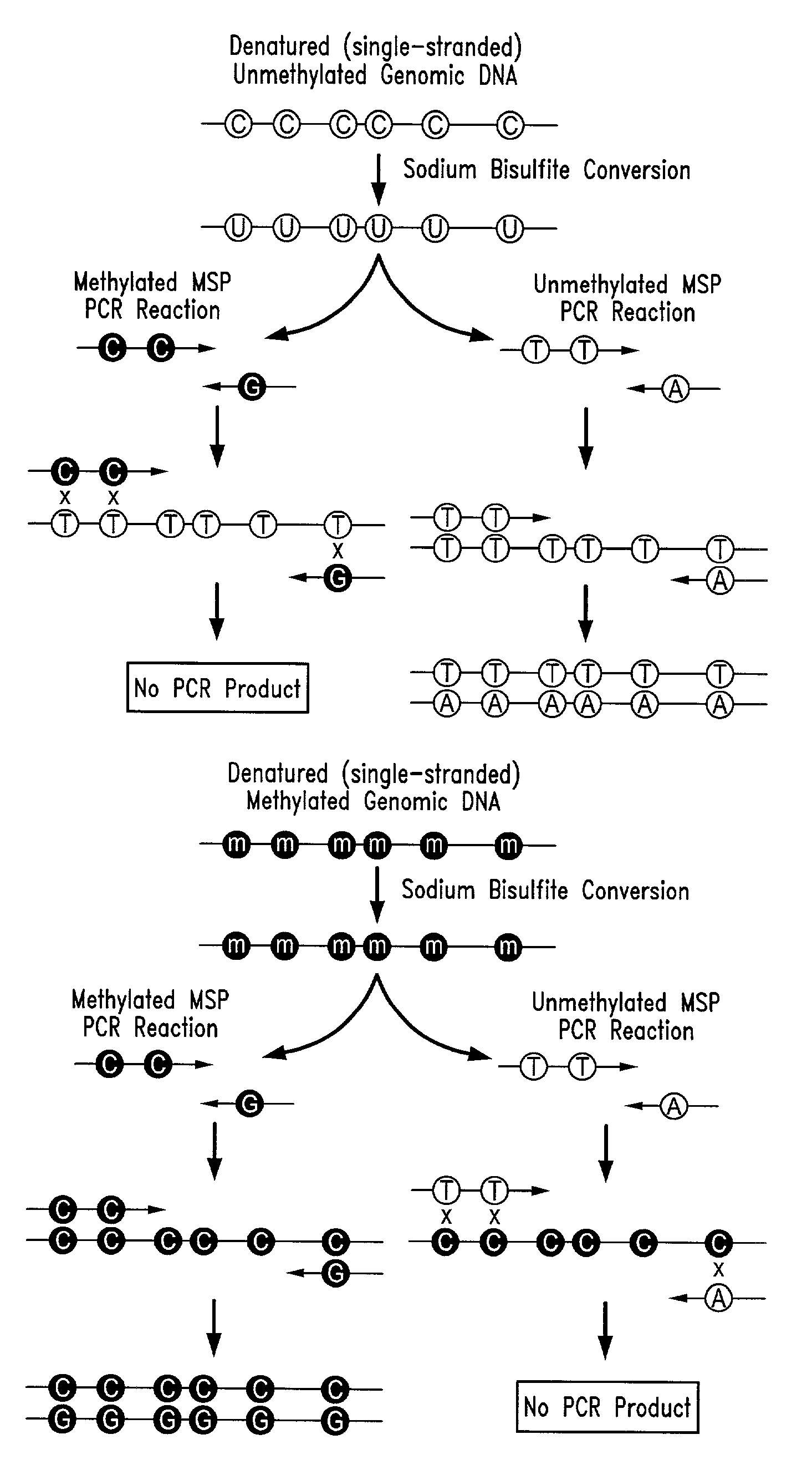
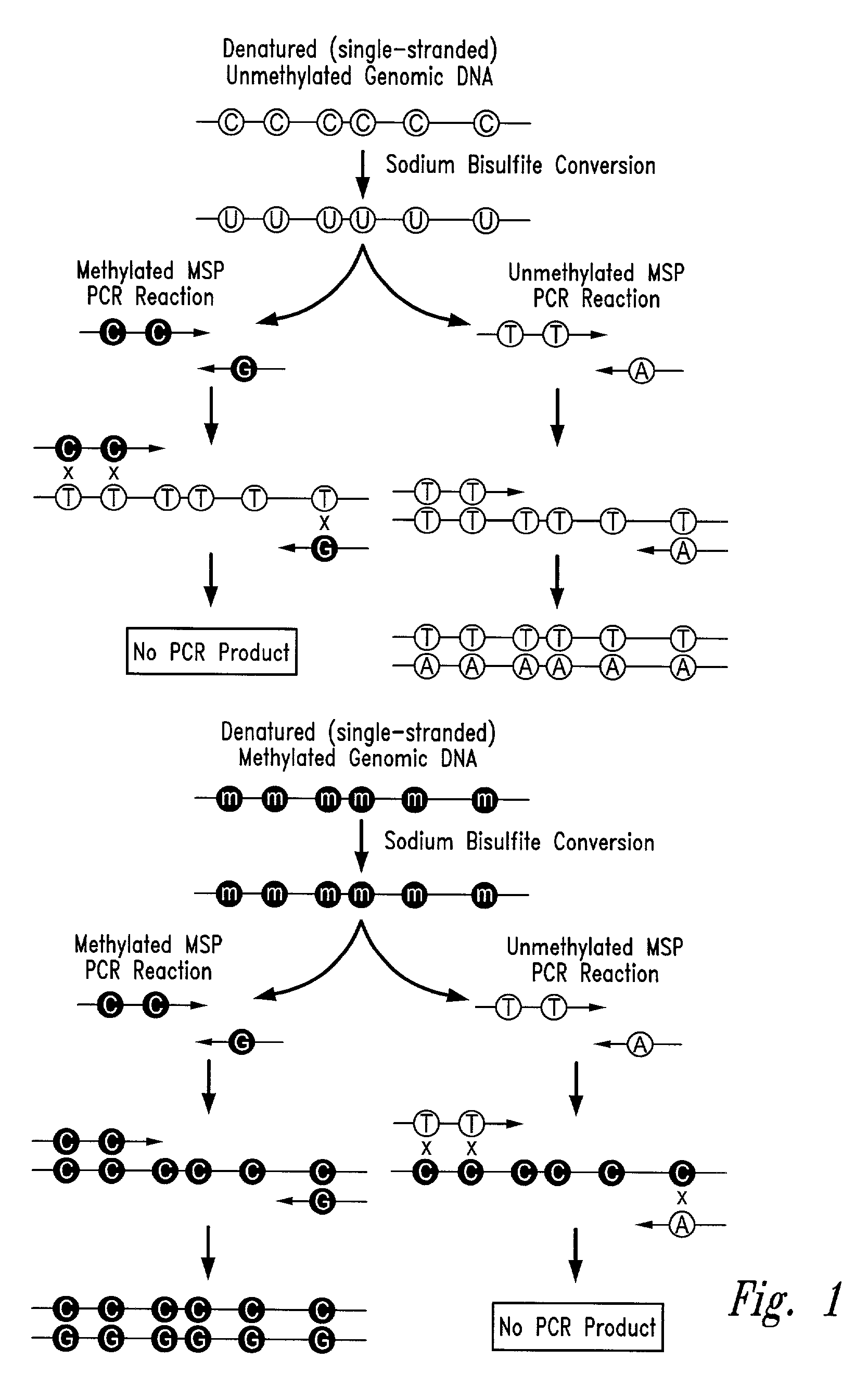
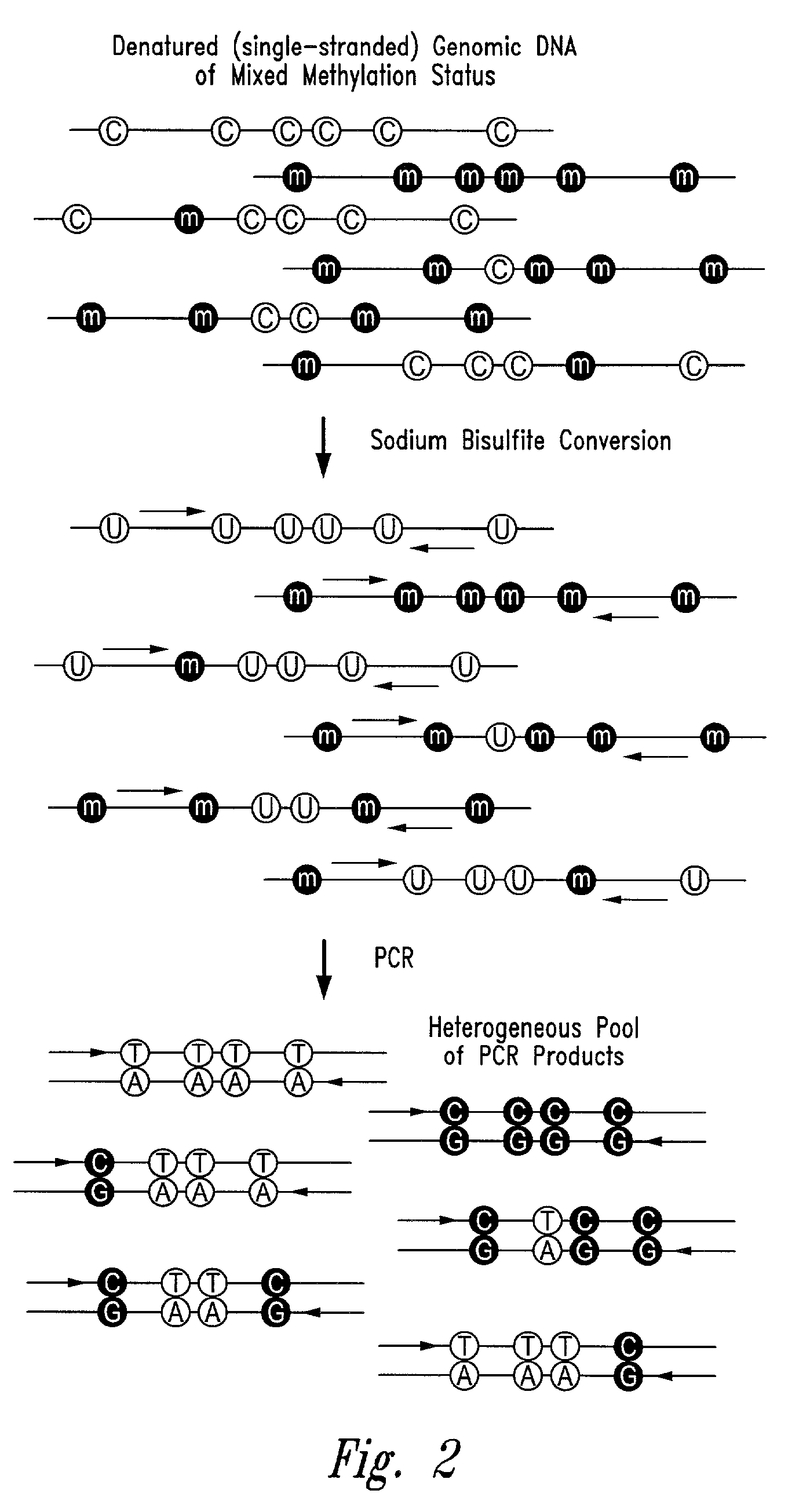
There is disclosed an improved high-throughput and quantitative process for determining methylation patterns in genomic DNA samples based on amplifying modified nucleic acid, and detecting methylated nucleic acid based on amplification-dependent displacement of specifically annealed hybridization probes. Specifically, the inventive process provides for treating genomic DNA samples with sodium bisulfite to create methylation-dependent sequence differences, followed by detection with fluorescence-based quantitative PCR techniques. The process is particularly well suited for the rapid analysis of a large number of nucleic acid samples, such as those from collections of tumor tissues.
Owner:UNIV OF SOUTHERN CALIFORNIA
Polymerase chain reaction (PCR) method for diagnosing human papillomavirus (HPV) and reagent kit thereof
InactiveCN101017141AEfficient, systematic, economical and simpleShorten the timeMicrobiological testing/measurementChemiluminescene/bioluminescenceHuman bodyHuman papillomavirus
This invention relates to one polymer enzyme linkage reaction fluorescence test method to dialogue dangerous human body nipple shape virus and to isolate DNA sample HPV gene type to test one set of dangerous HPV infection from patient by the method, wherein, it belongs to life science and biological technique. This invention agent case comprises one fluorescence meter PCR technique as base of multi-layer polymer enzyme reaction composed of multiple HPV positive lead object, reaction lead object and fluorescence detector.
Owner:GENETEL PHARMA SHENZHEN
crRNA (Clustered Regularly Interspaced Short Palindromic Repeat-Derived Ribonucleic Acid) for specifically detecting exon mutation of human KRAS genes #2 and #3 based on CRISPR technique
InactiveCN108893529AHigh sensitivityEasy to operateMicrobiological testing/measurementTemperature controlKRAS
The invention discloses crRNA (Clustered Regularly Interspaced Short Palindromic Repeat-Derived Ribonucleic Acid) for specifically detecting exon mutation of human KRAS genes #2 and #3 based on a CRISPR technique. The crRNA is characterized by comprising a CRISPR-Cas13a system as well as crRNA, wherein the crRNA can be combined with the CRISPR-Cas13a system; and the crRNA has a sequence format of5'-Cas13a protein anchoring sequence-crRNA guide sequence-3'. The crRNA has the advantages that according to detection results, whether the human KRAS gene have mutation or not can be judged intuitively through fluorescent reading, high-flux sequencing can be avoided, and the crRNA has the advantages of being low in cost, possible in multi-time detection, high in detection speed, possible in direct result analysis, and the like, and is applicable to large-scale clinical sample detection; the nucleic acid testing technique disclosed by the invention is different from a conventional detection technique which is based on PCR (Polymerase Chain Reaction) techniques, and no complex temperature control instrument or system is needed in a whole reaction process; and three steps of reactions are carried out in a same reaction system, so that operation procedures can be further simplified, and nucleic acid fragments with specific sequences can be detected within two hours.
Owner:江苏博嘉生物医学科技有限公司
Primer middle sequence interference PCR (Polymerase Chain Reaction) technology
InactiveCN103114131AHigh Sensitivity FeaturesQuantitatively accurateMicrobiological testing/measurementAgricultural scienceFluorescence
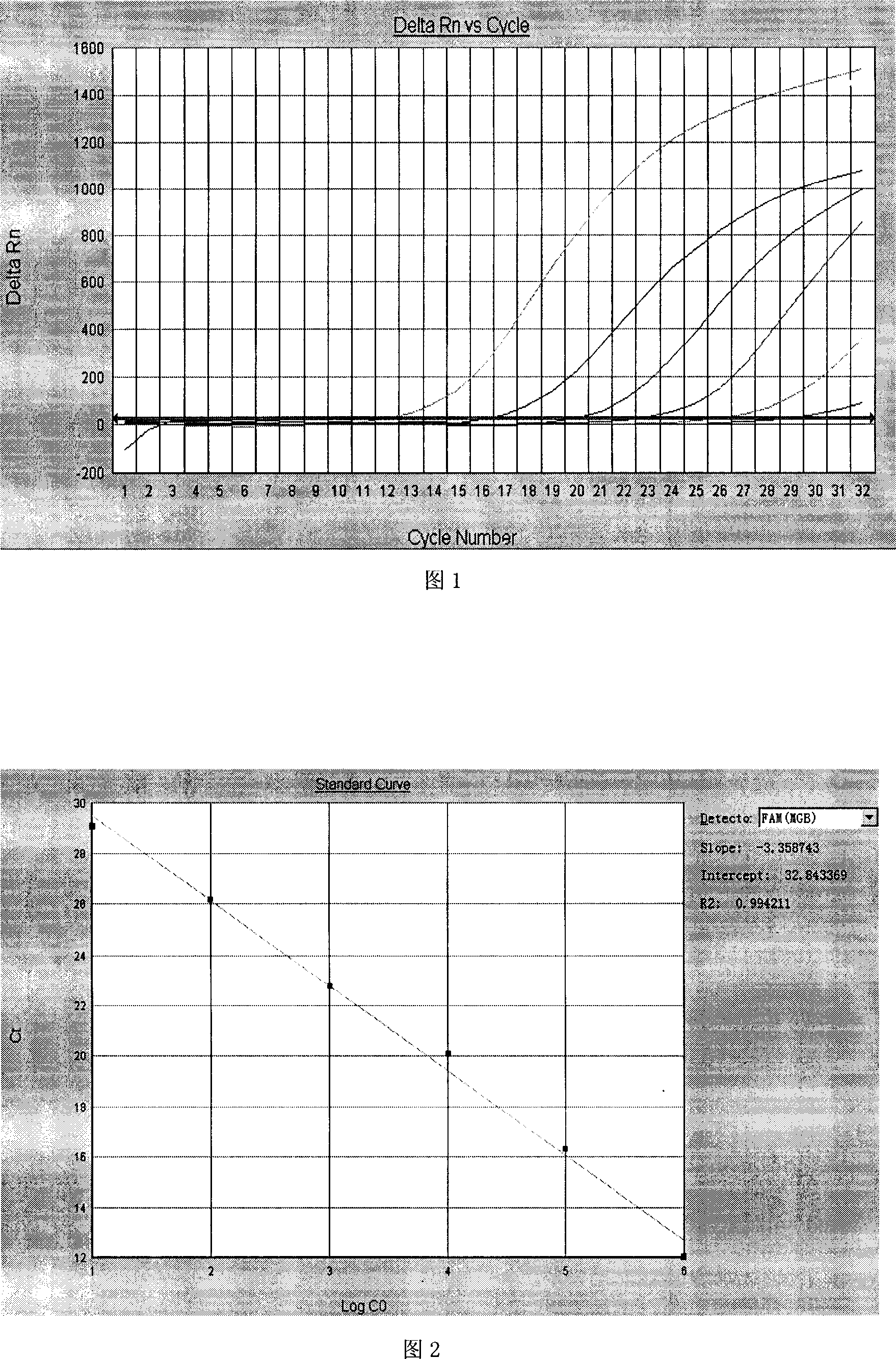
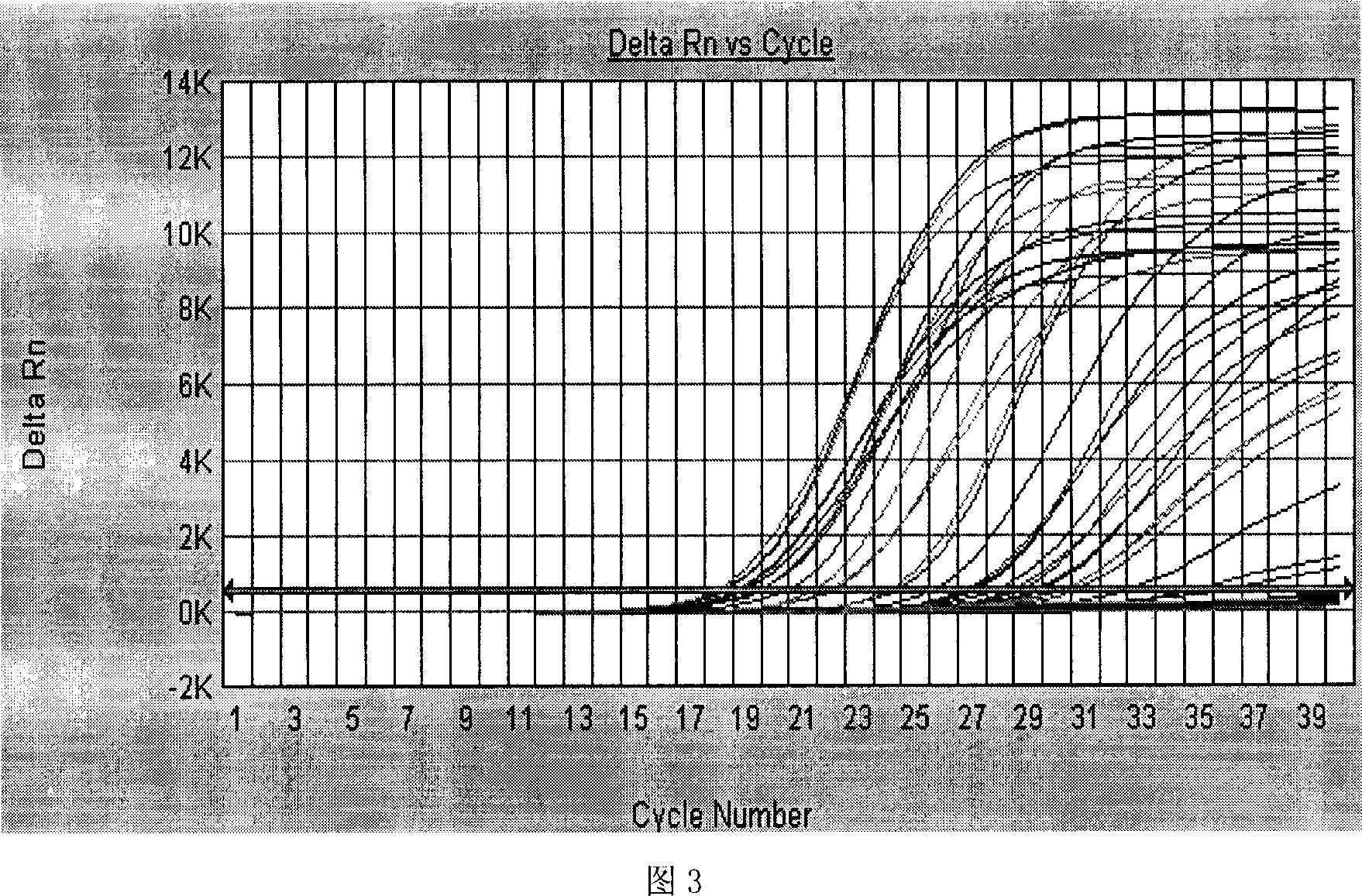
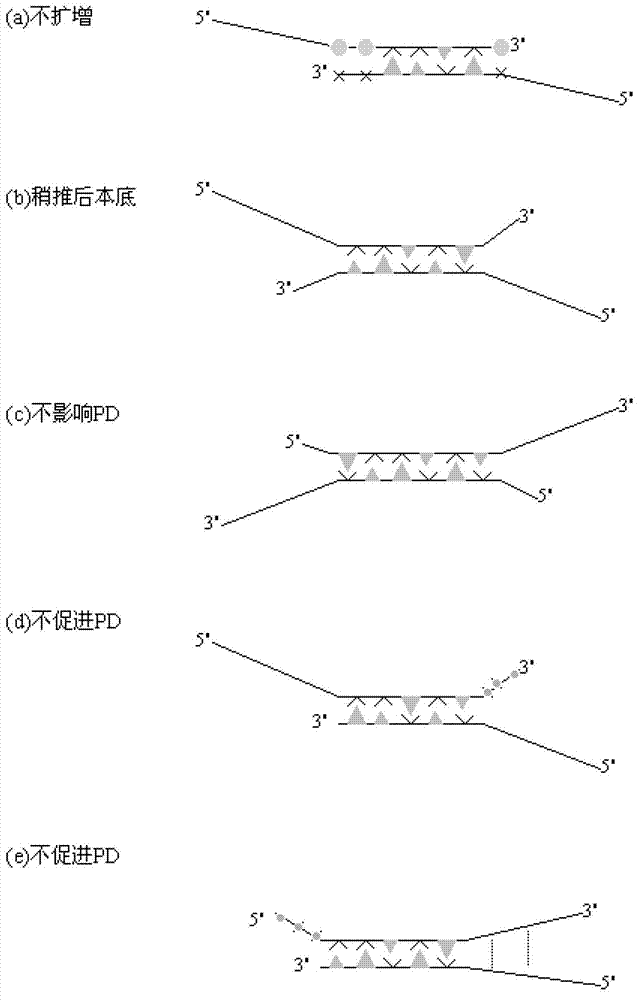
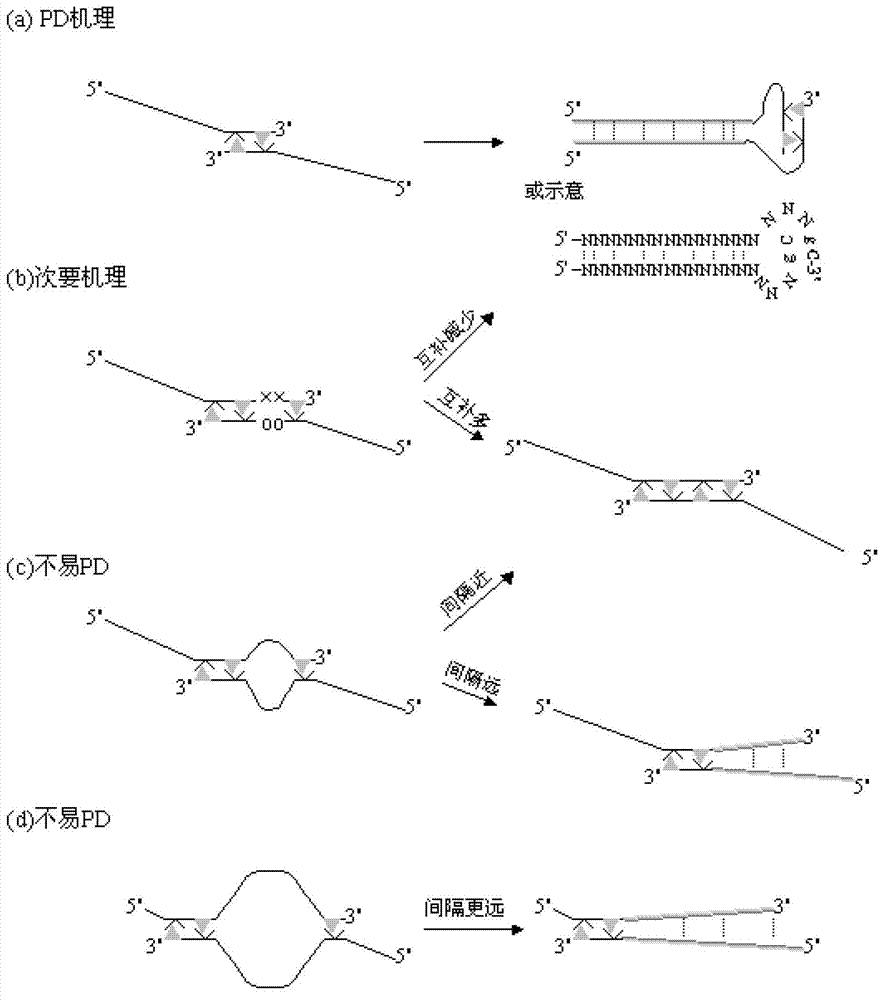
The invention relates to a primer middle sequence interference PCR (Polymerase Chain Reaction) technology. The improved PCR technology is characterized in that one segment of relatively non-complemented or same-sequence basic group primer molecules in the intermediate domain of primers perform the antisense interference inside and outside so as to competitively destroy the polymerization among the primers to selectively inhibit the primer dimer (PD) from being amplified. For the interference of the intermediate domain of the primers, based on the primers optimally selected by the conventional design principle, the technology that the intermediate domain (ID) of a pair of the primers are in parallel but are not complemented with each other or are in the same sequence or / and the technology that ID antisense oligonucleotides (Oligo) are added into the primers to perform the interference action or / and the Oligo antonymy is carried out in the primer molecules via the ID so as to perform the interference action are adopted, or the combined technology of the three types of the technologies is adopted. As a result, only the ID of the primers is interfered while the target specific amplification is not influenced; the combining force acting on the minority of base-group pairing hydrogen bonds at the tail end and the base-group hydrogen bonds outside the tail end due to the action of the primers is dispersed to a maximum extent, so that the PD is selectively inhibited. Therefore, the PD accumulation in the PCR system is avoided. If the mineral oil is additionally used, the sealed primers can slowly release the hot starting and the UDG pretreatment so as to prevent aerosol glue as a byproduct of the PCR system from causing the pollution. Consequently, the nucleic acid is amplified reliably and the real-time fluorescence PCR is quantified accurately.
Owner:珠海市坤元科技有限公司
Kit for diagnosing gene of Leber optic neuropathy in heredity and detecting method
InactiveCN1661116ASolve difficult collection problemsShorten the timeMicrobiological testing/measurementDiseaseLesion
A reagent kit for the gene diagnosis of Leber's hereditary optical nerve (LHON) lesion which is caused by the mutations on 3 primary pathogenic sites (G11778A, G3460A and T14484C) is disclosed. Its detecting process includes such steps as designing and synthesizing the site specific PCR primer, using whole blood as DNA template, PCR amplification, and detecting said mutant sites of LHON patient.
Owner:FUJIAN MEDICAL UNIV
Thalassemia gene detection method based on fluorescence labeling quantitation PCR (Polymerase Chain Reaction) technology
InactiveCN103255225ARealize detectionEnables fluorescence detectionMicrobiological testing/measurementFluorescenceThalassemia
The invention discloses a thalassemia gene detection method based on a fluorescence labeling quantitation PCR (Polymerase Chain Reaction) technology, and the detection method mainly comprises the following steps of: firstly designing a general fluorescence labeling primer; then designing an amplification primer aiming at a detection site; when the amplification primer is designed, tailing one primer of an amplification primer pair, wherein the tailing sequence is consistent to the sequence of the general fluorescence labeling primer; when PCR reaction is carried out, mixing a tailed primer group and the general fluorescence labeling primer to realize the fluorescence labeling of an amplification product; and after the PCR reaction is finished, placing PCR products into a sequencer, and confirming the lengths of different PCR products through a series of internal references of known molecular weight of a result by utilizing the Genemapper function of the sequencer to judge the genetype of a detected sample. The detection method disclosed by the invention can realize the detection of different genotypes of alpha thalassemia through a single tube.
Owner:钦州市妇幼保健院
Deafness susceptibility gene screen test kit
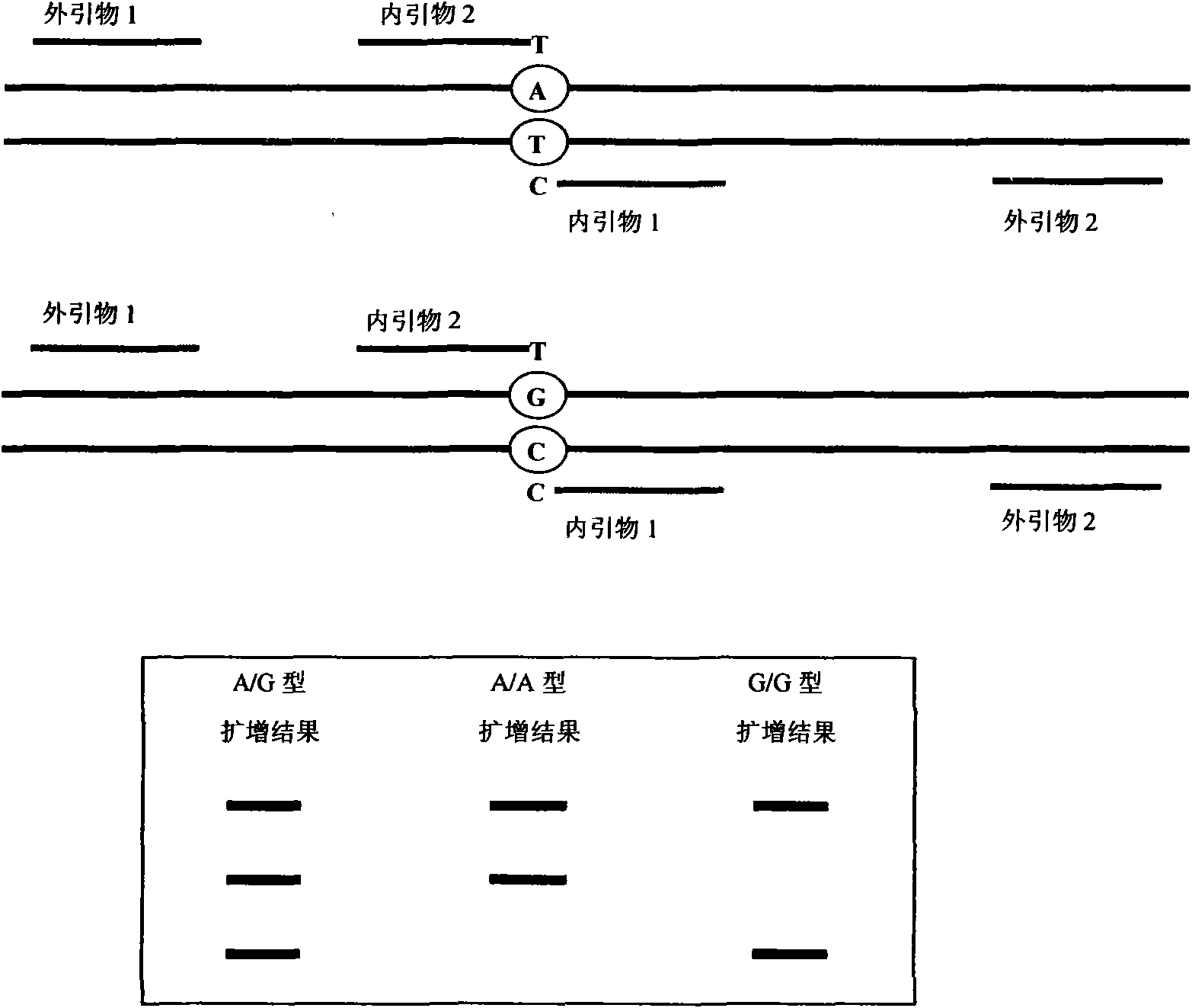
The invention relates to a hereditable deafness susceptibility gene screen test kit. The kit uses mutation from C to T of 1,494 locus of a 12s rRNA gene, mutation from A to G of 1,555 locus of the 12srRNA gene, mutation from A to G of IVS7(-2) locus of an SLC26A4 gene and C(+ / -) of 235 locus of a GJB2 gene as detecting objects, designs and optimizes a set of specific primers respectively againsteach locus to be tested according to the PCR technical principle of a tetra-primer amplification refractory mutation system, amplifies the whole set of the primers in the same reaction tube, and performs primary multiple PCR amplification and primary gel electrophoresis on the four reaction tubes to obtain gene types of four loci simultaneously.
Owner:GENERAL HOSPITAL OF PLA
Preparation method of virus-like particles (VLPs) of Chikungunya virus (CHIKV) and its application
InactiveCN102321639ANeutralizing activityViral antigen ingredientsInactivation/attenuationImmune effectsVirus-like particle
The invention relates to a preparation method of virus-like particles (VLPs) of Chikungunya virus (CHIKV). The method comprises the steps of: modifying genetic elements of a structural protein encoding gene C-E3-E2-6K-E1 of CHIKV, cloning the modified genetic elements into the expression vector of an insect cell, then transfecting the obtained recombined expression vector and baculovirus linear DNA respectively to an SF9 insect cell and making the cell secrete and express CHIKV VLPs. Additionally, the invention also makes preliminary studies on the immune effects of CHIKV VLPs and applicationof CHIKV VLPs in virus specific antibody detection, thus laying a foundation for research and preparation of immunological detection reagents and even vaccines based on CHIKV VLPs.
Owner:中国疾病预防控制中心病毒病预防控制所
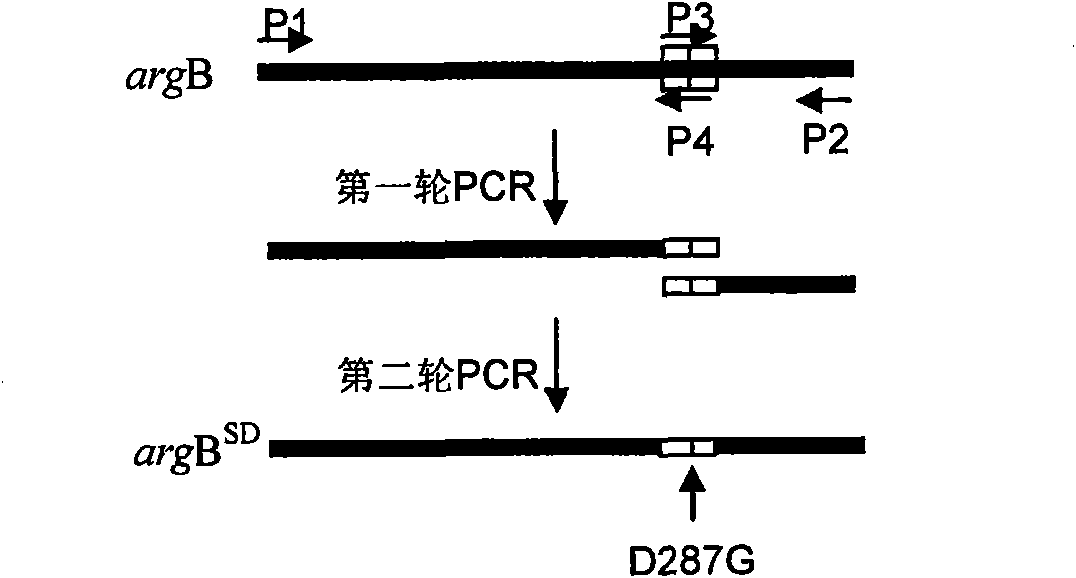
Method for improving yield of arginine by mutation of Corynebacterium crenatum N-acetyl glutamic acid kinase
The invention relates to a method for improving the yield of arginine by the mutation of Corynebacterium crenatum N-acetyl glutamic acid kinase. L-arginine is one of semi-essential basic amino acids for a human body, and has various particular physiology and pharmacology functions. High-yield arginine mutant strain C. crenatum SYPA5-5 is compounded into the arginine through a circulating way, and N-acetyl glutamic acid kinase (NAGK) is a key enzyme in the compounding way and is subject to the feedback inhibition of the product arginine. By using an overlapping PCR (Polymerase Chain Reaction) technique, a GAT (glycerol phosphate acyl transferase) codon used for coding aspartic acid is used for substituting a GGA (General Gonadotropic activity) codon used for coding glycine at the site 287 in the coding NAGK albumen, and the NAGK which has high activity and arginine with obviously reduced by the feedback inhibition can be obtained after the mutation. An argBSD gene is brought into the Corynebacterium crenatum of the high-yield arginine through pJCl-tac, and the expression volume of the key enzyme is further improved. The final acid yield is increased to 36.3g / L from original 28g / L, and the yield of the L-arginine is increased by 29.7 percent.
Owner:JIANGNAN UNIV
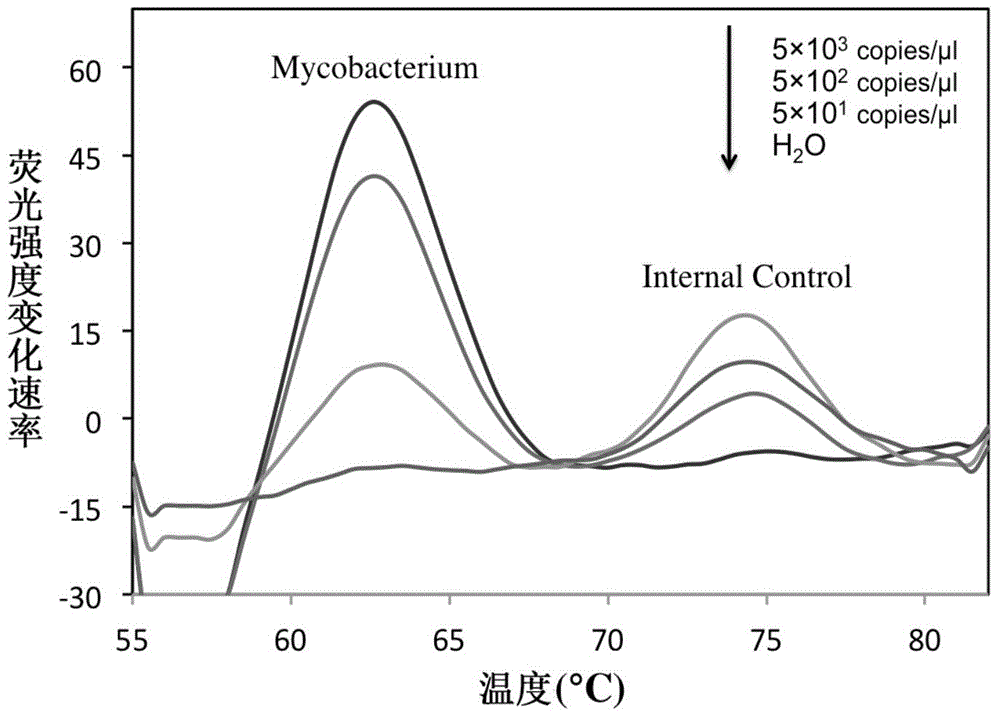
Rapid identification method and kit for MTBC (mycobacterium tuberculosis complex)
InactiveCN104561245AShorten diagnostic timeSimple and fast operationMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceRapid identification
The invention belongs to the field of a detection reagent, and relates to a detection and identification kit for MTBC (mycobacterium tuberculosis complex) and an identification method of the MTBC. According to the method, fluorescence probes and amplification primers for detecting the MTBC are designed through sequence alignment, mycobacterium and the MTBC specific SNP (single nucleotide polymorphism) loci on an rrs gene are detected based on a real-time fluorescent quantitative PCR (polymerase chain reaction) platform and with an asymmetric PCR amplification technology and a probe melting curve analysis technology, so that mycobacteria are identified, and the MTBC and NTM (nontuberculosis mycobacteria) are further identified. The method has the characteristics of convenience in operation, short detection time and high specificity and sensitivity.
Owner:FUDAN UNIV
Novel target gene for diagnosing and treating tongue squamous carcinoma and application thereof
ActiveCN105779618AHigh selectivitySimplify the process of quantitative detectionOrganic active ingredientsGenetic material ingredientsDiseaseCandidate Gene Association Study
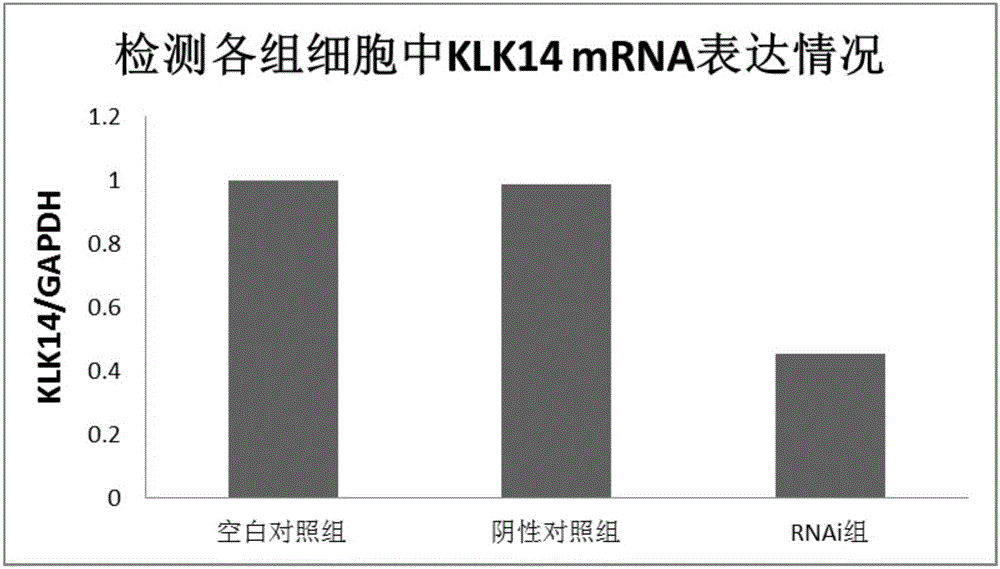
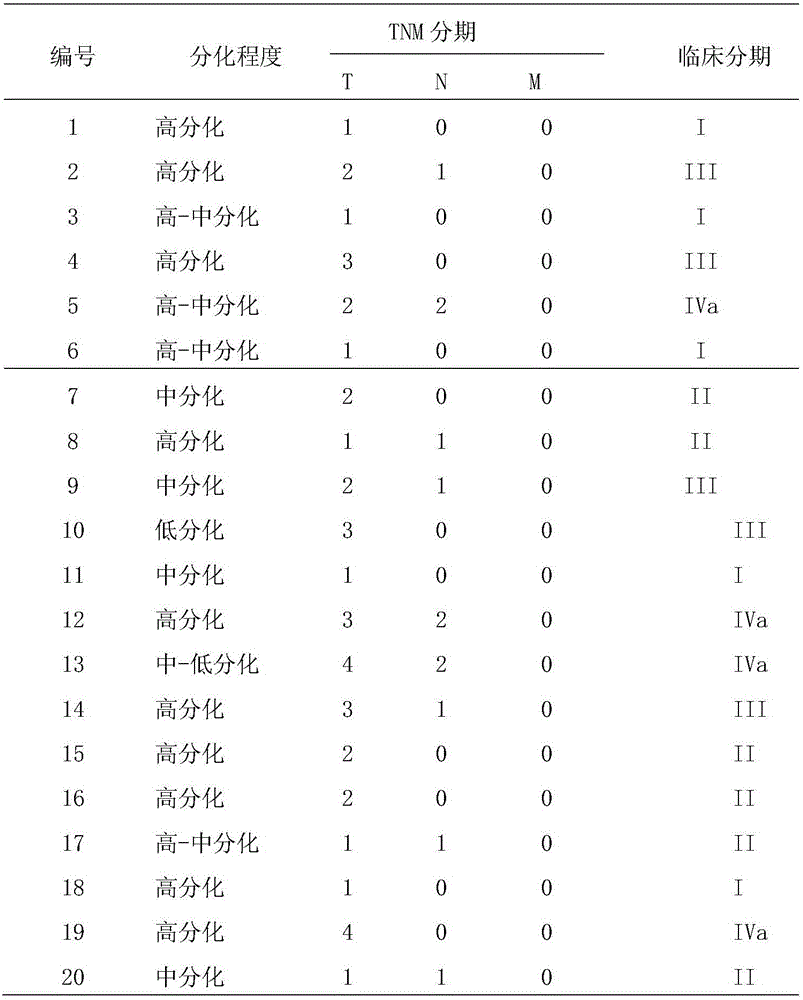
The invention provides a novel target gene for diagnosing and treating tongue squamous carcinoma and application thereof and particularly relates to application of a KLK14 gene and an expression product thereof to diagnosis and treatment of tongue squamous carcinoma. To research the occurrence and development mechanisms of tongue squamous carcinoma, search for an effective molecular target gene for diagnosing and treating tongue squamous carcinoma, promote early diagnosis, prevention and treatment of the disease and lower the death rate of tongue cancer, firstly, RNA-seq sequencing is utilized to detect differential expression genes of tongue squamous carcinoma, cancer branch and normal oral mucosa; secondly, a Real-time PCR technology is utilized to verify the sequencing result; then, an interference technology is utilized, and expression of the candidate gene KLK14 in tongue squamous carcinoma cells SCC15 is silenced. By means of the novel target gene for diagnosing and treating tongue squamous carcinoma and application thereof, an experimental foundation is laid for clinical application of the KLK14 gene to tongue squamous carcinoma, and a new target gene and theoretical basis are provided for early diagnosis and treatment of tongue squamous carcinoma.
Owner:THE SECOND XIANGYA HOSPITAL OF CENT SOUTH UNIV
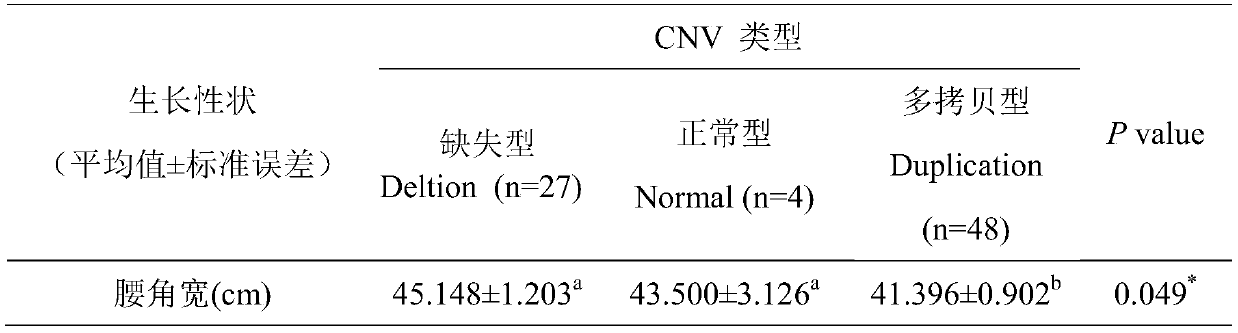
Method for quickly detecting cattle MLLT10 gene CNV marker and application of method
ActiveCN109943647AAccurate identificationLow costMicrobiological testing/measurementMarker-assisted selectionPcr ctpp
The invention discloses a method for quickly detecting a cattle MLLT10 gene CNV marker and application of the method. Based on the real-time fluorescent quantitation PCR technology, cattle genome DNAserves as a template, the cattle MLLT10 gene copy number variation area is amplified by using specific PCR primers, and partial fragments of the cattle BTF3 gene are amplified to serve as an internalreference, and finally, by using the 2-delta delta Ct method, the individual copy number variation type is computed and determined. The method lays a basis for the relationship between the cattle MLLT10 gene copy number variation and the growth trait, the detection method is simple and fast, the method can be used for accelerating cattle marker-assisted selection breeding work, and application andpopularization are facilitated.
Owner:NORTHWEST A & F UNIV
Multiple channel surface plasma resonant image sensor based on-chip PCR
InactiveCN1664560AIn line with the development trendImprove detection accuracyPolarisation-affecting propertiesScattering properties measurementsPrismSurface plasmonic resonance
The invention relates to a biosensor based on the surface plasma resonance principle, in particular to use the biology PCR technology combined with it. Which including the following steps: a) combining the SPR sample pond with the PCR reflecting pond and formatting the sensor reflecting pond; (b) the top of the sensor body is a rectangular reflecting sensor pond made up of the silicon chip, the lower is a cylinder prism, the two sides of the sensor body are set with the incident path, exit path and the computer plate connected with the end of the exit path;(c) the homogeneous light incidences, using the single color surface CCD to collect SPR picture and analyze through it, measuring the resonance and the change of it, which means the response of each channel. The invention has for characters: (a) measuring several samples once (b) measuring different characters of a sample once; (c) setting the consulting channel in the multi-channel and avoiding the effect of non-specific; (d) measuring the DNA molecule of low concentration.
Owner:NANKAI UNIV
Preparation method for microbial agent and application thereof
InactiveCN107058167APromote growthSpecies results are accurateBiocidePlant growth regulatorsGrowth plantMicrobial agent
The invention discloses a preparation method for a microbial agent. The method comprises the following steps: 1) taking soil of virgin forest in Changbai Mountain; 2) adopting sterilizing water for diluting the soil for 10 times and then uniformly mixing, adopting a gradient dilution separating method for preparing soil suspension and then respectively coating on five solid medium plates for culturing and selecting one plate with more single colonies as a statistic plate; 3) classifying the colonies on the statistic plate, counting and recording the quantity; 4) confirming three advanced strains according to a result of the step 3); 5) adopting a gram staining method for judging the classification of the colonies of the three advanced strains and then utilizing a PCR technique to expand the species of 16SrDNA identified strains; 6) storing the advanced strains; 7) culturing the three advanced strains, mixing at proper volume ratio and then centrifuging and suspending in the sterilized saline, thereby acquiring the microbial agent. The prepared microbial agent can be used for promoting the growth of flowers and plants. The method is simple, the operation is easy and the prepared microbial agent can obviously promote the plant growth.
Owner:HUBEI UNIV OF TECH
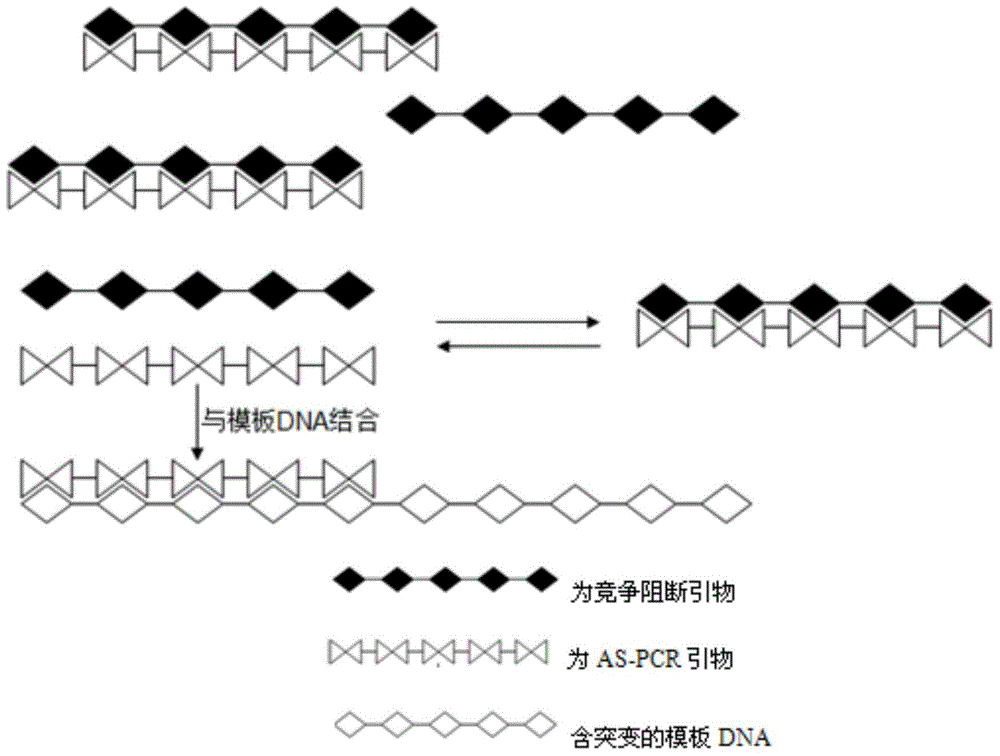
AS-PCR (allele-specific polymerase chain reaction) primer design method, gene mutation detection method and kit
InactiveCN104611427AStrong specificityImprove featuresMicrobiological testing/measurementDNA/RNA fragmentationPolymerase LOligonucleotide
The invention relates to the field of molecular biology, in particular to an AS-PCR (allele-specific polymerase chain reaction) primer design method, a gene mutation detection method and a kit. The AS-PCR primer design method comprises the following steps: (1), an AS-PCR primer is designed for a target sequence containing a to-be-detected allele mutation region; (2), a competition blocking primer is designed for the AS-PCR primer and adopts oligonucleotide in reverse complement with the AS-PCR primer. According to the gene mutation detection method, the AS-PCR primer and the competition blocking primer have a PCR amplification reaction. The kit comprises the AS-PCR primer, the competition blocking primer, a TaqMan probe, an internal control primer, an internal control probe, polymerase, dNTP (deoxynucleotide) and a buffer solution. The AS-PCR primer design method, the gene mutation detection method and the kit have the advantage that the difference of specificity of the AS-PCR primers on mutation sites due to strong and weak mismatch of the mutation sites is reduced effectively.
Owner:江苏宏泰格尔生物医学工程有限公司
Recombinant adenovirus of porcine reproductive and respirator syndrome virus and porcine Circovirus, and vaccine
InactiveCN1800375AHas the copy featureStable potencyViruses/bacteriophagesAntibody medical ingredientsEscherichia coliCircovirus
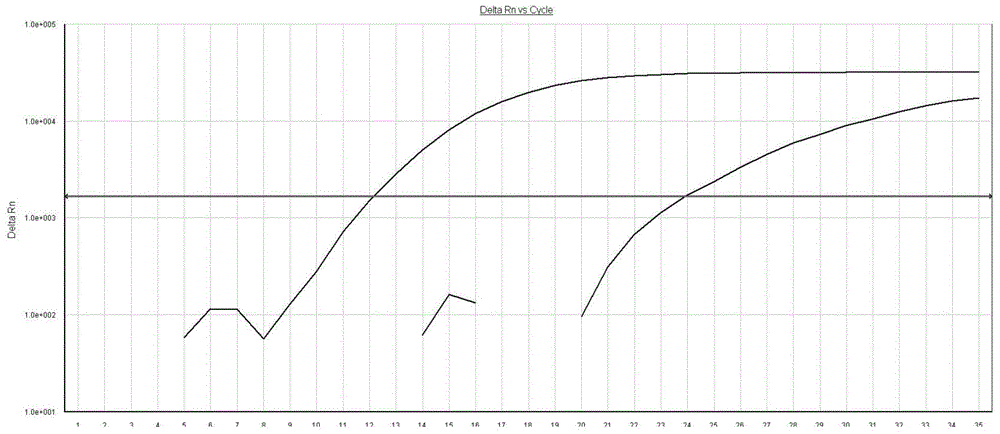
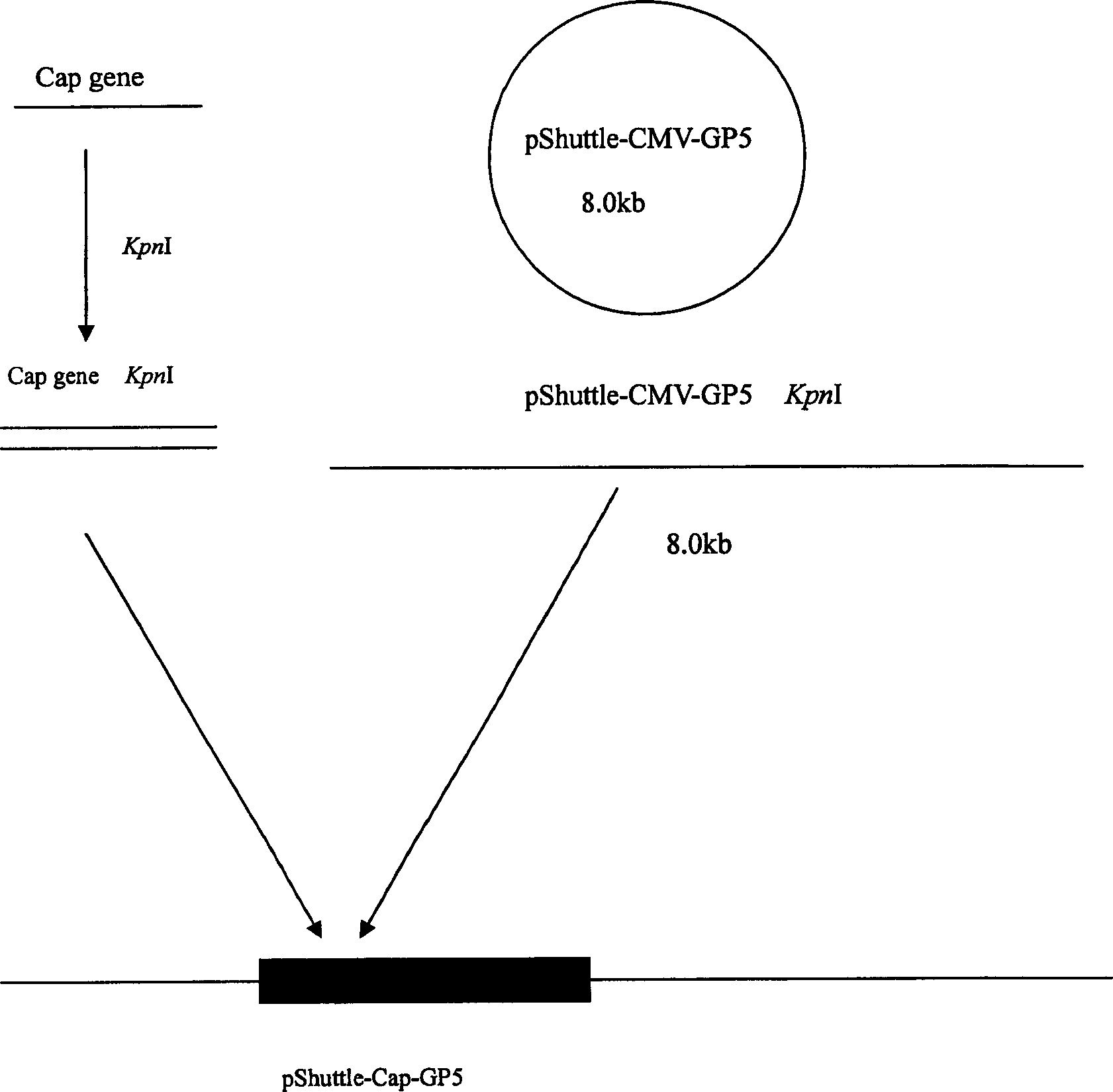
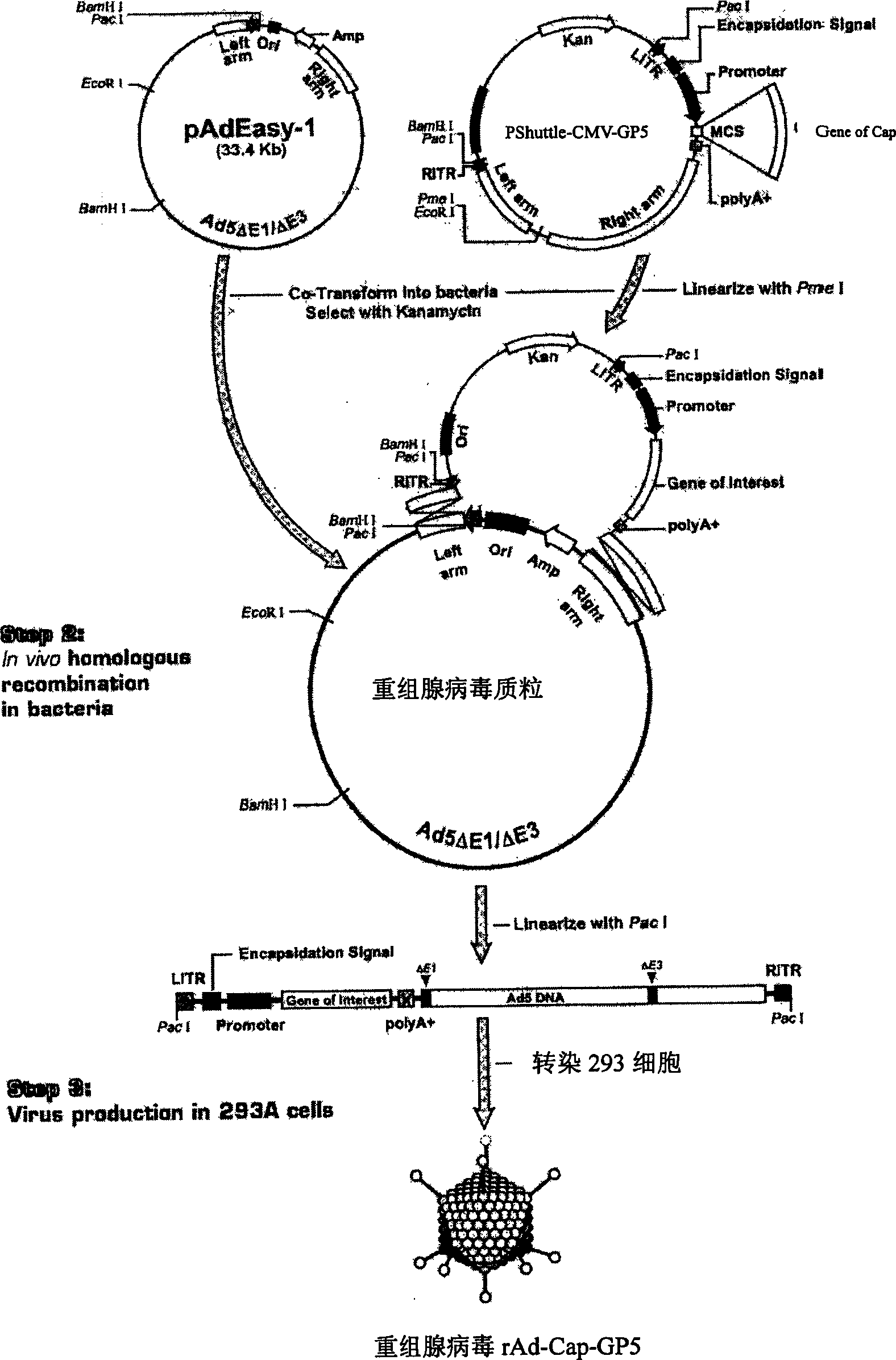
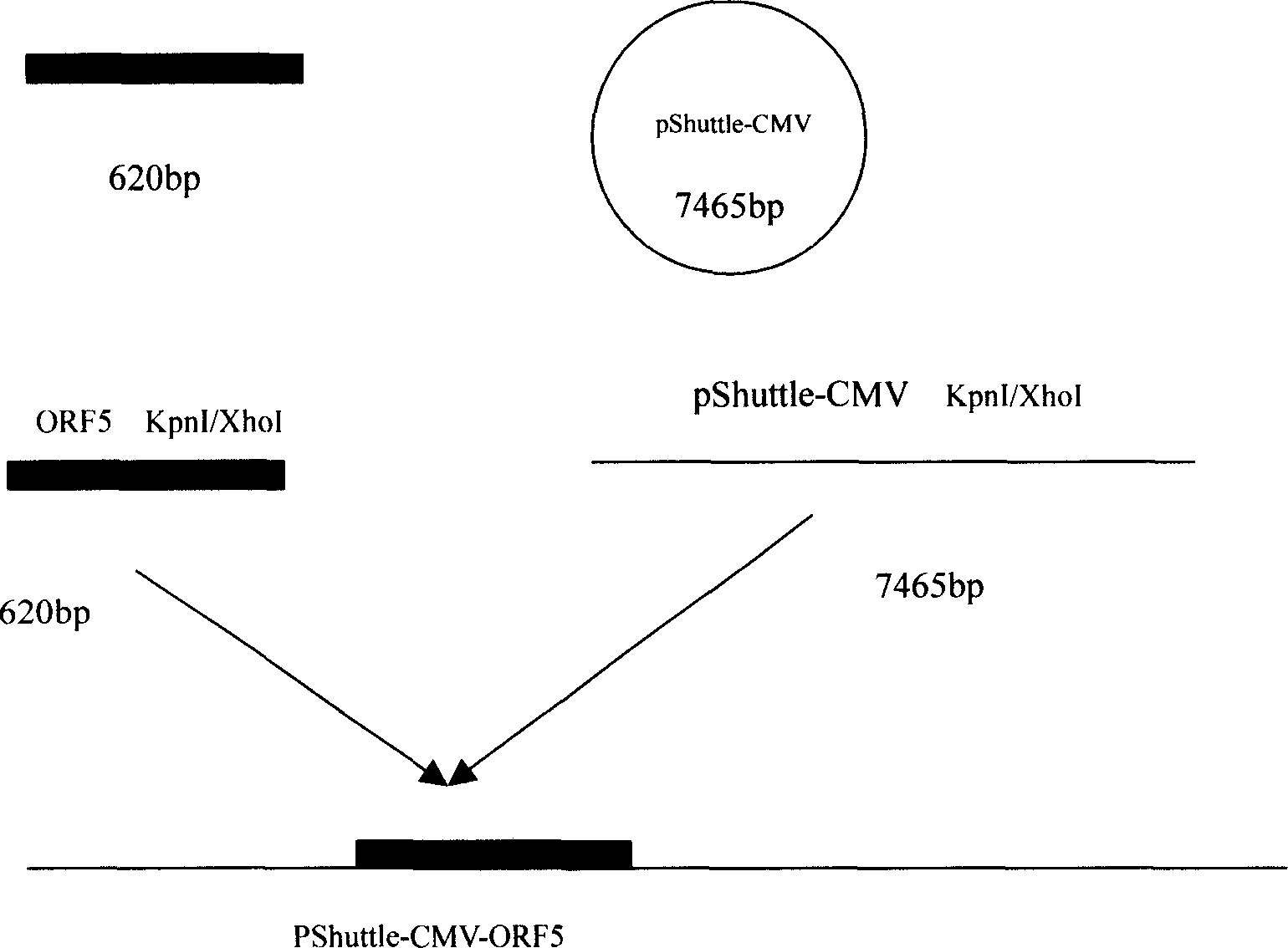
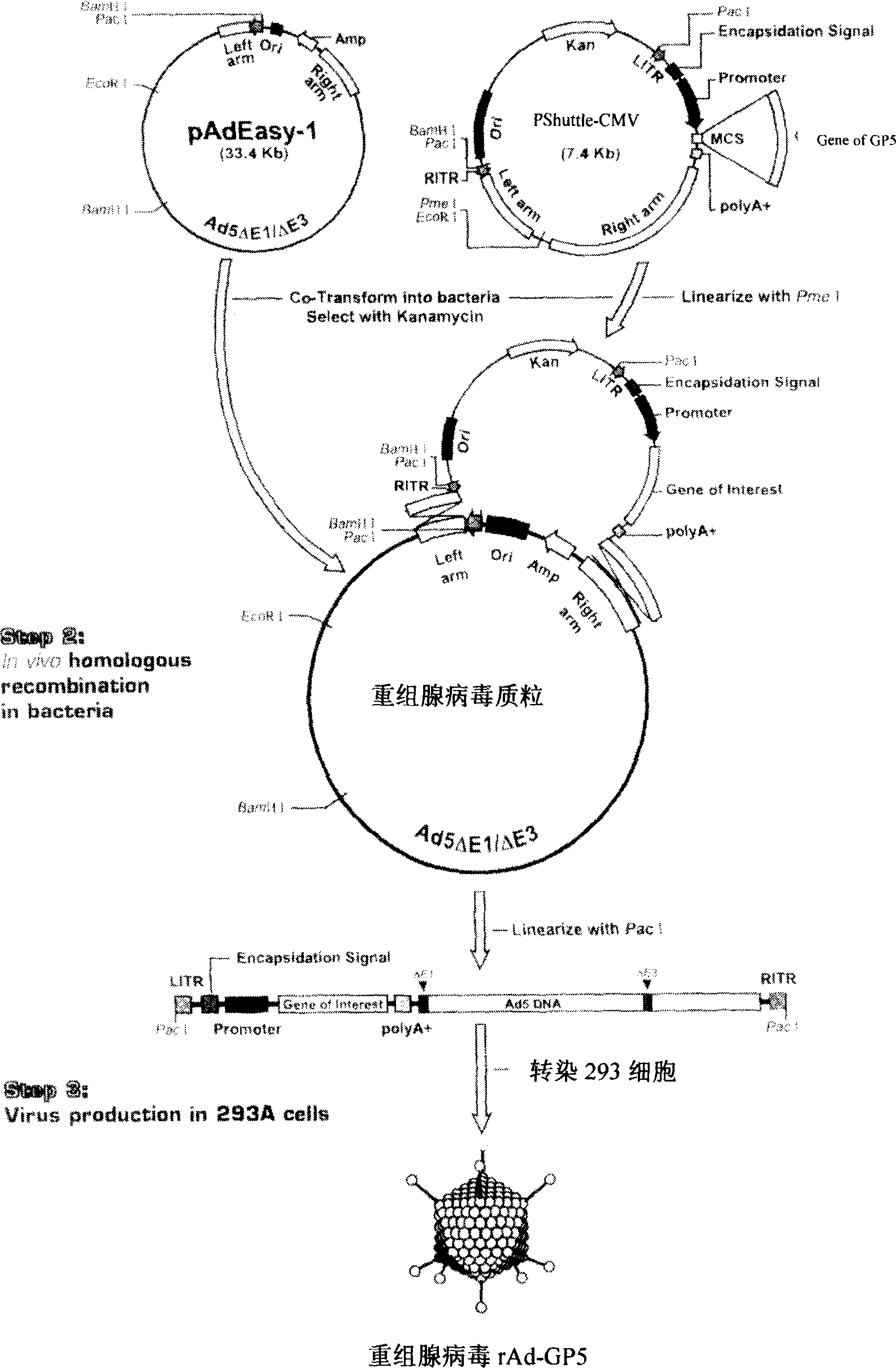
The invention relates to a pig breeding and breathing complex virus (PPRSV) and pig 2-type loop virus (PCV 2) series gene recombination adenovirus and vaccine in the field of high and new biotechnology. It uses PCR technology to clone the Cap protein gene into the plasmid carrier pShuttle-CMV-GP5 by open type reading rule and transforms tobacillus coli BJ5183strain with cage carrier pAdEasy-1 to capture the recombination plasmid; it uses recombination plasmid HEK293-A cell to capture recombination adenovirus and purifies it to express the recombination adenovirus rAd-Cap-GP5 of PRRSV GP5protein and PCV-2 Cap protein.
Owner:NANJING AGRICULTURAL UNIVERSITY
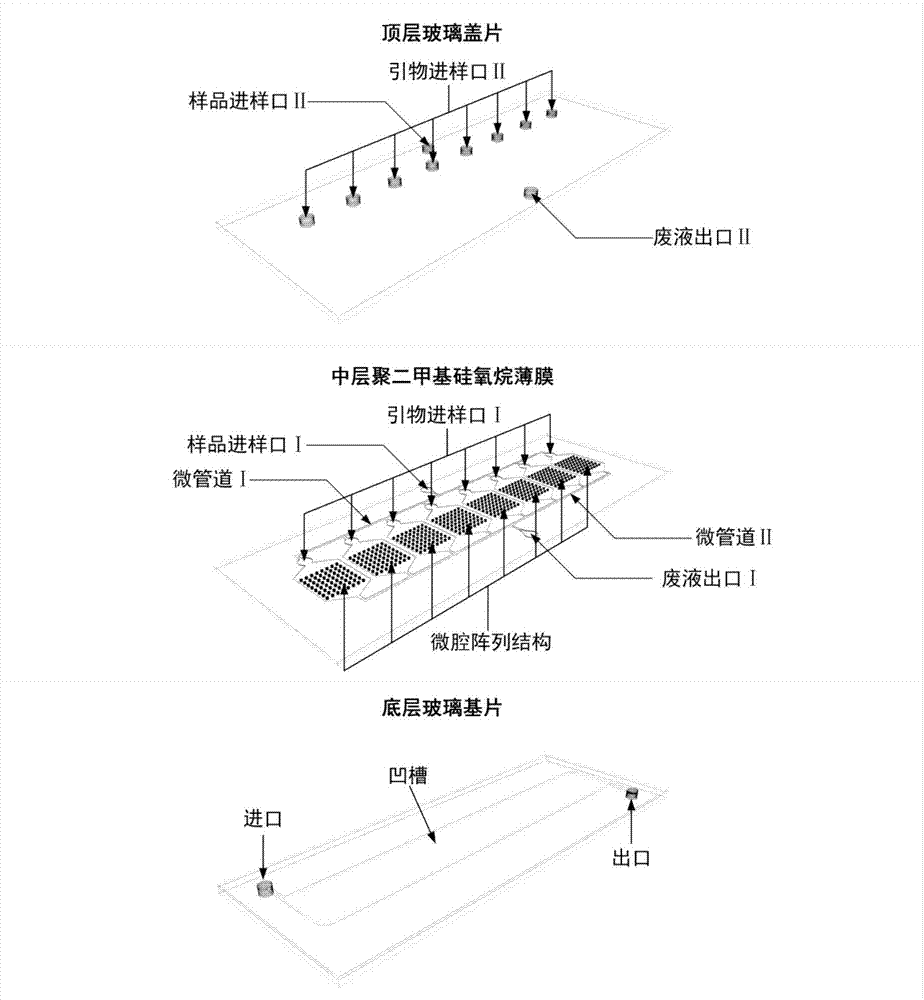
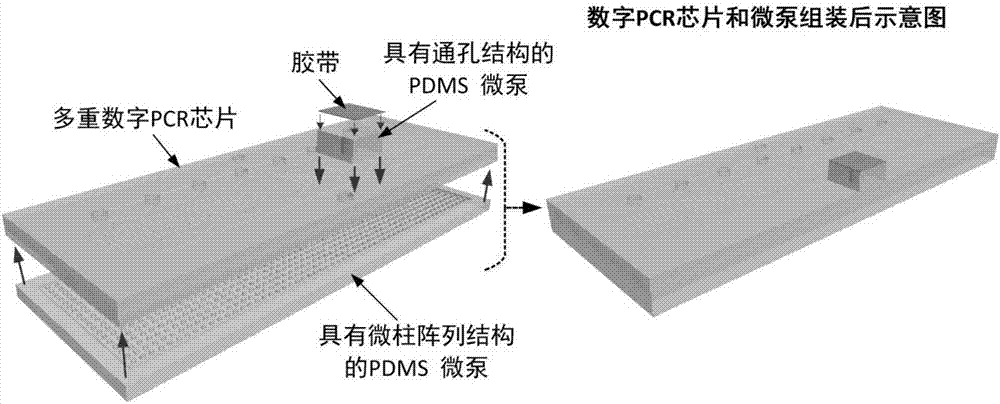
Integrated droplet microfluidic chip structure, preparation method and microfluidic chip assembly
PendingCN109825426AHighly integratedHigh degree of automationBioreactor/fermenter combinationsBiological substance pretreatmentsPositive pressureFluorescence
The invention provides an integrated droplet microfluidic chip structure. All functional modules of droplet generation, amplification and detection are integrated on the same microfluidic chip to achieve the whole enclosed process from the droplet generation to fluorescence detection. The invention also relates to an integrated droplet microfluidic chip structure preparation method and a microfluidic chip assembly. The structure is compatible with the positive pressure or negative pressure drive mode, the pressure response time is short, the rapid droplet generation can be achieved, and the sample preparation time is greatly reduced. Droplet generation oil is not required to be filled in advance, the operation is simple, and popularization and application in the technical field of digitalPCR are facilitated.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
Reagent kit for quantitatively assessing long-term recurrence risks of breast cancer
InactiveCN102586410AEasy to operateTest results are stableMicrobiological testing/measurementFluorescence/phosphorescenceGenomicsBreast cancer metastasis
The invention relates to the functional genomic and gene expression detection technology and the analysis technology, and discloses a reagent kit for quantitatively assessing long-term recurrence risks of breast cancer. Particularly, a type of genes capable of being used for breast cancer metastasis and prognostic molecular classification is screened within a human functional genome expression profile range, the detection technology is created, and the reagent kit is prepared and applied to breast cancer metastasis and prognostic assessment for a patient. The reagent kit for quantitatively assessing long-term recurrence risks of breast cancer comprises 21 pairs of primers, 21 specific taqman fluorescent probes, 10XRT-PCR (reverse transcription-polymerase chain reaction) buffer solution, 2.5mM of dNTP (diethyl-nitrophenyl thiophosphate) mixed liquor, reverse transcriptase, DNA (deoxyribose nucleic acid) polymerase, 10XPCR buffer solution and RNA (ribonucleic acid) enzyme inhibitor. The reverse transcription PCR technology is combined with the taqman fluorescent quantitative PCR technology, reverse transcription primers, real-time PCR primers and the taqman fluorescent probes are self-designed and optimized, reverse transcription PCR reagent and taqman fluorescent quantitative PCR reagent are integrated to prepare the detection reagent kit, operation is simple and fast, detection results are more stable, and detection cost is lower.
Owner:苏州科贝生物技术有限公司
Pig breeding and respiratory syndrome recombined adenovirus and vaccine
InactiveCN1554766ALittle changeChange propertiesGenetic material ingredientsInactivation/attenuationEscherichia coliBiotechnology
The present invention relates to pig reproduction and respiratory syndrome virus recombined adenovirus and vaccine, and belongs to the field of biological high-tech. Through RT-PCR process to proliferate whole PRRSV GP5 sequence, cloning the gene sequence to the shuttle vector pShuttle-CMV of adenovirus carrier system, cotransforming colibacillus BJ5183 strain together with the skeleton vector of adenovirus carrier system to obtain recombinant plasmid, transfecting HEK293-A cell to obtain recombinant adenovirus and plaque purification, and RT-PCR and indirect immunofluorescence technique inspection, the recombinant adenovirus rAd-GP5 expressing PRRSV GP5 protein is constituted. The recombinant adenovirus can set ahead the expression of PRRSV GP5 protein and raise the expression amount to simulate the immune protecting reaction of body effectively.
Owner:NANJING AGRICULTURAL UNIVERSITY
Specific molecular marker for acquiring genders of bighead carps and screening method and application thereof
ActiveCN106947827AOvercome operabilityTo overcome the disadvantages such as time-consumingMicrobiological testing/measurementDNA/RNA fragmentationGenomic sequencingScreening method
The invention discloses a specific molecular marker for acquiring the genders of bighead carps and a screening method and application thereof. The screening method comprises the following steps: firstly, performing reduced-representation sequencing on 5 female bighead carps and 5 male bighead carps by using a 2b-RAD technology to identify a male specific sequence; then, performing genome resequencing and genome assembly on one of the male bighead carps, comparatively searching 2b-RAD specific sequences of the male bighead carps in a male bighead carp genome to obtain a long-fragment male bighead carp specific sequence, and designing a male specific primer. By adopting the screening method, a male specific sequence and a gender specific primer of the bighead carps are screened for the first time, and a PCR (Polymerase Chain Reaction) technology for generic gender determination is built. The method has the advantages of low cost, easiness, rapidness and accuracy in operation, and the like, and a reliable technical measure is provided for the development of a gender marker and gender determination of the bighead carps. The method can be popularized and applied in gender marker development and gender determination research of other fishes.
Owner:INST OF AQUATIC LIFE ACAD SINICA
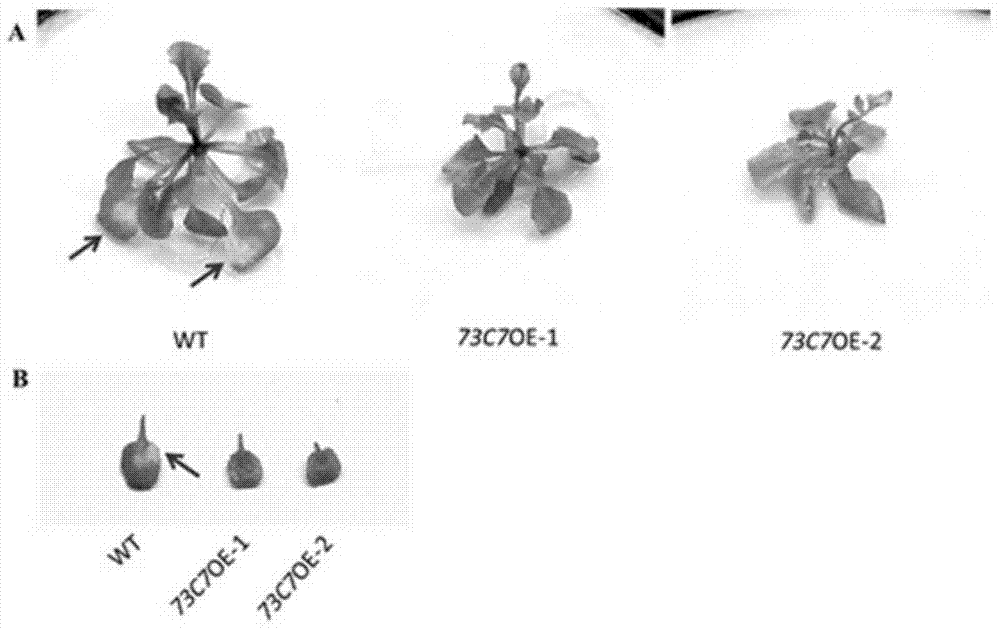
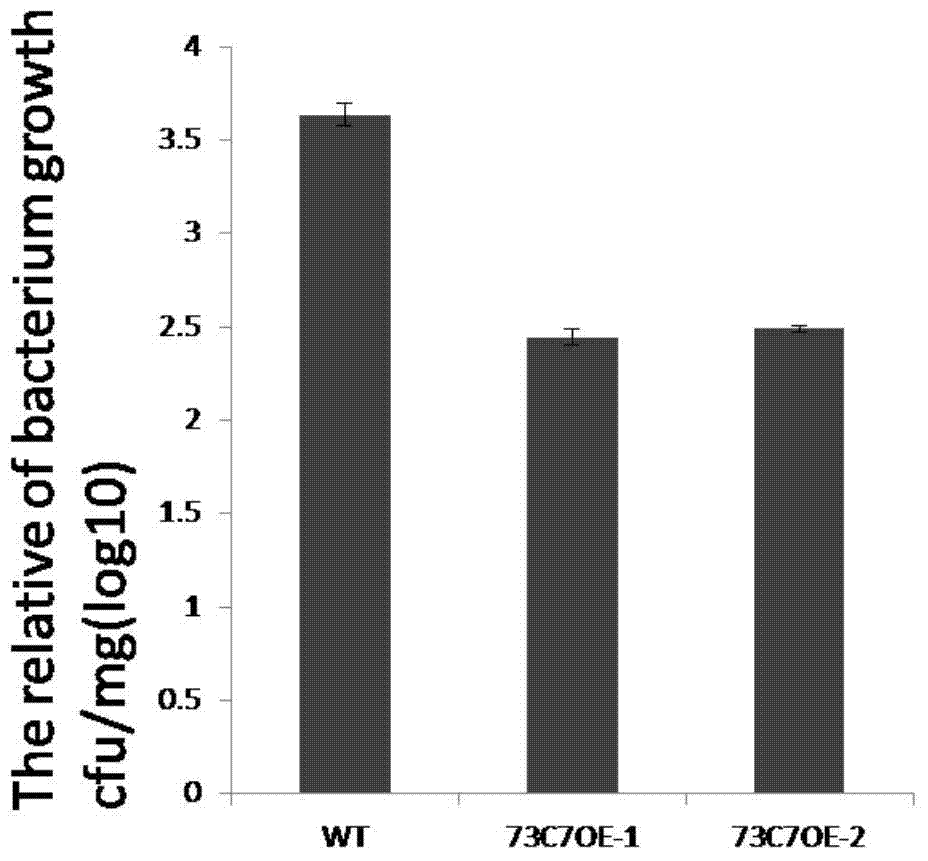
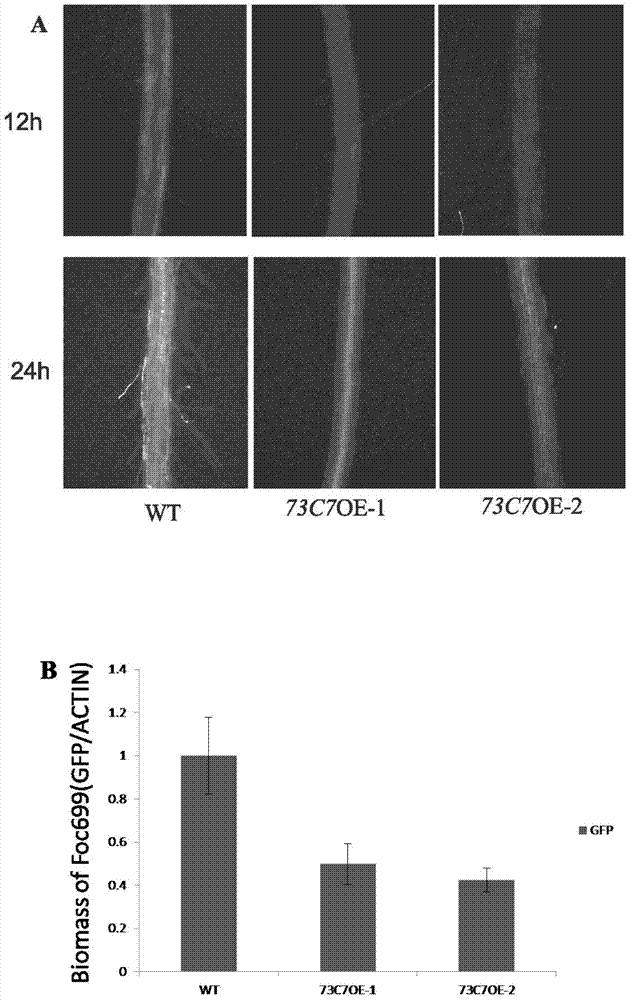
Application of Arabidopsis glycosyltransferase gene UGT73C7 in improving plant disease resistance
InactiveCN104845990AImprove disease resistanceIncrease varietyGenetic engineeringFermentationNucleotideArabidopsis
The invention discloses an application of Arabidopsis glycosyltransferase gene UGT73C7 in improving plant disease resistance, wherein the nucleotide sequence of the glycosyltransferase gene UGT73C7 is shown in SEQ ID No. 1 and is cloned by RT-PCR from Arabidopsis thaliana. The ene UGT73C7 is used to construct a plant overexpressing vector for plant transgene operation, so as to obtain the transgenic plants. The detection results show that the disease resistance of the transgenic plants is significantly improved, suggesting that new disease-resistant plants can be created after the implementation of the application disclosed by the invention, and the application can be used for the subsequent crop variety improvement and has great significance for China's agricultural production.
Owner:SHANDONG UNIV
Preparation method of biosensor with high sensitivity to heavy metal copper and product obtained thereby
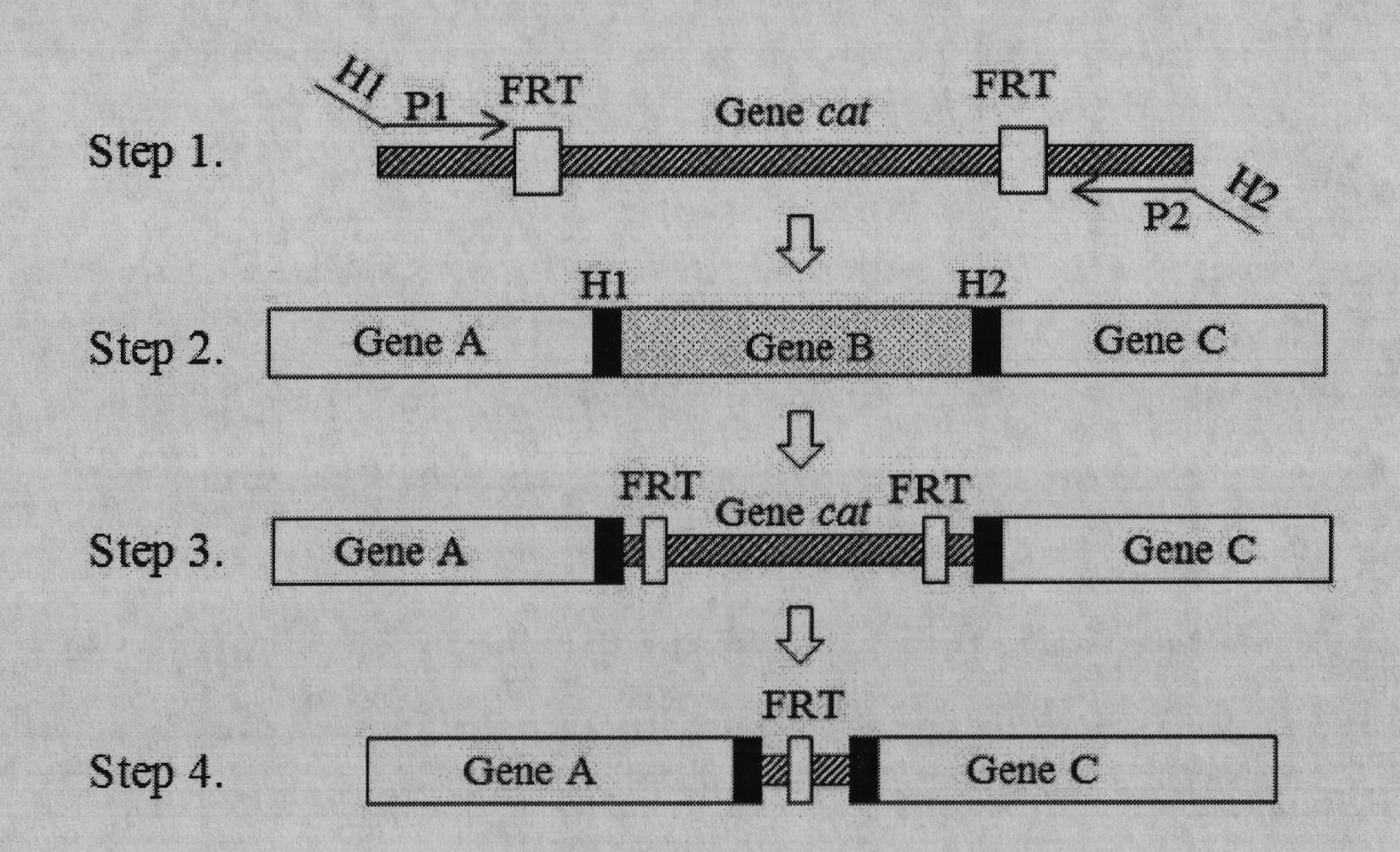
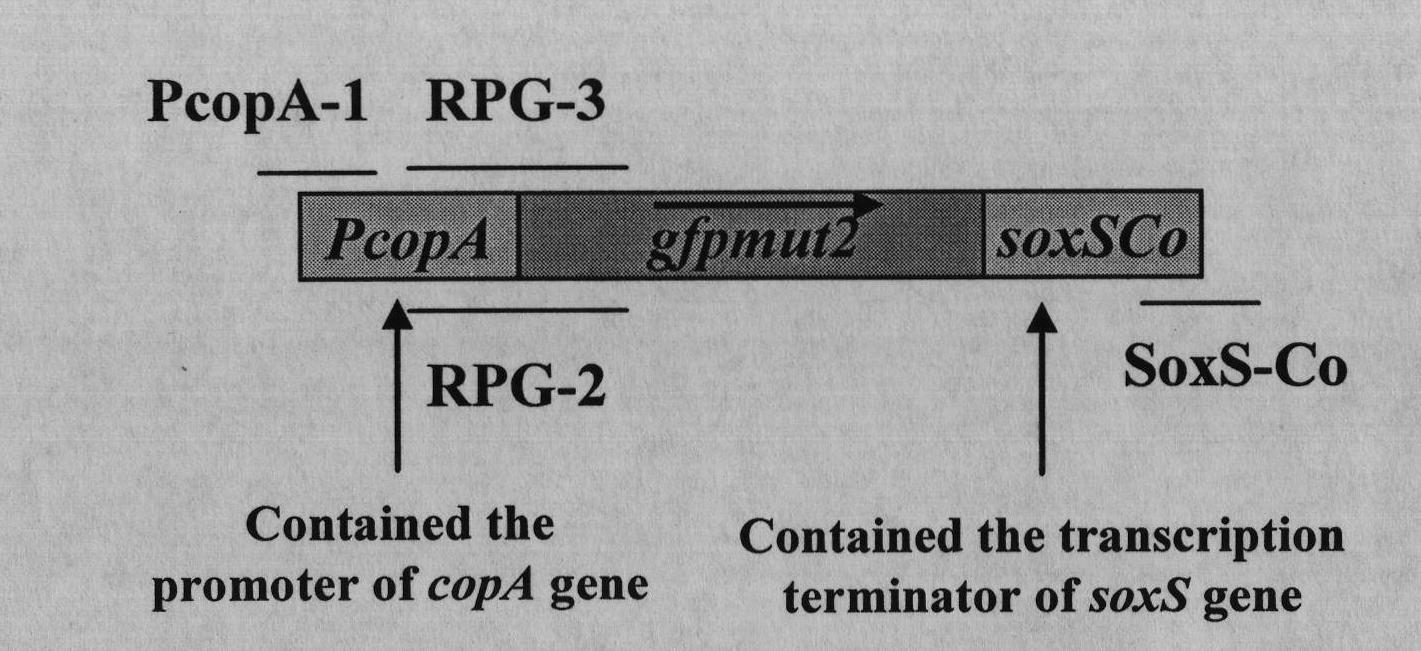
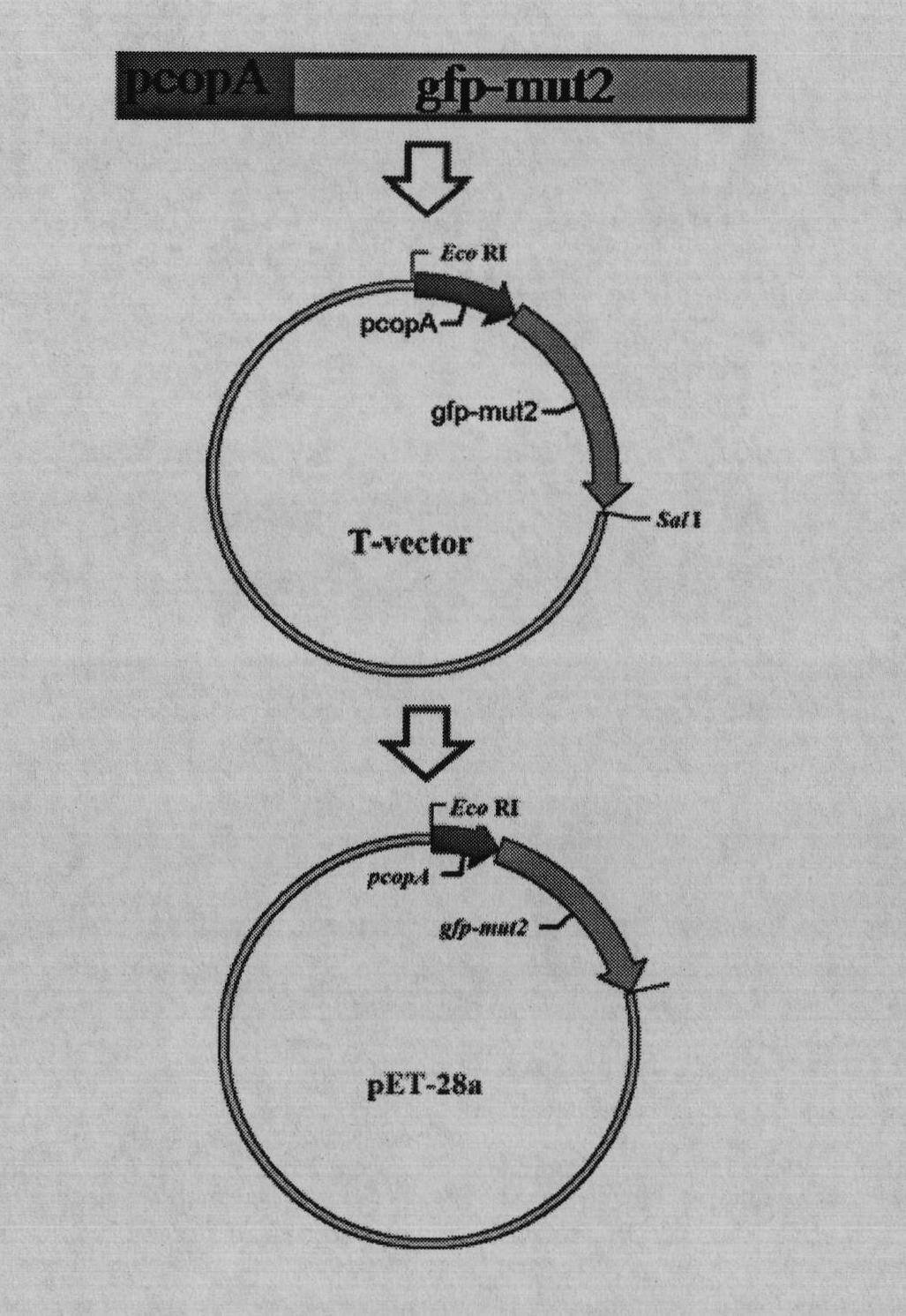
ActiveCN102417901AEasy to detectHigh sensitivityBacteriaVector-based foreign material introductionEscherichia coliDigestion
The invention relates to a preparation method of an Escherichia coli (E.coli) biosensor and a product obtained thereby. The preparation method comprises the following steps: (1) knocking out three genes, namely copA, cueO and cusA in a wild-type E.coli MC4100 genome by a Red recombination system; (2) constructing a fusion reporter gene PcopA::gfpmut2 by adopting a cross PCR (Polymerase Chain Reaction) technology; (3) cloning the segment obtained in step (2) to a pMD18-T vector; (4) carrying out double digestion on a PcopA::gfpmut2-T vector to obtain a target strip, and then connecting the target strip to a pET28a vector; and (5) converting a fusion reporter vector obtained in the step (4) to a deltacopA-deltacueO-deltacusA tri-gene mutant strain obtained in the step (1). In the preparation method, the three genes such as the copA, the cueO and the cusA are deleted from wild-type E.coli MC4100, and then the genes are converted to the fusion reporter vector PcopA::gfpmut2-pET28a so as to finally obtain the product. The product obtained by the method has the characteristics of high specificity, high sensitivity, high speed, high efficiency, high cost performance and the like, and is simple and convenient to use, thus being applicable to real-time field monitoring of copper ions.
Owner:WENZHOU MEDICAL UNIV
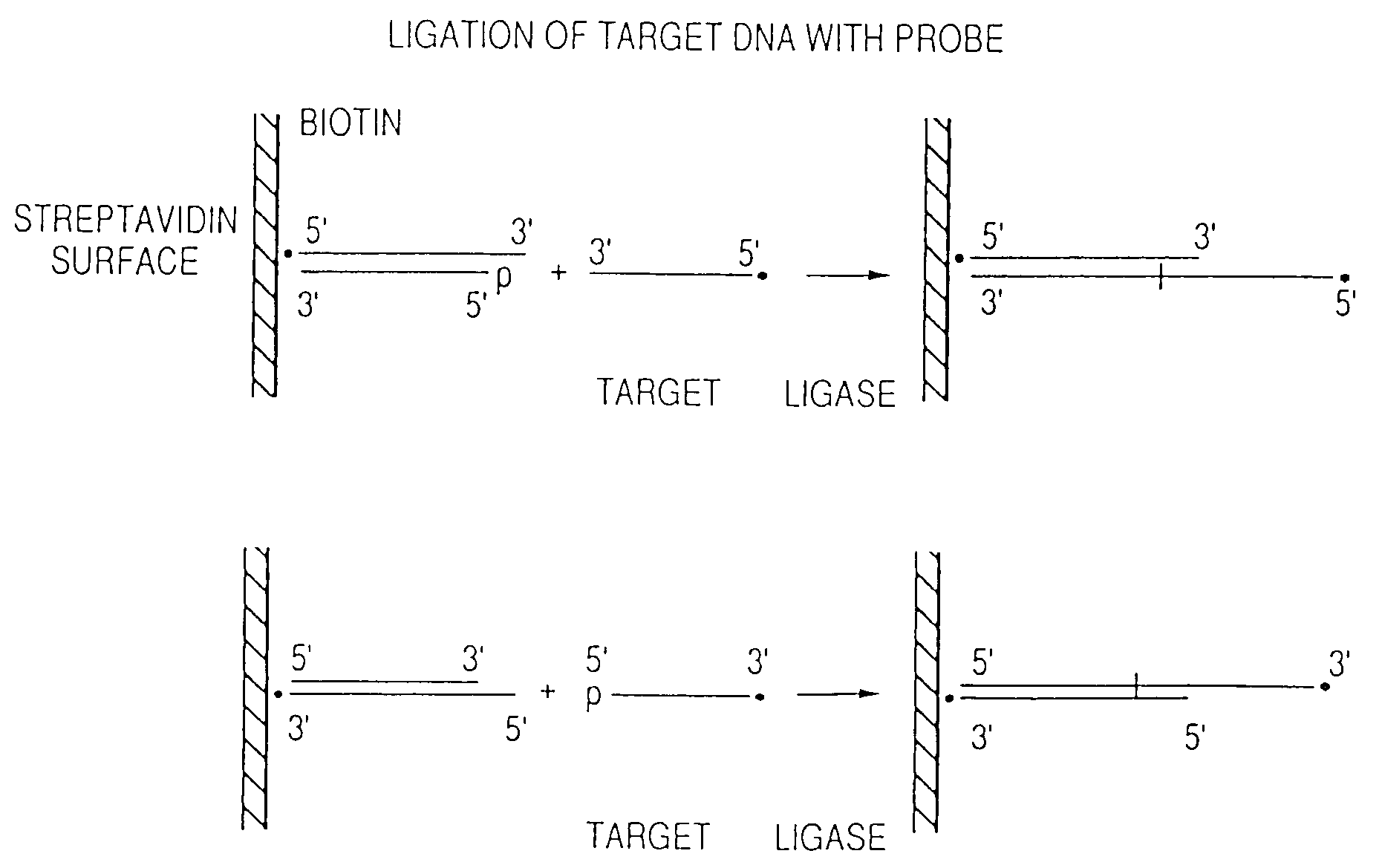
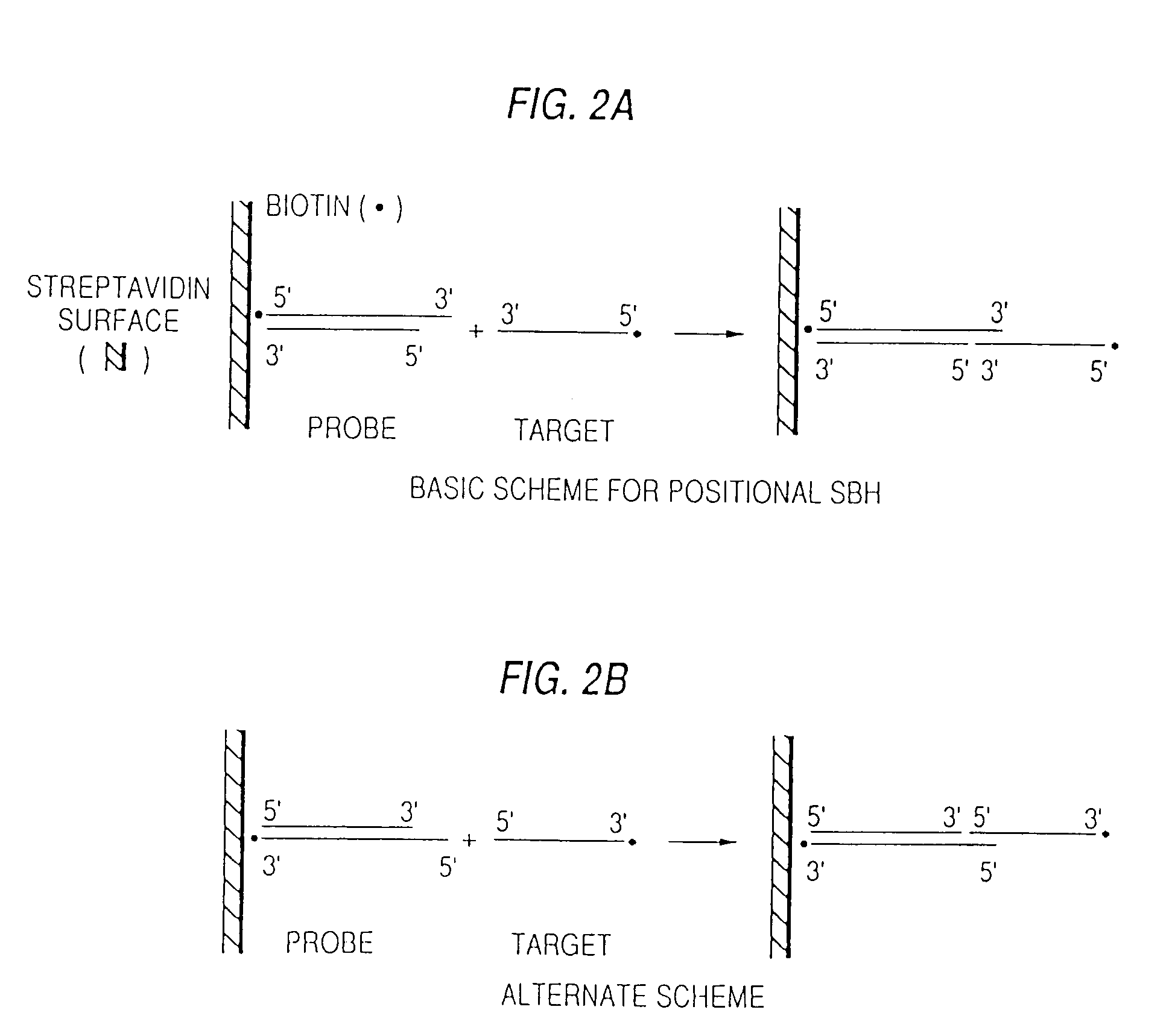
Arrays of probes for positional sequencing by hybridization
InactiveUS7319003B2Rapidly and accurately determining nucleotide sequenceBioreactor/fermenter combinationsSequential/parallel process reactionsPcr ctppBiology
This invention is directed to methods and reagents useful for sequencing nucleic acid targets utilizing sequencing by hybridization technology comprising probes, arrays of probes and methods whereby sequence information is obtained rapidly and efficiently in discrete packages. That information can be used for the detection, identification, purification and complete or partial sequencing of a particular target nucleic acid. When coupled with a ligation step, these methods can be performed under a single set of hybridization conditions. The invention also relates to the replication of probe arrays and methods for making and replicating arrays of probes which are useful for the large scale manufacture of diagnostic aids used to screen biological samples for specific target sequences. Arrays created using PCR technology may comprise probes with 5′- and / or 3′-overhangs.
Owner:TRUSTEES OF BOSTON UNIV
Bacillus licheniformis engineering bacteria for nattokinase production and method for producing nattokinase by using bacillus licheniformis engineering bacteria
ActiveCN104630124AAchieve secretory expressionSecreted expression highly mediatedBacteriaMicroorganism based processesBacillus licheniformisShuttle vector
Disclosed are bacillus licheniformis engineering bacteria for nattokinase production and a method for producing nattokinase by using the bacillus licheniformis engineering bacteria, which belong to the technical fields of microorganism genetic engineering and enzyme engineering. According to the invention, a nattokinase gene of bacillus subtilis MBS04-6 is obtained through PCR technology augmentation. Bacillus licheniformis BL10 is utilized as an expression host; promoter P43 of bacillus subtilis 168 is taken as a promoter; a signal peptide of extracellular serine protease Vpr of bacillus licheniformis WX-02 is taken a signal peptide; and a terminator sequence of alpha-amylase of bacillus licheniformis WX-02 is taken as a terminator. The expression elements are connected to shuttle vector pHY300PLK to form an expression vector, and the expression vector is transformed into the bacillus licheniformis BL10 to obtain the bacillus licheniformis engineering bacteria BL10 (pP43SNT) for nattokinase production. The bacterial strain has been deposited with the China Center for Type Culture Collection (CCTCC) at September 10th, 2013, and the accession number is CCTCC NO:M2013401. The invention also provides a method of the bacterial strain for producing nattokinase, and the maximum enzyme activity can reach 11.37 FU / mL in a liquid fermentation medium.
Owner:HUAZHONG AGRI UNIV
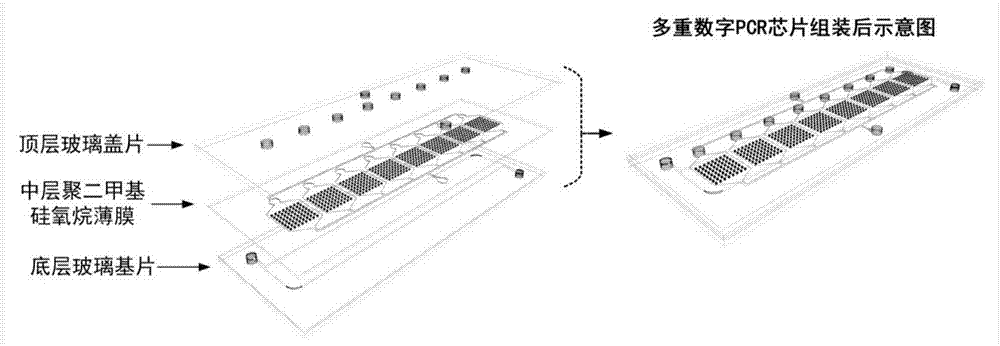
Multiple digital PCR chip and use method thereof
ActiveCN107262170ASimple and fast operationReduce complexityLaboratory glasswaresFluorescence/phosphorescencePcr chipEngineering
The invention relates to a multiple digital PCR chip and a use method thereof. The chip does not need to be driven by a precise micro-pump and be controlled by a complex micro-valve and does not need a complex macro-micro interface. The chip realizes simple, fast, low cost and large-scale autodecomposition and quantitative uniform distribution of samples. Compared with the existing commercial PCR chip, the chip greatly reduces the complexity and operating cost of the digital PCR system, simplifies the experimental operation and is free of professional training of an operator. The chip realizes the independence of each PCR reaction unit through microcavity arrays, is free of a surfactant and prevents the influence caused by an additive on the reaction system. Through integration of the isolated microcavity array zones and pre-deposition of primers corresponding to different specific target molecules in the microcavities in different zones, the chip can realize multiple detection through one step. The chip has the advantages of operation simpleness, low cost and strong multiple detection capacity and can promote development and wide application of the digital PCR technology.
Owner:CHONGQING UNIV
Method for obtaining sogatella furcifera carrying SRBSDV (southern rice black-streaked dwarf virus) and application of sogatella furcifera
InactiveCN102630640AOvercome technical bottlenecks that are difficult to preserve for a long timeRapid identificationHorticulture methodsRice cultivationDiseaseDiseased plant
The invention discloses a method for obtaining sogatella furcifera carrying SRBSDV (southern rice black-streaked dwarf virus) and an application sogatella furcifera. The method for obtaining the sogatella furcifera carrying the SRBSDV, comprises the following steps of: feeding the sogatella furcifera by utilizing rice infecting the SRBSDV or cryopreserved rice diseased plant so that the sogatella furcifera obtains the SRBSDV, transferring the infected sogatella furcifera to a healthy rice to be fed till passing through a circulation period of virus, and detecting the sogatella furcifera by utilizing an RT-PCR (reverse transcription-polymerase chain reaction) technology and a serology method to obtain the sogatella furcifera carrying the SRBSDV, wherein the carrier rate of the sogatella furcifera can reach more than 50% through a manual feeding manner based on the experiment. Through utilizing the method for obtaining the sogatella furcifera carrying the SRBSDV, the obtained sogatella furcifera carrying the SRBSDV can be applied to screening of SRBSDV-resisting monoclonal antibody to carry out a rice inoculation experiment to identify the resistance of the rice, screen disease-resistant variety and provide a service of the breeding for disease resistance. The invention can be further applied to researching mutual relation between the SRBSDV and a vector insect sogatella furcifera and provides powerful theoretical evidence for prevention and treatment on the viruses.
Owner:ZHEJIANG UNIV
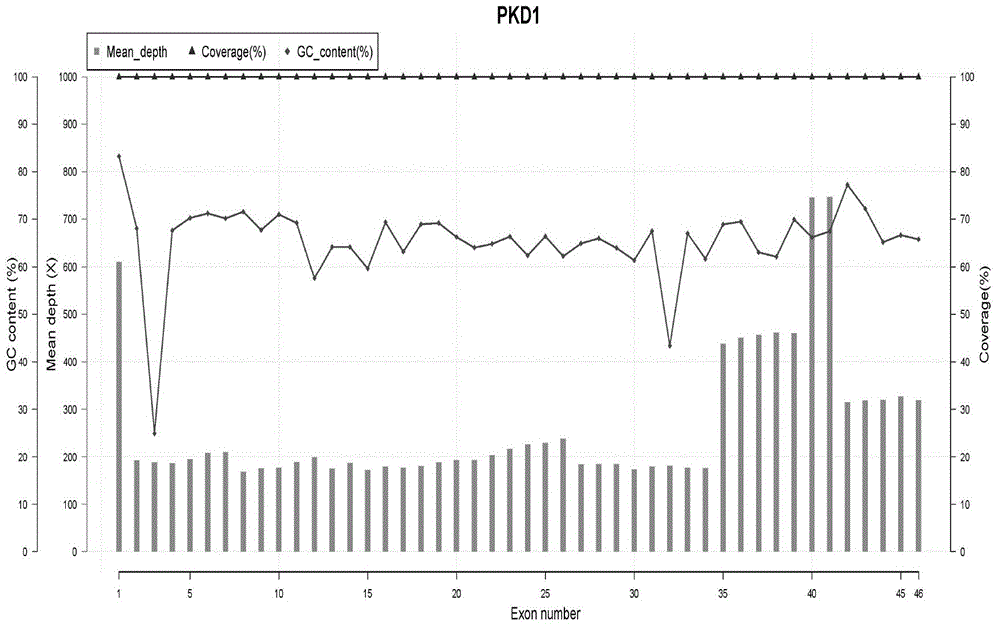
Amplimers, kit and method for detecting PKD1 gene mutation
ActiveCN104975081AAchieve separate markingAchieving Parallel SequencingMicrobiological testing/measurementDNA/RNA fragmentationPKD1Pcr ctpp
The present invention relates to a method for detecting PKD1 gene mutation. The method utilizes sequence information of the PKD1 gene, designs nine pairs of PCR tag primers, and uses a long-chain PCR technology for targeting amplification of PKD1 gene; while amplification products are labeled, PCR products are purified and mixed for direct construction of a single SMRTBell library; a PacBio RSII sequencer is employed for sequencing; and bioinformatic analysis is carried out to obtain PKD1 gene mutation information. The invention has the beneficial effects that a simplified pair of primers is designed and combined with tag primer technology to realize respective marking of PCR products of multiple DNA samples; the amplified PCR products do not need to perform knockout and can be directly used for the library construction, and the library construction does not need extra PCR, so that a plurality of samples are mixed into one library and processed simultaneously by a PacBio RSII sequencing library construction link, and the experimental operation is greatly simplified.
Owner:NANJING MATERNITY & CHILD HEALTH CARE HOSPITAL
Bacillus subtilis strain for excreting nattokinase at high efficiency and preparation technology of high-purity nattokinase
InactiveCN107058204AIncrease enzyme activityBacteriaMicroorganism based processesRestriction enzyme digestionDigestion
The invention relates to a bacillus subtilis strain for excreting nattokinase at high efficiency. An aprN gene of bacillus subtilis NK is amplified by a PCR (polymerase chain reaction) technique; codons of initial thirty amino acids are optimized according to the codon preference of the bacillus subtilis, a recombination expression plasmid pHT01-aprN is established, and the correctness is verified through the restriction enzymes digestion, PCR amplification and sequencing. The recombination expression plasmid pHT01-aprN is imported into the bacillus subtilis 168 by an electric shocking method, and is performed with resistance screening by adopting chloramphenicol, so as to obtain engineering bacteria. After the fermenting condition is optimized, and the expression is induced by IPTG, the highest enzyme activity in a shake culture fermenting liquid is 289U / ml; the highest enzyme activity in a high-density fermenting liquid is 7778U / ml; the highest enzyme activity of a nattokinase preparation prepared by affinity column chromatography is 981731U / g.
Owner:上海诺金科生物科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com